Abstract
COVID-19 causes neurological damage, systemic inflammation, and immune cell abnormalities. COVID-19-induced neurological impairment may be caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which directly infects cells of the central nervous system (CNS) and exerts toxic effects. Furthermore, SARS-CoV-2 mutations occur constantly, and it is not well understood how the infectivity of the virus to cells of the CNS changes as the virus mutates. Few studies have examined whether the infectivity of cells of CNS - neural stem/progenitor cells (NS/PCs), neurons, astrocytes, and microglia - varies among SARS-CoV-2 mutant strains. In this study, therefore, we investigated whether SARS-CoV-2 mutations increase infectivity to CNS cells, including microglia. Since it was essential to demonstrate the infectivity of the virus to CNS cells in vitro using human cells, we generated cortical neurons, astrocytes, and microglia from human induced pluripotent stem cells (hiPSCs). We added pseudotyped lentiviruses of SARS-CoV-2 to each type of cells, and then we examined their infectivity. We prepared three pseudotyped lentiviruses expressing the S protein of the original strain (the first SARS-CoV-2 discovered in the world), the Delta variant, and the Omicron variant on their envelopes and analyzed differences of their ability to infect CNS cells. We also generated brain organoids and investigated the infectivity of each virus. The viruses did not infect cortical neurons, astrocytes, or NS/PCs, but microglia were infected by the original, Delta, and Omicron pseudotyped viruses. In addition, DPP4 and CD147, potential core receptors of SARS-CoV-2, were highly expressed in the infected microglia, while DPP4 expression was deficient in cortical neurons, astrocytes, and NS/PCs. Our results suggest that DPP4, which is also a receptor for Middle East respiratory syndrome-coronavirus (MERS-CoV), may play an essential role in the CNS. Our study is applicable to the validation of the infectivity of viruses that cause various infectious diseases in CNS cells, which are difficult to sample from humans.
Keywords: Astrocyte, Cerebral organoid, COVID-19, hiPSC, Microglia, Neuron, NS/PC, Pseudotyped lentivirus, SARS-CoV-2
1. Introduction
COVID-19 has been reported to cause central nervous system (CNS) damage (Mao et al., 2020; Kase and Okano, 2021), with clinical findings including encephalitis, necrotizing encephalopathy (Poyiadji et al., 2020) and cognitive dysfunction (Becker et al., 2021). The clinical features are diverse, including brain fog (Centers for Disease Control and Prevention) (https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-care/post-covid-conditions.html) in those with long COVID. Although systemic inflammation and abnormal immune cell dynamics caused by COVID-19 have been associated with such neurological damage (Hojyo et al., 2020), it is not yet fully understood whether SARS-CoV-2 can directly infect cells of the nervous system.
RNA viruses such as SARS-CoV-2 are constantly mutating, and the clinical features of COVID-19 change slightly to reflect these mutations; however, it is not known how the infectivity of mutant viruses to cells of the CNS is altered. To verify the infectivity of mutant viruses, it is crucial to establish an in vitro system for studying viral infectivity.
Since CNS cells are composed of multiple cell types, not just neurons, it is necessary to prepare various cell types to verify whether the virus is directly causing neurological damage. However, since these CNS cells cannot be sampled directly from the human brain, establishing this type of system using conventional methods is challenging.
Recently, methods have been developed to induce not only neurons but also other CNS cells from human induced pluripotent stem cells (hiPSCs), and it is now possible to create brain organoids, a 3D culture technique. Here, we used the induction method developed by our group to generate cortical neurons (Sato et al., 2021), astrocytes (Leventoux et al., 2020), microglia (Sonn et al., 2022), and brain organoids from hiPSCs. We examined the difference in the infectivity among the original strain of SARS-CoV-2 and the Delta and Omicron variants.
2. Results
2.1. SARS-CoV-2 hardly infects cortical neurons
The cells that make up the brain include neurons, astrocytes, oligodendrocytes, microglia, blood vessels, pericytes, etc. We first focused on neurons.
Various reports have indicated that SARS-CoV-2 can directly infect neurons, while others report that it cannot directly infect neurons (Meinhardt et al., 2021; Andrews et al., 2022; Crunfli et al., 2022). Therefore, we generated cortical neurons from hiPSCs (Okita et al., 2011) and investigated SARS-CoV-2 infectivity.
Since there are several legal barriers to directly handling SARS-CoV-2 in Japan, we prepared a pseudotyped lentivirus of SARS-CoV-2. The S protein of SARS-CoV-2 was expressed in the lentivirus envelope instead of the G protein of the vesicular stomatitis virus (VSV-G), and its ability to infect CNS cells was evaluated.
To further evaluate changes in the ability of mutant viruses to infect CNS cells, three pseudotyped lentiviruses were prepared based on the first SARS-CoV-2 strain reported in the world (original strain) (Wu et al., 2020), the so-called Delta variant (B.1.617.2 strain) (Cherian et al., 2021; Planas et al., 2021), and the Omicron variant (B.1.1.529 strain) (Schmidt et al., 2022). Each SARS-CoV-2 strain or variant S protein was expressed in the envelope of these lentiviruses instead of VSV-G. The fluorescent protein reporter EGFP following the EF1a promoter was used as an indicator of infection. In addition, our group previously described the generation of cerebral cortical neurons, not pan-neurons, from hiPSCs (Sato et al., 2021), and we took an advantage of it in this study (Supplementary Fig. 1).
Supplementary Fig. 1.
