Abstract
Noroviruses (NoVs) are the primary cause of acute gastroenteritis worldwide. Histo-blood group antigens (HBGAs) are receptors or attachment factors that affect the prevalence and host susceptibility of NoVs. GII.6 NoV is one of the predominant genotypes in humans, which recognizes the type ABO secretor of HBGAs. However, the structural basis of GII.6 NoV's interaction with HBGAs receptors remains elusive. In this study, we investigated the binding features of the GII.6 strain to HBGAs using saliva- and glycan-ELISA assays and characterized the molecular basis of the GII.6 virus that recognizes H disaccharide. We showed that the GII.6 P domain recognized some A and O secretor's saliva samples, most B secretor's saliva samples, and H disaccharide antigen, but did not bind non-secretors’ saliva. Further, we determined the crystal structures of GII.6 and its complex with H disaccharides at 1.7 Å, revealing that the P domain of GII.6 shares the conventional binding interface and mode of GII HBGAs. Single residue mutations at the GII.6-H binding sites could inhibit the binding of GII.6 to HBGAs, demonstrating that the interaction residues were crucial in maintaining NoV-glycan integrity. Finally, structural and sequence analyses showed that the major residues of the GII.6-H interaction were conserved among NoVs in the GII genogroup. Taken together, our study characterized the functional and structural features of GII.6 that allow it to interact with HBGAs, and shed light on NoV evolution, epidemiology, and anti-viral drug development.
Keywords: Noroviruses (NoVs), Histo-blood group antigens (HBGAs), GII.6 P protein structure, H disaccharides
Highlights
-
•
We showed that GII.6 P domain recognized ABO secretors and H disaccharides antigen.
-
•
The native crystal structure of the GII.6 P domain and its interaction with H disaccharides had been characterized.
-
•
The GII.6 HBGAs binding sites were quite conserved among NoVs in the GII genogroup.
-
•
We characterized GII.6-H binding interaction residues that were crucial in maintaining NoVs-glycan integrity.
1. Introduction
Noroviruses (NoVs) belong to the Norovirus genus of the family Caliciviridae and are among the most important causes of acute gastroenteritis (Ahmed et al., 2014). Currently, there are no internationally approved drugs or vaccines for the specific treatment and prevention of human NoVs (HuNoVs). NoVs are genetically diverse and can be classified into ten genogroups (GI–GX) (Chhabra et al., 2019). Genogroup GII is the most prevalent and further into the most genotypes, accounting for more than 90% of NoV outbreaks.
NoVs are single-stranded, positive-sense RNA viruses encapsulated by the major capsid protein, VP1, which is assembled into the main structural protein. VP1 is composed of a shell (S) domain surrounding the viral RNA and a protruding (P) domain that can be further divided into two subdomains, P1 and P2 (Song et al., 2020). The P2 subdomain, which recognizes host receptors and plays an important role in virus-host interactions, exhibits more variable sequences (Cao et al., 2007). The P domain expressed in an E.coli system can form P particles and P dimer and is a very useful model for studying the receptor-binding function and structural basis of NoVs (Lindesmith et al., 2003; Cao et al., 2007; Tan and Jiang, 2012).
Histo-blood group antigens (HBGAs), which act as host attachment factors or susceptibility factors in HuNoVs, determine host susceptibility, host range, and the prevalence of NoV infection (Tan and Jiang, 2005, 2011, 2014). HBGAs are glycans containing fucose, which are mainly distributed on the surface of mucosal epithelial cells of the intestinal tract and are the determinants of various blood types, including ABO, and Lewis blood types. FUT2 codes for the alpha(1,2) fucosyltransferase that is responsible for the synthesis of the H antigen. Individuals with functional FUT2 called secretor. Nonfunctional FUT2s fail to present H antigens in secretions and on epithelial cells and are less susceptible to NoV infection, which are called non-secretors (Le Pendu, 2004).
The binding capability of NoVs to saliva sample of HBGAs has been reported to be positively correlated with its prevalence. As an example, several GII.4 strains, such as Dijon, Osaka, Sydney, and DenHaag, show a wide variety of HBGA-binding features and have thus had the highest prevalence over the past two decades (Rydell et al., 2009; De Rougemont et al., 2011; Morozov et al., 2018). In contrast, GI.1 does not bind HBGAs and thus has a lower prevalence (Huang et al., 2005). The GII.17 strain outbreak in 2014–2015 also showed a broader HBGA-binding spectrum, including ABO blood group types, no matter for secretors and non-secretors. However, prior to this outbreak, the earlier GII.17 was not prevalent and showed no binding capability to HBGAs (Sun et al., 2015; Jin et al., 2016; Qian et al., 2019).
The binding of various NoV strains to HBGAs has been clarified, including their binding patterns and terminal structures (Tan and Jiang, 2011; Cong et al., 2019; Qian et al., 2019). Many GII NoV strains, such as GII.4, GII.9, GII.10, GII.12, and GII.17, interact with HBGAs, mainly through α-Fuc (Chen et al., 2011; Frenck et al., 2012; Tan and Jiang, 2014). Compared to earlier variants, the GII.17 outbreak variant reconstructed its HBGA-binding site (HBS) by mutating 444 in VP1 to tyrosine to form a van der Waals bond, thus greatly enhancing HBGA binding capability (Qian et al., 2019). The HBS of the GII.2 BJSMQ outbreak strain, which has been prevalent in Chinese mainland in the last five years, has acquired stability and greater blood group antigen-binding capability via mutation of 382 in VP1 (Ao et al., 2018). Crystallological studies have also demonstrated that the emergence of an outbreak often results in changes in the molecular pattern of oligosaccharide binding, and these changes make the low HBGA binding intensity NoVs acquire the ability to cause outbreaks, supporting a close association between the molecular pattern of HuNoV binding to oligosaccharides and the virus’ prevalence (Qian et al., 2019).
Over the past two decades, GII.4 has been the dominant genotype causing HuNoV infection worldwide (Leshem et al., 2013), and in recent years, GII.17 and GII.2 have gradually become the main outbreak strains in many countries (Chan et al., 2015; Jin et al., 2016; Ao et al., 2017). GII.6 recognizes the type ABO secretor's saliva (Huo et al., 2017). The results from laboratory monitoring network in many countries showed that the epidemic of GII.6 was always secondary to those prevalent strains (Jin et al., 2020). However, the mechanism of GII.6 binding to HBGAs remains largely unknown.
In the present study, we elucidated the features of the GII.6 P domain in recognizing HBGAs, including ABO secretor's saliva samples and H disaccharides, by saliva and glycan-ELISA, and biolayer interferometry binding assays. We also elucidated the structural basis for the binding of GII.6 to H disaccharides and identified seven key amino acids required for HBGA receptor binding. Finally, we compared the structural features and binding capacity between GII.6 and the outbreak strain GII.17, and demonstrated that GII.6 showed a relatively low binding capacity. Our results thus revealed that the structural features and mechanism of recognition of HBGAs in GII.6 provided a mechanistic explanation for its prevalence in the population.
