Abstract
We have demonstrated previously by Western blotting that in naturally sensitized humans, the serum or salivary antibody response to Streptococcus mutans was directed predominantly to a protein antigen with a size of approximately 60-kDa. To identify this immunodominant antigen, specific serum antibodies were eluted from immunoblots and five positive clones with inserts ranging in length from 3 to 8 kb from identical chromosomal loci were obtained by screening a genomic expression library of Streptococcus mutans GS-5. Amino acid sequencing established the identity of this immunodominant antigen, a 60-kDa immunodominant glycoprotein (IDG-60), to be a cell wall-associated general stress protein GSP-781, which was originally predicted to have a molecular mass of approximately 45 kDa based on the derived nucleotide sequence. Discrepancy in the molecular mass was also observed in recombinant his-tagged IDG-60 (rIDG-60) expressed from Escherichia coli. Glycosylation, consisting of sialic acid, mannose galactose, and N-acetylgalactosamine, was detected by lectin binding to IDG-60 in cell wall extracts from S. mutans and rIDG-60 expressed in vivo or translated in vitro. Despite the presence of multiple Asn or Ser or Thr glycosylation sites, IDG-60 was resistant to the effect of N-glycosidase F and multiple O-glycosidase molecules but not to β-galactosidase. Insertional inactivation of the gene encoding IDG-60, sagA, resulted in a retarded growth rate, destabilization of the cell wall, and pleiomorphic cell shape with multifold ingrowth of cell wall. In addition, distinct from the parental GS-5 strain, the isogenic mutant GS-51 was unable to survive the challenge of low pH and high osmotic pressure or high temperature. Expression of the wild-type gene in trans within GS-51 from plasmid pDL277 complemented the growth defect and restored normal cell shape. These results suggested that IDG-60 is essential for maintaining the integrity of the cell wall and the uniformity of cell shape, both of which are indispensable for bacteria survival under stress conditions.
Streptococcus mutans is a primary pathogen of human dental caries in the oral cavity and occasionally causes infective endocarditis in patients with heart valve abnormalities (13, 17). The cell wall-associated proteins in this microorganism play an important role in bacterial adherence for colonization in distinct host compartments. On the other hand, the host immune response against S. mutans infection is induced by specific antibodies, either secretory immunoglobulin A (IgA) present in saliva or serum IgG in circulation, that recognize these bacterial proteins (28). Antibody-mediated protection is achieved through interference with adherence in situ or by enhanced bacterial clearance by phagocytic cells. Therefore, the identification and functional characterization of the cell wall-associated proteins in S. mutans may provide essential information for understanding the virulence mechanism and also for developing strategies for prevention of infection.
By analyzing the profiles of human salivary and serum antibodies to S. mutans antigens, we found several immunodominant antigens from cell surface protein extracts, but one protein with a size of approximately 60 kDa uniformly exhibited the strongest signals in Western blots probed with either salivary IgA or serum IgG from 157 volunteers (6). Predominant antibody responses to S. mutans antigens with sizes of approximately 60 kDa have been documented previously by other laboratories. One S. mutans surface antigen, named natural antigen, with a molecular mass of 60 kDa exhibited the strongest signal in Western blots detected by serum IgG from 20 adults (29). Dominant immunogenicity of this natural antigen was also demonstrated when Macaca fascicularis monkeys were infected with S. mutans (29). More recently, another surface antigen with glucan binding activity, GBP59, was found to be an immunodominant antigen recognized by salivary IgA from a limited number of adults and children (30). Immunization of rats with GBP59 could induce protective immunity against experimental dental caries (31). The lack of genetic information of either the natural antigen or GBP59 made the comparisons of these surface molecules impossible. Therefore, the immunodominant property of surface antigens with sizes of approximately 60 kDa in S. mutans appeared to be an interesting phenomenon in human populations of different origins, but the identity of these proteins is still not clear.
In the present report, we provide genetic and biological evidence to indicate that the immunodominant surface antigen, named IDG-60, is the general stress protein (GSP-781) of S. mutans reported recently by us (8). Interestingly, IDG-60 isolated from either S. mutans or recombinant E. coli undergoes posttranslational modification by glycosylation, which forms structural units intrinsically encoded by IDG-60. Functional characterization suggested that IDG-60 is essential for maintaining the integrity of the cell wall and uniformity of cell shape, which are indispensable for bacteria growing under stress. This is also the first finding of posttranslational modification by glycosylation in S. mutans.
MATERIALS AND METHODS
Bacteria and growth conditions.
S. mutans GS-5 and isogenic mutants of GS-5 were grown and maintained in brain heart infusion broth (BHI; Difco Laboratories, Inc., Detroit, Mich.) supplemented with erythromycin (10 μg/ml) or spectinomycin (100 μg/ml) when needed. Cell wall-associated proteins of S. mutans were prepared as described previously (6). E. coli JM109 was used as the plasmid host, and cultures were grown in Luria-Bertani (LB) medium supplemented with ampicillin (100 μg/ml) and/or agar (2%) as required. E. coli Strain XL1-Blue MRF′ used for the phage library and strain XLOLR used for phagemid recovery were grown and maintained according to the manufacturer's instructions (Stratagene, La Jolla, Calif.). E. coli host strain BL21(DE3) for recombinant His-tagged IDG-60 was grown and maintained according to the manufacturer's instructions (Novagen, Inc., Madison, Wisc.).
Immunological procedures.
One serum sample, no. 156, donated by a healthy young adult was selected for antibody elution and phage expression library screening. This antiserum recognized predominantly an antigen with a size of 60 kDa at a titer of 1:600 by Western blotting. The antibody directed specifically to this 60-kDa antigen was purified by methods developed in this laboratory. In brief, S. mutans cell wall-associated proteins were extracted, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and subsequently transferred electrophorectically to a Hybond-P membrane (Amersham, Buckinghamshire, United Kingdom). A portion of the transferred membrane was cut, and the 60-kDa antigen was detected by Western blotting using no. 156 serum. Regions on the remaining membrane corresponding to this 60-kDa protein were cut out and incubated with no. 156 serum for 30 min at 37°C. The bound antibody was washed phosphate-buffered saline (PBS; pH 7.2), eluted by adding 5 ml of glycine saline buffer (150 mM NaCl, 200 mM glycine [pH 2.8]), and shaken at room temperature for 30 min. The buffer was aspirated and neutralized with Tris buffer (pH 9.8), defined as “purified antibody,” and stored at 4°C. Western blotting confirmed the reactivity and specificity of the eluted antibody. Immunoprecipitation of IDG-60 from S. mutans cell wall extracts or phage lysate by serum or purified antibody was performed with protein A-Sepharose CL-4B following the manufacturer's instructions (Amersham Pharmacia Biotech AB, Uppsala, Sweden).
Genomic library and gene cloning.
Chromosomal DNA from S. mutans GS-5 was partially digested with Sau3AI and size fractionated by gel electrophoresis. Sections in the 4- to 12-kbp-size range were eluted from the gel, ligated into the BamHI-cut arms of lambda ZAP Express (Stratagene), packaged, and amplified according to the manufacturer's instructions. E. coli XL1 Blue cells were infected with the lambda ZAPII phage on L broth agar (Bacto-tryptone [10 g/liter], yeast extract [5 g/liter], NaCl [10 g/liter], maltose [2 g/liter], Bacto-agar [15 g/liter]) at 2,500 to 3,000 PFU/85-mm-diameter plate. Plaques were transferred to nitrocellulose filters (Millipore Corp., Bedford, Mass.) and incubated with 10 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 37°C for 4 h. These filters were blocked overnight at 4°C with bovine serum albumin (BSA; Sigma Chemical Co., St Louis, Mo.) and screened for positive reactions with the purified antibody detected by alkaline phosphatase-conjugated anti-human IgG (Sigma) with nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) (Stratagene). Positive plaques were transferred to 15-ml tubes containing 200 μl of SM buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.5], 10 mM MgSO4, 0.01% gelatin), and 2 or 3 drops of chloroform. Phage lysate was prepared as described below. The pBK-CMV phagemid containing hybridizing inserts were excised from the lambda ZAP Express vector as described by the manufacturer (Stratagene).
Protein extraction and analysis.
