Abstract
Background
Our study analyzed the immune infiltration of esophageal adenocarcinoma (EAC) tumor cells and identified long non-coding ribonucleic acid (lncRNA) genes to construct a prognostic model of EAC to evaluate the survival prognosis of patients and explore potential therapeutic targets.
Methods
The data of 89 patients with EAC, including 11 normal tissue samples and 78 EAC of tumor tissue samples, were downloaded from The Cancer Genome Atlas public database. Perl script and R software were used to run the code, conduct the statistical analysis, calculate the risk coefficients of the patients, and conduct the Cox regression analysis, immune-related lncRNA survival analysis, risk analysis, principal component analysis (PCA), and receiver operating characteristic (ROC) curve analysis.
Results
We screened and identified 19 prognostic biomarkers, including LINC01612, AC008443.2, and LINC02582, allocated the patients into high- and low-risk groups, and found significant differences in the prognosis between the high- and low-risk groups using the Kaplan-Meier survival analysis (P<0.001). A ROC curve was used to evaluate the feasibility of the prognostic model for EAC, and we found that the model had high predictability (area under the curve =0.964). A PCA analysis was performed of the complex transcriptome sequencing data and other cubes to transform the data into a 3-dimensional space constructed by feature vectors.
Conclusions
Our study effectively screened and identified the lncRNA genes related to the immune infiltration of EAC and successfully constructed a prognostic model. In total, 19 potential diagnostic and therapeutic target genes, including LINC01612, AC008443.2, and LINC02582, were identified that have certain significance in guiding the clinical treatment of EAC patients.
Keywords: Esophageal adenocarcinoma (EAC), immune infiltration, prognostic model, long non-coding ribonucleic acids (lncRNAs), prognostic risk
Highlight box.
Key findings
• Our study effectively screened and identified the lncRNA genes related to the immune infiltration of EAC and successfully constructed a prognostic model.
What is known and what is new?
• Immune infiltration plays important roles in various cancers.
• The lncRNA genes related to the immune infiltration of EAC.
What is the implication, and what should change now?
• The model may be useful in finding new potential therapeutic targets for EAC and providing personalized immunotherapy to EAC patients.
Introduction
Esophageal adenocarcinoma (EAC) is a common malignant tumor of the digestive tract (1-3). In terms of treatment and prognosis, immune-related genes (IRGs) are key factors in the development of the disease, and immune disorders in tumors are considered promoting factors in the process of tumor genesis and development. In recent years, immunotherapy has emerged as a promising potential treatment for EAC in addition to surgery and radiotherapy (4).
EAC tumor cells are rich in tumor antigens, including tumor-associated antigens and neoantigens, which have the ability to initiate dendritic cell-mediated tumor cytotoxic T lymphocytes early in cancer development (5). When EAC tumor cells begin to fight the immune system, they can acquire the ability to suppress anti-tumor immunity through immune checkpoints, secretory factors, and negatively modulated immune cells (6). These characteristics of IRGs may contribute to the diagnosis and individualized treatment of EAC (7). When the immune function is inhibited, cancer cells can proliferate and metastasize.
A study has reported a relationship between IRGs and the prognosis of EAC patients, but there is no systematic description or study of the IRGs in the tumor immune microenvironment and the prognosis of EAC patients (8). Moreover, their interactions are also need studied. Thus, a systematic description and analysis of the tumor immune microenvironment and the effect of IRGs on prognosis are necessary for EAC immunotherapy and patient prognosis (9).
In recent years, with the continuous development and promotion of tumor immune infiltration research, there have been a few reports on the establishment of IRG prognostic models for EAC (10). This study sought to successfully construct a prognostic model of immune-related EAC by analyzing EAC transcriptome data from The Cancer Genome Atlas (TCGA) public database and clinical follow-up data, and to screen and identify new biomarkers and prognostic long non-coding ribonucleic acids (lncRNAs) for EAC, thus providing potentially important therapeutic targets for EAC. We present the following article in accordance with the TRIPOD reporting checklist (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1279/rc).
