Abstract
The M protein is an important surface-located virulence factor of Streptococcus pyogenes, the group A streptococcus (GAS). Expression of M protein is primarily controlled by Mga, a transcriptional activator protein. A recent report suggested that the sag locus, which includes nine genes necessary and sufficient for production of streptolysin S, another GAS virulence factor, is also needed for transcription of emm, encoding the M protein (Z. Li, D. D. Sledjeski, B. Kreikemeyer, A. Podbielski, and M. D. Boyle, J. Bacteriol. 181:6019–6027, 1999). To investigate this in more detail, we constructed an insertion-deletion mutation in sagA, the first gene in the sag locus, in the M6 strain JRS4. The resulting strain, JRS470, produced no detectable streptolysin S and showed a drastic reduction in cell surface-associated M protein, as measured by cell aggregation and Western blot analysis. However, transcription of the emm gene was unaffected by the sagA mutation. Detailed analysis with monoclonal antibodies and an antipeptide antibody showed that the M protein in the sagA mutant strain was truncated so that it lacks the C-repeat region and the C-terminal domain required for anchoring it to the cell surface. This truncated M protein was largely found, as expected, in the culture supernatant. Lack of surface-located M protein made the sagA mutant strain susceptible to phagocytosis. Thus, although sagA does not affect transcription of the M6 protein gene, it is needed for the surface localization of this important virulence factor.
Streptococcus pyogenes (the group A streptococcus [GAS]) is a serious human pathogen capable of producing a wide variety of diseases. Such infections range from mild suppurative diseases, such as pharyngitis and pyoderma, to more severe and life-threatening invasive diseases, including myositis, necrotizing fasciitis, and the recently recognized and often fatal streptococcal toxic shock syndrome (for a recent review, see reference 11). The primary infections may also lead to serious sequelae such as acute rheumatic fever, glomerulonephritis, and reactive arthritis (6). It appears that many GAS strains can cause more than one of these diseases.
Different strains of GAS produce various virulence factors that are involved in the survival and persistence of this organism within its host. Among them, M protein, which appears as hair-like projections on the cell surface (52), is considered to be a major virulence factor. Probably the most important role of M protein in the virulence of the GAS is that it confers resistance to complement-mediated killing by polymorphonuclear leukocytes and macrophages (27), thus protecting the bacteria from phagocytosis. In addition, M protein is required for attachment of the GAS to keratinocytes and thus is likely to play a critical role in infections initiated at the skin surface. M protein also causes the GAS to aggregate when they attach to tonsillar epithelial cells (9), so it may also play a critical role in initiation of colonization of the respiratory mucosa.
The sequence and length of M proteins of different strains of GAS differ, and these diferences are used to classify strains of GAS serologically by M types. M proteins have a dimeric alpha-helical coiled-coil structure (43) and most have tandem direct sequence repeats responsible for the size variation observed (21). The N-terminal regions of M proteins are highly variable and are largely responsible for the antigenic variation. In contrast, the C-terminal part of the molecule is anchored to the GAS surface and is largely conserved. M proteins contain two to three 27-amino-acid C repeats that are highly conserved among strains of different serological M types (for reviews, see references 14, 15, and 48). These are followed by an LPXTG motif, a hydrophobic region, and a charged tail, which are required for anchoring the molecule to the bacterial cell surface (17). The M6 molecule, found on the strain used in our study, is the first M protein that was cloned and sequenced (20). In this protein, the variable N-terminal region is followed by five 14-amino-acid A repeats and about five 24-amino-acid B repeats. There is a pepsin hypersensitive site within the last B repeat of the M6 protein that separates the conserved region from the variable region of the molecule (20, 34).
The GAS also express a wide range of secreted and surface-attached proteases proposed to be involved in virulence. Among them is streptococcal cysteine protease, SpeB, which is secreted as a 40-kDa zymogen and autocatalytically cleaved into a 28-kDa active protease. The activated SpeB can cleave several host proteins, including extracellular matrix component (25) and plasma proteins such as fibrinogen (35). Furthermore, SpeB has been shown to process the M1 protein (46) as well as protein H (which binds to immunoglobulin) and the scpA-encoded C5a peptidase (4) that degrades this chemotactic member of the complement cascade. Streptokinase, another secreted serine protease with broad-spectrum activity, catalyzes the conversion of plasminogen to plasmin and is also thought to be an important virulence factor required for tissue invasion by the GAS (31).
The ability of GAS strains to invade and persist in different niches in the human host during various stages of disease is likely to depend on selective expression of virulence factors that are controlled by global regulatory mechanisms sensitive to environmental changes. A major component of this global regulatory circuit found in all GAS strains is the multiple gene regulator Mga, which activates expression of many virulence genes in an environmentally controlled fashion. These include emm (8, 10) and scpA (10, 50), and in strains where they are present, the emm-like genes (26, 44) sic (1), scl, and sof (37). Mga, which is also autoregulated, controls gene expression by direct interaction with the promoter region of the genes it regulates (36).
In addition to Mga, the recently described pel locus was found to influence expression of various GAS virulence factors, including M (28). This locus is identical to sagA, the first open reading frame (ORF) in the sag operon, which is required for streptolysin S (SLS) production. The sag operon consists of nine ORFs. The sagA gene encodes a putative 53-amino-acid product with sequence similarity to a bacteriocin and is believed to encode SLS. There is a putative promoter upstream of sagA and there are two potential rho-independent terminators between sagA and sagB and after the last ORF, sagI (40).
The molecular mechanism by which the sag locus modulates gene expression has not been determined. Previous investigations of the effect of sag mutations on expression of the emm gene gave different results. In one study, a mutant in which Tn916 was inserted in the presumed promoter region of sagA in strains of both serotypes M1 and M18 showed no differences in the amount of M protein compared to the isogenic wild-type strains (5). In contrast, a mutant in a serotype M49 strain in which Tn917 had inserted at the same location showed a drastic reduction of the emm transcript compared to the wild-type strain (28). This apparent difference and an interest in fully understanding the regulation of production of M protein prompted us to study the role of the sagA locus in the serotype M6 GAS strain JRS4. We found that although the level of emm transcript was the same in both the sagA insertion-deletion mutant and its isogenic wild type, the M protein was truncated at its C terminus and therefore was not anchored to the cell wall in the mutant strain. Thus, the sagA mutant strain was susceptible to phagocytosis.
MATERIALS AND METHODS
Bacterial strains and media.
