Summary
Background
Echocardiography (echo) based machine learning (ML) models may be useful in identifying patients at high-risk of all-cause mortality.
Methods
We developed ML models (ResNet deep learning using echo videos and CatBoost gradient boosting using echo measurements) to predict 1-year, 3-year, and 5-year mortality. Models were trained on the Mackay dataset, Taiwan (6083 echos, 3626 patients) and validated in the Alberta HEART dataset, Canada (997 echos, 595 patients). We examined the performance of the models overall, and in subgroups (healthy controls, at risk of heart failure (HF), HF with reduced ejection fraction (HFrEF) and HF with preserved ejection fraction (HFpEF)). We compared the models' performance to the MAGGIC risk score, and examined the correlation between the models’ predicted probability of death and baseline quality of life as measured by the Kansas City Cardiomyopathy Questionnaire (KCCQ).
Findings
Mortality rates at 1-, 3- and 5-years were 14.9%, 28.6%, and 42.5% in the Mackay cohort, and 3.0%, 10.3%, and 18.7%, in the Alberta HEART cohort. The ResNet and CatBoost models achieved area under the receiver-operating curve (AUROC) between 85% and 92% in internal validation. In external validation, the AUROCs for the ResNet (82%, 82%, and 78%) were significantly better than CatBoost (78%, 73%, and 75%), for 1-, 3- and 5-year mortality prediction respectively, with better or comparable performance to the MAGGIC score. ResNet models predicted higher probability of death in the HFpEF and HFrEF (30%–50%) subgroups than in controls and at risk patients (5%–20%). The predicted probabilities of death correlated with KCCQ scores (all p < 0.05).
Interpretation
Echo-based ML models to predict mortality had good internal and external validity, were generalizable, correlated with patients’ quality of life, and are comparable to an established HF risk score. These models can be leveraged for automated risk stratification at point-of-care.
Funding
Funding for Alberta HEART was provided by an Alberta Innovates - Health Solutions Interdisciplinary Team Grant no. AHFMRITG 200801018. P.K. holds a Canadian Institutes of Health Research (CIHR) Sex and Gender Science Chair and a Heart & Stroke Foundation Chair in Cardiovascular Research. A.V. and V.S. received funding from the Mitacs Globalink Research Internship.
Keywords: Echocardiography, Machine learning, Deep learning, Mortality, Heart failure, Prognostic models, Functional status
Abbreviations: Echo, Echocardiography; ML, Machine learning; HF, Heart failure; HFrEF, Heart failure with reduced ejection fraction; HFpEF, Heart failure with preserved ejection fraction; KCCQ, Kansas city cardiomyopathy questionnaire; AUROC, Area under the receiver-operating curve; PLAX, Parasternal long axis; Alberta HEART, Alberta Heart Failure Etiology and Analysis Research Team; MAGGIC, Meta-Analysis global group in chronic heart failure; CSS, Clinical summary score; OSS, Overall summary score; DNN, Deep neural networks; CNN, Convolution neural networks; IVS, Interventricular septal; LV, Left ventricular; LVIDs, Left ventricular internal dimension end-systolic; LVIDd, Left ventricular internal dimension end-diastolic; PWTDI, Pulsed-wave tissue Doppler imaging; ROC, Receiver operating characteristics; AUPRC, Area under the precision–recall curve; SHAP, SHapley additive exPlanations
Research in context.
Evidence before this study
All English-language articles in PubMed were screened from inception until May 2022 to identify studies that used “artificial intelligence” and “machine learning” (ML) for “mortality prediction” in different patient populations. Previous studies have suggested that echo-based deep learning models can be used to predict in-hospital and 1-year mortality in patients with suspected cardiovascular diseases. As an example, Ulloa Cerna et al. used convolutional neural networks to train on 812,278 echo videos from 34,362 patients and showed superior performance of deep learning models of echo videos in the prediction of one-year all-cause mortality compared to established clinical risk scores, cardiologists' clinical gestalt, or ML models based on human-crafted or electronic health records-driven parameters. However, none of these studies have examined the models’ performance in specific patient groups, such as those with heart failure (HF); modelled longer-term mortality beyond a 1-year time-period; or externally validated the models in independent cohorts.
Added value of this study
In this study, we developed deep learning- and gradient boosting-based models for 1-, 3- and 5-year mortality prediction using echo videos and expert-curated echo measurements from a cohort of patients with and without HF. We validated the models externally in an independent dataset captured in another country with predominantly distinct ethnicity, comorbidity burden, and outcomes. The deep learning models’ performance was better than or comparable to an established risk score [the Meta-Analysis Global Group in Chronic Heart Failure (MAGGIC) score] depending on HF subtype and follow-up period, as ResNet and CatBoost performed similarly across subgroups or showed better performance in HF with preserved ejection fraction (HFpEF), while MAGGIC score showed better performance in HF with reduced ejection fraction (HFrEF) compared to the HFpEF for the 1-and 3-year time-points. ResNet models predicted higher probability of death in HFpEF and HFrEF patients than in controls and at-risk patients. We also demonstrated the alignment of the deep learning-based mortality prediction probability with patient-reported functional status (measured by Kansas City Cardiomyopathy Questionnaire) at baseline.
Implications of all the available evidence
Echo-based machine-learning models can provide good-to-excellent prognostic information with respect to mortality outcomes. The deep-learning models can facilitate a fully automated decision support system that could be applied directly to images, prior to expert-curated annotations. The deep-learning models are generalizable, comparable in performance to established risk scores, and correlated with patient's quality of life measures. Our study findings support clinical implementation of point-of-care echo-based automated risk stratification systems.
Introduction
The global burden of heart failure (HF) is increasing over time due to ageing populations and it is associated with significant morbidity, mortality and healthcare resource utilization.1,2 Despite advancements in treatments over decades, the mortality rates remain high with a median survival of 5-years,2 with patients experiencing reduced functional status and quality of life.3 There is considerable interest in developing prognostic models to identify patients with HF who are at higher risk of adverse outcomes and could benefit from closer monitoring and more intense treatment.4
Echocardiography (echo) is the most common cardiac imaging modality employed in diagnosing and managing patients with HF.2 Previous studies have shown that deep learning models based on echo can be used to predict in-hospital and 1-year mortality in patients with suspected cardiovascular diseases.5,6 However, none of these earlier studies were focussed on patients with HF or have modelled long-term mortality beyond a 1-year time-period, and they have not been externally validated in independent cohorts. Therefore, their generalizability across ethnic, geographical location, operator and device-related variations remains to be tested. Furthermore, no previous study has examined how mortality predictions generated by machine learning (ML) algorithms align with patients-reported symptoms and functional status.
