Abstract
Viruses are ubiquitous in the gut of animals and play an important role in the ecology of the gut microbiome. The potential effects of these substances on the growth and development of the body are not fully known. Little is known about the effects of breeding environment on pig gut virome. Here, there are 3584 viral operational taxonomic units (vOTUs) longer than 5 kb identified by virus-enriched metagenome sequencing from 25 pig fecal samples. Only a small minority of vOTUs (11.16%) can be classified at the family level, and ∼50% of the genes could be annotated, supporting the concept of pig gut as reservoirs of substantial undescribed viral genetic diversity. The composition of pig gut virome in the six regions may be related to geography. There are only 20 viral clusters (VCs) shared among pig gut virome in six regions of Shanxi Province. These viruses rarely carry antibiotic resistance genes (ARGs). At the same time, they possess abundant auxiliary metabolic genes (AMGs) potentially involved in carbon, sulfur metabolism and cofactor biosynthesis, etc. This study has revealed the unique characteristics and potential function of pig gut DNA virome and established a foundation for the recognition of the viral roles in gut environment.
Keywords: Pig gut, Viral metagenome, Viral diversity, Antibiotic resistance genes, Auxiliary metabolic genes
1. Introduction
As livestock, pigs provide people with a large number of meat products. They are a major meat source worldwide and the backbone of Chinese livestock industry. The gut microbiome is a complex microecosystem [1], containing bacterial cells, viral particles, archaea and eukaryotes. Thanking the development of high-throughput sequencing technologies (such as whole-metagenomic sequencing or amplicon), the gut bacterial community has been well studied in recent years [[2], [3], [4], [5]]. As shown in previous studies, gut microbes play an important role in nutrient uptake and growth in pigs [6,7]. However, as another component of the gut microbial ecosystem, the pig gut viral community (gut virome) was poorly described [8].
The viral component of the microbiome is mainly composed of bacteriophages [9], which are thought to play crucial roles in shaping microbial communities through cell lysis and horizontal gene transfer. Understanding the spread of antibiotic resistance genes (ARGs) is critical to human health [10]. Although previous study have demonstrated that the pig gut virome carries very few ARGs, only a sample size of 14 was used, which is relatively small [11]. ARGs are frequently found in pig fecal samples [4], but insights into whether the pig gut virome carries ARGs are lacking. Besides, viruses are able to encode auxiliary metabolic genes (AMGs) that are beneficial to the host and promote the co-evolution of both [12]. At the same time, the abundance and community structure of host was affected. In recent years, auxiliary metabolic genes (AMGs) have been suggested to enhance host metabolism and promote the production of new viruses [13,14]. The role of viruses in different ecosystems, such as bovine rumen [15], human gut [16], mangrove soils [17], prawn-culture sediments [18], Mariana Trench sediments [19], permafrost [20], rhizosphere soils [21] and freshwater [22] had gradually been revealed. Analysis of virus-encoded AMGs showed that rumen viruses possess glycoside hydrolases (GHs) that increase the breakdown of complex carbohydrates, thereby increasing energy production [15]. Mangalea et al. suggest that Rheumatoid arthritis (RA) risk individuals have diverse phage communities that may alter intestinal metabolism through the introduction of phage-derived specific AMGs [16]. Viruses in mangrove sediments encode abundant auxiliary carbohydrate-activate enzyme (CAZyme) genes, most of which are necessary for the degradation of complex polysaccharides [17]. Chu et al. revealed that viral AMGs contribute to the enhancement of host responses to environmental gradients and drive the biogeochemical cycle of mariculture sediments [18]. However, there is currently limited knowledge about the viruses present in the gut of pigs, such as their identity, their survival strategy and if they contain novel AMGs.
Environmental factors have been identified as potential factors influencing viral communities [[23], [24], [25]]. Gu et al. found that six viromes from three saline lakes on Qinghai-Tibet Plateau were correlated with elevation, latitude, and concentrations of magnesium and calcium [23]. A study on the virome of the South China Sea found that surface viruses and deep viruses were affected by different environmental factors [24]. Surface viruses were affected by carbon flux and temperature. Deep viruses were associated with depth and salinity. Zuo et al. also found that the human gut DNA virome was affected by geography, diet, urbanization and ethnicity [25]. And geography had the strongest impact of the examined factors on the human gut virome variation [25]. However, other studies have reported opposite results [8,26]. Analysis of freshwater viral community from six different sites in the Yangtze River showed that viral community structure was independent of geographical location [26]. A study of pig virome across China also pointed out that the viral community structure was independent of geographical location [8]. In addition, farm management and biosafety are also potential factors affecting the viral community [8]. They found that administration, isolated housing, and hygiene of farms significantly affected the structure of pig viral community [8]. So, one focus of our study was to explore the influence of environmental factors on the pig gut virome.
Fecal samples were collected from 25 healthy pigs at six farms in Shanxi province. Shanxi Province in China is located between latitude 34°34′-40°44′ N and longitude 110°14′-114°33′ E. It is located in the mid-latitude inland and belongs to the temperate continental monsoon climate. Due to the influence of solar radiation, monsoon circulation and geographical factors, the climate of Shanxi is characterized by four distinct seasons and significant differences between the north and the south. The average annual temperature in Shanxi Province ranges from 4.2 °C to 14.2 °C, with an overall trend of increasing from north to south and decreasing from the basin to the mountains. Datong Lingqiu farm (DTLQ) is located in northern Shanxi. Taiyuan Qingxu farm (TYQX), Jinzhong Taigu farm (JZTG) and Jinzhong Shouyang farm (JZSY) are located in the central part of Shanxi. Changzhi Lucheng farm (CZLC) and Jincheng Zezhou farm (JCZZ) are located in the south of Shanxi (Fig. 1, Table S2). In this study, virus-enriched metagenome sequencing and virome-specific bioinformatics tools were used to investigate: (1) the effects of feeding environment on pig gut virome, (2) the viral communities possible roles in the pig gut.
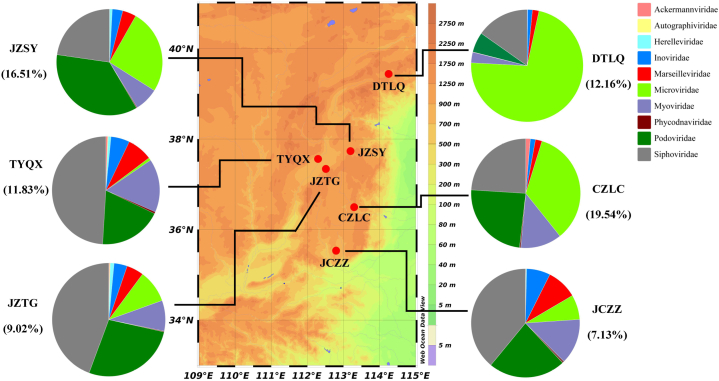
Fig. 1.
The geographical distribution of pig farms and the pig gut viral community structure of classified vOTUs in six regions of Shanxi Province, China. The pie charts show relative abundance on viral community composition, assessed for all vOTUs at the family level based on the results of CAT, vConTACT2 and mmseq2 “easy-taxonomy”. The proportion of annotated vOTUs in total vOTUs for each pig farm is marked below the farm name.
2. Method details
2.1. Experimental model and subject details
Fresh pig fecal samples were collected from six farms in the Shanxi province of China (Fig. 1, Table S2). These farms had the same feeding standards and were owned by the same company. Pigs are fed according to the pig nutritional requirements standards (GB/T 39,235–2020) (Table S14). There were 25 pig fecal samples were obtained and stored in a portable ice box. The quantity of feces collected from six farms was as follows: five fecal samples were collected from TYQX and four fecal samples were collected from each of the other five farms. Next, they were transferred the laboratory within 24 h. Samples were stored at −80 °C until DNA extraction. Our experiment was approved by the Institutional Animal Care and Use Committee of Shanxi Agricultural University to meet the requirements of animal ethical welfare. The project number is SXAU-EAW-2022P·CU.1,005,003.
2.2. Fecal VLP nucleic acid extraction
Fecal virus-like particles (VLPs) were extracted using the method described by Shkoporov et al. but with minor modifications [27]. Aliquots of 1.5 g of pig feces were resuspended in 30 mL of SM buffer [NaCl 5.8 g, MgSO4·7H2O 2.0 g, 1 mol/L Tris·HCl 50 mL (pH 7.0), 2% gelatin 5 mL, add pure water to 1 L, and store at 4 °C] and strongly vortexed for 5 min. The centrifuge tube was shaken on a shaker for 30 min. Tubes were then chilled on ice for 5 min prior to centrifugation at 5000 g in a centrifuge (Beckman) for 10 min at +4 °C. Supernatants were transferred to another 50 mL tube, and centrifugation was repeated once again. Supernatant was subsequently filtered twice through a 0.22-μm PES membrane. NaCl particles and PEG-8000 powders were then added to the filtrate to give a final concentration of 0.5 M and 10% w/v, respectively. After complete dissolution, samples were incubated overnight (24 h) at + 4 °C.
