Abstract
Form‐function relationships in mammalian feeding systems are active topics of research in evolutionary biology. This is due principally to their fundamental importance for understanding dietary adaptations in extinct taxa and macro‐evolutionary patterns of morphological transformations through changing environments. We hypothesize that three‐dimensional dental topographic metrics represent stronger predictors for dietary and other ecological variables than do linear measurements. To test this hypothesis, we measured three dental topographic metrics: Relief Index (RFI), Dirichlet Normal Energy (DNE), and Orientation Patch Count Rotated (OPCR) in 57 extant carnivoran species. Premolar and molar dental topographic indices were regressed against activity, diet breadth, habitat breadth, terrestriality, and trophic level variables within a phylogenetic framework. The results of this study showed significant correlations between RFI and the ecological variables diet breadth and trophic level. Weaker correlations are documented between OPCR and activity and between DNE and trophic level. Our results suggest that cusp height is strongly reflective of dietary ecology in carnivorans as a whole, and represents a proxy mainly for different degrees of hypercarnivory observed within this group of predatory mammals.
Keywords: Carnivora, dental topography, diet breadth, Dirichlet Normal Energy, Orientation Patch Count Rotated, Relief Index, teeth, terrestriality, trophic level
Dental topographic metrics represent stronger predictors for dietary and other ecological variables than do linear measurements. We measured three dental topographic metrics: Relief Index (RFI), Dirichlet Normal Energy (DNE), and Orientation Patch Count Rotated (OPCR) in 57 extant carnivoran species. Our results suggest that cusp height is strongly reflective of dietary ecology in carnivorans as a whole, and represents a proxy mainly for different degrees of hypercarnivory observed within this group of predatory mammals.
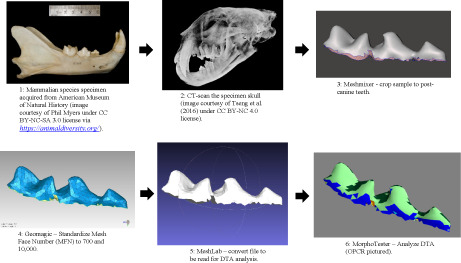
1. INTRODUCTION
Dietary habits and ecological traits are among some of the most commonly analyzed variables in comparative studies of mammalian diversity. Teeth are unique identifiers for these characteristics because the dentition is the only part of the skeleton directly exposed to and interacting with the external environment. Consequently, teeth are critical for understanding an individual's ecology and the direct interaction between organisms and their environment (Haeckel, 1866), particularly in determining feeding ecology. Additionally, teeth are ideal structures for determining rate and pattern of growth and development of mammals and other vertebrates, and the dentition also can be used to study both ontogenetic and phylogenetic patterns, as well as providing a rich source of shared‐derived (synapomorphic) phylogenetic traits for assisting taxonomic classification (Schwartz & Dean, 2000; Winchester et al., 2014).
As a quantitative method for characterizing ecological or dietary groupings using teeth, dental topographic analysis (DTA) metrics were first utilized to study and infer diet in fossil hominins. DTA subsequently has been expanded to include diet studies, wear stages, and lifestyle dental effects in a variety of both extinct and extant mammals (Reed, 1997; Thiery et al., 2019; Ungar & Williamson, 2000; Zuccotti et al., 1998).
Hopkins et al. (2021) demonstrated that using linear and area measurements of dental morphology in carnivoran mammals to interpret dietary ecology is susceptible to large errors as a function of limited dataset size and confounding phylogenetic signals. They also showed that phylogenetic non‐independence decreases the accuracy of diet predictions, and much of the information in dental morphology on dietary ecology cannot be decoupled from evolutionary constraints characteristic of particular clades due to ancestry. Thus, the utility of non‐dental cranial traits and linear dental morphology metrics in predicting or reconstructing diet in living and extinct mammals may rest on the use of multiple variables for more confident dietary inferences.
In an analysis of family‐level dental form‐function, Pérez‐Ramos et al. (2020) focused on Ursidae, and showed a phylogenetic signal associated with curvature and complexity in both living and extinct bears. Additionally, diets of ursids could be distinguished by DTA measurements. In an analysis of plant diets, Eronen et al. (2017) examined primates with specialized bamboo versus generalized fibrous plant diets and found that specialized fibrous (bamboo) diets correlated with extremely high complexity DTA value of molar surfaces.
Our study quantifies tooth morphology metrics of dental topography (Thiery et al., 2019; Ungar & Williamson, 2000; Zuccotti et al., 1998). Additionally, teeth are among the most commonly preserved body parts in the vertebrate fossil record, and thus are most useful for paleobiological studies, as soft tissue is rarely preserved and other hard tissues such as bones are not as frequently found in some environments or as precisely identifiable taxonomically as the more durable dental tissues (Shipman, 1981). Post‐canine mammalian dental morphology often has been used to infer behavior, diet, and ecology in extinct mammals (e.g., Berthaume et al., 2019, 2020). These correlations are especially strong in mammals that masticate food before ingestion (Lucas, 2004; Ungar, 2010). Therefore, not only can we predict dental morphology to be under strong selective pressure and tight genetic control, but we can also measure quantitative three‐dimensional dental topographic metrics to represent stronger predictors for dietary variables than linear measurements. We formulated and tested this form‐function hypothesis based on well‐defined dental topographic features (see next section).
1.1. Topographic analysis of tooth morphology
As introduced above, DTA is an important tool for quantification of tooth morphology. DTA has been applied extensively to mammalian dentitions to identify anatomical features, characterize worn teeth, and infer an organism's dietary grouping (Berthaume et al., 2020; Bunn et al., 2011). There are three principal dental topographic metrics: Relief Index (RFI), Orientation Patch Count Rotated (OPCR) (modified from Orientation Patch Count [OPC], see below), and Dirichlet Normal Energy (DNE).
Relief Index is a metric for determining overall tooth shape (Ungar & Williamson, 2000), calculated as the three‐dimensional dentition surface area divided by the two‐dimensional outline, multiplied by 100. RFI is documented to separate diets most effectively among these three metrics, distinguishing ecological insectivores (insect‐consumers) and folivores (leaf‐consumers) from frugivores (fruit‐consumers), in Euarchonta, a clade comprising extinct and extant primates, tree shrews (Scandentia), and colugos (Dermoptera) (Bunn et al., 2011).
Another common dental topographic metric is the OPC, which quantifies tooth surface complexity. If teeth are described as multi‐functional tools, OPC is the number of tools on a tooth's surface (Evans et al., 2007). OPC was originally measured by stacking a 2.5D grid over the surface of a dentition, thereby assigning the same number of dental surfaces to an equal number of polygons (Berthaume et al., 2019, 2020). The normal vector for each polygon was assigned, and polygons were distributed into “bins” based on the direction in which the normal vector pointed (anteriorly, anterior‐medially, medially, etc.). Polygons with three or more common edges were assigned to the same bin, and therefore formed a “patch”, with more complex teeth having more patches (Berthaume et al., 2019).
Currently, OPC is calculated by locating the orientations of grid points on a tooth in eight compass directions (Bunn et al., 2011). This measurement has been assessed and replicated in herbivores and hypercarnivores of two different orders in which species in the same dietary category possess similar OPC values (Evans et al., 2007). To improve the sensitivity of this metric, OPCR was introduced (Evans & Jernvall, 2009). This modified metric was applied to individual teeth instead of entire tooth rows and averaged eight OPC values for eight tooth orientations to account for variations in scan orientations (Berthaume et al., 2019). However, OPCR was not able to distinguish different primate dietary groups in Bunn et al.'s (2011) study. Additionally, OPCR has been considered an informative measure to understand diet preferences in extant “prosimian” primates and the extinct primatomorph class, plesiadapiformes, even with non‐trophic level diet categorizations (e.g., Boyer et al., 2010). In contrast, Berthaume et al. (2019) determined that OPCR does not predict diet in two primate groups analyzed: platyrrhines and prosimians.
A third parameter, DNE, was introduced as an orientation‐free dental metric quantifying tooth shape (molar), independent of position, orientation, scale, or landmarks (Bunn et al., 2011). Further, DNE was interpreted as surface “energy” and quantifies the deviation from a stable minimal state of energy, or planar form (Bunn et al., 2011). DNE allowed differentiation among Euarchonta species with diverse diets, such as insectivores and folivores (Bunn et al., 2011). In that same study, DNE was strongly associated with RFI, and weakly associated with OPCR; a conclusion was drawn to represent DNE as a variable reflecting the shape of the tools on the molar occlusal plane. High and steeply sloped cusps, as well as sharp shearing crests, all have high DNE values, whereas flatter more bulbous cusps produce smaller DNE values (Bunn et al., 2011). Based on these findings, DNE appears to be an informative metric to distinguish among insectivores, folivores, omnivores, and frugivores. Insectivores were reported as having the highest DNE, followed sequentially by folivores, omnivores, and then frugivores in extinct and extant primates within Euarchonta (Berthaume et al., 2020; Bunn et al., 2011).
Considered together, DNE, RFI, and OPCR are the most common dental topographic analytical metrics in recent quantitative tooth crown analyses, in preference to more localized measures such as shearing quotient and shearing ratio (Bunn et al., 2011). These three metrics can be applied reliably to both worn and unworn molar teeth, and are essentially free of errors introduced by manual selection of landmarks (Bunn et al., 2011). However, it is important to note that not only does digital model smoothing affect DNE, RFI, and OPCR, but if different dental models possess both smoothed and unsmoothed surfaces, then DNE and OPCR should not be compared (Berthaume et al., 2019). Based on the rigorous analyses and previously documented reliability of distinguishing dietary categories in other mammal groups, DNE, RFI, and OPCR are the three dental measurements applied in this study. As dental morphology is under tight genetic control, and tooth form generally correlates with diet preference, it also may be possible to infer selective adaptations between tooth form and diet function (Lucas, 2004).
