Abstract
The role of B cells in antitumor immunity is becoming increasingly appreciated, as B cell populations have been associated with response to immune checkpoint blockade (ICB) in patients with breast cancer and murine models of breast cancer. Deeper understanding of antibody responses to tumor antigens is needed to clarify the function of B cells in determining response to immunotherapy. We evaluated tumor antigen-specific antibody responses in patients with metastatic triple negative breast cancer treated with pembrolizumab following low-dose cyclophosphamide therapy using computational linear epitope prediction and custom peptide microarrays. We found that a minority of predicted linear epitopes were associated with antibody signal, and signal was associated with both neoepitopes and self-peptides. No association was observed between signal presence and subcellular localization or RNA expression of parent proteins. Patient-specific patterns of antibody signal boostability were observed that were independent of clinical response. Intriguingly, measures of cumulative antibody signal intensity relative to immunotherapy treatment showed that the one complete responder in the trial had the greatest increase in total antibody signal, which supports a potential association between ICB-dependent antibody boosting and clinical response. The antibody boost in the complete responder was largely driven by increased levels of IgG specific to a sequence of N-terminal residues in native Epidermal Growth Factor Receptor Pathway Substrate 8 (EPS8) protein, a known oncogene in several cancer types including breast cancer. Structural protein prediction showed that the targeted epitope of EPS8 was in a region of the protein with mixed linear/helical structure, and that this region was solvent-exposed and not predicted to bind to interacting macromolecules. This study highlights the potential importance of the humoral immune response targeting neoepitopes as well as self epitopes in shaping clinical response to immunotherapy.
Keywords: B-Lymphocytes; Breast Neoplasms; Antibody Specificity; Antigens, Neoplasm; Epitope Mapping
Background
There is a growing appreciation for the role of B cells in antitumor immunity. B cell functions include cytokine production, antigen presentation via MHC class-I and MHC class-II molecules to stimulate expansion of CD8+ and CD4+ T cells, and production of soluble antibody, which can opsonize/neutralize target antigen and facilitate antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity.1 B cells infiltrate tumors and associate with myeloid/lymphoid aggregates termed tertiary lymphoid structures (TLS),2–6 and promote antitumor immune responses through production of antigen-specific antibodies and via antigen presentation to promote expansion of CD4+ T cells.7 B cells are also a source of autoantibodies, which are self-targeting antibodies that result from a deficit in immunological tolerance, are characteristic of autoimmune diseases, and are observable in diverse cancer types.8–10 As potential diagnostic biomarkers, autoantibodies are detectable at early stages of cancer,11–13 and they may be involved in anti-tumor immune responses in certain types of cancer.14–17
We recently reported that increased B cell gene signature expression and B cell receptor diversity in pretreatment samples were associated with clinical response to immune checkpoint blockade (ICB) in triple negative breast cancer (TNBC).18 In murine models of TNBC, ICB response has been found to be dependent on B cell responses.19 Beyond TNBC, intratumoral presence of TLS have been associated with ICB response in various cancer types.2–6 In ovarian cancer, B-cell-derived IgA production promoted myeloid cell-dependent killing of ovarian cancer cells and antibody-dependent transcriptomic changes in cancer cells that sensitized them to T cell killing.20 B-cell-derived antibody responses have also been reported to promote the differentiation of neoantigen-specific CD4+ T cells, which can in turn enhance CD8+ T cell effector functionality through IL-21 production.21 Thus, there is a dynamic interplay between dendritic cells (DCs), T cells, and B cells within tumors, and further elucidation of the cellular and molecular mechanisms that govern this dynamic would be valuable for understanding immunotherapy response.
The goal of this study was to understand whether tumor-specific antibodies and autoantibodies could be discovered in TNBC patients treated with immunotherapy. To assess this, we used genomics data to predict linear epitopes in eleven patients for generation of custom peptide arrays, which were probed in a multiplex ELISA with patient plasma from two time points: pre-ICB and after two cycles of ICB. We found that a minority of predicted epitopes were associated with IgG antibody signal. Nuclear, cell surface and cytoplasmic locations were predominant in antibody-associated proteins, and RNA expression was not associated with antibody signal. For some patients, including the complete responder, the majority of peptides with antibody signal displayed increased signal after ICB treatment. Furthermore, ICB-dependent boost of both self-peptide-specific and mutated peptide-specific antibody signal was observed for some patients. A set of high-boosted epitopes that were self-specific in the complete responder was observed, and these epitopes were found in an N-terminal, surface-exposed region of the oncogenic Epidermal Growth Factor Receptor Pathway Substrate 8 (EPS8) protein. Together, these data offer an initial glimpse into the characteristics of antibody responses in TNBC patients treated with immunotherapy.