Methods
General information
We downloaded the EAC group gene transcription and clinical data from TCGA public database (https://portal.gdc.cancer.gov/). Based on the research inclusion and exclusion criteria, the data of 89 patients, including 11 normal healthy patients and 78 EAC patients, were downloaded. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Research methods
Perl scripts were used to conduct the statistical analysis of the downloaded lncRNA data and clinical data, and “Limma” package in R software (version 4.0.3) was then used to correct them. To examine differences in the overall survival (OS) of the high- and low-risk groups and identify the lncRNAs related to the prognosis of EAC patients, a survival curve for the EAC patients during the clinical follow-up period was generated using the “Survival” package. The risk curve was drawn by running the “PheatMap” package. In order to identify the lncRNAs were related to immune, the “BiocManager” package was downloaded and run to perform the principal component analysis (PCA) of the multi-dimensional and complex EAC gene data set and convert the data into a 3-dimensional space constructed by feature vectors. Finally, risk heat map was used to determine the high- and low-risk genes.
Statistical processing
SPSS software (version 23.0) and Bertel-Perl software (version 5.32.0.1) were used to screen the obtained data, and a Cox regression analysis and a PCA algorithm were used to analyze the differences in the lncRNAs related to immune infiltration in the tumor EAC tissues. All the data were statistically analyzed by R language software (version 4.0.3), and P<0.05 indicated statistically significant differences.
Results
Survival analysis
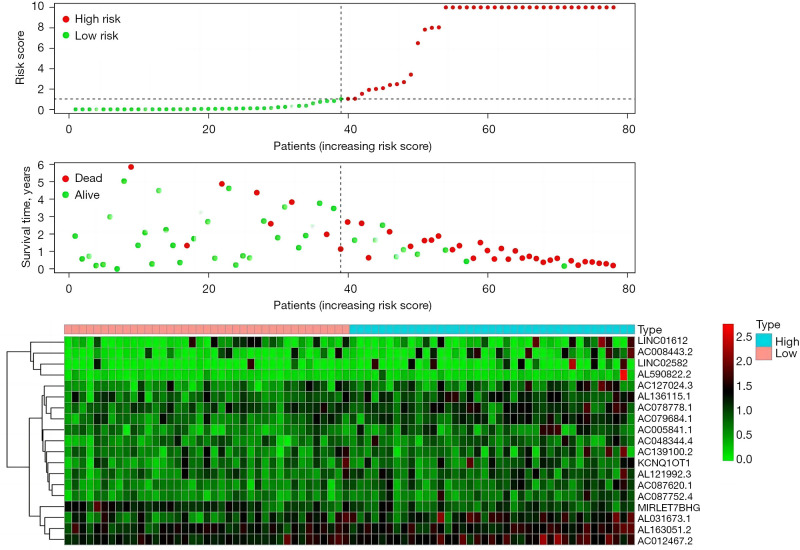
The follow-up data of 78 EAC patients were divided into high- and low-risk groups according to the expression of immune-prognostic lncRNAs. The 1-year survival rate (60%) and 3-year survival rate (10%) of the high-risk group were significantly lower than the 1-year survival rate (100%) and 3-year survival rate (90%) of the low-risk group (P<0.001) (Figure 1).
Figure 1.
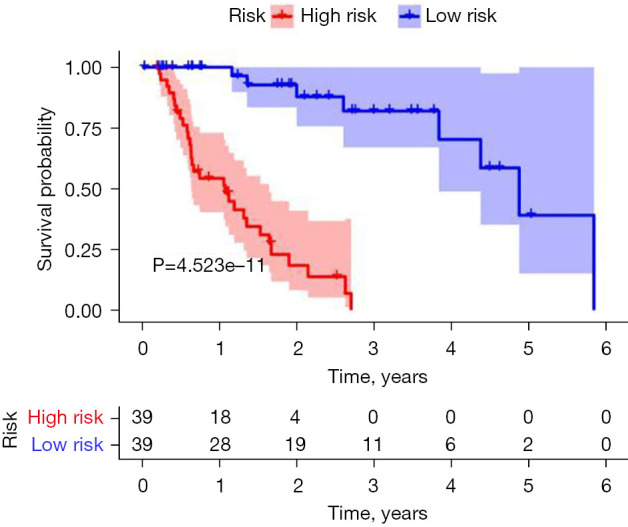
Kaplan-Meier survival analysis of EAC. EAC, esophageal adenocarcinoma.
