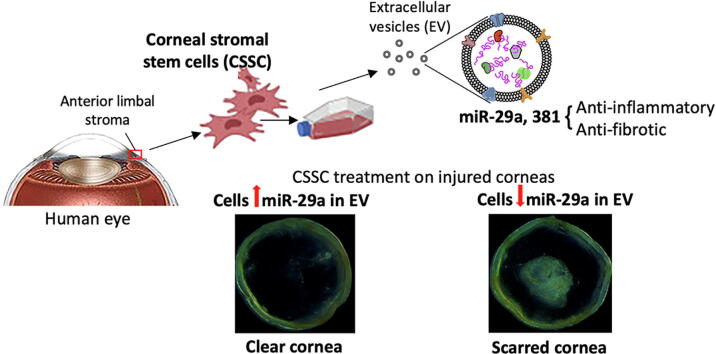
Graphical abstract
A schematic overview of this study showing corneal stromal stem cell (CSSC) isolation from anterior limbal stromal tissue, ex vivo cell culture, extraction of extracellular vesicles (EV) to identify the expression of miR-29a and 381-5p by Nanostring assay. The anti-inflammatory and anti-fibrotic effects were determined by in vitro and in vivo assays. Screening of miR-29a expression in CSSC-EV distinguished CSSC with good anti-scarring quality for cell-based therapy of corneal scarring after injury.
Keywords: Corneal blindness, Scarring, Stromal cell therapy, Stem cells, Extracellular vesicles, miR-29a
Highlights
-
•
Corneal stromal stem cell (CSSC) therapy rectifies corneal scarring, a cause of global blindness.
-
•
Cell potency and quality control are needed to assure cell product safety for patient use.
-
•
Transcriptomic assay found microRNAs enriched in extracellular vesicles (EV) of healing CSSC.
-
•
EV express miR-29a/381 to reduce inflammation and fibrosis, indicating anti-scarring potency.
-
•
High miR-29a levels in EV distinguished CSSC with good anti-scarring quality for clinical use.
Abstract
Introduction
Corneal blindness due to scarring is treated with corneal transplantation. However, a global problem is the donor material shortage. Preclinical and clinical studies have shown that cell-based therapy using corneal stromal stem cells (CSSCs) suppresses corneal scarring, potentially mediated by specific microRNAs transported in extracellular vesicles (EVs). However, not every CSSC batch from donors achieves similar anti-scarring effects.
Objectives
To examine miRNA profiles in EVs from human CSSCs showing “healing” versus “non-healing” effects on corneal scarring and to design a tool to select CSSCs with strong healing potency for clinical applications.
Methods
Small RNAs from CSSC-EVs were extracted for Nanostring nCounter Human miRNA v3 assay. MicroRNAs expressed > 20 folds in “healing” EVs (P < 0.05) were subject to enriched gene ontology (GO) term analysis. MiRNA groups with predictive regulation on inflammatory and fibrotic signalling were studied by mimic transfection to (1) mouse macrophages (RAW264.7) for M1 phenotype assay; (2) human corneal keratocytes for cytokine-induced fibrosis, and (3) human CSSCs for corneal scar prevention in vivo. The expression of miR-29a was screened in additional CSSC batches and the anti-scarring effect of cells was validated in mouse corneal wounds.
Results
Twenty-one miRNAs were significantly expressed in “healing” CSSC-EVs and 9 miRNA groups were predicted to associate with inflammatory and fibrotic responses, and tissue regeneration (P <10−6). Overexpression of miR-29a and 381-5p significantly prevented M1 phenotype transition in RAW264.7 cells after lipopolysaccharide treatment, suppressed transforming growth factor β1-induced fibrosis marker expression in keratocytes, and reduced scarring after corneal injury. High miR-29a expression in EV fractions distinguished human CSSCs with strong healing potency, which inhibited corneal scarring in vivo.
Conclusion
We characterized the anti-inflammatory and fibrotic roles of miR-29a and 381-5p in CSSCs, contributing to scar prevention. MiR-29a expression in EVs distinguished CSSCs with anti-scarring quality, identifying good quality cells for a scarless corneal healing.
Introduction
Corneal blindness is a leading cause of vision loss worldwide (World Report on Vision, WHO 2019; https://www.who.int/publications/i/item/world-report-on-vision). >200 million individuals have impaired vision, and about 4.5 million of them have a loss of corneal clarity [1]. In developing countries, it is estimated that 35 to 50% of blindness is due to corneal scarring [2]. The conventional treatment to replace scarred corneal tissues includes partial or full-thickness corneal transplantation using healthy donor corneas. As a matter of fact, corneal transplantation is one of the most frequently performed solid organ transplant surgeries globally. Despite advances in the field of keratoplasty, only 1 in 70 individuals with treatable corneal scarring can undergo surgery, due to a multitude of social, and economic issues, as well as, more importantly, a limited supply of transplantable donor corneas.
Different strategies have been attempted to treat corneal scarring and improve corneal clarity for light passage, including the prosthetic corneas and the use of biomaterials to replace the scarred tissues [3], [4], [5]. Since the discovery of corneal stromal stem cells (CSSCs) in 2005, stem cell therapy is an attractive approach to prevent or remediate corneal scarring [6], [7], [8]. The application of cells in a fibrin gel to corneal stromal wounds suppressed the injury-associated inflammation [9], reduced fibrosis (downregulated collagen III [Col3], α-smooth muscle actin [αSMA], and tenascin C [TNC]) and regenerated native-like stromal tissue architecture in animal models [10], [11], [12]. An interventional clinical trial (NCT02948023) using CSSC therapy in patients with corneal haze due to infection or after laser surgery or collagen crosslinking is underway in India, and preliminary results have demonstrated safety, scar regression and visual improvement [13]. Recently, we reported that the treatment with CSSCs having a reduced expression of Alix (an endosomal sorting complex required for transport [ESCRT]-associated gene) after small-interference RNA-mediated knockdown to block microRNA (miRNA) transport in the extracellular vesicles (EVs or exosomes), resulted in a loss of scar preventive and corneal regenerative functions [14]. These nanosized (typically 30 ∼ 150 nm in diameter) membrane-bound EVs contain diverse constituents as cargo molecules, including cytosolic and membrane proteins, amino acids, lipids, nucleic acids (namely mRNA, noncoding small RNA and DNA) and metabolites, which reflect their cell of origin [15]. The roles of EVs have been widely documented to mediate cell–cell communication in an autocrine and/or paracrine fashion, regulating a vast variety of cell and tissue processes, including development, immune responses, inflammation and infection, neurodegeneration, metabolic diseases and cancers [16], [17], [18]. Recent studies have further indicated a functional, targeted and mechanism-driven accumulation of specific components in EVs, suggesting that they could play roles to selectively induce specific signalling in recipient cells [19], [20], [21]. Likewise, specific soluble factors were identified in EVs from mesenchymal stem cells (MSCs) as active in tissue repair and regenerative process [22], [23]. Our previous finding has highlighted that certain miRNAs present in CSSC-derived EVs could play an essential role in the process by which CSSCs block scarring and initiate the regeneration of transparent corneal tissue [14]. In this study, we characterized the miRNA profile from EVs of human CSSC batches from different donor corneas using Nanostring nCounter microRNA profiler, followed by target gene search and enriched pathway analysis. Specific miRNA groups predicted to have significant association with tissue inflammation, and fibrosis were verified using in vitro assays of macrophage M1 phenotype transition and keratocyte fibrogenesis, as well as in vivo mouse corneal injury model.
Establishing standards for CSSC quality control and cell selection is necessary to assure the safety, quality and potency of cell products to be administered to patients. Though different studies used MSC markers (e.g. CD73, CD90 and CD105) to examine the cell homogeneity, owing to the mesenchymal nature of CSSCs [14], [24], [25], specific and reliable markers for cell identification are still missing. In addition, not every CSSC batches generated from different donor corneas achieve similar regenerative effect. Such variations in the healing efficiency is likely affected by their EV-containing miRNA content. Identification of specific miRNAs and their effects associated with scar prevention and corneal tissue healing (with respect to inflammation and fibrosis) would give us better understanding about the mechanisms of CSSC therapeutic action, facilitating the development of a cell-based treatment strategy for corneal wound management. This advance could transform the current treatment options and alleviate the heavy reliance on limited donor materials worldwide [26], [27]. The use of optimal CSSC batches would result in improved safety and efficacy, and the reduction of post-treatment complications [3]. Hence, in this study, we assessed a potential use of candidate miRNAs, by screening their expression in various CSSC cultures, to predict their anti-scarring potency, and the cells were functionally validated using in vivo corneal injury model.
Materials and methods
Ethics statements
The use of donor corneas followed the tenets of the Declaration of Helsinki and was approved by The University of Pittsburgh Institutional Review Board (IRB) and Committee for Oversight of Research and Clinical Training Involving Decedents (CORID), Protocol #161. Animal study was carried out in strict accordance with the guidelines for the Care and Use of Laboratory Animals of NIH and The Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. The protocol was approved by the Institutional Animal Care and Use Committee of The University of Pittsburgh (Protocol 18022511).
Donor corneas, CSSC isolation and culture
Human corneas, from de-identified donors younger than 70 years old, were obtained from the Centre for Organ Recovery and Education, Pittsburgh, PA (www.core.org) (donor information in Table S1). Corneal tissues were preserved in Optisol GS (Bausch & Lomb Inc., Rochester, NY) and used within 12 days post-enucleation. After clearing of corneal epithelium, iris and endothelium by scraping and rinses, the anterior limbal stroma (0.5–1 mm wide, 0.1 mm deep) was isolated for digestion using collagenase A (1 mg/ml, Roche, Indianapolis, IN) for 10 h at 37 °C, following our previous studies [10], [11]. Single cell suspension were cultured with stem cell growth medium (JM-H) containing 2% (vol/vol) pooled human serum (Innovative Res, Novi, MI), on culture surface pre-treated with FNC coating mix (Athena Enzyme Systems, Baltimore, MD). Cell colonies were expanded to passage 3 for experiments. To determine the cell homogeneity, flow cytometry with cell surface markers (CD73, 90 and 105) conventionally used to identify bone marrow mesenchymal stem cells (MSCs) was performed, as previously described [14].
In vitro anti-inflammatory activity of CSSC
1. Acute toll-like receptor (TLR)-mediated inflammatory assay - mouse RAW-BlueTM cells (InvivoGen, San Diego, CA), with chromosomal integration of SEAP reporter inducible by NF-kB and alkaline phosphatase (AP-1), were used in this study. They express pattern-recognition receptors, including most of TLRs, which play important roles in acute phase of corneal inflammation [28], [29]. RAW-blue cells were plated at 104 cells/cm2 in DMEM with 10% fetal bovine serum (FBS, Gibco) and 100 μg/ml Normocin (InvivoGen). The cultures were added with CMconc (500 μg protein; protocol referred to the following section) from different CSSC batches, in the presence of lipopolysaccharides (LPS, 20 ng/ml, Sigma) for 16 h. The culture supernatant was collected for SEAP reporter assay (Sigma-Aldrich), according to manufacturer’s instruction. In brief, the supernatant samples were heated to 65 °C for 30 min to inactivate endogenous AP, followed by incubation with QUANTI-BlueTM solution containing AP substrate for 4 h at 37 °C. Optical densities at excitation 620 nm with a reference wavelength of 420 nm were measured. Samples were run in triplicate. Mean and SD were calculated for samples treated or not with LPS and significance was determined by Mann-Whitney U test.