All GAS strains except JRS800 are derivatives of the M6 strain JRS4, a streptomycin-resistant derivative of D471 (49). JRS800 is a streptomycin-resistant derivative of M1 strain SF370 (13). GAS cultures were grown at 37°C without agitation in Todd-Hewitt broth with 0.2% yeast extract (THY). Escherichia coli XL1 Blue (Stratagene) was used as the host for plasmid construction and was grown in Luria broth (LB) with agitation. Antibiotic concentrations were as follows: chloramphenicol, 2 μg/ml for GAS and 20 μg/ml for E. coli; kanamycin, 300 μg/ml for GAS and 50 μg/ml for E. coli; spectinomycin, 50 μg/ml for both GAS and E. coli; erythromycin, 0.5 μg/ml for GAS and 500 μg/ml for E. coli; ampicillin, 100 μg/ml for E. coli.
To inhibit extracellular proteases, GAS strains were grown overnight, washed with saline, and reinoculated at a 1:100 dilution in fresh THY containing either 20 μM E64 (to inhibit cysteine protease; Sigma) as previously described (46, 51) or 10× complete protease inhibitor tablets (to inhibit serine, cysteine, and metalloproteases; Roche). Cells were grown to an optical density at 600 nm of 0.6 and collected for further analysis.
Inactivation of the sagA locus in the M6 GAS strain JRS4
The sagA gene was inactivated in JRS4 with a polar kanamycin resistance cassette (omega-Km2 [41]) containing the aphA3 gene. Since the sequence information is available from an M1 strain, upstream and downstream regions of the sagA locus were amplified from the chromosomal DNA of the M1 strain JRS800. Two primers, Kpn-Del5 (5′ agtgacggtaccCGCGCAGTAGGGATCAAGCGAGC) and Xho-Del5 (5′ agttgactcgagAAGGTTTACCTCCTTATCTAATAAG), were used to amplify 0.56 kb upstream of the sagA region (restriction sites are underlined and uppercase letters indicate sequence homology to the chromosome). Two primers, Bam-Del3 (5′ gcagttggatccTAATCTATTTAGCATCTCTATGTG) and Xba-Del3 (5′ gatctgtctagaGTCGACAATACTAGCTTGGAAGCC), were used to amplify 0.48 kb of the downstream region. These PCR products were digested with appropriate enzymes and cloned into pBluescriptII digested with KpnI and XhoI for the upstream fragment and with BamHI and XbaI for the downstream fragment to generate pJRS453 and pJRS454, respectively. An EcoRI fragment from pUC4ΩKm2 (41) containing the omega-Km2 cassette was then cloned into the EcoRI site of pJRS453 to generate pJRS459. An XbaI-PstI fragment from pJRS454 carrying the fragment downstream of sagA was cloned into XbaI-PstI-digested pJRS459 to create pJRS460. Finally, the KpnI-SacI fragment of pJRS460 containing the omega-Km2 cassette with fragments upstream and downstream of sagA was cloned (by blunting the SacI site) into KpnI-EcoRV sites of pJRS233 (42) to generate the gene replacement plasmid pJRS470. This plasmid contains a temperature-sensitive replication origin (42).
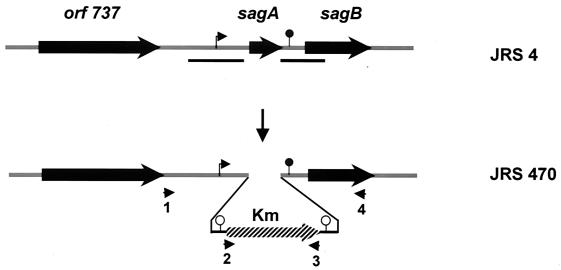
Plasmid pJRS470 was introduced into JRS4 and transformants were selected at 30°C, a temperature permitting plasmid replication. The chromosomal sagA locus was replaced with omega-Km2 by growing at the nonpermissive temperature (37°C) with selection for resistance to kanamycin following a previously described procedure (42) to create JRS470. The presence of a mutant sagA allele in the chromosome of JRS470 was confirmed by PCR analysis across the sagA locus with flanking primers (Fig. 1, primers 1 and 3, 2 and 4) and omega-Km2-specific primers (Fig. 1, primers 2 and 3) and by Southern hybridization analysis. The sagA locus was also deleted from JRS145, an isogenic M6 strain in which the cat86 gene replaces emm6 (7). This strain does not produce M protein (7).
FIG. 1.
Schematic diagram of sagA insertion-deletion mutant derivative of JRS4. The omega-Km2 cassette (hatched box) was used to replace the sagA locus to generate JRS470. Thick arrows indicate ORFs. The chromosomal segments used as the homologous region for gene replacement are shown by black lines below the diagram of JRS4. The putative promoter of the sag operon is indicated by a bent arrow and the putative transcriptional terminator is shown by a filled circle. Transcriptional terminators present in the omega-Km2 cassette are shown by open circles. Arrowheads represent primers (1 to 4) used to confirm the gene replacement event.
Preparation of whole-cell extract, cell wall, and supernatant proteins.
Overnight cultures were grown in THY, collected by centrifugation, and washed twice in saline. Cell density was adjusted to 5 cell units per ml with saline (1 cell unit per ml is equivalent to 1 ml of culture at an optical density at 600 nm of 2.0). Total cell extracts were prepared by lysing the cell suspension with a glass bead beater (BIO101) and were clarified by centrifugation. For cell wall preparations, cell amounts equivalent to 5 cell units were resuspended in 500 μl of 10 mM Tris-Cl (pH 8.0) containing 30% raffinose, 100 U of mutanolysin per ml, 1 mg of lysozyme (32) per ml, and complete protease inhibitor cocktail (Roche). Cells were digested for 3 h at 37°C with constant rotation and pelleted by centrifugation. The supernatant containing the cell wall fraction was used for further analysis. To obtain total proteins in the culture supernatant, cells were removed by centrifugation followed by filtration through a 0.2-μm-pore-size filter. Total proteins were obtained by precipitation with trichloroacetic acid (TCA; 20% [wt/vol] final concentration) on ice for 1 h, washed with acetone, and resuspended in water to 1/100 volume.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate–4 to 12% or 10% polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto a nitrocellulose membrane that had been blocked with 3% bovine serum albumin–0.1% Tween 20 in Tris-buffered saline for 1 h at room temperature. Monoclonal antibody (MAb) 10A11, 10B6, or 10F5 (23) diluted 1:2,000 or polyclonal antibody to the N-terminal 26 residues of mature M6 protein (23) diluted 1:1,000 was added and incubated for 1 h at room temperature. Filters were washed thoroughly with 0.1% Tween 20 in Tris-buffered saline, probed with anti-mouse (for monoclonal) or anti-rabbit (for polyclonal) immunoglobulin G alkaline phosphatase-conjugated secondary antibodies for 1 h, and washed again. Reactivity was detected using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) as substrates.