2. Materials and methods
2.1. GII.6 P domain expression and purification
The GII.6 P domain-encoding sequences (GenBank accession number: OL468714.1) were chemically synthesized by Genewiz (Suzhou, China). The cysteine-containing short peptide (CDCRGDCFC) was added to the C-terminal end of the P domain to stabilize P particle formation, and the wild-type P domain sequence was used for P dimer production (Tan et al., 2004; Tan and Jiang, 2011). For single GII.6 mutations, with the exception of the A358 mutant to G, GII.6 P domain mutants with T359, R360, D388, G449, G450, or Y451 single mutations to A were constructed using overlap PCR with the corresponding wild-type P domain as a template. The cDNA sequences encoding the wild-type (residues 224–551) and mutant GII.6 P domains were cloned into the pGEX-6P-1 expression vector and then transformed into BL21 (DE3) cells that were induced with 0.4 mmol/L IPTG. The GII.6 P particles and P dimer were purified using glutathione-Sepharose 4B to obtain the glutathione S-transferase (GST) fusion protein. The GST tag was cleaved using PreScission protease (Singh et al., 2016). The P dimer was further purified using Superdex 20016/600GL gel filtration chromatography with phosphate-buffered saline (PBS, pH 7.4). GII.17 KW308, and P[14] VP8∗ of rotavirus were used as positive controls (Chan et al., 2015; Sun et al., 2016a).
2.2. Glycan binding assay
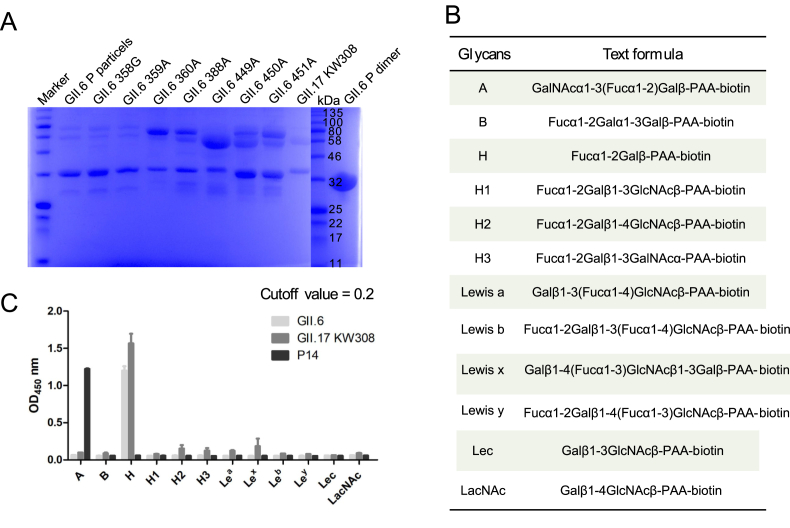
The glycan binding assay was used to test the binding of GII.6 P particles to oligosaccharides using enzyme-linked immunosorbent assay (ELISA) as previously described (Cong et al., 2019). Briefly, 20 μg/well of P particles was coated onto microtiter plates at 4 °C overnight. After blocking with 5% nonfat milk, 0.2 μg/well of biotin-labeled polyacrylamide (PAA)-conjugated oligosaccharides were added, including A trisaccharides (A), B trisaccharides (B), H disaccharides (H), H type 1 (H1), H type 2 (H2), H type 3 (H3), Lewis a (Lea), Lewis x (Lex), Lewis b (Leb), Lewis y (Ley), type I precursor (Lec), and type II precursor (LacNAc) (GlycoTech, USA). After incubation, horseradish peroxidase (HRP)-conjugated streptavidin (1:1500 in PBS) (Abcam, USA) was added. The 3,3′,5,5′-tetramethylbenzidine (TMB) kit (BD Biosciences, USA) was used to detect the HRP activity. The reaction was stopped by the addition of 1 mol/L phosphoric acid, and the absorbance was measured at 450 nm (OD450).
2.3. Saliva binding assay
A panel of well-characterized saliva samples with A, B, H, Lea, Leb, Lex, and Ley phenotypes and GII.6 P particles were used in the saliva binding assay as previously described (Cong et al., 2019). Microtiter plates were coated with different boiled saliva samples (1:1000) in PBS from ABO secretors and non-secretors. After blocking with 5% nonfat milk, 0.5 μg/well GII.6 P particles were incubated on a microtiter plate. Bound P particles were tested using a rabbit anti-P domain polyclonal antibody followed by an HRP-conjugated goat anti-rabbit IgG antibody. HRP activity was detected using a TMB kit, and absorbance was measured at OD450.
2.4. Biolayer interferometry binding analysis
The apparent binding affinity of GII.6 P particle and H disaccharides was measured by biolayer interferometry (BLI) using Octet RED96e. Biotin-labeled H disaccharides were immobilized onto streptavidin biosensors until saturation, typically 10 μg/mL for 2 min, in K buffer containing 0.02% Tween 20 and 0.1% bovine serum albumin (BSA) in PBS. The association and dissociation curves were measured. The data were normalized using Octet analysis software to calculate the apparent equilibrium dissociation constants for GII.6 P particles and oligosaccharides.
2.5. Protein crystallization
The GII.6 P dimer was concentrated to 3–5 mg/mL. Native crystal of the GII.6 P protein was grown using the sitting-drop vapor diffusion method by mixing protein solution with an equal volume of reservoir solution containing 0.2 mol/L calcium chloride dihydrate, 0.1 mol/L sodium acetate trihydrate (pH4.6), 20% (vol/vol) 2-propanol. GII.6 P dimer and H disaccharides (Dextra) were mixed at a 1:50 molar ratio and incubated for 5 h at 4 °C under the same conditions as the native protein. After incubation for 7 days at 18 °C, the native and complex crystals were transferred to a cryoprotectant containing mother liquor and 20% glycerol, and subsequently flash-frozen in liquid nitrogen.
2.6. Data collection and processing
X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF) BL19U and processed using HKL2000. Additional processing was performed using CCP4 software. The structure of the GII.6 P domain was determined using the molecular replacement module of PHASER with the GII.21 P structure (Protein Data Bank [PDB] code 4RLZ) as a search model. The model was further refined using phenix.refine in PHENIX (Adams et al., 2010; Afonine et al., 2012). The data collection and refinement statistics are summarized in Table 1. Structural analysis was performed using PyMOL software.
Table 1.
Crystallographic X-ray diffraction and refinement statistics of GII.6 and GII.6-H.
| Parametera | GII.6 | GII.6-H |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P212121 |
| Unit Cell dimensions | ||
| a, b, c (Å) | 62.444, 94.777, 109.010 | 62.631, 94.597, 108.968 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.00–1.70 (1.76–1.70) | 50.00–1.70 (1.76–1.70) |
| Rmerge (%)b | 0.100 (0.909) | 0.072 (0.599) |
| I/σI | 24.843 (2.345) | 37.9 (4.335) |
| Completeness (%) | 92.72 (65.99) | 99.28 (94.28) |
| Redundancy | 10.9 (9.3) | 12.3 (10.4) |
| Refinement | ||
| Resolution (Å) | 47.39–1.704 | 47.3–1.698 |
| No. reflections | 66,160 | 71,618 |
| Rwork/Rfree | 0.1725/0.1943 | 0.1427/0.1746 |
| No. atoms | 5378 | 5523 |
| Protein | 4668 | 4683 |
| Ligand/ion | 68 | |
| Water | 710 | 772 |
| B-factors | ||
| Protein | 17.63 | 18.63 |
| Water | 26.79 | 32.12 |
| Ligand/ion | 39.84 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | 0.006 |
| Bond angles (°) | 0.84 | 0.770 |
| Ramachandran plot | ||
| Favored (%) | 98.65 | 96.79 |
| Allowed (%) | 1.35 | 3.21 |
| Disallowed (%) | 0.00 | 0.00 |
Values in parentheses are given for the highest resolution shell.
