Abstract
Background
Phoebe goalparensis is an endemic forest species of North East India that belongs to Lauraceae family. P. goalparensis is used as timbers yielding plants for commercial importance in the local furniture markets of North East India. A rapid in vitro micropropagation protocol was established by using apical and axillary shoot tips on Murashige and Skoog medium with varied concentrations of plant growth regulators.
Results
In this study, 5.0 mg/l BAP augmented medium was chosen as the best for shoot multiplication of the plant. However, IBA (2.0 mg/l) was the most responsive for root induction. Moreover, 70% of root induction was recorded during rooting experiment and 80–85% survivability was observed during the acclimatization of this species. Clonal fidelity of P. goalparensis was determined with ISSR marker and it was observed that in vitro raised plantlets were polymonomorphic.
Conclusion
Hence, an efficient protocol with high proliferation and rooting was established for P. Goalparensis that could aid in massive propagation in future.
Keywords: Phoebe goalparensis, Micropropagation, Clonal fidelity, ISSR, Polymonomorphic
Background
Natural resources (Forest trees) are over-exploited due to the growth of population pressure. Nowadays, a noticeable decrease is observed in the natural evergreen forest tree population of Assam. According to Assam Forest Report, 32.5 thousand hectares of forest cover in Assam has been decreased during 2009–2012 due to indiscriminate and illicit felling by the villagers and unscrupulous traders.
Phoebe goalparensis Hutchinson which belongs to the family Lauraceae is one of the indigenous evergreen large forest trees of Assam. It constitutes excellent light-weight and one of the most valuable timbers of Assam, known as ‘Bonsum’ in trade. It is the principal source of Bonsum timber, which is used for building structure, planking and furniture and all kinds of cabinet work, and it resembles the same properties of Teak, and hence, it is known as ‘Assam Teak’ [1]. Though it flowers profusely during April to May, almost all the ripened fruits are eaten by birds and wild animals. Hence, availability of the mature seeds is scanty and fresh seeds have 35–70% germination in the soil but germination is inhibited due to its hydrophobic nature. Moreover, seeds are recalcitrant and, therefore, cannot be stored for a long period [2]. Germination starts after about 25 days of sowing and is completed in about 4 months. Seedlings become ready for plantation after about 6 months of transplanting. Direct sowing is not found successful for this species [3]. Therefore, the above information gives a critical bottleneck in the implementation of the conservation through conventional mode for large-scale regeneration of planting material of the target species.
Under this concept, micropropagation is reported to be an effective method for the conservation of important germplasms, which can produce large-scale plantlets within a short time frame without any genetic variation [4]. Various factors like plant growth regulator, media and temperature could affect the results of micropropagation. Therefore, molecular methods such as Random Amplified Polymorphic DNA (RAPD), Restriction Fragment Length Polymorphism (RFLP), Inter Simple Sequence Repeat (ISSR) and Simple Sequence Repeat (SSR) are used for the assessment of genetic homogeneity. In this study, in vitro regeneration protocol of P. goalparensis for rising propagule quality from elite plant source was first attempted to establish and proliferate, and the genetic fidelity of the obtained plants was assessed. The clonal planting stocks are necessary to ensure the performance of micropropagated plantlets of P. goalparensis. Moreover, it is the first attempt to evaluate the in vitro culture on clonal fidelity study of P. goalparensis. Therefore, an approach with biotechnological tools and techniques in regeneration of quality propagates and also for ex situ conservation with sustainable exploitation of this endemic species is highly essential.
Methods
Seedlings of P. goalparensis Hutchinson were collected from State Forest Department of Assam and established in the field of CSIR-NEIST Experimental farm for rising of in vitro culture as a mother plant (Fig. 1A). The apical and axillary shoot buds (3–4 cm) were excised from clone number Pg7 and cleaned smoothly under tap water for removal of dust and dart (Fig. 1B). Explants’ material was treated with 5% (v/v) Tween 20 solution for 1 h and 0.3% Bavistin for 30 min followed by rigorous cleaning with running water for 4–5 times and disinfected in 70% ethanol for 1 min, 0.1% Hgcl2 solution (v/v) for 8 min, and then flushed 3–4 times each in germfree double-distilled water.
Fig. 1.
In vitro propagation of Phoebe goalparensis. A Mother plant. B Preparation of explants material. C–F Emergence of shoot buds. G, H Shoot multiplication. I Root induction. J Hardening. K Acclimatization and soil transfer
Induction of in vitro shoot proliferation
Higher concentrations of TDZ causes no shoot proliferation, only lower concentration of it shows shoot proliferation [5]. Therefore, the surface-sterilized explants were cultured into BAP, Kin (0.5–5.0 mg/l) and TDZ (0.5–2.0 mg/l) enriched MS [6] medium for in vitro shoot proliferation augmented with 3% sucrose and 0.8% Agar powder from Hi Media Chemicals, India. All cultures were incubated under 16-h photoperiod at 24±2°C temperature with 35 μ mol m−2 s−1 of light power (cool white luminous light).
Root induction
Woody Plant Medium (WPM) of Lloyd and McCown [7] was used for rooting trials. Root induction experiment was conducted with various concentrations of auxins like NAA and IBA (0.5–5mg/l). The hardening procedure was involved by few steps process, i.e. shift of rooted shoot to MS basal liquid medium for 15–20 days, which was pursued by exhibiting the rooted shoots in half strength of MS basal liquid medium for another 15 days. Roots were in liquid medium so it was washed thoroughly in running tap water to remove the traces of MS liquid medium and caps of the flasks were also removed along with periodical rinsing with antifungal solution of Bavistin fungicide (0.2% solution). After that, plantlets were kept in filtered water for total of 30 days likely 15 days in culture room and subsequently another 15 days in ambient room temperature.
