Abstract
Virulence of Vibrio vulnificus correlates with changes in colony morphology that are indicative of a reversible phase variation for expression of capsular polysaccharide (CPS). Encapsulated variants are virulent with opaque colonies, whereas phase variants with reduced CPS expression are attenuated and are translucent. Using TnphoA mutagenesis, we identified a V. vulnificus CPS locus, which included an upstream ops element, a wza gene (wzaVv), and several open reading frames with homology to CPS biosynthetic genes. This genetic organization is characteristic of group 1 CPS operons. The wza gene product is required for transport of CPS to the cell surface in Escherichia coli. Polar transposon mutations in wzaVv eliminated expression of downstream biosynthetic genes, confirming operon structure. On the other hand, nonpolar inactivation of wzaVv was specific for CPS transport, did not alter CPS biosynthesis, and could be complemented in trans. Southern analysis of CPS phase variants revealed deletions or rearrangements at this locus. A survey of environmental isolates indicated a correlation between deletions in wzaVv and loss of virulent phenotype, suggesting a genetic mechanism for CPS phase variation. Full virulence in mice required surface expression of CPS and supported the essential role of capsule in the pathogenesis of V. vulnificus.
Vibrio vulnificus is indigenous to the estuarine environment and can produce rapidly fatal human infections associated with consumption of raw oysters. Pathogenesis of this gram-negative species involves a combination of host-pathogen interactions that are not completely understood. Predisposing host factors include iron overload, hepatic disease, and immune system dysfunction (3, 22, 45). This organism asymptomatically colonizes both fish (8) and shellfish (51) at relatively high levels (103 to 106 CFU/g of body weight) during warmer months, but mouse models, using exogenous iron, suggest that the infectious dose may be <10 CFU (48). Mortalities exceed 50% for septicemic patients, and V. vulnificus disease remains the leading cause of fatal infections associated with seafood consumption (37). Although symptoms of V. vulnificus septicemia resemble endotoxic shock, the lipopolysaccharide (LPS) of this organism is relatively inert, and the contribution of LPS to virulence remains unclear (24, 31). Conversely, expression of capsular polysaccharide (CPS) is clearly a prerequisite for virulence and correlates with lethality in mice (41, 54), resistance to phagocytosis (46) and complement-mediated lysis (40), cytokine induction (31), and opaque colonies. Phase variation to a phenotype with reduced CPS expression occurs at a frequency of about 10−4 and correlates with translucent colonies, increased serum sensitivity, and reduced virulence. CPS is a protective antigen in mice (9, 18), and its relationship to V. vulnificus disease was confirmed by the loss of virulence in acapsular transposon mutants (49, 52). CPS-independent virulence factors indicate that pathogenesis is multifactorial (23, 28, 42), but opaque and translucent phenotypes remain the most reliable predictors of virulence.
Polysaccharide capsules contribute to the virulence of many bacterial pathogens, and specific capsular types are often associated with systemic disease. CPS types in Escherichia coli have been grouped on the basis of their biochemistry, physiology, and genetics (30, 38), and these groups have been reviewed recently and restructured (47). Group 1 and colanic acid polysaccharides are primarily comprised of uronic acid sugars, are regulated by the rcs locus, and are generally induced at low temperatures (<20°C). Related structures are described for Klebsiella and Erwinia species. Group 1 operons are expressed as large transcripts and include unique, highly conserved transport genes such as wza. Group 2 polysaccharides are homopolymers of sialic acid, mapping to a different genetic locus from that of group 1, and homologues are described for Haemophilus and Neisseria species. Groups 2 and 3 share genomic loci and transport systems but differ in operon organization and relationship to CPS or 2-keto-3-deoxyoctulosonic acid (KDO) synthesis. Group 4 strains exhibit the “O-antigen capsule” or Klps associated with a lipid A core and map to the rfb locus.
The genetics of CPS biosynthesis and transport for V. vulnificus have not been defined, and the relationship among capsular types for this species and other CPS groups is unclear. V. vulnificus strains exhibit great diversity in CPS carbohydrate composition, but many contain uronic acid sugars similar to group 1 CPS (7, 33). An epimerase gene was recently reported to be required for CPS expression, but the relationship of this gene to a particular CPS locus was not demonstrated (55). We previously reported phenotypic analysis of CPS expression for several mutants that were either unable to synthesize CPS or that produced CPS but did not translocate capsule to the cell surface (50, 53). In the present study, we detail the genetics of these mutations to identify a CPS locus for V. vulnificus. Genes interrupted by transposon insertions were cloned and examined for sequence homology to other CPS transport and biosynthetic genes. A CPS transport gene, wzaVv, was identified and characterized by nonpolar mutagenesis in V. vulnificus. This gene previously has been shown to encode an outer membrane protein that forms a multimeric secretin-like complex for group 1 polysaccharide transport in E. coli (12). We examined wzaVv gene function in V. vulnificus, as well as the relationship of this gene to virulence and CPS phase variation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
V. vulnificus strains described in Table 1 were cultured in Luria broth (LB; Difco) or on LB agar at 37 or 30°C and were stored at −70°C in LB with 50% glycerol. When appropriate, antibiotics were added at the following concentrations: kanamycin (200 μg/ml), ampicillin (200 μg/ml), tetracycline (10 μg/ml), trimethoprim (50 μg/ml), and polymyxin (50 u/ml). E. coli was cultured as described above except that kanamycin (50 μg/ml) and tetracycline (50 μg/ml) were added at different concentrations.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Description |
|---|---|
| V. vulnificus strains | |
| M06-24/O,a M06-24/T, LC4/O, LC4/T, E4125/O, and E4125/T | Opaque- and translucent-phase variants of clinical isolates |
| 345/O and 345/T | Phase variants of environmental isolate |
| CVD752 | Acapsular TnphoA mutant of M06-24/O (insertion in wzaVv) |
| CVD737 | Acapsular TnphoA mutant of M06-24/O (insertion in ORF4) |
| M06-24/31T and M06-24/32T | Nonpolar mutant of M06-24/O (insertion in wzaVv) |
| Plasmids | |
| pACW15 | Partial wzaVv gene cloned into pBluescript II |
| pACW29 and pACW30 | pACW15 with in-frame or +1 nonpolar insertion in wzaVv |
| pACW31 and pACW32 | wzaVv gene from pACW29 and/or pACW30 cloned into pRK404 |
| pACW36 | Intact wzaVv gene cloned into pGEM-TEasy |
| pWZA1 | Intact wzaVv gene cloned into pRK404 |
| pWZA2 | Intact wzaVv gene with upstream sequences cloned into pRK404 |
| pR751 | IncP plasmid |
O, opaque; T, translucent.
