Abstract
Human induced pluripotent stem cell derived cardiomyocytes (hiPSC-CMs) are commonly used to model arrhythmogenic cardiomyopathy (ACM), a heritable cardiac disease characterized by severe ventricular arrhythmias, fibrofatty myocardial replacement and progressive ventricular dysfunction. Although ACM is inherited as an autosomal dominant disease, incomplete penetrance and variable expressivity are extremely common, resulting in different clinical manifestations. Here, we propose hiPSC-CMs as a powerful in vitro model to study incomplete penetrance in ACM. Six hiPSC lines were generated from blood samples of three ACM patients carrying a heterozygous deletion of exon 4 in the PKP2 gene, two asymptomatic (ASY) carriers of the same mutation and one healthy control (CTR), all belonging to the same family. Whole exome sequencing was performed in all family members and hiPSC-CMs were examined by ddPCR, western blot, Wes™ immunoassay system, patch clamp, immunofluorescence and RNASeq. Our results show molecular and functional differences between ACM and ASY hiPSC-CMs, including a higher amount of mutated PKP2 mRNA, a lower expression of the connexin-43 protein, a lower overall density of sodium current, a higher intracellular lipid accumulation and sarcomere disorganization in ACM compared to ASY hiPSC-CMs. Differentially expressed genes were also found, supporting a predisposition for a fatty phenotype in ACM hiPSC-CMs. These data indicate that hiPSC-CMs are a suitable model to study incomplete penetrance in ACM.
Abbreviations: ABC, active ß-catenin; ACM, arrhythmogenic cardiomyopathy; ASY, asymptomatic; BBB, bundle-branch block; CMs, cardiomyocytes; CTR, control; Cx43, connexin-43; DEGs, differentially expressed genes; GATK, Genome Analysis Toolkit; hiPSC, human induced pluripotent stem cell; ICD, implantable cardioverter-defibrillator; ID, intercalated disk; LBB, left bundle-branch block; MRI, magnetic resonance imagingmut, mutated; NSVT, non-sustained ventricular tachycardia; RV, right ventricle; wt, wild type
Keywords: Arrhythmogenic cardiomyopathy, Incomplete penetrance, Human induced pluripotent stem cell derived cardiomyocytes
Graphical Abstract
1. Introduction
Arrhythmogenic cardiomyopathy (ACM) is a genetic cardiac disorder, with an estimated prevalence in the general population ranging from 1:2000–1:5000 [1]. It is mainly caused by mutations affecting proteins of the intercalated disk (ID) [2], in particular desmosomal proteins [3]. PKP2 is one of the first identified [4] and most commonly mutated genes in ACM [2]. It encodes plakophilin-2, an armadillo protein, which interacts with plakoglobin, desmoplakin, desmoglien-2 and desmocollin-2 in the cardiac desmosome [5]. The loss of desmosomal integrity, due to mutated desmosomal proteins, with impaired cell-cell adhesion and intercellular gap widening [6], can lead to the disruption of mechanical function in ACM [7]. Growing evidence indicate that cell-cell junctions could have a role not only at mechanical level but also in the electrical propagation of action potential in the heart [8].
ACM is a major cause of sudden cardiac death, particularly in young adults, and it is associated with myocardial fibrofatty substitution and severe ventricular arrhythmias [1]. ACM is typically an autosomal dominant disorder with reduced penetrance and highly variable expressivity, indicating that additional determinants influence the phenotypic manifestation of a given disease-causative mutation [9]. A significant proportion of ACM patients (4–11%) carry multiple mutations in either the same (compound heterozygosity) or different genes (digenic heterozygosity). These patients show a clinically overt and often more severe phenotype, compared to single ACM mutation carriers [10], [11]. On the other hand, several family members, carrying ACM pathogenetic variants, do not develop the disease phenotype. This reduced penetrance depends on the family genotype, but it is relatively common in ACM (4–70%) [12], [13], [14], [15], [16]. Unfortunately, these individuals remain trapped in uncertainty about a possible future disease onset. As sudden cardiac death can be the first symptom of the disease, their clinical management remains very challenging. Over the past decades, significant progress has been made in identifying ACM related genes and their pathogenetic mechanisms [17], [18]. However, much less is known about the causes of incomplete penetrance, which cannot be investigated using heterologous expression systems and animal models [18], [19]. To elucidate the genetic determinants of incomplete penetrance in cardiac channelopathies and cardiomyopathies some studies focused on the coinheritance of functional variants that might influence the disease phenotype, by either enhancing or repressing the primary defect caused by the disease associated mutation [20]. Environmental factors, mutational heterogeneity, modifier genes, post-translational modifications, and epigenetic changes have been indicated as potential elements associated to incomplete penetrance and variable expressivity in ACM [9], [21]. Advances in human induced pluripotent stem cell (hiPSC) technology, including the improvement of protocols for their efficient differentiation into cardiomyocytes (CMs), have made possible to develop a new cellular model for studying several inherited cardiac diseases, reproducing the whole genetic background of the patient. While hiPSC-derived CMs (hiPSC-CMs) have previously been used as a cellular disease model to generate new insights into molecular mechanisms contributing to ACM pathogenesis [22], [23], this is the first report, in which they are used as a model for incomplete penetrance, specifically in ACM. Of note, hiPSC-CMs, hiPSC-derived retinal ganglion cells (hiPSC-RGCs) and hiPSC derived neural crest cells (hiPSC-NCCs), including autonomic and sensory neurons, have been successfully used to investigate reduced penetrance and variable expressivity, identifying different genetic modifiers in long QT Syndrome [24], [25], [26], [27], Leber's hereditary optic neuropathy [28] and familial dysautonomia [29].
Here, we tested hiPSC-CMs, obtained from an ACM family, whose members carry the same desmosomal PKP2 mutation but have different phenotypes, including either overt ACM or no disease manifestation.
2. Methods
2.1. Study design
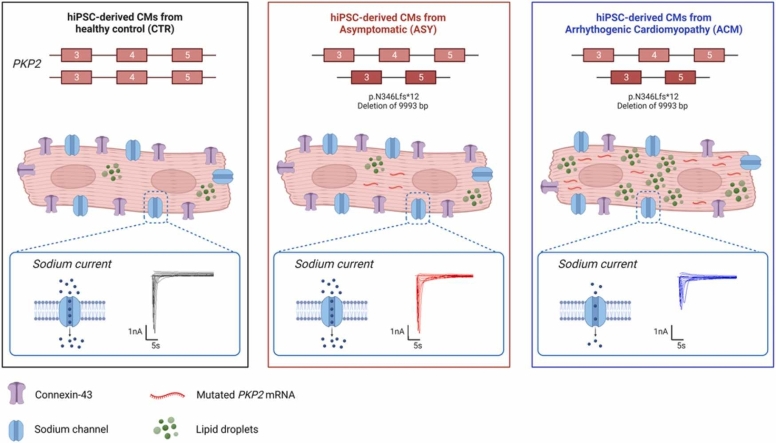
The overall objective of this study was to understand whether hiPSC-CMs are a suitable cellular model for studying the mechanisms of incomplete penetrance in ACM. We enrolled six individuals, belonging to an ACM family, identified and diagnosed according to 2010 Task Force criteria [30] by a clinical cardiac electrophysiologist who has been treating this family for 20 years (Fig. 1 and Table 1). Three of them are ACM patients, carrying a heterozygous truncating PKP2 mutation, two are asymptomatic (ASY) PKP2 carriers, and one is a healthy control (CTR). Six hiPSC lines were generated and differentiated into CMs. Several experiments were performed in these human CMs to recapitulate the genotype-phenotype discordance between ACM and ASY.
Fig. 1.
ACM family with the PKP2 mutation. CTR: internal healthy control. The red arrow indicates the proband. The red letters indicate the IDs assigned to each individual.
Table 1.
Clinical data of PKP2 mutation carriers.
| Subject | ID (#) | Major Criteria (M) | Minor Criteria (m) | Diagnosis and notes |
|---|---|---|---|---|
| I-1 | C (67 y) | 1) Family history and PKP2 mutation carrier | 1) Regional dyskinesia and mild RV dysfunction (2D echo+MRI) 2) > 500 ventricular extrasystoles per 24 h (Holter) |
ACM (1 M and 2 m) |
| I-2 | CTR (65 y) | - | - | -- |
| I-3 | G (66 y) | 1) Family history and PKP2 mutation carrier | - | (1 M) |
| I-4 | A (64 y) | 1) Family history and PKP2 mutation carrier 2) Regional dyskinesia, moderate RV dilatation and dysfunction (2D echo+MRI) |
1) Late potentials by SAECG | ACM (2 M and 1 m) (ICD) |
|
II-1 (Proband) |
E (32 y) | 1) Family history and PKP2 mutation carrier 2) Regional dyskinesia, moderate RV dilatation and dysfunction (2D echo+MRI) 3) Epsilon wave in right precordial leads 4) Inverted T waves in right precordial leads without RBB* 5) Non sustained ventricular tachycardia of LBB* * morphology with superior axis |
- | ACM (5 M) (ICD) |
| II-2 | D (24 y) | 1) Family history and PKP2 mutation carrier | - | (1 M) |
#: age at the time of blood sampling for cell reprogramming. RV: right ventricle; MRI: magnetic resonance imaging; *RBB: right bundle-branch block; * *LBB: left bundle-branch block; ICD implantable cardioverter defibrillator. CTR: internal healthy control.
2.2. Ethics approval and consent to participate
The present study complied with the principles outlined in the Declaration of Helsinki and it was approved by the Ethical Committee of the Province of Alto Adige/South Tyrol (Nr.1/2014, 12/03/2014). Blood samples were collected after patients and their family members gave their written informed consent.
2.3. Whole exome sequencing
For individuals I-1, I-2, II-1, and II-2 (Fig. 1), whole exome sequencing (WES) was performed on an Illumina HiSeq 2500 using the Nextera® Rapid Capture Exomes Kit v3. For individuals I-3 and I-4 (Fig. 1), WES was performed on an Illumina MiSeq using the Nextera® Rapid Capture Exomes Kit v2. To increase the number of reads available for analysis, the same pooled samples from individuals I-3 and I-4 were run four times on the MiSeq. For all samples, the resulting sequencing data was processed using the Genome Analysis Toolkit (GATK) v3.7 best practice recommendations [31], [32], [33]. Specifically, for each of the four MiSeq runs and the one HiSeq run, read quality was assessed with FastQC version 0.11.3 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Nextera adapters were trimmed with SeqPrep version 1.2 (https://github.com/jstjohn/SeqPrep). Trimmed reads were aligned to GRCh37 with BWA version 0.7.15 [34]. Quality of the resulting alignments was assessed with QualiMap v2.2.1 (http://qualimap.conesalab.org/). The four resulting bam files from individuals I-3 and I-4 were merged with picard tools 2.8.2 (https://broadinstitute.github.io/picard/). For each sample bam (the merged bam files for I-3 and I-4), duplicate marking with picard tools and indel realignment and base quality score recalibration with GATK v 3.7 was performed. Raw variant calling was made with the GATK HaplotypeCaller, followed by joint variant calling with GenotypeGVCFs of these six samples together with 57 WES samples unrelated to this study to increase the accuracy of the genotype calls. Variant quality score recalibration was performed, and variants were filtered based on a tranche sensitivity threshold of 0.99 for SNPs and indels. Variants were annotated using the Dintor software [35] and Ensembl v75.
