Abstract
The fast-developing synthetic biology (SB) has provided many genetic tools to reprogram and engineer cells for improved performance, novel functions, and diverse applications. Such cell engineering resources can play a critical role in the research and development of novel therapeutics. However, there are certain limitations and challenges in applying genetically engineered cells in clinical practice. This literature review updates the recent advances in biomedical applications, including diagnosis, treatment, and drug development, of SB-inspired cell engineering. It describes technologies and relevant examples in a clinical and experimental setup that may significantly impact the biomedicine field. At last, this review concludes the results with future directions to optimize the performances of synthetic gene circuits to regulate the therapeutic activities of cell-based tools in specific diseases.
Subject terms: Cancer, Drug delivery, Biomarkers, Genetic engineering
Introduction
Modern science is becoming highly multidisciplinary to promote the innovation of new theories and technologies. Synthetic biology (SB) is one of the fastest-growing and most dynamic multidisciplinary fields comprising different projects, approaches, and definitions.1–3 SB applies engineering principles to genetically design and transform cells or living organisms by constructing biological functional components, devices, and systems, creating or reinventing biological modules to meet human needs, or even creating novel biological systems.4 SB-inspired cell factory engineering for industrial applications, in a narrow concept, is about using renewable biomass resources as raw materials and synthetic assembly of existing components from microbial or animal cells to produce various products.5–7 Engineered and standardized functional modules are the strengths of SB. Appropriate interface design makes modules detachable, improving their stability and compatibility in different environments. Whole-genome synthesis, redesign, and reconstruction allow generating of artificial biomimetic/minimal cells, while atomic or module/domain-level protein engineering offers high specificity and controllability over more complex functions. Meanwhile, innovative modification and creative assembly of signaling and communication systems can further optimize cell engineering. In addition, mathematical models for functional module simulation and prediction help streamline circuit design for the development of invaluable research tools.8–10
Engineered cells can have various applications, including the production of industrial or pharmaceutical compounds and increasing agricultural productivity by alleviating pathogens and environmental challenges.11,12 SB-inspired cell engineering can be commercialized for medical and healthcare. The main goal of medical SB is to genetically modify cells and redesign/synthesize regulatory systems to assist in disease diagnosis and treatment.13,14 Such cell-based sensors and therapies could achieve more specific detection and treat diseases on a much broader scale.12,15,16 Medical SB utilizes rationally designed therapeutic gene circuits, which can be implanted into the human body via vectors to correct the targeted defect. SB-inspired cell engineering with efficient design, specificity, and control can maximize therapeutic effects while minimizing side effects.
There are several reviews on SB technologies for developing therapeutic strategies or improving clinical effectiveness; most of them covered different aspects or specific approaches of SB in medical applications (Fig. 1).17–26 The ongoing development of engineered cells-derived tools has attracted substantial public attention. Accordingly, this review summarizes the recent preclinical, clinical, and experimental data to discuss the benefits and limitations of SB techniques in cell engineering and biomedical research. Here, we elaborate on various SB-driven cell devices in diagnosis, treatment, and drug development. Also, we address the current and potential future challenges for SB and cell engineering in medical applications.
Fig. 1.
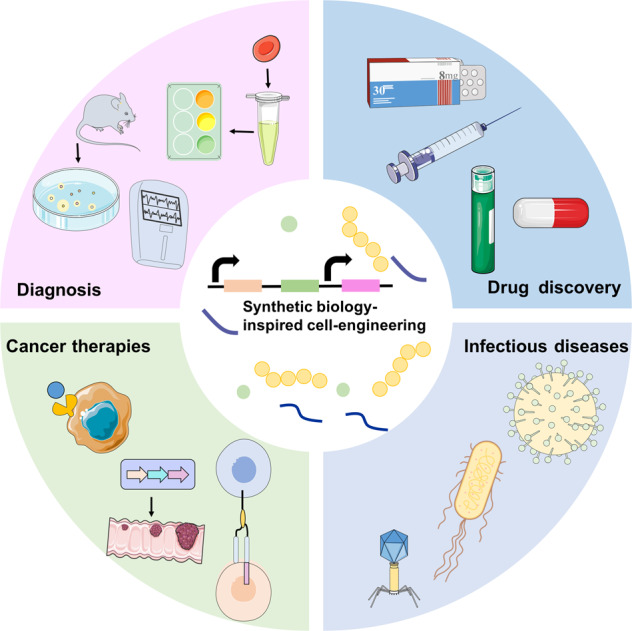
Synthetic biology-inspired cell engineering can be employed for various medical applications. Synthetic gene networks are uploaded into cells for disease diagnosis, cancer therapies, infectious diseases treatment, and drug discovery
Engineered cells for diagnosis
Accurate disease characterization and monitoring are critical for therapeutic planning and epidemiological records. However, most traditional diagnostic procedures involving pathological examination of biopsies cannot precisely detect the disease state in real-time and are time-consuming. Thus, there is an urgent need to enhance current medical diagnostics capabilities. SB-inspired cell engineering and programmable biosensors have focused on high-throughput biomolecular/pathogens identification and probing, which can improve clinical diagnosis and disease management protocols (Fig. 2 and Table 1).25,27,28 Here, we describe the emerging advances of SB-inspired cell engineering approaches in diagnostics that may speed up data acquisition with improved sensitivity, specificity, real-time quantification, and accuracy at a substantially lower cost.
Fig. 2.
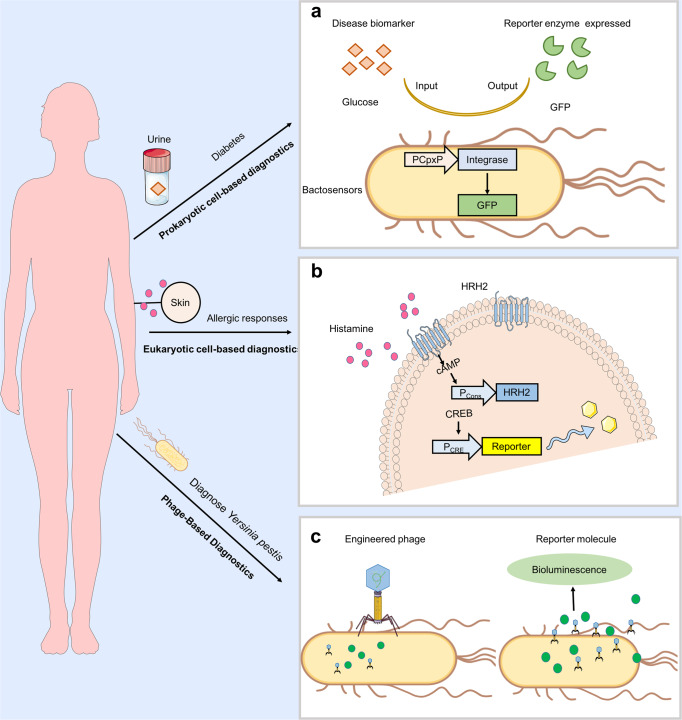
Synthetic biology platforms for diagnostics. a Schematic representation of engineered bacteria sense-and-respond systems adopt disease biomarkers (orange quadrilateral) to sense disease, in turn, produce a diagnostic readout such as β-lactamase and green fluorescent protein (GFP) (green Pacman). b Applications of mammalian synthetic biology in diagnostics. Monitoring of allergic immune by engineered mammalian cell encoding the histamine sense-and-response device. The constitutively expressed histamine-responsive receptor HRH2 sense histamine and triggers an endogenous signaling cascade leading to expression of a reporter protein. c Phage-based diagnostics rely on engineered phage can specific recognition of target bacterial species. Once the engineered phage bound to the target cells, the genome is injected into it then the phage replicate and reporter gene expressed
Table 1.
Advances in engineering cells for disease diagnosis
| Disease | Biomarker | Reporter | Diagnostics feature |
|---|---|---|---|
| Zinc deficiency | Zinc | Pigments | Zinc-responsive transcription factors Zur and ZntR are modules to respond extracellular concentrations of zinc37 |
| Diabetes | Glucose | Fluorescence | Glucose-responsive integrase expression cassette (PCpxP–integrase–pA) that expresses the green fluorescent protein (GFP) for glycosuria testing38 |
| Vibrio cholera infection | Cholera autoinducer 1 (CAI-1) | Colorimetric enzymatic reaction | CqsS-NisK quorum sensing block for cholera autoinducer 1 (CAI-1) detection44 |
| Staphylococcus aureus infection | Autoinducer peptide-I (AIP-I) | Yellow pigment p-nitrophenol | Agr quorum sensing (agrQS) from Staphylococcus aureus to detect autoinducer peptide-I (AIP-I)45 |
| Gut inflammation | Thiosulfate/tetrathionate | Colorimetric enzymatic reaction | Two-component signaling system ThsSR/TtrSR is engineered to detect thiosulfate/tetrathionate49,50 |
| Gut inflammation | Nitrate | Fluorescence | NarX-NarL two-component system in E. coli responds to nitrate51 |
| Liver tumors | Liver metastases | Bioluminescent | PROP-Z (programmable probiotics with lacZ) platform can report liver metastasis52 |
| Intestinal bleeding | Heme | Fluorescence | Probiotic biosensor based on the sensitivity to heme diagnosis of gastrointestinal bleeding in pigs53 |
| Allergic responses | Histamine | Citrine | Engineered HEK293 cells can sensing the extracellular histamine levels and triggering cAMP-induced SEAP (secreted alkaline phosphatase) expression65 |
| Hypercalcaemia | Calcium | Melanin | Engineered HEK293 cells monitor calcium levels and produce melanin visible through the skin66 |
| Yersinia pestis | Phage ΦA1122 with luxAB | Bioluminescence | Engineered a diagnostic phage for Yersinia pestis detection by bioluminescence reporter81 |
| Mycobacterium tuberculosis | Fluoromycobacteriophages | Bioluminescence | Microscopy-based method utilizing the reporter mycobacteriophage mCherrybombϕ for detection of M. tuberculosis82 |
| Listeria monocytogenes | Nanoluciferase (NLuc) -based phage | Bioluminescence | Engineered nanoluciferase (NLuc)-based phage reporters to identify Listeria monocyte83 |
Prokaryotic cell-based diagnostics
In the past several decades, bacteria-based biosensors have been extensively used in the food, environment, and medical industries.29,30 Bacterial biosensors can rapidly adapt to test environments to sense biological substances both in vitro and in vivo. Bacterial sensors are preferred for selectivity, sensitivity, automation, miniaturization, and accuracy.31 Engineered bacterial sensors have been successfully used for the analysis of environmental pollutants.32,33 Many bacterial sensors with specific and sensitive gene circuits for the identification of biological signals can be used to detect widespread clinically relevant biomarkers in vitro, such as in serum and urine.34 In particular, certain symbiotic bacteria that colonize specific tissues and organs are believed to be feasible candidates for cell engineering to serve as in vivo tools for real-time diagnosis.28,35,36
Most bacterial-based diagnostic biosensors produce an easily measurable output in the form of a pigment or fluorescent protein upon identification of a target signal.37,38 For instance, Zur and ZntR zinc-responsive transcription factors (TFs) modules respond to extracellular concentrations of zinc and produce visible pigments; Escherichia coli cells integrated with this circuit can sense serum zinc levels to identify zinc deficiency.37 Such a bacterial sensor impacts multiple metabolite production pathways through a single TF, reduces diagnostic time, and explains results through visual inspection. Similarly, the glucose-responsive integrase expression cassette (PCpxP–integrase-pA), which expresses the green fluorescent protein (GFP), is an ideal reporter to test glycosuria; hydrogel beads immobilized bacteria containing this module can evaluate urine glucose levels in diabetic patients with high sensitivity and specificity.38
Quorum sensing (QS) systems that determine population behavior are widely distributed among bacteria and have a long evolutionary history.39 QS signaling molecule profiling from specific pathogens are proper biomarkers for bacterial diagnosis; acyl-homoserine lactone (AHL), 2-heptyl-4-quinolone (HHQ), and acyl-homoserine lactones (acyl HSLs) have been used to identify P. aeruginosa infection.40–42 Importantly, QS signals can be directly monitored by genetically engineered non-pathogenic bacteria.43 Mao et al. reprogrammed the probiotic Lactococcus lactis by introducing a chimeric CqsS-NisK quorum sensing block for the detection of cholera autoinducer 1 (CAI-1) from Vibrio cholerae.44 This system can easily detect V. cholerae in fecal samples and intestinal environments. Another research group generated a probiotic Lactobacillus reuteri strain integrated with an agr quorum sensing (agrQS) biosensor from Staphylococcus aureus, which could identify quorum sensing molecule autoinducer peptide-I (AIP-I) from common pathogenic Staphylococcus sp with high sensitivity in the nanomolar range.45 Engineered probiotic bacteria integrated with a rational antimicrobial module can be novel therapeutic “robots” to track and eliminate pathogenic bacteria.
Two-component system (TCS), including a membrane-bound histidine kinase for signal sensing/transduction and a cognate intracellular response regulator to activate downstream gene expression, is a key mechanism of how bacteria perceive and respond to the environment.46 The well-defined TCSs have been used for diagnostic signal analysis.47 Holowko et al. integrated three V. cholerae-derived TCS elements CqsS, LuxU, and LuxO with a dCAS9-based GFP-reporter system into E. coli to construct a biosensor with a high sensitivity for QS-ligand cholera CAI-1 indicating the presence and proliferation of V. cholera.48 Previous studies engineered thiosulfate/tetrathionate-responsive E.coli strain by combining TCS sensors ThsSR/TtrSR with pre-installed reporter system or phage lambda cI/Cro-based memory system as potential therapeutics and diagnostics for inflammation49,50 Recently, a new TCS-based whole-cell biosensor was designed by adopting E. coli NarX-NarL to sense nitrate levels, a biomarker of gut inflammation.51 This highly specific nitrate-responsive genetic circuit allows non-invasive diagnosis of inflammation-related diseases.
In addition to in vitro diagnosis, engineered bacteria systems can also be exploited for the diagnosis of complex internal environments. Biomolecular monitoring of the gastrointestinal environment is often limited by access and complexity of the environment. Danino et al. developed a PROP-Z (programmable probiotics with lacZ) platform using a Nissle 1917 (EcN) derived bacterium with a series of expression cassettes. This orally administered probiotic can produce detectable signals in urine reliably indicating hepatic tumors.52 Another exquisitely engineered micro-bio-electronic device (IMBED) was developed by Mimee et al. They efficiently combined a bacterial-electronic system with SB circuits, electronic sensor, and wireless transmission platform to detect excess heme in blood samples offering rapid in situ diagnosis of gastrointestinal bleeding in pigs.53
Eukaryotic cell-based diagnostics
Microbial biosensors offer inexpensive and feasible disease detection strategies. Similar to engineered bacteria, eukaryotic cell-based biosensors have also been developed for real-time clinical applications.
The number of yeast/fungi-based diagnostic biosensors is growing. Instead of using optical signals such as fluorescence, luminescence, and colorimetry, many yeast-based biosensors utilize electrochemical elements (amperometry, potentiometry, conductometry, voltammetry) and growth indicating modules (HIS3, TRP1, LEU2).54–56 One of the advantages of eukaryotic cell biosensors is to incorporate G-protein-coupled receptors (GPCRs) which greatly expands their functional repertoire.57–59 Diagnostic yeast biosensors have often been used to detect the metabolic rate of oxygen, glucose, lactate, and ethanol through enzyme-catalyzed oxidation-reduction reactions; the generated electrical energy is measured by an implemented electrode which rapidly generates signals with high sensitivity and specificity. A d-lactate-selective biosensor was developed from the debris of the recombinant thermotolerant methylotrophic yeast H. polymorpha. This biosensor overproduces d-lactate: cytochrome c-oxidoreductase (DLDH) and has high selectivity and effective stability for d-lactate due to the deletion of cytochrome c-oxidoreductase (CYB2, responsible for l-lactate synthesis) in the C-105 strain genotype (gcr1 catX).60 The same research group further suggested that enriching the flavocytochrome b2 (FC b2) module incorporated H. polymorpha cells by enzyme-bound nanoparticles can improve amperometric current responses.61
Combining new ideas and technologies based on existing yeast-based biosensors stimulates more innovations. Holly V. Goodson’s research group designed an S. cerevisiae-based olfactory reporter by assembling the endogenous galactose-response system and the gene encoding acetyltransferase I (ATF1, isoamyl alcohol-converting enzyme). This system produces enough amount of isoamyl acetate (banana flavor) in response to galactose and is suitable for low-cost non-quantitative/semi-quantitative diagnosis.62 Furthermore, biological paper analytical devices (bioPADs) were created by embedding this bioengineered fluorescent yeast biosensor into a portable paper substrate. This simple but sensitive test strip can detect doxycycline in complex matrices such as in human urine and raw bovine serum.63 It is foreseeable that sustainable developments in fungal genome engineering would take yeast biosensors toward precise, accurate, and cost-effective editing.
