Abstract
Efficient utilization of heme as an iron (Fe) source by Bordetella avium requires bhuR, an Fe-regulated gene which encodes an outer membrane heme receptor. Upstream of bhuR is a 507-bp open reading frame, hereby designated rhuI (for regulator of heme uptake), which codes for a 19-kDa polypeptide. Whereas the 19-kDa polypeptide had homology to a subfamily of alternative sigma factors known as the extracytoplasmic function (ECF) sigma factors, it was hypothesized that rhuI encoded a potential in-trans regulator of the heme receptor gene in trans. Support for the model was strengthened by the identification of nucleotide sequences common to ECF sigma-dependent promoters in the region immediately upstream of bhuR. Experimental evidence for the regulatory activities of rhuI was first revealed by recombinant experiments in which overproduction of rhuI was correlated with a dramatically increased expression of BhuR. A putative rhuI-dependent bhuR promoter was identified in the 199-bp region located proximal to bhuR. When a transcriptional fusion of the 199-bp region and a promoterless lacZ gene was introduced into Escherichia coli, promoter activity was evident, but only when rhuI was coexpressed in the cell. Sigma competition experiments in E. coli demonstrated that rhuI conferred biological properties on the cell that were consistent with RhuI having sigma factor activity. Heme, hemoglobin, and several other heme-containing proteins were shown to be the extracellular inducers of the rhuI-dependent regulatory system. Fur titration assays indicated that expression of rhuI was probably Fur dependent.
Gene regulation in response to the local iron (Fe) concentration is a common adaptive response for nearly all pathogenic bacteria. Fe is an essential nutrient which must be obtained from the extracellular milieu, and many of the genes which are involved in acquisition of Fe are regulated in response to the metal (24). It has become clear, however, that some pathogenic bacteria recognize Fe starvation as a general signal to upregulate expression of virulence genes (24, 45). The Fe-associated induction of diphtheria toxin is one such example (41).
Of the various Fe-responsive regulatory factors, the global regulator Fur (Fe uptake regulator) (15) of Escherichia coli has been described in the greatest detail. Fur is a traditional repressor protein which, in the presence of Fe, binds to an operator sequence (Fur box) located upstream of a promoter. Binding of Fur to the Fur box blocks transcription by competing with RNA polymerase for promoter sequences (10). The activities of Fur, however, may extend beyond typical repressor functions. Fur was shown to activate the superoxide dismutase gene in E. coli (8). Functional Fur proteins have been identified in numerous bacteria (10). For many years, Fur was accepted as the major, if not sole, Fe-responsive regulator in prokaryotes. In contrast, the picture of bacterial Fe-dependent gene regulation has become increasingly complex, with a variety of other Fe-responsive regulatory proteins having been identified: Pseudomonas aeruginosa encodes PchR, an AraC-type regulator, which both activates and represses the expression of a siderophore receptor (17); irgA of Vibrio cholerae is upregulated by the LysR-type activator IrgB (13); Irr, a GntR-type regulator, has been shown to regulate heme biosynthesis in Bradyrhizobium japonicum in an Fe-dependent manner (14); and the diphtheria toxin genes of Corynebacterium diphtheriae are repressed under high-Fe conditions by DtxR (36).
Recently, a new family of Fe-responsive regulatory proteins which have characteristics of sigma factors and that respond indirectly to Fe was identified (21, 22, 37, 43). Sigma factors are small polypeptides which provide transcriptional specificity to RNA polymerase for promoter sequences (47). In contrast to other sigma factors, this new class of Fe-responsive sigma factors requires extracellular molecules to induce their biological activities (21, 22, 37, 43). To differentiate these inducible transcriptional regulators from other alternative sigma factors, these sigma factors have been designated extracytoplasmic function (ECF) sigma factors. Lonetto et al. described ECF sigma factors as a subfamily of ς70-type proteins which control an organism's response to the environment by regulating genes encoding adaptive proteins (26).
Expression of some ECF sigma factors has been shown to be directly controlled by Fur, and thus, indirectly by local Fe concentrations (28). A variety of Fur-dependent ECF sigma factors and the proteins which they regulate have been described. PupI in Pseudomonas putida regulates expression of the outer membrane receptor for the siderophore pseudobactin BN8 (21). PbrA in P. fluorescens regulates the transcription of genes for siderophore production and uptake, as well as the gene encoding a casein protease (37). In P. aeruginosa, PvdS, a virulence-associated ECF sigma factor, activates transcription of the genes for synthesis of the siderophore pyoverdin and regulates the expression of three other genes whose products control the expression of exotoxin A (22, 44). The paradigm for Fur-regulated ECF sigma factors, however, is FecI of E. coli for which the extracellular inducer is ferric dicitrate (32).
The E. coli fec system is comprised of two genetically linked polycistrons. The upstream operon encodes fecI and its response regulator, fecR. The downstream operon consists of five genes (fecABCDE) which encode the ferric dicitrate uptake machinery (9, 32). Upon Fe starvation, FecI and FecR are synthesized in a Fur-dependent manner (39). Binding of ferric dicitrate to the extracellular domain of FecA induces a signal which is transmitted to FecR, the FecA-dependent response regulator which is located in the plasma membrane (20). Once stimulated, FecR activates cytoplasmic FecI (30). Neither the composition of the signals nor the process by which the signal is transmitted from FecA to FecR or from FecR to FecI has been elucidated. Binding of activated FecI to core RNA polymerase subsequently promotes transcription of the fecA operon by directing the holoenzyme to a promoter located immediately upstream of fecA (31). The result of this regulatory cascade is a ferric dicitrate-dependent induction of fecA and its accessory genes fecBCDE.
Bordetella avium is a gram-negative bacillus which is the causative agent of coryza, an avian upper respiratory illness. Coryza has many symptomatic similarities to whooping cough, a human upper respiratory illness produced by Bordetella pertussis. We have identified BhuR, an Fe-regulated outer membrane receptor in B. avium, which is required for efficient utilization of heme and heme-containing proteins as sources of nutrient Fe (E. R. Murphy and T. D. Connell, unpublished data). We have identified two genes, rhuI and rhuR, which are located proximal to the bhuR gene in the B. avium chromosome. Experimental results presented here demonstrate that expression of BhuR is dependent upon RhuI and that RhuI activation requires extracellular heme or hemoprotein induction. These experimental results support the model that RhuI is a member of the ECF subfamily of ς70-type sigma factors. This is the first report of an ECF sigma factor which regulates a heme and hemoprotein utilization system and that responds to multiple inducers.
MATERIALS AND METHODS
Strains, antibiotics, and reagents.
Bacterial strains used in this study are listed in Table 1. B. avium strains were maintained on brain heart infusion (BHI) agar or in BHI broth (Difco Laboratories, Detroit, Mich.). E. coli strains were cultured on Luria-Bertani (LB) agar. BHI was supplemented with 144 μM FeSO4 for Fe-replete growth conditions. Fe stress conditions were achieved by supplementing BHI with 100 μM ethylenediamine di-o-hydroxyphenylacetic acid (EDDHA). Unless otherwise noted, the following antibiotics and concentrations were used: ampicillin (200 μg/ml), rifampin (10 μg/ml), streptomycin (200 μg/ml), tetracycline (10 μg/ml), kanamycin (50 μg/ml), and gentamicin (10 μg/ml). Antibiotics were obtained from Sigma Biochemicals (St. Louis, Mo.) and Amresco (Solon, Ohio). Biochemical reagents were purchased from Life Technologies, Inc. (Frederick, Md.) and Sigma Biochemicals. Restriction enzymes and DNA-modifying enzymes were obtained from Fermentas, Inc. (Hanover, Md.). Deionized water with an electrical resistance of >18 MΩ was used for all solutions.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| B. avium strains | ||
| 4169 | Wild type | 34 |
| 4169rif | Spontaneous Rifr mutant of 4169 | This study |
| Pho20 | 4169rif bhuR::TnphoA | 5 |
| Pho20(fur) | Pho20, fur gene interrupted by insertional integration of ptr5-1 | This study |
| E. coli strains | ||
| H1717 | Chromosomal lacZ under the control of a strong Fur-regulated promoter | 40 |
| MGIQ1 | K-12 derivative; lacIq1 Kanr | V. J. Hernandez |
| AF633 | MC4100 λφ(uspB-lacZ) | 11 |
| Plasmids | ||
| pCOS5 | Cosmid vector; Ampr Tetr | 4 |
| p1016 | 45-kbp fragment of 4169 chromosome encoding the rhuI-rhuR-bhuR locus ligated into pCOS5 | Murphy and Connell, unpublished |
| pMECA | Cloning vector, Ampr | W. A. Parrott |
| pERM1 | 3-kbp EcoRI fragment of p1016 ligated into pMECA | Murphy and Connell, unpublished |
| pBluescript KS | High-copy-number expression vector; Ampr | Stratagene |
| pAEK10 | 199-bp rhuR-bhuR intergenic region ligated into pBluescript KS | This study |
| pAEK13 | 142-bp rhuI promoter region ligated into pBluescript KS | This study |
| p1910 | Mobilizable ColE1 replicon; Ampr | S. Stibitz |
| ptr5-1 | Truncated fur with the ATG start codon mutated to TGA; mutant gene ligated into p1910 | 29 |
| pMGJ66 | lacIq1-containing plasmid; Tetr | M. G. Jobling |
| placI | lacIq1 allelle from pMGJ66 ligated into pUFR047 | This study |
| pET21a | Expression vector; Ampr | Novagen |
| pFI-7 | rhuI ORF ligated into pET21a | This study |
| pRK415 | Bordetella spp. expression vector, Tetr | 19 |
| pERM26 | rhuI ORF from pFI-7 ligated into pRK415 | This study |
| pERM26.4 | rhuI frameshifted at the XhoI site ligated into pRK415 | This study |
| pRK415Δ | Alpha peptide gene in pRK415 frameshifted at the XhoI site | This study |
| pRS415 | Promoter reporter vector; Ampr | 38 |
| pDJM31 | rhuR-bhuR intergenic region ligated into pRS415 | This study |
| pUFR047 | E. coli/Bordetella spp. shuttle vector; Ampr Gmr | 6 |
| pDJM41 | Fragment of pDJM31 containing the tandem transcription terminators, the rhuR-bhuR intergenic region, and the lacZYA locus ligated into pUFR047 | This study |
Construction of B. avium Pho20(fur).