Extraction and preparation of cell wall-associated proteins from S. mutans were described previously (6). S. mutans GS-5 was grown in BHI broth (Difco Laboratories). For extraction of cell wall-associated protein antigens (CWP-A), cells of streptococci from 20 liters of batch culture were washed extensively with 10 mM sodium phosphate buffer and incubated with 8 M urea extraction fluid for 1 h at 25°C. The extract was then dialyzed against 10 mM sodium phosphate buffer (pH 6.5) to remove the urea and subsequently concentrated by 60% (saturation) ammonium sulfate precipitation; the precipitate was dissolved and dialyzed against the same buffer containing 1 mM phenylmethylsulfonyl fluoride (PMSF).
E. coli XL1 Blue cells were infected with purified positive phage clones on L broth agar to give 5,000 PFU/85-mm-diameter plate and incubated at 42°C for 4 h. Fusion protein was induced by the addition of 1 ml of 10 mM IPTG/plate and incubation at 37°C for 3.5 h. The crude phage lysates were recovered by adding 0.5 ml of lysis buffer (25% β-mercaptoethanol, 10% SDS, 6 M urea, 300 mM Tris-HCl [pH 6.8]) and gently shaking the sealed plates at 37°C for 10 to 15 min. The phage lysate containing the fusion protein was centrifuged at 1,500 × g for 10 min to remove debris and stored at −20°C. The proteins were separated by SDS-PAGE followed by silver staining or transferred electrophorectically to a Hybond-P super membrane (Amersham). For Western blot analysis, the blotted membrane was exposed to human sera or purified antibody as described above.
Construction, expression, and purification of rIDG-60.
Recombinant IDG-60 (rIDG-60) expressed in E. coli was purified by chromatography on an Ni2+ affinity resin. The IDG-60 coding sequence, sagA, was digested with FspI and EcoRI from pZAP-781 and inserted into the PvuII-EcoRI sites of the plasmid pRSETA (Invitrogen, Carlsbad, Calif.). The resulting plasmid, pRSETA-sagA, expressing sagA with a deletion of its signal sequence (amino acids 1 to 32) replaced by an N-terminal 6-His tag, was introduced into E. coli BL21(DE3), which contains the T7 polymerase gene on the chromosome under the control of the lacUV5 promoter. E. coli harboring pRSETA-sagA was grown to an A550 of 0.4 to 0.5, and the T7 promoter was induced by the addition of lPTG to a final concentration of 2.0 mM. The cultures were grown for an additional 4 h and then harvested. The pellets were resuspended in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), and a cell lysate was prepared by disrupting the cells with sonication. The cell debris was removed by centrifugation at 15,000 × g for 30 min. Further steps in the purification of His-tagged rIDG-60 were performed according to the instruction manual provided by the manufacturers. Homogeneity of the purified proteins was confirmed by SDS-PAGE, followed by silver staining. Identity of the protein was analyzed by immunoblots with anti-6×His-tag monoclonal antibody (Qiagene) and amino acid sequence of the internal peptide fragments after in-gel digestion with trypsin as described below. The bands were analyzed with Electrophoresis Documentation and Analysis System 120 (Scientific Imaging Systems, Eastman Kodak Co., Rochester, N.Y.). Protein concentrations were determined using a modified method of Lowry et al. (18), with bicinchoninic acid (BCA) as the colorimetric detection reagent (BCA protein assay reagent; Pierce, Rockford, Ill.).
LC-MS-MS.
To confirm the identity of the authentic IDG-60 protein immunoprecipitated from cell wall extracts of S. mutans and rIDG-60 expressed from E. coli, IDG-60 or rIDG-60 was digested with trypsin and the amino acid composition of tryptic digest was determined by liquid chromatography-tandem mass spectrometry (LC-MS-MS) as described previously (33). For in-gel digestion, the gel piece containing IDG-60 or rIDG-60 was soaked in 25 mM NH4HCO3 for 10 min and then in 25 mM NH4HCO3–50% acetonitrile for 10 min. After being dried in a Speed-Vac (Savant), the gel was incubated in 100 μl of 2% β-mercaptoethanol–25 mM NH4HCO3 for 20 min at room temperature and in the dark. The same volume of 10% 4-vinylpyridine in NH4HCO3–50% acetonitrile was added for cysteine alkylation. After a 20-min incubation, the gel was soaked in 1 ml of 25 mM NH4HCO3 for 10 min. After being washed in 25 mM NH4HCO3–50% acetonitrile for 10 min, the gel was dried and then incubated with 25 mM NH4HCO3 containing 50 ng of modified trypsin (Promega, Madison, Wisc.) overnight for 18 h. The tryptic digest was removed from the gel by extraction with 200 μl of 0.1% formic acid. These two fractions were combined together, dried in a Speed-Vac, and then kept at −20°C for storage. The fraction was resuspended in 0.1% formic acid immediately before use. Samples were analyzed by SDS-PAGE with a 20% gel and LC-MS-MS analysis. Electrospray MS was performed using a Finnigan Mat LCQ ion-trap mass spectrometer interfaced with an ABI 140D HPLC (Perkin-Elmer). A PE Brownlee C18 column (150 by 0.5 mm; Perkin-Elmer) with mobile phases of A (0.1% formic acid in water) and B (0.085% formic acid in acetonitrile) was used. The peptides were eluted at a flow rate of 5 μl/min with an acetonitrile gradient, which consisted of 5 to 16% B in 5 min, 16 to 20% B in 40 min, and 20 to 65% B in 40 min. The spectra for the eluate were acquired as successive sets of three scan modes. The MS scan determines the intensity of the ions in the m/z range of 395 to 1,605, and a specific ion was selected for zoom scan and MS-MS scan. The acquired collision-in-dissociation (CID) spectra were interpreted using a Finnigan Corporation software package, the SEQUEST Browser. Those MS-MS scans that matched to peptide sequences with cleavage sites at lysine or arginine were considered significant and also were subjected to manual evaluation to confirm the SEQUEST results. The hypothetical m/z values of peptides were derived using the PEPSTAT algorithm in SEQUEST package. The EXPLORE program (Finnigan) was used to plot the ion tracings, to determine the retention time, and to calculate the peptide ion counts.
In vitro transcription and translation of rIDG-60.
Coupled transcription-translation of His-tagged sagA was conducted with the E. coli T7 S30 Extract System for Circular DNA (Promega) following the instruction of the manufacturer. Briefly, the S30 extract contains T7 RNA polymerase for transcription and all necessary components for translation. The translated products were precipitated by trichloroacetic acid before SDS-PAGE gel analysis. The in vitro-translated rIDG-60 was labeled with 1 μl of [35S]methionine (1,175 Ci/mmol) for detection or was not labeled for Western blotting and glycan staining.
Detection of glycoproteins and enzymatic deglycosylation.
IDG-60 from S. mutans and rIDG-60 purified from the expression system were subjected to SDS-PAGE and transferred to nitrocellulose for glycan detection. Glycan detection was performed with a digoxigenin glycan differentiation kit according to the protocol of the manufacturer (Roche Molecular Biochemicals, Mannheim, Germany). Briefly, digoxigenin-labeled lectins that selectively recognize terminal sugars and their linkages were reacted in separate reaction mixtures with the proteins transferred to nitrocellulose and the specificity of lectin binding was subsequently detected by polyclonal sheep antidigoxingenin Fab fragments conjugated with alkaline phosphatase. The lectins included Galanthus nivalis agglutinin (GNA) recognizing terminal mannose-linked α-(1–3), α-(1–6), or α-(1–2); Sambucus nigra agglutinin (SNA) recognizing sialic acid linked α-(2–6) to galactose; Maackia amurensis agglutinin (MAA) recognizing sialic acid linked α-(2–3) to galactose, peanut agglutinin (PNA) recognizing core disaccharide galactose β-(1–3)-N-acetylgalactosamine, and Datura stramonium agglutinin (DSA) recognizing Gal α-(1–4)-GlcNAc.
Analysis of glycan linkages of IDG-60 and rIDG-60 was performed with a panel of glycosidase enzymes. The proteins were incubated under denaturing conditions with endoglycosidases, N-glycosidase F (N-linked glycans), O-glycosidase (Galβ1-3GalNAc), and exoglycosidase, and β-galactosidase (β1-linked 1–4 terminal galactose) supplied in an enzymatic deglycosylation kit according to the protocol of the manufacturer (Bio-Rad Laboratories, Hercules, Calif.).
Inactivation and complementation of sagA.
To construct an sagA insertion mutant, a cassette encoding erythromycin resistance from pAMβ1 (5) was inserted into a unique BclI site of sagA in the plasmid pRSETA-sagA. The resulting plasmid was linearized and subsequently transformed in the S. mutans GS-5 strain to disrupt the corresponding chromosomal loci by double crossover. Isogenic mutants, GS-51, GS-52, and GS-53, defective in the expression of IDG-60, were verified at the genetic level by Southern blotting and at the protein level by the absence of reaction with purified antibody by Western blotting.