Based on the availability and compatibility of endogenous signal-transduction biomarkers, engineered mammalian cells are more suitable for certain diagnostics applications over bacterial and yeast biosensors.64 With the recent technological advances, a few mammalian cell-based diagnostics have been developed. For example, HEK293 cell-based biosensor with a synthetic histamine-responsive signal module (histamine receptor H2, HRH2) can detect allergies from blood samples of high-risk allergic patients by sensing the extracellular histamine levels and triggering the expression of cAMP-induced SEAP (secreted alkaline phosphatase).65 Such engineered mammalian cell systems can be used in high-throughput diagnostic applications such as testing novel drugs for allergic reactions. Many diseases are preceded by a period of asymptomatic pathogenesis and thus require reliable indirect molecular diagnosis methods. Accordingly, Martin Fussenegger’s research group designed a novel HEK293 cell for in vivo diagnostics by employing a calcium-sensing receptor constructed with a synthetic signaling pathway. In response to the increased calcium level in hypercalcemia, this system generates black pigment melanin on the skin via transgenic tyrosinase-mediated enzymatic reaction and thus is ideal for naked-eye detection.66
Except for epithelial cell lines, other mammalian cells including macrophages, neurons cell, mast cells, and B lymphocytes can also be exploited for biosensor applications. Based on the pattern of signal detection and test objective, a specific assay combined with proper cell lines can be used to assess pathogens, toxins, metabolic activity, or even cell proliferation.67 Monkey kidney cells (Vero cells) are sensitive to verotoxin Stx (Shiga-like toxins) due to their overproduced Stx receptor GB3 (globotriaosylceramide).68 Bioengineered Vero cells constructed in collagen matrix demonstrated better cell maturation and improved sensitivity to Stx and thus could detect Shiga-like toxin-producing pathogenic bacteria such as Salmonella, Listeria, Citrobacter, Serratia, and Hafnia.69 IgE-mediated vasoactive amines including histamine and serotonin degranulation are characteristics of basophil or mast cells, which make them a good choice for cell-based biosensor development.70 Jiang et al. reported an electrochemical sensor based on rat basophilic leukemia (RBL-2H3) cells exploited on folding paper-based equipment for casein detection.71 Certainly, engineered mammalian cell-based devices for therapeutic and diagnostic applications are a growing area of interest. The diverse cell behaviors in response to different compounds such as antibiotics, antibodies, and chemokines make mammalian cells attractive model for building diagnostic biosensors. Meanwhile, the complexity and packaging of eukaryotic genome have also to be considered when using them for such applications.
Phage-based diagnostics
Although cell-based biosensor diagnostics can fulfill most needs of clinical applications, the rational design of engineered phages promises some other advantages. Phages are viruses that infect bacteria by adhering to specific host cell surface receptors. A simple model for reporter phage design is that the presence of the correct epitope induces the transfer of the phage genome into bacteria, triggering bioluminescence from luciferase in the target bacteria.72,73 Bioluminescent reporter-based phages have been applied for the diagnosis of pathogens such as Bacillus anthracis,74,75 Vibrio parahaemolyticus,76 Salmonella typhimurium,77 pathogenic Escherichia coli,78,79 and Staphylococcus aureus.80 For example, an engineered Yersinia pestis phage was used for the detection of an etiologic agent in bubonic plague.81 A microscopy-based method in association with the fluoromycobacteriophage mCherrybombϕ was used to diagnose M. tuberculosis.82 Recently, Meile et al. engineered a set of nanoluciferase (NLuc)-based phage reporters to identify and differentiate Listeria monocytogenes in potentially contaminated milk, cold food, and other eatable samples.83 These NLuc-based reporter phages enabled rapid and ultrasensitive detection of one CFU of L. monocytogenes in 25 g of artificially contaminated natural food in less than 24 h.
Phages can also be exploited as bioelectrode elements by immobilizing them to an electrode surface or utilizing lytic phages to infect target bacteria to release bacterial cell contents such as adenosine triphosphate (ATP) for electrochemical detection.84 Niyomdecha et al. used an M13 bacteriophage-modified electrode for the detection of Salmonella spp.: this biosensor binds to target bacteria through the amino acid groups on the filamentous phage.85 Farooq et al. mobilized S. aureus-specific lytic phages in surface-modified bacterial cellulose (BC) matrix conducted by carboxylated multi-walled carbon nanotube (c-MWCNTs). The functionalized electrode could detect S. aureus through impedance measurements.86 Lately, gold electrodes were used as a transducer with chemically modified M13 phages to monitor caspase-3 activity in apoptotic HeLa cells with nanomole range sensitivity.87 Such bacterial cell and phage-based biosensors can effectively sense disease-associated biomarkers, paving the way for next-generation of medical diagnostics.
Engineered cell-based therapeutics for cancer
More than 19 million new cases of cancer are diagnosed every year, making it one of the most devastating diseases worldwide.88–90 Traditional cancer treatments such as surgical intervention, radiation, and chemotherapeutic drugs have several limitations, including high cytotoxicity and low selectivity.91–94 SB approaches for cancer treatment are the most vivid and promising trends that enable precise timing, location, and dosing of the drug. Accordingly, engineered mammalian and bacterial cells have been increasingly recognized as promising anticancer devices.95–97
Bacteria-derived tools for cancer treatment
Since some bacteria can colonize in a hypoxia intratumoral environment, human cancer can now be treated by SB-engineered bacteria.98–101 Certain bacterial species, including Streptococcus, Caulobacter, Escherichia, Bifidobacterium, and Salmonella, can automatically sense and accumulate in tumor tissues after external administration (injection or oral) to produce cytotoxic toxins or release delivery compounds.102 One of the major advantages of bacterial therapies for cancer is that they can be easily manipulated for the effective delivery of anticancer agents.103 Based on the reported studies, bacterial therapies for cancer majorly involves three types of mechanisms: expression of toxic proteins to directly kill cancer cells, immunotherapeutics that stimulate the immune system to kill cancer cells or the transfer of therapeutic drug delivery vehicles (Fig. 3 and Table 2).102,104,105
Fig. 3.
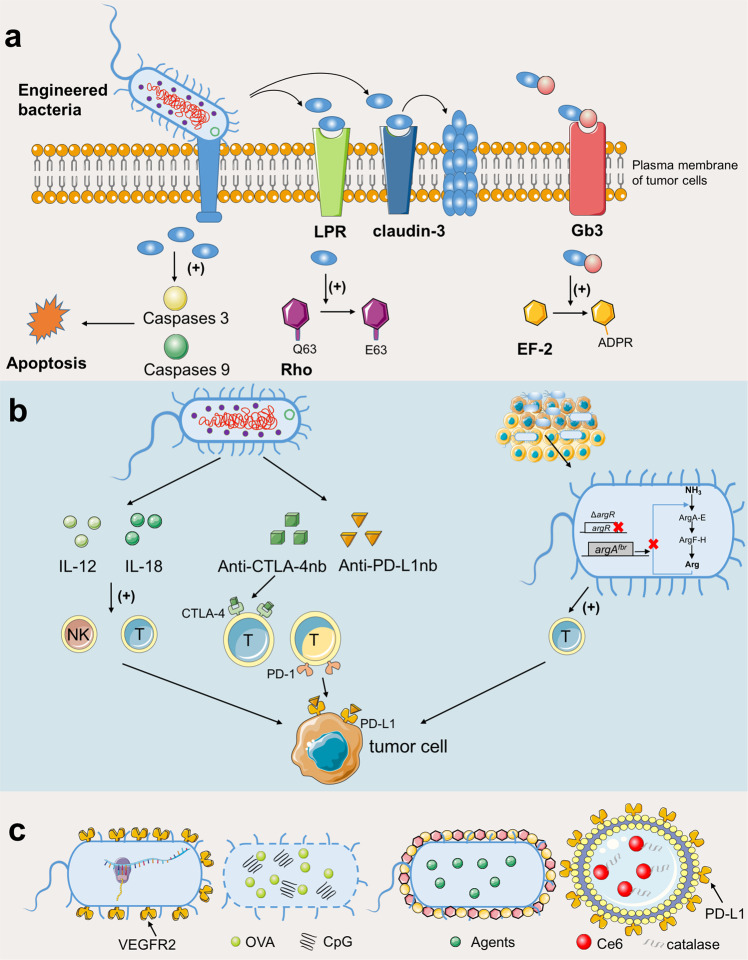
Engineered bacteria for cancer therapy. Current strategies can be divided into three types: a expression of toxic proteins to directly kill cancer cells, b immunotherapeutics, and c therapeutic drug delivery vehicles. a Bacteria could utilize secretion systems such as T3SS and T6SS to directly inject bacterial effector proteins into eukaryotic host cells, or secret effectors targeting receptors on cell surface. LRP laminin receptor precursor protein, EF-2 elongation factor 2. b Engineered bacteria could express cytokines or other chemical molecules to induce proper immune cells. IL-12 Interleukin 12, CTLA-4 cytotoxic T-lymphocyte-associated antigen-4, PD-1 programmed death-1, PD-L1 programmed cell death protein-ligand 1, nb nanobody. c Bacteria-based vaccine or drug delivery systems. VEGFR2 vascular endothelial growth factor 2, OVA ovalbumin
Table 2.
Bacteria-based therapies against cancer in clinical trials
| Type | Biological/tradename | Sponsor | NCT | Year | Phase | Status | Cancer type |
|---|---|---|---|---|---|---|---|
| Salmonella Typhimurium | VNP20009 | National Cancer Institute | NCT00004988 | 2008 | I | Completed | Cancer neoplasm, neoplasm, metastasis297 |
| VNP20009 | National Cancer Institute | NCT00006254 | 2013 | I | Completed | Unspecified adult solid tumor298 | |
| Salmonella-IL-2 | Masonic Cancer Center, University of Minnesota | NCT01099631 | 2020 | I | Completed | Solid tumor cancer299 | |
| VXM01/Avelumab | Vaximm GmbH | NCT03750071 | 2021 | I/II | Recruiting | Recurrent glioblastoma | |
| SGN1 | Guangzhou Sinogen Pharmaceutical Co., Ltd. | NCT05103345 | 2021 | I | Not yet recruiting | Advanced solid tumor | |
| Salmonella-IL-2 | Salspera LLC | NCT04589234 | 2022 | II | Recruiting | Metastatic pancreatic cancer | |
| VNP20009-M | Guangzhou Sinogen Pharmaceutical Co., Ltd. | NCT05038150 | 2022 | I | Not yet recruiting | Advanced solid tumor | |
| Escherichia coli | SYNB1891/Atezolizumab | Synlogic | NCT04167137 | 2021 | I | Recruiting | Metastatic solid neoplasm, lymphoma300 |
| VAX014 | Vaxiion Therapeutics | NCT03854721 | 2020 | I | Recruiting | Urothelial carcinoma of the urinary bladder301 | |
| Clostridium novyi-NT | Clostridium novyi-NT spores | BioMed Valley Discoveries, Inc. | NCT01118819 | 2016 | I | Terminated | Solid tumor malignancies |
| Clostridium novyi-NT spores | BioMed Valley Discoveries, Inc. | NCT01924689 | 2019 | I | Completed | Solid tumor malignancies302 | |
| Bifidobacterium longum | bacTRL-IL-12 | Iqvia Pty Ltd. | NCT04025307 | 2021 | I | Terminated | Solid tumors |
| APS001F/Flucytosine (5-FC)/10% maltose | Anaeropharma Science, Inc. | NCT01562626 | 2021 | I/II | Suspended | Tumors, neoplasms, cancer | |
|
Bifidobacterium animalis lactis |
EDP1503/Pembrolizumab | Evelo Biosciences, Inc. | NCT03775850 | 2021 | I/II | Completed |
Colorectal cancer metastatic, triple-negative breast cancer, non-small cell lung cancer303 Bladder cancer, gastroesophageal cancer, renal cell carcinoma, MSI-H |
| Listeria monocytogenes | ADU-623 | Providence Health & Services | NCT01967758 | 2018 | I | Completed | Astrocytic tumors304 |
| CRS-207 with Epacadostat | Aduro Biotech, Inc. | NCT02575807 | 2019 | I/II | Terminated | Platinum-resistant ovarian, fallopian, or peritoneal cancer | |
| ADXS31-142+ Pembrolizumab | Advaxis, Inc. | NCT02325557 | 2020 | I/II | Unknown | Prostate cancer | |
| CRS-207 in combination with chemotherapy | Aduro Biotech, Inc. | NCT01675765 | 2020 | I | Completed | Malignant pleural mesothelioma305 | |
| ADXS11-001 | Andrew Sikora | NCT02002182 | 2021 | II | Active, not recruiting | HPV-positive oropharyngeal cancer306 | |
| CRS-207 | Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins | NCT03190265 | 2022 | II | Active, not recruiting | Pancreatic cancer307 | |
| ADXS-503 with or without pembro | Advaxis, Inc. | NCT03847519 | 2022 | I/II | Recruiting | Metastatic non-small cell lung cancer | |
|
Multi-species/strain bacteria |
Dietary supplement: probiotic | Mayo Clinic | NCT03358511 | 2020 | Not applicable | Completed | Breast cancer308 |
| MRx0518 | 4D pharma plc | NCT03637803 | 2021 | I/II | Recruiting | Oncology, solid tumor, non-small cell lung cancer, renal cell carcinoma, melanoma, bladder cancer | |
| VE800 with Nivolumab | Vedanta Biosciences, Inc. | NCT04208958 | 2021 | I/II | Active, not recruiting | Metastatic cancer |
Natural bacterial toxins are emerging as potential anticancer agents to treat established solid tumors.104 Bacterial extract from Streptococcus pyogenes and Serratia marcescens was shown effective against sarcomas, melanomas, and myelomas, and named “Coley’s toxins” based on the name of the researcher.106,107 Since then, more bacterial toxins have been identified and tested for anticancer therapy.102,104 Cytolethal distending toxins from Salmonella typhi and the cycle-inhibiting factor (Cif) from enterohemorrhagic E. coli can inhibit cancer cell growth by blocking mitosis.108,109 The bifunctional Pseudomonas aeruginosa exotoxin T (ExoT) consists of a GTPase-activating protein (GAP) and an ADP-ribosyltransferase encoded from the N- and C-terminus, respectively.110,111 The GAP protein guides the mitochondrial dysfunction in the host cell through activating host caspases 3 and 9, while the ADP-ribosyltransferase domain upon activating the CT10 regulator of the kinase (Crk) leads to atypical anoikis apoptosis.112,113 Clostridium perfringens enterotoxin (CPE) can treat colorectal cancers by binding to claudin-3 on the tumor cell surface forming a multi-protein membrane pore complex, altering cellular osmotic equilibrium that leads to cell lysis.114,115 To improve the specific recognition of bacterial toxins for cancer cells, they can be reprogrammed either to conjugate with tumor surface antigens or ligands that selectively bind to the target cell receptor.104,116 For instance, P. aeruginosa exotoxin A (PE) was fused to anti-CD25 to form a novel recombinant toxin.117 Native diphtheria toxin (DT) was linked with the B-subunit of Shiga-like toxin (STXB) to improve its affinity for breast cancer cells and showed improved cytotoxicity against Gb3-expressing cancer cells in a dose-dependent manner.118 Besides, bacterial spores can also be exploited as anticancer agents as they can specifically germinate inside the necrotic/hypoxic tissues such as a solid tumor.119,120 Clostridium histolyticum spores showed considerable oncolysis and tumor regression effects in murine models.121 However, the original bacterial spores might not eliminate cancer cells and would require genetic modifications or combinational use with other anticancer strategies such as radiotherapy and chemotherapy.
Expression of exogenous cytotoxic proteins may trigger the host immune response raising safety concerns while engineered bacteria to induce specific cytokines can be an alternative anticancer strategy.102 Selective expression of cytokines can induce proper immune cells to clear tumors via multiple mechanisms with fewer side effects.122 Interleukin-2 (IL-2) and IL-18 are the most crucial components of cancer-related inflammation and participate in the immune regulation network. They activate the antitumor cytotoxicity of T cells and natural killer cells.123,124 Engineered Salmonella producing IL-2 and IL-18 was shown to inhibit tumor growth in animal experiments.125,126 Targeting programmed cell death protein-ligand 1 (PD-L1) and cytotoxic T-lymphocyte-associated protein-4 (CTLA-4) by immune-checkpoint inhibitors is a revolutionary cancer immunotherapy strategy but has side effects such as fatigue, skin rashes, and endocrine disorders.127,128 Increasing l-arginine levels in tumor tissue can activate the response of immune-checkpoint inhibitors. Engineered E. coli Nissle 1917 strain after accumulation in the tumor tissue converted ammonia to l-arginine recruiting tumor-infiltrating T cells showing a remarkable antitumor effect.129 Gurbatri et al. engineered a probiotic bacteria system to tightly control the release of nanobodies, which targeted PD-L1 and CTLA-4 and activated T cells.130 In another case, engineered Salmonella (△ppGpp) was utilized to secret the development endothelial locus 1 (Del-1) protein promoting the recruitment of M1 macrophage to tumors.131
The deoxygenated inflammatory microenvironment of a tumor is distinct from most other tissues and therefore allows selective accumulation of obligate anaerobes in the tumor for the delivery of bacteria-based vaccine or drug.102,105 Hu et al. used self-assembled synthetic nanoparticles coated attenuated bacteria to develop a novel oral DNA vaccine to express vascular VEGFR2 and specific antigens.132 This engineered bacteria could activate T cells and induce the production of cytokine for efficacious cancer immunotherapy. In 2017, researchers exploited demi-bacteria (DB) from Bacillus for the efficient accommodation of antigen and cytidine-phosphate-guanosine (CpG) adjuvants, inducing synergistic cellular and humoral immune responses in vivo.133 In combination with traditional chemotherapy and/or phototherapy, SB approaches can significantly promote the anticancer therapeutic outcome. For instance, attenuated Salmonella coated with degradable polydopamine was used for the delivery of photothermal agents to tumors.134 To efficaciously transport the chemotherapeutic drugs to the intestinal environment, Bacillus coagulans spores were modified with deoxycholic acid (DA) and then load them with drugs serving as autonomous nanoparticle (NP) producers.135 In addition, the bacteria-derived outer membrane vesicles (OMVs) have been explored as drug carriers in vaccine developments and cancer therapies.136,137 Based on oxygenated PDT and immunotherapy, Zhang et al. combined Chlorin e6 (Ce6) with negatively charged catalase (CAT) to form a molecular scaffold (CAT-Ce6) and transported it using aPDL1 modified (OMV-aPDL1) Salmonella OMVs to inhibit tumor growth.138
CAR-T-based cellular immunotherapy
Engineered cellular immunotherapy has great potential for advanced hematological malignancies and other solid tumors.139 Compared with traditional treatment and engineered bacterial and cell-based therapies have two major advantages: (1) a large number of basic research, clinical trials, and SB can make rapid development in this field, and (2) it offers long-term precise regulation and coordination of immune response in vivo. Current cancer cell therapy includes chimeric antigen receptor (CAR)-T cell, T-cell receptor (TCR) T cell, CAR-natural killer (NK)/NKT cells, tumor-infiltrating lymphocyte (TIL), choline transporter‐like proteins, etc.140 Among them, CAT-T-cell therapy is one of the most innovative gene therapies with the greatest breakthrough in the field.141,142 At present, five cell therapies have been approved by the Food and Drug Administration (FDA) in July 2021 after clinical trials and all of them are CAR-T-cell therapies (Table 3).140 Claudin18.2-specific CAR-T cells (CT041) showed good efficacy against digestive tract solid tumors with 48.6% objective remission rate and 73.0% disease control rate exceeding the best result of CAR-T-cell therapy for hematoma treatment (remission rate <30%).143 In the last decade, CAR has been developed to the fourth generation, the present CAR consists of a single-chain variable fragment (scFv), a flexible hinge, a transmembrane domain (CD8a), one or more co-stimulatory domains, and an intracellular CD3ζ domain from the T-cell receptor.139 Here, we discuss the main SB strategies for antitumor CAR engineering, including switchable control, logic gates, and armored CARs, and then discuss the future possible development of cell therapies from a SB perspective (Fig. 4).