Methods
Neoantigen peptide prediction
HLA major and minor class I alleles were determined from RNA expression data using OptiType V.1.3.122 via the authors’ published Docker container in RNA mode (--rna, as per https://github.com/FRED-2/OptiType). Annotated variant transcripts were created using ANNOVAR v2019Oct24,23 using their suggested single nucleotide polymorphism (SNP) filters: the Exome Aggregation Consortium repository (last updated May 16, 2019) and ANNOVAR’s modified dbSNP list, avnsp147. Functional prediction and annotation were done using the suggested dbNSFP V.3.0a24 database. NeoPredPipe V.4.025 was used to orchestrate the process of variant filtering, transcript annotation, and binding affinity calculations.
Peptide selection
Peptide candidates were generated from predicted variant protein sequences (arising from single nucleotide variants and inserstion/deletions; SNVs and INDELs, respectively) by iterating over possible 15mers including at least one variant amino acid in a sliding-window approach. For each predicted variant 15mer, a matched reference 15mer was produced as a control. Neoantigen peptides from frameshift or stop-loss mutations were discarded. Single amino acid substitutions were sorted based on their estimated amino acid exchangeability,26 with more dissimilar substitutions favored over more similar ones.
Peptide arrays
Neoepitopes and matched self-peptides were screened using peptide arrays printed by PEPperPRINT (Germany). Peptides were converted into two identical microarrays for each patient. The resulting arrays contained varying numbers of linear peptides (range: 2136–5498 total peptides, half of which were mutant peptides and the other half comprizing matched self-peptides) printed in duplicate, and were framed by additional HA (YPYDVPDYAP, 52 or 40 spots, respectively) and polio (KEVPALTAVETGAT, 52 or 38 spots, respectively) control peptides. Microarrays were prestained with the secondary and control antibodies (Goat anti-human IgG (Fc)-DyLight680 (0.1 µg/mL), Mouse monoclonal anti-HA (12CA5)-DyLight800 (0.1 µg/mL)) in incubation buffer (PBS, pH 7.4 with 0.05% Tween-20+10% Rockland blocking buffer MB-070) to investigate background interactions with linear peptides. Subsequent incubation of peptide microarrays with patient plasma samples of the respective patient at 1:20 dilution was followed by staining with secondary and control antibodies. Read-out was performed with an Innopsys InnoScan 710-IR Microarray Scanner at scanning gains of 50/10 (red/green). The additional HA peptides framing the peptide microarrays were simultaneously stained as internal quality control to confirm assay performance and peptide microarray integrity. Quantification of spot intensities and peptide annotation were based on 16-bit gray scale tiff files that exhibit a higher dynamic range than the 24-bit colorized tiff files. Microarray image analysis was done with PepSlide Analyzer. A software algorithm breaks down fluorescence intensities of each spot into raw, foreground and background signal, and calculates averaged median foreground intensities and spot-to-spot deviations of spot duplicates. A maximum spot-to-spot deviation of 40% was tolerated, otherwise the corresponding intensity value was zeroed. For analysis purposes, a background-corrected signal intensity of >500 F.U. (fluorescence units) and less than 2000 F.U. was denoted as weak signal, and signal intensity >2000 F.U. was denoted as moderate/strong signal (per recommendations from Pepperprint technical support team).
Results
A minority of predicted neoantigen-containing peptides were targeted by endogenous antibodies
We examined IgG reactivity to neoantigen-containing versus unmutated self linear epitopes from patients with TNBC who participated in a clinical trial to examine efficacy of PD-1 inhibition following cyclophosphamide treatment18 (figure 1, online supplemental tables 1 and 2). Of the 40 patients enrolled, 11 patients were chosen for evaluation based on differential response to ICB (1 with complete response, 4 with partial response and 6 with progressive disease; (CR, PR and PD, respectively); refer to online supplemental methods 1 for details on patient selection). The predicted peptides were prioritized/ranked (see online supplemental methods 1) and were printed on custom peptide arrays along with their corresponding normal self-peptides. Plasma drawn on cycle 1, day 1 (C1D1, prestudy) and cycle 3, day 1 (C3D1, after two completed cycles of pembrolizumab) was used to probe the arrays. Peptide spots that bind antibody would yield fluorescence signal after staining with a secondary antibody conjugated to a fluorescent label. As seen in the digitized scan images of representative microarrays (figure 1), a large majority of predicted peptides were not associated with IgG antibody binding, and this was true of both mutated and self-peptides. On average, 94.8% of predicted mutant peptides had signal denoted as very weak/noise (<500 F.U.), 3.66% had weak signal (>500 F.U. and <2000 F.U.), 1.24% had moderate signal (>2000 FU and <10 000 F.U.), and 0.26% had strong signal (>10 000 F.U.).
