Abstract
Monitoring the genetic structure of pathogen populations may be an economical and sensitive approach to quantify the impact of control on transmission dynamics, highlighting the need for a better understanding of changes in population genetic parameters as transmission declines. Here we describe the first population genetic analysis of the major human malaria parasites, Plasmodium falciparum (Pf) and Plasmodium vivax (Pv) following nationwide distribution of long-lasting insecticide treated nets (LLIN) in Papua New Guinea (PNG). Parasite isolates from pre- (2005-6) and post-LLIN (2010-2014) were genotyped using microsatellite markers. Despite parasite prevalence declining substantially (East Sepik Province: Pf=54.9-8.5%, Pv=35.7-5.6%, Madang Province: Pf=38.0-9.0%, Pv: 31.8-19.7%), genetically diverse and intermixing parasite populations remained. Pf diversity declined modestly post-LLIN relative to pre-LLIN (East Sepik: Rs = 7.1-6.4, He = 0.77-0.71; Madang: Rs= 8.2-6.1, He = 0.79-0.71). Unexpectedly, population structure present in pre-LLIN populations was lost post-LLIN, suggesting that more frequent human movement between provinces may have contributed to higher gene flow. Pv prevalence initially declined but increased again in one province, yet diversity remained high throughout the study period (East Sepik: Rs=11.4-9.3, He=0.83-0.80; Madang: Rs=12.2-14.5, He=0.85-0.88). Although genetic differentiation values increased between provinces over time, no significant population structure was observed at any time point. For both species, a decline in multiple infections and increasing clonal transmission and significant multilocus linkage disequilibrium (mLD) post-LLIN was a positive indicator of impact on the parasite population using microsatellite markers. These parameters may be useful adjuncts to traditional epidemiological tools in the early stages of transmission reduction.
Keywords: Plasmodium falciparum, Plasmodium vivax, malaria control, transmission dynamics, population genetics, microsatellite markers, Papua New Guinea
INTRODUCTION
Characterising pathogen transmission dynamics using population genomics is essential to guide containment efforts and to plan strategies for disease elimination (Grad & Lipsitch, 2014; Hedtke et al., 2019; Wlodarska, Johnston, Gardy, & Tang, 2015). Pathogen populations comprise genetically distinct individuals that are related to varying degrees due to the accumulation of genetic variation as they transmit from host to host. Genomic diversity within populations can thereby indicate the extent of transmission intensity, whilst that between populations determines their connectivity (gene flow) and is influenced by local selection and inbreeding. Measuring pairwise relationships between infections further identifies how infections are spreading from host to host within a population and allows epidemiological characteristics of transmission to be defined (e.g. endemic versus epidemic). Understanding how these population genetic parameters change under the pressure of control interventions is central to using genomic epidemiology as an effective tool to monitor pathogen transmission dynamics.
When utilising population genetics to measure transmission dynamics it is important to consider how genomic diversity is generated. Human malaria parasites acquire de novo mutations whilst replicating asexually and reassortment occurs through sexual recombination within the mosquito vector. However, the generation of novel recombinants occurs only if the mosquito has taken up multiple, genetically distinct clones in the blood meal, otherwise self-fertilization occurs, and progeny are clonal. Outcrossing is therefore dependent on the presence of multiple genetically distinct infections in the human host and increases with endemicity (Babiker et al., 1994; Paul et al., 1995). The population structure of the most virulent malaria parasite, Plasmodium falciparum is associated with transmission intensity (Anderson et al., 2000). At moderate to high transmission where multiclonal infections are frequently found, parasite populations are characterised by high diversity and a lack of population structure with low levels of linkage disequilibrium (LD) (Anderson et al., 2000; Gatei et al., 2010; Orjuela-Sanchez et al., 2013; Salgueiro, Vicente, Figueiredo, & Pinto, 2016; Schultz et al., 2010). At low transmission where multiclonal infections are less common, clonal transmission and inbreeding amongst closely related individuals is more common, resulting in lower overall diversity and high levels of LD, whilst population structure is more evident due to both lower gene flow between areas and within population transmission dynamics (Anderson et al., 2000; Branch et al., 2011; Chenet, Schneider, Villegas, & Escalante, 2012; Noviyanti et al., 2015). For P. vivax, also a significant human pathogen, relapsing infections and other unique features that enhance its transmission (Olliaro et al., 2016), result in a higher prevalence of multiclonal infections. Therefore, P. vivax populations are