Rmerge = Σhkl |I-<I>|/ΣhklI, where I is the intensity of unique reflection hkl and <I> is the average over symmetry-related observations of unique reflection hkl, hkl is the reflection indices.
3. Results
3.1. Glycan binding specificity of GII.6 P domain
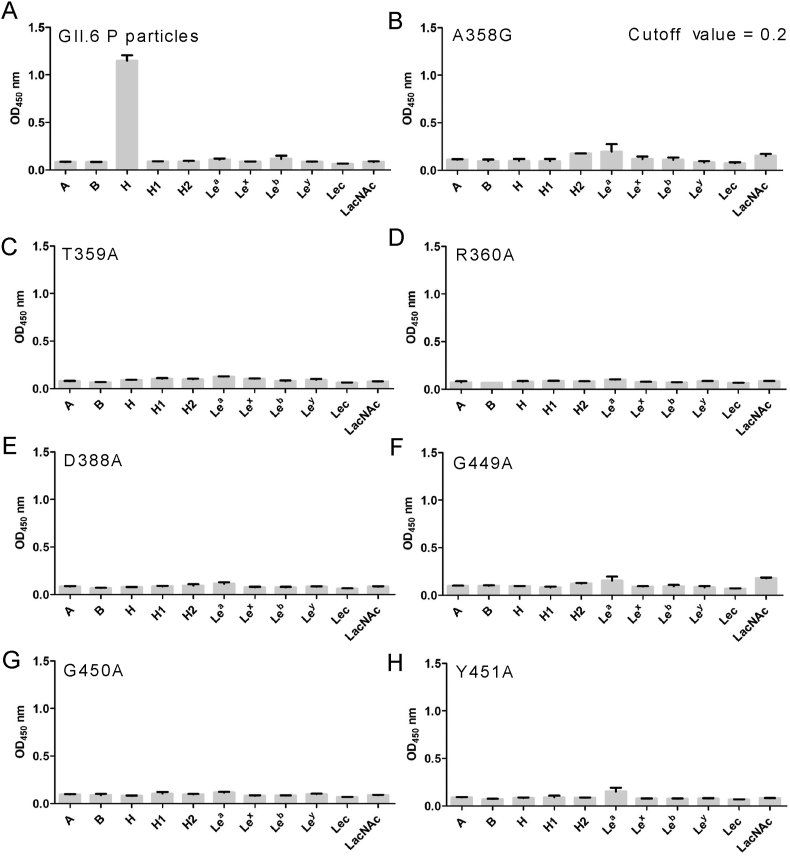
To determine the glycan-binding characteristics of GII.6, we expressed and purified fusion GST-labeled GII.6-GST P proteins with single mutations and obtained GII.6 P particles through 3C protease cleavage of GST protein (Fig. 1A). We also performed a glycan-binding assay with GII.6 P particles and synthetic HBGA oligosaccharides representing A, B, H, H1, H2, H3, Lea, Lex, Leb, Ley, Lec, and LacNAc (Fig. 1B). Using GII.17 KW308 and P14 as positive controls, we found that the GII.6 P particles bound strongly to H disaccharides and displayed no signals to other oligosaccharides (Fig. 1C).
Fig. 1.
Glycan-binding specificity of GII.6 P protein to synthetic HBGA oligosaccharides. A SDS-PAGE analysis of the P proteins of wild-type GII.6 virus and seven GII.6 mutants. B The glycans involved in this study. C The GII.6 P protein was tested for its binding to various glycans (x-axis) at 0.2 μg/well. The experiment was repeated twice independently and the data are expressed as means and SDs (error bars) for absorbance values.
3.2. GII.6 P domain interacted tightly with H disaccharides
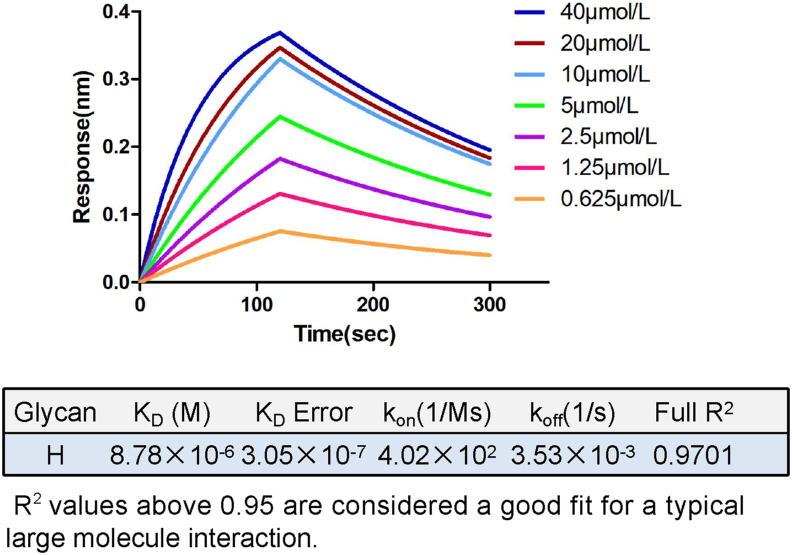
To determine the affinity between the GII.6 P particles and H disaccharides, we performed binding experiments using BLI. Our data showed that the P domain bound to H disaccharides with an affinity constant Kd of 8.78 μmol/L and rate constants Kon of 4.02 × 102 mol/L1·s−1 and Koff of 3.53 × 10−3 s−1 for association and dissociation, respectively (Fig. 2), indicating a tight interaction between the GII.6 P domain and H disaccharides.
Fig. 2.
BLI analysis of GII.6 P domain binding to H disaccharides. The P domain-H disaccharide association-dissociation curves were obtained through serial twofold dilutions of GII.6 P protein (0.625–40 μmol/L) plus buffer controls using Octet acquisition software. The sensograms for all concentrations are shown and labeled accordingly. The calculated KD, KD Error, Kon, Koff, and Full R2 are shown in a tabular form.