Clonal fidelity assessment of raised plantlets by ISSR
Clonal fidelity is a technique for determining the genetic stability of micropropagated plantlets before they are delivered to extensive plantations or other commercial applications. To assess the clonal fidelity, 12 in vitro raised plants were randomly selected along with its mother plant Pg7. ISSR was used to determine genetic stability and soma clonal variation of plantlets derived through micropropagation protocol. These plants were compared with the mother plant from which explants were taken. Total genomic DNA of the mother plant and in vitro raised plants was extracted from tender leaves using modified cetyltrimethyl ammonium bromide (CTAB) method [8] and quantified using Nano Drop spectrophotometer (G:BOX Syngene, UK). Initially, 25 ISSR (Genei) primers were screened and 10 ISSR primers were selected based on clear banding patterns. PCR amplification was performed in Thermal Cycler (Applied biosystem) with a total volume of 20μl reaction mixtures having 15ng/μl of DNA template, 10X PCR buffer, 2.5mM dNTPs, 5pmol primer, 1U Taq DNA polymerase (Sigma) and the final volume was adjusted with nuclease-free water. The PCR reaction programmes for ISSR were set at 94°C for 4 min followed by 35 cycles of 94°C for 30s, 90s at annealing temperature 45–50°C, according to primer’s Tm (melting temperature) then 2-min elongation at 72°C and 72°C final extension at 10 min. The amplified products were analysed in 1.5% agarose gel at 95V for 1.5h and visualized in the gel documentation system (G:BOX, Syngene, UK). Low range DNA ladder of 100bp to 1kb (Genei) was used as a standard for each marker analysis.
Data analysis
The experiments were conducted in completely randomized design in different replication in different experiments. Experimental results were analysed statistically using the techniques of analysis of variance for single-factor experiments. The significance of the treatment means differences were tested by the procedure of Duncan’s multiple range test [9].
The scoring of ISSR was done manually in a binary format (“1” for presence and “0” for absence of bands) and distinctive bands were calculated, whereas the unclear or faint bands were not considered for further analysis. To differentiate each marker, the parameters like the total number of bands, number of polymorphic and monomorphic bands, percentage of polymorphism, polymorphic information content (PIC), resolving power (RP) and marker index (MI) were evaluated using Microsoft Excel. The value of PIC was calculated as PIC=1−∑Pi2, where pi is the frequency of ith allele (band present) and summation extends over n alleles [10]. Resolving power (Rp) of each primer was estimated with Rp=∑Ib where Ib (informative fragments) = 1− [2 x (0.5−pi2)], where pi2 is the proportion of accession containing bands [11]. NTSYS (Numerical Taxonomy and Multivariate Analysis System) software v2.1 was used to generate the distance matrix and cluster analysis of the datasets [12]. A dendrogram representing the genetic association revealed by the similarity coefficient was generated using UPGMA with the Sequential agglomerative hierarchical and nested clustering method programme in NTSYS (SAHN).
Results
In vitro shoot regeneration
Among the various treatments, BAP (0.5–5.0 mg/l) enriched medium enhanced shoot bud proliferation in this species (Table 1). 1–3 numbers of shoot buds were induced from the cultured explants within 15–30 days of inoculation (Fig. 1C–F). 5.0 mg/l BAP augmented MS medium resulted in maximum number (5.3a) of shoot induction (Fig. 1G, H, Table 1). During 7–10 days of culture, shoot bud induction resulted. Lower conc. of BAP (0.5–1.0) reduced shoot bud induction. However, higher conc. of BAP resulted from an intervening callus phase at the basal portion of the explants. Effects of all three cytokinins i.e. BAP, Kin, and TDZ resulted in similar tendencies at lower and higher concentrations. But BAP manifested a notably higher effect on the number of shoot bud induction, and shoot length. MS basal medium was unresponsive towards shoot bud formation during the study.
Table 1.
Effect of cytokinins on shoot proliferation of Phoebe goalparensis
| Hormonal composition (mg/l) | Shoot number | Shoot length (cm) | Leaf number |
|---|---|---|---|
| Basal | 0.0 | 0.0 | 0.0 |
| BAP 0.5 | 2.7cd | 1.3fgh | 1.67fgh |
| BAP 1.0 | 4.0b | 2.17cdef | 2.33def |
| BAP 2.0 | 3.7bc | 3.27b | 3.67bc |
| BAP 5.0 | 5.3a | 4.8a | 5.3a |
| KIN 0.5 | 2.0d | 1.00h | 1.00h |
| KIN 1.0 | 2.3d | 1.97defg | 2.00efg |
| KIN 2.0 | 2.7cd | 2.5bcd | 2.67de |
| KIN 5.0 | 3.0bcd | 3.07bc | 4.3b |
| TDZ 0.5 | 2.0d | 1.83defgh | 1.67fgh |
| TDZ 1.0 | 2.3d | 2.33cde | 1.67fgh |
| TDZ 2.0 | 3.0bcd | 3.03bc | 3.67bc |
Means are from four repeated experiments. Each treatment consisted of three replicates. Means within a row followed by the same letters are not significant at P = 0.05 according to DMRT
Shoot induction acquired by Kin was found to be less effective than BAP. In this experiment, the role of Kin (1.0 and 5.0 mg/l) was not significant at all. Higher conc. of BAP (5.0 mg/l) and Kin (1.0 and 5.0 mg/l) did not show any significant result towards shoot/root formation.