For complementation test, the DNA fragment containing sagA and flanking sequences was digested from pZAP-781 and cloned into an E. colistreptococcus shuttle vector, pDL277 (3). The resultant plasmid, pDL-sag was transformed into GS-51, GS-52, or GS-53, and the transformants were selected on BHI agar plates with erythromycin and spectinomycin. Three complementing strains originating from individual sagA-defective strains, named GS-511, GS-521, and GS-531, respectively, were isolated, and the expression of IDG-60 in these strains was restored to thè level of that in their parental GS-5 strain. The characteristics of isogenic mutant strains and complementing strains were tested by a series of biochemical reactions and serological typing and showed no distinct changes from the wild-type strains.
General genetic manipulations.
Transformation of S. mutans was performed as described by Perry and Kuramitsu (25). DNA extraction and Southern blotting were performed as described previously (7). Automated DNA sequencing (ABI) was performed by the Molecular Biology Facility, College of Medicine, National Taiwan University, Taipei. Sequencing was carried out with custom oligonucleotides, and all sequencing was done in both directions.
Evaluation of adaptation to environmental stress.
Environmental adaptability of the wild-type (GS-5), isogenic mutants (GS-51, GS-52, and GS-53), and complementation strains (GS-511, GS-521, and GS-531) were evaluated by monitoring bacterial growth in Todd-Hewitt (TH) broth (Difco) containing 50 mM sodium acetate at appropriate pH values or the indicated concentrations of NaCl. Temperature sensitivity was determined from 37 to 42°C. To initiate the growth experiments, bacteria grown to early log phase were inoculated into 3 ml of TH broth with or without the indicated modification. Bacterial growth was monitored by measurement of the optical density at 550 nm (OD550). The number of surviving cells was determined for each mutant strain tested and compared with that of the corresponding wild-type strain.
Scanning electron microscopy.
S. mutans cells were fixed by the addition of glutaraldehyde to a final concentration of 2% (wt/vol) in 0.1 M sodium cacodylate buffer (SCB) (pH 6.8). After this first fixation, cells were rinsed with 0.1 M SCB and fixed for 1 h in 1% (wt/vol) osmium tetroxide in 0.1 M SCB. The samples were then washed twice with 0.1 M SCB, were dehydrated with acetone, and were critical-point dried by the CO2 method of Anderson (2). The dehydrated samples were coated with gold (SC-502, Fison, United Kingdom) and examined using a vacuum sputter coater TopCon ABT-60 scanning electron microscope operating at 10 kV.
TEM.
For transmission electron microscopy (TEM), bacteria grown in BHI in early stationary phase were fixed in 2.5% glutaldehyde in PBSC (0.1 M sodium buffer [pH 7.2 to 7.4], sucrose, and calcium) for 2 to 3 h at room temperature with shaking. Samples were washed with PBSC for 30 min twice and postfixed for 2 h at 4°C in 1% (wt/vol) OsO4 at room temperature. After being washed with distilled water, samples were prestained in uranyl acetate in 50% methanol for 15 min at room temperature. Dehydration was performed in upgraded ethanol, and then the samples were embedded in Spurr's resin (32). Thick sections (5 μm) stained with toluidine blue were examined first; subsequently, ultrathin sections were mounted on gold grid, which was restained by uranyl acetate and lead citrate. Samples were analyzed with a model EM109 electron microscope (Zeiss).
Nucleotide sequence accession number.
The complete sequence of sagA has been deposited in GenBank under the accession number AF338445. It is an apparent homolog of an unpublished sequence of Enterococcus faecium sagA (secreted antigen) reported in GenBank (AF242196).
RESULTS
Antibody elution and identification of IDG-60.
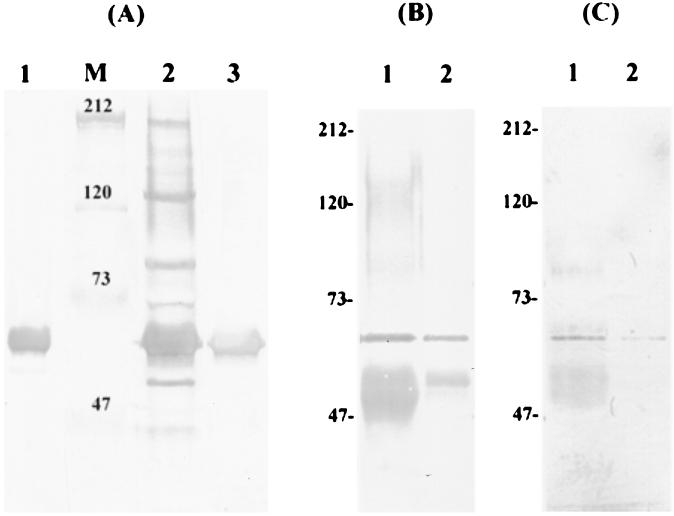
Previous screening of serum and salivary antibody profiles against S. mutans indicated that, in naturally sensitized humans, the strongest response was directed against a cell wall-associated protein with a size of approximately 60 kDa, among a number of antigens detectable by Western blotting (6). A total of 156 individuals tested were classified on the basis of the number and relative intensity of the bands detectable on immunoblots into categories of either high or low responders. Regardless of the tested antibody being a high responder or a low responder, this 60-kDa cell wall-associated protein was the immunodominant antigen (Fig. 1A, lanes 1 and 2). Interestingly, in some low responders, the 60-kDa protein appeared to be the only antigen detectable by serum antibodies at a titer of 1:600. These observations prompted us to initiate studies to isolate the corresponding gene and to analyze the biological function of this protein.
FIG. 1.
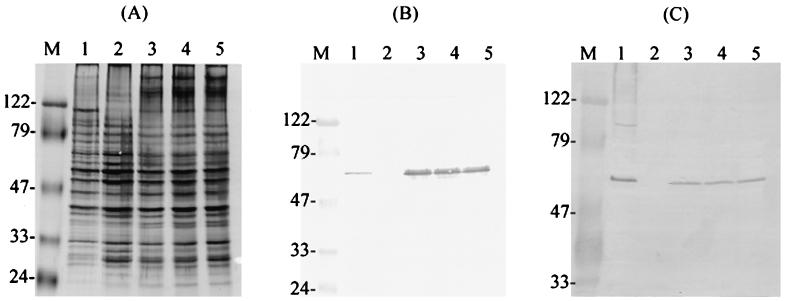
Immunoblot analysis of IDG-60. (A) Representative immunoblot analysis of serum IgG to CWP-A of S. mutans GS-5 from a low responder (no. 156) (lane 1) and a high responder (lane 2). At a titer of 1/600, IDG-60 exhibited the strongest reaction in both cases and was the only antigen detectable in this low responder. The purified anti-IDG-60 exhibited a single band on immunoblot (lane 3). The IDG-60 in the CWP-A fraction of S. mutans GS-5 was immunoprecipitated by human serum no. 156 (lane 1) or purified anti-IDG-60 (lane 2) and subsequently detected on immunoblots by purified anti-IDG-60 antibody (B) or by digoxigenin-labeled lectins (SNA and DSA) (C). The prestained high-molecular-mass standards in kilodaltons are indicated to the left (lane M).
