Graphical abstract
Keywords: Pathogenic bacteria, Deep sea, Sediment, Mouse, Gut microbiota, Alien species invasion
Highlights
-
•
The bacteria from 9 of 106 deep-sea sediment samples could alter the gut bacterial communities of mice and induce glucose metabolism deterioration, liver damage and inflammatory symptom, thus yielding alien species invasion of deep-sea bacteria into mammal gut microbiota.
-
•
Bacillus cereus DP040, a bacterium isolated from deep-sea sediment DP040, could invade the gut microbiota of mice to change the gut microbial structure, leading to inflammatory symptom of mice.
-
•
The deep-sea sediments containing the bacteria destroying the health of mice were distributed in hydrothermal vent, ocean basin and hadal trench of the Indian Ocean, the Atlantic Ocean and the Pacific Ocean. Deep sea may be an important origin of potential pathogenic bacteria.
Abstract
Introduction
Deep sea has numerous bacteria which dominate in the biomass of deep-sea sediments. Some deep-sea bacteria may possess the capacity to destroy mammal health by the alteration of gut microbiota, acting as potential pathogens.
Objectives
Pathogenic bacteria are great threats to human health. However, the ultimate origin of pathogenic bacteria has not been intensively explored. In this study, therefore, the influence of deep-sea bacteria on the gut microbiota was evaluated on a global scale.
Methods
The bacteria isolated from each of 106 deep-sea sediment samples were transplanted into mice in our study to assess the infectiousness of deep-sea bacteria.
Results
The results showed that some bacteria from deep sea, an area that has existed since the earth was formed, could proliferate in mouse gut. Based on the infectious evaluation of the bacteria from each of 106 deep-sea sediments, the bacteria isolated from 13 sediments invaded the gut bacterial communities of mice, leading to the significant alteration of mouse gut microbiota. Among the 13 deep-sea sediments, the bacteria isolated from 9 sediments could destroy mouse health by inducing glucose metabolism deterioration, liver damage and inflammatory symptom. As an example, a bacterium was isolated from deep-sea sediment DP040, which was identified to be Bacillus cereus (termed as Bacillus cereus DP040). Bacillus cereus DP040 could invade the gut microbiota of mice to change the gut microbial structure, leading to inflammatory symptom of mice. The deep-sea sediments containing the bacteria destroying the health of mice were distributed in hydrothermal vent, mid-ocean ridge and hadal trench of the Indian Ocean, the Atlantic Ocean and the Pacific Ocean.
Conclusion
Our findings demonstrate that deep sea is an important origin of potential pathogenic bacteria and provide the first biosecurity insight into the alien species invasion of deep-sea bacteria into mammal gut microbiota.
Introduction
The human gut microbiota consists of several trillion microbes, which has been found to be one of the most important factors affecting human health. [1] With the fast technological development, the understanding of the composition of gut microbiota, as well as their functions and roles in human health and diseases, has been advanced in the past decade. [2] Gut microbes can ferment non-digestible polysaccharides, thereby producing short-chain fatty acids and other metabolites to achieve evolutionarily conserved roles in the metabolism, immunity, development and behavior of the host. [3], [4] The most abundant bacteria in gut microbiota include Bacteroidales S24-7 group_norank, Lactobacillus, Faecalibaculum, Alloprevotella, Bacteroides, Lachnospiraceae NK4A136 group, Lachnospiraceae_uncultured, Escherichia- Shigella and Enterorhabdus, while the core microbiota are Bacteroidales S24-7 group_norank, Lactobacillus, Alloprevotella, Bacteroides, Lachnospiraceae NK4A136 group, Lachnospiraceae_uncultured, Alistipes and Ruminiclostridium. [5] The change of the composition of gut microbiota has been directly implicated in the etiopathogenesis of a number of pathological states, such as obesity, inflammation, colorectal cancer and autism. [6] Some intrinsic and extrinsic factors can change the composition of gut microbiota. Studies have shown that diet, which counts as a host-extrinsic factor, has a dominating role in shaping the structure of gut microbiota. [7] Host-intrinsic factors, such as genetics, mostly cause differences between individuals. The invasion of pathogenic bacteria is one of the most frequent causes of food-borne gastroenteritis. In this context, alien bacterial species that can alter gut microbiota may cause human diseases. However, the role of alien bacterial species that originate from inaccessible areas for human being, such as deep sea, has not been explored.
As the largest biome on the earth, deep sea is rich in microorganisms. [8] In the deep-sea ecosystems, the microbes can serve important ecological functions, such as organic matter re-mineralization, nutrient cycling and the transfer of energy to higher trophic levels, [9], [10], [11], [12] thus microorganisms are the most important players in the biogeochemical cycles in deep sea. Besides, deep-sea microbes are broadly regarded as important resources for drugs, rare-earth elements and other applications. [13], [14], [15], [16] Due to the importance of deep-sea microbes for researches and applications, more and more deep-sea samples, especially deep-sea sediments, have been brought into the land. The deep-sea samples, which are not properly treated for biosafety before being brought into the land, contain a large number of microbes, most of which are unidentified and may affect human health by changing the composition of gut microbiota after intake. The analysis of deep-sea samples reveals the presence of the bacteria with phenotypic and genotypic similarities to the human pathogenic bacteria, such as Vibrio [17], [18] and Bacillus toyonensis. [19] The genomes of these deep-sea bacteria contain many homologs of virulence genes that are prevalent in the pathogenic bacteria of humans and other animals. These deep-sea-originated microbes may become invasive species in human gut microbiota. Up to date, however, the biosafety of deep-sea samples has not been evaluated.
To evaluate the biosafety of deep-sea microbes to mammal health by altering the composition of gut microbiota, the bacteria isolated from deep-sea sediments were transplanted into mice in this investigation. Since the microbial communities in ocean waters are affected by currents, [20] the microbial communities of ocean waters do not accurately represent their deep-sea counterparts. Therefore the deep-sea sediments were used in this study. The results indicated that the gut microbiota of mice was seriously changed by the bacteria from some of the deep-sea sediments, leading to the damage of mouse health.
Materials and methods
Deep-sea sediment samples
Deep-sea sediment samples were collected in the Pacific Ocean, the Atlantic Ocean and Indian Ocean during the 22th, 26th, 30th, 34th, 39th, 40th and 45th cruises of oceanic vessel No.1 (Dayang No. 1) geomicrobiology cruises of China from 2010 to 2018 (Table S1). The sampling environments included hydrothermal vent (66 sediment samples), cold seep (8 sediment samples), ocean basin (20 sediment samples), hadal trench (8 sediment samples) and mid-ocean ridge (4 sediment samples). The depth of sampling stations ranged from 1,100 to 6,682 m with an average of 3,241 m. All samples were sealed into boxes in situ that were prefilled with sterilized sea water to minimize contamination. After arriving on deck, the surfaces of samples were removed using sterile shovels to further prevent mixing with seawater. To ensure most of the bacteria were survival, the sediment samples were immediately stored at 20 °C until the bacteria were isolated.
Isolation of bacteria from deep-sea sediments
Each sediment sample (5 g) was vibrated with glass beads and 5 mL of SM buffer (10 mM Tris-HCl, 100 mM NaCl, 10 mM MgSO4, pH7.5) for 30 min. After centrifuge at 1000 × g for 7 min, the supernatant was collected. Then 5 mL of fresh SM buffer was added to the same sediment and vibrated for 30 min for the second time. This process was repeated for 9 times. All of the supernatants were pooled together, followed by centrifuge at 5000 × g for 20 min. Subsequently the bacteria were collected and dissolved in sterilized water for later use. The isolated bacteria were examined using transmission electron microscope.
Samples from the mice treated with deep-sea bacteria
A total of 6 ICR (Institute of Cancer Research) female and 6 ICR male mice (8 weeks old) were randomly divided into two groups. The 8-week-old mice are considered to be adult mice, which can receive oral gavage. Each group contained 3 female mice and 3 male mice. To eliminate possible effects of bacteria in the environment on the gut microbiota of mice, all mice were kept in a sterilized container and fed them with sterilized water and food. Both groups of mice were raised in a sterilized condition for 3 days to stabilize their gut microbiota. One group of the mice (experimental mice) were fed with the isolated deep-sea bacteria (3 × 105) dissolved in sterilized water, while the mice of another group (control mice) were fed with sterilized water. After feeding with deep-sea bacteria or sterilized water everyday for 3 days, both groups of the mice were raised for 4 days in a sterilized condition to stabilize their gut microbiota. To evaluate the influence of deep-sea bacteria on the mouse gut microbiota, the body weight of mice was measured and the feces of mice were collected at Day 3 and Day 10. At Day 10, all the mice were sacrificed. The blood and intestinal tissues of mice were collected for later use.
Sequencing of bacterial 16S rRNA
Genomic DNA of bacteria was extracted from fecal samples using the FastDNA® SPIN kit (MP Biomedicals, USA) following the manufacturer’s protocols. The extracted DNA was used for the amplification of bacterial 16S rDNA with gene-specific primers (515F, 5′-GTGCCAGCMGCCGCGG-3′; 907R, 5′-CCGTCA ATTCMTTTRAGTTT-3′) (M = A/C; R = A/G). Subsequently the libraries were generated using NEB Next®Ultra™DNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendation. The libraries were sequenced on an Illumina MiSeq platform.