On the following day, the samples were centrifuged at 5000 g for 20 min at + 4 °C to collect viral particles (Fig. S1). Supernatant was removed, and tubes were left in inverted position on paper towels for 5 min to remove last traces of supernatant. Viral particles were then resuspended in 250 μL of SM buffer. To ensure that the collected virus is not disturbed by free DNA in the environment, DNase I is added to pellets at 37 °C for 2 h to remove free DNA [17]. Extra DNase Ⅰ was deactivated at 70 °C for 10 min.
2.3. Viral DNA extraction
The viral DNA was extracted by using Nucleic acid extraction reagent (magnetic bead method, 502-B Type) according to the manufacturer's protocols (GENFINE). The extracted DNA was measured concentration using Qubit 3 Fluorometer and gel electrophoresis.
2.4. Shotgun sequencing of viromes
At the beginning, we were worried that the concentration of viral DNA extracted was too low to complete the construction of the library. Therefore, samples 3 and 4 from each farm were mixed and sequenced separately, including DTLQ3_4, TYQX3_4, JZSY3_4, JZTG3_4, CZLC3_4, JCZZ3_4. Since DTLQ samples 1 and 2 had less sample mass, they were mixed together and sequenced. The remaining 14 samples were sequenced separately, including TYQX1, TYQX2, TYQX4, TYQX6, JZTG1, JZTG5, JZSY1, JZSY2, CZLC1, CZLC5, JCZZ2, JCZZ3, JCZZ4, JCZZ6 (Table S2). We are using the same batch of samples. The 14 fecal samples were sequenced separately to explore individual differences in pig gut virome. All viral DNA samples were subjected to shotgun metagenomic sequencing by using the Illumina Novaseq6000 platform. Libraries were prepared with a fragment length of approximately 350 bp. Paired-end reads were generated using 150 bp in the forward and reverse directions.
2.5. Pig gut DNA virome assembly and identification
The quality control of raw reads was performed using fastp (0.19.7 --cut_tail -W 4 -M 20 -n 5 -c -l50 -w 3) [28]. Then bwa (0.7.17-r1194-dirty) was used to map reads to the pig reference genome (Sus_scrofa.Ss-crofa11.1. dna.toplevel.fa.gz) for removing host contaminated reads [29]. Each sample was individually assembled using MEGAHIT (v1.2.9) with the options “--min-count 2 --k-min 27 --k-max 87 --k-step 10 --min-contig-len 500”. The unassembled reads underwent a second round of assembly and were then combined with the first round of assembly [5,30]. A downstream analysis was performed for longer than 5 kb contigs and they were extracted by seqkit (2.2.0) [31]. After that, viral contigs were identified by Yan et at.‘s protocol [32]. Longer than 5 kb contigs were identified as viruses when they satisfied one of the following conditions: 1, use VIBRANT's default parameter to identify viruses (-virome mode -l 5000) [33]. 2, the number of viral genes is more than microbial genes based on searching against the CheckV marker gene set [34]. 3, P < 0.01 and score >0.9 in the DeepVirFinder, a virus sequence-based k-mers word frequency and machine learning method for virus sequence recognition [35] (an upgraded version of VirFinder) [36]. Viral contigs were pairwise blasted and the highly consistent viruses with 95% nucleotide identity and 80% coverage of the sequence were further clustered into viral operational taxonomic units (vOTUs) using CD-hit-est [37]. The obtained vOTUs were evaluated for the completeness of genomes and filtered out host-contaminated regions in viral contigs by CheckV [34].
2.6. Calculating relative abundances and taxonomic annotation of virome
To calculate the relative abundance of viral communities in each farm, an index of 3584 vOTUs was built using Bowtie2 [38]. Next, clean reads in each sample were mapped each vOTU using SAMtools (v1.9) [39]. Then, the CheckM (v1.2.0) was used to get the number of reads mapped in each viral contig [40]. Finally, Reads per Kilobase per million Mapped Reads (RPKM) was used as the relative abundance of the viral contigs.
Classification annotations in viral contigs at the family level use the following three methods: First, (1) CAT based on the lowest common ancestor (LCA) algorithm [41]. (2) vConTACT2 based on gene-sharing networks [42]. (3) Use NCBI nr database (June 2022) to extract the virus sequences corresponding to the international Committee on Taxonomy of Viruses (ICTV), using mmseqs2 “easy-taxonomy” mode to annotate viral contigs (https://github.com/apcamargo/ictv-mmseqs2-protein-data-base). Pig gut virome was compared with the reference virus genome (virus RefSeq201) using vConTACT2 [42] (--rel-mode ‘Diamond’ --pcs-mode MCL --vcs-mode ClusterONE). At the same time, each farm pig gut virome was also compared using the same way. The final results were presented using Cytoscape v3.9.1 software (http://cytoscape.org/), using an edge-weighted spring embedded model, which allows shared viral clusters (VCs) to be displayed in a closer manner [43].
2.7. Phylogenetic analyses of terL gene
The large terminase subunit gene terL is a key feature of Caudovirales (or tailed dsDNA phages). It is highly conserved in phage phylogeny [17,44]. As a result, it was utilized to construct a genetic tree to evaluate the diversity of pig gut Caudovirales viruses. Pfam domains of TerL proteins of pig gut viral contigs were searched against the Pfam 33.1 database [45] using the hmmsearch program in the HMMER3 package (E-value 1e-5) [46] using the method described by Ma et al. [47], and four domains, including Terminase_1 (PF03354), Terminase_3 (PF04466), Terminase_GpA (PF05876) and Terminase_6 (PF03237). The TerL proteins of viral references were download form NCBI Viruse database (https://www.ncbi.nlm.nih.gov/labs/virus/) in May 2022, including “TerL”, “terminase large subunit” and “large terminase subunit” proteins. TerL protein sequences from pig gut virome and viral reference were aligned using MUSCLE v3.8.31 (-Super5) [48]. Gaps were trimmed using trimAl v.1.4 (-gappyout) [49]. Phylogenetic reconstruction was performed using the Fasttree (-wag) program [50]. The tree was manually edited by iTOL v6 [51].
2.8. Virus-host prediction
Host CRISPR-spacers match was used to predict viral hosts. Spacers come from two parts. First, CRT [52] and PILER-CR [53] were used to identify spacers (https://git-hub.com/snayfach/MGV/tree/master/crispr_spacers) from 24 pig gut metagenomic datas (Bingzhen Ji et al., unpublished data) and 11,804 spacers were got from microbial hosts. Besides, 720,391 spacers were predicted using CRISPRFinder and PILER-CR from bacterial and archaea sequences [54]. The viral sequences were matched spacers using balstn (-task blastn-short -evalue 1e-5 -outfmt 6 -dust no -word_size 18). And final results were filtered with bit score ≥45 [55,56]. CAT [41] was used to classify contig of pig gut microbial hosts. And only contigs with annotated results were retained. Finally, if a bacterium produced the highest number of spacers hits, it was considered as the primary host.
2.9. Functional profiles and identification of viral AMGs
Antibiotic resistance genes were identified using four tools: (1) ARG Resfinder [57] (E-value 1e-5), (2) the NCBI AMRfinder tool v3.10.24 using default options [58], (3) the Resistance Gene Identifier (RGI) v5.2.1, CARD v3.2.4 [59], (4) ABRicate (https://github.com/tsee-mann/abricate). 5300 viral contigs were verified for the presence or absence of ARGs before virus clustering. VIBRANT and NCBI nt databases were used to determine the source of these contigs. VIBRANT was also used to determine whether the contigs were linear or circular. All virus-encoded proteins were blasted against the eggNOG database v5.0.2 [60] using a python script, emapper. py v2.1.7, with the parameters --matrix BLOSUM62 --evalue 1e-5 --dbtype seqdb -m diamond [61]. And the clusters of orthologous groups (COGs) information for each protein was assigned and count the number of categories.