The allelic-exchange construction ptr5-1 (29) was used to engineer a fur mutation in the B. avium strain Pho20. ptr5-1 contains a C-terminally truncated fur whose ATG start codon has been mutated to a TGA nonsense codon. The plasmid was conjugated into Pho20, and the transconjugants were plated on BHI agar containing ampicillin. Since ptr5-1 does not replicate in B. avium, plating transconjugants on ampicillin-containing medium selects for a plasmid integration at the chromosomal fur. Integration of ptr5-1 causes a gene duplication in which one copy of the fur gene is truncated and the other copy is substituted at the ATG start codon with a translation termination codon (TGA). The genotype of the fur mutant was confirmed by PCR analysis and Southern hybridization (data not shown).
Construction of pERM26.
The 507-bp rhuI open reading frame (ORF) was amplified by PCR from the plasmid pERM1 (Murphy and Connell, unpublished) using synthetic oligonucleotides which were homologous to the 3′ terminus (5′-CTTGCAACATATGTTCTCTGCTTCCCAGG-3′ [NdeI site underlined]) and the 5′ terminus (5′-GGAATTCTCATGACGCATCCATCACCAGA −3′ [EcoRI site underlined]) of the gene. The 3′ primer incorporated an ATG codon in place of the wild-type GTG codon at the start of rhuI to maximize expression of the cloned gene in E. coli. (PCR was done with a DNA thermal cycler [model 480; Perkin-Elmer, Norwalk, Conn.]. PCR was performed in a solution of 1× PCR amplification buffer [Life Technologies], 1 μM (each) oligonucleotides, 25 μM (each) deoxynucleoside triphosphates, 2.5 mM MgCl2, and 5% dimethyl sulfoxide. Thirty cycles of PCR were done, with one cycle consisting of denaturation at 92°C for 1 min, annealing at 45°C for 1 min, and extension at 72°C for 6 min.) The resulting product was directionally cloned into pET21a (Novagen, Madison, Wis.) at the NdeI/EcoRI sites, which placed rhuI under control of the vector's T7 promoter, producing the plasmid pFI-7. To overexpress RhuI in strains lacking the T7 polymerase, a fragment containing the rhuI ORF and the pET21a-encoded ribosomal binding sequence was isolated by digestion of pFI-7 with EcoRI and XbaI. The 520-bp fragment was directionally cloned into pRK415 (19), which placed the gene under the control of the lac promoter. This plasmid was designated pERM26.
To assure that results acquired with pERM26 were due to expression of the RhuI protein and not to the presence of extra copies of the rhuI gene, a frameshift was engineered in the rhuI gene of pERM26. Digestion with XhoI (restriction site located at nucleotide 274 of the rhuI ORF) was used to linearize the plasmid, and the overhanging ends were repaired using Klenow polymerase. The linearized and blunted DNA was religated, transformed into E. coli, and the rhuI gene in the plasmid (pERM26.4) was sequenced to confirm that the gene had been shifted out of its proper reading frame.
Measurement of alkaline phosphatase activity.
Expression of the bhuR::phoA fusion of B. avium Pho20 was determined using a modified p-nitrophenyl phosphate assay (NPP) (35). Pho20 and Pho20 containing various plasmids were cultured to stationary phase in Fe-limited BHI broth. Fe-stressed cells were used to inoculate a secondary culture in BHI broth containing 144 μM FeSO4 or BHI broth containing 100 μM EDDHA. At stationary phase, 1 ml of the secondary culture was centrifuged at 3,000 × g for 5 min to pellet the bacteria. Cells were resuspended in 1 ml of 1 M Tris (pH 8.0), after which the optical density at 600 nm (OD600) of the cell suspension was recorded. The substrate NPP was added to the cell suspension (100 μl of 4-mg/ml NPP) and the OD at 420 and 550 nm (OD420/550) was recorded at 30- to 90-min intervals over a period of 8 h using a Beckman DU 640B spectrophotometer (Fullerton, Calif.). A final OD420/550 was recorded after 24 h. Alkaline phosphatase activity was calculated by use of the following formula (35): activity = {[(OD420 − OD550)/0.0162]/OD600}. Activity is reported as the mean from triplicate assays and is representative of at least two separate experiments.
Measurement of β-galactosidase activity.
Expression of the lacZ reporter gene in pRS415-derived plasmids (38) was determined using a combination of two previously described methods (2, 27). Primary overnight cultures were used to inoculate a 3-ml secondary culture. For experiments in E. coli, secondary cultures were induced after a 2-h incubation at 37°C with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and incubated for an additional 2 h. For experiments in B. avium, the secondary culture was incubated at 37°C for 16 h. After the appropriate incubation, bacteria in 1 ml of culture were pelleted by centrifugation for 5 min at 3,000 × g. The cell pellet was resuspended in 1 ml of Z buffer (60 mM Na2HPO4 · 7H2O, 40 mM NaH2PO4 · H2O, 10 mM KCl, 1 mM MgSO4 · 7H2O, 38 mM β-mercaptoethanol), and the OD600 was adjusted to 0.2 to 0.7. Four hundred microliters of cell suspension, diluted with 400 μl of Z buffer, were permeabilized with 45 μl of 0.1% sodium dodecyl sulfate (SDS) and 90 μl of chloroform and incubated at 30°C for 15 min. o-Nitrophenyl-β-d-galactopyranoside (ONPG) was added (160 μl of a 4-mg/ml solution) to the permeabilized cells, and the reaction was incubated at 30°C until yellow color developed. The reaction was stopped by the addition of 400 μl of 1 M Na2CO3. The reactions were centrifuged briefly to pellet debris, and the OD420/550 of the solution was recorded. β-galactosidase activity was calculated by use of the formula: {1,000[OD420 − 1.75(OD550]}/(t)(V)(OD600), where t is the time of reaction (in minutes) and V is the volume (in milliliters) of cell suspension used in the reaction. Relative enzymatic activity is reported as the mean of triplicate assays and was derived from at least two separate experiments.
Outer membrane protein isolation.
B. avium outer membranes were isolated by a modified version of the protocol of Leyh and Griffith (23). Bacterial strains were cultured at 37°C to stationary phase in BHI broth supplemented with 100 μM EDDHA. The stationary-phase cultures were used to inoculate 25- to 500-ml secondary cultures in BHI supplemented with appropriate Fe supplements. After 24 h of incubation at 37°C in a shaking incubator, cells were pelleted by centrifugation for 10 min at 5,000 × g, resuspended in 20 ml of ice-cold 10 mM HEPES (pH 7.4) supplemented with the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) (0.1 mM), and frozen overnight at −80°C to facilitate cell disruption. After thawing on ice, the cells were sonicated (four 1-min pulses) at 50% duty cycle using a sonifier (Branson Ultrasonics Corp., Dansbury, Conn.) fitted with a microtip. The lysed cells were incubated on ice for 1 h, and a second sonication was performed (two 1-min pulses). Unbroken cells were removed from the lysate by centrifugation (3,000 × g for 20 min). The cleared lysate was centrifuged at 100,000 × g for 1 h to pellet total membranes which were subsequently treated with a solution of 10 mM HEPES (pH 7.4), 1% (wt/vol) N-lauroylsarcosine (Sarkosyl) for 1 h at room temperature by gentle agitation to dissolve cytoplasmic membranes. After centrifugation at 100,000 × g for 60 min, the lysate was treated a second time with the Sarkosyl solution. Outer membranes which remained in the lysate were pelleted by centrifugation as described above, resuspended in 200 μl of deionized water, and stored at −80°C. Total protein concentrations of the outer membranes were determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, Calif.) by comparison to a bovine serum albumin standard. Proteins were separated on SDS–8.75% polyacrylamide gels and visualized by staining with Coomassie brilliant blue.
