Abstract
The lipopolysaccharide (LPS)-binding protein (LBP) has a concentration-dependent dual role in the pathogenesis of gram-negative sepsis: low concentrations of LBP enhance the LPS-induced activation of mononuclear cells (MNC), whereas the acute-phase rise in LBP concentrations inhibits LPS-induced cellular stimulation. In stimulation experiments, we have found that LBP mediates the LPS-induced cytokine release from MNC even under serum-free conditions. In biophysical experiments we demonstrated that LBP binds and intercalates into lipid membranes, amplified by negative charges of the latter, and that intercalated LBP can mediate the CD14-independent intercalation of LPS into membranes in a lipid-specific and temperature-dependent manner. In contrast, prior complexation of LBP and LPS inhibited binding of these complexes to membranes due to different binding of LBP to LPS or phospholipids. This results in a neutralization of LPS and, therefore, to a reduced production of tumor necrosis factor by MNC. We propose that LBP is not only present as a soluble protein in the serum but may also be incorporated as a transmembrane protein in the cytoplasmic membrane of MNC and that the interaction of LPS with membrane-associated LBP may be an important step in LBP-mediated activation of MNC, whereas LBP-LPS complexation in the serum leads to a neutralization of LPS.
Human lipopolysaccharide (LPS)-binding protein (LBP) is a serum glycoprotein belonging to a family of lipid-binding proteins which includes bactericidal/permeability-increasing protein (BPI), phospholipid ester transfer protein, and cholesterol ester transfer protein (1, 18, 36). It consists of 456 amino acid residues preceded by a hydrophobic signal sequence of 25 residues (31). LBP is synthesized by hepatocytes (26) and intestinal epithelial cells (42) and is present in normal serum at concentrations of 5 to 10 μg/ml, rising up to 200 μg/ml 24 h after induction of an acute-phase response (35). This rise in LBP levels is caused by transcriptional activation of the LBP gene mediated by interleukin-1 (IL-1) and IL-6 (17). LBP has a concentration-dependent dual role: low concentrations of LBP enhance the LPS-induced activation of mononuclear cells (MNC), whereas the acute-phase rise in LBP concentrations inhibits LPS-induced cellular stimulation (20). LBP binds a variety of LPS (endotoxin) chemotypes from rough and smooth strains of gram-negative bacteria and even lipid A, the lipid moiety of LPS (37, 38). The LPS molecules, components of the outer membrane of gram-negative bacteria, are important mediators in the pathogenesis of gram-negative sepsis and septic shock (25). Because the lipid A moiety has been shown to be responsible for the biological activity of LPS in most in vivo and in vitro test systems, it has been termed the endotoxic principle of LPS (27).
LPSs activate monocytes and macrophages to secrete inflammatory cytokines (tumor necrosis factor alpha [TNF-α] and IL-1, etc.) and other potent mediators (32) by an intracellular signal amplification pathway. These mediators, in turn, act on additional target cells to produce cardiovascular shock, multisystem organ failure, and septic shock (6, 13), one of the major causes of death in intensive care units. Specific cellular responses in organisms are generally mediated by receptors. For endotoxin recognition, a binding protein/receptor system has been postulated that involves LBP, the membrane bound and soluble CD14 molecules, members of the family of Toll-like receptors (32, 39), and a K+ channel (5, 24).
LBP increases the capacity of LPS to induce cytokine release by mononuclear phagocytes (8, 15), and neutralization of LBP with rabbit anti-LBP antibodies (Abs) prevents binding of LPS to monocytes (15) and protects mice from lethal endotoxemia (11). The important role of LBP in LPS-induced cell activation has been underlined by the observation that blood from mice with a targeted deletion of the LBP gene was hyporesponsive to LPS by at least 1,000-fold (48). In these mice, a transfer of LPS to CD14 was not observed (16). It was shown recently, using reconstituted planar membranes, that LBP intercalates in a directed manner and transmembranously into bilayers composed of an extracellular leaflet with a negative surface charge density. LPS and lipid A were shown to bind to LBP on both sides of the membrane, and binding at the extracellular side led to a conformational change of the protein or a change of its orientation in the membrane (14). Moreover, it has been shown that LBP transfers phospholipids to LPS micelles (50). It has been shown that an interaction of LPS with membrane-associated LBP is more likely to occur than the function of LBP as a shuttle protein bringing LPS to the cell surfaces independent of CD14 (31). For the subsequent signal transduction, binding of complexes of LPS and LBP to CD14 is necessary (46). Therefore, the formation of microdomains of LBP, CD14, and other proteins involved in LPS signaling, e.g., the Toll-like receptors or ion channels, is likely.
In contrast to the enhancement of the LPS-induced activation of MNC by LBP, an LPS-neutralizing effect has also been observed. At high concentrations, LBP inhibits LPS-mediated cytokine release and prevents hepatic failure, resulting in a significantly decreased mortality rate in LPS-challenged mice as well as in a murine model of bacteremia (20). It has been shown that LBP knockout mice were more susceptible to the lethal effects of infection with live bacteria than healthy mice (16). Furthermore, LBP has been found to mediate LPS transfer to reconstituted high density lipoprotein (HDL) and to low density lipoprotein, attenuating its stimulatory effects (40, 47). LBP facilitates binding of a series of phosphatidylinosides and phosphatidylserine to membrane-bound CD14 (43), resulting in an inhibition of the LPS-induced response in monocytes.
LBP has often been compared to BPI. Both proteins bind LPS, and a sequence comparison for human LBP and BPI revealed 44% amino acid identity (31). BPI has been found on the cell surface of human peripheral blood monocytes (7). In contrast to BPI, LBP has no effect on the viability of gram-negative bacteria at concentrations at which BPI is very effective (36), and the effects of LBP and BPI on LPS-induced cytokine release from mononuclear phagocytic cells are counteractive (8).
In this work, we focused on the differences leading to the dual role of LBP in neutralization of LPS and enhancement of LPS-mediated activation of MNC. To this end, we performed experiments to determine the LPS-induced TNF-α production by MNC. These data were correlated with those obtained from biophysical experiments on the binding of LBP to phospholipids and on CD14-independent binding and intercalation of LBP and the LBP-mediated LPS binding to phospholipids. For these experiments, surface plasmon resonance (SPR) and fluorescence resonance energy transfer (FRET) techniques were utilized.
MATERIALS AND METHODS
Lipids and other chemicals.
Deep rough mutant (Re) LPS from Escherichia coli strain F515 (F515 LPS) (chemical structure according to references 28 and 45) was extracted by the phenol/chloroform/petroleum ether method (9), purified, lyophilized, and transformed into the triethylamine salt form. For the preparation of the aggregates, LPSs were suspended in buffer (100 mM KCl, 5 mM HEPES, pH 7) by thorough vortexing and temperature cycled at least twice between 4 and 56°C. Each cycle was followed by intense vortexing for a few minutes, and then the LPSs were stored at 4°C for at least 12 h before measurement.