Cloning, PCR, and DNA sequence analysis.
V. vulnificus strains and recombinant plasmids are detailed in Table 1. DNA that flanked transposon insertions in V. vulnificus CVD737 and CVD752 (49) was cloned, using the kanamycin resistance marker in TnphoA. Plasmid clones were constructed in pBR325 and transformed into E. coli DH5α (Gibco-BRL) by standard methods (39). These recombinant plasmids were isolated by polyethylene glycol precipitation and were sequenced by cycle sequencing using dideoxy chain termination on an automated sequencer (Applied Biosystems, Inc.). These sequences were used to derive oligonucleotide primers (UMB Biopolymer Laboratory) for PCR amplification of DNA in order to recover intact parental DNA. DNA (100 ng) from V. vulnificus M06-24/O was amplified by PCR using Taq polymerase (Promega) or High Fidelity polymerase (Boehringer Mannheim) on a thermocycler (MJ Research) under the following conditions: incubation at 92°C for 5 min followed by 35 cycles of 92°C for 1 min, 57° to 60°C for 2 min, and 72°C for 2 min with a final 10-min extension at 72°C. The region encompassing the wzaVv gene was amplified by forward primer 712A (5′ ATT CCG TGA CCG ATT GAG CGT 3′), 712C (5′ TGC AGC AAG CCA TTA GAG CT 3′), or 712K (5′ CCA GCA ACT TAC GTT CAC TT3′) and by reverse primer 752J (5′ GCA GTA GAA GAT ACA CCT AGG 3′). PCR products were gel purified by Micropure separators (Amicon) and cloned into either T/A vector pGEM-TEasy (Promega), pBluescript II (Stratagene), or into pRK404 (10) for complementation studies. Plasmid DNA or gel-purified PCR products were sequenced as described above. DNA from multiple isolates of plasmid clones was sequenced in both directions. Sequence identity searches and alignments were done with the TFASTA, FASTA, or PILEUP program (GCG Wisconsin Package) or BLAST (National Center for Biotechnology Information).
Southern analysis and colony blots.
Chromosomal DNA was extracted with Qiamp Tissue Extraction kits (Qiagen) and digested with restriction enzymes according to the manufacturer's recommendations (Promega). DNA was visualized on 0.5 to 1.0% agarose gels with ethidium bromide and transferred to Zetaprobe GT nylon membranes (Bio-Rad) using alkaline transfer in 0.4 M NaOH. A NotI digest of pACW36, which included all of the wzaVv gene, was gel purified as described above and labeled with 32P by random priming (Amersham). Membranes were hybridized with this probe in phosphate buffer with 7% sodium dodecyl sulfate, washed under stringent conditions at 65°C in SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with decreasing amounts of sodium dodecyl sulfate, and visualized by autoradiography.
Environmental isolates (n = 107) from our culture collection and that of M. Tamplin were confirmed as V. vulnificus using the species-specific VVAP oligonucleotide probe and colony blot hybridization (50). VVAP probe-positive strains were assayed with wzaVv-specific oligonucleotide probe (712A, described above) using the same methodology. Chi-square analysis with one degree of freedom was performed to test the null hypothesis that the frequency of wzaVv-negative strains was significantly different in translucent-phase versus opaque-phase environmental V. vulnificus isolates.
Nonpolar mutagenesis of V. vulnificus wza.
Clones for marker exchange were derived from pACW15 and constructed by insertion of kanamycin resistance cassettes into a unique SphI site in wzaVv. These cassettes, previously shown to introduce nonpolar mutations in the ipa genes of Shigella flexneri (25), are on SmaI fragments of 850 or 851 bp and include start and stop codons and a ribosomal binding site but lack transcriptional terminators. Cassettes were inserted into the cloned wzaVv gene either in frame or with a +1 orientation (pACW29 and pACW30, respectively). EcoRI fragments containing the interrupted wzaVv genes from these plasmids were subsequently cloned (pACW31 and pACW32) into a broad-host-range vector (pRK404) and were transformed into conjugation-competent E. coli 17S for conjugation into V. vulnificus. Crossover events were facilitated by the introduction of another plasmid (pR751) from the same incompatibility group (IncP) with dual selection for the inserted kanamycin resistance marker and trimethoprim resistance from pR751. About 50% of the resulting transconjugates were positive for the crossover events, as determined by acquisition of translucence. Insertions were confirmed by PCR and Southern analysis. Phenotypes of V. vulnificus strains were confirmed by examining their ability to bind CPS-specific monoclonal antibody, as determined by enzyme-linked immunosorbent assay, immunoelectron microscopy, and flow cytometry, as previously described (52). The nonpolar mutant was able to synthesize CPS but did not exhibit surface expression, whereas the transposon mutants showed no evidence of CPS synthesis.
Complementation of wzaVv mutants.
The intact wzaVv gene, with or without upstream sequences, was recovered by PCR amplification using conditions described above and primer pair 712K and 752J or 712C and 752J, respectively. PCR products were first cloned into pGEMT-Easy (Promega) in E. coli JM109, and then NsiI fragments, containing the wzaVv, were further subcloned into the PstI site of pRK404 in conjugation-competent E. coli 17S to yield pWZA1 (without upstream sequences) or pWZA2 (with upstream sequences). Filter matings of V. vulnificus M06-24/31T and either E. coli 17S (pWZA1), E. coli 17S (pWZA2), or E. coli 17S (pRK404) were performed overnight at 37°C, and transconjugates were plated to LB agar with tetracycline and polymyxin B. Filter matings were also performed with V. vulnificus CVD752 and E. coli 17S (pWZA2). Complementation was detected by recovery of opaqueness, and CPS expression of transconjugates was confirmed by the ability to resist the lytic effects of complement as previously described (49). Briefly, bacteria were grown to log phase and inoculated into either fresh or heat-inactivated (56°C for 1 h) human sera at a concentration of about 107 CFU/ml. Sera and cells were incubated for 2 h at 37°C, and survival was determined by standard plate counts using serial dilutions in phosphate-buffered saline (PBS), and colony morphology was recorded.
Virulence assay.