Accordingly, the objectives of our study were to develop echo-based deep learning models to predict patients' all-cause mortality at 1-, 3- and 5-years, and to test the models' generalizability in an external cohort of patients from a different country with predominantly distinct ethnicity, comorbidity burden, and outcomes. We further tested the validity of the models by: examining the models' predicted probability of death in subgroups of patients across the HF class spectrum; comparing the models' performance against an established risk score (the Meta-Analysis Global Group in Chronic Heart Failure (MAGGIC) risk score); and examining the correlation between the models’ predicted probability of death and patient-reported measures of symptoms and functional status as measured by the Kansas City Cardiomyopathy Questionnaire (KCCQ).
Methods
Training and internal validation dataset
We have followed the PRIME checklist for standardized reporting of cardiovascular imaging-related machine learning investigations to report our modelling methods and results.7 The models were trained and internally tested on a part of a dataset from the Mackay Memorial hospital in Taiwan (referred subsequently as the Mackay dataset).8 Among the various echo views, we selected the parasternal long axis (PLAX) view for modeling based on an earlier study which showed that the PLAX view is the most relevant for mortality prediction.6 The Mackay data included echos of healthy participants, patients with comorbidities without prevalent HF, and patients with HF (both inpatients and outpatients). The echos were performed during an annual cardiovascular health check-up at an outpatient clinic. We included 6083 echos with PLAX views (both videos and expert-curated echo measurements) from 3626 patients with at least 1-year follow-up on mortality status.
External validation dataset
The models were externally tested on 997 echos with a PLAX view from 595 patients collected as part of the Alberta Heart Failure Etiology and Analysis Research Team (Alberta HEART) Study.9 The Alberta HEART study was a prospective study that enrolled healthy controls, patients with comorbidities at risk of HF, patients with HF with preserved ejection fraction (HFpEF), and patients with HF with reduced ejection fraction (HFrEF) in the province of Alberta, Canada. The supplementary material provides more information on the Alberta and Mackay datasets.
We calculated the ‘Meta-Analysis Global Group in Chronic Heart Failure’10 (MAGGIC) risk score at baseline. The score is based on 13 predictor variables: age, sex, body mass index, systolic blood pressure, ejection fraction, creatinine, current smoker, diabetes, chronic obstructive pulmonary disease, New York Heart Association class, HF duration >18 months, β-blocker use, and angiotensin-converting enzyme inhibitor use. MAGGIC scores range from 0 to 40, with higher scores indicating higher risk status.
As part of the Alberta HEART study, patients completed the KCCQ at baseline, which collects information on several domains including physical function, symptoms, self-efficacy, social limitation, and quality of life.11 Responses are transformed into a clinical summary score (CSS) based on symptoms and physical function; and an overall summary score (OSS) incorporating all five above-mentioned domains. Both summary scores range from 0 to 100 with higher scores indicating better functional status.
Ethics
Data from Mackay Memorial Hospital was retrospectively identified. A waiver of consent was obtained from the Mackay Memorial Hospital institutional review board. All participants in Alberta HEART study signed informed consent, and the study was approved by the Health Research Ethics Boards at the University of Alberta, Covenant Health and the University of Calgary (Pro00117313). After consent, patients were enrolled and had a comprehensive clinical exam, and protocolized echo.
Echo selection
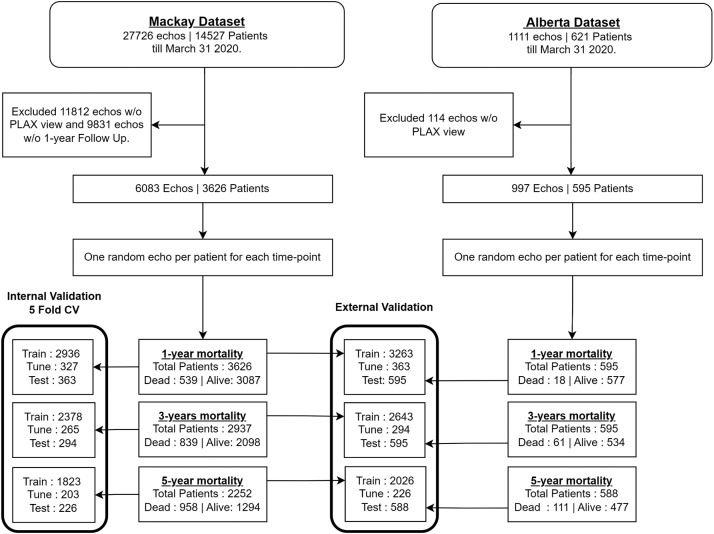
Number of days until death was calculated for each echo video based on the ‘death date minus the date of image acquisition’, and was used to assign ‘dead’ and ‘alive’ classes for modeling 1-, 3-, and 5-year time points. The Mackay dataset did not have fixed study duration or study end date, whereas in the Alberta HEART dataset, follow-up as of March 31, 2020 was available via linkage to provincial insurance and vital status registry data. Therefore, whenever the date of death was not available in the Mackay dataset, the last confirmed date of being alive was obtained from their last visit information. Echo videos were then used for modeling only if the confirmed ‘alive’ duration (last visit date minus the echo acquisition date) was greater than the respective 1-year, 3-year, or 5-year period (Fig. 1). For modeling each time point, only one echo video per patient was chosen randomly among the possible candidates. We made sure that all echo videos belonging to a particular patient were included exclusively either in the training or in the test set, so that none of the echo videos acquired in any of visits of a test patient are included in the training set, or vice versa.
Fig. 1.
Study flow chart showing the sample sizes used for training and testing prediction models. CV: cross-validation; PLAX: parasternal long axis; w/o: without.
Video preprocessing
Each video varied from 2 to 10 s in duration with a frame rate of 30–80 frames per second, covering 2-3 heart cycles. To ensure consistency, all the videos were converted to a constant frame rate of 30 frames per second. Videos were cleaned to remove texts and other patient information, and then down-sampled using cubic interpolation, and standardized to the same frame size of 112 × 122 (original dimensions depended on machine used for echo acquisition–GE Medical Systems Ultrasound: 640 × 432, Philips iE33 Ultrasound: 816 × 608). Given a large number of frames available per video, we divided the original videos to 16 sub-clips, and then, the first frame from each sub-clip was taken such that input for our deep learning model was consistently 16 frames. This choice of preprocessing was based on previous studies, which also used the equally spaced frames that provided uniform sampling along the temporal dimension.12, 13, 14, 15
Prediction models
We used supervised learning with CatBoost and ResNet-based deep learning algorithms to train our models to predict the probability of patients dying within 1-year, 3-year, and 5-year following the acquisition date of the echo. We developed six models using two algorithms for each of the three time points. CatBoost was chosen as it provides state-of-art performance for structured tabular data with fast training time, supports categorical and missing values intrinsically, and also provides explainability and visualization functions.16 For deep learning, we used ResNet architecture based on its successful performance in previous studies with similar datasets.17 Our ResNet models use non-annotated echo PLAX views, whereas the CatBoost models use patient characteristics and manually-acquired measurements. The models were implemented using PyTorch 1.8.1 and CatBoost 0.26.1 in Python 3.8.6.