3.3. Saliva binding specificity of GII.6 P domain
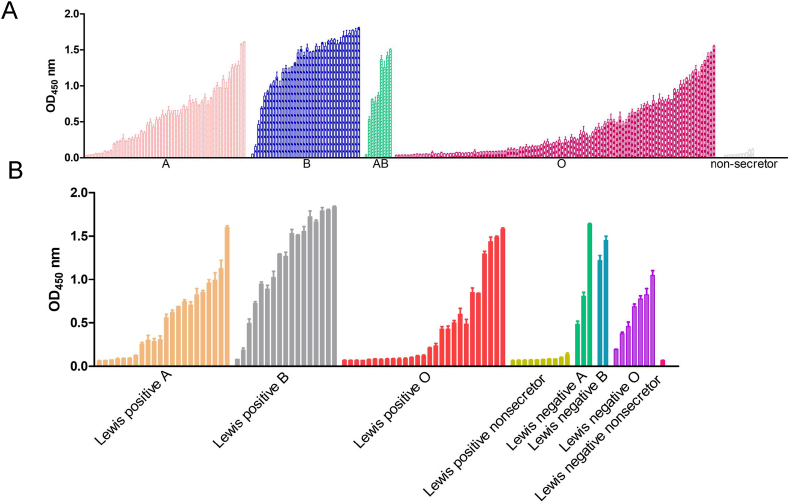
To further assess the specificity of the binding of the GII.6 P domain to HBGAs, we chose saliva samples that contained both ABO secretors and non-secretors. Our data showed that the GII.6 P protein selectively bound to the secretor's saliva samples, including type A, B, AB, and O, but not to non-secretor's saliva samples (Fig. 3A). The results of the Lewis classification method showed that GII.6 binds to the saliva samples of Lewis-positive A/B/O and Lewis-negative A/B/O, but not to Lewis-positive non-secretor or Lewis-negative non-secretor (Fig. 3B). As expected, the GST protein, as a negative control, did not show any binding activity.
Fig. 3.
Saliva-binding specificity of GII.6 P protein. A Saliva-based binding assay showed the binding signals (y-axis) of the GII.6 protein (0.5 μg/well) to type secretors (A, B, O), and non-secretor's saliva samples (x-axis). B Saliva-based binding assay of GII.6 P protein to Lewis positive A/B/O/non-secretor and Lewis negative A/B/O/non-secretor's saliva samples.
3.4. Crystal structure of the GII.6 P dimer in complex with H disaccharides
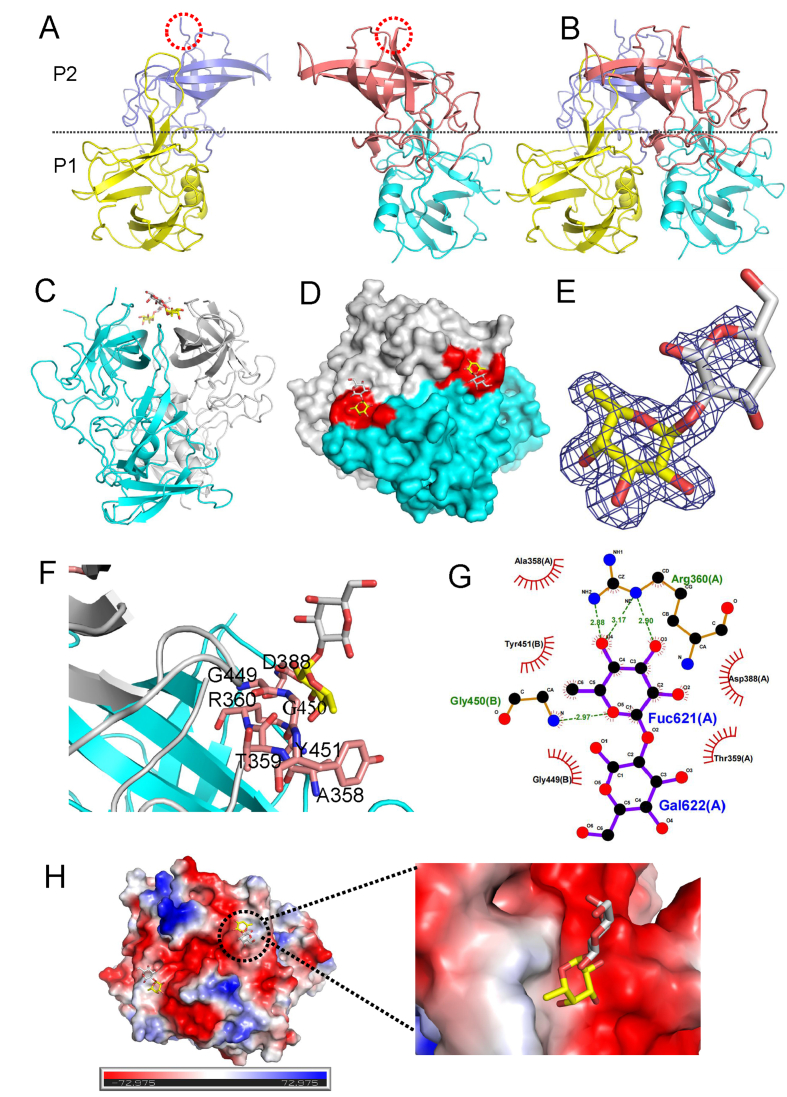
To further understand the structural basis of the interaction between the GII.6 P dimer and H disaccharides, we determined the X-ray structure of the GII.6 P dimer and its complex with H disaccharides at 1.7 Å (Fig. 4). Both the GII.6 P dimer and its complex residues were clearly defined, except for 15 amino acids from 293 to 307, that showed no discernible electron density, suggesting a disorder of flexibility in this loop (Fig. 4A and B). Besides, we observed that the H disaccharide binding sites were located at the interface between the two P monomers (Fig. 4C and D). H disaccharides were visible in the (2mFo-DFc) omit the difference electron density map, and two sugar rings were fitted into the map (Fig. 4E). Six residues from the P2 domain were involved in binding to the α-Fuc of H disaccharides via hydrogen bonding and hydrophobic interactions (Fig. 4F). The α-Fuc interacted with the A chain of R360 and the B chain of G450 through hydrogen bonds, while A358, T359, and D388 of the A chain and Y451 of the B chain formed hydrophobic interactions with Fuc to support the binding outcomes. In contrast, the other saccharide, -galactose (-Gal), pointed away from the surface of the P dimer, and only a hydrophobic interaction was formed with G449 of the B chain (Fig. 4G). It also showed that α-Fuc was bound to the electronegative glycan-binding site (Fig. 4H).
Fig. 4.
Crystal structures of GII.6 P dimer in complex with H disaccharides. A The structures of the two native GII.6 P protein monomers. A red dashed line shows the missing loop. B The structure of the native GII.6 P dimer. P1: yellow and cyan; P2: slate and salmon. The dashed line marks the boundary between the P1 and the P2 subdomains. C The cartoon represents the complex formed by GII.6 P dimer in complex with H disaccharides. D The surface of the GII.6 P dimer with H disaccharides. Cyan and gray, the two monomers of the GII.6 P dimer. E The mesh map of H disaccharides was contoured at 1 (blue) around the selection site, with a coverage of 1.6 Å radius. F The network of interactions between GII.6 and H disaccharides. The amino acids involved in the interaction are shown as purple sticks. Fuc, yellow; Gal, gray. G Schematic diagram of the GII.6-H disaccharide interaction. The details of the interaction of GII.6-H mediated by hydrogen bonds and hydrophobic contacts were analyzed using LIGPLOT. Hydrogen bonds, green dashed lines; hydrophobic contacts, red arcs. Black, red, and blue represent carbon, oxygen, and nitrogen atoms, respectively. H The electrostatic surface potentials of the glycan binding sites of GII.6 P dimer. GII.6 P dimers are shown in surface representation. The blue and red color indicates the positive and negative electrostatic surface potentials, respectively. The glycan binding sites are framed by the dashed line boxes. A large view of the electrostatic surface potentials of the glycan binding sites is shown on the right.