Direct binding assay on whole cells for fibrinogen, fibronectin, and M6 antibody.
GAS strains grown overnight were collected by centrifugation and washed twice with 0.9% saline. Cell density was adjusted to 2.5 cell units/ml and serial dilutions were made in saline. Aliquots of 5 μl were spotted onto nitrocellulose membranes and air dried, and nonspecific sites were blocked with 3% bovine serum albumin–0.1% Tween 20 in phosphate-buffered saline at room temperature for 1 h. Digoxigenin (DIG)-labeled fibrinogen or fibronectin was added to the membrane to a final concentration of 1 μg/ml and incubated at room temperature for 1 h. Membranes were washed twice with 0.05% Tween 20 in phosphate-buffered saline. Binding of ligand to the cell wall was detected using alkaline phosphatase-conjugated anti-DIG antibody with nitroblue tetrazolium and BCIP as substrates. Detection of M6 protein on the cell surface was performed as described above using MAb 10A11.
RNA slot blot.
GAS strains were cultured in THY and growth was monitored using a Klett-Summerson colorimeter with a red filter. Total RNA was isolated from samples using the FastaPrep system (BIO 101) and treated with DNase I to remove residual DNA as described previously (12). RNA was assayed on a Zeta probe membrane (Bio-Rad) by slot blot as previously described (12). DNA probes were prepared by PCR amplification using the JRS4 chromosomal DNA as template. Primer pairs used for probes were emm (5′GAGTGTAATAGGGGCAGGA3′ and 5′AGTTTCCTTCATTGGTGCT3′), rpsL (5′gccgaattcGAATGTAGATGCCTACAATTAACCA3′ and 5′cccaagcttTTTACGACTCATTTCTCTTTATCCC3′), and gyrA (5′GATCTGCAGGAAGAAGAAGATGTTTTGATTAC3′ and 5′GTCATCCTGACCGCTTGTCAAAAGG3′) (lowercase letters indicate nonhomologous bases). PCR fragments were labeled with [α-32P]dATP by random priming using the DECAprimeII kit (Ambion). RNA blots were analyzed with a phosphorimager (Molecular Dynamics).
Phagocytosis in human blood.
Resistance to phagocytosis was determined with the bactericidal assay as previously described (43). Briefly, freshly drawn human blood from a nonimmune individual was mixed with heparin (10 U/ml) and the plasma was obtained by centrifugation. GAS strains were grown to mid-logarithmic phase in THY and diluted to 100 to 200 CFU/ml. A mixture of 0.1 ml of THY, 0.1 ml of diluted bacterial cultures (approximately 10 to 20 bacteria), and 0.4 ml of either human blood or plasma (control for anti-M antibodies) was incubated at 37°C for 3 h with constant rotation. CFU were counted by the pour plate method with THY agar.
RESULTS
Construction of a sagA insertion-deletion mutant in the serotype M6 GAS strain JRS4.
To elucidate the role of sagA in the expression of M protein by an M6 GAS strain, the chromosomal sagA locus was replaced by insertion of a kanamycin gene. The DNA regions upstream and downstream of sagA were amplified separately from the chromosome of the sequenced M1 GAS strain JRS800, since the sequence of this region of JRS4 is not known. The omega-Km2 cassette was cloned between these DNA fragments in the temperature-sensitive shuttle vector pJRS233 (42) to construct plasmid pJRS470. This plasmid was used to replace the wild-type sagA locus of strain JRS4 with omega-Km2, following a double recombination event, to create strain JRS470 (Fig. 1). The same plasmid was used to replace the sagA gene in strain JRS145 (7), a derivative of JRS4 that contains cat86 in place of emm under the emm gene promoter, to create strain JRS480. Both strain constructions were confirmed by PCR using primers that flank the sagA gene (Fig. 1) and by Southern blot analysis.
Since the sag locus is required for expression of SLS, cell-associated SLS activity was assayed (28) in JRS4, JRS470, and JRS145. As expected, JRS4 and JRS145 hemolyzed sheep erythrocytes while JRS470 did not (data not shown).
M protein expression in the sagA mutant
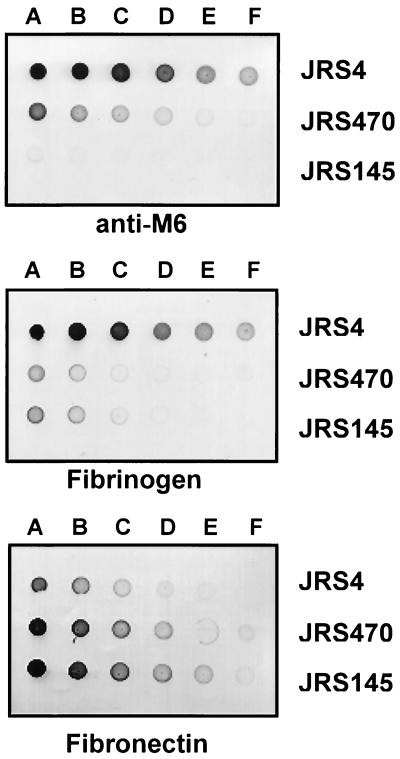
The simplest test for the presence of M protein on the GAS surface is the clumping of the cells in undisturbed liquid cultures. Although normal clumping was observed for JRS4, no clumping was seen for JRS145 or JRS470, suggesting that, like the M− strain, the sagA mutant strain lacks surface M protein. This was confirmed by spotting whole cells onto nitrocellulose membranes and developing the membranes with MAb 10A11, which recognizes an epitope in the B repeat region of the M6 protein (24), and with DIG-labeled fibrinogen, which binds M protein (53, 54) (Fig. 2). As a control, DIG-labeled fibronectin (which binds a different cell surface protein) was also used to develop the whole-cell dot blots (Fig. 2). The wild-type parent strain, JRS4, bound anti-M antibody, fibrinogen, and fibronectin, as expected. The emm deletion strain JRS145 bound fibronectin but did not bind anti-M protein antibody and showed reduced binding of fibrinogen, consistent with the probable presence of one or more fibrinogen binding proteins in addition to the M protein on the surface of this strain (29). The sagA mutant strain JRS470 bound fibronectin, showed reduced fibrinogen binding, and bound significantly less anti-M6 antibody than the parent strain. This suggests that JRS470 has much less M protein on its surface than the parent. (The increased binding of fibronectin by JRS145 and JRS470 compared to JRS4 is probably due to interference by M protein with the fibronectin binding of protein F.)