Based on the findings discussed above, we hypothesized that carnivoran dental topographic metrics of tooth surface energy or sharpness (DNE) and tooth complexity (OPCR) would be related to multiple ecological variables studied, but no ecological trait would be related to relative cusp height (RFI). Tooth form is expected to correlate closely with dietary preference, as seen in previous studies and because the dentition is among the first organs to contact food (Bunn et al., 2011; Eronen et al., 2017; Pérez‐Ramos et al., 2020). Furthermore, DTA values are expected to correlate to the feeding ecology and life history traits of diet breadth, habitat breadth, and trophic level (based on dietary pressure placed specifically on the dentition), but not to locomotor traits that are less directly linked to feeding ecology and dental morphology (such as activity cycle or terrestriality).
2. MATERIALS AND METHODS
We sampled one individual for each of 57 extant carnivoran species representing all 16 modern families of Carnivora (Table S1). For consistency among specimens, we focused our analyses on the left lower postcanine teeth. The 57 specimens were from the American Museum of Natural History (AMNH) Mammalogy Collections, augmented with datasets from the literature, preferentially analyzing adult, wild‐caught skulls. These specimens were scanned in the AMNH Microscopy and Imaging Facility (MIF) via high‐resolution X‐ray micro‐computed tomography (μCT) using a GE v|tome|x system, with voxel resolutions set between ~30 and 140 μm, scaled to specimen size (larger specimens had image datasets with larger voxel sizes to yield resolution comparable to smaller specimens). The image data then were segmented using Avizo Lite (Version 9.0) (Thermo Fisher Scientific) to produce triangular mesh models that contained different quantities of mesh face number (MFN) that form the surface of a tooth model (Boyer, 2008). These models were uploaded into Autodesk Meshmixer, 2017 (Version 3.5.474; Autodesk, Inc.) to crop out all features except the lower left postcanine teeth and exported as *.ply files. Each *.ply file was then cropped, resampled, and smoothed (Berthaume et al., 2019) in Geomagic Wrap (Version 2017; 3D Systems). Figure 1 represents an illustrated workflow using the specimen example of Panthera pardus.
FIGURE 1.
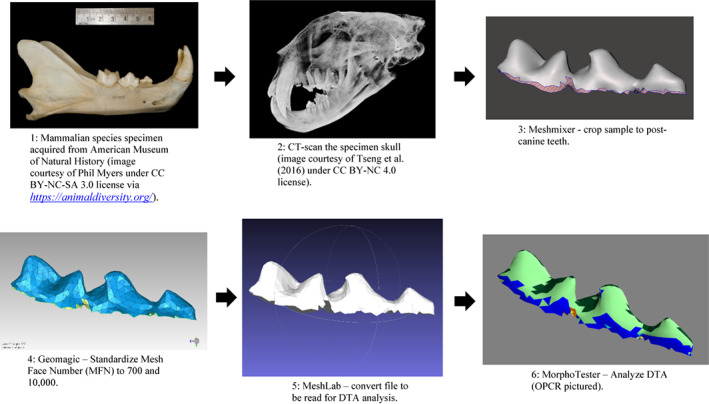
Illustrated research workflow using example specimen, Panthera pardus.
Two methods used by Berthaume et al. (2019) for cropping out all other tooth structure excluding the enamel are: basin cut‐off (BCO) and entire enamel cap (EEC). BCO isolates the enamel cap above the lowest point on the central basin, whereas EEC includes the entire outer surface of the enamel cap (Berthaume et al., 2019). The EEC method, which produces values for the whole tooth shape, was reported to differ significantly in dental metrics from BCO, which includes significant portions of the root surfaces (Berthaume et al., 2018; Boyer, 2008; Winchester, 2016). In this study, we use only the EEC cropping method of Berthaume et al. (2019) to analyze each specimen consistently. Cropped files from Geomagic Wrap were opened in MeshLab version 2016.12 (Cignoni et al., 2008), inspected for any errors such as holes in the surface of the dentition or incompletely scanned areas, then uploaded into MorphoTester 1.0.1 software (Winchester, 2016) and analyzed to determine DNE, RFI, and OPCR (Boyer et al., 2014) metrics with methods described in Berthaume et al. (2019).
In addition to analyses of the tooth mesh models, generated using the method described above, we followed the Pampush et al. (2016, 2019) protocol to standardize MFN to adjust for mesh sensitivity differences among species. Models were loaded into Geomagic Wrap 2017 to standardize the mesh number. As in Berthaume et al. (2019), cropping followed by resampling was used to examine the effect of mesh density on dental topographic metrics. DNE and OPCR, for example, are sensitive to triangle count, whereas RFI is reportedly not as sensitive (Winchester, 2016). However, at high mesh resolutions DNE is independent of triangle count, due to the lack of sharp edges or bumpy surfaces being factored into the calculation (Berthaume et al., 2019). In addition, at high mesh resolutions RFI will not be sensitive to triangle count, due to small changes in crown occlusal surface area not being dependent on overall dentition height (Lazzari & Guy, 2014). OPCR is most sensitive to changes in mesh resolution, as determined by the number of triangles that point in different orientations (Berthaume et al., 2019). To combat variable counts of tooth mesh number in studies, Berthaume et al. (2019) reported that teeth are commonly resampled at a constant triangle count. During the computed tomography (CT) data collection stage, larger teeth may have higher voxel resolution, translating to higher mesh counts, as opposed to smaller teeth that typically are not scanned at as high relative resolution and thus might have fewer mesh counts making up a comparable feature (Skinner & Gunz, 2010). Models used in this study had markedly different mesh counts, with the highest mesh count at 21,278 in the spotted‐necked otter (Lutra maculicollis) and the lowest mesh count at 683 in the aardwolf (Proteles cristatus; Table 1) (Werdelin et al., 2021). In previously reported DNE calculations, most teeth are standardized arbitrarily to 10,000 triangles (Bunn et al., 2011; Winchester et al., 2014), while other studies uniformly simplified the mesh counts to 22,000 and 55,000 triangles, respectively (Guy et al., 2013, 2015). The study by Berthaume et al. (2019) stated that the effects of cropping and analysis of triangle counts were most variable in low triangle counts (under 210), and the final mesh number count for a low triangle count did not reflect actual tooth surface area and size measurements; all triangle counts in this study lie above that threshold.
TABLE 1.
Species and dental topographic measurement dataset.
| Species | Clade | MFN 10k | DNE 10k | RFI 10k | OPCR 10k | MFN 700 | DNE 700 | RFI 700 | OPCR 700 | Tooth # (all postcanine) | Premolar # (total) | Molar # (total) | Missing teeth |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinonyx jubatus | Feliformia | 9998 | 912.012 | 7.706 | 169.125 | 699 | 321.662 | 7.626 | 44.75 | 3 | 2 | 1 | 0 |
| Ailuropoda melanoleuca | Caniformia | 9999 | 478.686 | 7.519 | 71.25 | 698 | 216.84 | 7.717 | 54.875 | 6 | 3 | 3 | 0 |
| Ailurus fulgens | Caniformia | 9998 | 300.266 | 4.336 | 64.125 | 700 | 156.569 | 4.42 | 46.125 | 6 | 4 | 2 | 0 |
| Aonyx capensis | Caniformia | 9998 | 369.401 | 2.2 | 154.125 | 698 | 186.015 | 2.233 | 62 | 3 | 3 | 2 | 2 |
| Aonyx cinerea | Caniformia | 9998 | 461.541 | 7.148 | 59.125 | 698 | 254.235 | 7.3 | 72 | 5 | 3 | 2 | 0 |
| Arctonyx collaris | Caniformia | 9999 | 271.044 | 9.158 | 56.125 | 698 | 163.039 | 9.154 | 53.25 | 5 | 3 | 2 | 0 |