Construction of PbhuR reporters.
A 200-bp DNA fragment containing the 3′ terminus of rhuR, the 102-bp intergenic region, and the 5′ terminus of bhuR was amplified by PCR from the plasmid pERM1 (Murphy and Connell, unpublished). Synthetic oligonucleotides pAD3-29E (5′-GGAATTCGCCGTATTGGATCGT-3′ [the EcoRI site underlined]) and pAD3-30.1B (5′-CGGGATCCTCACGTGAGAACAGACGA-3′ [the BamHI site underlined and the stop codon in bold type]) were used as primers for the reaction. (PCR was performed in a solution of 1× PCR amplification buffer, 10% dimethyl sulfoxide, 1 μM (each) oligonucleotides, and 50 μM (each) deoxynucleoside triphosphates. Thirty cycles of PCR were done, with one cycle consisting of denaturation at 94°C for 1 min, annealing at 50°C for 1 min, and extension at 72°C for 1 min.) A translational stop codon (TGA) was incorporated at the 3′ end of the fragment to prevent formation of a BhuR-LacZ translational fusion. The amplified fragment was directionally cloned into the promoter reporter vector pRS415 (38) at the EcoRI and BamHI sites, a position that was 5′ to a promoterless lacZYA reporter, to engineer pDJM31. Four tandem transcription terminators are situated in the vector at a region directly upstream of the insertion site to block readthrough transcription from vector promoters.
For examination of the promoter activity in B. avium, the 8.5-kbp region containing the multiple terminators, PbhuR, and the lacZYA reporter was removed by digestion of pDJM31 with PstI and SalI, and ligated into pUFR047 (6), a Bordetella shuttle vector. The plasmid was designated pDJM41.
Determination of Fur binding sites.
The Fur titration assay (FURTA) was used to determine whether promoter sequences were capable of binding Fur. E. coli strain H1717, a strain harboring a wild-type fur gene, was engineered so as to include a chromosomal lacZ reporter gene under the control of a Fur-regulated promoter (40). Transformation of H1717 with a high-copy-number vector bearing a DNA sequence capable of binding Fur elicits an increase in β-galactosidase activity due to titration of Fur from the chromosomal reporter. A DNA fragment encoding the rhuR-bhuR intergenic region was isolated from pDJM31 by double digestion with EcoRI/BamHI and ligated into the EcoRI/BamHI sites of the high-copy-number vector pBluescript KS (Stratagene, La Jolla, Calif.) to produce pAEK10. The rhuI promoter region was amplified by PCR from pERM1 (Murphy and Connell, unpublished) using the synthetic oligonucleotides pfrgI-5E (5′-GGAATTCTGACCTCGCCTGAGCCT-3′ [EcoRI site underlined]) and pfrgI-3B (5′-CGGGATCCTCATGGGAAGCAGAGAA-3′ [BamHI site underlined]) as primers. (PCR was performed in a solution of 1× Extensor Hi-Fidelity PCR Master Mix, buffer 2 [Marsh Bioproducts, Rochester, N.Y.], and 0.1 μM (each) oligonucleotides. Thirty cycles of PCR were done, with one cycle consisting of denaturation at 94°C for 1 min, annealing at 45°C for 30 s, and extension at 72°C for 45 s). To construct pAEK13, the amplified 158-bp fragment was digested with EcoRI and BamHI and ligated into the high-copy-number vector pBluescript KS.
After the plasmids were transferred into E. coli H1717, stationary-phase cultures of the transformants were used to inoculate a secondary culture in LB supplemented with 144 μM FeSO4 to activate the Fur repressor. The secondary culture was incubated at 37°C until mid-log phase at which time the β-galactosidase activity of the cells was determined by a modified version of the Miller assay (2, 27) as described above.
Nucleotide sequencing and sequence analysis.
Nucleotide sequencing was performed by the CAMBI Nucleotide Sequencing Facility at The University at Buffalo, The State University of New York, and by the Biopolymer Facility at Roswell Park Cancer Institute (Buffalo, N.Y.). Sequence analysis was performed using the Wisconsin Package version 9.0 software package (Genetics Computer Group, Madison, Wis.) and ClustalW (http://www.ebi.ac.uk/clustalw).
RESULTS
Fur regulation of bhuR.
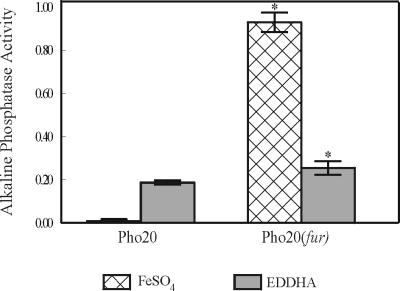
In initial efforts to identify Fe-regulated outer membrane proteins of B. avium, a TnphoA-derived library was screened for mutants which expressed Fe-dependent PhoA fusion proteins (5). Pho20, one of the seven mutants which were identified, was shown to have a TnphoA insertion in a 2,567-bp ORF which was designated bhuR (for Bordetella heme uptake receptor). Subsequent analysis demonstrated that bhuR encodes an outer membrane protein with significant homology to heme and hemoglobin receptors of other species (Murphy and Connell, unpublished). Recently, Vanderpool and Armstrong reported that the bhuR homolog in B. pertussis is necessary for iron acquisition from heme and heme complexes (42). Expression of bhuR was shown to be Fe responsive in that, compared to expression of the bhuR::phoA fusion in Pho20 cells cultured in Fe-replete BHI broth, the reporter fusion in cells cultured under Fe stress was stimulated at least 54-fold (Fig. 1). To assess the contribution of Fur to the regulation of bhuR, the nucleotide sequence directly upstream of bhuR was analyzed by the FURTA (40). This region exhibited a weak but significant titration activity for the Fur repressor of E. coli (32.2 ± 1.8 versus 24.6 ± 2.3 Miller units for the vector control). Upon close inspection of the region, a nucleotide sequence with weak homology to the E. coli consensus Fur box (7) was identified (8 of 19 nucleotides) (Fig. 2C). Little is known about the Fur repressor of B. avium (29). Thus, B. avium Fur may recognize significantly different nucleotide sequences than does E. coli Fur. To further investigate the influence of Fur in the regulation of bhuR, a derivative of Pho20 was engineered in which fur was genetically inactivated using a strategy similar to that used to produce the fur mutant of B. avium Pho6 (29). As expected, Pho20(fur) cultured in Fe-replete conditions exhibited derepressed expression of the bhuR::phoA translation fusion (Fig. 1). However, expression in Fe-replete conditions was not equivalent to expression in Fe-limiting conditions. Rather, expression of the bhuR::phoA fusion of Pho20(fur) was greater in Fe-replete conditions (Fig. 1). This discrepancy in Fur dependency suggested that bhuR is regulated only partly by Fur and that additional regulatory factors are likely involved in expression of the gene.
FIG. 1.
Fur-dependent regulation of bhuR::phoA in B. avium Pho20. A fur knockout in Pho20 was engineered by integration of ptr5-1 (29) into the fur gene of Pho20. Pho20 and Pho20(fur) were cultured in Fe-replete and Fe-stressed BHI broth. Expression of the bhuR::phoA reporter in each strain was determined by measuring alkaline phosphatase activity using a colorimetric assay (35). Activity is reported as micromoles of NPP hydrolyzed per minute per OD600 unit. The mean ± 1 standard deviation from the mean (indicated by the error bar) is shown for each value. Asterisks indicate that the Pho20(fur) values are significantly different (P < 0.05) from the values for Pho20 cultured under identical conditions, as determined by the Student t test.
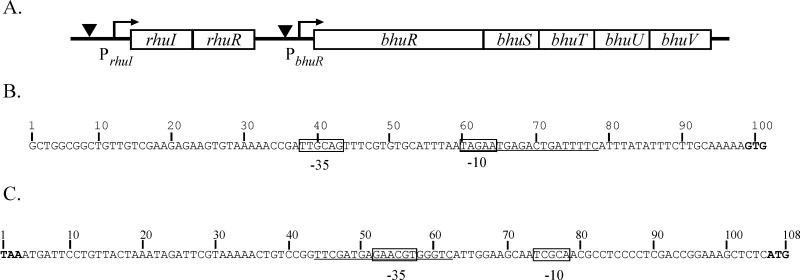
FIG. 2.