Data analysis
Sequences analysis was performed by UPARSE software package using the UPARSE-OTU and UPARSE-OTUref algorithms. For each sample, about 12 Mb raw sequences of bacterial 16S rRNA gene were obtained. After removal of the primers, the spacers, the low-quality fragments and the sequences shorter than 50 bp, the remaining sequences were further denoised and screened for chimeric sequences with the pre.cluster command and chimera.uchime command in Mothur. Sequences with ≥ 97% similarity were assigned to the same operational taxonomic units (OTUs). One representative sequence for each OTU was picked and the uclust was used to annotate taxonomic information. Community richness, community diversity and rarefaction curve were analyzed using mothur.
Analysis of blood routine, alanine aminotransferase (ALT) and blood glucose (GLU)
Blood samples were collected from mice’s eye sockets with anticoagulant [136 mM ethylene diamine tetraacetic acid (EDTA)-Na2]. Blood routine test was performed using anticoagulated whole blood on Sysmex xt-2000i whole blood cell analyzer (Sysmex, Japan). The serum was subjected to ALT and GLU test on Roche cobas c311 fully automatic biochemical analyzer (Roche, Switzerland).
Detections of IL (interleukin)-25 expression level and tuft cells
The intestinal tissue samples were harvested from small intestines of mice. The expression level of IL25 was quantified by qPCR. Briefly the total RNAs, extracted from tissue samples using RNA isolation kit (Ambion, USA), were reversely transcribed with PrimeScript RT reagent kit (TaKaRa, Japan). Quantitative real-time PCR was performed using 2 × ChamQ SYBR qPCR Master Mix (Vazyme, USA). The PCR reaction mixture (10 μL) contained Rox reference Dye, cDNA, ChamQ SYBR qPCR Master Mix (Vazyme) and primers (IL25, 5′-CTAACCTGCTCCAGTCAGCC- 3′ and 5′-CACCTAATCTGGGTCGCTCC-3′; GAPDH, 5′-GGTATCGTGGAAGGA CTCATGAC-3′ and 5′ATGCCAGTGAGCTTCCCGTTCAG-3′). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was included for normalization. The 2-(△△Ct) method was used to calculate the relative fold change of mRNA expression.
To detect tuft cells, the intestinal tissue samples of mice were fixed in 4% paraformaldehyde solution and stored at 4 °C. Subsequently the sample was dehydrated with 10% sucrose solution before embedding in paraffin. The paraffin block was sliced with a frozen microtome and placed on clean glass slides. After drying, the sample was washed with xylene for 15 min, anhydrous ethanol for 10 min, 85% alcohol for 5 min and 75% alcohol for 5 min. Then, the tissue sections were heated in a repair box filled with EDTA antigen-repair buffer (pH9.0) (Wuhan Servicebio Technology, China). The slides were washed with phosphate buffer saline (PBS), followed by incubation with primary antibody (Abcam, UK) overnight at 4 °C and then with fluorescence-labeled secondary antibody (Abcam, UK) for 50 min at room temperature. To detect tuft cells, the antibody against double cortin-like kinase 1 (DCLK1), a marker of tuft cells, was used. The nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI). Subsequently the sample was observed under fluorescence microscope. The antibodies used were purchased from a commercial company (Abcam, UK).
Taxonomic classification and phylogenetic tree construction
Bacteria were identified using the deposited reference sequences in the National Center for Biotechnology Information-Basic Local Alignment Search Tool (NCBI-BLAST) server (https://blast.ncbi.nlm.nih.gov/blast/Blast.cgi). The BLAST results of the identified bacteria were combined and aligned by the neighbour-joining method with gap corrections. The phylogenetic tree, including the identified bacteria, was annotated and visualized as an unrooted tree using Blast Tree View (https://www.ncbi.nlm.nih.gov/blast/treeview/treeView.cgi).
Bacterial diversity analysis
The alpha diversity of mouse gut bacteria was evaluated using the online Mothur software (https://www.mothur.org/wiki). The alpha diversity included Chao, Simpson and Shannon indices.
Principal co-ordinates analysis
The weighted unifrac algorithm was applied for principal co-ordinates analysis at the OTU level to analyze beta diversity of bacteria. The v egan of package R (version 3.4.4) (https://www.r-project.org/ Mingkebio, China) was used.
Correlation analysis
Correlation analysis was performed using the online OmicStudio tools (https://www.omicstudio.cn). Based on Spearman's rank correlation coefficient, the vegan of package R (Version 3.6.1) was used.
Isolation and identification of deep-sea bacteria
A deep-sea sediment was suspended in liquid enterobacteria enrichment broth (Hope Bio, China) or beef extract peptone medium (0. 3% beef extract, 1% peptone, 0.5% NaCl). After incubation at 37 °C for 3 h, the liquid was plated onto agar media (liquid medium containing 1.5% agar) and cultured for 12 h at 37 °C. Subsequently the colonies were subjected to bacterial 16S gene sequencing. The bacteria, after purification, were identified based on their 16S gene sequences. Transmission electron microscope was used to examine the purity and morphology of the isolated bacteria.
Genome sequencing and sequence analysis
High-quality DNA was extracted from cultured bacteria using Qiagen kit. DNA was sequenced using Oxford Nanopore Technology on PromethION sequencer. A total of 2 Gb raw sequences were generated. After quality control, the sequencing data were assembled using unicycler (version 0.4.8), Pilon (version 1.23) and NextPolish (version v1.4.13). Coding genes, tRNA genes and rRNA genes were predicted by prodial, tRNAscan-SE (version 2.0) and RNAmmer (version 1.2), respectively. The other non-coding RNAs were predicted by searching the RFAM database (https://rfam.xfam.org/) in Infernal (version 1.1.3). The proteins were annotated using Interproscan (version 5.25–64.0), TIGRFAMs (https://tigrfams. jcvi.org/cgi-bin/index.cgi), Pfam (https://pfam.xfam.org/), gene ontology (GO) (https://geneontology.org/), the Kyoto encyclopedia of genes and genomes (KEGG) (https://www.kegg.jp/kegg/), Refseq (https://www.ncbi.nlm.nih.gov/refseq/) and the clusters of orthologous groups (COG).
Statistical analysis
The numerical data were analyzed by one-way analysis of variance. The difference between different treatments was analyzed using Student’s t test. All experiments were performed three times and the data were presented as mean ± standard deviation.
Ethics statement
All experiments involving animals were conducted according to the ethical policies and procedures approved by the ethics committee of Zhejiang University, China (Approval no. 14843).
Results
Bacterial community in deep-sea sediments
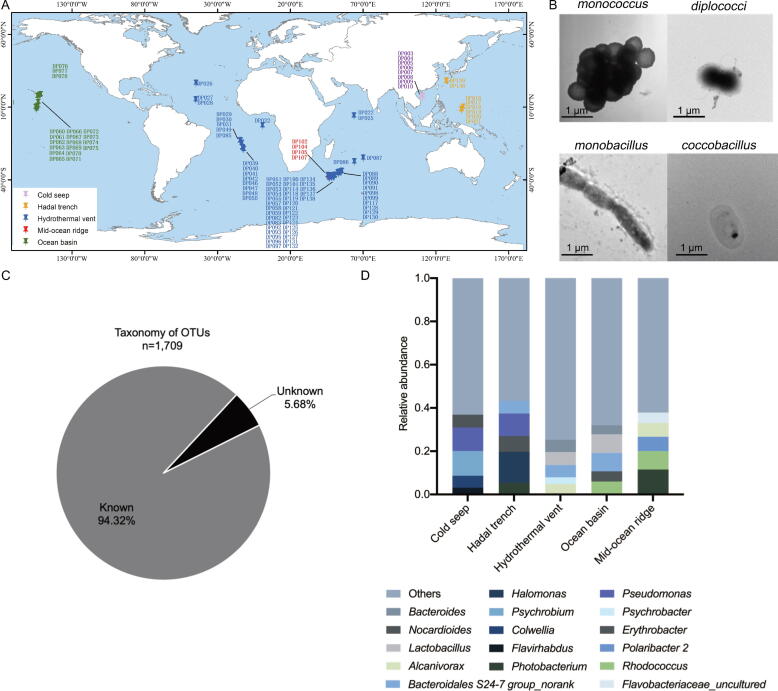
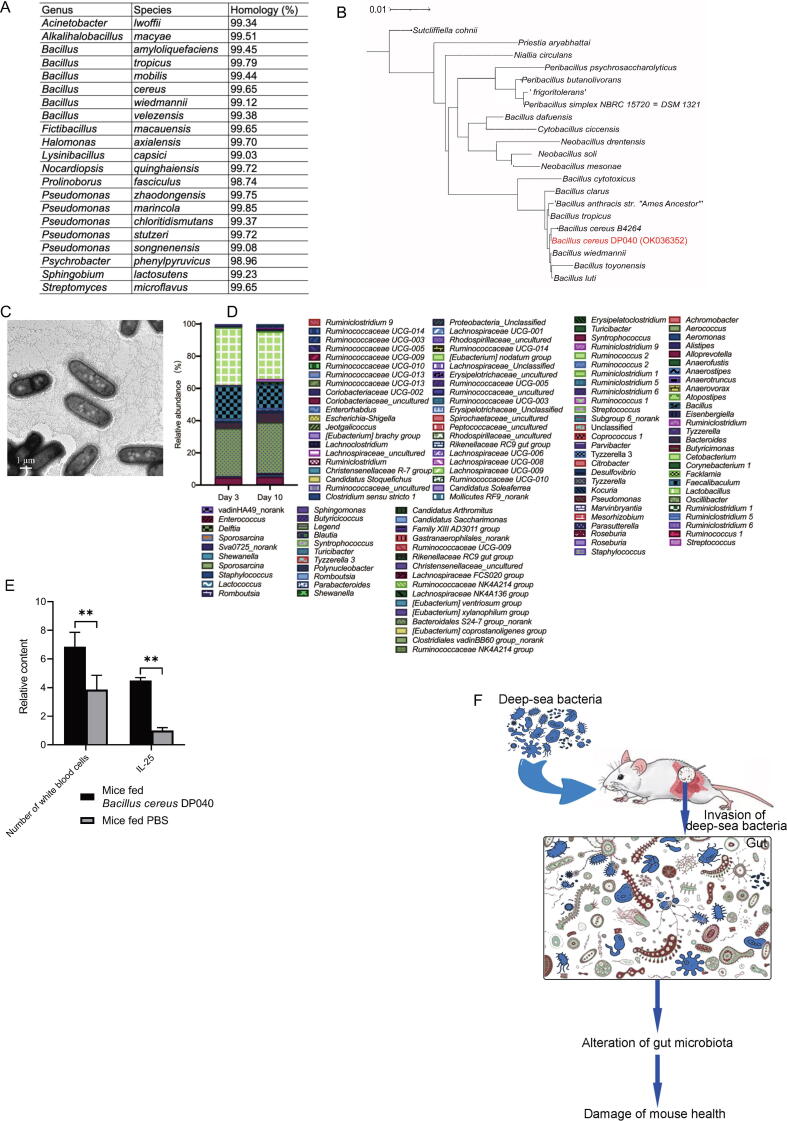
To identify the bacterial community of deep-sea sediment samples from different environments, the mixed bacteria (total bacterial community) were isolated from each of 106 deep-sea sediments which were collected from five deep-sea environments including hydrothermal vent, ocean basin, cold seep, mid-ocean ridge and hadal trench (Fig. 1A and Table S1). The transmission electron microscopy data demonstrated that the isolated deep-sea bacteria were spherical-shaped or rod-shaped with an average diameter of 0.4–1 μm (Fig. 1B).