FIG. 2.
Expression of surface proteins in the sagA mutant. Twofold serial dilutions starting with 2.5 cell units/ml (A to F) from overnight cultures of the wild type (JRS4), the sagA mutant (JRS470), and the M deletion mutant strain (JRS145) were spotted onto a nitrocellulose membrane and probed with anti-M6 antibody (MAb 10A11), DIG-labeled fibrinogen, and DIG-labeled fibronectin as indicated.
Transcription of the emm gene in the sagA mutant.
To measure transcription of emm, two different approaches were taken. In the first, cat86 was used as a reporter for the promoter of emm6. Strain JRS145 and its sagA insertion-deletion derivative JRS480 contain cat86 in place of emm under the Pemm promoter. The ability of these strains to grow in the presence of increasing amounts of chloramphenicol was measured in liquid cultures to monitor the expression of chroramphenicol acetyltransferase. The MIC for both strains was 80 ± 10 μg/ml, indicating that there is no difference in cat86 expression from Pemm between the wild type and the sagA mutant.
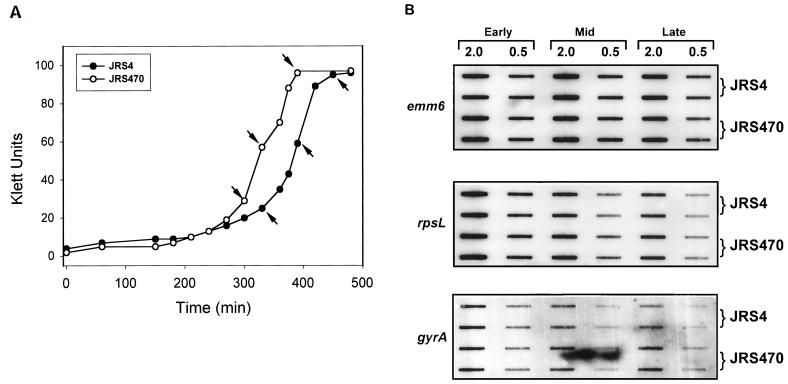
To confirm this observation, the amount of emm transcript was measured directly by RNA hybridization to a PCR-derived emm probe at three different stages in the growth cycle (Fig. 3A). To assure that equal amounts of mRNA from each strain were loaded on the gel, blots were also probed with rpsL and with gyrA as controls. When the sagA mutant strain JRS470 was compared with its parent, JRS4, there were no significant differences in the amount of emm gene transcript at any stage of growth (Fig. 3B). Therefore, the sagA locus does not affect transcription of emm in this M6 GAS strain.
FIG. 3.
Transcription of the emm gene in the wild-type and sagA mutant strains at different stages of growth. (A) Growth curve indicating the times of RNA isolation at early and mid-log phases of growth and at the transition to the stationary phase of growth (arrows). (B) Hybridization of specific DNA probes internal to the genes indicated to RNA isolated at the stages of growth indicated in panel A. On each filter, 2.0 and 0.5 μg of RNA was applied in vertically arranged duplicates.
The M6 protein is truncated in the sagA mutant.
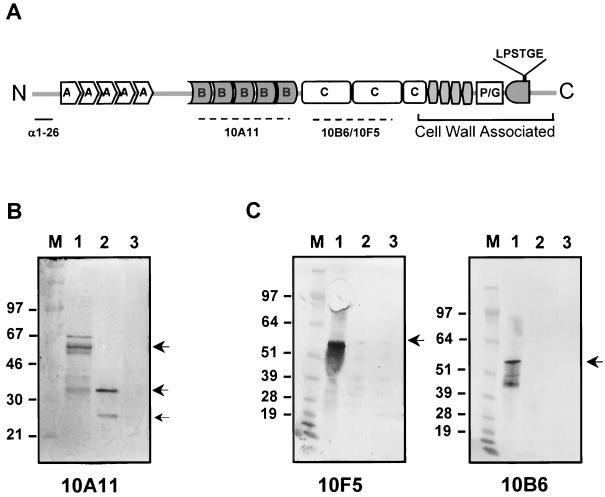
Since we have found that the emm transcript is produced at the wild-type level but the amount of M6 protein on the cell surface is reduced, we wished to determine whether the M protein was translated and was stable. To investigate this, we used four different anti-M antibodies that recognize specific regions of the M6 protein (Fig. 4A). Whole-cell protein extracts from both wild-type and mutant strains were separated by SDS-PAGE and probed first with MAb 10A11, which recognizes an epitope in the B repeat region of the M6 molecule (24). The multiple banding pattern characteristic of full-length M6 protein (between ≈50 and ≈66 kDa) was observed in the whole-cell extracts of wild-type strain JRS4 (Fig. 4B, lane 1). (Since the M protein is covalently attached to the cell wall, the physical disruption of the wall used in this work probably results in cell wall fragments remaining attached to the M protein, as has been found using phage lysin extraction of M [16, 23].) In contrast, the extract from the sagA mutant strain (JRS470) showed two smaller bands (with apparent molecular masses of ≈35 and ≈28 kDa) that reacted with MAb 10A11 (Fig. 4B, lane 2). This indicates that the M6 protein is truncated in this mutant strain. JRS145, which contains no emm6 gene, was included as a negative control (Fig. 4B, lane 3).
FIG. 4.
Analysis of the M protein of the wild type and the sagA mutant. (A) Schematic diagram of the structure of the M6 protein indicating the location of the epitopes recognized by the antibodies used. Repeat regions are shown by capital letters and the cell-associated region and cell wall anchoring signals are indicated. (B and C) Western blot analysis of whole-cell extracts separated by SDS–10% (B) or 4 to 12% (C) PAGE. The antibodies used are indicated below the gels. Lanes contain extracts from JRS4 (lanes 1), JRS470 (lanes 2), and JRS145 (lanes 3). M, Rainbow (B; Amersham) or See Blue (C; Invitrogen) molecular mass marker. Bands that reacted with anti-M antibody are indicated by arrows.