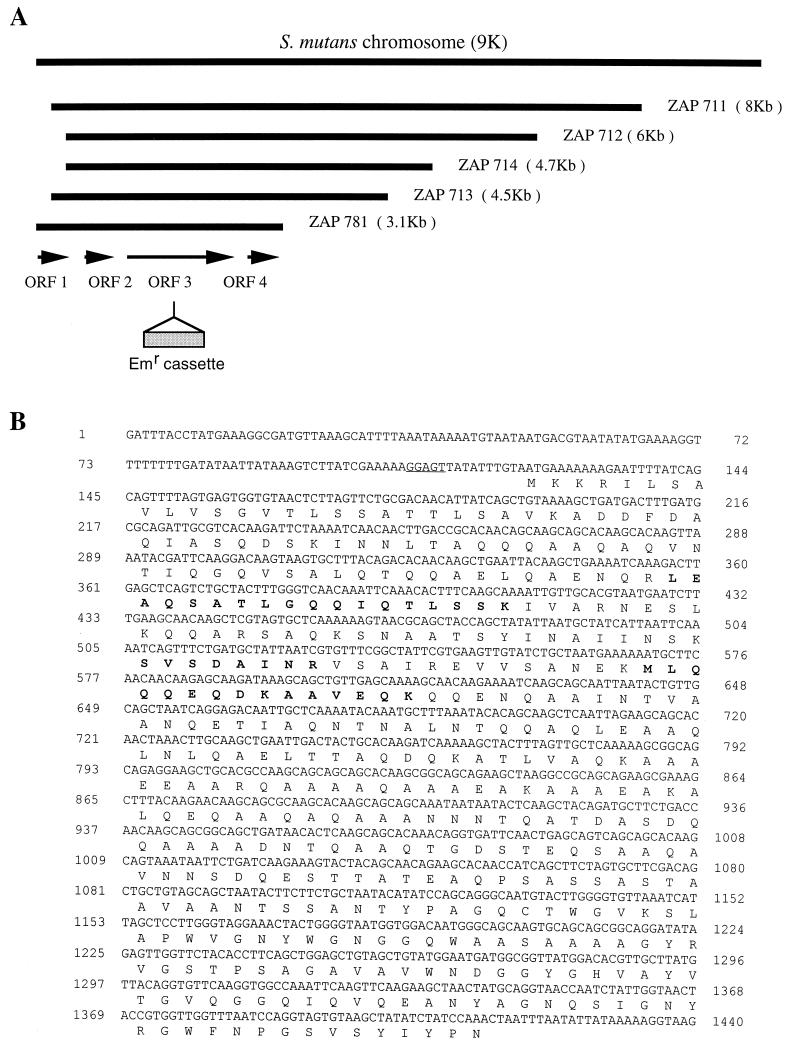
Specific anti-60-kDa antibody with high affinity was purified by elution from the immunoblots and recognized specifically the protein with a size of approximately 60 kDa on immunoblots (Fig. 1A, lane 3). Subsequently, the purified antibody was used for screening positive clones from a phage expression library of S. mutans GS-5. Five positive clones were obtained after screening >105 PFU on 85-mm-diameter plates. Western blot analysis of the phage lysates confirmed that each of these clones contained an open reading frame which expressed a 60-kDa protein. The expression of this protein from these clones was independent of the induction by IPTG, which specifically drives the expression of inserted DNA fragments from the T7 promoter located at one arm of the lamda phage vector. This result indicated that the 60-kDa protein was transcribed from its own promoter and also that all five clones contained the complete open reading frame of the targeted protein. Nucleotide sequences of all five clones from both directions of the phage revealed an identical chromosome location but different sizes, from 3.1 to 8 kb, compared with the nucleotide sequence of the S. mutans genome released from the University of Oklahoma's Advanced Center for Genome Technology (OU-ACGT). One of the clones (pZAP-781), smallest in size, was completely sequenced for both DNA strands and found to contain four potential open reading frames (Fig. 2). Southern blot analysis confirmed that pZAP-781 contained DNA originating from S. mutans GS-5 and also the expected restriction patterns by sequence analysis.
FIG. 2.
Maps of positive phage clones and sequence analysis of IDG-60. (A) Schematic representation of a portion of the S. mutans chromosome deduced form the OU-ACGT (www.genome.ou.edu) and individual phage clones. The relative position and size of each phage clone was mapped to the chromosome after nucleotide sequence analysis of each clone from both ends. For insertional inactivation of sagA, an erythromycin cassette (as indicated) was inserted in the sagA in plasmid pRSETA-sag for allelic exchange. (B) Nucleotide sequence of the sagA gene and the deduced amino acid sequence. The putative Shine-Dalgarno sequence is underlined. The amino acid sequence of peptide confirmed by LC-MS-MS is bold faced.
Nucleotide and amino acid sequence analysis.
Computer analysis of the sequence obtained for pZAP-781 revealed four complete open reading frames, ORF1, ORF2, ORF3, and ORF4. These ORFs and portions of the sequenced 3,120-bp fragments are depicted in Fig. 2. The deduced amino acid sequence of ORF1 and ORF2 defined two small polypeptides of 155 amino acids (12 kDa) and 168 amino acids (13 kDa), which share homology (∼50%) to the cell shape-determining proteins MreC and MreD of Bacillus subtilis, respectively (15). Sequences similar to these two and to ORF3 were also found in a cluster in E. faecium (GenBank AF242196), and ORF3 was designated sagA, the name given the corresponding gene in E. faecium. ORF3 encodes a protein of 431 amino acids (45 kDa), which is a general stress protein, GSP-781, recently identified by our laboratory by differential display reverse transcription-PCR (8). This protein was named IDG-60 for immunodominant glycoprotein with a size of 60 kDa in the present report. IDG60 shares approximately 65% identity with PscB, a protein most recently identified from group B streptococcus (27), and approximately 45% identity with another secreted protein, Usp45, of unknown functions from Lactococcus lactis (36). The polypeptide from ORF4 is composed of 129 amino acids (11 kDa) and shares more than 90% homology with phosphoribosyl PPi synthetase (prs) from Listeria monocytogenes (10).
Because Western blot analysis of the phage lysates from all five positive clones (ZAP711 to ZAP781 [Fig. 2]) showed positive identification of IDG-60 and because two of the clones do not contain the complete coding sequences of ORF1, these results indicated that the coding sequence for IDG-60 was located in ORF2, ORF3, or ORF4, but none of them encodes a protein with a size of 60 kDa predicted from the deduced amino acid sequences. To confirm the identity of the IDG-60, the protein was immunoprecipitated from the cell wall extracts of S. mutans GS-5 by the purified anti-IDG-60 antibody, separated on SDS-PAGE, and confirmed by its molecular weight and a positive reaction with the antiIDG-60 antibody (Fig. 1B). The internal amino acid sequence determined by LC-MS-MS confirmed that IDG-60 is encoded by ORF3, and the sequence obtained is shown in Fig. 2. To test the hypothesis that the discrepancy in the predicted versus the observed molecular weight is due to posttranslational modification by glycosylation, glycoprotein staining was performed by digoxigenin-labeled lectins. As shown in Fig. 1C, IDG-60 immunoprecipitated by either serum or purified anti-IDG-60 antibody gave a positive reaction to the glycoprotein staining. Therefore, the immunodominant antigen IDG-60 located on the cell wall of S. mutans is a general stress protein with posttranslational modification by glycosylation.
rIDG-60 purification and glycoprotein composition analysis.
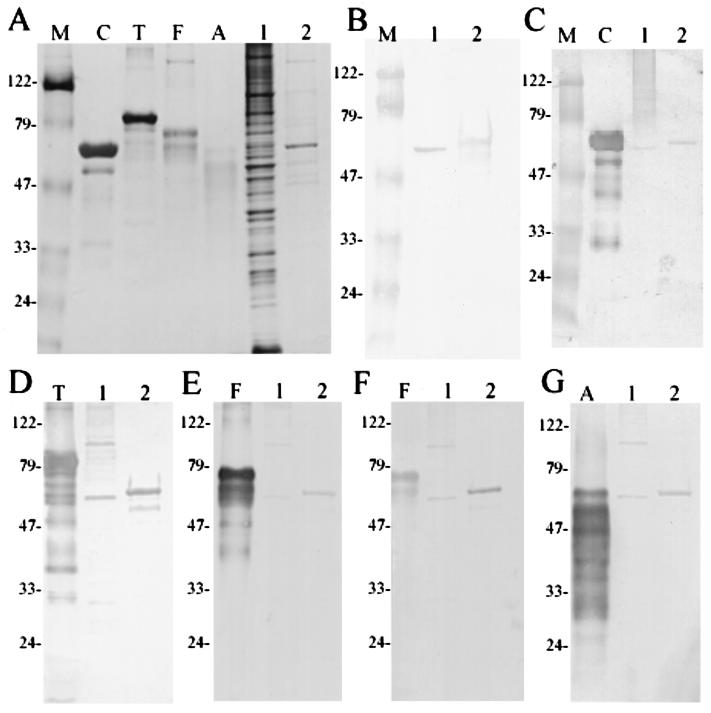
Previous results from Western blot analysis indicated that IDG-60 constituted only a minor fraction in the cell wall-associated protein extracts concentrated from a large batch (20 liters) of cultures. To obtain purified IDG-60 and to determine if glycosylation is restricted to its native host, rIDG-60 with a His tag was expressed and purified by affinity chromatography. After purification and separation by SDS-PAGE, only rIDG-60 with a molecular mass of approximately 60 kDa was detected (Fig. 3A, lane 2). The identity of rIDG-60 was confirmed by a positive response to the purified anti-IDG-60 antibody and by internal amino acid sequence using LC-MS-MS after in-gel digestion with trypsin. To confirm if nonglycosylated rIDG-60 existed and if it exhibited a different binding affinity with the Ni2+ affinity resin, crude extracts of E. coli expressing rIDG-60 or eluents from different elution conditions were analyzed. None of them exhibited positive bands of the predicted 45-kDa protein by Western blotting with anti-His tag monoclonal antibody. The glycosylation of rIDG-60 was subsequently confirmed by lectin staining as well (see below). Therefore, only the glycosylated rIDG-60 could be identified when it was expressed in E. coli. These results indicated that the glycosylation of IDG-60 was an intrinsic property determined by the protein and that this function is retained across species boundaries.
