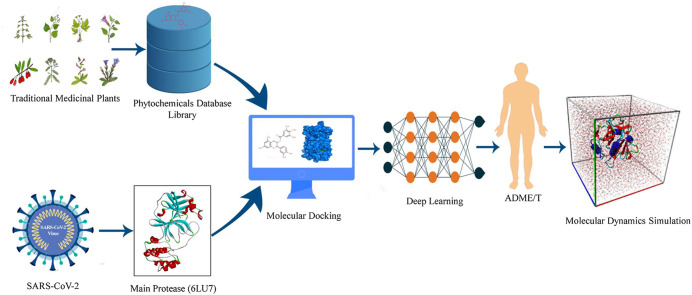
Abstract
Highly transmissive and rapidly evolving Coronavirus disease-2019 (COVID-19), a viral disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), triggered a global pandemic, which is one of the most researched viruses in the academia. Effective drugs to treat people with COVID-19 have yet to be developed to reduce mortality and transmission. Studies on the SARS-CoV-2 virus identified that its main protease (Mpro) might be a potential therapeutic target for drug development, as this enzyme plays a key role in viral replication. In search of potential inhibitors of Mpro, we developed a phytochemical library consisting of 2431 phytochemicals from 104 Korean medicinal plants that exhibited medicinal and antioxidant properties. The library was screened by molecular docking, followed by revalidation by re-screening with a deep learning method. Recurrent Neural Networks (RNN) computing system was used to develop an inhibitory predictive model using SARS coronavirus Mpro dataset. It was deployed to screen the top 12 compounds based on their docked binding affinity that ranged from −8.0 to −8.9 kcal/mol. The top two lead compounds, Catechin gallate and Quercetin 3-O-malonylglucoside, were selected depending on inhibitory potency against Mpro. Interactions with the target protein active sites, including His41, Met49, Cys145, Met165, and Thr190 were also examined. Molecular dynamics simulation was performed to analyze root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (RG), solvent accessible surface area (SASA), and number of hydrogen bonds. Results confirmed the inflexible nature of the docked complexes. Absorption, distribution, metabolism, excretion, and toxicity (ADMET), as well as bioactivity prediction confirmed the pharmaceutical activities of the lead compound. Findings of this research might help scientists to optimize compatible drugs for the treatment of COVID-19 patients.
Keywords: Deep learning, Catechin gallate, Main protease, Molecular docking, SARS-CoV-2
Graphical abstract
1. Introduction
The COVID-19 pandemic, the extremely contagious viral infection induced by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been posing serious threats to the world in a multitude of ways. As of February 2023, there have been 677.74 million confirmed cases of COVID-19 along with 6.78 million deaths, reported by the World Health Organization (WHO). Although significant advancements in clinical research have improved our knowledge of SARS-CoV-2 and COVID-19 management, there is growing worried about how to prevent the dissemination of this virus and its mutations. SARS-CoV-2 is still causing havoc around the globe and the advent of mutant variations is primarily to blame for the second or third wave of breakouts of this viral illness that are currently affecting more than 230 countries [1]. SARS-CoV-2 is a new beta coronavirus that belongs to the identical subgenus (SARS-CoV, 2002) and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV, 2013), and their previous mortality rates were up to 10% and 35%, respectively [2]. Throughout this pandemic, many SARS-CoV-2 variants have been identified among which merely a few are recognized as variants of concern by the WHO due to their effect on the world's public health such as Alpha (B.1.1.7) in the United Kingdom, Beta (B.1.351) in South Africa, Gamma (P.1) in Brazil, Delta (B.1.617.2) in India and Omicron (B.1.1.529) in South Africa [1].
SARS-CoV-2 is a single-stranded positive-sense RNA virus containing ∼30,000 nucleotides and 11 protein-coding genes, with 12 expressed proteins [[3], [4], [5]]. Among these few encoded proteins, is the Mpro, a cysteine protease that mediates the maturation cleavage of polyproteins at the time of viral replication [6,7]. The Mpro is a homodimer that consists of three domains (domain I, II, and III), however, is situated in the region adjacent to domains I and II and the protomers. They play a significant role in the formation of the substrate-binding site and bind to each other through their N-terminal residues 1 through 7, which are positioned between domains II and III [[8], [9], [10], [11], [12]]. The Mpro offers a promising target for the development of broad-spectrum anti-coronaviral therapeutic agents due to its highly conserved three-dimensional structure [13].
All coronaviruses share the Mpro protein [14], which is essential for the maturation of both the Mpro itself and other significant polyproteins [7]. Additionally, it plays a crucial role in how viruses enter host cells; when this enzyme is inhibited, the viral entrance and subsequent infection are halted [15]. Moreover, it also plays a crucial role in the proteolytic release of enzymes necessary for viral replication, such as nsp 13, which acted as both an RNA helicase and an NTPase [16,17]. These crucial roles of the viral protease enzyme, Mpro make them an intriguing therapeutic target for reducing illnesses associated with coronaviruses [5,16]. Mpro is highly conserved among various coronaviruses and due to its relatively conserved nature, structurally tailored broad-spectrum medications substantially block the Mpro of coronavirus [18,19]. Similar to how SARS-CoV-2 Mpro has garnered considerable interest in creating medications to combat the existing COVID-19 pandemic.
Structure-based drug design strategies became widespread to explicate antiviral activity through active constituents present in traditional medicinal plants. Several natural products have been identified as adjuvant therapy for COVID-19 treatment, direct antiviral therapy [20], as well as immune enhancer [21]. Studies both in silico and in vitro/in vivo showed that numerous types of inhibitors effectively bound to and inhibited SARS-CoV-2 Mpro [22]. Several studies demonstrated that some potential inhibitors such as carmofour, ebselen, cinanserin, famotidine, nelfinavir, lopinavir, and ritonavir bind with SARS-CoV-2 Mpro and inhibit viral replication, thereby reducing viral activity through the inhibition of Mpro [[23], [24], [25], [26]]. These inhibitors are unlikely to have any adverse side effects or toxicity because humans do not have any proteases with a similar cleavage specificity [[27], [28], [29]]. Using molecular dynamics simulations, the binding characteristics of the Delta and Omicron (BA.1) variants were investigated at the interface of the spike protein receptor binding domain and human angiotensin-converting enzyme-2 ectodomain and compared the molecular modeling systems with others [30].
In addition, a number of commonly used medications, such as valrubicin (antitumor), colistin (antibiotic), bepotastine (prescribed for rhinitis), icatibant (used to treat hereditary angioedema), perphenazine (antipsychotic), and caspofungin (antifungal), also bind to the Mpro even though they are more tolerant to mutation than lopinavir/ritonavir, suggesting potential drug candidates [26]. In another investigation, the FDA-approved antiviral medications tipranavir, lopinavir-ritonavir, and raltegravir were found to bind to the SARS-CoV-2 Mpro active site with robust, persistent, and versatile affinity [31]. Therefore, the replication process of the virus can be inhibited by searching for a potential inhibitor against SARS-CoV-2 Mpro.
Drug development activities are achieved efficiently in the burgeoning field of computational modeling. For successful binding to the SARS-CoV-2 Mpro active site, researchers have examined new and well-known antiviral substances such as vaniprevir, sovaprevir, boceprevir, glecaprevir, simeprevir, danoprevir, paritaprevir, and grazoprevir those firmly adhere to SARS-CoV-2 Mpro [32]. Medicinal plants are renowned as complementary approaches in which antiviral natural products have been established as safe and effective drugs [33]. Furthermore, among other things, the best medicine can be predicted with the help of computational drug design tools. Even drug development depends on safety measures regarding toxicity in the human body through a series of clinical trials, however, computer-based toxicity detection is also facilitated as it can be done before development. Therefore, a number of studies focused on the in-silico design of effective medications targeting SARS-CoV-2 Mpro is continuously rising. However, given the potential drawbacks of passing clinical studies, the therapeutic efficacy of these desired medications is debatable. In this study, computer-aided drug design is used for its more time and cost-efficient way of developing novel target drug candidates for inhibiting SARS-CoV-2 Mpro activity, in turn, the severity and mortality due to SARS-CoV-2 will decrease.
2. Materials and methods
2.1. Retrieval and protein preparation
The three-dimensional crystal structure of SARS-CoV-2 Mpro with N3 inhibitor (PDB ID: 6LU7) was retrieved in pdb format from the Protein Data Bank. Then the protein structure was prepared by removing all heteroatoms and water molecules using PyMol and Discovery Studio client. The clean protein structure was prepared by optimizing important factors like missing hydrogen bonds, side-chain geometry, and improper bond order were energy minimized using the GROMOS 43B1 force field [34] in Swiss-PDB Viewer.
2.2. Preparation of ligand
Initially, 104 Korean medicinal plants and their 2431 phytochemicals, identified by GC-MS, were listed (Supplementary Table 01) through a rigorous literature review. Then the compounds were downloaded in 3D SDF format from the PubChem Database. Energy minimization of the ligand was utilized using the mmff94 force field and the steepest descent optimization algorithm steps 2000 in PyRx software before performing molecular docking analysis.
2.3. Molecular docking study
Docking is a technique used to predict the binding interaction of a drug with its target molecule. The AutoDock Vina virtual screening tool was used to perform molecular docking analysis to determine the specific residues in the target protein that interact with the particular ligands [35]. For docking analysis, AutoDock vina was used to convert pdb into a pdbqt format to input protein and ligands. Re-docking is the process of removing the ligand molecule from the receptor model. Therefore, to verify the docking studies, 6LU7 was re-docked with its crystallographic inhibitor N3 and compared the differences between the re-docking pose and conformation of the ligand in the crystal structure. This protein structure has several cavity sites and inhibitor N3 bound to at site 1 (X: 11.90, Y: 20.33, Z: 58.94) to the main protease. Consequently, site 1 was selected as an inhibition site. In this study, docking was used to understand the binding of ligands to the active sites of the Mpro protein. To dock the compounds against 6LU7, the center of the grid box was kept at X: 26.294, Y: 12.6009, Z: 58.9473, and dimensions were kept at X: 51.3565, Y: 66.9335, and Z: 59.6050 Å. The binding affinity of the ligand was by kcal/mol as a unit for negative binding score [36]. The compounds’ docking results were calculated and ranked based on their highest negative values, representing the highest binding affinities [35,37]. The interactions of drug molecules with the Mpro active site residues were confirmed by examining the docking and re-docking results at each docking position.