Genetic organization of the rhuIR-bhuRSTUV locus of B. avium. (A) Schematic of the organization of the rhu-bhu locus in B. avium. rhuI and rhuR are homologous to genes encoding ECF sigma factor regulatory systems. bhuR has been shown to encode an outer membrane heme receptor (Murphy and Connell, unpublished). bhuSTUV are homologous to accessory genes of other known heme and hemoglobin receptors. Arrows denote putative promoter sequences. Inverted triangles represent putative Fur binding sites. The map is not drawn to scale. (B) Nucleotide sequence 5′ of the rhuI ORF contains sequences homologous to ς70-type promoters. The putative −35 and −10 regions of the PrhuI promoter are boxed. Underlined sequences share 13 of 19 nucleotides with the consensus E. coli Fur operator sequence (7). The GTG start codon of rhuI is indicated in bold type. (C) Nucleotide sequence of the 102-bp rhuR-bhuR intergenic region contains sequences homologous to ECF-dependent promoters. Nucleotides are numbered from the TAA stop codon of rhuR (bold) to the ATG start codon of bhuR (bold). Putative −35 and −10 regions of the RhuI-dependent bhuR promoter are boxed. Nucleotide sequences which have identity at 8 of 19 nucleotide positions with the E. coli consensus Fur operator sequence (Fur box) (7) are underlined.
rhuI encodes a putative ECF sigma factor.
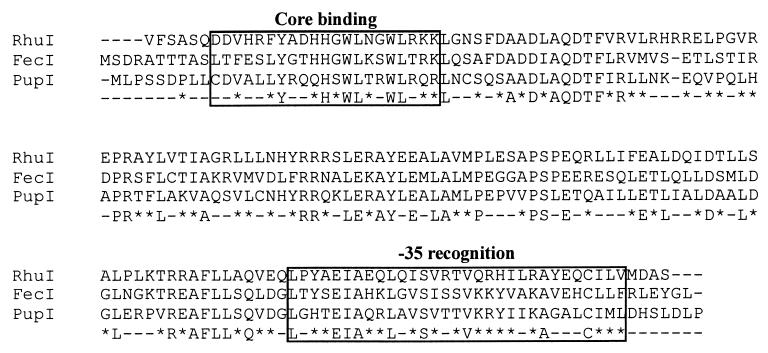
Genes encoding regulatory proteins are often found in close proximity to the genes which are regulated. Cosmid clone p1016 contains an approximately 45-kbp segment of the B. avium chromosome within which is encoded bhuR (Murphy and Connell, unpublished). To search for regulatory elements, nucleotide sequence was obtained from the region upstream of the bhuR gene in p1016. Analysis of the nucleotide sequences revealed two ORFs located 102 bp upstream of bhuR (Fig. 2A). The distal 507-bp ORF, designated rhuI (for regulator of heme uptake), codes for a polypeptide of 19.4 kDa. Overlapping the 3′ terminus of rhuI by 4 bp and located proximal to bhuR was a second ORF of 923 bp which encodes a predicted polypeptide of 33.6 kDa. The second ORF was designated rhuR. BLAST searches revealed that RhuI shares 56% amino acid similarity to the Fe-regulated ECF sigma factor FecI of E. coli (32) and 57% amino acid similarity to the Fe-regulated ECF sigma factor PupI of P. putida (21) (Fig. 3). Similar database searches for polypeptides with homology to RhuR revealed that the predicted polypeptide has 38% amino acid similarity to the antisigma factors FecR of E. coli (32) and 42% amino acid similarity to PupR of P. putida (21). Both antisigma factors are intimately involved in the Fe-dependent regulatory activities of FecI and PupI, respectively. Based on the similar genetic organizations of the fec, pup, and bhu loci (Fig. 2), we hypothesized that rhuI and rhuR are likely Fe-dependent regulators of bhuR expression.
FIG. 3.
Comparison of the amino acid sequence of RhuI to those of two Fur-regulated ECF sigma factors. The predicted amino acid sequence of RhuI was aligned with the amino acid sequences of FecI of E. coli (1) and PupI of P. putida (21) using ClustalW software (http://www.ebi.ac.uk/clustalw/). Regions 2.1 and 4.2, regions important for the function of ECF sigma factors, are boxed, and their functions, core binding and −35 recognition, respectively, are noted. Amino acids are designated by the single-letter code. Dashes in the three sequences represent gaps introduced to maximize alignment. The consensus sequence for the three sequences is shown below the three sequences; in the consensus sequence, positions with amino acid similarity are indicated by asterisks, and positions with nonidentity are designated with dashes. The putative start codon (GTG) of rhuI codes for valine.
Fur regulation of rhuI expression.
Fur has been shown to regulate both fecI and pupI, and Fur binding sites have been identified in the promoters of those genes (21, 43). To investigate whether rhuI has similar Fur-dependent regulatory features, the region upstream of rhuI was analyzed for Fur binding sequences. Nucleotide sequence analysis revealed a region located 20 bp upstream of the rhuI translational start codon in which 13 of 19 positions were identical to the consensus Fur box of E. coli (7) (Fig. 2B). Additionally, nucleotide sequences consistent with −35 (TTGCAG) and −10 (TAGAA) regions of ς70-dependent promoters (47) were also identified upstream of rhuI (Fig. 2B). The putative Fur box overlaps the predicted −10 sequence. To determine whether the region contained a functional Fur binding box, a 142-bp region encompassing the predicted promoter, the Fur box, and the first 20 bp of rhuI was analyzed by FURTA (40). As expected, the 142-bp region titrated Fur from the Fur-regulated chromosomal reporter in the E. coli FURTA strain (78.7 ± 4.4 versus 24.6 ± 2.3 Miller units for the vector control). These data indicated that the region contains a strong Fur binding site and supported the hypothesis that the rhuI gene of B. avium is Fur regulated.
Expression of bhuR is dependent upon rhuI .
To test the hypothesis that RhuI is an Fe-dependent ECF sigma factor regulating bhuR expression, we took advantage of the observation that overexpression of a sigma factor commonly upregulates expression from its respective sigma factor-dependent promoter (21, 46). This effect was observed in P. putida, in which overexpression of pupI stimulated expression from the pupB promoter (21). When pERM26, a pRK415-derived plasmid that expresses rhuI from a strong constitutive promoter, was introduced by conjugation into B. avium Pho20, a 22.5-fold induction of the bhuR::phoA gene over that of the control strain was observed when both strains were cultured in Fe-limited broth (3.087 versus 0.137 U) (Table 2). B. avium Pho20(pERM26) cultured in Fe-replete broth also exhibited rhuI-dependent expression of the fusion reporter (1.127 versus 0.003 U) (Table 2). Thus, the local availability of Fe did not appear to exert a dramatic effect on expression of the bhuR::phoA gene in the mutant strain. The greater expression of the bhuR::phoA reporter in Fe-stressed cells is likely attributable to the direct influence of Fur on the bhuR promoter. In Fe-replete cells, Fur would be expected to exert a negative influence on expression from the bhuR promoter by binding to the Fur box in the rhuIR-bhuR intergenic region; in Fe-stressed cells, the RhuI-dependent bhuR promoter would be derepressed.
TABLE 2.
RhuI upregulates bhuR::phoA expression in B. avium Pho20a
| Plasmid | RhuI expression | Alkaline phosphatase activityb
|
|
|---|---|---|---|
| Fe-replete | Fe-limited | ||
| pRK415 | − | 0.003 (0.0) | 0.137 (0.0) |
| pERM26 | + | 1.127 (0.1)* | 3.087 (0.1)* |
Plasmids were conjugated into B. avium Pho20, which contains a bhuR::phoA translational fusion reporter in the bhu locus. Cells were cultured in Fe-replete BHI broth (100 μM EDDHA and 144 μM FeSO4) or in Fe-limited BHI broth (100 μM EDDHA).
Alkaline phosphatase activity was determined by the colorimetric NPP assay (35). The mean and standard deviation (in parentheses) are shown for each value. Activity is reported as micromoles of NPP hydrolyzed per minute per OD600 unit. Asterisks indicate the values that are significantly different (P < 0.05) from the value for the vector control when each strain is cultured under identical conditions, as determined by the Student t test.