Fifty percent lethal dose (LD50) determinations in C57BL/6 female mice (Charles River) were conducted with or without exogenous iron supplement as previously described (48). Strains were inoculated from single colonies into LB and were cultured overnight at 30°C. Fresh LB culture was seeded at a 1:100 dilution from the corresponding overnight culture and incubated at 30°C for 2 to 3 h. Bacterial cells were collected by centrifugation and washed once with PBS. Bacterial cell concentrations were estimated by determining the optical density at 600 nm of washed cells and were confirmed by plate counts. Groups of mice (n = 5) were injected intraperitoneally with serial log dilutions of each bacterial strain (0.5 ml/mouse) and PBS (0.2 ml) with or without ferric ammonium citrate (80 mg); the control group was given PBS alone. Lethality was observed at 24 h, and calculations were determined according to the method of Reed and Muench (34).
RESULTS
Identification of CPS locus for V. vulnificus M06-24.
A CPS locus for V. vulnificus was characterized using previously described TnphoA mutants, CVD752 and CVD737, which are acapsular and unable to synthesize CPS (49, 52). The present study details the genetic analysis of these strains. DNA sequences for regions flanking the insertion sites in both CVD752 and CVD737 exhibited open reading frames (ORFs) with DNA identity and amino acid similarity to genes previously described for CPS biosynthetic and transport function (Table 2). Only ORF 1 (ORF1) from CVD 752 did not show significant homology to GenBank sequences. ORF2 was homologous to the conserved gene family, wza, which encodes outer membrane proteins involved in CPS transport. We obtained the complete sequence for this gene (GenBank accession number AY055488), which we have designated wzaVv according to recently described nomenclature (35). The remaining partial ORFs from CVD752 (ORF3) or CVD737 (ORF4 and ORF5 from the 3′ end of the insertion site) also showed homology to known CPS biosynthetic genes.
TABLE 2.
Comparison of V. vulnificus DNA and deduced amino acid sequences to potential homologues for CPS biosynthesis and transport
| V. vulnificus ORF | Homologue (accession no.) | Source | Function | % DNA identity (no. of bases aligned) | % Amino acid identity | % Amino acid similarity |
|---|---|---|---|---|---|---|
| wzaVv | wzaEc (AF104912) | E. coli | Transport | 64 (1,237) | 61 | 76 |
| ORF3 | rfbO (X60665) | Salmonella enterica | Biosynthesis | 59 (438) | 15 | 62 |
| ORF4 | capM (U10927) | Staphylococcus aureus | Biosynthesis | 54 (312) | 28 | 66 |
| ORF5 | capD (U10927) | S. aureus | Biosynthesis | 57 (480) | 36 | 76 |
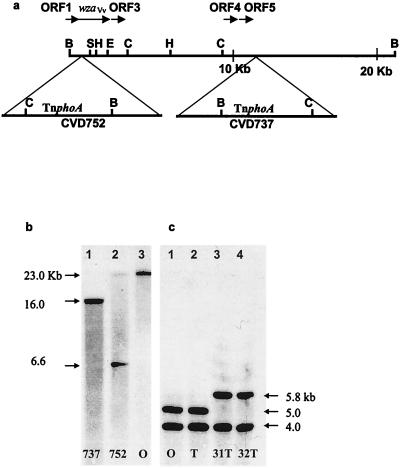
Both transposon insertions localized to the same 23-kb BamHI fragment, and a partial map of the CPS locus is shown in Fig. 1a. Southern analysis with a wza probe indicated that both mutants exhibited novel BamHI restriction sites that were not present in the parent strain, V. vulnificus M06-24/O (Fig. 1b), or in the phase variant, V. vulnificus M06-24/T (not shown). These sites were introduced presumably from the BamHI site in TnphoA. Restriction fragment analysis of these digests, as well as of other restriction enzyme digests (not shown), indicated that the transposon insertion in V. vulnificus CVD737 was approximately 10 kb downstream of the insertion in CVD752. Thus, both acapsular transposon mutants exhibited independent insertions at the same locus, disrupting genes related to either CPS transport or biosynthesis.
FIG. 1.
Analysis of the V. vulnificus CPS genetic locus. (a) Restriction map for ClaI (C), HindIII (H), SphI (S), and EcoRI (E) sites within the 23-kb BamHI (B) fragment of the V. vulnificus CPS genetic locus. Relative placement and direction of transcription (horizontal arrows) of ORFs and wzaVv are shown. Location of 7.7-kb TnphoA (with a BamHI site at 5 kb) in V. vulnificus CVD737 and CVD752 is indicated by triangles. (b) Southern analysis for BamHI restriction digests of the following V. vulnificus strains: CVD737, lane 1; CVD752, lane 2; and M06-24/O, lane 3. DNA was hybridized with the wzaVv gene probe as described in text. Approximate size of bands (arrows) was determined by extrapolation from known size markers of lambda DNA HindIII digests. CVD737 digests probed with wzaVv show a single band of ca. 16 kb as a product of the new BamHI site introduced 11 kb downstream from the 5′ BamHI site with an additional 5 kb from TnphoA. The insertion in CVD752 occurred within wzaVv 1.6 kb from the 5′ BamHI site to produce 2 bands of ca. 6.6 and 23.4 kb. (c) HindIII restriction digests of chromosomal DNA from the following V. vulnificus strains are shown: lane 1, M06-24/O; lane 2, M06-24/T; lane 3, M06-24/31T; and lane 4, M06-24/32T. DNA was hybridized with a wzaVv probe as described in the text, and the approximate sizes of bands, as determined by comparison to known markers of lambda DNA HindIII digests, are indicated by arrows.
Upstream regulatory sequences for V. vulnificus wza.
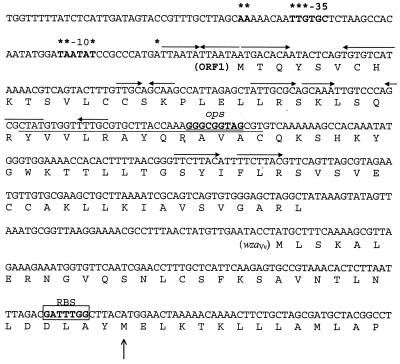
The wzaVv gene is characteristic of group 1 CPS operons. Other features common to these operons were identified upstream of this gene in V. vulnificus. As shown in Fig. 2, ORF1 contains a genetic element, ops, which is located at the 5′ end of all group 1 CPS operons (47). This element is generally found within a larger JUMPstart (just upstream of many polysaccharides) sequence (16). The upstream region also included a putative promoter containing all eight of the most conserved bases described for strong E. coli promoters (15), i.e., the “−10” and “−35” regions and a potential RNA start site. No ribosomal binding site was identified for ORF1; however, a possible ribosomal binding site was found for the wzaVv gene, which preceded the second Met (amino acid 33) of the deduced amino acid sequence, corresponding to the translational start for other wza homologues.