2. Chronic pro-inflammatory osteoclastogenesis assay – inflammatory bone resorption capacity by osteoclast formation was observed at 3 to 7 days after induction by RANK ligand (RANK-L) peptide in RAW264.7 cells [30]. In this assay, mouse macrophage RAW264.7 cells (American Type Cell Collection, Manassas, VA) at passages below 25th were plated at 104 cells/cm2 in DMEM/F12 with 5% FBS in a 6-well plate. The cells were treated with RANK-L peptide (50 ng/ml, Sigma-Aldrich) and Concanavalin A (20 μg/ml, ConA, Sigma-Aldrich), in the presence of native or heat-denatured CMconc (500 μμg protein). After 5 days, cells were harvested in RLT lysis buffer (Qiagen) for total RNA extraction, followed by qPCR for osteoclast markers: tartrate-resistant alkaline phosphatase (ACP5), cathepsin K (CTSK) and matrix metalloproteinase 9 (MMP9) genes (primer information in Table S2) as an indicator of NFkB activation. Delta Ct was determined by comparison with housekeeping 18S.
Mouse corneal injury model and cell treatment
The c57bl/6 mice at 6–8 weeks old were anesthetized by intraperitoneal ketamine (50 mg/kg) and xylazine (5 mg/kg) injection [11]. Right eyes received topical proparacaine hydrochloride (0.5%, Alcaine, Alcon, Fort Worth, TX) for local analgesia. After saline rinses, the corneal epithelium (2 mm diameter) was removed using high speed rotation of Algerbrush II (Accutome Inc, Malvern, PA, US) and scraped with a surgical blade #15. The basement membrane and anterior stromal layers were damaged by a second Algerbrush burring. After normal saline rinses and briefly drying with a sterile cotton spear, the wounded stromal surface was overlaid with one μl of CSSC (5x104 cells) in fibrinogen (37 μg/ml, Sigma-Aldrich), followed by 0.5 μl thrombin (100 U/ml, Sigma-Aldrich). The fibrin gel formed within a minute and the treated eye received topical gentamicin (0.3%, Genoptic, USP, Rockville, MD).
Assessment of corneal scarring
Before and immediately after injury, and at time interval, cross-sectional corneal structure were scanned (4 mm diameter) with Spectral Domain Ophthalmic Coherence Tomography (SD-OCT, Envisu R2210, Leica [Bioptigen], Morrisville, NC). Images were processed with NIS-Elements software (Nikon, Melville, NY) in a masked fashion. Central corneal thickness (CCT) was measured as the mean of 3 measurements obtained at the centre (0 mm) and at 0.5 mm on either side [31]. At day 10 post-treatment, mice were sacrificed with overdose of sodium pentobarbital, followed by cervical dislocation. All corneas were isolated under dissecting microscopy, imaged and the scar area was determined in a masked fashion using the Fiji open-source image analysis software package (https://fiji.sc/) and MetaMorph 7.7.3 (Molecular Devices Inc., San Jose, CA) [10]. Threshold images were generated after digitally removing the corneal epithelium. Naïve corneas were used to set the threshold value for scar volume measurement. Statistical analyses were performed with Prism 7 (GraphPad Prism) using paired Student’s t-test.
Mouse corneas, total RNA isolation and qPCR analysis
Corneas were isolated and placed in RLT reagent, and disrupted with MagNA Lyser green beads in a MagNA Lyser Instrument (Roche). The lysates were processed using QiaShredder (Qiagen), and total RNA was extracted using RNeasy Mini kit (Qiagen) with on-column RNase-free DNase digestion (Qiagen), per manufacturer’s instruction. RNA was quantified using NanoDrop One (Thermo Fisher) and 500 ng RNA was reverse-transcribed to cDNA using SuperScript III RT-PCR kit (Thermo Fisher) and random hexanucleotide primers. Target gene expression was performed with specific primers (Table S2) using SYBR Green Real-Time Master Mix (Life Technologies, Carlsbad, CA) in a QuantStudio 3 Real-Time PCR System (Applied Biosystems). Experiments were done in triplicate and relative RNA abundance was assayed by the 2-ΔΔCt method after normalization with housekeeping 18S gene and fold changes were expressed as mean ± SD. Significance was determined by non-parametric Mann-Whitney U test. Primer validation by agarose gel electrophoresis and melting curves is shown in Supplementary Fig. S9.
CSSC conditioned media, EV isolation and characterization
CSSCs at passage 3 (30 ∼ 40 population doublings) were grown to about 50% confluence, washed and replenished with JM-H containing human serum that was pre-cleared of particulate material (including EVs) by centrifugation at 100,000 g for 18 h. Conditioned media (CM) was collected after 4 days. It was firstly spun at 400 g for 5 min to remove cellular debris. The supernatant was then concentrated by MicroCon centrifugal filter (YM-10 membrane, Millipore) at 12,000 g for 40 min at 4 °C to achieve about 20x concentration. The total protein content of CM concentrate (CMconc) was quantified using Pierce BCA Protein Assay kit (Thermo Fisher Sci.). EVs were precipitated from CMconc by polymer exclusion using Total Exosome Purification Reagent (Invitrogen), and harvested by centrifugation at 10,000 g [14]. The pellet was resuspended in sterile PBS containing 50 mM trehalose (Sigma) (PBST) with gentle shaking at 4 °C overnight. Samples were centrifuged at 10,000 g, at 4 °C for 1 h, and the supernatant was further centrifuged at 100,000 g for 3 h. The pellet containing EVs was resuspended in PBST overnight at 4 °C. All relevant data of our experiments was submitted to the EV-TRACK knowledgebase (EV-TRACK ID: EV220192) [32].
Characterization assays: (1) EV size profiling by a tunable resistive pulse sensing (TRPS) assay using a qNano Gold System (IZON Sci Ltd) [33]. Two different nanopores, NP100 (50–330 nm) and NP400 (185–1100 nm) (IZON Sci Ltd) were used to target EVs < 0.5 μm in size. The assay procedure followed manufacturer’s instruction. Sample suspensions were diluted with Solution Q (qEV Electrolyte; IZON Sci Ltd) and concentration was adjusted as required to target a particle rate of 1,000–2,000/min. Samples were filtered with a 0.22 μm syringe filter before being analysed with NP200 (85–500 nm) or NP400. Samples and calibration particles measurements were run under the same conditions, and a minimum of 1,000 particles were recorded. Data obtained were analysed using Izon Control Suite software (Izon Control Suite v.3.2.2.268; Izon Sci Ltd). The data sets were filtered to analyse specific populations.
(2) EV marker expression by flow cytometry. The expression of EV-specific membrane antigens CD9, 29, and 81, and the low or absent expression of hematopoietic CD34, 45, and lymphocytic marker CD107a were analysed with a FACSAria flow cytometer (BD Bioscience, San Jose, CA) at the Flow Cytometry core facility of The Eye and Ear Institute of Pittsburgh [34] (Table S2). The LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Life Technologies, Carlsbad, CA) was used to exclude dead cells. Data were analysed by using FlowJo, LLC (v10.0.7; Ashland, OR).
RNA isolation from EVs and microRNA profiling by Nanostring platform
Purified EVs were lysed with RLT buffer and total RNA, including small RNAs, were extracted using miRNeasy kit. MicroRNA profiling of EV-RNA fractions (300 ng RNA) were conducted on 8 CSSC batches, all cultured under same protocol. The miRNA profiling was performed using a Nanostring nCounter microRNA profiler (Human v3 miRNA assay, Nanostring Tech Inc., Seattle, WA), which screens for a collection of 800 human miRNA species referenced from miRBase. The miRNA data was normalized using a global method, which normalizes to total counts of the expressed miRNA targets across all samples using nSolver software (NanoString Tech Inc.). Mean fold changes and statistical differences between the abundance of miRNA from 3 EV samples derived from CSSC batches with strong in vitro anti-inflammatory effects and in vivo anti-scarring effects on corneas, and miRNA from another 3 EV samples from CSSC batches without corrective effects, were determined using one-sample t-test. Differentially expressed miRNAs were identified with P values < 0.05 and fold changes > 20.
MicroRNA database search and enriched pathway analyses
Target gene prediction for each miRNA was performed using online TargetScan v7.2 (https://www.targetscan.org/vert_80/) and miRDB/miRTarget (https://www.mirdb.org/). The target genes from each platform were ranked according to the confidence level and the top 50 genes occurring commonly in both platforms were selected for enriched pathway analyses. Since one or several miRNAs could mediate similar gene regulatory effects, the gene lists from different candidate miRNAs were grouped and imported to DAVID (Database for Annotation, Visualization and Integration Discovery) v6.8 (https://david.ncifcrf.gov/) for gene ontology (GO) term analysis. The non-coding genes were selected out, and genes present in genome were identified and examined for over-representation in particular signalling pathways, biological processes and phenotypes. P < 10-6 (Bonferroni adjustment) was considered to be statistically significant.
MicroRNA mimic transfection and gene expression
The regulatory functions of target miRNA groups on inflammation were assessed by transfecting their respective mimics (Table S2) into mouse RAW264.7 cells using Lipofectamine RNAiMax (3 μl per 10 pmol RNA; Thermo Fisher), before or after treatment of LPS (50 ng/ml) or IL4 (20 ng/ml), followed by RNA analysis for M1 pro-inflammatory genes, including inducible nitric oxide synthase (iNOS), monocyte chemoattractant protein (MCP1) and C-X-C motif chemokine ligand 10 (CXCL10). Alternatively, miRNA regulation on corneal cell fibrosis was examined by transfecting mimics to primary human corneal stromal keratocytes (CSKs) [35], followed by treatment with recombinant human TGFβ1 (0.25 ng/ml, Sigma-Aldrich) and L-ascorbate 2-phosphate (0.5 mM) for 7 days [36]. RNA analysis of fibrosis markers, including Col3A1, FN, SPARC, and αSMA were performed by qPCR.
Statistics
All experiments were done in triplicate and the animal number was 6 or more in each group, unless stated otherwise. Data were presented as mean ± SD. Mean value was compared using unpaired two-tailed Student’s t-test or ANOVA with a post hoc Bonferroni test using GraphPad Prism 7. Non-parametric comparison was done by Mann-Whitney U test. P < 0.05 was considered statistically significant.