To check for variants relevant for ACM, the obtained call set was filtered for variants present in the affected individuals I-1, I-4, II-1 and absent in the unaffected individuals I-2, I-3, and II-2 accounting for both a dominant and a recessive effect. Variants with missing genotype data in one or more samples were discarded. Variants were further filtered to (i) include only variants of “high” or “moderate” impact as defined by Ensembl (https://grch37.ensembl.org/info/genome/variation/prediction/predicted_data.html), (ii) that had a minor allele frequency< 0.01 in the ExAC database, (iii) and were present in one of the 22 selected known ACM causing genes (Supplementary Table S1) [3] or in 839 candidate genes implicated in sudden cardiac death, adipogenesis, Wnt/βcatenin, Hippo and TGFβ pathways (Supplementary Table S2) [36].
2.4. Generation of hiPSCs
Peripheral blood mononuclear cells, isolated from the six family members, were amplified and reprogrammed into hiPSCs using a protocol established in our laboratory [37]. Two different clones were generated for the healthy intrafamilial control (CTR, I-2, Fig. 1). hiPSCs were then characterized according to standard procedures including karyotyping, mutation sequencing, STR analysis, pluripotency marker expression and differentiation towards the three germ layers [38].
2.5. hiPSC culture and in vitro cardiac differentiation
All hiPSC lines were cultured on Matrigel® (Corning) coated plates in StemMACS ™ iPS-Brew XF medium (Miltenyi Biotec) according to the manufacturer’s protocol. When cells reached a confluence of 80–90% they were detached using TrypLE™ Express enzyme (Thermo Fisher Scientific), centrifuged at 200 g for 5 min, resuspended in the culture medium supplemented with 10 μM of Rock Inhibitor (Y-27632, Biomed Reagent) and plated in a new Matrigel® coated plate. The day after, Rock Inhibitor was removed by changing medium. Cells were kept at 37 °C in 5% CO2 and routinely tested for mycoplasma and karyotype.
Cardiac differentiation was induced in cells seeded on Matrigel® coated 6-well plates, at 70–80% of confluence using the PSC Cardiomyocyte Differentiation Kit (Thermo Fisher Scientific), based on the protocol described by Burridge et al. [39]. Briefly, cells were cultured for two days in Medium A (to stimulate the Wnt pathway), then for additional two days in Medium B (to suppress the Wnt pathway) and subsequently in Maintenance (M) medium. The beating monolayers appeared after 10–12 days and were then cultured for a total of 30 days, refreshing the M medium three times per week.
The beating monolayers were then dissociated using the Multi tissue dissociation kit 3 (Miltenyi Biotec). CMs were purified from non-CMs cells using the PSC-Derived Cardiomyocyte Isolation Kit (Miltenyi Biotec) and magnetically enriched on the MACS Manual Separators (Miltenyi Biotec).
2.6. Culture and treatment of hiPSC-CMs
Purified hiPSC-CMs were maintained in culture for additional 30 days in basal and adipogenic media in Matrigel® coated 12-well plates. Basal medium was composed of High Glucose DMEM (Gibco), 2% Hyclone Fetal Bovine Defined (GE Healthcare Life Sciences), 1% non-essential Amino Acids (Gibco), 1% Penicillin/Streptomycin (Gibco) and 0,09% β-mercapto-ethanol (Gibco). Adipogenic Medium, composed of basal medium supplemented with 50 μg/ml Insulin (Sigma-Aldrich), 0.5 μM Dexamethasone (Sigma-Aldrich), 0.25 mM 3-isobutyl-1 methylxanthine (IBMX) (Sigma-Aldrich), 200 μM Indomethacin (Sigma Aldrich) and 5 μM Rosiglitazone (Vinci-Biochem), was used to induce adipogenic differentiation [22]. The culture media were refreshed twice a week.
2.7. Droplet digital PCR (ddPCR)
Total RNA was extracted using Direct-zol RNA Kit (Zymo Research) and then reverse transcribed to cDNA using the SuperScript VILO cDNA Synthesis Kit (Invitrogen), following the manufacturer’s instructions. The reaction mixture for the ddPCR (20 μl/reaction) contained 2 × ddPCR Supermix for Probe (no dUTP) (Bio-Rad), 20 × primer/probe assay for each target, 1 ng of cDNA and water up to the final volume. A multiplex reaction was set up in order to simultaneously detect both, the mutated and wildtype PKP2 mRNA, in the same well-reaction, by using the following 20 × primer/probe assays: the custom made assay for the specific detection of mutated PKP2 (Bio-Rad: C-PKP2_EX3–5 (FAM); (Forward Primer: GAGAGAAGCACTTTCACTG; Reverse Primer: CAGAAGCTTGAGGATGC; Probe: CCAGCTGGGGTTAACC)), the commercial assays for the wildtype PKP2, on exon 4, (Bio-Rad: dCNS128841330 (High HEX)) and the reference HPRT1 gene (Bio-Rad: dHsaCPE5192872 (Low HEX)). To generate the droplets, 20 μl of the reaction mix and 70 μl of oil Droplet Generation oil for Probes (Bio-Rad) were respectively loaded into separate wells of DG8 cartridges (Bio-Rad) which were then inserted into the QX200™ Droplet Generator (Bio-Rad). Subsequently, 40 μl of the resulting droplets emulsion was transferred to a 96-well PCR semi-skirted plate (Bio-Rad). The PCR reaction was performed using a GeneAmp™ PCR System 9700 (Applied Biosystems). The protocol conditions consisted of 95 °C for 10 min, (94 °C for 30 s, 57 °C for 2 min) × 40 cycles, 98 °C for 10 min and 4 °C for the storage. Amplification signals were read using the QX200™ Droplet Reader (Bio-Rad) and analyzed using the QuantaSoft software (Bio-Rad).
2.8. Western blot and Wes™ immunoassay system
hiPSC-CMs were lysed using RIPA lysis buffer, composed of 10 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Igepal CA630 (NP-40), 1% sodium deoxycholate (NaDoc), 0.1% SDS (Sodium Dodecyl Sulfate), 1% Glycerol, supplemented with protease and phosphatase inhibitors (Complete Tablets, Mini EASYpack, Roche). The protein concentration was quantified using Pierce™ BCA Protein Assay Kit (Thermo Scientific). Total proteins (15 ug) were separated by SDS-PAGE on precast gradient (4–12%) gels (Invitrogen) and then transferred to a nitrocellulose membrane (Bio-Rad). The blotted membranes were blocked in PBS 0.1% Tween 5% skim milk for 1 h at room temperature (RT) and then incubated overnight at 4 °C with the following primary antibodies: mouse anti-plakophilin2 (PROGEN 651101, 1:100) (against the C-terminal of the protein), mouse anti-plakophilin2 (BD Biosciences, 610788, 1:1000) (against the N-terminal of the protein), rabbit anti active ß-catenin (Cell Signaling, #8814, 1:1000) and mouse anti-GAPDH (Santa Cruz Biotechnology, sc-32233, 1:5000). After washing, the membrane was incubated with goat HRP-conjugated anti-mouse and anti-rabbit secondary antibodies in blocking solution at RT for 1 h. Detection was performed using the enhanced chemiluminescence system (ECL, Pierce™ ECL Western Blotting Substrate kit). Images were acquired with the ChemiDoc MP Imaging System (Bio-rad) and quantified using Image Lab software 5.2.1 (Bio-Rad).
Capillary Western immunoassay was performed on a Protein Simple Wes™ system according to the manufacturer's instructions using a 12–240 kDa and 66–440 kDa Separation Module (Bio-Techne R&D Systems) and the Anti-Rabbit and the Anti-Mouse Detection Modules (Bio-Techne R&D Systems). Protein lysates were diluted with 0.1X Sample Buffer to an appropriate concentration (from 1 to 0.5 μg/μl), then mixed with 5X Fluorescent Master Mix and heated at 95 °C for 5 min. The samples and all the proper reagents (ladder, blocking antibody diluent, primary and HRP-conjugated secondary antibodies, and the luminol-peroxide mixture) were loaded into the prefilled plates with stacking and separation matrixes. Instrument default settings were used: separation at 375 V for 25 min, blocking for 5 min, primary and secondary antibody incubation for 30 min. High dynamic range (HDR) function was applied for the luminol/peroxide chemiluminescence detection. Compass software was used to process and analyze all data results. The following primary antibodies were used: mouse anti-plakophilin2 (BD Biosciences 610788) at several dilutions (1:50; 1:25; 1:10), rabbit anti-connexin43 (Abcam, ab11370, 1:200), mouse anti-desmoplakin (Santa Cruz Biotechnology, sc-390975, 1:50), rabbit anti-desmoglein2 (Abcam, ab150372, 1:200), mouse anti-GSK-3α/β (Santa Cruz Biotechnology, sc-7291, 1:50), rabbit anti-cyclin-D1 (Cell Signaling, #2922, 1:50) and rabbit anti-HSP90 as reference housekeeping (Cell Signaling, #4874, 1:200).
2.9. Electrophysiology
After enrichment step, 1–2 × 105 hiPSC-CMs were plated on fibronectin-coated (38 μg/ml, Corning) FluoroDishes (World Precision Instruments, Inc. – tissue culture dish with cover glass bottom) in RPMI 1640 medium (with/L-Glutamine, Gibco, USA) supplemented with B27 (Life Technologies) and Penicilin/Streptomicin (Euroclone). Electrophysiological recordings were performed on single cardiomyocytes, identified based on their typical morphology, 7 days after the plating. Data were collected from a minimum of three independent differentiations per line.
For the sodium current (INa) measurement, the bath solution contained (mmol/l): 140 NaCl, 5.4 KCl, 1.8 CaCl2, 1.2 MgCl2, 10 glucose, 5 HEPES, pH 7.3 (NaOH). The pipette solution contained 10 mM HEPES, 126 mM KCl, 6 mM NaCl, 1.2 mM MgCl2, 5 mM EGTA, 11 mM glucose and 1 mM MgATP, pH 7.4 (KOH).