Phosphatidylcholine (PC) and phosphatidylglycerol (PG) from egg, sphingomyelin (SM) and phosphatidylserine (PS) from bovine brain, and phosphatidylethanolamine (PE) from E. coli were from Avanti Polar Lipids (Alabaster, Ala.). All phospholipids were used without further purification. For preparation of the membranes from the phospholipid mixture resembling the composition of the cytoplasmic membrane of macrophages (PL), PC, PE, SM, and PS were mixed in a molar ratio of 38.1:27.3:19.4:15.2 (2, 19).
The fluorescent dyes N-(7-nitro-2,1,3-benzoxadiazol-4-yl)-PE (NBD-PE) and N-(rhodamine B sulfonyl)-PE (Rh-PE) were purchased from Molecular Probes (Eugene, Oreg.).
Proteins and antibodies.
Recombinant human LBP (456-amino-acid holoprotein rLBP50) in 10 mM HEPES, pH 7.5, was prepared according to the method described in reference 34. The monoclonal mouse anti-mouse LBP Ab biG 33 immunoglobulin G1, which is cross-reactive with human LBP, was obtained from Biometec (Greifswald, Germany).
Stimulation of human MNC by LPS.
In experiments aiming at the determination of the cytokine-inducing capacity of LPS, human MNC were stimulated by LPS, and TNF-α production of the cells was determined in the supernatant.
For the isolation of MNC, heparinized blood (20 IU/ml) from healthy donors was processed directly by mixing with an equal volume of Hanks' balanced salt solution and centrifugation on a Ficoll density gradient for 40 min (21°C, 500 × g). The interphase layer of MNC was collected and washed twice in serum-free Hanks' balanced salt solution and once in serum-free RPMI 1640 containing 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. The cells were resuspended in serum-free medium, and the number of cells was adjusted to 5 × 106 cells per ml. For stimulation experiments, 200 μl of MNC per well (106 cells/well) was transferred to 96-well culture plates. LPS was serially diluted in serum-free RPMI 1640 and added to the cultures (20 μl per well). The cultures were incubated for 4 h at 37°C and 5% CO2. Supernatants were collected after centrifugation of the culture plates for 10 min at 400 × g and stored at −20°C until determination of the cytokine content.
The immunological determination of TNF-α in the cell supernatant was determined in a sandwich enzyme-linked immunosorbent assay as recommended by the manufacturer and described elsewhere (10). Ninety-six-well plates (Greiner, Solingen, Germany) were coated with a monoclonal Ab against TNF-α (clone 6b; Intex AG, Muttenz, Switzerland). Cell culture supernatants and the standard (recombinant TNF-α [rTNF-α]; Intex) were diluted with buffer. After exposure to appropriately diluted test samples and serial dilutions of standard rTNF-α, the plates were exposed to peroxidase-conjugated rabbit anti-rTNF-α Ab. The plates were shaken for 16 to 24 h at 4°C. For the removal of free Ab, the plates were washed six times in distilled water. Subsequently, the color reaction was started by the addition of tetramethylbenzidine-H2O2 in an alcoholic solution, and after 5 to 15 min, it was stopped by the addition of 1 M sulfuric acid. In the color reaction, the substrate is cleaved enzymatically, and the product is measured photometrically. This was done on an enzyme-linked immunosorbent assay reader (Rainbow; Tecan, Crailsham, Germany) at a wavelength of 450 nm, and the values were related to the standard. The TNF-α concentration was determined in duplicate at two different dilutions, and the values were averaged.
FRET.
The FRET technique was used as a probe dilution assay (30, 33) to obtain information on the intercalation of LBP and LPS into liposomes made from various phospholipids. For the FRET experiments, phospholipid liposomes were double labeled with NBD-PE and Rh-PE. The fluorescent dyes were dissolved together with PC, PG, SM, PS, PE, or mixtures of these phospholipids in chloroform in molar ratios of 100:1:1 (lipid/NBD-PE/Rh-PE). The solvent was evaporated under a stream of nitrogen, and the lipids were resuspended in bathing solutions with 100 mM KCl and 5 mM HEPES at pH 7.0, mixed thoroughly, and sonicated with a Branson sonicator for 1 min (1 ml of solution). Subsequently, the preparation was cooled for 30 min at 4°C, heated for 30 min at 56°C, and recooled to 4°C. Preparations were stored at 4°C overnight prior to measurement. A preparation of 900 μl of the double-labeled lipid liposomes (0.1 mM) at 37°C was excited at 470 nm (excitation wavelength of NBD-PE), and the intensities of the emission light of the donor NBD-PE (531 nm) and acceptor Rh-PE (593 nm) were measured simultaneously on the fluorescence spectrometer SPEX F1T11 (SPEX Instruments, Edison, N.J.). LBP, Ab, and LPS aggregates were added after 50, 100, and 150 s. Intercalation could be detected as a change in fluorescence intensity as a function of time (increase of the donor signal, decrease of the acceptor signal). For the quantitative analysis of FRET data obtained from experiments using liposomes composed of different lipids, the dilution effects (influence of non-membrane-active solutions on the emission intensities) for each type of lipid were determined in control experiments, and the donor signal was corrected correspondingly. This method allows for the comparison of effects induced by LBP and LPS in liposomes composed of different lipids. The increases in the corrected donor intensities at t = 5 min (at which point the intensities have nearly reached constant values) were depicted graphically (see Fig. 3 and 4). For a qualitative analyses of experiments utilizing liposomes composed of the same type of lipid in each case, the quotient of the intensities of the donor dye and the acceptor dye were plotted against time (hereafter designated the FRET signal) (see Fig. 5).
FIG. 3.
Changes in the donor emission intensity in FRET experiments upon addition of 10 μg of LBP (180 nM) (A) or additional F515 LPS (10 μM) (B) to double-labeled liposomes composed of different phospholipids (10 μM). PS-PC and PS-PE are equimolar mixtures, and PL is composed of PC, PE, SM, and PS in molar ratios of 38.1:27.3:19.4:15.2. Changes in donor intensity are plotted against the average number of net negative charges per lipid molecule.
FIG. 4.
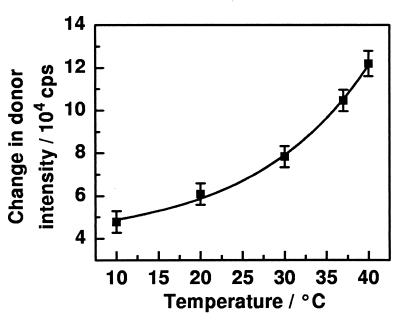
Changes in the donor emission intensity in FRET experiments upon addition of LBP (180 nM) and F515 LPS (10 μM) to double-labeled liposomes composed of PS (10 μM) to determine dependence on buffer temperature.
FIG. 5.
