Abstract
Aphid species (Insecta, Hemiptera) are economically important invasive pest throughout the world, though their identification is intricate due to tiny size and inconspicuous nature of morphology. Mitochondrial cytochrome c oxidase I (mtCOI) region has been proven to be a standard barcode to identify the diverse array of insect groups. Isolation of good quality DNA is a fundamental first step in insect DNA barcoding which is obtained by standardizing the DNA isolation method. In this study, we demonstrate a modified CTAB method for the isolation of DNA to maximize the quality and yield from small aphids. This method will help the researchers to efficiently isolate DNA from small aphid and the method can be utilized for other small insects as well. We evaluated the quality of the isolated DNA and the mtCOI gene region were subjected to PCR amplification. Further, the gene segment was sequenced and gene annotation was done by NCBI BLAST program through which the insect was found to be Aphis gossypii. This study provides a set of molecular tools that can be used for identification of insect at species level through DNA barcoding and biodiversity analysis.
-
•
Detailed method to maximize quality and quantity of genomic DNA isolated from aphids.
-
•
Molecular identification of aphids using mtCOI gene amplification and sequence validation.
-
•
First report on Aphis gossypii infecting Solanum trilobatum provides insights of pest identification and management.
Keywords: Aphid, biodiversity; COI; DNA barcoding; Insect DNA; Modified CTAB; PCR
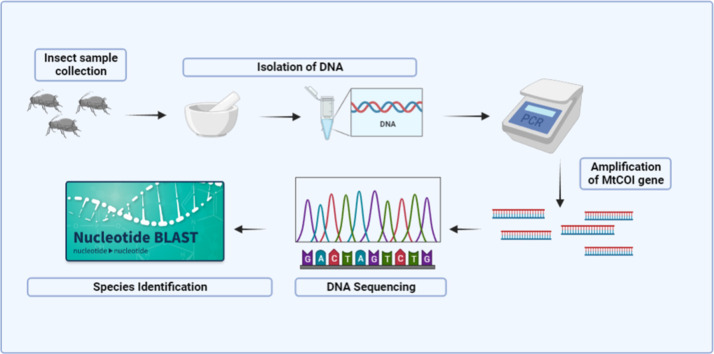
Method name: DNA Extraction and PCR amplification of mtCOI gene
Graphical abstract
Specifications table
| Subject area: | Biochemistry, Genetics and Molecular Biology |
| More specific subject area: | Molecular identification of Aphids |
| Name of your method: | DNA Extraction and PCR amplification of mtCOI gene |
| Name and reference of original method: | M. Salah, Aljanabi, I. Martinez, Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques, Nuc. Acids Res. 25 (1997) 4692-4693. 10.1093/nar/25.22.4692. |
| Resource availability: | A. gossypii MtCOI gene sequence (NCBI Submission ID: A. gossypii: 2636395) |
Method details
Background
Aphids (Hemiptera: Aphididae), are a polyphagous insect cause heavy crop loss by virus transmission and by colonizing the leaves and stem of the plant. Aphids have become a worldwide threat to many agricultural crops [1,2]. In India, 530 species of medicinal plants belonging to 117 plant families were infested by 428 species/sub-species of aphids. Study further reveals that among the aphids, A. gossypii complex is highly observed species in 224 plant species [3]. Due to its sucking nature, aphids cause direct physical damage by sucking the amino acid and carbohydrate from plant phloem which leads to significant yield reduction [4]. Crop loss due to aphid infestation and control measures are calculated around US$200 million annually [5]. Proper identification of insect species is necessary for the integrated pest management and for early detection [6].
Traditional morphological identification of aphids needs the examination and analysis of various features viz., characteristics of antennae, siphunculi and caudas, even though several keys are exist, morphological differentiation of aphids is difficult due to the notable similarities between species and intraspecific variation [7]. Aphid's tiny size is another drawback for morphological identification. In order to overcome difficulties in morphological examination, molecular method of identification have been employed for easy identification of economically relevant aphid species. DNA barcoding has been projected as a standardized molecular identification of insect species in any life stages [8]. Molecular characterization of insect was done by amplifying the mitochondrial gene called cytochrome c oxidase 1 COI. Several research studies reported that the region of 710bp was successfully used as a standard barcode for the identification of insect pests [9,10]. mtCOI region has high mutation speed and highly conserved through which insect species and subspecies can be easily identified.