FIG. 3.
Glycoprotein detection of IDG-60 and purified rIDG-60 by specific lectin binding. (A) SDS-PAGE analysis of glycoproteins and CWP-A of S. mutans GS-5 after silver staining. Lanes: (C) carboxypeptidase Y (63 kDa); T, transferrin (80 kDa); F, fetuin (68 kDa); A, asialofetuin (61 kDa); 1, CWP-A of GS-5; 2, purified rIDG-60 (8 μg loaded); and M; molecular mass standards (in kilodaltons). IDG-60 in CWP-A or purified rIDG-60 was detected by Western blotting with purified anti-IDG-60 antibody (B) or detected for binding with specific lectins with known glycoproteins as positive controls. The following digoxigenin-labeled lectins were used: GNA (C), SNA (D), MAA (E), PNA (F) and DSA (G). Specific binding of lectins to carbohydrate moieties were detected by antidigoxigenin Fab fragments conjugated with alkaline phospatase.
To determine the monosaccharide composition, glycan staining was performed with a set of labeled lectins that specifically recognize particular terminal monosaccharides and their linkage to protein. Both authentic IDG-60 found in crude extracts of S. mutans and purified rIDG-60 were recognized by all five lectins tested (Fig. 3C to G). These lectin interactions suggested that the monosaccharides terminally linked to the glycan of IDG-60 include mannose, sialic acid, and galatose. Positive reactions with SNA and MAA (Fig. 3C and D) suggested that sialic acid is terminally linked as α(2–6) and α(2–3) to galactose. Positive reactions with PNA suggested the the presence of the disaccharide galactose-β-(1–3)-N-acetylgalactosamine, which usually forms the core unit of O glycan. Finally, positive reactions with DSA suggested the existence of both galactose-β(1–4)-N-acetylglucosamine and individual N-acetylglucosamine residues with O-glycosidic links to serine or threonine (11).
Glycosidase analysis and in vitro translation of IDG-60.
One of our primary interests was to determine if the immunodominant characteristics of IDG-60 were determined by the glycans and/or the protein backbone. To achieve this goal and also to determine whether the glycosylation is N linked and/or O linked, the removal of monosaccharides from the authentic protein or rIDG-60 was attempted with different kinds of glycosidases suggested by the carbohydrate composition and linkage analysis determined by specific lectin staining. Incubation of authentic IDG-60 in crude extracts from S. mutans or purified rIDG-60 with endoglycosidase N-glycosidase F (N-linked glycans) or O-glycosidase did not result in deglycosylation as determined by a reduction in both the electrophoretic mobilities and intensities of the proteins as determined by Western blot analysis with purified anti-IDG-60 antibody. Nevertheless, the authentic IDG-60 was sensitive to the activity of an exoglycosidase, β-galactosidase, which cleaved the β-linked 1–4 terminal galactose (data not shown). This result is consistent with the lectin binding staining assay using DSA, which binds specifically to the β-linked 1–4 terminal galactose (Fig. 3G). In addition, the sensitivity of IDG-60 to β-galactosidase provided direct evidence of the covalent linkage of terminal galactose in IDG-60.
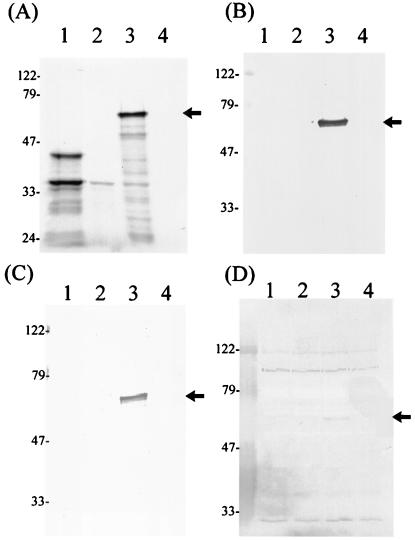
To test if nonglycosylated IDG-60 could be obtained in a cell-free system using an in vitro translation system designed for prokaryotes, the plasmid pRSETA-sagA, expressing IDG-60 with an N-terminal 6-His tag, was incubated in vitro with the S30 extract, which contains T7 RNA polymerase for transcription and all necessary components for translation from E. coli. The identity of the translated product was confirmed by positive reaction with either anti-His antibody or purified anti-IDG-60 antibody (Fig. 4B and C). Surprisingly, similar to the in vivo-expressed product, in vitro-translated IDG-60 exhibited a molecular mass of 60 kDa only and a positive, but weak, binding to the tested lectin (Fig. 4D). These results indicated that glycosylation of IDG-60 functions in vitro in a cell-free system, even though at a much lower efficiency than that which occurred in vivo inside the bacterial host.
FIG. 4.
Detection of rIDG-60 translated in vitro. The plasmid pRSETA-sag was incubated in vitro with the S30 extract, which contains T7 RNA polymerase for transcription and all necessary components for translation from E. coli. (A) Autoradiograph of translational products labeled with [35S]methionine. Lanes 1, positive control proteins, PinPoint/CAT fusion protein (39 kDa) and β-lactamase (28 kDa); 2, plasmid control with pRSETA only; 3, IDG-60 (60 kDa); and 4, negative control without plasmid DNA. The identity of rIDG-60 was confirmed by immunoblots with either anti-his monoclonal antibody (B) or purified anti-IDG-60 antibody (C). Glycosylation was confirmed by lectin binding (D)., IDG-60 is marked by arrow head.
Growth characteristics of sagA mutant and complementation strains.
Transcriptional analysis by Northern blots reported previously (8) confirmed that sagA is transcribed monocistronically in S. mutans and transcription at the mid-logarithmic-growth phase was enhanced by stress treatment such as low pH, high osmolarity, and high temperature. To analyze the function of IDG60 in S. mutans, we constructed an isogenic mutant by insertion inactivation using a 1.2-kb erythromycin cassette introduced by allelic exchange. Despite the essential role of IDG-60 in S. mutans (see below) and the retarded growth rate of the isolated mutants, successful inactivation of sagA was achieved in S. mutans GS-5, and three mutants, GS-51, GS-52, and GS-53 were isolated. All three mutants exhibited positive reaction with anti-serotype c antiserum (7) and GS-51 was selected for further analysis. Southern blot analysis confirmed the predicted integration and disruption of sagA as indicated by the shifting of a HindIII-digested fragment from the length of 2.2 to 3.3 kb. Western blotting using purified anti-IDG-60 IgG showed a strong band in the cell wall extracts of parental GS-5 but not in the extracts of its isogenic mutant GS-51 (Fig. 5B, lanes 1 and 2). The GS-51 mutants exhibited poor growth in BHI agar or TH broth with a doubling time over 10 h for growth anaerobically in the TH broth. In addition, sagA mutant GS-51 tends to aggregate and grows in clumps. When examined with a light microscope, GS-51 lost the ability to retain the crystal violet staining typical for gram-positive cell wall. To confirm the observed phenotypic changes were entirely due to the defect in sagA rather than pleiotropic effects through other mutations that may have occurred during selection of mutants, complementation of the sagA defective strain GS-51 was carried out by transformation with streptococcus-E. coli shuttle vector pDL277 carrying wild-type copy of sagA. The resultant transformants, GS-511, GS-512, and GS-513 restored the expression of IDG-60 (Fig. 5B) and exhibited phenotypic characteristics, such as growth rate, positive gram staining, etc., indistinguishable from those of the parental GS-5 strain.
FIG. 5.
