Abstract
Crimean-Congo hemorrhagic fever virus (CCHFV) is a biosafety level-4 (BSL-4) pathogen that causes Crimean-Congo hemorrhagic fever (CCHF) characterized by hemorrhagic manifestation, multiple organ failure and high mortality rate, posing great threat to public health. Despite the recently increasing research efforts on CCHFV, host cell responses associated with CCHFV infection remain to be further characterized. Here, to better understand the cellular response to CCHFV infection, we performed a transcriptomic analysis in human kidney HEK293 cells by high-throughput RNA sequencing (RNA-seq) technology. In total, 496 differentially expressed genes (DEGs), including 361 up-regulated and 135 down-regulated genes, were identified in CCHFV-infected cells. These regulated genes were mainly involved in host processes including defense response to virus, response to stress, regulation of viral process, immune response, metabolism, stimulus, apoptosis and protein catabolic process. Therein, a significant up-regulation of type III interferon (IFN) signaling pathway as well as endoplasmic reticulum (ER) stress response was especially remarkable. Subsequently, representative DEGs from these processes were well validated by RT-qPCR, confirming the RNA-seq results and the typical regulation of IFN responses and ER stress by CCHFV. Furthermore, we demonstrate that not only type I but also type III IFNs (even at low dosages) have substantial anti-CCHFV activities. Collectively, the data may provide new and comprehensive insights into the virus-host interactions and particularly highlights the potential role of type III IFNs in restricting CCHFV, which may help inform further mechanistic delineation of the viral infection and development of anti-CCHFV strategies.
Keywords: Crimean-Congo hemorrhagic fever virus (CCHFV), Biosafety level-4 (BSL-4) pathogen, Bunyavirus, Transcriptome, Type III interferon, ER stress, Virus-host interaction, Antiviral therapeutics
Highlights
-
•
Transcriptomic analysis of CCHFV-infected HEK293 cells identified 496 DEGs.
-
•
The DEGs were involved in host processes including defense response, stress response, metabolism, etc.
-
•
Therein, a significant up-regulation of type III IFN pathway and ER stress response was highlighted and validated by qPCR.
-
•
Antiviral assays showed that type III IFNs (even at low dosages) have substantial anti-CCHFV activities.
1. Introduction
Crimean Congo hemorrhagic fever virus (CCHFV), a member of the Orthonairovirus genus in Nairoviridae family of Bunyavirales order, is a tick-born virus classified as biosafety level-4 (BSL-4) pathogen (Abudurexiti et al., 2019). As one of the most dangerous pathogens, CCHFV infection in humans can cause a severe acute infectious disease called Crimean-Congo hemorrhagic fever (CCHF) with case mortality rates of up to 60% (Hawman and Feldmann, 2018). Routes of CCHFV transmission mainly include tick (particularly Hyalomma spp.) bites and human-to-human or animal-to-human contacts (Bente et al., 2013). CCHFV spreads widely in about 50 countries throughout Africa, Asia and Europe with more than 1000 cases reported every year (Leblebicioglu et al., 2017; Nasirian, 2020). However, no FDA-approved antiviral drugs or vaccines are currently available (Dai S. Y. et al., 2021b). Moreover, the knowledge regarding CCHFV-host interactions is poorly understood. Thus, CCHFV has been included in the list of the top 10 priority pathogens urgently requiring research and development efforts by the World Health Organization (WHO) (Mazzola and Kelly-Cirino, 2019).
CCHFV is tri-segmented RNA virus with a genome containing large (L), medium (M) and small (S) segment. The L segment codes RNA-dependent RNA polymerase (RdRp) and S codes nucleocapsid protein (N) as well as non-structural protein (NSs), respectively, while the M segment encodes envelope glycoproteins (Gn/Gc) and non-structural glycoproteins (Bente et al., 2013). During viral infection, host often initiates defense responses such as immune response, cell stress response as well as apoptosis for protection. Likewise, upon CCHFV infection, several cellular or viral factors were reported to modulate the viral infection or host response by involvement in virus-host interactions. The cytoskeleton component actin and cellular chaperon heat shock protein 70 family may interact with CCHFV N protein and modulate virus replication (Surtees et al., 2016). Host aquaporin 6, a water channel facilitating fluxes of water and small solutes across membranes, showed possible protective role upon CCHFV challenge (Molinas et al., 2016). On the other hand, CCHFV may have evolved strategies conflicting with some cellular responses. One example is that CCHFV may delay type I interferon (IFN) pathway possibly by interfering with the activation of IFN-regulatory factor 3 (IRF-3) (Andersson et al., 2008). Besides, the OTU domain-containing RdRp of CCHFV showed deconjugating activity toward ubiquitinated and ISGylated proteins, thus inhibiting Ub and ISG15 dependent innate immunity pathway (Frias-Staheli Natalia et al., 2007a). Additionally, the NSs protein of CCHFV was demonstrated to induce apoptosis by both mitochondrial death pathway and death receptor pathway (Barnwal et al., 2016). In spite of these studies, the global profile of host cell response upon the virus infection remains to be further investigated.
RNA sequencing (RNA-Seq) technology is a recently developed high-throughput approach which enables us to get comprehensive measurement of host transcriptome upon virus infection (Stark et al., 2019). In this study, we analyzed the transcriptome profile of CCHFV-infected human kidney HEK293 cells by RNA-seq. A total of 496 differentially expressed genes (DEGs) following CCHFV infection were identified, with 361 up-regulated and 135 down-regulated. Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses showed that most of the DEGs were involved in the processes concerning defense to virus (particularly type III IFN pathways) and response to stimulus (including ER-stress response). Further, the regulation of transcripts related to IFN responses and ER stress was validated by reverse transcription quantitative PCR (RT-qPCR), corroborating the RNA-seq results and the typical stimulation of the biological processes by CCHFV infection. Moreover, antiviral testing demonstrated the substantial inhibitory effects of not only type I but also type III IFNs against CCHFV replication. Collectively, this study provides a new global cellular transcriptional profile of CCHFV infection and broadens the understanding of the CCHFV-host interactions, which perhaps will benefit the development of antiviral intervention strategies.
2. Materials and methods
2.1. Cell and virus
HEK293 (human embryonic kidney) and Vero (African green monkey kidney) cells were cultured in minimum Eagle's medium (MEM) supplemented with 10% fetal bovine serum (FBS) at 37 °C under 5% CO2. Huh7 (human liver cancer cells), HUVEC (human umbilical vein endothelial cells), and A549 (adenocarcinomic human alveolar basal epithelial cells) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. CCHFV (strain YL16070, CSTR: 16533.06. IVCAS 6.6329) was propagated in the monolayer of Vero cells as described previously (Guo et al., 2017; Dai S. et al., 2021a). The titer was determined in Vero cells by 50% tissue culture infectious dose (TCID50) method (Ning et al., 2017; Min Yuan-Qin et al., 2020a; Dai S. et al., 2021a). All CCHFV infection experiments were conducted in the National High-level Biosafety Laboratory, Wuhan, Chinese Academy of Sciences.
2.2. CCHFV infection and sample preparation
HEK293 cells with 60% confluence were seeded into 6-well plate, followed by mock or CCHFV (0.3 MOI) incubation at 4 °C for 1 h. After 3 times of washing with PBS, the cells were incubated at 37 °C with complete medium for 24 h. Then the cells were lysed with TRIzol for the following RNA extraction. Three biological replicates were prepared for each group. RNA qualities were evaluated with labchip and Nanodrop.
2.3. Generation of cDNA library and sequencing
Polyadenylated mRNAs were enriched by magnetic oligo (dT) beads from high qualified RNA prepared above. The first strand cDNA was synthetized using random primers, followed by conversion into double stranded cDNA and purification with AMPure XP beads. After poly (A) repair and 5′-adaptor ligation, the second cDNA containing uracil (U) was digested by USER enzyme for the preparation of specific cDNA library following quality evaluation with Qubit3.0 and size detection with Qsep100. Library sequencing was conducted on Illumina HiSeqTM2000 by following the manufacturer's instructions.
2.4. Sequence analysis and annotation
RNA sequencing quality analysis was performed using FastQC. Clean reads were extracted from raw reads using Fastp V0.20.0 and indexed to homo sapiens genome Homo_sapiens.GRCh38.98 as reference. By using Hisat2, the clean reads were mapped to the reference genome. Analysis of gene expression levels was calculated using the fragments per kilobase of exon per million mapped reads (FPKM) method.
2.5. DEG analysis and functional enrichment analysis
DEG analysis between mock- and CCHFV-infected samples was based on the gene transcription level by DESeq2 using Negative Binomial distribution as model for read counts. DEGs were identified based on the criterions that |log2 (fold change) | > 1 and P-value < 0.05. Identified DEGs were summarized in Supplementarty Table S1 and S2. GO analysis for DEGs were performed based on GO databases and KEGG databases using the R package clusterProfiler. Functional enrichment analysis was performed by using PANTHER16.0. Dotplot of DEGs from top 15 significant GO terms categorized into biological process and cellular component were presented by R package ggplot2. Heatmap of DEGs was shown by TBtools. Hypergeometric distribution test was used to calculate the enrichment for GO terms and KEGG pathways and p-value less than 0.05 was considered significantly enriched.
2.6. Validation of DEGs by RT-qPCR
Representative DEGs (IFIT1, IFITM1, ISG15, MXA, OAS1, IFN-β, IFNλ1, IFNλ2, IFNλ3, HSPA5 and XBP1) were confirmed by quantification of relative mRNA levels with human GAPDH mRNA levels serving as control. Briefly, the samples were prepared the same as those used for sequencing with three biological repeats. Total RNAs were extracted with TRIzol reagent followed by cDNA synthesis with PrimeScript™ RT reagent Kit with gDNA Eraser kit (TaKaRa). Then, quantitative real-time PCR analyses were performed as described previously (Ning et al., 2015; Ning et al., 2019; Min Y. Q. et al., 2020b; Mo et al., 2020; Feng et al., 2021). Primers used in the study were listed in Supplementarty Table S3. Note that due to the high sequence identities of IFNλ2 (IL-28A) and IFNλ3 (IL-28B/C), a pair of primers targeting both were used to detect the total IL-28 mRNAs in qPCR. Relative mRNA levels were calculated by 2-ΔΔCT method (Livak and Schmittgen, 2001).