For deep neural networks (DNN), we used modified 3D convolution neural networks (CNN), i.e. (2 + 1)D CNN (ResNet) with 31 million parameters, to extract the spatial and temporal features from the echo videos.18 The model was trained using the AdamW optimizer and Cross-Entropy as loss function.19 The learning rate was scheduled with an initial learning rate of 0.0005, for 25 maximum of epochs, and patience of 10 epoch interval. If validation loss in the tuning set continued to increase for an interval of 10 epochs, then the learning rate was reduced by a factor of 0.1 and the best model weights (the weights prior to the interval) were used.
We also trained a CatBoost model using 18 patient characteristics.16 These included two demographic features (age and sex) and the following 16 human expert-curated echo measurements.9 Interventricular septal (IVS) thickness, left ventricular (LV) posterior wall thickness at end-diastole, LV internal dimension both end-systolic and end-diastolic (LVIDs and LVIDd), LV end-diastolic volume, LV end-systolic volume, heart rate, deceleration time, isovolumic relaxation time, mitral valve E wave velocity (cm/s), mitral valve A wave velocity (cm/s), pulsed-wave Tissue Doppler imaging (PWTDI) for lateral and septal mitral annulus e’ velocity, left atrial max volume, tricuspid regurgitation velocity and LV ejection fraction. The hyperparameters for CatBoost–tree depth, and L2 regularization terms were tuned based on grid-search within the training sets. CatBoost models were learnt for a maximum of 600 epochs, and the learning process was stopped if validation loss in the tuning set did not reduce for 100 epochs. Class weights were enabled to ensure that both classes are given equal weights during gradient updates of the training process for both models. The video dataset used for ResNet was complete without any missingness, but echo measurements used for training and testing CatBoost models had missing values (reported in Table 1). We did not perform any data imputations, as patterns of missingness can have relevant information for prediction tasks and can be handled inherently by CatBoost. We used 10% of training data as a model tuning set for both ResNet and CatBoost models. The optimal cut-points for binarizing the predicted probabilities were estimated using the Youden index based on the training receiver operating characteristics (ROC) curve, such that the sum of the sensitivity and specificity is maximized.
Table 1.
Baseline characteristics of subjects in development and validation datasets.
| Patient Features | Description | Mackay dataset (n = 3626) |
Alberta dataset (n = 595) |
||||
|---|---|---|---|---|---|---|---|
| Missing | Median [Q1, Q3] | Mean (SD) | Missing | Median [Q1, Q3] | Mean (SD) | ||
| Gender | Female (n%) | 490 | 1247 (34.4) | 0 | 252 (42.4) | ||
| Male (n%) | 2379 (65.6) | 343 (57.6) | |||||
| Age | (in years) | 785 | 67.0 [53.0, 78.0] | 65.6 (15.4) | 0 | 67.0 [59.0, 75.0] | 66.3 (11.7) |
| LVEF | Left ventricular ejection fraction | 691 | 62.9 [55.0, 68.1] | 59.7 (12.4) | 34 | 58.6 [46.0, 65.6] | 54.6 (14.9) |
| IVS | Interventricular septal thickness | 551 | 9.7 [8.7, 11.0] | 11.3 (12.3) | 10 | 10.5 [9.0, 12.0] | 10.6 (2.4) |
| LVPW | Left ventricular posterior wall thickness at end-diastole (dcm) | 553 | 9.7 [8.8, 11.0] | 9.9 (1.9) | 10 | 10.0 [8.7, 11.8] | 10.2 (2.2) |
| LVIDd | Left ventricular internal dimension end-diastolic (dcm) | 552 | 47.1 [44.0, 50.3] | 47.0 (11.3) | 11 | 48.0 [43.0, 54.7] | 49.4 (9.2) |
| LVIDs | Left ventricular internal dimension end-systolic (dcm) | 552 | 31.0 [28.0, 35.0] | 32.7 (7.5) | 20 | 32.0 [26.4, 40.0] | 34.6 (11.4) |
| LVEDV | Left ventricle end-diastolic volume (mL) | 614 | 98.0 [80.3, 118.2] | 102.9 (36.1) | 35 | 105.8 [80.2, 145.9] | 120.6 (58.1) |
| LVESV | Left ventricle end-systolic volume (mL) | 611 | 35.9 [27.9, 51.0] | 46.3 (30.3) | 31 | 42.0 [29.1, 70.2] | 60.0 (48.2) |
| HR | Heart rate | 463 | 70.0 [62.0, 83.0] | 74.1 (18.3) | 3 | 65.0 [57.0, 73.0] | 66.6 (14.1) |
| DT | Deceleration time (ms) | 613 | 206.0 [170.0, 240.0] | 204.1 (77.0) | 412 | 250.0 [190.0, 300.0] | 252.2 (105.9) |
| IVRT | Isovolumic relaxation time (ms) | 638 | 89.0 [70.0, 100.0] | 89.5 (38.6) | 360 | 100.0 [80.0, 120.0] | 106.9 (51.5) |
| E | Mitral valve E wave velocity (cm/s) | 597 | 72.0 [57.6, 90.0] | 75.8 (28.1) | 336 | 77.3 [62.0, 96.2] | 84.9 (59.2) |
| A | Mitral valve A wave velocity (cm/s) | 574 | 40.0 [31.0, 71.2] | 51.8 (32.5) | 369 | 77.5 [61.5, 92.0] | 77.0 (28.8) |
| PWTDI lateral e’ | Pulsed-wave Tissue Doppler imaging lateral e’ velocity (cm/s) | 2369 | 8.9 [7.0, 11.0] | 9.1 (3.0) | 80 | 8.5 [6.5, 11.0] | 9.5 (6.5) |
| PWTDI septal e’ | Pulsed-wave Tissue Doppler imaging septal e’ velocity (cm/s) | 2694 | 7.5 [6.2, 9.1] | 7.9 (5.2) | 72 | 8.3 [5.9, 11.8] | 9.7 (5.4) |
| LA MAX Volume | Left atrial max volume (mlˆ3) | 3119 | 28.0 [22.4, 35.8] | 30.4 (11.1) | 366 | 62.3 [47.2, 87.0] | 69.6 (32.0) |
| TR Velocity | Tricuspid regurgitation velocity (m/s) | 3068 | 2.1 [1.9, 2.3] | 2.1 (0.4) | 302 | 2.1 [1.5, 2.6] | 2.0 (0.8) |
A: mitral valve A wave velocity; DT: deceleration time; E: mitral valve E wave velocity; HR: heart rate; IVRT: isovolumic relaxation time; IVS: interventricular septal thickness; LA: left atrial; LV: left ventricular; LVEDV: left ventricular end diastolic volume; LVEF: left ventricular ejection fraction; LVESV: left ventricular end systolic volume; LVIDd: left ventricular internal dimension at the end of diastole; LVIDs: left ventricular internal dimension at the end of systole; n: number; PWTDI: pulsed-wave Tissue Doppler imaging for mitral annulus e’ velocity; Q1: first quartile; Q3: third quartile; SD: standard deviation; TR: tricuspid regurgitation.