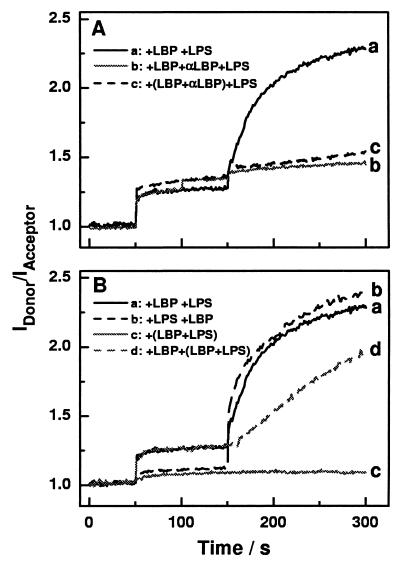
Time kinetics of changes in the quotient of the donor and acceptor emission intensities (IDonor/IAcceptor) in FRET experiments upon addition of LBP (200 nM), F515 LPS (10 μM), and anti-LBP Ab (20 μl, 1 mg/ml) to double-labeled PS liposomes (10 μM) at various time points and in various combinations (A) Addition of LBP at t = 50 s and F515 LPS at t = 150 s (trace a); addition of LBP at t = 50 s, anti-LBP Ab at t = 100 s, and F515 LPS at t = 150 s (trace b); addition of preincubated LBP and anti-LBP Ab at t = 50 s and F515 LPS at t = 150 s (trace c). (B) Addition of LBP at t = 50 s and F515 LPS at t = 150 s (trace a); addition of F515 LPS at t = 50 s and LBP at t = 150 s (trace b); addition of preincubated LBP and F515 LPS at t = 50 s (trace c); addition of LBP at t = 50 s and preincubated LBP and F515 LPS at t = 150 s (trace d).
SPR experiments.
An SPR technique (23) was used as a binding assay to detect the interaction of LBP and LPS with immobilized liposomes made from PS. First, a C1 sensor chip (Biacore AB, Uppsala, Sweden) was pretreated with 20 μl of a 4-μg/ml polylysine (Sigma Chemical Co., St. Louis, Mo.) solution to obtain a positively charged chip surface. Then a 10 μM suspension of PS liposomes was injected to obtain an immobilized lipid matrix for interaction experiments with LBP and LPS. LBP, LPS, and a preincubated (15 min, 37°C) mixture of LBP and LPS were added at concentrations of 100 nM, 10 μM, and 100 nM plus 10 μM, respectively. The running buffer was 100 mM KCl and 5 mM HEPES at pH 7.0, and experiments were performed at 25 or 37°C at a flow rate of 10 μl/min in a BIACORE 3000.
RESULTS
Influence of LBP on the LPS-induced TNF-α production by MNC under serum-free conditions.
As a measure of the biological activity of LPS, its ability to induce TNF-α in human MNC was determined. TNF-α, together with IL-1 and IL-6, is one of the important mediators induced by endotoxin.
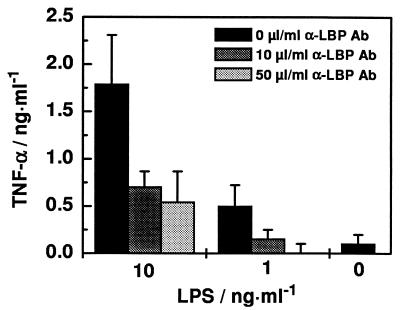
To investigate whether the LBP-mediated activation of MNC by LPS is dependent on the presence of free LBP in the serum, we washed MNC three times in serum-free medium to remove serum LBP as completely as possibly. The addition of 1 and 10 ng of LPS per ml clearly led to a concentration-dependent increase of TNF-α production (Fig. 1) which was also observed in lower amounts when the cells were washed four or five times in serum-free medium. This TNF-α production could be reduced or even completely inhibited by preincubating the MNC with the monoclonal anti-LBP Ab prior to LPS addition (Fig. 1).
FIG. 1.
Effects of monoclonal anti-LBP Ab biG 33 immunoglobulin G1 on LPS-stimulated TNF-α production by MNC under serum-free conditions. MNC (5 × 106 cells/ml) were simulated with 0, 1, or 10 ng of LPS per ml from Re mutant strain E. coli F515 in the absence of Ab or in the presence of 10 or 50 μl of Ab (1 mg/ml).
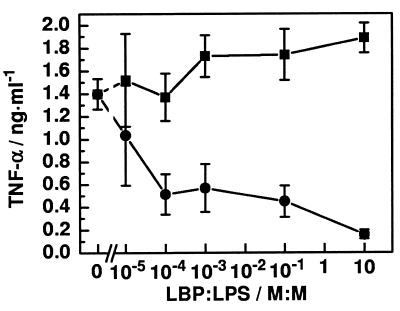
To investigate whether the sequence of addition of LBP and LPS plays a role in the function of LBP, we applied LBP at different concentrations and LPS at identical concentrations (1 ng of LPS per ml = 400 pM) in different sequences to serum-free MNC. Under both conditions, an increase in LPS-induced TNF-α release from MNC was observed with increasing amounts of LBP (Fig. 2); no significant difference resulted from the different sequences of LPS and LBP addition (data not shown). In contrast, the addition of a preincubated mixture of LBP and LPS (15 min, 37°C) to the MNC led to a significantly lower level of TNF-α production than did the absence of additional LBP. A TNF-α concentration comparable to that of unstimulated MNC was reached at a molar ratio of 10:1 (LBP/LPS) of the preincubated complexes. LPS-induced IL-6 release was also reduced in a dose-dependent way when LPS was preincubated with LBP (data not shown). Qualitatively identical results were obtained at an LPS concentration of 10 ng/ml (data not shown).
FIG. 2.
Influence of LPS, LBP, and LPS/LBP complexes on the TNF-α production of MNC under serum-free conditions. The F515 LPS concentration (1 ng/ml = 400 pM) and number of cells (5 × 106 cells/ml) were identical in all experiments. LBP was either added subsequently (■) or as preincubated (15 min, 37°C) LPS/LBP complexes (●) at the indicated LBP/LPS molar ratios.
Intercalation of LBP and LPS into phospholipid membranes.
FRET spectroscopy was used to investigate the intercalation of LBP into phospholipid membranes and its ability to mediate endotoxin intercalation into the respective bilayers. Liposomes prepared of PC, PS, PG, PL, and mixtures of PS and PC (PS-PC) and PS and PE (PS-PE) were labeled with the donor and acceptor dyes. In Fig. 3, the difference between changes in the donor intensity after the addition of 10 μl of buffer and the addition of LBP (10 μl, 180 nM) (Fig. 3A) or LBP (10 μl, 180 nM) and LPS (100 μl, 10 μM) (Fig. 3B) to the various liposome suspensions (10 μM) is plotted against the average number of negative charges per lipid molecule. In the absence of LPS, a linear correlation between these two parameters was observed (Fig. 3A). Even for the electrically neutral PC liposomes, a slight increase in the donor intensity is observed. In contrast, the LBP-mediated intercalation of LPS into the liposomes is not linearly correlated with the number of charges per lipid molecule (Fig. 3B): a change in donor intensity after subsequent addition of LPS to liposomes in the presence of LBP was observed for PS but not for PG liposomes. Moreover, higher increases in the donor intensity were obtained with PS-PE or PL liposomes than with PS-PC liposomes. A slight but significant increase in the donor signal was also observed after the addition of LBP and LPS to PC liposomes.