2.7. Measurement of anti-CCHFV activities of IFNs
The following IFNs were used in anti-CCHFV testing: recombinant human IFN-α (300-02AA), IFN-β (300–02BCE), and IFNλ1 (300-02L) were all purchased from Peprotech. IL-28B/IFNλ3 (5259-IL-025) was purchased from R&D Systems.
HEK293 cells (∼4 × 105 cells/well) were seeded into 24-well plates overnight. Then the cells were pretreated with increasing amounts of either IFNλ1 (0, 1, 10, 100 ng/mL) or IFNλ3 (0, 1, 10, 100 ng/mL), or with IFN-α (100 U/mL) or IFN-β (100 ng/mL). After 24 h, the IFN-containing culture medium was removed, followed by CCHFV infection (0.1 MOI) in fresh medium without IFNs. At 24 h post-infection (hpi), cells were lysed with TRIzol for RNA extraction. Viral RNAs and representative ISGs (OAS1, ISG15, MxA, IFIT1 and IFITM1) were respectively quantified by qRT-PCR with 2-ΔΔCT method. Similarly, anti-CCHFV activities of IFNλ1 or IFNλ3 (10 ng/mL) were also measured in Huh7 cells.
2.8. Quantification and statistical analysis
Graphs were drawn by GraphPad Prism 8 software. All data are shown as means ± SD. Statistical analyses were performed using SPSS19.0 with Student's t-test for the comparison between two groups. Differences were considered statistically significant if P-value < 0.05. Significance levels are: ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
3. Results
3.1. RNA-seq data analysis
To identify the host transcriptome profile of CCHFV infection, mRNAs extracted from mock-infected or CCHFV-infected HEK293 cells in three independent experiments were subjected to sequencing on the Illumina HiSeqTM2000 platform. Totally, 159, 414, 742 and 169, 649, 726 raw sequencing reads were obtained from the mock and infected samples, respectively. After quality analysis of the sequencing results using FastQC by removing low-quality sequences. We obtained clean reads 158, 701, 660 in mock groups and 168, 830, 014 in infected groups, with the average clean Q20 and Q30 rates were 98.15% and 94.40%, respectively (Table 1). All of the reads were mapped to the homo sapiens genome by Hisat2 and the average mapped read rate was 97.09%, in which the non-unique mapped read rate and unique mapped read rate were 8.78% and 91.22%, respectively (Table 2), indicating high quality of the sequencing data.
Table 1.
Summary of RNA-seq data.
| Samples | Raw reads | Clean reads | Clean rate (%) | Q20 (%) | Q30 (%) | GC content (%) |
|---|---|---|---|---|---|---|
| M1 | 55,308,480 | 55,068,456 | 99.57 | 98.21 | 94.47 | 45.91 |
| M2 | 48,780,100 | 48,544,258 | 99.52 | 97.96 | 94.01 | 45.31 |
| M3 | 55,326,162 | 55,088,946 | 99.57 | 98.33 | 94.75 | 46.22 |
| V1 | 56,045,550 | 55,760,164 | 99.49 | 97.97 | 94.05 | 45.49 |
| V2 | 60,323,304 | 60,001,316 | 99.47 | 98.02 | 94.18 | 45.48 |
| V3 | 53,280,872 | 53,068,534 | 99.60 | 98.38 | 94.91 | 46.66 |
M1, M2, and M3: three biological replicates of the mock-infected samples; V1, V2, and V3: three biological replicates of the CCHFV-infected samples.
Table 2.
Alignment of RNA-seq reads between CCHFV-infected and mock-infected samples.
| Samples | Total reads | Mapped reads | Non-unique Mapped reads |
Unique Mapped reads |
|---|---|---|---|---|
| M1 | 55,068,456 (100%) | 53,528,015 (97.20%) | 4,580,558 (8.56%) | 48,947,457 (91.44%) |
| M2 | 48,544,258 (100%) | 47,126,425 (97.08%) | 4,116,553 (8.74%) | 43,009,872 (91.26%) |
| M3 | 55,088,946 (100%) | 53,595,907 (97.29%) | 4,690,044 (8.75%) | 48,905,863 (91.25%) |
| V1 | 55,760,164 (100%) | 54,031,155 (96.90%) | 4,833,614 (8.95%) | 49,197,541 (91.05%) |
| V2 | 60,001,316 (100%) | 58,162,671 (96.94%) | 5,194,280 (8.93%) | 52,968,391 (91.07%) |
| V3 | 53,068,534 (100%) | 51,556,131 (97.15%) | 4,511,142 (8.75%) | 47,044,989 (91.25%) |
M1, M2, and M3: three biological replicates of the mock-infected samples; V1, V2, and V3: three biological replicates of the CCHFV-infected samples.
3.2. DEGs upon CCHFV infection
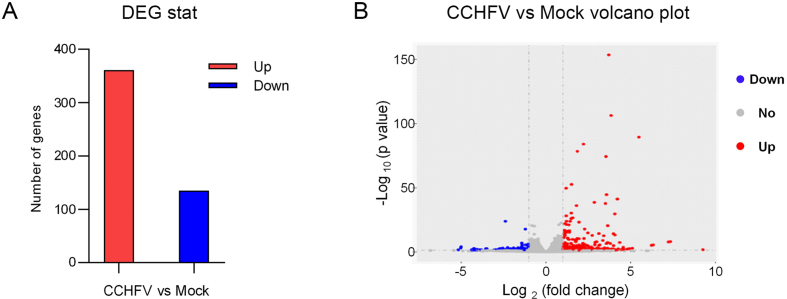
Gene expression levels were evaluated by read number mapping to each gene with featureCounts V1.5.0. Then the fragments per kilobase of exon per million mapped reads (FPKM) method was used to analysis the gene expression levels. The analysis of DEGs was performed with DESeq2 using Negative Binomial distribution as models for read counts. DEGs were identified following the criterions with fold change >2, P-value < 0.05 and biological replicates ≥ 3. Finally, a total of 496 DEGs with 361 promoted DEGs and 135 suppressed DEGs were identified in CCHFV-infected cells (Fig. 1A and B, Supplementary Table S1 and S2). It should be pointed out that among the 496 DEGs, only 366 genes were noted with name while the left 130 ones were unnoted. The fold changes in upregulated genes ranged from 614-folds to 2-folds and the downregulated genes changed from 36-folds to 2-folds. Among the upregulated genes, 135 genes were increased about 4 folds, and 74 genes were upregulated ∼8 folds. As for the downregulated genes, the mRNA expression levels of 51 genes were decreased over 4 times.
Fig. 1.
Analysis of differentially expressed genes (DEGs) in mock- and CCHFV-infected samples. A Number of DEGs between CCHFV-treated and mock groups. Red represents up-regulated genes and blue represents down-regulated genes. The y-axis indicates the number of genes. B Volcano plots of the distribution of DEGs. The x-axis indicates the Log2(fold change) of DEGs. The y-axis represents the statistical significance, -Log10(p-value). Red, blue, and gray dots represent up-regulated genes, down-regulated genes, and not significantly regulated genes, respectively. Dashed lines indicate the cutoff values, |log2(fold change)| = 1 and P = 0.05.
3.3. GO and KEGG pathway enrichment and interaction network analyses based on the identified DEGs reveal important aspects of CCHFV infection biology
To get a whole picture of the DEGs during CCFHV infection, regulated host genes were firstly categorized by the classification of molecular function, biological process, and cellular component, according to the GO enrichment analysis with PANTHER.
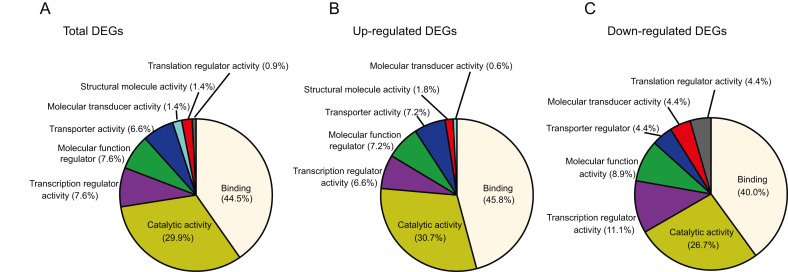
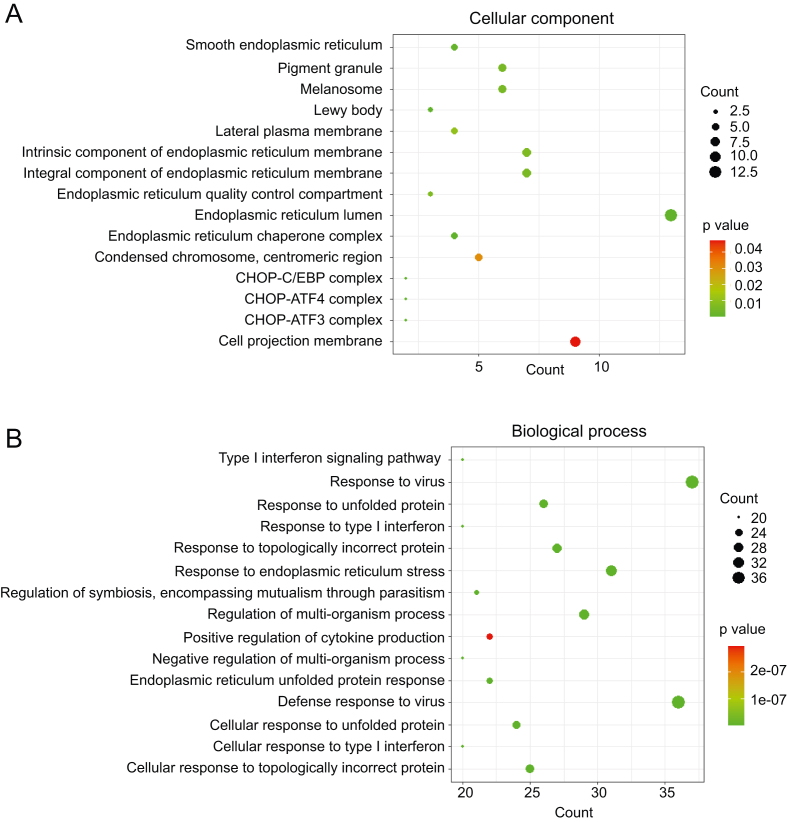
The primary molecular functions of the DEGs are mainly enriched into binding (44.5%), catalytic activity (29.9%), transcription regulator activity (7.6%), molecular function regulation (7.6%), transporter activity (6.6%), molecular transducer activity (1.4%), and structural molecule activity (1.4%) (Fig. 2A). The distribution patterns of the up-regulated or down-regulated genes are similar to those of the whole DEGs (Fig. 2B and C). As for the cellular component, DEGs were roughly related to endoplasmic reticulum chaperone complex, melanosome, pigment granule, lewy body core and protein complexes (Fig. 3A and Fig. 4). Based on the biological processes, the GO terms were mainly summarized into the following categories, namely, defense response to virus, response to stress, interferon signaling pathway, regulation of viral process, immune response, metabolism, stimulus, apoptosis and protein catabolic process (Figs. 3B and 4).