Evaluation and visualization
To evaluate the performance of our models, we used 5-fold cross-validation within the Mackay dataset for internal validation, and then retrained the model on the entire Mackay dataset and evaluated it on the Alberta HEART dataset for external validation. We reported the following performance metrics–area under the ROC curve (AUROC, also known as C-index), area under the precision–recall curve (AUPRC), F1 Score, Specificity, Recall, Precision and Accuracy. The performance scores were compared between models by bootstrapping 10,000 replicates of AUROC in the external validation dataset with random replacement sampling. The mean of pairwise differences between the model performances was estimated based on the bootstrap point estimate and 95% confidence intervals (CI). If the 95% CI contained zero, then the performance differences were considered not to be significant at the 0.05 level. We used this method to compare the AUROC performances of our models with that of the MAGGIC risk score.
We calculated the observed mortality rate at 1-year, 3-year and 5-year among Alberta HEART patients across tertiles of predicted probability of death from the ResNet models. We examined the mean predicted probabilities from the ResNet models as well as the AUROC performances across the patient subgroups (HFpEF, HFrEF, at risk, and controls). Lastly, we examined the correlation (spearman) between the predicted probabilities of death from the ResNet model with the baseline functional status scores–KCCQ CSS and KCCQ OSS.
We used GradCAM to visualize the gradient activation maps that contributed towards the model's prediction of a particular class.20 The gradients were pooled frame-wise and channel-wise and multiplied with the corresponding weights to get weighted activation channels. We represented these in an image superimposed with the first frame of the echo video to indicate regions that played a key role in the prediction. Also, we used SHAP21 (SHapley Additive exPlanations) to identify the echo measurements that were key contributors of average mortality prediction in the CatBoost models.
Role of the funders
The study was funded by general research funds available to Dr. Kaul as part of her Canadian Institutes of Health Research (CIHR) Sex and Gender Science Chair and a Heart & Stroke Foundation Chair in Cardiovascular Research. These agencies had no input into the study design, data collection, analysis, interpretation of data, writing of the report, or decision regarding publication.
Results
Table 1 shows patients’ demographic and echo characteristics in the Mackay and Alberta HEART cohorts. The mean age of participants in the Mackay cohort was 65.6 ± 15.4 years, while it was 66.3 ± 11.4 years in the Alberta HEART cohort. In the Mackay dataset, 34.4% were women, while the proportion was 42.4% in the Alberta HEART cohort.
Internal validation
The models were trained on 3626 patients with confirmed mortality status at 1-year in the Mackay cohort (Fig. 1). At 1-year, 539 (14.9%) had died. The 3-year and 5-year models were trained on 2937 and 2252 patients, respectively. Mortality rates in the 3- and 5-year cohorts were 28.6% and 42.5%, respectively. Table 2 and Fig. 2 show the models’ performances with 5-fold cross-validation in the Mackay data. The AUROC ranged from 85% for 1-year to 92% for 5-years Supplementary Table S1 shows the results for each fold of 5-fold cross-validation.
Table 2.
Model performance in the internal validation cohort (Mackay dataset) and the external validation cohort (Alberta HEART dataset).
| Timeline | Model | AUROC | AUPRC | F1 Score | Specificity | Recall | Precision | Accuracy | Observed |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Dead | Alive | |||||||||
| Mackay cohort (internal validation) | ||||||||||
| 1-year | ResNet | 0.85 ± 0.02 | 0.46 ± 0.04 | 0.52 ± 0.02 | 0.82 ± 0.02 | 0.73 ± 0.03 | 0.41 ± 0.02 | 0.80 ± 0.01 | 54 | 309 |
| CatBoost | 0.85 ± 0.00 | 0.48 ± 0.01 | 0.48 ± 0.01 | 0.80 ± 0.04 | 0.73 ± 0.02 | 0.34 ± 0.01 | 0.74 ± 0.01 | 54 | 309 | |
| 3-years | ResNet | 0.89 ± 0.01 | 0.75 ± 0.03 | 0.72 ± 0.03 | 0.84 ± 0.02 | 0.80 ± 0.06 | 0.66 ± 0.03 | 0.83 ± 0.01 | 84 | 210 |
| CatBoost | 0.88 ± 0.00 | 0.69 ± 0.01 | 0.70 ± 0.01 | 0.74 ± 0.22 | 0.79 ± 0.01 | 0.61 ± 0.01 | 0.80 ± 0.00 | 84 | 210 | |
| 5-years | ResNet | 0.91 ± 0.01 | 0.86 ± 0.01 | 0.81 ± 0.02 | 0.82 ± 0.01 | 0.85 ± 0.04 | 0.79 ± 0.03 | 0.83 ± 0.01 | 94 | 143 |
| CatBoost | 0.92 ± 0.00 | 0.89 ± 0.00 | 0.81 ± 0.01 | 0.84 ± 0.04 | 0.82 ± 0.06 | 0.78 ± 0.05 | 0.83 ± 0.01 | 94 | 143 | |
| Alberta heart cohort (external validation) | ||||||||||
| 1-year | ResNet | 0.82 | 0.13 | 0.16 | 0.81 | 0.61 | 0.09 | 0.81 | 18 | 577 |
| CatBoost | 0.78 | 0.24 | 0.14 | 0.17 | 0.96 | 0.12 | 0.94 | 18 | 577 | |
| MAGGIC | 0.82 | 0.16 | 0.07 | 0.19 | 1 | 0.37 | 0.21 | 18 | 577 | |
| 3-years | ResNet | 0.82 | 0.32 | 0.37 | 0.75 | 0.72 | 0.25 | 0.75 | 61 | 534 |
| CatBoost | 0.73 | 0.23 | 0.18 | 0.15 | 0.95 | 0.24 | 0.87 | 61 | 534 | |
| MAGGIC | 0.78 | 0.37 | 0.21 | 0.20 | 0.97 | 0.12 | 0.28 | 61 | 534 | |
| 5-years | ResNet | 0.78 | 0.43 | 0.49 | 0.74 | 0.68 | 0.38 | 0.73 | 111 | 477 |
| CatBoost | 0.75 | 0.42 | 0.27 | 0.18 | 0.96 | 0.51 | 0.81 | 111 | 477 | |
| MAGGIC | 0.79 | 0.50 | 0.36 | 0.22 | 0.96 | 0.22 | 0.36 | 111 | 477 | |
Model performance in the internal validation cohort is generated based on 5-fold cross-validation. For MAGGIC, we used the cut point score of 12 that was previously used in literature for dichotomizing the predictions.34 AUROC: area under the receiver operating characteristic curve; AUPRC: area under the precision–recall curve.