2.4. Re-screening using deep learning
The compounds with above −8.0 kcal/mol binding affinity were selected for re-evaluation with deep learning method using Recurrent Neural Networks (RNN) to predict models using the CHEMBL3927 dataset consisting of inhibitors for Mpro of SARS-CoV-1. The DeepScreeing server was used for this process and the server shows the average and median area under the ROC curve value of 0.86, and 0.89 under several constructed deep-learning models indicating good performance in screening [38]. PubChem fingerprint was used by PaDEL to generate 881 molecular fingerprints [39]. The hyperparameters for this developed model were manually set like learning rate (0.0001), number of neurons (2000,700,200), and hidden layer (3). The activation function and sigmoid function were used in hidden and output layers, respectively. The predicted pIC50 score was calculated using this equation.
| pIC50 = −log10IC50 (M) = 9 − log10IC50 (nM) |
This model was employed to re-screen the top docked 12 compounds and presented in Table 1 .
Table 1.
Binding affinity, Predicted IC50 score and non-bond interactions between SARS-CoV-2 main protease and the top twelve compounds.
| Compound name and CID | 2D Structure | Predicted pIC50 score | Binding affinity (kcal/mol) | Residue in contact | Interaction type | Bond distance in Å |
|---|---|---|---|---|---|---|
| Catechin gallate 6419835 | fx1 | 4.7227 | −8.5 | THR190 | Hydrogen Bond | 2.19304 |
| ARG188 | Hydrogen Bond | 2.97179 | ||||
| HIS163 | Hydrogen Bond | 1.68962 | ||||
| SER144 | Hydrogen Bond | 2.21275 | ||||
| PRO168 | Carbon Hydrogen Bond | 2.67908 | ||||
| CYS145 | Pi-Sulfur | 5.40505 | ||||
| MET165 | Pi-Sulfur | 5.40764 | ||||
| HIS41 | Pi-Pi Stacked | 4.73238 | ||||
| MET49 | Pi-Alkyl | 5.26499 | ||||
| Quercetin 3-O-malonylglucoside 5282159 | fx2 | 4.7172 | −8.2 | ASN238 | Hydrogen Bond | 1.88031 |
| ASP197 | Hydrogen Bond | 2.75917 | ||||
| LYS137 | Hydrogen Bond | 2.49689 | ||||
| THR199 | Hydrogen Bond | 2.72559 | ||||
| TYR239 | Hydrogen Bond | 2.15711 | ||||
| LEU272 | Carbon Hydrogen Bond | 2.6859 | ||||
| TYR237 | Pi-Pi T-shaped | 5.05381 | ||||
| Rutin 5280805 |
fx3 | 4.6303 | −8.9 | GLY143 | Hydrogen Bond | 2.25632 |
| GLU166 | Hydrogen Bond | 2.31189 | ||||
| PHE140 | Hydrogen Bond | 2.90128 | ||||
| ARG188 | Hydrogen Bond | 2.12475 | ||||
| THR26 | Carbon Hydrogen Bond | 1.84333 | ||||
| GLN189 | Carbon Hydrogen Bond | 2.90367 | ||||
| ASN142 | Carbon Hydrogen Bond | 2.74658 | ||||
| CYS145 | Pi-Sulfur | 4.9642 | ||||
| MET165 | Pi-Sulfur | 5.16804 | ||||
| Isoquercitin 10813969 | fx4 | 4.6179 | −8.6 | ARG188 | Hydrogen Bond | 2.40792 |
| THR190 | Hydrogen Bond | 2.12519 | ||||
| HIS163 | Hydrogen Bond | 2.05993 | ||||
| GLN192 | Hydrogen Bond | 2.42437 | ||||
| GLU166 | Carbon Hydrogen Bond | 2.22159 | ||||
| PRO168 | Carbon Hydrogen Bond | 2.88806 | ||||
| CYS145 | Pi-Sulfur | 5.81078 | ||||
| MET165 | Pi-Sulfur | 5.57255 | ||||
| HIS41 | Pi-Pi Stacked | 4.9543 | ||||
| MET49 | Pi-Alkyl | 4.94603 | ||||
| Hyperoside 5281643 | fx5 | 4.6179 | −8.6 | SER144 | Hydrogen Bond | 2.84073 |
| LEU141 | Hydrogen Bond | 2.56989 | ||||
| ARG188 | Carbon Hydrogen Bond | 2.83337 | ||||
| CYS145 | Pi-Sulfur | 5.06909 | ||||
| HIS41 | Pi-Pi T-shaped | 4.59608 | ||||
| MET49 | Pi-Alkyl | 5.04361 | ||||
| Quercetin-3-D-Xyloside 5320863 | fx6 | 4.6179 | −8.5 | LYS137 | Hydrogen Bond | 2.73634 |
| ASN238 | Hydrogen Bond | 2.30343 | ||||
| GLU290 | Hydrogen Bond | 2.36161 | ||||
| THR199 | Carbon Hydrogen Bond | 2.24907 | ||||
| ASP289 | Pi-Anion | 3.65208 | ||||
| LEU286 | Pi-Alkyl | 4.87541 | ||||
| Cacticin 5318644 | fx7 | 4.6171 | −8.4 | THR190 | Hydrogen Bond | 2.73319 |
| ARG188 | Hydrogen Bond | 2.42079 | ||||
| MET165 | Hydrogen Bond | 2.30981 | ||||
| GLN189 | Carbon Hydrogen Bond | 2.62132 | ||||
| GLU166 | Carbon Hydrogen Bond | 2.4619 | ||||
| THR190 | Pi-Donor Hydrogen Bond | 2.7937 | ||||
| MET49 | Alkyl | 4.15532 | ||||
| HIS41 | Pi-Alkyl | 3.72376 | ||||
| Narcissoside 5481663 | fx8 | 4.595 | −8.7 | GLY143 | Hydrogen Bond | 2.25307 |
| GLU166 | Hydrogen Bond | 2.35394 | ||||
| LEU141 | Hydrogen Bond | 2.24284 | ||||
| THR190 | Hydrogen Bond | 2.27931 | ||||
| THR26 | Hydrogen Bond | 2.31897 | ||||
| HIS41 | Hydrogen Bond | 2.82795 | ||||
| GLN189 | Carbon Hydrogen Bond | 2.76494 | ||||
| PHE140 | Carbon Hydrogen Bond | 2.5103 | ||||
| THR26 | Carbon Hydrogen Bond | 3.09029 | ||||
| CYS145 | Pi-Sulfur | 4.84568 | ||||
| MET165 | Pi-Sulfur | 5.1467 | ||||
| 3,4-Dicaffeoylquinic acid5281780 | fx9 | 4.5823 | −8.2 | LEU271 | Hydrogen Bond | 2.30224 |
| ALA285 | Hydrogen Bond | 2.78731 | ||||
| LEU272 | Hydrogen Bond | 2.31922 | ||||
| ASN238 | Hydrogen Bond | 2.75446 | ||||
| THR199 | Hydrogen Bond | 2.11357 | ||||
| ASP197 | Hydrogen Bond | 2.91729 | ||||
| ARG131 | Hydrogen Bond | 2.35626 | ||||
| LYS137 | Hydrogen Bond | 2.12948 | ||||
| MET276 | Pi-Alkyl | 5.48501 | ||||
| LEU287 | Pi-Alkyl | 4.78701 | ||||
| luteolin-7-O-rutinoside 14032966 | fx10 | 4.4184 | −8.5 | THR199 | Hydrogen Bond | 2.18848 |
| ASN133 | Hydrogen Bond | 2.8903 | ||||
| ASP197 | Hydrogen Bond | 2.38578 | ||||
| TYR237 | Carbon Hydrogen Bond | 2.50399 | ||||
| ASP197 | Carbon Hydrogen Bond | 2.45529 | ||||
| LEU272 | Pi-Alkyl | 5.11035 | ||||
| LEU287 | Pi-Alkyl | 5.38559 | ||||
| Cynaroside 5280637 | fx11 | 4.4176 | −8.2 | MET165 | Hydrogen Bond | 2.7413 |
| ARG188 | Hydrogen Bond | 2.68482 | ||||
| GLU166 | Hydrogen Bond | 2.39233 | ||||
| HIS163 | Hydrogen Bond | 3.08162 | ||||
| THR26 | Hydrogen Bond | 2.57113 | ||||
| SER144 | Carbon Hydrogen Bond | 2.4719 | ||||
| CYS145 | Pi-Sulfur | 4.70184 | ||||
| Cosmosiin 5280704 | fx12 | 4.3704 | −8.0 | THR199 | Hydrogen Bond | 2.42785 |
| LEU287 | Hydrogen Bond | 2.14488 | ||||
| THR199 | Hydrogen Bond | 2.65619 | ||||
| LEU286 | Carbon Hydrogen Bond | 3.09014 | ||||
| TYR237 | Pi-Pi T-shaped | 4.72839 | ||||
| Inhibitor N3 146025593 | fx13 | 4.690 | −8.1 | GLN189 | Carbon Hydrogen Bond | 2.48559 |
| GLU166 | Hydrogen Bond | 1.93628 | ||||
| GLY143 | Hydrogen Bond | 1.86317 | ||||
| HIS172 | Carbon Hydrogen Bond | 2.39426 | ||||
| HIS41 | Pi-Alkyl Bond | 4.3148 | ||||
| MET165 | Carbon Hydrogen Bond | 2.72823 | ||||
| MET49 | Alkyl | 4.66427 | ||||
| ALA191 | Pi-Alkyl | 4.53889 | ||||
| PRO168 | Pi-Alkyl | 4.84869 | ||||
| LEU167 | Alkyl | 5.46094 | ||||
| THR190 | Hydrogen Bond | 1.79854 | ||||
| HIS164 | Hydrogen Bond | 2.95502 | ||||
| PHE140 | Hydrogen Bond | 2.4722 | ||||
| ASN142 | Carbon Hydrogen Bond | 2.8316 |
CID; PubChem CID, pIC50 score in nM.