| Atilax paludinosus | Feliformia | 9999 | 586.433 | 3.41 | 164.875 | 698 | 302.172 | 3.438 | 77.625 | 5 | 3 | 2 | 0 |
| Bassariscus astutus | Caniformia | 9999 | 365.483 | 8.899 | 38.125 | 699 | 187.814 | 9.022 | 48.625 | 5 | 4 | 2 | 1 |
| Callorhinus ursinus | Caniformia | 9998 | 245.685 | 2.684 | 65.625 | 698 | 150.662 | 2.758 | 62.625 | 5 | 4 | 1 | 0 |
| Canis lupus | Caniformia | 10,000 | 481.171 | 7.774 | 89.625 | 699 | 272.544 | 7.957 | 65.75 | 7 | 4 | 3 | 0 |
| Civettictis civetta | Feliformia | 9999 | 243.672 | 4.471 | 42.125 | 698 | 148.853 | 4.558 | 47.75 | 6 | 4 | 2 | 0 |
| Crocuta crocuta | Feliformia | 9999 | 334.153 | 6.286 | 67 | 698 | 171.888 | 6.379 | 48.375 | 4 | 3 | 1 | 0 |
| Cryptoprocta ferox | Feliformia | 9998 | 359.447 | 4.299 | 82.125 | 698 | 204.977 | 4.416 | 58.375 | 4 | 3 | 1 | 0 |
| Cynogale bennettii | Feliformia | 9999 | 794.515 | 3.087 | 138.125 | 699 | 377.092 | 3.178 | 68.5 | 6 | 4 | 2 | 0 |
| Eira barbara | Caniformia | 10,000 | 354.739 | 2.865 | 82.5 | 700 | 122.855 | 2.911 | 46.875 | 4 | 2 | 2 | 1 |
| Enhydra lutris | Caniformia | 9999 | 201.457 | 2.555 | 111.375 | 699 | 133.173 | 2.597 | 56.5 | 4 | 3 | 1 | 1 |
| Erignathus barbatus | Caniformia | 9999 | 183.71 | 2.922 | 74.75 | 697 | 107.169 | 2.982 | 46.25 | 5 | 5 | 0 | 0 |
| Eupleres goudotii | Feliformia | 9998 | 539.481 | 2.654 | 190.5 | 698 | 307.119 | 2.725 | 84 | 6 | 4 | 2 | 0 |
| Felis silvestris | Feliformia | 10,000 | 295.737 | 4.059 | 62.625 | 698 | 184.879 | 4.131 | 54.75 | 3 | 2 | 1 | 0 |
| Fossa fossana | Feliformia | 9998 | 615.905 | 3.054 | 163.25 | 698 | 318.72 | 3.173 | 77.25 | 6 | 4 | 2 | 0 |
| Galidictis fasciata | Feliformia | 9998 | 528.274 | 7.788 | 84.25 | 696 | 269.046 | 7.971 | 66.5 | 5 | 3 | 2 | 0 |
| Genetta johnstoni | Feliformia | 9999 | 530.568 | 3.231 | 173.25 | 698 | 315.658 | 3.359 | 73.625 | 6 | 4 | 2 | 0 |
| Gulo gulo | Caniformia | 9998 | 642.8 | 5.069 | 171.875 | 698 | 218.873 | 5.12 | 68.625 | 6 | 4 | 2 | 0 |
| Herpestes javanicus | Feliformia | 9998 | 702.737 | 5.082 | 147.5 | 698 | 373.223 | 5.214 | 79.75 | 6 | 4 | 2 | 0 |
| Lutra maculicollis | Caniformia | 10,000 | 542.674 | 7.214 | 114 | 699 | 272.381 | 7.486 | 66.125 | 5 | 3 | 2 | 0 |
| Hydrurga leptonyx | Caniformia | 10,002 | 772.541 | 3.155 | 165.125 | 699 | 352.076 | 3.238 | 82.5 | 5 | 5 | 0 | 0 |
| Lontra canadensis | Caniformia | 9998 | 512.178 | 2.841 | 165.25 | 698 | 257.217 | 2.956 | 72.75 | 5 | 3 | 2 | 0 |
| Lontra felina | Caniformia | 10,000 | 459.788 | 6.304 | 103 | 699 | 240.146 | 6.459 | 59.25 | 5 | 3 | 2 | 0 |
| Lontra longicaudis | Caniformia | 9999 | 650.266 | 6.725 | 119.5 | 699 | 280.348 | 6.906 | 71 | 5 | 3 | 2 | 0 |
| Lutra lutra | Caniformia | 9999 | 468.35 | 6.936 | 88.375 | 699 | 239.916 | 7.024 | 66.125 | 5 | 3 | 2 | 0 |
| Lutrogale perspicillata | Caniformia | 9999 | 552.091 | 5.418 | 139 | 698 | 273.206 | 5.518 | 73.875 | 5 | 3 | 2 | 0 |
| Lycaon pictus | Caniformia | 9999 | 481.114 | 7.013 | 114 | 698 | 264.737 | 7.107 | 57.125 | 6 | 4 | 2 | 1 |
| Mellivora capensis | Caniformia | 9998 | 316.738 | 2.882 | 103.625 | 698 | 101.389 | 2.901 | 40.125 | 4 | 3 | 1 | 0 |
| Mephitis mephitis | Caniformia | 10,000 | 367.923 | 3.882 | 107.125 | 699 | 221.932 | 3.935 | 63 | 5 | 3 | 2 | 0 |
| Mungos mungo | Caniformia | 9999 | 313.556 | 2.861 | 134.875 | 699 | 178.482 | 2.964 | 65.75 | 5 | 3 | 2 | 0 |
| Mustela frenata | Caniformia | 10,001 | 469.33 | 2.837 | 138.75 | 699 | 220.935 | 2.897 | 63.5 | 5 | 3 | 2 | 0 |
| Mydaus javanensis | Caniformia | 9998 | 522.531 | 2.559 | 179 | 698 | 202.628 | 2.601 | 66 | 3 | 1 | 2 | 3 |
| Nandinia binotata | Feliformia | 10,000 | 268.888 | 5.365 | 74.875 | 697 | 172.846 | 5.479 | 62.25 | 6 | 4 | 2 | 0 |
| Neofelis nebulosa | Feliformia | 10,001 | 226.012 | 4.249 | 86.25 | 700 | 126.006 | 4.283 | 39.875 | 2 | 1 | 1 | 1 |
| Odobenus rosmarus | Caniformia | 10,000 | 307.126 | 2.429 | 66.375 | 699 | 211.413 | 2.478 | 43.5 | 3 | 3 | 0 | 0 |
| Panthera pardus | Feliformia | 9999 | 330.939 | 5.224 | 77.375 | 697 | 153.304 | 5.327 | 43.875 | 3 | 2 | 1 | 0 |
| Paradoxurus hermaphroditus | Feliformia | 9999 | 914.83 | 2.555 | 112.375 | 700 | 463.81 | 2.643 | 66 | 5 | 3 | 2 | 1 |
| Hyaena brunnea | Feliformia | 9999 | 231.389 | 2.417 | 75 | 697 | 118.426 | 2.444 | 49.25 | 4 | 3 | 1 | 0 |
| Phoca vitulina | Caniformia | 9999 | 261.887 | 2.486 | 78.5 | 699 | 123.133 | 2.525 | 45.875 | 3 | 3 | 0 | 2 |
| Potos flavus | Caniformia | 9999 | 144.464 | 3.392 | 26.375 | 698 | 89.102 | 3.427 | 27.375 | 4 | 2 | 2 | 1 |
| Prionodon linsang | Feliformia | 9999 | 500.401 | 6.108 | 115.625 | 696 | 280.909 | 6.274 | 64.5 | 5 | 4 | 1 | 1 |
| Procyon lotor | Caniformia | 9999 | 354.128 | 3.565 | 84.625 | 697 | 186.486 | 3.598 | 59.375 | 6 | 4 | 2 | 0 |
| Proteles cristatus | Feliformia | 9997 | 152.268 | 4.529 | 91.375 | 699 | 62.37 | 4.538 | 23 | 3 | 2 | 1 | na |
| Pteronura brasiliensis | Caniformia | 9999 | 538.663 | 7.145 | 96.75 | 699 | 257.744 | 7.293 | 74.375 | 5 | 3 | 2 | 0 |
| Spilogale putorius | Caniformia | 9998 | 684.021 | 3.22 | 161.5 | 698 | 326.143 | 3.371 | 78.5 | 5 | 3 | 2 | 0 |
| Suricata suricatta | Feliformia | 9998 | 284.636 | 3.415 | 95.375 | 699 | 180.386 | 3.458 | 63.375 | 5 | 3 | 2 | 0 |
| Taxidea taxus | Caniformia | 10,001 | 291.772 | 2.537 | 105 | 699 | 184.045 | 2.567 | 66.25 | 4 | 3 | 1 | 1 |
| Tremarctos ornatus | Caniformia | 10,000 | 257.361 | 2.471 | 104.75 | 700 | 110.657 | 2.501 | 43.75 | 4 | 2 | 2 | 3 |
| Ursus arctos | Caniformia | 9999 | 390.217 | 2.42 | 136.75 | 699 | 202.01 | 2.49 | 68.375 | 6 | 3 | 3 | 0 |
| Viverra tangalunga | Feliformia | 9997 | 378.789 | 3.885 | 94.25 | 697 | 194.221 | 3.964 | 68.625 | 6 | 4 | 2 | 0 |
| Vulpes vulpes | Caniformia | 10,000 | 699.159 | 2.64 | 267.375 | 699 | 350.194 | 2.669 | 83.5 | 7 | 4 | 3 | 0 |
| Zalophus californianus | Caniformia | 9999 | 275.467 | 2.912 | 73.25 | 697 | 159.604 | 2.987 | 53.875 | 4 | 4 | 0 | 1 |
Note: Values of columns include mesh face number (MFN), Dirichlet Normal Energy (DNE), Relief Index (RFI), and Orientation Patch Count Rotated (OPCR) each at 700 MFN and 10,000 MFN counts. Tooth # presents the number of analyzed teeth, and missing teeth lists any teeth that are missing from the analyzed specimen's mandible, and therefore not included in the overall analysis.
To assess the effect of different standardized MFNs, we generated two additional uniformly standardized datasets of all tooth mesh models at 700 and 10,000 mesh counts, respectively. We repeated 10 iterations of surface refinement (triangle subdivision) in Geomagic Wrap, then down‐sampled specimens that originally were over 10,000 mesh count to the targets of 10,000 (Pampush et al., 2016, 2019) and 700 mesh counts. These computer‐altered specimens were then saved in the same *.ply file format as the original meshes. We then noted the MFN under each model dataset/sampling regime and repeated the associated DNE, RFI, and OPCR analyses for each of the two resampled model datasets (see Table 1 for results).