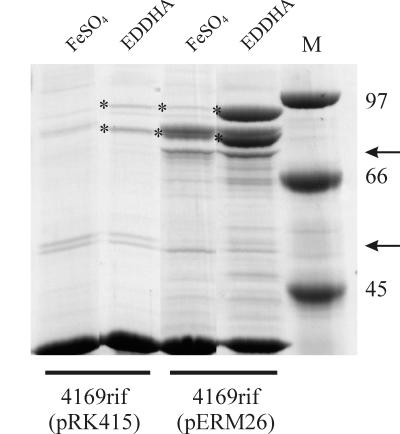
To complement the experiments performed with B. avium Pho20, pERM26 was also introduced into 4169rif, a spontaneous rifampin-resistant mutant of B. avium which contains a wild-type chromosomal copy of bhuR. The outer membrane of Fe-stressed B. avium contains at least four major Fe-regulated proteins (5) (Fig. 4). Two of these Fe-regulated proteins (93 and 91.5 kDa) have been determined by amino acid sequence analysis to be encoded by bhuR (Murphy and Connell, unpublished). Analysis of proteolytic fragments of both polypeptides indicated that the 91.5-kDa polypeptide is likely a cleavage product of the 93-kDa BhuR protein. In B. avium 4169rif(pERM26), both the 93- and 91.5-kDa proteins were upregulated (Fig. 4). Furthermore, the level of expression of BhuR in 4169rif(pERM26) when cultured in Fe-replete (Fig. 4, FeSO4) broth was equivalent to the level of expression of BhuR in the vector control when those cells were cultured under Fe stress (Fig. 4, EDDHA). These data indicated that constitutive expression of rhuI in B. avium was correlated with increased expression of BhuR, yet constitutive expression of rhuI did not absolutely abrogate the regulatory effects of Fe on expression of BhuR in 4169rif(pERM26). In comparison to Fe-replete 4169rif(pERM26), Fe-stressed 4169rif(pERM26) cells produced significantly higher levels of BhuR (Fig. 4). We attributed this difference to the influence of Fur on the bhuR promoter. Nonetheless, the data obtained from the RhuI overexpression experiments strongly indicated that the Fe-responsive regulation of bhuR is dependent, at least in part, on RhuI.
FIG. 4.
Overexpression of RhuI upregulates expression of BhuR. B. avium 4169rif containing either pERM26 (rhuI overexpressing plasmid) or pRK415 (vector control) were cultured in Fe-replete broth (BHI plus 144 μM FeSO4) or in Fe-stressed broth (BHI plus 100 μM EDDHA). Outer membrane proteins from each strain were resolved on an SDS–8.75% polyacrylamide gel and stained with Coomassie brilliant blue. Each lane was loaded with 8 μg of total protein. The positions (in kilodaltons) of molecular mass standards (M) are shown to the right of the gel. The two forms of the Fe-regulated BhuR outer membrane protein of 4169rif (93 and 91.5 kDa) are denoted by asterisks. Arrows denote the positions of the 74- and 50-kDa outer membrane proteins which are influenced by RhuI overexpression.
The regulatory role of rhuI may not be limited to bhuR expression. Production of two other outer membrane proteins was influenced either negatively or positively by RhuI (Fig. 4). While synthesis of a 74-kDa protein was upregulated by RhuI overexpression, production of a 50-kDa protein was downregulated. The roles of these proteins in Fe acquisition have yet to be determined.
RhuI dependence of the bhuR promoter.
In the paralogous fec (9) and pup (21) systems, the ECF sigma factor-dependent promoter is located directly upstream of the genes encoding the regulated receptors (fecA and pupB). Nucleotide sequence analysis was performed to ascertain whether promoter sequences were evident within the region located immediately upstream of bhuR. Although nucleotide sequences similar to the consensus ς70 promoter sequence were absent, nucleotide sequences consistent with an ECF sigma factor-responsive promoter (28) were detected within the 102-bp rhuR-bhuR intergenic region (Fig. 2C). A conserved −35 site (5′-GAACGT-3′) was located 48 bp upstream of the ATG start codon of bhuR, and a sequence with similarity to the consensus −10 region (5′-TCGCA-3′) was identified at a site 16 bp downstream of the putative −35 region. To determine whether the intergenic region encoded a promoter which responds to RhuI, a 199-bp fragment containing the intergenic region, 32 bp of rhuR, and 48 bp of bhuR obtained from pERM1 by PCR was ligated directionally into the promoter probe vector pRS415 at a site immediately upstream of a promoterless lacZYA reporter operon (38). Four transcriptional terminators located in a tandem array upstream of the insertion site in pRS415 efficiently isolated the DNA fragment containing the putative bhuR promoter (PbhuR) and the lacZYA reporter from plasmid-encoded promoters. pDJM31 was transformed into E. coli MGIQ1, a strain encoding the lacIq1 allele (a gift from V. J. Hernandez). To demonstrate that a RhuI-dependent promoter was located within the intergenic region, MGIQ1(pDJM31) was subsequently transformed with the RhuI-expressing plasmid pERM26. Cells induced for expression of RhuI by addition of IPTG to the culture medium were assayed for production of β-galactosidase activity as an indirect measure of promoter activity. Upon induction of rhuI by IPTG, MGIQ1(pDJM31, pERM26) exhibited a 62-fold increase in β-galactosidase activity (3,069.8 versus 49.1 Miller units) when the strain was cultured in Fe-replete broth compared to the vector control (Table 3). In the absence of IPTG induction, lower but still significant differences in RhuI-induced promoter activity (37-fold) were evident (1,706.8 versus 46.5 Miller units) (Table 3). This activity was likely a result of leaky expression of the plasmid-encoded rhuI gene. To further demonstrate that expression of the RhuI polypeptide was needed for bhuR promoter induction, the rhuI gene in pERM26 was mutated by frameshifting the gene at a single codon. Introduction of this plasmid, pERM26.4, into MGIQ1(pDJM31) did not stimulate expression of the reporter gene (data not shown). Taken together, these data support the hypothesis that the 102-bp region located immediately upstream to bhuR encompasses a promoter (PbhuR) whose activity is responsive in trans to stimulation by RhuI.
TABLE 3.
The rhuR-bhuR intergenic region exhibits RhuI-dependent promoter activitya
| Plasmids | RhuI expression | Growth conditionb | β-Galactosidase activity (Miller units)a
|
|
|---|---|---|---|---|
| −IPTG | +IPTG | |||
| pDJM31 + pRK415Δ | − | FeSO4 | 46.5 (1.7) | 49.1 (0.5) |
| EDDHA | 119.0 (4.2)† | 124.0 (5.5)† | ||
| pDJM31 + pERM26 | + | FeSO4 | 1,706.8 (52.3) | 3,069.8 (112.6)* |
| EDDHA | 2,260.0 (142.4)*† | 3,958.1 (186.2)*† | ||
Reporter and expression plasmids were cotransformed into E. coli MGIQ1, an engineered K-12 strain that encodes a chromosomal lacIq1 allele (V.J. Hernandez). Cells were induced at mid-log stage for 2 h by adding IPTG to a final concentration of 1 mM to the culture broth.
Primary cultures were incubated overnight in LB supplemented with 100 μM EDDHA. The Fe-stressed overnight cultures were used to inoculate secondary cultures under the indicated conditions. FeSO4, LB supplemented with 100 μM EDDHA and 144 μM FeSO4; EDDHA, LB supplemented with 100 μM EDDHA.
Reporter activity was determined by a modified version of the Miller assay (2, 27). The mean and standard deviation (in parentheses) are shown for each value. Values that are significantly different (P < 0.05) from the value for the vector control, as determined by the Student t test, are indicated by asterisks. Values that are significantly different (P < 0.05) from the value for the Fe-replete strain, as determined by the Student t test, are indicated by daggers.
To assess the contribution of Fur to the regulation of bhuR, the RhuI-dependent PbhuR reporter was analyzed in E. coli for responsiveness to local Fe availability. Consistent with Fur regulation, Fe-limited cells expressed a higher level of β-galactosidase activity than did cells cultured in Fe-replete conditions. Expression of the reporter was independent of RhuI or IPTG (Table 3). Compared to activity in Fe-replete cells, reporter activity increased by 76% when E. coli MGIQ1(pDJM31, pERM26) cells were Fe stressed. Unexpectedly, the reporter responded weakly in the absence of RhuI (Table 3), which indicated that PbhuR is responsive to an unknown E. coli sigma factor. This responsiveness was not influenced by the endogenous E. coli fec system; Fe-dependent PbhuR activity was unaffected by adding citrate to the culture (data not shown). It was clear from these experiments that PbhuR in E. coli is regulated in a direct manner by RhuI and Fur.
Competition of ςS-dependent transcription by RhuI.
Although homology comparisons are useful in predicting the molecular role of a new protein, functional assays are required before a biochemical role can be assigned to any protein. In the case of sigma factors, gel shift experiments and coimmunoprecipitations using core RNA polymerase are routinely used to characterize their biological activities. Significant amounts of purified proteins, however, are required for both assays. Various attempts to purify properly folded, recombinant RhuI were unsuccessful (data not shown). As an alternative to gel shift and coimmunoprecipitation experiments, a genetic model of sigma factor competition for RNA polymerase was utilized to demonstrate that RhuI had biological properties which were consistent with a sigma factor (11). Transcription of uspB, a gene which encodes a universal stress protein, is controlled by ςS, a stationary-phase sigma factor (12). When ς70 was overexpressed in the cell, transcription from the uspB promoter was significantly decreased due to competition between ς70 and ςS for binding to core RNA polymerase (11). When pERM26 was transformed into AF633, an engineered strain of E. coli (11) which contains a transcriptional fusion of the uspB promoter and a β-galactosidase reporter gene, and the recombinant strain was IPTG induced for high-level expression of RhuI, a 24% decrease in PuspB activity was observed (114.0 versus 86.7 Miller units) (Table 4). No significant difference in PuspB activity (109.2 versus 114.0 Miller units) was observed in cells when IPTG was omitted from the culture (Table 4). The RhuI-dependent decrease in PuspB activity was not associated with general disruption of cellular metabolism, since both AF633(pERM26, placI) and AF633(pRK415, placI) grew in culture broth with equal proficiency and to similar cell densities (data not shown). Thus, these experimental results were consistent with a model in which the sigma factor RhuI successfully competed in vivo with ςS for binding to core RNA polymerase.