Risk curve analysis
The “Pheatmap” package in R software was used to arrange and collate the downloaded data from TCGA, calculate the risk coefficients of the EAC patients, calculate the distribution matrix of the survival and death of the patients, analyze the differential expression of lncRNAs in an immune infiltration-related prognosis heatmap, and successfully build the EAC prognostic model. We found that as the expression level of the lncRNAs (the following 19 high-risk lncRNAs) increased, the prognosis of patients worsened. Further, we also found that the high expression of lncRNA may be related to prognosis and high mortality rates for EAC patients. The following 19 high-risk lncRNAs were identified: LINC01612, AC008443.2, LINC02582, AL590822.2, AC127024.3, AL136115.1, AC078778.1, AC079684.1, AC005841.1, AC048344.4, AC139100.2, KCNQ1OT1, AL121992.3, AC087620.1, AC087752.4, MIRLET7BHG, AL031673.1, AL163051.2, and AC012467.2 (Figure 2).
Figure 2.
Survival prognostic risk curve, death case distribution matrix, and risk heat map of the 78 EAC patients. EAC, esophageal adenocarcinoma.
ROC curve analysis of prognostic model
A prognostic model of EAC was successfully constructed through TCGA public database, and a receiver operating characteristic (ROC) curve was used to verify the predictive value of the model. We found that the area under the curve (AUC) was significant >0.7 (AUC =0.964), indicating that compared to other clinically relevant indicators, this model has a high prediction accuracy and has certain value in guiding risk assessments used to determine the prognosis of EAC patients (Figure 3).
Figure 3.
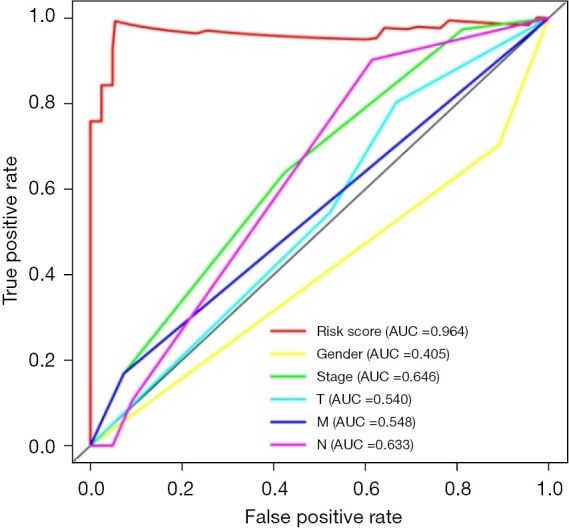
ROC curve of the prognostic model of EAC associated with immune infiltration. ROC, receiver operating characteristic; AUC, area under the curve; EAC, esophageal adenocarcinoma.
PCA analysis
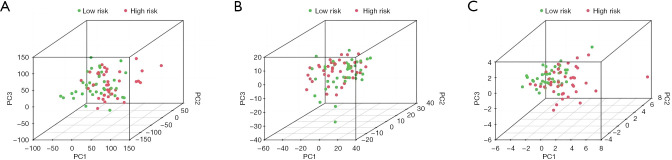
A PCA analysis was performed of these complex transcriptome data. Figure 4A shows, all the lncRNA gene sets related to EAC. Figure 4B shows the part of the gene sets related to immune infiltration. Figure 4C shows the lncRNA gene sets with prognostic value of the first 2 gene sets (Figure 4).
Figure 4.