Results
Characterization of EVs derived from human CSSCs exhibiting anti-scarring effect on corneal injury and batch variation from different donors
The homogeneity of human CSSC cultures was determined by flow cytometry with cell surface MSC markers (CD73, 90 and 105) (Fig. S1). Cultures with > 90% CD-positive viable cells and negligible CD31 (vascular endothelial marker) were used in the experiments. EVs were prepared from CM of qualified human CSSC cultures following our reported protocols [14]. The collected EVs were spherical in morphology under transmission electron microscopy (Fig. S2A). Size measurement by a TRPS assay using qNano Gold System showed the peak size of EVs was around 60–135 nm (Fig. S2B shows the representative peak sizes at 67 and 88 nm). According to MISEV2018, human CSSC-derived EVs fell within the size range of small and/or medium EVs [37]. By flow cytometry, the samples expressed positive EV markers (CD9, 29, 81) but not negative markers (CD34, 45, HLA-G and HLA-DR) (Fig. S2C and Fig. S3).
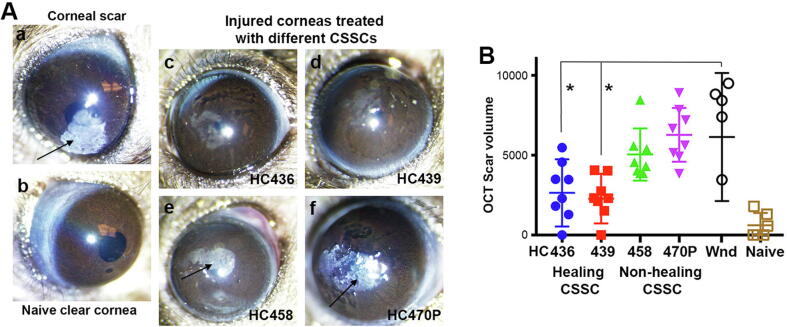
Injured mouse corneas (n = 36) were treated with human CSSCs (5x104 cells) loaded in fibrin gel, or untreated as wound control. After 10 days, the wounded corneas (n = 6) without treatment developed significant scarring (Fig. 1A, a). Cell treatment with 4 different CSSC batches modulated scar severity (Fig. 1A, c to f), as described previously [14]. From serial OCT images, the scar volume changed after cell treatments, as compared to wound controls (Fig. 1B). Treatment with HC436 and 439 significantly reduced the scar severity and volume (n = 6 per treatment group) (Fig. 1B) (referred as “healing” CSSCs), whereas HC458 and 470P had reduced efficacy versus scar progression (referred as “non-healing” CSSCs).
Fig. 1.
Differential anti-scarring effect of treatment with CSSCs from different donors. (A) Macroscopic images of mouse eyes under diffuse light illumination revealed opaque scars (arrows) in wound control (a) and corneas treated by HC458 and 470P (non-healing CSSC) (e and f). In contrast, relatively clear corneas were resulted after treatment with HC436 and 439 (healing CSSC) (c and d), as compared to naïve cornea (b). (B) Scar volume measurement from serial transverse OCT images showed a significant scar reduction in corneas treated with healing CSSC, when compared to wound controls (*P < 0.05, paired t-test). Injured mouse corneas treated with non-healing CSSC exhibited scar development similar to wound control.
CSSC-conditioned media (CM) exhibited differential anti-inflammatory effect
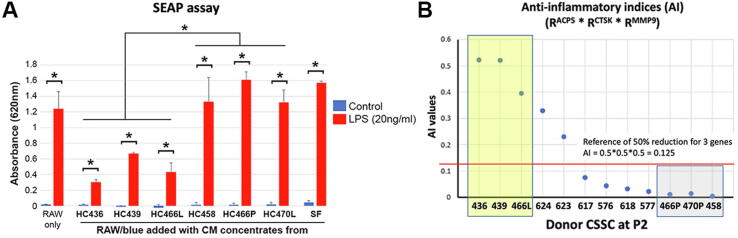
Next, we determined whether the different anti-scarring effect from CSSCs was related to their capability to suppress acute and chronic inflammatory reactions. In acute reaction, SEAP reporter assay with mouse RAW-BlueTM cultures was used to measure toll-like receptor (TLR)-mediated inflammatory responses. In RAW-BlueTM cultures, the addition of CMconc from HC436, 439 and 466L (“healing” CSSC) significantly suppressed the upregulation of SEAP reporter expression induced by LPS, when compared to CMconc from HC466P, 470P and 458 (“non-healing” CSSC) (P < 0.05; Mann-Whitney U test) (Fig. 2A). Controls without CMconc treatment or with stromal fibroblast (SF)-derived CMconc had elevated SEAP reporter expression. Similar anti-inflammatory effect was found in chronic inflammatory osteoclastogenesis assay. Mouse RAW264.7 cells were treated with RANK-L and ConA for 5 days to induce osteoclast phenotype with upregulated expression of ACP5, CTSK and MMP9 genes (Fig. S4). This induction was reduced after cells were incubated with an increasing amount of CSSC (HC436)-derived CM concentrate (CMconc, 20 to 500 μg protein). This finding indicated a dose-dependent suppressive effect of CSSC secretome (containing EVs) on induced inflammation. The test was performed using 12 different CSSC batches (donor information in Table S1) to check their anti-inflammatory capability. After RANK-L/ConA-treated cells were incubated with native or heat-denatured CMconc (500 μg protein), the ratios of reduction (R) for ACP5, CTSK and MMP9 expression were calculated (Table S3). The anti-inflammatory index (AI), represented as RACP5 × RCTSK × RMMP9, was calculated for each CSSC batch. As shown in Fig. 2B, higher AI values were found for HC436, 439 and 466L, of which the cell batches suppressed corneal scarring in vivo (Fig. 1 and Fig. S5). In contrast, low AI values for HC466P, 470P and 458 were consistent with their null effect on corneal scar correction. These results clearly distinguished HC436, 439 and 466L with relatively strong healing potential from the non-healing HC466P, 470P and 458. Figure S5 shows the in vivo corrective effect of cell treatment on mouse corneal scarring. Clear corneas were recovered with HC436, 439 and 466L, whereas corneas treated with HC466P, 470P and 458 developed intense scarring. Similar strong scarring was found in untreated wound corneas and with fibrin only. This finding demonstrated that healing CSSCs had higher anti-inflammatory activity than the non-healing cells.
Fig. 2.
Anti-inflammatory efficiency of CSSC-derived CM. (A) SEAP reporter assay confirmed the inflammation suppression efficiency of CMconc from cells selected from A. The incubation of CMconc from HC436, 439, and 466L (high AI) significantly suppressed the upregulation of SEAP reporter expression induced by LPS (20 ng/ml), when compared to CMconc from HC466P, 470P, and 458 (low AI) (*P < 0.05; Mann-Whitney U test). Controls without CM treatment or with stromal fibroblast (SF)-derived CMconc had strong SEAP reporter expression. (B) Suppression efficiency of osteoclast marker expression (ACP5, CTSK, and MMP9) after adding CM concentrates (CMconc) from different CSSC strains to RAW cells induced by RANK-L/ConA treatment. Anti-inflammatory index (AI) was calculated as a multiplication of rates of reduction (R) of ACP5, CTSK, and MMP9. The red line indicates the reference AI representing 50% reduction in each gene. The yellow box contains 3 CSSC strains (HC436, 439, 466L) with high AI, and the grey box has cells of low AI (HC466P, 470P, 458). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
MicroRNA profiling of “healing” versus “non-healing” CSSC-derived EVs and enriched pathway analysis.
To uncover specific miRNAs in association with anti-inflammatory and anti-scarring effects of CSSC-EVs, we performed Nanostring analysis using small RNA isolated from “healing” EVs of HC436, 439, and 466L, versus “non-healing” EVs of HC458, 466P, and 470P. RNA libraries were constructed without polyA selection so that the sequence analysis included both mRNA and small RNA populations. After data filtering, and map reading by nSolver analysis software, a total of 59 candidate miRNAs were differentially expressed in CSSC-EVs showing “healing” versus “non-healing” effects (volcano plot in Fig. S6) (orange dots > 10-fold; black dots > 20-fold; all P < 0.05). Table S4 lists miRNAs with expression > 10-fold (P < 0.05) in “healing” EVs. Complete Nanostring data is listed in Supplementary Table S7.
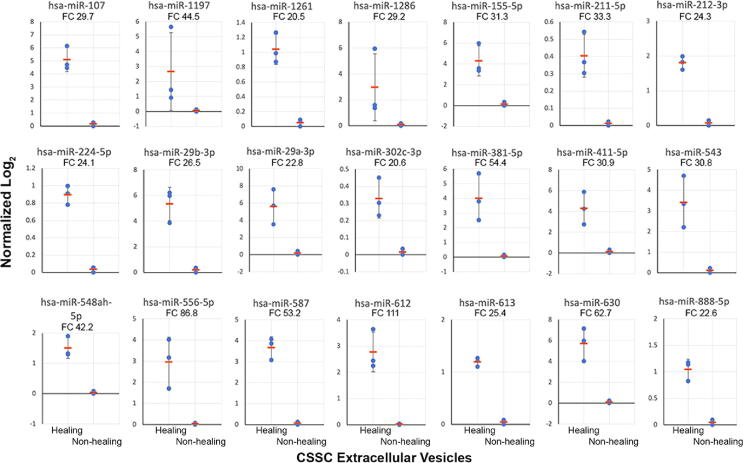
A total of 21 miRNAs with > 20-fold expression in the “healing” EVs were identified (Fig. 3). Among them, 5 were expressed > 50-fold higher in the “healing” than “non-healing” EVs (miR-612 [111-fold], 556-5p [87-fold], 630 [63-fold], 381-5p [54-fold], and 587 [53-fold]). Also, 11 miRNAs (miR-29a, 29b, 107, 155, 211-5p, 212, 224, 411, 302c, 381-5p, and 543) were reported in association with tissue fibrosis and inflammation by regulating different signalling pathways in fibroblast growth and collagen synthesis (Table S6). Five miRNAs (miR-548ah, 888, 1197, 1261, and 1286), were not narrated previously with specific functions. We hypothesized that one or more of these miRNAs participate(s) in the anti-scarring effect and the regenerative capability delivered by the “healing” CSSC-EVs.
Fig. 3.
Nanostring miRNA analysis of CSSC-EVs. Twenty-one miRNAs expressed with > 20-fold higher (P < 0.05) in “healing” compared to “non-healing” EVs. Data were normalized to the gTotal intensity signals and log2-transformed. Fold change (FC) of each miRNA is indicated.
In order to assess if any of these miRNAs regulate tissue healing, we performed target gene search for each miRNA, randomly grouped one to multiple gene lists for pathway analysis, and identified miRNA groups with significant association to tissue inflammation and fibrosis-related pathways. Using TargetScan and miRDB/miRTarget for target gene search, the gene lists were generated. The top 50 common target genes for each miRNA were sorted and the gene list of different miRNAs were grouped for functional annotation analysis using DAVID bioinformatics. A total of 9 miRNA groups showed enriched GO terms significantly linked to the inflammatory and immune pathways (including T-cell antigen receptor signalling, toll-like receptor cascade, focal adhesion/Akt/mTOR signalling), fibrosis (TET signalling, integrin and collagen modification), senescence and autophagy, as well as tissue regeneration (mesodermal commitment, pluripotency) (Table 1). All groups contained miR-29, together with miRNAs that were previously reported in regulating tissue fibrosis and inflammation.