The increase in donor intensity after the addition of LBP (5.5 μl, 100 nM) did not depend significantly on temperature (data not shown), but the further addition of LPS (100 μl, 10 μM) led to changes in the donor intensity which increased exponentially with increasing temperature (Fig. 4).
To gain further insight into the influence of the monoclonal anti-LBP Ab on the interaction of LBP with the target membranes, experiments with PS liposomes were performed. Below we designate the quotient of the intensities of the donor dye and the acceptor dye as the FRET signal. The addition of LBP (180 nM) to double-labeled PS liposomes (10 μM) at t = 50 s led to an increase in the FRET signal, and the addition of LPS (10 μM) at t = 150 s led to a further increase (Fig. 5A). When anti-LBP Ab (20 μl at t = 100 s) was added after the first addition of LBP (at t = 50 s), the FRET signal increased slightly, and the effect of the LPS addition (at t = 150 s) was completely abolished (Fig. 5A). To investigate whether the Ab can inhibit the intercalation of LBP into the PS liposomes, we preincubated LBP and the Ab for 15 min at 37°C and then added these complexes to the PS liposomes. Under these conditions, the FRET signal increased slightly compared to that seen with LBP alone, and the effect of subsequently added LPS was completely abolished again (Fig. 5A).
To investigate the role of uncomplexed and LPS-complexed LBP in the interaction with membranes, LBP and LPS were added successively or as a preincubated mixture to double-labeled PS liposomes. The results are shown in Fig. 5. The successive addition of LBP (180 nM) and LPS (10 μM) resulted in nearly identical changes to those described above (Fig. 5A). The inverse sequence of addition led only to a slight increase in the FRET signal after LPS addition, which is indicative for a negligible interaction between LPS and PS liposomes (Fig. 5B). The further addition of LBP caused an increase in the FRET signal to a value (2.4) comparable to that of the experiment in which 180 nM LBP and 10 μM LPS were added. In contrast to the effects of uncomplexed LBP and LPS, the addition of preincubated LPS/LBP complexes did not change the FRET signal (Fig. 5B). When LBP was added to PS liposomes prior to the addition of LPS/LBP complexes, an increase in the FRET signal was observed (Fig. 5B) which was, however, smaller than that obtained after the addition of LPS alone (Fig. 5B).
Binding of LBP and endotoxin to immobilized lipids.
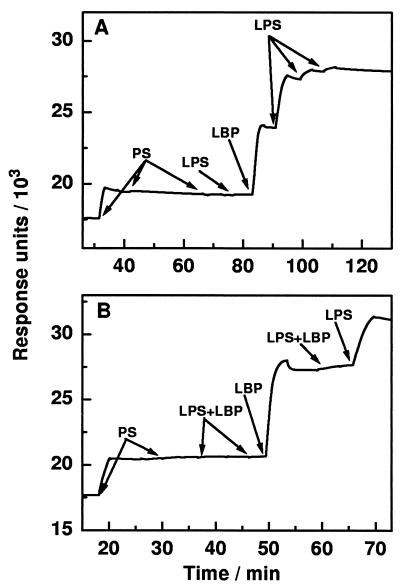
SPR experiments were performed to investigate the binding of LBP and LPS to immobilized liposomes, and the results are summarized in Fig. 6. From both panels of the figure it can clearly be seen that the first injection of PS liposomes (20 μl, 10 μM) led to an increase in response units, indicating binding of the liposomes to the polylysine-covered surface of the chip. Further injections of PS liposomes (20 and 40 μl) did not increase the signal, leading to the conclusion that the first addition led to complete coverage of the surface with PS. The subsequent addition of LPS (40 μl, 10 μM) did not lead to its binding to the PS surface (Fig. 6A). In contrast, injection of LBP (40 μl, 100 nM) resulted in its binding to the PS surface, and this could not be reversed by washing the surface with running buffer. Subsequent injection of 10 μM LPS suspension in steps of 40 μl led to a saturable binding to the PS/LBP surface in the absence of unbound LBP (Fig. 6A). The absence of unbound LBP is guaranteed by the continuous flow of LBP-free buffer solution over the surface. Comparable masses of LBP and LPS bound to the chip, and therefore, more than a 20 fold-higher number of LPS molecules bound to the PS/LBP surface than of LBP molecules to the PS surface (based on the relative molecular masses of LPS and LBP). In contrast to this observation, LPS/LBP complexes (40-μl complex of 10 μM LPS and 100 nM LBP preincubated for 15 min at 37°C) did not bind to the PS surface (Fig. 6B). Injection of LBP (40 μl, 100 nM) led to the previously described binding of LBP to the PS surface; however, the further addition of LPS/LBP complexes (40 μl) did not increase the response. Thus, it may be concluded that the LPS/LBP complexes do not bind either to PS or to PS/LBP surfaces. Binding of LBP and LBP-mediated binding of LPS to a PC surface was also observed, but the effects were reduced compared to their binding to PS surfaces (data not shown).
FIG. 6.
Time kinetics of changes in the response units in SPR experiments upon injection of LBP and LPS. Injections are marked in the diagram, and the following concentrations were used: CPS = 10 μM, CF515 LPS = 10 μM, and CLBP = 100 nM. For preincubated (15 min at 37°C) complexes of LPS/LBP, 10 μM F515 LPS and 100 nM LBP were used.
All effects observed in the FRET and SPR experiments depend on the concentrations of LBP and LPS and/or the molar ratio of LBP/LPS used. The data shown in Fig. 3 to 6 represent the results with the best visible effects.
DISCUSSION
In earlier investigations into the influence of LBP on the LPS-mediated activation of MNC, a dual role was observed. It was the aim of this study to elucidate the different mechanistic principles leading to the enhancement of LPS-mediated activation of MNC and the inhibition of this effect at higher LBP concentrations (20).
LBP-mediated activation of MNC.
LPS is a potent inducer of TNF-α production by MNC, even under serum-free conditions (15, 29). This was verified by our data shown in Fig. 1. Our observation of a suppression of TNF-α release by anti-LBP Ab under serum-free conditions provides strong evidence that LBP is still present (Fig. 1). The presence of LBP may be explained in two ways: washing of MNC does not completely remove serum LBP or LBP is present in a membrane-bound state on the surface of MNC. There is considerable evidence in the literature supporting the proposed binding of LBP to the cytoplasmic membrane of MNC: (i) binding of LPS to monocytes in serum-free media independent of CD14 was observed (15), possibly mediated by LBP, (ii) LBP interacts with various phospholipids (14, 30, 50), and (iii) BPI, a protein with high sequence homology to LBP, has been detected on the surface of monocytes (7). These findings stimulated us to investigate the interaction between LBP and phospholipid membranes utilizing different biophysical techniques and also standard biological assays for determination of TNF-α production.
