Abstract
Shigella strains are in reality clones of Escherichia coli and are believed to have emerged relatively recently (G. M. Pupo, R. Lan, and P. R. Reeves, Proc. Natl. Acad. Sci. USA 97:10567–10572, 2000). There are 33 O-antigen forms in these Shigella clones, of which 12 are identical to O antigens of other E. coli strains. We sequenced O-antigen gene clusters from Shigella boydii serotypes 4, 5, 6, and 9 and also studied the O53- and O79-antigen gene clusters of E. coli, encoding O antigens identical to those of S. boydii serotype 4 and S. boydii serotype 5, respectively. In both cases the S. boydii and E. coli O-antigen gene clusters have the same genes and organization. The clusters of both S. boydii 6 and S. boydii 9 O antigens have atypical features, with a functional insertion sequence and a wzx gene located in the orientation opposite to that of all other genes in S. boydii serotype 9 and an rmlC gene located away from other rml genes in S. boydii serotype 6. Sequences of O-antigen gene clusters from another three Shigella clones have been published, and two of them also have abnormal structures, with either the entire cluster or one gene being located on a plasmid in Shigella sonnei or Shigella dysenteriae, respectively. It appears that a high proportion of clusters coding for O antigens specific to Shigella clones have atypical features, perhaps indicating recent formation of these gene clusters.
Lipopolysaccharide (LPS) is a key component of the outer membranes of gram-negative bacteria. It comprises three distinct regions: lipid A, an oligosaccharide core, and, commonly, a repeat unit polysaccharide O antigen. The O antigen is one of the most variable cell constituents, with variation in the types of sugars present, their arrangement within the O unit, and the linkages between O units. The highly variable nature of the O antigen provides the basis for serotyping, and 187 O-antigen forms (serotypes) have been recognized in Escherichia coli (including Shigella strains) (11, 23, 25).
The genes for O-antigen synthesis are normally in a gene cluster which maps between galF and gnd in E. coli and Salmonella enterica. The differences between the many forms of O antigen are almost entirely due to genetic variation in this gene cluster. It has been proposed that inter- and intraspecies lateral transfer of O-antigen genes played an important role in redistributing the polymorphic forms (e.g., references 20, 24, 51, and 53). In regard to the origin of the polymorphism, it has been found that new forms can be formed by homologous recombination or recombination mediated by a transposable element (e.g., references 8, 15, 52, 53, and 57).
The O antigen is on the cell surface and appears to be a major target of both the immune system and bacteriophages, which must apply intense selection. Selection is probably a major factor in the origin and maintenance of the high level of variation. Each strain expresses only one O-antigen form, and the variation is thought to allow each of the various clones of a species to present a surface that offers a selective advantage in the niche occupied by that clone. It has been estimated that a selective advantage of only 0.1% for one O antigen over another in a given niche is more than sufficient to maintain different alleles in different clones (45).
Analysis of sequence variation in housekeeping genes showed that most of the 46 Shigella serotypes fall into three clusters within E. coli, with five outlier strains (see reference 43). It is important to note that although 46 Shigella serotypes are recognized, there are only 33 distinct O antigens, the others being modifications that in E. coli or S. enterica would not be given separate status. There are only two distinct O-antigen forms for the 14 Shigella flexneri serotypes (see reference 43), and also Shigella boydii serotype 15 and Shigella dysenteriae serotype 2 have identical O antigens (11). Most O-antigen variation is in clusters 1 and 2, with 19 and 7 O-antigen forms, respectively (43). Based on sequence diversity, it was estimated that strains within these two clusters diverged over 50,000 to 270,000 years.
Of the 33 O-antigen forms found in the two clusters, 12 are identical to other known E. coli O antigens and 21 are unique to Shigella clones. This determination is based on cross-reactions summarized by Ewing (11). In many cases the conclusions have been confirmed by other structure data or the extensive restriction fragment length polymorphism analysis of the O-antigen gene cluster reported by Coimbra et al. (6), although there are a few discrepancies that might lead to minor adjustments when they are resolved. If the unique forms were gained in Shigella rather than lost by other E. coli strains, the 21 new O antigens gained by Shigella clones in the last 50,000 to 270,000 years represent 11% of the total number of E. coli O antigens, a very rapid expansion by interspecies lateral transfer.
To start analysis of this phenomenon, we sequenced gene clusters for S. boydii O antigens 4, 5, 6, and 9. S. boydii O antigens 4 and 6 are in cluster 1, while O antigens 5 and 9 are in cluster 2. S. boydii O antigens 4 and 5 are identical to O antigens 53 and 79, respectively, of traditional E. coli strains, and we also studied the O-antigen gene clusters for these E. coli O antigens.
MATERIALS AND METHODS
Bacterial strains.
S. boydii strains LSPQ2428 (type 4), LSPQ3686 (type 5), LSPQ3687 (type 6), and LSPQ3482 (type 9) were kindly provided by J. Lefebvre of the Canadian National Laboratory for Enteric Pathogens, Ste-Anne-de- Bellevue, Ontario, Canada, where they are used as reference strains (21). Their antigens were confirmed in our laboratory by agglutination using antisera obtained from Denka Seiken Co. Ltd., Tokyo, Japan. E. coli Bi 7327-41(O53:H3) and E. coli E49(O79:H40), the O53 and O79 type strains, were from the Institute of Medical and Veterinary Science, Adelaide, Australia. Plasmids were maintained in E. coli K-12 strain JM109.
Construction of a random DNase I bank for sequencing DNA fragments.