Root induction
After 10 days of inoculation, root initiation was noticed in IBA (2.0 mg/l) enriched WPM medium and at this conc. the root number (6.67a), length of roots (2.70a) and root induction percentage (70%) were also higher in comparison with other conc. (Fig. 1I). But, in 4.0 mg/l IBA containing WPM medium rooting was noticed after 20 days of inoculation with retarded root growth. In case of NAA-enriched WPM medium root initiation was delayed by 25–30 days and root growth was reduced than IBA. Moreover, IAA was not at all responsive for root induction of this species. At the higher conc. of IBA (4.0–5.0mg/l), reduced percentage of rooting was observed in this species (Table 2).
Table 2:
Effect of auxins on in vitro root induction of Phoebe goalparensis
| Hormonal composition (mg/l) | Root number | Root length (cm) | Root induction % | |
|---|---|---|---|---|
| NAA | IBA | |||
| 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 0.5 | 1.00g | 0.40e | 20 | |
| 1.0 | 1.67fg | 0.93cde | 41 | |
| 2.0 | 3.67bc | 1.70b | 50 | |
| 3.0 | 3.33bcd | 1.17bcd | 45 | |
| 4.0 | 2.33ef | 1.07bcde | 40 | |
| 5.0 | 1.33fg | 0.50de | 32 | |
| 0.5 | 1.33fg | 1.17bcd | 30 | |
| 1.0 | 3.67bc | 1.70b | 45 | |
| 2.0 | 6.67a | 2.70a | 70 | |
| 3.0 | 4.33b | 1.76b | 55 | |
| 4.0 | 3.00cde | 1.33bc | 50 | |
| 5.0 | 2.00fg | 1.00bcde | 35 | |
Means are from four repeated experiments. Each treatment consisted of three replicates. Means within a row followed by the same letters are not significant at P = 0.05 according to DMRT
Hardening plantlets of P. goalparensis were shifted to polythene sleeves containing 1:1:2 soil: sand: cow-dung mixture for acclimatization and retained in a net house for 30–40 days prior to field transfer (Fig. 1J, K).
Clonal fidelity of tissue culture raised plantlets by ISSR
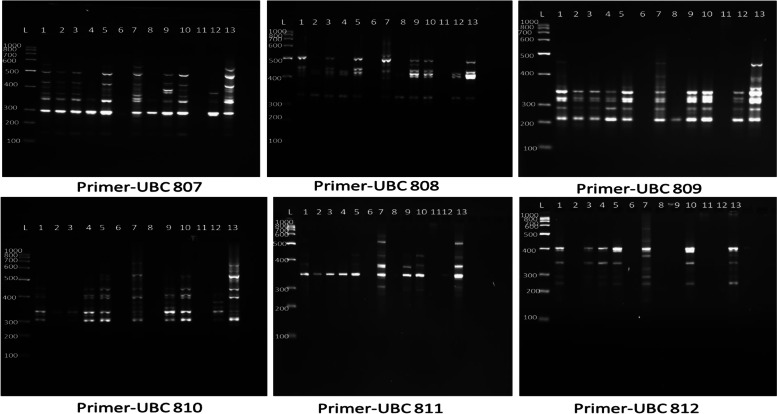
ISSR markers are the direct reflection of abundances and distribution of microsatellite repeat in the genome. Out of 25 screened ISSR primers, 10 anchored ISSR primers produced 73 clear bands with 13 DNA samples of micropropagated plantlets (Fig. 2). The number of amplified fragments varied from 3 to 11, with an average of 7.30 bands per primer (Table 3). Among 73 bands, 62 bands were polymorphic and the average polymorphic percentage was 85.40% across all the plantlets. Highest percentage of polymorphism (100%) was found with the primer numbers UBC812, UBC818 and UBC834, and lowest percentage (71.43%) was found in 2 primers, i.e. UBC808 and UBC809. The PIC value ranged from 0.45 (UBC807) to 0.79 (UBC812), with a mean of 0.63 per loci. The MI and Rp of the ISSR varied from 0.28 to 0.98 and 17.5 to 18.75 and the maximum MI and Rp were found in UBC810 and UBC807.
Fig. 2.
ISSR Products of Phoebe goalparensis amplified along with ISSR marker. A ISSR Primer-807. B ISSR Primer- 808. C ISSR Primer-809. D ISSR Primer-810. E ISSR Primer-811. F ISSR Primer-812
Table 3.