Interaction between LBP and phospholipid matrices.
The SPR experiments clearly demonstrate binding of LBP to immobilized PS (Fig. 6A) and, to a lesser degree, to PC (data not shown) which could not be reversed by washing with buffer. Furthermore, the FRET data provide strong evidence for an intercalation of LBP into differently composed phospholipid liposomes with a dependence on the average number of net negative charges per lipid molecule and even into zwitterionic PC liposomes (Fig. 3A). Thus, the existence of negatively charged lipids, or of other negatively charged constituents in the outer membrane leaflet of MNC, is not a prerequisite but may enhance the membrane intercalation of LBP. For this reason, in further experiments negatively charged PS liposomes were used to obtain signals significantly above the detection limit of the FRET system.
In this context it is noteworthy that the macrophage cytoplasmic membrane contains negatively charged lipids, mainly PS and cardiolipin (2, 19); however, the distribution to the two leaflets is not well known. LBP shares 44% amino acid sequence identity with BPI (31), which has been detected on the cytoplasmic membrane of human peripheral blood monocytes (7) in tight association. In studies using reconstituted membranes, negative charges are responsible for the binding and intercalation of the polycationic BPI (44) which may be represented by anionic lipids or even other negatively charged constituents present in the cytoplasmic membrane. The intercalation of LBP into the lipid matrix is temperature independent (data not shown) and cannot be inhibited by the monoclonal anti-LBP Ab (Fig. 5A).
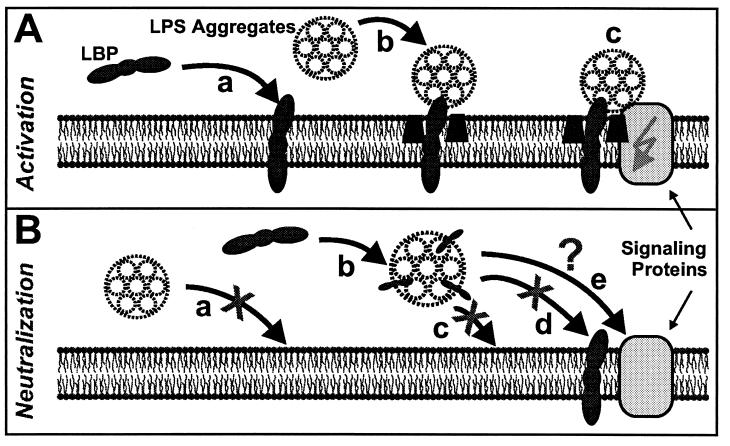
In experiments using reconstituted planar bilayers, it was previously shown that for symmetric PS membranes, LBP intercalates transmembranously and in a directed orientation into bilayers (14), as shown schematically in Fig. 7A. Binding and intercalation of LBP into LPS aggregates were also observed (12, 38) (Fig. 7B).
FIG. 7.
Cartoon of proposed mechanisms of interaction between LBP, LPS, and phospholipid membranes independent of membrane-bound CD14. (A) Interactions that may be involved in the LPS-induced activation of MNC: intercalation of LBP into the membrane (a), LBP-mediated binding and/or intercalation of LPS (different scales were used for the cartoons of LPS aggregates in solution and in membrane-intercalated LPS) (b), and activation of MNC by signaling proteins such as Toll-like receptors or ion channels (c). (B) Interactions that may cause neutralization of LPS by LBP: LPS aggregates do not bind to or intercalate into lipid membranes (a), LBP intercalates into LPS aggregates (b), and LPS/LBP complexes do not bind to or intercalate into lipid membranes (c) or bind to LBP-doped membranes (d), whereas binding to other components of MNC is still possible (e).
LBP-mediated intercalation of LPS.
For an elucidation of the biological function of LBP, the characterization of the interaction of this protein with LPS is important. It is well known that LBP binds to a variety of LPS chemotypes from rough and smooth strains of gram-negative bacteria and to lipid A (37, 38) and mediates the intercalation of LPS from LPS aggregates, which do not interact with lipids by themselves (Fig. 7B), into phospholipid liposomes (30, 49) (Fig. 7A). We show here in SPR experiments that more than 10 LPS molecules bind to one LBP molecule associated with immobilized PS. It is rather unlikely that one LBP molecule binds such a number of LPS molecules directly because the proposed LPS binding site of LBP is too small. Thus, we propose that LBP binds to individual LPS molecules within an LPS aggregate or that LBP transfers a number of LPS molecules into the target membrane.
The LBP-mediated intercalation of LPS into phospholipid matrices depends significantly on the lipid composition of these matrices, which may result from different geometries of the lipid headgroups and/or different phase states (fluidity of the acyl chains). An influence of the phase state of LPS and/or the lipid matrix can also be derived from the exponential increase of LBP-mediated LPS intercalation in dependence on temperature (Fig. 4). However, a difference cannot be distinguished between the influence of the fluidity of the PS matrix and that of Re LPS. Both lipids have a phase transition temperature (Tc) within the experimental temperature range of 10 to 40°C (PS, Tc, ≈ 18°C; Re LPS, Tc ≈ 36°C) and, therefore, undergo significant changes in their fluidities.
The monoclonal anti-LBP Ab does not influence the intercalation of LBP into PS liposomes (Fig. 5A), but it nearly completely inhibits the LBP-mediated LPS intercalation. It made no difference whether LBP was preincubated with the Ab or if the Ab was even bound to LBP already intercalated into PS liposomes. These data demonstrate that the Ab used in our experiments can interact with intercalated LBP and subsequently inhibit interaction with LPS. This mechanism may thus explain the observed inhibition of LPS-induced MNC activation.
Thus, LBP binds and intercalates into phospholipid membranes and mediates binding of more than 10 Re LPS molecules to each of these LBP/lipid complexes. Both of these phenomena vary in their intensity, depending on the lipid composition. Inhibition of LPS-induced TNF-α production by MNC by the anti-LBP Ab may be interpreted as an inhibition of LBP-mediated intercalation of LPS into the lipid matrix as confirmed by FRET experiments. Thus, we propose that the interaction of LPS with membrane-associated LBP can be an important step in LBP-mediated activation of MNC by LPS (Fig. 7A). This concept of membrane-bound LBP would explain the observation that LPS binds to monocytes in serum-free media independent of CD14 (15). However, the concentration of LBP on the cytoplasmic membrane of MNC and the concentration required for cell activation are unknown. For subsequent signal transduction, the binding of LPS/LBP complexes to CD14 has been shown to be a necessary step (46). Therefore, the formation of microdomains (41) of LBP, CD14, and other proteins involved in LPS signaling, such as the Toll-like receptors or ion channels, is likely to occur.