TABLE 4.
RhuI competes with ςS for binding to E. coli RNA polymerasea
| Plasmid | RhuI expression | β-Galactosidase activity (Miller units)b
|
|
|---|---|---|---|
| −IPTG | +IPTG | ||
| pRK415 | − | 109.2 (3.9) | 100.3 (3.9) |
| pERM26 | + | 114.0 (4.9) | 86.7 (1.6)* |
Plasmids were transformed into E. coli AF633 which contains a lacZYA reporter operon under the control of the ςS-dependent uspB promoter (11). Expression of a sigma factor other than ςS in this strain results in a decreased expression of the LacZ reporter. All plasmids were cotransformed with placI as a source of the LacIq1 repressor. Cells were induced for RhuI expression by the addition of IPTG to a final concentration 0.1 mM at the time of inoculation.
Reporter activity was determined by a modified version of the Miller assay (2, 27). The mean and standard deviation (in parentheses) are shown for each value. The asterisk indicates that the value is significantly different from the value for the uninduced control (P < 0.05), as determined by the Student t test.
Induction of RhuI-dependent expression of bhuR by heme and hemoglobin.
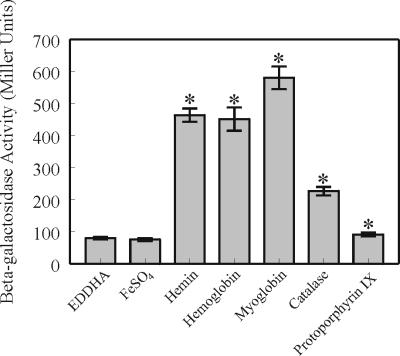
In genetic systems regulated by ECF sigma factors, maximal activation of ECF sigma factors was promoted by binding of the cognate extracellular ligands to the surface-exposed receptors (16, 21). In E. coli, ferric dicitrate, the molecule that is bound by FecA receptor, is the inducer for the fecABCDE polycistron (16). The inductive signal is transmitted from FecA to FecI via the cytoplasmic membrane protein FecR (30). Similarly, the siderophore pseudobactin was shown to induce the activity of PupI in P. putida via PupB, the outer membrane pseudobactin receptor (21). Prior studies in our laboratory demonstrated that B. avium requires BhuR for the efficient acquisition of heme, hemoglobin, myoglobin, and catalase as sources of nutrient Fe (Murphy and Connell, unpublished). To test whether heme and hemoproteins are the cognate inducers for RhuI-dependent transcription of bhuR, a PbhuR reporter plasmid (pDJM41) was engineered in pUFR047 (6) for use in B. avium. When B. avium 4169rif(pDJM41) was cultured in BHI broth which had been supplemented with FeSO4, β-galactosidase activity from the PbhuR-lacZYA reporter fusion was negligible (Fig. 5). Addition of 0.16 μM hemin to the Fe-replete culture had no effect on the basal reporter activity (data not shown). However, addition of hemin to BHI broth which had been reduced for free Fe by supplementation with EDDHA stimulated a sevenfold increase in reporter activity over that observed with cells cultured in BHI supplemented with EDDHA alone (Fig. 5). A similar experiment was performed to investigate whether the heme-containing molecules hemoglobin, myoglobin, and catalase would induce PbhuR. Addition of these three hemoproteins (hemoglobin, myoglobin, and catalase) to the Fe-stressed cultures resulted in a significant induction of PbhuR in all cases (Fig. 5). Since Fe uptake systems in other bacteria often require that the ferromolecules are complexed with Fe, protoporphyrin IX, the Fe-free precursor of heme, was also tested for inductive potential. Unexpectedly, protoporphyrin IX elicited a weak potential for induction of PbhuR (Fig. 5, Protoporphyrin IX). This slight inductive capacity of the molecule was attributed to a weak binding affinity of the outer membrane receptor BhuR for protoporphyrin IX. It is not yet clear whether the hemoproteins per se are responsible for induction, as it is feasible that heme is released from the molecules by an undescribed cell surface or extracellular degradase. Preliminary experiments, however, revealed that neither culture supernatants nor intact B. avium cells had hemoglobin degradative activity (T. D. Connell, unpublished data). Nonetheless, these experiments firmly established that free heme and several heme-containing molecules induced expression of bhuR in a RhuI-dependent manner.
FIG. 5.
Hemin and hemoproteins induce expression from the bhuR promoter. B. avium 4169rif(pDJM41) was cultured in BHI broth supplemented with 100 μM EDDHA in combination with each of the following molecules: FeSO4, hemin, turkey hemoglobin, myoglobin, catalase, or protoporphyrin IX. Each of the molecules was used at a concentration such that the total Fe concentration was 5 μM. β-Galactosidase activity was determined by using a modified version of the Miller assay (2, 27). The mean ± 1 standard deviation from the mean (indicated by the error bar) is shown for each value. Asterisks indicate the values that are significantly different (P < 0.05) from the value for EDDHA alone, as determined by the Student t test.
Heme induction of PbhuR requires expression of BhuR.
For E. coli and P. aeruginosa, the FecA and PupB outer membrane receptors, respectively, are integral components of the ligand-responsive signaling cascades (16, 21). A reasonable hypothesis for B. avium was that BhuR is the ligand sensor for induced expression of bhuR. To test this hypothesis, pDJM41 was conjugated into B. avium Pho20. In contrast to B. avium 4169rif(pDJM41) (Fig. 5), Pho20(pDJM41) was unresponsive to heme in either Fe-replete or Fe-stressed media (data not shown). These data supported a model in which BhuR is an essential participant in the heme-sensing pathway of B. avium.
DISCUSSION
Prokaryotic regulatory systems evolved in response to evolutionary pressures that selected for pathways to minimize wasteful expenditures of energy. The evolutionary advantage to the bacterium is that only the most optimal metabolic pathway(s) will be expressed at any one moment and under any particular environmental condition. Ideally, bacteria would downregulate genes encoding specific nutrient uptake and utilization pathways when the nutrient is available at high concentrations in the local environment. Conversely, the bacterium would be expected to upregulate these genes when it is starved for that particular nutrient. This predicted pattern of gene expression is evident in the expression of bacterial systems for Fe acquisition. In response to local Fe concentrations, bacteria tightly regulate those genes which encode for Fe acquisition proteins. Fur, an Fe-responsive global regulator, has been shown to be the major regulatory molecule for a variety of bacterial Fe acquisition systems. In the case of bacterial pathogens, however, more-complicated regulatory mechanisms for mediating expression of genes for Fe acquisition are often encountered. While Fe starvation is a sufficient stimulus for some pathogens to upregulate transport systems, other bacteria do not respond to free Fe but require induction by particular Fe-containing molecules (14, 21, 43). One of the most efficient bacterial systems to control expression of Fe acquisition machinery, the ECF sigma factor mechanism, utilizes both inducer-activated and Fur-regulated mediators. In our model, the RhuI/RhuR ECF regulatory system exerts both positive and negative control on expression of the bhu heme utilization regulon in B. avium. When Fe is limiting, repression of the Fur-dependent rhuIR polycistron is released and the regulatory proteins RhuI and RhuR are synthesized. Upon appropriate indirect stimulation by extracellular heme, these regulatory proteins promote increased expression of bhuR. When Fe is abundant, expression of rhuI and rhuR, and thus, bhuR is repressed, most likely in a Fur-dependent manner.
Regulated expression of the heme utilization system by B. avium requires both transcriptional activation and ligand sensing. In this regard, BhuR is an essential component of the regulatory cascade for heme acquisition. Our current model is that BhuR is simultaneously a heme receptor and a heme response sensor. Under Fe stress, low but adequate amounts of BhuR are produced to prime the cell for heme-dependent induction of the rhuIR/bhuR regulon. When heme or other heme-containing molecules become locally available, binding of the ligand(s) to BhuR initiates the regulatory cascade to dramatically upregulate expression of bhuR in a RhuI-dependent manner. The independent, but converging fur and rhu regulatory systems, therefore, tightly regulate transcription of bhuR. As a result, biosynthetic energy is conserved until the cell experiences both Fe stress and the presence of heme or heme-containing molecules in the local environment.