Fig. 2.
Molecular function analysis of DEGs. DEGs were submitted to PANTHER to analyze the molecular functional distribution. The total (A), up-regulated (B), and down-regulated DEGs (C) categorized according to the molecular function are shown. Color represents different functions and the area size is positively correlated to the proportion of the DEGs with indicated functions.
Fig. 3.
Dotplot of GO enrichment analysis based on the identified DEGs. Top 15 significant GO terms categorized into cellular components (A) and biological processes (B) are respectively shown. The y-axis indicates GO terms and the x-axis represents the counts of DEGs mapping to the corresponding GO terms.
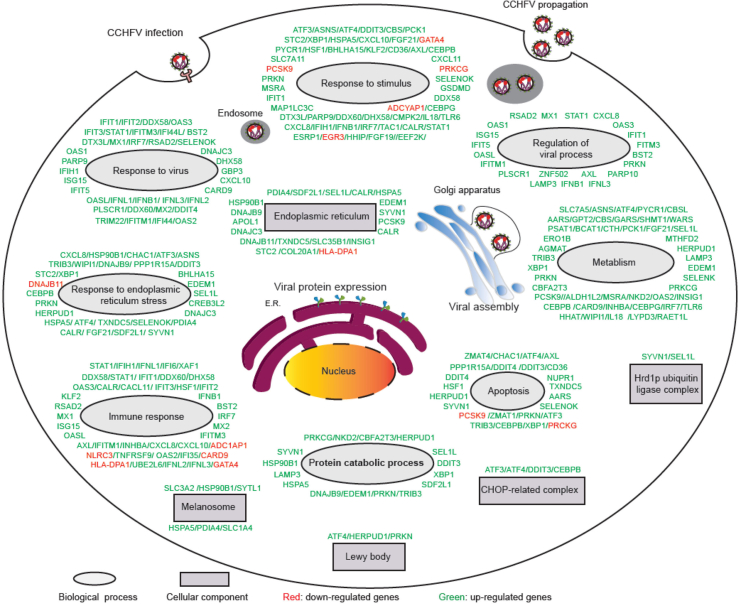
Fig. 4.
Global view of DEGs regulated by CCHFV infection. The DEGs were categorized according to the biological process and cellular component enrichment analyses and respectively illustrated in the cell model for global view presentation. The virus-cell interplays could take place in various steps of the viral life cycle, such as entry, replication/transcription, viral protein production, assembly, and release.
Furthermore, KEGG analysis on the DEGs revealed that innate immune antiviral signaling pathways, protein processing in endoplasmic reticulum, cysteine and methionine metabolism, and viral related infections ranked at top 15 significantly enriched pathways (Supplementary Fig. S1).
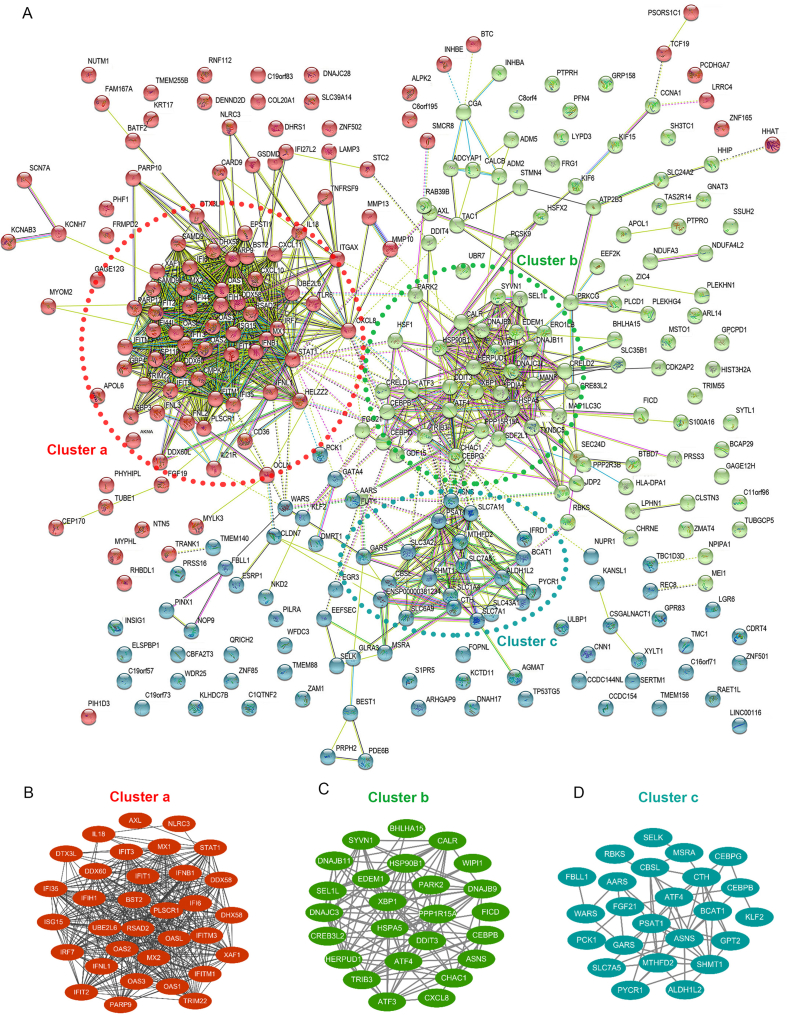
To view the potential connection network of the DEGs, the experimentally determined and predicted interactions were analyzed by submitting the DEGs into STRING database and visualized by Cytoscape (Fig. 5). In total, 295 nodes with 1324 interactions (edges) are represented in the protein-protein interaction (PPI) network (Fig. 5A). Consistent with the GO and KEGG analyses, most of the DEGs in the interaction network could be clustered into three categories, i.e., the IFN immune pathway, responses to stimuli (especially ER-stress response), and cellular metabolism (Fig. 5B–D), indicating that these are likely the significant aspects of CCHFV infection and CCHFV-cell interplays. Therefore, they are further elucidated in the following.
Fig. 5.
Protein-protein interaction analysis of DEGs. A Potential protein-protein interaction network was analyzed by STRING. Nodes represent the proteins and the biological relationship between two nodes is represented as an edge. Genes/proteins involved in the following biological processes, IFN pathway (Cluster a), ER-stress (Cluster b) and metabolism (Cluster c), are notably clustered in the interaction network and further illustrated in (B), (C), and (D), respectively.