Detection of IDG-60 in sagA mutant and complementation strains. (A) Analysis of crude extracts of CWP-A from GS-5 (lane 1), GS-51 (lane 2), GS-511 (lane 3), GS-512 (lane 4), and GS-513 (lane 5) by SDS-PAGE and silver staining. The total amounts of proteins loaded in each lane were nearly identical. (B) Immunoblot of IDG-60 by purified antibody. A single band of expected mobility was present in the wild-type, GS-5 (lane 1), and complementation strains (lanes 3, 4, and 5) but absent from extracts of sagA mutant (lane 2). (C) Glycoprotein detection by lectin binding. Positive reaction was detected in the wild-type, GS-5 (lane 1), and complementation strains (lanes 3, 4, and 5) but not in the sagA mutant (lane 2).
Since previous results indicated that IDG-60 may function as a general stress protein, the growth characteristics of the parental GS-5 strain and mutant GS-51 and GS-511 strains were challenged with different stress conditions and their survival rates were determined. Strain GS-511 grew at the same rate as the wild-type GS-5 strain at pH 7.5 and exhibited similar resistance to the challenge of low pH (5.5), high osmolarity (400 mM NaCl), and high temperature (42°C). But strain GS-51, lacking expression of IDG-60, could not survive any of the challenges list above. These results in conjunction with our earlier transcriptional analysis confirmed that IDG-60 is a general stress protein which is essential for survival of S. mutans encountering environmental stress.
Microscopic analysis of IDG-60.
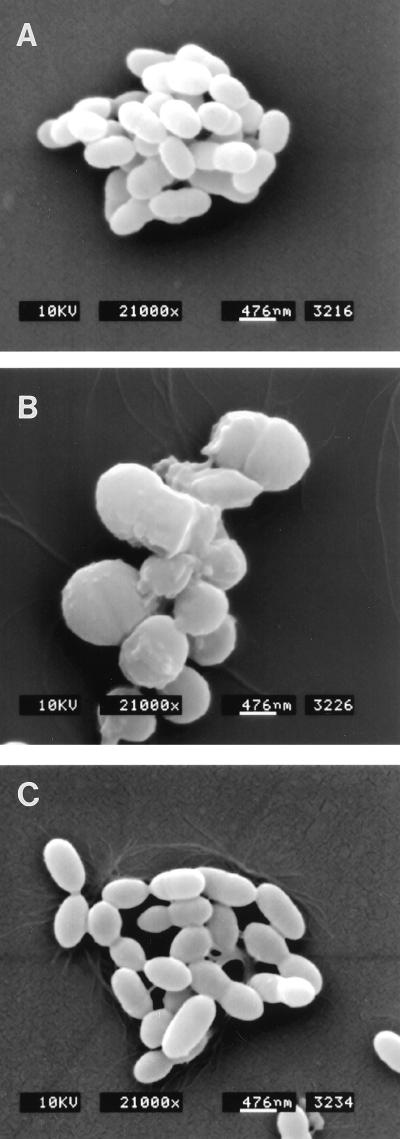
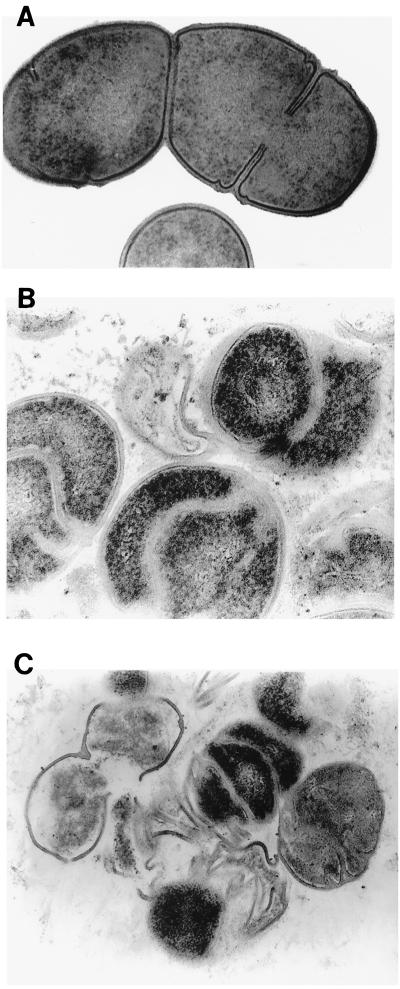
The failure of the GS-51 mutant to retain crystal violet and the increased sensitivity to osmotic pressure led us to hypothesize that the role of IDG-60 is to maintain integrity of the cell wall. Scanning electron microscopy examination of the wild-type GS-5 strain showed characteristics typical of S. mutans as short rods (Fig. 6A). However, the GS-51 mutant lacking expression of IDG-60 exhibited pleimorphic cell shape. The cells of the GS-51 mutant acquired irregular shapes, with a general increase in the size to magnitudes different from those of GS-5 (Fig. 6B). Complementation of the normal copy of sagA restored the shape of GS-511 indistinguishable from that of the wild type GS-5 strain (Fig. 6C). To analyze the changes in the structure of the cell wall in more detail, stationary cells of GS-5 and GS-51 were fixed and examined by TEM. As depicted in Fig. 7, the wild-type GS-5 strain showed a typical cell wall with condensed structure and with typical septum formation (Fig. 7A). However, in the GS-51 mutant, the cell wall was less condensed and revealed several layers of ingrowth of cell wall distinct from septum formation during cell division (Fig. 7B). In addition, the cell wall of the GS-51 mutant appeared to be fragile, because rupture of the cell wall in this mutant, but not in the parental GS5 strain, was easily observed by mild sonication that is normally applied to disrupt the streptococcus chains (Fig. 7C). Taken together, these results suggest that IDG-60 is required for maintaining the integrity of the S. mutans cell wall and cell shape which, in turn, are essential for withstanding stress conditions such as osmotic pressure.
FIG. 6.
Scanning electron microscopic analysis of S. mutans GS-5, sagA mutant strain GS-51 and complementation strain GS-511. Cells of GS-5 are short rods with uniform size and shape (A), GS-51 cells are irregular and exhibit enlarged size of various magnitude (B), and shape or size of GS-511 is indistinguishable from GS-5 (C).
FIG. 7.
Transmission electron analysis of GS-5 (A) and its isogenic mutant GS-51 (B and C). GS-5 has a typical cell wall with condensed structure and septum formation (A), while mutant GS-51 exhibits polymorphism in cell wall orientation and shows multilayer ingrowth of cell wall distinct from septum formation found in GS-5 (B). The cell wall of GS-51 is fragile and lysed cells or cells with disruption in the cell walls can be easily observed by mild sonication, which normally disrupts the streptococcal chains without damaging the intact cell wall (C). Original magnification, ×50,000.
DISCUSSION
Independent approaches using completely different strategies conducted in our laboratory led to the identification of IDG-60. The first approach was to identify stress-responsive genes in S. mutans by differential display RT-PCR of RNA from cells exposed to different stress conditions, including low pH, high osmolarity, and saliva or plasma components, etc. We have recently reported the identification of several acid stress proteins which were induced or repressed at the transcriptional level by challenge at pH of 5.5 (8). RNA of one protein, GSP-781, was up-regulated significantly when bacteria were exposed to high osmolarity and temperature in addition to low pH, and therefore, GSP-781 is a general stress protein. The second approach was to identify the immunodominant antigens located on the cell wall of S. mutans, which are important for host immune response to S. mutans infections. By using purified antibody eluted from human donors, we identified IDG-60 (this study), and the nucleotide sequence of this protein revealed its identity to GSP-781. The results of the present study provide additional information to elucidate the posttranslational modification as well as the functional role of this interesting protein.
Glycoproteins of prokaryotes have been known for two decades, but information about their biological function is still limited. There are now an increasing number of glycoproteins identified from both archaebacteria and eubacteria. Based on their cellular localization, prokaryotic glycoproteins have been classified as crystalline surface layer (S-layer) membrane-associated and surface-associated glycoproteins, as well as secreted glycoproteins and exoenzymes (for a review, see reference 21). Regardless of their distribution and localization, the glycan structure of prokaryotic glycoproteins differs considerably from that of eukaryotic glycoconjugates in terms of sugar composition and glycosidic linkages. S-layer glycans of eubacteria, for example, are often composed of linear or branched charbohydrate chains linked via common N-glycosidic linkage to asparagine or O-glycosidic linkage to serine or threonine, although carbohydrates O linked to tyrosine have been identified in Paenibacillus alvei (1, 22) and Thermoanaerobacter thermohydrosulfuricus (4, 23). Such distinct characteristics in structure and linkages may account for the observed lack of response of prokaryotic glycoproteins to the action of endoglycosidases, which usually are utilized for the deglycosylation of eukaryotic glycoconjugates. IDG-60 of S. mutans, for example, is resistant to the action of both N-glycosidase F and O-glycosidase, and therefore, additional experiments are required to confirm the glycosidic linkages in this protein. It has been shown that a glycoprotein from Streptococcus sanguis, platelet aggregation-associated protein (PAAP), contains an N-glycosidic linkage between N-acetylglucosamine and asparagines (9).