Result of different genetic expressions of Phoebe goalparensis showing the percentage of monomorphism and Polymorphism, PIC, EMR and MI
| Sl No. | Primer | Sequences | Total no. of bands | Total no. of monomorphic bands | Total No. of polymorphic bands | Percentage of polymorphism | Polymorphic information content | EMR | MI |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 807 | AGAGAGAGAGAGAGAGT | 8 | 2 | 6 | 75 | 0.45 | 4.5 | 2.04 |
| 2 | 808 | AGAGAGAGAGAGAGAGC | 7 | 2 | 5 | 71.43% | 0.54 | 3.6 | 1.97 |
| 3 | 809 | AGAGAGAGAGAGAGAGG | 7 | 2 | 5 | 71.43% | 0.54 | 3.6 | 1.95 |
| 4 | 810 | GAGAGAGAGAGAGAGAT | 11 | 1 | 10 | 90.90% | 0.61 | 9.1 | 5.58 |
| 5 | 811 | GAGAGAGAGAGAGAGAC | 5 | 1 | 4 | 80% | 0.75 | 3.2 | 2.43 |
| 6 | 812 | GAGAGAGAGAGAGAGAA | 6 | 0 | 6 | 100% | 0.79 | 6 | 4.75 |
| 7 | 818 | CACACACACACACACAG | 3 | 0 | 3 | 100% | 0.66 | 3 | 1.99 |
| 8 | 826 | ACACACACACACACACC | 8 | 1 | 7 | 87.5% | 0.60 | 6.12 | 3.72 |
| 9 | 834 | AGAGAGAGAGAGAGAGYT | 9 | 0 | 9 | 100% | 0.73 | 9 | 6.59 |
| 10 | 836 | AGAGAGAGAGAGAGAGYA | 9 | 2 | 7 | 77.78% | 0.67 | 5.44 | 3.67 |
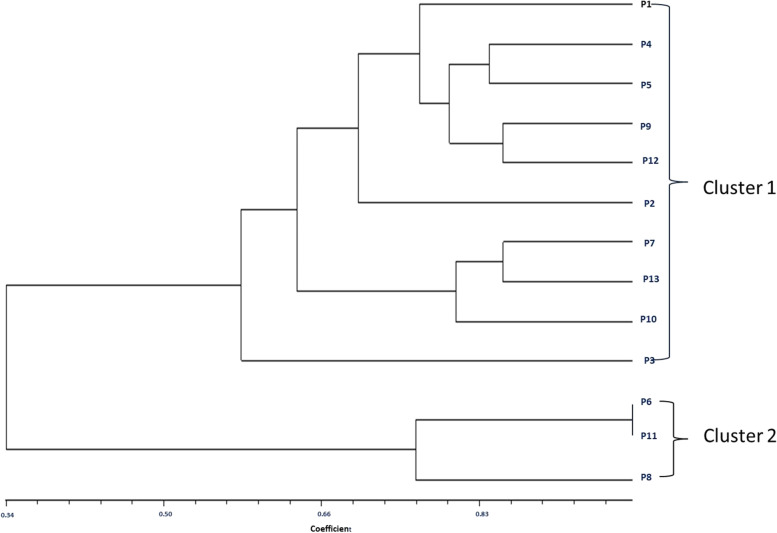
The genetic similarities among the species of P. goalparensis were estimated according to the ISSR data. Jaccard’s coefficient showed that there were two closely related accessions, i.e. P6-P11 with the highest similarity index 0.9863014, and two distance-related accessions, i.e. P5-P11 with the lowest similarity index 0.1369863 respectively (Table 4). Cluster analysis was performed with NTSys using UPGMA method and two major clusters were generated (Fig. 3). Cluster 1 includes 10 genotypes with samples P1, P4, P5, P9, P12, P2, P7, P13, P10 and P3. Cluster 2 consists of 3 genotypes with P6, P11 and P8 samples.
Table 4.
Similarity matrix of 12 in vitro raised plantlets and mother plant (Pg7) of P. goalparensis
| P1 | P2 | P3 | P4 | P5 | P6 | P7 | P8 | P9 | P10 | P11 | P12 | P13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 1.0000000 | ||||||||||||
| P2 | 0.6438356 | 1.0000000 | |||||||||||
| P3 | 0.6164384 | 0.6164384 | 1.0000000 | ||||||||||
| P4 | 0.7534247 | 0.7260274 | 0.5342466 | 1.0000000 | |||||||||
| P5 | 0.8082192 | 0.6712329 | 0.5342466 | 0.8356164 | 1.0000000 | ||||||||
| P6 | 0.1780822 | 0.4520548 | 0.5616438 | 0.2876712 | 0.1506849 | 1.0000000 | |||||||
| P7 | 0.7671233 | 0.4931507 | 0.6027397 | 0.6027397 | 0.7123288 | 0.2191781 | 1.0000000 | ||||||
| P8 | 0.4109589 | 0.6575342 | 0.6301370 | 0.4931507 | 0.3561644 | 0.7671233 | 0.2876712 | 1.0000000 | |||||
| P9 | 0.7808219 | 0.7260274 | 0.5342466 | 0.7808219 | 0.8082192 | 0.2328767 | 0.6849315 | 0.4657534 | 1.0000000 | ||||
| P10 | 0.7397260 | 0.5205479 | 0.6575342 | 0.6849315 | 0.7397260 | 0.3013699 | 0.8082192 | 0.3698630 | 0.7671233 | 1.0000000 | |||
| P11 | 0.1643836 | 0.4383562 | 0.5479452 | 0.2739726 | 0.1369863 | 0.9863014 | 0.2054795 | 0.7534247 | 0.2191781 | 0.2876712 | 1.0000000 | ||
| P12 | 0.7123288 | 0.7397260 | 0.5205479 | 0.8219178 | 0.7671233 | 0.3287671 | 0.5890411 | 0.5068493 | 0.8493151 | 0.6986301 | 0.3150685 | 1.0000000 | |
| P13 | 0.6986301 | 0.4520548 | 0.6164384 | 0.5342466 | 0.6986301 | 0.2328767 | 0.8493151 | 0.3013699 | 0.5616438 | 0.7945205 | 0.2191781 | 0.5479452 | 1.0000000 |
Fig. 3.