To account for data non‐independence and different degrees of evolutionary relatedness between species, a molecular phylogeny was generated based on data from Upham et al. (2019). DTA measurements were quantitatively compared to the ecological characteristics of species in this study using pairwise phylogenetically informed regression analyses. All possible pairings of DTA (n = 3 variables) to ecological (n = 5 variables) traits were regressed in order to assess the degree to which each DTA variable correlates with ecological ones; no interactions among multiple DTA and/or ecological variables were tested. The five ecological variables were activity cycle, diet breadth, habitat breadth, terrestriality/arboreality, and trophic level, with ecological data from the PanTHERIA database (Jones et al., 2009) (Table 2). Diet breadth is defined as the number of dietary groupings, from among these classifications of food sources: vertebrate, invertebrate, fruit, flower/nectar/pollen, leaves/branches/bark, seeds, grass, and roots/tubers. Habitat breadth is the number of habitat layers a species populates, and is characterized as scansorial, aquatic, fossorial (under‐ground dwelling), or ground‐dwelling. Trophic level for each species is defined as herbivore, omnivore, or carnivore. Activity is categorized as nocturnal only, nocturnal/crepuscular (twilight; also includes cathemeral or equally distributed in the 24‐h daily cycle), or crepuscular or diurnal/crepuscular. Degrees of terrestriality are determined from the PanTHERIA database, defined as a qualitative or quantitative time measure of the dominant percentage of time spent by a species in a specific habitat, and further subdivided into either as fossorial (or ground‐dwelling), scansorial (both above‐ground and ground‐dwelling), semi‐aquatic, or fully aquatic (Jones et al., 2009). Scansorial was defined as ground‐dwelling species who have the additional lifestyle ability to climb and live in an arboreal setting (Nakagawa et al., 2007). In this study, we also use this term to define a group of mammals which are ground‐dwelling, but spend a significant amount of time above‐ground in trees, but are not exclusively an arboreal‐dwelling taxa. For the variables analyzed, only the bearded seal (Erignathus barbatus) and smooth‐coated otter (Lutrogale perspicillata) did not have complete life history information for activity cycle in the PanTHERIA database, and therefore these two species were excluded from the analysis for that variable.
TABLE 2.
Species and ecology/life history values dataset.
| Species | Clade | Activity (1: Nocturnal; 2: Nocturnal/crepuscular; 3: Diurnal/crepuscular) | Diet breadth (from 1 [narrowest] to 6 [broadest]) | Habitat breadth (from 1 [narrowest] to 3 [broadest]) | Terrestriality (1: Fossorial; 2: Scansorial; 3: Semi‐aquatic; 4: Fully aquatic) | Trophic level (1: Herbivore; 2: Omnivore; 3:Carnivore) | Family |
|---|---|---|---|---|---|---|---|
| Acinonyx jubatus | Feliformia | 3 | 1 | 1 | 1 | 3 | Felidae |
| Ailuropoda melanoleuca | Caniformia | 2 | 3 | 1 | 1 | 1 | Ursidae |
| Ailurus fulgens | Caniformia | 2 | 3 | 1 | 1 | 1 | Ailuridae |
| Aonyx capensis | Caniformia | 2 | 6 | 2 | 3 | 2 | Mustelidae |
| Aonyx cinerea | Caniformia | 1 | 2 | 2 | 3 | 3 | Mustelidae |
| Arctonyx collaris | Caniformia | 1 | 6 | 1 | 1 | 2 | Mustelidae |
| Atilax paludinosus | Feliformia | 2 | 6 | 2 | 3 | 2 | Herpestidae |
| Bassariscus astutus | Caniformia | 1 | 6 | 1 | 1 | 2 | Procyonidae |
| Callorhinus ursinus | Caniformia | 2 | 6 | 2 | 4 | 2 | Otariidae |
| Canis lupus | Caniformia | 2 | 1 | 1 | 1 | 3 | Canidae |
| Civettictis civetta | Feliformia | 1 | 6 | 1 | 1 | 2 | Viverridae |
| Crocuta crocuta | Feliformia | 1 | 1 | 1 | 1 | 3 | Hyaenidae |
| Cryptoprocta ferox | Feliformia | 2 | 2 | 3 | 2 | 3 | Eupleridae |
| Cynogale bennettii | Feliformia | 2 | 6 | 2 | 3 | 2 | Viverridae |
| Eira barbara | Caniformia | 2 | 2 | 2 | 2 | 2 | Mustelidae |
| Enhydra lutris | Caniformia | 3 | 6 | 1 | 4 | 2 | Mustelidae |
| Erignathus barbatus | Caniformia | na | 2 | 1 | 4 | na | Phocidae |
| Eupleres goudotii | Feliformia | 2 | 6 | 1 | 1 | 2 | Eupleridae |
| Felis silvestris | Feliformia | 2 | 1 | 1 | 1 | 3 | Felidae |
| Fossa fossana | Feliformia | 1 | 6 | 2 | 2 | 3 | Eupleridae |
| Galidictis fasciata | Feliformia | 2 | 2 | 1 | 1 | 3 | Eupleridae |
| Genetta johnstoni | Feliformia | 1 | 1 | 2 | 3 | 3 | Viverridae |
| Gulo gulo | Caniformia | 1 | 1 | 1 | 1 | 3 | Mustelidae |
| Herpestes javanicus | Feliformia | 3 | 6 | 1 | 1 | 2 | Herpestidae |
| Lutra maculicollis | Caniformia | 2 | 1 | 2 | 3 | 3 | Mustelidae |
| Hydrurga leptonyx | Caniformia | 2 | 6 | 2 | 4 | 2 | Phocidae |
| Lontra canadensis | Caniformia | 2 | 1 | 2 | 3 | 3 | Mustelidae |
| Lontra felina | Caniformia | 3 | 2 | 2 | 3 | 3 | Mustelidae |
| Lontra longicaudis | Caniformia | 2 | 2 | 3 | 3 | 3 | Mustelidae |
| Lutra lutra | Caniformia | 1 | 1 | 2 | 3 | 3 | Mustelidae |
| Lutrogale perspicillata | Caniformia | na | 2 | 2 | 3 | 3 | Mustelidae |
| Lycaon pictus | Caniformia | 3 | 1 | 1 | 1 | 3 | Canidae |
| Mellivora capensis | Caniformia | 2 | 6 | 1 | 1 | 2 | Mustelidae |
| Mephitis mephitis | Caniformia | 1 | 6 | 1 | 1 | 2 | Mephitidae |
| Mungos mungo | Caniformia | 3 | 1 | 1 | 1 | 3 | Herpestidae |
| Mustela frenata | Caniformia | 2 | 1 | 2 | 2 | 3 | Mustelidae |
| Mydaus javanensis | Caniformia | 1 | 6 | 1 | 1 | 2 | Mephitidae |
| Nandinia binotata | Feliformia | 1 | 2 | 2 | 2 | 1 | Nandiniidae |
| Neofelis nebulosa | Feliformia | 2 | 1 | 2 | 2 | 3 | Felidae |
| Odobenus rosmarus | Caniformia | 2 | 6 | 2 | 4 | 2 | Odobenidae |
| Panthera pardus | Feliformia | 2 | 1 | 1 | 1 | 3 | Felidae |
| Paradoxurus hermaphroditus | Feliformia | 1 | 6 | 1 | 1 | 2 | Viverridae |
| Hyaena brunnea | Feliformia | 1 | 6 | 2 | 1 | 2 | Hyaenidae |
| Phoca vitulina | Caniformia | 2 | 1 | 2 | 4 | 3 | Phocidae |
| Potos flavus | Caniformia | 1 | 4 | 2 | 2 | 2 | Procyonidae |
| Prionodon linsang | Feliformia | 1 | 6 | 1 | 1 | 2 | Prionodontidae |
| Procyon lotor | Caniformia | 2 | 6 | 2 | 2 | 2 | Procyonidae |
| Proteles cristatus | Feliformia | 1 | 1 | 1 | 1 | 3 | Hyaenidae |
| Pteronura brasiliensis | Caniformia | 3 | 1 | 2 | 3 | 3 | Mustelidae |
| Spilogale putorius | Caniformia | 1 | 6 | 1 | 1 | 2 | Mephitidae |
| Suricata suricatta | Feliformia | 3 | 1 | 1 | 1 | 3 | Herpestidae |
| Taxidea taxus | Caniformia | 1 | 1 | 1 | 1 | 3 | Mustelidae |
| Tremarctos ornatus | Caniformia | 2 | 5 | 1 | 1 | 2 | Ursidae |
| Ursus arctos | Caniformia | 2 | 6 | 1 | 1 | 2 | Ursidae |
| Viverra tangalunga | Feliformia | 1 | 6 | 1 | 1 | 2 | Viverridae |
| Vulpes vulpes | Caniformia | 1 | 1 | 1 | 1 | 3 | Canidae |
| Zalophus californianus | Caniformia | 2 | 6 | 2 | 4 | 2 | Otariidae |
Note: Values of columns include species' clade and family of specimens, and the numerically quantified life history variables as extracted from the PanTHERIA dataset.
Statistical analyses were performed using R v4.0.4 and RStudio v1.3.959 (R Core Team, 2020) with the following packages: ape (Paradis & Schliep, 2019), nlme (Pinheiro et al., 2022), geiger (Pennell et al., 2014), caper (David et al., 2018; Orme et al., 2013), and picante (Kembel et al., 2010). Phylogenetic generalized least squares (PGLS) analysis was used in the analysis of correlations between dental topographic metrics and the five life history traits examined, to account for the effect of covariance with phylogeny, as data from evolutionarily more closely related species need to be controlled for their sharing more similar traits due solely to shared ancestry (Mundry, 2014). A phylogenetic signal test using the Kmult statistic was used to determine the influence of phylogenetic relatedness on the three DTA metrics and ecological variables (Münkemüller et al., 2012).
3. RESULTS
The first set of analyzed data for DNE, RFI, OPCR, and MFN was compared to the five life history traits (activity, diet breadth, habitat breadth, terrestriality, and trophic level), with phylogenetic relationships taken into account using PGLS. The first data set used original, unadjusted MFN proportional to CT scan resolution (Table A1), before adjusting to standardized resolutions of 700 and 10,000 mesh counts, as described above.
We examined tooth size allometry using Autodesk Meshmixer to fill sample teeth, and subsequently rendered and analyzed volume of the dentition with regard to DNE, RFI, and OPCR. Using tooth volume as a measure of size, no DTA variables examined correlated to the measured analyses (Table 3).
TABLE 3.
Tooth allometry compared to volume of post‐canine teeth using high and low MFN (mesh face number) with statistical significances.
| Index | Mesh face number (MFN) | p‐Values |
|---|---|---|
| DNE | 10,000 Triangles | 0.35 |
| 700 Triangles | 0.54 | |
| OPCR | 10,000 Triangles | 0.38 |
| 700 Triangles | 0.05 | |
| RFI | 10,000 Triangles | 0.81 |
| 700 Triangles | 0.72 |
Abbreviations: DNE, Dirichlet Normal Energy; OPCR, Orientation Patch Count Rotated; RFI, Relief Index.