For viral AMGs identification, the following two methods were mainly used: (1) CAZymes from these viral open reading frames (ORFs) were identified on the dbCAN web server based on CAZyme family-specific HMMs [62]. ORFs related to carbohydrate metabolism were compared with NCBI nr database and Pfam database to determine the best annotation. (2) To identify downstream AMGs, CheckV [34] was used to remove the host contaminated region of each viral contig. DRAM-v (DRAM-v.py) [63] was uesd to annotate pig gut vOTUs and recover their putative AMGs. Due to DRAM-v required VirSorter2 (v2.2.3) [64] output, 3119 viral contigs with high viral scores (>0.5) were selected throuth VirSorter2 (--prep-for-dramv). Next, the default parameters of DRAM-v were used to identify viral AMGs. From the DRAM-v output, putative AMGs with auxiliary scores <4 and gene descriptions were retained. As a complement, the default parameters of the VIBRANT were used to identify viral AMGs [33]. For VIBRANT results, those annotated as “energy metabolism” of those annotations were considered to be a potential AMGs [19]. A manual calibration method was used for getting high confidence AMGs based on DRAM-v and VIBRANT outputs [65]. As previously reported [66,67], to ensure the reliability of viral AMGs, the genes related to nucleotide metabolism, organic nitrogen, ribosomal proteins and glycosyltransferases were not considered as viral AMGs. Furthermore, if a putative AMG was located between two viral hallmark genes or viral genes, or be close to viral hallmark gene or viral gene, the gene was considered to be a high confidence viral AMGs for the subsequent analysis [19]. For identification of virus genomic context, genes adjacent to AMG were searched to the UniProt virus database for getting annotations (2022.3 release, -e 1e-5). For these AMGs, the conserved domains of viral AMGs were identified by NCBI CD-search tool [68] and the Phyre2 online was used to search the protein structural homology [69].
Previous studies have shown that viral AMGs can be derived by host horizontal gene transfer [70]. Thereby phylogenetic trees of virus and reference microbial hosts AMGs can be constructed to predict host of these viruses [17,71,72]. For getting the relevant reference sequences of the relevant species, the amino acid sequences of AMGs were searched against the NCBI nr database (online comparison) (-e 1e-5 and bitscore >50) [17,72]. To establish an AMG phylogenetic tree, the amino acid sequences from the reference sequences and viral AMG sequence were blasted by MEGA11 [73]. And a neighbor-joining tree was built and NO. Of Bootstrap Replications was 1000.
3. Quantification and statistical analysis
The α-diversity (Simpson's H and Shannon's H) statistics were performed using vegan (v 2.5–6) in Rstudio (v February 1, 5001; R Stats v 3.6.1), based on the relative abundances of vOTUs. Viral communities of PCoA and CCA used https://bioincloud.tech/. Viral genome quality assessment used GraphPad Prism8. The sankey diagram of virus host prediction and AMGs gene cluster were made by https://www.chiplot.online/
4. Results
4.1. An overview of six pig gut viromes
Fecal samples from pigs were collected in six different regions of Shanxi Province. All pig farms are shown in Fig. 1. The biological repeats for each region were as follows: DTLQ = 2, JZTG = 3, JZSY = 3, TYQX = 5, CZLC = 3 and JCZZ = 5. Prior to metagenomic sequencing, the PEG8000+NaCl physical precipitation method was used for VLP enrichment. The sequencing volume of each library is approximately 1G. And twenty-one viral metagenomic libraries were obtained after Illumina sequencing, containing approximately 21G bases raw data (a total of 120, 009, 418 raw paired-end reads [150 bp each-end]) (Table S2). After quality control, six viromes contained a total of approximately 75 million paired clean reads. A total of 17,790 contigs longer than 5 kb were assembled by MEGAHIT. The combination of the prediction outputs of VIBRANT, DeepVirFinder and CheckV [[33], [34], [35]] resulted in the detection of 5300 viral contigs longer than 5 kb (Fig. S2A). Based on 95% nucleotide similarity and 80% coverage, 3584 vOTUs were obtained (Fig. S2A). There were a total of 365 viral contigs that had more than 50% completeness, with 143 of them having more than 90% completeness (Table S3, Fig. S2B). There were 1264 viral contigs that were more than 10 kb and 3 viral contigs more than 100 kb. The longest contig was 156,869 bp. A total of 90 provirus sequences were found by CheckV [34], ranging from 90 bp to 51,316 bp (Table S3).
4.2. Pig gut viral community structure
In order to explore the difference of pig gut viral composition among six regions of Shanxi, the composition of annotated vOTUs at the family level was compared in each region. The vOTUs were annotated using CAT, vContact2 and a custom mmseqs2 viral protein database [41,42] and represented an average relative abundance of 12.70% at the family level. (Fig. 1). Siphoviridae (15.21%∼49.03%) and Podoviridae (5.81%∼35.78%) were the abundant families of these pig gut viromes. However, the Microviridae [74], a nonenveloped ssDNA viruses, occupied a high proportion (72.41% and 34.45%) in the DTLQ and CZLC. The Phycodnaviridae, a Nucleocytoplasmic large DNA viruses, were also discovered in all samples based on mmseqs2 “easy-taxonomy”. There was a similar viral composition between TYQX and JZTG, while the other four pig gut viromes had a big difference. Inoviridae were also prevalent in the pig gut virome from six regions, which was consistent with previous studies [10]. In addition, the composition of gut virome of each pig was also individual specific at the family level (Fig. S3).
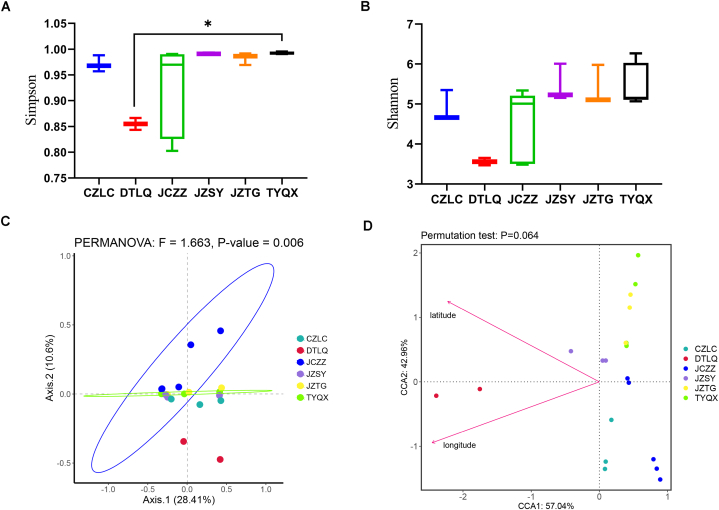
Simpson and Shannon indexes were analyzed to explore whether there was a difference in the alpha diversity of pig gut DNA viromes in six different regions. The Simpson index of the pig gut virome from DTLQ was significantly lower than that virome from TYQX (p < 0.05) (Ordinary one-way ANOVA, Fig. 2A). In addition, the Shannon index of six pig gut DNA viromes was not significantly different. (Kruskal-Wallis test, Fig. 2B). Pig gut viromes in Shanxi Province were related to geography by principal-coordinate analysis (PCoA) (Bray-Curtis distance, Fig. 2C). The pig gut viromes from TYQX, JZTG, JZSY and CZLC were very similar. While JCZZ and DTLQ's pig gut viromes showed significant variation from the pig gut viromes in other four regions respectively (p < 0.01) (Fig. 2C). Canonical Correspondence analysis (CCA) analysis was also used to explore the association between pig gut viral communities and environmental factors. Taking into account consistent feeding standards, we tentatively propose that geographical location could be a factor impacting the pig gut viral community, despite the small sample size. (p < 0.1) (Fig. 2D).
Fig. 2.
A comparison of the pig gut viral community diversity in six regions of Shanxi Province and their relationship with geography. Simpson (A) and Shannon (B) indices of the viral community diversity from six regions of Shanxi Province. (C) PCoA of pig gut viromes from six regions. (D) CCA of the relationship between pig gut viral communities and geographical locations. Asterisks denote significance, with * indicating p < 0.05.