A variety of polysaccharides are present in prokaryotic cell walls, usually attached to lipids or peptidoglycan. In S. mutans, the polysaccharides in the cell wall responsible for the serotype-specific reactions have been well characterized. This serotype-specific antigen is composed primarily of backbone structures of 1,2- and 1,3-linked rhamnose with glucose side chains unique to different serotypes of mutans streptococci (16, 26). The seroptype-specific polysaccharides are immunodominant antigens; that is, they can elicit predominant antibody responses that specifically recognize and differentiate the structural details of the side chains on the cell walls of different serotypes of mutans streptococci (16). Although the biosynthetic pathway of these seroptype-specific carbohydrate polymers has recently been elucidated (34, 35), the linkage and the components to which these polysaccharides attach are still not clear. The results of the present study indicated that glycan of IDG-60 may not be related to the serotype-specific carbohydrates and may represent a distinct entity on the cell wall of S. mutans. Evidence in support of this conclusion is derived from the finding that the GS-51 mutant lacking IDG-60 still exhibits serotype-specific carbohydrate antigens on the cell wall identified by the serotype c-specific antiserum from our laboratory (8). Based on the lectin staining (Fig. 3), there is more than one glycosylated protein detectable in the cell wall extracts of S. mutans. A glycoprotein with a molecular mass higher than that of IDG-60 exhibited positive reactions with the lectins of SNA, MAA, PNA, and DSA (Fig. 3). Identification of this second glycoprotein is currently underway in our laboratory.
Even though the glycosidic linkage and structural details of IDG-60 merit further investigation, the results of the present study provide direct evidence to elucidate the biological function of this glycoprotein in S. mutans. By using periodic acid-Schiff staining and lectin binding, the presence of glycoproteins has been demonstrated among cell wall antigens from other streptococci, including S. sanguis (9) and Streptococcus pyogenes (14). It was hypothesized that carbohydrates in these proteins may bind to the inner surface of the bacterial membrane, facilitating the secretion and subsequent incorporation of the protein into the cell wall (9). Therefore, glycosylation of prokaryotic proteins may function in a manner similar to that in the eukaryotes for targeting proteins for secretion from either the cell or organelles (24). Insertional inactivation of sagA in conjunction with microscopic analyses confirmed that cell wall-associated glycoprotein IDG-60 plays an essential role in maintaining the integrity of the cell wall and cell shape. An analogous functional role of glycoprotein in shape determination has been reported in an archaeobacteria, Halobacteri Salinarium (20). Upon removal of the Glc-Asn-linked glycan from an S-layer glycoprotein, the rod-shaped halobacteria change their shape to round spheres.
Based on the results of primary amino acid sequence comparison, IDG-60 was found to share 64.7% identity with the recently reported PscB, a protein required for cell wall separation from group B streptococci (27). Insertional inactivation of pcsB resulted in a mutant strain, Sep1, that exhibited phenotyic characteristics similar to the IDG-60-defective mutant of S. mutans GS-51. First, Spe1 exhibited a drastically reduced growth rate compared to that of the parental streptococcal strain and showed an increased susceptibility to osmotic pressure. Secondly, the TEM picture of Spe1 revealed multifold ingrowth of the cell wall, which was hypothesized to be due to the formation of multiple septa. This latter finding led the group to hypothesize that PscB was a murein hydrolase which splits the peptidoglycan septum during cell division (12). But no murein hydrolase activity could be demonstrated in vitro for PscB. Distinct from Spe1, GS-51 exhibited enlarged cell shape with pleiomorphic cell shapes from spherical to square-like shapes. Although GS-51 had the tendency to aggregate in broth culture, the cell division appeared to be normal in this mutant strain. In addition, the ingrowth of cell wall is distinct from the septa normally found in the plane of cell division because the former contains new newly synthesized peptidoglycan which appears as an electron-dense layer by scanning electron microscopy (Fig. 7A). The fact that the cell wall of GS-51 was fragile and easily disrupted by mild sonication lead us to hypothesize that the multifold ingrowth of cell wall observed in GS-51 by TEM might be a compensation for the fragile cell wall lacking IDG-60 in order to maintain normal function in resisting osmotic pressure and other stresses.
IDG-60 also shares 45% identity with a secreted protein, Usp45, from Lactococcus latis with no previously known biological significance (37). The nucleotide sequence of usp45 revealed an ORF of 1,383 bp encoding a protein of 461 amino acids with an estimated molecular mass of 45 kDa. Interestingly, Western blot analysis of extracellular proteins of L. lactis with anti-Usp45 antibodies revealed a protein with a size of 60 kDa (36). In addition, expression of usp45 in E. coli also resulted in a recombinant Usp45 protein with a molecular mass of 60 kDa. No commentary or discussion was made on the discrepancy of the observed molecular mass of Usp45 in that report. But glycosylation of heterogeneous recombinant proteins in E. coli has been documented recently with proteins from Ehrlichia. Two proteins, P120 (120 kDa) and P140 (140 kDa), were immunodominant antigens from E. chaffeensis and E. cani, cani respectively (38, 39). The predicted molecular mass of the P120 recombinant protein is 67 kDa, and that of the P140 is 57 kDa. However, the molecular masses of both recombinant proteins observed were 1.6 times larger than the protein size predicted by the amino acid sequence (19). Carbohydrates of recombinant P120 and P140 were subsequently demonstrated by biotin labeling of the oxidized sugars.
Similarly, rIDG-60 was also glycosylated when expressed in E. coli and the results of lectin binding assays suggestèd that glycosylation appeared to be indistinguishable from that in the authentic host S. mutans. In addition, our results also indicated that glycosylation of IDG-60 can proceed in vitro in a cell free system. These results indicate that the signals for glycosylation is intrinsically encoded and can cross species boundaries.
Characterization of eubacterial glycoproteins has revealed diverse carbohydrate structures compared to eukaryotes, which have more conserved glycan structures. The main problem is the lack of sufficient structural information to draw a picture of the general architecture of the prokaryotic glycoproteins. Homologous serine- and threonine-rich motifs were identified in the repeat regions of E. chaffeensis P120, E. cani P140, and Streptococcus parasanguis Fap1, which is a fimbria-associated adhesin glycoprotein (19, 37). No direct repeats were found in IDG-60 and IDG-60 and share no homology with P120, P140, or Fap1. Nevertheless, a common characteristic shared by these glycoproteins and IDG-60 was their immunoreactivity. All of them are immunodominant antigens and immunoreactivity of both the native and recombinant counterparts is reported to be strong. As shown in the present study, the nonglycosylated form of IDG-60 was not found when expressed in vivo in E. coli or translated in vitro. These results suggest that glycosylation might be essential for maintaining proper structure or folding of IDG-60. The role that carbohydrates play in recognition of these proteins, during the immune response remains to be determined.
ACKNOWLEDGMENTS
We thank P. E. Kolenbrander for plasmid pDL277 and Gallen Wang for technique assistance in SEM. We thank Sheng-Chung Lee, Graduate Institute of Molecular Biology, National Taiwan University, Taipei, for assistance with LC/MS/MS. We thank Tim J. Harrison, Reader in Molecular Virology, University Department of Medicine, Royal Free Hospital School of Medicine, London, United Kingdom for his kind review and help in the preparation of the manuscript.
This work was supported in part by the National Science Council (grants NSC 90-2320-B-002-134 and 90-2314-B-002M05) and the National Health Research Institute (grant NHRI-ZX90-8814BC).