The giant panda (Ailuropoda melanoleuca) is classified as a ground‐dwelling herbivore according to PanTHERIA, and is a clear outlier for the ground‐dwelling taxa with its high RFI values. Our analyses suggest that giant panda postcanine teeth possess a very high RFI value (RFI = 7.52), exceeding those of all other ground‐dwelling carnivores (RFI ≤ 6.5). Pérez‐Ramos et al. (2020) found similar DTA measurements, and concluded that the giant panda shows the highest values of tooth metric RFI indices due to their extremely specialized diet of consuming highly fibrous, strictly bamboo‐based, vegetation (requiring shearing that would more closely resemble carnivores than omnivores or other herbivores) (Eronen et al., 2017; Figueirido et al., 2014). All other relatively high RFI values are observed in ground‐dwelling ecological carnivores (broad‐striped Malagasy mongoose [Galidictis fasciata], cheetah [Acinonyx jubatus], African wild dog [Lycaon pictus], wolf [Canis lupus], ringtail [Bassariscus astutus], hog badger [Arctonyx collaris]), as well as the semi‐aquatic species: spotted‐necked otter (Lutra maculicollis), Asian small‐clawed otter (Aonyx cinerea), and giant otter (Pteronura brasiliensis), and include representatives from both Caniformia and Feliformia clades. The highest RFI value is 9.158 at 10,000 MFN in Ar. collaris, which is classified as a ground‐dwelling omnivore. Both Ar. collaris and Ai. melanoleuca have the highest RFI values despite being partially or completely herbivorous, compared to high RFI values otherwise being limited only to taxa classified as ground‐dwelling carnivores in the PanTHERIA dataset. The lowest RFI value observed is 2.2 at 10,000 MFN in the African clawless otter (Ao. capensis), classified as a semi‐aquatic omnivore in the PanTHERIA database. While both extremes in the range of RFI values occur in individual ground‐dwelling omnivore species, higher value RFI is seen in specifically ground‐dwelling mammals Ar. collaris and Ai. melanoleuca, while the lowest RFI value is observed in the semi‐aquatic species Ao. capensis. DNE and OPCR are not significantly related to any of the life history traits analyzed in this study, and cannot be relied on to characterize activity, diet breadth, habitat breadth, terrestriality, or trophic level.
We also analyzed DNE, measuring surface curvature. The range of values at 10,000 MFN found the lowest DNE (144.5) in the kinkajou (Potos flavus) and the highest DNE (914.83) in the Asian palm civet (Paradoxurus hermaphroditus) (Table 1). The kinkajou (Po. flavus) has the smallest value of OPCR (measuring tooth surface complexity), at 26.38 with an MFN of 10,000. The largest reported OPCR is for the red fox (Vulpes vulpes), which has a considerably higher OPCR than any other reported value, at 267.38 with 10,000 MFN.
We statistically analyzed the effects of phylogenetic relatedness and ecological variables using the K phylogenetic signal test (Tables 4 and 5). We tested for phylogenetic signal with and without pinnipeds, due to the clear environmental homogeneity for these marine carnivorans, as shown in Casares‐Hidalgo et al. (2019). We also conducted all comparative analyses using alternative branch length configuration with nodal calibration points from Tseng and Flynn (2018) (Tables 6 and 7).
TABLE 4.
PGLS analysis with pinnipeds included, p‐values calculated with standardized triangle counts (both at 10,000 and 700 MFN) as compared to ecological or life history variables.
| Index | Mesh face number (MFN) | Activity | Diet breadth | Habitat breadth | Terrestriality | Trophic level |
|---|---|---|---|---|---|---|
| DNE | 10,000 Triangles | 0.93 | 0.94 | 0.19 | 0.62 | 0.50 |
| 700 Triangles | 0.90 | 0.87 | 0.34 | 0.90 | 0.47 | |
| OPCR | 10,000 Triangles | 0.20 | 0.27 | 0.79 | 0.54 | 0.54 |
| 700 Triangles | 0.24 | 0.55 | 0.15 | 0.73 | 0.74 | |
| RFI | 10,000 Triangles | 0.08 | 0.01 | 0.53 | 0.13 | 0.02 |
| 700 Triangles | 0.09 | 0.01 | 0.55 | 0.11 | 0.01 |
Note: Values of rows include mesh face number (MFN), Dirichlet Normal Energy (DNE), Relief Index (RFI), and Orientation Patch Count Rotated (OPCR).
Abbreviation: PGLS, phylogenetic generalized least squares.
TABLE 5.
PGLS analysis with pinnipeds excluded, p‐values calculated with standardized triangle counts (both at 10,000 and 700 MFN) as compared to ecological or life history variables.
| Index | Mesh face number (MFN) | Activity | Diet breadth | Habitat breadth | Terrestriality | Trophic level |
|---|---|---|---|---|---|---|
| DNE | 10,000 Triangles | 0.56 | 0.59 | 0.30 | 0.66 | 0.49 |
| 700 Triangles | 0.51 | 0.55 | 0.49 | 0.94 | 0.40 | |
| OPCR | 10,000 Triangles | 0.11 | 0.44 | 0.90 | 0.45 | 0.49 |
| 700 Triangles | 0.11 | 0.99 | 0.20 | 0.67 | 0.72 | |
| RFI | 10,000 Triangles | 0.13 | 0.01 | 0.57 | 0.14 | 0.01 |
| 700 Triangles | 0.14 | 0.01 | 0.59 | 0.15 | 0.01 |
Note: Values of rows include mesh face number (MFN), Dirichlet Normal energy (DNE), Relief Index (RFI), and Orientation Patch Count Rotated (OPCR).
Abbreviation: PGLS, phylogenetic generalized least squares.
TABLE 6.
PGLS analysis using the Upham et al. (2019) phylogeny but including nodal calibration from Tseng and Flynn (2018) analysis with pinnipeds, p‐values calculated with standardized triangle counts (both at 10,000 and 700 MFN) as compared to ecological or life history variables.
| Mesh face number (MFN) | Activity | Diet breadth | Habitat breadth | Terrestriality | Trophic level |
|---|---|---|---|---|---|
| 10,000 Triangles | 0.37 | 0.19 | 0.16 | 0.48 | 0.50 |
| 700 Triangles | 0.61 | 0.18 | 0.37 | 0.86 | 0.13 |
| 10,000 Triangles | 0.04 | 0.53 | 0.71 | 0.50 | 0.55 |
| 700 Triangles | 0.20 | 0.55 | 0.19 | 0.60 | 0.40 |
| 10,000 Triangles | 0.06 | 0.01 | 0.61 | 0.07 | 0.001 |
| 700 Triangles | 0.06 | 0.01 | 0.63 | 0.07 | 0.001 |
Note: Values of rows include mesh face number (MFN), Dirichlet Normal energy (DNE), Relief Index (RFI), and Orientation Patch Count Rotated (OPCR).
Abbreviation: PGLS, phylogenetic generalized least squares.
TABLE 7.
PGLS analysis using the Upham et al. (2019) phylogeny but including nodal calibration from Tseng and Flynn (2018) analysis with pinnipeds excluded, p‐values calculated with standardized triangle counts (both at 10,000 and 700 MFN) as compared to ecological or life history variables.
| Index | Mesh face number (MFN) | Activity | Diet breadth | Habitat breadth | Terrestriality | Trophic level |
|---|---|---|---|---|---|---|
| DNE | 10,000 Triangles | 0.23 | 0.14 | 0.20 | 0.47 | 0.04 |
| 700 Triangles | 0.41 | 0.11 | 0.44 | 0.83 | 0.13 | |
| OPCR | 10,000 Triangles | 0.03 | 0.47 | 0.71 | 0.37 | 0.37 |
| 700 Triangles | 0.18 | 0.41 | 0.21 | 0.58 | 0.17 | |
| RFI | 10,000 Triangles | 0.09 | 0.01 | 0.71 | 0.10 | 0.001 |
| 700 Triangles | 0.10 | 0.01 | 0.73 | 0.10 | 0.001 |
Note: Values of rows include mesh face number (MFN), Dirichlet Normal energy (DNE), Relief Index (RFI), and Orientation Patch Count Rotated (OPCR).
Abbreviation: PGLS, phylogenetic generalized least squares.
Consequently, when pinnipeds are included in both calibrated phylogenetic trees, our analyzes show that RFI is the only DTA variable that is significantly correlated with the ecological variables of diet breadth and trophic level. For p‐values of RFI using the phylogeny of Upham et al. (2019), RFI is significantly related to diet breadth at 700 MFN (p = 0.01) and 10,000 MFN (p = 0.01), as well as to trophic level at 700 MFN (p = 0.01) and 10,000 MFN (p = 0.02). Evaluating further, applying the phylogeny of Upham et al. (2019), with nodes adjusted with calibrations of Tseng and Flynn (2018), provides highly similar p‐values, with RFI significantly related to diet breadth at 700 MFN (p = 0.01) and 10,000 MFN (p = 0.01), as well as to trophic level at 700 MFN (p = 0.001) and 10,000 MFN (p = 0.001).
Analyses excluding pinnipeds yield more significant results. Assessing against the Upham et al. (2019) phylogeny alone, RFI is significantly related to diet breadth at 700 MFN (p = 0.01) and 10,000 MFN (p = 0.01), as well as to trophic level at 700 MFN (p = 0.01) and 10,000 MFN (p = 0.01). However, the additional nodal analysis including the calibrations of Tseng and Flynn (2018) shows more significant variables when pinnipeds are excluded: in that case, DNE is correlated to trophic level at 10,000 MFN (p = 0.04), OPCR is significantly related to activity at 700 MFN (p = 0.03), and to RFI and diet breadth at 700 MFN and 10,000 MFN (p = 0.01), and RFI is significantly correlated with trophic level at 700 MFN and 10,000 MFN (p = 0.001).