3.3.1. IFN signaling pathways induced by CCHFV
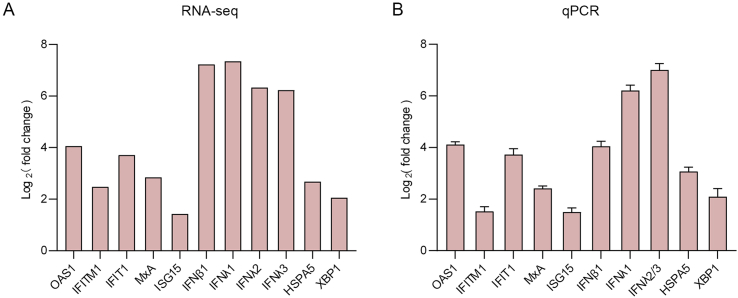
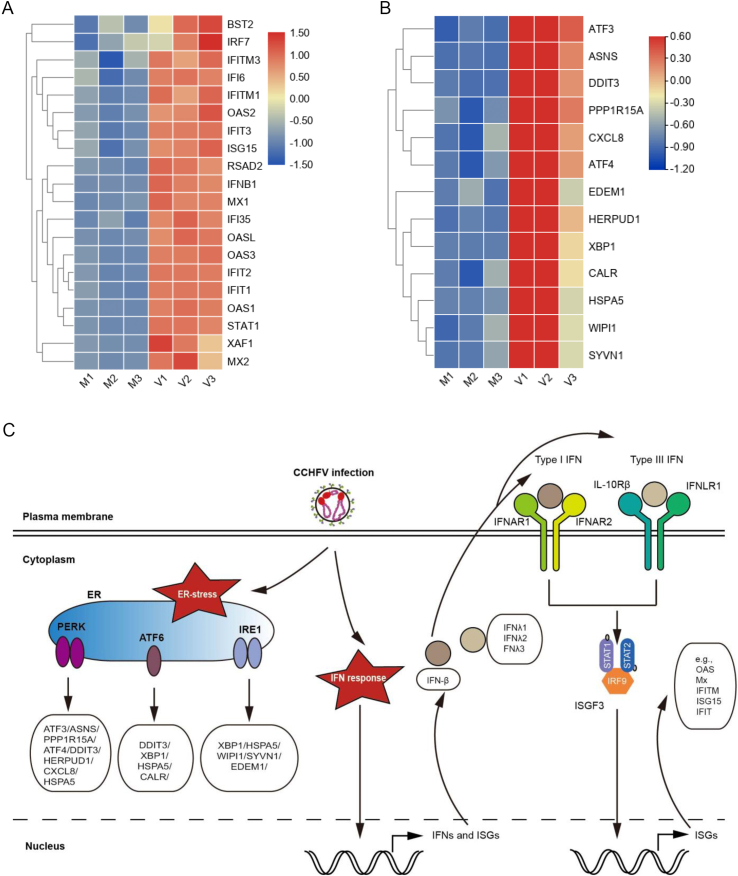
The antiviral IFN system plays pivotal roles in protecting the host from virus infection and pathogenicity. Although both type I and III IFNs are ubiquitously expressed and recognized as “antiviral IFNs”, type I IFNs contain many members including 13 subtypes of IFN-α, one type of IFN-β, as well as IFN-ε, IFN-ω, etc., while type III IFNs consists of only four homologs, IFNλ1, IFNλ2, IFNλ3 and IFNλ4. Type I IFNs (especially IFN-β and IFN-α) are extensively induced by numerous viruses (Stanifer et al., 2019); however, the reports on induction of type III IFNs by viruses are relatively limited. In the present transcriptome profile of CCHFV infection, one type I IFN, IFN-β (∼150.4 folds), and intriguingly, three type III IFNs, IFNλ1 (∼163.0 folds), IFNλ2 (∼80.5 folds) and IFNλ3 (∼75.1 folds), were dramatically stimulated (as top 5 up-regulated genes) (Supplementarty Table S1). Moreover, many IFN-induced ISGs, such as MxA (∼7.2 folds), ISG15 (∼2.7 folds), OAS1 (∼16.7 folds), IFIT1 (∼13.1 folds) and IFITM1 (∼5.6 folds) were also significantly elicited by CCHFV infection (Supplementarty Table S1, Fig. 7A). These findings suggest that CCHFV triggers robust antiviral IFN (especially type III IFN) responses and ISG expression which, in turn, would restrict viral replication and spread. Consistent with the GO, KEGG and PPI analyses, the data indicate that innate immune responses mediated by IFNs and particularly the type III IFNs are likely the pivotal aspect of virus-host interactions upon CCHFV infection (Fig. 6C).
Fig. 7.
Validation of representative DEGs involved in the IFN antiviral pathways and ER-stress responses by RT-qPCR. A RNA-seq results of DEGs related to IFN response and ER stress. B Induction of the representative DEGs were verified by RT-qPCR. HEK293 cells infected with CCHFV or mock infected were subjected to RT-qPCR analysis for the relative expression of the indicated DEGs at 24 h post infection (hpi). GAPDH was used as an internal control.
Fig. 6.
CCHFV-stimulated IFN response and ER-stress pathways. Heatmap of DEGs related to IFN responses (A) and ER-stress (B) upon CCHFV infection were clustered and visualized by TBtools. C A model depicting the three branches of ER-stress and the two phases of IFN responses (IFN induction and IFN signaling) stimulated by CCHFV.
3.3.2. Response to stimulus upon CCHFV infection
Based on the transcriptome profile obtained here, CCHFV infection likely induced notable ER stress, oxidative stress, and biotic stimuli. As an important cellular organelle, endoplasmic reticulum (ER) is the major site responsible for protein synthesis, folding and sorting. The unfolded protein response (UPR) would be activated by ER homeostasis disturbance such as ER stress (Jheng et al., 2014). In our results, following CCHFV infection, X box binding protein 1 (Xbp-1), a marker of inositol-requiring protein 1 (IRE1) branch upon UPR induction, was upregulated ∼4.3 folds. Another protein involved in ER, HSPA5 [Heat Shock Protein Family A (Hsp70) Member 5], was induced almost 8.4 folds in activating transcriptional factor 6 (ATF6) branch of UPR. Besides, other ER-stress involved genes such as homocysteine-inducible endoplasmic reticulum stress-inducible ubiquitin-like domain member 1 (HERPUD1), DNA damage inducible transcript 3 (DDIT3), and cation transport regulator-like protein 1 (CHAC1) that is a novel gene identified in activating transcription factor 4 (ATF4) arm of the PKR-like ER protein kinase (PERK) branch of the UPR were upregulated ranking at the top 3 with the induction folds of 12.5, 10.2 and 11.9, respectively (Supplementary Table S1 and Fig. 6B). These data suggest that upon CCHFV infection, three UPRs pathways mediated by IRE1, ATF6 and PERK were likely all stimulated (Fig. 6C).
3.3.3. Metabolism changes during CCHFV infection
Many viruses, such as hepatitis C virus, human cytomegalovirus, influenza virus, and rhinovirus, have been documented to manipulate the cellular metabolism process for their use (Syed et al., 2010; McArdle et al., 2011; Vijayan and Hahm, 2014; Gualdoni et al., 2018). However, whether and how CCHFV regulates host cell metabolism remains unclear. In the present study, many cellular metabolism-related proteins seem to be significantly modulated. For example, fibroblast growth factor 21 (FGF21) which controls the process of energy metabolism through glucose and lipid metabolism was upregulated about 8.7 folds (Salminen et al., 2017). TRIB3 (tribbles homolog 3), a stress-inducible gene responding to oxidative and endoplasmic reticulum (ER) stress, was elevated ∼5.4 folds (Qian et al., 2008). The protein interactions referring to the metabolism were analyzed and shown in Fig. 5D. These results indicate that CCHFV infection may also modulate cellular metabolism processes especially those regarding cellular synthesis or catabolic process of cytokines, lipoproteins and amino acids.
3.3.4. Other biological processes regulated by CCHFV infection
In addition, several other biological processes, such as apoptosis and proteasomal degradation, also seemed to be regulated upon CCHFV infection (as summarized in the global view of the DEGs in Fig. 4). For example, ChaC glutathione specific gamma-glutamylcyclotransferase 1 (CHAC1), a proapoptotic γ-glutamyl cyclotransferase that depletes glutathione, was upregulated ∼12 folds (Kumar et al., 2012). Another gene transcription factor-4 (ATF4), a downstream molecular of PERK-eIF2-ATF4 axis pathway promoting apoptosis, was upregulated about 2.2 folds. CCAAT/enhancer binding protein beta (CEBPB) which participates in endoplasmic reticulum-mediated apoptosis was increased about 4.68 folds (Pellerito et al., 2010). These indicate that CCHFV infection probably regulated apoptosis that could be an act of host protection against viral infection. Furthermore, several proteins involved in proteasomal protein catabolic process were also differently regulated. Suppressor/enhancer of Lin-12-like (Sel1L) and selenoprotein K (SELENOK) were upregulated about 4 folds. These two proteins are involved in the endoplasmic reticulum (ER)-associated degradation (ERAD) (Sugimoto et al., 2017; Addinsall et al., 2018). Also, parkin RBR E3 ubiquitin protein ligase (PRKN) was elevated about 3.5 times. PRKN can mediate the targeting of substrate proteins for proteasomal degradation, and was also reported to be associated with Parkinson disease and mitophagy (Yamada et al., 2019). Together, the data indicate that apoptosis and proteasomal degradation are likely regulated by CCHFV infection as well.
3.4. Validation of DEGs by quantitative RT-PCR
Next, a series of representative DEGs including IFNs (IFN-β, IFNλ1, IFNλ2, and IFNλ3), typical IFN-inducible antiviral effectors (such as MxA, OAS1, ISG15, IFITM1, and IFIT1), and ER-stress pathway-related genes (like HSPA5 and XBP1) were selected for quantitative RT-qPCR analyses. As shown in Fig. 7, the quantitative results in HEK293 cells by qPCR were well in accordance with the RNA-Seq data, which further validates the transcriptome analysis and corroborates the induction of IFN immune responses and ER-stress pathway by CCHFV. Additionally, qPCR quantification results show that the induction of type III IFNs, IFNλ1 (∼74.4 folds) and IFNλ2/3 (∼129.9 folds), seems to be higher than that of IFN-β (∼16.7 folds) (Fig. 7B). Further, induction of all these genes by CCHFV infection was also observed in human lung A549 cells (Supplementary Fig. S2A). However, no notable up-regulation of IFN-β was found in human liver Huh7 cells, and HSPA5 and XBP1 were not markedly induced in either Huh7 or HUVEC (human blood vessel endothelial) cells (Supplementary Figs. S2B and S2C), whereas evident induction of type III IFNs and ISGs was detected in all these cell types tested (Fig. 7 and Supplementary Figs. S2A–2C), suggesting general stimulation of type III IFN responses by CCHFV in various cells.
