Abstract
Background
Studies for developing diagnostics and treatments for infectious diseases usually require observing the onset of infection during the study period. However, when the infection base rate incidence is low, the cohort size required to measure an effect becomes large, and recruitment becomes costly and prolonged. We developed a model for reducing recruiting time and resources in a COVID-19 detection study by targeting recruitment to high-risk individuals.
Methods
We conducted an observational longitudinal cohort study at individual sites throughout the U.S., enrolling adults who were members of an online health and research platform. Through direct and longitudinal connection with research participants, we applied machine learning techniques to compute individual risk scores from individually permissioned data about socioeconomic and behavioral data, in combination with predicted local prevalence data. The modeled risk scores were then used to target candidates for enrollment in a hypothetical COVID-19 detection study. The main outcome measure was the incidence rate of COVID-19 according to the risk model compared with incidence rates in actual vaccine trials.
Results
When we used risk scores from 66,040 participants to recruit a balanced cohort of participants for a COVID-19 detection study, we obtained a 4- to 7-fold greater COVID-19 infection incidence rate compared with similar real-world study cohorts.
Conclusion
This risk model offers the possibility of reducing costs, increasing the power of analyses, and shortening study periods by targeting for recruitment participants at higher risk.
Keywords: COVID-19, Clinical trials, Risk modeling
1. Introduction
The costs of recruiting large numbers of participants for clinical trials can be high. The power of clinical trials also can depend on the number of “rare events” observed (such as COVID-19 infections), which often takes long periods to accrue. Efforts have therefore been attempted to reduce costs, increase the power of analyses, and shorten study periods by targeting participants at higher risk (those more exposed to infection) during recruitment [1]. Many prospective incidence trials already use basic demographics and health state-based approach to define populations at increased risk, which helps to figure out how vaccines work in the defined population (high-risk population), with the caveat of potentially making data less-generalizable.
We present an enrichment approach based on connection with members of the Evidation health and research platform [2]. This reward platform encourages users to develop healthy habits—such as walking, meditating, and logging meals—and incentivizes them to participate in research by completing surveys and sharing data from commercial-grade wearable sensors [3,4]. For example, the application has been used since 2017 for voluntary monitoring of annual influenza cases [5]. We therefore had access to a large pool of potential study participants we could easily survey.
Our modeling approach applied machine-learning techniques to compute individual risk scores from socioeconomic and behavioral data, in combination with predicted local prevalence data. The modeled risk scores were then used to target candidates for enrollment.
2. Material and methods
2.1. Design
We leveraged our longitudinal relationship with members of the Evidation platform and applied machine-learning modeling of COVID-19 risk to determine and recruit a high-risk study population. The study protocol was approved by the WCB Institutional Review Board (#20202887), and participants gave written consent to take part in the study.
The machine-learning model quantified risk using participants’ locations, occupations, and behavior, given that these features likely affected exposure level to SARS-CoV-2. The initial training phase for the model was 49 days, during which we followed >100,000 members to characterize those who contracted COVID-19.
We measured enrichment in terms of the incidence of COVID-19 infection using this enhanced selection process versus infection incidence in the control groups of 3 COVID-19 vaccine trials [[6], [7], [8]] and another cohort generated via the Evidation platform but not using a precision recruitment approach [9]. Because each study had different enrollment dates and demographics, we normalized each study's incidence by matching dates and demographics of the comparator study to the US incidence.
Specifically, for this study, we launched a Risk of Occupational Exposure to COVID-19 deep-labeling survey on June 15, 2020. This survey, which collected demographic, socioeconomic, and behavioral data on potential high-risk populations, had received 128,629 responses at the time of analysis. Short follow-up surveys were sent to respondents to this survey who indicated they had not had a diagnosis or symptoms of COVID-19 as of August 3, 2020, to determine whether any individuals had received a diagnosis (and date of diagnosis) within ∼2 months since completing the initial survey. Of the 94,700 who were sent the follow-up survey, 66,040 (69.7%) responded, and 514 (0.8%) indicated they had received a COVID-19 diagnosis in the interim.