LPS neutralization by LBP.
In contrast to the enhancement of LPS-induced activation of MNC by LBP, an LPS-neutralizing effect has also been characterized. At high concentrations, LBP inhibits LPS-mediated cytokine release and prevents hepatic failure, resulting in a significantly decreased mortality rate in LPS-challenged mice as well as in a murine model of bacteremia (20). Furthermore, LBP is essential for the survival of an intraperitonally induced Salmonella enterica serovar Typhimurium infection as shown in experiments using LBP−/− knockout mice (16). It was previously found that LBP mediates LPS transfer to reconstituted HDL, attenuating its stimulatory effects (47). This transfer may represent one possible mechanism for neutralization of LPS by LBP, and our data also support a second mechanism, the complexation of LBP and LPS.
The subsequent addition of LBP and LPS, or vice versa, to MNC induced an LBP concentration-dependent increase in TNF-α production as shown in Fig. 2 and as previously published by other groups (8, 15). Complexation of LPS and LBP (by preincubation for 15 min at 37°C) prior to application, however, led to a decrease of TNF-α (and also IL-6) production with increasing LBP concentration (Fig. 2). These results are indicative of a direct LPS-neutralizing effect of LBP independent of other components like HDL or low density lipoprotein. As can be seen from Fig. 2, a very low LBP/LPS molar ratio of 1:104 affects the biological activity of LPS. This can be explained by two possible effects. First, since LPS is diluted from a stock solution down to the final concentration, it exists as aggregates. Therefore, the number of effective LPS molecules at the aggregates' surfaces is much smaller than the total number of molecules in the aggregates, and thus, the binding of LBP to one of the effective LPS surface molecules leads to the binding of an aggregate. Second, electron micrographs of freeze fractured phospholipid liposomes showed that LBP causes a cross-linking of the liposomes, thus reducing the number of single aggregates (data not shown) and, with that, the number of effective molecules at the surface.
To confirm our hypothesis that the interaction of LPS with membrane-associated LBP is an important step in LBP-mediated activation of MNC, the neutralizing effect of LBP has to be understood. To this end we performed FRET and SPR experiments to elucidate the missing interaction of the LPS/LBP complexes with lipid membranes. The FRET data clearly document that LPS/LBP complexes do not intercalate into PS liposomes (Fig. 5B), whereas the data on the intercalation of these complexes into PS liposomes containing intercalated LBP do not allow unequivocal conclusions because the presence of free LBP in the buffer cannot be excluded in these experiments. Due to the removal of all unbound components, more precise data on the binding of LPS/LBP complexes to lipids can be obtained from SPR experiments. These experiments show that LPS/LBP complexes do not bind to immobilized PS liposomes (Fig. 7B) or to LBP intercalated into the PS membranes (Fig. 6B and 7B). Both of these experiments provide important information: (i) LBP inhibits the binding of LPS aggregates to LBP intercalated in the target membrane, (ii) LPS inhibits the binding of LBP to the target membrane, and (iii) the binding of LPS to LBP is different from that of phospholipids to LBP. Thus, we propose that LBP contains two binding domains, one exclusively for phospholipids and one for LPS and phospholipids. Based on the observation that the double mutant Glu-94/Glu-95 of the N-terminal domain of LBP is completely lacking LPS transfer and cell stimulatory activity (21), it is likely that a tip on the N-terminal domain is responsible for LPS binding (4).
Summarizing, complexation of LPS and LBP prior to the binding of LPS to membrane-associated LBP results in LPS neutralization and, thus, to inhibition of MNC activation. These results do not exclude binding of LPS/LBP complexes to other proteins such as CD14 (Fig. 7B). However, from the fact that no TNF-α production was induced by the LPS/LBP complexes, it may be presumed that even if the complexes bind to other components, LPS is neutralized by LBP.
In this work, we focused on understanding the different mechanisms underlying the dual role of LBP in neutralizing LPS and enhancing the LPS-mediated activation of MNC. We provide evidence that LBP mediates the LPS-induced cytokine release of MNC under serum-free conditions. LBP binding and intercalation into lipid membranes is enhanced by negatively charged components, e.g., lipids, and intercalated LBP can mediate the intercalation of LPS into membranes. In contrast, prior complexation of LBP and LPS causes inhibition of the binding of these complexes to the membrane due to different binding between LBP and LPS and LBP and phospholipids. This results in a neutralization of LPS and, with that, in a reduction of TNF-α production by MNC. We thus propose that LBP is located in strong association with the cytoplasmic membrane of MNC, as has been shown for BPI (7), or even intercalated into it and that interaction of LPS with the membrane-associated LBP may be an important step in LBP-mediated activation of MNC by LPS, whereas binding of LPS to serum LBP provokes neutralization of LPS. In in vitro (in the presence of serum) and in particular in vivo experiments, this clear distinction between the two roles of LBP cannot be elaborated because the effects are concentration dependent and a variety of further proteins and other serum constituents, such as soluble CD14 (3), BPI (8), CAP18 (22), and lipoproteins (47), enhance or suppress LPS-induced activation of MNC.
ACKNOWLEDGMENTS
We gratefully acknowledge the technical assistance of Christine Hamann.
This work was financially supported by the Deutsche Forschungsgemeinschaft (SFB 367, project B8) and the Federal Ministry of Education, Science, Research, and Technology (BMBF, 01 KI 9851, Project A6). T.G. acknowledges a fellowship of the Sparkassenstiftung Schleswig-Holstein.