To determine whether the region of M6 missing from the protein extracted from JRS470 is the C-terminal half of the molecule, we used two MAbs, 10F5 and 10B6, that recognize epitopes within the C repeat region (Fig. 4A). Both antibodies reacted with the intact M6 protein in the JRS4 extract, and both failed to recognize the truncated M6 protein present in the mutant strain (Fig. 4C), implying that the C-terminal half of M6 is not present in the protein extracted from the sagA mutant. The presence of the N-terminal portion of M6 in the truncated protein of the sagA mutant was confirmed using an antibody raised against the first 26 amino acids of the mature protein (see below).
Localization of the truncated M6 protein.
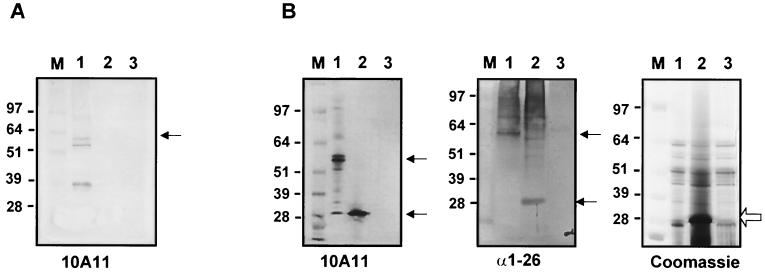
M proteins are transported through the cytoplasmic membrane of GAS by the Sec system, which cleaves the 42-amino-acid N-terminal signal peptide, and is anchored to the GAS cell wall through a well-conserved LPXTG motif present in the C-terminal region of the molecule, distal to the C repeats (17) (Fig. 4A). Because the truncated M6 protein in the extract of the sagA mutant lacks the C-terminal half of the molecule, it seemed likely that this protein would not be anchored to the cell wall. To investigate this, cell wall-associated proteins were prepared from both wild-type and mutant strains and analyzed by Western blotting with MAb 10A11, specific for the B repeats. As expected, the truncated M6 protein seen in whole-cell extracts of the sagA mutant was absent from the cell wall fraction but intact M protein was present in the cell wall preparation from the wild-type parent (Fig. 5A, compare lanes 1 and 2).
FIG. 5.
Localization of M6 protein in the mutant and wild-type strains. Analysis of cell wall fractions (A) or concentrated culture supernatants (B) from various GAS strains separated on SDS–4 to 12% polyacrylamide gels and probed with MAb 10A11 or anti-amino acids 1 to 26 (α1-26) or stained with Coomassie blue, as indicated below each panel. Lanes contain samples from JRS4 (lanes 1), JRS470 (lanes 2), and JRS145 (lanes 3). M, See Blue (Invitrogen) molecular mass markers whose sizes are indicated on the left of each panel. Bands that reacted with anti-M antibody are indicated by arrows.
To determine whether the truncated M6 protein has an intact N terminus and whether it is transported through the cell membrane and secreted outside the cell, concentrated culture supernatant proteins from the wild type and the sagA mutant were reacted with antibody specific for the B repeats (MAb 10A11) and with antibody made against the N-terminal 26 amino acids of the mature M6 molecule (23). Both antibodies recognized mature M6 protein secreted from the wild-type strain (Fig. 5B, lanes 1) and no bands were detectable in either the culture supernatant or cell wall extracts from the strain with no M protein gene (Fig. 5, lanes 3), as expected. In the culture supernatant of the sagA mutant, both antibodies recognized an ≈28-kDa band (Fig. 5B, lanes 2). This indicates that the truncated M6 protein of the mutant strain contains the N terminus and that it is released outside the cell. Coomassie blue staining of the same culture supernatants showed that the sagA mutant produced a large amount of a protein that migrates to the location of the truncated M6 protein, suggesting that a large amount of truncated M6 is produced and accumulates in the supernatant.
Truncation is not caused by external proteases.
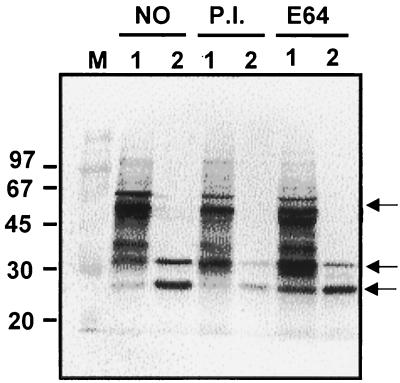
Many strains of GAS produce and secrete abundant quantities of proteases, including the speB-encoded cysteine protease SpeB, which has been shown to remove the C-terminal fragment from the M1 protein (4, 46). Therefore, we investigated the possible role extracellular proteases might have in the truncation of the M6 protein in the sagA mutant strain. To inhibit SpeB, cells were grown in the presence of E64, a cysteine protease inhibitor, and to inhibit other proteases, a commercial mixture of inhibitors of both serine and cysteine proteases were added to the medium during growth. When total cell lysates from these cultures were analyzed by Western blot with MAb 10A11, we found that addition of protease inhibitors had no effect on the truncation of M protein produced in the sagA mutant strain (Fig. 6). The extract from the sagA mutant strain (JRS470) showed the same two smaller bands of ≈35 and ≈28 kDa as those seen in the absence of any protease inhibitor (compare Fig. 6, lanes 1 and 2, to Fig. 4B, lanes l and 2). The reduction of intensity of these two bands in protease inhibitor samples is probably due to the difference in the amount of sample present (Fig. 6, lanes 1). The protease inhibitor(s) was determined to be active in the growth medium by demonstrating inhibition of hydrolysis of casein on plates (an activity associated with SpeB [33, 46]) using the M1 strain JRS800 known to produce large amounts of protease (data not shown).
FIG. 6.
M proteins in cell extracts grown in the presence or absence of protease inhibitors. The wild-type (lane 1) and sagA mutant (lane 2) strains were grown in the absence of protease inhibitor (NO), the presence of complete protease inhibitor (P.I.), or with cysteine protease inhibitor E64. Whole-cell extracts were separated on SDS–10% polyacrylamide gels and reacted with MAb 10A11. M, molecular mass marker (Rainbow).
The sagA mutant is sensitive to phagocytosis.