2.2. Risk modeling
We then created a machine-learning model using labeling responses from the initial survey, which performed better than chance at identifying who would receive a subsequent diagnosis. This model incorporated predictions of COVID-19 local prevalence (using generalized additive models [GAMS]) as a variable, along with socioeconomic and behavioral data from the initial survey. Using random forests, the model was trained on respondents to the second survey, with the outcome variable being whether they had contracted COVID-19 during the 49-day follow-up period. See the Appendix for detailed descriptions of the modeling process.
The trained model was then used to calculate a risk score for each respondent to the initial survey. Persons with the highest risk scores were primarily targeted for recruitment and were selected to generate a dataset with balanced demographic variables (eg, age, sex, and ethnicity).
2.3. Comparison with previous studies
To compare our findings with enrollment in the other studies, we first calculated the incidence rate as the number of confirmed COVID-19 cases per 1000 person-years of follow-up for our cohort and each comparison cohort. Only the person-days at risk of contracting COVID-19 were considered, and we excluded all days occurring after vaccination or contracting COVID-19. With this method, breakthrough infections were not included, therefore providing a conservative estimate.
To calculate the US-matched incidence for our cohort, we used individual-level data from the Centers for Disease Control and Prevention (CDC) describing all confirmed COVID-19 cases in the U.S. (These data likely underestimate the true number of cases.) We aggregated counts by date and by sex by age group.
For each comparator study, we calculated the US incidence of COVID-19 during the study period for each demographic group, using 2019 US Census data for the size of each demographic group in the U.S.[10] To calculate the final US-matched incidence, we measured the proportion of each demographic group in the comparator study, and then took the weighted average of the US incidences across demographic groups (weighted by proportions of these groups in the comparator study). By dividing each study incidence by the US-matched incidence, we ensured that our findings were not biased by differences in the study period or demographics.
We calculated 95% confidence intervals for the incidence in each study and the ratios using the exact method (Poisson distribution).
3. Results
From candidates with the highest risk scores, we recruited a demographically balanced cohort of 840 participants and followed them from November 5, 2020 to April 15th, 2021. The total follow-up time to reported infection or vaccination was 141.2 person-years, and 104 participants (12.3%) developed confirmed COVID-19 infection.
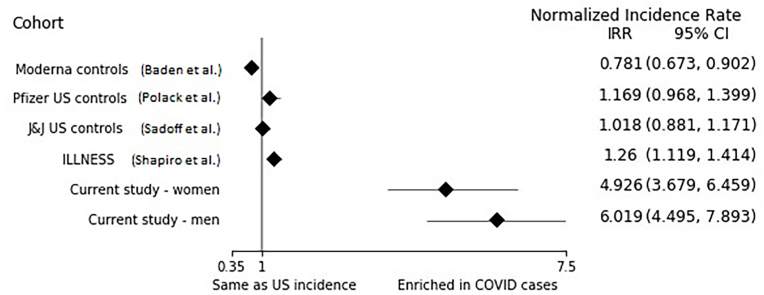
Comparing our model with recruitment in other studies, we observed 4- to 7-fold greater detection of COVID-19 cases after accounting for differences in study periods and numbers of COVID-19 cases in the U.S. at those times (Fig. 1). The normalized incidence rate for our study was 4.93 (CI [3.68–6.46]) for women and 6.02 (CI [4.50–7.89]) for men. See Appendix Table 1 for the data listings for each study included in the analysis. This supplementary table also shows the raw numbers of infections in the control groups of the comparison trials (between 1% and 2.2% of participants got infected in each of the comparison groups vs. 12.3% when we use our precision recruitment approach).
Fig. 1.
Covid-19 incidence rate in each cohort normalized by US incidence rate matched for time, age, and sex.