4. DISCUSSION
Our dental topographic data analyses using digital tooth models of almost five dozen carnivoran species indicate that cusp height is closely related to dietary ecology (diet breadth and trophic level, as categorized in the PanTHERIA database). Relationships that became more weakly correlated when pinnipeds are excluded from the analyses include tooth complexity relative to activity and surface energy relative to trophic level. No other life history, dietary grouping, or ecological trait examined is significantly correlated with DNE, OPCR, or RFI values. The observation of RFI (cusp height) relating to dietary breadth and trophic level are intuitive, based on the fact that gripping and cutting actions require more sharply angled tools to efficiently chew and rip food in carnivores (Lucas, 2004).
Within our dataset, species with the highest RFI values are Ar. collaris (RFI = 9.158) classified with broadest diet breadth and omnivore trophic level, B. astutus (RFI = 8.899) which is classified as broad diet breadth and omnivore trophic level. Species with the lowest RFI values are Ao. capensis (RFI = 2.20) classified with broadest diet breadth and omnivore trophic level, Ursus arctos (RFI = 2.42) which is classified as broad diet breadth and omnivore trophic level, Hyaena brunnea (RFI = 2.417) classified with broadest diet breadth and omnivore trophic level, and Odobenus rosmarus (RFI = 2.429) classified with broadest diet breadth and omnivore trophic level. Therefore, species shown with the highest and lowest recorded RFI values are considered species with broad diets and dietary omnivores.
Focusing on DNE, species with highest DNE counts are Ac. jubatus (DNE = 912.01) and Pa. hermaphroditus (DNE = 914.83), which have a trophic level of carnivore and omnivore, respectively. Lowest values of DNE represented are Po. flavus (DNE = 144.464) and Pr. cristatus (DNE = 152.268) which are defined as an omnivore and carnivore, respectively. Looking at general trends, species have weak correlations of trophic level assocations with DNE values.
Finally, highest valued species of OPCR are species Eupleres goudotii (OPCR = 109.5) with an activity of nocturnal/crepuscular, and V. vulpes (OPCR = 267.375) with a nocturnal activity. Lowest values of OPCR are species: B. astutus (38.125) and Civettictis civetta (OPCR = 42.125) both defined as having nocturnal activity. Generally speaking, species with nocturnal activity levels are more likely to be correlated to OPCR.
Looking past dentition, many non‐dental cranial features have been compared to ecological traits, such as orbit orientation and skull shape. Orbit orientation was compared to visual strategy, habitat, and substrate preference in carnivoran mammals by Casares‐Hidalgo et al. (2019). Their results showed phylogenetic influence on the relationship between only orbital orientation and substrate preference. Casares‐Hidalgo et al. (2019) looked further into the results of their study and removed pinnipeds due to environmental homogeneity. The association between orbit orientation and substrate preference was found to be no longer significant. Casares‐Hidalgo et al. (2019) concluded that orbit orientation was phylogenetically correlated only to substrate preference, but only when pinnipeds were included in the sample; once pinnipeds were removed, no significant relationship was observed. In Tseng and Flynn (2018), cranial landmarks were used to capture cranial anatomical shape, and above‐ground dwelling (terrestriality/arboreality) and monthly precipitation as well as temperature were shown to be significantly correlated to skull shape in species with different dietary breadths, and to vary across feeding categories; no diet‐specific signals were detected from overall skull shape or biomechanical measures. In addition, Tseng and Flynn (2018) also determined that cranial shape is significantly correlated to dietary groupings especially distinguishing taxa with narrow versus generalist diets.
In Tseng and Flynn's (2015) analyses, diet and skull stiffness distinguished omnivores and invertebrate/vertebrate feeders in Carnivora. A majority of skulls demonstrated increasing efficiency and force output from anterior to posterior, however, hypercarnivores had differential force optimization of anterior teeth for killing (felids) and posterior teeth for crushing (canids). Overall, general cranial shape differentiation is closely linked to input load and strain energy gradients within feliforms and caniforms, and feeding behaviors cannot be determined based on mechanical efficiency and strain energy alone, so that multiple factors are needed to assess diet based on cranial form (Tseng & Flynn, 2015).
Previous analyses suggest that dietary ecological similarities may yield functional morphological convergence in carnivorans in select cases (e.g., Ailuropoda melanoleuca and Ailurus fulgens), but there is no broad support for such convergences across extant carnivores as a whole (Tamagnini et al., 2021). Tamagnini et al. (2021) found that convergent evolution was seen in cranial shapes only, and almost never in mandibular shapes of any dietary class. Further studies support this finding that mandibular shape is a conserved feature, and convergent functional morphological evolution in carnivoran skulls may not reflect dietary ecology alone (Meloro et al., 2015; Tseng & Flynn, 2018).
Based on our initial hypotheses, it was unexpected to find RFI primarily related to the life history traits studied, with DNE and OPCR being only weakly correlated to ecology when pinnipeds were excluded. Most previous studies showed correlations between diet and dentition, but few also analyzed mammalian trophic level. Pérez‐Ramos et al. (2020) compared the DTA indices of extinct cave bears to all living bear species, demonstrating that RFI values did not distinguish between dietary groupings, feeding behavior, or phylogeny. However, Pérez‐Ramos et al. (2020) also concluded that DNE and OPCR distinguished dietary groupings and feeding behavior, but also that these metrics were influenced by phylogeny, as were dental topographic curvature and complexity, each showing phylogenetic structure. Additionally, tooth curvature and complexity (DNE and OPCR) values were found by Pérez‐Ramos et al. (2020) to be highest in giant pandas, which eat large amounts of tough, high‐fiber vegetation: bamboo. Highest values of DNE and OPCR were observed among mainly herbivorous bears, with them having the second highest values among omnivores; the lowest values were found in faunivores and folivores‐frugivores (Pérez‐Ramos et al., 2020). Our findings indicate that although family‐level data does suggest that DTA is useful in dietary inferences, it is difficult to comparably distinguish dietary categories using DTA when these same metrics are assessed across all Carnivora rather than only in ursids. This could be due to convergence of similar features different species of Carnivora, which may be expanded within a single family and lead to specialization of omnivore or carnivoran diet. However, for omnivoran diets, a similarly functioning dentition as a hypercarnivore may assist with acquiring food in diverse landscapes. Overall, stronger dietary and dental signals within a single family could improve with hunting/foraging in a specific niche instead of a stronger signal across multiple families of different niches.
Pineda‐Munoz et al. (2017) sampled 134 extant mammal species (28 marsupial species from the order Diprotodontia, and 106 placental species from the orders Carnivora, Primates, and Rodentia) and determined that no single dental metric discriminates one individual diet category from the others in pairwise comparisons across that broad taxonomic sample, and that only OPCR had any significant correlation with any dietary category, differentiating dietarily carnivorous from non‐carnivorous species.
Other ecological and paleontological studies documented a strong correlation between dietary grouping and tooth molar shape, particularly in Primates (Cuozzo & Sauther, 2006; M'Kirera & Ungar, 2003). Two lemur species, the white‐footed sportive lemur (Lepilemur leucopus) and the ring‐tailed lemur (Lemur catta), were observed at Beza Mahafaly Special Reserve, Madagascar for environmental effects on their dentitions. Removal of ground plants by modern anthropogenic activity has led to severe tooth wear and tooth loss in the opportunistically omnivorous ring‐tailed lemurs, but not as severely in sympatric tamarind‐fruit consuming, scansorial sportive lemurs (Cuozzo & Sauther, 2012). These observations support the description of dental ecology by Cuozzo and Sauther (2012) and provide evidence supporting environmental influences on the teeth of sympatric species on ecological timescales. By contrast, in our study, dietary signals in dental topographic features may not be as obvious within the carnivorans analyzed herein.
The lack of consistent and significant correlations between some of the indices we analyzed (OPCR, DNE) and ecological and environmental characteristics may be attributed to (1) the actual absence of correlations; (2) limited categorizations in the PanTHERIA database, which provides only generalized dietary categories that may be mismatched with the fine‐scale correlations that DTA indices capture; or (3) a combination of both phenomena. At this time, no database provides detailed, comprehensive carnivoran diet and life history traits. Carnivoran tooth crowns have a wide range of cusp shapes and varieties, but such fine scale variability is perhaps more detailed than the broad and limited ecological categories currently available can reflect when analyzing much more precise dental metric traits.
In terms of potential limitations of this study, scan resolution may be considered to be a complication, as each scan was not made at the same resolution, given large variations in absolute size among taxa. Additionally, each specimen was not at the same age/ontogenetic stage, resulting in models scanned with variable amounts of heavy pre‐existing dental wear or either traumatically or congenitally missing teeth. While trying to account for dissimilar dental topography by standardizing the mesh resolution of the MFN at 700 or 10,000, the scan surface was modified automatically by the software algorithm without regard for preferential preservation of anatomically important feature information during these resampling methods. Each of these factors may play a role in possibly increasing the noise‐to‐signal ratio of diet‐informative DTA data.