Table 3.
Currently CAR-T products approved by FDA for cancer therapy
| Product name | Target | Sponsor | First time offered to market | Approved countries | Vector | Cancer type |
|---|---|---|---|---|---|---|
| Kymriah (tisagenlecleucel) | CD19 | Novartis | September 30, 2017 | USA (2017), EU (2018), Canada (2019), Japan (2019) | Lentivirus | B-cell acute lymphoblastic leukemia309 |
| Yescarta (axicabtagene ciloleucel) | CD19 | Kite Pharma, Inc./Gilead Sciences lnc. | October 18, 2017 | USA (2017), EU (2018), Canada (2019) | Retrovirus | Relapsed/refractory diffuse large B-cell lymphoma, relapsed/refractory primary mediastinal B-cell lymphoma, relapsed/refractory transformed follicular lymphoma, relapsed/refractory high-grade B-cell lymphoma310 |
| Tecartus (Brexucabtagene autoleucel) | CD19 | Kite Pharma, Inc./Gilead Sciences lnc. | October 28, 2020 | USA (2020), EU (2020) | Retrovirus | Relapsed/refractory mantle cell lymphoma311 |
| Breyanzi (lisocabtagene ciloleucel) | CD19 | Juno Therapeutics, Inc/BMS | February 5, 2021 | USA (2021) | Lentivirus | Relapsed/refractory diffuse large B-cell lymphoma, relapsed/refractory primary mediastinal B-cell lymphoma, relapsed/refractory transformed follicular lymphoma, relapsed/refractory high-grade B-cell lymphoma312 |
| Abecma (idecabtagene vicleucel) | BCMA | Celgene Corporation/BMS | March 26, 2021 | USA (2021) | Lentivirus | Multiple myeloma313 |
| Carvykti (Ciltacabtagene autoleucel) | BCMA | Legendbiotech, Johnson & Johnson | February 28, 2022 | USA (2022) | Lentivirus | Recurrent or refractory multiple myeloma314 |
Fig. 4.
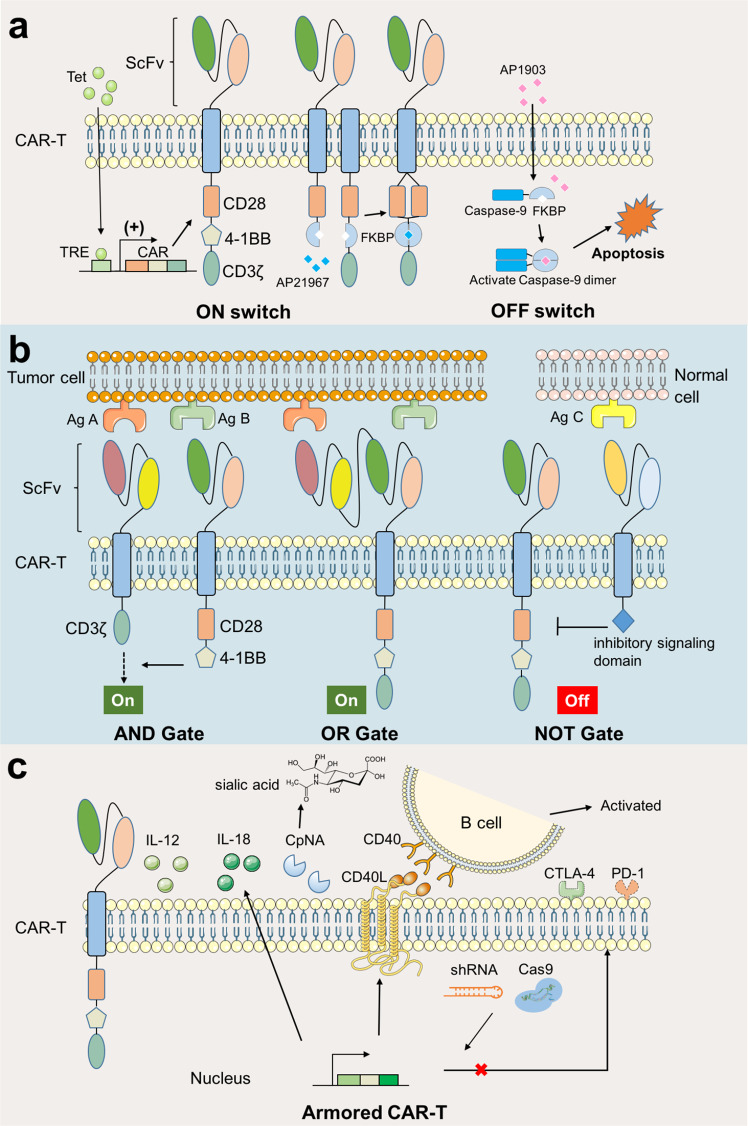
Synthetic Biology in CAR-T Cell Engineering for cancer therapy. Current strategies can be divided into three types: a Switchable CAR, b Logic gates, and c Armored CAR. a Switchable CAR includes ON switches and OFF switches. In ON switches, exogenous molecules could activate the expression of CAR elements or the recombination for activate state. In OFF switches, the exogenous molecules induce the suicide genes such as iCasp9 for controlled elimination. Tet tetracycline, TRE tetracycline response element, FKBP a rapamycin-binding protein, AP21967 rapamycin analog, AP1903 dimer inducers. b Logic gates includes AND Gate, NOT Gate, and OR Gate. The activation of CAR depends on the number and type of antigens from tumor cells. Ag antigen. c As for Armored CAR-T, CAR-T cells are engineered to expression extracellular proteins including IL-12, IL-18, other enzymes or transmembrane ligands or receptors to enhance immune response. Intracellularly engineered CAR-T cell is another powerful strategy, such as inhibiting the transcription of immune-checkpoint molecules by shRNA or CRISPR-Cas9. IL-12 Interleukin 12, CpNA C. perfringens neuraminidase, CD40 cluster of differentiation 40, CD40L CD40 ligand, shRNA short-hairpin RNAs, CTLA-4 cytotoxic T-lymphocyte-associated antigen-4, PD-1 programmed death-1
Engineered switches are powerful control systems that minimize the life-threatening toxicities and other unfavorable side effects of CAR-T cells and have two types, named ON and OFF switches.26,144 A good example is the tetracycline regulatory platform, in which, exogenous tetracycline or doxycycline triggers the time- and dose-dependent expression of CAR components. The co-stimulatory domain could also be separated from the CAR structure and fused with small molecules binding domain for specific recognition and recombination. In addition, the antigen binding region can be engineered to recognize multiple tumor antigens, such as leucine zippers, unique epitopes, or chemical tags.145 In contrast to an ON switch, an OFF switch senses inhibitory signals to stop CAR T-cell activation. The drug-inducible caspase-9 (iCasp9) kill switch is one of the most successful designs, which has been used in several clinical trials for CAR T-cell therapies.146 The iCasp9 switch consists of a domain encoding chemically inducible dimerization system derived from FKBP12 (a rapamycin-binding protein) and split caspase-9 protein; the small-molecule AP1903 can induce the dimerization of FKBP12-caspase-9 causing apoptosis. CAR-T cells carrying the iCasp9 switch have shown effective curative effects in several clinical trials.147,148 There are other non-caspase strategies, such as combining MyD88 and CD40, based on the FKBP12/AP1903 dimerization system, that can overcome the limitation of elimination from the body.149
Considering the sophisticated tumor microenvironment and nonspecific antigens on tumor cells, more precise treatments are necessary to balance safety and efficacy. Improved genetic logic circuits, including AND Gate, NOT Gate, and OR Gate, were designed to solve these problems.144 All of the antigens present in AND Gate are needed for CAR-T-cell activation. Another common strategy is to separate and fuse the activation signal (CD3ζ) and the co-stimulatory domain to two different antigen-recognizing receptors, such as SynNotch system,150 bispecific tandem CAR (TanCAR)151 and masked CAR (mCAR),152 however, this also has the disadvantage of slow activation kinetics. Following the principle “A and not B”, NOT Gate can restrict off-target attacks of T cells reducing unintended immune attacks; however, this has not been validated in clinical trials so far. Alternatively, the purpose of OR Gate is to overcome antigen heterogeneity and antigen escape, thus multiple-targeting CAR-T therapy is developed by fusing different CAR-T cells or generating specific T cells with different CAR suits. The other two alternative strategies are combining different antigen targeting adapters and generation of bispecific CAR-T cells consisting of two or more different scFvs; however, the concerns of potential cytokine release syndrome, neurotoxicity, and B-cell aplasia remain.144
Another strategy for enhancing the antitumor efficacy of the CAR-T cells is to delete or express immunologic factors rather than CAR, named “Armored” CAR-T cells.153,154 Engineered CAR-T cells can secrete extracellular cytokines (including IL-12,155 IL-15,156 and IL-18157), antibodies, and bacterial enzymes increasing the prodrug concentration at the disease site.158 C. perfringens neuraminidase (CpNA) can remove sialic acid residues from the target cells. CpNA-restrained CAR-T cells exhibited superior effector function and cytotoxicity in vitro, showing enhanced antitumor efficacy in a Nalm-6 xenograft model of leukemia, xenograft model of glioblastoma, and syngeneic model of melanoma.159 Enabled CAR-T cells with additional natural or genetically modified transmembrane proteins can have more physiological functions such as signal transduction, recognition, and attachment. Kuhn et al. developed CAR-T cells to produce CD40L, a TNF-associated activation protein (TRAP), to recruit and activate immune effectors.160 Moreover, engineered CAR-T cells disturbing the intracellular transduction of inhibitory signaling pathways is another powerful strategy to inhibit the expression of immune-checkpoint molecules, including short-hairpin RNAs or CRISPR-Cas9.161,162
Although CAR-T therapy offers spectacular therapeutic opportunities against cancer, it has certain limitations such as tumor antigen escape relapse lowering the therapeutic efficacy. To conquer such challenges, novel strategies to optimize the CAR-T cells are needed for increased safety.
Certainly, the developments in SB will continue to promote the progress of tumor treatments; designing subtle gene circuits and finding novel biological components and the right targets could be the main endeavors. Meanwhile, controlling biosafety and reducing in vivo toxicity will also be necessary to balance cost and effectiveness.
Engineered cell-based therapeutics for infectious diseases
Infectious diseases are the major reasons for death and disability worldwide.163 Pathogenic infections including those from bacteria, fungi, and viruses are treated by conventional antibiotic therapy, however, many antibiotics have side effects causing intestinal flora disorder, hearing impairment, nephrotoxicity, and allergic reactions. Moreover, the emergence of multidrug-resistant (MDR) bacteria urgently demands new treatment strategies.164,165 The developing SB could also provide efficient, accurate, and cost-effective tools to treat infectious diseases. Here, we mainly discuss novel strategies against infectious diseases from the SB perspective.
SB tools against bacteria
Bacteria evolved with a variety of molecular weapons to kill or interfere with competitors, such as bacteriocins, effector proteins, and other chemicals.166–168 Thus, engineered bacteria can be exploited for novel antimicrobial biotherapeutics. Jana et al. introduced the arabinose-induced Type VI Secretion System (T6SS) into Vibrio natriegens, a safe bacterium, to manipulate the effector against competitors.169 Ting et al. developed programmed inhibitor cells (PICs), a new system expressing surface nanobodies to mediate antigen-specific cell-cell adhesion, exerting T6SS antimicrobial activity directly against selected bacteria.170 Bacteria use chemical language, termed quorum sensing (QS), to communicate in the community and regulate cell density.168 Probiotic E. coli designed to produce AI-2, which is naturally produced by Vibrio cholera, caused significant growth inhibition of V. cholerae and increased the survival rate of infected animals.171 Engineered Δalr ΔdadX E. coli Nissle 1917 strain can produce Dispersin B, Pyocin S5, Lysin E7, and sense autoinducer 3OC12HSL secreted by P. aeruginosa.172 In the presence of 3OC12HSL, this E. coli strain triggers E7 lysozyme-mediated self-lysis releasing Pyocin S5 and Dispersin B that kills P. aeruginosa. In another case, attenuated human lung pathogen Mycoplasma pneumonia was reformed to secrete anti-biofilm microbial enzymes that degraded Staphylococcus aureus biofilm both in in vitro and in vivo.173 Lysostaphin-producing Staphylococcus simulans could treat methicillin-susceptible and methicillin-resistant Staphylococcus aureus infections.174 Although bacteria can be easily tailored to combat microbial pathogens, their use is limited by the on/off switch of effector release, potential toxic effects in human cells, and long-term stability.175 A new research direction has evolved combining Computer-Based Frameworks and SB for the discover of potential antimicrobial peptide (AMP).176
Producing selective endolysins (lysins) to break bacterial cells, bacteriophages are one of the most effective therapeutic approaches against both Gram-positive and Gram-negative bacteria.177,178 Furthermore, engineered bacteriophage may overcome the limitations of frequent escape of bacterial hosts by modifying cell-wall related receptors, CRISPR-Cas disruption, or restriction-modification systems. E. coli bacteriophage T7 encoding depolymerase Dispersin B (T7DspB) could specifically hydrolyze polymeric b-1,6-N-acetyl-D-glucosamine (PNAG) of bacterial biofilms reducing biofilm formation of E. coli.179 Masuda et al. generated recombinant lnqQ-T7 phage by integrating lnqQ (structure gene of Leaderless bacteriocins, LLB, lacticin Q) into the T7 phage genome, which significantly inhibited lacticin Q-susceptible Bacillus coagulans and E. coli.180 Hsu et al. engineered λ phage to express transcriptional repressor of Shiga toxin (Stx), which successfully reduced Shiga toxin production in the E. coli population colonizing the intestinal tract model.181 The phages can also effectively transport molecules (such as proteins, peptides, DNA, RNA, and drugs) to targeted bacteria. Staphylococcus aureus phage encoding an antimicrobial small acid-soluble spore protein (SASP) was developed to kill S. aureus.182 This engineered phage (SASPject PT1.2) showed 100% activity against the 225 geographically diverse S. aureus isolates. However, bacteriophage treatment will inevitably lead to bacteriophage resistance and serious side effects such as the release of bacterial endotoxins. Also, bacteriophage is highly specific and attacks limited bacteria. In future, SB and genome engineering can expand the scope of phage therapy.182
SB tools against viral infection
SB-based tools can also contribute to the quick diagnosis, therapy, and prevention of viral infections.26 Wei et al. engineered Lactobacillus jensenii, a commensal bacterium that colonizes the cervicovaginal mucosa, to prevent HIV transmission; the engineered strain secretes two-domain CD4 (2D CD4) proteins,183 which associate with HIV type 1 (HIV-1) gp120 and inhibit HIV-1 invasion. Similarly, the probiotic bacterium E. coli Nissle 1917 was designed to secret HIV-gp41-hemolysin peptides, which blocks HIV fusion and subsequent invasion.184 This engineered strain could continuously and stably colonize the colon and cecum and released anti-HIV peptides for several months in mice. Furthermore, a genetically modified Lactobacillus acidophilus ATCC 4356 expressing human CD4 receptors on its surface can adsorb HIV-1 particles.185 The results showed that CD4-carrying bacteria could capture HIV-1 particles and attenuate infection efficiency in vitro. Martins et al. developed a potential microfluidic attenuator containing engineered E. coli to trap Ebola viruses in human blood.186 Along these lines, incorporating engineered probiotics into the vaginal microbiota can inhibit the spread of viruses.