Isolation of high quality and quantity DNA from small insects is often difficult and fundamental first step in insect barcoding also in invasion studies, phylogenetic analysis and evolutionary genetics. Generally, organic extraction is commonly followed for isolating DNA from varied insect orders by considering cost effectiveness and high yield [11]. DNA isolation from small insects using commercial kits are high cost hence rapid, cost-effective method for DNA isolation from small insects is necessary to obtain a good quality DNA for further molecular studies. Organic extraction is convenient by following steps including addition of CTAB (Cetyltrimethylammonium Bromide) extraction buffer to break the insect cuticle, protein separation by phenol-chloroform and DNA precipitation by isopropanol/ethanol precipitation. Also this method is mostly preferred in many laboratories due to easy availability of laboratory chemicals and high quantity of DNA [12]. In this study, modified CTAB method was followed to isolate the aphid DNA by following short incubation time after the addition of lysis buffer and the usage of β- mercaptoethanol was avoided by considering the health concern like severe skin and eye irritation and destructive to respiratory track and central nervous system. Also, PCR sensitivity of isolated DNA was checked by amplifying the mtCOI region using PCR. The present study reports a cheap, simple and rapid method for isolating DNA from small insects and molecular identification of aphid in S. trilobatum. COI gene amplification and sequence analysis confirmed that the insect belongs to the genus Aphis and the species of gossypii. A. gossypii species reported in many agricultural plants [13,14] however, in this study first time reported in S. trilobatum.
Insect sample
The aphids were found in the leaves and stem of the S. trilobatum plants in the area of Maduravoyal, Tiruvallur, Tamil Nadu, India. Several colonization was observed in the leaves which further affects the growth of the plants. As, S. trilobatum is a medicinal plant, leaves are the main source which are heavily colonized by aphids (Fig.1). Adult aphids such as A. gossypii were collected using tweezers and stored in 70% ethanol. Ethanol was added to avoid bacterial or fungal DNA contamination during DNA isolation. The tubes were stored at 4 °C until further studies.
Fig. 1.
Identification of Aphids in the leaves of S. trilobatum plant.
Reagents and solutions
The reagents and chemicals used are CTAB lysis buffer [5% CTAB, 100 mM Tris-HCI (pH 8.0), 0.5 M sodium chloride, 10 mM ethylenediaminetetraacetic acid (EDTA)], chloroform: isoamylalcohol (24:1), isopropanol, 75% ethanol, Tris EDTA (TE) buffer (10 mM Tris-HCl, 1 mM disodium EDTA, pH 8.0).
Procedure for isolation of DNA from Aphid sample

Determination of quantity and purity of isolated DNA
DNA sample was separated in 1% agarose gel containing ethidium bromide (EtBr) and visualized under UV transilluninator. Also, the DNA sample was diluted 20 times with ddH2O and quantified using a spectrophotometer (Amersham biosciences).
Validation of DNA quality and integrity using PCR amplification and sequencing
Folmer fragment of the 5′ region of COI was amplified using polymerase chain reaction (PCR) by using forward primer LCO (5′- GGTCAACAAATCATAAAGATATTGG-3′) and reverse primer HCO (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) [15]. PCR reaction was performed in a total volume of 25 µL reaction mixture containing 1 µL template DNA (50–150 ng), 4.7 µL 10X PCR buffer (contains 25mM magnesium chloride), 1 µL of forward and reverse primer (10 µmol each), 2 µL dNTP mixture (0.2 mmol/L each of dATP, dCTP, dGTP, and dTTP), 0.3 µL Taq DNA polymerase (3U/µl), and final volume made with 16 µL distilled water. The PCR mixture was processed in a Thermal Cycler (eppendorf) with 35 cycles, each cycle consists of 3 min of pre-denaturation at 94 °C, followed by 30 cycles of amplification (40 sec of denaturation at 94 °C; 1 min of annealing at 47 °C for COI; 45 s of extension at 72 °C), and final extension at 72 °C for 10 min. The amplified PCR products were separated on 1.2% agarose gel/ EtBr (0.5 µg/mL). Sequencing of COI gene was performed by Barcode Biosciences, (Bangalore, Karnataka) by following Sanger method of sequencing.