Table 1.
Nine groups of miRNAs and their predicted significant pathways. P < 10-6 indicates statistical significance. Group 10 is the control to negative actions.
| miRNA Groups | Predicted Enriched Pathways | P values | |
|---|---|---|---|
| 1 | 29 | Collagen chain trimerization | 5.50E-18 |
| Collagen biosynthesis and modifying enzymes | 6.20E-16 | ||
| Integrins in angiogenesis | 2.75E-14 | ||
| β1 integrin cell surface interactions | 4.28E-14 | ||
| ECM organization | 1.12E-13 | ||
| miRNA targets in ECM and membrane receptors | 7.61E-08 | ||
| Binding and uptake of ligands by scavenger receptors | 3.21E-06 | ||
| 2 | 29 + 107 + 155 | Collagen chain trimerization | 6.42E-13 |
| Integrins in angiogenesis | 9.52E-13 | ||
| Collagen biosynthesis and modifying enzymes | 6.50E-11 | ||
| Syndecan-1-mediated signalling events | 8.10E-10 | ||
| β1 integrin cell surface interactions | 1.23E-09 | ||
| ECM organization | 2.52E-07 | ||
| Focal adhesion-PI3K-Akt-mTOR-signalling pathway | 3.42E-07 | ||
| 3 | 29 + 107 + 155 + 381 | Integrins in angiogenesis | 5.55E-13 |
| Collagen chain trimerization | 7.81E-12 | ||
| Collagen biosynthesis and modifying enzymes | 7.55E-10 | ||
| Syndecan-1-mediated signalling events | 5.79E-09 | ||
| β1 integrin cell surface interactions | 1.10E-08 | ||
| Focal adhesion-PI3K-Akt-mTOR-signalling pathway | 1.03E-06 | ||
| ECM organization | 4.30E-06 | ||
| Signalling regulating pluripotency of stem cells | 6.82E-06 | ||
| 4 | 29 + 107 + 155 + 543 | Integrins in angiogenesis | 1.55E-12 |
| Collagen chain trimerization | 1.83E-11 | ||
| Syndecan-1-mediated signalling events | 4.32E-10 | ||
| Collagen biosynthesis and modifying enzymes | 1.74E-09 | ||
| β1 integrin cell surface interactions | 2.32E-08 | ||
| Mesodermal commitment pathway | 5.13E-07 | ||
| Focal Adhesion-PI3K-Akt-mTOR-signalling pathway | 5.19E-07 | ||
| ECM organization | 2.09E-06 | ||
| 5 | 29 + 381 | Collagen chain trimerization | 4.20E-15 |
| Integrins in angiogenesis | 2.02E-13 | ||
| Collagen biosynthesis and modifying enzymes | 4.53E-13 | ||
| β1 integrin cell surface interactions | 1.48E-11 | ||
| Syndecan-1-mediated signalling events | 1.57E-11 | ||
| ECM organization | 5.88E-10 | ||
| Focal Adhesion-PI3K-Akt-mTOR-signalling pathway | 8.17E-10 | ||
| miRNA targets in ECM and membrane receptors | 1.64E-06 | ||
| 6 | 29 + 543 | Collagen chain trimerization | 2.51E-14 |
| Integrins in angiogenesis | 1.19E-12 | ||
| Syndecan-1-mediated signalling events | 1.20E-12 | ||
| Collagen biosynthesis and modifying enzymes | 2.65E-12 | ||
| β1 integrin cell surface interactions | 7.12E-11 | ||
| ECM organization | 4.81E-10 | ||
| Focal adhesion-PI3K-Akt-mTOR-signalling pathway | 6.84E-10 | ||
| TET1,2,3 and TDG demethylate DNA | 6.14E-07 | ||
| Senescence and autophagy in cancer | 1.65E-06 | ||
| Protein alkylation leading to liver fibrosis | 7.22E-06 | ||
| 7 | 29 + 155 | Integrins in angiogenesis | 8.04E-15 |
| Collagen chain trimerization | 8.36E-15 | ||
| Collagen biosynthesis and modifying enzymes | 8.95E-13 | ||
| Syndecan-1-mediated signalling events | 2.69E-11 | ||
| β1 integrin cell surface interactions | 2.71E-11 | ||
| ECM organization | 1.38E-09 | ||
| Focal adhesion-PI3K-Akt-mTOR-signalling pathway | 1.91E-09 | ||
| miRNA targets in ECM and membrane receptors | 2.26E-06 | ||
| 8 | 29 + 155 + 381 + 543 | Integrins in angiogenesis | 3.18E-14 |
| Collagen chain trimerization | 1.16E-11 | ||
| Syndecan-1-mediated signalling events | 2.87E-10 | ||
| Collagen biosynthesis and modifying enzymes | 1.12E-09 | ||
| β1 integrin cell surface interactions | 1.56E-08 | ||
| Focal adhesion-PI3K-Akt-mTOR-signalling pathway | 4.57E-08 | ||
| PDGFR-β signalling pathway | 4.91E-07 | ||
| ECM organization | 1.21E-06 | ||
| Protein alkylation leading to liver fibrosis | 9.21E-06 | ||
| 9 | 29 + 381 + 543 | Integrins in angiogenesis | 6.22E-13 |
| Collagen chain trimerization | 1.13E-10 | ||
| Syndecan-1-mediated signalling events | 2.19E-09 | ||
| Collagen biosynthesis and modifying enzymes | 1.02E-08 | ||
| β1 integrin cell surface interactions | 1.13E-07 | ||
| Mesodermal commitment pathway | 4.32E-07 | ||
| Focal adhesion-PI3K-Akt-mTOR-signalling pathway | 1.08E-06 | ||
| PDGFR-β signalling pathway | 3.94E-06 | ||
| Signalling regulating pluripotency of stem cells | 8.83E-06 | ||
| 10 | 107 + 155 + 381 + 543 | Mesodermal commitment pathway | 3.50E-07 |
| (serve as neutral reference) | PDGFR-β signalling pathway | 4.79E-06 | |
Functional validation of miR-29a and 381-5p for anti-inflammatory activity in vitro
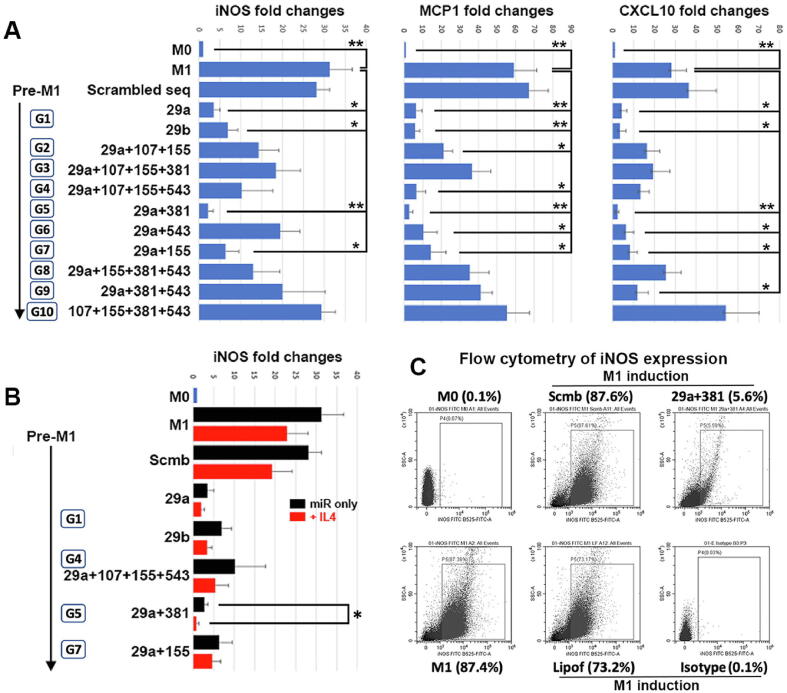
To validate which miRNA groups regulated inflammatory activities as predicted earlier, RAW264.7 cells were transfected with the respective miRNA mimics before LPS treatment to induce M1 phenotype. qPCR results showed that LPS treatment significantly induced the expression of M1 markers (iNOS, MCP1 and CXCL10), compared to M0 cells (Fig. 4). In triplicate study using cells transfected with miRNA mimics, M1 marker expression was generally reduced. All 3 markers were significantly downregulated in cells transfected with miR-29a, 29b, 29a + 381 (*P < 0.05 and **P < 0.01; Mann-Whitney U test), and with miR-29a + 155 (P < 0.05) (Fig. 4A). Compared to M1 cells without transfection, the expression of iNOS was reduced by a mean of 89% after miR-29a transfection, 78.2% with miR-29b, 93.2% with miR-29a + 381 and 79.8% with miR-29a + 155; MCP1 expression was suppressed by 89.3% with miR-29a transfection, 90% with miR-29b, 95.5% with miR-29a + 381, and 75.9% with miR-29a + 155; and CXCL10 was down-regulated by 84.3% with miR-29a transfection, 88.1% with miR-29b, 92% with miR-29a + 381, and 70.4% with miR-29a + 155. M1 cells transfected with scrambled sequences showed high inflammatory marker expression, similar to control M1.
Fig. 4.
In vitro anti-inflammatory assay of mouse RAW264.7 cells after miRNA mimic transfection and LPS treatment. (A) RAW cells were treated with LPS to M1 phenotype followed by transfection with 9 different groups of miRNA mimics (G1 to 9) or one control group (G10). Compared to non-transfected M1 cells, ectopic expression of different miRNA groups differentially reduced pro-inflammatory marker (iNOS, MCP1 and CXCL10) expression (*P < 0.05, **P < 0.01; Mann-Whitney U test). Transfection of miR-29a and 381 (G5) consistently inhibited all 3 marker expressions. (B) miR-29a and 381 expression augmented iNOS suppression after cells were treated with IL4 to revert the M1 phenotype (*P < 0.05). Transfection of other miRNA mimics also downregulated iNOS expression but was insignificant. (C) Flow cytometry assay showing miR29a and 381 transfection reduced iNOS positive M1 cells, when compared to M1 cells without transfection, transfected with scrambled sequences, or with lipofectamine only.