REFERENCES
- 1.Agellon L B, Quinet E M, Gillette T G, Drayna D T, Brown M L, Tall A R. Organization of the human cholesteryl ester transfer protein gene. Biochemistry. 1990;29:1372–1376. doi: 10.1021/bi00458a004. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez E, Ruiz-Gutierrez V, Santa M C, Machado A. Age-dependent modification of lipid composition and lipid structural order parameter of rat peritoneal macrophage membranes. Mech Ageing Dev. 1993;71:1–12. doi: 10.1016/0047-6374(93)90030-u. [DOI] [PubMed] [Google Scholar]
- 3.Bazil V, Horejsi V, Baudys M, Kristofova H, Strominger J L, Kostka W, Hilgert I. Biochemical characterization of a soluble form of the 53-kDa monocyte surface antigen. Eur J Immunol. 1986;16:1583–1589. doi: 10.1002/eji.1830161218. [DOI] [PubMed] [Google Scholar]
- 4.Beamer L J, Carroll S F, Eisenberg D. The BPI/LBP family of proteins: a structural analysis of conserved regions. Protein Sci. 1998;7:906–914. doi: 10.1002/pro.5560070408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blunck R, Scheel O, Müller M, Brandenburg K, Seitzer U, Seydel U. New insights into endotoxin-induced activation of macrophages: involvement of a K+ channel in transmembrane signaling. J Immunol. 2001;166:1009–1015. doi: 10.4049/jimmunol.166.2.1009. [DOI] [PubMed] [Google Scholar]
- 6.Bone R C. The pathogenesis of sepsis. Ann Intern Med. 1991;115:457–469. doi: 10.7326/0003-4819-115-6-457. [DOI] [PubMed] [Google Scholar]
- 7.Dentener M A, Francot G J M, Buurman W A. Bactericidal/permeability-increasing protein, a lipopolysaccharide-specific protein on the surface of human peripheral blood monocytes. J Infect Dis. 1996;173:232–235. doi: 10.1093/infdis/173.1.252. [DOI] [PubMed] [Google Scholar]
- 8.Dentener M A, Von Asmuth E J U, Francot G J M, Marra M N, Buurman W A. Antagonistic effects of lipopolysaccharide binding protein and bactericidal/permeability-increasing protein on lipopolysaccharide-induced cytokine release by mononuclear phagocytes: competition for binding to lipopolysaccharide. J Immunol. 1993;151:4258–4265. [PubMed] [Google Scholar]
- 9.Galanos C, Lüderitz O, Westphal O. A new method for the extraction of R lipopolysaccharides. Eur J Biochem. 1969;9:245–249. doi: 10.1111/j.1432-1033.1969.tb00601.x. [DOI] [PubMed] [Google Scholar]
- 10.Gallati H. Interferon: a greatly simplified immuno enzyme determination with two monoclonal antibodies. J Clin Chem Clin Biochem. 1982;20:907–914. [PubMed] [Google Scholar]
- 11.Gallay P, Heumann D, Le Roy D, Barras C, Glauser M P. Lipopolysaccharide-binding protein as a major plasma protein responsible for endotoxemic shock. Proc Natl Acad Sci USA. 1993;90:9935–9938. doi: 10.1073/pnas.90.21.9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazzano-Santoro H, Mészáros K, Birr C, Carroll S F, Theofan G, Horwitz A H, Lim E, Aberle S, Kasler H, Parent J B. Competition between rBPI23, a recombinant fragment of bactericidal/permeability-increasing protein, and lipopolysaccharide (LPS)-binding protein for binding to LPS and gram-negative bacteria. Infect Immun. 1994;62:1185–1191. doi: 10.1128/iai.62.4.1185-1191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glauser M P, Zanetti G, Baumgartner J D, Cohen J. Septic shock: pathogenesis. Lancet. 1991;338:732–736. doi: 10.1016/0140-6736(91)91452-z. [DOI] [PubMed] [Google Scholar]
- 14.Gutsmann T, Haberer N, Seydel U, Wiese A. Interaction between lipopolysaccharide (LPS), LPS-binding protein (LBP), and planar membranes. Biol Chem. 2001;382:425–434. doi: 10.1515/BC.2001.052. [DOI] [PubMed] [Google Scholar]
- 15.Heumann D, Gallay P, Barras C, Zaech P, Ulevitch R J, Tobias P S, Glauser M P, Baumgartner J D. Control of lipopolysaccharide (LPS) binding and LPS-induced tumor necrosis factor secretion in human peripheral blood monocytes. J Immunol. 1992;148:3505–3512. [PubMed] [Google Scholar]
- 16.Jack R S, Fan X, Bernheiden M, Rune G, Ehlers M, Weber A, Kirsch G, Mentel R, Fürli B, Freudenberg M, Schmitz G, Stelter F, Schütt C. Lipopolysaccharide-binding protein is required to combat a murine Gram-negative bacterial infection. Nature. 1997;389:742–745. doi: 10.1038/39622. [DOI] [PubMed] [Google Scholar]
- 17.Kirschning C, Unbehaun A, Lamping N, Pfeil D, Herrmann F, Schumann R R. Control of transcriptional activation of the lipopolysaccharide binding protein (LBP) gene by proinflammatory cytokines. Cytokines Cell Mol Ther. 1997;3:59–62. . (Erratum, 3:137.) [PubMed] [Google Scholar]
- 18.Kirschning C J, Au-Young J, Lamping N, Reuter D, Pfeil D, Seilhamer J J, Schumann R R. Similar organization of the lipopolysaccharide-binding protein (LBP) and phospholipid transfer protein (PLTP) genes suggests a common gene family of lipid-binding proteins. Genomics. 1997;46:416–425. doi: 10.1006/geno.1997.5030. [DOI] [PubMed] [Google Scholar]
- 19.Kröner E E, Peskar B A, Fischer H, Ferber E. Control of arachidonic acid accumulation in bone marrow-derived macrophages by acyltransferases. J Biol Chem. 1981;256:3690–3697. [PubMed] [Google Scholar]
- 20.Lamping N, Dettmer R, Schroeder N W J, Pfeil D, Hallatschek W, Burger R, Schumann R R. LPS-binding protein protects mice from septic shock caused by LPS or gram-negative bacteria. J Clin Investig. 1998;101:2065–2071. doi: 10.1172/JCI2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamping N, Hoess A, Yu B, Park T C, Kirschning C J, Pfeil D, Reuter D, Wright S D, Herrmann F, Schumann R R. Effects of site-directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of lipopolysaccharide (LPS)-binding protein on binding and transfer of LPS and subsequent immune cell activation. J Immunol. 1996;157:4648–4656. [PubMed] [Google Scholar]
- 22.Larrick J W, Hirata M, Zheng H, Zhong J, Bolin D, Cavaillon J-M, Warren H S, Wright S C. A novel granulocyte-derived peptide with lipopolysaccharide-neutralizing activity. J Immunol. 1994;152:231–240. [PubMed] [Google Scholar]
- 23.Malmqvist M. Biospecific interaction analysis using biosensor technology. Nature. 1993;361:186–187. doi: 10.1038/361186a0. [DOI] [PubMed] [Google Scholar]
- 24.Maruyama N, Yasunori K, Yamauchi K, Aizawa T, Ohrui T, Nara M, Oshiro T, Ohno L, Tanura G, Shimura S, Saschi H, Tahishima T, Shirato K. Quinine inhibits production of tumor necrosis factor-α from human alveolar macrophages. Am J Respir Cell Mol Biol. 1994;10:514–520. doi: 10.1165/ajrcmb.10.5.8179913. [DOI] [PubMed] [Google Scholar]