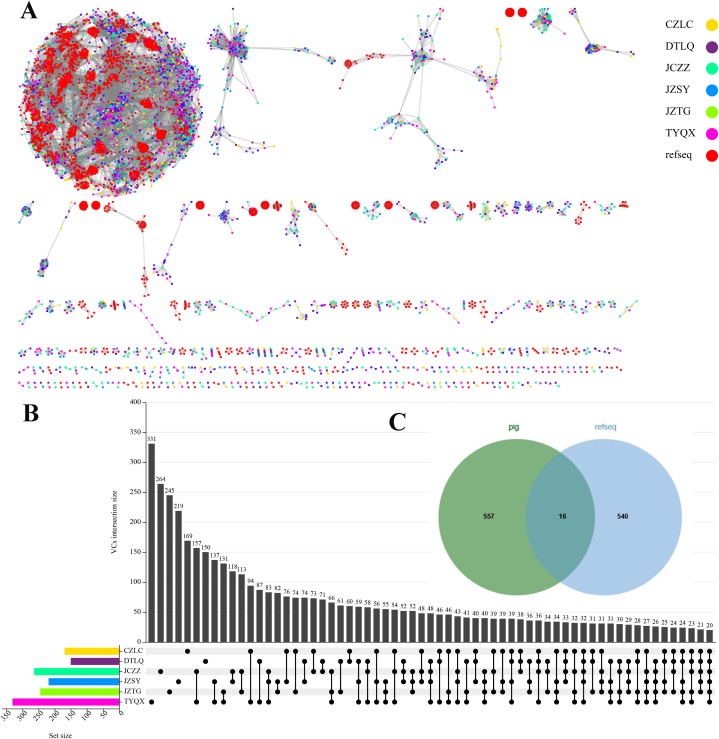
In order to compare the composition of pig gut virome in different regions of Shanxi and the differences between the discovered pig gut virome and the reference database, a gene sharing network was constructed using vContact2 [42]. There were only 20 viral clusters (VCs, with a very high probability of the same genus in a VC) shared by the viruses in the six regions of Shanxi Province, suggesting that there were significant differences among different regions of pig gut virome (Fig. 3A and B). Only 16 VCs were shared between the pig gut and viral RefSeq201, suggesting that there's still a lot of dark matter in the pig gut virome. (Fig. 3C). Interestingly, Oengus-like Phages were also found at VC152_0 b y cluster analysis of vContact2 (Table S5). It is a particularly large one of 171 elements, 170 of which originated from pig gut viral contigs. Of note, one reference phage genome was related to them. It is temperate phage Oengus (Oengus-like Phages) infected the Firmicute Faecalibacterium prausnitzii [75].
Fig. 3.
Comparison of pig gut virome based on gene sharing network in six regions of Shanxi Province. (A) Gene sharing network diagram of pig gut virome in six regions of Shanxi Province and reference genome. (B) The upset graph shows the differences in the composition of pig gut virome based on virus clusters in six regions of Shanxi Province. (C) The venn diagram shows the differences in the composition of the pig gut virome and the reference genome based on virus clusters.
4.3. Phylogenetic tree of terL gene
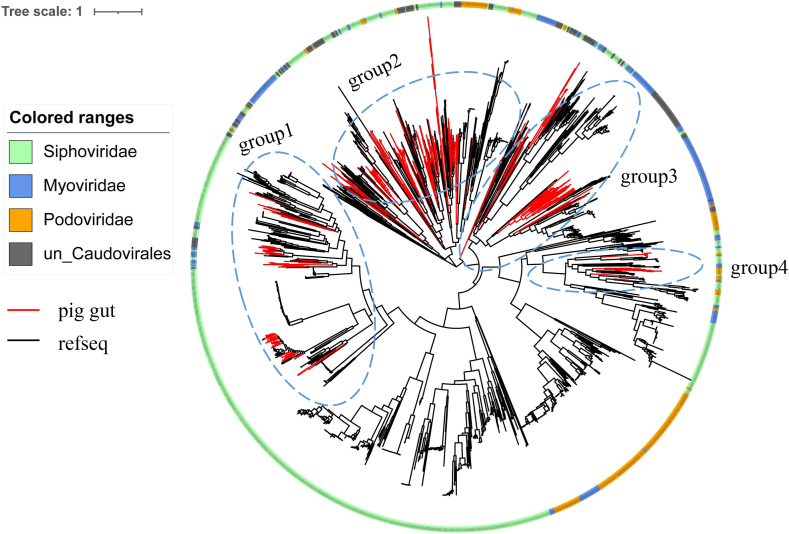
Caudovirales viruses are the most abundant viruses in pig gut, and the terL gene is highly conserved among these viruses. Phylogenetic analysis of TerL proteins was used to assess the diversity and genetic distance of pig gut virome. A total of 319 pig gut terL genes and 1586 reference terL genes were used to construct a phylogenetic tree. To date, although Caudovirales have been well studied, there were still phylogenetic distances between pig gut Caudovirales sequences and the known reference viral sequences. Pig gut Caudovirales sequences are divided into four groups (Fig. 4). As shown in Fig. 4, most of the viral sequences were Siphoviridae (n = 1154), followed by Myoviridae (n = 324) and Podoviridae (n = 274). In addition, similar results were obtained for pig gut Caudovirales sequences. Most of the pig gut Caudovirales sequences were Siphoviridae (n = 154), followed by Myoviridae (n = 37) and Podoviridae (n = 11). The clades of pig gut Caudovirales sequences enriched previously uncharacterized diversity and defined new branches of Caudovirales. There is a new branch of viruses that contains one reference protein, thermus phage P23-45 (NC_009803.1) and 43 pig gut viruses. It suggested that these pig gut phages had the ability to infect thermus bacteria.
Fig. 4.
Phylogenetic tree of terL genes of Caudovirales from pig gut virome and reference viral sequences. The red lines represent pig gut virome sequences and the black lines represent reference viral sequences.
4.4. Host prediction of viruses
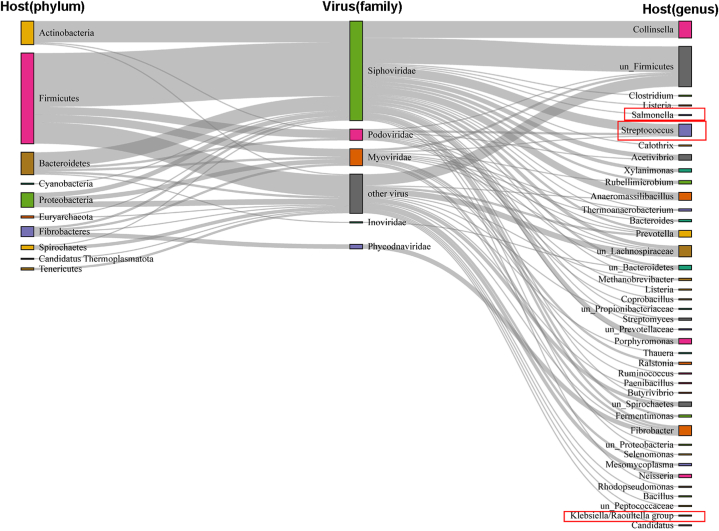
By host CRISPR-spacers match, 154 viruses of the 3584 vOTUs were able to assign the potential hosts at the phylum level (10 phyla) and the genus level (40 genera) (Fig. 5). For host prediction of viruses, the use of the bulk metagenomes from the same pig farm could improve the rate of viral host prediction, as compared to use reference genomes from bacteria and archaea alone. It was consistent with previous results [18]. Most viruses had a narrow host range, which only infected one phyla. Only five viruses were able to infect two phyla, one virus can infect three phyla, which is consistent with previous results [18,66,76,77]. The number of hosts for these viruses was Firmicutes (n = 81), Actinobacteria (n = 21) and Bacteroidetes (n = 20), followed by Proteobacteria (n = 13) at the phyla level. Interestingly, there were three viruses associated with two archaea phyla, including Euryarchaeota and Candidatus Thermoplasmatota. These results illustrated that pig gut virome was strongly associated with microbial hosts, which in turn affected the gut environment.
Fig. 5.
Host prediction of pig gut virome. Sankey diagram shows that the association between pig gut virome and host based on host CRISPR-spacers match. The red boxes are the pathogenic bacteria.
4.5. ARGs annotation of pig gut virome
ARGs were detected in 5300 viral contigs using different antibiotic resistance gene annotation software. The search against Resfinder [57] revealed that one viral contig encoded one putative ARG (e = 8.12e-11). In order to further verify whether the viral contigs carried ARGs, AMRfinder, RGI and ABRicate were used [58,59]. Surprisingly, only one contig was detected in four softwares (Table 1). It encoded a blaVIM-48-1 gene. The origins of this contig were determined using VIBRANT software and NCBI nt database (August 2022) [33]. Although this contig was annotated as a virus by the result of VIBRANT. After alignmenting with the NCBI nt database, this contig turned out to be 99.77% idential to Sphingobium sp. TKS plasmid pTK5 (Accession CP005089.1). In addition, this ∼6.5 kb contig was linear and contained a 49 kb deletion compared to plasmid pTK5. So, it was rather a plasmid fragment than a virus. In summary, only one contig (0.019%) out of 5300 contigs, or below 0.0179 per Mb of assembled viral contigs, may carry ARG. It was revealed that the pig gut virome rarely encodes ARGs.
Table 1.