We next investigated if the transfection of these miRNA groups augmented the anti-inflammatory action of IL-4, which reverted M1 phenotype shown by reduced iNOS expression. RAW cells transfected with miRNA mimics were treated with LPS for 6 h, followed by cell harvest or IL-4 treatment for another 12 h. Ectopic expression of miRNA mimics which showed downregulated M1 phenotype in previous section caused further iNOS suppression after IL-4 treatment (Fig. 4B). Among them, cells with miR-29a + 381 transfection significantly down-regulated iNOS expression by a mean of 72.2% (P < 0.05) (n = 3 tests). On the other hand, RAW M0 cells were transfected with miRNA mimics for 6 h, followed by IL-4 (20 ng/ml) treatment for another 12 h. Cells transfected with miR mimics showed insignificant upregulation of Arg1, M2 phenotypic marker (Fig. S7). The expression of Arg1 was induced by IL-4 treatment towards similar levels as cells without miRNA mimic transfection. This indicated that miR-29a + 381 expression in RAW cells suppressed pro-inflammatory M1, but not necessarily promoting M2 phenotype.
By flow cytometry, the population of iNOS-expressing RAW cells was substantially reduced by miR-29a + 381 transfection (5.6% positive cells) after LPS treatment, compared to controls (non-transfected M1 cells was 87.4%, scrambled sequence-transfected M1 cells 87.6% and lipofectamine-treated M1 cells 73.2%) (Fig. 4C). Our finding showed that the expression of miR29a and 381-5p reduced the inflammatory response of mouse macrophages.
Validation of miR-29a and 381-5p for anti-fibrotic activity in vitro
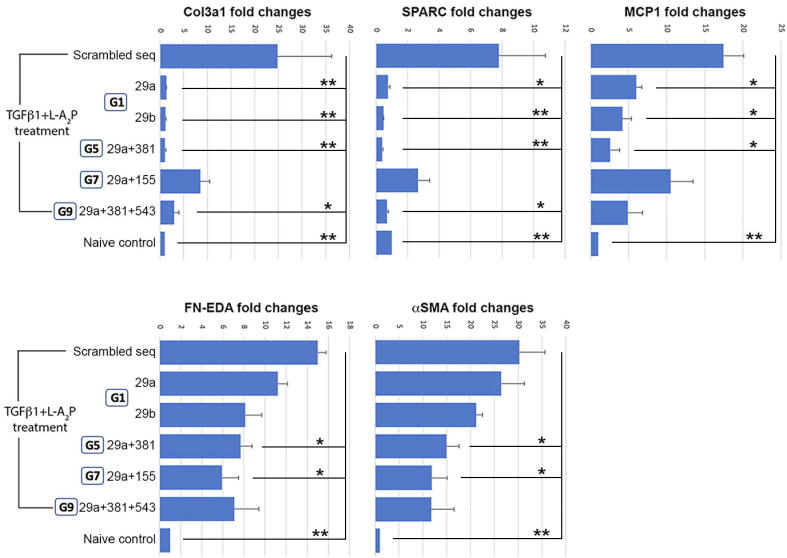
Primary human CSKs were treated with recombinant human TGFβ1 for 7 days in low serum culture to induce fibrogenesis [36]. CSKs transfected with miRNA mimics showed reduced fibrosis gene expression, when compared to cells with scrambled sequences (Fig. 5). In triplicate study, the expression of Col3A1 was significantly reduced by a mean of 92.2% after miR-29a transfection, 93.2% with miR-29b, and 93.4% with miR-29a + 381 (P < 0.01), and 88.2% with miR-29a + 381 + 543 (P < 0.05). SPARC expression also was suppressed by 92.8% with miR-29b transfection, and 93.1% with miR-29a + 381 (P < 0.01), 90.3% with miR-29a and 89.5% with miR-29a + 381 + 543 (P < 0.05). For MCP1, the expression dropped by 64.7% with miR-29a transfection, 76.5% with miR-29b, and 85.3% with miR-29a + 381 (P < 0.05). FN expression decreased after miR-29a + 381 (48%) and miR-29a + 155 transfection (60.3%) (P < 0.05). Similarly, αSMA reduction was lower when transfected with miR-29a + 381 (50.2%) and miR-29a + 155 (59.6%) (P < 0.05), but not significant after miR29a or miR-29b transfection. Among these miRNA groups, miR-29a + 381 transfection consistently and significantly downregulated most fibrosis gene expression.
Fig. 5.
In vitro anti-fibrosis assay of human corneal stromal keratocytes transfected with miRNA mimics. Cells transfected with miRNA groups showing significant anti-inflammatory activity in the previous assay were treated by TGFβ1 and ascorbate to undergo fibrogenesis. Compared to cells with scrambled sequences, various miRNA expressions downregulated fibrosis gene expression. Among them, miR-29a and 381 consistently and significantly suppressed Col3A1, SPARC, MCP1, FN-EDA and αSMA (*P < 0.05, **P < 0.01; Mann-Whitney U test).
Validation of in vivo anti-scarring effect of CSSCs transfected with miR-29a and 381-5p mimics
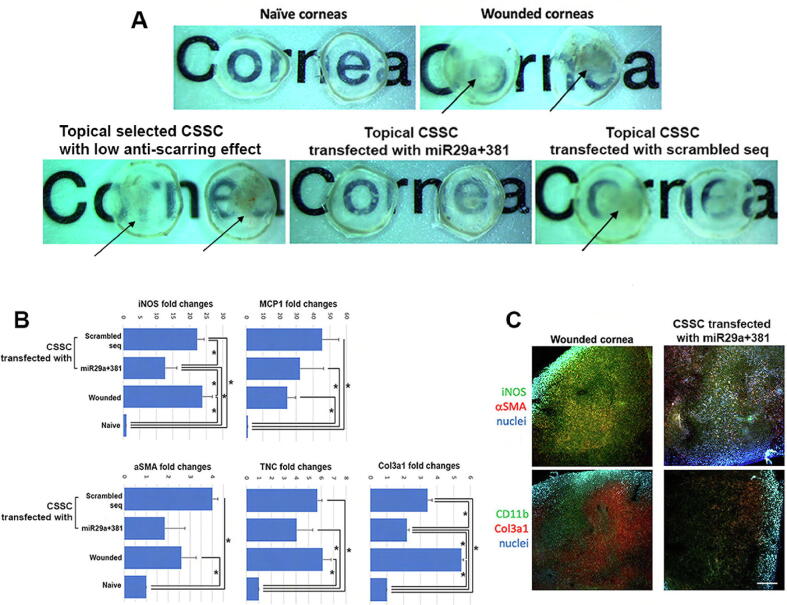
CSSCs (HC458 and HC470P previously shown to have poor anti-scarring effect on corneal wounds, Supplementary Fig. S5) were transfected with miR-29a + 381 mimics, or scrambled sequences for 48 h. Cells were collected and applied topically to mouse corneal stromal wounds (n = 6 per group; 5x104 cells each treatment). After 10 days, the untreated injured corneas and corneas receiving non-transfected CSSC developed intense scarring, compared to naïve clear corneas (Fig. 6A). Corneas treated with miR-29a + 381 transfected cells showed reduced scarring and only mild haziness, while those treated with cells having scrambled sequences were blurred with heavy scarring, similar as wound controls. The expression of inflammatory and fibrosis genes were markedly upregulated after wounding and in injured corneas treated with cells containing scrambled sequences (Fig. 6B). Treatment with miR-29a + 381 transfected cells showed a repressed upregulation of these markers. Compared to wound controls, the expression of iNOS was reduced by a mean of 48% (P < 0.05), αSMA by 29.2%, TNC by 35.3% and Col3A1 by 61.4% (P < 0.05) (single cornea per sample, a total of 3 corneas per cell treatment). Similar results were shown by immunostaining on wholemount samples and the expression of iNOS, αSMA and Col3A1 in mouse corneas treated with miR-29a + 381 transfected CSSCs were generally downregulated (Fig. 6C). These results illustrate that CSSC overexpressing miR-29a and 381-5p have corrective effect on corneal scarring.
Fig. 6.
In vivo anti-scarring test of human CSSCs transfected with miR-29a and 381 mimics. Topical CSSC treatment was performed on mouse anterior stromal wounds and corneas were harvested at day 7. (A) Wound control and corneas receiving selected CSSCs previously shown to have low anti-scarring effects. CSSCs transfected with scrambled sequences showed intense scarring (arrows), while corneas with cells transfected with miR-29a and 381 mimics had inhibited scar formation. (B) By qPCR, the expression of inflammatory (mouse iNOS and MCP1) and fibrosis genes (mouse αSMA, TNC, and Col3A1) were downregulated in corneas treated with cells having miR-29a and 381 transfection (*P < 0.05; Mann-Whitney U test). (C) Wholemount immunofluorescence demonstrated the reduced expression of mouse iNOS, αSMA, CD11b and Col3A1 in mouse corneas having CSSC with miR29a and 381 expression when compared to wound controls. Scale bar: 50 μm.
Screening of miR29a and 381-5p expression in CSSC EV fractions to predict treatment outcome on corneal scarring in a mouse model of corneal scarring.
CM was collected from an additional 8 primary CSSC batches (donor information in Table S1), and total EV-RNA was extracted for the screening of miR-29a and 381 expression by qPCR. To determine target miRNA expression precisely, a robust normalization with uniformly expressed EV-containing miRNA is necessary. However, there is no consensus on reference miRNA for the abundance normalization of EV miRNAs. Hence we first determined a CSSC EV miRNA normalizer using our Nanostring data. Based on the number of reads in gTotal signals, the expression reads of miR-16 was invariant between “healing” and “non-healing” EV batches (each n = 3) (Table S5). Other miRNAs, which have been reported as housekeeping in cells, showed abundance variation with diverse fold differences among EV samples. Hence, we selected miR-16 as a miRNA normalizer to evaluate miRNA abundance inside EVs.
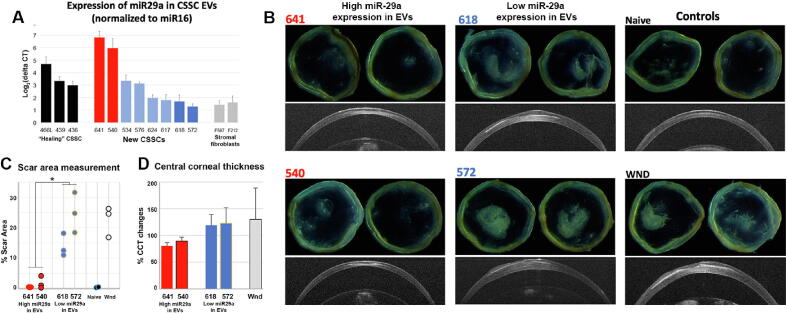
The normalized expression of miR-29a was upregulated in EV fractions of HC540 and 641, and was low in HC572 and 618 (Fig. 7A). It was 5.9-fold (in HC540) and 6.8-fold (in HC641) higher than the respective miR-16 levels. Similarly upregulated miR-29a expression was found in CSSC (HC436, 439 and 466L) that previously reported to have good healing effects. In contrast, narrow fold changes of miR-29a to miR-16 was detected in HC572 (1.3-fold) and HC618 (1.7-fold), and this level was similar to SFs (F215 and F587). On the other hand, miR-381-5p was generally expressed in most CSSC batches, but negligibly detectable in SFs (Fig. S8). Different to miR-29a, we hardly observed any pattern of miR-381 expression that could indicate their regenerative potency of CSSCs.