3.5. Validation of the HBGA binding interface
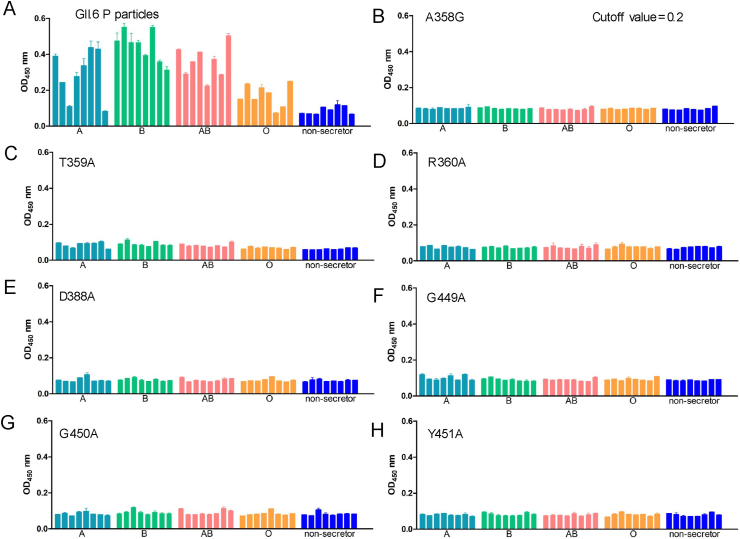
To investigate the residues that were critical for GII.6 binding to HBGAs, we constructed seven GII.6 mutants using single-point mutagenesis, except for A mutated to G at 358, other residues including T359, R360, D388, G449, G450 and Y451, which were mutated to A. The results from saliva or glycan-ELISA showed that compared to the wild-type P particle of GII.6 virus that bound to ABO secretor saliva samples and H disaccharides (Fig. 5A and Fig. 6A), no signal representing different HBGA types for the binding activity of the seven GII.6 mutants was detected (Fig. 5B–H and Fig. 6B–H). This showed that mutants with a mutation to alanine or glycine at any of these amino acids affected the overall conformation and completely prevented the binding of H disaccharide antigen and ABO secretors’ saliva. These results demonstrate that the A358, T359, R360, D388, G449, G450, and Y451 residues of GII.6 are the key amino acids required for binding to HBGAs and play an important role in the structural and functional integrity of the HBGAs binding interface.
Fig. 5.
Saliva-binding assay of wild-type and various mutant P particles with single amino acid mutations at the HBGA-binding interface of GII.6 virus. A Saliva binding of wild-type GII.6 P particles with a panel of oligosaccharides representing different HBGA, secretor ABO, and non-secretor's saliva samples. B–H Saliva binding of seven mutant P particles with single amino acid mutations at the HBGA-binding interface with the same panel of saliva as wild-type P particles. The Y axes indicate the binding signals in optical densities at OD450, while the X-axes indicate different saliva samples.
Fig. 6.
Glycan-binding assay of wild-type and various mutant P particles with single amino acid mutations at the HBGA-binding interface of GII.6 virus. A Glycan binding of wild-type GII.6 P particles with a panel of oligosaccharides representing different HBGAs. B–H Glycan binding of seven mutant P particles with single amino acid mutations at the HBGA-binding interface with the same panel of oligosaccharides. The concentrations of the P particles were 20 μg/well, while the oligosaccharides for plate coating were 0.2 μg/well. Y axes indicate the binding signals in optical densities at OD450, while the X-axes indicate different oligosaccharides.
3.6. Sequence alignment and structural comparison of GII.6 and different GII genotypes
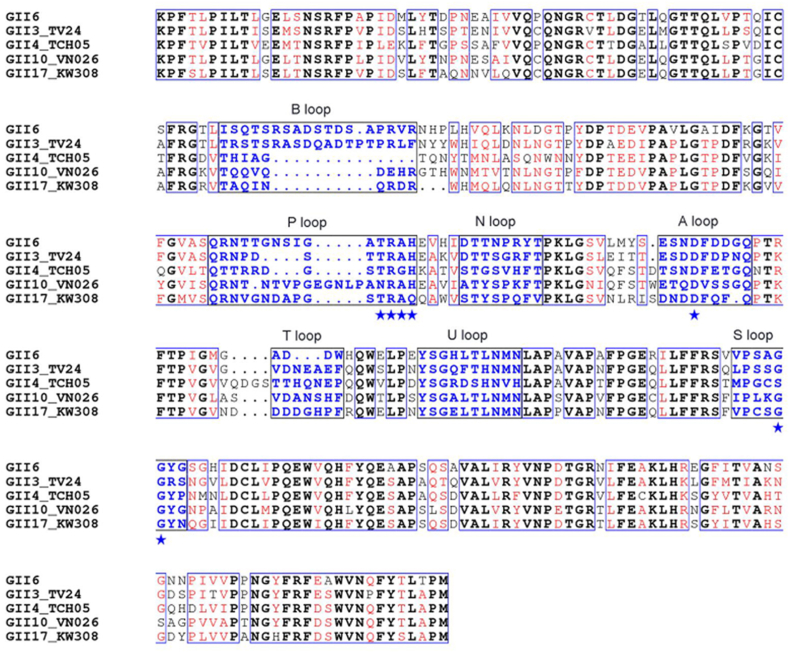
To explore similarities in sequence and structure in GII.6 and other GII genotypes, GII.6 and other four GII genotypes with well-defined P structures, including GII.3 TV24, GII.4 TCH05, GII.10 VN026, and GII.17 KW308 were aligned using structure-based sequence analysis (Fig. 7). A structure-based amino acid multiple sequence alignment of the P domain showed that GII.6 shares 66.8%, 56.4%, 64.1%, and 65.8% sequence similarity with GII.3 TV24, GII.4 TCH05, GII.10 VN026, and GII.17 KW308, respectively. The GII.6 P protein has the three conventional GII HBGA binding sites and shows structural features similar to those of GII.3 TV24, GII.4 TCH05, GII.10 VN026, and GII.17 KW308 with Cα atoms r.m.s.d. = 0.61 Å, 0.68 Å, 0.49 Å, and 0.60 Å, respectively. Overall, similar to other GII genotypes, the sequence and structure of GII.6 P domain were relatively conserved. However, significant sequence differences were observed at the top surface of the P2 subdomain. Except for GII.3 in the same branch, for the B loop, an insert of approximately nine amino acids was observed in the sequence of GII.6, whereas this loop was a disorder of flexibility where electron density was not discernible. A deletion of three amino acids was observed in the sequence of the T loop. The A, B, P, and T loops had the lowest sequence identity and structural superimposition. The N, U, and S loops of GII.6 were more conserved than those of the other GII genotypes. These differences in sequences, surface structures, and conformations of the GII.6 P dimer also contribute to the similar HBGA-binding features of other GII genotypes.
Fig. 7.