Fig. 1.
Bacterial community in deep-sea sediments. (A) Distribution of deep-sea sediments. (B) Representative images of bacteria isolated from deep-sea sediments. The bacteria were observed with transmission electron microscopy Scale bar, 1 μm. (C) Pie diagram of known and unknown taxonomy of OTUs in deep-sea sediment samples at the genus level. (D) The dominant genera of bacteria in different deep-sea geographical environments.
Based on the 16S rRNA gene sequencing in our laboratory (GenBank accession no PRJNA721272), the bacterial communities of 106 deep-sea sediments contained 1,709 operational taxonomic units (OTUs) at the genus level. Among them, 94.32% OTUs were matched with the known bacteria, while 5.68% OTUs were unclassified (Fig. 1C). Of the known genera, the most abundant bacterium was Bacteroidales S24-7 group_norank, followed by Pseudomonas, Lactobacillus, Halomonas and Rhodococcus. The dominant genera were different in different geographical environments (Fig. 1D).
Effects of deep-sea bacteria on gut microbiota of mice
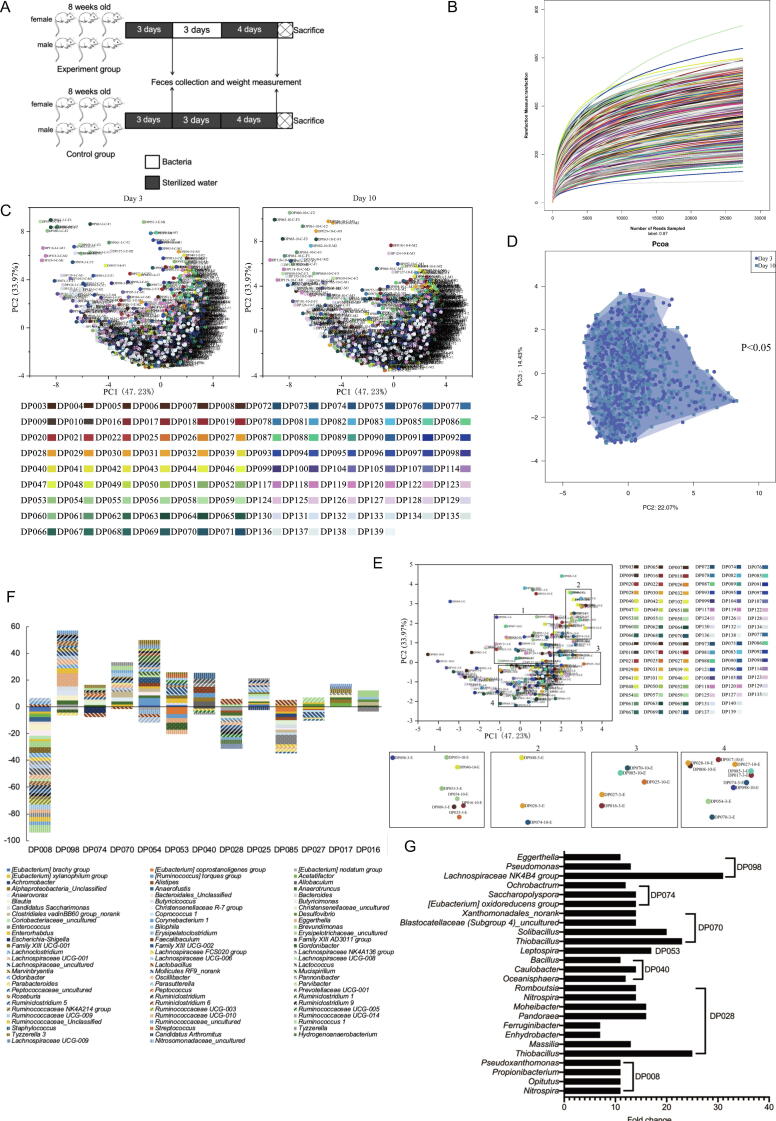
To evaluate the effects of deep-sea bacteria on the microbiota of mammalian gut, the ICR (Institute of Cancer Research) mice were fed with the mixed bacteria (total bacterial community) isolated from each of 106 deep-sea sediments (experiment group) or sterilized water (control group) (Fig. 2A). A total of 106 experiment groups and the corresponding 106 control groups were performed. At Day 3 and Day 10, the mouse feces of experiment and control groups were collected and subject to the analysis of gut microbiota (Fig. 2A). The sequencing analysis of bacterial 16S rRNA gene revealed a total of 12,280,349 clean reads and identified 4,043 OTUs (Table S2) (GenBank accession no. PRJNA721276). Based on OTUs with 97% similarity, the rarefaction curves of all samples were approaching plateaus (Fig. 2B). At the same time, the coverage of sequencing for all samples ranged from 99.2% to 99.9% (Table S2). These results showed that the sequencing data represented the bacterial communities of mouse gut microbiota which could be used for the further analysis. Based on Shannon, Simpson and Chao indices, the mouse gut bacteria possessed high diversities (Table S2).
Fig. 2.
Effects of deep-sea bacteria on gut microbiota of mice. (A) A flow diagram of the experiments. Male and female mice (8 weeks old) were fed with sterilized water in a sterile environment for 3 days, followed by feeding with the isolated deep-sea bacteria or sterilized water for 3 days. After maintenance of mice for further 4 days, the mice were sacrificed. Fecal sample collection and weight measurement of mice were conducted at Day 3 and Day 10. (B) Rarefaction curves of the bacterial 16S rRNA genes for all samples. (C) Principal co-ordinates analysis of the mouse gut bacterial communities of 106 experiment groups and 106 control groups at Day 3 (left panel) and Day 10 (right panel). For each of experiment groups or control groups, 3 female and 3 male mice were used. (D) Principal co-ordinates analysis of the gut microbiota of the control mice (control group). The bacterial communities of the feces of 106 control groups at Day 3 and Day 10 were analyzed with principal co-ordinates analysis. Each dot represented the bacterial community of mouse at Day 3 (blue) or Day 10 (green). (E) Principal co-ordinates analysis of the bacterial communities of the fecal samples of the mice treated with the bacteria isolated from 106 deep-sea sediments. The mice were fed with the bacteria from one of 106 deep-sea sediment samples, followed by the analysis of gut microbiota. Each dot represented an experiment group at Day 3 or Day 10. The enlarged images of the samples with significant alteration (p < 0.05) were shown at the lower panel. “E” indicated the fecal samples of experiment group. (F) The bacteria significantly increased or decreased in the gut microbiota of the mice treated with the bacteria isolated from deep-sea sediments. Based on the sequencing data of bacterial 16S rRNA gene, the mouse gut bacteria with significant fold change were indicated at the genus level. The deep-sea-originated bacteria were shown. (G) The deep-sea-originated bacteria in the gut microbiota of mice. After treatment of deep-sea bacteria, 24 bacteria appeared in the gut microbiota of mice. These 24 bacteria were absent in the gut microbiota of the control mice, showing they originated from deep-sea sediments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The principal co-ordinates analysis showed that the bacterial communities of mouse gut between six repeats (3 female and 3 male mice) of each treatment of 106 experiment groups or 106 control groups were similar to each other (Fig. 2C), indicating that the data were reliable and could be used for further analysis. At the same time, there was no significant difference of mouse gut bacterial communities between control groups at Day 3 and Day 10 (Fig. 2D), showing that the bacterial communities of mouse gut were stable.