2.5. ADME/T and bioactivity analysis
The ADME/T were predicted using SwissADME, pKCSM, and ProTox-II server using SMILES (simplified molecular-input line-entry system) specification as an input syntax of the compounds. Pharmacokinetic and pharmacodynamic properties related to the metabolism, absorption, distribution, excretion, and toxicity of the compounds were evaluated. Molinspiration server was used to evaluate the biological activities of the compounds. Here, bioactivity score >0 is regarded as a biologically active compound, −5.0 to <0 as a moderately active compound, and <-5.0 as a biologically inactive compound.
2.6. Molecular dynamics simulation study
Molecular Dynamics (MD) simulation helps to determine and validate the structural flexibility and entropic effects of protein-ligand complexes in an artificial environment. Prior to performing the production simulations, energy minimization and equilibration simulations were conducted to remove bad contacts between the solute and solvent water molecules [40]. The solute and lipid chains were subjected to a harmonic restraint in the first step of energy minimization, and the second step was to allow all atoms to move freely [41]. The YASARA dynamics software was used in a molecular dynamics simulation to validate the predictions from the docking study [42]. In this simulation study, the AMBER14 force field [43] was utilized, which is widely used in macromolecular systems. The Mpro protein was used as a control system to compare the structural integrity against the complexes. The particle mesh Ewald method [44] was used to calculate long-range electrostatic interactions, and 8 Å cutoff radius was used to analyze short-range van der Waals and Coulomb interactions [45]. The total environmental condition of the system was set to a temperature of 298 K, pH 7.4, and 0.9% NaCl. The steepest descent algorithm was used to minimize the system [35]. A Monte Carlo barostat and a Langevin thermostat were used to maintain the pressure and temperature, respectively [46]. The MD simulation trajectories were saved after every 100 ps using a time step of 1.25 fs [47]. To ensure the simulation convergence, multiple parallel trajectories were conducted for each complex [40]. The MD simulation study was conducted for 100 ns to analyze the root mean square deviation (RMSD), the root mean square fluctuation (RMSF), the radius of gyration (RG), the solvent accessible surface area (SASA), and the number of hydrogen bonds [45,[48], [49], [50]]. The MD plots were visualized by Matplotlib.
3. Results and discussion
Drug discovery against viruses facilitated the successful identification of potential inhibitors by rational screening and understanding the bio-physio-chemical basis of inhibitor binding mechanism through molecular dynamics simulations as well as a series of conformational and toxicity analyses [51]. Molecular docking has been widely used to predict the bioactive compounds from plants to develop drugs against target proteins that cause infection by viruses. Mpro is the important therapeutic target of a virus as it cleaves the polyprotein of the virus that is crucial in viral transcription and replication that maintains the viral life cycle [23,52,53]. Therefore, this study aimed at the molecular docking and MD simulation approaches using Korean medicinal plants to predict their inhibitory role against virus infection. The target protein of this study has well-defined active sites that help in designing novel drugs against coronaviruses [54]. The binding propensity of the compounds against Mpro was calculated and ranked based on the AutoDock Vina docking score. Lesser binding affinity was considered to be a good docking score for screening the compound library. The lowest affinity from several binding poses of each compound was selected because it indicates the lowest energy release for bond formation [55].
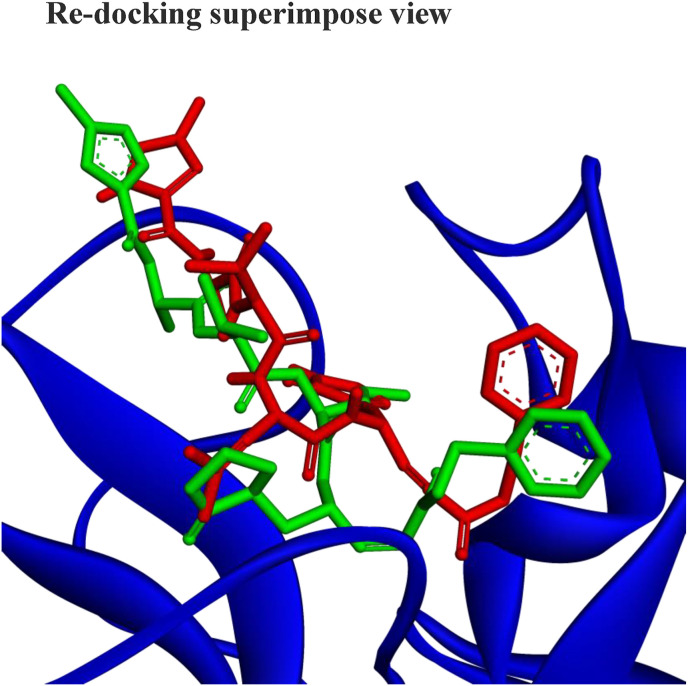
At first, the native inhibitor N3 of COVID-19 Mpro has been docked in order to legitimatize the virtual molecular docking accuracy and then re-docked. The re-docking of inhibitor N3 exhibited a binding affinity of −8.1 kcal/mol against the inhibition site of the target protein (Table 1). Further, validated the re-docking results of their conformational superimposition of ligand binding modes in the target protein (Fig. 1 ). The RMSD value between the re-docked and co-crystal native ligand: pose of inhibitor N3 was 0.037 Å which was within 2 Å indicating effective docking protocol. This conformational superimposition of ligands RMSD value represented good reproduction of the correct pose mostly with identical inhibition sites and residues. The inhibitors of the molecules in the active pocket of Mpro of the protein and their interaction type are shown in Fig. 2 .
Fig. 1.
Three-dimensional view of the best re-docking pose and the conformational superposition of ligands (inhibitor N3) and the RMSD value of 0.037 Å.
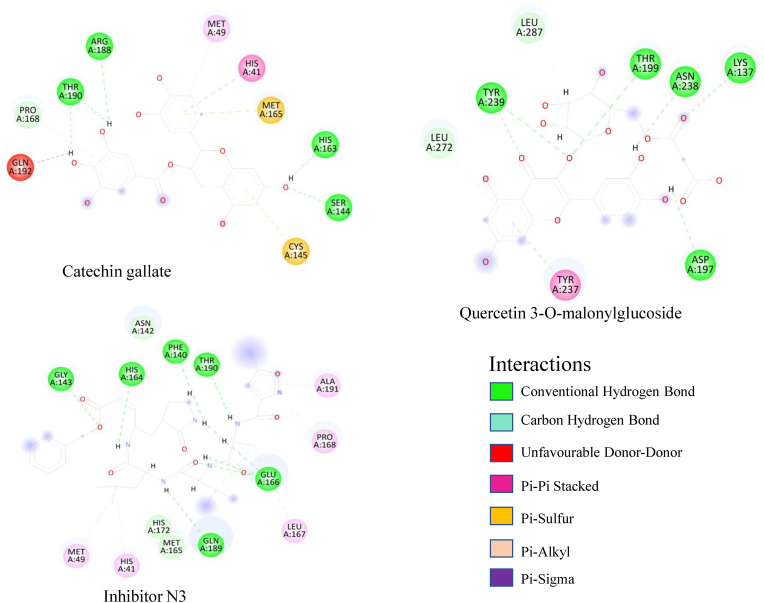
Fig. 2.
Molecular docking interaction and two-dimensional (2D) binding mode of selected ligands in SARS-CoV-2 main protease active site (PDB ID: 6LU7).
After docking of the phytochemicals of Korean medicinal plants, their lower binding affinity than inhibitor N3 was analyzed. Based on the lowest affinity, the top 12 compounds, i.e. Catechin gallate, Cynaroside, Cosmosiin, Isoquercitrin, Rutin, Hyperoside, Isochlorogenic acid b, Quercetin 3-O-malonylglucoside, Cacticin, Narcissoside, Guaijaverin, Luteolin-7-O-Rutinoside were selected which showed negative binding affinity (kcal/mol) of −8.5, −8.2, 8.0, −8.6, −8.9, −8.6, −8.2, −8.2, −8.4, −8.7, −8.5, −8.5, respectively (Table 1).
Other molecular properties and 2D structure of the predicted compounds are presented in Table 1. Further, catechin gallate and quercetin 3-O-malonylglucoside had been indented as top potential candidates those had binding affinities of −8.5 and −8.2, kcal/mol (Fig. 3 ). Moreover, their other physio-chemical properties, solubility, drug-likeness, etc. have been demonstrated in Table 2 , and their non-bonded interactions within the active and catalytic sites of the Mpro have been illustrated in Fig. 4 . Re-docking performed the N3 inhibitor against 6LU7, which was used as a positive reference molecule. The top two lead compounds exhibited notable docking conformations with a binding affinity of more than −8.1 kcal/mol (binding affinity of N3) in the active site of a target protein. The protein-ligand interaction profile and binding affinity between the top two compounds and the inhibitor N3 in the binding site of Mpro, and the comparative docking score are presented in Table 1. Several natural compounds like theaflavin, quercetin, withanone, dihydrowithaferin, and caffeic acid phenethyl ester have been reported as potential inhibitors of SARS-CoV-2 [51,56,57].