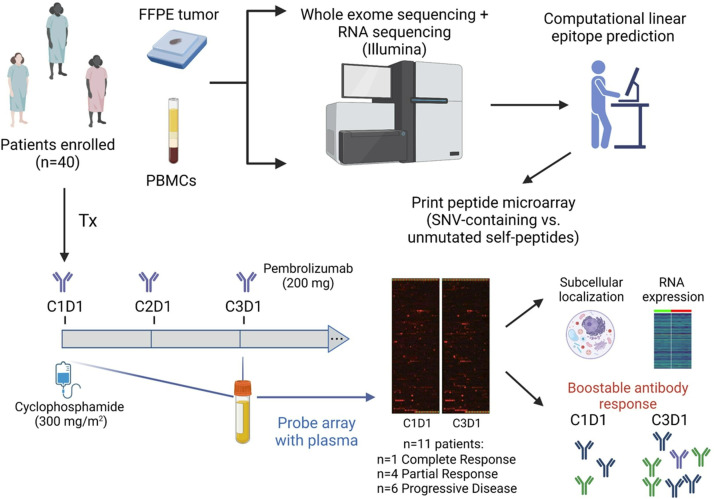
Figure 1.
Experimental approach to examine neoantigen-specific antibody responses. Patients with metastatic triple negative breast cancer underwent a clinical trial to examine the efficacy of regulatory T cell depletion with cyclophosphamide plus PD-1 inhibition with pembrolizumab.18 Eleven patients from this cohort were selected based on clinical response for analysis of tumor antigen-specific antibody responses via multiplex ELISA (peptide arrays). Downstream analyses included examination of protein subcellular localization and RNA expression, as well as antibody boostability relative to immunotherapy treatment. Antibody responses were seen at baseline, and in some patients, increased after therapy. FFPE, formalin-fixed paraffin-embedded; SNV, single nucleodie variant; PBMCs, peripheral blood mononuclear cells.
jitc-2022-005848supp001.pdf (2.1MB, pdf)
Subcellular distribution and RNA expression analysis of peptides and associated antibody signal
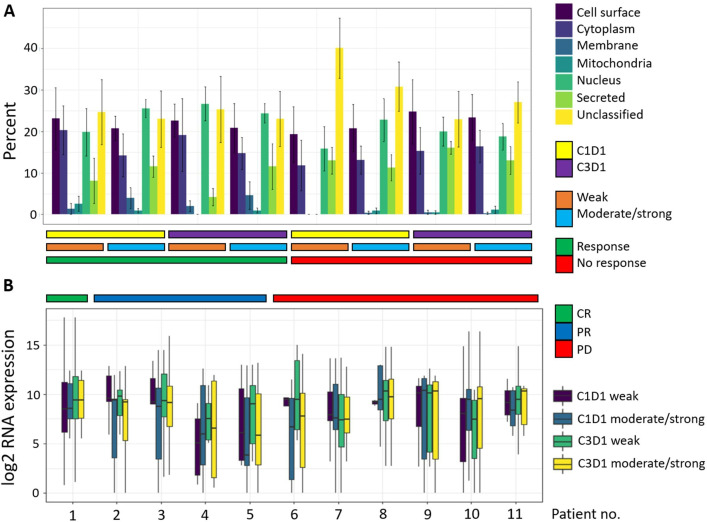
We sought to understand whether there were any subcellular locations enriched for antibody signal-associated peptides. In order to focus on antibody signal with putative biological function, we compared antibody signal that was weak but above background noise (>500 F.U. and <2000 F.U.) to antibody signal that was at least moderate in intensity (>2000 F.U.). Subcellular localization analysis showed that peptides associated with antibody signal were most often derived from nuclear proteins, with cell surface and cytoplasmic subcellular locations also being more highly represented relative to membrane, mitochondrial and secreted proteins (figure 2A). Additionally, there was no subcellular distribution difference between peptides associated with weak versus stronger signal intensity (figure 2A). We were also interested in determining if parent proteins of peptides associated with stronger antibody signal intensity would have increased tumor RNA expression levels, but no such difference was observed (figure 2B).
Figure 2.