FIG. 2.
The 5′ region of the CPS locus for V. vulnificus. Single-letter deduced amino acid sequence is given below nucleotide coding region. Putative promoter region is shown in boldface, and locations of −10 and −35 sites are indicated above coding sequence with asterisks over highly conserved nucleotides (16). Inverted repeats are indicated by horizontal arrows. The JUMPstart region is underlined, and the ops element is doubly underlined. Possible ribosomal binding site (RBS) for wzaVv is boxed and followed by the second Met (vertical arrow), which aligns translation start site with other wza homologues.
Functional analysis of V. vulnificus wzaVv as CPS transport gene.
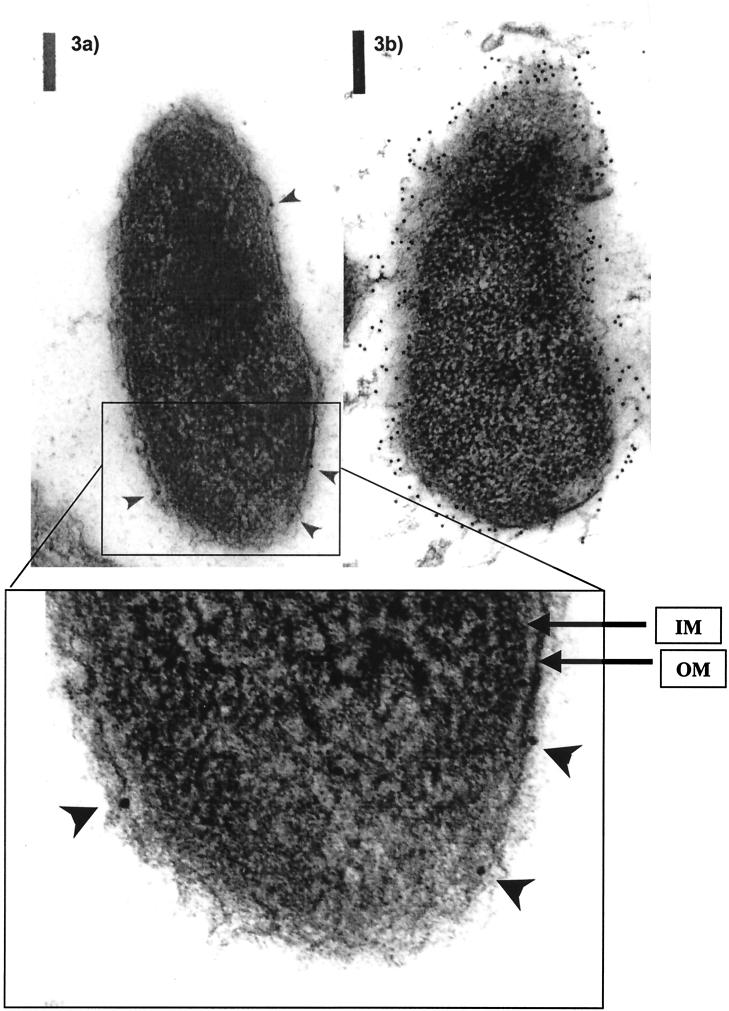
To determine the function of the wzaVv gene product, nonpolar mutations were introduced at the SphI site (Fig. 1a) of wzaVv and recombined into the chromosome of V. vulnificus M06-24/O. Insertions of the nonpolar kanamycin cassette into mutant strains M06-24/31T and M06-24/32T were confirmed by increased sizes of a HindIII fragments in mutants (5.8 kb) compared to the parental strain (5.0 kb), as indicated by Southern analysis (Fig. 1c) and by sequencing of PCR products spanning the insertion sites. Mutants were translucent and did not revert to opaque colonies when grown with or without antibiotic selection. In a previous study, CPS expression was examined in detail for V. vulnificus M06-24/O and its derivatives, using a combination of enzyme-linked immunosorbent assay, flow cytometry, and immunoelectron microscopy (52). Those results demonstrated that the nonpolar mutant was able to synthesize CPS but did not translocate the polysaccharide to the cell surface. This strain differed from the TnphoA mutants, which do not synthesize the polysaccharide. Closer examination of electron micrographs confirmed that CPS was detected consistently in the cytoplasm and occasionally in the periplasm of M06-31T but was not seen beyond the outer membrane (Fig. 3a). These results differed from those for the wild-type encapsulated strain, where label was clearly detected beyond the outer membrane (Fig. 3b).
FIG. 3.
Immunoelectron micrograph of V. vulnificus wza mutant. Bacterial thin sections of wild-type wzaVv mutant M06-24/31T (A) and M06-24/O (B) were immunolabeled using V. vulnificus CPS-specific monoclonal antibody (7/G4-D2) and were visualized by gold-labeled secondary goat anti-mouse immunoglobulin A conjugate as previously described (53). Inner membrane (IM) and outer membrane (OM) are shown for inset, and arrows indicate gold-labeled CPS for wzaVv mutant M06-24/31T. Bar = 1 μm.
Introduction of the intact wzaVv gene with the upstream sequences (pWZA2) into M06-24/31T complemented the mutation, and all transconjugates had the opaque colonies of the parental phenotype. However, conjugation of the cloned wzaVv gene without these upstream sequences (pWZA1) into M06-24/31T did not complement the mutation, and transconjugates had translucent colonies. To confirm complementation and CPS expression in M06-31T (pWZA2), resistance to the lytic effects of complement was examined in this strain. The nonpolar mutant M06-24/31T was extremely sensitive to complement and exhibited a >5.0-log-CFU/ml reduction in normal serum in comparison to heat-inactivated serum. However, complement sensitivity of M06-24/31T containing the pWZA2 was similar to that of the encapsulated parental strain, with only 0.1- and 0.2-log reductions in number of CFU per milliliter, respectively. Conjugation of pWZA2 into transposon mutant CVD752 or of the vector only into M06-24/31T did not complement in trans, yielding translucent transconjugates.
Transport of CPS to cell surface increases virulence of V. vulnificus in mice.