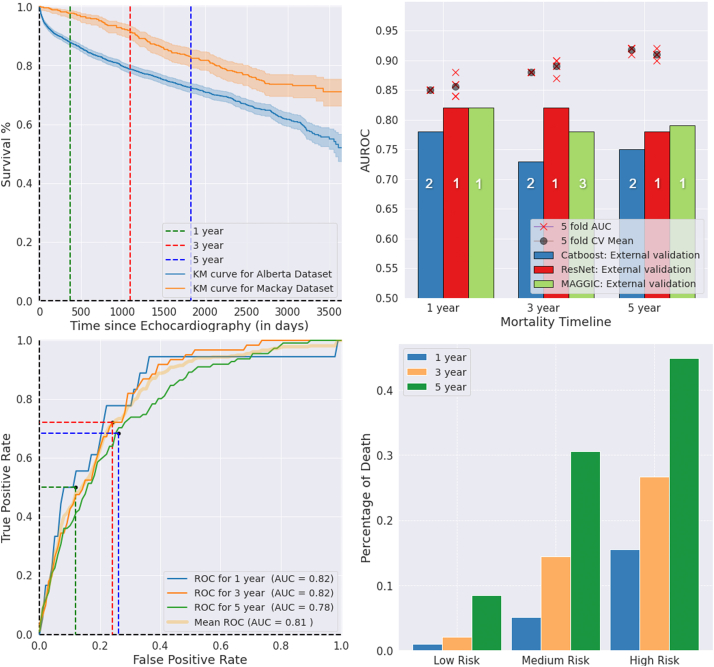
Fig. 2.
Top Left: Kaplan–Meier curve for the development and external validation datasets. Top Right: ResNet and CatBoost model performances in internal 5-fold CV and external validation for 1-year, 3-year and 5-year mortality. Bottom Left: ROC curves ResNet mortality prediction in the external validation dataset with optimal cut-points based on training Youden index. Bottom Right: Percentage of observed deaths for risk groups based on predicted probability (binned by 33 percentile cutpoints) by ResNet models for 1-year, 3-year and 5-year mortality in the external validation dataset. AUROC: Area under the receiver operating curve; AUC: area under the curve; CV: cross-validation; KM: Kaplan–Meier; ROC: receiver operating curve. In the top right panel, ranking (with ties) of the models are indicated on the bars, based on the bootstrap significance testing of pairwise differences between the models' external performances.
External validation
As of March 31, 2020, 1- and 3-year mortality status was available for all 595 patients, while 5-year mortality status was available for 588 patients, in the Alberta HEART Study (Fig. 1). At 1-, 3-, and 5-years, 18 (3.0%), 61 (10.3%), and 111 (18.7%) of the patients had died. Supplementary Tables S2–S4 provide baseline characteristics of subjects who were alive versus dead in the 1-, 3-, and 5-years mortality periods. The externally validated AUROC scores ranged from 78% to 82% for ResNet models, and 73%–78% for CatBoost models (Table 2). ResNet models achieved significantly better performance than CatBoost models at all three time-points (bootstrap significance test, p < 0.05, Fig. 2). Also, optimal threshold cut-points selected based on the development dataset showed a better balance between recall and specificity (hence, better F1 scores) for the ResNet models than for CatBoost models. Fig. 2 shows the Kaplan–Meier curves for the development and external datasets, internal 5-fold cross-validation and external validation AUROC for the models, ROC curves and the percentage of observed deaths in risk groups based on tertiles of prediction probabilities for the ResNet models in the external validation. Supplementary Fig. S1 shows the Precision-Recall curves for ResNet mortality prediction, number of patients and percentage of observed deaths in predicted risk groups for CatBoost and ResNet models in the external validation.
ResNet model performance relative to MAGGIC risk score and in patient subgroups
AUROC performance for the MAGGIC risk score ranged from 78% to 82% for the three time-points on the external validation cohort (Table 2, Fig. 2). The performance of the ResNet model was statistically comparable to the MAGGIC risk score for 1-year mortality (AUROC = 82% for both, mean and 95% CI of ResNet–MAGGIC performance difference = −0.11% (−3.40%, 3.06%)); significantly better for 3-year mortality (ResNet AUROC = 82% vs. MAGGIC AUROC = 78%, mean and 95% CI of ResNet–MAGGIC performance difference = 3.59% (1.13%, 6.03%)); and comparable for 5-year mortality (ResNet AUROC = 78% vs. MAGGIC AUROC = 79%, mean and 95% CI of ResNet–MAGGIC performance difference = −1.29% (−3.19%, 0.58%), Supplementary Fig. S2).
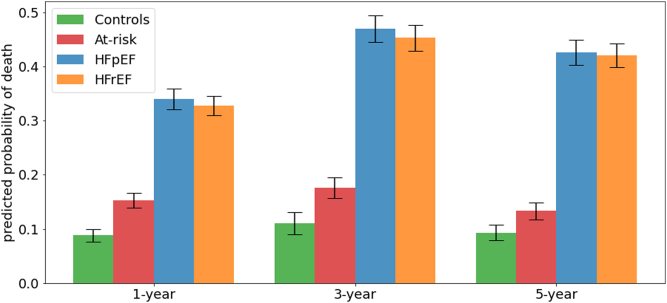
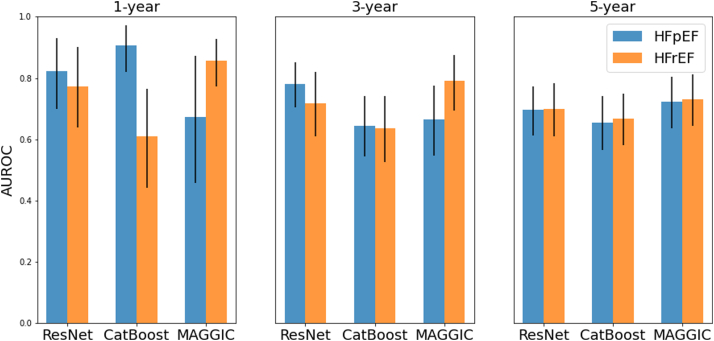
The ResNet models, while being agnostic to HF status, predicted higher probability of death among patients with HFpEF and HFrEF (30%–50%) than among healthy control and patients at risk of HF (5%–20%), thus providing additional evidence for the validity of our models (Fig. 3). The performance of the models for each outcome in HFrEF and HFpEF subgroups are reported in Fig. 4. MAGGIC score showed better performance in HFrEF compared to the HFpEF for the 1- and 3-year time-points, while ResNet and CatBoost performed similarly across subgroups or showed better performance in HFpEF. Models performed similarly between HFrEF and HFpEF subgroups for the 5-year time-point.
Fig. 3.
Bar plots showing mean and standard errors of mean for ResNet predicted probabilities of mortality between patient groups based on HF risk and cardiac function in the external validation dataset. HF: heart failure; HFrEF: HF with reduced ejection fraction; HFpEF: HF with preserved ejection fraction.
Fig. 4.
Bar plots showing mean and 95% CI of AUROC model performances for HFrEF and HFpEF subgroups. HF: heart failure; HFrEF: HF with reduced ejection fraction; HFpEF: HF with preserved ejection fraction; AUROC: Area under the receiver operating curve; CI: Confidence Interval.