Subcellular protein localization and RNA expression do not associate with antibody signal or response group. (A) Subcellular localization of parent proteins of self/mutant peptide pairs subset based on associated antibody signal level (weak vs moderate/strong), and further categorized by treatment time point and response. Peptide pairs were considered to have moderate/strong signal if either peptide of the pair (self or mutant) had >2000 F.U. signal intensity. Peptide pairs were considered to have weak signal if both peptides of the pair (self and mutant) had signal intensity >500 F.U. and <2000 F.U. Error bars represent SE (n=5 for responders; n=6 for non-responders). Subcellular localization was determined using the R Bioconductor package SubCellBarCode (https://bioconductor.org/packages/release/bioc/html/SubCellBarCode.html) and the R package UniprotR (https://rdrr.io/cran/UniprotR/). Each gene was assigned a subcellular annotation based on consensus between the outputs of these two R packages. If there was no consensus, then the subcellular annotation attained using SubCellBarCode was used as this method is based on subcellular confirmation using mass spec data from 5 cell lines. For annotation of cell surface proteins, the Cancer Surfaceome Atlas (doi: 10.1038/s43018-021-00282-w) was used. (B) Distribution of RNA expression of parent proteins categorized according to antibody signal level, treatment time point and response. CR, complete response; PR, partial response; PD, progressive disease.
Immunogenomic correlates of antibody response
We examined correlations between tissue-derived immunogenomics features and metrics of antibody response. Pretreatment tumor immune gene signatures (IGS) negatively associated with antibody responses included IGS reflective of Th1 cells, mast cells, and an ICB-response signature; conversely, positive correlation was noted with an EMT signature (online supplemental figure 1). Associations of antibody responses to measures of T and B cell repertoire diversity were also observed (online supplemental figures 2–4). In pretreatment tumors, a positive correlation was seen between self-specific antibody signal at C3D1 and IGH/IGK species diversity (online supplemental figure 2). In peripheral blood, a negative association was noted between measures of T cell diversity and antibody responses both before and on-treatment; positive associations were also observed between antibody responses and IGH/IGL evenness (online supplemental figures 3 and 2). Further analysis examining putative associations between antibody responses and predicted neoantigen load, including traditional and alternative neoantigen sources (eg, SNV, InDel, Virus/ERV, CTA/Self-antigens, fusions, and splice variants), was performed. Positive correlation between the number of predicted InDel-associated MHC class I-restricted neoantigens and metrics of antibody response was noted, with the strongest correlation observed with respect to total specific antibody signal (mutant-specific plus self-specific antibody level) at C3D1 (online supplemental figure 5). Negative correlation was observed between self-specific antibody level at C3D1 level and splice variant-derived MHC class I antigens (online supplemental figure 5).
Observation of patient-specific patterns of antibody boostability
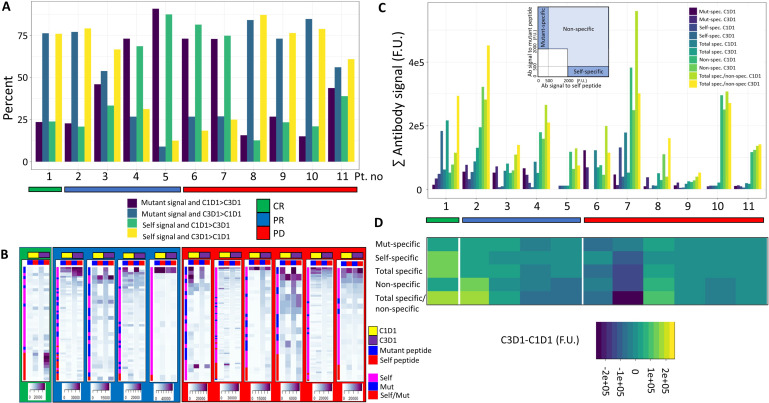
We next examined the relationship between pembrolizumab treatment and boostability of antibody signal (eg, increase in antibody signal between pretreatment and post-treatment time points). ICB-dependent boost of both self-peptide-specific and mutated peptide-specific antibody signal was observed for some patients, but this was not significantly associated with clinical response class (figure 3, online supplemental figure 6). Patients 1 (CR) and 2 (PR), which were the patients with the highest tumor mutation burden, exhibited strong ICB-dependent antibody boost. The boost observed with patient 1 was largely driven by an increase in antibody signal to self-specific peptides (figure 3B–D), although mutated peptides were also boosted. Alternatively, the boost observed with patient 2 was driven by an increase in non-specific antibody signal (eg, antibody response to both mutant and matched self-peptides). Of note, signal boost was not associated with clinical response to checkpoint inhibition, although the magnitude of boost was generally higher in responders than in non-responders. To guage the absolute value of antibody signal boost relative to ICB treatment, we calculated the difference between total antibody signal at C3D1 and C1D1 (figure 3C,D). Interestingly, patient 1 exhibited the strongest absolute signal intensity difference relative to immunotherapy treatment. Together, these data suggest a possible association between ICB-dependent antibody boostability and clinical response.
Figure 3.