Recently, COVID-19 (SARS-CoV-2) pandemic became a serious concern for public health safety,187,188 urgently requiring an efficient vaccine. Chen et al. engineered yeast to stably express the SARS-CoV-2 receptor binding domain (RBD) without impacting protein structure and functionality, which can be a promising candidate vaccine for COVID-19.189 Furthermore, the yeast expressed RBD combined with the 3M-052-alum provoked powerful immune response against SARS-CoV-2 in rhesus monkeys.190 Apart from recombinant proteins, nucleic acid-based (such as messenger RNA) vaccines are potential candidates against SARS-CoV-2. mRNA vaccines can transform host cells into “protein factories”, providing in situ the protein for a balanced immune response for both cellular and humoral immunity. In 2021, clinical trials demonstrated that 2 doses of mRNA vaccine BNT162b2 successfully induced a persistent germinal center B-cell response and robust humoral immunity. This made BNT162b2 to be the first licensed mRNA vaccine against COVID-19 by FDA for people over 16 years of age. Although a considerable number of mRNA vaccines have recently been developed, including PTX-COVID-19-B, SYS6006, and mRNA-LNP, they have several limitations including allergic reactions, uncertain immunity length, and difficult storage.191 Notably, SB tools also can help predict and identify the emerging SARS-CoV-2 variants. Based on reverse genetics, Thao et al. developed a rapid and robust yeast-based synthetic genomics platform for the genetic reconstruction of SARS-CoV-2, which can help generate and functionally characterize evolving viral variants in real-time.192 Scientists have used yeast screening to identify antibody escape mutants of SARS-CoV-2 S RBD to provide new therapeutic strategies against SARS-CoV-2. Importantly, viruses are evolutionarily autonomous; the emergence of new mutations led to the rapid global evolution of influenza viruses.193,194 Accordingly, the virus evolutionary genetic algorithm should be applied for the development of next-generation SB tools.
Engineered cell-based therapeutics for imbalanced flora-related diseases
The human body is inhabited by a multitude of microbial communities, which co-evolve with human hosts and are essential for their health.195–197 It is now well-established that human microbiome disturbance can lead to many chronic diseases, such as colorectal cancer (CRC), autism, and diabetes mellitus (DM).72,198–201 Microbial population modulation is a new treatment strategy for intestinal and neurodegenerative diseases (Fig. 5).105
Fig. 5.
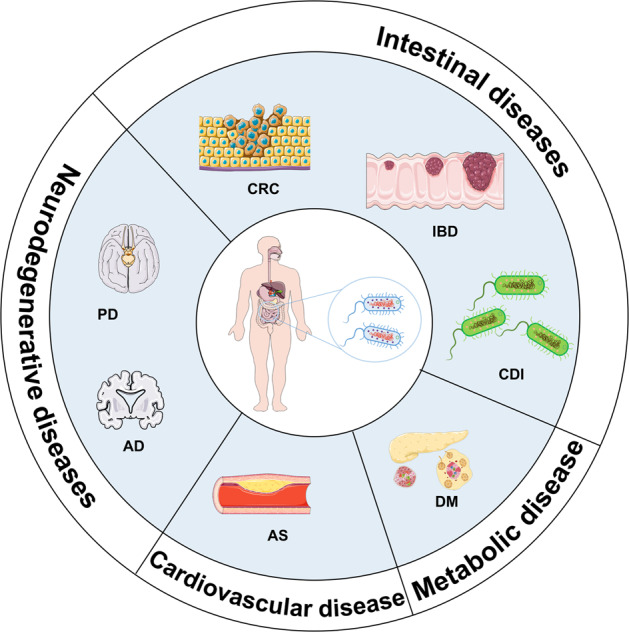
Microbial modulation for disease treatment. AD Alzheimer’s disease, PD Parkinson’s disease, CRC colorectal cancer, DM diabetes mellitus, CDI Clostridium difficile infection, IBD inflammatory bowel disease, AS atherosclerosis
Intestinal diseases
Clostridium difficile infection (CDI), which causes gut dysbiosis, is a major cause of morbidity and death worldwide.202,203 Supplementation with Firmicutes, which convert primary bile acids to secondary bile acids, can significantly lower the growth and virulence of C. difficile by competitively colonizing the infected gut.204 Microbiome product Seres SER-262, which was isolated from healthy donors, could eradicate C. difficile infection. Altered composition, function, and metabolites of the intestinal flora have been associated with the progression of inflammatory bowel disease (IBD).205 Traditional IBD treatment is limited by side effects and complications. Lee et al. developed a hyaluronic acid-bilirubin nanodrug (HABN) which after accumulation restored the epithelial barrier alleviating inflamed colonic epithelia by restoring gut microbiota balance.206 Whelan et al. engineered the probiotic bacterium E.coli Nissle 1917 for the treatment of IBD, which secretes nematode immunomodulator cystine in the intestine.207 This modified probiotic significantly inhibited gut inflammation in murine acute colitis and can potentially treat other similar diseases. A Lactobacillus probiotic was engineered to produce anti-inflammatory molecules, in particular IL-10, in response to murine colitis.208 An engineered E. coli, named EcNL4, improved intestinal microbiota and thereby colitis in mice through the sustainable release of (R)-3-hydroxybutyric acid (3HB).209 An E. coli strain, named SYNB1618, expressing the phenylalanine ammonia-lyase and L-amino acid deaminase genes was engineered to consume phenylalanine in the gut. Overall, the above studies indicated that engineered bacteria can be potentially used to treat rare metabolic diseases.
Neurodegenerative and other diseases
Neurodegenerative diseases such as Parkinson’s disease (PD), Alzheimer’s disease (AD), and amyotrophic lateral sclerosis (ALS) have similar features.210 Emerging evidence closely associates gut dysbiosis with the development of neurodegenerative diseases through the gut–microbiota–brain axis.211,212 Wu et al. showed that Enterobacteria infection strongly exacerbated neurodegeneration by promoting the recruitment of immune hemocytes to the brain.213 Being functionally related to the brain immune system, the intestine has been termed the “second brain”. A study showed that oral administration of engineered MG136-pMG36e-GLP-1 strain derived from Lactococcus lactis MG1363 continuously produced Glucagon-like peptide-1 (GLP-1), which dramatically reduced lipopolysaccharide (LPS)-induced memory impairment and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP)-induced motor dysfunction, and the abundance of Enterococcus and Proteus pathogens in mice.214,215 This strain could be a novel intervention in AD and PD.214–221 Notably, probiotics can compete with pathogenic bacteria for nutrients and binding sites and secrete/produce bacterio-toxins which repress the invasion and adhesion of pathogens.218 Oral bacteriotherapy with SLAB51 probiotic and p62 (SQSTM1)-engineered lactic acid bacteria inhibited the progression of AD, indicating the positive effect of the probiotic intervention in neuronal diseases.219 So far, Lactobacillus and Bifidobacterium strains are the most promising candidates for improving cognitive functioning by reducing the levels of oxidative and inflammatory biomarkers in AD.220
Engineered commensal bacteria also have significant potential in other chronic diseases, i.e., DM and Atherosclerosis (AS). Duan et al. engineered Lactobacillus gasseri ATCC 33323 to secrete GLP-1 (1–37) to stimulate the conversion of intestinal epithelial cells into insulin-secreting cells in a diabetic rat model. They showed that rats fed daily with engineered bacteria developed insulin-producing cells in the upper intestine, sufficient enough to ameliorate hyperglycemia.214 A recent study showed that an engineered strain MG1363-pMG36e-GLP-1 (M-GLP-1) dramatically decreased body weight and improved glucose intolerance in high-fat diet (HFD) obese mice.215 May-Zhang et al. developed a novel E. coli Nissle 1917 strain expressing N-acyl phosphatidylethanolamines, which are precursors of endogenous lipid satiety factor N-acylethanolamides; the strain significantly reduced the levels of adiposity, hepatic triglycerides, and fatty acid synthesis lowering serum cholesterol levels and extent of necrosis in atherosclerotic mice.217
Similar to chemical drugs or cell-based therapies, the safety, tolerability, and efficacy of engineered therapeutic microbes for chronic disease therapies have attracted widespread attention.221 The long-term persistence, potential sensitization, and influence of engineered probiotics on native gut microbiota are yet to be completely comprehended. With the advances in SB tools and massive clinical trial data, this field is anticipated to develop therapies for many chronic diseases in the future.
Synthetic cells in drug discovery and development
Microorganisms, plants, marine resources, insects, and even mammals have always been important sources of pharmaceuticals/natural compounds. In nature, such compounds exist in low concentrations and are often difficult to synthesize due to their structural complexity. Advances in SB allow the production of bioactive compounds in non-natural hosts, which has revolutionized the pharmaceutical industry. Using computational and experimental tools, SB can help design chassis cells as novel drug screening platforms or as biofactories for the production of difficult bioactive compounds (Fig. 6).
Fig. 6.
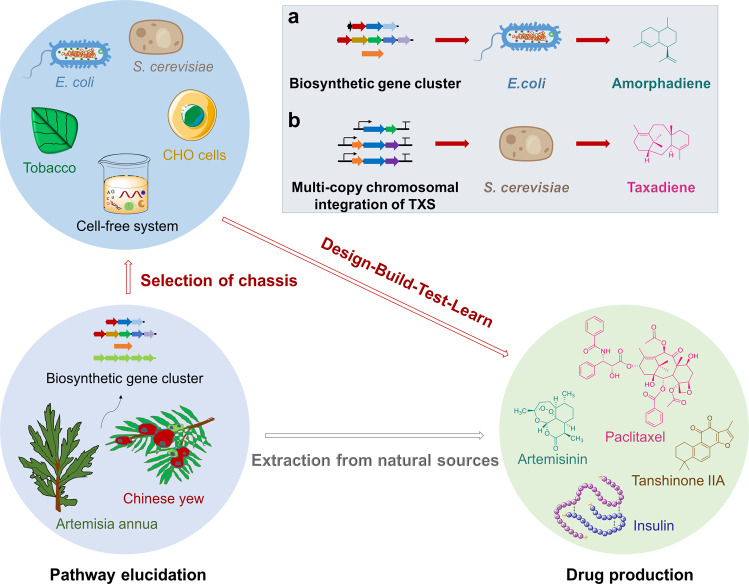
Synthetic biology facilitates drug production in diverse chassis systems. Outlined scheme of the typical pipeline of drug development consisting of pathway elucidation, chassis selection and drug production. Gray arrow indicates extraction of bioactive compounds from natural sources, red arrows indicate general synthetic biology pathways. a Synthetic biology production of amorphadiene in E. coli chassis. b Synthetic biology production of taxadiene in S. cerevisiae chassis
SB in drug discovery
E. coli is the best-studied prokaryotic model organism with properties of a well-defined genetic background, rapid reproduction, low cost, and high levels of foreign protein expression. Therefore, it serves as an important component of high-throughput drug screening platforms. For example, multidrug-resistant E. coli strains are utilized to identify novel antimicrobial compounds.222 Male et al. developed a high-throughput gene coding screening platform to screen a SICLOPPS library of cyclic hexapeptides in E. coli that could inhibit the protein interaction between the human CMG2 receptor and the Bacillus anthracis protective antigen (PA).223 Van der Donk and co-workers generated a bicyclic peptide library in E. coli by combining the generation of a lanthipeptide library with a bacterial reverse two-hybrid peptide library (RTHS), and identified interaction inhibitors for the HIV p6 protein and UEV domain of the human TSG101 protein.224
E. coli lack the regulation mechanisms of eukaryotic gene expression and therefore is not suitable for screening bioactive compounds against eukaryotic targets.225 Saccharomyces cerevisiae is a commonly used eukaryotic model organism for the expression of active eukaryotic proteins undergoing post-translational modifications, including disulfide bond formation and glycosylation. In addition, S. cerevisiae can also tolerate a wide range of pH and high osmotic pressure, which makes it a robust organism in practice.226 S. cerevisiae strains, such as a yeast strain expressing human PARP1, have been used to screen inhibitors of enzymes involved in cancer progression.227 A yeast-based chemogenomic platform was used to identify the interaction between the chemotherapeutic agent methotrexate and the protein Dfr1 using haploinsufficiency profiling.228
The complexity of mammalian cells limits their engineering as host cells. However, recent developments in SB are changing the situation. For example, CRISPR-Cas9 technology was applied to create a high-throughput platform of primary human CD4+ T cells for the functional analysis of host factors in HIV infection and pathogenesis.229 Mammalian cells, such as Chinese hamster ovary (CHO) cell lines, are used for primary screening and production of complicated biopharmaceuticals. We will now discuss gene circuits based on mammalian cells. The mammalian cells have a high cost of production and also suffer from the risk of contamination from human-specific pathogens.
Synthetic gene circuits offer a huge opportunity for advanced drug discovery. Antimicrobial resistance (AMR) is emerging as a serious health threat, which can possibly be addressed by synthetic biology. Synthetic mammalian gene circuits can be used for the screening of novel antimicrobial compounds. Streptogramin-inducible (PipON) and streptogramin-repressible (PipOFF) mammalian gene-regulation systems were constructed based on the pristmamycin-responsive interaction between pristinamycin-induced protein (Pip) and its ptr promoter (Pptr) target sequence.230 The Streptomyces coelicolor Pip was adapted to modulate reporter gene expression (SEAP, secreted alkaline) in Chinese Hamster Ovary (CHO) cells in response to streptogramin antibiotics. This screening allowed the identification of new bioavailable and more sensitive antibiotics.231 Another example is the discovery of new anti-tuberculosis drugs. Ethylthionamide repressor (EthR)-based genetic circuit screening was used to identify 2-phenylethyl-butyrate, an anti-tuberculosis drug candidate against multidrug-resistant M. tuberculosis.232,233
SB in natural drug production
Natural products are a valuable source of pharmaceuticals; however, they are difficult to synthesize and exist in low amounts in natural hosts. Rapid advances in SB now allow the production of high-value natural medicines and precursor compounds (Table 4).
Table 4.
Advances in the research of high-value natural drugs and precursor compounds
| Target drugs | Host | Products | Achievements |
|---|---|---|---|
| Artemisinin | E. coli | Amorphadiene | First production of artemisinin precursors in a microbial system239 |
| S. cerevisiae | Artemisinic acid | The synthesis of artemisinic acid in S. cerevisiae was achieved240 | |
| N. tabacum and P. patens | Artemisinin | Production of artemisinin in model plants242,243 | |
| Paclitaxel | E. coli | Taxadiene | The first report of taxadiene production in E. coli250 |
| E. coli | Taxadiene | Taxadiene accumulation to 1 g/L.251 | |
| S. cerevisiae | Taxadiene | Taxadiene accumulation to 129 ± 15 mg/L in S. cerevisiae252 | |
| E. coli and S. cerevisiae | Oxygenated taxanes | Combining the strengths of E. coli and S. cerevisiae produced 33 mg/L oxygenated taxanes254 | |
| N. benthamiana | Taxadiene and taxadiene-5α-ol | The first heterologous synthesis of taxadiene-5α-ol in a plant chassis255 | |
| Tanshinones | S. cerevisiae | Miltiradiene | A high yield of miltiradiene (365 mg/L) was obtained in yeast257 |
| S. cerevisiae | Miltiradiene | A high yield of miltiradiene (3.5 g/L) was obtained in bioreactor.258 | |
| Hairy roots | Tanshinone | Enhancement of tanshinone content and antioxidant activity by hairy root metabolic engineering for the first time259 | |
| Opioids | S. cerevisiae | Thebaine and hydrocodone | Total biosynthesis of opioids in yeast260 |
| Cannabinoids | S. cerevisiae | Cannabidiolic acid etc. | Total biosynthesis of cannabinoids in yeast261 |
| Insulin | E. coli | Insulin A and B chains | Insulin was first produced in E. coli263 |
| S. cerevisiae | Proinsulin | S. cerevisiae-based expression system is efficient and secret human insulin265 | |
| Recombinant protein vaccines | E. coli | Trumenba® | Recombinant protein vaccine involving E. coli expression system270 |
| S. cerevisiae | Gardasil | Recombinant protein vaccine involving yeasts expression system271 | |
| CHO | ZF2001 | Recombinant protein vaccine involving mammalians cell expression system272 | |
| Sf9 insect cells | Recombinant COVID-19 vaccine | Recombinant protein vaccine involving Insect cells expression system273 | |
| AMPs | E. coli | GKY20 | High expression level in E. coli by inclusion body formation275 |
| E. coli | Melittin etc. | Expression in E. coli using CaM as a carrier protein276 | |
| P. pastoris | Apidaecin | Super heterologous expression and secretion of apidaecin in yeast278 | |
| Valinomycin | Cell-free system | Valinomycin | The first NRP that was fully biosynthesized in vitro using an cell-free system282 |
| Nisin | Cell-free system | Nisin | Cell-free biosynthesis of nisin was achieved by a CFPS platform284 |
Semi-synthetic artemisinin is a successful case of SB-based synthesis. Artemisinin, a highly effective antimalarial drug, is a sesquiterpene lactone, which is naturally obtained from Artemisia annua (Asteraceae).234 Artemisinin and its derivatives also have anticancer,235 anti-inflammation,236 antiviral,237 and anti-SARS-CoV-2 activity.238 Initially, Martin et al. attempted the production of artemisinin precursors in microbial systems; a mevalonate pathway from S. cerevisiae and a synthetic amorpha-4,11-diene synthase (ADS) were heterologously expressed in E. coli, producing amorphadiene (Fig. 6a).239 Synthesis of artemisinic acid in S. cerevisiae was achieved using an engineered mevalonate pathway, amorphadiene synthase, and a novel CYP450 from the eukaryotic origin, all of which could not be perfectly expressed in E. coli; the synthesized artemisinic acid was transported out of the engineered yeast simplifying purification.240 A complete biosynthetic pathway, including a plant dehydrogenase and a second cytochrome, allowed the production of artemisinic acid in engineered S. cerevisiae at a fermentation titer of 25 g/L; singlet oxygen was used to convert artemisinic acid to artemisinin.241 Artemisinin was also produced in two model plants, tobacco (Nicotiana benthamiana) and moss (Physcomitrella patens).242,243
Paclitaxel (taxol), a diterpenoid secondary metabolite derived from the stem bark of Chinese yew (Taxus brevifolia),244 is widely used to treat breast,245 ovarian,246 pancreatic,247 lung,248 and prostate cancers.249 T. brevifolia resources are scarce and have paclitaxel in very low amounts. The chemical synthesis of paclitaxel is complex and costly. Therefore, the industrialized production of paclitaxel by attempted using the tools of SB. Taxadiene, an important intermediate in the paclitaxel biosynthesis pathway, was obtained for the first time by overexpressing DXP synthase, IPP isomerase, GGPP synthase, and taxane synthase genes in E. coli at a titer close to 0.5 mg/L.250 Multiple module metabolic engineering (MMME) in E. coli was used to divide the paclitaxel biosynthetic pathway into an upstream module (containing dxs, idi, ispD, ispF genes) for IPP and DMAPP synthesis and a downstream module (containing GGPPS, TXS genes) for the production of GGPP and taxadiene. The conditions for the optimal balance of the two pathway modules were determined by a systematic multivariate search, resulting in a taxadiene yield of ~1 g/L, which is the highest yield reported so far.251 Since several CYP450s (membrane-bound proteins) are involved in the biosynthesis of paclitaxel, eukaryotic yeasts are more suitable for the heterologous production of paclitaxel and its intermediates. In an S. cerevisiae system, multi-copy chromosomal integration of taxadiene synthase (TXS) harboring fusion solubility tags improved taxadiene titers (Fig. 6b). The effects of the selected promoter, Mg2+ concentration, TXS truncation length, chromosomal gene copy, and cultivation temperature were evaluated; a maximum taxadiene titer of 129 ± 15 mg/L was achieved after strain optimization.252 Recently, a statistical definitive screening design approach combined with the state-of-the-art high-throughput micro-bioreactors resulted in a taxane titer of 229 mg/L.253 A co-cultivation method for the production of oxygenated taxanes was established in a synthetic consortium combining the strengths of E. coli and S. cerevisiae, which produced 33 mg/L of oxygenated taxanes.254 Taxadiene synthase, taxadiene-5α-hydroxylase, and CYP450 reductase were introduced in plant N. benthamian to achieve high-level production of taxadiene and taxadiene-5α-ol using a chloroplastic compartmentalized metabolic engineering strategy combined with enhanced isoprenoid precursors.255 Although many excellent attempts have been made, more efforts are needed to optimize the complete biosynthetic pathway of paclitaxel for its industrial production.