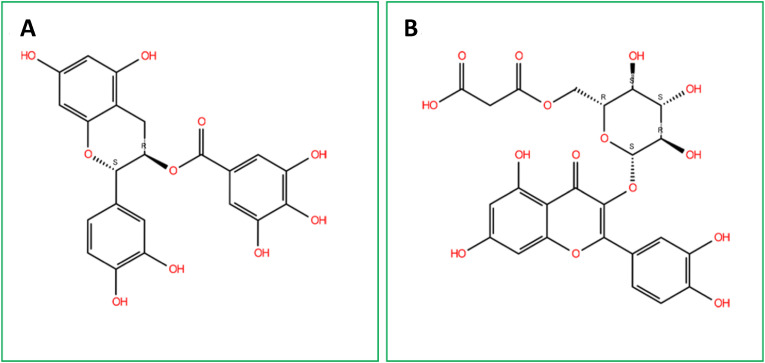
Fig. 3.
2D chemical structure of (A) Catechin gallate and (B) Quercetin 3-O-malonylglucoside. The structures were drawn using the 2D sketcher (Beta) software.
Table 2.
Pharmacological profiles of the top two potential candidates derived from the SwissADME and pKCSM webservers.
| Properties | Catechin gallate | Quercetin 3-O-malonylglucoside | |
|---|---|---|---|
| Physico-chemical Properties | MW (g/mol) | 442.37 g/mol | 550.42 g/mol |
| Heavy atoms | 32 | 39 | |
| Aro. Atoms | 18 | 16 | |
| Rotatable bonds | 4 | 8 | |
| H-bond acceptors | 10 | 15 | |
| H-bond donors | 7 | 8 | |
| TPSA (Å2) | 177.14 | 253.88 | |
| Lipophilicity | Log Po/w (Cons) | 1.25 | −0.67 |
| Water Solubility | Log S (ESOL) | Soluble | Soluble |
| Pharmacokinetics | GI absorption | Low | Low |
| BBB permeant | No | No | |
| P-GP substrate | No | Yes | |
| Drug likeness | Lipinski | Yes | No |
| Medi. Chemistry | Synth. accessibility | 4.16 | 5.63 |
Fig. 4.
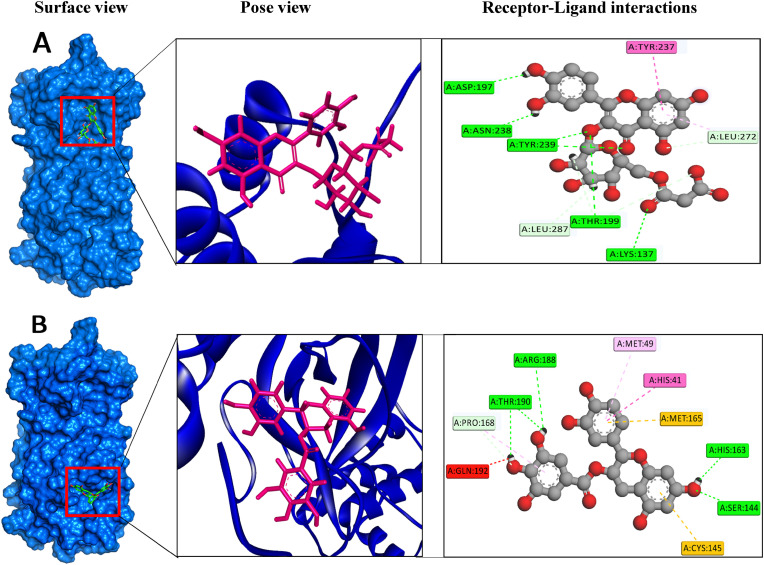
Binding modes for the selected compounds. The figures illustrate non-bonded interactions of the docked complexes for the top two compounds within the active and catalytic sites of the main protease. (A) Quercetin 3-O-malonylglucoside, and (B) Catechin gallate.
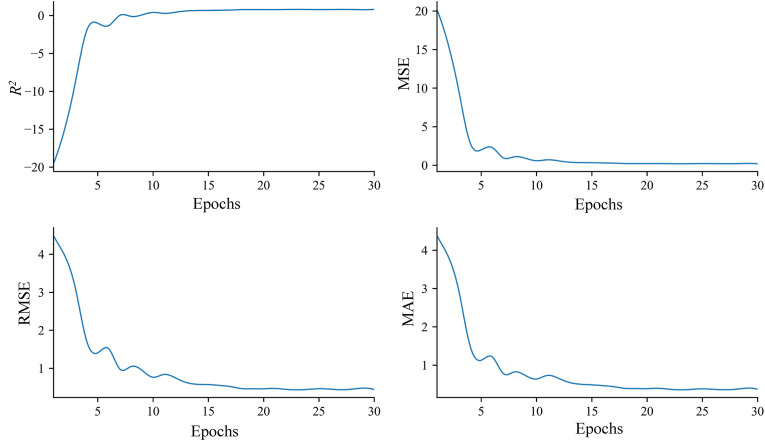
The Deep learning method uses various artificial neural network frameworks on the dataset to predict molecular properties and bioactivities from large libraries unitizing different chemical, topological, and biological data patterns of compounds [[58], [59], [60]]. It was performed to screen selected compounds based on inhibitory activities against the target protein. Compounds with inhibitory properties against Mpro of SARS-CoV-1 were used for building many regression models based on IC50. Evaluation matrices, i.e., coefficient of determination (R2), mean square error (MSE), root mean square error (RMSE), and mean absolute error (MAE) were measured to evaluate the performance and quality of the selected model. These parameters represent the fitness of the regression model and the errors between predicted and actual data [61,62]. The values of R2, MSE, RMSE, and MAE were 0.81, 0.26, 0.51, and 0.41, respectively (Fig. 5 ). High R2 value showed that the model had higher fitness and a good performance against the test data. Small MSE, RMSE, and MAE values indicated the higher accuracy of the model [63]. Selected compounds were screened in the regression model against the biologically active inhibitors for Mpro of SARS-CoV-1. Both catechin gallate and quercetin 3-O-malonylglucoside showed pIC50 values of 4.723 and 4.717, respectively, demonstrating that these two compounds were biologically active and able to inhibit the protein.
Fig. 5.
Regression model accuracy plots showing various statistical parameters of the deep learning model. Upper left plot showed R2 value of 0.81 at epoch 30. Upper right plot showed MSE value of 0.26 at epoch 30. Lower left plot denoted RMSE value of 0.51 at epoch 30. Lower right plot showed MAE value of 0.41 at epoch 30.
It is reported that Mpro of SARS-CoV-2 has three domains consisting of residues 8–101, 102–184, and 201–303 [51,64]. Among these residues, Cys145-His41 acts as a catalytic dyad between domains I and II, and is responsible for protease activity and substrate binding [[65], [66], [67], [68]]. In domain I, residue Phe140, His172, Asn142, His163, Glu166, and Leu141 are involved in substrate binding. Residue Asp187, Met165, Tyr54, Met49, and His41 in domain II and residue Gln192, Met165, Gln189, Phe185, and Leu167 in domain III are also important for substrate binding [69,70]. In the activity of the protein, the large number of hydrogen bonds between ligands and residues signifies a strong binding affinity [49]. Post-dock analysis showed Catechin gallate had a better binding affinity with the protein because of having more electrostatic force than the Quercetin 3-O-malonylglucoside. Catechin gallate had no interaction with domain III of the protein. It formed four conventional hydrogen bonds with residue Thr190, Arg188, His163, and Ser144 residue creating strong affinity on the domain I and II. Two non-covenant pi-sulfur bonds were formed with Cys145 and Met165 residue. Pi-Pi Stacked bond with residue His41 interacting with both aromatic rings and pi-alkyl bond with residue Met49 was also observed, that created a pi-electron cloud over the alkyl group. Carbon-hydrogen bonds with Pro168 residue were also formed, which gave this compound higher interaction affinity with the protein (Fig. 4). The residues Gln189 and Leu455 have already been found to be potent drug binding sites [51,71]. Catechin gallate complex interactions also match with another research by Mousavi, 2021 [72]. Thus, Catechin gallate formed chemical bonds with Cys145-His41 catalytic dyad residues and interacted with Met165 and His163, which play an important role in substrate binding.
Polyphenol flavonoids show activity against various human viruses like herpes simplex, hepatitis B, HIV, hepatitis C, influenza, dengue, chikungunya, and zika. Studies were conducted on Camellia sinensis (tea) which demonstrated that catechins and gallate derivatives including epicatechin 3-O-caffeoate, etc-pyrrolidinone C, etc-pyrrolidinone D, epigallocatechin gallate, epicatechin gallate, and gallocatechin-3-gallate have inhibition properties against Mpro in intracellular and extracellular inhibition assays. Catechin gallate also had stable complex formation with the viral protein due to hydrogen bonds and non-covalent interactions [64,[73], [74], [75]]. Various studies showed that Catechin gallate has better binding energy compared to Michael acceptor inhibitors known as N3, which have a half-maximal inhibitory concentration (IC50) value of 125 μM against SARS-CoV-2 Mpro. Its derivatives, like gallo catechin gallate with two carboxy group replacements, also have the potential to decrease infection of SARS-CoV-2 omicron mutant [76].
Similarly, Quercetin 3-O-malonylglucoside formed five conventional hydrogen bonds with residue Asn238, Asp197, Lys137, Thr199, and Tyr239. Leu272 residue formed a carbon-hydrogen bond and Tyr237 formed a pi-pi t-shaped bond with the compound. The majority of chemical interactions were found with domain III of Mpro and no bond was formed with the first domain that consists of residue 8–101. Lys137 was the only residue that formed a hydrogen bond with the compound. Quercetin 3-O-malonylglucoside bind at Asp197 within a loop among the domains. This analysis suggests that both compounds have strong and stable binding with Mpro because of the multiple numbers of bonds that were formed during docking and Catechin gallate formed bonds with catalytic and binding residues of the target protein.