The results of principal co-ordinates analysis indicated that among 106 deep-sea sediments, the bacteria from 13 sediments significantly altered the bacterial communities of gut microbiota of mice (Fig. 2E). These deep-sea sediments included DP008, DP016, DP017, DP025, DP027, DP028, DP040, DP053, DP054, DP070, DP074, DP085 and DP098.
Among 13 deep-sea sediments, the bacteria from DP008 significantly decreased or increased the relative abundance of the mouse gut bacteria belonging to 30 genera or 7 genera (Fig. 2F and Table S3). The bacteria from DP054 led to significant decrease or increase of the mouse gut bacteria belonging to 4 or 21 genera (Fig. 2F and Table S3). The bacteria from the remaining 11 deep-sea sediments also significantly changed the bacterial communities of gut microbiota of mice (Fig. 2F and Table S3). In addition, it is interesting that 24 bacteria, which were absent in the gut microbiota of the control mice, were presented in the gut microbiota of the mice treated with the bacteria from 7 deep-sea sediments (Fig. 2G), indicating that these bacteria were originated from deep sea. These 24 bacteria included Nitrospira, Opitutus, Propionibacterium, Pseudoxanthomonas, Lachnospiraceae NK4B4 group, Pseudomonas, Eggerthella, [Eubacterium] oxidoreducens group, Saccharopolyspora, Ochrobactrum, Thiobacillus, Solibacillus, Blastocatellaceae (Subgroup 4) uncultured, Xanthomonadales_norank, Leptospira, Oceanisphaera, Caulobacter, Bacillus, Massilia, Enhydrobacter, Ferruginibacter, Pandoraea, Moheibacter and Romboutsia. A total of 50 OTUs from deep sea were identified to be these 24 bacteria. These data demonstrated that the deep-sea bacteria could invade the gut microbiota of mice to change the bacterial community of mouse gut.
Collectively, the bacteria from 13 of 106 deep-sea sediments invaded the bacterial communities of gut microbiota of mice, leading to the significant alteration of mouse gut microbiota.
Influence of deep-sea bacteria on the health of mice
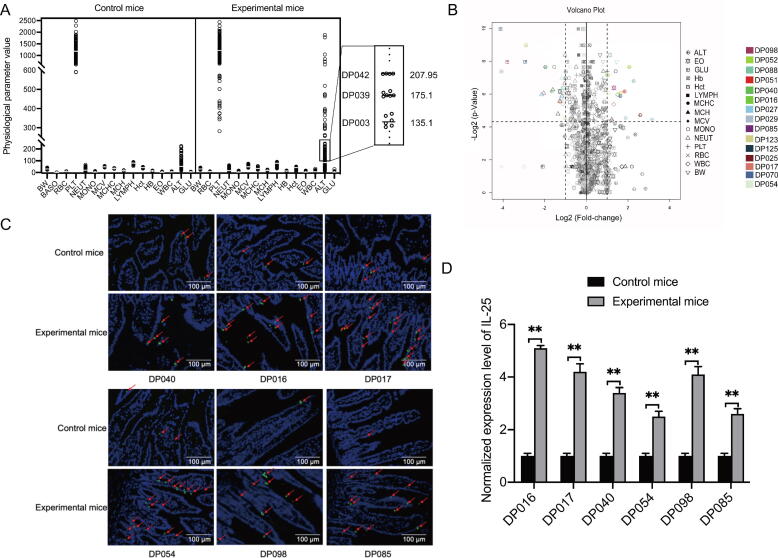
To reveal the effects of deep-sea bacteria on the health of mice, the blood and gut tissue samples of experiment groups and control groups (Fig. 2A) were collected. The mice of experiment groups fed the isolated bacteria from each of 106 deep-sea sediments, while the mice of control groups fed water. The blood samples were subjected to the detections of blood routine, alanine aminotransferase (ALT) and blood glucose (GLU), and the intestinal tissue samples were analyzed to detect the expression level of IL-25 and Tuft cells. These physiological parameters could indicate liver disease, obesity or/and colitis. For each of 106 treatments (bacteria or water), there was no difference of physiological parameters between 6 mouse individuals of experiment or control mice (Fig. 3A), indicating that the data were reliable.
Fig. 3.
Influence of deep-sea bacteria on the health of mice. (A) Scatter plots of the value of 16 physiological parameters of control mice and experimental mice. Both control mice and experimental mice consisted of 3 female and 3 male ICR mice. Each of the experimental mice was fed with the bacteria isolated from a deep-sea sediment, while the control mice were fed with sterilized water. In total, 106 deep-sea sediments were used. Subsequently the physiological parameters of the experimental and control mice were examined. Each dot represented the mean of a physiological parameter of 6 mice (3 female and 3 male mice). As an example, ALT of experimental mice was enlarged. In the enlarged image, each line indicated the mean (on the right) of 6 ALT values of the mice treated with the bacteria from a deep-sea sediment (on the left). As shown in the enlarged image, the ALT values of 6 mice were close to each other, showing that there was no difference of physiological parameters between 6 mouse individuals. BW, body weight; BASO, basophilic granulocyte; RBC, red blood cell; PLT, platelet; NEUT, neutrophil; MONO, monocyte; MCV, mean corpuscular volume; MCHC, mean corpuscular hemoglobin concentration; MCH, mean corpuscular hemoglobin; LYMPH, lymphocyte; Hct, hematocrit; Hb, hemoglobin; EO, eosinophil; WBC, white blood cell, ALT, alanine aminotransferase; GLU, glucose. (B) Volcano plots showing statistical significance (y axis) versus fold change (x axis) of each physiological parameter of mice after administration of the bacteria from each deep-sea sediment sample. The colored points above the horizontal dashed line indicated the significant changes with p < 0.05 (lower line) and p < 0.01 (upper line). The percentage of BASO was 0% in most of the samples. Therefore it was not shown. (C) Influence of deep-sea bacteria on the number of tuft cells in the small intestine of mice. The small intestines of control and experimental mice were labeled with the antibody against DCLK1 (green), the marker protein of tuft cells. The nuclei were stained with DAPI (blue). The tuft cells were indicated with arrows. Scale bar, 100 μm. (D) The expression level of IL-25 in the small intestine tissues of control and experimental mice. Quantitative real-time PCR was used to examine the expression profiles of IL-25 (**, p < 0.01). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The results showed that 7 of 15 physiological parameters showed no significant difference between 106 experiment groups and control groups (Fig. 3B). These physiological parameters included basophilic granulocyte (BASO), red blood cell (RBC), erythrocyte mean corpuscular volume (MCV), erythrocyte mean corpuscular hemoglobin concentrate (MCHC), erythrocyte mean corpuscular hemoglobin (MCH), hematocrit (Hct) and hemoglobin (Hb). Of these parameters, BASO, an effector cell of allergy, was not affected by the isolated deep-sea bacteria, indicating that the deep-sea bacteria did not trigger allergic symptoms of mammals.
The bacteria isolated from each of 9 deep-sea sediments (DP040, DP054, DP098, DP016, DP017, DP027, DP070, DP025 and DP085) significantly increased or decreased 9 physiological parameters, including BW, PLT, NEUT, MONO, LYMPH, EO, WBC, ALT and GLU. Of the bacteria isolated from 106 deep-sea sediments, the bacteria from 5 sediments (DP029, DP070, DP098, DP123 and DP125) significantly decreased the glucose (GLU) content of mice, while the bacteria from the remaining sediments had no effect on the mouse GLU level (Fig. 3B and Table S4). At the same time, the bacteria isolated from 2 of 5 sediments, including DP098 and DP070, significantly decreased the body weight of mice (Fig. 3B and Table S4), indicating that these deep-sea bacteria could cause glucose metabolism deterioration of mice. Based on the detection of alanine aminotransferase (ALT), the ALT levels of the mice fed the bacteria isolated from 8 deep-sea sediments (DP025, DP027, DP029, DP051, DP052, DP088, DP123 and DP125) were significantly increased (Fig. 3B and Table S4). Among 8 sediments, the bacteria from 2 sediments (DP025 and DP027) significantly decreased the mouse body weight and gut bacterial communities (Fig. 3B and Table S4), suggesting that the bacteria from deep-sea sediments DP025 and DP027 caused liver damage of mice through dysbiosis of gut microbiota.
The number of white blood cells (WBC) in the mice fed the bacteria isolated from 3 deep-sea sediments (DP040, DP016 and DP017) was significantly increased compared with that of the control mice (Fig. 3B and Table S4). The percentage of monocytes (MONO) in the mice fed the bacteria from sediment DP085 was significantly increased (Fig. 3B and Table S4). For sediments DP054 and DP098, the isolated bacteria significantly increased the percentage of mouse neutrophil (NEUT) (Fig. 3B and Table S4). At the same time, the mice fed the bacteria from sample DP098 showed a significant weight loss. However, the percentage of lymphocyte (LYMPH) in the mice fed the bacteria from sediments DP040, DP016, DP017, DP085, DP054 and DP098 was not altered. These data indicated that the bacteria from deep-sea sediments DP040, DP016, DP017, DP085, DP054 and DP098 caused inflammation of mice.