Fig. 7.
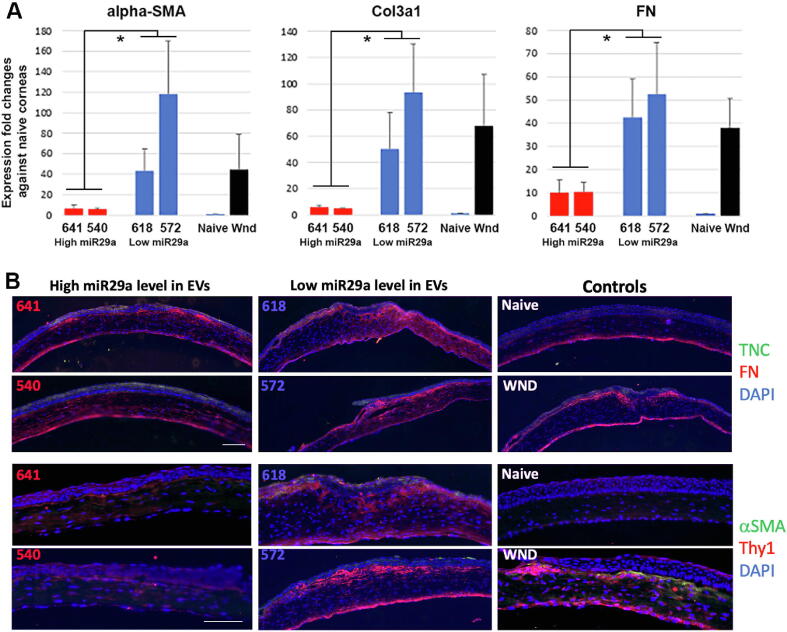
Quality test of miR-29a expression in human CSSC-EVs and functional validation of anti-scarring efficiency. (A) Compared among known healing CSSCs (HC436, 439 and 466L) and stromal fibroblasts (F587 and F212) without non-healing function on corneal wounds, new CSSCs were distinguished with higher miR-29a expression (HC540 and 641; red bars) and low miR-29a expression (HC572 and 618; dark blue bars) in the collected EVs. (B) Isolated corneas after cell treatment showed different degrees of scar formation at day 10. In contrast to naïve corneas, wound control and corneas treated with HC618 and 572 (low miR-29a expression in EVs) developed intense scarring. Treatment with HC540 and 641 (high miR-29a expression) had relatively much reduced scarring. OCT images showed thin corneas after HC540 and 641 treatment, but not for treatment with HC618 and 572, and wound controls. (C) Scar area measurement illustrated a significant scar reduction after treatment with HC540 and 641, compared to corneas treated with HC618 and 572, as well as wound controls (n = 3 corneas; *P < 0.05; paired t-test). (D) Percentage changes of mean central corneal thickness (CCT) compared to pre-operative level (n = 6 corneas). CCT of HC641 and 540-treated corneas were lower than that of HC618 and 572-treated and wound controls. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
When topically applied to mouse corneal stromal wounds (n = 6 per cell treatment), HC540 and 641 (with higher miR-29a expression in EVs) (5x104 cells per treatment) nearly prevented corneal scarring at day 10 (Fig. 7B). Corneas treated with HC618 and 572 (with low miR-29a expression) developed intense scarring, similar to wound control. The percentage of scar area of HC540- and 641-treated corneas was significantly smaller than HC618- and 572-treated, and wounded corneas (P < 0.05, ANOVA; n = 3 corneas) (Fig. 7C). From serial OCT images, the mean central corneal thickness (CCT) of HC540- and 641-treated corneas stayed at normal range similar to naïve controls (n = 6 each group) (Fig. 7D). In contrast, HC618- and 572-treated corneas were thicker.
We studied the fibrosis marker expression to assess the treatment outcome on corneal scar progression. Using qPCR, the expression of Col3A1, FN, and αSMA were markedly upregulated 10 days after wounding, similar to our previous report [11]. Mouse corneas treated with HC540 and 641 (with higher miR-29a expression) showed inhibited upregulation of these genes, compared to wound controls (Fig. 8A). This suppressive effect was significantly weaker after treatment with HC618 and 572 (lower miR-29a expression) (P < 0.05; ANOVA), and this was indistinguishable from the wound controls. Immunofluorescence also showed intense expression of mouse FN, αSMA and CD90/Thy1 in the anterior stromal region of HC618- and 572-treated, and wound control corneas (n = 3 corneas per treatment group) (Fig. 8B). Reduced signals of these markers were detected in HC540- and 641-treated corneas. While TNC was expressed in the regenerated corneal epithelia, moderate signals were also found in the anterior stroma of HC618- and 572-treated, and wound corneas, but not in HC540- and 641 corneas. Hence, our results demonstrated that CSSCs with higher miR-29a expression in CM had strong anti-scarring potency and regenerated clear corneas after injury.
Fig. 8.
Anti-scarring phenotypes of mouse corneas treated with human CSSCs after quality test by miR-29a expression in EVs. (A) By qPCR, injured corneas treated with HC540 and 641 (high miR-29a expression) had significantly downregulated expression of fibrosis genes (αSMA, Col3A1 and FN), when compared to corneas after HC618 and 572 cell treatment (low miR-29a expression) and wound controls (triplicate run with single cornea each sample; *P < 0.05; Mann-Whitney U test). (B) Immunofluorescence showed suppressed expression of fibrosis markers (FN, αSMA and CD90/Thy1) in mouse corneas treated by HC540 and 641 cells (n = 3 each). In contrast, wounded corneas without treatment or treated with HC618 and 572 showed stronger immunoreactive signals. Scale bars: 20 μm.
Discussion
In this study, we demonstrated that the healing effect exhibited by human CSSCs on corneal stromal injury was related to the paracrine actions mediated by a differential expression of miRNAs in EVs produced by cells. Among 21 miRNAs with upregulated expression (>20 folds) in the healing EVs, miR-29a and 381 were shown to reduce LPS-stimulated M1 pro-inflammatory response in mouse macrophages and suppress TGFβ1-caused fibrotic reaction of human stromal keratocytes. These in vitro effects were validated by the in vivo anti-scarring outcome in a mouse model of corneal stromal injury. The EV expression of miR-29a was further shown to be a valid quantitative tool for quality testing of CSSCs. Selection of cells with high miR-29a expression in EVs indicated strong healing potency that could achieve the clinical efficacy in corneal stromal regeneration, and restore corneal transparency.
Scarring of corneal stroma is an intrinsic response to trauma, inflammation or infection. It disrupts vision, causing corneal blindness in millions of people worldwide [1], [2]. The primary clinical approach to restore vision is by surgical replacement of scarred tissue with clear donor corneas through transplantation. Recent studies have reported cell-based therapeutic approaches using either keratocytes from central stroma or their progenitors (CSSCs from the limbal stroma) to reduce scar formation and progression [10], [31]. As a progenitor cell type, CSSCs possess the property of MSC-like phenotype with the expression of stem cell markers (including ABCG2, nestin, CD73, CD90) and clonal expansion in vitro [8]. CSSC cultures with high homogeneity (with > 90% CD73/90/105-positive viable cells and negligible CD31) were used in our experiments. In low-mitogen condition supplemented with insulin and ascorbate, CSSCs expressed an array of keratocyte-specific markers, ALDH3A1 and keratocan [6] and produced significant amounts of ECM with a deposition of aligned collagen fibrils, similar to that seen in corneal stroma in vivo [38], [39]. These beneficial features lead to regenerating clear corneas in pre-clinical models of corneal wounding. Both topical application and stromal injection of CSSCs remodelled the defective stromal structure, replaced the fibrotic ECM proteins (like Col3A1, hyaluronan and FN, produced by the activated stromal fibroblasts), and restored the native stromal ECM, resulting in better light passage with reduced deviation and scattering [7], [10], [11], [40]. Moreover, the anti-scarring effect of CSSCs was shown to be associated with significant reduction of corneal inflammation (inhibiting neutrophil infiltration) [9]. Using both toll-like receptor (TLR)-mediated inflammatory and osteoclastogenesis assays, we demonstrated the acute and chronic anti-inflammatory effects by CSSCs. Similar to other MSCs, human CSSCs when administered to mouse corneal stroma did not elicit xenogeneic T-cell-mediated immune rejection, demonstrating its immunomodulatory function which can enhance the therapeutic efficacy and stability [7]. Preliminary results from an ongoing clinical trial in India using CSSC therapy on patients with corneal scarring showed scar regression and visual improvement [13].
In generating a clinical grade product, cell manufacturing under GMP regulatory framework must ensure the product safety. Another aspect of evaluation is the product potency, which is a quantitative measure of how the product achieves a defined biological effect. Such quantitative evaluations provide the means to control product quality and may relate to, but not exactly define the clinical efficacy of the product. The selection of proper CSSC batches with good regenerative potential has been challenging due to the batch-to-batch variability of cells generated from different donor corneas. The variability also occurs in the scale-up manufacturing process due to differences in equipment, reagents and supplies. In this study, we report the product testing with a quantitative measure of miR-29a expression in CSSC secretome containing EVs. MicroRNAs are a class of evolutionarily conserved small non-coding RNA molecules (19–24 nucleotides) that control gene expression at post-transcriptional level by repressing target mRNAs [41]. Our previous study identified that the presence of miRNAs in CSSC EVs was related to their anti-scarring effect in vivo [14].
This report uncovered miRNAs specific to CSSC EVs with regenerative potential typified by suppressing inflammation and blocking fibrosis/scarring during corneal healing. Using the Nanostring nCounter Human v3 miRNA Expression assay screening 800 miRNAs, we found 59 miRNAs which were differentially expressed by>10 folds in the healing EVs and 21 of them had > 20 folds of difference (Table S6). The analysis was based on the assumption that not every miRNA identified is involved in regulating inflammatory and fibrotic reactions. Some miRNAs can be redundant, while miRNAs with relevant activity may work in tandem to regulate multiple genes. We therefore employed a bioinformatic approach with target gene search and enriched pathway prediction to identify single or groups of miRNAs that potentially regulate pathways of inflammation, and/or fibrosis. Different online databases are available for target gene search; however, each uses its proprietary algorithm in calculating the binding score to a target sequence and thus has limitations. We employed more than 1 platform to increase the confidence in hits identified independently while compensating for the limitations of each. From TargetScan and miRDB/miRTarget, we obtained respective gene lists and the top 50 genes shared by both platforms were listed for each miRNA. The gene lists of different miRNAs were then combined and imported for enriched GO term analysis using DAVID bioinformatics.