Chromosomal DNA used as the template for PCR was prepared using a Wizard DNA preparation kit from Promega. Long PCR was carried out using the Expand Long Template PCR System from Roche, and products were subjected to DNase I digestion and cloned into pGEM-T to make banks for sequencing by using the method described previously (54).
Sequencing and analysis.
The DNA template for sequencing was prepared using a 96-well-format plasmid DNA miniprep kit from Advanced Genetic Technologies, and sequencing was performed with an Applied Biosystem 377 automated DNA sequencer. Sequence data were assembled using the Phred/Phrap package of the University of Washington Genome Center, and the sequence annotation was done using the program Artemis from the Sanger Centre. Further analysis was undertaken using programs available through the Australian National Genomic Information Service at The University of Sydney. Sequence comparisons were analyzed using the MULTICOMP package (48), which gives pairwise comparisons of DNA and amino acid sequences.
Nucleotide sequence accession numbers.
The DNA sequences of S. boydii 4, 5, 6, and 9 O-antigen gene clusters have been deposited in GenBank under accession numbers AF402312 to AF402315, respectively. The DNA sequences of segments of the E. coli O53 and O79 gene clusters have been deposited in GenBank under accession numbers AF409075 to AF409080.
RESULTS AND DISCUSSION
Sequences of O-antigen gene clusters for S. boydii O antigens 4, 5, 6, and 9.
The O-antigen gene clusters from S. boydii strains of O-antigen types 4, 5, 6, and 9 were PCR amplified using primers #1523 (5′-ATTGTGGCTGCAGGGATCAAAGAAATC) and #1524 (5′-TAGTCXCGCTGNGCCTGXATYAXGTTZGC), which bind to the 5′ end of the upstream galF gene and the 3′ end of the downstream gnd gene, respectively. To limit the effect of PCR errors, 10 individual PCR products were pooled before we made the bank for each gene cluster.
For S. boydii O antigens 4, 5, 6, and 9, sequences of 10,551 bp (10 genes), 13,116 bp (13 genes), 12,611 bp (11 genes), and 8,829 bp (9 genes), respectively, were found between galF and gnd (Fig. 1). The nucleotide and amino acid sequences were used to search available databases for indication of possible function.
FIG. 1.
O-antigen gene clusters of E. coli S. boydii serotypes 4, 5, 6, and 9. All genes are transcribed in the direction from galF to gnd, except for the wzx gene in O6. The G+C content is given above each gene.
The four gene clusters are very similar to those for most other E. coli O antigens, with nucleotide sugar biosynthesis genes, wzx, wzy, and sugar transferase genes found in each. The O-antigen chain length determinant gene (wzz) is generally located outside of the main O-antigen gene cluster in E. coli (4, 5) and was not found in any of the four newly sequenced gene clusters.
The galF and gnd genes of these four S. boydii strains are typical E. coli genes.
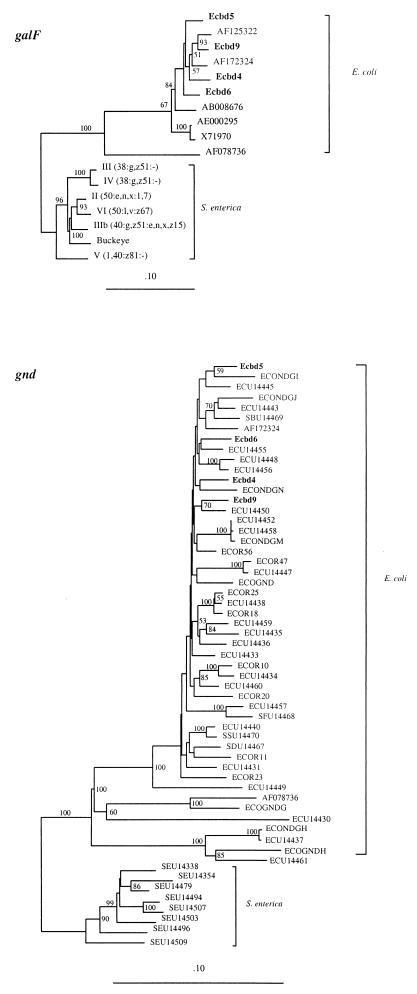
DNA from positions 1 to 765 encodes most of GalF (from amino acid [aa] 44 to the C′ terminus) in each of the four sequences. The last 1,218 bp of each sequence encodes part of Gnd (from aa 1 to 406). We compared these sequences with those of all the known galF and gnd genes from E. coli and S. enterica, and trees for the two genes are shown in Fig. 2. For both trees, genes from E. coli and S. enterica strains form separate groups and the genes from these four S. boydii strains are within the E. coli group.
FIG. 2.
Phylogenetic trees for the galF and gnd genes generated by the neighbor-joining method, including sequences from the four S. boydii strains (Ecbd4, Ecbd5, Ecbd6, and Ecbd9). For gnd genes from S. enterica and gnd and galF genes from other E. coli strains, GenBank accession numbers are used. galF sequences from S. enterica strains are unpublished data (R. Lan, D. M. Ryan, and P. R. Reeves), and serovar names are given. The values adjacent to the nodes indicate percentages of 1,000 bootstrap trees that contain the node. Only those greater than 50% are shown.
Nucleotide sugar biosynthesis genes.
O-antigen gene clusters generally contain three classes of (i) genes for synthesis of nucleotide sugar precursors such as dTDP-rhamnose, (ii) genes for transfer of sugars to build the O unit, and (iii) genes which carry out specific assembly or processing steps in the conversion of the O unit to the O antigen as part of complete LPS, such as the O-antigen flippase gene (wzx) and the O-antigen polymerase gene (wzy) (see reviews by Reeves [46, 47] and Whitfield [56]).