PCA analysis of the lncRNAs included in the prognostic model of EAC. (A) All genes PCA; (B) immune lncRNA PCA; (C) risk gene PCA. PCA, principal component analysis; lncRNAs, long non-coding ribonucleic acids; EAC, esophageal adenocarcinoma.
Discussion
Esophageal carcinoma is a malignant tumor of the digestive tract with a poor prognosis, and its pathology is mainly esophageal squamous carcinoma and EAC (11-14). In Southeast Asia, including China, Japan and South Korea, esophageal squamous cell carcinoma is common (15). In North America and Europe, including the United States, Canada and the United Kingdom, the incidence of EAC is high (16). However, in recent years, the morbidity and mortality of EAC in China has shown an increasing trend, which has aroused widespread concern (17-19).
Despite continued advances in its diagnosis and treatment, the prognosis of EAC patients remains poor (20). Thus, we need to identify the molecular mechanisms of EAC pathogenesis and biomarkers for use in prognostic risk assessments (21). There is a growing body of evidence that immune infiltration plays an important role in tumor cell proliferation, invasion, and metastasis, and cannot be ignored (21,22). Thus, we need to explore the tumor cells involved in immune signaling and the occurrence and development of EAC. This will help us in specific targeting loci treatment prognosis and survival risk model building. There are many EAC-related biomarkers, but there are few reports on the establishment and analysis of lncRNA immune-prognostic models (23).
In this study, gene transcription data sets, the associations between prognostic risk and immune infiltration, lncRNA were screened and identified. Ultimately, 19 potential prognostic lncRNAs were screened and identified, and survival, risk prediction, and PCA analyses were performed using these lncRNAs. The EAC patients were divided into high- and low-risk groups, and a differential analysis was conducted to identify important prognostic indicators (24). Models constructed based on immune infiltration and prognostic risk coefficients are effective at assessing disease status and determining treatment options for EAC patients (25). These 19 lncRNAs were used to construct a Cox regression risk model, which could predict the prognosis of high-risk and low-risk patients. The accuracy of the test model was analyzed using a ROC curve.
In recent years, a large number of studies have explored the prognosis of different diseases in order to guide treatment (26-30). The prognostic model of esophageal cancer was constructed based on the characteristics of messenger RNA, and the role of specific immunotherapy was highlighted. Our findings may lead to the development of new immunotherapies for esophageal cancer. The latest checkpoint inhibitors and related targeting sites for esophageal cancer and have thus provided a research basis for the discovery of new targeted therapeutic sites for EAC. One study retrospectively analyzed the differences in immune cell infiltration and functional phenotypes in the tumor cells of 72 EAC patients, and found that the cytokine staining had a mixture functions of pro-inflammatory and anti-inflammatory properties in EAC tumors tissue and found that memory cytotoxic T cells were rich in memory cytotoxic T cells (26). This result will greatly promote the study of the EAC tumor immune microenvironment and immune cell reaction mechanism (31).
Limitation
We did not validate the representative lncRNA related to immune infiltration by experiments.
Conclusions
This study established an EAC prognostic model based on the immune infiltration that can accurately predict the survival prognosis of EAC patients. In addition, the model may be useful in finding new potential therapeutic targets for EAC and providing personalized immunotherapy to EAC patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: The study was supported by Yunnan Province High-level Health and Family Planning Personnel Training Project (Yunwei Science and Education Development [2017] No. 14); Basic Research Joint Special General Project of Yunnan Provincial Local Universities (Part) (Nos. 2018FH001-076, 2018FH001-080); The 8th Research Project of Education and Teaching Reform of Dali University (Special Medical Education Reform Project, Nos. 2022JGYX08-01, 2022JGYX08-02).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1279/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-22-1279/coif). The authors have no conflicts of interest to declare.