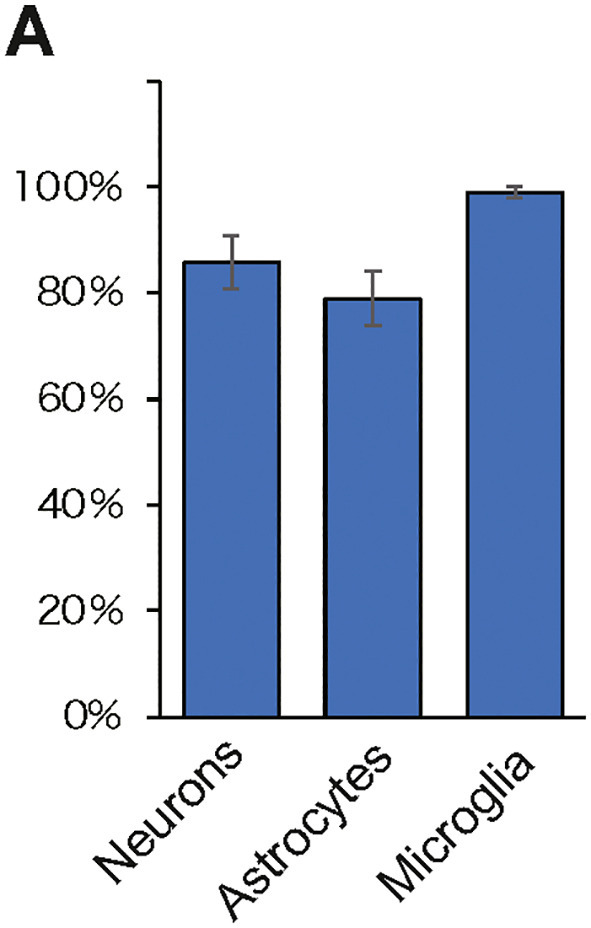
Induction efficiency of each cell induced from hiPSCs.
(A) Induction efficiency of neurons, astrocytes, and microglia induced from hiPSCs is shown as a percentage of 100.
Neurons: # of β-IIItubulin positive cells/Hoechst.
Astrocytes: # of GFAP positive cells /Hoechst.
Microglia: # of Iba1positive cells/Hoechst.
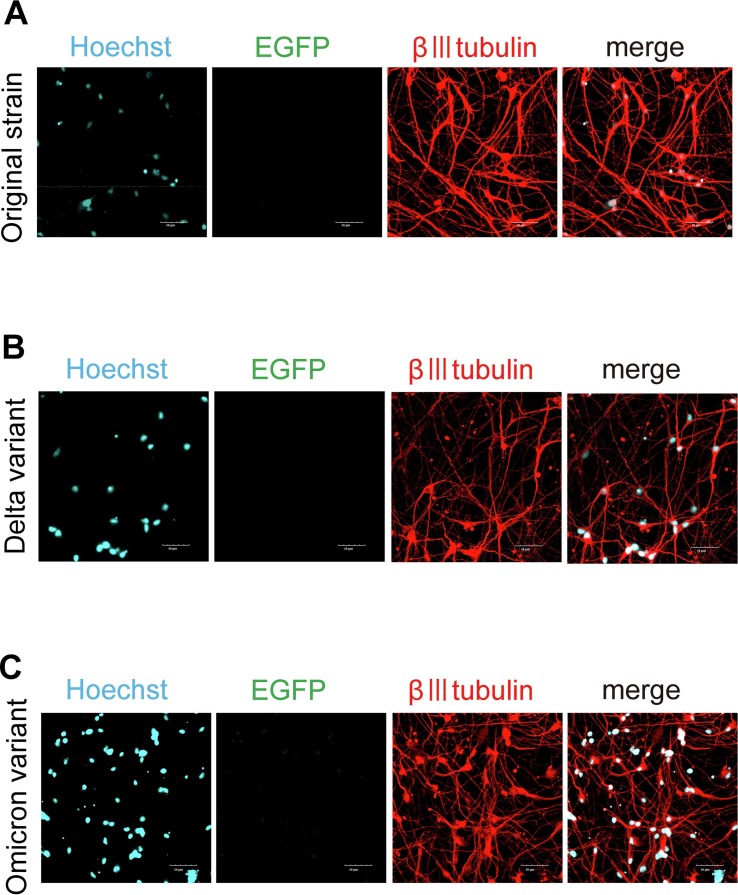
Neurons generated from hiPSCs were infected with these pseudotyped lentiviruses (MOI: 3). The medium was changed to virus-free medium the next day, and samples were fixed and immunocyto-stained after two days of culture (Fig. 1 ). No β-III-tubulin-positive neurons expressing EGFP were found in the samples (Figs. 1A-1C).
Fig. 1.
SARS-CoV-2 pseudotyped lentivirus did not infect β-III tubulin-positive neurons.
(A) Pseudotyped lentivirus of the original SARS-CoV-2 strain was added to hiPSC-derived neurons and stained with β-III tubulin (red), a neuronal marker. No expression of EGFP, a reporter protein that indicates the establishment of infection, was observed. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
(B) Pseudotyped lentivirus of the Delta variant was added to hiPSC-derived neurons and stained with β-III tubulin (red), a neuronal marker. No expression of EGFP, a reporter protein that indicates the establishment of infection, was observed. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
(C) Pseudotyped lentivirus of the Omicron variant was added to hiPSC-derived neurons and stained with β-III tubulin (red), a neuronal marker. No expression of EGFP, a reporter protein that indicates the establishment of infection, was observed. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
The original strain of SARS-CoV-2 and the Delta and Omicron variants had difficulty in infecting cortical neurons.
2.2. SARS-CoV-2 hardly infects astrocytes
Astrocytes act as supportive cells for neurons, and recently astrocytes have recently been recognized as as a vital role player in the healthy functioning of neuronal networks (Sofroniew, 2015). Some reports have linked astrocytes to disease (Kidana et al., 2018), so we examined the ability of each lentivirus to infect astrocytes.