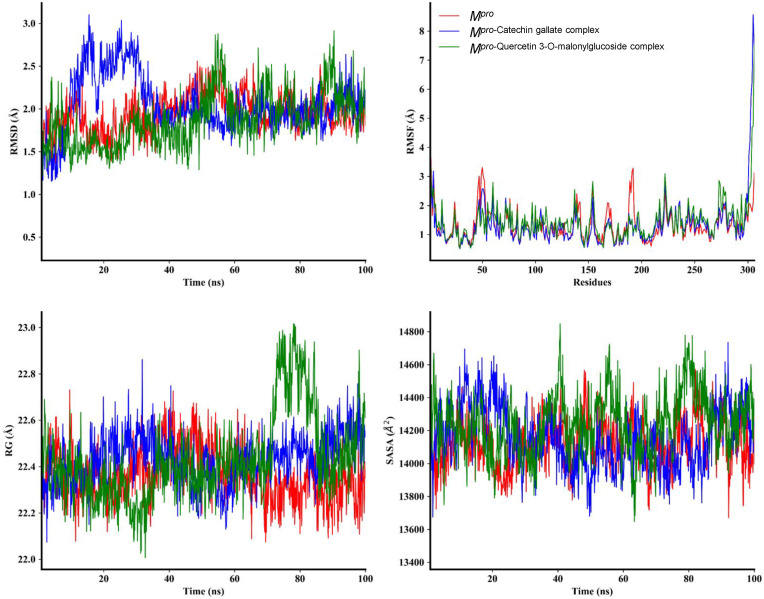
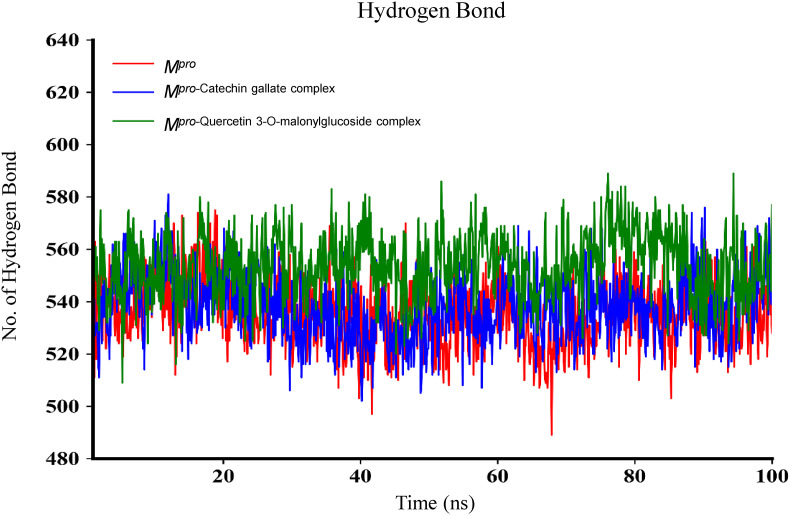
Molecular dynamics (MD) simulation is vital for post-dock analysis to investigate time-dependent stability and atom movements of biological compounds [77]. MD simulation was performed for 100 ns to understand the structural behavior, binding mechanism, and structural flexibility of Mpro [77,78], Mpro-Catechin gallate complex, and Mpro-Quercetin 3-O-malonylglucoside complex. The comparison between initial and final structural behavior of the complexes has been demonstrated in Fig. 6 . The parameters computed and analyzed after 100 ns dynamics trajectory were root-mean-square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (RG), solvent accessible surface area (SASA) (Fig. 7 ) as well as hydrogen bonding (Fig. 8 ), those were measured as because such stabilities are essential to obtain good binding affinities [79]. RMSDs of Cα atoms of these compounds were performed and it was observed that Mpro had a minor fluctuation from 45 ns to 67 ns but afterward it remained at an equilibrium state. As for Mpro-Catechin gallate complex, 8 ns–35 ns showed a high fluctuation, after that it also reached a stable position. Mpro-Quercetin 3-O-malonylglucoside complex showed stability at first but after 45 ns it had several fluctuations. Nevertheless 92 ns afterward it again showed stability. These two complexes showed stability regarding the reference protein at the end.
Fig. 6.
Comparing between initial and final structures of the complexes acquired from MD simulations.
Fig. 7.
Analysis of RMSD, RMSF, RG, and SASA of unligated Mpro and Mpro- Catechin gallate/Quercetin 3-O-malonylglucoside complex. The MD simulations for each system were performed for 100 ns. These MD trajectories were analyzed with the aid of RMSD, RMSF, RG and SASA. RMSD analysis for the alpha carbon atoms, RMSF for the flexibility analysis of amino acids residues, RG for the degree of rigidity and compactness analysis, and SASA for the protein volume with expansion analysis.
Fig. 8.
Determination of Hydrogen bond analysis of unligated Mpro and Mpro- Catechin gallate/Quercetin 3-O-malonylglucoside complex. The MD simulation for the system was performed for 100 ns.
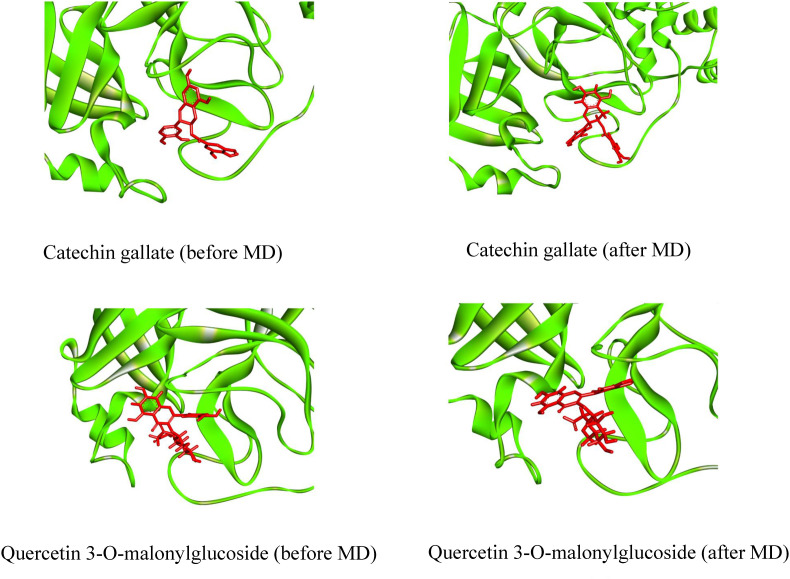
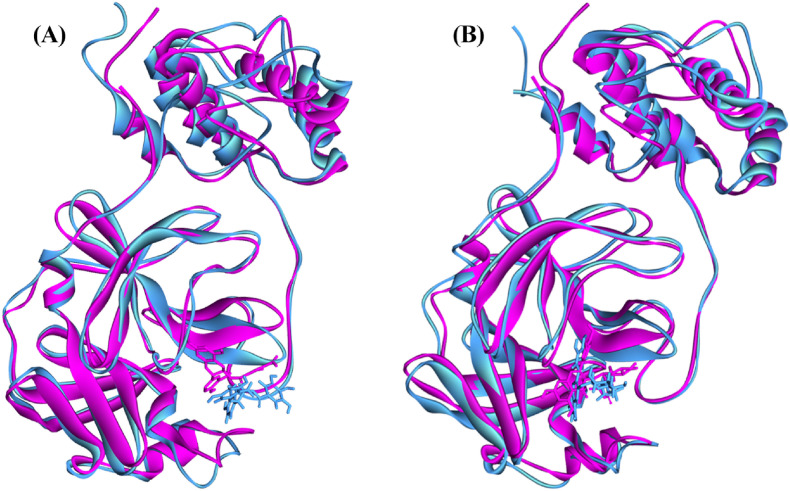
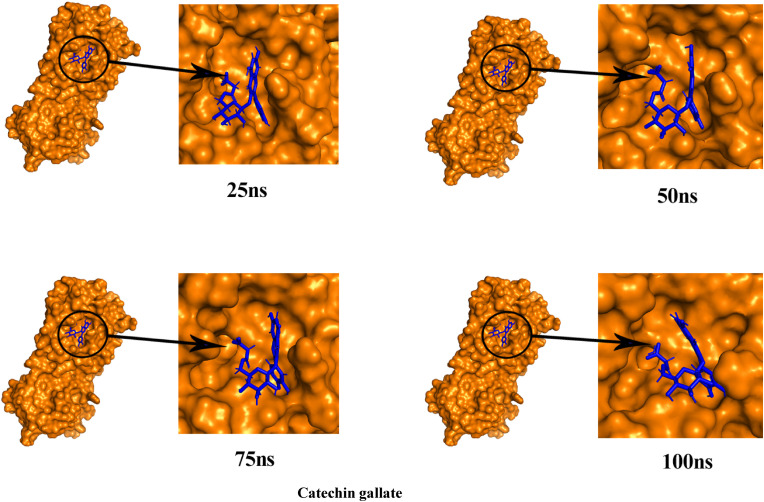
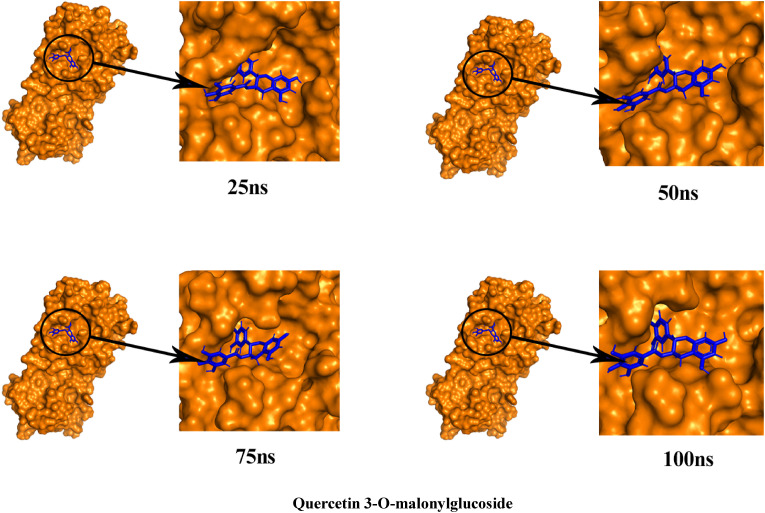
To better understand the structural compactness, SASA was examined [78]. Mpro-Catechin gallate complex showed two higher trajectories at the beginning of the simulation compared to the Mpro. However, both showed relatively similar trajectories near the end demonstrating that these complexes had slightly lower compactness. In contrast, Mpro-Quercetin 3-O-malonylglucoside complex showed higher trajectories after 7 ns for 10 ns (Fig. 7). Despite of this the latter complex showed high RG values between 72 ns and 85 ns, both of them showed comparatively alike plot indicating these complexes somewhat had loosed protein structure packaging [80] that supports SASA result analysis. The reference protein Mpro and the other two complexes showed higher fluctuation in RMSF analysis over a similar range of residues, e.g. 45–56, 132–143, 222–230, and 272–279, which strongly suggest homogeneous protein residue flexibility [81]. The hydrogen bonds between intermolecular binding demonstrated that the Mpro only and Mpro-Catechin gallate had similar hydrogen bond numbers between intra-molecules but Mpro-Quercetin 3-O-malonylglucoside complex showed a higher number of hydrogen bindings than these two molecules donating its strong stability (Fig. 8). Analysis from these various results indicates that despite having minor conformational changes due to MD simulation, these two complexes have similar stability like Mpro reference protein. The superimposition of the structures between pre- and post-molecular dynamics simulation of the structures of Catechin gallate and Quercetin 3-O-malonylglucoside also was performed to determine changes in the binding cavity (Fig. 9 ). After the post-dock analysis showed their structural change in every 25 ns, surface view of Mpro-Catechin gallate and Mpro-Quercetin 3-O-malonylglucoside complexes in MD simulation. The snapshot was taken from 25, 50, 75, and 100 ns, respectively, shown in Fig. 10, Fig. 11 .