(English Language Editor: L. Huleatt)
References
- 1.Conroy MJ, Kennedy SA, Doyle SL, et al. A study of the immune infiltrate and patient outcomes in esophageal cancer. Carcinogenesis 2021;42:395-404. 10.1093/carcin/bgaa101 [DOI] [PubMed] [Google Scholar]
- 2.Jammula S, Katz-Summercorn AC, Li X, et al. Identification of Subtypes of Barrett's Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data. Gastroenterology 2020;158:1682-97.e1. Erratum in: Gastroenterology 2021;161:1727. 10.1053/j.gastro.2020.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gokon Y, Fujishima F, Taniyama Y, et al. Immune microenvironment in Barrett's esophagus adjacent to esophageal adenocarcinoma: possible influence of adjacent mucosa on cancer development and progression. Virchows Arch 2020;477:825-34. 10.1007/s00428-020-02854-0 [DOI] [PubMed] [Google Scholar]
- 4.Lagisetty KH, McEwen DP, Nancarrow DJ, et al. Immune determinants of Barrett's progression to esophageal adenocarcinoma. JCI Insight 2021;6:e143888. 10.1172/jci.insight.143888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi L, Huang P, Zou X, et al. Integrative stemness characteristics associated with prognosis and the immune microenvironment in esophageal cancer. Pharmacol Res 2020;161:105144. 10.1016/j.phrs.2020.105144 [DOI] [PubMed] [Google Scholar]
- 6.Strizova Z, Snajdauf M, Stakheev D, et al. The paratumoral immune cell signature reveals the potential for the implementation of immunotherapy in esophageal carcinoma patients. J Cancer Res Clin Oncol 2020;146:1979-92. 10.1007/s00432-020-03258-y [DOI] [PubMed] [Google Scholar]
- 7.Zhang P, Liu M, Cui Y, et al. Microsatellite instability status differentially associates with intratumoral immune microenvironment in human cancers. Brief Bioinform 2021;22:bbaa180. 10.1093/bib/bbaa180 [DOI] [PubMed] [Google Scholar]
- 8.Porter RJ, Murray GI, Brice DP, et al. Novel biomarkers for risk stratification of Barrett's oesophagus associated neoplastic progression-epithelial HMGB1 expression and stromal lymphocytic phenotype. Br J Cancer 2020;122:545-54. 10.1038/s41416-019-0685-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hynes CF, Kwon DH, Vadlamudi C, et al. Programmed Death Ligand 1: A Step Toward Immunoscore for Esophageal Cancer. Ann Thorac Surg 2018;106:1002-7. 10.1016/j.athoracsur.2018.05.002 [DOI] [PubMed] [Google Scholar]
- 10.Dong Z, Wang J, Zhang H, et al. Identification of potential key genes in esophageal adenocarcinoma using bioinformatics. Exp Ther Med 2019;18:3291-8. 10.3892/etm.2019.7973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajendra S, Sharma P, Gautam SD, et al. Association of Biomarkers for Human Papillomavirus With Survival Among Adults With Barrett High-grade Dysplasia and Esophageal Adenocarcinoma. JAMA Netw Open 2020;3:e1921189. 10.1001/jamanetworkopen.2019.21189 [DOI] [PubMed] [Google Scholar]
- 12.Zhao M, Wang J, Yuan M, et al. Multivariate gene expression-based survival predictor model in esophageal adenocarcinoma. Thorac Cancer 2020;11:2896-908. 10.1111/1759-7714.13626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Liang D, Pierzynski JA, et al. D-mannose: a novel prognostic biomarker for patients with esophageal adenocarcinoma. Carcinogenesis 2017;38:162-7. 10.1093/carcin/bgw207 [DOI] [PubMed] [Google Scholar]
- 14.Hedner C, Borg D, Nodin B, et al. Expression and Prognostic Significance of Human Epidermal Growth Factor Receptors 1 and 3 in Gastric and Esophageal Adenocarcinoma. PLoS One 2016;11:e0148101. 10.1371/journal.pone.0148101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battaglin F, Naseem M, Puccini A, et al. Molecular biomarkers in gastro-esophageal cancer: recent developments, current trends and future directions. Cancer Cell Int 2018;18:99. 10.1186/s12935-018-0594-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin DC, Dinh HQ, Xie JJ, et al. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut 2018;67:1769-79. 10.1136/gutjnl-2017-314607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang S, Dai Y, Mei Y, et al. Clinical significance and biological role of cancer-derived Type I collagen in lung and esophageal cancers. Thorac Cancer 2019;10:277-88. 10.1111/1759-7714.12947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao S, Li Y, Lu Z, et al. Survival-associated alternative splicing signatures in esophageal carcinoma. Carcinogenesis 2019;40:121-30. 10.1093/carcin/bgy123 [DOI] [PubMed] [Google Scholar]