- 25.Morrison D C, Ryan J L. Endotoxins and disease mechanisms. Annu Rev Med. 1987;38:417–432. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 26.Ramadori G, Meyer zum Buschenfelde K H, Tobias P S, Mathison J C, Ulevitch R J. Biosynthesis of lipopolysaccharide-binding protein in rabbit hepatocytes. Pathobiology. 1990;58:89–94. doi: 10.1159/000163569. [DOI] [PubMed] [Google Scholar]
- 27.Rietschel E T, Brade H, Brade L, Brandenburg K, Schade U, Seydel U, Zahringer U, Galanos C, Luderitz O, Westphal O. Lipid A, the endotoxic center of bacterial lipopolysaccharides: relation of chemical structure to biological activity. Prog Clin Biol Res. 1987;231:25–53. [PubMed] [Google Scholar]
- 28.Rietschel E T, Brade L, Lindner B, Zähringer U. Biochemistry of lipopolysaccharides. In: Morrison D C, Ryan J L, editors. Bacterial endotoxic lipopolysaccharides. I. Molecular biochemistry and cellular biology. Boca Raton, Fla: CRC Press, Inc.; 1992. pp. 3–41. [Google Scholar]
- 29.Schromm A B, Brandenburg K, Loppnow H, Zähringer U, Rietschel E T, Carroll S F, Koch M H J, Kusumoto S, Seydel U. The charge of endotoxin molecules influences their conformation and interleukin-6 inducing capacity. J Immunol. 1998;161:5464–5471. [PubMed] [Google Scholar]
- 30.Schromm A B, Brandenburg K, Rietschel E T, Flad H-D, Carroll S F, Seydel U. Lipopolysaccharide binding protein (LBP) mediates CD14-independent intercalation of lipopolysaccharide into phospholipid membranes. FEBS Lett. 1996;399:267–271. doi: 10.1016/s0014-5793(96)01338-5. [DOI] [PubMed] [Google Scholar]
- 31.Schumann R R, Leong S R, Flaggs G W, Gray P W, Wright S D, Mathison J C, Tobias P S, Ulevitch R J. Structure and function of lipopolysaccharide binding protein. Science. 1990;249:1429–1431. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 32.Schumann R R, Rietschel E T, Loppnow H. The role of CD14 and lipopolysaccharide-binding protein (LBP) in the activation of different cell types by endotoxin. Med Microbiol Immunol (Berlin) 1994;183:279–297. doi: 10.1007/BF00196679. [DOI] [PubMed] [Google Scholar]
- 33.Struck D K, Hoekstra D, Pagano R E. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 34.Theofan G, Horwitz A H, Williams R E, Liu P-S, Chan I, Birr C, Carroll S F, Mészáros K, Parent J B, Kasler H, Aberle S, Trown P W, Gazzano-Santoro H. An amino-terminal fragment of human lipopolysaccharide-binding protein retains lipid A binding but not CD14-stimulatory activity. J Immunol. 1994;152:3623–3629. [PubMed] [Google Scholar]
- 35.Tobias P S, Mathison J, Mintz D, Lee J D, Kravchenko V, Kato K, Pugin J, Ulevitch R J. Participation of lipopolysaccharide-binding protein in lipopolysaccharide-dependent macrophage activation. Am J Respir Cell Mol Biol. 1992;7:239–245. doi: 10.1165/ajrcmb/7.3.239. [DOI] [PubMed] [Google Scholar]
- 36.Tobias P S, Mathison J C, Ulevitch R J. A family of lipopolysaccharide binding proteins involved in responses to gram-negative sepsis. J Biol Chem. 1988;263:13479–13481. [PubMed] [Google Scholar]
- 37.Tobias P S, Soldau K, Ulevitch R J. Isolation of a lipopolysaccharide-binding acute phase reactant from rabbit serum. J Exp Med. 1986;164:777–793. doi: 10.1084/jem.164.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tobias P S, Soldau K, Ulevitch R J. Identification of a lipid A binding site in the acute phase reactant lipopolysaccharide binding protein. J Biol Chem. 1989;264:10867–10871. [PubMed] [Google Scholar]
- 39.Ulevitch R J, Tobias P S. Recognition of gram-negative bacteria and endotoxin by the innate immune system. Curr Opin Immunol. 1999;11:19–22. doi: 10.1016/s0952-7915(99)80004-1. [DOI] [PubMed] [Google Scholar]
- 40.Van Lenten B J, Fogelman A M, Haberland M E, Edwards P A. The role of lipoproteins and receptor-mediated endocytosis in the transport of bacterial lipopolysaccharide. Proc Natl Acad Sci USA. 1986;83:2704–2708. doi: 10.1073/pnas.83.8.2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 42.Vreugdenhil A C, Dentener M A, Snoek A M, Greve J W, Buurman W A. Lipopolysaccharide binding protein and serum amyloid A secretion by human intestinal epithelial cells during the acute phase response. J Immunol. 1999;163:2792–2798. [PubMed] [Google Scholar]
- 43.Wang P Y, Kitchens R L, Munford R S. Phosphatidylinositides bind to plasma membrane CD14 and can prevent monocyte activation by bacterial lipopolysaccharide. J Biol Chem. 1998;273:24309–24313. doi: 10.1074/jbc.273.38.24309. [DOI] [PubMed] [Google Scholar]
- 44.Wiese A, Brandenburg K, Lindner B, Schromm A B, Carroll S F, Rietschel E T, Seydel U. Mechanisms of action of the bactericidal/permeability-increasing protein BPI on endotoxin and phospholipid monolayers and aggregates. Biochemistry. 1997;36:10301–10310. doi: 10.1021/bi970176m. [DOI] [PubMed] [Google Scholar]
- 45.Wiese A, Münstermann M, Gutsmann T, Lindner B, Kawahara K, Zähringer U, Seydel U. Molecular mechanisms of polymyxin B-membrane interactions: direct correlation between surface charge density and self-promoted uptake. J Membr Biol. 1998;162:127–138. doi: 10.1007/s002329900350. [DOI] [PubMed] [Google Scholar]
- 46.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 47.Wurfel M M, Kunitake S T, Lichenstein H, Kane J P, Wright S D. Lipopolysaccharide (LPS)-binding protein is carried on lipoproteins and acts as a cofactor in the neutralization of LPS. J Exp Med. 1994;180:1025–1035. doi: 10.1084/jem.180.3.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wurfel M M, Monks B G, Ingalls R R, Dedrick R L, Delude R, Zhou D, Lamping N, Schumann R R, Thieringer R, Fenton M J, Wright S D, Golenbock D. Targeted deletion of the lipopolysaccharide (LPS)-binding protein gene leads to profound suppression of LPS responses ex vivo, whereas in vivo responses remain intact. J Exp Med. 1997;186:2051–2056. doi: 10.1084/jem.186.12.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wurfel M M, Wright S D. Lipopolysaccharide-binding protein and soluble CD14 transfer lipopolysaccharide to phospholipid bilayers—preferential interaction with particular classes of lipid. J Immunol. 1997;158:3925–3934. [PubMed] [Google Scholar]
- 50.Yu B, Hailman E, Wright S D. Lipopolysaccharide binding protein and soluble CD14 catalyze exchange of phospholipids. J Clin Investig. 1997;99:315–324. doi: 10.1172/JCI119160. [DOI] [PMC free article] [PubMed] [Google Scholar]