Since it is difficult to collect astrocytes directly from the human brain, we induced and created astrocytes from hiPSCs for our experiments. Our group previously described the highly efficient generation of astrocytes from hiPSCs (Leventoux et al., 2020), and we used this technology (Supplementary Fig. 1). As in the neuron experiments, each pseudotyped lentivirus was added to astrocytes (MOI: 3), and the expression levels of the astrocytic markers GFAP and EGFP were checked after the samples were fixed. The experiment results showed no cells expressing EGFP in GFAP positive cells after the treatment of any pseudotyped lentivirus (Fig. 2A-2C).
Fig. 2.
Pseudotyped lentivirus of SARS-CoV-2 did not infect astrocytes.
(A) Although the pseudotyped lentivirus of the original SARS-CoV-2 strain was added, the reporter EGFP was not expressed in GFAP (red)-positive cells, an astrocyte marker. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
(B) Although pseudotyped lentivirus of the Delta variant was added, the reporter EGFP was not expressed in GFAP (red)-positive cells, an astrocyte marker. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
(C) Although pseudotyped lentivirus of the Omicron variant was added, the reporter EGFP was not expressed in GFAP (red)-positive cells, an astrocyte marker. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
2.3. SARS-CoV-2 hardly infects NS/PCs within cerebral organoids
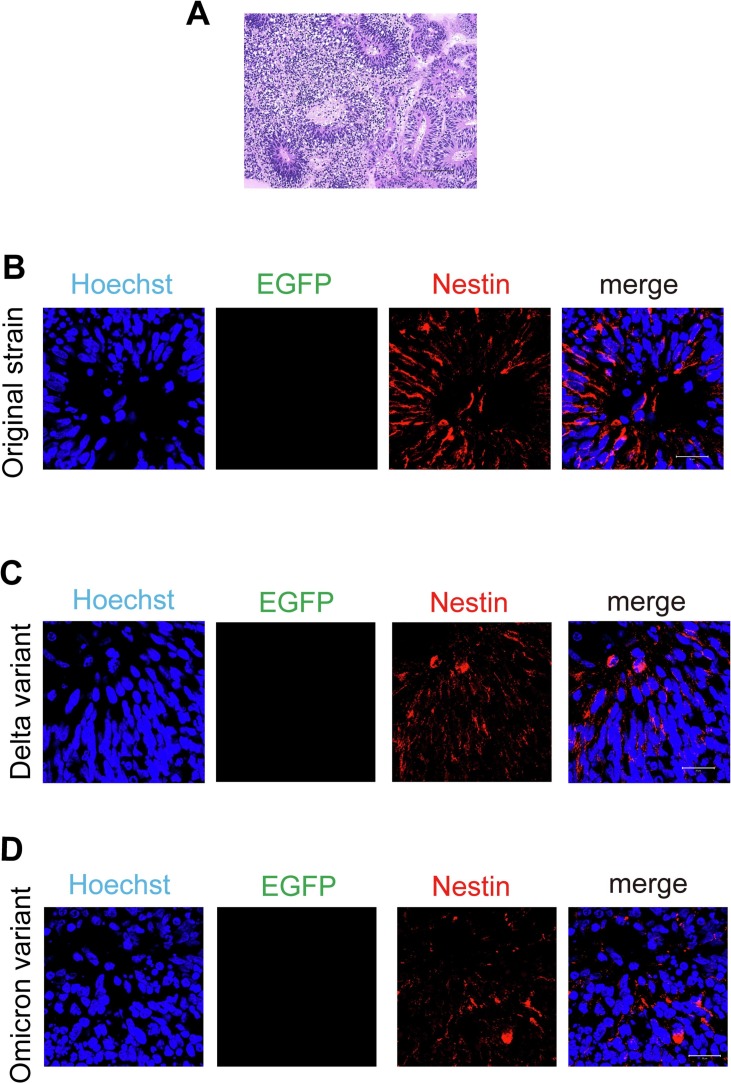
Next, cerebral organoids were prepared in 3D culture (Fig. 3A) to examine the infectivity of each variant of lentivirus to NS/PCs. Brain organoids were cultured for 30 days, and then paraffin sections were prepared and immunostained.
Fig. 3.
Pseudotyped lentivirus of SARS-CoV-2 did not infect NS/PCs in cerebral organoids.
(A) Cerebral organoids were prepared to examine the infectivity of pseudotyped lentivirus of SARS-CoV-2 to NS/PCs. A representative image of paraffin sections stained with hematoxylin-eosin. Scale bar: 100 μm.
(B) EGFP, a reporter of the pseudotyped lentivirus of the original strain of SARS-CoV-2, was not detected in nestin-positive cells (red), a marker for NS/PCs. Nuclei were counterstained with Hoechst (blue). Scale bar: 20 μm.
(C) EGFP, a reporter of the pseudotyped lentivirus of the Delta variant, was not detected in nestin-positive cells (red), a marker for NS/PCs. Nuclei were counterstained with Hoechst (blue). Scale bar: 20 μm.
(D) EGFP, a reporter of the pseudotyped lentivirus of the Omicron variant, was not detected in nestin-positive cells (red), a marker for NS/PCs. Nuclei were counterstained with Hoechst (blue). Scale bar: 20 μm.
Nestin (Kase et al., 2019) was used as a marker for NS/PCs to determine whether they were infected with each lentivirus, but EGFP was not expressed in nestin-positive cells (Fig. 3B-3D).
2.4. SARS-CoV-2 efficiently infects microglia
Finally, we investigated the ability of the virus to infect microglia. Microglia can also be generated from hiPSCs (Sonn et al., 2022) (Supplementary Fig. 1). Then, unlike in the experiments with neurons, astrocytes and NS/PCs, EGFP, the reporter of the pseudotyped lentiviruses, was expressed in Iba1-positive microglia. Almost all Iba1-positive cells were infected by the pseudotyped lentiviruses of the original strain, Delta variant, and Omicron variant, with no difference in infectivity among the mutant strains (Figs. 4A-4C).
Fig. 4.
Microglia derived from hiPSCs were infected with pseudotyped lentiviruses of SARS-CoV-2.