Sequence alignment of the P domain of GII.6 with other GII strains based on the secondary structure. P domains of GII.6 were aligned with GII.3 TV24, GII.4 TCH05, GII.10 VN026 and GII.17 KW308. Identical residues are black characters, while similar residues are shown in red characters. The conserved amino acid residues forming the conventional GII-HBGA binding interface are indicated by blue stars. The seven surface loops that constitute the HBGA-binding interface are indicated by blue typeface. The letters in black indicate the identical amino acid sequences, while the red letters indicate the similar amino acids among the five NoVs. Blue boxes frame both identical and similar residues.
3.7. Comparison of GII.6 and GII.17 genotypes using structure superimposition and HBGA-binding
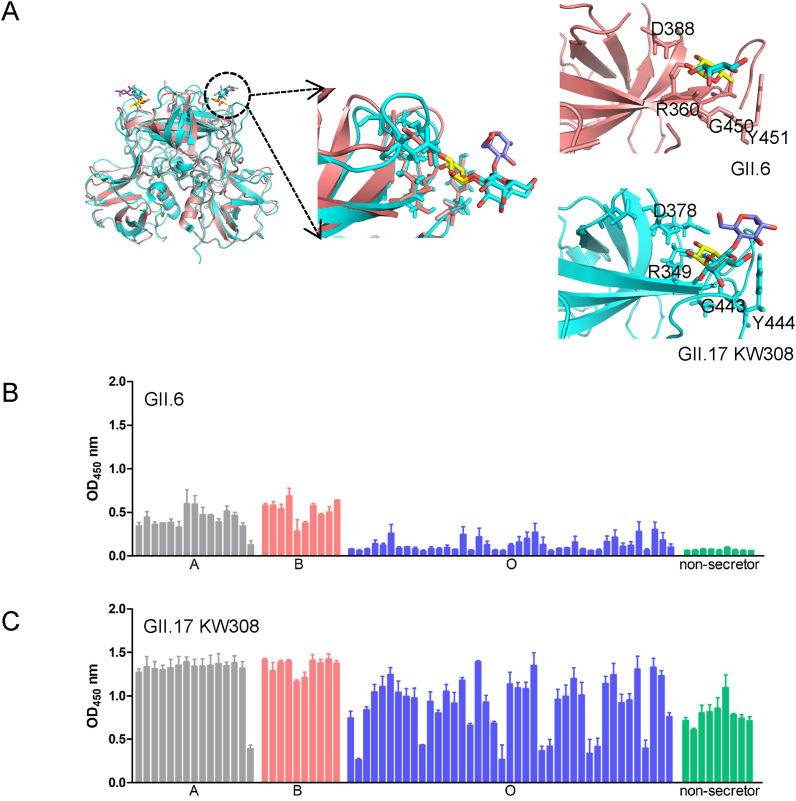
To compare the HBGA-binding ability and structural features of GII.6 and the outbreak strain, we selected GII.17 KW308 as the representative of the outbreak/epidemic strain, overlapped the structures of these two strains and compared their binding abilities through ELISA of saliva samples. Structural superimposition indicated that the binding sites of GII.6 in R360, D388, G450, and Y451 were the same as those of R349, D378, G443, and Y444 in GII.17 KW308, respectively (Fig. 8A). The saliva binding assay showed that all binding signals of GII.6 (Fig. 8B) in secretor and non-secretor's saliva samples were lower than those of GII.17 KW308 (Fig. 8C).
Fig. 8.
Structural superimposition and comparison of the saliva-binding of GII.6/GII.17. A A cartoon of structure superimposition of GII.6 and GII.17 (PDB: 5lkg). The same interaction residues of GII.6 (salmon) and GII.17 (cyan) are shown in stick in the right panel. Yellow, Fuc; Gal, cyan; Gla, purple. Black, red, and blue in glycans represent carbon, oxygen, and nitrogen atoms, respectively. B–C Saliva-ELISA assays show the binding signals of the GII.6 (B) and GII.17 (C) protein (0.5 μg/well) to type secretors (A, B, O) and non-secretor saliva samples. The experiment was repeated twice independently, and the data are mean and standard deviation (error bars) absorbance values.
4. Discussion
HBGAs act as host susceptibility factors or receptors and play an important role in the infection and prevalence of HuNoVs. A large number of studies have investigated the relationship between HBGA binding features and the prevalence of various HuNoVs, such as GII.4, GII.17, GII.2 (Tan and Jiang, 2014; Chan et al., 2015; Ao et al., 2018). Although GII.6 virus exhibits a comparatively high epidemic according to many previous studies, its binding features to HBGAs remain incompletely understood. In the present study, we characterized the binding specificity and structural features of HuNoVs GII.6 P protein binding to HBGAs. We demonstrated that GII.6 P particles selectively bind ABO secretor's saliva samples and H disaccharides, but do not bind to non-secretor's saliva samples. More importantly, we solved the crystal structures of the complex of GII.6 P protein-H disaccharide complex, which revealed that the GII.6 P protein has a conventional GII genogroup HBGA-binding interface. Using a point mutagenesis strategy, we identified seven important amino acids at the HBGA-binding interface of the GII.6 P protein that critically contribute to the interaction between the GII.6 P protein and H disaccharides. Lastly, we demonstrated that the binding capacity of GII.6 was lower than that of the outbreak strain GII.17, by comparing saliva binding features. Our findings depicted an important role for the structural features of GII.6 in recognizing HBGAs and provided a structural basis for the interaction between the GII.6 P domain and H disaccharides.
The degree and mode of salivary binding are important for NoV prevalence, as the high epidemic intensities of GII.4 and GII.17 are attributed to their strong binding capability to most ABO secretors and non-secretors (Huang et al., 2005; Jin et al., 2016). The ABO secretor types in the population are far more common than non-secretors (Storry and Olsson, 2009), and our data showed that the GII.6 P protein binds to ABO secretor's saliva samples, but does not bind to non-secretor's saliva samples. This difference may explain the relatively lower prevalence of GII.6, compared to epidemic strains. O secretor is the most common HBGA type, followed by types A and B secretors (Storry and Olsson, 2009). Here, we showed that GII.6 bind strongest to samples of B secretor, then A type, and O type the least. The degree of GII.6 recognizes to ABO secretor's saliva samples, may explain why the prevalence of GII.6 was lower than that of the outbreak strains. However, whether there are other reasons for the incomplete binding of GII.6 to ABO secretor's saliva samples needs to be further explored. One unexpected finding in this study is that GII.6 does not appear to bind to A and B oligosaccharides as is shown in previous studies (Sun et al., 2016b; Cong et al., 2019). There are two possible explanations for this finding. One is that GII.6 may indeed bind to A and B, but the binding might be too weak to be detected in the glycan-ELISA assay. The other is that saliva has a relatively complicated composition and may contain molecules that have not yet been identified. GII.6 bind to H disaccharide in glycan-ELISA, not to H1, H2, and H3 trisaccharides. H epitope exists on glycoproteins, glycolipids or secreted oligosaccharides in form of type 1/2/3/4 chains. It is also a puzzling that the protein did not recognize the trisaccharides with H antigen on type 1, 2, and 3 chains. We speculate that the possible reason is that they have weak interactions compared with the disaccharide and are not recognized. H4 may be a receptor, however, we cannot get H4 glycan from commercial company or other methods. These possibilities need to be further investigated in future studies.