3.5. Antiviral activities of IFN-β and type III IFNs against CCHFV
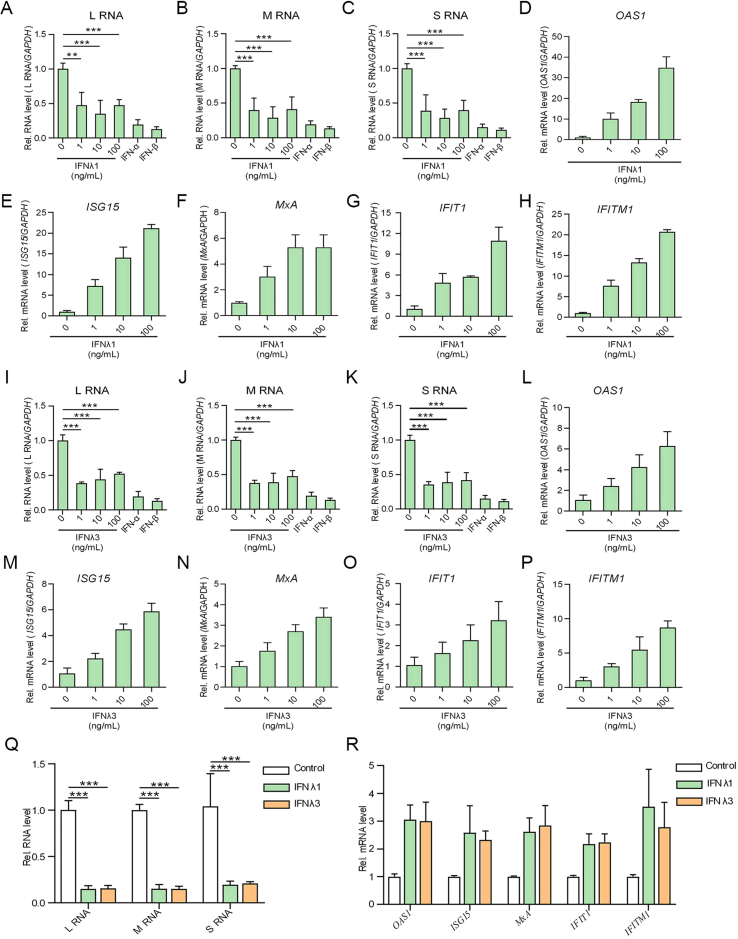
Given the marked IFN responses upon CCHFV infection revealed by the transcriptome and qPCR analyses, we then investigated the anti-CCHFV effects of the IFNs (especially type III IFNs). HEK293 cells were pretreated with different concentrations of type III IFNs (IFNλ1 or IFNλ3) or high concentrations of type I IFNs (100 U/mL IFN-α or 100 ng/mL IFN-β) and then infected with CCHFV (MOI = 0.1), followed by qPCR quantitative analyses of the viral RNA levels. Note that IFN-α here was used as a control as its antiviral activity against CCHFV had been shown in A549 and Huh7 cell lines (Bordi et al., 2015), although no significant induction of IFN-α was detected in the RNA-seq analysis. As shown in Fig. 8, treatments with the tested IFNs all exhibited evident anti-CCHFV activities. Interestingly, the low concentration of type III IFNs showed antiviral activities against CCHFV comparable with the high-concentration groups (Fig. 8A–C and I–K). Further, induction of several representative ISGs including OAS1, ISG15, MxA, IFIT1 and IFITM1 further confirmed the antiviral responses triggered by the type III IFNs in HEK293 cells (Fig. 8D–H and L-P). Additionally, significant anti-CCHFV activities of the type III IFNs were also observed in Huh7 cells, along with induction of ISGs (Fig. 8Q and R).
Fig. 8.
Inhibitory effects of type I and III IFNs on CCHFV infection. A–P HEK293 cells were pretreated with the indicated concentrations of IFNλ1 (A–H) or IFNλ3 (I–P), or 100 U/mL IFN-α or 100 ng/mL IFN-β for 24 h, followed by CCHFV infection and RT-qPCR detection at 24 hpi. CCHFV RNA levels (A–C and I–K) and representative ISGs in type III IFN-treated cells (D–H and L–P) were analyzed. (Q and R) Anti-CCHFV activities of type III IFNs in Huh7 cells. Huh7 cells were pretreated with IFNλ1 and IFNλ3 (10 ng/mL) for 24 h, followed by CCHFV infection and RT-qPCR analyses of the viral RNA (Q) or ISG mRNA (R) levels as above. Data are shown as means ± SD, n ≥ 3. ∗∗P < 0.01; ∗∗∗P < 0.001.
4. Discussion
CCHFV is a BSL-4 pathogen threatening public health in about 50 countries of Asia, Africa, southern Europe, and the Middle East (Nasirian, 2020). Elucidation of virus-host interactions is urgently needed for not only the understanding of the viral infection and pathogenesis but also the design of antiviral therapies. However, as one of the most dangerous human pathogens, experimental handling of live CCHFV is strictly restricted in high-containment laboratories, slowing down the virological studies including animal and cell infection model development and optimization (Hawman and Feldmann, 2018). HEK293 is a well-recognized human cell model derived from kidney and permissive to CCHFV infection (Foldes et al., 2020; Dai S. et al., 2021a). Moreover, clinical observations have shown that CCHFV has tropism to kidney tissue (Ardalan et al., 2006; Deveci et al., 2013; Khazaei et al., 2018; Foldes et al., 2020). Additionally, HEK293 is a most often chosen model for omics study because of its availability of annotated human omics databases to conduct gene identification and function assignment and ready validation of the omics results by further experimental analysis with it. Thus, we here establish a cellular transcriptome profile of CCHFV infection in HEK293 cells by using RNA-seq technology. In total, 496 DEGs, including 361 up-regulated and 135 down-regulated genes, were identified in CCHFV-infected cells. These DEGs were involved in multiple biological processes potentially linked with CCHFV infection. Particularly, type III IFN and ER stress responses were regulated significantly. Furthermore, RT-qPCR validates these marked regulations by CCHFV. Antiviral assays show that not only type I but also type III IFNs exhibit anti-CCHFV activities. Collectively, the current study may shed lights on some significant aspects of CCHFV-host interactions, which may benefit further mechanistic delineation of the viral infection and pathogenicity and inform future development of anti-CCHFV strategies. The representative aspects of CCHFV-cell interplays highlighted by this study are further discussed as follows.
4.1. Transcriptome profiling of bunyavirus infections
To date, few studies were reported regarding to the transcriptome profiling upon bunyavirus infections. Previously, two groups analyzed the transcriptome changes upon Rift Valley fever virus (RVFV, Phlebovirus genus, Phenuiviridae family, Bunyavirales order) infection by RNA-seq. Popova et al. showed that RVFV infection seemed to alter many pathways, such as G2/M DNA damage checkpoint, antiviral response, integrin-linked kinase (ILK) signaling, and mitochondrial dysfunction in human small airway epithelial cells (HSAECs) (Popova et al., 2010). In the study by Havranek et al., it was noted that host splicing pattern, immune response, and cell proliferation were changed in response to RVFV infection in HEK293 cells (Havranek et al., 2019). Furthermore, RNA-seq analysis of bovine cell infection with Schmallenberg virus (SBV), another member of Orthobunyavirus in Peribunyaviridae family of Bunyavirales order showed that NSs-deleted virus stimulated dramatic transcription of antiviral ISGs suggesting the critical role of NSs in shutting down host immune response (Blomström et al., 2015; Abudurexiti et al., 2019). We here analyzed the effect of CCHFV infection on host transcriptome by RNA-seq in HEK293 cells that could be considered as a priority human cell model for omics analyses of CCHFV infection. Overall, in comparison with the previously reported bunyaviruses, CCHFV infection results in not only some similar transcriptional changes in the immune response but also unique regulations such as the remarkable induction of type III IFNs, ER stress as well as the possible apoptosis (further discussed below). Recently, Arnold et al. utilized a targeted gene panel to analyzed transcriptional changes in whole blood samples of virus-infected cynomolgus macaques and Kozak et al. characterized dual gene expression regulations in hepatocarcinoma cell lines (Kozak et al., 2020; Arnold et al., 2021). Common immune stimulations including type I IFN and ISG expression were observed, while the responses of type III IFNs and ER stress were not uncovered in these studies (Kozak et al., 2020; Arnold et al., 2021). Aside from omics analysis, no further experimental validations were conducted in both studies (Kozak et al., 2020; Arnold et al., 2021). However, it is still likely suggestive of the dependence of transcriptional responses on the host cell types or viral strains. Besides, technical platforms, sequencing depth, and experimental settings may also lead to differences of obtained RNA-seq results. Indeed, in our study, we observed that type III IFN responses were generally induced by CCHFV in various human cells, while IFN-β and ER-stress related genes were not notably stimulated in some cell types (Supplementary Fig. S2). Additionally, CCHFV causes lethal disease with severe clinical manifestations in humans but is usually nonpathogenic in wild or domestic animals (Ergonul, 2006; Öncü, 2013). In view of this, experimental investigations in human-derived cells might be particularly informative. Further comparative transcriptomic analyses in more other human cell types or even in clinical patients are merited. During the revision of our manuscript, Neogi et al. reported a system-wide network-based system biology analysis of peripheral blood mononuclear cells (PBMCs) from CCHF patients and interestingly, their results also highlight notable changes of metabolism (Neogi et al., 2022). Apart from IFN responses and ER stress, metabolism process is the third enriched network regulated by CCHFV infection according to our data (Fig. 5). The consistent findings further underline the metabolic changes of particular importance involved in CCHFV infection and also support the potential value and significance of studies based on cell infection models like CCHFV-infected HEK293 in the present study. In addition, the data from whole blood or PBMCs, which both are mixtures of cell populations and even remain to be fully characterized in infectivity of CCHFV, are convoluted to be interpreted and depend on cultured cell infection model for validation and detailed depiction. Temporal analysis of transcription which is not achieved in our study due to the restrictions of using high-level biosafety laboratory especially during the current COVID-19 pandemic may be also informative and merited to be conducted for characterizing the kinetics of viral regulation of host cells in the future. Importantly, by combination of in vivo and in vitro study models, further experimental validations of the CCHFV-regulated cellular processes revealed by the transcriptome profiling is more urgently required to help deepen our understanding of this life-threatening pathogen and design specific antiviral therapeutics.
4.2. Antiviral IFN responses upon CCHFV infection
Antiviral IFN system driven by type I and III IFNs is the key component of host innate immune defense against viral infection. Upon virus infection, cellular pattern-recognition receptors (PRRs) can recognize pathogen-associated molecular patterns (PAMPs) produced by virus infection and trigger IFN expression. Secreted type I and III IFNs bind to their receptors on the cell surface and direct the following JAK-STAT signaling cascades, resulting in IFN-stimulated genes (ISGs) expression and establishing antiviral state of host cell (Maher et al., 2008; Ivashkiv and Donlin, 2014). The transcriptome profiling and qPCR analysis of our study show that upon CCHFV infection, IFNλ1, IFNλ2, IFNλ3, and IFN-β, together with many ISGs, were significantly upregulated, indicating systematic response of IFN signaling against CCHFV. Moreover, two ISGs, RIG-I (a main PRR sensing CCHFV infection) and IRF7 (an important transcription factor stimulating IFN expression) that can act as positive feedback regulation factors of IFN signaling cascades were also elevated (∼4.6 and 2.3 folds, respectively), which could further amplify the antiviral IFN responses.