Tanshinones, including tanshinone I, tanshinone IIA, tanshinone IIB, dihydrotanshnone I, and cryptotanshinone, are bioactive diterpenoid compounds produced from the roots of Salvia miltiorrhiza Bunge (Danshen in Chinese), which have a wide range of pharmacological activities including antibacterial, antioxidant, anti-inflammatory, cardiovascular protective, and antineoplastic properties.256 Modularized pathway engineering strategies were applied to create SmCPS and SmKSL fusion proteins with adjacent active sites and fused BTS1 and ERG20 to produce a high yield of miltiradiene (365 mg/L) in S. cerevisiae.257 By increasing the carbon flux toward GGPP and optimizing the gene module of miltiradiene synthases, the production of miltiradiene was further improved in S. cerevisiae, reaching 3.5 g/L in a 5-L bioreactor.258 Hairy roots are also an attractive system for drug production. Three genes, HMGR, DXS, and GGPPS, involved in the biosynthesis pathway for tanshinone were introduced in Salvia miltiorrhiza hairy root for a high-yield production of tanshinone with higher antioxidant activity.259
There have been several efforts for the heterologous production of small-molecule natural drugs based on SB approaches. The full biosynthesis of the opioid compounds thebaine and hydrocodone starting from sugar was engineered in yeast.260 The complete biosynthesis of the major cannabinoids, cannabigerolic acid, Δ9-tetrahydrocannabinolic acid, cannabidiolic acid, Δ9-tetrahydrocannabivarinic acid, and cannabidivarinic acid, starting from the simple sugar galactose was accomplished in S. cerevisiae.261
SB in recombinant protein and nucleic acid production
Protein drugs mainly include recombinant protein drugs, monoclonal antibodies, and recombinant protein vaccines. Compared with small-molecule drugs, protein drugs have high activity, strong specificity, low toxicity, clear biological function, and favorable clinical application.262 In this section, recent examples of recombinant protein and nucleic acid production will be presented; an overview of typical examples is presented in Table 4.
Recombinant human insulin was the first recombinant protein drug. It was produced by the expression of chemically synthesized cDNAs encoding for the insulin A and B chains separately in E. coli and the disulfide bonds were formed under optimal conditions to obtain bioactive insulin.263 A new and more efficient method for human insulin production was developed using a PCR-based cloning strategy that optimized human insulin expression in E. coli using the pET21b expression vector. It eliminated the use of affinity tags, tedious insulin renaturation, inclusion body recovery steps, and the expensive enzymatic cleavage of C-peptide.264 An insulin secretion expression system in S. cerevisiae has been developed, which expresses a cDNA encoding for a proinsulin-like molecule. ThreonineB30 was deleted for the fusion of proinsulin molecule with the S. cerevisiae α-factor prepro-peptide. This was later replaced by human proinsulin C-peptide with a small C-peptide. The purified proinsulin is subsequently converted into human insulin by tryptic transpeptidation.265 Recombinant human insulin has also been successfully produced in transgenic animals and plants, such as mice,266 Arabidopsis thaliana,267 and cucumber.268
The production of monoclonal antibodies (mAbs) has taken tremendous attention in the pharmaceutical industry. A modular synthetic biology approach is proposed to rationally engineer E. coli according to three functional modules to facilitate high-titer production of immunoglobulin G (IgG).269 Bacteria, yeast, insect cells, and mammalian cells are used to express recombinant protein vaccines. Trumenba®, produced in E. coli, was developed by Pfizer and uses two variants of the meningococcal factor H-binding protein as antigens.270 Quadrivalent HPV vaccine, Gardasil, is the first commercially available FDA-licensed HPV vaccine; the L1 gene of four human papillomaviruses is expressed in S. cerevisiae and is used with an aluminum adjuvant.271 Most current COVID-19 recombinant protein vaccines are expressed in mammalian cell culture-based expression systems. ZF2001, consisting of tandemly repeated RBD-dimers and aluminum adjuvants, and the RBD-sc-dimers of MERS-CoV and SARS-CoV-2 both were produced at a high yield in an industry-standard CHO cell system.272 The West China Hospital of Sichuan University has developed a recombinant COVID-19 vaccine using baculovirus as a vector; the RBD region of the SARS-CoV-2 spike protein receptor binding domain is expressed in Sf9 insect cells, which showed effective immunogenicity in phase I and phase II clinical trials.273 According to WHO, there were 54 recombinant COVID-19 protein vaccine candidates in the clinical phase as of June 17, 2022.
Peptide natural products exhibit diverse biological activities, however, the toxicity, limited bioavailability, and difficult mass production hinder their development as therapeutic agents. Natural peptides can be biologically synthesized with the help of SB,274 including recombinant production in bacteria, fungi, and cell-free systems. One way to reduce host toxicity is to produce peptides in E. coli as inclusion bodies. The antimicrobial short peptide (AMP) GKY20 derived from the C-terminus of human thrombin was fused with the C-terminus of Onconase for very high expression as inclusion bodies.275 Fusion proteins also have reduced toxicity for the heterologous expression host. Calmodulin (CaM), a common calcium-sensitive protein in eukaryotes, consists of two independent target binding domains with malleable methionine-rich interaction surfaces that can accommodate many basic and hydrophobic residues.276 CaM can be used as a universal carrier protein to express various types of AMPs in E. coli, such as melittin, fowlicidin-1, tritrpticin, indolicidin, puroindoline A peptide, and human β-defensin 3 (HBD-3). This strategy effectively masks the toxic activity of many AMPs and protects them from degradation during expression and purification.276 Recently, E. coli MSI001, a lethal strain, was isolated from mice; the strain was used for screening bacteria-binding heptapeptides based on an integrative bioinformatics approach, phage display technology, and high-throughput sequencing.277 Yeast is another promising cell factory for the production of peptides. Apidaecins, a series of small and proline-rich antimicrobial peptides act as a defense system against many drug-resistant bacteria. Apidaecin expression in Pichia pastoris was improved after the selection of a mutant strain that limits the loss of the integrated plasmid after induction.278 Additionally, P. pastoris has been used to produce AMPs such as cecropin279 and defensins.280 Cell-free systems provide another solution for the production of natural peptides. Valinomycin is a non-ribosomal peptide with antibacterial, antiviral, and antitumor activities.281 A highly efficient cell-free platform was developed for the rapid, in vitro total biosynthesis of valinomycin with a yield close to 30 mg/L; the cell-free protein synthesis system (CFPS) was coupled with cell-free metabolic engineering system (CFME) by mixing two enzyme-enriched cell lysates to perform the two-stage biosynthesis.282 Other natural peptide products, RiPPs, were also synthesized in cell-free systems. Nisin, a lantipeptide has antibacterial activity and is widely used in the food industry. The cell-free biosynthesis of nisin was achieved by the complete reconstitution of its pathway in a CFPS. The same system can also be used for the genome mining of nisin analogs, screening of mutants, and guiding the overproduction of lanthipeptides in vivo.283 In addition, four different lasso peptides were synthesized using cell-free systems, including known examples such as burhizin, capistruin, and fusilassin and predicted lariat peptides such as ellulassin from Thermobifida halotolerans. This system can rapidly generate and characterize novel lasso peptide variants.284
Nucleic acid vaccines are based on either DNA or mRNA and are amenable to SB design. The target nucleic acid is inserted into human cells, which then produce copies of viral protein. Most of the nucleic acid vaccines against SARS-CoV-2 encode the virus’s spike protein.285 INO-4800, a DNA vaccine candidate targeting the SARS-CoV-2 spike antigen,286 is currently in phase 3 trials. BNT162b2 and mRNR-1273 are the approved mRNA vaccines, both of these LNP-formulated nucleoside-modified mRNA vaccines encode PS2 spike protein of SARS-CoV-2.287 There are 16 DNA-based and 37 RNA-based vaccine candidates of SARS-CoV-2 in the clinical phase, together accounting for about 32% of the total vaccine candidates.
With the advancement in SB, bacteria can also be used as active drugs. An engineered E. coli (EcNL4) was shown to improve gut microbiome and ameliorate colitis in mice.209 A combination therapy of low-dose anti-CD3 with a clinical-grade self-containing Lactococcus lactis (appropriate for human DM application, secreting human proinsulin and interleukin-10) cured 66% of mice with new-onset diabetes.288 A Salmonella typhimurium strain was engineered to secrete Vibrio vulnificus flagellin B (FlaB) in tumor tissues for cancer immunotherapy.289 Virus-based vaccines, such as polio and measles vaccines, are created utilizing an inactivated or weakened pathogen while maintaining the antigenic properties285 Recently, Beijing Sinovac’s inactivated SARS-CoV-2 vaccine has been approved.
Conclusions and prospects
From bacterial-based biosensors and drug development to efficient treatments against pathogens, previously untreatable cancers, and other intractable human pathologies, SB has opened up various possibilities.290 For instance, there are currently seven cancer cell therapies through clinical trials that were approved by the FDA by July 2022.291 Despite these successes, SB-inspired cell engineering in medicine still faces many challenges, such as the lack of available sensors for essential biomarkers. Current microbial diagnostic and theragnostic approaches mostly rely on the existing sensors while numerous proteins with potential applications remain to be fully elucidated.38 Genome editing and manufacturing is a costly and lengthy process. Also, the therapeutic efficacy and technical difficulties limit their clinical applications.292 In addition, precise control of cellular output in engineered cells is very challenging, including the issues of bacteria diffusing to other tissues or being quickly eliminated by the host immune system. CAR-T-cell therapy is hampered by persistent infusion into patients and susceptibility to tumor antigen escape.293 Thus, additional efforts are required to integrate biological, chemical, and mechanical aspects for next-generation cell engineering.
Due to its dynamic multidisciplinary nature comprising different projects, approaches, and definitions, SB is evolving with advancements in biology, pharmacology, materials science, physics, and computational biology.294 The novel synthetic gene circuits combined with “on/off switches”, suicide gene systems, or CRISPR-Cas9 can create more precise control strategies for diagnostics and cell therapeutics. CRISPR-Cas9 significantly eases genome editing at targeted position(s).162 The iCasp9 kill switch has been licensed for more than 20 clinical trials including CAR-T cell and other type cell therapies and may help overcome limitations of currently approved CAR-T-cell therapies.295 In addition, non-biological factors have also been incorporated in SB approaches, such as temperature, light, rays, and nanomaterials. The integration of chemical biotechnology such as nanomaterial-coated bacteria provides tremendous promise for efficient transport, long-term storage, and spatial-temporal control in host.26 Gujrati et al. engineered bacteria to secrete melanin-containing OMVs with high photothermal conversion efficiency for photoacoustic imaging and photothermal therapy of 4T1 breast tumor.296 Moreover, the tools of computational biology such as machine learning can change the approach of SB by improving the search for biological components, optimizing design, and reforming/creating novel elements.
Taken together, recent advances in SB-related cellular diagnostics and therapeutics can significantly transform healthcare paradigms. Currently approved CAR-T therapies and upcoming clinical trials of other engineered cell therapies also inspire the rapid expansion of SB. SB-based biofactories can contribute to the screening and development of new and affordable medicines, including recombinant vaccines. In light of this, SB-inspired cell engineering will enter a new perspective and expand its application for various disease treatments.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 81871615).
Author contributions
RB and HW conceived, supervised, and revised the paper; NZ, YS, and XX participated in different parts of writing and organized figures and formatted the paper; ZZ, CD, and CN collected related literature.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ninglin Zhao, Yingjie Song, Xiangqian Xie.
Contributor Information
Huan Wang, Email: wanghuan@nju.edu.cn.
Rui Bao, Email: baorui@scu.edu.cn.