The role of CPS expression in virulence was examined using a mouse model with or without exogenous iron. As shown in Table 3, LD50 determinations in mice (n = 5) agreed with previously published data (50). As expected, translucent-phase variant V. vulnificus M06-24/T was less virulent than the encapsulated M06-24/O and acapsular TnphoA mutant V. vulnificus CVD752 was less virulent than all other strains, with no deaths observed at the highest concentration of bacteria inoculated (108). The LD50 was lower for all strains following iron injections but was dramatically reduced to one bacterium for the opaque, encapsulated isolate. The LD50 for the wzaVv mutant without iron was equivalent to the translucent variant but was intermediate between the translucent-phase variant and the acapsular transposon mutant for iron-treated mice.
TABLE 3.
Virulence of V. vulnificus strains
| V. vulnificus strain | LD50
|
|
|---|---|---|
| With Fe | Without Fe | |
| M06-24/O | <10 | 4.0 × 103 |
| M06-24/T | 1.2 × 104 | 1.9 × 107 |
| CVD752 | 4.0 × 105 | >108 |
| M06-24/31T | 1.2 × 105 | 1.6 × 107 |
Sequence comparisons of wza homologues.
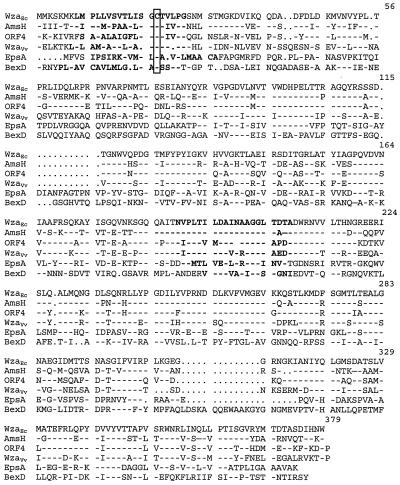
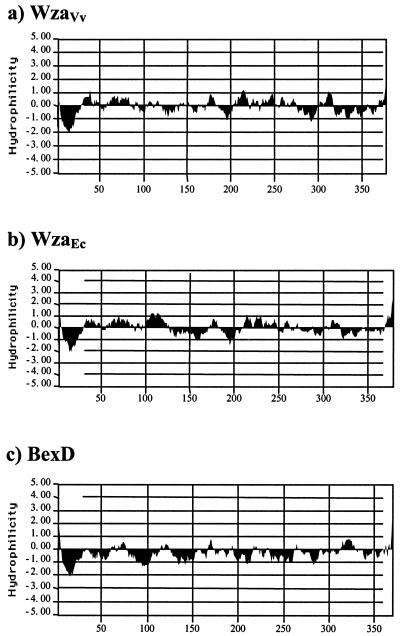
The wza gene family has been reported (47) to include the E. coli wza for group 1 and colanic acid CPS (44), amsH of Erwinia amylovora (6), ORF4 of Klebsiella pneumoniae (1), epsA of Pseudomonas solanacearum (17), and exoF of Rhizobium meliloti (36). Deduced amino acid sequence alignments indicated that homologues from V. vulnificus, E. coli, K. pneumoniae, and E. amylovora are more closely related to each other than those from P. solanacearum (Fig. 4) or R. meliloti (not shown). The former group shared exact conservation of sequence for 163 of 381 (43%) residues. On the other hand, EpsA shared only 58 of these residues (15%), and alignment of ExoF of R. meliloti was much closer to that of E. coli KpsD by BLAST analysis than to that of Wza homologues (not shown). At amino acid 112, EpsA also exhibited a 12-amino-acid insertion region that was not present in the other sequences. Other putative outer membrane transport systems include those from group 2 CPS, such as BexD from Haemophilus influenzae (19) and CtrA from Neisseria meningitidis (13, 14), which are more closely related to each other than to Wza homologues. Interestingly, BexD also contained an 8-amino-acid insertion found at the same location as the one in EpsA. Differences between group 1 and group 2 homologues were reflected in hydropathy analyses of the deduced amino acid sequences (Fig. 5). BexD, which has been characterized as a porin with multiple membrane-spanning regions, was considerably more hydrophobic than the Wza homologues.
FIG. 4.
Comparison of possible Wza homologues. Deduced amino acid sequences from E. coli WzaEc, E. amylovora AmsH, K. pneumoniae ORF4, V. vulnificus WzaVv, P. solanacearum EpsA, and H. influenzae BexD are shown. Identity is indicated by dashes, and gaps are indicated by periods. Hydrophobic regions are in boldface, and conserved cysteines at the end of the leader sequences are boxed.
FIG. 5.
Hydropathy comparison of deduced amino acid sequences for CPS outer membrane transport. Hydrophilicity plots of the deduced amino acid sequences of E. coli WzaEc, V. vulnificus WzaVv, and H. influenzae BexD are shown and were determined using the algorithm of Kyte and Doolittle (21).
Conservation and phase variation of wzaVv sequences among V. vulnificus strains.
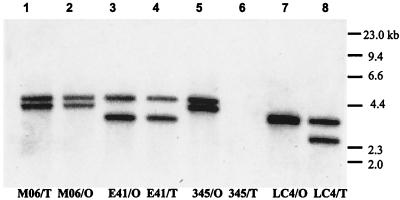
The wza gene of E. coli is highly conserved among different group 1 K serotypes (47). Therefore, isolates of V. vulnificus from clinical and environmental sources were examined for the presence and diversity of wzaVv. Chromosomal DNA from the opaque- or translucent-phase variants of V. vulnificus strains M06-24, LC4, 345, and E4125 was amplified by PCR, using primers derived from sequences flanking the wzaVv gene. PCR products of identical size were observed for all strains except the translucent-phase variant V. vulnificus 345/T (the only environmental isolate), which did not amplify with these or nested primers. Southern analysis confirmed deletion of wzaVv in V. vulnificus 345/T (Fig. 6), and restriction fragment length polymorphism was observed among all strains. No differences in restriction fragment lengths were detected between phase variants of either V. vulnificus M06-24 or E4125, but a deletion or restriction site polymorphism was seen for the translucent-phase variant of strain LC4.
FIG. 6.
Southern analysis of wzaVv from opaque- and translucent-phase variants. HindIII digests of chromosomal DNA extracted from the following strains are shown: lane 1, M06-24/O; lane 2, M06-24/T; lane 3, E4125/O; lane 4, E4125/T; lane 5, 345/O; lane 6, 345/T; lane 7, LC4/O; and lane 8, LC4/T. Size markers for a lambda DNA HindIII digest are indicated.