Standard patch-clamp recording techniques were used to measure INa in the whole-cell configuration at 37 °C in a heated chamber (TC-344 C (Dual Automatic Temperature Controller, Warner Instruments) with a bath solution perfusion system (VC-6 six channel valve controller, Warner Instruments) of bathing solution mounted on an inverted Eclipse-Ti microscope (Nikon, Tokyo, Japan). Patch electrodes were pulled from borosilicate glass capillaries (Sutter Instruments) using a P-1000 Next Generation Micropipette Puller (Sutter Instrument, USA). Pipette resistance ranged from 3.0 to 5.5 MΩ. Signals were acquired at 20 kHz and filtered at 3 kHz with the Multiclamp 700B amplifier and Digidata 1440B digitizer hardware as well as pClamp10.7 software (Axon Instruments, Molecular Devices, Sunnyvale, CA) in standard setup. Cardiomyocytes were held at − 80 mV. Only cells with good access resistance were included in the analysis. Current run-down effects were carefully monitored overtime.
Current-voltage characteristics (IV characteristics) of INa were measured using 400 ms depolarizing steps with an interval of 10 mV from a holding potential to 60 mV. Current densities were calculated by dividing current amplitude by cell Cm. The conductance (G) was calculated and then the activation curve G/Gmax was fitted with the Boltzmann function (I=((A1-A2)/(1 +exp(V-V1/2)/dV)) + A2) where A1 is the initial value, A2 is the final value, V1/2 is the half-activation voltage, dV/k is a slope factor.
Steady-state inactivation was measured by applying a double pulse protocol, the pre-pulse was from holding potential using 500 ms depolarizing steps with an interval of 10 mV from − 120–70 mV, the test pulse was to − 20 mV taking 50 ms. Steady-state inactivation values were fitted to an equation of the form I/ Imax = (I0 + 1/(1 + exp((V-V1/2)/dV))), where I is the current amplitude measured during the first test pulse, Imax is the maximum current amplitude measured during the second test pulse, V1/2 is the half-maximum inactivation voltage, dV/k is a slope factor, and I0 is the non-inactivating part of the current.
Recovery from inactivation was assessed with a double pulse protocol, the pre-pulse was from holding potential to − 20 mV taking 1000 ms, the test pulse was to 20 mV taking 50 ms with a changing time interval between them (24 steps, Δt = 5 ms). Peak current amplitude was fit to the following equation: Itest/Ipre-pulse= 1/exp(t/τrec) where Itest and Ipre-pulse represent the amplitudes of the pre-pulse and test pulse, and τrec represent the recovery time constant. The protocol for measuring recovery from inactivation is used to characterize the ratio of available channels for next stimulus.
2.10. Immunofluorescence and confocal imaging
After 60 days from induced cardiac differentiation, hiPSC-CMs were washed with PBS then fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton-X100 for 10 min, incubated for 1 h in blocking solution (5% goat serum or 5% bovine serum albumin) at room temperature (RT), and incubated over night with primary antibodies at 4 °C. The day after, cells were washed three times with PBS and incubated for 1 h at 37 °C with goat anti mouse or anti rabbit secondary antibodies coupled to Alexa fluorescent dyes (Thermo Fisher Scientific; dilution 1:1000). The following primary antibodies were used: mouse anti sarcomeric α-actinin (Sigma-Aldrich; A7732, 1:250), mouse anti-plakophilin2 (PROGEN, 651101, 1:100) and rabbit anti-troponin I (Abcam ab47003, 1:100). Nuclei were stained with DAPI (Invitrogen, Carlsbad, California, USA). Intracellular lipid droplet accumulation was evaluated using BODIPY 493/503 assay (dilution 0.1 µg/ml in PBS; Thermo Fisher Scientific, Waltham, Massachusetts, USA) incubated for 20 min at RT. The images were acquired using confocal microscopy (Leica Microsystem CMs GmBH Type: TCSSP8X) and analyzed with the ImageJ software. Intensity fluorescence was normalized on nuclei number.
The sarcomeric organization expressed as sarcomere alignment index based on the regularity and periodicity of Z-bands was evaluated on hiPSC-CMs stained for sarcomeric α-actinin. The analysis of sarcomere alignment index was analyzed using a plugin based on Fast Fourier Transform (FFT) algorithm (Fiji Software, ImageJ) as previously described [40], [41]. Briefly, the periodicity of Z-bands was reproduced by the peak components of the power spectrum curve and the index of sarcomere alignment was obtained by normalizing the area under the first order peak for the total area of the power spectrum profile.
2.11. Transcriptome analysis
Total RNA was extracted from hiPSC-CMs using the Direct-zol RNA Kit (Zymo Research). Concentration and purity of isolated RNA were assessed by Nanodrop and Qubit, and integrity was evaluated using the Experion RNA StdSens Analysis Kit (Bio-Rad). High-quality RNA was used for subsequent library preparation. Libraries were prepared for 500 ng of RNA from each sample using the QuantSeq 3’ mRNA-Seq Library Prep FWD (Lexogen). Samples were sequenced on a HiSeq2500 platform according to the SR1000 protocol. Reads were aligned using STAR (version 2.5) against the Homo sapiens GRCh38 genome assembly with recommended options and thresholds. HTSeq-count (version 0.9) was used to generate raw gene counts. All subsequent analysis was performed in R (version 4.2.1). Annotation of genes was performed using the ensembldb [42] package (version 2.21.2) based on annotations from Ensembl release 90. The data set was pre-filtered by removing genes with an average read count below 64 in all sample groups reducing the data set from initially 58,302 genes to 11,457. The data set consisted of gene expression profiles of hiPSC-CMs from three ACM patients (two males, one female) and two asymptomatic individuals (two females) cultivated either in basal culture medium or adipogenic medium. The DESeq2 [43] package (version 1.36.0) was used for differential expression analysis. No adjustment for the present sex-bias was performed because of the collinearity with the contrasts of interest (ACM against ASY). Genes differentially expressed due to the sex difference were identified in a separate analysis by comparing the two male ACM samples against the one female ACM sample. This information was used to flag genes found to be significant in the main comparisons of ACM vs ASY samples. Raw p-values from the linear models were adjusted for multiple hypothesis testing using the method from Benjamini and Hochberg. Genes with an adjusted p-value smaller than 0.05 and an absolute log2 fold change value larger than 0.7 were considered significant. The RNA-seq raw data were deposited at the Gene Expression Omnibus (Accession Number GSE222793).
2.12. Statistical analysis
OriginPro version 2021 (OriginLab Corporation, Northampton, MA, USA.) was used for electrophysiology data analyses. For all other experiments, statistical analyses were performed using Prism 5 (Graphpad software, Inc., CA, USA).
One-way, two-way ANOVA and Kruskal-Wallis ANOVA were used to compare continuous variables with normal and non-normal distributions, respectively. For parametric data one-way, two-way ANOVA with Bonferroni post-test for multiple comparisons was performed. For non-parametric data, the Kruskal-Wallis test with Dunn’s multiple comparisons post-test was used. If not otherwise indicated data are shown as mean± SEM.
3. Results
3.1. Family history, clinical data and genetic screening
The family enrolled in this study was evaluated after proband E (Fig. 1, II-1) was diagnosed with ACM at the age of 22 years. She showed regional dyskinesia, moderate right ventricle (RV) dilatation and dysfunction, epsilon wave, inverted T waves without right bundle-branch block (BBB) and non-sustained ventricular tachycardia (NSVT) of left BBB morphology with superior axis (Table 1). At the age of 24 years, the patient received an implantable cardioverter-defibrillator (ICD) for primary prevention. She experienced several appropriate ICD discharges. Diagnostic genetic analysis revealed a heterozygous deletion of the entire exon 4 in the PKP2 gene. Familial screening identified four additional PKP2 mutation carriers. The father C (Fig. 1, I-1) and the uncle A (Fig. 1, I-4) (Table 1) were diagnosed with ACM, at the age of 66 years and 63 years, respectively, whereas the sister D (Fig. 1, II −2) and the aunt G (Fig. 1, I-3) were completely asymptomatic (Table 1). The proband’s mother CTR (Fig. 1, I-2) was negative for the PKP2 mutation and did not show any sign of heart disease.
We have defined the breakpoints of the PKP2 mutation within intron 3 and intron 4, indicating a deletion of 9993 bp (g.22512_32505del; c.1035–5203_1171–13298del; p.Asn346Leufs*12) (Supplementary Fig. S1A and S1B) [38]. At protein level, this deletion is predicted to insert a premature stop codon within the first ARM repeat, leading to a truncated protein (Supplementary Fig. S1C). Whole exome sequencing was performed to discover additional mutations that might act as second hits together with the PKP2 mutation. Sequencing data from all family members passed quality controls and were retained for analysis. All samples achieved a mean coverage of 55X (20X–95X) with 75% (41–95%) of the exonic target region covered by at least 20X across all samples. The GATK variant calling pipeline resulted in 117,832 retained variants in the six samples. Overall, 119 variants matched the required genotype pattern (present in I-1, I-4, II-1 and absent in I-2, I-3, II-2). Of these, no rare variants in known ACM genes and candidate genes (Supplementary Table S1 and S2) had a consequence of “high” or “moderate” impact as defined by Ensembl.
3.2. Mutated PKP2 mRNA is more expressed in ACM than ASY hiPSC-CMs
Using ddPCR it was tested whether there were differences in the expression of the wild type (wt) (Green high HEX signal in Fig. 2A) and mutated (mut) PKP2 (Blue FAM signal in Fig. 2A) alleles among CTR, ASY and ACM hiPSC-CMs. As expected, half the amount of wtPKP2 mRNA was found in ACM and ASY compared with CTR hiPSC-CMs (2.25 ± 0.27 vs 4.29 ± 0.70 wtPKP2/HPRT1; p = 0.029) (2.08 ± 0.38 vs 4.29 ± 0.70 wtPKP2/HPRT1; p = 0.031) (Fig. 2B). Of note, the mutPKP2 mRNA was significantly more expressed in ACM than in ASY iPSC-CMs (0.33 ± 0.04 vs 0.11 ± 0.06 mutPKP2/HPRT1; p = 0.029) (Fig. 2C).
Fig. 2.
Analysis of wild type (wt) and mutated (mut) PKP2 mRNA expression in CTR, ASY and ACM hiPSC-CMs. A) Representative outcome of the ddPCR assay showing mutPKP2 (FAM in blue), wtPKP2 (High HEX in green) and HPRT1 (Low HEX in green) expression. CN: ddPCR negative control B) Graph indicates half amount of wtPKP2 mRNA in both ACM and ASY compared to CTR hiPSC-CMs. *p = 0.029 vs ACM; *p = 0.031 vs ASY. C) Graph indicates a significantly higher expression of mutPKP2 mRNA in ACM compared to ASY hiPSC-CMs. *p = 0.029 vs ASY. All dots in the B and C plots correspond to independent cardiomyogenic differentiations: N = 5 for CTR, N = 6 for ASY and N = 11 ACM. The black and gray circles referred to the 2 different hiPSC clones of the internal healthy CTR. Orange (G) and red (D) squares for the 2 ASY hiPSC. Dark purple (A), blue (E) and light blue (C) triangles for the 3 ACM hiPSC. Non parametric test: Kruskal-Wallis test followed by multiple comparisons.