Boostability of antibody response relative to ICB treatment. (A) Percentage of mutant or self-peptides that had associated antibody signal (>500 F.U. at either C1D1 or C3D1) and greater signal relative to the other time point. (B) Heatmap depiction of antibody signal relative to treatment time point. Signal was ranked according to intensity of signal to C3D1 mutant peptide. A peptide pair was included if there was any signal >2000 F.U. for either self or mutant peptide at either C1D1 or C3D1 time point. Colored sidebar denotes whether antibody signal was associated with mutant peptide, self peptide or both. (C) Absolute antibody signal (summed) categorized by signal specificity, treatment time point and response. Inlay depicts signal thresholds that were used to denote specificity classes. (D) ICB-associated boostability difference in antibody signal, which is derived by subtracting C1D1 antibody signal from C3D1 signal for respective specificity classes. ICB, immune checkpoint blockade; CR, complete response; PR, partial response; PD, progressive disease.
Boostable self-specific antibodies to EPS8 dominate antibody response in complete responder
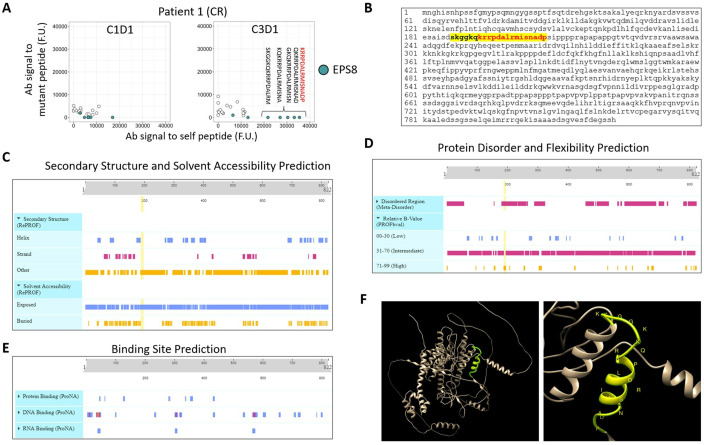
We next focused on the ICB-dependent boost that was observed in the complete responder to try and determine if there was something particular about this boost that associated with clinical response. A strong boost to EPS8 self-peptides was seen (figure 4A), and these peptides were located within an N-terminal region of the protein corresponding to residues S187-P107 (figure 4B). This region was predicted to have a mixed linear/helical structure, be surface-exposed/solvent-accessible, and to be disordered/flexible (figure 4C–F). These physical properties may contribute to a potentiated B cell response against these epitopes.
Figure 4.
Complete responder displayed ICB-dependent boostability of both mutant and self-peptides, with particularly strong boost of IgG specific to native EPS8 peptides. (A) Relative antibody signal to self- vs mutant peptides at C1D1 and C3D1. Annotation shows ICB-dependent boost of antibody signal to self-EPS8 peptides. Peptides with greatest boost corresponding to S187-P107 residues are shown. (B) Primary protein structure of EPS8, with S187-P107 highlighted. The primary structure for EPS8 protein was input into PredictProtein (https://predictprotein.org) to query secondary structure and solvent accessibility (C), protein disorder and flexibility (D), and macromolecular binding site predictions (E). (F) Tertiary structure of EPS8 as predicted by Alphafold Protein Structure Database (https://alphafold.com), with S187-P107 highlighted in yellow. Three-dimensional structure was visualized and annotated using Chimera V.1.16 software (https://www.cgl.ucsf.edu/chimera/). ICB, immune checkpoint blockade; EPS8, Epidermal Growth Factor Receptor Pathway Substrate 8; CR, complete response.
Discussion
T cells dominate our concept of ‘The Cancer Immunity Cycle’27 for good historical reasons: they kill tumor cells directly, have efficacy in adoptive transfer, and associate with clinical response to immune checkpoint inhibition. That said, studies from our group and others over the past five years have implicated the B cell arm of the adaptive immune system in tumor control. In breast cancer specifically, B cell population features consistent with antigen-driven clonal expansion associate with improved survival and response to immunotherapy. Thus, there is a need to discover the action(s) of tumor antigen-specific B cells in antitumor immunity in breast cancer.
We have taken a step in that direction by measuring tumor antigen-specific antibodies in the plasma of patients with breast cancer treated with immunotherapy. There is evidence that antitumor antibodies can be important to achieve curative responses in large established murine tumors,28 and deeper understanding of the intricacies of antigen-specific B cell responses is necessary. Importantly, this study establishes that ICB can boost antibody responses to neoantigens, thus providing rationale for combining B cell-epitope-targeting vaccine strategies with ICB. Moreover, in the case of the complete responder, we observed a boostable antibody response targeting EPS8 protein, which is a known oncogene potentiating growth/survival (eg, mTOR/PI3K/AKT/EGFR signaling) in multiple cancer types including breast cancer.29 This finding highlights the potential utility of immunotherapeutic strategies aimed at boosting antibody responses to tumor-associated antigens. It has been previously shown that elevated levels of autoantibodies targeting HER2 in breast cancer were significantly associated with increased recurrence-free survival in multivariable models that included clinicopathological characteristics,15 thus supporting a putative relationship between autoantibody levels and protective antitumor responses.