(A) Microglia derived from hiPSCs (Iba1-positive cells) were infected with a pseudotyped lentivirus of the original SARS-CoV-2 strain and expressed the reporter EGFP. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
(B) Microglia derived from hiPSCs (Iba1-positive cells) (red) were infected with a pseudotyped lentivirus of the Delta variant and expressed the reporter EGFP. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
(C) Microglia derived from hiPSCs (Iba1-positive cells) were infected with a pseudotyped lentivirus of the Omicron variant and expressed the reporter EGFP. Nuclei were counterstained with Hoechst (blue). Scale bar: 50 μm.
2.5. DPP4 may play a pivotal role as a receptor for SARS-COV-2 in CNS cells
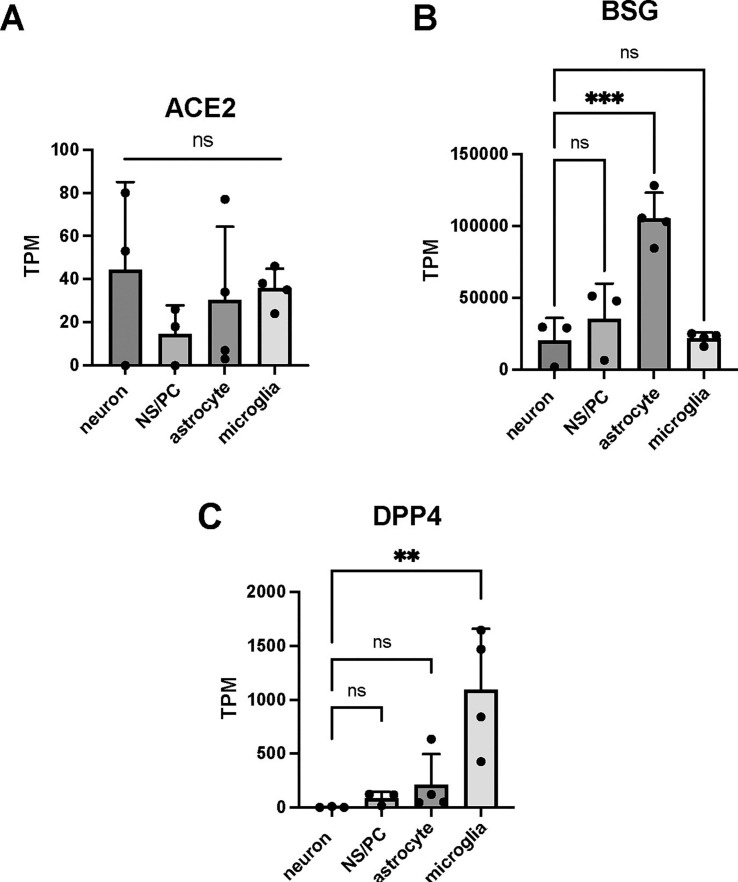
Then, we examined why each virus was able to infect only microglia. Andrews et al. examined the infectivity of SARS-CoV-2 in cerebral organoids and reported that although SARS-CoV-2 does not easily infect neurons, the virus does infect astrocytes with high expression of dipeptidyl peptidase 4 (DPP4) and CD147 (Andrews et al., 2022). Furthermore, although angiotensin converting enzyme 2 (ACE2) is not entirely absent in the CNS, its expression levels are low, leading the researchers to conclude that DPP4 and CD147 are the core receptors.
In our experimental system, we examined the expression of ACE2, DPP4, and CD147 at the transcriptome level and found that ACE2 was expressed in neurons, NS/PCs, astrocytes, and microglia at a very low level (Fig. 5A). Basigin (BSG), the gene encoding CD147, was expressed at high levels in all these types of cells, with average TPM values exceeding approximately 20,000 (Fig. 5B). However, DPP4 was significantly higher in microglia than in other cells. Its expression level was comparatively lower in nonmicroglial cells (Fig. 5C). These results suggest that DPP4 is an essential factor in whether SARS-CoV-2 infection is viable in cells of the CNS.
Fig. 5.
ACE2 expression is low in neurons, NS/PCs, astrocytes, and microglia derived from hiPSCs, and DPP4 may play an important role as a core receptor.
Bulk RNA-seq data on the expression level (TPM) of ACE2 (A), BSG (B) and DPP4 (C) in neurons, NP/SCs, astrocytes and microglia.
TPM of DPP4 in neurons: 0, 1, 8.
ns, not significant; ** p < 0.01; *** p < 0.001. All data are expressed as the mean ± SD. SD, standard deviation.
3. Discussion
COVID-19 causes CNS damage, and it is crucial to verify whether SARS-CoV-2 directly infects CNS cells and exerts toxicity to develop a therapeutic strategy.
Many studies have been performed on autopsy brains, 2D cultures, and organoids, and many of these studies have reported negative results regarding the direct infectivity of SARS-CoV-2 to neurons. Our current results support this conclusion. The brain organoids used in the paper by Andrew et al. (Andrews et al., 2022) did not infect NS/PCs or neurons, and this was also true for the neurons and brain organoids we generated. However, it was not well understood whether mutations in the viral S protein alter its ability to infect CNS cells, including microglia.
As we have previously reported, the receptor for SARS-CoV-2, ACE2, is expressed in neurons, albeit to a low degree and in varying amounts in different brain regions (Kase and Okano, 2020). Low ACE2 expression is also found in astrocytes and even microglia. Andrews et al. demonstrated DDP4 and CD147 as candidate receptors for SARS-CoV-2 in the CNS (Andrews et al., 2022). Considering the high expression of DPP4 and CD147 in our microglia and the inability of pseudotyped lentiviruses to infect neurons, astrocytes, and NS/PCs with low DPP4 expression, it is likely that DPP4, which functions as a receptor for MERS-CoV, also plays a crucial role as a receptor for SARS-CoV-2 in the CNS.