The one ARG-positive contig is derived from pig gut virome.
| Contig_ID | ARG Resfinder name | Resfinder (E-val) | AMRfinder | RGI | ABRicate | Contig properties |
|||
|---|---|---|---|---|---|---|---|---|---|
| Phage VIBRANT | best BLASTn hit | Size,nt | RPKM in viromes | ||||||
| k87_1425 | blaVIM-48-1 | 8.12e-11 | No | No | No | Yes | TKS plasmid PTK5 (complete) (99.77%) 53,908 nt | 6448 nt | 194.33 |
4.6. Functional annotation and identification of AMGs from pig gut virome
In order to explore functional diversity of pig gut virome, the predicted ORFs of viral contigs were annotated by searching against the clusters of orthologous groups (COGs) in the eggNOG database. Only 26.22% of the viral open reading frames (ORFs) (13,746 out of 52,425) were assigned to some COG classes (Table S8). 51.43% of the viral ORFs (7069 out of 13,746) were “function unknown”, indicating the presence of a large number of unidentified viral genes in the pig gut virome (Fig. S4). Although the ORFs of annotated viruses are widely distributed across most COG function categories, most of the ORFs are related to conventional viral functions, primarily “replication, recombination, and repair”, “cell wall/be/envelope biogenesis”, “transcription” and “nucleotide transport and metabolism” (Fig. S4, Table S8). These functions are essential for viral reproduction and morphogenesis.
A large number of CAZymes genes in pig gut virome were identified using the dbCAN server. These CAZymes genes mainly included Glycoside Hydrolases (GHs), Carbohydrate Esterases (CEs), Auxiliary Activities (AAs), Glycosyl-Transferases (GTs) and Carbohydrate-Binding Modules (CBMs) (Fig. S5). 52.74% of CAZymes genes (346 out of 656) were GHs (Fig. S5, Table S7). Viral AMGs were also identified by viral protein annotation using VIBRANT and DRAM-v software, with manual curation. These AMGs were mainly involved in carbohydrate metabolism, sulfur metabolism and cofactor biosynthesis. Finally, 16 AMGs had strongly supported (>98% confidence) structures predictions as enzymes involved in carbon metabolism, sulfur metabolism, cofactor biosynthesis. 93.75% of them (15 out of 16) possessed the conserved functional domains (Table S12).
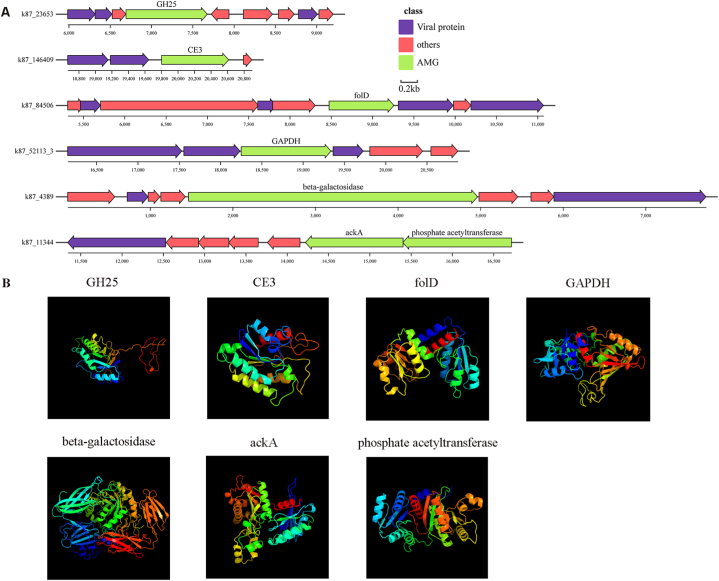
In general, AMGs of seven viruses were involved in carbon metabolism, including the genes of glycoside hydrolases (GH25), Carbohydrate Esterases (CE3), methenyltetrahydrofolate cyclohydrolase (folD), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), acetate kinase (ackA), phosphate acetyltransferase and beta-galactosidase (Fig. 6A). Protein tertiary structure of seven viral AMGs associated with carbon metabolism was shown in Fig. 6B. Viral GH25, CE3 and GAPDH may be involved in the breakdown of complex polysaccharides in pig gut. Besides, beta-galactosidase was involved in galactose metabolism and contributed to energy production. The acetate kinase (ackA)–phosphate acetyltransferase pathway controled acetyl phosphate levels. Viral ackA was predicted to convert acetate to acetyl phosphate. And viral phosphate acetyltransferase was hypothesized to convert acetyl phosphate to acetyl-CoA [78]. So, the viral ackA and phosphate acetyltransferase potentially participated in inorganic carbon fixation. Viral folD genes and a large number of glycine hydroxymethyltransferase (glyA) genes in pig gut virome suggested that there was a reprogramming of carbon metabolism to folate metabolism, similar to what had been observed in the rumen of bovine [15].
Fig. 6.
Genomic context and protein tertiary structure of AMGs related to carbon metabolism. (A) Genomic context of viral contigs. AMGs are in green, viral genes are in blue, and non-virus-like genes are in pink. (B) Protein tertiary structure of viral AMGs.
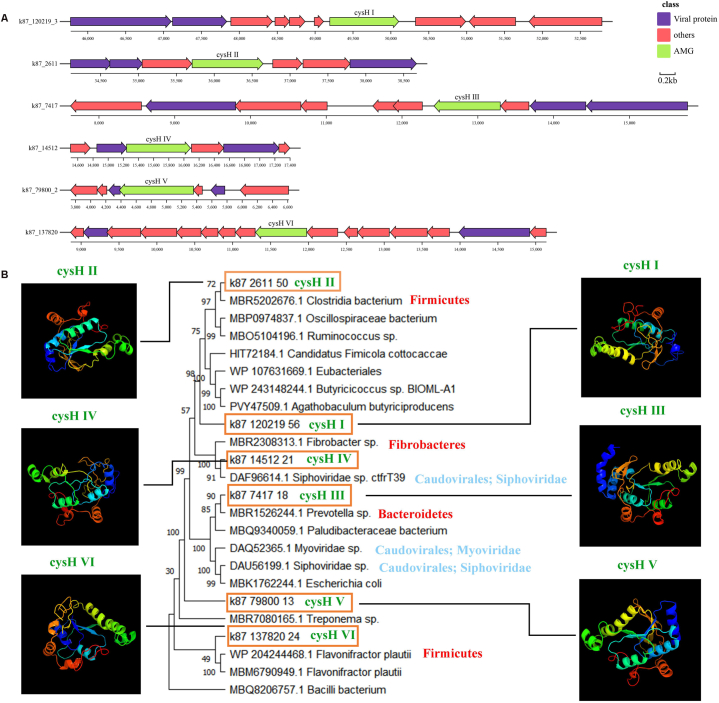
In addition, seven viral AMGs involved in sulfur metabolism were identified too. Among them, six viral AMGs (cysH Ⅰ to Ⅵ) encoded phosphoadenosine phosphoreductases (cysH) genes (Fig. 7A), involved in assimilative sulfate reduction [79]. The remaining one was viral cytochrome subunit of sulfide dehydrogenase (fccA) gene, which can serve as the electron transport subunit of sulfide dehydrogenase (Table S12). As for the viral AMGs involved in cofactor biosynthesis, two viral genes encoding cobaltochelatases (CobS and CobT) participate in cobalamin (vitamin B12) biosynthesis (Table S12).
Fig. 7.
Genomic context, protein tertiary structure and phylogenetic tree of viral cysH gene. (A) Genomic context of viral contigs containing cysH gene. AMGs are in green, viral genes are in blue, and non-virus-like genes are in pink. (B) Tertiary structure and phylogenetic tree of viral cysH gene. It contains 6 pig gut virome cysH genes and 18 reference sequences, 3 of which are reference viral sequences.
Viral AMGs usually come from the host via horizontal gene transfer [70]. Those viruses encoding AMGs were predicted potential hosts based on AMG phylogenetic trees. The host of seven AMGs associated with carbon metabolism was predicted (Figure S6, S7 and S8). For example, phylogenetic analysis showed that viral GH25 clustered with GH25 from Firmicutes and Bacteroidetes (Fig. S6A). It indicated that this AMG was possibly derived from phages infecting these bacteria. Besides, six cysH gene involved in sulful metabolism may be related to Frimicutes, Fibrobacteres and Bacteroidetes (Fig. 7B).