Figure 3 shows the structures of the four O antigens. UDP-Glc and UDP-GlcNAc are synthesized by housekeeping genes located outside of the O-antigen gene cluster in E. coli. UDP-GlcA is synthesized from UDP-Glc by UDP-glucose-6-dehydrogenase (Ugd). ugd is located outside of the O-antigen gene cluster between the gnd and the his operons in E. coli (5).
FIG. 3.
O-antigen repeat units of E. coli and S. boydii O4 (29), O5 (28), O6 (9), and O9 (27). Gal, galactose; Glc, glucose; GlcA, glucorunic acid; GlcNAc, N-acetylglucosamine; Man, mannose; Rha, rhamnose.
We expect genes for the synthesis of dTDP-rhamnose from glucose-1-phosphate in the gene clusters of S. boydii O antigens 4, 5, and 9. Four genes from each of the three gene clusters were identified as rmlB (dTDP-glucose-4,6-dehydratase), rmlD (dTDP-l-rhamnose synthase), rmlA (glucose-1-phosphate thymidyl transferase), and rmlC (dTDP-4-keto-6-deoxyglucose-3,5-epimerase) by their high levels of identity to many rml genes.
Genes for the synthesis of GDP-mannose are expected in the gene clusters of S. boydii O antigens 5 and 6. manB and manC, coding for phosphomannomutase and GDP-mannose pyrophosphosphorylase, are identified from both gene clusters based on their high levels of identity to other GDP-mannose synthesis genes. GDP-mannose is synthesized from fructose-6-phosphate by products of manA, manB, and manC, and manA maps as an individual gene not associated with polysaccharide gene clusters due to its role in mannose catabolism in E. coli and S. enterica (37).
wzx and wzy genes.
Presumptive wzx genes were first identified as encoding a potential integral inner membrane protein with 12 predicted transmembrane segments. Wzx proteins can be difficult to identify with confidence by sequence searches as sequence identity levels are low (49), and motif searches are often more convincing. Each of the putative Wzx proteins was grouped using the BLOCKMAKER program (14) with known or putative Wzx proteins, and this analysis revealed motifs which are conserved among this group of proteins. The consensus sequence was used to run the program PSI-BLAST (2) to search the Genpept database; the input Wzx proteins and many other distantly related Wzx proteins but no other proteins were retrieved (E value ≤ 4e × 10−6) after several iterations, confirming the designation.
The genes we consider to be wzy encode proteins having 10 or 11 predicted transmembrane segments with a large periplasmic loop, a characteristic topology for O-antigen polymerases (36). Each of these proteins was grouped with known or putative Wzy proteins, and motifs were generated and used to search databases as described above for Wzx. Only Wzy proteins were retrieved after two iterations (E value ≤ 3e × 10−10), confirming the designation.
Putative transferase genes.
Based on the O-antigen structures (Fig. 3), we expect 5, 6, 5, and 4 sugar transferases for S. boydii O antigens 4, 5, 6, and 9, respectively, including one to add the first sugar to the carrier lipid undecaprenol phosphate (UndP). It has been shown that WecA transfers GlcNAc phosphate or GalNAc phosphate to UndP to initiate oligosaccharide unit synthesis in E. coli strains with GlcNAc or GalNAc as the first O-unit sugar (1, 3). WecA also initiates enterobacterial common antigen synthesis by transfer of GlcNAc phosphate, and the wecA gene is located within the enterobacterial common antigen gene cluster in E. coli (5). Thus, WecA is the first transferase for the four S. boydii O antigens, and we expect to find four, five, four, and three additional transferase genes in gene clusters for S. boydii O antigens 4, 5, 6, and 9, respectively.
S. boydii O4.
WbdS shows 50% similarity to Cps2T (WchF), a putative sugar transferase of Streptococcus pneumoniae serotype 2 (16). WbdG shares 49.5% similarity with WaaK, an N-acetylglucosamine transferase involved in the synthesis of the oligosaccharide core of LPS in S. enterica (30). WbdE and WbdF do not share similarity with any known proteins. Because two more transferases are needed for the synthesis of the O4 unit, we assume that wbdF and wbdG are also transferase genes.
S. boydii O5.
WbdT and WbdX share 49 and 54% similarity with Cps14I (WchL; N-acetylglucosaminyl transferase) and Cps14J (WchM; galactosyl transferase), respectively, of S. pneumoniae serotype 14 (19). WbdU, WbdV, and WbdW do not share similarity with any known proteins, and it was presumed that they are the three additional transferases.
S. boydii O6.
WbaT shares 49% similarity with WaaB, a galactosyl transferase catalyzing the galactosyl 1–6 glucose linkage in the oligosaccharide core of S. enterica (13, 50). We suggest that wbaT is the galactosyl transferase gene for the α-galactosyl 1-6 mannose linkage. WbaX and WbaY share 50.8 and 52.5% similarity with WbaW and WbdC, respectively. WbaW is a mannosyl transferases catalyzing α-mannosyl 1-2 mannose linkages in the S. enterica C2 O antigen (26). WbdC in the E. coli O9a antigen gene cluster is also a mannosyl transferase, which puts mannose onto the pyrophosphorylundecaprenol-linked glucose via a 1,3 linkage (17). We propose that WbaX and WbaY are mannosyl transferases for the α-mannosyl 1-2 mannose and the α-mannosyl 1-3 N-acetylgalactosamine linkages, respectively, in S. boydii O antigen 6 (Fig. 3). WbaS shares 49% similarity with AceP of Acetobacter xylinum, a β-d-1,6 glucosyl transferase catalyzing a β-glucosyl 1-6 α-glucose linkage (10). We propose that WbaS is the remaining transferase, responsible for the α glucuronic acid 1-4 β-galactose linkage.