Microglia, the only immunocompetent cells in the brain parenchyma, have been reported to be abnormally activated by COVID-19 (Kishimoto-Urata et al., 2022). They showed that SARS-CoV-2 causes abnormal microglial activation in hamsters, but it was not known whether it infects microglia. We have shown that SARS-CoV-2 is cable of directly infecting human microglia.
Nevertheless, if SARS-CoV-2 can directly infect microglia, it can be expected to cause abnormal microglial activation more directly. Jeong et al. (2022) reported on a microglial cell line and showed that SARS-CoV-2 could infect microglia and even cause cell death. In our study, we demonstrated that even SARS-CoV-2 variants (Delta and Omicron variants) can infect microglia and that infection efficiency is unchanged.
Although it is generally believed that viral diseases are spread by mutants that are more infectious and less virulent to humans, this experiment suggests thatSARS-CoV-2 can infect microglia since its emergence. This may be why many COVID-19-related CNS disorders were reported at the time of the emergence of SARS-CoV-2.
3.1. Limitations of the study
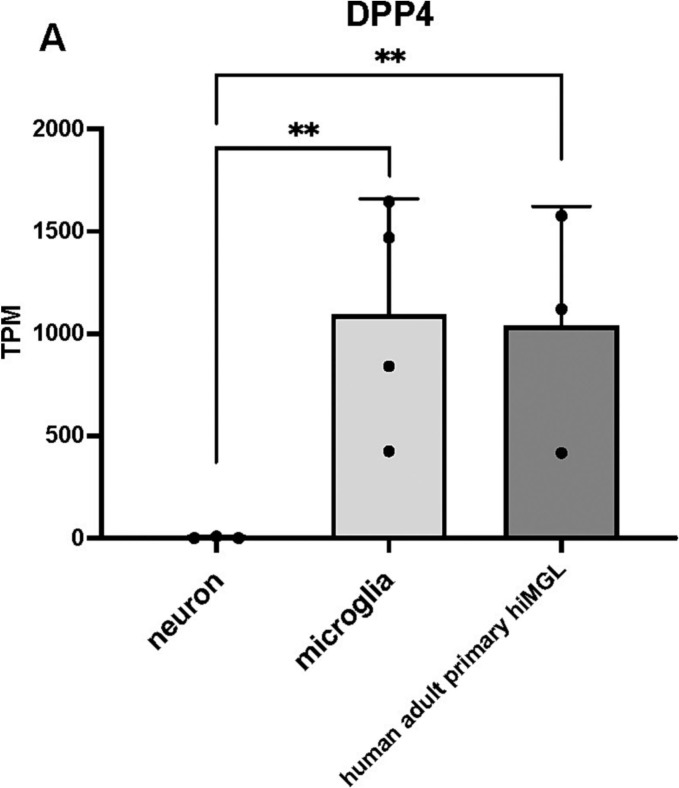
Using cortical slice cultures and brain organoids, Andrews et al. (Andrews et al., 2022) reported that astrocytes are the main target of SARS-CoV-2 infection. Nevertheless, our generated astrocytes were not infected by the pseudotyped lentiviruses and had low DPP4 expression. The reason for this difference may be the difference in the method used to generate the astrocytes. Because the technique we used to induce astrocytes from hiPSCs without using transcription factors may not fully reflect all the characteristics of astrocytes in the human brain. Since astrocytes are generated without the introduction of transcription factors in our method, it is possible that the expression of DPP4 and other factors may not correspond to the actual expression of astrocytes in the human brain. On the other hand, microglia were generated by introducing transcription factors and we compared our microglia with RNA-seq data from adult human microglia; DPP4 expression was comparable (Fig. 6A).
Fig. 6.
DPP4 was low in neurons but highly expressed by microglia.
(A) Bulk RNA-seq data indicated that DPP4 was expressed in human adult primary microglia, consistent with our microglia derived from hiPSCs. ns, not significant; ** p < 0.01; *** p < 0.001. All data are expressed as the mean ± SD. SD, standard deviation.
In addition, Andrew et al. (Andrews et al., 2022) used the SARS-CoV2 virus itself, whereas we used a pseudotyped lentivirus, which may explain the difference in experimental results. Since pseudotyped lentiviruses were used in our experiments, we were able to verify the change in infection efficiency with mutation but not the change in virulence.
4. Materials and methods
4.1. Culture of undifferentiated hiPSCs
The hiPSC lines (CiRA; Kyoto University) were cultured with mitomycin C-treated SNL murine fibroblast feeder cells in standard human embryonic stem cell (hESC) medium (Reprocell, RCHEMD001) containing 0.5% penicillin–streptomycin (Nacalai Tesque, 26,253–84) and 4 ng/mL fibroblast growth factor 2 (FGF-2) (PeproTech, 100-18B) in an atmosphere containing 5% CO2. We always make sure that the morphological characteristics of the colony are maintained.
4.2. Formation of neurons
hiPSCs (201B7 line) were pretreated for 6 days with 3 μM SB431542 (Tocris, 301,836–41-9) and 150 nM LDN193189 (StemRD, 1,062,368–24-4). The cells were then dissociated and seeded at a density of 1 × 105 cells per milliliter in ultralow-attachment culture dishes (Corning) in neuronal induction medium consisting of medium hormone mix (MHM) supplemented with 2% B27 supplement without vitamin A (Thermo Fisher, 17,504–044), 20 ng/mL FGF-2, 10 μM Y27632 (Nacalai Tesque, 08945–71), 1 μM retinoic acid (RA; Sigma, R2625-1G), 3 μM CHIR 99021 (Reprocell, 04–0004) and 10 μM SB431542 (Calbiochem, 301,836–41-9) in a hypoxic and humidified atmosphere (4% O2, 5% CO2) for 6 days. The formed neurospheres were passaged by dissociation into single cells and then cultured in slightly modified neuronal induction medium, MHM supplemented with 2% B27 without vitamin A, 20 ng/mL FGF-2, 10 μM Y27632, and 1 μM RA for 6 days under 4% O2 (hypoxic) conditions.