Autoantibodies may be associated with clinical outcomes to cancer therapy. For example, lower baseline and greater increase in autoantibody levels during the course of ICB treatment were associated with development of immune-related adverse events (irAEs),30 and increased severity of irAEs has been found to be related to specific autoantibody profiles.31 Although irAEs are harmful, such ICB-associated B cell-driven autoimmune responses may have beneficial sequelae.17 Studies of melanoma patients treated with ICB found associations between therapy response/survival and serum IgG/autoantibody levels.17 32 33 Interestingly, ICB-dependent expansion of CD21lo memory B cells and CD27+CD38+ plasmablasts preceded and associated with the development of irAEs in melanoma patients, and CD21lo B cells exhibited increased levels of IFNG signaling and B cell activation, as well as increased clonality in some patients.34 Plasmablasts are known to secrete autoantibodies,35 and CD21lo memory B cells potentially contribute to autoreactive T cell expansion via autoantigen presentation.36 Notably, a significant association between plasmablast levels and ICB response was shown in previous analysis of TNBC patients from our study cohort.18 It is thus possible that this population contributed to both autoantibody and neoantigen-specific antibody production, which warrants further investigation. While the above studies provide evidence of a positive relationship between autoantibodies and ICB response/survival, other studies have not found this. No association was found between median baseline autoantibody levels and disease recurrence in a prospective study of melanoma patients treated with ICB in the adjuvant setting, although an autoantibody signature that predicted recurrence-free survival and irAE development with high accuracy was reported.37 The authors posited that the immunogenicity of specific autoantigens, such as those included in their recurrence signature, may possess superior predictive power relative to total autoantibody levels.37 Lack of relationship between pretreatment and post-ICB autoantibody levels and clinical endpoints has also been described in a pan-cancer study,38 although the authors assayed presence of a prescribed and limited panel of autoantigens and they did not examine autoantibody boostability relative to response. With regard to chemotherapy associations with autoantibodies, a reduction of autoantibody levels was observed in a breast cancer study after treatment with different combinations of chemotherapy, radiation, and hormonal therapy.39 Conversely, a case study of a colon cancer patient treated with a FOLFIRI/cetuximab regimen showed increased IgM autoantibody levels on treatment that were boostable with further treatment cycles.40 Chemotherapy-induced boosting of antibody titer is a possible but unlikely explanation of our data, as patients in the study cohort were pretreated with cyclophosphamide prior to ICB, which is known to deplete both T and B cells.41
The present study is limited in important ways. Due to the relative lack of responders in the study cohort (n=6 responders out of n=40 patients enrolled; 1 PR had data that was unusable due to low signal-to-noise ratio), this study is not powered to associate antibody signal with clinical response or genomics and immunogenomics features between patient groups. It also lacks power to interrogate the relative frequency of mutation-specific, unmutated self-specific and cross-reactive antibodies in the treated population. Metrics of IgG abundance/boostability were not corrected for TMB, and such analysis may yield further insight in later studies with larger cohorts. The framework of this analysis does not provide evidence that these antibody signals are associated with tumor growth, cytotoxicity, support of T cells, or other biological function(s). It is possible that the observed antibody boostability is merely a surrogate of T-cell expansion that may or may not have a therapeutic effect. Even if the observed antibody responses are real, we do not know how generalizable antibody production is across patients or tumor types. An additional deficit of this study is that it is limited in antibody discovery, as we have neither predicted nor measured conformational B cell epitopes, which may be the dominant epitopes in the system. It is unclear how these antibodies, or others targeting autoantigens, contribute to immunotherapy response.
In summary, we have performed an initial analysis of antibody responses in TNBC patients treated with pembrolizumab following cyclophosphamide. We have found both tumor neoantigen-specific, self-specific and non-specific antibodies that increased in signal after two cycles of pembrolizumab therapy. Similar studies with increased power to delineate the characteristics and contribution of ICB-dependent antibody responses to clinical response are warranted.
Acknowledgments
The authors thank Ken Fowler and Karen McKinnon with the Immune Monitoring and Genomics Facility (IMGF) at UNC-Chapel Hill for their assistance with this study. We also thank the patients in this study and their families, without whom this study would not have been possible.
Footnotes
Twitter: @SeroBMT1, @BenjaminGVincen
EDR and MGW contributed equally.
Contributors: EDR and BGV conceived the study. MGW performed in silico peptide predictions. EDR performed data analysis and figure preparation. EDR and BGV wrote the manuscript. WB and SPV performed in silico neoantigen predictions. All authors contributed to the review of the manuscript.