Numerous viruses have been reported to stimulate type I IFN induction, while reports concerning type III IFN production in response to virus infection are limited (Ank et al., 2006; Vitour et al., 2014; Killip et al., 2015; Huang et al., 2017). Interestingly, here we observed that CCHFV infection triggers significant induction of three types of IFN-λs (all ranked top 5 up-regulated genes together with IFN-β), revealing a distinct IFN induction pattern and highlighting the notable involvement of type III IFNs in CCHFV-host interactions. Further, we also demonstrated the substantial anti-CCHFV activities of type III IFNs in HEK293 cells even at low application dosages. Additionally, higher concentration (e.g. 100 ng/mL) appeared not to increase the antiviral efficacy, which suggests that the low-dosage treatments may have resulted in a plateau of antiviral effect and is reminiscent of the previously reported complex antagonistic capabilities of CCHFV (Frias-Staheli N. et al., 2007b; Andersson et al., 2008; Fajs et al., 2014). Besides, some of the ISGs that have significant anti-CCHFV activity (e.g. MxA) (Andersson et al., 2004) may have reached an expression plateau as well in type III IFN treatment of lower concentration as seen in Fig. 8F, which may also explain the less obvious dose-dependent manner of the antiviral efficiency. Despite the similar induction pathway, type III IFNs are believed to have different kinetics in action in comparison with type I IFNs: the former stimulates ISGs in a relatively slower but much more sustained pattern (Pervolaraki et al., 2018; Sun et al., 2018; Ye et al., 2019). Moreover, administration of type III IFNs as antiviral agents is likely much safer with fewer side effects due to their more specific and local actions at epithelial cells and mucosal barriers (Kotenko and Durbin, 2017; Ye et al., 2019). Thus, further testing of the inhibitory effects of type III IFNs on CCHFV replication (especially in animal infection models) is warranted to develop anti-CCHFV therapies in the future.
4.3. ER-stress induced by CCHFV infection
The ER is an organelle with multiple biological functions which are of significance for the correct protein modification (Jheng et al., 2014). Retaining of incorrect proteins in ER will stimulate UPR. The UPR consists of PERK, IRE1 and ATF6 pathways (Tirasophon et al., 1998; Haze et al., 1999; SOOD et al., 2000). Many viruses, including DNA and RNA viruses were reported to induce ER-stress and UPR pathways during infection. Virus-induced ER stress and UPR are either beneficial or detrimental for virus replication. For example, transmissible gastroenteritis virus (TGEV), a member belonging to alphacoronavirus, triggers all the three UPR pathways, while the replication of TGEV was suppressed by the PERK-eIF2α axis-mediated UPR pathway (Xue et al., 2018). In contrast, lymphocytic choriomeningitis virus (LCMV), a member with bi-segmented negative sense RNA genome newly defined to family Arenaviridae, order Bunyavirales, activates the ATF6 branch of UPR pathway to benefit viral replication and cell viability (Pasqual et al., 2011). A recent quantitative proteomic study demonstrated that severe fever with thrombocytopenia syndrome virus (SFTSV), another bunyavirus (Dabie bandavirus species, Bandavirus genus, Phenuiviridae family of Bunyavirales order), appeared to activate all the main three UPR responses with two of the pathways favoring SFTSV infection (Zhang et al., 2019; Kuhn et al., 2020). As for CCHFV, preliminary studies revealing CCHFV infection-triggered ER-stress, especially the UPR in Huh7 cells, are in line with the investigation of IRE1-mediated splicing of XBP1 transcript (Fraisier et al., 2014). In agreement with their results, we observed the upregulation of XBP1, a hallmark of UPR stimulation. To be more specific, all the three arms of UPR (ATF6-/PERK-/IRE1-mediated unfolded protein responses) were likely stimulated according to our transcriptomic analysis here, reflecting their potential roles in CCHFV infection. Nonetheless, the mechanisms underlying the regulation of the UPR pathways by CCHFV remain to be clearly unraveled. Further, whether and how these ER-stress responses affect CCHFV infection and pathogenicity are also merited to be determined in the future study.
4.4. Cross talks of cellular biological processes in response to CCHFV infection
As discussed above, IFN immune response, UPR, and apoptosis regulation are marked biological processes induced by CCHFV infection. Previous studies have shown that UPR promotes phosphorylation-dependent ubiquitination and degradation of IFNAR1, highlighting the link with immune response, especially the type I IFN pathway (Liu et al., 2009). Therefore, it is of interest to investigate the connection between the two responses during CCHFV infection. Further, viral infection induces UPR signaling, while prolonged and irremediable ER-stress could trigger apoptosis in PERK-eIF2α-ATF-CHOP (C/EBP-homologous protein) mediated pathway (Abudurexiti et al., 2019). As a cellular parasite, CCHFV utilizes the host ER to produce large amounts of viral glycoproteins (Serretiello et al., 2020), thus likely leading to misfolded and unfolded protein accumulation in the ER lumen. To alleviate this stress, UPR then could be initiated during CCHFV infection. In terms of extensive UPR, the intrinsic apoptotic cascade-related proteins such as CHOP-ATF3, CHOP-ATF4 and CHOP-C/EBP complexes would be upregulated as seen in the present transcriptomic data, perhaps leading to the intrinsic apoptosis (Liu et al., 2016). Additionally, immune responses including the IFN pathways could also contribute to the apoptosis regulation. In our study, many genes involved in the three processes (immune response, UPR and apoptosis) were significantly stimulated (Fig. 4). The result agrees with the finding that CCHFV infection of Huh7 cells induced ER-stress and modulated apoptosis involving mitochondrial and death receptor pathways as reported by Rodrigues et al. (2012). Thus, there would be complicated cross talks among these processes during CCHFV infection. It will be interesting to investigate whether and how CCHFV balances these pathways to create an environment favorable for its replication or manipulates them for its pathogenesis.
5. Conclusions
CCHFV is a high-virulent BSL-4 pathogen posing a severe threat to worldwide public health. In the current study, we establish the cellular transcriptome profile of CCHFV infection in human kidney HEK293 cells by RNA-seq and identify 496 DEGs, including 361 up-regulated and 135 down-regulated genes. The data suggest that multiple biological processes including defense response to virus, response to stress, regulation of viral process, immune response, metabolism, stimulus, apoptosis and protein catabolic process are likely implicated in the cellular responses upon CCHFV infection. Among them, type III IFN pathways as well as ER stress responses are particularly regulated and highlighted by qPCR analyses, indicating their potential roles in CCHFV-cell interplays. Furthermore, antiviral assays demonstrate that not only type I but also type III IFNs exhibit notable anti-CCHFV activities. These findings provide new insights into CCHFV-host interactions and may help advance the understanding of viral infection and pathogenesis, which perhaps will eventually inform the development of medical countermeasures for disease prevention and treatment.
Data availability
All data generated or analyzed in the current study are included in this published article and its supplementary materials.
Ethics statement
All experiments involving infectious CCHFV were conducted in the National High-level Biosafety Laboratory, Wuhan, Chinese Academy of Sciences (CAS), and was approved by the biosafety committee of Wuhan Institute of Virology, CAS.
Author contributions
Qiong Mo: conceptualization, methodology, investigation, data curation, formal analysis, software, validation, writing – original draft. Kuan Feng: investigation, data curation, validation. Shiyu Da: investigation. Qiaoli Wu: investigation. Zhong Zhang: investigation. Ashaq Ali: methodology, formal analysis. Fei Deng: conceptualization, project administration, resources, software, supervision. Hualin Wang: conceptualization, funding acquisition, project administration, resources, software, supervision. Yun-Jia Ning: conceptualization, methodology, data curation, funding acquisition, project administration, resources, software, supervision, visualization, writing – original draft, writing – review & editing. All the authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFA0507202), the National Natural Science Foundation of China (32170171, 31870162, and 82161138003), and the Youth Innovation Promotion Association of Chinese Academy of Sciences. We thank the Institutional Center for Shared Technologies and Facilities of Wuhan Institute of Virology, CAS for technical assistance and the National Virus Resource Center (Wuhan Institute of Virology) for virus resources. We are also grateful to the running team of National Biosafety Laboratory, Wuhan, Chinese Academy of Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2022.09.002.
Contributor Information
Fei Deng, Email: df@wh.iov.cn.
Hualin Wang, Email: h.wang@wh.iov.cn.