4.3. Formation of astrocytes
In this study, the previously reported astrocyte differentiation protocol from feeder-free hiPSCs was used (Leventoux et al., 2020). On differentiation day ∼0, hiPSCs were cultured with mitomycin-C-treated SNL murine fibroblast feeder cells on 0.1% gelatin-coated culture dishes. hiPSCs were cultured using human ES medium, which contained KnockOut™ serum replacement (final 20%, KSR, Gibco, Japan), 200 mM l-glutamine (final 1%, Gibco), nonessential amino acids (final 0.8%, NEAA, Nacalai, Japan), 2-mercaptoethanol (final 0.1 mM, Sigma, USA), fibroblast growth factor 2 (final 4 ng/mL, FGF2, PeproTech, Japan), and penicillin–streptomycin at 37 °C in a humidified atmosphere of 3% CO2. On day 0 of astrocyte differentiation, iPSCs were removed from feeder cells with a dissociation solution (0.25% trypsin, 100 μg/mL collagenase IV (Invitrogen, USA), 1 mM CaCl2, and 20% KSR) and cultured in FGF2-free human ES medium with 10 μM Y-27632 (Nacalai, Japan) to form embryoid bodies (EBs) at 37 °C in a humidified atmosphere of 3% CO2. On days 1–3 of differentiation, to enhance the differentiation to the neural lineage, the medium was changed to EB medium (DMEM/F12 containing 5% KSR, 1% l-glutamine, 0.8% NEAA, 0.1 mM 2-mercaptoethanol and penicillin–streptomycin) with 3 μM dorsomorphin (Sigma), 3 μM SB431542 (Sigma), and 3 μM CHIR99021 (Focus Biomolecules, USA). On day 4 of differentiation, the medium was changed to EB medium with only 1 μM retinoic acid (Sigma). On days 7, 10, and 13 of differentiation, the medium was changed to EB medium with 1 μM retinoic acid and 1 μM purmorphamine (Cayman, Japan), and the culture was maintained until day 16. On day 16 of differentiation, the EBs were dissociated into single cells using TrypLE Select (Thermo Fisher Scientific, USA), and the dissociated cells were cultured as a cell suspension with 1–4 × 105 cells/mL cell density in neurosphere medium, which contained media hormone mix (MHM) supplemented with B-27 (final 2%, Nacalai), NEAA (final 0.8%), purmorphamine (final 1 μM), FGF2 (final 20 ng/mL), and epidermal growth factor (final 10 ng/mL, EGF, PeproTech, USA) in a humidified atmosphere of 5% CO2. On days 17–31 of differentiation, the supplemented neurosphere medium was changed every 4–6 days to form spheres until day 32. On differentiation day 32, the primary neurospheres were dissociated as described above and cultured with the same cell density in supplemented neurosphere medium without purmorphamine. On days 33–47 of differentiation, the culture was maintained until day 48. On day 48 of differentiation, the secondary neurospheres were dissociated in the same way as on days 16 and 32 and plated at a density of 4–10 × 105 cells/well in 6-well plates coated with 0.5% growth-factor-reduced Matrigel (Corning, USA) with iPSC-derived astrocyte differentiation medium, which consisted of MHM medium containing B-27 (final 2%, Nacalai), NEAA (final 0.8%), brain-derived neurotrophic factor (final 10 ng/mL, BDNF, R&D, USA), and glial cell-derived neurotrophic factor (final 10 ng/mL, GDNF, Alamone, Israel) in a humidified atmosphere of 5% CO2. On days 49–76 of differentiation, the medium was changed to fresh medium every 4–7 days until day 77. On differentiation day 77, the iPSC-derived astrocytes were dissociated using Accutase (Nacalai), and the dissociated cells were plated at a density of 1 × 106 cells/dish in 100-mm dishes coated with 0.05% Matrigel in iPSC-derived astrocyte differentiation medium. On days 78–97 of differentiation, the plated cells were dissociated and cultured as described above once a week (at least twice during days 78–97). On differentiation day 98, iPSC-derived astrocytes were dissociated, used for experiments and stored in serum-free CellBanker2 (Zenoaq, Japan) at −180 °C.
4.4. Formation of microglia
Details of microglial differentiation from hiPSCs are summarized in Sonn et al., 2022. Briefly, GeneJuice (Sigma–Aldrich, 70,967) and Opti-MEM medium (Gibco™, 31,985,070) were used to establish a doxycycline-inducible PU.1-overexpression clone of hiPSCs. Then, the generation of microglial cells from iPSCs was divided into two stages. First, hematopoietic progenitor cells were generated from hiPSCs (hiHPCs; ∼day 18) in modified StemPro™-34 SFM (1×) (Gibco™, 10,639,011) medium supplied with multiple cytokines and chemicals (BMP4 (PEPROTECH), CHIR99021 (Focus Biomolecules), VEGF (Thermo Fisher Scientific), FGF2 (PEPROTECH), SCF (PEPROTECH), IL-3 (PEPROTECH), IL-6 (PEPROTECH), and IWR-1e (Thermo Fisher Scientific). Doxycycline (1 μg/mL) was added to the medium during days 6 ∼ 18.
Second, microglia-like cells were differentiated from hiHPCs (day 19∼) in modified DMEM/F12 (1:1, Gibco™) supplied with a cocktail of five cytokines (100 ng/mL IL-34, 50 ng/mL TGFβ1, 25 ng/mL M-CSF, 100 ng/mL CD200, and 100 ng/mL CX3CL1, all from PEPROTECH). Induced microglia were used for analysis on day 21.