To further evaluate the influence of deep-sea bacteria on gut inflammation, the number of tuft cells and the level of interleukin (IL)-25 of the small intestine of the mice fed the bacteria from DP040, DP016, DP017, DP085, DP054 and DP098, the parameters of inflammatory symptom, were examined. Compared to the control groups, the tuft cell hyperplasia was observed in the small intestine of experiment groups (Fig. 3C), indicating that the bacteria from DP040, DP016, DP017, DP085, DP054 and DP098 promoted inflammation of mice. At the same time, IL-25, secreted by tuft cells, [21] was significantly upregulated in experiment groups (Fig. 3D). These results demonstrated that the bacteria from deep-sea sediments DP040, DP016, DP017, DP085, DP054 and DP098 could induce an inflammatory symptom.
Collectively, these findings revealed that deep-sea bacteria did not trigger allergic symptoms of mammals, but some deep-sea bacteria could destroy mouse health by inducing glucose metabolism deterioration, liver damage or/and inflammatory symptom.
Relationship between mouse health and mouse gut bacterial communities mediated by deep-sea bacteria
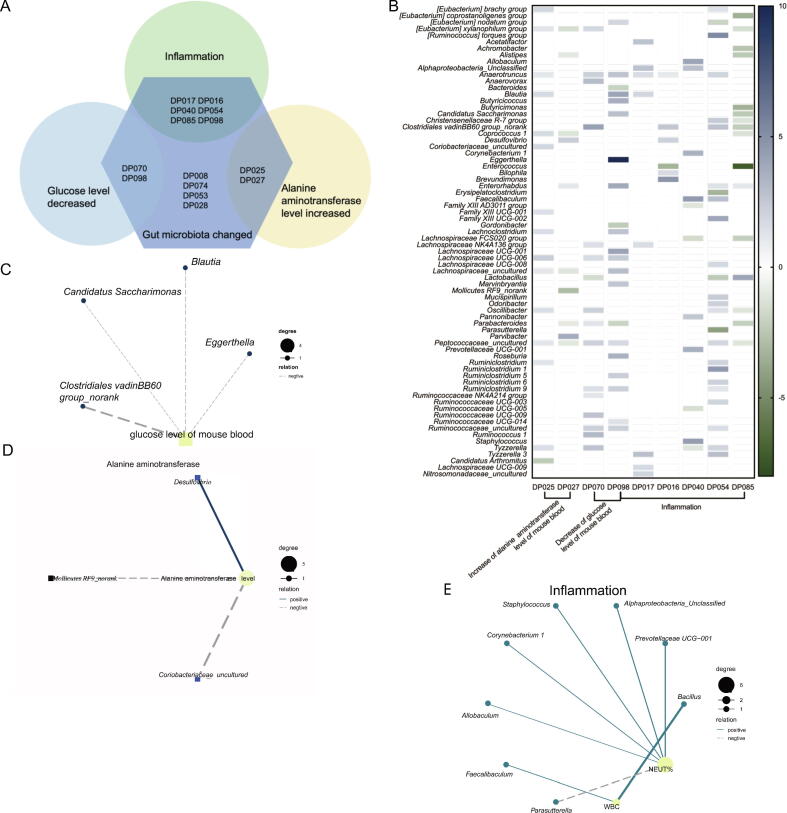
To explore whether the health damage of mice treated with deep-sea bacteria resulted from the changes of mouse gut bacteria, the relationship between mouse gut bacterial communities and mouse health mediated by deep-sea bacteria was characterized. The results showed that the bacteria from 9 deep-sea sediments (DP017, DP016, DP025, DP027, DP040, DP054, DP070, DP085 and DP098) significantly altered the mouse gut bacterial communities, leading to the damage of mouse health (Fig. 4A). The bacteria from DP070 and DP098 significantly decreased the glucose level of mouse blood, while the bacteria from DP025 and DP027 significantly increased the alanine aminotransferase level of mouse blood (Fig. 4A). The bacteria from DP016, DP017, DP040, DP054, DP085 and DP098 caused intestinal inflammation of mice by altering the mouse gut bacterial communities (Fig. 4A). However, the bacteria from 6 deep-sea sediments (DP029, DP051, DP052, DP088, DP123 and DP125) changed the physiological parameters of mice, but did not alter the mouse gut microbiota.
Fig. 4.
Relationship between mouse health and mouse gut bacterial communities mediated by deep-sea bacteria. (A) The deep-sea bacteria significantly changing the mouse gut bacterial communities and destroying the mouse health. Among 106 deep-sea sediments, the bacteria from each of 9 sediments significantly altered the gut bacterial communities of mice, leading to inflammation, the decrease of glucose level and/or the increase of alanine aminotransferase. (B) Influence of the invasion of deep-sea bacteria on the gut microbiota and health of mice. The increase and decrease of the mouse gut bacteria, mediated by the bacteria from deep-sea sediments, led to the decrease of the glucose level of mouse blood, the increase of the alanine aminotransferase level of mouse blood or/and the intestinal inflammation of mice. The increase or decrease level of bacteria was shown using fold-change value. Only the bacteria with fold-change ≥ 2 were indicated. (C) Correlation analysis of the bacterial community and the glucose level of mouse blood. The correlation analysis was conducted using Spearman's rank correlation coefficient. (D) Correlation analysis of the bacterial community and the alanine aminotransferase level of mouse blood. (E) Correlation analysis of the bacterial community and the inflammatory parameters of mice.
The results showed that the bacteria from DP070 and DP098 significantly changed the abundance of 31 bacteria of mouse gut microbiota at the genus level, resulting in the decrease of the glucose level of mouse blood (Fig. 4B). To reveal the origins of the 31 bacteria, they were compared with the bacterial community of mouse gut microbiota (GenBank accession no. PRJNA721276) and the bacterial communities of deep-sea sediments (GenBank accession no PRJNA721272). It was shown that 3 bacteria were only found in the mouse gut bacterial community, while 27 bacteria were simultaneously found in the mouse gut bacterial community and the deep-sea sediments (Table 1). Interestingly, the bacterium Eggerthella only existed in deep-sea sediments (Table 1). The correlation analysis demonstrated that based on the relative abundance of bacteria, 4 of 31 bacteria at the genus (Blautia, Candidatus Saccharimonas and Clostridiales vadinBB60 group_norank) were negatively correlated with the glucose level of mouse blood, and no bacterium was positively correlated with the glucose level of mouse blood (Fig. 4C).
Table 1.
The origins of the altered bacteria in the gut microbiota of mice treated with the isolated bacteria from deep-sea sediments.
| Effect | Origin | Bacteria at the genus level |
|---|---|---|
| the decrease of the glucose level of mouse blood | Mouse gut bacterial community | Candidatus Saccharimonas, Lachnospiraceae UCG-006, Gordonibacter |
| Deep-sea sediments | Eggerthella | |
| Mouse gut bacterial community and deep-sea sediments | Enterorhabdus, Lachnospiraceae UCG-001, Ruminiclostridium 5, Marvinbryantia, Butyricicoccus, [Eubacterium] xylanophilum group, Peptococcaceae_uncultured, Clostridiales vadinBB60 group_norank, Lachnoclostridium, Blautia, Roseburia, Ruminococcus 1, [Eubacterium] nodatum group, Anaerovorax, Oscillibacter, Ruminococcaceae NK4A214 group, Ruminococcaceae UCG-009, Tyzzerella, Lachnospiraceae NK4A136 group, Ruminococcaceae UCG-014, Lachnospiraceae_uncultured, Anaerotruncus, Ruminiclostridium 9, Ruminococcaceae_uncultured, Lactobacillus, Parabacteroides, Bacteroides | |
| the increase of the alanine aminotransferase level of mouse blood | Mouse gut bacterial community | Candidatus Arthromitus, Desulfovibrio, Family XIII UCG-001, [Eubacterium] xylanophilum group, Blautia, Parvibacter |
| Mouse gut bacterial community and deep-sea sediments | Peptococcaceae_uncultured, Enterorhabdus, Coriobacteriaceae_uncultured, Lachnoclostridium, Ruminiclostridium, [Eubacterium] brachy group, Coprococcus 1, Parabacteroides, Tyzzerella, Lachnospiraceae_uncultured, Alistipes, Oscillibacter, Anaerotruncus, Mollicutes RF9_norank, Lachnospiraceae UCG-006 | |
| intestinal inflammation of mice | Mouse gut bacterial community | Acetatifactor, Bilophila, Eggerthella, Gordonibacter, Lachnospiraceae UCG-006, Lachnospiraceae UCG-009 |
| Mouse gut bacterial community and deep-sea sediments | Achromobacter, Alistipes, Allobaculum, Alphaproteobacteria_Unclassified, Anaerotruncus, Bacteroides, Blautia, Brevundimonas, Butyricicoccus, Butyricimonas, Candidatus Saccharimonas, Christensenellaceae R-7 group, Clostridiales vadinBB60 group_norank, Coprococcus 1, Corynebacterium 1, Desulfovibrio, Enterococcus, Enterorhabdus, Erysipelatoclostridium, Faecalibaculum, Family XIII AD3011 group, Family XIII UCG-002, Lachnoclostridium, Lachnospiraceae FCS020 group, Lachnospiraceae NK4A136 group, Lachnospiraceae UCG-001, Lachnospiraceae UCG-008, Lachnospiraceae_uncultured, Lactobacillus, Marvinbryantia, Mucispirillum, Nitrosomonadaceae_uncultured, Odoribacter, Oscillibacter, Pannonibacter, Parabacteroides, Parasutterella, Peptococcaceae_uncultured, Prevotellaceae UCG-001, Roseburia, Ruminiclostridium, Ruminiclostridium 1, Ruminiclostridium 5, Ruminiclostridium 6, Ruminiclostridium 9, Ruminococcaceae UCG-005, Ruminococcaceae UCG-014, Ruminococcaceae_uncultured, Staphylococcus, Tyzzerella, Tyzzerella 3, [Eubacterium] brachy group, [Eubacterium] coprostanoligenes group, [Eubacterium] nodatum group, [Eubacterium] xylanophilum group, [Ruminococcus] torques group | |
| Deep-sea sediments | Bacillus |
When the mice were treated with the bacteria from DP025 and DP027, the abundance of 21 bacteria of mouse gut microbiota at the genus level was significantly altered and the alanine aminotransferase level of mouse blood was significantly increased (Fig. 4B). Among the 21 bacteria, 6 bacteria existed in the mouse gut microbiota, while the remaining 15 bacteria could be found in the deep-sea sediments and the mouse gut microbiota (Table 1). The correlation analysis revealed that of the 21 bacteria, Desulfovibrio was positively correlated with the alanine aminotransferase level of mouse blood, while Coriobacteriaceae_uncultured and Mollicutes RF9_norank were negatively correlated with the alanine aminotransferase level of mouse blood (Fig. 4D).