The best-documented function of M proteins is prevention of phagocytosis of GAS strains by polymorphonuclear leukocytes in human blood (27). Since there is little M6 protein on the surface of the sagA mutant strain JRS470 and the protein it releases into the culture supernatant is truncated, we investigated whether the sagA mutation rendered the strain susceptible to phagocytosis. We compared survival and growth of the mutant with its parent, JRS4, in whole blood and plasma (Table 1). As expected, the doubling time of JRS4, 25 min, was the same in whole blood and plasma, an indication of phagocytosis resistance. In contrast, both the M− control strain JRS145 and the sagA mutant strain JRS470 grew well in plasma but were phagocytized by the cells in whole blood (Table 1). This is in agreement with the above results indicating the lack of M protein on the surface of the sagA mutant.
TABLE 1.
Phagocytosis of GAS in human blood
| Strain | Relevant genotype | Amount of GAS (CFU) ina:
|
||
|---|---|---|---|---|
| Inoculum | Whole blood | Plasma | ||
| JRS4 | emm6 (wild type) | 1.65 × 102 ± 0.11 × 102 | 1.28 × 104 ± 0.05 × 104 | 2.15 × 104 ± 0.03 × 104 |
| JRS145 | Δemm6 | 1.89 × 102 ± 0.13 × 102 | 0.75 × 101 ± 0.43 × 101 | 2.94 × 104 ± 0.25 × 104 |
| JRS470 | emm6 (wild type) ΔsagA | 0.94 × 102 ± 0.11 × 102 | 9.50 × 101 ± 0.50 × 101 | 2.07 × 104 ± 0.17 × 104 |
The ability of GAS to survive in fresh human blood was measured as described in the text. The numbers reported are the averages of two assays.
DISCUSSION
SLS activity produces one of the defining phenotypes of GAS, and the genes that are necessary and sufficient to produce this hemolysin, the sag genes, have been found in all GAS strains studied (40). The major transcript from this operon was found to be 450 bp long, which corresponds to the size predicted for a message that starts at the sag promoter and ends at the terminator motif located between sagA and sagB. However, reverse transcription-PCR analysis showed that the sag genes downstream of sagA are cotranscribed as a polycistronic message together with sagA, indicating read-through transcription from the sagA promoter through the terminator sequence following sagA (40). In this study, we used a sagA insertion-deletion mutant in which the additional terminators surrounding the omega-Km2 (41) cassette are inserted between the sagA promoter and the sagB gene. This mutant is therefore expected to be polar on all the downstream genes in the sag operon. Three of these genes, sagG, sagH, and sagI, encode putative ABC transporter proteins, which may be involved in exporting SagA and/or other molecules across the cytoplasmic membrane. Another gene, sagB, encodes a protein with a high degree of homology to immunity proteins often involved in inhibition of bacteriocin action. The other genes downstream of sagA (sagC, sagD, and sagF) have little or no homology to any known genes, but they encode proteins with putative membrane-spanning domains and thus their products may be surface located. Because the insertion-deletion mutant we studied should show reduced expression of all the genes in the sag operon, the effect on M6 protein that we observed could be due to loss of function of any of the genes in the operon.
The relationship between SLS production and the expression of various virulence factors has been studied in GAS strains of several different serotypes. In an M12 strain, a Tn916 insertion mutant that lacked SLS activity had wild-type levels of both M and T antigen (39). In an M3 strain, inactivation of SLS by a Tn916 insertion was associated with an altered growth requirement, but M protein was not affected (30). Unfortunately, in both cases, the locations of the Tn916 insertions were not mapped and thus the reduction of SLS activity may have been an indirect effect of insertion in some unknown regulatory locus.
More recently, Tn916 insertion mutants that mapped to the sagA promoter region were isolated in strains of both serotypes M1 and M18. These mutants were reported to retain wild-type levels of M protein, as determined by Western blot analysis with a MAb specific for the C-terminal region of the molecule (5). In contrast, in a serotype M49 strain, a Tn917 insertion mutant in the sagA promoter region showed virtually no emm gene transcription (28). However, in our study with a strain of M6 serotype we found an unaltered level of emm transcript but a greatly reduced amount of full-length M protein in the sagA insertion-deletion mutant we constructed. Combined, these results indicate that the sagA locus regulates M expression differently in different GAS strains.
At this time, we can only speculate about the molecular mechanisms by which the sag locus regulates the M protein. In the M6 strain we studied, the sag locus mutation led to truncation of the M protein. It seems likely, therefore, that either sagA or one of the other sag genes directly or indirectly decreases the expression of a protease or proteases that truncate the M6 molecule. Although the sequence of the sagA gene implies that SagA is a secreted peptide, it has been suggested that SagA may act as a pleiotropic transcriptional regulator (28). In that case, it is possible that SagA represses transcription of the protease(s) in the M6 strain. In the strain(s) in which transcription of emm is reduced in the sag mutant, it is possible that the hypothetical GAS protease(s) repressed by the sag locus may cleave the activator of the emm promoter, Mga. The sequence of Mga differs among the strains investigated. Thus, a cleavage site for the proposed protease may be present in the divergent Mga of the M49 strain but absent from the mga gene in the other strains, including that of serotype M6.
Although it seems unusual that the amount of M protein on the surface of sag mutants of different GAS strains is reduced for different reasons, there is precedent for different regulatory mechanisms controlling production of other secreted GAS proteins. For example, the speB gene is carried by all GAS strains, but the degree of SpeB expression varies greatly in strains of different serotypes (22, 51) and this results from the different actions of different transcriptional regulators. In a strain of M49 serotype, the speB gene is negatively regulated by Nra (38), whereas in an M6 serotype strain, RofA, an Nra homolog (3), has no effect on speB transcription. Furthermore, Nra also acts as a negative regulator of transcription of prtF (47), a gene encoding protein F, whereas RofA acts as a positive regulator of prtF transcription (18, 19). Taken together, these observations indicate that regulators act on similar genes differently in different strains.
In addition to a decrease in M protein expression, the M49 sagA mutant strain showed reduced amounts of streptokinase (encoded by ska) and the cysteine protease encoded by speB. However, the ska transcript remained at wild-type levels while the transcript levels for speB and emm were reduced (28). Thus, the mechanism of regulation of surface and secreted proteins by sagA may vary even within a single strain.
We found that in an M6 strain, a sagA mutation causes truncation of the M protein, which results in the loss of the domains of the protein required for anchoring to the GAS cell surface. To produce the larger (35 kDa) truncated protein found in the whole-cell extract, cleavage probably occurs at a site that separates the C repeats from the B repeats, since this truncated species failed to react with antibody specific for the C repeat region but did react with the B repeat-specific antibody. The location of this cleavage may coincide with the pepsin-sensitive site in the M6 molecule (20). It seems likely that this region of the protein is more exposed and thus more sensitive to proteolysis. Consistent with cleavage that removes the C repeats and the C-terminal region of the M6 protein is the result that the truncated M protein is not anchored to the cell wall.