Method validation
Validation of modified CTAB method by COI gene amplification
In this study, modified CTAB method was effectively extracted genomic DNA from single aphid. The evaluation of integrity of the genomic DNA showed clear DNA bands on 1% agarose gel (Fig. 2) which proves that no DNA degradation had occurred during the isolation process. Spectrophotometric analysis reveals that A260/A280 ratios were greater than 1.7. The DNA concentration was measured as 120 ng/µL and yield recovery was ∼2000 ng. Usage of toxic solvents (β- mercaptoethanol and phenol) was eliminated in this method. LCO/HCO forward and reverse primer was successfully utilized for the COI gene amplification and intact/clear band was observed. The PCR product was resolved on 1.2% agarose gel and 1Kb DNA marker was used for identifying the size of CO1 gene amplicon. The amplification was visible at 709 bp (Fig. 3).
Fig. 2.
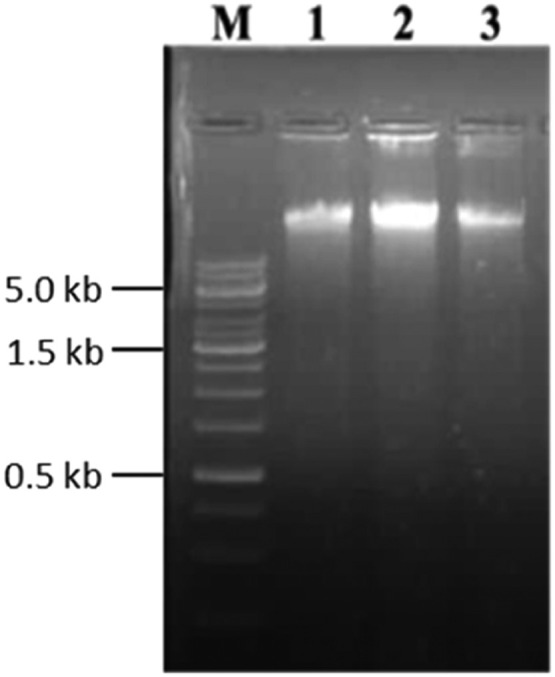
Genomic DNA isolation from Aphid using modified CTAB method Lane M. 1Kb DNA Marker, Lane 1-3. Aphids DNA with replications.
Fig. 3.
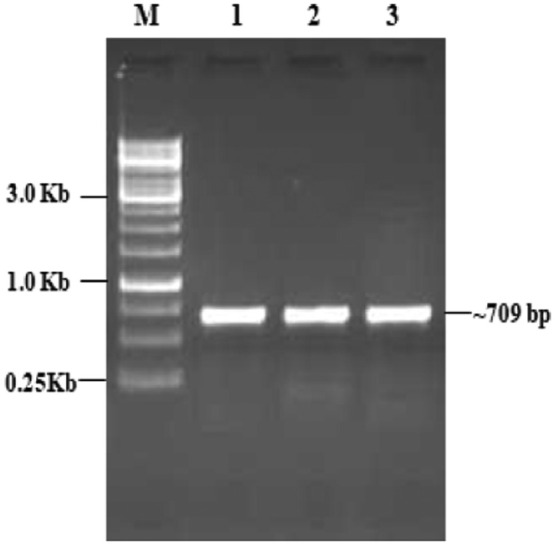
PCR amplification of CO1 gene using DNA isolated from aphid using modified CTAB Method. Lane M. 1Kb DNA Marker, Lane 1-3. Aphids CO1 gene amplified as replications.