3.3. Plakophillin-2 protein haploinsufficiency
Considering the identified differences in PKP2 mRNA expression among the three groups, we next evaluated the wt and mut plakophillin-2 protein expression.
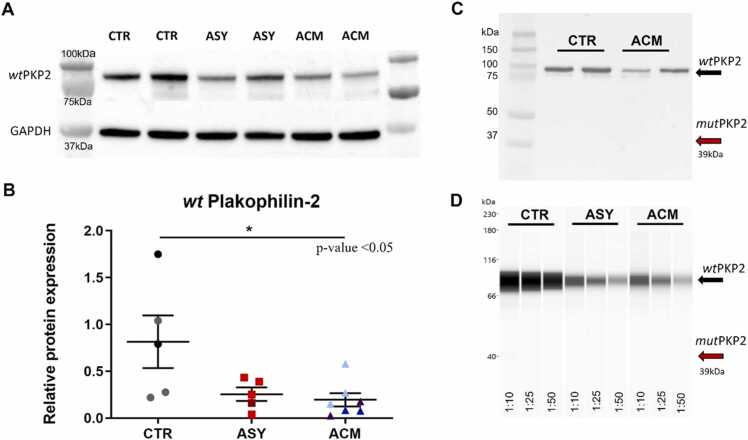
We confirmed a reduction of wt plakophillin-2 protein in ACM and ASY compared to CTR hiPSC-CMs (0.19 ± 0.07 vs 0.81 ± 0.28 wtPKP2/GAPDH; p = 0.016), (0.25 ± 0.07 vs 0.81 ± 0.28 wtPKP2/GAPDH; p = 0.11) (Fig. 3A, B). On the contrary, the mutant protein, which is supposed to be higher expressed in ACM hiPSC-CMs, was not detectable by western blot (Fig. 3C). We further try to identify the mut plakophillin-2 protein by Wes™ system, with a much more concentrated primary PKP2 antibody (1:10; 1:25; 1:50), but still, it was not detectable neither in ACM hiPSC-CMs nor in ASY hiPSC-CMs (Fig. 3D).
Fig. 3.
Analysis of wild type (wt) and mutated (mut) plakophillin-2 protein expression in hiPSC-CMs. A-B) Western blot representative bands and densitometry graph show half of wt plakophillin-2 protein in both mutation carriers compared to CTR, *p = 0.016 vs ACM. All dots in the B plot correspond to independent cardiomyogenic differentiations: N = 5 for CTR, N = 5 for ASY and N = 7 ACM. The black and gray circles referred to the 2 different hiPSC clones of the internal healthy CTR. Orange (G) and red (D) squares indicate the 2 ASY hiPSC. Dark purple (A), blue (E) and light blue (C) triangles point to the 3 ACM hiPSC. Non parametric test: Kruskal-Wallis test followed by multiple comparisons. C-D) The predicted mutant protein was not detectable neither by western blot nor by Wes™ system. 1:10, 1:25, 1:50 are antibody dilutions used against the N-terminal of plakophillin-2.
These results indicate a clear condition of haploinsufficiency for the plakophillin-2 protein in both ACM and ASY hiPSC-CMs. Immunofluorescence analysis revealed that the expressed protein was similarly present at the cell membrane in CTR, ASY and ACM hiPSC-CMs, with no significant changes in its localization (Fig. 4).
Fig. 4.
Plakophillin-2 membrane localization in CTR, ASY and ACM hiPSC-CMs. Sarcomeric marker troponin I is shown in (red) and desmosomal plakophillin-2 is highlighted in green. DAPI was used to stain nuclei (blue).
3.4. Area composita and gap junction analysis
Considering the multiple interactions of the plakophillin-2 at intercalated disks, we further analyzed the expression of desmoglein-2, desmoplakin, active ß-catenin (ABC) proteins of the area composita, and connexin-43 (Cx43), the main protein of the gap junction, in CTR, ASY and ACM hiPSC-CMs.
Desmoplakin and desmoglein-2 proteins were similarly expressed in the three groups (Fig. 5A-B). In contrast, active ß-catenin resulted to be expressed at a lower level in both ACM (0.80 ± 0.14 vs 1.36 ± 0.08; p = 0.02) and ASY (0.75 ± 0.10 vs 1.36 ± 0.08; p = 0.007) compared to CTR hiPSC-CMs (Fig. 5C). Finally, Cx43 protein was significantly reduced in ACM compared to ASY iPSC-CMs (0.63 ± 0.12 vs 1.91 ± 0.46; p = 0.02) and CTR iPSC-CMs (0.63 ± 0.12 vs 1.95 ± 0.62; p = 0.01) (Fig. 5D). We also analyzed GSK-3β and cyclin-D1 protein expression, to test possible alterations in the Wnt/ß-catenin signaling pathway, but we did not find any difference among the three groups (Fig. 5E-F).
Fig. 5.
Area composita, gap junction proteins and Wnt/ß-catenin signaling markers in CTR, ASY and ACM hiPSC-CMs. A-B) Desmoplakin and desmolgein-2 relative protein expression. C) Active ß-catenin (ABC) showing a lower expression in both mutation carriers compared to CTR hiPSC-CMs. *p = 0.007 vs ASY hiPSC-CMs, *p = 0.02 vs ACM hiPSC-CMs. D) Cx43 showing a lower expression in ACM hiPSC-CMs. *p = 0.02 vs ASY hiPSC-CMs, *p = 0.01 vs CTR hiPSC-CMs. E-F) GSK-3β and cyclin-D1 relative proteins expression. Dots in all plots correspond to independent cardiomyogenic differentiations: N = 5–7 for CTR, N = 5–9 for ASY and N = 5–8 ACM. The black and gray circles referred to the 2 different hiPSC clones of the internal CTR. Orange (G) and red (D) squares indicate the 2 ASY hiPSC. Dark purple (A), blue (E) and light blue (C) triangles point to the 3 ACM hiPSC. Non parametric test: Kruskal-Wallis test followed by multiple comparisons.
3.5. Sodium current impairment in ACM hiPSC-CMs
Whole-cell sodium currents in CTR, ASY and ACM hiPSC-CMs were recorded using patch-clamp techniques, as shown by the representative traces in Fig. 6A. ACM hiPSC-CMs showed a significantly altered overall sodium current density compared to CTR and ASY hiPSC-CMs (CTR: −68.32 ± 4.93 pA/pF, N = 28 vs. ASY: −69.22 ± 4.40 pA/pF, N = 37 vs. ACM: −49.61 ± 2.83 pA/pF, N = 44; Fig. 6B) (two-way ANOVA, p (model) = <0.001, p (factor voltage)< 0.001, p (group)< 0.001). However, the peak recorded at − 20 mV was not significantly different among the groups (Table 2, one-way ANOVA, p = 0.078). We did not observe significant changes in activation evaluated as comparison of the half-activation voltage V1/2 (Table 2, one-way ANOVA, p = 0.317) and slope factor k among groups (Table 2, one-way ANOVA, p = 0.052). Concomitantly, we did not observe any significant difference in steady-state inactivation among groups when we compared the half maximum-inactivation voltage V1/2 (Table 2, one-way ANOVA, p = 0.148) and slope factor k (Table 2, one-way ANOVA, p = 0.528). Nevertheless, a significant difference in the kinetic of the sodium current recovery was observed (Fig. 6C). The time constant τ appeared in fact significantly higher in ACM hiPSC-CMs when compared to control and asymptomatic groups (Table 2, one-way ANOVA, p = 0.0005).
Fig. 6.
The sodium current IV characteristics elicited from CTR, ASY and ACM hiPSC-CMs. A) Representative traces of currents elicited by selected voltages from control, asymptomatic and patient/ACM hiPSC-CMs (black-control, red-asymptomatic, blue-patient/ACM). B) The current was normalized to cell capacitance to give current density of groups (control-black square, asymptomatic-red dot, patient-blue triangle) displayed as mean value with SEM for every applied voltage. C) The % of recovered sodium current is calculated as the ratio of current amplitude of pre-pulse to test pulse (Itest/Ipre-pulse). The scatter plot represents the mean values of Itest/Ipre-pulse with SEM in selected time intervals (control-black square, asymptomatic-red dot, patient-blue triangle). The insets (in B and C) display the stimulation waveform. Data were collected from 7, 9, 7 independent experiments for CTR, ASY and ACM, respectively. The number of cells analyzed per each group is: 98 for CTR, 116 for ASY and 124 for ACM. Parametric test: ANOVA followed by multiple comparisons.
Table 2.
The biophysical parameters of sodium channels in control, asymptomatic and patient hiPSC-derived cardiomyocytes.
| Parameter | CTR | ASY | ACM | p-value (ANOVA) |
|---|---|---|---|---|
| Peak INa(at − 20 mV) | ||||
| pA/pF n |
-164.73 ± 22.75 28 |
-176.37 ± 20.29 37 |
-123.40 ± 12.77 44 |
n.s (0.078) |
| Recovery from inactivation | ||||
| τ (ms) n |
16.67 ± 1.55 12 |
15.74 ± 1.20 15 |
21.17 ± 1.26 21 |
0.0005 |
| Activation | ||||
| V1/2 (mV) k n |
-34.57 ± 1.26 2.09 ± 0.21 29 |
-31.57 ± 1.72 2.55 ± 0.17 42 |
-32.21 ± 0.91 2.74 ± 0.17 53 |
n.s (0.317) n.s (0.052) |
| Steady-state inactivation | ||||
| V1/2 (mV) k n |
-58.31 ± 3.59 7.27 ± 0.92 5 |
-63.86 ± 1.94 7.13 ± 0.53 9 |
-63.98 ± 1.19 8.50 ± 1.07 13 |
n.s (0.148) n.s (0.528) |
Peak INa (pA/pF) was calculated as current density a − 20 mV in selected groups. Activation and steady-state inactivation parameters were determined by data fitting to selected functions (see Methods). V1/2 (mV) is half-maximal voltage for activation or steady-state inactivation, k is the slope factor, n is the number of cells per group. Recovery from inactivation data was fitted to an exponential function to obtain the time constant τ. Values are expressed as mean± SEM.
Since the beat rate could be affected in ACM hiPSC-CMs, we have manually analyzed the beats per seconds (bps) in different recordings of hiPSC-CMs after 60 days in basal condition. The 3 groups showed a similar mean beat rate of about 0.4–0.6 bps (Supplementary Fig. S2). Moreover, the analysis of action potential of ACM hiPSC-CMs, under unstimulated conditions, did not reveal the presence of early afterdepolarizations (EADs) and delayed afterdepolarizations (DADs) (data not shown).