Yun-Jia Ning, Email: nyj@wh.iov.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Abudurexiti A., Adkins S., Alioto D., Alkhovsky S.V., Avšič-Županc T., Ballinger M.J., Bente D.A., Beer M., Bergeron É., Blair C.D., et al. Taxonomy of the order bunyavirales: update 2019. Arch. Virol. 2019;164:1949–1965. doi: 10.1007/s00705-019-04253-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addinsall A.B., Wright C.R., Andrikopoulos S., van der Poel C., Stupka N. Emerging roles of endoplasmic reticulum-resident selenoproteins in the regulation of cellular stress responses and the implications for metabolic disease. Biochem. J. 2018;475:1037–1057. doi: 10.1042/BCJ20170920. [DOI] [PubMed] [Google Scholar]
- Andersson I., Karlberg H., Mousavi-Jazi M., Martinez-Sobrido L., Weber F., Mirazimi A. Crimean-Congo hemorrhagic fever virus delays activation of the innate immune response. J. Med. Virol. 2008;80:1397–1404. doi: 10.1002/jmv.21222. [DOI] [PubMed] [Google Scholar]
- Andersson I., Bladh L., Mousavi-Jazi M., Magnusson K.E., Lundkvist A., Haller O., Mirazimi A. Human mxa protein inhibits the replication of crimean-Congo hemorrhagic fever virus. J. Virol. 2004;78:4323–4329. doi: 10.1128/JVI.78.8.4323-4329.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. Lambda interferon (ifn-lambda), a type iii ifn, is induced by viruses and ifns and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardalan M.R., Tubbs R.S., Chinikar S., Shoja M.M. Crimean-Congo haemorrhagic fever presenting as thrombotic microangiopathy and acute renal failure. Nephrol. Dial. Transplant. 2006;21:2304–2307. doi: 10.1093/ndt/gfl248. [DOI] [PubMed] [Google Scholar]
- Arnold C.E., Shoemaker C.J., Smith D.R., Douglas C.E., Blancett C.D., Graham A.S., Minogue T.D. Host response transcriptomic analysis of crimean-Congo hemorrhagic fever pathogenesis in the cynomolgus macaque model. Sci Rep-UK. 2021;11 doi: 10.1038/s41598-021-99130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwal B., Karlberg H., Mirazimi A., Tan Y.-J. The non-structural protein of crimean-Congo hemorrhagic fever virus disrupts the mitochondrial membrane potential and induces apoptosis. J. Biol. Chem. 2016;291:582–592. doi: 10.1074/jbc.M115.667436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bente D.A., Forrester N.L., Watts D.M., McAuley A.J., Whitehouse C.A., Bray M. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res. 2013;100:159–189. doi: 10.1016/j.antiviral.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Blomström A.L., Gu Q., Barry G., Wilkie G., Skelton J.K., Baird M., McFarlane M., Schnettler E., Elliott R.M., Palmarini M., Kohl A. Transcriptome analysis reveals the host response to schmallenberg virus in bovine cells and antagonistic effects of the nss protein. BMC Genom. 2015;16:324. doi: 10.1186/s12864-015-1538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi L., Lalle E., Caglioti C., Travaglini D., Lapa D., Marsella P., Quartu S., Kis Z., Arien K.K., Huemer H.P., Meschi S., Ippolito G., Di Caro A., Capobianchi M.R., Castilletti C. Antagonistic antiviral activity between ifn-lambda and ifn-alpha against lethal crimean-Congo hemorrhagic fever virus in vitro. PLoS One. 2015;10 doi: 10.1371/journal.pone.0116816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Wu Q., Wu X., Peng C., Liu J., Tang S., Zhang T., Deng F., Shen S. Differential cell line susceptibility to crimean-Congo hemorrhagic fever virus. Front. Cell. Infect. Microbiol. 2021;11:648077. doi: 10.3389/fcimb.2021.648077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S.Y., Deng F., Wang H.L., Ning Y.J. Crimean-Congo hemorrhagic fever virus: current advances and future prospects of antiviral strategies. Viruses-Basel. 2021;13 doi: 10.3390/v13071195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveci K., Uysal E.B., Kaya A., Sancakdar E., Alkan F. Evaluation of renal involvement in children with crimean-Congo hemorrhagic fever. Jpn. J. Infect. Dis. 2013;66:493–496. doi: 10.7883/yoken.66.493. [DOI] [PubMed] [Google Scholar]
- Ergonul O. Crimean-Congo haemorrhagic fever. Lancet Infect. Dis. 2006;6:203–214. doi: 10.1016/S1473-3099(06)70435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajs L., Resman K., Avsic-Zupanc T. Crimean-Congo hemorrhagic fever virus nucleoprotein suppresses ifn-beta-promoter-mediated gene expression. Arch. Virol. 2014;159:345–348. doi: 10.1007/s00705-013-1816-2. [DOI] [PubMed] [Google Scholar]
- Feng K., Min Y.Q., Sun X.L., Deng F., Li P.Q., Wang H.L., Ning Y.J. Interactome profiling reveals interaction of sars-cov-2 nsp13 with host factor stat1 to suppress interferon signaling. J. Mol. Cell Biol. 2021;13:760–762. doi: 10.1093/jmcb/mjab068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldes K., Aligholipour Farzani T., Ergunay K., Ozkul A. Differential growth characteristics of crimean-Congo hemorrhagic fever virus in kidney cells of human and bovine origin. Viruses. 2020;12:685. doi: 10.3390/v12060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisier C., Rodrigues R., Vu Hai V., Belghazi M., Bourdon S., Paranhos-Baccala G., Camoin L., Almeras L., Peyrefitte C.N. Hepatocyte pathway alterations in response to in vitro crimean Congo hemorrhagic fever virus infection. Virus Res. 2014;179:187–203. doi: 10.1016/j.virusres.2013.10.013. [DOI] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R., Schmaljohn C.S., Lenschow D.J., Snijder E.J., García-Sastre A., Virgin H.W. Ovarian tumor domain-containing viral proteases evade ubiquitin- and isg15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Staheli N., Giannakopoulos N.V., Kikkert M., Taylor S.L., Bridgen A., Paragas J., Richt J.A., Rowland R.R., Schmaljohn C.S., Lenschow D.J., Snijder E.J., Garcia-Sastre A., Virgin HWt. Ovarian tumor domain-containing viral proteases evade ubiquitin- and isg15-dependent innate immune responses. Cell Host Microbe. 2007;2:404–416. doi: 10.1016/j.chom.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdoni G.A., Mayer K.A., Kapsch A.M., Kreuzberg K., Puck A., Kienzl P., Oberndorfer F., Frühwirth K., Winkler S., Blaas D., Zlabinger G.J., Stöckl J. Rhinovirus induces an anabolic reprogramming in host cell metabolism essential for viral replication. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E7158–e7165. doi: 10.1073/pnas.1800525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R., Shen S., Zhang Y., Shi J., Su Z., Liu D., Liu J., Yang J., Wang Q., Hu Z., Zhang Y., Deng F. A new strain of crimean-Congo hemorrhagic fever virus isolated from xinjiang, China. Virol. Sin. 2017;32:80–88. doi: 10.1007/s12250-016-3936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havranek K.E., White L.A., Lanchy J.M., Lodmell J.S. Transcriptome profiling in rift valley fever virus infected cells reveals modified transcriptional and alternative splicing programs. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawman D.W., Feldmann H. Recent advances in understanding crimean-Congo hemorrhagic fever virus. F1000Res. 2018;7 doi: 10.12688/f1000research.16189.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze K., Yoshida H., Yanagi H., Yura T., Mori K. Mammalian transcription factor atf6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B., Li J., Zhang X., Zhao Q., Lu M., Lv Y. Rig-1 and mda-5 signaling pathways contribute to ifn-β production and viral replication in porcine circovirus virus type 2-infected pk-15 cells in vitro. Vet. Microbiol. 2017;211:36–42. doi: 10.1016/j.vetmic.2017.09.022. [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Donlin L.T. Regulation of type i interferon responses. Nat. Rev. Immunol. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jheng J.-R., Ho J.-Y., Horng J.-T. ER stress, autophagy, and RNA viruses. Front. Microbiol. 2014;5:388. doi: 10.3389/fmicb.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaei Z., Darvishi I., Amiri M., Sohrabivafa M., Kamran S. Crimean Congo hemorrhagic fever: a brief report regarding kidney involvement. J. Ren. Inj. Prev. 2018;7:129–131. [Google Scholar]
- Killip M.J., Fodor E., Randall R.E. Influenza virus activation of the interferon system. Virus Res. 2015;209:11–22. doi: 10.1016/j.virusres.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotenko S.V., Durbin J.E. Contribution of type iii interferons to antiviral immunity: location, location, location. J. Biol. Chem. 2017;292:7295–7303. doi: 10.1074/jbc.R117.777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak R.A., Fraser R.S., Biondi M.J., Majer A., Medina S.J., Griffin B.D., Kobasa D., Stapleton P.J., Urfano C., Babuadze G., Antonation K., Fernando L., Booth S., Lillie B.N., Kobinger G.P. Dual rna-seq characterization of host and pathogen gene expression in liver cells infected with crimean-Congo hemorrhagic fever virus. PLoS Neglected Trop. Dis. 2020;14 doi: 10.1371/journal.pntd.0008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Adkins S., Alioto D., Alkhovsky S.V., Amarasinghe G.K., Anthony S.J., Avšič-Županc T., Ayllón M.A., Bahl J., Balkema-Buschmann A., et al. 2020 taxonomic update for phylum negarnaviricota (riboviria: orthornavirae), including the large orders bunyavirales and mononegavirales. Arch. Virol. 2020;165:3023–3072. doi: 10.1007/s00705-020-04731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Tikoo S., Maity S., Sengupta S., Sengupta S., Kaur A., Bachhawat A.K. Mammalian proapoptotic factor chac1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012;13:1095–1101. doi: 10.1038/embor.2012.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblebicioglu H., Ozaras R., Sunbul M. Crimean-Congo hemorrhagic fever: a neglected infectious disease with potential nosocomial infection threat. Am. J. Infect. Control. 2017;45:815–816. doi: 10.1016/j.ajic.2016.05.039. [DOI] [PubMed] [Google Scholar]