Additional environmental isolates were examined for the presence of wzaVv, based on hybridization with an oligonucleotide probe. The majority (88%) of 97 opaque-phase and presumably encapsulated isolates were positive for the wzaVv probe, indicating a high degree of conservation for this gene among V. vulnificus environmental isolates. However, a significantly greater percentage (50%) of translucent-phase strains (n = 10) than of opaque-phase strains were negative for wza (P = 0.007, Fisher's two-tailed exact test), suggesting a higher deletion rate for translucent-phase than for opaque-phase isolates.
DISCUSSION
These data represent the first description of a CPS operon in V. vulnificus. CPS-associated homologues were identified by transposon mutagenesis and shown to localize to the same 23-kb chromosomal fragment. Genetic organization at this locus indicated homology to the group 1 and colanic acid CPS operons. In E. coli, an 8-bp ops element always follows the promoter of group 1 and colanic acid CPS operons and is contained within a larger 39-bp consensus region termed JUMPstart. This element exhibits nonrandom distribution throughout the chromosome and functions to suppress transcriptional polarity, greatly enhancing the activity of both homologous and heterologous promoters (26). It is orientation dependent and requires cotranscription by the promoter. Within ORF1 of the V. vulnificus sequence, we identified an ops element contained in a region with 67% homology to the consensus for JUMPstart. This element in V. vulnificus was downstream of a putative promoter, which showed genetic structure similar to the E. coli colanic acid CPS promoter (44). Distances between promoters and ops elements were almost identical for V. vulnificus and E. coli (123 and 124 bp, respectively), and both regions contained multiple inverted repeats that may serve as recognition sites for DNA-binding proteins. Further, this promoter preceded the CPS transport gene, wzaVv, which is usually the first gene in group 1 operons.
The presence of wza is a criterion for group 1 CPS operon classification (47), and mutational analysis of wzaVv was consistent with identification of a V. vulnificus CPS operon. Mutations in this gene in E. coli specifically disrupt transport and surface expression of CPS (11). However, a single-transposon insertion in this gene in V. vulnificus CVD752 completely eliminated biosynthetic function as well. The polar effects of TnphoA insertions have been established for a number of species, including vibrios (5); therefore, loss of CPS biosynthesis in CVD752 is due presumably to an inability to transcribe downstream biosynthetic genes within the CPS operon. On the other hand, nonpolar mutagenesis of this gene in V. vulnificus M06-31T specifically eliminated transport function without disturbing CPS biosynthesis, and the nonpolar mutation of wzaVv was complemented in trans, whereas the introduction of the cloned gene did not complement the TnphoA mutation in CVD752. Furthermore, a complementation was achieved with a plasmid construct that included both the cloned wzaVv and its upstream promoter region; however, a similar construct, lacking the upstream DNA, did not complement. Although confirmation will require more extensive analysis, we predict that transcription of the V. vulnificus CPS operon begins upstream of wzaVv.
Deduced amino acid sequences support a specific outer membrane function for transport of CPS by Wza protein homologues (11), and studies with E. coli recently confirmed that Wza proteins form multimeric secretins exposed on the cell surface (12). TnphoA mutagenesis of the V. vulnificus wzaVv gene was consistent with an outer membrane location for the gene product. TnphoA lacks a signal sequence and will not produce functional enzyme without a fusion to an exported or secreted gene product. TnphoA insertion into the wzaVv gene in V. vulnificus CVD752 resulted in a strain that acquired alkaline phosphatase activity (49). However, the transposon insertion into cytoplasmic, biosynthetic homologues in CVD737 did not produce enzymatic activity. These results indicate that wzaVv provides the signal sequence for the export of the TnphoA fusion product through the inner membrane, supporting a periplasmic or outer membrane location for wza.
Group 2 inner membrane transport genes are well defined, but homologues have not been found in group 1 CPS operons. Thus, the mechanism for coordination of inner and outer transport in group 1 is unknown. The genetic analysis of V. vulnificus CPS mutants reported here, in the context of previously detailed phenotypic examination (52), demonstrated specific outer membrane transport function for wzaVv; the nonpolar mutant was able to synthesize CPS but lacked cell surface expression. Micrographs of this mutant differed greatly from E. coli mutants deficient in either inner membrane or periplasmic transport. Deletion of group 2 CPS inner membrane transport genes, kpsM and kpsT, produced strains that accumulated intact CPS in the cytoplasm, sequestering the polymers in discrete electron-lucent spaces just below the inner membrane (4, 29). Strains deficient in the periplasmic protein KpsD accumulated CPS in the periplasm and exhibited electron-lucent spaces that expanded the periplasm (53). Micrographs of the nonpolar wzaVv mutant indicated accumulation of CPS in the cytoplasm, with relatively little material in the periplasm and no surface CPS. Zones of adhesion or “Bayer junctions” have been proposed to span both membranes and form junctions for CPS transport that bypasses the periplasm altogether (2). The presence of CPS in the periplasm of the wzaVv mutant would argue against this hypothesis, but lack of periplasmic CPS accumulation does suggest coordination of inner and outer membrane transport.
Analyses of deduced amino acid sequences of group 1 wza homologues indicated that they encode proteins distinct from those described for group 2 CPS transport and are likely to differ in origin and structure (35). We present evidence that Wza homologues lack the multiple hydrophobic regions characteristic of the porin structure of group 2 homologues (i.e., BexD) and agree that the transport mechanisms of groups 1 and 2 are not closely related. However, we also found striking diversity among strains that are generally classified within group 1. Sequence alignments showed that Wza homologues from V. vulnificus, E. coli, K. pneumoniae, and E. amylovora share much greater overall conservation of sequence than those of P. solanacearum and R. meliloti. All Wza homologues include a lipoprotein signal peptidase II recognition site with a conserved cysteine (amino acid 22 of the E. coli Wza), characteristic of outer membrane proteins (6, 32, 43), but the former group has a common threonine at the membrane-sorting signal site that was not found in the latter species. In addition, both of the latter strains contain insertions not found in the other sequences. Therefore, we suggest that the grouping of homologues from P. solanacearum and R. meliloti within the wza gene family requires further examination.