A total of 9 miRNA groups showed enriched GO terms significantly linked to inflammatory and immune pathways, fibrosis, senescence and autophagy, as well as tissue regeneration (Table 1). Importantly, all groups contained miR-29, which is known as a “master fibro-miRNA” regulator, with pivotal roles in regulating organ fibrosis, including cardiac, hepatic, lung fibrosis, systemic sclerosis and keloid [42]. The human miR-29 family contains 3 members, miR-29a, b and c, encoded by 2 gene clusters in chr. 7q32.3 and chr. 1q32.2, respectively. They share common seed sequences and are predicted to target largely overlapping sets of genes, including at least 16 ECM genes as direct targets. They are the collagen isoforms, laminin γ1, fibrillin 1, elastin, MMP-2, and integrin β1 [43], [44], [45]. Ectopic knockdown of miR-29 in canine atrial fibroblasts significantly upregulated Col1A1, 3A1 and FN [46], but did not affect αSMA expression or myofibroblasts [47]. Hence, miR-29 could negatively regulate fibrosis by targeting collagen matrix synthesis rather than by inhibiting myofibroblasts. The miR-29 family is an ECM homeostatic modulator. MiR-29a-3p was the most deregulated miRNA in Fuchs’ corneal dystrophy endothelium. Downregulated miR-29a increased collagen I, IV and laminin mRNA and protein levels [48]. Moreover, TGFβ2 differentially regulated miR-29a and 29b expression related to ECM synthesis in the trabecular meshwork [49]. Since miR-29a is significantly enriched in the cytoplasm where EVs originate, while 29b is more localized inside the nucleus [50], we studied miR-29a (i.e., EV-bearing) for its anti-inflammatory and anti-fibrotic effect in vitro and in vivo. Remarkably, our results showed that co-expressing miR-29a and 381-5p in mouse RAW macrophages significantly suppressed pro-inflammatory iNOS, MCP1 and CXCL10 expression after LPS treatment (reduced M1 induction), and further downregulated iNOS expression after IL-4 treatment (reverted M1 phenotype), indicating the anti-inflammatory effects. However, the indifferent Arg1 expression after IL-4 treatment has suggested that M2 phenotype was not necessarily induced in the transfected RAW cells. When human CSKs were treated with TGFβ1 to induce fibrogenesis, the transfection of miR-29a + 381 significantly reduced fibrosis gene expression (Col3A1, SPARC and FN). Intriguingly, the expression of αSMA was also downregulated. This could be related to the positive feedback loop in the expression of myofibroblast-related AOC3 (amine oxidase, copper containing 3, a cell surface marker to distinguish viable cells) and Nkx3-2 (also Bapx1, a direct target of miR-381-5p) [51]. Nkx3-2 forms complex with HDAC1 and Smad1/4 in a BMP-dependent manner and acts downstream of TGFβ1/2 to regulate fibrosis gene expression [52]. This transcription factor was transiently expressed at early corneal injury in mice when fibrosis was initiated (unpublished data). MiR-381 targeting on Nkx3-2 probably affects the viability of myofibroblasts and their differentiation, providing additive action to negatively regulate scar formation [53]. The exact connection between miR-381 and myofibroblasts that create the scar niche remains to be elucidated. The regulatory effect on tissue inflammation by miR-381, via its direct targeting on IκBα, was also reported to affect NFkB signalling, hence regulating pro-inflammatory TNFα, IL-6 and COX-2 expression [54]. CSSCs over-expressing miR-29a and 381-5p were further shown to suppress corneal scar formation in vivo, indicating their functionality in blocking fibrosis. Whether such therapeutic effect can be achieved by EVs enriched with miR-29a and 381-5p expression and the dosage effect of miRNA quantity in EVs will be of interest to investigate in ongoing studies.
Screening specific miRNA expression in secretome or EVs is highly feasible during the scale-up cell manufacturing as in-process testing or at the end of production prior to the release of cell product for clinical use (lot release testing). Since cell therapy can be developed as patient-specific, there is usually a limited quantity of final cell product. Use of secretome for product testing can avoid this limitation and will not affect the quantity and clinical effectiveness of the cell product. Moreover, there are practical advantages of target miRNA expression over microarrays for potency testing. The array platforms are still evolving and results are not always comparable among platforms. The array analysis also involves multiple steps, including RNA isolation, amplification, fluorescence labelling, hybridization and data analysis with expertise, which takes much longer working time than simple qPCR for target miRNAs. This may delay the product release for patient use. Target miRNA identification for screening purpose can also be developed as tailored chips or specific qPCR kits for multiple lot testing. Our work demonstrated that miR-29a expression in EV fractions distinguished CSSCs with different healing potencies. This could be due to more EVs containing miR-29a or increased miR29a copy number in each EV, which was not investigated in the present study. Treatment using CSSCs with higher miR-29a expression significantly reduced corneal scar formation and fibrosis gene expression, resulting in clear corneas and normal corneal thickness. Mouse corneas treated with cells having lower miR-29a expression developed intense scarring, similar to wound controls. This miR-29a expression assay thus can be a suitable quality test for CSSCs prior to clinical application. Further investigations on miR-29a and other cell markers may result in development of quantitative assays to predict the clinical efficacy of cell therapy. However, the accuracy of miR-29a expression is hindered by a lack of standard reference miRNAs in EVs for qPCR normalization. In cell and tissue samples, there are “housekeeping miRNAs” with stable expressions, such as U6, RNU44, RNU48, let-7, and they have been extensively used for normalizing target miRNA quantification. Instead, they are not suitable for normalizing circulating or secretory miRNAs [55], [56], [57]. Their expression is highly variable in plasma and sera of healthy individuals and patients with cancers or fibrosis. From our Nanostring data, most reported “housekeeping” miRNAs showed variation in abundance, based on their number of reads in gTotal signals (Table S5), and this may be due to tissue specificity. Only miR-16 showed invariant readings between “healing” and “non-healing” EVs. It has been described as a stable control for miRNA expression analysis from blood samples of cancer patients and healthy subjects [58], [59]. The approach using exogenous miRNA (like ath-miR-159a of plant origin) as spike-in control and normalization reference is useful to control the technical biases related to sample preparation, and quantification of miRNA copy number, however biological variables, such as EV size and cargo density, may not be elucidated [56].
Conclusions
Our study, an endeavour to characterize the miRNA profile of EVs from CSSCs, demonstrated the utility of miR-29a as a screening tool to identify CSSCs with anti-scarring quality, as validated by in vivo corneal injury model. These findings would facilitate the production of GMP grade CSSC batches with uniform regenerative quality for clinical trials.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank K Davoli for histological processing of corneal samples; K Lathrop for guidance in microscopy; J Patel for EV assay development; AJ St Leger, PhD for flow cytometry; I Khandaker, PhD for image analysis; D Dhaliwal, MD, LAc for intellectual discussion and I Billig, PhD for project coordination. We also thank RM Shanks, PhD and SK Swamynathan, PhD for reading and providing comments on the manuscript. This work was supported by NIH Grant RO1 EY016415 (JF, YD); Stein Innovator Award\Inside Ophthalmology from Research to Prevent Blindness (JF, GY, YD); Immune Transplant and Therapy Centre of UPMC (JS, GY). Additional funds for the department were provided by Hillman Foundation, Eye and Ear Foundation of Pittsburgh, Louis J Fox Centre for Vision Restoration, and unrestricted funds from Research to Prevent Blindness and National Eye Institute [P30, EY008098]. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Ethics approval
The use of donor corneas followed the tenets of the Declaration of Helsinki and was approved by The University of Pittsburgh Institutional Review Board (IRB) and Committee for Oversight of Research and Clinical Training Involving Decedents (CORID), Protocol #161. Animal study was carried out in strict accordance with the guidelines for the Care and Use of Laboratory Animals of NIH and The Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research. The protocol was approved by the Institutional Animal Care and Use Committee of The University of Pittsburgh (Protocol 18022511).
Author contributions
Conception and design: GY, JF, MF. Collection and assembly of data, data analysis: GY, TY, MG, MS, MF, ER. Financial support: GY, JF, YD, JS. Manuscript writing: GY, MF, VJ, JS. All authors approved the manuscript.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.05.008.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data Availability.
All data are included in the text and supplementary materials.