- Liu J., HuangFu W.C., Kumar K.G., Qian J., Casey J.P., Hamanaka R.B., Grigoriadou C., Aldabe R., Diehl J.A., Fuchs S.Y. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type i interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Shi Q., Song X., Wang Y., Wang Y., Song E., Song Y. Activating transcription factor 4 (atf4)-atf3-c/ebp homologous protein (chop) cascade shows an essential role in the er stress-induced sensitization of tetrachlorobenzoquinone-challenged pc12 cells to ros-mediated apoptosis via death receptor 5 (dr5) signaling. Chem. Res. Toxicol. 2016;29:1510–1518. doi: 10.1021/acs.chemrestox.6b00181. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−δδct method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maher S.G., Sheikh F., Scarzello A.J., Romero-Weaver A.L., Baker D.P., Donnelly R.P., Gamero A.M. Ifn-α and ifn-λ differ in their antiproliferative effects and duration of jak/stat signaling activity. Cancer Biol. Ther. 2008;7:1109–1115. doi: 10.4161/cbt.7.7.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzola L.T., Kelly-Cirino C. Diagnostic tests for crimean-Congo haemorrhagic fever: a widespread tickborne disease. BMJ Global Health. 2019;4 doi: 10.1136/bmjgh-2018-001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle J., Schafer X.L., Munger J. Inhibition of calmodulin-dependent kinase kinase blocks human cytomegalovirus-induced glycolytic activation and severely attenuates production of viral progeny. J. Virol. 2011;85:705–714. doi: 10.1128/JVI.01557-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y.-Q., Ning Y.-J., Wang H., Deng F. A rig-i-like receptor directs antiviral responses to a bunyavirus and is antagonized by virus-induced blockade of trim25-mediated ubiquitination. J. Biol. Chem. 2020;295:9691–9711. doi: 10.1074/jbc.RA120.013973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y.Q., Shi C., Yao T., Feng K., Mo Q., Deng F., Wang H., Ning Y.J. The nonstructural protein of guertu virus disrupts host defenses by blocking antiviral interferon induction and action. ACS Infect. Dis. 2020;6:857–870. doi: 10.1021/acsinfecdis.9b00492. [DOI] [PubMed] [Google Scholar]
- Mo Q., Xu Z., Deng F., Wang H., Ning Y.-J. Host restriction of emerging high-pathogenic bunyaviruses via mov10 by targeting viral nucleoprotein and blocking ribonucleoprotein assembly. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009129. e1009129-e1009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinas A., Mirazimi A., Holm A., Loitto V.M., Magnusson K.-E., Vikström E. Protective role of host aquaporin 6 against hazara virus, a model for crimean–Congo hemorrhagic fever virus infection. FEMS Microbiol. Lett. 2016;363 doi: 10.1093/femsle/fnw058. [DOI] [PubMed] [Google Scholar]
- Nasirian H. New aspects about crimean-Congo hemorrhagic fever (cchf) cases and associated fatality trends: a global systematic review and meta-analysis. Comp. Immunol. Microbiol. Infect. Dis. 2020;69:101429. doi: 10.1016/j.cimid.2020.101429. [DOI] [PubMed] [Google Scholar]
- Neogi U., Elaldi N., Appelberg S., Ambikan A., Kennedy E., Dowall S., Bagci B.K., Gupta S., Rodriguez J.E., Svensson-Akusjarvi S., Monteil V., Vegvari A., Benfeitas R., Banerjea A., Weber F., Hewson R., Mirazimi A. Multi-omics insights into host-viral response and pathogenesis in crimean-Congo hemorrhagic fever viruses for novel therapeutic target. Elife. 2022;11:e76071. doi: 10.7554/eLife.76071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y.-J., Feng K., Min Y.-Q., Deng F., Hu Z., Wang H. Heartland virus nss protein disrupts host defenses by blocking the tbk1 kinase-irf3 transcription factor interaction and signaling required for interferon induction. J. Biol. Chem. 2017;292:16722–16733. doi: 10.1074/jbc.M117.805127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y.-J., Feng K., Min Y.-Q., Cao W.-C., Wang M., Deng F., Hu Z., Wang H. Disruption of type i interferon signaling by the nonstructural protein of severe fever with thrombocytopenia syndrome virus via the hijacking of stat2 and stat1 into inclusion bodies. J. Virol. 2015;89:4227. doi: 10.1128/JVI.00154-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning Y.-J., Mo Q., Feng K., Min Y.-Q., Li M., Hou D., Peng C., Zheng X., Deng F., Hu Z., Wang H. Interferon-γ-directed inhibition of a novel high-pathogenic phlebovirus and viral antagonism of the antiviral signaling by targeting stat1. Front. Immunol. 2019;10:1182. doi: 10.3389/fimmu.2019.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öncü S. Crimean-Congo hemorrhagic fever: an overview. Virol. Sin. 2013;28:193–201. doi: 10.1007/s12250-013-3327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqual G., Burri D.J., Pasquato A., de la Torre J.C., Kunz S. Role of the host cell's unfolded protein response in arenavirus infection. J. Virol. 2011;85:1662–1670. doi: 10.1128/JVI.01782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerito O., Calvaruso G., Portanova P., De Blasio A., Santulli A., Vento R., Tesoriere G., Giuliano M. The synthetic cannabinoid win 55,212-2 sensitizes hepatocellular carcinoma cells to tumor necrosis factor-related apoptosis-inducing ligand (trail)-induced apoptosis by activating p8/ccaat/enhancer binding protein homologous protein (chop)/death receptor 5 (dr5) axis. Mol. Pharm. 2010;77:854. doi: 10.1124/mol.109.062257. [DOI] [PubMed] [Google Scholar]
- Pervolaraki K., Talemi S.R., Albrecht D., Bormann F., Bamford C., Mendoza J.L., Garcia K.C., McLauchlan J., Hofer T., Stanifer M.L., Boulant S. Differential induction of interferon stimulated genes between type i and type iii interferons is independent of interferon receptor abundance. PLoS Pathog. 2018;14:e1007420. doi: 10.1371/journal.ppat.1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova T.G., Turell M.J., Espina V., Kehn-Hall K., Kidd J., Narayanan A., Liotta L., Petricoin E.F., 3rd, Kashanchi F., Bailey C., Popov S.G. Reverse-phase phosphoproteome analysis of signaling pathways induced by rift valley fever virus in human small airway epithelial cells. PLoS One. 2010;5:e13805. doi: 10.1371/journal.pone.0013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B., Wang H., Men X., Zhang W., Cai H., Xu S., Xu Y., Ye L., Wollheim C.B., Lou J. Trib3 [corrected] is implicated in glucotoxicity- and endoplasmic reticulum-stress-induced [corrected] beta-cell apoptosis. J. Endocrinol. 2008;199:407–416. doi: 10.1677/JOE-08-0331. [DOI] [PubMed] [Google Scholar]
- Rodrigues R., Paranhos-Baccalà G., Vernet G., Peyrefitte C.N. Crimean-Congo hemorrhagic fever virus-infected hepatocytes induce er-stress and apoptosis crosstalk. PLoS One. 2012;7 doi: 10.1371/journal.pone.0029712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A., Kaarniranta K., Kauppinen A. Regulation of longevity by fgf21: interaction between energy metabolism and stress responses. Ageing Res. Rev. 2017;37:79–93. doi: 10.1016/j.arr.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Serretiello E., Astorri R., Chianese A., Stelitano D., Zannella C., Folliero V., Santella B., Galdiero M., Franci G., Galdiero M. The emerging tick-borne crimean-Congo haemorrhagic fever virus: a narrative review. Trav. Med. Infect. Dis. 2020;37 doi: 10.1016/j.tmaid.2020.101871. [DOI] [PubMed] [Google Scholar]
- Sood R., Porter A.C., Ma K., Quilliam L.A., Wek R.C. Pancreatic eukaryotic initiation factor-2α kinase (pek) homologues in humans, drosophila melanogaster and caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem. J. 2000;346:281–293. [PMC free article] [PubMed] [Google Scholar]
- Stanifer M.L., Pervolaraki K., Boulant S. Differential regulation of type i and type iii interferon signaling. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R., Grzelak M., Hadfield J. Rna sequencing: the teenage years. Nat. Rev. Genet. 2019;20:631–656. doi: 10.1038/s41576-019-0150-2. [DOI] [PubMed] [Google Scholar]
- Sugimoto T., Ninagawa S., Yamano S., Ishikawa T., Okada T., Takeda S., Mori K. Cell Struct Funct; 2017. Sel1l-dependent Substrates Require Derlin2/3 and Herp1/2 for Endoplasmic Reticulum-Associated Degradation. (advpub) [DOI] [PubMed] [Google Scholar]
- Sun Y., Jiang J., Tien P., Liu W., Li J. Ifn-lambda: a new spotlight in innate immunity against influenza virus infection. Protein Cell. 2018;9:832–837. doi: 10.1007/s13238-017-0503-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surtees R., Dowall S.D., Shaw A., Armstrong S., Hewson R., Carroll M.W., Mankouri J., Edwards T.A., Hiscox J.A., Barr J.N. Heat shock protein 70 family members interact with crimean-Congo hemorrhagic fever virus and hazara virus nucleocapsid proteins and perform a functional role in the nairovirus replication cycle. J. Virol. 2016;90:9305–9316. doi: 10.1128/JVI.00661-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed G.H., Amako Y., Siddiqui A. Hepatitis c virus hijacks host lipid metabolism. Trends Endocrin Met. 2010;21:33–40. doi: 10.1016/j.tem.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirasophon W., Welihinda A.A., Kaufman R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayan M., Hahm B. Influenza viral manipulation of sphingolipid metabolism and signaling to modulate host defense system. Sci. Tech. Rep. 2014;2014:793815. doi: 10.1155/2014/793815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitour D., Doceul V., Ruscanu S., Chauveau E., Schwartz-Cornil I., Zientara S. Induction and control of the type i interferon pathway by bluetongue virus. Virus Res. 2014;182:59–70. doi: 10.1016/j.virusres.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Fu F., Ma Y., Zhang X., Li L., Feng L., Liu P. The perk arm of the unfolded protein response negatively regulates transmissible gastroenteritis virus replication by suppressing protein translation and promoting type i interferon production. J. Virol. 2018;92 doi: 10.1128/JVI.00431-18. e00431–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Dawson T.M., Yanagawa T., Iijima M., Sesaki H. Sqstm1/p62 promotes mitochondrial ubiquitination independently of pink1 and prkn/parkin in mitophagy. Autophagy. 2019;15:2012–2018. doi: 10.1080/15548627.2019.1643185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L., Schnepf D., Staeheli P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat. Rev. Immunol. 2019;19:614–625. doi: 10.1038/s41577-019-0182-z. [DOI] [PubMed] [Google Scholar]
- Zhang L.K., Wang B., Xin Q., Shang W., Shen S., Xiao G., Deng F., Wang H., Hu Z., Wang M. Quantitative proteomic analysis reveals unfolded-protein response involved in severe fever with thrombocytopenia syndrome virus infection. J. Virol. 2019;93 doi: 10.1128/JVI.00308-19. e00308–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in the current study are included in this published article and its supplementary materials.