Findings in our study are consistent with those using only linear and area measurements for dental topography (Hopkins et al., 2021), in terms of phylogenetic influence on dental morphological data. Our observations suggest that more work is needed to test the conventional interpretation that an extinct animal's teeth are a direct and reliable line of evidence for inferring their diet (Friscia et al., 2007; Pineda‐Munoz et al., 2017; Van Valkenburgh, 1989). One source of form‐function complexity may lie in regional variations in the development of the dentition. For example, specific signaling molecules determine molar size along a tooth row in Mammalia (Kavanagh et al., 2007), and canines generally vary more in length and width than other teeth (Dayan et al., 2002; Reuter et al., 2021; Szuma, 2000). Interactions among factors such as diet, genetics, and sexual dimorphism likely influence carnivoran intraspecific tooth‐size variation (Reuter et al., 2021), to the degree that occlusion‐driven functional demands may not constitute the principal covariation with DTA. It follows that more studies detailing a broader range of dentition patterns and measurements, and standardizing scan resolution, wear/age stage, and other attributes, may be needed to further understand patterns and effects of anatomical variation and to improve form‐function inferences from the fossil record (Reuter et al., 2021). Another challenge that remains in applying DTA within a broader phylogenetic framework is homology versus analogy of tooth surfaces used in different analyses across distantly related taxa, potentially limiting the utility of composite, meta‐analyses that leverage the broad range of mammalian taxa already studied using DTA (Berthaume et al., 2019).
5. CONCLUSIONS
Results of our study of lower postcanine teeth in 57 species of modern Carnivora, representing all 16 extant families, show that the RFI dental topographic metric significantly correlates with diet breadth and trophic level, suggesting an association between steeper cusp height and distinct dietary ecology. Other dental topographic metrics such as OPCR and DNE correlate, but only weakly, to activity and trophic level in carnivoran data partitions which exclude the marine pinnipeds.
Given these results, it is clear that cusp height strongly reflects mammalian diet. Analyzing a larger suite of carnivorans, as well as sampling more broadly across the mammal tree of life, should lead to even more conclusive studies on if or how dental topographic metrics relate to life history traits. Previous studies showed strong predilections of DNE and OPCR for distinguishing dietary groupings in primates, ursid carnivorans, and dietarily carnivorous versus non‐carnivorous mammal species. Surface curvature as related to trophic level may have a secondary influence on the shape of teeth in addition to the cusp height, as found for carnivoran trophic levels. Similarly, surface complexity has been shown to relate strongly to highly fibrous diets, and has a relatively smaller influence in carnivoran mammals in our analyses. Whereas cusp height has been quantified as a primary descriptor for carnivoran dietary ecology in our study, previous studies supported the relationship that OPCR and DNE are influenced by ecological variables more strongly than what we observed in our analysis.
We show that quantitative 3D topographic metrics are subject to phylogenetic influence and form‐function complexity, as linear metrics also are. Our analyses also paint a more nuanced picture of how dental morphology may have limited utility in dietary inferences of certain feeding ecologies, such as carnivory. Ecological and life history data currently available in large composite databases provide only crude dietary categorizations of Carnivora species, and more specific diet groupings would benefit the life history classification of extant species and reconstruction of diet in fossil taxa.
AUTHOR CONTRIBUTIONS
Concept and design: Emily Waldman, Z. Jack Tseng. Data acquisition: Emily Waldman, Z. Jack Tseng, John J. Flynn. Data analysis and interpretation: Emily Waldman, Z. Jack Tseng, John J. Flynn. Drafting of the manuscript: Emily Waldman, Z. Jack Tseng, John J. Flynn, Yoly Gonzalez. Editing and approval of the article: Emily Waldman, Z. Jack Tseng, John J. Flynn, Yoly Gonzalez.
OPEN RESEARCH BADGES
This article has earned Open Data, Open Materials and Preregistered Research Design badges. Data, materials and the preregistered design and analysis plan are available in this paper.
Supporting information
Table S1.
ACKNOWLEDGMENTS
Support for this project was provided by US National Science Foundation DEB 1257572 (to JJF, ZJT), DBI 2128146 (to ZJT) and the Frick Fund (Division of Paleontology, American Museum of Natural History). We thank the AMNH Department of Mammalogy for loans of specimens and the AMNH MIF (Microscopy and Imaging Facility) for overseeing the CT scanning system used in this study, and we especially acknowledge the assistance of M. Hill Chase, C. Grohé, and E. Westwig.
APPENDIX A.
TABLE A1.
p‐Values using unedited triangle count mesh face numbers (MFN) from species with various CT‐scanned triangle counts compared to ecological or life history variables.
| Index | Dataset | Activity | Diet breadth | Habitat breadth | Terrestriality | Trophic level |
|---|---|---|---|---|---|---|
| DNE | Raw | 0.99 | 0.55 | 0.12 | 0.18 | 0.99 |
| OPCR | Raw | 0.4 | 0.42 | 0.83 | 0.62 | 0.73 |
| RFI | Raw | 0.04 | 0.08 | 0.33 | 0.02 | 0.06 |
Note: Values of rows include mesh face number (MFN), Dirichlet Normal energy (DNE), Relief Index (RFI), and Orientation Patch Count Rotated (OPCR).
Abbreviation: CT, computed tomography.
Waldman, E. , Gonzalez, Y. , Flynn, J.J. & Tseng, Z.J. (2023) Dental topographic proxies for ecological characteristics in carnivoran mammals. Journal of Anatomy, 242, 627–641. Available from: 10.1111/joa.13806
DATA AVAILABILITY STATEMENT
This paper includes new data but no conflict is preventing from sharing this data.
REFERENCES
- Berthaume, M.A. , Delezene, L.K. & Kupczik, K. (2018) Dental topography and the diet of Homo naledi . Journal of Human Evolution, 118, 14–26. [DOI] [PubMed] [Google Scholar]
- Berthaume, M.A. , Lazzari, V. & Guy, F. (2020) The landscape of tooth shape: over 20 years of dental topography in primates. Evolutionary Anthropology: Issues, News, and Reviews, 29(5), 245–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthaume, M.A. , Winchester, J. & Kupczik, K. (2019) Effects of cropping, smoothing, triangle count, and mesh resolution on 6 dental topographic metrics. PLoS One, 14(5), e0216229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer, D.M. (2008) Relief index of second mandibular molars is a correlate of diet among prosimian primates and other euarchontan mammals. Journal of Human Evolution, 55(6), 1118–1137. [DOI] [PubMed] [Google Scholar]
- Boyer, D.M. , Evans, A.R. & Jernvall, J. (2010) Evidence of dietary differentiation among late Paleocene–early Eocene plesiadapids (Mammalia, Primates). American Journal of Physical Anthropology, 142(2), 194–210. [DOI] [PubMed] [Google Scholar]
- Boyer, D.M. , Kaufman, S. , Gunnell, G.F. , Rosenberger, A.L. & Delson, E. (2014) Managing 3D digital data sets of morphology: MorphoSource is a new project‐based data archiving and distribution tool. American Journal of Physical Anthropology, 153, 84. [Google Scholar]
- Bunn, J.M. , Boyer, D.M. , Lipman, Y. , St Clair, E.M. , Jernvall, J. & Daubechies, I. (2011) Comparing Dirichlet normal surface energy of tooth crowns, a new technique of molar shape quantification for dietary inference, with previous methods in isolation and in combination. American Journal of Physical Anthropology, 145(2), 247–261. [DOI] [PubMed] [Google Scholar]
- Casares‐Hidalgo, C. , Pérez‐Ramos, A. , Forner‐Gumbau, M. , Pastor, F.J. & Figueirido, B. (2019) Taking a look into the orbit of mammalian carnivorans. Journal of Anatomy, 234, 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cignoni, P. , Callieri, M. , Corsini, M. , Dellepiane, M. , Ganovelli, F. & Ranzuglia, G. (2008) MeshLab: an open‐source mesh processing tool . Eurographics Italian Chapter Conference. Available at: https://www.meshlab.net [Accessed 1st February 2020].
- Cuozzo, F.P. & Sauther, M.L. (2006) Severe wear and tooth loss in wild ring‐tailed lemurs (Lemur catta): a function of feeding ecology, dental structure, and individual life history. Journal of Human Evolution, 51(5), 490–505. [DOI] [PubMed] [Google Scholar]
- Cuozzo, F.P. & Sauther, M.L. (2012) What is dental ecology? American Journal of Physical Anthropology, 148(2), 163–170. [DOI] [PubMed] [Google Scholar]
- David, O. , Freckleton, R. , Thomas, G. , Petzoldt, T. , Fritz, S. , Isaac, N. et al. (2018) Caper: comparative analyses of phylogenetics and evolution in R . Available at: https://cran.r‐project.org/web/packages/caper/index.html [Accessed 1st March 2021].