4.5. Formation of cerebral organoids
Cerebral organoids were generated based on the protocol of the STEMdiff Cerebral Organoid Kit (STEMCELL Technologies, ST-08570). On day 0, hiPSCs (414C2) were suspended at a density of 9.0 × 104/mL in EB Formation Medium (with Y27632 (Nacalai Tesque, 08945–71)). A total of 100 μL of cell suspension was seeded in a 96-well ultralow-attachment plate (9000 cells/well) and incubated in a humidified atmosphere of 5% CO2 at 37 °C. On days 2 and 4, 100 μL of EB Formation Medium (without Y27632) was added. On day 5, the solution was replaced with induction medium, and organoids were cultured for 48 h. From day 7, organoids were embedded in Matrigel Matrix (Cornig, 356,230). On day 7, the solution was replaced with Expansion Medium, and the organoids were incubated for 72 h. From day 10, plates were shaken on an orbital shaker at 70 rpm, and organoids were cultured in maturation medium. The medium was changed every 4 days until day 30.
4.6. Pseudotyped lentivirus production
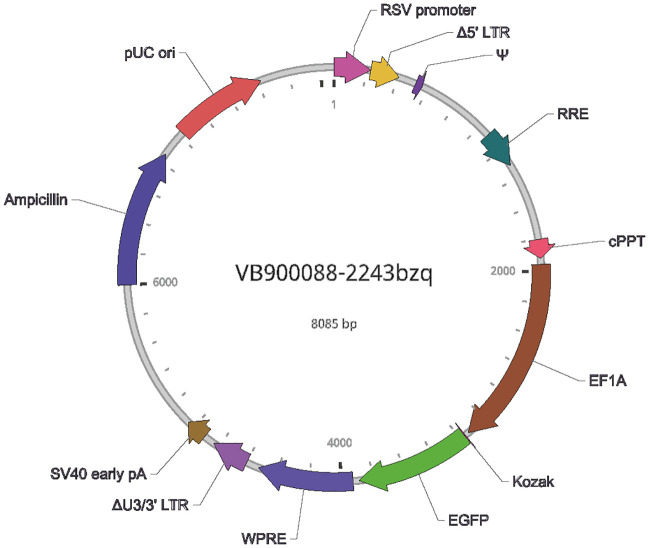
SARS-CoV-2_S pseudotyped lentivirus, SARS-CoV-2_S (B.1.617.2 var./Delta) pseudotyped lentivirus, and SARS-CoV-2_S (B.1.1.529, Omicron) pseudotyped lentivirus were synthesized by VectorBuilder. EGFP was inserted under the EF1a promoter as a fluorescent reporter (pLV-EF1a-EGFP). Instead of the VSV-G envelope, the S protein of the coronavirus is expressed, a virus that can mimic the manner in which SARS-Co-2 infects cells. It does not transduce the S-protein coding gene into the host cells. See Supplementary Fig. 2 for the vector map.
Supplementary Fig. 2.
Vector map of pseudotyped lentivirus.
4.7. Immunostaining
For immunocytochemistry, samples were plated onto poly-L-ornithine/fibronectin-coated chamber slide glasses or 12-well plates and fixed in 4% PFA/PBS for 30 min at room temperature. The samples were rinsed with PBS three times and permeabilized with 0.3% Triton X-100/PBS for 5 min at room temperature. After blocking with Blocking One (Nacalai Tesque, 03953–95) for 15 min at room temperature, the slides were incubated at 4 °C overnight with the following antibodies: mouse anti-β-III tubulin (Sigma–Aldrich, T8660-2ML; 1:500), mouse anti-nestin (Merck Millipore, MAB5326; 1:500), anti-GFAP, and rabbit anti-Iba1 (FUJIFILM, 019–19,741; 1:500). After washing three times with PBS, the samples were incubated with secondary antibodies conjugated to Alexa 488 (Thermo Fisher Scientific, A-11034), Alexa 555 (Thermo Fisher Scientific, A-21424; 1:500), Alexa 555 (Thermo Fisher Scientific, A-21434; 1:500) or Alexa 555 (Thermo Fisher Scientific, A-21428; 1:500) for 60 min at room temperature and then subjected to nuclear counterstaining with Hoechst 33258 (Sigma–Aldrich, B2883; 10 μg/mL). The samples were analyzed with a FLUOVIEW FV3000 (OLYMPUS) and LSM700 (Carl Zeiss).
At least 100 cells were observed, and this was done for n = 3. All microglia were infected with pseudotyped lentivirus, but no other neurons, astrocytes, or NS/PCs were infected at all, so no quantitative graphs were made.
4.8. Bulk mRNA-seq analysis
For mRNA-seq analysis (in Fig. 5, Fig. 6), we included data from previous studies deposited in the GEO (https://www.ncbi.nlm.nih.gov/geo/) and DDBJ (https://www.ddbj.nig.ac.jp/) databases as follows: neurons and NS/PCs in GSE196144, astrocytes in GSE161024, and microglia in GSE178284. mRNA-seq (FASTQ file format) data were quality checked, and low-quality reads (score < 30), adapter sequences, and overrepresented sequences such as those with a poly-A chain were trimmed using Trim Galore! (ver.0.4.0). The remaining reads were mapped to the Homo sapiens (hg19) genome using HISAT2 (ver. 2.2.2.0) (Kim et al., 2019), and the output file (BAM file format) was summarized using StringTie (ver. 2.1.5).
4.9. Statistical analysis
For each statistical analysis, three or four independent experiments were carried out. Statistical significance was determined with one-way ANOVA followed by the Tukey–Kramer test. Throughout the study, p values <0.05 were considered to indicate statistical significance. The following significance thresholds were used throughout the study: ** p < 0.01 and *** p < 0.001. The values in the bar and line graphs represent the means ± SDs in Fig. 5, Fig. 6, and ns means not significant. GraphPad Prism Version 9.4.1 (MDF) was used for the analysis.
The following are the supplementary data related to this article.
Funding
This work was supported by grants from SENSHIN Medical Research Foundation (to YK) and from the Mitsubishi Foundation (to HO).