Funding: Merck Sharp & Dohme, a subsidiary of Merck & Co., Rahway, New Jersey, USA (MSD) provided financial support (#OTSP58116) for the clinical trial under which samples from this study were acquired. This work was also supported by Susan G. Komen for the Cure (BVG, #CCR17483467), V Foundation for Cancer Research (BGV, #T2018-009), and UNC University Cancer Research Fund (MGW, JSS, BGV).
Competing interests: BGV declares consulting fees from GeneCentric Therapeutics, not relevant to this work. JSS receives funding from MSD, GSK, and Carisma, is a scientific consultant for PIQUE Therapeutics and has filed IP for the use of STING agonists to enhance CAR T cell for breast cancer. The other authors have no conflicts requiring disclosure.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the University of North Carolina at Chapel Hill Institutional Review Board, #16-1025. Participants gave informed consent to participate in the study before taking part.
References
- 1. LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood 2008;112:1570–80. 10.1182/blood-2008-02-078071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cabrita R, Lauss M, Sanna A, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561–5. 10.1038/s41586-019-1914-8 [DOI] [PubMed] [Google Scholar]
- 3. Helmink BA, Reddy SM, Gao J, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vanhersecke L, Brunet M, Guégan J-P, et al. Mature tertiary lymphoid structures predict immune checkpoint inhibitor efficacy in solid tumors independently of PD-L1 expression. Nat Cancer 2021;2:794–802. 10.1038/s43018-021-00232-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petitprez F, de Reyniès A, Keung EZ, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577:556–60. 10.1038/s41586-019-1906-8 [DOI] [PubMed] [Google Scholar]
- 6. Meylan M, Petitprez F, Becht E, et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity 2022;55:527–41. 10.1016/j.immuni.2022.02.001 [DOI] [PubMed] [Google Scholar]
- 7. Laumont CM, Banville AC, Gilardi M, et al. Tumour-infiltrating B cells: immunological mechanisms, clinical impact and therapeutic opportunities. Nat Rev Cancer 2022;22:414–30. 10.1038/s41568-022-00466-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan EM. Autoantibodies as reporters identifying aberrant cellular mechanisms in tumorigenesis. J Clin Invest 2001;108:1411–5. 10.1172/JCI14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan EM, Zhang J. Autoantibodies to tumor-associated antigens: reporters from the immune system. Immunol Rev 2008;222:328–40. 10.1111/j.1600-065X.2008.00611.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Jonge H, Iamele L, Maggi M, et al. Anti-cancer auto-antibodies: roles, applications and open issues. Cancers (Basel) 2021;13:813. 10.3390/cancers13040813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chapman C, Murray A, Chakrabarti J, et al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol 2007;18:868–73. 10.1093/annonc/mdm007 [DOI] [PubMed] [Google Scholar]
- 12. Wang X, Yu J, Sreekumar A, et al. Autoantibody signatures in prostate cancer. N Engl J Med 2005;353:1224–35. 10.1056/NEJMoa051931 [DOI] [PubMed] [Google Scholar]
- 13. Zhong L, Peng X, Hidalgo GE, et al. Antibodies to HSP70 and HSP90 in serum in non-small cell lung cancer patients. Cancer Detect Prev 2003;27:285–90. 10.1016/s0361-090x(03)00097-7 [DOI] [PubMed] [Google Scholar]
- 14. Montgomery RB, Makary E, Schiffman K, et al. Endogenous anti-HER2 antibodies block HER2 phosphorylation and signaling through extracellular signal-regulated kinase. Cancer Res 2005;65:650–6. 10.1158/0008-5472.650.65.2 [DOI] [PubMed] [Google Scholar]
- 15. Tabuchi Y, Shimoda M, Kagara N, et al. Protective effect of naturally occurring anti-HER2 autoantibodies on breast cancer. Breast Cancer Res Treat 2016;157:55–63. 10.1007/s10549-016-3801-4 [DOI] [PubMed] [Google Scholar]
- 16. Chen WS, Haynes WA, Waitz R, et al. Autoantibody landscape in patients with advanced prostate cancer. Clinical Cancer Research 2020;26:6204–14. 10.1158/1078-0432.CCR-20-1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zitvogel L, Perreault C, Finn OJ, et al. Beneficial autoimmunity improves cancer prognosis. Nat Rev Clin Oncol 2021;18:591–602. 10.1038/s41571-021-00508-x [DOI] [PubMed] [Google Scholar]