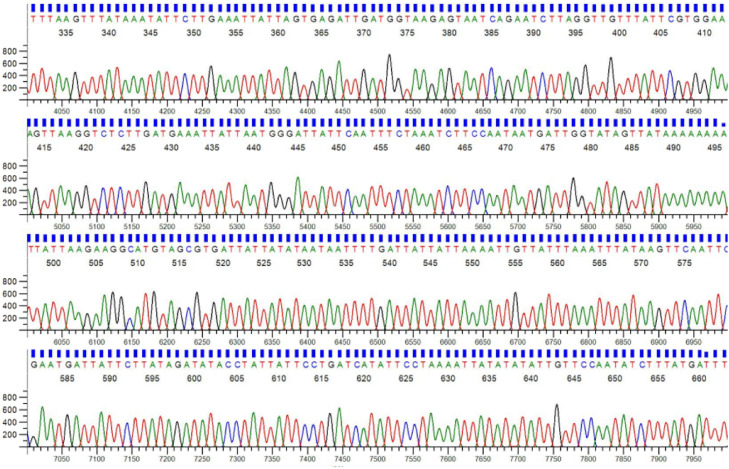
The results shows that DNA samples isolated by modified CTAB method successfully utilized by the LCO/HCO primers in the PCR reaction and showed clear and thick band of expected amplicon size 709 bp in the gel which indicates the modified method provided good quality DNA with sufficient quantity. COI gene was sequenced using Sanger sequencing method and good chromatogram was obtained (Fig. 4). Finally, the COI gene sequence was blasted against the National Centre for Biotechnology Information (NCBI) database and the sequence showed 99% similarity with previously reported sequence of A. gossypii. Sequence analysis revealed that collected aphidbelongs to the genus Aphis and species gossypii.
Fig. 4.
Chromatogram of a fragment of the COI gene.
DNA barcoding is a recent taxonomic method that uses a short region of a gene as a marker in an organism's DNA to identify the particular species. The main principle in DNA barcoding technique is the standardization of DNA isolation and PCR reaction for mtCOI gene amplification [16]. To reduce the complications and easing the work of the researchers, organism may be identified by standardized molecular technique by adopting standardized DNA isolation procedure [17]. Sometimes, DNA barcoding of the tiny insects may be failed due to the difficulty in isolating the DNA. Still, many successful manual methods were optimised for getting high quantity and high yield of insect DNA from tiny insect specimen [18]. Aphids are considered to be a serious agricultural insect due to the nature of spreading plant virus and related diseases (Black man and Eastop.2000). Rose plants are severely infested by rose aphids further stops the growth of leaf bud, flower bud and twigs which cause high annual losses [19]. In North America, soybean aphid (Aphis glycines) was noted as a exotic pest and cause considerable economic losses every year [20].
Conclusion
In conclusion, it is quite sensitive and challenging to obtain clean DNA from soft-bodied insects like aphids, we have developed modified CTAB method for DNA isolation from aphids. This method yields good quality and quantity of DNA, also with good stability in storage at 4°C for a short period. This method of DNA extraction could be a feasible option for cost effective and time saving to screen large populations. Easy amplification of COI gene in aphid DNA indicates that the proposed modified DNA isolation method is reliable for molecular identification of agricultural important aphids. First time, COI gene marker was successfully used for molecular identification of aphid in S. trilobatum plant. NCBI BLAST analysis revealed that the aphid belongs to the species gossypii. Accurate identification of insects will pave a way for insect pest management.
Ethics statements
We have used Aphids for our experimental studies, and we have followed the safety and laboratory guidelines for Aphids handling as per the institutional norms.
Declaration of Competing Interest
Please tick the appropriate statement below (please do not delete either statement) and declare any financial interests/personal relationships which may affect your work in the box below.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Please declare any financial interests/personal relationships which may be considered as potential competing interests here.
Acknowledgments
The authors sincerely acknowledge the Department of Biotechnology, Vels Institute of Science, Technology and Advanced Studies (VISTAS), for providing the laboratory and research facilities.
Contributor Information
M. Suganthi, Email: sugenthi88@gmail.com.
Senthilkumar Palanisamy, Email: mpsenthilkumar@gmail.com.
Data availability
Data will be made available on request.