3.6. Higher lipid accumulation and sarcomere disarray in ACM than asymptomatic hiPSC-CMs
No differences in lipid accumulation and sarcomeric organization were observed among CTR, ASY and ACM hiPSC-CMs in basal medium (Fig. 7). However, when cells were grown in adipogenic conditions, ACM hiPSC-CMs underwent significant higher lipid accumulation and sarcomere disorganization than CTR hiPSC-CMs (1580 ± 219 vs 615 ± 199 intensity/nuclei; p = 0.0351) (0.019 ± 0.001 vs 0.028 ± 0.003 sarcomere alignment index; p = 0.039) and ASY hiPSC-CMs (1580 ± 219 vs 689 ± 146 intensity/nuclei; p = 0.005) (0.019 ± 0.001 vs 0.027 ± 0.001 sarcomere alignment index; p = 0.010) (Fig. 7).
Fig. 7.
Lipid accumulation and sarcomere organization in CTR, ASY and ACM hiPSC-CMs, in basal and adipogenic conditions. A) Representative images showing BODIPY to stain neutral lipids (green) and sarcomeric marker α-Actinin (red). B) Bodipy quantification. C) Sarcomere alignment index. Dots in all plots correspond to N = 4 for CTR, N = 4–8 for ASY and N = 7–16 ACM independent cardiomyogenic differentiations, with 5–6 available microscopy fields for each differentiation. The black and gray circles referred to the 2 different hiPSC clones of the internal CTR. Orange (G) and red (D) squares indicate the 2 ASY hiPSC. Dark purple (A), blue (E) and light blue (C) triangles point to the 3 ACM hiPSC. Non parametric test: Kruskal-Wallis test followed by multiple comparisons.
3.7. Differentially expressed genes between ACM and ASY hiPSC-CMs in basal and adipogenic conditions
We next aimed to identify possible differences in the transcriptome of ACM and ASY hiPCS-CMs. Thus, we compared the gene expression profile of ACM cells to the one of ASY cells cultured in either basal or adipogenic medium (full results in Supplementary Table S3). In addition, we have provided the results of gene expression comparison between the 2 groups, for the subset of known and candidate ACM genes (Supplementary Tables S4-S5).
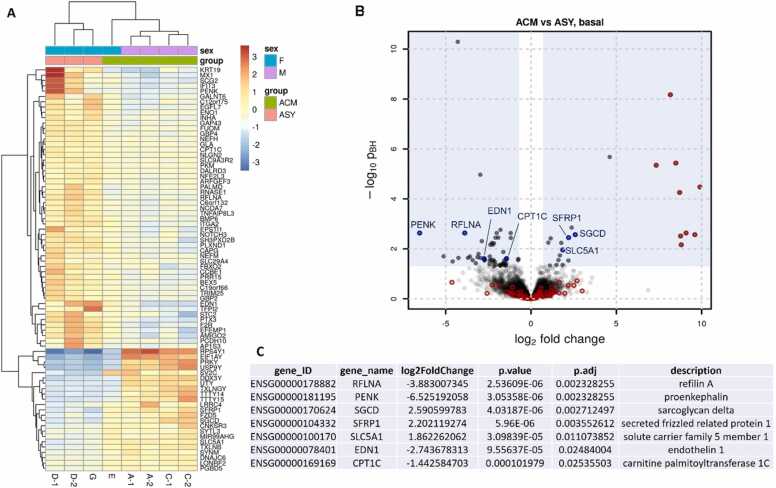
In basal conditions 76 genes were differentially expressed between ACM and ASY hiPSC-CMs (Fig. 8), including a few genes involved in cardiac function. In particular RFLNA, PENK, EDN1, and CPT1C were significantly downregulated, while SGCD, SFRP1 and SLC5A1 were significantly upregulated in ACM compared to ASY iPSC-CMs (Fig. 8).
Fig. 8.
Differential gene expression between ACM and ASY hiPSC-CM in basal condition. A) Heatmap of relative expression (compared to the average of the gene in all samples) of genes found to be significantly differentially expressed between the compared samples (ACM vs ASY). Two replicates were considered for the analysis, excluded for G and E cell lines (see Methods section). B) Volcano plot representing the differential expression results with the log2 differential expression of each gene on the x-axis against its significance expressed as the -log10 adjusted p-value. The blue shaded area defines the used significance criteria. Genes highlighted in red were also found significantly differentially expressed between ACM samples of male and female study participants and thus most likely represent sex-based differential expression. These genes were then excluded in the final comparison analysis between ACM and ASY hiPSC-CM. C) Selected genes by function (also indicated in blue in the volcano plot).
In adipogenic conditions, only 18 genes were differentially expressed (Fig. 9). Among them, PENK was downregulated, similarly to what observed in basal conditions, while NRAP, OSGIN1 and NMRK2 were upregulated (Fig. 9).
Fig. 9.
Differential gene expression between ACM and ASY hiPSC-CM in adipogenic condition. A) Heatmap of relative expression (compared to the average of the gene in all samples) of genes found to be significantly differentially expressed between the compared samples (ACM vs ASY). Two replicates were considered for the analysis, excluded for G and E cell lines (see Methods section). B) Volcano plot representing the differential expression results with the log2 differential expression of each gene on the x-axis against its significance expressed as the -log10 adjusted p-value. The blue shaded area defines the used significance criteria. Genes highlighted in red were also found significantly differentially expressed between ACM samples of male and female study participants and thus most likely represent sex-based differential expression. These genes were then excluded in the final comparison analysis between ACM and ASY hiPSC-CM. C) Selected genes by function (also indicated in blue in the volcano plot).
4. Discussion
Multiple risk factors contribute to ACM pathogenesis, such as intense physical activity [44], but the genetic predisposition remains the main underlying cause of this disease [14], with about 60% of ACM patients being carriers of a causative mutation [9]. Several clinical and genetic evaluations of probands family members have revealed incomplete penetrance, identifying relatives carrying the same mutation but remaining completely asymptomatic [12], [16].
Here we describe a family that typically represents the incomplete penetrance in ACM, composed of three affected individuals, carrying a heterozygous truncating PKP2 mutation, and two family members with the same mutation but without clinical manifestation. Excluding sex and age, since the proband is a young female and the other 2 ACM patients are males of older age, another plausible explanation for this different phenotype could be an additional pathogenetic variant segregating only in the three ACM patients. Our analysis did not identify any mutation in either 22 ACM genes or 839 candidate genes. Nevertheless, we cannot exclude the presence of variants in modifier genes not included in the candidate gene panel or variants in non-coding regions, which cannot be identified with this sequencing approach.
While important progress has been done in identifying ACM related genes, much less is known about the causes of incomplete penetrance. Over the past years, patient hiPSC-CMs were used to model ACM which faithfully reproduce in vitro the disease phenotype [22], [23]. Others have demonstrated that hiPSC-CMs, hiPSC-RGCs and hiPSC-NCCs can be used to study the molecular mechanisms of variable expressivity and incomplete penetrance in the long QT syndrome [24], [25], [26], [27], in Leber's hereditary optic neuropathy [28] and in familial dysautonomia [29], demonstrating that this technology is contributing to a better knowledge of the molecular pathogenesis of different diseases and has the potential to improve the risk stratification in patients, paving the way for a more personalized medicine.
Here, we have examined ACM and ASY hiPSC-CMs at both molecular and functional levels.
Starting from the truncating PKP2 mutation, involving the exon 4 of the gene, we have hypothesized a nonsense-mediated mRNA decay, as previously described for other truncating PKP2 mutations, leading to protein haploinsufficiency [45], [46]. In line with this previous evidence, our data confirm a lower expression of wt plakophilin-2 protein and the absence of the predicted mutant protein in both ACM and ASY hiPSC-CMs (Fig. 3). On the contrary, the mut PKP2 mRNA was expressed at a significantly higher level in ACM than in ASY hiPSC-CMs (Fig. 2). Interestingly, differential expression of wild-type and mutant mRNAs was previously reported to affect penetrance in hypertrophic cardiomyopathy, as described in [47]; however, in this case authors were able to detect an unequal expression of the corresponding proteins by mass-spectrometry. The different sensitivity of this method, compared to standard western blot and capillary western immunoassay technology used here, may explain why we were unable to identify the mutant protein, which, if present, is expected to be expressed at a much lower amount than the wt protein. In addition, the mutated mRNA may have a function per se, as it is increasingly being reported for these small molecules in heart diseases [48]. The actual role of this mutated mRNA in ACM hiPSC-CMs remains to be investigated.
As mentioned, ACM is a disease of IDs [2], important cellular structures for the mechanical and electrical coupling between neighboring cardiac cells. Desmosomes, adherens junctions, gap junctions and sodium channels are not independent units, localized at the IDs, but they mutually interact [49], [50]. In this study, expression analysis for desmosomal (desmoplakin, desmoglein-2, Fig. 5A-B; plakophillin-2, Fig. 3), adherens junction (active ß-catenin, Fig. 5C) and gap junction (Cx43, Fig. 5F) proteins revealed that the mechanical components in ACM and ASY hiPSC-CMs are similarly affected with a reduced amount of active ß-catenin and plakophillin-2 proteins, without major changes in plakophillin-2 protein localization (Fig. 4). On the other hand, Cx43 was significantly lower expressed in ACM than in ASY hiPSC-CMs indicating a likely cell-cell electrical coupling alteration specifically in ACM cells. Interestingly, lower Cx43 protein expression, with unchanged desmoglein2 and desmoplakin, is consistent with findings in other ACM hiPSC-CMs and patient heart tissue [23], [51], [52], [53]. Since we did not performed electron microscopy experiments we cannot exclude changes in the IDs ultrastructure in ACM and ASY hiPSC-CMs. However, it was previously reported that ACM hiPSC-CMs with an altered expression of desmosomal related proteins showed a structural distortion of desmosomes [23]. Moreover, these data are in line with previous literature reporting a Cx43 remodeling caused by inhibition of plakophilin-2 expression in cardiac cells [54], [55]. In addition, it has been demonstrated that decreased Cx43 expression is associated with a lower membrane Nav1.5 trafficking and a reduced sodium current [51], [56], [57], suggesting a functional link between these two important players in electrical signal propagation. These observations point to a close relationship between Cx43 and Nav1.5, suggesting that altered expression and function of these proteins promote conduction disturbances and make the heart highly prone to arrhythmias. In line with this evidence, whole-cell patch clamp experiments revealed that average sodium current density in ACM hiPSC-CMs was significantly reduced when compared to ASY hiPSC-CMs (Fig. 6A, B). Also, a slower recovery from inactivation in ACM compared to ASY cells was observed (Fig. 6C). Significantly decreased sodium current density is often described in studies from animals with reduced abundance of plakophilin-2 [58] or from other ACM hiPSC-CMs [59]. A negative shift in steady-state inactivation and a slower recovery from inactivation was also observed in PKP2-heterozygous-null murine hearts [58], whereas no alterations in gating modulation of sodium channel were observed in ACM hiPSC-CMs [59]. Overall, our findings suggest a partial impairment of cardiac sodium current in ACM hiPSC-CMs, possibly linked to the reduced amount of Cx43 protein, whereas the electrophysiological phenotype of the sodium current in ASY hiPSC-CMs was similar to CTR hiPSC-CMs. It remains to be understood whether ion channel dysfunction may also affect conduction velocity or signal propagation in ACM hiPSC-CMs. Further investigation will be necessary to clarify this aspect.