S. boydii O9.
WbgS shares 55% similarity also with AceP of A. xylinum, and we suggest that WbgS catalyzes the α glucosyl 1-4 glucuronic acid linkage in S. boydii O9. WbgR shares 47% similarity with CpsI, an N-acetylglucosaminyl transferase of S. pneumoniae serotype 14 (19). WbgQ shares 58% similarity with WcgB, a putative glycosyltransferase of Bacteroides fragilis (7). We suggest that WbgR and WbgO are the two remaining transferases.
In summary, we have found, in each of the four S. boydii O-antigen gene clusters, all genes expected for the synthesis and processing of the O unit. There is also an insertion (IS) sequence in the S. boydii O6 gene cluster, and this will be discussed below.
The rml genes of S. boydii O antigens 4, 5, and 9.
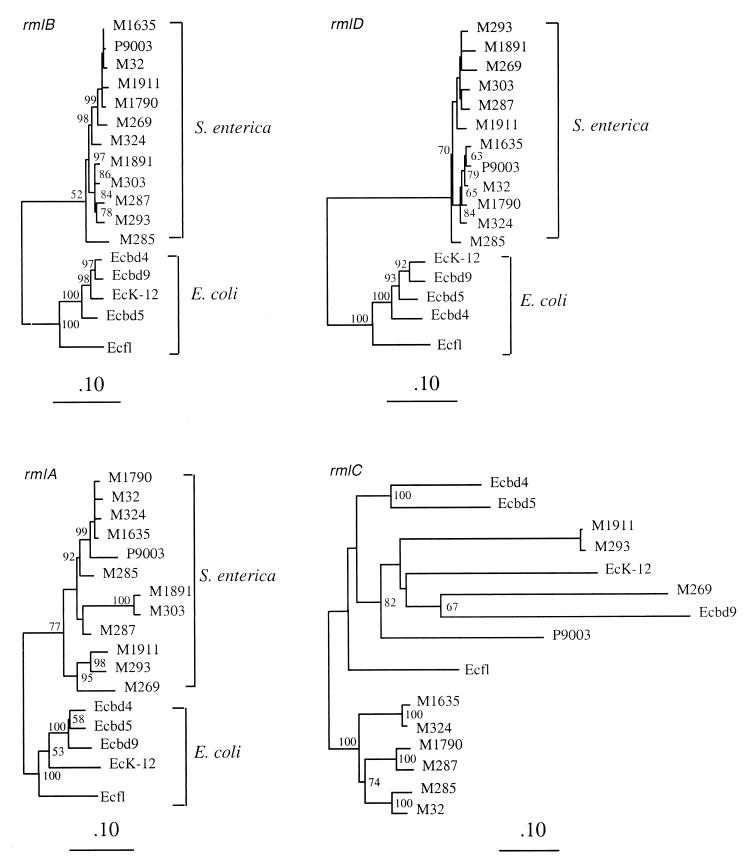
Three of the S. boydii gene clusters include the rml gene set. Rhamnose is widely distributed in O antigens of gram-negative bacteria. The four rml genes are usually grouped together; they have been identified in a range of species and are clearly homologous, although the gene order may be different in different species (24). Many polysaccharide gene clusters have a cassette structure with a central set of varied serotype-specific genes flanked by genes widely present in that class of gene clusters. In E. coli and S. enterica, the four rml genes are generally clustered in the order rmlB rmlD rmlA rmlC at the 5′ end of the O-antigen gene cluster (24). We found the same gene order in the three S. boydii O-antigen gene clusters except that in the O9 gene cluster rmlC was not found immediately downstream of rmlA but was separated by four genes (Fig. 1). DNA from positions 1 to 4048, containing the galF, rmlB, rmlD, and rmlA genes, shows identity levels ranging from 95.1 to 95.5% in pairwise comparisons among the three S. boydii gene clusters. These DNA fragments share 92 and 86% identity with corresponding genes from E. coli K-12 (GenBank accession number D90842) and Flexneri 2a (GenBank accession number SFRF BAJ), respectively.
Phylogenetic trees of the rml genes of the three S. boydii strains, E. coli K-12 (GenBank accession number D90842), S. flexneri 2a (GenBank accession number SFRFBAJ), and 12 S. enterica strains (24) were constructed using the neighbor-joining method (Fig. 4). The rmlB, rmlD, and rmlA genes of the three S. boydii strains were grouped with those of other E. coli strains and separated from the S. enterica genes.
FIG. 4.
Phylogenetic trees for the rmlB, rmlD, rmlA, and rmlC genes generated by the neighbor-joining method. Sequences used include those from three S. boydii strains (Ecbd4, Ecbd5, and Ecbd9), E. coli K-12 (EcK-12), and Flexneri 2a (Ecfl), and 12 S. enterica strains (laboratory names are used as in reference 24). The values adjacent to the nodes indicate percentages of 1,000 bootstrap trees that contain the node. Only those greater than 50% are shown.