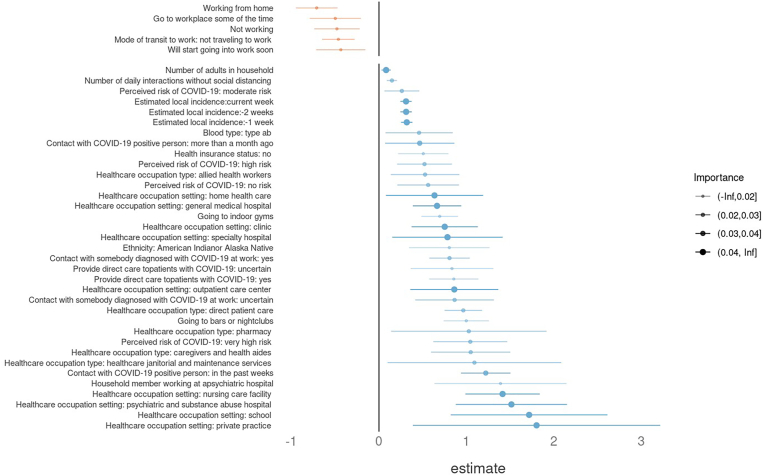
Fig. 2 shows the variables most important to the prediction of COVID-19 infection. The top features related to the number of potentially risky contacts (household size and residential situation), location (living in a city with numerous COVID-19 cases at recruitment) and working in a risky occupation (healthcare workers). See the Appendix for more details about these features. The features that were least important related to non-healthcare work settings (hospitality, public transit, agriculture, self-employed, etc.)
Fig. 2.
The 20 Most Important Groups of Predictors of Risk Ranked by Random Forest Feature Importance. Sub-predictors from the same group (e.g., Healthcare occupation) have been separated and rearranged for visual clarity. Positive coefficients that are statistically significant (risk increasing) are in blue, and negative ones are in orange. More detailed descriptions of each factor and variable are available in the Appendix. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
We compared a precision recruitment approach to that of another internal study [9] and external vaccine trials [[6], [7], [8]] while taking into account that these trials happened at different times and with different sample demographics. We used calculations of matched US incidences (in time, age, and sex) for each trial to properly make those comparisons. The comparisons showed a substantial enrichment factor of 4–7 times the incidence of COVID-19 infections. This means that our precision recruitment approach could be applied to reduce the needed sample size of future trials by this factor, or shorten the trials’ duration by the same factor. Beyond the benefit of reducing the cost of trials, this precision recruitment approach could be of great utility in situations of emergency, such as during a pandemic.
Although our risk model showed up to a 7-fold increase in recruitment possibility, we can improve it further through including additional variables that might be relevant, such as information about contacts with other people, especially with school-age children. We also could improve the training process and GAM models.
The current analysis has several limitations. For some of the datasets [[6], [7], [8]], we had access only to publicly available data. For example, we had no access to data about each participant's start and end dates in the trial. Therefore, when computing the matched US incidence for each trial (matched in time, age, and sex), we used the overall study start and end dates of each trial. This does not consider variability in when each participant started and ended the trial. If a study had very gradual or slow enrollment, its incidence might have differed substantially from the US-matched incidence over its duration.
Additionally, some studies tallied only events happening 14 days after the second vaccination dose/placebo. Given that we lacked individual-level data for when each participant received their second dose, we could not account for that variability. Instead, as shown in Appendix Table 1, we recalculated the US-matched incidences after removing the initial days corresponding to the length of its vaccination protocol plus 14 days (instead of using the full length of each study). This attempt to count from a later start date resulted in a higher US-matched incidence and therefore even lower comparative incidence ratios for the vaccine trials. Thus, our current estimate of enrichment compared with other studies is a lower bound on the actual enrichment. Our comparisons to US-matched incidences are imperfect (because of lack of individual-level data) but consistent across studies.
Whether the findings obtained on the recruited population would be generalizable to the full population is not guaranteed when using a technique for precision recruitment as described. For a vaccine trial this should be true, given that our population was not biologically different in demographics or comorbidities. The difference with the full population relates only to greater exposure to COVID-19 (through location, occupation, or behavior) rather than different biological susceptibility to it. Therefore, we do not expect any conclusion made regarding vaccine efficacy to be affected by our precision recruitment procedure, and results should be generalizable to other populations.