References
- 1.Furlan L., Pozzebon A., Duso C. An update of the worldwide integrated assessment (WIA) on systemic insecticides. part 3: alternatives to systemic insecticides. Environ. Sci. Pollut. Res. 2021;28:11798–11820. doi: 10.1007/s11356-017-1052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo K., Zhao H., Wang X., Kang Z. Prevalent pest management strategies for grain aphids: opportunities and challenges. Front. Plant. Sci. 2022 doi: 10.3389/fpls.2021.790919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R., Ruhi A., Sharmila P. Records of aphids infesting Indian medicinal plants. J. Aphidol. 2003;17:1–58. [Google Scholar]
- 4.Louis J., Singh V., Shah J. Arabidopsis thaliana-Aphid interaction. Arab. Book. 2012;10:e0159. doi: 10.1199/tab.0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jangra S., Ghosh A. Rapid and zero-cost DNA extraction from soft-bodied insects for routine PCR-based applications. PLOS One. 2022;17(7) doi: 10.1371/journal.pone.0271312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller G.L., Foottit R.G. Vol. 20. Blackwell Publishing; 2017. The taxonomy of crop pests: the aphids. (Insect Biodiversity: Science and Society). [DOI] [Google Scholar]
- 7.Pereira G.B., Valente V., Queiroz M.S., Vianna M.C., Paco-Larson M.L. Overexpression of kermit/dGIPC is associated with lethality in drosophila melanogaster. Braz. J. Med. Biol. Res. 2011;44(4):283–290. doi: 10.1590/s0100-879x2011007500022. [DOI] [PubMed] [Google Scholar]
- 8.Floyd R.M., Wilson J.J., Hebert P.D.N. Insect Biodiversity: Science and Society. Blackwell Publishing; 2009. DNA barcodes and insect biodiversity. [DOI] [Google Scholar]
- 9.Kinyanjui G., Khamis F.M., Ombura F.L.O., Kenya E.U., Ekesi S., Mohamed S.A. Infestation levels and molecular identification based on mitochondrial COI barcode region of five invasive gelechiidae pest species in Kenya. J. Eco. Entomol. 2019;112(2):872–882. doi: 10.1093/jee/toy357. [DOI] [PubMed] [Google Scholar]
- 10.Suganthi M., Chandrashekara K.N., Arvinth S., Raj Kumar R. Molecular characterization of tea mosquito bug from tea growing regions of India. Mitochondrial DNA A DNA Mapp. Seq. Anal. 2016;27(5):3504–3506. doi: 10.3109/19401736.2015.1066369. [DOI] [PubMed] [Google Scholar]
- 11.Asghar U., Malik M.F., Anwar F., Javed A., Raza A., Asghar U. DNA extraction from insects by using different techniques: a review. Adv. Entomol. 2015;3:132–138. [Google Scholar]
- 12.Milligan B.G. Molecular Genetic Analysis of Population: A Practical Approach. 2nd ed.2nd ed. John Wiley & Sons, Ltd; 1998. Total DNA isolation; pp. 29–69. Hoelzel AR, editor. [Google Scholar]
- 13.S. Garima, N.P. Singh, S. Rajendra, Food plants of a major agricultural pest Aphis gossypii Glover (Homoptera: Aphididae) from India: An updated checklist, Int. J. Life Sci. Biotechnol. Pharma Res. 3 (2) 1-26.
- 14.Wang L., Zhang S., Luo Y., Wang Y., Lv M., Zhu Z., Li H., Cui J. Identification of Aphis gossypii Glover (Hemiptera: Aphididae) Biotypes from different host plants in North China. PLOS One. 2016;11(1) doi: 10.1371/journal.pone.0146345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folmer O., Black M., Hoeh W. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Marine Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 16.Hollingsworth P.M., Graham S.W., Little D.P. Choosing and using a plant DNA barcode. PLOS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seifert K.A., Samson R.A., Dewaard J.R., Houbraken J., Levesque C.A., Moncalvo J.M., LouisSeize G., Herbert P.D. Prospects for fungus identification using COI DNA barcodes: with Penicillium as a test case study. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3901–3906. doi: 10.1073/pnas.0611691104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salah M., Aljanabi I.M. Universal and rapid salt-extraction of high-quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi S., Lokeshwari D., Krishna Kumar N.K., Manjunatha H., Verghese A., Jalali K. Wahlgreniella nervata (Hemiptera: Aphididae), a new pest of Rose in India. Fla. Entomol. 2014;97:162–167. [Google Scholar]
- 20.Tilmon K.J., Hodgson E.W., O Neal M.E., Ragsdale D.W. Biology of the Soybean Aphid, Aphis glycines (Hemiptera: Aphididae) in the United States. J. Int. Pest Manag. 2011;2(2):A1–A7. doi: 10.1603/IPM10016. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.