Although there are two different species of truncated M6 protein in the whole-cell extract of the sagA mutant strain, of about 35 and 28 kDa, the larger truncated species is absent from the culture supernatant. This suggests that the 35-kDa protein is further processed during export across the cell membrane. The second proteolytic event that generates the 28-kDa truncated species probably occurs within the B repeats, since this species reacted with both antibodies specific for the N terminus of the M6 molecule and with MAb 10A11 (Fig. 5). Because M protein migrates anomalously in SDS-PAGE (21), the location of the 28-kDa band cannot be used to accurately localize the site of cleavage.
It seems highly probable that an endogenous GAS protease is involved in production of the truncated M6 proteins found in the sagA mutant strain. The presence of two different truncated products, one of which appears to be produced during transport through the cell membrane, suggests the possibility that more than one protease may be involved in the truncation. An intracellular protease probably cleaves mature M protein to produce the 35-kDa product while another protease further processes it to generate a 28-kDa protein found predominantly in the supernatant.
In GAS, SpeB is the major secreted and cell-associated protease. Furthermore, SpeB has been found to process the M1 molecule by removing its N-terminal region as well as by cleaving internal regions (4, 46). However, SpeB is probably not involved in either of the truncations of M6 protein resulting from the sagA mutation for at least three reasons. First, in the strain we studied, there is very little SpeB expressed (12, 51). Second, in a serotype M49 strain, the sagA mutant down-regulates SpeB activity instead of increasing it (28). And third, the presence of E64, a cysteine protease-specific inhibitor that reduces SpeB activity, had no effect on the presence of the truncated M6 products in a cell wall extract. Thus, it appears that proteases other than SpeB are involved in the truncation.
The results presented in this study emphasize the importance of the sag locus in virulence of the GAS. The reduction in virulence in a subcutaneous inoculation murine model of a sagA insertion mutant may result from a direct and/or indirect effect of sagA on virulence (5). It is likely that SLS plays an important role in dissemination of GAS during infection, but because of the pleiotropic nature of mutations in this gene it is not yet possible to test this idea directly. The M protein has long been considered the major virulence determinant of GAS because of its role in protecting the bacteria from phagocytosis and its role in attachment to initiate infection and it has been demonstrated to be important for virulence in a subcutaneous inoculation murine model (2). Mutation in the sag locus in several strains results in a lack of M protein on their surfaces, although the mechanism leading to this phenotype appears to be different in different strains of GAS and is not yet understood. We have shown that for an M6 strain, the polar sagA mutation demonstrates proteolytic truncation of the M protein that prevents its attachment to the surface of GAS. This causes a loss of the ability of the bacterium to resist phagocytosis and thus should have a significant effect on the virulence of the strain. Further understanding of the complex functions of the sag locus in different GAS strains should greatly enhance our knowledge of the pathogenesis of this heterogeneous bacterial species.
ACKNOWLEDGMENT
We thank Vince Fischetti for providing anti-M6 polyclonal and monoclonal antibodies and Tim Barnett for helping with plasmid construction and for helpful discussion.
This work was supported by NIH grant R37-AI20723.
REFERENCES
- 1.Akesson P, Sjoholm A G, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh C D, Warren H B, Carey V J, Wessels M R. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Investig. 1998;102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckert S, Kreikemeyer B, Podbielski A. Group A streptococcal rofA gene is involved in the control of several virulence genes and eukaryotic cell attachment and internalization. Infect Immun. 2001;69:534–537. doi: 10.1128/IAI.69.1.534-537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berge A, Bjorck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J Biol Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- 5.Betschel S D, Borgia S M, Barg N L, Low D E, De Azavedo J C. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bronze M S, Dale J B. The reemergence of serious group A streptococcal infections and acute rheumatic fever. Am J Med Sci. 1996;311:41–54. doi: 10.1097/00000441-199601000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Caparon M G, Geist R T, Perez-Casal J, Scott J R. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J Bacteriol. 1992;174:5693–5701. doi: 10.1128/jb.174.17.5693-5701.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caparon M G, Stephens D S, Olsen A, Scott J R. Role of M protein in adherence of group A streptococci. Infect Immun. 1991;59:1811–1817. doi: 10.1128/iai.59.5.1811-1817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 11.Cunningham M W. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Federle M J, McIver K S, Scott J R. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferretti J J, McShan W M, Ajdic D, Savic D J, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov A N, Kenton S, Lai H S, Lin S P, Qian Y, Jia H G, Najar F Z, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton S W, Roe B A, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischetti V A, Jones K F, Hollingshead S K, Scott J R. Structure, function, and genetics of streptococcal M protein. Rev Infect Dis. 1988;10(Suppl. 2):S356–S359. doi: 10.1093/cid/10.supplement_2.s356. [DOI] [PubMed] [Google Scholar]
- 16.Fischetti V A, Jones K F, Scott J R. Size variation of the M protein in group A streptococci. J Exp Med. 1985;161:1384–1401. doi: 10.1084/jem.161.6.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischetti V A, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- 18.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an m gamma delta-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 19.Granok A B, Parsonage D, Ross R P, Caparon M G. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J Bacteriol. 2000;182:1529–1540. doi: 10.1128/jb.182.6.1529-1540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollingshead S K, Fischetti V A, Scott J R. Complete nucleotide sequence of type 6 M protein of the group A streptococcus. Repetitive structure and membrane anchor. J Biol Chem. 1986;261:1677–1686. [PubMed] [Google Scholar]
- 21.Hollingshead S K, Fischetti V A, Scott J R. Size variation in group A streptococcal M protein is generated by homologous recombination between intragenic repeats. Mol Gen Genet. 1987;207:196–203. doi: 10.1007/BF00331578. [DOI] [PubMed] [Google Scholar]
- 22.Hytonen J, Haataja S, Gerlach D, Podbielski A, Finne J. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol Microbiol. 2001;39:512–519. doi: 10.1046/j.1365-2958.2001.02269.x. [DOI] [PubMed] [Google Scholar]