Funding
This project was funded in whole or in part with federal funds from the Department of Health and Human Services (HHS), Office of the Assistant Secretary for Preparedness and Response, Biomedical Advanced Research and Development Authority (BARDA) [contract number 75A50120C00091]. Additional support was provided by an appointment to the BARDA Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the HHS. The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author contributions
Aziz Mezlini: investigation, writing - original draft, writing - review & editing. Eamon Caddigan: investigation; Allison Shapiro: investigation. Ernesto Ramirez: conceptualization, methodology, investigation, writing - original draft, writing - review & editing. Helena M. Kondow-McConaghy: investigation, writing - original draft, writing - review & editing. Justin Yang: investigation, writing - original draft, writing - review & editing. Kerry DeMarco: investigation, writing - original draft, writing - review & editing. Pejman Naraghi-Arani: conceptualization, methodology, investigation. Luca Foschini: conceptualization, methodology, investigation, writing - original draft, writing - review & editing.
Data sharing statement
The datasets analyzed in this study are not publicly available but can be shared for scientific collaboration upon publication by contacting the corresponding author.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:AMM, EC, AS, ER, and LF are employees of Evidation Health, Inc., developers of the Evidation health and research platform.
Acknowledgements
This manuscript is not an endorsement of any technology or platform. The authors thank Patricia French of Left Lane Communications for editing and formatting assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.conctc.2023.101113.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Park T. Behind Covid-19 vaccine development. 2021. https://news.mit.edu/2021/behind-covid-19-vaccine-development-0518
- 2.Evidation health, Inc. 2022. https://www.evidation.com
- 3.Deering S., Grade M.M., Uppal J.K., Foschini L., Juusola J.L., Amdur A.M., Stepnowsky C.J. Accelerating research with technology: rapid recruitment for a large-scale web-based sleep study. J. Med. Internet Res. Protoc. 2019;8 doi: 10.2196/10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar S., Tran J.L., Lee W., Bradshaw B., Foschini L., Juusola J. Longitudinal data from activity trackers show that those with greater inconsistency in activity levels are more likely to develop more severe depression. Value Health. 2018;21:S191. http://www.valueinhealthjournal.com/article/S1098301518315857/pdf [Google Scholar]
- 5.Konty K.J., Bradshaw B., Ramirez E., Lee W.-N., Signorini A., Foschini L. Influenza surveillance using wearable mobile health devices. Online J. Public Health Inform. 2019;11:e249. doi: 10.5210/ojphi.v11i1.9758. [DOI] [Google Scholar]
- 6.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., Fierro C., Schwartz H., Neuzil K., Corey L., Gilbert P., Janes H., Follmann D., Marovich M., Mascola J., Polakowski L., Ledgerwood J., Graham B.S., Bennett H., Pajon R., Knightly C., Leav B., Deng W., Zhou H., Han S., Ivarsson M., Miller J., Zaks T., COVE Study Group Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/nejmoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. C4591001 clinical trial group, safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/nejmoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., Offergeld K., Scheper G., Taylor K.L., Robb M.L., Treanor J., Barouch D.H., Stoddard J., Ryser M.F., Marovich M.A., Neuzil K.M., Corey L., Cauwenberghs N., Tanner T., Hardt K., Ruiz-Guiñazú J., Le Gars M., Schuitemaker H., Van Hoof J., Struyf F., Douoguih M. ENSEMBLE study group, safety and efficacy of single-dose Ad26.COV2.S vaccine against covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/nejmoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro A., Marinsek N., Clay I., Bradshaw B., Ramirez E., Min J., Trister A., Wang Y., Althoff T., Foschini L. Characterizing COVID-19 and influenza illnesses in the real world via person-generated health data. Patterns. 2020;2 doi: 10.1016/j.patter.2020.100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.U.S. Census Bureau Age and sex composition in the United States: 2019. 2020. https://www.census.gov/data/tables/2019/demo/age-and-sex/2019-age-sex-composition.html Accessed.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.