Author contributions
YK obtained the funding, designed this research, investigated all experiments, analyzed all results, and wrote the original draft. IS created the hiPSC-derived microglia, analyzed the RNA-seq data, and wrote the original draft. MG created the cerebral organoids, performed immunostaining experiments, and wrote the original draft. RM created the hiPSC-derived astrocytes and wrote the original draft, and TS created the hiPSC-derived neurons. HO obtained the funding, edited the paper, and supervised the project. All authors approved the final manuscript.
Consent for publication
All authors have consented to the submission of the manuscript to the journal.
Ethics approval
Ethical approval of the present study was obtained from the Ethics Committee of Keio University School of Medicine (Approval Number: 20130146).
Declaration of Competing Interest
HO is a compensated scientific consultant for San Bio Co., Ltd.; RMiC; and K Pharma, Inc. The other authors have no potential conflicts of interest to declare.
Acknowledgments
We thank Professor Shinya Yamanaka at the Center for iPS Cell Research and Application (CiRA), Kyoto University, for providing us with human iPSC lines.
This research was conducted as a part of the Keio Donner Project against COVID-19.
Data availability
GEO (https://www.ncbi.nlm.nih.gov/geo/) and DDBJ (https://www.ddbj.nig.ac.jp/) databases as follows: neurons and NS/PCs in GSE196144, astrocytes in GSE161024, and microglia in GSE178284. Further information and requests for resources and reagents should be directed to and will be fulfilled by Hideyuki Okano (hidokano@keio.jp).
References
- Andrews M.G., Mukhtar T., Eze U.C., et al. Tropism of SARS-CoV-2 for human cortical astrocytes. Proc. Natl. Acad. Sci. U. S. A. 2022;119(30) doi: 10.1073/pnas.2122236119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.H., Lin J.J., Doernberg M., et al. Assessment of cognitive function in patients after COVID-19 infection. JAMA Netw. Open. 2021;4(10) doi: 10.1001/jamanetworkopen.2021.30645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., et al. SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. Published 2021 Jul 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunfli F., Carregari V.C., Veras F.P., et al. Morphological, cellular, and molecular basis of brain infection in COVID-19 patients. Proc. Natl. Acad. Sci. U. S. A. 2022;119(35) doi: 10.1073/pnas.2200960119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojyo S., Uchida M., Tanaka K., et al. How COVID-19 induces cytokine storm with high mortality. Inflamm Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong G.U., Lyu J., Kim K.D., et al. SARS-CoV-2 infection of microglia elicits Proinflammatory activation and apoptotic cell death. Microbiol. Spectr. 2022;10(3) doi: 10.1128/spectrum.01091-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase Y., Okano H. Expression of ACE2 and a viral virulence-regulating factor CCN family member 1 in human iPSC-derived neural cells: implications for COVID-19-related CNS disorders. Inflamm. Regen. 2020;40:32. doi: 10.1186/s41232-020-00143-6. Published 2020 Sep 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase Y., Okano H. Neurological pathogenesis of SARS-CoV-2 (COVID-19): from virological features to clinical symptoms. Inflamm Regen. 2021;41(1):15. doi: 10.1186/s41232-021-00165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kase Y., Otsu K., Shimazaki T., Okano H. Involvement of p38 in age-related decline in adult neurogenesis via modulation of Wnt signaling. Stem Cell Rep. 2019;12(6):1313–1328. doi: 10.1016/j.stemcr.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidana K., Tatebe T., Ito K., et al. Loss of kallikrein-related peptidase 7 exacerbates amyloid pathology in Alzheimer’s disease model mice. EMBO Mol. Med. 2018;10(3) doi: 10.15252/emmm.201708184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto-Urata M., Urata S., Kagoya R., et al. Prolonged and extended impacts of SARS-CoV-2 on the olfactory neurocircuit. Sci. Rep. 2022;12(1):5728. doi: 10.1038/s41598-022-09731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventoux N., Morimoto S., Imaizumi K., et al. Human astrocytes model derived from induced pluripotent stem cells. Cells. 2020;9(12):2680. doi: 10.3390/cells9122680. Published 2020 Dec 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., et al. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China [published online ahead of print, 2020 Apr 10] JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. e201127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt J., Radke J., Dittmayer C., et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021;24(2):168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K.I., et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Poyiadji N., Shahin G., Noujaim D., Stone M., Patel S., Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: CT and MRI features [published online ahead of print, 2020 Mar 31] Radiology. 2020:201187. doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Imaizumi K., Watanabe H., Ishikawa M., Okano H. Generation of region-specific and high-purity neurons from human feeder-free iPSCs. Neurosci. Lett. 2021;746:135676. doi: 10.1016/j.neulet.2021.135676. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Muecksch F., Weisblum Y., et al. Plasma neutralization of the SARS-CoV-2 omicron variant. N. Engl. J. Med. 2022;386(6):599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M.V. Astrocyte barriers to neurotoxic inflammation [published correction appears in Nat Rev Neurosci. 2015 Jun;16(6):372] Nat. Rev. Neurosci. 2015;16(5):249–263. doi: 10.1038/nrn3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonn I., Honda-Ozaki F., Yoshimatsu S., Morimoto S., Watanabe H., Okano H. Single transcription factor efficiently leads human induced pluripotent stem cells to functional microglia. Inflamm. Regen. 2022;42(1):20. doi: 10.1186/s41232-022-00201-1. Published 2022 Jul 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China [published correction appears in nature. 2020 Apr;580(7803):E7] Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GEO (https://www.ncbi.nlm.nih.gov/geo/) and DDBJ (https://www.ddbj.nig.ac.jp/) databases as follows: neurons and NS/PCs in GSE196144, astrocytes in GSE161024, and microglia in GSE178284. Further information and requests for resources and reagents should be directed to and will be fulfilled by Hideyuki Okano (hidokano@keio.jp).