Previous studies have used crystallography to clarify the binding features of various NoV strains to HBGAs and have demonstrated genome-specific binding patterns (Tan and Jiang, 2011; Cong et al., 2019; Qian et al., 2019). The binding pocket of most GII genotypes of HuNoVs that interacts with HBGAs is alpha fucose (α-Fuc), which is located at the interface of the two P monomers (Chen et al., 2011; Frenck et al., 2012; Tan and Jiang, 2014). Notably, GII.13/21 harbors a unique binding pattern, in which the binding pocket interacts with β-Gal and has a low prevalence (Cong et al., 2019). The binding pockets of the outbreak strains GII.4 and GII.17 are located between the two P monomers and mainly recognize α-Fuc (Frenck et al., 2012; Chan et al., 2015). Our data showed that GII.6 recognizes α-Fuc of HBGAs through the conventional binding pattern of GII genotypes that occur between two P monomers, similar to the epidemic strains GII.4 or GII.17. At the same time, further structure-based analysis revealed that GII.6 shares good similarity in sequence structure features and conservation of HBGA binding sites with conventional GII genotypes. Therefore, GII.6 may be the dominant epidemic strain with structural features similar to those of outbreak strains.
NoVs have many genotypes and show diversity in their recognition of HBGAs. Overall, the GII.6 P domain is relatively conserved in terms of sequence and structure compared to other GII genotypes. The N, U, and S loops of GII.6 are relatively more conserved than those of other GII genotypes. However, the A, B, P, and T loops have the least sequence identity and structural superimposition, especially in the P2 subdomain, where insertions and deletions of amino acids are found in the B and T loops, respectively. In addition, owing to the insertion of amino acids in the B loop in GII.6 and its complex structures, we did not observe electron density in these regions. Our homology modeling data showed that GII.6 and the outbreak strain GII.17 KW308 exhibit similar binding pockets and amino acids that interact with glycans, including R360, D388, G450, and Y451 in GII.6, and R349, D378, G443, and Y444 in GII.17 KW308. However, GII.6 showed a narrow and weak binding pattern compared to GII.17 KW308 in the saliva ELISA using the same samples, this difference in binding ability, strength and structural data may explain why GII.6 has not become an epidemic strain. Notably, though, the outbreak strain GII.2 has an A/B secretor's saliva binding spectrum, which is relatively narrow compared to GII.6 (Ao et al., 2018). These data also urge a focus on NoV non-structural proteins, such as polymerase, which could also serve as a driving force in causing outbreaks. HuNoVs can gain the capability to target new populations and lead to an epidemic of acute gastroenteritis or a worldwide outbreak (Chan et al., 2015; Jin et al., 2016; Ao et al., 2018). Thus, the GII.6 genotype will require continuous monitoring since structural mutations may lead to a wider range of HBGA-binding ability in epidemic genotypes.
The GII.6 P domain interacts with HBGAs through seven amino acids: A358, T359, R360, D388, G449, G450, and Y451. Our mutational binding results showed that mutations in each of the seven amino acids affect the activity of the GII.6 P domain. They inhibit the formation of hydrogen bonds or the hydrophobicity of the GII.6 P protein and HBGAs, suggesting that these amino acids are critical for the stability and integrity of HBGA-binding pockets and may also serve as an important target for structure-based drug design. Moreover, understanding the GII.6-HBGA interaction could also help with continuous monitoring and control of potential outbreaks caused by new mutant GII.6 strains.
5. Conclusions
We have comprehensively evaluated the HBGA binding specificity of the GII.6 P domain and elucidated the structural basis for the GII.6 P domain and H disaccharides, which is similar to the outbreak strains GII.17 and GII.4, and determined the conventional GII-HBGA binding interface. Moreover, we identified the interaction sites between the GII.6 P domain and HBGAs that are necessary for the P protein to recognize HBGAs. Our findings expand the understanding of NoV host interaction, evolution, and epidemiology, which may facilitate the development of strategies for the control and prevention of HuNoVs.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The structures of GII.6 and GII.6-H P domains have been deposited under PDB codes 7YQB and 7YQG, respectively.
Ethics statement
The saliva samples for this research were approved by the meeting of ethics committee of the National Institute for Viral Disease Control and Prevention, China CDC.
Author contributions
Xin Cong: conceptualization, methodology, writing-original draft, writing-review and editing. Hanbo Li: data curation, methodology, writing-original draft. Xiaoman Sun: methodology. Jianxun Qi: data Curation. Qing Zhang: methodology. Zhaojun Duan, Yong Xu and Wenlan Liu: conceptualization, writing-review and editing.
Conflict of interest
The authors declared no conflict of interests.
Acknowledgements
We acknowledge George F. Gao at the Institute of Microbiology, Chinese Academy of Sciences, for his assistance in the X-ray data collection and processing. This research was supported by grants from the National Natural Science Foundation of China (no. 32100111, 21934005), Guangdong Basic and Applied Basic Reuter Foundation (no. 2019A1515110220), China Postdoctoral Science Foundation (no. 2020M682900) and Shenzhen High-level Hospital Construction Fund.
Contributor Information
Zhao-jun Duan, Email: zhaojund@126.com.
Yong Xu, Email: xuyong_2000@tom.com.
Wen-lan Liu, Email: wlliu@szu.edu.cn.