In addition to the altered electrophysiological phenotype, fibro-fatty replacement is the other pathological hallmark of ACM. Induction of a cell adult-like metabolism is essential to establish an ACM disease model using patient hiPSC-CMs [22], [23]. In fact, a much higher lipid accumulation can be induced in ACM hiPSC-CMs compared to CTR hiPSC-CMs when exposed to adipogenic stimuli [22], [23]. Consistently, we observed that ACM hiPSC-CMs showed higher lipid accumulation compared both to CTR and ASY hiPSC-CMs, under adipogenic conditions (Fig. 7). As well, sarcomere disorganization has been observed in ACM hiPSC-CMs, as a potential secondary effect caused by the increased steric hindrance of lipid droplets (Fig. 7), which localize close to T-tubules and between myofibrils in human CMs [60]. Moreover, our transcriptome data suggest a higher predisposition to a fatty phenotype in ACM hiPSC-CMs compared to ASY hiPSC-CMs, already in basal condition. Indeed, the SFRP1 gene was upregulated in ACM hiPSC-CMs compared to ASY hiPSC-CMs and its constitutive expression was described to promote adipogenesis and lipid accumulation [61]. In line with our results, this gene was previously reported to show a higher expression level in the RV of ACM patients compared with RV samples from healthy myocardium [62]. SFRP1 is an important player in the inhibition of both canonical and noncanonical Wnt signaling pathways [63]. In our model, active ß-catenin, GSK-3ß and cyclin-D1 protein expression was not different between ACM and ASY hiPSC-CMs, suggesting that canonical Wnt signaling was not involved in the incomplete penetrance mechanism. However, we cannot exclude the involvement of the noncanonical Wnt pathways [63]. Barandon et al., demonstrated that disruption of a ß-catenin independent Wnt pathway, due to the overexpression of SFRP1, inhibited GSK-3ß phosphorylation as well as protein kinase C epsilon activation in cardiomyocytes [64]. In line with these findings, GSK-3β phosphorylation, mTOR pathway activation, and cardioprotection were impaired in transgenic hearts overexpressing SFRP1 [65]. Back to the fatty phenotype, the CPT1C gene, which encodes the carnitine palmitoyltransferase 1 C protein shows a decreased expression in ACM hiPSC-CMs compared to ASY hiPSC-CMs. This enzyme catalyzes carnitinylation of fatty acids to be transported into mitochondria for β-oxidation. A recent paper described an increased lipid accumulation in catecholamine-treated hiPSC-CMs from a patient with Takotsubo cardiomyopathy showing a higher expression of the lipid importer CD36 and a decreased expression of the CPT1C lipid translocase compared to control hiPSC-CMs [66]. Moreover, we have previously demonstrated that when ACM hIPSC-CMs were cultured in adipogenic medium showed an increased amount of membrane CD36 and a higher lipid accumulation compared with CTR hIPSC-CMs [67]. Here, in the same adipogenic conditions, we have checked the CD36 gene expression in ACM and ASY hiPSC-CMs. Although the CD36 expression is higher in ACM compared to ASY hiPSC-CMs, this difference is not statistically significant (Supplementary Table S3), probably due to the low available number of replicates. Nevertheless, the differentially expressed genes (DEGs) identified between ACM and ASY hiPSC-CMs are in line with findings mentioned above, showing a higher lipid accumulation in ACM hiPSC-CMs. We can therefore speculate that ACM hiPSC-CMs could have a lower ability to translocate lipids from the cytoplasm into the mitochondria compared to the ASY hiPSC-CMs. Of note, the possible role of mitochondrial dysfunction in ACM was also indicated by Perez-Hernandez et al., showing transcriptional downregulation of the electron transport chain proteins with increased oxidant production, as an early event in the molecular pathophysiology of the disease [68]. In addition, we have recently reported in ACM cardiac stromal cells (CStCs) a higher mitochondrial oxidative stress compared to CTR CStCs [69]. However, for a fully comprehension of the role of mitochondria on ACM, further specific studies are necessary.
Other DEGs between ACM and ASY hiPSC-CMs are RFLNA and SGCD, which are both related to actin-binding protein filamin C, previously associated with ACM [70], [71], [72]. Moreover, a low expression of endothelin 1 protein, encoded by EDN1 gene, was previously described to cause cardiac dysfunction [73]. PENK gene encodes the proenkephalin protein, which is proteolytically processed to generate the opioid receptor agonists enkephalins. Enkephalins have been shown to attenuate contraction in isolated cardiomyocytes by suppressing the L-type Ca2 + current [74]. It is thus conceivable that ACM hiPSC-CMs with a decreased level of PENK expression have a lower ability to regulate calcium homeostasis. Interestingly, cardiac-specific NRAP overexpression causes right ventricular dysfunction in mice [75]. Of note, specific NRAP staining was only observed at the intercalated disks in adult mouse hearts [76] and a homozygous truncating mutation in this gene was identified in a patient with dilated cardiomyopathy [77]. Moreover, OSGIN1 gene encodes an oxidative stress response protein, reported to be induced by oxidized phospholipids and fatty acids [78], [79]. Lastly, NMRK2 gene encodes nicotinamide riboside kinase 2 interestingly reported to be an AMPK-PPARα–responsive gene that is induced by energy stress and NAD+ depletion in cardiac cells [80].
In summary, we have found that ACM hiPSC-CMs express a significantly higher amount of mutated PKP2 mRNA than ASY hiPSC-CMs. Even if no differences were detected for plakophilin-2 and β-catenin proteins, the SFRP1 gene was differentially expressed between ACM and ASY hiPSCM-CMs, thus suggesting that the Wnt pathway might have a role in the disease incomplete penetrance, although not through a direct action on β-catenin activation. Compared to ASY, ACM hiPSC-CMs showed a lower expression of Cx43 associated with an altered sodium current density, a higher lipid accumulation and sarcomere disorganization. The present study focused on the main ACM hallmarks in order to elucidate differences between ACM and ASY hiPSC-CMs; although we are aware that the spectrum of molecular mechanisms underlying incomplete penetrance in ACM is much broader, this is behind the scope of this work and requires further investigation.
Taken together, these data provide evidence that hiPSC-CMs show discordant molecular and functional features between ACM patients and ASY cases, therefore supporting their use as a patient based cellular model to gain insight into the molecular mechanisms of ACM incomplete penetrance. However, there are still many open questions that must be addressed to provide a robust set of knowledge, that are needed, before implementation of this cellular approach as a platform for a personalized medicine, in a larger scale. It would be really relevant to study further ACM families showing incomplete penetrance related to mutations in other known ACM genes. In addition, it would be also important to generate hiPSC lines from young asymptomatic individuals in parallel with clinical follow-up, to monitor over time some possible early cellular changes that may precede clinical manifestation. Only after these further studies it might make sense to move to a more clinical use of this system to try to stratify disease risk in familial mutation carriers. The founding of laboratories specialized in these specific techniques could become an important new resource for having support in clinical diagnosis, prognosis and patient management, reducing in the future the contemporary high costs and times needed for this kind of analysis.
Funding
This study was supported by the Department of Innovation, Research and University of the Autonomous Province of Bolzano-South Tyrol (Italy) and by the project INCardio (ITAT1047 grant) funded by the European Regional Development Fund and Interreg V-A Italy-Austria 2014–2020.
CRediT authorship contribution statement
Marzia De Bortoli: Conceptualization, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing, Project administration. Viviana Meraviglia: Formal analysis, Investigation, Methodology, Writing – review & editing. Katarina Mackova: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Laura S. Frommelt: Investigation, Methodology, Writing – review & editing. Eva Konig: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. Johannes Rainer: Data curation, Formal analysis, Methodology, Visualization. Chiara Volani: Investigation, Methodology, Writing – review & editing. Patrizia Benzoni: Formal analysis. Maja Schlittler: Investigation, Methodology, Writing – review & editing. Giada Cattelan: Investigation, Methodology, Writing – review & editing. Benedetta Maria Motta: Investigation, Methodology. Volpato Claudia: Investigation, Methodology. Werner Rauhe: Resources, Writing – review & editing. Andrea Barbuti: Supervision, Writing – review & editing. Serena Zacchigna: Supervision, Writing – review & editing. Peter P. Pramstaller: Supervision, Writing – review & editing. A. Rossini: Conceptualization, Project administration, Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
The authors thank the family members recruited for this study and Drs. Veronica De Sanctis, Roberto Bertorelli and Silvano Piazza (University of Trento, CIBIO, NGS Core Facility) for library generation and NGS sequencing on HiSeq2500. The authors also acknowledge the contribution of Drs. Marcelo D. Rosato-Siri, Rita Chiarelli, Nadia Bernardi during the initial phase of this project. The authors thank the Department of Innovation, Research and University of the Autonomous Province of Bozen/Bolzano for covering the Open Access publication costs.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.02.029.
Contributor Information
Marzia De Bortoli, Email: Marzia.DeBortoli@eurac.it, marzia.debortoli@eurac.edu.
Viviana Meraviglia, Email: v.meraviglia@lumc.nl.
Katarina Mackova, Email: Katarina.Mackova@eurac.edu.
Laura S. Frommelt, Email: LauraSophie.Frommelt@eurac.edu.
Eva König, Email: Eva.Koenig@eurac.edu.
Johannes Rainer, Email: Johannes.Rainer@eurac.edu.
Chiara Volani, Email: Chiara.Volani@eurac.edu.
Patrizia Benzoni, Email: patrizia.benzoni@unimi.it.
Maja Schlittler, Email: Maja.Schlittler@eurac.edu.
Giada Cattelan, Email: Giada.Cattelan@eurac.edu.
Benedetta M. Motta, Email: bmotta@unisa.it.
Claudia Volpato, Email: Claudia.Volpato@eurac.edu.
Werner Rauhe, Email: werraubz@yahoo.it.
Andrea Barbuti, Email: andrea.barbuti@unimi.it.
Serena Zacchigna, Email: Serena.Zacchigna@icgeb.org.
Peter P. Pramstaller, Email: Peter.Pramstaller@eurac.edu.