Consensus tree of Phoebe goalparensis accession developed on the basis of their banding patterns with ISSR Markers
Discussion
Micropropagation provides a feasible substitute to seed propagation as it entitles rapid propagation of elite stock cultivars in a fairly short duration of time. For raising of quality plant material, genetic consistency of micropropagated plants is a prerequisite factor. It is well known that adenine uses its ontogenesis effects on growth and organ formation of plant tissues by determining nucleic acid metabolism [13]. Efficacy of BAP towards shoot multiplication was investigated in various plant species such as Eucalyptus [14], Psidium guava [15], Syzygiumcumini [16], Bambusanutans [17], Bambusabalcooa [18], Chlorophytumborivilianum [19], Calotropisprocera [20], Artemisia arborescens [21], Boerhaaviadiffusa [22], Andrographisalata [23] and in cucurbits also [24, 25]. In our study, among the three cytokinins BAP, Kin and TDZ, BAP was the most superior, followed by Kin and TDZ. This finding was in accordance with Curcuma caesia, which initiated BAP was more suitable for shoot proliferation than Kin and TDZ [26, 27]. Similarly, BAP was mentioned as the most potent cytokinin for shoot induction and multiplication. The efficacy of BAP over other types of cytokinin in shoot multiplication might be due to the ability of the plant to metabolize BAP more readily as nucleotides, and ribosides stability is present naturally in BAP [28]. In contrast to our results, the regeneration potentiality of Kin is at least a fold and a half more than the percentage of regeneration using the other three types of cytokinin. The varied physiological response of using Kin than the other types of cytokinin was reported earlier. In Cucumissativus, Kin gives less than twofold of increasing number of shoots/explant than the other types of cytokinin with 1.0 mg/1 concentration. When 2.0 mg/1 concentration of TDZ was used, the regenerates using Kin, with a concentration of 1.0 mg/l, is more by 1.7 fold, which differed from our result [29].
It is prevalent that auxins play a pivotal role in the assurance of rooting capacity, which is crucial for vegetative propagation. The inclusion of exogenous auxin resolves the issue of difficult to root woody plant species and has been described to be imperative for root formation in various species like Oak [30], Chestnut [31] and Poplar [32]. However, the refreshing effect of IBA on rooting was earlier described in Excoecariaagallocha [33], Meliaazedarach [34], Bambusa vulgaris [35] and Eucalyptus globulus [36]. Contrary to our study, efficacy of NAA towards promoting root induction was well explained earlier [37–39]. Moreover, prior studies reported that high concentration of auxin inhibits shoot growth along with root elongation and stimulates cell differentiation [40].
In contrast, genetic instability occurs in the in vitro regenerated plants (somaclonal variation) due to use of hyper-optimum potency of growth regulators and continuous sub culturing. ISSR marker was used in order to detect genetic stability of in vitro raised 12 plantlets of P. goalparensis with the mother plant (Pg7). Allelic composition of 12 tissue culture raised plantlets and its mother plant were polymonomorphic. This result suggested somaclonal variation in P. goalparensis, which interfered with the integrity of the regenerated clonal plantlets. Somaclonal variation amid in vitro propagation may occur from pre-existing variations, the type of explant accustomed, the concentration and type of growth regulator in the medium, the number and duration of subcultures, effect of stress, genotype and the method of propagation espoused. The concentration of synthetic plant growth regulators in the medium is also associated with somaclonal variation [41]. Moreover, the preparation of many explants from only one donor plant augments the chances of variation in cultures [42].
The results of the ISSR marker system in the present study revealed the genetic variability among the in vitro raised plantlets of P. goalparensis. PCR contours of the 12 micropropagated plantlets for 11 ISSR primers exhibit polymorphism among themselves and with the mother plant. Bhatia et al. estimated clonal variation among the Gerbera plants by using ISSR marker regenerated from leaf explants [43]. The exact cause of variation might be due to explant source or due to mode of plant regeneration [44], media composition, or culture conditions and sub- or supra-optimal levels of plant phytohormones [45]. Still, somaclonal variation was reported for a numerous of in vitro derived plant species, such as Populus deltoides [46], Populus tremuloides [47], Prunus persica [48], Actinidia deliciosa [49], grapevine cv. Crimson Seedless [50], Solanum tuberosum [51], etc.
Unlike our findings, genetic stability was reported in several cases, namely, Picea mariana plants regenerated from somatic embryogenesis [52], micropropagated shoots of Pinus thunbergii Parl [44]., adventitious shoots of Pinus taeda [53], axillary bud proliferation of chestnut rootstock hybrids [54] and tissue culture-derived plantlets of Almond [55] and sugarcane [56].
Conclusion
A high shoot multiplication rate for rooting and reproducible protocol for micropropagation of P. goalparensis was established. This study indicates that 5mg/l BAP augmented medium was the best for shoot multiplication and ISSR analysis accessed the genetic stability of in vitro raised 12 plantlets and suggests that the genetic variability among the in vitro raised plantlets of P. goalparensis could be used in the future for large-scale propagation.