There is convincing comparative evidence that RhuI is a new member of the ECF subfamily of sigma factors. RhuI has significant amino acid homology to FecI (32) and PupI (21), two well-described ECF sigma factors, particularly in regions 2.1, 4.2, and 3 (Fig. 3). Each of those domains is known to be important for sigma factor function (25). Region 2.1 has been implicated in RNA polymerase core binding, region 4.2 has been implicated in −35 region recognition (Fig. 3), and region 3 has the potential to form a helix-turn-helix motif which is thought to contribute DNA-binding activity to the protein (25). RhuI has no detectable homology to a domain (region 1) that is found in most ς70-type sigma factors but which is absent in ECF sigma factors (26). Additional evidence to support inclusion of RhuI into the ECF sigma factor subfamily is provided by analysis of the genetic structure of the putative RhuI-dependent bhuR promoter. Nucleotide sequences that are reminiscent of other ECF sigma factor-dependent promoters are observed within the putative −35 region (GAACGT) of PbhuR (Fig. 2C). An adenine dinucleotide at the second and third positions, the guanosine at the 5′ position, and the thymidine at the 3′ position are present in most promoters that are regulated by ECF sigma factors (26). Conversely, the putative −10 regions of ECF-dependent promoters are not highly conserved with the exception of a relative abundance of thymidine nucleotides (26). Since a species may produce multiple ECF sigma factors (3, 28), a system to discriminate between different promoters is a logical necessity. From the patterns of sequence conservation, it is reasonable to propose that the −35 region of ECF-dependent promoters provides broad specificity for all ECF sigma factors, while narrow specificity of the promoters for their cognate ECF sigma factors is likely to be mediated by the divergence in the −10 sequences. Evidence for this model was demonstrated in Bacillus subtilis in which the −10 regions of the ECF sigma ςX- and ςW-dependent promoters were genetically switched. In these mutants, the sigma specificity of the promoters was shown to be dependent upon the respective −10 regions (33).
Promoter reporter experiments performed in E. coli established that the rhuR-bhuR intergenic region had promoter activity and that the promoter was responsive to RhuI. In control experiments, however, PbhuR was weakly active in the absence of RhuI (Table 3, pDJM31 and pRK415Δ). We hypothesize that an endogenous and unknown E. coli sigma factor interacts with PbhuR. Subsequent experiments confirmed that the hypothesized sigma factor is not FecI, an endogenous ECF sigma factor of E. coli, since conditions known to induce FecI transcriptional activity did not increase expression from PbhuR (data not shown). The responsiveness of PbhuR to the hypothetical sigma factor is apparently species specific, as a similar Fe dependency was not evident in promoter experiments performed in B. avium 4169rif(pDJM41). Specifically, PbhuR in 4169rif(pDJM41) was unresponsive to Fe limitation in the absence of heme or other extracellular inducers (Fig. 5).
Expression of PbhuR was elicited in E. coli without concurrent expression of RhuR, the putative antisigma factor of RhuI. We concluded from those data that, under those experimental conditions, RhuR must not be essential for RhuI activation. At first glance, these data supported the traditional model that RhuR is a member of the sequestering subfamily of antisigma factors. One would predict that in B. avium, RhuR, induced during Fe stress, would sequester RhuI and, thereby, limit expression of PbhuR, yet the converse was observed. In promoter experiments performed in B. avium, expression of BhuR::PhoA in Fe-starved Pho20(pERM26) exceeded expression of BhuR::PhoA in Fe-replete Pho20(pERM26) cells (Table 2). The explanation for this somewhat perplexing observation may reside in copy number considerations; i.e., the single chromosomal copy of rhuR likely produced insufficient amounts of RhuR to sequester the prodigious amounts of recombinant RhuI that was expressed from pERM26. Although most antisigma factors are negative regulators (18), positive regulation by antisigma factors has been documented (30), and RhuR may be of this latter class. Experiments to examine the character of RhuR in RhuI stimulation or activation are in progress.
Overexpression of RhuI evoked a dramatic increase in the amount of BhuR in the outer membrane of B. avium 4169rif (Fig. 4). Both the 93-kDa form of the protein and the smaller 91.5-kDa form of BhuR were upregulated in response to constitutive expression of RhuI. Fe availability, however, altered the stoichiometric expression of the two forms. Whereas the 93-kDa form of BhuR predominated in the outer membrane of Fe-stressed 4169rif(pRK415), the 91.5-kDa form was present only in low amounts (Fig. 4). In contrast, in Fe-stressed 4169rif(pERM26) in which RhuI was overexpressed, the two forms of BhuR were synthesized in essentially equivalent amounts (Fig. 4). In Fe-replete cells overexpressing RhuI, the smaller form of BhuR predominated. This pattern of expression suggested that the processing of BhuR from the 93-kDa form to the 91.5-kDa form may be a regulated event in response to Fe concentration. Furthermore, processing of BhuR may be influenced, directly or indirectly, by overexpression of RhuI. Neither the functional significance of the two forms of BhuR in the outer membrane of B. avium nor the role of RhuI in synthesis of the two forms has been elucidated, but the observations are consistent with a model in which expression of activity of a specific protease is controlled by RhuI.
In E. coli and P. aeruginosa, the ECF sigma factor regulons are activated only after binding of the pertinent extracellular inducers to their cognate receptors (16, 21). We hypothesized a similar model for BhuR regulation in that RhuI activity in B. avium is induced by the presence of extracellular heme. Promoter reporter experiments using B. avium 4169rif(pDJM41) confirmed that the bhuR promoter responded to heme (Fig. 5). To our surprise, promoter activity was also induced by the heme-containing proteins hemoglobin, myoglobin, and catalase. Each of these molecules was shown to induce expression from PbhuR in an RhuI-dependent manner (Fig. 5). The level of Fe responsiveness of the bhuR promoter was dependent upon the context in which it was presented. While the bhuR::phoA reporter gene in B. avium Pho20(pRK415) was responsive to Fe stress (Table 2), the plasmid-encoded PbhuR in 4169rif (pDJM41) was unresponsive to Fe limitation unless inducer was present (Fig. 5). This seemingly paradoxical observation is resolved by considering the differences between the two reporter systems. In Pho20, the bhuR::phoA reporter gene resides on the chromosome. In contrast, the PbhuR element in 4169rif(pDJM41) is encoded on an extrachromosomal element (i.e., pDJM41). The most likely source of the cis-acting Fe-dependent regulation of bhuR::phoA is the Fur box located immediately upstream of rhuI. The data are consistent with the following model. Under Fe stress, the Fur-dependent rhuIR promoter in strain Pho20 (and in the wild-type strain 4169rif) is derepressed, enabling RNA polymerase to transcribe rhuI and rhuR. At an unknown frequency, a longer transcript is produced by readthrough transcription when RNA polymerase continues to transcribe beyond rhuIR and into bhuR. Biologically, such inefficient transcriptional termination would be adaptive, in that small amounts of BhuR would be produced in the absence of heme induction during Fe stress. That small amount of BhuR in the outer membrane would thus prime the cell for detection of heme should the inducer become available. At that time, binding of inducer to the limited number of receptors would initiate the RhuIR-BhuR regulatory cascade, thus stimulating a dramatic increase in BhuR synthesis. The model, however, does not resolve the conundrum of why Pho20(fur) does not exhibit equivalent expression of bhuR::phoA regardless of Fe availability (Fig. 1). We hypothesize that B. avium has a third regulatory mechanism for the expression of bhuR that has yet to be identified. Transcriptional experiments are ongoing in B. avium to investigate the role of readthrough transcription under various conditions of Fe availability in bhuR expression.
Recently, three ORFs, designated hurI, hurR, and bhuR, with sequence homology to the rhuI, rhuR, and bhuR genes of B. avium have been identified in B. pertussis (42). While the structural sequences of the three genes of B. pertussis were highly homologous to their respective partners in B. avium, there was significant divergence in the nucleotide sequences of the rhuR-bhuR intergenic regions. Experiments similar to those detailed in this study performed in our laboratory demonstrated that the rhuR-bhuR intergenic region of B. pertussis has promoter activity that is responsive to RhuI of B. avium (data not shown). These experiments provide convincing evidence that B. pertussis expresses a rhuI-dependent heme-inducible system for heme uptake that is functionally similar to that expressed by B. avium. Examining the incomplete genomes of Bordetella parapertussis and Bordetella bronchiseptica by using the Sanger Center database (Bordetella sequencing group of the Sanger Center [http://www.sanger.ac.uk/Projects]) revealed rhuIR sequences in these species as well. Additionally, BhuR in B. bronchiseptica has been shown to be heme induced (42). Future experiments will investigate whether the rhuIR (hurIR) and bhuR genes of B. pertussis, B. parapertussis, and B. bronchiseptica are the functional equivalents of the respective genes in B. avium.
ACKNOWLEDGMENTS
This study was supported in part by funds made available to T.D.C. from the School of Medicine and Biomedical Sciences at The University of Buffalo, The State University of New York. A.E.K. and E.R.M. were supported by an NIH training grant (grant AIO7614) awarded to the Witebsky Center for Microbial Pathogenesis and Immunology. A.E.K. was partly supported by a Presidential Fellowship administered by the University of Buffalo Office of the Provost.