We recently studied the rml genes in S. enterica strains that varied in O antigens and subspecies (24). It was found that the 5′ end of the rml gene set, including rmlB, rmlD, and most of rmlA, is subspecies specific and has a level of variation comparable to that of housekeeping genes but that the 3′ end, including part of rmlA and all of rmlC, is much more varied and O-antigen specific (24). Extensive recombination in the gene set, probably related to O-antigen transfer between subspecies, was also evident (24). It was concluded that recombination in rml genes plays a role in mediating the transfer of the central serotype-specific genes, which are located downstream of the rmlC gene in S. enterica (24). With one exception discussed below, the rml gene set of E. coli is in the same position as in S. enterica. The rmlB, rmlD, and rmlA genes have many characteristics of housekeeping genes: the variation within E. coli is very similar to that of the adjacent gnd and galF genes (see above) and comparable to variation in housekeeping genes in general. Also as observed above, the E. coli and S. enterica genes form separate groups in the phylogenetic tree, with levels of divergence similar to those for housekeeping genes of these species. The rmlC gene is quite different, as it is much more varied in E. coli than the rmlB, rmlD, and rmlA genes. It appears that, as proposed for S. enterica (24), the rmlC genes found in E. coli were those associated with the O-antigen-specific transferase genes at the time of transfer to the species but that the rmlB, rmlD, and rmlA genes have been in E. coli for a long time and presumably have become part of the particular gene clusters where we now find them by recombination, probably during the transfer of O antigens within E. coli.
It is interesting that the rmlC gene of S. boydii serotype 9 is located four genes downstream of the rmlBDA region (Fig. 1) and is also the most divergent (Fig. 4). It is highly likely that this gene and the four genes upstream of it were recently introduced into the O9 gene cluster by recombination involving at one end one of the first three rml genes.
Anomalies in the S. boydii 6 O-antigen gene cluster.
The S. boydii O6 gene cluster has an IS sequence (positions 9724 to 11036) which shares 97.9% sequence identity with IS629 of S. sonnei (35). The IS sequence interrupts an open reading frame of 984 bp (positions 9421 to 11737). The N-terminal half (including amino acids encoded by DNA on both sides of the IS) of the protein encoded by this open reading frame shares 47% similarity to the entire Gpt protein, a purine phosphoribosyltransferase in Thermus flavus (38). The fact that neither half of this gene has any indels or stop codons indicates that the IS element was inserted recently.
The wzx gene of S. boydii O6 is located at the 3′ end of the gene cluster in the opposite orientation to that of the other O-antigen genes (Fig. 1). All previously described E. coli and S. enterica O-antigen gene clusters have their genes transcribed in the same direction (see http://www.angis.su.oz.au/BacPolGenes/welcome/html), and this is the first exception to this general observation. It may indicate that this gene was introduced to its current position very recently. It is worth noting that this wzx gene is located adjacent to the gene mutated by the IS insertion, and it may further indicate that the region including wzx and its flanking DNA was assembled recently.
O-antigen genes of S. boydii O4 and O5 are almost identical to those of E. coli O53 and O79, respectively.
S. boydii O4 and O5 antigens are identical to O antigens 53 and 79 of traditional E. coli strains (11). We carried out adjacent-gene PCR for all O-antigen genes on the type strains for E. coli O53 and O79, with PCR primers based on the O-antigen sequences (including the flanking galF and gnd genes) of S. boydii O4 and O5, respectively. We included the two S. boydii strains, and the E. coli O53 and O79 strains gave the same PCR results as the S. boydii O4 and O5 strains, respectively. This showed that the S. boydii O4 and O5 gene clusters have the same genes in the same order as those of the E. coli O53 and O79 gene clusters, respectively. We sequenced three PCR products from each of the O53 and O79 strains. DNAs of the E. coli O53 type strain and the corresponding regions in S. boydii O4 from positions 5630 to 6408, 8808 to 9373, and 9909 to 10393 share 99.8, 99.8, and 100% identity, respectively. DNAs of the E. coli O79 type strain and the corresponding regions in S. boydii O5 from positions 6426 to 6854, 7166 to 7664, and 13683 to 14138 share 98.4, 99.2, and 99.3% identity, respectively.
As described above, Shigella strains are considered to be clones of E. coli based on a comparison of housekeeping genes (42, 43). We show here that, when Shigella strains and other E. coli strains share an O antigen, the antigen genes show very high levels of identity, as is usual for strains of the same species.
We can compare the difference between typical and Shigella strains of E. coli with the difference between E. coli and S. enterica in similar ways. There are three O antigens common to E. coli and S. enterica, although they have different names in the two species. E. coli O111 and S. enterica O35 are one such pair. We recently sequenced the S. enterica O35 gene cluster and compared it with that of E. coli O111: the two gene clusters have the same genes and gene order, with DNA identity levels ranging from 88.3 to 78.2% between corresponding genes (55). In this case the divergence is comparable with that in housekeeping genes and consistent with the hypothesis that the two gene clusters evolved from a gene cluster present in their common ancestor (55).
The intergenic regions between galF and rmlB in S. boydii O4, O5, and O9 strains.
The intergenic regions between galF and the first O-antigen gene, rmlB (positions 766 to 1137 in the S. boydii O4, O5, and O9 gene clusters), show 97.6 to 98.9% identity among S. boydii strains, 95.6 to 97% identify between S. boydii strains and K-12, and 81.9 to 82.4% identity between S. boydii and S. flexneri strains. The intergenic regions between galF and the first O-antigen gene (wbdH) are 540 and 519 bp in length, respectively, in S. enterica O35 and E. coli O111 and share much less DNA identity at 64% than do coding regions. Again, the Shigella and E. coli strains are clearly within one species, with much less difference than for the two well-differentiated species E. coli and S. enterica.
Expansion of O-antigen diversity in Shigella strains.