Loss of virulence in acapsular transposon mutants presumably results from decreased anticomplement and antiphagocytic protection. The virulence of genetically undefined translucent-phase variants, expressing patchy and incomplete capsules, was intermediate between opaque-phase variants and acapsular mutants (49, 52). Thus, residual CPS expression by translucent-phase variants may confer some degree of protection and implies a correlation of virulence with the amount of CPS expressed. Alternatively, a small number of cells within the translucent-phase population may revert to the opaque-phase phenotype and function to lower the LD50, especially in the presence of exogenous iron. Interestingly, the wzaVv nonpolar transport mutant, which lacks surface CPS and does not revert to the opaque phase, was also more virulent than the acapsular transposon mutants. Thus, in vivo release of intracellular polymer from the wzaVv mutant may contribute directly to the disease process in a manner that is independent of its surface properties. Recent observations have indicated that, during infections in mice, V. vulnificus CPS elicits inflammatory host cytokines, which may contribute to pathogenesis by inducing toxic shock (31). Therefore, virulence analysis of V. vulnificus strains was consistent with an inflammatory role for CPS; however, full virulence required complete encapsulation of the bacteria.
Phase variation of CPS expression is common and has been attributed to insertional inactivation of CPS genes by insertion elements (27) or deletion mutations (20). Deletions or rearrangements of wzaVv were more frequent among V. vulnificus that were translucent than among those that were opaque, suggesting that wzaVv is a possible target for phase variation. However, translucent-phase V. vulnificus strains with intact wzaVv genes, as well as wzaVv-negative opaque-phase isolates, were observed and indicated that other mechanisms for phase variation exist. These data also raise the question of whether multiple CPS operons may exist for V. vulnificus strains, as they do for E. coli. The previously reported V. vulnificus epimerase gene was required for CPS expression, but operon structure was not demonstrated (55). This gene showed extensive identity to a sequence found in the V. cholerae O139 CPS operon, which is not related to group 1 operons and appears to be derived from an insertion of CPS genes into a LPS locus. We are presently evaluating the linkage of the epimerase gene to the group 1 operon described here, as well as the possibility of multiple CPS operons.
In summary, this study identified a group 1-like CPS operon for V. vulnificus. Experimental analysis provided evidence to support the role of the wza gene family in outer membrane transport of bacterial polysaccharides. Despite great variation in CPS composition within this species, this transport gene was highly conserved among different strains of V. vulnificus, and naturally occurring deletions observed in wzaVv may represent at least one of the mechanisms responsible for phase variation. Finally, animal studies confirmed the essential contribution of capsule expression as a virulence determinant for V. vulnificus.
ACKNOWLEDGMENTS
Funding was provided in part by a Merit Review grant from the Department of Veterans Affairs and by a grant from the United States Department of Agriculture (2001-35201-09954).
We thank Anne Sill for the statistical analysis and Lynne Ensor for electron micrographs.
REFERENCES
- 1.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayer M, Bayer M H. Biophysical and structural aspects of the bacterial capsule. ASM News. 1994;60:192–198. [Google Scholar]
- 3.Blake P A, Merson M H, Weaver R E, Hollis D G, Heublein P C. Disease caused by a marine Vibrio. Clinical characteristics and epidemiology. N Engl J Med. 1979;300:1–5. doi: 10.1056/NEJM197901043000101. [DOI] [PubMed] [Google Scholar]
- 4.Bronner D, Seiberth V, Pazzani C, Roberts S, Boulnois G J, Jann B, Jann K. Expression of the capsular K5 polysaccharide of Escherichia coli: biochemical and electron microscopic analyses of mutants with defects in region 1 of the gene cluster. J Bacteriol. 1993;175:5984–5992. doi: 10.1128/jb.175.18.5984-5992.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown R C, Taylor R K. Organization of tcp, acf, and toxT genes within a ToxT-dependent operon. Mol Microbiol. 1995;16:425–439. doi: 10.1111/j.1365-2958.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- 6.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 7.Bush C A, Patel P, Gunawardena S, Powell J, Joseph A, Johnson J A, Morris J G. Classification of Vibrio vulnificus strains by the carbohydrate composition of their capsular polysaccharides. Anal Biochem. 1997;250:186–195. doi: 10.1006/abio.1997.2219. [DOI] [PubMed] [Google Scholar]
- 8.DePaola A, Capers G M, Alexander D. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast Appl Environ Microbiol. 1994;60:984–988. doi: 10.1128/aem.60.3.984-988.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devi S J, Hayat U, Powell J L, Morris J G., Jr Preclinical immunoprophylactic and immunotherapeutic efficacy of antisera to capsular polysaccharide-tetanus toxoid conjugate vaccines of Vibrio vulnificus. Infect Immun. 1996;64:2220–2224. doi: 10.1128/iai.64.6.2220-2224.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ditta G, Schmidhauser T, Yakobson E, Lu P, Liang X W, Finlay D R, Guiney D, Helinski D R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985;13:149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- 11.Drummelsmith J, Whitfield C. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30) Mol Microbiol. 1999;31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 12.Drummelsmith J, Whitfield C. Translocation of group 1 capsular polysaccharide to the surface of Escherichia coli requires a multimeric complex in the outer membrane. EMBO J. 2000;19:57–66. doi: 10.1093/emboj/19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frosch M, Edwards U, Bousset K, Krausse B, Weisgerber C. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 14.Frosch M, Muller D, Bousset K, Muller S. Conserved outer membrane protein of Neisseria meningitidis involved in capsule expression. Infect Immun. 1992;60:798–803. doi: 10.1128/iai.60.3.798-803.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gentz R, Bujard H. Promoters recognized by Escherichia coli RNA polymerase selected by function: highly efficient promoters from bacteriophage T5. J Bacteriol. 1985;164:70–77. doi: 10.1128/jb.164.1.70-77.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobbs M, Reeves P R. The JUMPstart sequence: a 39 bp element common to several polysaccharide gene clusters. Mol Microbiol. 1994;12:855–856. doi: 10.1111/j.1365-2958.1994.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Schell M. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol. 1995;16:677–689. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 18.Kreger A S, Gray L D, Testa J. Protection of mice against Vibrio vulnificus disease by vaccination with surface antigen preparation and anti-surface antigen antisera. Infect Immun. 1984;45:537–543. doi: 10.1128/iai.45.3.537-543.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroll J S, Loynds B, Brophy L M, Moxon E R. The bex locus in encapsulated Haemophilus influenzae: a chromosomal region involved in capsule polysaccharide export. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 20.Kroll J S, Moxon E R, Loynds B M. An ancestral mutation enhancing the fitness and increasing the virulence of Haemophilus influenzae type b. J Infect Dis. 1993;168:172–176. doi: 10.1093/infdis/168.1.172. [DOI] [PubMed] [Google Scholar]