- 18. Anders CK, Woodcock MG, Van Swearingen AED, et al. Evaluating the efficacy of a priming dose of cyclophosphamide prior to pembrolizumab to treat metastatic triple negative breast cancer. J Immunother Cancer 2022;10:e003427. 10.1136/jitc-2021-003427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hollern DP, Xu N, Thennavan A, et al. B cells and T follicular helper cells mediate response to checkpoint inhibitors in high mutation burden mouse models of breast cancer. Cell 2019;179:1191–206. 10.1016/j.cell.2019.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Biswas S, Mandal G, Payne KK, et al. IgA transcytosis and antigen recognition govern ovarian cancer immunity. Nature 2021;591:464–70. 10.1038/s41586-020-03144-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cui C, Wang J, Fagerberg E, et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell 2021;184:6101–18. 10.1016/j.cell.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Szolek A, Schubert B, Mohr C, et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 2014;30:3310–6. 10.1093/bioinformatics/btu548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu X, Wu C, Li C, et al. DbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 2016;37:235–41. 10.1002/humu.22932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schenck RO, Lakatos E, Gatenbee C, et al. NeoPredPipe: high-throughput neoantigen prediction and recognition potential pipeline. BMC Bioinformatics 2019;20:264. 10.1186/s12859-019-2876-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yampolsky LY, Stoltzfus A. The exchangeability of amino acids in proteins. Genetics 2005;170:1459–72. 10.1534/genetics.104.039107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39:1–10. 10.1016/j.immuni.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 28. Moynihan KD, Opel CF, Szeto GL, et al. Eradication of large established tumors in mice by combination immunotherapy that engages innate and adaptive immune responses. Nat Med 2016;22:1402–10. 10.1038/nm.4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C, Liang Z, Huang W, et al. EPS8 regulates cellular proliferation and migration of breast cancer. Int J Oncol 2015;46:205–14. 10.3892/ijo.2014.2710 [DOI] [PubMed] [Google Scholar]
- 30. Ghosh N, Postow M, Zhu C, et al. Lower baseline autoantibody levels are associated with immune-related adverse events from immune checkpoint inhibition. J Immunother Cancer 2022;10:e004008. 10.1136/jitc-2021-004008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gowen MF, Giles KM, Simpson D, et al. Baseline antibody profiles predict toxicity in melanoma patients treated with immune checkpoint inhibitors. J Transl Med 2018;16:82. 10.1186/s12967-018-1452-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Diem S, Fässler M, Bomze D, et al. Immunoglobulin G and subclasses as potential biomarkers in metastatic melanoma patients starting checkpoint inhibitor treatment. J Immunother 2019;42:89–93. 10.1097/CJI.0000000000000255 [DOI] [PubMed] [Google Scholar]
- 33. de Moel EC, Rozeman EA, Kapiteijn EH, et al. Autoantibody development under treatment with immune-checkpoint inhibitors. Cancer Immunol Res 2019;7:6–11. 10.1158/2326-6066.CIR-18-0245 [DOI] [PubMed] [Google Scholar]
- 34. Das R, Bar N, Ferreira M, et al. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J Clin Invest 2018;128:715–20. 10.1172/JCI96798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stathopoulos P, Kumar A, Nowak RJ, et al. Autoantibody-producing plasmablasts after B cell depletion identified in muscle-specific kinase myasthenia gravis. JCI Insight 2017;2:17. 10.1172/jci.insight.94263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reincke ME, Payne KJ, Harder I, et al. The antigen presenting potential of cd21low B cells. Front Immunol 2020;11:535784. 10.3389/fimmu.2020.535784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johannet P, Liu W, Fenyo D, et al. Baseline serum autoantibody signatures predict recurrence and toxicity in melanoma patients receiving adjuvant immune checkpoint blockade. Clin Cancer Res 2022;28:4121–30. 10.1158/1078-0432.CCR-22-0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barth DA, Stanzer S, Spiegelberg J, et al. Evaluation of autoantibodies as predictors of treatment response and immune-related adverse events during the treatment with immune checkpoint inhibitors: a prospective longitudinal pan-cancer study. Cancer Med 2022;11:3074–83. 10.1002/cam4.4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans RL, Pottala JV, Nagata S, et al. Longitudinal autoantibody responses against tumor-associated antigens decrease in breast cancer patients according to treatment modality. BMC Cancer 2018;18:119. 10.1186/s12885-018-4022-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Broecker F, Shanin E, Lysov N, et al. Chemotherapy-induced, broadly reactive autoantibodies in a colon cancer patient. Cureus 2022;14:e31954. 10.7759/cureus.31954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chetchotisakd P, Anunnatsiri S, Nanagara R, et al. Intravenous cyclophosphamide therapy for anti-IFN-gamma autoantibody-associated Mycobacterium abscessus infection. J Immunol Res 2018;2018:6473629. 10.1155/2018/6473629 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005848supp001.pdf (2.1MB, pdf)
Data Availability Statement
Data are available in a public, open access repository.