In the mice treated with the bacteria from DP016, DP017, DP040, DP054, DP085 and DP098, the abundance of 63 bacteria of mouse gut microbiota at the genus level was significantly changed, resulting in intestinal inflammation of mice (Fig. 4B). Of the 63 bacteria, 6 bacteria, including Acetatifactor, Bilophila, Eggerthella, Gordonibacter, Lachnospiraceae UCG-006, were found in the mouse gut microbiota, while a bacterium Bacillus only existed in the deep-sea sediments (Table 1). The remaining 56 bacteria co-existed in the deep-sea sediments and the mouse gut microbiota (Table 1). The correlation analysis showed that among the 63 bacteria, Bacillus, Prevotellaceae UCG-001, Alphaproteobacteria_Unclassified, Staphylococcus, Corynebacterium 1, Faecalibaculum and Allobaculum were positively correlated with inflammation, whereas Parasutterella was negatively correlated with inflammation (Fig. 4E).
Collectively, these results indicated that the deep-sea bacteria could invade mouse gut microbiota, leading to the alteration of gut bacterial community and further the damage of mouse health such as inflammation, liver damage and decreased blood glucose level.
Characterization of the deep-sea bacteria destroying mouse health
To identify the deep-sea bacteria that could invade the gut microbiota of mouse to destroy mouse health, the bacteria were isolated from deep-sea sediments. Based on the screening of bacteria from deep-sea sediments DP016, DP017, DP025, DP027, DP040, DP054, DP070, DP085 and DP098, many bacterial colonies were observed. More than 70 colonies were selected randomly for the bacterial 16S rRNA gene sequencing. A total of 21 bacterial species were identified (Fig. 5A). Among these bacteria, a bacterium from DP040, identified to be Bacillus cereus (termed as Bacillus cereus DP040) based on the bacterial 16S rRNA gene sequence (GenBank accession number OK036352) (Fig. 5B), existed only in the gut microbiota of the mice treated with deep-sea bacteria, but not in the gut microbiota of control mice or experimental mice before treatment (Fig. 2F and Table 1), suggesting that Bacillus cereus DP040 might play important roles in destroying mouse health. Therefore Bacillus cereus DP040 was further characterized. The transmission electron microscopic data showed that Bacillus cereus DP040 was rod-shaped (Fig. 5C).
Fig. 5.
Characterization of the deep-sea bacteria destroying mouse health. (A) The bacteria isolated from deep-sea sediments. All bacteria were isolated using beef extract peptone medium or enterobacteria enrichment broth at 37 °C. The bacteria were identified based on the bacterial 16S rRNA gene sequencing. (B) Phylogenetic tree of Bacillus cereus isolated from deep-sea sediment DP040. The tree was constructed using the neighbor-joining analysis of a distance matrix obtained from a multiple-sequence alignment. Scale bar represented the genetic distance (1 substitution per 100 nucleotides). (C) Image of Bacillus cereus DP040. The bacteria were observed under a transmission electron microscope. Scale bar, 500 nm. (D) Influence of Bacillus cereus DP040 on the gut bacterial communities of mice. The mice (8 weeks old) were fed with sterilized water in a sterile environment for 3 days to stabilize the mouse gut bacterial communities. Then 3 male and 3 female mice were fed with the purified deep-sea Bacillus cereus DP040 for 3 days. After maintenance of the mice for further 4 days, the mice were sacrificed. At Day 3 and Day 10, the fecal samples were collected and subjected to the bacterial 16S rRNA gene sequencing. (E) Detection of inflammatory symptom of the mice treated with Bacillus cereus DP040. Mice were fed Bacillus cereus DP040 or PBS. Seven days later, the number of white blood cells (WBC) and Il-25 level of mice were examined (**, p < 0.01). (F) Model for the invasion of deep-sea bacteria into mouse gut microbiota. The bacteria from deep sea invaded the gut microbiota of mice to alter the mouse gut microbiota, leading to the damage of mouse health.
As reported, Bacillus cereus, a Gram-positive bacterium, is an important opportunistic human pathogen that can be well adapted for growth in the intestinal tract of mammals, leading to food poisoning. [22] Therefore the influence of deep-sea Bacillus cereus on mouse gut microbiota and health was explored. The results indicated that the contents of 14 bacterial genera, including Candidatus Soleaferrea, Oscillibacter, Roseburia, Tyzzerella, Lachnospiraceae NK4A136 group, Ruminiclostridium 9, Anaerotruncus, Lachnospiraceae_uncultured, [Eubacterium] nodatum group, Christensenellaceae_uncultured, Anaerovorax, Candidatus Saccharimonas, [Eubacterium] brachy group and Ruminococcaceae UCG-014, were significantly increased in the gut microbiota of the mice treated with Bacillus cereus DP040 (6 × 10 [8] bacteria/mouse) (Fig. 5D). At the same time, the number of white blood cells (WBC) and Il-25 level of the mice fed Bacillus cereus DP040 were significantly increased compared with the controls (Fig. 5E), which were consistent with the previous data (Fig. 3B, 3D and Table S4).
Taken together, these results indicated that some bacteria in deep-sea sediments, such as Bacillus cereus DP040, could invade the gut microbiota of mice to change its structure, leading to the health damage of mice (Fig. 5F).
Genomic characterization of deep-sea Bacillus cereus DP040
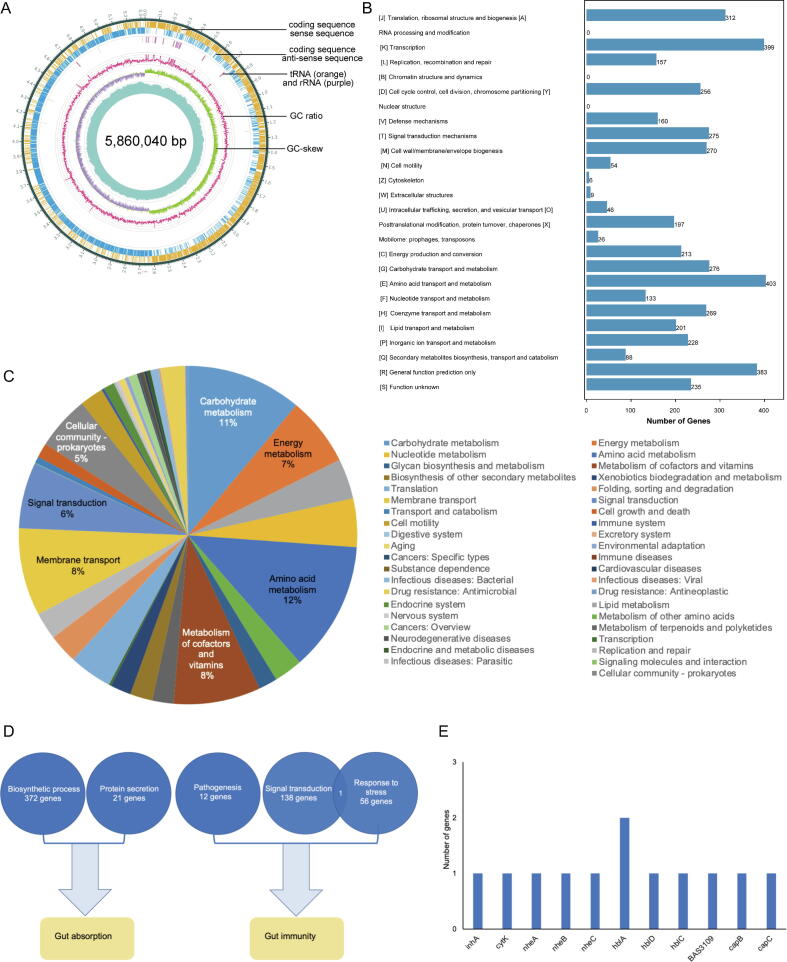
To further characterize the bacteria isolated from deep-sea sediment sample DP040, the whole genome of Bacillus cereus DP040 was sequenced. The results showed that Bacillus cereus DP040 contained a 5,860,040-bp genome with an average GC content of 34% (Fig. 6A and Table 2) (GenBank accession number PRJNA786723). Bacillus cereus DP040 shared 73.6% identity of genome sequence with the known Bacillus cereus. The Bacillus cereus DP040 genome contained 5,694 open reading frames (ORFs), ranging from 93 bp to 15,033 bp in length with an average length of 854.23 bp (Table 2). Of these ORFs, 4,068 ORFs were annotated into 26 categories in the Clusters of Orthologous Group (COG) database (Fig. 6B). The largest COG group was the [E] group (amino acid transport and metabolism), followed by the [K] group (transcription). Based on KEGG analysis, 2,552 of 5,694 genes were involved in amino acid metabolism (12%), carbohydrate metabolism (11%), metabolism of cofactors and vitamins (8%), membrane transport (8%) and energy metabolism (7%) (Fig. 6C).