- 19.Tian S, Wang C, Zhang J, et al. The cox-filter method identifies respective subtype-specific lncRNA prognostic signatures for two human cancers. BMC Med Genomics 2020;13:18. 10.1186/s12920-020-0691-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao D, He S, Harada K,et al. Integrated genomic profiling and modelling for risk stratifica-tion in patients with advanced oesophagogastric adenocarcinoma. Gut. 2020 Dec 17:gutjnl-2020-322707. [DOI] [PMC free article] [PubMed]
- 21.Kelly RJ. Immunotherapy for Esophageal and Gastric Cancer. Am Soc Clin Oncol Educ Book 2017;37:292-300. 10.1200/EDBK_175231 [DOI] [PubMed] [Google Scholar]
- 22.Wu L, Zhong Y, Wu D, et al. Immunomodulatory Factor TIM3 of Cytolytic Active Genes Affected the Survival and Prognosis of Lung Adenocarcinoma Patients by Multi-Omics Analysis. Biomedicines 2022;10:2248. 10.3390/biomedicines10092248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu L, Zheng Y, Ruan X, et al. Long-chain noncoding ribonucleic acids affect the survival and prognosis of patients with esophageal adenocarcinoma through the autophagy pathway: construction of a prognostic model. Anticancer Drugs 2022;33:e590-603. 10.1097/CAD.0000000000001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L, Zhong Y, Yu X, et al. Selective poly adenylation predicts the efficacy of immunotherapy in patients with lung adenocarcinoma by multiple omics research. Anticancer Drugs 2022;33:943-59. 10.1097/CAD.0000000000001319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Ling X, Fang C, et al. Identification and validation of an eight-lncRNA signature that predicts prognosis in patients with esophageal squamous cell carcinoma. Cell Mol Biol Lett 2022;27:39. 10.1186/s11658-022-00331-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng D, Xia S, Hu S, et al. miR-887-3p Inhibits the Progression of Colorectal Cancer via Downregulating DNMT1 Expression and Regulating P53 Expression. Comput Intell Neurosci 2022;2022:7179733. 10.1155/2022/7179733 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Chen Y, Wang J, Zhang X, et al. Correlation between apparent diffusion coefficient and pathological characteristics of patients with invasive breast cancer. Ann Transl Med 2021;9:143. 10.21037/atm-20-7746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qi A, Li Y, Yan S, et al. Effect of postoperative chemotherapy on blood glucose and lipid metabolism in patients with invasive breast cancer. Gland Surg 2021;10:1470-7. 10.21037/gs-21-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He XC, Chen HY, Qiu Y, et al. Associations of iron status with breast cancer risk factors in adult women: Findings from National Health and Nutrition Examination Survey 2017-2018. J Trace Elem Med Biol 2021;68:126867. 10.1016/j.jtemb.2021.126867 [DOI] [PubMed] [Google Scholar]
- 30.Qi A, Li Y, Sun H, et al. Incidence and risk factors of sexual dysfunction in young breast cancer survivors. Ann Palliat Med 2021;10:4428-34. 10.21037/apm-21-352 [DOI] [PubMed] [Google Scholar]
- 31.Zhang H, Pan E, Zhang Y, et al. LncRNA RPL34-AS1 suppresses the proliferation, migration and invasion of esophageal squamous cell carcinoma via targeting miR-575/ACAA2 axis. BMC Cancer 2022;22:1017. 10.1186/s12885-022-10104-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as