Alessandra Rossini, Email: Alessandra.Rossini@eurac.edu.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- 1.Corrado D., Basso C., Judge D.P. Arrhythmogenic cardiomyopathy. Circ Res. 2017;121(7):784–802. doi: 10.1161/CIRCRESAHA.117.309345. [DOI] [PubMed] [Google Scholar]
- 2.Calore M., Lorenzon A., De Bortoli M., Poloni G., Rampazzo A. Arrhythmogenic cardiomyopathy: a disease of intercalated discs. Cell Tissue Res. 2015;360(3):491–500. doi: 10.1007/s00441-014-2015-5. [DOI] [PubMed] [Google Scholar]
- 3.James C.A., Jongbloed J.D.H., Hershberger R.E., Morales A., Judge D.P., Syrris P., et al. International evidence based reappraisal of genes associated with arrhythmogenic right ventricular cardiomyopathy using the clinical genome resource framework. Circ Genom Precis Med. 2021;14(3) doi: 10.1161/CIRCGEN.120.003273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerull B., Heuser A., Wichter T., Paul M., Basson C.T., McDermott D.A., et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36(11):1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 5.Chen X., Bonne S., Hatzfeld M., van Roy F., Green K.J. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta -catenin signaling. J Biol Chem. 2002;277(12):10512–10522. doi: 10.1074/jbc.M108765200. [DOI] [PubMed] [Google Scholar]
- 6.Basso C., Czarnowska E., Della Barbera M., Bauce B., Beffagna G., Wlodarska E.K., et al. Ultrastructural evidence of intercalated disc remodelling in arrhythmogenic right ventricular cardiomyopathy: an electron microscopy investigation on endomyocardial biopsies. Eur Heart J. 2006;27(15):1847–1854. doi: 10.1093/eurheartj/ehl095. [DOI] [PubMed] [Google Scholar]
- 7.Mezzano V., Sheikh F. Cell-cell junction remodeling in the heart: possible role in cardiac conduction system function and arrhythmias? Life Sci. 2012;90(9–10):313–321. doi: 10.1016/j.lfs.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mezzano V., Pellman J., Sheikh F. Cell junctions in the specialized conduction system of the heart. Cell Commun Adhes. 2014;21(3):149–159. doi: 10.3109/15419061.2014.905928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel V., Asatryan B., Siripanthong B., Munroe P.B., Tiku-Owens A., Lopes L.R., et al. State of the art review on genetics and precision medicine in arrhythmogenic cardiomyopathy. Int J Mol Sci. 2020;21(18):6615. doi: 10.3390/ijms21186615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauce B., Nava A., Beffagna G., Basso C., Lorenzon A., Smaniotto G., et al. Multiple mutations in desmosomal proteins encoding genes in arrhythmogenic right ventricular cardiomyopathy/dysplasia. Heart Rhythm. 2010;7(1):22–29. doi: 10.1016/j.hrthm.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 11.Xu T., Yang Z., Vatta M., Rampazzo A., Beffagna G., Pilichou K., et al. Compound and digenic heterozygosity contributes to arrhythmogenic right ventricular cardiomyopathy. J Am Coll Cardiol. 2010;55(6):587–597. doi: 10.1016/j.jacc.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quarta G., Muir A., Pantazis A., Syrris P., Gehmlich K., Garcia-Pavia P., et al. Familial evaluation in arrhythmogenic right ventricular cardiomyopathy: impact of genetics and revised task force criteria. Circulation. 2011;123(23) doi: 10.1161/CIRCULATIONAHA.110.976936. 2701-2109. [DOI] [PubMed] [Google Scholar]
- 13.Chivulescu M., Lie Ø.H., Popescu B.A., Skulstad H., Edvardsen T., Jurcut R.O., Haugaa K.H. High penetrance and similar disease progression in probands and in family members with arrhythmogenic cardiomyopathy. Eur Heart J. 2020;41(14):1401–1410. doi: 10.1093/eurheartj/ehz570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groeneweg J.A., Bhonsale A., James C.A., te Riele A.S., Dooijes D., Tichnell C., et al. Clinical presentation, long-term follow-up, and outcomes of 1001 arrhythmogenic right ventricular dysplasia/cardiomyopathy patients and family members. Circ Cardiovasc Genet. 2015;8(3):437–446. doi: 10.1161/CIRCGENETICS.114.001003. [DOI] [PubMed] [Google Scholar]
- 15.Protonotarios N., Anastasakis A., Antoniades L., Chlouverakis G., Syrris P., Basso C., et al. Arrhythmogenic right ventricular cardiomyopathy/dysplasia on the basis of the revised diagnostic criteria in affected families with desmosomal mutations. Eur Heart J. 2011;32(9):1097–1104. doi: 10.1093/eurheartj/ehr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma A., Bosman L.P., Tichnell C., Nanavati J., Murray B., Nonyane B.A.S., et al. Arrhythmogenic right ventricular cardiomyopathy prevalence and arrhythmic outcomes in at-risk family members: a systematic review and meta-analysis. Circ Genom Precis Med. 2022;15(3) doi: 10.1161/CIRCGEN.121.003530. [DOI] [PubMed] [Google Scholar]
- 17.Gerull B., Brodehl A. Insights into genetics and pathophysiology of arrhythmogenic cardiomyopathy. Curr Heart Fail Rep. 2022;18(6):378–390. doi: 10.1007/s11897-021-00532-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Austin K.M., Trembley M.A., Chandler S.F., Sanders S.P., Saffitz J.E., Abrams D.J., Pu W.T. Molecular mechanisms of arrhythmogenic cardiomyopathy. Nat Rev Cardiol. 2019;16(9):519–537. doi: 10.1038/s41569-019-0200-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerull B., Brodehl A. Genetic animal models for arrhythmogenic cardiomyopathy. Front Physiol. 2020;11:624. doi: 10.3389/fphys.2020.00624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giudicessi J.R., Ackerman M.J. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res. 2013;161(1):1–14. doi: 10.1016/j.trsl.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sen-Chowdhry S., Syrris P., Pantazis A., Quarta G., McKenna W.J., Chambers J.C. Mutational heterogeneity, modifier genes, and environmental influences contribute to phenotypic diversity of arrhythmogenic cardiomyopathy. Circ Cardiovasc Genet. 2010;3(4):323–330. doi: 10.1161/CIRCGENETICS.109.935262. [DOI] [PubMed] [Google Scholar]
- 22.Kim C., Wong J., Wen J., Wang S., Wang C., Spiering S., et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494(7435):105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspi O., Huber I., Gepstein A., Arbel G., Maizels L., Boulos M., Gepstein L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ Cardiovasc Genet. 2013;6(6):557–568. doi: 10.1161/CIRCGENETICS.113.000188. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y.K., Sala L., Mura M., Rocchetti M., Pedrazzini M., Ran X., et al. MTMR4 SNVs modulate ion channel degradation and clinical severity in congenital long QT syndrome: insights in the mechanism of action of protective modifier genes. Cardiovasc Res. 2021;117(3):767–779. doi: 10.1093/cvr/cvaa019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chai S., Wan X., Ramirez-Navarro A., Tesar P.J., Kaufman E.S., Ficker E., et al. Physiological genomics identifies genetic modifiers of long QT syndrome type 2 severity. J Clin Invest. 2018;128(3):1043–1056. doi: 10.1172/JCI94996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Brink L., Brandão K.O., Yiangou L., Blanch-Asensio A., Mol M.P.H., Mummery C.L., et al. The linkage phase of the polymorphism KCNH2-K897T influences the electrophysiological phenotype in hipsc models of LQT2. Front Physiol. 2021;12 doi: 10.3389/fphys.2021.755642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shah D., Prajapati C., Penttinen K., Cherian R.M., Koivumäki J.T., Alexanova A., et al. hiPSC-derived cardiomyocyte model of LQT2 syndrome derived from asymptomatic and symptomatic mutation carriers reproduces clinical differences in aggregates but not in single cells. Cells. 2020;9(5):1153. doi: 10.3390/cells9051153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang T.C., Yarmishyn A.A., Yang Y.P., Lu P.C., Chou S.J., Wang M.L., et al. Mitochondrial transport mediates survival of retinal ganglion cells in affected LHON patients. Hum Mol Genet. 2020;29(9):1454–1464. doi: 10.1093/hmg/ddaa063. [DOI] [PubMed] [Google Scholar]
- 29.Zeltner N., Fattahi F., Dubois N.C., Saurat N., Lafaille F., Shang L., et al. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat Med. 2016;22(12):1421–1427. doi: 10.1038/nm.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marcus F.I., McKenna W.J., Sherrill D., Basso C., Bauce B., Bluemke D.A., et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J. 2010;31(7):806–814. doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma. 2013;43(1110):11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weichenberger C.X., Blankenburg H., Palermo A., D'Elia Y., König E., Bernstein E., Domingues F.S. Dintor: functional annotation of genomic and proteomic data. BMC Genom. 2015;16:1081. doi: 10.1186/s12864-015-2279-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao S., Puthenvedu D., Lombardi R., Chen S.N. Established and emerging mechanisms in the pathogenesis of arrhythmogenic cardiomyopathy: a multifaceted disease. Int J Mol Sci. 2020;21(17):6320. doi: 10.3390/ijms21176320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meraviglia V., Zanon A., Lavdas A.A., Schwienbacher C., Silipigni R., Di Segni M., et al. Generation of induced pluripotent stem cells from frozen buffy coats using non-integrating episomal plasmids. J Vis Exp. 2015;100 doi: 10.3791/52885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meraviglia V., Cattelan G., De Bortoli M., Motta B.M., Volpato C., Frommelt L.S., et al. Vol. 55. 2021. Generation and characterization of three human induced pluripotent stem cell lines (EURACi007-A, EURACi008-A, EURACi009-A) from three different individuals of the same family with arrhythmogenic cardiomyopathy (ACM) carrying the plakophillin2 p.N346Lfs*12 mutation. (Stem Cell Res). [DOI] [PubMed] [Google Scholar]
- 39.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., et al. Chemically defined generation of human cardiomyocytes. Nat Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pioner J.M., Racca A.W., Klaiman J.M., Yang K.C., Guan X., Pabon L., et al. Isolation and mechanical measurements of myofibrils from human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rep. 2016;6(6):885–896. doi: 10.1016/j.stemcr.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrantini C., Coppini R., Sacconi L., Tosi B., Zhang M.L., Wang G.L., et al. Impact of detubulation on force and kinetics of cardiac muscle contraction. J Gen Physiol. 2014;143(6):783–797. doi: 10.1085/jgp.201311125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rainer J., Gatto L., Weichenberger C.X. ensembldb: an R package to create and use Ensembl-based annotation resources. Bioinformatics. 2019;35(17):3151–3153. doi: 10.1093/bioinformatics/btz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beffagna G., Sommariva E., Bellin M. Mechanotransduction and adrenergic stimulation in arrhythmogenic cardiomyopathy: an overview of in vitro and in vivo models. Front Physiol. 2020;11 doi: 10.3389/fphys.2020.568535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerull B., Heuser A., Wichter T., Paul M., Basson C.T., McDermott D.A., et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36(11):1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- 46.Rasmussen T.B., Nissen P.H., Palmfeldt J., Gehmlich K., Dalager S., Jensen U.B., et al. Truncating plakophilin-2 mutations in arrhythmogenic cardiomyopathy are associated with protein haploinsufficiency in both myocardium and epidermis. Circ Cardiovasc Genet. 2014;7(3):230–240. doi: 10.1161/CIRCGENETICS.113.000338. [DOI] [PubMed] [Google Scholar]