We thank Jason P. Folster for critical review of the manuscript.
REFERENCES
- 1.Angerer A, Enz S, Ochs M, Braun V. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. FecI belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol Microbiol. 1995;18:163–174. doi: 10.1111/j.1365-2958.1995.mmi_18010163.x. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1995. Saccharomyces cerevisiae; pp. 13–30. [Google Scholar]
- 3.Bibb M J, Molle V, Buttner M J. ςBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor. J Bacteriol. 2000;182:4606–4616. doi: 10.1128/jb.182.16.4606-4616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connell T D, Martone A J, Holmes R K. A new mobilizable cosmid vector for use in Vibrio cholerae and other gram-negative bacteria. Gene. 1995;153:85–87. doi: 10.1016/0378-1119(94)00804-2. [DOI] [PubMed] [Google Scholar]
- 5.Connell T D, Dickenson A, Martone A J, Militello K T, Filiatraut M J, Hayman M L, Pitula J. Iron starvation of Bordetella avium stimulates expression of five outer membrane proteins and regulates a gene involved in acquiring iron from serum. Infect Immun. 1998;66:3597–3605. doi: 10.1128/iai.66.8.3597-3605.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFeyter R, Kado C I, Gabriel D W. Small, stable shuttle vectors for use in Xanthomonas. Gene. 1990;88:65–72. doi: 10.1016/0378-1119(90)90060-5. [DOI] [PubMed] [Google Scholar]
- 7.deLorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubrac S, Touati D. Fur-positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J Bacteriol. 2000;182:3802–3808. doi: 10.1128/jb.182.13.3802-3808.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enz S, Braun V, Crosa J H. Transcription of the region encoding the ferric dicitrate-transport system in Escherichia coli: similarity between promoters for fecA and for extracytoplasmic function sigma factors. Gene. 1995;163:13–18. doi: 10.1016/0378-1119(95)00380-o. [DOI] [PubMed] [Google Scholar]
- 10.Escolar L, Perez-Martin J, deLorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farewell A, Kvint K, Nystrom T. Negative regulation by RpoS: a case of sigma factor competition. Mol Microbiol. 1998;29:1039–1051. doi: 10.1046/j.1365-2958.1998.00990.x. [DOI] [PubMed] [Google Scholar]
- 12.Farewell A, Kvint K, Nystrom T. uspB, a new ςS-regulated gene in Escherichia coli which is required for stationary-phase resistance to ethanol. J Bacteriol. 1998;180:6140–6147. doi: 10.1128/jb.180.23.6140-6147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldberg M B, Boyko S A, Calderwood S B. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc Natl Acad Sci USA. 1991;88:1125–1129. doi: 10.1073/pnas.88.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamza I, Qi Z, King N D, O'Brian M R. Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum. Microbiology. 2000;146:669–676. doi: 10.1099/00221287-146-3-669. [DOI] [PubMed] [Google Scholar]
- 15.Hantke K. Regulation of ferric iron transport in Escherichia coli K-12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 16.Harle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995;14:1430–1438. doi: 10.1002/j.1460-2075.1995.tb07129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrichs D E, Poole K. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol. 1996;178:2586–2592. doi: 10.1128/jb.178.9.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes K T, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 19.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 20.Kim I, Stiefel A, Plantor S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 21.Koster M, van Klompenburg W, Bitter W, Leong J, Weisbeek P. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 1994;13:2805–2813. doi: 10.1002/j.1460-2075.1994.tb06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leoni L, Orsi N, deLorenzo V, Visca P. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1481–1491. doi: 10.1128/jb.182.6.1481-1491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leyh R, Griffith R W. Characterization of the outer membrane proteins of Bordetella avium. Infect Immun. 1992;60:958–964. doi: 10.1128/iai.60.3.958-964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Litwin C M, Calderwood S B. Role of iron in regulation of virulence genes. Clin Microbiol Rev. 1993;6:137–149. doi: 10.1128/cmr.6.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lonetto M, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial ς factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller J H. A short course in bacterial genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. Procedures for working with lac; pp. 72–74. [Google Scholar]
- 28.Missiakas D, Raina S. The extracytoplasmic function sigma factors: role and regulation. Mol Microbiol. 1998;28:1059–1066. doi: 10.1046/j.1365-2958.1998.00865.x. [DOI] [PubMed] [Google Scholar]
- 29.Murphy E R, Dickenson A, Militello K T, Connell T D. Genetic characterization of wild-type and mutant fur genes of Bordetella avium. Infect Immun. 1999;67:3160–3165. doi: 10.1128/iai.67.6.3160-3165.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ochs M, Veitinger S, Kim I, Welz D, Angerer A, Braun V. Regulation of citrate-dependent iron transport of Escherichia coli: FecR is required for transcription activation by FecI. Mol Microbiol. 1995;15:119–132. doi: 10.1111/j.1365-2958.1995.tb02226.x. [DOI] [PubMed] [Google Scholar]
- 31.Ochs M, Angerer A, Enz S, Braun V. Surface signaling in transcriptional regulation of the ferric citrate transport system of Escherichia coli: mutational analysis of the alternative sigma factor FecI supports its essential role in fec transport gene transcription. Mol Gen Genet. 1996;250:455–465. doi: 10.1007/BF02174034. [DOI] [PubMed] [Google Scholar]
- 32.Pressler U, Staudenmaier H, Zimmerman L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–2724. doi: 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu J, Helmann J D. The −10 region is a key promoter specificity determinant for the Bacillus subtilis extracytoplasmic-function ς factors ςX and ςW. J Bacteriol. 2001;183:1921–1927. doi: 10.1128/JB.183.6.1921-1927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rimler R B, Simmons D G. Differentiation among bacteria isolated from turkeys with coryza (rhinotracheitis) Avian Dis. 1983;27:491–500. [PubMed] [Google Scholar]
- 35.Russo T A, Singh G. An extraintestinal, pathogenic isolate of Escherichia coli (O4/K54/H5) can produce a group 1 capsule which is divergently regulated from its constitutively produced group 2: K54 capsular polysaccharide. J Bacteriol. 1993;175:7617–7623. doi: 10.1128/jb.175.23.7617-7623.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitt M P, Holmes R K. Iron-dependent regulation of diphtheria toxin and siderophore expression by the cloned Corynebacterium diphtheriae repressor gene dtxR in C. diphtheriae C7 strains. Infect Immun. 1991;59:1899–1904. doi: 10.1128/iai.59.6.1899-1904.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sexton R, Gill P R, Callanan M J, O'Sullivan D J, Dowling D N, O'Gara F. Iron-responsive gene expression in Pseudomonas fluorescens M114: cloning and characterization of a transcription-activating factor, PbrA. Mol Microbiol. 1995;15:297–306. doi: 10.1111/j.1365-2958.1995.tb02244.x. [DOI] [PubMed] [Google Scholar]
- 38.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 39.Stiefel A, Mahren S, Ochs M, Schindler P T, Enz S, Braun V. Control of the ferric citrate transport system of Escherichia coli: mutations in region 2.1 of the FecI extracytoplasmic-function sigma factor suppress mutations in the FecR transmembrane regulatory protein. J Bacteriol. 2001;183:162–170. doi: 10.1128/JB.183.1.162-170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. . (Erratum, 240:271.) [DOI] [PubMed] [Google Scholar]
- 41.Tai S P, Holmes R K. Iron regulation of the cloned diphtheria toxin promoter in Escherichia coli. Infect Immun. 1988;56:2430–2436. doi: 10.1128/iai.56.9.2430-2436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanderpool C K, Armstrong S K. The Bordetella bhu locus is required for heme iron utilization. J Bacteriol. 2001;183:4278–4287. doi: 10.1128/JB.183.14.4278-4287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Hove B, Staudenmaier H, Braun V. Novel two-component transmembrane transcription control: regulation of iron dicitrate transport in Escherichia coli K-12. J Bacteriol. 1990;172:6749–6758. doi: 10.1128/jb.172.12.6749-6758.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasil M L, Ochsner U A, Johnson Z, Colmer J A, Hamood A N. The Fur-regulated gene encoding the alternative sigma factor PvdS is required for iron-dependent expression of the LysR-type regulator PtxR in Pseudomonas aeruginosa. J Bacteriol. 1998;180:6784–6788. doi: 10.1128/jb.180.24.6784-6788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vasil M L, Ochsner U A. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 46.Wei Z M, Beer S V. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J Bacteriol. 1995;177:6201–6210. doi: 10.1128/jb.177.21.6201-6210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wosten M M. Eubacterial sigma-factors. FEMS Microbiol Rev. 1998;22:127–150. doi: 10.1111/j.1574-6976.1998.tb00364.x. [DOI] [PubMed] [Google Scholar]