References
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., Mccoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C., Richardson J.S., Terwilliger T.C., Zwart P.H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S.M., Hall A.J., Robinson A.E., Verhoef L., Premkumar P., Parashar U.D., Koopmans M., Lopman B.A. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect. Dis. 2014;14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Y., Cong X., Jin M., Sun X., Wei X., Wang J., Zhang Q., Song J., Yu J., Cui J., Qi J., Tan M., Duan Z. Genetic analysis of reemerging GII.P16-GII.2 Noroviruses in 2016-2017 in China. J. Infect. Dis. 2018;218:133–143. doi: 10.1093/infdis/jiy182. [DOI] [PubMed] [Google Scholar]
- Ao Y., Wang J., Ling H., He Y., Dong X., Wang X., Peng J., Zhang H., Jin M., Duan Z. Norovirus GII.P16/GII.2–Associated gastroenteritis, China, 2016. Emerg. Infect. Dis. 2017;23:1172–1175. doi: 10.3201/eid2307.170034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Lou Z., Tan M., Chen Y., Liu Y., Zhang Z., Zhang X.C., Jiang X., Li X., Rao Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan M.C., Lee N., Hung T.N., Kwok K., Cheung K., Tin E.K., Lai R.W., Nelson E.A., Leung T.F., Chan P.K. Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat. Commun. 2015;6 doi: 10.1038/ncomms10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Tan M., Xia M., Hao N., Zhang X.C., Huang P., Jiang X., Li X., Rao Z. Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra P., De Graaf M., Parra G.I., Chan M.C., Green K., Martella V., Wang Q., White P.A., Katayama K., Vennema H., Koopmans M.P.G., Vinje J. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019;100:1393–1406. doi: 10.1099/jgv.0.001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X., Sun X.M., Qi J.X., Li H.B., Chai W.G., Zhang Q., Wang H., Kong X.Y., Song J., Pang L.L., Jin M., Li D.D., Tan M., Duan Z.J. GII.13/21 Noroviruses recognize glycans with a terminal beta-galactose via an unconventional glycan binding site. J. Virol. 2019;93:e00723–19. doi: 10.1128/JVI.00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rougemont A., Ruvoen-Clouet N., Simon B., Estienney M., Elie-Caille C., Aho S., Pothier P., Le Pendu J., Boireau W., Belliot G. Qualitative and quantitative analysis of the binding of GII.4 norovirus variants onto human blood group antigens. J. Virol. 2011;85:4057–4070. doi: 10.1128/JVI.02077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenck R., Bernstein D.I., Xia M., Huang P., Zhong W., Parker S., Dickey M., Mcneal M., Jiang X. Predicting susceptibility to norovirus GII.4 by use of a challenge model involving humans. J. Infect. Dis. 2012;206:1386–1393. doi: 10.1093/infdis/jis514. [DOI] [PubMed] [Google Scholar]
- Huang P., Farkas T., Zhong W., Tan M., Thornton S., Morrow A.L., Jiang X. Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 2005;79:6714–6722. doi: 10.1128/JVI.79.11.6714-6722.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y., Zheng L., Chen X., Ge L., Wang Y. Expression and characterization of the major capsid protein derived from a GII.6 norovirus strain isolated in China. Microb. Pathog. 2017;105:131–137. doi: 10.1016/j.micpath.2017.02.025. [DOI] [PubMed] [Google Scholar]
- Jin M., Wu S., Kong X., Xie H., Fu J., He Y., Feng W., Liu N., Li J., Rainey J.J., Hall A.J., Vinje J., Duan Z. Norovirus outbreak surveillance, China, 2016-2018. Emerg. Infect. Dis. 2020;26:437–445. doi: 10.3201/eid2603.191183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M., Zhou Y.K., Xie H.P., Fu J.G., He Y.Q., Zhang S., Jing H.B., Kong X.Y., Sun X.M., Li H.Y., Zhang Q., Li K., Zhang Y.J., Zhou D.Q., Xing W.J., Liao Q.H., Liu N., Yu H.J., Jiang X., Tan M., Duan Z.J. Characterization of the new GII.17 norovirus variant that emerged recently as the predominant strain in China. J. Gen. Virol. 2016;97:2620–2632. doi: 10.1099/jgv.0.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pendu J. Histo-blood group antigen and human milk oligosaccharides: genetic polymorphism and risk of infectious diseases. Adv. Exp. Med. Biol. 2004;554:135–143. doi: 10.1007/978-1-4757-4242-8_13. [DOI] [PubMed] [Google Scholar]
- Leshem E., Wikswo M., Barclay L., Brandt E., Storm W., Salehi E., Desalvo T., Davis T., Saupe A., Dobbins G., Booth H.A., Biggs C., Garman K., Woron A.M., Parashar U.D., Vinje J., Hall A.J. Effects and clinical significance of GII.4 Sydney norovirus, United States, 2012-2013. Emerg. Infect. Dis. 2013;19:1231–1238. doi: 10.3201/eid1908.130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindesmith L., Moe C., Marionneau S., Ruvoen N., Jiang X., Lindblad L., Stewart P., Lependu J., Baric R. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 2003;9:548–553. doi: 10.1038/nm860. [DOI] [PubMed] [Google Scholar]
- Morozov V., Hanisch F.G., Wegner K.M., Schroten H. Pandemic GII.4 Sydney and epidemic GII.17 Kawasaki308 Noroviruses display distinct specificities for histo-blood group Antigens leading to different transmission vector dynamics in Pacific oysters. Front. Microbiol. 2018;9:2826. doi: 10.3389/fmicb.2018.02826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Song M., Jiang X., Xia M., Meller J., Tan M., Chen Y., Li X., Rao Z. Structural adaptations of Norovirus GII.17/13/21 lineage through two distinct evolutionary paths. J. Virol. 2019;93:e01655–18. doi: 10.1128/JVI.01655-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydell G.E., Nilsson J., Rodriguez-Diaz J., Ruvoen-Clouet N., Svensson L., Le Pendu J., Larson G. Human noroviruses recognize sialyl Lewis x neoglycoprotein. Glycobiology. 2009;19:309–320. doi: 10.1093/glycob/cwn139. [DOI] [PubMed] [Google Scholar]
- Singh B.K., Leuthold M.M., Hansman G.S. Structural constraints on human Norovirus binding to histo-blood group Antigens. mSphere. 2016;1 doi: 10.1128/mSphere.00049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Takai-Todaka R., Miki M., Haga K., Fujimoto A., Ishiyama R., Oikawa K., Yokoyama M., Miyazaki N., Iwasaki K., Murakami K., Katayama K., Murata K. Dynamic rotation of the protruding domain enhances the infectivity of norovirus. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storry J.R., Olsson M.L. The ABO blood group system revisited: a review and update. Immunohematol. 2009;25:48–59. [PubMed] [Google Scholar]
- Sun C., Schattgen S.A., Pisitkun P., Jorgensen J.P., Hilterbrand A.T., Wang L.J., West J.A., Hansen K., Horan K.A., Jakobsen M.R., O'hare P., Adler H., Sun R., Ploegh H.L., Damania B., Upton J.W., Fitzgerald K.A., Paludan S.R. Evasion of innate cytosolic DNA sensing by a gammaherpesvirus facilitates establishment of latent infection. J. Immunol. 2015;194:1819–1831. doi: 10.4049/jimmunol.1402495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Guo N., Xu Z., Li D., Duan Z.J. P[14] Rotavirus VP8∗ protein specifically binds to A type histo-blood group antigen. Chin. J. Exp. Clin. Virol. 2016;30:315–318. [Google Scholar]
- Sun X., Li D., Peng R., Guo N., Jin M., Zhou Y., Xie G., Pang L., Zhang Q., Qi J., Duan Z.J. Functional and structural characterization of P[19] rotavirus VP8∗ interaction with histo-blood group Antigens. J. Virol. 2016;90:9758–9765. doi: 10.1128/JVI.01566-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Hegde R.S., Jiang X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 2004;78:6233–6242. doi: 10.1128/JVI.78.12.6233-6242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Jiang X. Norovirus and its histo-blood group antigen receptors: an answer to a historical puzzle. Trends Microbiol. 2005;13:285–293. doi: 10.1016/j.tim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Tan M., Jiang X. Norovirus–host interaction: multi-selections by human histo-blood group antigens. Trends Microbiol. 2011;19:382–388. doi: 10.1016/j.tim.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Jiang X. Norovirus P particle: a subviral nanoparticle for vaccine development against norovirus, rotavirus and influenza virus. Nanomedicine. 2012;7:889–897. doi: 10.2217/nnm.12.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M., Jiang X. Histo-blood group antigens: a common niche for norovirus and rotavirus. Expet Rev. Mol. Med. 2014;16:e5. doi: 10.1017/erm.2014.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request. The structures of GII.6 and GII.6-H P domains have been deposited under PDB codes 7YQB and 7YQG, respectively.