Fig. 6.
Genomic characterization of deep-sea Bacillus cereus DP040. (A) Circos plot showing the Bacillus cereus DP040 genome. The coding sequence, tRNA (orange), rRNA (purple) were integrated to draw the circos plot of the nuclear genome of Bacillus cereus DP040. (B) Functional classifications of the genes encoded by Bacillus cereus DP040 based on the Clusters of Orthologous Group (COG) database. (C) The percentage of the genes assigned in KEGG categories. (D) The genes assigned with functions that affected the functions of gut. (E) The identified virulence factors in Bacillus cereus DP040 genome. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 2.
Characteristics of the Bacillus cereus DP040 genome.
| Characteristics | Bacillus cereus DP040 |
|---|---|
| Length (bp) | 5,860,040 |
| GC content | 35.11% |
| No. of plasmid | 1 |
| No. of coding genes | 5,694 |
| Ns (%) | 0 |
| No. of non-coding RNA | 278 |
| No. of ribosomal RNA | 42 |
| No. of transfer RNA | 108 |
Based on the genes encoded by Bacillus cereus DP040, Bacillus cereus DP040 might affect the gut absorption and immunity. A total of 372 genes participated in the biosynthetic process and 21 genes were involved in the protein secretion (Fig. 6D), which could affect the absorption ability of animal gut. There were 12, 138 and 56 genes related to pathogenesis, signal transduction and responses to drug, antibiotic, heat and other types of stresses, respectively (Fig. 6D), which played important roles in immunity. These results indicated that the deep-sea Bacillus cereus DP040 had the capacity to affect the intestinal functions of mice. The analysis of virulence factors of Bacillus cereus DP040 genome revealed that 11 genes, including cytK and hblA, could trigger immune responses of mice to destroy mouse’s health (Fig. 6E).
Collectively, these findings revealed that Bacillus cereus DP040 could encode the proteins to alter the gut absorption and immunity of mice, thus having the ability to destroy mouse’s health.
Distribution of deep-sea sediments containing the bacteria destroying the mouse health
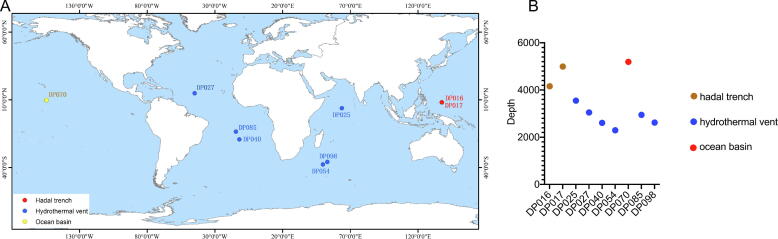
To reveal the distribution of the deep-sea sediments containing the bacteria harmful to the health of mice, the deep-sea sediment samples were analyzed. The results showed that among 106 deep-sea sediment samples, the bacteria from each of 9 sediments (DP016, DP017, DP025, DP027, DP040, DP054, DP070, DP085 and DP098) could destroy the health of mice via altering the structure of mouse gut microbiota. These 9 deep-sea sediment samples were distributed in the Indian Ocean (DP025, DP054, DP085 and DP098), the Atlantic Ocean (DP027 and DP040) and the Pacific Ocean (DP016, DP017 and DP070) (Fig. 7A). Of the 9 samples, 6 samples (DP025, DP027, DP040, DP054, DP085 and DP098) came from hydrothermal vents, 1 sample (DP070) was from ocean basins, and 2 (DP016 and DP017) were hadal trench samples (Fig. 7A). The 9 sediments were mainly distributed at about 10°north.
Fig. 7.
Distribution of deep-sea sediments containing the bacteria destroying the mouse health. (A) Distribution of deep-sea sediments containing the bacteria destroying the mouse health. (B) The sampling depth of deep-sea sediments containing the bacteria destroying the mouse health.
The results showed that the depth of the 9 sediment samples ranged from 2295 m to 5192 m with an average depth of 3490.44 m (Fig. 7B). Four samples (DP040, DP054, DP085 and DP098) were distributed between 2000 and 3000 m (Fig. 7B).
Taken together, these results demonstrated that among 106 deep-sea sediment samples, 9 samples containing the bacteria destroying the health of mice were distributed in hydrothermal vent, ocean basin and hadal trench of the Indian Ocean, the Atlantic Ocean and the Pacific Ocean.
Discussion
Pathogens, including bacteria, viruses, fungi and microscopic protozoans, possess an inherent capacity to cause diseases in hosts. [23] The human diseases caused by pathogens include but not limited to enteritis, [24] hepatitis, [25] pneumonia, [26] cholera and gastritis. [27], [28] As one of the main pathogens, bacteria have evolved to exploit humans as a rich source of nutrients to support bacterial survival and replication, [29] therefore colonizing and causing clinical symptoms. The sources of pathogenic bacteria can be foods, animals and even environments. [30], [31], [32] In recent years, environments have become the important sources of pathogenic bacteria, such as waters and soils in which the bacterial pathogens can cause sepsis, traumatic infection, gastroenteritis and even death. [33], [34] Up to date, however, whether there exist pathogenic bacteria in deep sea, one of the less explored and extreme environments on the earth, has not been explored. In this study, based on the characterization of the bacteria isolated from 106 deep-sea sediments on a global scale, the bacteria from 9 sediments of three oceans (the Indian Ocean, the Atlantic Ocean and the Pacific Ocean) could destroy mouse health via altering the mouse gut microbiota, suggesting the existence of potential pathogenic bacteria in deep sea. As an example, our findings showed that Bacillus cereus DP040, which was isolated from the deep-sea sediment, had the ability to invade mouse gut microbiota, leading to the damage of mouse health. Our study clearly demonstrates for the first time that the alien species invasion of potential pathogenic bacteria from deep sea is a great threat to mammal health.
Deep sea, covering more than 50% of the planet’s surface and 66% of global sea-floor area, [35] has existed for more than 4.2 billion years. [36] Deep sea, one of the most ancient and extreme environments on the earth, is characterized by high pressure, lack of light, extreme oxygen and salinity concentration, along with extreme temperature. [37] In the deep-sea ecosystems, there are abundant biological communities, of which microbes are the most abundant.[38] Among the deep-sea microorganisms that can be isolated and cultured, bacteria are a main type of microbes, most of which belong to Gamma-proteobacteria subgroup, including Shewanella, Moritella, Psychromonas, Photobacterium and Colwellia. Our findings revealed that 9 deep-sea sediment samples from hydrothermal vent, ocean basin and hadal trench of the Indian Ocean, the Atlantic Ocean and the Pacific Ocean contained the potential pathogenic bacteria that were harmful to mice. These deep-sea sediments were mainly distributed in hydrothermal vents. The potential pathogenic bacteria from deep sea could invade the gut microbiota of mice to alter the gut bacterial community and further to destroy the mouse health. The numerous adaptation genes and virulence factors encoded by deep-sea bacteria enabled the potential pathogenic bacteria from deep sea to quickly proliferate in the mouse gut, thus leading to alien species invasion of deep-sea bacteria into mouse gut microbiota. Due to their proliferation in mouse gut, the deep-sea bacteria altered the mouse gut microbiota and further resulted in the damage of mouse health, such as intestinal inflammation, liver damage and decrease of blood glucose level. The underlying mechanism of the invasion of deep-sea bacteria into mammal gut microbiota merited to be explored in the future work. At the same time, the biosecurity of deep-sea bacteria needed to be further investigated.
Compliance with ethics requirements
All Institutional and National Guidelines for the care and use of animals (fisheries) were followed.
CRediT authorship contribution statement
Mengqi Chu: Conceptualization, Methodology, Validation, Formal analysis, Writing – original draft, Visualization, Supervision. Xiaobo Zhang: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This work was supported by China Ocean Mineral Resources R & D Association (DY135-B-04) and the National Key Research and Development Program of China (2018YFC0310703).
Disclosure statement
There is no conflict of interest that I should disclose.