References
- 1.Cameron, D. E., Bashor, C. J. & Collins, J. J. A brief history of synthetic biology. Nat. Rev. Microbiol.12, 381–390 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Shapira, P., Kwon, S. & Youtie, J. Tracking the emergence of synthetic biology. Scientometrics112, 1439–1469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng, F. & Ellis, T. The second decade of synthetic biology: 2010–2020. Nat. Commun.11, 5174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie, M., Haellman, V. & Fussenegger, M. Synthetic biology-application-oriented cell engineering. Curr. Opin. Biotechnol.40, 139–148 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Schwille, P. & Diez, S. Synthetic biology of minimal systems. Crit. Rev. Biochem. Mol. Biol.44, 223–242 (2009). [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., Krasnogor, N. & Schmidt, M. For the sake of the Bioeconomy: define what a synthetic biology chassis is! N. Biotechnol.60, 44–51 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Polizzi, K. M. What is synthetic biology? Methods Mol. Biol.1073, 3–6 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Purnick, P. E. & Weiss, R. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol.10, 410–422 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y. Y., Galloway, K. E. & Smolke, C. D. Synthetic biology: advancing biological frontiers by building synthetic systems. Genome Biol.13, 240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh, V. Recent advancements in synthetic biology: current status and challenges. Gene535, 1–11 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Khalil, A. S. & Collins, J. J. Synthetic biology: applications come of age. Nat. Rev. Genet.11, 367–379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruder, W. C., Lu, T. & Collins, J. J. Synthetic biology moving into the clinic. Science333, 1248–1252 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Wang, F., Zhao, D. & Qi, L. S. Application of genome engineering in medical synthetic biology. Sheng Wu Gong. Cheng Xue Bao.33, 422–435 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Courbet, A., Renard, E. & Molina, F. Bringing next-generation diagnostics to the clinic through synthetic biology. EMBO Mol. Med.8, 987–991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng, A. A. & Lu, T. K. Synthetic biology: an emerging engineering discipline. Annu. Rev. Biomed. Eng.14, 155–178 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Abil, Z., Xiong, X. & Zhao, H. Synthetic biology for therapeutic applications. Mol. Pharm.12, 322–331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien, T., Doshi, A. & Danino, T. Advances in bacterial cancer therapies using synthetic biology. Curr. Opin. Syst. Biol.5, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bober, J. R., Beisel, C. L. & Nair, N. U. Synthetic biology approaches to engineer probiotics and members of the human microbiota for biomedical applications. Annu. Rev. Biomed. Eng.20, 277–300 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemire, S., Yehl, K. M. & Lu, T. K. Phage-based applications in synthetic biology. Annu. Rev. Virol.5, 453–476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ozdemir, T., Fedorec, A. J. H., Danino, T. & Barnes, C. P. Synthetic biology and engineered live biotherapeutics: toward increasing system complexity. Cell Syst.7, 5–16 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Wu, M. R., Jusiak, B. & Lu, T. K. Engineering advanced cancer therapies with synthetic biology. Nat. Rev. Cancer19, 187–195 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Romanowski, S. & Eustaquio, A. S. Synthetic biology for natural product drug production and engineering. Curr. Opin. Chem. Biol.58, 137–145 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Cubillos-Ruiz, A. et al. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov.20, 941–960 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Lenneman, B. R. et al. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol.68, 151–159 (2021). [DOI] [PubMed] [Google Scholar]
- 25.McNerney, M. P. et al. Theranostic cells: emerging clinical applications of synthetic biology. Nat. Rev. Genet.22, 730–746 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan, X., Letendre, J. H., Collins, J. J. & Wong, W. W. Synthetic biology in the clinic: engineering vaccines, diagnostics, and therapeutics. Cell184, 881–898 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amrofell, M. B., Rottinghaus, A. G. & Moon, T. S. Engineering microbial diagnostics and therapeutics with smart control. Curr. Opin. Biotechnol.66, 11–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slomovic, S., Pardee, K. & Collins, J. J. Synthetic biology devices for in vitro and in vivo diagnostics. Proc. Natl Acad. Sci. USA112, 14429–14435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gui, Q. et al. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors17, 1623 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali, A. A., Altemimi, A. B., Alhelfi, N. & Ibrahim, S. A. Application of biosensors for detection of pathogenic food bacteria: a review. Biosensors10, 58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su, L., Jia, W., Hou, C. & Lei, Y. Microbial biosensors: a review. Biosens. Bioelectron.26, 1788–1799 (2011). [DOI] [PubMed] [Google Scholar]
- 32.van der Meer, J. R. & Belkin, S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat. Rev. Microbiol.8, 511–522 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Liu, L., Bilal, M., Duan, X. & Iqbal, H. M. N. Mitigation of environmental pollution by genetically engineered bacteria—current challenges and future perspectives. Sci. Total Environ.667, 444–454 (2019). [DOI] [PubMed] [Google Scholar]
- 34.Hicks, M., Bachmann, T. T. & Wang, B. Synthetic biology enables programmable cell-based biosensors. Chemphyschem21, 132–144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soleimany, A. P. & Bhatia, S. N. Activity-based diagnostics: an emerging paradigm for disease detection and monitoring. Trends Mol. Med.26, 450–468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei, T. Y. & Cheng, C. M. Synthetic biology-based point-of-care diagnostics for infectious disease. Cell Chem. Biol.23, 1056–1066 (2016). [DOI] [PubMed] [Google Scholar]
- 37.McNerney, M. P. et al. Dynamic and tunable metabolite control for robust minimal-equipment assessment of serum zinc. Nat. Commun.10, 5514 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Courbet, A. et al. Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med.7, 289ra283 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Hooshangi, S. & Bentley, W. E. From unicellular properties to multicellular behavior: bacteria quorum sensing circuitry and applications. Curr. Opin. Biotechnol.19, 550–555 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Kumari, A., Pasini, P. & Daunert, S. Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem.391, 1619–1627 (2008). [DOI] [PubMed] [Google Scholar]
- 41.Capatina, D. et al. Analytical methods for the characterization and diagnosis of infection with Pseudomonas aeruginosa: a critical review. Anal. Chim. Acta1204, 339696 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Li, Y. et al. Advanced detection and sensing strategies of Pseudomonas aeruginosa and quorum sensing biomarkers: a review. Talanta240, 123210 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Stephens, K. & Bentley, W. E. Synthetic biology for manipulating quorum sensing in microbial consortia. Trends Microbiol.28, 633–643 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Mao, N., Cubillos-Ruiz, A., Cameron, D. E. & Collins, J. J. Probiotic strains detect and suppress cholera in mice. Sci. Transl. Med.10, eaao2586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lubkowicz, D. et al. Reprogramming probiotic Lactobacillus reuteri as a biosensor for Staphylococcus aureus derived AIP-I detection. ACS Synth. Biol.7, 1229–1237 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Beier, D. & Gross, R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol.9, 143–152 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Lazar, J. T. & Tabor, J. J. Bacterial two-component systems as sensors for synthetic biology applications. Curr. Opin. Syst. Biol.28, 100398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holowko, M. B., Wang, H., Jayaraman, P. & Poh, C. L. Biosensing Vibrio cholerae with genetically engineered Escherichia coli. ACS Synth. Biol.5, 1275–1283 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Daeffler, K. N. et al. Engineering bacterial thiosulfate and tetrathionate sensors for detecting gut inflammation. Mol. Syst. Biol.13, 923 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riglar, D. T. et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol.35, 653–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woo, S. G. et al. A designed whole-cell biosensor for live diagnosis of gut inflammation through nitrate sensing. Biosens. Bioelectron.168, 112523 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Danino, T. et al. Programmable probiotics for detection of cancer in urine. Sci. Transl. Med.7, 289ra284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mimee, M. et al. An ingestible bacterial-electronic system to monitor gastrointestinal health. Science360, 915–918 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan, C. et al. Measurement of biodegradable substances using the salt-tolerant yeast Arxula adeninivorans for a microbial sensor immobilized with poly(carbamoyl) sulfonate (PCS) Part I: construction and characterization of the microbial sensor. Biosens. Bioelectron.14, 131–138 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Shimomura-Shimizu, M. & Karube, I. Yeast based sensors. Adv. Biochem. Eng. Biotechnol.117, 1–19 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Sun, L., Liao, K. & Wang, D. Effects of magnolol and honokiol on adhesion, yeast-hyphal transition, and formation of biofilm by Candida albicans. PLoS ONE10, e0117695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ostrov, N. et al. A modular yeast biosensor for low-cost point-of-care pathogen detection. Sci. Adv.3, e1603221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobson, K. A. New paradigms in GPCR drug discovery. Biochem. Pharmacol.98, 541–555 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miao, Y. & McCammon, J. A. G-protein coupled receptors: advances in simulation and drug discovery. Curr. Opin. Struct. Biol.41, 83–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smutok, O. V. et al. d-lactate-selective amperometric biosensor based on the cell debris of the recombinant yeast Hansenula polymorpha. Talanta125, 227–232 (2014). [DOI] [PubMed] [Google Scholar]
- 61.Karkovska, M., Smutok, O., Stasyuk, N. & Gonchar, M. L-lactate-selective microbial sensor based on flavocytochrome b2-enriched yeast cells using recombinant and nanotechnology approaches. Talanta144, 1195–1200 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Miller, R. A. et al. “Scentsor”: a whole-cell yeast biosensor with an olfactory reporter for low-cost and equipment-free detection of pharmaceuticals. ACS Sens.5, 3025–3030 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller, R. A. et al. Development of a paper-immobilized yeast biosensor for the detection of physiological concentrations of doxycycline in technology-limited settings. Anal. Methods12, 2123–2132 (2020). [Google Scholar]
- 64.Lienert, F., Lohmueller, J. J., Garg, A. & Silver, P. A. Synthetic biology in mammalian cells: next generation research tools and therapeutics. Nat. Rev. Mol. Cell Biol.15, 95–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auslander, D. et al. A designer cell-based histamine-specific human allergy profiler. Nat. Commun.5, 4408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tastanova, A. et al. Synthetic biology-based cellular biomedical tattoo for detection of hypercalcemia associated with cancer. Sci. Transl. Med.10, eaap8562 (2018). [DOI] [PubMed] [Google Scholar]
- 67.Banerjee, P. & Bhunia, A. K. Mammalian cell-based biosensors for pathogens and toxins. Trends Biotechnol.27, 179–188 (2009). [DOI] [PubMed] [Google Scholar]
- 68.Menge, C. Molecular biology of Escherichia coli shiga toxins’ effects on mammalian cells. Toxins12, 345 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.To, C. Z. & Bhunia, A. K. Three dimensional vero cell-platform for rapid and sensitive screening of shiga-toxin producing Escherichia coli. Front. Microbiol.10, 949 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carlos, D. et al. Histamine modulates mast cell degranulation through an indirect mechanism in a model IgE-mediated reaction. Eur. J. Immunol.36, 1494–1503 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Jiang, D. et al. A novel electrochemical mast cell-based paper biosensor for the rapid detection of milk allergen casein. Biosens. Bioelectron.130, 299–306 (2019). [DOI] [PubMed] [Google Scholar]
- 72.Schofield, D. A., Sharp, N. J. & Westwater, C. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage2, 105–283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lu, T. K., Bowers, J. & Koeris, M. S. Advancing bacteriophage-based microbial diagnostics with synthetic biology. Trends Biotechnol.31, 325–327 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Sharp, N. J., Vandamm, J. P., Molineux, I. J. & Schofield, D. A. Rapid detection of Bacillus anthracis in complex food matrices using phage-mediated bioluminescence. J. Food Prot.78, 963–968 (2015). [DOI] [PubMed] [Google Scholar]
- 75.Sharp, N. J., Molineux, I. J., Page, M. A. & Schofield, D. A. Rapid detection of viable Bacillus anthracis spores in environmental samples by using engineered reporter phages. Appl. Environ. Microbiol.82, 2380–2387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peng, Y. et al. Application of the VPp1 bacteriophage combined with a coupled enzyme system in the rapid detection of Vibrio parahaemolyticus. J. Microbiol. Methods98, 99–104 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Kim, S., Kim, M. & Ryu, S. Development of an engineered bioluminescent reporter phage for the sensitive detection of viable Salmonella typhimurium. Anal. Chem.86, 5858–5864 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Oosterik, L. H. et al. Bioluminescent avian pathogenic Escherichia coli for monitoring colibacillosis in experimentally infected chickens. Vet. J.216, 87–92 (2016). [DOI] [PubMed] [Google Scholar]
- 79.Kim, J., Kim, M., Kim, S. & Ryu, S. Sensitive detection of viable Escherichia coli O157:H7 from foods using a luciferase-reporter phage phiV10lux. Int. J. Food Microbiol.254, 11–17 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Rees, J. C., Pierce, C. L., Schieltz, D. M. & Barr, J. R. Simultaneous identification and susceptibility determination to multiple antibiotics of Staphylococcus aureus by bacteriophage amplification detection combined with mass spectrometry. Anal. Chem.87, 6769–6777 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vandamm, J. P. et al. Rapid detection and simultaneous antibiotic susceptibility analysis of Yersinia pestis directly from clinical specimens by use of reporter phage. J. Clin. Microbiol.52, 2998–3003 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rondon, L. et al. Fluoromycobacteriophages can detect viable Mycobacterium tuberculosis and determine phenotypic rifampicin resistance in 3-5 days from sputum collection. Front. Microbiol.9, 1471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meile, S. et al. Engineered reporter phages for rapid bioluminescence-based detection and differentiation of viable listeria cells. Appl. Environ. Microbiol.86, e00442–00420 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu, J., Chau, Y. & Lee, Y. K. Phage-based electrochemical sensors: a review. Micromachines10, 855 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Niyomdecha, S. et al. Phage-based capacitive biosensor for Salmonella detection. Talanta188, 658–664 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Farooq, U. et al. High-density phage particles immobilization in surface-modified bacterial cellulose for ultra-sensitive and selective electrochemical detection of Staphylococcus aureus. Biosens. Bioelectron.157, 112163 (2020). [DOI] [PubMed] [Google Scholar]
- 87.Shin, J. H. et al. Electrochemical detection of caspase-3 based on a chemically modified M13 phage virus. Bioelectrochemistry145, 108090 (2022). [DOI] [PubMed] [Google Scholar]
- 88.Hu, L. et al. Study on the conduction analysis and blocking intervention scheme of emotional disorders between cancer patients and their families. Comput. Math. Methods Med.2022, 4820090 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Qiu, X., Zhang, K., Zhang, Y. & Sun, L. Benefit finding and related factors of patients with early-stage cancer in China. Int. J. Environ. Res. Public Health19, 4284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cao, W. et al. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin. Med. J.134, 783–791 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu, D.-Y. et al. Drug combination in clinical cancer treatments. Rev. Recent Clin. Trials12, 202–211 (2017). [DOI] [PubMed] [Google Scholar]
- 92.Bell Gorrod, H. et al. A review of survival analysis methods used in NICE technology appraisals of cancer treatments: consistency, limitations, and areas for improvement. Med Decis. Mak.39, 899–909 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Wilt, T. J. et al. Systematic review: comparative effectiveness and harms of treatments for clinically localized prostate cancer. Ann. Intern. Med.148, 435–448 (2008). [DOI] [PubMed] [Google Scholar]
- 94.Marin, J. J. et al. Importance and limitations of chemotherapy among the available treatments for gastrointestinal tumours. Anticancer Agents Med. Chem.9, 162–184 (2009). [DOI] [PubMed] [Google Scholar]
- 95.Kalos, M. & June, C. H. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity39, 49–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choe, J. H., Williams, J. Z. & Lim, W. A. Engineering T cells to treat cancer: the convergence of immuno-oncology and synthetic biology. Annu. Rev. Cancer Biol.4, 121–139 (2020). [Google Scholar]
- 97.Chakravarti, D. & Wong, W. W. Synthetic biology in cell-based cancer immunotherapy. Trends Biotechnol.33, 449–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jain, R. K. & Forbes, N. S. Can engineered bacteria help control cancer? Proc. Natl Acad. Sci. USA98, 14748–14750 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Anderson, J. C., Clarke, E. J., Arkin, A. P. & Voigt, C. A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol.355, 619–627 (2006). [DOI] [PubMed] [Google Scholar]
- 100.Lee, C.-H. Engineering bacteria toward tumor targeting for cancer treatment: current state and perspectives. Appl. Microbiol. Biotechnol.93, 517–523 (2012). [DOI] [PubMed] [Google Scholar]
- 101.Yin, T. et al. Engineering bacteria and bionic bacterial derivatives with nanoparticles for cancer therapy. Small18, 2104643 (2022). [DOI] [PubMed] [Google Scholar]
- 102.Forbes, N. S. Engineering the perfect (bacterial) cancer therapy. Nat. Rev. Cancer10, 785–794 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mengesha, A. et al. Potential and limitations of bacterial-mediated cancer therapy. Front. Biosci.12, 3880–3891 (2007). [DOI] [PubMed] [Google Scholar]
- 104.Weerakkody, L. R. & Witharana, C. The role of bacterial toxins and spores in cancer therapy. Life Sci.235, 116839 (2019). [DOI] [PubMed] [Google Scholar]
- 105.Li, Z. et al. Chemically and biologically engineered bacteria‐based delivery systems for emerging diagnosis and advanced therapy. Adv. Mater.33, 2102580 (2021). [DOI] [PubMed] [Google Scholar]
- 106.Wiemann, B. & Starnes, C. O. Coley’s toxins, tumor necrosis factor and cancer research: a historical perspective. Pharmacol. Ther.64, 529–564 (1994). [DOI] [PubMed] [Google Scholar]