5. Discussion
5.1. Novelty and predominant families of viruses in pig gut
Although some progress has been made in understanding the pig gut virome [8,10,80], our study found that the pig gut still contains a lot of “viral dark matter” and the role of viruses in pig gut needs further exploration. The lack of classification at the family level for most viral contigs found in the pig gut, as well as the lack of homology for more than half of viral ORFs in public databases, suggests that these viruses are novel. There are two primary reasons for this: (1): most viral sequences share little homology with current reference databases [81]. (2) Viral genomes lack universal marker genes that could aid in their taxonomic assignment. Although much research has been conducted on Caudovirales, there is still a great deal of unknown information about them. The Caudovirales in pig gut virome were characterized by phylogenetic tree and gene-sharing network. The four clades of Caudovirales were identified by using the terL gene. These clades have the potential to be new viruses in phylogeny. The novelty was also confirmed at the approximate genera level based on gene-sharing network. It is a lower taxonomic resolution (i.e. a higher taxonomic rank) compared to the vOTU level, which can overcome the virome individuality and allow cohort comparisons [66].
Caudovirales is the dominant viral group among the viruses that have been classified in a variety of environments, including rumen, mangrove sediments and human gut [15,17,56], similar results were obtained with pig gut. Siphoviridae accounted for the largest proportion in pig gut virome and also represented a large proportion of viruses in human gut [56]. A global survey of ocean viral genomes showed that the Siphoviridae is the third most common family of viruses [82]. However, Microviridae has the highest proportion in DTLQ and CZLC (Fig. 1). There are three reasons for this situation: (1): it may be due to the individual specific of pig gut virome (Fig. S3). This is also seen in the human gut virome [56]. (2): it is possible that exaggerated PCR amplification bias during viral DNA preparation [83]. (3): it is possible that the limited classification of viral contigs is impacting the composition of pig gut virome at the family level. Besides, our study found that inoviruses and Oengus-like phages were rich in pig gut virome, which is consistent with previous research [10].
5.2. Effects of environmental factors on pig gut virome
The gut microbiome is influenced by a variety of factors, such as dietary patterns [84] and living conditions [85], and the environment plays a greater role in shaping the gut microbiome than the host's genetic background [86]. The composition of viral communities in the human gut is greatly influenced by geography [25,[87], [88], [89]]. Although previous study have shown that the viral composition of each pig farm is not affected by geographical location [8]. However, our experiment controlled the same feeding standard and designed a good model for further exploring the potential impact of environmental factors on pig gut virome. Through PCoA analysis, we found that the gut viral communities of pig farms in different locations in Shanxi present regional differences. Although, according to CCA analysis, geographical location did not significantly affect the composition of the pig gut viruses. The viral community in the DTLQ pig farm still showed a relatively large separation trend from the other five areas (Fig. 2D). It may be due to the unique viral community structure that was formed by environmental factors. The Microviridae bacteriophages accounted for a large proportion of the gut viruses in DTLQ pig farms and also showed its unique viral community structure (Fig. 1). In fact, geography can affect both altitude and latitude, which in turn can impact temperature. This, in turn, may play a role in viral diversity [23]. Although the geographical location did not have an impact on the composition of pig gut virome dramatically, the temperature difference among northern, central and southern Shanxi may have been a contributing factor to the variation of pig gut virome. At the same time, we are aware that, limited by our sample size, it is insufficient to answer the question of environmental influences on pig gut viruses. In future studies, we will increase the number of samples and environmental parameters investigated to further explore the influence of environmental factors on the pig gut viruses.
5.3. Potential value of pig gut viruses for phage therapy
It is believed that antibiotic resistance in bacteria is caused by intensive pig farming [91,92]. The use of phage therapy to fight drug-resistant bacteria has regained popularity in recent years [93]. Although bacteriophages themselves can promote the horizontal transfer of antibiotic resistance genes [11], our conclusion offered prospects for the application of bacteriophages. Here, phages encode few antibiotic resistance genes were proved in pig gut. Only 1 contig (0.019%) out of 5300 viral contigs or below 0.0179 per Mb of assembled viral contigs was considered as ARG-containing contig in pig gut (Table 1). None of the ARG-containing contig appears to belong to active phage, which is consistent with previous research [10]. It indicates that the feasibility and safety of phage therapy with pig gut viruses. In addition, previous study have shown that phage-mediated transfer of ARGs is overestimated [11]. Experimental testing was also needed to test whether a predicted ORF is an ARG. When using a phage in practice, pre-experiments need to be done to fully study the dynamic curve of specific bacteria and its phage to evaluate the application value of the phage.
Furthermore, some pathogenic bacteria were found based on host CRISPR-spacers match, such as Streptocuss, Klebsiella and Salmonella (Fig. 5). It revealed that the abundant phages in the pig gut have the potential to kill pathogenic bacteria. These results suggest that viruses from pig guts could be used as new antimicrobial agents. The majority of pig gut phages infect Firmicutes and Bacteroidetes, both of which are abundant in the gut and highly adapted to its environment. Compared to a previous study [8], Fibrobacteres, Spirochaetes, Cyanobacteria and Candidatus Thermoplasmatota were found to have a relationship with pig gut phages for the first time (Fig. 5). This enriches the contact between pig gut viruses and microbial hosts. To sum up, these host-virus linkages may be important for designing phage therapies or understanding host-virus co-evolutionary dynamics in the future.
5.4. Viral AMGs and their potential functions
To the best of our knowledge, this is the first time that AMGs have been extracted from pig gut viruses. Based on the annotation results of VIBRANT and DRAM-v, a number of AMGs involved in carbon metabolism, sulfur metabolism, and cofactor biosynthesis were identified (Figs. 6 and 7 and Table S12).
Viral AMGs for carbon metabolisms have been extensively investigated in many environments, such as the marine, rumen and mangrove soil [15,17,94]. A majority of them are involved in central carbon metabolism to promote viral replication [15,17]. According to previous sduty from the marine environment, central carbon AMGs shift host microbial metabolism to mimic a state of starvation [94]. Virally encoded glycogen synthase interferes with host glycolysis and forces host metabolism away from amino acid biosynthesis and towards pathways that support phage replication [15,17,94]. Because complex carbohydrates or polysaccharides are highly difficult to degrade, AMGs associated with carbon metabolism in many environments may contribute to viral reproduction and host metabolism [15,17]. It is believed that pig feedstuff often contains starch, non-starch polysaccharide, crude fiber and other nutrients that are difficult to digest and absorb. It is suspected that viruses that are related to carbon metabolism paly a role in the digestion and absorption of pig gut. The viral GH25, CE3 and beta-galactosidase may contribute to the degradation of polysaccharides. AckA and phosphate acetyltransferase work together to generate acetyl-CoA, which can then be used by the virus for energy during replication. In addition, similar to rumen viruses [15], we found that pig gut viruses have extensive reprogramming of carbon metabolism toward folate biosynthesis. Tetrahydrofolate (THF) can serve as a donor of one-carbon unit and enable various functions in three different states: 5-methyl-THF is important for the reaction of homocysteine to methionine, 5,10-methylene-THF was needed for the conversion of deoxyuridylate (dUMP) to deoxythymidine monophosphate (dTMP), and 10-formyl-THF was a formyl donor in de novo purine biosynthesis reactions [95,96]. Viral fold gene was detected and found to be responsible for the interconversion of 5,10-methylene-THF and 10-formyl-THF. The viral glyA gene was also found in the pig gut, encoding the reversible interconversion of serine to glycine and THF to 5,10-methylene-THF. Besides, the phylogenetic analysis of AMGs showed that viral CAZyme genes are diverse and probably derive from phages infecting distinct hosts of different phyla, suggesting that auxiliary carbohydrate metabolic genes may be widespread in pig gut viruses.
So far, some progress has been made in the study of virus-associated sulfur metabolism in human and environmental systems [97,98]. Sulfur metabolism has been found to potentially affect human gut homeostasis [97]. The cysH gene can be involved in the assimilation reduction of sulfate, which is required for cysteine and methionine synthesis [79,99]. In studies of phage infection of Burkholderia, it has been proposed that cysH can act as a fitness factor during sulfur-limiting periods [100]. Here, a large number of viral AMGs genes that assist sulfur metabolism, including cysH and fccA genes (Fig. 7 and Table S12) were found in our study. Therefore, the viral cysH gene may help pig gut viruses adapt to complex gut environment and affect gut homeostasis. The host of the virus encoding the cysH gene was compared between pig gut and the Mariah Trench [19]. There was a significant difference between the two environments. Through data analysis, we preliminarily come to this conclusion: viruses containing the cysH gene can infect Firmicutes in the pig gut, but this is not observed in the Mariah Trench. At the same time, some differences were found based on the homologous structure comparison of the proteins. We hypothesized that it might be caused by the difference between the gut environment and the Marine environment, suggesting that pig gut might be a unique environment for viruses to inhabit. In addition, fccA gene can also participate in sulfate assimilation and reduction, and similar results have been found in mariculture sediments [18].