Acknowledgements
The authors are thankful to Dr. G. N. Sastry, Director, CSIR-North-East Institute of Science and Technology, Jorhat, Assam, India, for providing facilities.
Authors’ contributions
Dr. KDM, Dr. BG, Ms. GB, Ms. MH, Ms. TT and Ms. KB designed and performed the experiment, analysed the data and wrote the manuscript. Dr. KDM and Mr. HL had contributed to the conception and design of the work. Dr. SPS contributed to the conception and design of the work and also provided technical support and drafted the manuscript. The authors read and approved the final manuscript.
Funding
A financial grant to KDM from Department of Biotechnology (DBT), Govt. of India, New Delhi, in the form of BioCare project (SAN No.102/IFD/SAN/3384/2014-15 dated Nov 19, 2014) is gratefully acknowledged.
Availability of data and materials
The data are available within the manuscript and also accessible from the corresponding author upon request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Given by all authors.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kanjilal UN, Kanjilal PC, De RN, Das A. Flora of Assam. Vol. IV. Nyctaginaceae to Cycadaceae. Flora of Assam. Vol. IV. Nyctaginaceae to Cycadaceae. 1940. [Google Scholar]
- 2.Berjak P, Pammenter N. Implications of the lack of desiccation tolerance in recalcitrant seeds. Front Plant Sci. 2013;4:478. doi: 10.3389/fpls.2013.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kundu M, Chanda S, Kachari J. Germination and storage behaviour of Phoebe goalparensis Hutch seeds. Seed SciTechnol. 2003;31(3):659–666. doi: 10.15258/sst.2003.31.3.15. [DOI] [Google Scholar]
- 4.Sudha CG, Seeni S. In vitro multiplication and field establishment of Adhatodabeddomei CB Clarke, a rare medicinal plant. Plant Cell Rep. 1994;13(3):203–207. doi: 10.1007/BF00239893. [DOI] [PubMed] [Google Scholar]
- 5.Pai SR, Desai NS. Effect of TDZ on various plant cultures. Thidiazuron: From urea derivative to plant growth regulator; 2018. pp. 439–454. [Google Scholar]
- 6.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 7.Lloyd G, McCown B. Commercially-feasible micropropagation of mountain laurel, Kalmia latifolia, by use of shoot-tip culture. Comb proc Int Plant Propag Soc. 1980;30:421–427. [Google Scholar]
- 8.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem bull. 1987;19(1):11–15. [Google Scholar]
- 9.Duncan DB. Multiple ranges and multiple F test. Biometrics. 1955;11:1–42. doi: 10.2307/3001478. [DOI] [Google Scholar]
- 10.Liu B, Larsson L, Franssens V, Hao X, Hill SM, Andersson V, Hoglund D, Song J, Yang X, Oling D, Grantham J. Segregation of protein aggregates involves actin and the polarity machinery. Cell. 2011;147(5):959–961. doi: 10.1016/j.cell.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Prevost A, Wilkinson MJ. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. TheorAppl Genet. 1999;98(1):107–112. doi: 10.1007/s001220051046. [DOI] [Google Scholar]
- 12.Rohlf FJ. NTSyS-p.c. Numerical Taxonomy and Multivariate Analysis System (Version 2.0) Exeter Software Publishers Ltd., Setauket; 1998. [Google Scholar]
- 13.NowakowskaK PA, Siedlecka E, Pacholczak A. The effect of cytokinins on shoot proliferation, biochemical changes and genetic stability of Rhododendron ‘Kazimierzodnowiciel’ in the in vitro cultures. Plant Cell Tiss Organ Cult. 2021;4:1–0. doi: 10.1007/s11240-021-02206-z. [DOI] [Google Scholar]
- 14.Gupta PK, Mehta UJ, Mascarenhas AF. A tissue culture method for rapid clonal propagation of mature trees of Eucalyptus torelliana and Eucalyptus camaldulensis. Plant Cell Rep. 1983;2(6):296–299. doi: 10.1007/BF00270185. [DOI] [PubMed] [Google Scholar]
- 15.Amin MN, Jaiswal VS. Rapid clonal propagation of guava through in vitro shoot proliferation on nodal explants of mature trees. Plant Cell Tiss Organ Cult. 1987;9(3):235–243. doi: 10.1007/BF00040809. [DOI] [Google Scholar]
- 16.Yadav U, Lal M, Jaiswal VS. In vitro micropropagation of the tropical fruit tree Syzygium cuminii L. Plant Cell Tiss Organ Cult. 1990;21(1):87–92. doi: 10.1007/BF00034498. [DOI] [Google Scholar]
- 17.Yasodha R, Sumathi R, Malliga P, Gurumurthi K. Genetic enhancement and mass production of quality propagules of Bambusa nutans and Dendrocalamus membranaceous. Indian Forester. 1997;123(4):303–306. [Google Scholar]