Association with patient-reported quality of life
Overall, we observed a modest negative correlation between ResNet's predicted probability of death and patient-reported functional status measures (Supplementary Fig. S3). The correlation between increasing predicted probability of mortality at 1-, 3-, and 5-year and baseline KCCQ-CSS scores was −0.35, −0.28, and −0.33 (p < 0.05). Similarly, the correlation between predicted probability of mortality and baseline KCCQ-OSS scores was −0.34 for 1-year; −0.29 for 3-year; and −0.33 for 5-year, respectively (p < 0.05). When we examined average KCCQ-CSS and KCCQ-OSS scores across groups based on tertiles of prediction probabilities from the ResNet models, we found that the difference in average scores in the low-risk group compared to the medium and high risk groups were substantially higher than the minimal clinically important difference of 5 points (Supplementary Table S5).22
Visualization
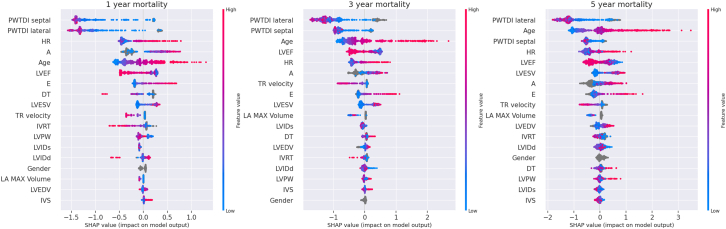
Supplementary Fig. S4 shows the Grad-CAM map for the ResNet model overlaid on the first frame of the video for anatomical reference (for representative cases with at least 90% prediction probability in their respective class), with areas of highest importance around the left atrium, or mitral and aortic valves of the heart.20 SHAP analysis revealed that, in general, lateral mitral annulus e’ velocity, septal mitral annulus e’ velocity, age, heart rate, mitral valve A wave velocity and ejection fraction were the top contributors of mortality prediction in the CatBoost models (Fig. 5).
Fig. 5.
SHAP feature importance analysis for 1 year, 3-year and 5 year CatBoost models (left to right). On the X-axis, the features are listed in decreasing order of their importance (top to bottom). The color represents the feature value–red for high values and blue for low values. For the features such as PWTDI lateral e’ velocity, PWTDI septal e’ velocity and ejection fraction–low (blue) values contribute towards high Shapley values i.e. towards mortality (class ‘dead’). On the other hand, for age, heart rate, mitral valve A wave velocity–high (red) values contribute towards mortality. A: mitral valve A wave velocity; DT: deceleration time; E: mitral valve E wave velocity; EF: ejection fraction; HR: heart rate; IVRT: Isovolumic relaxation time; IVS: interventricular septal thickness; LA: left atrial; LVEDV: left ventricular end diastolic volume; LVESV: left ventricular end systolic volume; LVIDd: left ventricular internal dimension at the end of diastole; LVIDs: left ventricular internal dimension at the end of systole; LVPW: left ventricular posterior wall thickness; PWTDI: pulsed-wave Tissue Doppler imaging for mitral annulus e’ velocity; TR: tricuspid regurgitation.
Discussion
Our study demonstrates that echo-based ML models can provide good-to-excellent prognostic information with respect to 1-, 3-, and 5-year mortality. Our deep-learning models can facilitate a fully automated decision support system that could be applied directly to images, prior to expert-curated annotations. In addition to deep-learning (ResNet) models based on echo videos, we developed gradient boosting (CatBoost) models based on echo measurements. We found both types of ML models were generalizable, comparable in performance to established risk scores, and correlated with patient's quality of life measures. Our findings suggest echo-based automated systems could be used to facilitate risk stratification at point-of-care.
Increasingly, ML based models are being considered to assist with both the diagnosis and prognostication of patients with suspected HF. Traditional clinical prediction models for patient survival based on detailed demographic, clinical, medication, and biomarker data have shown performance with discrimination indices of ∼80% in limited, single centre patient population.23 Several ML models using multimodal clinical or imaging data have performed better than traditional regression models in predicting clinical events such as mortality.5,24 Few ML studies have combined echo measurements with demographic,5 clinical data from electronic health records25 and/or other variables such as electrocardiogram, and blood markers24 for predicting mortality. However, these models rely on operator-crafted echo measurements which were sometimes extracted from echo reports using text mining approaches.
Detailed review of echo videos and providing precise measurements can be cumbersome and time consuming,26 which presents an opportunity for unbiased and automated pattern recognition using deep learning techniques. Deep learning has the potential to manage the complexity of tasks and assist the physicians in arriving at efficient clinical decisions, directly using echo videos without the need of intermediate steps for expert-guided annotations. The performance of our deep learning model was better than MAGGIC at 3-year time-point and comparable to the MAGGIC risk score for other two time-points. However, it is noteworthy that calculating MAGGIC score for a patient requires an extensive set of characteristics including ejection fraction, laboratory tests, comorbidities, smoking status, activity class and medication details that may be expensive to operationalize in terms of time, effort, resources and trained personnel to be made readily available at the point of risk assessment. Further, MAGGIC risk score and the ML models (ResNet and CatBoost) showed differential performances between the HFpEF and HFrEF subgroups, particularly for 1- and 3-year mortality, generating the hypothesis that echo-based deep learning models might be a better fit than MAGGIC for prognostication across HF subgroups.
Previously, Ulloa Cerna et al. used convolutional neural networks to train on 812,278 echo videos from 34,362 patients and showed superior performance of deep learning models of annotation-free echo videos in the prediction of one-year all-cause mortality compared to established clinical risk scores, cardiologists’ clinical gestalt, or ML models based on human-crafted or electronic health records-driven parameters.6 We extend this observation to longer time periods and also provided further validation on an independent dataset acquired from a completely different geographical location, and from patients with different ethnicity, clinical characteristics, and outcome rates than the development dataset.
We observed significant correlation between our ML-based mortality predictions and patient-reported functional status as measured by the KCCQ. The KCCQ has been shown to provide important prognostic information with respect to clinical outcomes, and the U.S. Food and Drug Administration has recently approved its use as a measure of patient-reported functional status and quality of life outcomes in clinical trials.10 In general, we found that the difference in baseline KCCQ-CSS and KCCQ-OSS across patients grouped according to tertiles of predicted probability of death from our ResNet models was higher than the minimal clinically important difference of 5 points.3,22,27 This observation needs to be confirmed in future prospective studies, but has the potential to help clinicians and researchers in identifying those at risk of reduced functional status and providing opportunities to improve quality of life.
Echo is one of several imaging modalities that are used for diagnosing cardiovascular disease and provide useful prognostic information. For example, cardiac magnetic resonance (CMR) imaging provides valuable data regarding cardiac structural and functional abnormality. Several previous studies have shown models based on CMR-related features to have moderate to excellent performance (c-indices: 0.62–0.86) for predicting adverse events.28, 29, 30, 31, 32, 33, 34 However, CMR is a costly procedure that is not widely available. In contrast, echo is a more widely available, less costly and invasive procedure that provides valuable information on cardiac anatomy and function. These factors, combined with its diagnostic and prognostic utility, suggest that echo would be a good choice for the development of an end-to-end artificial intelligence-augmented prognostic application.