REFERENCES
- 1.Altman E, Brisson J-R, Messner P, Sleytr U B. Structure of the glycan chain from the surface layer glycoprotein of Bacillus alvei CCM 2051. Biochem Cell Biol. 1991;69:72–78. doi: 10.1139/o91-010. [DOI] [PubMed] [Google Scholar]
- 2.Anderson T F. Techniques for the preservation of three-dimensional structure in preparing specimens for the electron microscope. Trans N Y Acad Sci. 1951;13:130–134. [Google Scholar]
- 3.Aspiras M B, Kazmerzak K M, Kolenbrander P E, McNab R, Hardegen N, Jenkinson H F. Expression of green fluorescent protein in Streptococcus gordonii DL1 and its use as a species-specific marker in coadhesion with Streptococcus oralis 34 in saliva-conditioned biofilms in vitro. Appl Environ Microbiol. 2000;66:4074–4083. doi: 10.1128/aem.66.9.4074-4083.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock K, Schuster-Kolbe J, Altman E, Allmaier G, Stahl B, Christian R, Sleytr U B, Messner P. Primary structure of the O-glycosidically linked glycan chain of the crystalline surface layer glycoprotein of Thermoanaerobacter thermohydrosulfuricus L111–69. Galactosyl tyrosine as a novel linkage unit. J Biol Chem. 1994;269:7137–7144. [PubMed] [Google Scholar]
- 5.Brehm J, Salmond G, Minton N. Sequence of the adenine methylase gene of the Streptococcus faecalis plasmid pAMβ1. Nucleic Acids Res. 1987;15:3177. doi: 10.1093/nar/15.7.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chia, J. S., W. C. Chang, C. S. Yang, and J. Y. Chen. Salivary and serum antibody response to Streptococcus mutans antigens in humans. Oral Microbiol. Immunol. 15:131–138. [DOI] [PubMed]
- 7.Chia J S, Hsu T Y, Teng L J, Chen J Y, Hahn L J, Yang C S. Glucosyltransferase gene polymorphism among Streptococcus mutans strains. Infect Immun. 1991;59:1656–1660. doi: 10.1128/iai.59.5.1656-1660.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chia J S, Lee Y Y, Huang P T, Chen J Y. Identification of stress-responsive genes in Streptococcus mutans by differential display reverse transcription-PCR. Infect Immun. 2001;69:2493–2501. doi: 10.1128/IAI.69.4.2493-2501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson P R, Herzberg M C. Evidence for the covalent linkage of carbohydrate polymers to a glycoprotein from Streptococcus sanguis. J Biol Chem. 1993;268:23780–23783. [PubMed] [Google Scholar]
- 10.Gouin E, Mengaud J, Cossart P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect Immun. 1994;62:3550–3553. doi: 10.1128/iai.62.8.3550-3553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart G W, Holt G D, Haltiwanger R S. Nuclear and cytoplasmic glycosylation: novel saccharide linkages in unexpected places. Trends Biochem Sci. 1989;13:380–384. doi: 10.1016/0968-0004(88)90179-x. [DOI] [PubMed] [Google Scholar]
- 12.Holtje J-V. From growth to autolysis: the murein hydrolses in Escherichia coli. Arch Microbiol. 1995;164:243–254. doi: 10.1007/BF02529958. [DOI] [PubMed] [Google Scholar]
- 13.Horaud T, Delbos F. Viridans streptococci in infective endocarditis: species distribution and susceptibility to antibiotics. Eur Heart J. 1984;5(Suppl. C):39–44. doi: 10.1093/eurheartj/5.suppl_c.39. [DOI] [PubMed] [Google Scholar]
- 14.Kanaoka M, Fukita Y, Taya K, Kawanaka C, Negoro T, Agui H. Antitumor activity of streptococcal acid glycoprotein produced by Streptococcus pyogenes Su. Jpn J Cancer Res. 1987;78:1409–1414. [PubMed] [Google Scholar]
- 15.Levin P A, Margolis P S, Setlow P L, Losick R, Sun D. Identification of Bacillus subtilis genes for septum placement and shape determination. J Bacteriol. 1992;174:6717–6728. doi: 10.1128/jb.174.21.6717-6728.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linzer R, Reddy M S, Levin M J. Immunological aspects of serotype carbohydrate antigens of Streptococcus mutans. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunobiology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publisher; 1986. pp. 29–38. [Google Scholar]
- 17.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 19.McBride J W, Yu X J, Walker D H. Glycosylation of homologous immunodominant proteins of Ehrlichia chaffeensis and Ehrlichia canis. Infect Immun. 2000;68:13–18. doi: 10.1128/iai.68.1.13-18.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mescher M F, Strominger J L. Structural (shape-maintaining) role of the cell surface glycoprotein of Halobacterium salinarium. Proc Natl Acad Sci USA. 1976;73:2687–2691. doi: 10.1073/pnas.73.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messner P. Bacterial glycoproteins. Glycoconjugate J. 1997;14:3–11. doi: 10.1023/a:1018551228663. [DOI] [PubMed] [Google Scholar]
- 22.Messner P, Christian R, Neuninger C, Schulz G. Similarity of “core” structures in two different glycans of tyrosine eubacterial S-layer glycoproteins. J Bacteriol. 1995;177:2188–2193. doi: 10.1128/jb.177.8.2188-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messner P, Christian R, Kolbe J, Schulz G, Sleytr U B. Analysis of a novel linkage unit of O-linked carbohydrates from the crystalline surface layer glycoprotein of Clostridium thermohydrosulfuricum S102–70. J Bacteriol. 1992;174:2236–2240. doi: 10.1128/jb.174.7.2236-2240.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Reilly M A, Nogee L, Whitsett J A. Differential effects of glucocorticoid on expression of surfactant protein in a human lung adenocarcinoma cell line. Biochem Biophys Acta. 1988;969:176–184. doi: 10.1016/0167-4889(88)90179-6. [DOI] [PubMed] [Google Scholar]
- 25.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prichard D G, Gregory R L, Michalek S M, McGhee J R. Biochemical aspects of serotype carbohydrate antigens of Streptococcus mutans. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunobiology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publisher; 1986. pp. 39–49. [Google Scholar]
- 27.Reinscheid D J, Gottschalk B, Schubert A, Eikmanns B J, Chhatwal G S. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J Bacteriol. 2001;183:1175–1183. doi: 10.1128/JB.183.4.1175-1183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russell M W, Hajishengallis G, Childers N K, Michalek S M. Secretory immunity in defense against cariogenic mutans streptococci. Caries Res. 1999;33:4–15. doi: 10.1159/000016490. [DOI] [PubMed] [Google Scholar]
- 29.Russell R R B, Abdulla E, Gilpin M L, Smith K. Characterization of Streptococcus mutans surface antigens. In: Hamada S, Michalek S M, Kiyono H, Menaker L, McGhee J R, editors. Molecular microbiology and immunobiology of Streptococcus mutans. Amsterdam, The Netherlands: Elsevier Science Publisher; 1986. pp. 61–70. [Google Scholar]
- 30.Smith D J, Akita H, King W F, Taubman M A. Purification and antigenecity of a novel glucan-binding protein of Streptococcus mutans. Infect Immun. 1994;62:2545–2552. doi: 10.1128/iai.62.6.2545-2552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith D J, Taubman M A. Experimental immunization with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries. Infect Immun. 1996;62:2545–2552. doi: 10.1128/iai.64.8.3069-3073.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spurr A R. A low-viscosity epoxy resin embedding medium for electron microscope. J Ultrastruc Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 33.Tsay Y G, Wang Y-H, Chiu C-M, Shen B-J, Lee S-C. A strategy for identification and quantitation of phosphopeptides by liquid chromatography/tandem mass spectrometry. Anal Biochem. 2000;287:55–64. doi: 10.1006/abio.2000.4837. [DOI] [PubMed] [Google Scholar]
- 34.Tsukioka Y, Yamashita Y, Oho T, Nakano Y, Koga T. Biological function of the dTDP-rhamnose synthesis pathway in Streptococcus mutans. J Bacteriol. 1997;179:1126–1134. doi: 10.1128/jb.179.4.1126-1134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsukioka Y, Yamashita Y, Nakano Y, Oho T, Koga T. Identification of a fourth gene involved in the dTDP-rhamnose synthesis in Streptococcus mutans. J Bacteriol. 1997;179:4411–4414. doi: 10.1128/jb.179.13.4411-4414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Asseldonk M, Rutten G, Oteman M, Siezen R J, de Vos M W, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990;95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, Mintz P, Ladha M, Fives-Taylor P M. Isolation and charaterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- 38.Yu X J, Crocquet-Valdes P, Walker D H. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene. 1997;184:149–154. doi: 10.1016/s0378-1119(96)00586-0. [DOI] [PubMed] [Google Scholar]
- 39.Yu X J, McBride J W, Diaz C M, Walker D H. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J Clin Microbiol. 2000;38:369–374. doi: 10.1128/jcm.38.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]