Shigella has 33 distinct O-antigen forms, of which 12 are also found in E. coli and 21 are unique to Shigella strains. It has been shown that Shigella strains evolved recently within E. coli and proposed that the Shigella strains obtained these 21 unique O-antigen forms since the Shigella mode of pathogenicity arose in E. coli (43). In this study, by analyzing sequences of two newly sequenced and two previously sequenced (see below) gene clusters encoding O antigens unique to Shigella, we obtained evidence suggesting that the expansion of O-antigen diversity occurred by at least two means: by obtaining new clusters from other species and by modifying E. coli O-antigen gene clusters.
S. boydii O-antigens 6 and 9 are unique to Shigella, and gene clusters for these two O antigens are atypical, with O6 having an IS and a gene in the wrong orientation and O9 having the rmlC gene separated from other rml genes. These atypical features may indicate that the O6 and O9 gene clusters were assembled recently whereas those for S. boydii O4 and O5, also found in other E. coli strains, are quite typical. The gene clusters for two other O antigens unique to Shigella have been sequenced, and both S. sonnei (18) and S. dysenteriae 1 (51) have atypical features. The S. sonnei O-antigen gene cluster is on a plasmid, and we have shown that this gene cluster was recently transferred from Plesiomonas shigelloides (51). We have also shown that S. sonnei once had a normal chromosomal O-antigen gene cluster which has undergone a major deletion, presumably after transfer of the plasmid-borne O-antigen genes (20). One of the O-antigen genes of S. dysenteriae 1 is also located on a plasmid, but in this case the other O-antigen genes are on the chromosome between galF and gnd (18). Most of the S. dysenteriae 1 chromosomal O-antigen gene cluster has been sequenced, and all necessary genes were identified (18). However, examination of the published sequence revealed a mutated glycosyl transferase gene at the 3′ end of the gene cluster (unpublished observation), indicating that the original S. dysenteriae 1 gene cluster lost at least one gene, presumably after gaining the plasmid-borne gene. Again we are probably seeing an early stage in the origin of a new O antigen, with all the genes present and expressing but not yet assembled into a single gene cluster.
The S. flexneri 2a O-antigen gene cluster has also been sequenced (31–33, 44) and shows all the typical features of an E. coli O-antigen gene cluster: the wzz gene is between ugd and the his operon; all other genes, including wzx and wzy, are located between galF and gnd on the chromosome; and the rml gene set is in the usual location. S. flexneri serotypes 1 through 5 have a common basic O antigen, and this is also present in E. coli O13. This gene cluster with those of S. boydii O4 and O5 make three clusters for O antigens also found in other E. coli strains and hence presumably have been recently acquired by transfer within E. coli in the broad sense.
We now have sequences for seven Shigella O-antigen gene clusters. Four of the seven are unique to Shigella strains and, as discussed above, all have atypical features. In contrast, the three genes encoding O antigens also found in traditional E. coli strains are all typical for the species. Based on this small number, it seems that there is a correlation between whether a Shigella O antigen also occurs in traditional E. coli strains and the likelihood of it having atypical features. Gene clusters with atypical features encode O-antigen forms not found in traditional E. coli strains, with the evidence suggesting that most arose within E. coli by reassortment of genes but that S. sonnei acquired the entire O-antigen gene cluster from outside.
We can only speculate on the reason for the rapid expansion of O-antigen forms in Shigella strains. It has been observed that some E. coli O-antigen forms are disproportionately represented in pathogenic clones and concluded that the specificity of an O antigen is important in determining pathogenicity (39–41). It has also been shown that the virulence of S. flexneri is reduced if the O antigen is changed (12), and isogenic S. enterica serovar Typhimurium strains with antigen O4 are more virulent than those in which the O4 antigen has been experimentally replaced with antigen O9 (34). There is thus considerable support for the concept that O-antigen specificity is important for host colonization, at least for pathogenic strains. It is possible that the great diversification of O antigens in the three clusters of Shigella strains (see reference 43) by phage-encoded modification in cluster 3 and by the gain of new antigenic forms in clusters 1 and 2 is related to their development of intracellular invasion properties in relatively recent times. One can speculate that the O antigens previously in E. coli were not ideal for strains with the intracellular mode of colonization, providing strong selection for the modification of existing E. coli O antigens and the gain of others from other species.
Comments on nomenclature for Shigella and E. coli.
In this paper we add further support for the widely accepted view that Shigella and E. coli are really one species. Indeed, in discussing specific genes we treat those of Shigella and E. coli as genes of one species, and as we develop a better understanding of diversity within this species, it becomes very confusing to continue with the current terminology, which gives us phylogenetic trees with data from five named species intermingled. We draw attention to the urgent need to develop a new nomenclature that reflects evolutionary relationships as was done for Salmonella with adoption of the name S. enterica (20).
ACKNOWLEDGMENTS
We thank J. Lefebvre for kindly supplying S. boydii strains.
This study was supported by the Australian Research Council.