Fig. 9.
The superimposed between pre- and post-molecular dynamics simulation structures of (A) Catechin gallate and (B) Quercetin 3-O-malonylglucoside. The pink color denotes the pre-molecular dynamics structure, and the sky color denotes the post-molecular dynamics structure.
Fig. 10.
The surface view of Catechin gallate and Mpro complex in MD simulation. The snapshot was taken from 25, 50, 75, and 100 ns, respectively.
Fig. 11.
The surface view of Quercetin 3-O-malonylglucoside and Mpro complex in MD simulation. The snapshot was taken from 25, 50, 75, and 100 ns, respectively.
Biologically active compounds act on proteins, subsequently causing physiological effects directly. The potential biological activities of the compounds were assessed by investigating G protein-coupled receptor (GPCR) ligand activity, ion channel inhibitor, enzyme inhibition, protease inhibition, kinase inhibition, and nuclear receptor ligand activity (Table 3 ). Catechin gallate exhibited positive biological activity against all of these biological mechanisms, whereas Quercetin 3-O-malonylglucoside showed moderate activity in ion channel, kinase and protease inhibition. Quercetin 3-O-malonylglucoside showed a better result as an enzyme inhibitor, but Catechin gallate outperformed all other biological receptors.
Table 3.
Biological activities of the screened hit phytochemicals.
| Compounds | GPCR ligand | Ion channel inhibitor | Kinase inhibitor | Nuclear receptor ligand | Protease inhibitor | Enzyme inhibitor |
|---|---|---|---|---|---|---|
| Catechin gallate | 0.17 | 0.02 | 0.05 | 0.34 | 0.13 | 0.25 |
| Quercetin 3-O-malonylglucoside | 0.04 | −0.21 | −0.02 | 0.14 | −0.03 | 0.35 |
Activities were calculated from the Molinospiration chemoinformatics software package. Here bioactivity score >0.0; biologically active, between −5.0 – 0.0; moderately active, and <-5.0; biologically inactive.
Safety issues, including toxicities and adverse drug effects, are always an essential prerequisite that should be evaluated during drug development. Even though the clinical trial is obligatory, computational predictive models built for various toxicities also facilitated the progress of drug design [82]. Computational methods to investigate toxicities and adverse drug effects have shown significant advantages since they are accurate and can be done before a compound is synthesized [83]. Toxicity assessment and pharmacokinetic characteristics are required to assess the effectiveness and level of indemnification of the lead compounds [84]. These two lead compounds were further analyzed with the pharmacological activity that is an essential indicator for disposition within the human body with ADME/T technology whose intensive application has been reviewed recently [82]. Furthermore, Catechin gallate and Quercetin 3-O-malonylglucoside were evaluated by examining Central Nervous System (CNS) permeability, hepatotoxicity, p-glycoprotein inhibition, and Cytochrome P450 (CYP) inhibition, among several other aspects. CNS permeability refers to the capacity of a drug to penetrate the semipermeable blood-brain barrier, which is meant to protect the CNS from potentially harmful compounds. Permeability of CNS higher than −4 is thought to reflect the ability to pass across the blood-brain barrier. The molecular weights of Quercetin 3-O-malonylglucoside and Catechin gallate were 550.42 and 442.37 g/mol, respectively. None showed inhibitory properties against cytochromes p450 (CYPs) thus, these compounds would be metabolized without having increased side effect risks or possible toxicity [85]. Catechin gallate is in agreement with Lipinski and Ghoses' [86] rule of drug-likeness, however Quercetin 3-O-malonylglucoside fails in their filter. Consequently, Catechin gallate showed better results in physiochemical properties, water solubility, lipophilicity, pharmacokinetics, and toxicity parameters concerning drug-likeness compared to Quercetin 3-O-malonylglucoside.
Moreover, the majority of the potential drug candidates fail clinical trials due to the toxicity they produce which makes it an important filter for the drug design process [87]. Various toxicity properties e.g., hepatotoxicity, carcinogenicity, immunotoxicity, mutagenicity, cytotoxicity, oral rat acute toxicity, LD50 value, and toxicity class of the selected two compounds have been assessed (Table 4 ). These machine learning methods have been applied to construct classifiers and regression models to predict LD50 or their toxic categories [88,89]. Both of the compounds were inactive in organ toxicity endpoints. Predicted LD50 of Quercetin 3-O-malonylglucoside was 5000 mg kg−1, and for Catechin gallate, it was 1000 mg kg−1 making them nearly non-toxic to biological systems. Catechin gallate exhibited comparatively better favorable pharmacokinetics, biological activities, and toxicity properties than Quercetin 3-O-malonylglucoside, which indicates it has a minimal likelihood to fail during clinical trials.
Table 4.
Various toxicity properties of the selected two compounds.
| Endpoint | Target | Catechin gallate | Quercetin 3-O-malonylglucoside |
|---|---|---|---|
| Organ toxicity and toxicity endpoints | Hepatotoxicity | Inactive | Inactive |
| Carcinogenicity | Inactive | Inactive | |
| Immunotoxicity | Inactive | Inactive | |
| Mutagenicity | Inactive | Inactive | |
| Cytotoxicity | Inactive | Inactive | |
| LD50 (mg kg−1) | 1000 | 5000 | |
| Toxicity class | 4 | 5 |
This study provides feature learning and artificial neural network-based deep structured learning using artificial intelligence that supports the previous studies on SARS-CoV-2 Mpro and Catechin gallate. Cheminformatics-based bioactivity prediction that employed a quantitative structure–activity relationship model on main human enzymes also holds up these findings and data. In our investigation and analysis, we predicted a potential inhibitor, Catechin gallate, against Mpro from a large phytochemical library through molecular docking screening and molecular dynamics simulation. We investigated their pharmaceutical profile as a possible drug candidate against SARS-CoV-2. Our findings might help to understand the potential drug design based on their inhibitory activity of a target protein.
4. Conclusion
This study revealed that collected phytochemicals from Korean traditional medicinal plants bind with Mpro with different poses. Catechin gallate and quercetin 3-O-malonylglucoside were selected through molecular docking and deep learning method based on binding affinity and inhibitory potency. They were further subjected to post-dock analysis that showed these compounds were stable forming various electrostatic forces like H-bond, pi-bond, and C–H bond, while catechin gallate formed important bonds with catalytic residues. MD simulation method also validated the stability, compactness, flexibility, binding energy, and residue fluctuations of the selected compounds. These were further investigated with ADME/T pharmacokinetic properties and bioactivity properties, denoting that these bioactive dispositions of the compounds inside the body as a chemical would be safe without causing toxicity or carcinogenicity. These analyses demonstrated that Catechin gallate might be a potential drug candidate against SARS-CoV-2. Our identification also might help to advancement of knowledge of Korean traditional medicinal plants.