6. Conclusions
The complex gut microbial ecosystem of pig has a significant impact on body health and productivity through the breakdown and digestion of feed into available energy. Despite the growing understanding of the functions and importance of viruses in other ecosystems, little is known regarding the roles and dynamics of pig gut viruses. In this study, for the first time, the differences in pig gut virome from six regions of Shanxi Province are revealed. The pig gut virome is individual specific. At the same time, there is a potential relationship between the pig gut virome and geography. Phylogenetic tree analysis of terL gene has expanded our understanding of pig gut virome. There was little resemblance between pig gut virome and viral RefSeq based on gene-sharing network. In addition, the potential function of pig gut virome were investigated. Pig gut virome encoded almost no ARGs and contained abundant AMGs related to carbon and sulfur metabolism. In conclusion, our study provides new insights into the differences of pig gut virome in different regions and the viral communities possible roles in the pig gut.
Author contribution statement
Junjun Qin: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Bingzhen Ji: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Yijia Ma: Contributed reagents, materials, analysis tools or data.
Xin Liu: Contributed reagents, materials, analysis tools or data.
Tian Wang: Contributed reagents, materials, analysis tools or data.
Guiming Liu: Contributed reagents, materials, analysis tools or data.
Bugao Li: Analyzed and interpreted the data.
Guoliang Wang: Conceived and designed the experiments; Analyzed and interpreted the data.
Pengfei Gao: Conceived and designed the experiments; Analyzed and interpreted the data.
Funding statement
This work was funded by the Financial Special Foundation from Beijing Academy of Agriculture and Forestry Sciences (no. CZZJ202202), Basic Re-search Project of Shanxi Province (Grant No. 202203021211268), Shanxi Agricultural University 2022 Industry Leading Project (CYYL22-13), Open Fund of Shanxi Provincial Key Laboratory of livestock and poultry Genetic Resources Exploration and Precision Breeding and the Natural Science Foundation of Shanxi Province, China.
Data availability statement
Data associated with this study has been deposited at The raw reads for 21 viral metagenomes are stored in the NCBI Sequence Read Archive (SRA) with accession numbers PRJNA888429.
Declaration of interest's statement
The authors declare no conflict of interest.
Additional information
Supplementary content related to this article has been published online at [URL].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e14020.
Contributor Information
Junjun Qin, Email: junjun_qin@163.com.
Guoliang Wang, Email: wangglpro@163.com.
Pengfei Gao, Email: gpf800411@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Lynch S.V., Pedersen O. The human intestinal microbiome in health and disease. N. Engl. J. Med. 2016;375(24):2369–2379. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Turnbaugh P.J., Ley R.E., Mahowald M.A., et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 3.Qin J., Li R., Raes J., et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao L., Estellé J., Kiilerich P., et al. A reference gene catalogue of the pig gut microbiome. Nature Microbiology. 2016;1 doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 5.Chen C., Zhou Y., Fu H., et al. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021;12(1):1106. doi: 10.1038/s41467-021-21295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patil Y., Gooneratne R., Ju X.-H. Interactions between host and gut microbiota in domestic pigs: a review. Gut Microb. 2020;11(3):310–334. doi: 10.1080/19490976.2019.1690363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia B., Wu W., Zhang L., et al. Gut microbiota mediates the effects of inulin on enhancing sulfomucin production and mucosal barrier function in a pig model. Food Funct. 2021;12(21):10967–10982. doi: 10.1039/d1fo02582a. [DOI] [PubMed] [Google Scholar]
- 8.He B., Gong W., Yan X., et al. Viral metagenome-based precision surveillance of pig population at large scale reveals viromic signatures of sample types and influence of farming management on pig virome. mSystems. 2021;6(3) doi: 10.1128/mSystems.00420-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shkoporov A.N., Hill C. Bacteriophages of the human gut: the "known unknown" of the microbiome. Cell Host Microbe. 2019;25(2):195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Billaud M., Lamy-Besnier Q., Lossouarn J., et al. Analysis of viromes and microbiomes from pig fecal samples reveals that phages and prophages rarely carry antibiotic resistance genes. ISME Communications. 2021;1(1):55. doi: 10.1038/s43705-021-00054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enault F., Briet A., Bouteille L., et al. Phages rarely encode antibiotic resistance genes: a cautionary tale for virome analyses. ISME J. 2017;11(1):237–247. doi: 10.1038/ismej.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison E., Brockhurst M.A. Ecological and evolutionary benefits of temperate phage: what does or doesn't kill you makes you stronger. Bioessays : News and Reviews In Molecular, Cellular and Developmental Biology. 2017;39(12) doi: 10.1002/bies.201700112. [DOI] [PubMed] [Google Scholar]
- 13.Breitbart M. Marine viruses: truth or dare. Ann. Rev. Mar. Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- 14.Rohwer F., Thurber R.V. Viruses manipulate the marine environment. Nature. 2009;459(7244):207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 15.Anderson C.L., Sullivan M.B., Fernando S.C. Dietary energy drives the dynamic response of bovine rumen viral communities. Microbiome. 2017;5(1):155. doi: 10.1186/s40168-017-0374-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangalea M.R., Paez-Espino D., Kieft K., et al. Individuals at risk for rheumatoid arthritis harbor differential intestinal bacteriophage communities with distinct metabolic potential. Cell Host Microbe. 2021;29(5) doi: 10.1016/j.chom.2021.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin M., Guo X., Zhang R., et al. Diversities and potential biogeochemical impacts of mangrove soil viruses. Microbiome. 2019;7(1):58. doi: 10.1186/s40168-019-0675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu Y., Zhao Z., Cai L., et al. Viral diversity and biogeochemical potential revealed in different prawn-culture sediments by virus-enriched metagenome analysis. Environ. Res. 2022;210 doi: 10.1016/j.envres.2022.112901. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J., Jing H., Wang Z., et al. Novel viral communities potentially assisting in carbon, nitrogen, and sulfur metabolism in the upper slope sediments of Mariana Trench. mSystems. 2022;7(1) doi: 10.1128/msystems.01358-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emerson J.B., Roux S., Brum J.R., et al. Host-linked soil viral ecology along a permafrost thaw gradient. Nature Microbiology. 2018;3(8):870–880. doi: 10.1038/s41564-018-0190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bi L., Yu D.-T., Du S., et al. Diversity and potential biogeochemical impacts of viruses in bulk and rhizosphere soils. Environ. Microbiol. 2021;23(2):588–599. doi: 10.1111/1462-2920.15010. [DOI] [PubMed] [Google Scholar]
- 22.Labbé M., Girard C., Vincent W.F., et al. Extreme viral partitioning in a marine-derived high arctic lake. mSphere. 2020;5(3) doi: 10.1128/mSphere.00334-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu C., Liang Y., Li J., et al. Saline lakes on the Qinghai-Tibet Plateau harbor unique viral assemblages mediating microbial environmental adaption. iScience. 2021;24(12) doi: 10.1016/j.isci.2021.103439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y., Wang L., Wang Z., et al. Metagenomic analysis of the diversity of DNA viruses in the surface and deep sea of the South China sea. Front. Microbiol. 2019;10:1951. doi: 10.3389/fmicb.2019.01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo T., Sun Y., Wan Y., et al. Human-gut-DNA virome variations across geography, ethnicity, and urbanization. Cell Host Microbe. 2020;28(5):741–751 e744. doi: 10.1016/j.chom.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Lu J., Yang S., Zhang X., et al. Metagenomic analysis of viral community in the Yangtze River expands known eukaryotic and prokaryotic virus diversity in freshwater. Virol. Sin. 2022;37(1):60–69. doi: 10.1016/j.virs.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shkoporov A.N., Ryan F.J., Draper L.A., et al. Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome. 2018;6(1):68. doi: 10.1186/s40168-018-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S., Zhou Y., Chen Y., et al. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li D., Luo R., Liu C.-M., et al. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods (San Diego, CA, U. S.) 2016:102. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 31.Shen W., Le S., Li Y., et al. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan Q., Wang Y., Chen X., et al. Characterization of the gut DNA and RNA viromes in a cohort of Chinese residents and visiting Pakistanis. Virus Evolution. 2021;7(1):veab022. doi: 10.1093/ve/veab022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kieft K., Zhou Z., Anantharaman K. VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome. 2020;8(1):90. doi: 10.1186/s40168-020-00867-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayfach S., Camargo A.P., Schulz F., et al. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat. Biotechnol. 2021;39(5):578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren J., Song K., Deng C., et al. Identifying viruses from metagenomic data using deep learning. Quantitative Biology (Beijing, China) 2020;8(1):64–77. doi: 10.1007/s40484-019-0187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren J., Ahlgren N.A., Lu Y.Y., et al. VirFinder: a novel k-mer based tool for identifying viral sequences from assembled metagenomic data. Microbiome. 2017;5(1):69. doi: 10.1186/s40168-017-0283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu L., Niu B., Zhu Z., et al. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Handsaker B., Wysoker A., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parks D.H., Imelfort M., Skennerton C.T., et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Von Meijenfeldt FaB., Arkhipova K., Cambuy D.D., et al. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 2019;20(1):217. doi: 10.1186/s13059-019-1817-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bin Jang H., Bolduc B., Zablocki O., et al. Taxonomic assignment of uncultivated prokaryotic virus genomes is enabled by gene-sharing networks. Nat. Biotechnol. 2019;37(6):632–639. doi: 10.1038/s41587-019-0100-8. [DOI] [PubMed] [Google Scholar]