Data availability statement
The data that support the findings of this study are openly available in NCBI database at https://www.ncbi.nlm.nih.gov, reference number PRJNA721276.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Rosshart S.P., Vassallo B.G., Angeletti D., Hutchinson D.S., Morgan A.P., Takeda K., et al. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171(5):1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmidt T.S.B., Raes J., Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172(6):1198–1215. doi: 10.1016/j.cell.2018.02.044. PMID: 29522742. [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed F., Roswall J., Peng Y., Feng Q., Jia H., Kovatcheva-Datchary P., et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Kho Z.Y., Lal S.K. The human gut microbiome - a potential controller of wellness and disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. PMID: 30154767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slobodkina G.B., Mardanov A.V., Ravin N.V., Frolova A.A., Chernyh N.A., Bonch-Osmolovskaya E.A., et al. Respiratory ammonification of nitrate coupled to anaerobic oxidation of elemental sulfur in deep-sea autotrophic thermophilic bacteria. Front Microbiol. 2017;8:87. doi: 10.3389/fmicb.2017.00087. PMID: 28194142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kinross J.M., Darzi A.W., Nicholson J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011;3(3):14. doi: 10.1186/gm228. PMID: 21392406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. PMID: 21850056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salazar G., Cornejo-Castillo F.M., Benítez-Barrios V., Fraile-Nuez E., Álvarez-Salgado X.A., Duarte C.M., et al. Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 2016;10(3):596–608. doi: 10.1038/ismej.2015.137. PMID: 26251871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corinaldesi C. New perspectives in benthic deep-sea microbial ecology. Front Mar Sci. 2015;2 doi: 10.3389/fmars.2015.00017. [DOI] [Google Scholar]
- 10.Wu W.F., Wang F.P., Li J.H., Yang X.W., Xiao X., Pan Y.X. Iron Reduction and mineralization of deep-sea iron reducing bacterium shewanella piezotolerans WP3 at elevated hydrostatic pressures. Geobiology. 2013;11(6):593–601. doi: 10.1111/gbi.12061. PMID: 24102974. [DOI] [PubMed] [Google Scholar]
- 11.Scott K.M., Sievert S.M., Abril F.N., Ball L.A., Barrett C.J., Blake R.A., et al. The genome of deep-sea vent chemolithoautotroph thiomicrospira crunogena XCL-2. PLoS Biol. 2006;4(12):e383. doi: 10.1371/journal.pbio.0040383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruff S.E., Felden J., Gruber-Vodicka H.R., Marcon Y., Knittel K., Ramette A., et al. In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J. 2019;13(1):197–213. doi: 10.1038/s41396-018-0263-1. PMID: 30154496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J.L., Liu H.X., Chen Y.C., Tan H.B., Guo H., Xu L.Q., et al. Highly substituted benzophenone aldehydes and eremophilane derivatives from the deep-sea derived fungus Phomopsis lithocarpus FS508. Mar Drugs. 2018;16(9):329. doi: 10.3390/md16090329. PMID: 30208615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He T., Jin M., Xu C., Ma Z., Wu F., Zhang X. The homeostasis-maintaining metabolites from bacterial stress response to bacteriophage infection suppress tumor metastasis. Oncogene. 2018;37(43):5766–5779. doi: 10.1038/s41388-018-0376-z. PMID: 29925861. [DOI] [PubMed] [Google Scholar]
- 15.Takaya Y., Yasukawa K., Kawasaki T., Fujinaga K., Ohta J., Usui Y., et al. The tremendous potential of deep-sea mud as a source of rare-earth elements. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-23948-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang B., Huang J., Zhou X., Lin X., Liu J., Liao S., et al. The fungal metabolites with potential antiplasmodial activity. Curr Med Chem. 2018;25(31):3796–3825. doi: 10.2174/0929867325666180313105406. PMID: 29532754. [DOI] [PubMed] [Google Scholar]
- 17.Hasan N.A., Grim C.J., Lipp E.K., Rivera I.N.G., Chun J., Haley B.J., et al. Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species. Proc Natl Acad Sci U S A. 2015;112(21) doi: 10.1073/pnas.1503928112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasa A., Auguste M., Lema A., Oliveri C., Borello A., Taviani E., et al. A deep-sea bacterium related to coastal marine pathogens. Environ Microbiol. 2021;23(9):5349–5363. doi: 10.1111/1462-2920.15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo J.C., Long H., Zhang J., Zhao Y., Sun L. Characterization of a deep sea bacillus toyonensis isolate: genomic and pathogenic features. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.629116. PMID: 33777842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hays G.C. Ocean currents and marine life. Curr Biol. 2017;27(11):R470–R473. doi: 10.1016/j.cub.2017.01.044. PMID: 28586681. [DOI] [PubMed] [Google Scholar]
- 21.von Moltke J., Ji M., Liang H.E., Locksley R.M. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529(7585):221–225. doi: 10.1038/nature16161. PMID: 26675736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enosi Tuipulotu D., Mathur A., Ngo C., Man S.M. Bacillus cereus: epidemiology, virulence factors, and host-pathogen interactions. Trends Microbiol. 2021;29(5):458–471. doi: 10.1016/j.tim.2020.09.003. PMID: 33004259. [DOI] [PubMed] [Google Scholar]
- 23.Méthot P.O., Alizon S. What is a pathogen? toward a process view of host-parasite interactions. Virulence. 2014;5(8):775–785. doi: 10.4161/21505594.2014.960726. PMID: 25483864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Costa D., Iraola G. Pathogenomics of emerging Campylobacter Species. Clin Microbiol Rev. 2019;32(4):e00072–e118. doi: 10.1128/CMR.00072-18. PMID: 31270126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinstone S.M. History of the discovery of Hepatitis A virus. Cold Spring Harb Perspect Med. 2019;9(5) doi: 10.1101/cshperspect.a031740. PMID: 29712682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang T., Du Z., Zhu F., Cao Z., An Y., Gao Y., et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395(10228) doi: 10.1016/S0140-6736(20)30558-4. PMID: 32171074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothenbacher F.P., Zhu J. Efficient responses to host and bacterial signals during Vibrio cholerae colonization. Gut Microbes. 2014;5(1):120–128. doi: 10.4161/gmic.26944. PMID: 24256715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamboj A.K., Cotter T.G., Oxentenko A.S. Helicobacter pylori: the past, present, and future in management. Mayo Clin Proc. 2017;92(4):599–604. doi: 10.1016/j.mayocp.2016.11.017. PMID: 28209367. [DOI] [PubMed] [Google Scholar]
- 29.Passalacqua K.D., Charbonneau M.-E., O’Riordan M.X.D., Kudva I.T., Cornick N.A. Bacterial metabolism shapes the host-pathogen interface. Microbiol Spectr. 2016;4(3) doi: 10.1128/microbiolspec.VMBF-0027-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raudales R., Parke J.L., Guy C.L., Fisher P.R. Control of waterborne microbes in irrigation: a review. Agric Water Manage. 2014;143:9–28. doi: 10.1016/j.agwat.2014.06.007. [DOI] [Google Scholar]
- 31.Umesha S., Manukumar H.M. Advanced molecular diagnostic techniques for detection of food-borne pathogens: current applications and future challenges. Crit Rev Food Sci Nutr. 2018;58(1):84–104. doi: 10.1080/10408398.2015.1126701. PMID: 26745757. [DOI] [PubMed] [Google Scholar]
- 32.Liu H., Whitehouse C.A., Li B. Presence and persistence of salmonella in water: the impact on microbial quality of water and food safety. Front Public Health. 2018;6:159. doi: 10.3389/fpubh.2018.00159. PMID: 29900166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen J.E., Wang R., Shen R.-F., Wu W.W., Keller J.E., Brüggemann H. Comparative pathogenomics of Clostridium tetani. PLoS ONE. 2017;12(8):e0182909. doi: 10.1371/journal.pone.0182909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leng F., Lin S., Wu W., Zhang J., Song J., Zhong M. Epidemiology, pathogenetic mechanism, clinical characteristics, and treatment of vibrio vulnificus infection: a case report and literature review. Eur J Clin Microbiol Infect Dis. 2019;38(11):1999–2004. doi: 10.1007/s10096-019-03629-5. PMID: 31325061. [DOI] [PubMed] [Google Scholar]
- 35.Woolley S.N., Tittensor D.P., Dunstan P.K., Guillera-Arroita G., Lahoz-Monfort J.J., Wintle B.A., et al. Deep-sea diversity patterns are shaped by energy availability. Nature. 2016;533(7603):393–396. doi: 10.1038/nature17937. PMID: 27193685. [DOI] [PubMed] [Google Scholar]
- 36.Daniel I., Oger P., Winter R. Origins of life and biochemistry under high-pressure conditions. Chem Soc Rev. 2006;35(10):858–875. doi: 10.1039/b517766a. PMID: 17003893. [DOI] [PubMed] [Google Scholar]
- 37.Kamjam M., Sivalingam P., Deng Z., Hong K. Deep sea actinomycetes and their secondary metabolites. Front Microbiol. 2017;8:760. doi: 10.3389/fmicb.2017.00760. PMID: 28507537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierella Karlusich J.J., Ibarbalz F.M., Bowler C. Phytoplankton in the Tara ocean. Ann Rev Mar Sci. 2020;12:233–265. doi: 10.1146/annurev-marine-010419-010706. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in NCBI database at https://www.ncbi.nlm.nih.gov, reference number PRJNA721276.