Author contributions
AH; Conceptualization, methodology, formal analysis, investigation, validation, writing – original draft, MNM, KN; Conceptualization, methodology, investigation, validation, writing –review & editing, MFR; Validation, funding acquisition, review, editing, MER, MSR , MOF; Formal analysis, review & editing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known conflict of interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors wish to thank the Molecular Genetics Lab. Dept. of Genetic Engineering and Biotechnology, University of Rajshahi, Bangladesh, for providing all facilities during the research.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2023.106785.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Di Napoli R. 2022. Features, Evaluation, and Treatment of Coronavirus (COVID-19) Statpearls [Internet] [PubMed] [Google Scholar]
- 2.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S., Yuen K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.-J., Shieh C.-K., Gorbalenya A.E., V Koonin E., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M.M.C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziebuhr J., Snijder E.J., Gorbalenya A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 8.Anand K., Palm G.J., Mesters J.R., Siddell S.G., Ziebuhr J., Hilgenfeld R. Structure of coronavirus main proteinase reveals combination of a chymotrypsin fold with an extra α-helical domain. EMBO J. 2002;21:3213–3224. doi: 10.1093/emboj/cdf327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 10.Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou C.-Y., Chang H.-C., Hsu W.-C., Lin T.-Z., Lin C.-H., Chang G.-G. Quaternary structure of the severe acute respiratory syndrome (SARS) coronavirus main protease. Biochemistry. 2004;43:14958–14970. doi: 10.1021/bi0490237. [DOI] [PubMed] [Google Scholar]
- 12.Hsu M.-F., Kuo C.-J., Chang K.-T., Chang H.-C., Chou C.-C., Ko T.-P., Shr H.-L., Chang G.-G., Wang A.H.-J., Liang P.-H. Mechanism of the maturation process of SARS-CoV 3CL protease*[boxs] J. Biol. Chem. 2005;280:31257–31266. doi: 10.1074/jbc.M502577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morse J.S., Lalonde T., Xu S., Liu W.R. Learning from the past: possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden F.G., Turner R.B., Gwaltney J.M., Chi-Burris K., Gersten M., Hsyu P., Patick A.K., Smith G.J., III, Zalman L.S. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain R., Mujwar S. Repurposing metocurine as main protease inhibitor to develop novel antiviral therapy for COVID-19. Struct. Chem. 2020;31:2487–2499. doi: 10.1007/s11224-020-01605-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 17.Su H., Yao S., Zhao W., Li M., Liu J., Shang W., Xie H., Ke C., Gao M., Yu K. BioRxiv; 2020. Discovery of Baicalin and Baicalein as Novel, Natural Product Inhibitors of SARS-CoV-2 3CL Protease in Vitro. [Google Scholar]
- 18.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV Mpro with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Islam M.T., Sarkar C., El‐Kersh D.M., Jamaddar S., Uddin S.J., Shilpi J.A., Mubarak M.S. Natural products and their derivatives against coronavirus: a review of the non‐clinical and pre‐clinical data. Phytother Res. 2020;34:2471–2492. doi: 10.1002/ptr.6700. [DOI] [PubMed] [Google Scholar]
- 21.Shah M.A., Rasul A., Yousaf R., Haris M., Faheem H.I., Hamid A., Khan H., Khan A.H., Aschner M., Batiha G.E. Combination of natural antivirals and potent immune invigorators: a natural remedy to combat COVID‐19. Phytother Res. 2021;35:6530–6551. doi: 10.1002/ptr.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma M., Prasher P., Mehta M., Zacconi F.C., Singh Y., Kapoor D.N., Dureja H., Pardhi D.M., Tambuwala M.M., Gupta G. Drug Development Research; 2020. Probing 3CL Protease: Rationally Designed Chemical Moieties for COVID-19. [DOI] [PubMed] [Google Scholar]
- 23.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 24.Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., Drosten C., Bai D., He X., Ludewig B. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortega J.T., Serrano M.L., Jastrzebska B. Class AG protein-coupled receptor antagonist famotidine as a therapeutic alternative against SARS-CoV2: an in silico analysis. Biomolecules. 2020;10:954. doi: 10.3390/biom10060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu X., Wang X.-J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J. Genetics Genom. 2020;47:119. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281:4085–4096. doi: 10.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mengist H.M., Dilnessa T., Jin T. Structural basis of potential inhibitors targeting SARS-CoV-2 main protease. Front. Chem. 2021;9 doi: 10.3389/fchem.2021.622898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adegbola P.I., Semire B., Fadahunsi O.S., Adegoke A.E. Molecular docking and ADMET studies of Allium cepa, Azadirachta indica and Xylopia aethiopica isolates as potential anti-viral drugs for Covid-19. VirusDisease. 2021;32:85–97. doi: 10.1007/s13337-021-00682-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pitsillou E., Liang J.J., Beh R.C., Hung A., Karagiannis T.C. Molecular dynamics simulations highlight the altered binding landscape at the spike-ACE2 interface between the Delta and Omicron variants compared to the SARS-CoV-2 original strain. Comput. Biol. Med. 2022;149 doi: 10.1016/j.compbiomed.2022.106035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar Y., Singh H., Patel C.N. Silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing. J. Infect. Public Health. 2020;13:1210–1223. doi: 10.1016/j.jiph.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bafna K., Krug R.M., Montelione G.T. ChemRxiv; 2020. Structural Similarity of SARS-CoV2 Mpro and HCV NS3/4A Proteases Suggests New Approaches for Identifying Existing Drugs Useful as COVID-19 Therapeutics. [Google Scholar]
- 33.Liu Q., Kwan K., Cao T., Yan B., Ganesan K., Jia L., Zhang F., Lim C., Wu Y., Feng Y. Broad‐spectrum antiviral activity of Spatholobus suberectus Dunn against SARS‐CoV‐2, SARS‐CoV‐1, H5N1, and other enveloped viruses. Phytother Res. 2022;36:3232–3247. doi: 10.1002/ptr.7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaplan W., Littlejohn T.G. Swiss-PDB viewer (deep view) Briefings Bioinf. 2001;2:195–197. doi: 10.1093/bib/2.2.195. [DOI] [PubMed] [Google Scholar]
- 35.Arshia A.H., Shadravan S., Solhjoo A., Sakhteman A., Sami A. De novo design of novel protease inhibitor candidates in the treatment of SARS-CoV-2 using deep learning, docking, and molecular dynamic simulations. Comput. Biol. Med. 2021;139 doi: 10.1016/j.compbiomed.2021.104967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murugesan S., Kottekad S., Crasta I., Sreevathsan S., Usharani D., Perumal M.K., Mudliar S.N. Targeting COVID-19 (SARS-CoV-2) main protease through active phytocompounds of ayurvedic medicinal plants–Emblica officinalis (Amla), Phyllanthus niruri Linn.(Bhumi Amla) and Tinospora cordifolia (Giloy)–A molecular docking and simulation study. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Z., Du J., Fang J., Yin Y., Xu G., Xie L. 2019. DeepScreening : a Deep Learning-Based Screening Web Server for Accelerating Drug Discovery; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yap C.W.E.I. 2010. Software News and Update PaDEL-Descriptor : an Open Source Software to Calculate Molecular Descriptors and Fingerprints. [DOI] [PubMed] [Google Scholar]
- 40.ting Fu T., Tu G., Ping M., xun Zheng G., yuan Yang F., yi Yang J., Zhang Y., jun Yao X., wei Xue W., Zhu F. Subtype-selective mechanisms of negative allosteric modulators binding to group I metabotropic glutamate receptors. Acta Pharmacol. Sin. 2021;42:1354–1367. doi: 10.1038/s41401-020-00541-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue W., Wang P., Tu G., Yang F., Zheng G., Li X., Li X., Chen Y., Yao X., Zhu F. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys. Chem. Chem. Phys. 2018;20:6606–6616. doi: 10.1039/c7cp07869b. [DOI] [PubMed] [Google Scholar]
- 42.Krieger E., Darden T., Nabuurs S.B., Finkelstein A., Vriend G. Making optimal use of empirical energy functions: force‐field parameterization in crystal space. Proteins: Struct., Funct., Bioinf. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- 43.Case D.A., Cheatham T.E., III, Darden T., Gohlke H., Luo R., Merz K.M., Jr., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krieger E., Nielsen J.E., Spronk C.A.E.M., Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 45.M. Dutta, A.M. Tareq, A. Rakib, S. Mahmud, S.A. Sami, J. Mallick, M.N. Islam, M. Majumder, Z. Uddin, A. Alsubaie, A.S.A. Almalki, M.U. Khandaker, D.A. Bradley, S. Rana, T. Bin Emran, Molecular Dynamics Simulations, (2021).