A few limitations are noteworthy in interpreting the results of the current study. First, some echo measurement features used for CatBoost modeling were missing for several patients. Although this situation mimics realistic scenarios of data availability in the clinics, missingness can pose an unseen challenge to generalizability of models. Second, deep learning models are generally regarded as black boxes due to their complexity and inability to indicate human-identifiable patterns. Even though we have attempted to identify areas of importance using GradCAM, which pointed towards anatomically important parts of the heart such as the left atrium, or mitral and aortic valves, in general, it failed to detect distinct, clinically-useful patterns. We also have reported interpretability for CatBoost models with the SHAP method, which has highlighted a few echo measurement parameters as key contributors of mortality. Third, we used uniform sampling of echo images, and have not explored all the alternative ways of preprocessing echo data. However, we have used statistically appropriate methods of model development and evaluation, given our preprocessing pipeline. Fourth, we developed prognostic models based primarily on echo data/images. Our models did not account for other factors that may be associated with mortality outcomes, such as duration of disease, treatment strategies, as well as sociodemographic factors such as race/ethnicity. Furthermore, variables needed to calculate the MAGGIC score and data on patient reported functional status were only available in the Alberta dataset and not in the Mackay dataset. Future studies are needed to examine the extent to which the addition of these factors improves models’ prognostic performance. Fifth, we did not have data on the cause of death, and were therefore unable to verify whether the models performed better at predicting cardiovascular death compared to all-cause death. Lastly, this study provides a proof of concept that needs careful consideration including wider scale generalizability of these models before clinical implementation.
In conclusion, both ResNet and CatBoost models provided good-to-excellent prognostic performance for 1-, 3- and 5-year mortality prediction tasks in the internal and external validation cohorts. ResNet models provided higher performance than the CatBoost models in the external validation, suggesting better generalizability of prognostication performance with the deep learning models compared to CatBoost models. Furthermore, our models that were agnostic to clinical status of patients, provided clinically meaningful prediction probabilities that correlated with patients' clinical severity, as measured by patients’ risk of HF, ejection fraction, and functional status. These models can be leveraged for echo-based automatic risk stratification at the individual and population level, and can potentially guide the downstream management.
Contributors
Akshay Valsaraj - Data curation, formal analysis, investigation, methodology, software, validation, visualization, writing – original draft.
Sunil Vasu Kalmady - Conceptualization, methodology, project administration, supervision, writing – original draft, writing – review & editing.
Vaibhav Sharma - Data curation, investigation, software.
Matthew Frost - Project administration, resources, software, supervision, validation.
Weijie Sun - Formal analysis, validation, visualization, writing – review & editing.
Nariman Sepehrvand - Conceptualization, supervision, writing – review & editing.
Marcus Ong - Writing – review & editing.
Cyril Equibec - Writing – review & editing.
Jason R. B. Dyck - Writing – review & editing.
Todd Anderson - Writing – review & editing.
Harald Becher - Writing – review & editing.
Sarah Weeks - Writing – review & editing.
Jasper Tromp - Writing – review & editing.
Chung-Lieh Hung - Writing – review & editing.
Justin A. Ezekowitz - Writing – review & editing.
Padma Kaul - Conceptualization, funding acquisition, project administration, supervision, writing – review & editing.
All authors have read and approved the final version of the manuscript. Akshay Valsaraj and Matthew Frost have accessed and verified the underlying data.
Data sharing statement
Both the Alberta HEART and Mackay data may be accessible upon request and after approval of a proposal, with a signed data access agreement.
Code availability statement
The codebase for training and evaluating the CatBoost and deep learning models, and, also, for generating figures in this paper will be available at: https://doi.org/10.6084/m9.figshare.21620877.
Declaration of interests
J.T. reports consulting or speaker fees from Daiichi-Sankyo, Boehringer Ingelheim, Roche Diagnostics and Us2.ai, owns patent US-10702247-B2 unrelated to the present work. M.F., M.O. and C.E. report owning stock or stock options from Us2.ai.
J.E. reports research grants from Bayer, Merck & Co, Novo Nordisk, Cytokinetics, Applied Therapeutics, American Regent, US2.ai, Canadian Institutes of Health Research, Heart and Stroke Foundation, Weston Foundation; consulting fees from AstraZeneca, Boehringer Ingelheim, Novo Nordisk, Otsuka, Bayer, Novartis; participation on a Data Safety Monitoring Board or Advisory Board for Cardiac Sarcoidosis Randomized Trial (CHASM-CS-RCT); leadership or fiduciary role in the Canadian Heart Failure Society. C-L.H. reports honoraria for lectures or presentations as a speaker with AstraZeneca, Boehringer Ingelheim, Sanofi and Bayer Pharma; participation in Advisory Board activity in AstraZeneca, Boehringer Ingelheim, Sanofi and Bayer Pharma. All other authors report no conflicts of interest.
Acknowledgments
Funding for Alberta HEART was provided by an Alberta Innovates - Health Solutions Interdisciplinary Team Grant no. AHFMR ITG 200801018. P.K. holds a Canadian Institutes of Health Research (CIHR) Sex and Gender Science Chair and a Heart & Stroke Foundation Chair in Cardiovascular Research. A.V. and V.S. received funding from the Mitacs Globalink Research Internship.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104479.