- 18.Mudoi KD, Borthakur M. In vitro micropropagation of Bambusa balcooa Roxb. through nodal explants from field-grown culms and scope for upscaling. CurrSci. 2009;10:962–966. [Google Scholar]
- 19.Khatri P, Rana JS, Sindhu A, Jamdagni P. Effect of additives on enhanced in-vitro shoot multiplication and their functional group identification of Chlorophytum borivilianum Sant. Et Fernand. SN ApplSci. 2019;1(9):1–10. doi: 10.1007/s42452-019-1118-8. [DOI] [Google Scholar]
- 20.Sethy R, Kullu B. Micropropagation of ethnomedicinal plant Calotropis sp. and enhanced production of stigmasterol. Plant Cell Tissue Organ Cult. 2022;149:147–158. doi: 10.1007/s11240-022-02231-6. [DOI] [Google Scholar]
- 21.Riahi L, Chograni H, Ben RF, Ben RM, Masmoudi AS, Cherif A. Efficient in vitro regeneration of the endangered species Artemisia arborescens L. through direct organogenesis and impact on secondary metabolites production. Hortic Environ Biotechnol. 2022;63:439–450. doi: 10.1007/s13580-021-00400-8. [DOI] [Google Scholar]
- 22.Majumder S, Biswas A, Rahman MM. In vitro propagation and cytological analysis of Boerhaaviadiffusa L. - An important medicinal plant species of Bangladesh. Am J Plant Sci. 2021;12:1347–1360. doi: 10.4236/ajps.2021.129095. [DOI] [Google Scholar]
- 23.Kadapatti SS, Murthy HN (2021) Rapid plant regeneration, analysis of genetic fidelity, and neoandrographolide content of micropropagated plants of Andrographis alata (Vahl) Nees. J Genet EngBiotechnol:19–20 [DOI] [PMC free article] [PubMed]
- 24.Saha S, Mori H, Hattori K. Synergistic effect of kinetin and benzyl adenine plays a vital role in high frequency regeneration from cotyledonary explants of bottle gourd (Lagenaria siceraria) in relation to ethylene production. Breed Sci. 2007;57:197–202. doi: 10.1270/jsbbs.57.197. [DOI] [Google Scholar]
- 25.Ganasan K, Huyop F. In vitro regeneration of Citrullus lanatuscv. Round dragon. J BiolSci. 2010;10:131–137. doi: 10.3923/jbs.2010.131.137. [DOI] [Google Scholar]
- 26.Fong YM, Sani HB. Studies on micropropagation of Curcuma caesia Roxb. (Kunyit Hitam) Int J InnovSci EngTechnol. 2019;6:101–106. [Google Scholar]
- 27.Haida Z, Sinniah UR, Nakasha JJ, Hakiman M. Shoot Induction, Multiplication, Rooting and Acclimatization of Black Turmeric (Curcuma caesia Roxb.): an important and endangered Curcuma species. Horticulturae. 2022;8:740. doi: 10.3390/horticulturae8080740. [DOI] [Google Scholar]
- 28.Khajuria AK, Bisht NS, Bhagat N. In vitro organogenesis and plant regeneration of Thymus serpyllum L.: An important aromatic medicinal plant. In Vitro Cell DevBiol Plant. 2020;56:652–661. doi: 10.1007/s11627-020-10094-9. [DOI] [Google Scholar]
- 29.Abu-Romman S, Al-Hadid EKA, Arabiyyat AR (2015) Kinetin is the most effective cytokinin on shoot multiplication from cucumber. J Agri Sci 7(10) ISSN 1916-9752 E-ISSN 1916-9760
- 30.Chalupa V. In vitro propagation of oak (Quercus robur L.) and linden (Tilia cortdata Mill.) Biol Plant. 1984;26:374–377. doi: 10.1007/BF02898577. [DOI] [Google Scholar]
- 31.Mato MC, Vieitez AM. Changes in auxin protectors and IAA oxidases during the rooting of chestnut shoots in vitro. Physiol Plant. 1986;66:491–494. doi: 10.1111/j.1399-3054.1986.tb05956.x. [DOI] [Google Scholar]
- 32.Hausman JF, Kevers C, Gaspar T. Involvement of putrescine in the inductive rooting phase of poplar shoots in vitro. Physiol Plant. 1994;92:201–206. doi: 10.1111/j.1399-3054.1994.tb05327.x. [DOI] [Google Scholar]
- 33.Rao CS, Eganathan P, Anand A, Balakrishna P, Reddy TP. Protocol for in vitro propagation of Excoecariaagallocha L., a medicinally important mangrove species. Plant Cell Rep. 1998;17(11):861–865. doi: 10.1007/s002990050498. [DOI] [PubMed] [Google Scholar]
- 34.Thakur R, Rao PS, Bapat VA. In vitro plant regeneration in Melia azedarach L. Plant Cell Rep. 1998;18(1):127–131. doi: 10.1007/s002990050544. [DOI] [Google Scholar]
- 35.Ndiaye A, Diallo MS, Niang D, Gassama-Dia YK (2006) In vitro regeneration of adult trees of Bambusa vulgaris. Afr J Biotechnol 5(13). 10.5897/AJB06.149
- 36.Fogaça CM, Fett-Neto AG. Role of auxin and its modulators in the adventitious rooting of Eucalyptus species differing in recalcitrance. Plant Growth Regul. 2005;45:1–10. doi: 10.1007/s10725-004-6547-7. [DOI] [Google Scholar]
- 37.Nisa C, Rodinah (2005) Tissue culture of several banana (Musa paradisiaca L.) with NAA and kinetin treatment. Bioscientiae 2:23-36.