REFERENCES
- 1.Alexander D C, Valvano M A. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3398–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amor P A, Whitfield C. Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his-region containing gene clusters for multiple cell-surface polysaccharides. Mol Microbiol. 1997;26:145–161. doi: 10.1046/j.1365-2958.1997.5631930.x. [DOI] [PubMed] [Google Scholar]
- 4.Bastin D A, Brown P K, Haase A, Stevenson G, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerisation resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 5.Berlyn M K B. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coimbra R S, Grimont F, Lenormand P, Burguiere P, Beutin L, Grimont P A. Identification of Escherichia coli O-serogroups by restriction of the amplified O-antigen gene cluster (rfb-RFLP) Res Microbiol. 2000;151:639–654. doi: 10.1016/s0923-2508(00)00134-0. [DOI] [PubMed] [Google Scholar]
- 7.Comstock L E, Coyne M J, Tzianbos A O, Kasper D L. Interstrain variation of the polysaccharide B biosynthesis locus of Bacteroides fragilis: characterization of the region from strain 638R. J Bacteriol. 1999;181:6192–6196. doi: 10.1128/jb.181.19.6192-6196.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curd H, Liu D, Reeves P R. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998;180:1002–1007. doi: 10.1128/jb.180.4.1002-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dmitriev B A, Backinowsky L V, Lvov V L, Knirel Y A, Kochetkov N K. The structure of the chemical repeating-unit of the O-specific polysaccharide chain of Shigella boydii 6 lipopolysaccharide. Carbohydr Res. 1975;41:329–333. doi: 10.1016/s0008-6215(00)87035-4. [DOI] [PubMed] [Google Scholar]
- 10.Edwards K J, Jay A J, Colquhoun I J, Morris V J, Gasson M J, Griffin A M. Generation of a novel polysaccharide by inactivation of the aceP gene from the acetan biosynthetic pathway in Acetobacter xylinum. Microbiology. 1999;145:1499–1506. doi: 10.1099/13500872-145-6-1499. [DOI] [PubMed] [Google Scholar]
- 11.Ewing W H. Edwards and Ewing's identification of the Enterobacteriaceae. Amsterdam, The Netherlands: Elsevier Science Publishers; 1986. [Google Scholar]
- 12.Gemski P J, Sheahan D G, Washington O, Formal S B. Virulence of Shigella flexneri hybrids expressing Escherichia coli somatic antigens. Infect Immun. 1972;6:104–111. doi: 10.1128/iai.6.2.104-111.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff S, Henikoff J G, Alford W J, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs M, Reeves P R. Genetic organisation and evolution of Yersinia pseudotuberculosis 3,6-dideoxyhexose biosynthetic genes. Biochim Biophys Acta. 1995;1245:273–277. doi: 10.1016/0304-4165(95)00126-3. [DOI] [PubMed] [Google Scholar]
- 16.Iannelli F, Pearce B J, Pozzi G. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol. 1999;181:2652–2654. doi: 10.1128/jb.181.8.2652-2654.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kido N, Torgov V I, Sugiyama T, Uchiya K, Sugihara H, Komatsu T, Kato N, Jann K. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP-binding cassette transport system. J Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klena J D, Schnaitman C A. Function of the rfb gene cluster and the rfe gene in the synthesis of O antigen by Shigella dysenteriae 1. Mol Microbiol. 1993;9:393–402. doi: 10.1111/j.1365-2958.1993.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 19.Kolkman M A B, van der Zeijst B A M, Nuijten P J M. Functional analysis of glycosyltransferases encoded by the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae serotype 14. J Biol Chem. 1997;272:19502–19508. doi: 10.1074/jbc.272.31.19502. [DOI] [PubMed] [Google Scholar]
- 20.Lai V, Wang L, Reeves P R. Escherichia coli clone Sonnei (Shigella sonnei) had a chromosomal O-antigen gene cluster prior to gaining its current plasmid-borne O-antigen genes. J Bacteriol. 1998;180:2983–2986. doi: 10.1128/jb.180.11.2983-2986.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefebvre J, Gosselin F, Ismail J, Lorange M, Lior H, Woodward D. Evaluation of commercial antisera for Shigella serogrouping. J Clin Microbiol. 1995;33:1997–2001. doi: 10.1128/jcm.33.8.1997-2001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Minor L, Popoff M Y. Designation of Salmonella enterica sp. nov., nom. rev., as the type and only species of the genus Salmonella. Int J Syst Bacteriol. 1987;37:465–468. [Google Scholar]
- 23.Le Minor L, Richard C. Méthod de laboratoire pour l'identification des entérobactéries. Paris, France: Institut Pasteur; 1993. pp. 72–78. [Google Scholar]
- 24.Li Q, Reeves P R. Genetic variation of dTDP-l-rhamnose pathway genes in Salmonella enterica. Microbiology. 2000;146:2291–2307. doi: 10.1099/00221287-146-9-2291. [DOI] [PubMed] [Google Scholar]
- 25.Lior H. Classification of Escherichia coli. In: Gyles C L, editor. Escherichia coli in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1994. pp. 31–72. [Google Scholar]
- 26.Liu D, Haase A M, Lindqvist L, Lindberg A A, Reeves P R. Glycosyl transferases of O-antigen biosynthesis in Salmonella enterica: identification and characterization of transferase genes of groups B, C2, and E1. J Bacteriol. 1993;175:3408–3413. doi: 10.1128/jb.175.11.3408-3413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.L'vov V L, Musina L I, Shashkov A S, Ermakov G P, Dmitriev B A. Antigenic polysaccharides of bacteria of the genus Shigella. Determination of the structure of polysaccharide chains of Shigella boydii type 9 lipopolysaccharide and detection of unusually high molecular weight glycolipid. Bioorg Khim. 1987;13:1245–1255. [PubMed] [Google Scholar]
- 28.L'vov V L, Shashkov A S, Knirel Y A, Arifulina A E, Senchenkova S N, Yakovlev A V, Dmitriev B A. Structure of the O-specific polysaccharide chain of Shigella boydii type 5 lipopolysaccharide: a repeated study. Carbohydr Res. 1995;279:183–192. doi: 10.1016/0008-6215(95)00276-6. [DOI] [PubMed] [Google Scholar]