- 43.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adriaenssens E.M., Cowan D.A. Using signature genes as tools to assess environmental viral ecology and diversity. Appl. Environ. Microbiol. 2014;80(15):4470–4480. doi: 10.1128/AEM.00878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mistry J., Chuguransky S., Williams L., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49(D1):D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 47.Ma Y., You X., Mai G., et al. A human gut phage catalog correlates the gut phageome with type 2 diabetes. Microbiome. 2018;6(1):24. doi: 10.1186/s40168-018-0410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price M.N., Dehal P.S., Arkin A.P. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bland C., Ramsey T.L., Sabree F., et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinf. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edgar R.C. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinf. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shmakov S.A., Sitnik V., Makarova K.S., et al. The CRISPR spacer space is dominated by sequences from species-specific mobilomes. mBio. 2017;8(5) doi: 10.1128/mBio.01397-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nayfach S., Páez-Espino D., Call L., et al. Metagenomic compendium of 189,680 DNA viruses from the human gut microbiome. Nature Microbiology. 2021;6(7):960–970. doi: 10.1038/s41564-021-00928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shkoporov A.N., Clooney A.G., Sutton T.D.S., et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527–541 e525. doi: 10.1016/j.chom.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 57.Bortolaia V., Kaas R.S., Ruppe E., et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feldgarden M., Brover V., Haft D.H., et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019;63(11) doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alcock B.P., Raphenya A.R., Lau T.T.Y., et al. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020;48(D1):D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huerta-Cepas J., Szklarczyk D., Heller D., et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huerta-Cepas J., Forslund K., Coelho L.P., et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34(8):2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin Y., Mao X., Yang J., et al. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–W451. doi: 10.1093/nar/gks479. Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaffer M., Borton M.A., Mcgivern B.B., et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48(16):8883–8900. doi: 10.1093/nar/gkaa621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo J., Bolduc B., Zayed A.A., et al. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9(1):37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pratama A.A., Bolduc B., Zayed A.A., et al. Expanding standards in viromics: in silico evaluation of dsDNA viral genome identification, classification, and auxiliary metabolic gene curation. PeerJ. 2021;9 doi: 10.7717/peerj.11447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li Z., Pan D., Wei G., et al. Deep sea sediments associated with cold seeps are a subsurface reservoir of viral diversity. ISME J. 2021;15(8):2366–2378. doi: 10.1038/s41396-021-00932-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ter Horst A.M., Santos-Medellín C., Sorensen J.W., et al. Minnesota peat viromes reveal terrestrial and aquatic niche partitioning for local and global viral populations. Microbiome. 2021;9(1):233. doi: 10.1186/s40168-021-01156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu S., Wang J., Chitsaz F., et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kelley L.A., Mezulis S., Yates C.M., et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sullivan M.B., Lindell D., Lee J.A., et al. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4(8):e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roux S., Brum J.R., Dutilh B.E., et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537(7622):689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- 72.Edwards R.A., Mcnair K., Faust K., et al. Computational approaches to predict bacteriophage-host relationships. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2016;40(2):258–272. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013;30(5):1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- 74.Doore S.M., Fane B.A. The microviridae: diversity, assembly, and experimental evolution. Virology. 2016;491:45–55. doi: 10.1016/j.virol.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 75.Cornuault J.K., Petit M.-A., Mariadassou M., et al. Phages infecting Faecalibacterium prausnitzii belong to novel viral genera that help to decipher intestinal viromes. Microbiome. 2018;6(1):65. doi: 10.1186/s40168-018-0452-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., et al. Uncovering Earth's virome. Nature. 2016;536(7617):425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 77.Roux S., Hallam S.J., Woyke T., et al. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife. 2015:4. doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ingram-Smith C., Martin S.R., Smith K.S. Acetate kinase: not just a bacterial enzyme. Trends Microbiol. 2006;14(6):249–253. doi: 10.1016/j.tim.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 79.Bick J.A., Dennis J.J., Zylstra G.J., et al. Identification of a new class of 5'-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J. Bacteriol. 2000;182(1):135–142. doi: 10.1128/jb.182.1.135-142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shkoporov A.N., Stockdale S.R., Lavelle A., et al. Viral biogeography of the mammalian gut and parenchymal organs[J] Nature Microbiology. 2022;7(8):1301–1311. doi: 10.1038/s41564-022-01178-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aggarwala V., Liang G., Bushman F.D. Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mobile DNA. 2017;8:12. doi: 10.1186/s13100-017-0095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gregory A.C., Zayed A.A., Conceição-Neto N., et al. Marine DNA viral macro- and microdiversity from Pole to Pole. Cell. 2019;177(5) doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parras-Moltó M., Rodríguez-Galet A., Suárez-Rodríguez P., et al. Evaluation of bias induced by viral enrichment and random amplification protocols in metagenomic surveys of saliva DNA viruses. Microbiome. 2018;6(1):119. doi: 10.1186/s40168-018-0507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nelson T.M., Rogers T.L., Carlini A.R., et al. Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 2013;15(4):1132–1145. doi: 10.1111/1462-2920.12022. [DOI] [PubMed] [Google Scholar]
- 85.Amato K.R., Yeoman C.J., Kent A., et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes. ISME J. 2013;7(7):1344–1353. doi: 10.1038/ismej.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rothschild D., Weissbrod O., Barkan E., et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 87.Monaghan T.M., Sloan T.J., Stockdale S.R., et al. Metagenomics reveals impact of geography and acute diarrheal disease on the Central Indian human gut microbiome. Gut Microb. 2020;12(1) doi: 10.1080/19490976.2020.1752605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rampelli S., Turroni S., Schnorr S.L., et al. Characterization of the human DNA gut virome across populations with different subsistence strategies and geographical origin. Environ. Microbiol. 2017;19(11):4728–4735. doi: 10.1111/1462-2920.13938. [DOI] [PubMed] [Google Scholar]
- 89.Holtz L.R., Cao S., Zhao G., et al. Geographic variation in the eukaryotic virome of human diarrhea. Virology. 2014;468–470:556–564. doi: 10.1016/j.virol.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu Y.-G., Johnson T.A., Su J.-Q., et al. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. U. S. A. 2013;110(9):3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang R., Van Dorp L., Shaw L.P., et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 2018;9(1):1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kortright K.E., Chan B.K., Koff J.L., et al. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe. 2019;25(2):219–232. doi: 10.1016/j.chom.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 94.Hurwitz B.L., U'ren J.M. Viral metabolic reprogramming in marine ecosystems. Curr. Opin. Microbiol. 2016;31:161–168. doi: 10.1016/j.mib.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 95.De Crécy-Lagard V., El Yacoubi B., De La Garza R.D., et al. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genom. 2007;8:245. doi: 10.1186/1471-2164-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13(8):572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kieft K., Breister A.M., Huss P., et al. Virus-associated organosulfur metabolism in human and environmental systems. Cell Rep. 2021;36(5) doi: 10.1016/j.celrep.2021.109471. [DOI] [PubMed] [Google Scholar]
- 98.Kieft K., Zhou Z., Anderson R.E., et al. Ecology of inorganic sulfur auxiliary metabolism in widespread bacteriophages. Nat. Commun. 2021;12(1):3503. doi: 10.1038/s41467-021-23698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Woods D.E., Jeddeloh J.A., Fritz D.L., et al. Burkholderia thailandensis E125 harbors a temperate bacteriophage specific for Burkholderia mallei. J. Bacteriol. 2002;184(14):4003–4017. doi: 10.1128/JB.184.14.4003-4017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Summer E.J., Gonzalez C.F., Bomer M., et al. Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. J. Bacteriol. 2006;188(1):255–268. doi: 10.1128/JB.188.1.255-268.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at The raw reads for 21 viral metagenomes are stored in the NCBI Sequence Read Archive (SRA) with accession numbers PRJNA888429.