- 38.Karjadi AK, Buchori A. Effect of NAA and BAP on the growth of garlic meristem tissue on medium B5. J Hort. 2007;17:217–223. [Google Scholar]
- 39.Sirchl MHT, Kadir MA, Aziz MA, Rashid AA, Rapat A, Javadi MB. Amelioration of mangosteen micro propagation through leaf and seed segments (Garcinia mangostana L.) Afr J Biotechnol. 2008;7:2025–2029. doi: 10.5897/AJB2008.000-5051. [DOI] [Google Scholar]
- 40.Harahap F, Poerwanto R, Suharsono SC, Rahayu S. In vitro growth and rooting of mangosteen (Garcinia mangostana L.) on medium with different concentrations of plant growth regulator. Hayati. J Biosci. 2014;21(4):151–158. doi: 10.4308/hjb.21.4.151. [DOI] [Google Scholar]
- 41.Martin KP, Pachathundikandi SK, Zhang CL, Slater A, Madassery J. RAPD analysis of a variant of banana (Musa sp.) cv. Grande Naine and its propagation viashoot tip culture. In Vitro Cell Dev Biol Plant. 2006;42:188–192. doi: 10.1079/IVP2005736. [DOI] [Google Scholar]
- 42.Kunitake H, Koreeda K, Mii M. Morphological and cytological characteristics of protoplast-derived plants of statice (Limonium perezii Hubbard) Scientia Horticulturae. 1995;60:305–312. doi: 10.1016/0304-4238(94)00713-P. [DOI] [Google Scholar]
- 43.Bhatia A, Pathak H, Jain N, Singh PK, Singh AK. Global warming potential of manure amended soils under rice–wheat system in the Indo-Gangetic plains. Atmos Environ. 2005;39(37):6976–6984. doi: 10.1016/j.atmosenv.2005.07.052. [DOI] [Google Scholar]
- 44.Goto S, Thakur RC, Ishii K. Determination of genetic stability in long-term micropropagated shoots of Pinus thunbergii Parl. using RAPD markers. Plant Cell Rep. 1998;18(3):193–197. doi: 10.1007/s002990050555. [DOI] [PubMed] [Google Scholar]
- 45.Bednarek PT, Orłowska R. Plant tissue culture environment as a switch-key of (epi) genetic changes. Plant Cell Tiss Organ Cult. 2020;140(2):245–257. doi: 10.1007/s11240-019-01724-1. [DOI] [Google Scholar]
- 46.Rani V, Parida A, Raina SN. Random amplified polymorphic DNA (RAPD) markers for genetic analysis in micropropagated plants of Populus deltoides Marsh. Plant Cell Rep. 1995;14(7):459–462. doi: 10.1007/BF00234055. [DOI] [PubMed] [Google Scholar]
- 47.Rahman M, Rajora O. Microsatellite DNA somaclonal variation in micropropagated trembling aspen (Populus tremuloides) Plant Cell Rep. 2001;20(6):531–536. doi: 10.1007/s002990100365. [DOI] [Google Scholar]
- 48.Hashmi G, Huettel R, Meyer R, Krusberg L, Hammerschlag F. RAPD analysis of somaclonal variants derived from embryo callus cultures of peach. Plant Cell Rep. 1997;16(9):624–627. doi: 10.1007/BF01275503. [DOI] [PubMed] [Google Scholar]
- 49.Palombi MA, Damiano C (2001) Use of different molecular markers, RAPDs and SSRs, to investigate clonal stability in micropropagated kiwifruit (Actinidia deliciosa A. Chev.). Acta Hortic:609–614. 10.17660/ActaHortic.2001.546.85
- 50.Nookaraju A, Agrawal DC. Genetic homogeneity of in vitro raised plants of grapevine cv. Crimson Seedless revealed by ISSR and microsatellite markers. S Afr J Bot. 2012;78:302–306. doi: 10.1016/j.sajb.2011.08.009. [DOI] [Google Scholar]
- 51.Fossi M, Amundson K, Kuppu S, Britt A, Comai L. Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 2019;180(1):78–86. doi: 10.1104/pp.18.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Isabel N, Tremblay L, Michaud M, Tremblay FM, Bousquet J. RAPDs as an aid to evaluate the genetic integrity of somatic embryogenesis-derived populations of Picea mariana (Mill.) BSP. Theor Appl Genet. 1993;86(1):81–87. doi: 10.1007/BF00223811. [DOI] [PubMed] [Google Scholar]
- 53.Tang W. In vitro regeneration of loblolly pine and random amplified polymorphic DNA analyses of regenerated plantlets. Plant Cell Rep. 2001;20(2):163–168. doi: 10.1007/s002990000297. [DOI] [PubMed] [Google Scholar]
- 54.Carvalho CH, Zehr UB, Gunaratna N, Anderson J, Kononowicz HH, Hodges TK, Axtell JD. Agrobacterium-mediated transformation of sorghum: factors that affect transformation efficiency. Genet Mol Biol. 2004;27(2):259–269. doi: 10.1590/S1415-47572004000200022. [DOI] [Google Scholar]
- 55.Martins M, Sarmento D, Oliveira MM. Genetic stability of micropropagated almond plantlets, as assessed by RAPD and ISSR markers. Plant Cell Rep. 2004;23(7):492–496. doi: 10.1007/s00299-004-0870-3. [DOI] [PubMed] [Google Scholar]
- 56.Thorat AS, Sonone NA, Choudhari VV, Devarumath RM, Babu KH. Plant regeneration from cell suspension culture in Saccharum officinarum L. and ascertaining of genetic fidelity through RAPD and ISSR markers. 3 Biotech. 2017;7(1):1–2. doi: 10.1007/s13205-016-0579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available within the manuscript and also accessible from the corresponding author upon request.