- 107.McCarthy, E. F. The toxins of William B. Coley and the treatment of bone and soft-tissue sarcomas. Iowa Orthop. J.26, 154 (2006). [PMC free article] [PubMed] [Google Scholar]
- 108.Guerra, L., Cortes-Bratti, X., Guidi, R. & Frisan, T. The biology of the cytolethal distending toxins. Toxins3, 172–190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Taieb, F., Nougayrède, J.-P. & Oswald, E. Cycle inhibiting factors (cifs): cyclomodulins that usurp the ubiquitin-dependent degradation pathway of host cells. Toxins3, 356–368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goldufsky, J. et al. Pseudomonas aeruginosa exotoxin T induces potent cytotoxicity against a variety of murine and human cancer cell lines. J. Med. Microbiol.64, 164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Balachandran, P. et al. The ubiquitin ligase Cbl-b limits Pseudomonas aeruginosa exotoxin T-mediated virulence. J. Clin. Investig.117, 419–427 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shafikhani, S. H., Morales, C. & Engel, J. The Pseudomonas aeruginosa type III secreted toxin ExoT is necessary and sufficient to induce apoptosis in epithelial cells. Cell. Microbiol.10, 994–1007 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wood, S. J. et al. Pseudomonas aeruginosa ExoT induces mitochondrial apoptosis in target host cells in a manner that depends on its GTPase-activating protein (GAP) domain activity. J. Biol. Chem.290, 29063–29073 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Veshnyakova, A. et al. On the interaction of Clostridium perfringens enterotoxin with claudins. Toxins2, 1336–1356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kominsky, S. L. et al. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am. J. Pathol.164, 1627–1633 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moghadam, Z. M. et al. Designing and analyzing the structure of DT-STXB fusion protein as an anti-tumor agent: an in silico approach. Iran. J. Pathol.14, 305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang, H. et al. Expression, purification, and characterization of an immunotoxin containing a humanized anti-CD25 single-chain fragment variable antibody fused to a modified truncated Pseudomonas exotoxin A. Protein Expr. Purif.58, 140–147 (2008). [DOI] [PubMed] [Google Scholar]
- 118.Mohseni, Z. et al. Potent in vitro antitumor activity of B-subunit of Shiga toxin conjugated to the diphtheria toxin against breast cancer. Eur. J. Pharmacol.899, 174057 (2021). [DOI] [PubMed] [Google Scholar]
- 119.Patyar, S. et al. Bacteria in cancer therapy: a novel experimental strategy. J. Biomed. Sci.17, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barbé, S., Van Mellaert, L. & Anné, J. The use of clostridial spores for cancer treatment. J. Appl. Microbiol.101, 571–578 (2006). [DOI] [PubMed] [Google Scholar]
- 121.Yaghoubi, A. et al. The use of Clostridium in cancer therapy: a promising way. Rev. Med. Microbiol.33, 121–127 (2022). [Google Scholar]
- 122.Barbé, S. et al. Secretory production of biologically active rat interleukin-2 by Clostridium acetobutylicum DSM792 as a tool for anti-tumor treatment. FEMS Microbiol.246, 67–73 (2005). [DOI] [PubMed] [Google Scholar]
- 123.Nelson, B. H. IL-2, regulatory T cells, and tolerance. J. Immunol.172, 3983–3988 (2004). [DOI] [PubMed] [Google Scholar]
- 124.Dinarello, C. A., Novick, D., Kim, S. & Kaplanski, G. Interleukin-18 and IL-18 binding protein. Front. Immunol.4, 289 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Saltzman, D. A. et al. Attenuated Salmonella typhimurium containing interleukin-2 decreases MC-38 hepatic metastases: a novel anti-tumor agent. Cancer Biother. Radiopharm.11, 145–153 (1996). [DOI] [PubMed] [Google Scholar]
- 126.Loeffler, M., Le’Negrate, G., Krajewska, M. & Reed, J. C. IL-18-producing Salmonella inhibit tumor growth. Cancer Gene Ther.15, 787–794 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ariyarathna, H. et al. Increased programmed death ligand (PD-L1) and cytotoxic T-lymphocyte antigen-4 (CTLA-4) expression is associated with metastasis and poor prognosis in malignant canine mammary gland tumours. Vet. Immunol. Immunopathol.230, 110142 (2020). [DOI] [PubMed] [Google Scholar]
- 128.Zhang, H. et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res.40, 1–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Canale, F. P. et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature598, 662–666 (2021). [DOI] [PubMed] [Google Scholar]
- 130.Gurbatri, C. R. et al. Engineered probiotics for local tumor delivery of checkpoint blockade nanobodies. Sci. Transl. Med.12, eaax0876 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tian, S., Lin, G., Piao, L. & Liu, X. Del-1 enhances therapeutic efficacy of bacterial cancer immunotherapy by blocking recruitment of tumor-infiltrating neutrophils. Clin. Transl. Oncol.24, 244–253 (2022). [DOI] [PubMed] [Google Scholar]
- 132.Hu, Q. et al. Engineering nanoparticle-coated bacteria as oral DNA vaccines for cancer immunotherapy. Nano Lett.15, 2732–2739 (2015). [DOI] [PubMed] [Google Scholar]
- 133.Ni, D. et al. Biomimetically engineered demi‐bacteria potentiate vaccination against cancer. Adv. Sci.4, 1700083 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang, W. et al. Mussel‐inspired polydopamine: the bridge for targeting drug delivery system and synergistic cancer treatment. Macromol. Biosci.20, 2000222 (2020). [DOI] [PubMed] [Google Scholar]
- 135.Song, Q. et al. A probiotic spore‐based oral autonomous nanoparticles generator for cancer therapy. Adv. Mater.31, 1903793 (2019). [DOI] [PubMed] [Google Scholar]
- 136.Zhang, Y. et al. Design of outer membrane vesicles as cancer vaccines: a new toolkit for cancer therapy. Cancers11, 1314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gujrati, V. et al. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano.8, 1525–1537 (2014). [DOI] [PubMed] [Google Scholar]
- 138.Zhang, J. et al. Self-assembly catalase nanocomplex conveyed by bacterial vesicles for oxygenated photodynamic therapy and tumor immunotherapy. Int. J. Nanomed.17, 1971 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Akhoundi, M. et al. CAR T cell therapy as a promising approach in cancer immunotherapy: challenges and opportunities. Cell Oncol.44, 495–523 (2021). [DOI] [PubMed] [Google Scholar]
- 140.Upadhaya, S. et al. The clinical pipeline for cancer cell therapies. Nat. Rev. Drug Discov.20, 503–504 (2021). [DOI] [PubMed] [Google Scholar]
- 141.Wang, Z., Wu, Z., Liu, Y. & Han, W. New development in CAR-T cell therapy. J. Hematol. Oncol.10, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sengsayadeth, S., Savani, B. N., Oluwole, O. & Dholaria, B. Overview of approved CAR‐T therapies, ongoing clinical trials, and its impact on clinical practice. EJHaem3, 6–10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Qi, C. et al. Claudin18.2-specific CAR T cells in gastrointestinal cancers: phase 1 trial interim results. Nat. Med.28, 1189–1198 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang, C., Zhuang, Q., Liu, J. & Liu, X. Synthetic biology in chimeric antigen receptor T (CAR T) cell engineering. ACS Synth. Biol.11, 1–15 (2022). [DOI] [PubMed] [Google Scholar]
- 145.Rafiq, S., Hackett, C. S. & Brentjens, R. J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol.17, 147–167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu, S., Yi, M., Qin, S. & Wu, K. Next generation chimeric antigen receptor T cells: safety strategies to overcome toxicity. Mol. Cancer18, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Andrea, A. E., Chiron, A., Bessoles, S. & Hacein-Bey-Abina, S. Engineering next-generation CAR-T cells for better toxicity management. Int. J. Mol. Sci.21, 8620 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chang, P. S. et al. Manufacturing CD20/CD19-targeted iCasp9 regulatable CAR-TSCM cells using qCART, the Quantum pBac-based CAR-T system. Preprint at https://www.biorxiv.org/content/10.1101/2022.05.03.490475v2 (2022). [DOI] [PMC free article] [PubMed]
- 149.Prinzing, B. et al. MyD88/CD40 signaling retains CAR T cells in a less differentiated state. JCI Insight5, e136093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Moghimi, B. et al. Preclinical assessment of the efficacy and specificity of GD2-B7H3 SynNotch CAR-T in metastatic neuroblastoma. Nat. Commun.12, 1–15 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hegde, M. et al. Tandem CAR T cells targeting HER2 and IL13Rα2 mitigate tumor antigen escape. J. Clin. Investig.126, 3036–3052 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Han, X. et al. Masked chimeric antigen receptor for tumor-specific activation. Mol. Ther.25, 274–284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yeku, O. O. & Brentjens, R. J. Armored CAR T-cells: utilizing cytokines and pro-inflammatory ligands to enhance CAR T-cell anti-tumour efficacy. Biochem. Soc. Trans.44, 412–418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yeku, O. O. et al. Armored CAR T cells enhance antitumor efficacy and overcome the tumor microenvironment. Sci. Rep.7, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chmielewski, M. & Abken, H. CAR T cells transform to trucks: chimeric antigen receptor-redirected T cells engineered to deliver inducible IL-12 modulate the tumour stroma to combat cancer. Cancer Immunol. Immunother.61, 1269–1277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lanitis, E. et al. Optimized gene engineering of murine CAR-T cells reveals the beneficial effects of IL-15 coexpression. J. Exp. Med.218, e20192203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hu, B. et al. Augmentation of antitumor immunity by human and mouse CAR T cells secreting IL-18. Cell Rep.20, 3025–3033 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vincent, R. & Danino, T. CAR-T cells SEAK help from enzymes. Nat. Chem. Biol.18, 122–123 (2022). [DOI] [PubMed] [Google Scholar]
- 159.Durgin, J. S. et al. Enhancing CAR T function with the engineered secretion of C. perfringens neuraminidase. Mol. Ther.30, 1201–1214 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kuhn, N. F. et al. CD40 ligand-modified chimeric antigen receptor T cells enhance antitumor function by eliciting an endogenous antitumor response. Cancer Cell35, 473–488.e476 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Jafarzadeh, L. et al. Targeted knockdown of Tim3 by short hairpin RNAs improves the function of anti-mesothelin CAR T cells. Mol. Immunol.139, 1–9 (2021). [DOI] [PubMed] [Google Scholar]
- 162.Li, C., Mei, H. & Hu, Y. Applications and explorations of CRISPR/Cas9 in CAR T-cell therapy. Brief. Funct. Genomics19, 175–182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lindahl, J. F. & Grace, D. The consequences of human actions on risks for infectious diseases: a review. Infect. Ecol. Epidemiol.5, 30048 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Van Duin, D. & Paterson, D. L. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect. Dis. Clin. North Am.30, 377–390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chaoui, L., Mhand, R., Mellouki, F. & Rhallabi, N. Contamination of the surfaces of a health care environment by multidrug-resistant (MDR) bacteria. Int. J. Microbiol.2019, 3236526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McDermott, J. E. et al. Computational prediction of type III and IV secreted effectors in gram-negative bacteria. Infect. Immun.79, 23–32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pinaud, L., Sansonetti, P. J. & Phalipon, A. Host cell targeting by enteropathogenic bacteria T3SS effectors. Trends Microbiol.26, 266–283 (2018). [DOI] [PubMed] [Google Scholar]
- 168.Sperandio, V. et al. Bacteria–host communication: the language of hormones. Proc. Natl Acad. Sci. USA100, 8951–8956 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jana, B., Keppel, K. & Salomon, D. Engineering a customizable antibacterial T6SS‐based platform in Vibrio natriegens. EMBO Rep.22, e53681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ting, S.-Y. et al. Targeted depletion of bacteria from mixed populations by programmable adhesion with antagonistic competitor cells. Cell Host Microbe28, 313–321.e316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Duan, F. & March, J. C. Engineered bacterial communication prevents Vibrio cholerae virulence in an infant mouse model. Proc. Natl Acad. Sci. USA107, 11260–11264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hwang, I. Y. et al. Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun.8, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Matteau, D. & Rodrigue, S. An engineered Mycoplasma pneumoniae to fight Staphylococcus aureus. Mol. Syst. Biol.17, e10574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jayakumar, J., Kumar, V., Biswas, L. & Biswas, R. Therapeutic applications of lysostaphin against Staphylococcus aureus. J. Appl. Microbiol.131, 1072–1082 (2021). [DOI] [PubMed] [Google Scholar]
- 175.Jana, B., Keppel, K. & Salomon, D. Engineering a customizable antibacterial T6SS-based platform in Vibrio natriegens. EMBO Rep.22, e53681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Torres, M. D. et al. Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano.15, 2143–2164 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fenton, M., McAuliffe, O., O’Mahony, J. & Coffey, A. Recombinant bacteriophage lysins as antibacterials. Bioeng. Bugs1, 9–16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ajuebor, J. et al. Bacteriophage endolysins and their applications. Sci. Prog.99, 183–199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Meile, S. et al. Engineering therapeutic phages for enhanced antibacterial efficacy. Curr. Opin. Virol.52, 182–191 (2022). [DOI] [PubMed] [Google Scholar]
- 180.Masuda, Y. et al. Construction of leaderless-bacteriocin-producing bacteriophage targeting E. coli and neighboring Gram-positive pathogens. Microbiol. Spectr.9, e00141–00121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hsu, B. B., Way, J. C. & Silver, P. A. Stable neutralization of a virulence factor in bacteria using temperate phage in the mammalian gut. Msystems5, e00013–e00020 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cass, J., Barnard, A. & Fairhead, H. Engineered bacteriophage as a delivery vehicle for antibacterial protein, SASP. Pharmaceuticals14, 1038 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Chang, T. L.-Y. et al. Inhibition of HIV infectivity by a natural human isolate of Lactobacillus jensenii engineered to express functional two-domain CD4. Proc. Natl Acad. Sci. USA100, 11672–11677 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rao, S. et al. Toward a live microbial microbicide for HIV: commensal bacteria secreting an HIV fusion inhibitor peptide. Proc. Natl Acad. Sci. USA102, 11993–11998 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wei, W. et al. Blocking HIV-1 infection by chromosomal integrative expression of human CD4 on the surface of Lactobacillus acidophilus ATCC 4356. J. Virol.93, e01830–01818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Martins, D. P. et al. Computational models for trapping ebola virus using engineered bacteria. IEEE/ACM Trans. Comput Biol. Bioinform.15, 2017–2027 (2018). [DOI] [PubMed] [Google Scholar]
- 187.Wang, H. et al. Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020–21. Lancet399, 1513–1536 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ciotti, M. et al. The COVID-19 pandemic. Crit. Rev. Clin. Lab Sci.57, 365–388 (2020). [DOI] [PubMed] [Google Scholar]
- 189.Chen, W. H. et al. Genetic modification to design a stable yeast-expressed recombinant SARS-CoV-2 receptor binding domain as a COVID-19 vaccine candidate. Biochim Biophys. Acta Gen. Subj.1865, 129893 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pino, M. et al. A yeast expressed RBD-based SARS-CoV-2 vaccine formulated with 3M-052-alum adjuvant promotes protective efficacy in non-human primates. Sci. Immunol.6, eabh3634 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Turner, J. S. et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature596, 109–113 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Thi Nhu Thao, T. et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature582, 561–565 (2020). [DOI] [PubMed] [Google Scholar]
- 193.Xue, K. S., Moncla, L. H., Bedford, T. & Bloom, J. D. Within-host evolution of human influenza virus. Trends Microbiol.26, 781–793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Koonin, E. V., Dolja, V. V. & Krupovic, M. The logic of virus evolution. Cell Host Microbe30, 917–929 (2022). [DOI] [PubMed] [Google Scholar]
- 195.Shafquat, A., Joice, R., Simmons, S. L. & Huttenhower, C. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol.22, 261–266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Berrilli, F., Di Cave, D., Cavallero, S. & D’Amelio, S. Interactions between parasites and microbial communities in the human gut. Front. Cell. Infect. Microbiol.2, 141 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Robinson, C. J., Bohannan, B. J. & Young, V. B. From structure to function: the ecology of host-associated microbial communities. Microbiol. Mol. Biol. Rev.74, 453–476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Rosenfeld, C. S. Microbiome disturbances and autism spectrum disorders. Drug Metab. Dispos.43, 1557–1571 (2015). [DOI] [PubMed] [Google Scholar]
- 199.Genua, F. et al. The role of gut barrier dysfunction and microbiome dysbiosis in colorectal cancer development. Front. Oncol.11, 626349 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sharma, S. & Tripathi, P. Gut microbiome and type 2 diabetes: where we are and where to go? J. Nutr. Biochem.63, 101–108 (2019). [DOI] [PubMed] [Google Scholar]
- 201.Sikalidis, A. K. & Maykish, A. The gut microbiome and type 2 diabetes mellitus: discussing a complex relationship. Biomedicines8, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bien, J., Palagani, V. & Bozko, P. The intestinal microbiota dysbiosis and Clostridium difficile infection: is there a relationship with inflammatory bowel disease? Ther. Adv. Gastroenterol.6, 53–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Antharam, V. C. et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol.51, 2884–2892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Engevik, M. A. et al. Human Clostridium difficile infection: altered mucus production and composition. Am. J. Physiol. Gastrointest. Liver Physiol.308, G510–G524 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Loh, G. & Blaut, M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut microbes3, 544–555 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lee, Y. et al. Hyaluronic acid–bilirubin nanomedicine for targeted modulation of dysregulated intestinal barrier, microbiome and immune responses in colitis. Nat. Mater.19, 118–126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Whelan, R. A. et al. A transgenic probiotic secreting a parasite immunomodulator for site-directed treatment of gut inflammation. Mol. Ther.22, 1730–1740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Carroll, I. M. et al. Anti-inflammatory properties of Lactobacillus gasseri expressing manganese superoxide dismutase using the interleukin 10-deficient mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol.293, G729–G738 (2007). [DOI] [PubMed] [Google Scholar]
- 209.Yan, X. et al. Construction of a sustainable 3-hydroxybutyrate-producing probiotic Escherichia coli for treatment of colitis. Cell. Mol. Immunol.18, 2344–2357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Eisen, A. & Calne, D. Amyotrophic lateral sclerosis, Parkinson’s disease and Alzheimer’s disease: phylogenetic disorders of the human neocortex sharing many characteristics. Can. J. Neurol. Sci.19, 117–123 (1992). [PubMed] [Google Scholar]