- Dayan, T. , Woll, D. & Simberloff, D. (2002) Variation and covariation of skulls and teeth: modern carnivores and the interpretation of fossil mammals. Paleobiology, 28(4), 508–526. [Google Scholar]
- Eronen, J.T. , Zohdy, S. , Evans, A.R. , Tecot, S.R. , Wright, P.C. & Jernvall, J. (2017) Feeding ecology and morphology make a bamboo specialist vulnerable to climate change. Current Biology, 27(21), 3384–3389.e2. [DOI] [PubMed] [Google Scholar]
- Evans, A.R. & Jernvall, J. (2009) Patterns and constraints in carnivoran and rodent dental complexity and tooth size. Journal of Vertebrate Paleontology, 29, 24A. [Google Scholar]
- Evans, A.R. , Wilson, G.P. , Fortelius, M. & Jernvall, J. (2007) High‐level similarity of dentitions in carnivorans and rodents. Nature, 445(7123), 78–81. [DOI] [PubMed] [Google Scholar]
- Figueirido, B. , Tseng, Z.J. , Serrano‐Alarcón, F.J. , Martín‐Serra, A. & Pastor, J.F. (2014) Three‐dimensional computer simulations of feeding behavior in red and giant pandas relate skull biomechanics with dietary niche partitioning. Biology Letters, 10, 20140196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friscia, A.R. , Van Valkenburgh, B. & Biknevicius, A.R. (2007) An ecomorphological analysis of extant small carnivorans. Journal of Zoology, 272(1), 82–100. [Google Scholar]
- Guy, F. , Gouvard, F. , Boistel, R. , Euriat, A. & Lazzari, V. (2013) Prospective in (primate) dental analysis through tooth 3D topographical quantification. PLoS One, 8(6), e66142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, F. , Lazzari, V. , Gilissen, E. & Thiery, G. (2015) To what extent is primate second molar enamel occlusal morphology shaped by the enamel‐dentine junction? PLoS One, 10(9), e0138802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel, E. (1866) Generelle Morphologie der Organismen. Allgemeine Grundzü ge der organischen Formen‐Wissenschaft, mechanisch begrü ndet durch die von Charles Darwin reformierte Deszendenz‐Theorie. Band. I. Allgemeine Anatomie der Organismen. Berlin: Georg Reimer. [Google Scholar]
- Hopkins, S.S.B. , Price, S.A. & Chiono, A.J. (2021) Influence of phylogeny on the estimation of diet from dental morphology in the Carnivora. Paleobiology, 2(892), 1–16. [Google Scholar]
- Jones, K.E. , Bielby, J. , Cardillo, M. , Fritz, S.A. , O'Dell, J. , Orme, C.D.L. et al. (2009) PanTHERIA: a species‐level database of life history, ecology, and geography of extant and recently extinct mammals. Ecological archives E090‐184. Ecology, 90(9), 2648. [Google Scholar]
- Kavanagh, K.D. , Evans, A.R. & Jernvall, J. (2007) Predicting evolutionary patterns of mammalian teeth from development. Nature, 449(7161), 427–432. [DOI] [PubMed] [Google Scholar]
- Kembel, S. , Cowan, P.D. , Helmus, M.R. , Cornwell, W.K. , Morlon, H. , Ackerly, D.D. et al. (2010) Picante: R tools for integrating phylogenies and ecology. Bioinformatics, 26, 1463–1464. [DOI] [PubMed] [Google Scholar]
- Lazzari, V. & Guy, F. (2014) Quantitative three‐dimensional topography in taxonomy applied to the dental morphology of catarrhines. BMSAP, 26(3), 140–146. [Google Scholar]
- Lucas, P.W. (2004) Dental functional morphology: how teeth work. New York: Cambridge University Press. [Google Scholar]
- Meloro, C. , Clauss, M. & Raia, P. (2015) Ecomorphology of Carnivora challenges convergent evolution. Organisms Diversity and Evolution, 15, 711–720. [Google Scholar]
- M'Kirera, F. & Ungar, P.S. (2003) Occlusal relief changes with molar wear in Pan troglodytes and Gorilla . American Journal of Primatology, 60(2), 31–41. [DOI] [PubMed] [Google Scholar]
- Mundry, R. (2014) Statistical issues and assumptions of phylogenetic generalized least squares. In: Garamszegi, L. (Ed.) Modern phylogenetic comparative methods and their application in evolutionary biology. Berlin, Heidelberg: Springer, pp. 131–153. [Google Scholar]
- Münkemüller, T. , Lavergne, S. , Bzeznik, B. , Dray, S. , Jombart, T. , Schiffers, K. et al. (2012) How to measure and test phylogenetic signal. Methods in Ecology and Evolution, 3, 743–756. [Google Scholar]
- Myers, P. (2014) Lower_lateral.jpg. Animal diversity web . Available at: https://animaldiversity.org/collections/contributors/phil_myers/ADW_mammals/specimens/Carnivora/Felidae/Panthera_pardus/lower_lateral/ [Accessed 1st March 2021].
- Nakagawa, M. , Miguchi, H. , Sato, K. & Sakai, S. (2007) Population dynamics of arboreal and terrestrial small mammals in a tropical rainforest, Sarawak, Malaysia. The Raffles Bulletin of Zoology, 55(2), 389–395. [Google Scholar]
- Orme, D. , Freckleton, R.P. , Thomas, G.H. , Petzoldt, T. , Fritz, S.A. & Isaac, N. (2013) CAPER: comparative analyses of phylogenetics and evolution in R. Methods in Ecology and Evolution, 3, 145–151. [Google Scholar]
- Pampush, J.D. , Crowell, J. , Karme, A. , Macrae, S.A. , Kay, R.F. , Ungar, P. et al. (2019) Comparing dental topography software using platyrrhine molars. American Journal of Physical Anthropology, 169(1), 179–185. [DOI] [PubMed] [Google Scholar]
- Pampush, J.D. , Winchester, J.M. , Morse, P.E. , Vining, A.Q. , Boyer, D.M. & Kay, R.F. (2016) Introducing molaR: a new R package for quantitative topographic analysis of teeth (and other topographic surfaces). Journal of Mammalian Evolution, 23(4), 397–412. [Google Scholar]
- Paradis, E. & Schliep, K. (2019) Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics, 35(3), 526–528. [DOI] [PubMed] [Google Scholar]
- Pennell, M.W. , Eastman, J.M. , Slater, G.J. , Brown, J.W. , Uyeda, J.C. , FitzJohn, R.G. et al. (2014) Geiger v2. 0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics, 30(15), 2216–2218. [DOI] [PubMed] [Google Scholar]
- Pérez‐Ramos, A. , Romero, A. , Rodriguez, E. & Figueirido, B. (2020) Three‐dimensional dental topography and feeding ecology in the extinct cave bear. Biology Letters, 16(12), 20200792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda‐Munoz, S. , Lazagabaster, I.A. , Alroy, J. & Evans, A.R. (2017) Inferring diet from dental morphology in terrestrial mammals. Methods in Ecology and Evolution, 8(4), 481–491. [Google Scholar]
- Pinheiro, J. , Bates, D. , & R Core Team . (2022). nlme: linear and nonlinear mixed effects models. R package version 3.1‐161. https://CRAN.R‐project.org/package=nlme
- R Core Team . (2020) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: http://www.r‐project.org/index.html [Google Scholar]
- Reed, D. (1997) Contour mapping as a new method for interpreting diet from tooth morphology. American Journal of Physical Anthropology, 31, 517–535. [Google Scholar]
- Reuter, D.M. , Hopkins, S.S.B. & Davis, E.B. (2021) Carnivoran intraspecific tooth‐size variation shows heterogeneity along the tooth row and among species. Journal of Mammalogy, 102(1), 236–249. [Google Scholar]
- Schwartz, G.T. & Dean, C. (2000) Interpreting the hominid dentition: ontogenetic and phylogenetic aspects. In: O'Higgins, R. & Cohen, M. (Eds.) Development, growth and evolution: implications for the study of the hominid skeleton. London: Academic Press, pp. 207–233. [Google Scholar]
- Shipman, P. (1981) Life history of a fossil: an introduction to taphonomy and paleoecology. Cambridge: Harvard University Press. [Google Scholar]
- Skinner, M.M. & Gunz, P. (2010) The presence of accessory cusps in chimpanzee lower molars is consistent with a patterning cascade model of development. Journal of Anatomy, 217(3), 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuma, E. (2000) Variation and correlation patterns in the dentition of the red fox from Poland. Annales Zoologici Fennici, 37, 113–127. [Google Scholar]
- Tamagnini, D. , Meloro, C. , Raia, P. & Maiorano, L. (2021) Testing the occurrence of convergence in the cranio‐mandibular shape evolution of living carnivorans. Evolution, 75(7), 1738–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery, G. , Guy, F. & Lazzari, V. (2019) A comparison of relief estimates used in three‐dimensional dental topography. American Journal of Physical Anthropology, 170(2), 260–274. [DOI] [PubMed] [Google Scholar]
- Tseng, Z.J. & Flynn, J.J. (2015) Are cranial biomechanical stimulation data linked to known diets in extant taxa? A method for applying diet‐biomechanics linkage models to infer feeding capability of extinct species. PLoS One, 10(4), e0124020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, Z.J. & Flynn, J.J. (2018) Structure‐function covariation with nonfeeding ecological variables influences evolution of feeding specialization in Carnivora. Science Advances, 4(2), eaao5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng, Z.J. , Su, D.F. , Wang, X. , White, S.C. & Ji, X. (2017) Feeding capability in extinct giant Siamogale melilutra and comparative mandibular biomechanics of living Lutrinae. Scientific Reports, 7, 15225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungar, P. & Williamson, M. (2000) Exploring the effects of tooth wear on functional morphology: a preliminary study using dental topographic analysis. Palaeontologia Electronica, 3(1), 1–18. [Google Scholar]
- Ungar, P.S. (2010) Mammal teeth: origin, evolution, and diversity. Journal of Mammalogy, 92(5), 1138–1140. [Google Scholar]
- Upham, N.S. , Esselstyn, J.A. & Jetz, W. (2019) Inferring the mammal tree: species‐level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biology, 17(12), e3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Valkenburgh, B. (1989) Carnivore dental adaptations and diet: a study of trophic diversity within guilds. In: Gittleman, J.L. (Ed.) Carnivore Behavior, Ecology, and Evolution. Boston: Springer, pp. 410–436. [Google Scholar]
- Werdelin, L. , Kitchener, A.C. , Abramov, A. , Vernon, G. & Linh San, E.D.O. (2021) The scientific name of the aardwolf is Proteles cristatus . African Journal of Wildlife Research, 51(1), 149–152. 10.3957/056.051.0149. [DOI] [Google Scholar]
- Winchester, J.M. (2016) MorphoTester: an open source application for morphological topographic analysis. PLoS One, 11(2), e0147649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester, J.M. , Boyer, D.M. , St Clair, E.M. , Gosselin‐Ildari, A.D. , Cooke, S.B. & Ledogar, J.A. (2014) Dental topography of platyrrhines and prosimians: convergence and contrasts. American Journal of Physical Anthropology, 153(1), 29–44. [DOI] [PubMed] [Google Scholar]
- Zuccotti, L.F. , Williamson, M.D. , Limp, W.F. & Ungar, P.S. (1998) Modeling primate occlusal topography using geographic information systems technology. American Journal of Physical Anthropology, 107(1), 137–142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Data Availability Statement
This paper includes new data but no conflict is preventing from sharing this data.