- 29.L'vov V L, Tochtamysheva N V, Dmitriev B A, Kochetkov N K, Hofman I L. Bacterial antigenic polysaccarides. X. The structure of polysaccharide chain of Shigella boydii type 4 lipopolysaccharide. Bioorg Khim. 1980;6:1842–1850. [Google Scholar]
- 30.MacLachlan P R, Kadam S K, Sanderson K E. Cloning, characterization, and DNA sequence of the rfaLK region for lipopolysaccharide synthesis in Salmonella typhimurium LT2. J Bacteriol. 1991;173:7151–7163. doi: 10.1128/jb.173.22.7151-7163.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macpherson D F, Manning P A, Morona R. Characterization of the dTDP-rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol. 1994;11:281–292. doi: 10.1111/j.1365-2958.1994.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 32.Macpherson D F, Manning P A, Morona R. Genetic analysis of the rfbX gene of Shigella flexneri. Gene. 1995;155:9–17. doi: 10.1016/0378-1119(94)00918-i. [DOI] [PubMed] [Google Scholar]
- 33.Macpherson D F, Morona R, Beger D W, Cheah K-C, Manning P A. Genetic analysis of the rfb region of Shigella flexneri encoding the Y serotype O-antigen specificity. Mol Microbiol. 1991;5:1491–1499. doi: 10.1111/j.1365-2958.1991.tb00795.x. [DOI] [PubMed] [Google Scholar]
- 34.Mäkelä P H, Valtonen V V, Valtonen M. Role of O-antigen (lipopolysaccharide) factors in the virulence of Salmonella. J Infect Dis. 1973;128(Suppl.):S84–S85. doi: 10.1093/infdis/128.supplement_1.s81. [DOI] [PubMed] [Google Scholar]
- 35.Matsutani S, Ohtsubo H, Maeda Y, Ohtsubo E. Isolation and characterization of IS elements repeated in the bacterial chromosome. J Mol Biol. 1987;196:445–455. doi: 10.1016/0022-2836(87)90023-4. [DOI] [PubMed] [Google Scholar]
- 36.Morona R, Mavris M, Fallarino A, Manning P A. Characterisation of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. [Google Scholar]
- 38.Nishiyama M, Kukimoto M, Beppu T, Horinouchi S. An operon encoding aspartokinase and purine phosphoribosyltransferase in Thermus flavus. Microbiology. 1995;141:1211–1219. doi: 10.1099/13500872-141-5-1211. [DOI] [PubMed] [Google Scholar]
- 39.Ø F, Ørskov I. Special Escherichia coli serotypes among enterotoxigenic strains from diarrhoea in adults and children. Med Microbiol Immunol. 1976;162:73–80. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- 40.Ørskov F, Ørskov I. Special Escherichia coli serotypes from enteropathies in domestic animals and man. Fortschr Vetmed. 1979;529:7–14. [Google Scholar]
- 41.Ørskov I, Ørskov F. Special O:K:H serotypes among enterotoxigenic Escherichia coli strains from diarrhoea in adults and children. Med Microbiol Immunol. 1977;103:99–110. doi: 10.1007/BF02121318. [DOI] [PubMed] [Google Scholar]
- 42.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pupo G M, Lan R, Reeves P R. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajakumar K, Jost B H, Sasakawa C, Okada N, Yoshikawa M, Adler B. Nucleotide sequence of the rhamnose biosynthetic operon of Shigella flexneri 2a and role of lipopolysaccharide in virulence. J Bacteriol. 1994;176:2362–2373. doi: 10.1128/jb.176.8.2362-2373.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reeves P R. Variation in O antigens, niche specific selection and bacterial populations. FEMS Microbiol Lett. 1992;100:509–516. doi: 10.1111/j.1574-6968.1992.tb14085.x. [DOI] [PubMed] [Google Scholar]
- 46.Reeves P R. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 47.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. In: Neuberger A, van Deenen L L M, editors. Bacterial cell wall. New comprehensive biochemistry. Vol. 27. Amsterdam, The Netherlands: Elsevier Science Publishers; 1994. pp. 281–314. [Google Scholar]
- 48.Reeves P R, Farnell L, Lan R. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. CABIOS. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 49.Reeves P R, Hobbs M, Valvano M, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C, Rick P. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 50.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shepherd J G, Wang L, Reeves P R. Comparison of O-antigen gene clusters of Escherichia coli (Shigella) Sonnei and Plesiomonas shigelloides O17: Sonnei gained its current plasmid-borne O-antigen genes from P. shigelloides in a recent event. Infect Immun. 2000;68:6056–6061. doi: 10.1128/iai.68.10.6056-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Evolutionary relationship among rfb gene clusters synthesizing mannose homopolymer as O-specific polysaccharides in Escherichia coli and Klebsiella. Gene. 1997;198:111–113. doi: 10.1016/s0378-1119(97)00300-4. [DOI] [PubMed] [Google Scholar]
- 53.Sugiyama T, Kido N, Kato Y, Koide N, Yoshida T, Yokochi T. Generation of Escherichia coli O9a serotype, a subtype of E. coli O9, by transfer of the wb∗ gene cluster of Klebsiella O3 into E. coli via recombination. J Bacteriol. 1998;180:2775–2778. doi: 10.1128/jb.180.10.2775-2778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Reeves P R. Organization of Escherichia coli O157 O-antigen gene cluster and identification of its specific genes. Infect Immun. 1998;66:3545–3551. doi: 10.1128/iai.66.8.3545-3551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Reeves P R. The Escherichia coli O111 and Salmonella enterica O35 gene clusters: gene clusters encoding the same colitose-containing O antigen are highly conserved. J Bacteriol. 2000;182:5256–5261. doi: 10.1128/jb.182.18.5256-5261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitfield C. Biosynthesis of lipopolysaccharide O-antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 57.Xiang S H, Hobbs M, Reeves P R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994;176:4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]