- 211.Sarkar, S. R. & Banerjee, S. Gut microbiota in neurodegenerative disorders. J. Neuroimmunol.328, 98–104 (2019). [DOI] [PubMed] [Google Scholar]
- 212.Chidambaram, S. B. et al. Gut dysbiosis, defective autophagy and altered immune responses in neurodegenerative diseases: tales of a vicious cycle. Pharmacol. Ther.231, 107988 (2021). [DOI] [PubMed] [Google Scholar]
- 213.Wu, S.-C., Cao, Z.-S., Chang, K.-M. & Juang, J.-L. Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat. Commun.8, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Duan, F. F., Liu, J. H. & March, J. C. Engineered commensal bacteria reprogram intestinal cells into glucose-responsive insulin-secreting cells for the treatment of diabetes. Diabetes64, 1794–1803 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wang, L. et al. Engineered bacteria of MG1363-pMG36e-GLP-1 attenuated obesity-induced by high fat diet in mice. Front. Cell. Infect. Microbiol.11, 595575 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fang, X. et al. Therapeutic effect of GLP-1 engineered strain on mice model of Alzheimer’s disease and Parkinson’s disease. AMB Express10, 80 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.May-Zhang, L. S. et al. Administration of N-acyl-phosphatidylethanolamine expressing bacteria to low density lipoprotein receptor(-/-) mice improves indices of cardiometabolic disease. Sci. Rep.9, 420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Feyisetan, O., Tracey, C. & Hellawell, G. O. Probiotics, dendritic cells and bladder cancer. BJU Int.109, 1594–1597 (2012). [DOI] [PubMed] [Google Scholar]
- 219.Cecarini, V. et al. Neuroprotective effects of p62 (SQSTM1)-engineered lactic acid bacteria in Alzheimer’s disease: a pre-clinical study. Aging12, 15995 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Den, H., Dong, X., Chen, M. & Zou, Z. Efficacy of probiotics on cognition, and biomarkers of inflammation and oxidative stress in adults with Alzheimer’s disease or mild cognitive impairment—a meta-analysis of randomized controlled trials. Aging12, 4010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hwang, I. Y. & Chang, M. W. Engineering commensal bacteria to rewire host-microbiome interactions. Curr. Opin. Biotechnol.62, 116–122 (2020). [DOI] [PubMed] [Google Scholar]
- 222.Sadaka, C., Damborg, P. & Watts, J. L. High-throughput screen identifying the thiosemicarbazone NSC319726 compound as a potent antimicrobial lead against resistant strains of Escherichia coli. Biomolecules8, 166–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Male, A. L. et al. Targeting Bacillus anthracis toxicity with a genetically selected inhibitor of the PA/CMG2 protein-protein interaction. Sci. Rep.7, 3104–3112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yang, X. et al. A lanthipeptide library used to identify a protein-protein interaction inhibitor. Nat. Chem. Biol.14, 375–380 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Li, C. Q. et al. Recent advances in the synthetic biology of natural drugs. Front. Bioeng. Biotechnol.9, 691152 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Baghban, R. et al. Yeast expression systems: overview and recent advances. Mol. Biotechnol.61, 365–384 (2019). [DOI] [PubMed] [Google Scholar]
- 227.Perkins, E. et al. Novel inhibitors of Poly(ADP-ribose) polymerase PARP1 and PARP2 identified using a cell-based screen in yeast. Cancer Res.61, 4175–4183 (2001). [PubMed] [Google Scholar]
- 228.Birrell, G. W. et al. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl Acad. Sci. USA98, 12608–12613 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hultquist, J. F. et al. CRISPR-Cas9 genome engineering of primary CD4(+) T cells for the interrogation of HIV-host factor interactions. Nat. Protoc.14, 1–27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fussenegger, M. et al. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol.18, 1203–1208 (2000). [DOI] [PubMed] [Google Scholar]
- 231.Aubel, D. et al. Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiot.54, 44–55 (2001). [DOI] [PubMed] [Google Scholar]
- 232.Baulard, A. R. et al. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem.275, 28326–28331 (2000). [DOI] [PubMed] [Google Scholar]
- 233.Weber, W. et al. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc. Natl Acad. Sci. USA105, 9994–9998 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tu, Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med.17, 1217–1220 (2011). [DOI] [PubMed] [Google Scholar]
- 235.Li, D., Zhang, J. & Zhao, X. Mechanisms and molecular targets of artemisinin in cancer treatment. Cancer Invest.39, 675–684 (2021). [DOI] [PubMed] [Google Scholar]
- 236.Cheong, D. H. J., Tan, D. W. S., Wong, F. W. S. & Tran, T. Anti-malarial drug, artemisinin and its derivatives for the treatment of respiratory diseases. Pharmacol. Res.158, 104901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Efferth, T. Beyond malaria: the inhibition of viruses by artemisinin-type compounds. Biotechnol. Adv.36, 1730–1737 (2018). [DOI] [PubMed] [Google Scholar]
- 238.Uckun, F. M., Saund, S., Windlass, H. & Trieu, V. Repurposing anti-malaria phytomedicine artemisinin as a COVID-19 drug. Front. Pharmacol.12, 649532 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Martin, V. J. et al. Engineering a mevalonate pathway in E. coli for production of terpenoids. Nat. Biotechnol.21, 796–802 (2003). [DOI] [PubMed] [Google Scholar]
- 240.Ro, D. K. et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature440, 940–943 (2006). [DOI] [PubMed] [Google Scholar]
- 241.Paddon, C. J. et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature496, 528–532 (2013). [DOI] [PubMed] [Google Scholar]
- 242.Farhi, M. et al. Generation of the potent anti-malarial drug artemisinin in tobacco. Nat. Biotechnol.29, 1072–1074 (2011). [DOI] [PubMed] [Google Scholar]
- 243.Khairul Ikram, N. K. B. et al. Stable production of the antimalarial drug artemisinin in the moss Physcomitrella patens. Front. Bioeng. Biotechnol.5, 47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Wani, M. C. et al. Plant antitumor agents. VI. Isolation and structure of taxol, a novel antileukemic and antitumor agent from Taxus brevifolia. J. Am. Chem. Soc.93, 2325–2327 (1971). [DOI] [PubMed] [Google Scholar]
- 245.Abu Samaan, T. M. et al. Paclitaxel’s mechanistic and clinical effects on breast cancer. Biomolecules9, 789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bookman, M. A. Optimal primary therapy of ovarian cancer. Ann. Oncol.27(Suppl 1), i58–i62 (2016). [DOI] [PubMed] [Google Scholar]
- 247.Lemstrova, R., Melichar, B. & Mohelnikova-Duchonova, B. Therapeutic potential of taxanes in the treatment of metastatic pancreatic cancer. Cancer Chemother. Pharmacol.78, 1101–1111 (2016). [DOI] [PubMed] [Google Scholar]
- 248.Joshi, M., Liu, X. & Belani, C. P. Taxanes, past, present, and future impact on non-small cell lung cancer. Anticancer Drugs25, 571–583 (2014). [DOI] [PubMed] [Google Scholar]
- 249.Zhu, M. L. et al. Tubulin-targeting chemotherapy impairs androgen receptor activity in prostate cancer. Cancer Res.70, 7992–8002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Huang, Q., Roessner, C. A., Croteau, R. & Scott, A. I. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorg. Med. Chem.9, 2237–2242 (2001). [DOI] [PubMed] [Google Scholar]
- 251.Ajikumar, P. K. et al. Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science330, 70–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Nowrouzi, B. et al. Enhanced production of taxadiene in Saccharomyces cerevisiae. Microb. Cell Fact.19, 200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Walls, L. E., Martinez, J. L. & Rios-Solis, L. Enhancing Saccharomyces cerevisiae taxane biosynthesis and overcoming nutritional stress-induced pseudohyphal growth. Microorganisms10, 163 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Zhou, K., Qiao, K., Edgar, S. & Stephanopoulos, G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat. Biotechnol.33, 377–383 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Li, J. et al. Chloroplastic metabolic engineering coupled with isoprenoid pool enhancement for committed taxanes biosynthesis in Nicotiana benthamiana. Nat. Commun.10, 4850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Xu, J. et al. Ethnopharmacology, phytochemistry, and pharmacology of Chinese Salvia species: a review. J. Ethnopharmacol.225, 18–30 (2018). [DOI] [PubMed] [Google Scholar]
- 257.Zhou, Y. J. et al. Modular pathway engineering of diterpenoid synthases and the mevalonic acid pathway for miltiradiene production. J. Am. Chem. Soc.134, 3234–3241 (2012). [DOI] [PubMed] [Google Scholar]
- 258.Hu, T. et al. Engineering chimeric diterpene synthases and isoprenoid biosynthetic pathways enables high-level production of miltiradiene in yeast. Metab. Eng.60, 87–96 (2020). [DOI] [PubMed] [Google Scholar]
- 259.Kai, G. et al. Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng.13, 319–327 (2011). [DOI] [PubMed] [Google Scholar]
- 260.Galanie, S. et al. Complete biosynthesis of opioids in yeast. Science349, 1095–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Luo, X. et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature567, 123–126 (2019). [DOI] [PubMed] [Google Scholar]
- 262.Leader, B., Baca, Q. J. & Golan, D. E. Protein therapeutics a summary and pharmacological classification. Nat. Rev. Drug Discov.7, 21–39 (2008). [DOI] [PubMed] [Google Scholar]
- 263.Chance, R. E. & Frank, B. H. Research, development, production, and safety of biosynthetic human insulin. Diabetes Care16, 133–142 (1993). [DOI] [PubMed] [Google Scholar]
- 264.Govender, K. et al. A novel and more efficient biosynthesis approach for human insulin production in Escherichia coli (E. coli). AMB Express10, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kjeldsen, T. Yeast secretory expression of insulin precursors. Appl. Microbiol. Biotechnol.54, 277–286 (2000). [DOI] [PubMed] [Google Scholar]
- 266.Bucchini, D. et al. Pancreatic expression of human insulin gene in transgenic mice. Proc. Natl Acad. Sci. USA83, 2511–2515 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Nykiforuk, C. L. et al. Transgenic expression and recovery of biologically active recombinant human insulin from Arabidopsis thaliana seeds. Plant Biotechnol. J.4, 77–85 (2006). [DOI] [PubMed] [Google Scholar]
- 268.Abookazemi, K., Jalali, J., Mohebodini, M. & Vaseghi, A. Transfer of human proinsulin gene into Cucumber (Cucumis sativus L.) via agrobacterium method. Genetika49, 717–728 (2017). [Google Scholar]
- 269.Zhang, J. et al. Synthetic sRNA-based engineering of Escherichia coli for enhanced production of full-length immunoglobulin G. J. Biotechnol.15, e1900363 (2020). [DOI] [PubMed] [Google Scholar]
- 270.Shirley, M. & Dhillon, S. Bivalent rLP2086 vaccine (Trumenba(R)): a review in active immunization against invasive meningococcal group B disease in individuals aged 10–25 years. BioDrugs29, 353–361 (2015). [DOI] [PubMed] [Google Scholar]
- 271.Claudia Figueiredo, M. Vaccination against human papillomavirus. Einstein11, 547–549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Dai, L. et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell182, 722–733.e711 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Meng, F. Y. et al. Safety and immunogenicity of a recombinant COVID-19 vaccine (Sf9 cells) in healthy population aged 18 years or older: two single-center, randomised, double-blind, placebo-controlled, phase 1 and phase 2 trials. Signal Transduct. Target Ther.6, 271 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhao, C. et al. Conformational remodeling enhances activity of lanthipeptide zinc-metallopeptidases. Nat. Chem. Biol.18, 724–732 (2022). [DOI] [PubMed] [Google Scholar]
- 275.Pane, K. et al. Rational design of a carrier protein for the production of recombinant toxic peptides in Escherichia coli. PLoS ONE11, e0146552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Ishida, H. et al. Overexpression of antimicrobial, anticancer, and transmembrane peptides in Escherichia coli through a calmodulin-peptide fusion system. J. Am. Chem. Soc.138, 11318–11326 (2016). [DOI] [PubMed] [Google Scholar]
- 277.Zhang, X. et al. Identification of heptapeptides targeting a lethal bacterial strain in septic mice through an integrative approach. Signal Transduct. Target Ther.7, 245 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Chen, X. et al. High-level heterologous production and functional secretion by recombinant Pichia pastoris of the shortest proline-rich antibacterial honeybee peptide Apidaecin. Sci. Rep.7, 14543 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Guo, C. et al. Secretion and activity of antimicrobial peptide cecropin D expressed in Pichia pastoris. Exp. Ther. Med.4, 1063–1068 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wang, A. et al. High level expression and purification of bioactive human alpha-defensin 5 mature peptide in Pichia pastoris. Appl. Microbiol. Biotechnol.84, 877–884 (2009). [DOI] [PubMed] [Google Scholar]
- 281.Huang, S. et al. The nonribosomal peptide valinomycin: from discovery to bioactivity and biosynthesis. Microorganisms9, 780 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhuang, L. et al. Total in vitro biosynthesis of the nonribosomal macrolactone peptide valinomycin. Metab. Eng.60, 37–44 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Liu, R. et al. A cell-free platform based on nisin biosynthesis for discovering novel lanthipeptides and guiding their overproduction in vivo. Adv. Sci.7, 2001616 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Si, Y. et al. Cell-free biosynthesis to evaluate lasso peptide formation and enzyme-substrate tolerance. J. Am. Chem. Soc.143, 5917–5927 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Callaway, E. The race for coronavirus vaccines: a graphical guide. Nature580, 576–577 (2020). [DOI] [PubMed] [Google Scholar]
- 286.Tebas, P. et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: a preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine31, 100689 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Bandaru, R. et al. Clinical progress of therapeutics and vaccines: rising hope against COVID-19 treatment. Process Biochem.118, 154–170 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Takiishi, T. et al. Reversal of diabetes in NOD mice by clinical-grade proinsulin and IL-10-secreting lactococcus lactis in combination with low-dose anti-CD3 depends on the induction of Foxp3-positive T Cells. Diabetes66, 448–459 (2017). [DOI] [PubMed] [Google Scholar]
- 289.Zheng, J. H. et al. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med.9, 1–10 (2017). [DOI] [PubMed] [Google Scholar]
- 290.Tew, D. Synthetic biology and healthcare. Emerg. Top. Life Sci.3, 659–667 (2019). [DOI] [PubMed] [Google Scholar]
- 291.Sun, W., Jiang, Z., Jiang, W. & Yang, R. Universal chimeric antigen receptor T cell therapy—the future of cell therapy: a review providing clinical evidence. Cancer Treat. Res. Commun.33, 100638 (2022). [DOI] [PubMed] [Google Scholar]
- 292.Foss, D. V., Hochstrasser, M. L. & Wilson, R. C. Clinical applications of CRISPR-based genome editing and diagnostics. Transfusion59, 1389–1399 (2019). [DOI] [PubMed] [Google Scholar]
- 293.McNerney, M. P., Watstein, D. M. & Styczynski, M. P. Precision metabolic engineering: the design of responsive, selective, and controllable metabolic systems. Metab. Eng.31, 123–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Santos-Moreno, J. & Schaerli, Y. Using synthetic biology to engineer spatial patterns. Adv. Biosyst.3, e1800280 (2019). [DOI] [PubMed] [Google Scholar]
- 295.Ohta, K., Sakoda, Y. & Tamada, K. Novel technologies for improving the safety and efficacy of CAR-T cell therapy. Int. J. Hematol. (2022). [DOI] [PubMed]
- 296.Gujrati, V. et al. Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun.10, 1114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Low, K. B. et al. Lipid A mutant Salmonella with suppressed virulence and TNFα induction retain tumor-targeting in vivo. Nat. Biotechnol.17, 37–41 (1999). [DOI] [PubMed] [Google Scholar]
- 298.Afkhami-Poostchi, A., Mashreghi, M., Iranshahi, M. & Matin, M. M. Use of a genetically engineered E. coli overexpressing β-glucuronidase accompanied by glycyrrhizic acid, a natural and anti-inflammatory agent, for directed treatment of colon carcinoma in a mouse model. Int. J. Pharm.579, 119159 (2020). [DOI] [PubMed] [Google Scholar]
- 299.Broadway, K. M., Suh, S., Behkam, B. & Scharf, B. E. Optimizing the restored chemotactic behavior of anticancer agent Salmonella enterica serovar Typhimurium VNP20009. J. Biotechnol.251, 76–83 (2017). [DOI] [PubMed] [Google Scholar]
- 300.Janku, F. et al. Abstract CT110: intratumoral injection of SYNB1891, a synthetic biotic designed to activate the innate immune system, demonstrates target engagement in humans including intratumoral STING activation. Cancer Res.81, CT110 (2021). [Google Scholar]
- 301.Schöder, H. et al. Considerations on integrating prostate-specific membrane antigen positron emission tomography imaging into clinical prostate cancer trials by National Clinical Trials Network Cooperative Groups. J. Clin. Oncol.40, 1500–1505 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Roberts, N. J. et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med.6, 249ra111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.McHale, D. et al. P-325 Oral delivery of a single microbial strain, EDP1503, induces anti-tumor responses via gut-mediated activation of both innate and adaptive immunity. Ann. Oncol.31, S195 (2020). [Google Scholar]
- 304.Jackson, C. M., Lim, M. & Drake, C. G. Immunotherapy for brain cancer: recent progress and future promiseimmunotherapy for brain cancer. Clin. Cancer Res.20, 3651–3659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Le, D. T., Dubenksy, T. W. Jr. & Brockstedt, D. G. Clinical development of Listeria monocytogenes-based immunotherapies. Semin. Oncol.39, 311–322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Hanna, G. J., Adkins, D. R., Zolkind, P. & Uppaluri, R. Rationale for neoadjuvant immunotherapy in head and neck squamous cell carcinoma. Oral. Oncol.73, 65–69 (2017). [DOI] [PubMed] [Google Scholar]
- 307.Uzunparmak, B. & Sahin, I. H. Pancreatic cancer microenvironment: a current dilemma. Clin. Transl. Med.8, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Chervin, C. S. & Gajewski, T. Microbiome-based interventions: therapeutic strategies in cancer immunotherapy. Immunooncol. Technol.8, 12–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Morrow, T. Novartis’s Kymriah: harnessing immune system comes with worry about reining in costs. Manag Care26, 28–30 (2017). [PubMed] [Google Scholar]
- 310.Strimvelis, K. & Yescarta, L. Gene therapy’s next installment. Nat. Biotechnol.37, 697 (2019). [DOI] [PubMed] [Google Scholar]
- 311.Springer, G. & Facharztmagazine, R. Tecartus®-chance auf dauerhaftes ansprechen beim mantelzelllymphom. Info. Onkol.24, 76–77 (2021). [Google Scholar]
- 312.Jaklevic, M. C. CAR-T therapy is approved for non-Hodgkin lymphoma. J. Am. Med. Assoc.325, 1032 (2021). [DOI] [PubMed] [Google Scholar]
- 313.CRS, C. R. S., NT, N. T. & Lymphohistiocytosis, H. US Food and Drug Administration approves Bristol Myers Squibb’s and bluebird bio’s Abecma (idecabtagene vicleucel), the first anti-BCMA CAR T cell therapy for relapsed or refractory multiple myeloma. Abecma is a first-in-class BCMA-directed personalized immune cell therapy delivered as a one-time infusion for triple-class exposed patients with multiple survival. 5, 11–12 (2021).
- 314.Borogovac, A. et al. Safety and feasibility of outpatient chimeric antigen receptor (CAR) T-cell therapy: experience from a tertiary care center. Bone Marrow Transplant.57, 1025–1027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]