- 21.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 22.Linkous D A, Oliver J D. Pathogenesis of Vibrio vulnificus. FEMS Microbiol Lett. 1999;174:207–214. doi: 10.1111/j.1574-6968.1999.tb13570.x. [DOI] [PubMed] [Google Scholar]
- 23.Litwin C M, Rayback T W, Skinner J. Role of catechol siderophore synthesis in Vibrio vulnificus virulence. Infect Immun. 1996;64:2834–2838. doi: 10.1128/iai.64.7.2834-2838.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McPherson V L, Watts J A, Simpson L M, Oliver J D. Physiological effects of the lipopolysaccharide of Vibrio vulnificus on mice and rats. Microbios. 1991;67:141–149. [PubMed] [Google Scholar]
- 25.Menard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nieto J M, Bailey M J A, Hughes C, Koronakis V. Suppression of transcription polarity in the Escherichia coli haemolysin operon by a short upstream element shared by polysaccharide and DNA transfer determinants. Mol Microbiol. 1996;19:705–713. doi: 10.1046/j.1365-2958.1996.446951.x. [DOI] [PubMed] [Google Scholar]
- 27.Ou J T, Baron L S, Rubin F A, Kopecko D J. Specific insertion and deletion of insertion sequence 1-like DNA element causes the reversible expression of the virulence capsular antigen Vi of Citrobacter freundii in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:4402–4405. doi: 10.1073/pnas.85.12.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paranjpye R N, Lara J C, Pepe J C, Pepe C M, Strom M S. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect Immun. 1998;66:5659–5668. doi: 10.1128/iai.66.12.5659-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavelka M S, Jr, Hayes S F, Silver R P. Characterization of KpsT, the ATP-binding component of the ABC-transporter involved with the export of capsular polysialic acid in Escherichia coli K1. J Biol Chem. 1994;269:20149–20158. [PubMed] [Google Scholar]
- 30.Pearce R, Roberts I S. Cloning and analysis of gene clusters for production of the Escherichia coli K10 and K54 antigens: identification of a new group of serA-linked capsule gene clusters. J Bacteriol. 1995;177:3992–3997. doi: 10.1128/jb.177.14.3992-3997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell J L, Wright A C, Wasserman S W, Hone D M, Morris J G., Jr Release of tumor necrosis factor alpha in response to Vibrio vulnificus capsular polysaccharide using in vivo and in vitro models. Infect Immun. 1997;65:3713–3718. doi: 10.1128/iai.65.9.3713-3718.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy G P, Hayat U, Abeygunawardana C, Fox C, Wright A C, Manevel D R, Jr, Bush C A, Morris J G., Jr Purification and determination of the structure of capsular polysaccharide of Vibrio vulnificus M06-24. J Bacteriol. 1992;174:2620–2630. doi: 10.1128/jb.174.8.2620-2630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reed L J, Meunch H. A simple method of estimating the fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 35.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 36.Reuber T L, Walker G C. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 37.Rippey S R. Infectious diseases associated with molluscan shellfish consumption. Clin Microbiol Rev. 1994;7:419–425. doi: 10.1128/cmr.7.4.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts I S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch I F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Shinoda S, Kobayashi M, Yamada H, Yoshida S, Ogawa M, Mizuguchi M. Inhibitory effect of capsular antigen of Vibrio vulnificus on bactericidal activity of human serum. Microbiol Immunol. 1987;31:393–401. doi: 10.1111/j.1348-0421.1987.tb03102.x. [DOI] [PubMed] [Google Scholar]
- 41.Simpson L M, White V K, Zane S F, Oliver J D. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect Immun. 1987;55:269–272. doi: 10.1128/iai.55.1.269-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Starks A M, Schoeb T R, Tamplin M L, Parveen S, Doyle T J, Bomeisl P E, Escudero G M, Gulig P A. Pathogenesis of infection by clinical and environmental strains of Vibrio vulnificus in iron-dextran-treated mice. Infect Immun. 2000;68:5785–5793. doi: 10.1128/iai.68.10.5785-5793.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stout V. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J Bacteriol. 1996;178:4273–4280. doi: 10.1128/jb.178.14.4273-4280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strom M S, Paranjpye R N. Epidemiology and pathogenesis of Vibrio vulnificus. Microb Infect. 2000;2:177–188. doi: 10.1016/s1286-4579(00)00270-7. [DOI] [PubMed] [Google Scholar]
- 46.Tamplin M L, Specter S, Rodrick G E, Friedman H. Vibrio vulnificus resists phagocytosis in the absence of serum opsonins. Infect Immun. 1985;49:715–718. doi: 10.1128/iai.49.3.715-718.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitfield C, Roberts I S. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31:1307–1319. doi: 10.1046/j.1365-2958.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 48.Wright A C, Simpson L M, Oliver J D. The role of iron in the pathogenesis of Vibrio vulnificus. Infect Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wright A C, Simpson L M, Oliver J D, Morris J G., Jr Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect Immun. 1990;58:1769–1773. doi: 10.1128/iai.58.6.1769-1773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wright A C, Miceli G A, Landry W L, Christy J B, Watkins W D, Morris J G. Rapid identification of Vibrio vulnificus on nonselective media with an alkaline phosphatase-labeled oligonucleotide probe. Appl Environ Microbiol. 1993;59:541–546. doi: 10.1128/aem.59.2.541-546.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wright A C, Hill R T, Johnson J A, Colwell R R, Morris J G., Jr Distribution of V. vulnificus in the Chesapeake Bay. Appl Envron Microbiol. 1996;62:717–724. doi: 10.1128/aem.62.2.717-724.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wright A C, Powell J L, Tanner M K, Ensor L A, Karpas A B, Morris J G, Jr, Sztein M B. Differential expression of Vibrio vulnificus capsular polysaccharide. Infect Immun. 1999;67:2250–2257. doi: 10.1128/iai.67.5.2250-2257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wunder D E, Aaronson W, Hayes S F, Bliss J M, Silver R P. Nucleotide sequence and mutational analysis of the gene encoding KpsD, a periplasmic protein involved in transport of polysialic acid in Escherichia coli K1. J Bacteriol. 1994;176:4025–4033. doi: 10.1128/jb.176.13.4025-4033.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida S I, Ogawa M, Mizuguchi Y. Relation of capsular materials and colony opacity to Vibrio vulnificus. Infect Immun. 1985;47:446–451. doi: 10.1128/iai.47.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zuppardo A B, Siebeling R J. An epimerase gene essential for capsule synthesis in Vibrio vulnificus. Infect Immun. 1998;66:2601–2606. doi: 10.1128/iai.66.6.2601-2606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]