- 23.Jones K F, Hollingshead S K, Scott J R, Fischetti V A. Spontaneous M6 protein size mutants of group A streptococci display variation in antigenic and opsonogenic epitopes. Proc Natl Acad Sci USA. 1988;85:8271–8275. doi: 10.1073/pnas.85.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones K F, Khan S A, Erickson B W, Hollingshead S K, Scott J R, Fischetti V A. Immunochemical localization and amino acid sequences of crossreactive epitopes within the group A streptococcal M6 protein. J Exp Med. 1986;164:1226–1238. doi: 10.1084/jem.164.4.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapur V, Majesky M W, Li L L, Black R A, Musser J M. Cleavage of interleukin 1 beta (IL-1 beta) precursor to produce active IL-1 beta by a conserved extracellular cysteine protease from Streptococcus pyogenes. Proc Natl Acad Sci USA. 1993;90:7676–7680. doi: 10.1073/pnas.90.16.7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kihlberg B M, Cooney J, Caparon M G, Olsen A, Bjorck L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb Pathog. 1995;19:299–315. doi: 10.1016/s0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 27.Lancefield R C. Current knowledge of type specific M antigens of group A streptococci. J Immunol. 1962;89:307–313. [PubMed] [Google Scholar]
- 28.Li Z, Sledjeski D D, Kreikemeyer B, Podbielski A, Boyle M D. Identification of pel, a Streptococcus pyogenes locus that affects both surface and secreted proteins. J Bacteriol. 1999;181:6019–6027. doi: 10.1128/jb.181.19.6019-6027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limbago B, McIver K S, Penumalli V, Weinrick B, Scott J R. Restoration of Mga function to a Streptococcus pyogenes strain (M Type 50) that is virulent in mice. Infect Immun. 2001;69:1215–1220. doi: 10.1128/IAI.69.2.1215-1220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Sela S, Cohen G, Jadoun J, Cheung A, Ofek I. Insertional inactivation of streptolysin S expression is associated with altered riboflavin metabolism in Streptococcus pyogenes. Microb Pathog. 1997;22:227–234. doi: 10.1006/mpat.1996.0107. [DOI] [PubMed] [Google Scholar]
- 31.Lottenberg R, DesJardin L E, Wang H, Boyle M D. Streptokinase-producing streptococci grown in human plasma acquire unregulated cell-associated plasmin activity. J Infect Dis. 1992;166:436–440. doi: 10.1093/infdis/166.2.436. [DOI] [PubMed] [Google Scholar]
- 32.Lukomski S, Nakashima K, Abdi I, Cipriano V J, Ireland R M, Reid S D, Adams G G, Musser J M. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon W R, Gibson C M, Caparon M G. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manjula B N, Fischetti V A. Tropomyosin-like seven residue periodicity in three immunologically distinct streptococcal M proteins and its implications for the antiphagocytic property of the molecule. J Exp Med. 1980;151:695–708. doi: 10.1084/jem.151.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuka Y V, Pillai S, Gubba S, Musser J M, Olmsted S B. Fibrinogen cleavage by the Streptococcus pyogenes extracellular cysteine protease and generation of antibodies that inhibit enzyme proteolytic activity. Infect Immun. 1999;67:4326–4333. doi: 10.1128/iai.67.9.4326-4333.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIver K S, Thurman A S, Scott J R. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J Bacteriol. 1999;181:5373–5383. doi: 10.1128/jb.181.17.5373-5383.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLandsborough L A, Cleary P P. Insertional inactivation of virR in Streptococcus pyogenes M49 demonstrates that VirR functions as a positive regulator of streptococcal C5a peptidase and M protein in OF+ strains. Dev Biol Stand. 1995;85:149–152. [PubMed] [Google Scholar]
- 38.Molinari G, Rohde M, Talay S R, Chhatwal G S, Beckert S, Podbielski A. The role played by the group A streptococcal negative regulator Nra on bacterial interactions with epithelial cells. Mol Microbiol. 2001;40:99–114. doi: 10.1046/j.1365-2958.2001.02373.x. [DOI] [PubMed] [Google Scholar]
- 39.Nida K, Cleary P P. Insertional inactivation of streptolysin S expression in Streptococcus pyogenes. J Bacteriol. 1983;155:1156–1161. doi: 10.1128/jb.155.3.1156-1161.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nizet V, Beall B, Bast D J, Datta V, Kilburn L, Low D E, De Azavedo J C. Genetic locus for streptolysin S production by group A streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Casal J, Price J A, Maguin E, Scott J R. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol Microbiol. 1993;8:809–819. doi: 10.1111/j.1365-2958.1993.tb01628.x. [DOI] [PubMed] [Google Scholar]
- 43.Phillips G N, Flicker P F, Cohen C, Manjula B N, Fischetti V A. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci USA. 1981;78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Podbielski A, Woischnik M, Leonard B A, Schmidt K H. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 46.Raeder R, Woischnik M, Podbielski A, Boyle M D. A secreted streptococcal cysteine protease can cleave a surface-expressed M1 protein and alter the immunoglobulin binding properties. Res Microbiol. 1998;149:539–548. doi: 10.1016/s0923-2508(99)80001-1. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen M, Eden A, Bjorck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect Immun. 2000;68:6370–6377. doi: 10.1128/iai.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott J R. The M protein of group A streptococcus: evolution and regulation. In: Iglewski B M, Clark V L, editors. The bacteria: molecular basis of bacterial pathogenesis. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–203. [Google Scholar]
- 49.Scott J R, Guenthner P C, Malone L M, Fischetti V A. Conversion of an M-group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J Exp Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson W J, LaPenta D, Chen C, Cleary P P. Coregulation of type 12 M protein and streptococcal C5a peptidase genes in group A streptococci: evidence for a virulence regulon controlled by the virR locus. J Bacteriol. 1990;172:696–700. doi: 10.1128/jb.172.2.696-700.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson M D, Scaramuzzino D A, Sjobring U, Olsen A, Frank C, Bessen D E. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol Microbiol. 2000;38:242–253. doi: 10.1046/j.1365-2958.2000.02144.x. [DOI] [PubMed] [Google Scholar]
- 52.Swanson J, Hsu K C, Gotschlich E C. Electron microscopic studies on streptococci. I. M antigen. J Exp Med. 1969;130:1063–1091. doi: 10.1084/jem.130.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitnack E, Beachey E H. Antiopsonic activity of fibrinogen bound to M protein on the surface of group A streptococci. J Clin Investig. 1982;69:1042–1045. doi: 10.1172/JCI110508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitnack E, Beachey E H. Biochemical and biological properties of the binding of human fibrinogen to M protein in group A streptococci. J Bacteriol. 1985;164:350–358. doi: 10.1128/jb.164.1.350-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]