- 47.Tripathi S., Schultz I., Becker E., Montag J., Borchert B., Francino A., et al. Unequal allelic expression of wild-type and mutated β-myosin in familial hypertrophic cardiomyopathy. Basic Res Cardiol. 2011;106(6):1041–1055. doi: 10.1007/s00395-011-0205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shah A.M., Giacca M. Small non-coding RNA therapeutics for cardiovascular disease. Eur Heart J. 2022:ehac463. doi: 10.1093/eurheartj/ehac463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiss E., Fischer C., Sauter J.M., Sun J., Ullrich N.D. The structural and the functional aspects of intercellular communication in iPSC-cardiomyocytes. Int J Mol Sci. 2022;23(8):4460. doi: 10.3390/ijms23084460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moise N., Struckman H.L., Dagher C., Veeraraghavan R., Weinberg S.H. Intercalated disk nanoscale structure regulates cardiac conduction. J Gen Physiol. 2021;153(8) doi: 10.1085/jgp.202112897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noorman M., Hakim S., Kessler E., Groeneweg J.A., Cox M.G., Asimaki A., et al. Remodeling of the cardiac sodium channel, connexin43, and plakoglobin at the intercalated disk in patients with arrhythmogenic cardiomyopathy. Heart Rhythm. 2013;10(3):412–419. doi: 10.1016/j.hrthm.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inoue H., Nakamura S., Higo S., Shiba M., Kohama Y., Kondo T., et al. Modeling reduced contractility and impaired desmosome assembly due to plakophilin-2 deficiency using isogenic iPS cell-derived cardiomyocytes. Stem Cell Rep. 2022;17(2):337–351. doi: 10.1016/j.stemcr.2021.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fidler L.M., Wilson G.J., Liu F., Cui X., Scherer S.W., Taylor G.P., Hamilton R.M. Abnormal connexin43 in arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 mutations. J Cell Mol Med. 2009;13(10):4219–4228. doi: 10.1111/j.1582-4934.2008.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oxford E.M., Musa H., Maass K., Coombs W., Taffet S.M., Delmar M. Connexin43 remodeling caused by inhibition of plakophilin-2 expression in cardiac cells. Circ Res. 2007;101(7):703–711. doi: 10.1161/CIRCRESAHA.107.154252. [DOI] [PubMed] [Google Scholar]
- 55.Agullo-Pascual E., Reid D.A., Keegan S., Sidhu M., Fenyö D., Rothenberg E., Delmar M. Super-resolution fluorescence microscopy of the cardiac connexome reveals plakophilin-2 inside the connexin43 plaque. Cardiovasc Res. 2013;100(2):231–240. doi: 10.1093/cvr/cvt191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agullo-Pascual E., Lin X., Leo-Macias A., Zhang M., Liang F.X., Li Z., et al. Super-resolution imaging reveals that loss of the C-terminus of connexin43 limits microtubule plus-end capture and NaV1.5 localization at the intercalated disc. Cardiovasc Res. 2014;104(2):371–381. doi: 10.1093/cvr/cvu195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jansen J.A., Noorman M., Musa H., Stein M., de Jong S., van der Nagel R., et al. Reduced heterogeneous expression of Cx43 results in decreased Nav1.5 expression and reduced sodium current that accounts for arrhythmia vulnerability in conditional Cx43 knockout mice. Heart Rhythm. 2012;9(4):600–607. doi: 10.1016/j.hrthm.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cerrone M., Noorman M., Lin X., Chkourko H., Liang F.X., van der Nagel R., et al. Sodium current deficit and arrhythmogenesis in a murine model of plakophilin-2 haploinsufficiency. Cardiovasc Res. 2012;95(4):460–468. doi: 10.1093/cvr/cvs218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khudiakov A., Zaytseva A., Perepelina K., Smolina N., Pervunina T., Vasichkina E., et al. Sodium current abnormalities and deregulation of Wnt/β-catenin signaling in iPSC-derived cardiomyocytes generated from patient with arrhythmogenic cardiomyopathy harboring compound genetic variants in plakophilin 2 gene. Biochim Biophys Acta Mol Basis Dis. 2020;1866(11) doi: 10.1016/j.bbadis.2020.165915. [DOI] [PubMed] [Google Scholar]
- 60.Sulkin M.S., Yang F., Holzem K.M., Van Leer B., Bugge C., Laughner J.I., Green K., Efimov I.R. Nanoscale three-dimensional imaging of the human myocyte. J Struct Biol. 2014;188(1):55–60. doi: 10.1016/j.jsb.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lagathu C., Christodoulides C., Tan C.Y., Virtue S., Laudes M., Campbell M., et al. Secreted frizzled-related protein 1 regulates adipose tissue expansion and is dysregulated in severe obesity. Int J Obes. 2010;34(12):1695–1705. doi: 10.1038/ijo.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaertner A., Schwientek P., Ellinghaus P., Summer H., Golz S., Kassner A., et al. Myocardial transcriptome analysis of human arrhythmogenic right ventricular cardiomyopathy. Physiol Genom. 2012;44(1):99–109. doi: 10.1152/physiolgenomics.00094.2011. [DOI] [PubMed] [Google Scholar]
- 63.Hsueh Y.C., Hodgkinson C.P., Gomez J.A. The role of Sfrp and DKK proteins in cardiomyocyte development. Physiol Rep. 2021;9(3) doi: 10.14814/phy2.14678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barandon L., Dufourcq P., Costet P., Moreau C., Allières C., Daret D., et al. Involvement of FrzA/sFRP-1 and the Wnt/frizzled pathway in ischemic preconditioning. Circ Res. 2005;96(12):1299–1306. doi: 10.1161/01.RES.0000171895.06914.2c. [DOI] [PubMed] [Google Scholar]
- 65.Vigneron F., Dos Santos P., Lemoine S., Bonnet M., Tariosse L., Couffinhal T., et al. GSK-3β at the crossroads in the signalling of heart preconditioning: implication of mTOR and Wnt pathways. Cardiovasc Res. 2011;90(1):49–56. doi: 10.1093/cvr/cvr002. [DOI] [PubMed] [Google Scholar]
- 66.Borchert T., Hübscher D., Guessoum C.I., Lam T.D., Ghadri J.R., Schellinger I.N., et al. Catecholamine-dependent β-adrenergic signaling in a pluripotent stem cell model of Takotsubo Cardiomyopathy. J Am Coll Cardiol. 2017;70(8):975–991. doi: 10.1016/j.jacc.2017.06.061. [DOI] [PubMed] [Google Scholar]
- 67.Sommariva E., Stadiotti I., Casella M., Catto V., Dello Russo A., Carbucicchio C., et al. Oxidized LDL-dependent pathway as new pathogenic trigger in arrhythmogenic cardiomyopathy. EMBO Mol Med. 2021;13(9) doi: 10.15252/emmm.202114365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pérez-Hernández M., van Opbergen C.J.M., Bagwan N., Vissing C.R., Marrón-Liñares G.M., Zhang M., et al. Loss of nuclear envelope integrity and increased oxidant production cause DNA damage in adult hearts deficient in PKP2: a molecular substrate of ARVC. Circulation. 2022;146(11):851–867. doi: 10.1161/CIRCULATIONAHA.122.060454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Volani C., Pagliaro A., Rainer J., Paglia G., Porro B., Stadiotti I., et al. GCN5 contributes to intracellular lipid accumulation in human primary cardiac stromal cells from patients affected by Arrhythmogenic cardiomyopathy. J Cell Mol Med. 2022;26(13):3687–3701. doi: 10.1111/jcmm.17396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oz S., Yonath H., Visochyk L., Ofek E., Landa N., Reznik-Wolf H., et al. Reduction in Filamin C transcript is associated with arrhythmogenic cardiomyopathy in Ashkenazi Jews. Int J Cardiol. 2020;317:133–138. doi: 10.1016/j.ijcard.2020.04.005. [DOI] [PubMed] [Google Scholar]
- 71.Brun F., Gigli M., Graw S.L., Judge D.P., Merlo M., Murray B., et al. FLNC truncations cause arrhythmogenic right ventricular cardiomyopathy. J Med Genet. 2020;57(4):254–257. doi: 10.1136/jmedgenet-2019-106394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hall C.L., Gurha P., Sabater-Molina M., Asimaki A., Futema M., Lovering R.C., et al. RNA sequencing-based transcriptome profiling of cardiac tissue implicates novel putative disease mechanisms in FLNC-associated arrhythmogenic cardiomyopathy. Int J Cardiol. 2020;302:124–130. doi: 10.1016/j.ijcard.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hathaway C.K., Grant R., Hagaman J.R., Hiller S., Li F., Xu L., et al. Endothelin-1 critically influences cardiac function via superoxide-MMP9 cascade. Proc Natl Acad Sci USA. 2015;112(16):5141–5146. doi: 10.1073/pnas.1504557112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao R.P., Pepe S., Spurgeon H.A., Capogrossi M.C., Lakatta E.G. Opioid peptide receptor stimulation reverses beta-adrenergic effects in rat heart cells. Am J Physiol. 1997;272(2Pt2):H797–H805. doi: 10.1152/ajpheart.1997.272.2.H797. [DOI] [PubMed] [Google Scholar]
- 75.Lu S., Crawford G.L., Dore J., Anderson S.A., Despres D., Horowits R. Cardiac-specific NRAP overexpression causes right ventricular dysfunction in mice. Exp Cell Res. 2011;317(8):1226–1237. doi: 10.1016/j.yexcr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu S., Borst D.E., Horowits R. N-RAP expression during mouse heart development. Dev Dyn. 2005;233(1):201–212. doi: 10.1002/dvdy.20314. [DOI] [PubMed] [Google Scholar]
- 77.Truszkowska G.T., Bilińska Z.T., Muchowicz A., Pollak A., Biernacka A., Kozar-Kamińska K., et al. Homozygous truncating mutation in NRAP gene identified by whole exome sequencing in a patient with dilated cardiomyopathy. Sci Rep. 2017;7(1):3362. doi: 10.1038/s41598-017-03189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li R., Chen W., Yanes R., Lee S., Berliner J.A. OKL38 is an oxidative stress response gene stimulated by oxidized phospholipids. J Lipid Res. 2007;48(3):709–715. doi: 10.1194/jlr.M600501-JLR200. [DOI] [PubMed] [Google Scholar]
- 79.Yan X., Lee S., Gugiu B.G., Koroniak L., Jung M.E., Berliner J., Cheng J., Li R. Fatty acid epoxyisoprostane E2 stimulates an oxidative stress response in endothelial cells. Biochem Biophys Res Commun. 2014;444(1):69–74. doi: 10.1016/j.bbrc.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Diguet N., Trammell S.A.J., Tannous C., Deloux R., Piquereau J., Mougenot N., et al. Nicotinamide riboside preserves cardiac function in a mouse model of dilated cardiomyopathy. Circulation. 2018;137(21):2256–2273. doi: 10.1161/CIRCULATIONAHA.116.026099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material