- 46.Tu G., Fu T., Yang F., Yang J., Zhang Z., Yao X., Xue W., Zhu F. Understanding the polypharmacological profiles of triple reuptake inhibitors by molecular simulation. ACS Chem. Neurosci. 2021;12:2013–2026. doi: 10.1021/acschemneuro.1c00127. [DOI] [PubMed] [Google Scholar]
- 47.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahmud S., Parves M.R., Riza Y.M., Sujon K.M., Ray S., Tithi F.A., Zaoti Z.F., Alam S., Absar N. Exploring the potent inhibitors and binding modes of phospholipase A2 through in silico investigation. J. Biomol. Struct. Dyn. 2020;38:4221–4231. doi: 10.1080/07391102.2019.1680440. [DOI] [PubMed] [Google Scholar]
- 49.Khan M.A., Mahmud S., Alam A.S.M.R.U., Rahman M.E., Ahmed F., Rahmatullah M. Comparative molecular investigation of the potential inhibitors against SARS-CoV-2 main protease: a molecular docking study. J. Biomol. Struct. Dyn. 2021;39:6317–6323. doi: 10.1080/07391102.2020.1796813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Islam M.J., Parves M.R., Mahmud S., Tithi F.A., Reza M.A. Assessment of structurally and functionally high-risk nsSNPs impacts on human bone morphogenetic protein receptor type IA (BMPR1A) by computational approach. Comput. Biol. Chem. 2019;80:31–45. doi: 10.1016/j.compbiolchem.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Parida P.K., Paul D., Chakravorty D. The natural way forward: molecular dynamics simulation analysis of phytochemicals from Indian medicinal plants as potential inhibitors of SARS‐CoV‐2 targets. Phytother Res. 2020;34:3420–3433. doi: 10.1002/ptr.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J., Kenward C., Worrall L.J., Vuckovic M., Gentile F., Ton A.-T., Ng M., Cherkasov A., Strynadka N.C.J., Paetzel M. X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation. Nat. Commun. 2022;13:1–13. doi: 10.1038/s41467-022-32854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vuong W., Fischer C., Khan M.B., van Belkum M.J., Lamer T., Willoughby K.D., Lu J., Arutyunova E., Joyce M.A., Saffran H.A. Improved SARS-CoV-2 Mpro inhibitors based on feline antiviral drug GC376: structural enhancements, increased solubility, and micellar studies. Eur. J. Med. Chem. 2021;222 doi: 10.1016/j.ejmech.2021.113584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.V Stoddard S., Stoddard S.D., Oelkers B.K., Fitts K., Whalum K., Whalum K., Hemphill A.D., Manikonda J., Martinez L.M., Riley E.G. Optimization rules for SARS-CoV-2 Mpro antivirals: ensemble docking and exploration of the coronavirus protease active site. Viruses. 2020;12:942. doi: 10.3390/v12090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue Q., Liu X., Russell P., Li J., Pan W., Fu J., Zhang A. Evaluation of the binding performance of flavonoids to estrogen receptor alpha by Autodock, Autodock Vina and Surflex-Dock. Ecotoxicol. Environ. Saf. 2022;233 doi: 10.1016/j.ecoenv.2022.113323. [DOI] [PubMed] [Google Scholar]
- 56.Lung J., Lin Y., Yang Y., Chou Y., Shu L., Cheng Y., Te Liu H., Wu C. The potential chemical structure of anti‐SARS‐CoV‐2 RNA‐dependent RNA polymerase. J. Med. Virol. 2020;92:693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robinson M.C., Glen R.C. Validating the validation: reanalyzing a large-scale comparison of deep learning and machine learning models for bioactivity prediction. J. Comput. Aided Mol. Des. 2020;34:717–730. doi: 10.1007/s10822-019-00274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walters W.P., Barzilay R. Applications of deep learning in molecule generation and molecular property prediction. Acc. Chem. Res. 2020;54:263–270. doi: 10.1021/acs.accounts.0c00699. [DOI] [PubMed] [Google Scholar]
- 60.Kim J., Park S., Min D., Kim W. Comprehensive survey of recent drug discovery using deep learning. Int. J. Mol. Sci. 2021;22:9983. doi: 10.3390/ijms22189983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joshi T., Joshi T., Pundir H., Sharma P., Mathpal S., Chandra S. Predictive modeling by deep learning, virtual screening and molecular dynamics study of natural compounds against SARS-CoV-2 main protease. J. Biomol. Struct. Dyn. 2021;39:6728–6746. doi: 10.1080/07391102.2020.1802341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nand M., Maiti P., Joshi T., Chandra S., Pande V., Kuniyal J.C., Ramakrishnan M.A. Virtual screening of anti-HIV1 compounds against SARS-CoV-2: machine learning modeling, chemoinformatics and molecular dynamics simulation based analysis. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-77524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chicco D., Warrens M.J., Jurman G. The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ. Comp. Sci. 2021;7:e623. doi: 10.7717/peerj-cs.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ghosh R., Chakraborty A., Biswas A., Chowdhuri S. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors–an in silico docking and molecular dynamics simulation study. J. Biomol. Struct. Dyn. 2021;39:4362–4374. doi: 10.1080/07391102.2020.1779818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou X., Zhong F., Lin C., Hu X., Zhang Y., Xiong B., Yin X., Fu J., He W., Duan J. Structure of SARS-CoV-2 main protease in the apo state. Sci. China Life Sci. 2021;64:656–659. doi: 10.1007/s11427-020-1791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kneller D.W., Phillips G., O'Neill H.M., Jedrzejczak R., Stols L., Langan P., Joachimiak A., Coates L., Kovalevsky A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020;11:3202. doi: 10.1038/s41467-020-16954-7. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferreira J.C., Fadl S., Villanueva A.J., Rabeh W.M. Catalytic dyad residues His41 and Cys145 impact the catalytic activity and overall conformational fold of the main SARS-CoV-2 protease 3-chymotrypsin-like protease. Front. Chem. 2021;9:491. doi: 10.3389/fchem.2021.692168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Narayanan A., Toner S.A., Jose J. Structure-based inhibitor design and repurposing clinical drugs to target SARS-CoV-2 proteases. Biochem. Soc. Trans. 2022;50:151–165. doi: 10.1042/BST20211180. [DOI] [PubMed] [Google Scholar]
- 69.Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Coronavirus Disease 2019 (COVID-19) Springer; 2020. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pp. 23–31. [Google Scholar]
- 70.Khan S., Fakhar Z., Hussain A., Ahmad A., Jairajpuri D.S., Alajmi M.F., Hassan M.I. Structure-based identification of potential SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2022;40:3595–3608. doi: 10.1080/07391102.2020.1848634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.ul Qamar M.T., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharmaceut. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.S.S. Mousavi, A. Karami, T.M. Haghighi, S.G. Tumilaar, Fatimawali, Idroes, R. In silico evaluation of Iranian medicinal plant phytoconstituents as inhibitors against main protease and the receptor-binding domain of SARS-CoV-2. Molecules. 26 (2021) 5724. [DOI] [PMC free article] [PubMed]
- 73.Peterson L. SSRN; 2020. COVID-19 and Flavonoids, Silico Molecular Dynamics Docking to the Active Catalytic Site of SARS-CoV and SARS-CoV-2 Main Protease. [Google Scholar]
- 74.Liu S.-Y., Wang W., Ke J.-P., Zhang P., Chu G.-X., Bao G.-H. Discovery of Camellia sinensis catechins as SARS-CoV-2 3CL protease inhibitors through molecular docking, intra and extra cellular assays. Phytomedicine. 2022;96 doi: 10.1016/j.phymed.2021.153853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mahmud S., Biswas S., Paul G.K., Mita M.A., Promi M.M., Afrose S., Hasan M.R., Zaman S., Uddin M.S., Dhama K. Plant-based phytochemical screening by targeting main protease of SARS-CoV-2 to design effective potent inhibitors. Biology. 2021;10:589. doi: 10.3390/biology10070589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hanai T. Quantitative in silico analysis of SARS-CoV-2 S-RBD omicron mutant transmissibility. Talanta. 2022;240 doi: 10.1016/j.talanta.2022.123206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi K.-E., Kim J.-M., Rhee J., Park A.K., Kim E.-J., Kang N.S. Molecular Dynamics Studies on the structural characteristics for the stability prediction of SARS-CoV-2. Int. J. Mol. Sci. 2021;22:8714. doi: 10.3390/ijms22168714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh V.K., Chaurasia H., Kumari P., Som A., Mishra R., Srivastava R., Naaz F., Singh A., Singh R.K. Design, synthesis, and molecular dynamics simulation studies of quinoline derivatives as protease inhibitors against SARS-CoV-2. J. Biomol. Struct. Dyn. 2021:1–24. doi: 10.1080/07391102.2021.1946716. [DOI] [PubMed] [Google Scholar]
- 79.Dutta Dubey K., Kumar Tiwari R., Prasad Ojha R. Recent advances in protein− ligand interactions: molecular dynamics simulations and binding free energy. Curr. Comput. Aided Drug Des. 2013;9:518–531. doi: 10.2174/15734099113096660036. [DOI] [PubMed] [Google Scholar]
- 80.Mosquera-Yuqui F., Lopez-Guerra N., Moncayo-Palacio E.A. Targeting the 3CLpro and RdRp of SARS-CoV-2 with phytochemicals from medicinal plants of the Andean Region: molecular docking and molecular dynamics simulations. J. Biomol. Struct. Dyn. 2022;40:2010–2023. doi: 10.1080/07391102.2020.1835716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mohapatra R.K., Dhama K., El–Arabey A.A., Sarangi A.K., Tiwari R., Bin Emran T., Azam M., Al-Resayes S.I., Raval M.K., Seidel V. Repurposing benzimidazole and benzothiazole derivatives as potential inhibitors of SARS-CoV-2: DFT, QSAR, molecular docking, molecular dynamics simulation, and in-silico pharmacokinetic and toxicity studies. J. King Saud Univ. Sci. 2021;33 doi: 10.1016/j.jksus.2021.101637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang H., Sun L., Li W., Liu G., Tang Y. In silico prediction of chemical toxicity for drug design using machine learning methods and structural alerts. Front. Chem. 2018;6:30. doi: 10.3389/fchem.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Segall M.D., Barber C. Addressing toxicity risk when designing and selecting compounds in early drug discovery. Drug Discov. Today. 2014;19:688–693. doi: 10.1016/j.drudis.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 84.Williams J.K., Wang B., Sam A., Hoop C.L., Case D.A., Baum J. Molecular dynamics analysis of a flexible loop at the binding interface of the SARS‐CoV‐2 spike protein receptor‐binding domain, Proteins. Struct. Funct. Bioinformat. 2022;90:1044–1053. doi: 10.1002/prot.26208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Esteves F., Rueff J., Kranendonk M. The central role of cytochrome P450 in xenobiotic metabolism—a brief review on a fascinating enzyme family. J. Xenobio. 2021;11:94–114. doi: 10.3390/jox11030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.El-Banna A.A., Darwish R.S., Ghareeb D.A., Yassin A.M., Abdulmalek S.A., Dawood H.M. Metabolic profiling of Lantana camara L. using UPLC-MS/MS and revealing its inflammation-related targets using network pharmacology-based and molecular docking analyses. Sci. Rep. 2022;12:1–17. doi: 10.1038/s41598-022-19137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Swain S.S., Hussain T. Role of bioinformatics in early drug discovery: an overview and perspective. Computat. BioInformat.: Multidisc. Appl. 2021:49–67. [Google Scholar]
- 88.Lei T., Li Y., Song Y., Li D., Sun H., Hou T. ADMET evaluation in drug discovery: 15. Accurate prediction of rat oral acute toxicity using relevance vector machine and consensus modeling. J. Cheminf. 2016;8:1–19. doi: 10.1186/s13321-016-0117-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu Y., Pei J., Lai L. Deep learning based regression and multiclass models for acute oral toxicity prediction with automatic chemical feature extraction. J. Chem. Inf. Model. 2017;57:2672–2685. doi: 10.1021/acs.jcim.7b00244. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.