Appendix A. Supplementary data
References
- 1.Virani S.S., Alonso A., Benjamin E.J., et al. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- 2.McDonagh T.A., Metra M., Adamo M., et al. 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2021;42:3599–3726. doi: 10.1093/eurheartj/ehab368. [DOI] [PubMed] [Google Scholar]
- 3.Butler J., Khan M.S., Mori C., et al. Minimal clinically important difference in quality of life scores for patients with heart failure and reduced ejection fraction. Eur J Heart Fail. 2020;22:999–1005. doi: 10.1002/ejhf.1810. [DOI] [PubMed] [Google Scholar]
- 4.Pocock S.J., Ariti C.A., McMurray J.J.V., et al. Predicting survival in heart failure: a risk score based on 39 372 patients from 30 studies. Eur Heart J. 2013;34:1404–1413. doi: 10.1093/eurheartj/ehs337. [DOI] [PubMed] [Google Scholar]
- 5.Kwon J.-M., Kim K.-H., Jeon K.-H., Park J. Deep learning for predicting in-hospital mortality among heart disease patients based on echocardiography. Echocardiography. 2019;36:213–218. doi: 10.1111/echo.14220. [DOI] [PubMed] [Google Scholar]
- 6.Ulloa Cerna A.E., Jing L., Good C.W., et al. Deep-learning-assisted analysis of echocardiographic videos improves predictions of all-cause mortality. Nat Biomed Eng. 2021;5:546–554. doi: 10.1038/s41551-020-00667-9. [DOI] [PubMed] [Google Scholar]
- 7.Sengupta P.P., Shrestha S., Berthon B., et al. Proposed requirements for cardiovascular imaging-related machine learning evaluation (PRIME): a checklist. JACC Cardiovasc Imaging. 2020;13:2017–2035. doi: 10.1016/j.jcmg.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuo J.-Y., Chang S.-H., Sung K.-T., et al. Left ventricular dysfunction in atrial fibrillation and heart failure risk. ESC Heart Fail. 2020;7:3694–3706. doi: 10.1002/ehf2.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ezekowitz J.A., Becher H., Belenkie I., et al. The Alberta Heart Failure Etiology and Analysis Research Team (HEART) study. BMC Cardiovasc Disord. 2014;14:91. doi: 10.1186/1471-2261-14-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spertus J.A., Jones P.G., Sandhu A.T., Arnold S.V. Interpreting the Kansas city cardiomyopathy questionnaire in clinical trials and clinical care: JACC state-of-the-art review. J Am Coll Cardiol. 2020;76:2379–2390. doi: 10.1016/j.jacc.2020.09.542. [DOI] [PubMed] [Google Scholar]
- 11.Sepehrvand N., Savu A., Spertus J.A., et al. Change of health-related quality of life over time and its association with patient outcomes in patients with heart failure. J Am Heart Assoc. 2020;9 doi: 10.1161/JAHA.120.017278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yudistira N., Kurita T. Gated spatio and temporal convolutional neural network for activity recognition: towards gated multimodal deep learning. EURASIP J Image Video Process. 2017;2017 doi: 10.1186/s13640-017-0235-9. [DOI] [Google Scholar]
- 13.Wang L., Xiong Y., Wang Z., et al. Computer vision – ECCV 2016. Springer International Publishing; 2016. Temporal segment networks: towards good practices for deep action recognition; pp. 20–36. [Google Scholar]
- 14.Avendi M. Packt Publishing Limited; 2020. PyTorch Computer vision cookbook: over 70 recipes to master the art of computer vision with deep learning and PyTorch 1. x. [Google Scholar]
- 15.Wu Z., Xiong C., Ma C.-Y., Socher R., Davis L.S. 2019 IEEE/CVF conference on computer vision and pattern recognition (CVPR), Long Beach, CA, USA. 2019. Adaframe: adaptive frame selection for fast video recognition; pp. 1278–1287. [Google Scholar]
- 16.Prokhorenkova L., Gusev G., Vorobev A., Dorogush A.V., Gulin A. Proceedings of the 32nd international conference on neural information processing systems (NIPS'18) Curran Associates Inc; Red Hook, NY, USA: 2018. CatBoost: unbiased boosting with categorical features; pp. 6639–6649. [Google Scholar]
- 17.Ouyang D., He B., Ghorbani A., et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature. 2020;580:252–256. doi: 10.1038/s41586-020-2145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara K., Kataoka H., Satoh Y. Proceedings of the IEEE international conference on computer vision (ICCV) Workshops. 2017. Learning spatio-temporal features with 3D residual networks for action recognition. [Google Scholar]
- 19.Loshchilov I., Hutter F. International Conference on Learning Representations; 2019. Decoupled weight decay regularization. [Google Scholar]
- 20.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. 2017 IEEE international conference on computer vision (ICCV) 2017. Grad-CAM: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 21.Lundberg S.M., Lee S.-U. Proceedings of the 31st international conference on neural information processing systems (NIPS'17) Curran Associates Inc; Red Hook, NY, USA: 2017. A unified approach to interpreting model predictions; pp. 4768–4777. [Google Scholar]
- 22.Spertus J., Peterson E., Conard M.W., et al. Monitoring clinical changes in patients with heart failure: a comparison of methods. Am Heart J. 2005;150:707–715. doi: 10.1016/j.ahj.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Codina P., Lupón J., Borrellas A., et al. Head-to-head comparison of contemporary heart failure risk scores. Eur J Heart Fail. 2021;23:2035–2044. doi: 10.1002/ejhf.2352. [DOI] [PubMed] [Google Scholar]
- 24.Tse G., Zhou J., Woo S.W.D., et al. Multi-modality machine learning approach for risk stratification in heart failure with left ventricular ejection fraction ≤ 45. ESC Heart Fail. 2020;7:3716–3725. doi: 10.1002/ehf2.12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samad M.D., Ulloa A., Wehner G.J., et al. Predicting survival from large echocardiography and electronic health record datasets: optimization with machine learning. JACC Cardiovasc Imaging. 2019;12:681–689. doi: 10.1016/j.jcmg.2018.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furiasse N., Thomas J.D. Automated algorithmic software in echocardiography: artificial intelligence? J Am Coll Cardiol. 2015;66:1467–1469. doi: 10.1016/j.jacc.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Butler J., Shahzeb Khan M., Lindenfeld J., et al. Minimally clinically important difference in health status scores in patients with HFREF vs HFPEF. JACC Heart Fail. 2022;10(9):651–661. doi: 10.1016/j.jchf.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Romano S., Judd R.M., Kim R.J., et al. Feature-tracking global longitudinal strain predicts mortality in patients with preserved ejection fraction: a multicenter study. JACC Cardiovasc Imaging. 2020;13:940–947. doi: 10.1016/j.jcmg.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pezel T., Unterseeh T., Kinnel M., et al. Long-term prognostic value of stress perfusion cardiovascular magnetic resonance in patients without known coronary artery disease. J Cardiovasc Magn Reson. 2021;23:43. doi: 10.1186/s12968-021-00737-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano S., Judd R.M., Kim R.J., et al. Feature-tracking global longitudinal strain predicts death in a multicenter population of patients with ischemic and nonischemic dilated cardiomyopathy incremental to ejection fraction and late gadolinium enhancement. JACC Cardiovasc Imaging. 2018;11:1419–1429. doi: 10.1016/j.jcmg.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanov A., Mohamed A., Asfour A., et al. Right atrial volume by cardiovascular magnetic resonance predicts mortality in patients with heart failure with reduced ejection fraction. PLoS One. 2017;12 doi: 10.1371/journal.pone.0173245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pezel T., Garot P., Kinnel M., et al. Prognostic value of stress cardiovascular magnetic resonance in asymptomatic patients without known coronary artery disease. Eur Radiol. 2021;31:6172–6183. doi: 10.1007/s00330-021-08078-3. [DOI] [PubMed] [Google Scholar]
- 33.Gulati A., Japp A.G., Raza S., et al. Absence of myocardial fibrosis predicts favorable long-term survival in new-onset heart failure. Circ Cardiovasc Imaging. 2018;11 doi: 10.1161/CIRCIMAGING.118.007722. [DOI] [PubMed] [Google Scholar]
- 34.Rohen F.M., de Ávila D.X., Cabrita Lemos C.M., et al. The MAGGIC risk score in the prediction of death or hospitalization in patients with heart failure: comparison with natriuretic peptides. Rev Port Cardiol. 2022;S0870-2551(22):363–368. doi: 10.1016/j.repc.2021.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.