References
- 1.Flaxman S.R., Bourne R.R.A., Resnikoff S., Ackland P., Braithwaite T., Cicinelli M.V., et al. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Global Health. 2017;5:e1221–e1234. doi: 10.1016/S2214-109X(17)30393-5. [DOI] [PubMed] [Google Scholar]
- 2.Whitcher J.P., Srinivasan M., Upadhyay M.P. Corneal blindness: a global perspective. Bull World Health Organ. 2001;79:214–221. [PMC free article] [PubMed] [Google Scholar]
- 3.Fuest M., Yam G.H., Peh G.S., Mehta J.S. Advances in corneal cell therapy. Regen Med. 2016;11(6):601–615. doi: 10.2217/rme-2016-0054. [DOI] [PubMed] [Google Scholar]
- 4.Riau A.K., Liu Y.C., Yam G.H.F., Mehta J.S. Stromal keratophakia: corneal inlay implantation. Prog Retin Eye Res. 2020;75:100780. doi: 10.1016/j.preteyeres.2019.100780. [DOI] [PubMed] [Google Scholar]
- 5.El Zarif M., Alió J.L., Alió del Barrio J.L., Abdul Jawad K., Palazón-Bru A., Abdul Jawad Z., et al. Corneal stromal regeneration therapy for advanced keratoconus: long-term outcomes at 3 years. Cornea. 2021;40(6):741–754. doi: 10.1097/ICO.0000000000002646. [DOI] [PubMed] [Google Scholar]
- 6.Du Y., Funderburgh M.L., Mann M.M., SundarRaj N., Funderburgh J.L. Multipotent stem cells in human corneal stroma. Stem Cells. 2005;23(9):1266–1275. doi: 10.1634/stemcells.2004-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y., Carlson E.C., Funderburgh M.L., Birk D.E., Pearlman E., Guo N., et al. Stem cell therapy restores transparency to defective murine corneas. Stem Cells. 2009;27:1635–1642. doi: 10.1002/stem.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Funderburgh J.L., Funderburgh M.L., Du Y. Stem cells in the limbal stroma. Ocul Surf. 2016;14(2):113–120. doi: 10.1016/j.jtos.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hertsenberg A.J., Shojaati G., Funderburgh M.L., Mann M.M., Du Y., Funderburgh J.L., et al. Corneal stromal stem cells reduce corneal scarring by mediating neutrophil infiltration after wounding. PLoS ONE. 2017;12(3) doi: 10.1371/journal.pone.0171712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu S., Hertsenberg A.J., Funderburgh M.L., Burrow M.K., Mann M.M., Du Y., et al. Human limbal biopsy–derived stromal stem cells prevent corneal scarring. Sci Transl Med. 2014;6(266) doi: 10.1126/scitranslmed.3009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khandaker I., Funderburgh J.L., Geary M.L., Funderburgh M.L., Jhanji V., Du Y., et al. A novel transgenic mouse model for corneal scar visualization. Exp Eye Res. 2020;200:108270. doi: 10.1016/j.exer.2020.108270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weng L., Funderburgh J., Khandaker I., Geary M.L., Basu R., Funderburgh M.L., et al. The anti-scarring effect of corneal stromal stem cell therapy is mediated by transforming growth factor β3. Eye Vis. 2020;7:52. doi: 10.1186/s40662-020-00217-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basu S., Damala M., Tavakkoli F., Mitragotri N., Singh V. Human limbus-derived mesenchymal/stromal stem cell therapy for superficial corneal pathologies: two-year outcomes. Invest Ophthalmol Vis Sci. 2019;60:4146. [Google Scholar]
- 14.Shojaati G., Khandaker I., Funderburgh M.L., Mann M.M., Basu R., Stolz D.B., et al. Mesenchymal stem cells reduce corneal fibrosis and inflammation via extracellular vesicle-mediated delivery of miRNA. Stem Cells Trans Med. 2019;8:1192–1201. doi: 10.1002/sctm.18-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phinney D.G., Pittenger M.F. Concise review: MSC-derived exosomes for cell-free therapy. Stem Cells. 2017;35:851–858. doi: 10.1002/stem.2575. [DOI] [PubMed] [Google Scholar]
- 17.Doyle L.M., Wang M.Z. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8:727. doi: 10.3390/cells8070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai J., Su Y., Zhong S., Cong L.i., Liu B., Yang J., et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5(1):145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossetti C., Iraci N., Mercer T.R., Leonardi T., Alpi E., Drago D., et al. Extracellular vesicles from neural stem cells transfer IFNg via Ifngr1 to activate Stat1 signaling in target cells. Mol Cell. 2014;56:193–204. doi: 10.1016/j.molcel.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vojtech L., Woo S., Hughes S., Levy C., Ballweber L., Sauteraud R.P., et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res. 2014;42(11):7290–7304. doi: 10.1093/nar/gku347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi D., Montermini L., Kim D.K., Meehan B., Roth F.P., Rak J. The impact of oncogenic EGFRvIII on proteome of extracellular vesicles released from glioblastoma cells. Mol Cell Proteomics. 2018;17:1948–1964. doi: 10.1074/mcp.RA118.000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nikfarjam S., Rezaie J., Zolbanin N.M., Jafari R. Mesenchymal stem cell derived-exosomes: a modern approach in translational medicine. J Trans Med. 2020;18:449. doi: 10.1186/s12967-020-02622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao T., Sun F., Liu J., Ding T., She J., Mao F., et al. Emerging role of mesenchymal stem cell-derived exosomes in regenerative medicine. Curr Stem Cell Res Ther. 2019;14(6):482–494. doi: 10.2174/1574888X14666190228103230. [DOI] [PubMed] [Google Scholar]
- 24.Liu X.-N., Chen Y., Wang Y. Corneal stromal mesenchymal stem cells: reconstructing a bioactive cornea and repairing the corneal limbus and stromal microenvironment. Int J Ophthalmol. 2021;14(3):448–455. doi: 10.18240/ijo.2021.03.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashmani K., Branch M.J., Sidney L.E., Dhillon P.S., Verma M., McIntosh O.D., et al. Characterization of corneal stromal stem cells with the potential for epithelial transdifferentiation. Stem Cell Res Ther. 2013;4(3):75. doi: 10.1186/scrt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mansoor H., Ong H.S., Riau A.K., Stanzel T.P., Mehta J.S., Yam G.H. Current trends and future perspective of mesenchymal stem cells and exosomes in corneal diseases. Int J Mol Sci. 2019;20:2853. doi: 10.3390/ijms20122853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jhanji V., Billig I., Yam G.H. Cell-free biological approach for corneal stromal wound healing. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.671405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu F.S., Hazlett L.D. Toll-like receptors and the eye. Invest Ophthalmol Vis Sci. 2006;47:1255–1263. doi: 10.1167/iovs.05-0956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bognar E., Sarszegi Z., Szabo A., Debreceni B., Kalman N., Tucsek Z., et al. Antioxidant and anti-inflammatory effects in RAW264.7 macrophages of malvidin, a major red wine polyphenol. PLoS ONE. 2013;8(6):e65355. doi: 10.1371/journal.pone.0065355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakanishi-Matsui M., Yano S., Matsumoto N., Futai M. Lipopolysaccharide induces multinuclear cell from RAW264.7 line with increased phagocytosis activity. Biochem Biophys Res Commun. 2012;425(2):144–149. doi: 10.1016/j.bbrc.2012.07.050. [DOI] [PubMed] [Google Scholar]
- 31.Yam G.-F., Fuest M., Yusoff N.Z.B.M., Goh T.-W., Bandeira F., Setiawan M., et al. Safety and feasibility of intrastromal injection of cultivated human corneal stromal keratocytes as cell-based therapy for corneal opacities. Invest Ophthalmol Vis Sci. 2018;59(8):3340. doi: 10.1167/iovs.17-23575. [DOI] [PubMed] [Google Scholar]
- 32.Van Deun J., Mestdagh P., Agostinis P., Akay Ö., Anand S., Anckaert J., et al. EV-TRACK: transparent reporting and centralizing knowledge in extracellular vesicle research. Nat Methods. 2017;14(3):228–232. doi: 10.1038/nmeth.4185. [DOI] [PubMed] [Google Scholar]
- 33.Almizraq R.J., Holovati J.L., Acker J.P. Characteristics of extracellular vesicles in red blood concentrates change with storage time and blood manufacturing method. Transfus Med Hemother. 2018;45:185–193. doi: 10.1159/000486137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dos Santos A., Balayan A., Funderburgh M.L., Ngo J., Funderburgh J.L., Deng S.X. Differentiation capacity of human mesenchymal stem cells into keratocyte lineage. Invest Ophthalmol Vis Sci. 2019;60(8):3013. doi: 10.1167/iovs.19-27008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yam G.-F., Yusoff N.Z.B.M., Kadaba A., Tian D., Myint H.H., Beuerman R.W., et al. Ex vivo propagation of human corneal stromal “activated keratocytes” for tissue engineering. Cell Transplant. 2015;24(9):1845–1861. doi: 10.3727/096368914X685069. [DOI] [PubMed] [Google Scholar]
- 36.Karamichos D., Guo X.Q., Hutcheon A.E., Zieske J.D. Human corneal fibrosis: an in vitro model. Invest Ophthalmol Vis Sci. 2010;51:1382–1388. doi: 10.1167/iovs.09-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Ves. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Y., SundarRaj N., Funderburgh M.L., Harvey S.A., Birk D.E., Funderburgh J.L. Secretion and organization of a cornea-like tissue in vitro by stem cells from human corneal stroma. Invest Ophthalmol Vis Sci. 2007;48(11):5038. doi: 10.1167/iovs.07-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu J., Du Y., Mann M.M., Yang E., Funderburgh J.L., Wagner W.R. Bioengineering organized, multilamellar human corneal stromal tissue by growth factor supplementation on highly aligned synthetic substrates. Tissue Eng Part A. 2013;19(17-18):2063–2075. doi: 10.1089/ten.tea.2012.0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shojaati G., Khandaker I., Sylakowski K., Funderburgh M.L., Du Y., Funderburgh J.L. Compressed collagen enhances stem cell therapy for corneal scarring. Stem Cells Trans Med. 2018;7:487–494. doi: 10.1002/sctm.17-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng Z., He Y., Yang X., Shi H., Shi A.o., Lu L., et al. MicroRNA-29: A crucial player in fibrotic disease. Mol Diagn Ther. 2017;21(3):285–294. doi: 10.1007/s40291-016-0253-9. [DOI] [PubMed] [Google Scholar]
- 43.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Nat Acad Sci (USA) 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Taylor N.E., Lu L., Usa K., Cowley A.W., Ferreri N.R., et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55(4):974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwon J.J., Factora T.D., Dey S., Kota J. A systematic review of miR-29 in cancer. Mol Ther Oncolytics. 2019;12:173–194. doi: 10.1016/j.omto.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dawson K., Wakili R., Ördög B., Clauss S., Chen Y.u., Iwasaki Y., et al. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. 2013;127(14):1466–1475. doi: 10.1161/CIRCULATIONAHA.112.001207. [DOI] [PubMed] [Google Scholar]
- 47.Qin W., Chung A.C., Huang X.R., Meng X.M., Hui D.S., Yu C.M., et al. TGFb/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol. 2011;22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matthaei M., Hu J., Kallay L., Eberhart C.G., Cursiefen C., Qian J., et al. Endothelial cell microRNA expression in human late-onset Fuchs' dystrophy. Invest Ophthalmol Vis Sci. 2014;55(1):216. doi: 10.1167/iovs.13-12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Villarreal G., Jr., Oh D.J., Kang M.H., Rhee D.J. Coordinated regulation of extracellular matrix synthesis by the microRNA-29 family in the trabecular meshwork. Invest Ophthalmol Vis Sci. 2011;52:3391–3397. doi: 10.1167/iovs.10-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang H.W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315(5808):97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 51.Hsia L.T., Ashley N., Ouaret D., Wang L.M., Wilding J., Bodmer W.F. Myofibroblasts are distinguished from activated skin fibroblasts by the expression of AOC3 and other associated markers. Proc Nat Acad Sci (USA) 2016;113:E2162–E2171. doi: 10.1073/pnas.1603534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim D.W., Lassar A.B. Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol Cell Biol. 2003;23(23):8704–8717. doi: 10.1128/MCB.23.23.8704-8717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishida W., Nakamura M., Mori S., Takahashi M., Ohkawa Y., Tadokoro S., et al. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277(9):7308–7317. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]
- 54.Xu Z., Dong D., Chen X., Huang H., Wen S. MicroRNA-381 negatively regulates TLR4 signaling in A549 cells in response to LPS stimulation. BioMed Res Int. 2015;2015:1–8. doi: 10.1155/2015/849475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benz F., Roderburg C., Vargas Cardenas D., Vucur M., Gautheron J., Koch A., et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med. 2013;45(9):e42. doi: 10.1038/emm.2013.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marabita F., de Candia P., Torri A., Tegnér J., Abrignani S., Rossi R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief Bioinform. 2016;17(2):204–212. doi: 10.1093/bib/bbv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Donati S., Ciuffi S., Brandi M.L. Human circulating miRNAs real-time qRT-PCR-based analysis: an overview of endogenous reference genes used for data normalization. Int J Mol Sci. 2019;20:4353. doi: 10.3390/ijms20184353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McDermott A.M., Kerin M.J., Miller N. Identification and validation of miRNAs as endogenous controls for RT-PCR in blood specimens for breast cancer studies. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0083718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Müller V., Gade S., Steinbach B., Loibl S., von Minckwitz G., Untch M., et al. Changes in serum levels of miR-21, miR-210, and miR-373 in HER2-positive breast cancer patients undergoing neoadjuvant therapy: a translational research project within the Geparquinto trial. Breast Cancer Res Treat. 2014;147(1):61–68. doi: 10.1007/s10549-014-3079-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the text and supplementary materials.