often characterised by high genetic diversity, even at low transmission (Ferreira et al., 2007; Fola et al., 2017; Gunawardena, Ferreira, Kapilananda, Wirth, & Karunaweera, 2014; Noviyanti et al., 2015; Waltmann et al., 2018). In the South West Pacific region, a modest decline in diversity and increasing population structure occurs with the eastward decline in transmission (Fola et al., 2017; Koepfli et al., 2013; Waltmann et al., 2018). LD and pockets of clonal P. vivax transmission have been observed in several studies, suggesting increasingly focal transmission as malaria rates decline (Abdullah et al., 2013; Batista, Barbosa, Da Silva Bastos, Viana, & Ferreira, 2015; Chenet et al., 2012; Delgado-Ratto et al., 2016; Ferreira et al., 2007; Imwong et al., 2007; Iwagami et al., 2012; Noviyanti et al., 2015; Orjuela-Sanchez et al., 2013). Comparative analyses show P. vivax has a higher effective transmission intensity (Hofmann et al., 2017; Lin et al., 2010; Robinson et al., 2015) and higher diversity than P. falciparum due to a longer association with humans and fewer population bottlenecks (Chenet et al., 2012; Gilabert et al., 2018; Hupalo et al., 2016; Jennison et al., 2015; Liu et al., 2014; Loy et al., 2017; Neafsey et al., 2012; Noviyanti et al., 2015; Orjuela-Sanchez et al., 2013; Pava et al., 2017). P. vivax is more resilient to control efforts and thus may be less likely to show changes in parasite population structure than P. falciparum (Barry, Waltmann, Koepfli, Barnadas, & Mueller, 2015; Cornejo & Escalante, 2006; Feachem et al., 2010; Liu et al., 2014; Neafsey et al., 2012; Oliveira-Ferreira et al., 2010; Waltmann et al., 2015). No studies have yet investigated the impact of intensified control on the population genetics of sympatric P. vivax and P. falciparum populations.
The worldwide scale up of malaria control since the early 2000s, has reduced transmission dramatically around the world. Indeed, between 2010 and 2016, disease incidence declined by 18% and mortality by 32% (WHO, 2017, 2019). The incidence of clinical cases and infection prevalence remain the mainstay of malaria surveillance however population genetic surveillance has emerged as a promising and high-resolution approach for malaria monitoring (Arnott, Barry, & Reeder, 2012; Barry et al., 2015; Dalmat, Naughton, Kwan-Gett, Slyker, & Stuckey, 2019; Koepfli & Mueller, 2017; malEra Consultative Group on Monitoring & Surveillance, 2011). Specifically, these approaches identify local transmission dynamics (e.g. endemic, epidemic, imported infections), connectivity between parasite populations in different endemic areas (Anderson et al., 2000; Fola et al., 2017; Noviyanti et al., 2015; Vardo-Zalik et al., 2013; Waltmann et al., 2018) and “sources and sinks”, which together could help to design targeted control interventions (Auburn & Barry, 2017; Barry et al., 2015; Koepfli & Mueller, 2017). Population genetic surveys could also identify local drivers contributing to sustained transmission such as particular human social and economic interactions (Barry et al., 2015; Delgado-Ratto et al., 2016; Koepfli & Mueller, 2017). While parasite population genetics and genomics is becoming more popular and accessible, the impact on control programs has been limited, and to date few studies have systematically assessed the long-term impact of malaria control using these approaches (Bei et al., 2018; Chenet, Taylor, Blair, Zuluaga, & Escalante, 2015; R. F. Daniels et al., 2015; Gatei et al., 2010; Gunawardena et al., 2014; Nkhoma et al., 2013; Vardo-Zalik et al., 2013). Moreover, it is not clear how long transmission needs to be disrupted, or to which extent prevalence should be reduced, before changes in parasite population structure can be seen. A better understanding of the impact of malaria control interventions on P. falciparum and P. vivax population structure is urgently required to capitalise on the potential of genomic surveillance for malaria control and elimination.
Population genetic surveys using panels of well-validated neutral microsatellite markers (Anderson et al., 2000; Imwong et al., 2007; Karunaweera, Ferreira, Hartl, & Wirth, 2006) were conducted on the north coast of Papua New Guinea before the intensification of malaria control (2005/2006). P. vivax showed higher genetic diversity and a lack of population structure yet there was significant population structure of P. falciparum populations (Jennison et al., 2015; Koepfli et al., 2013; Schultz et al., 2010; Waltmann et al., 2018). Significant inbreeding (mLD) was not observed for sub-populations of either species, confirming high levels of outcrossing and endemic transmission (Jennison et al., 2015). Since that time, PNG has implemented an intensified control program including the free nationwide distribution of Long Lasting Insecticide Treated Nets (LLIN). This resulted in a significant decline in infections across the country including the north coast provinces previously covered in our population genetic surveys (Arnott et al., 2013; Barry et al., 2013; Hetzel et al., 2016; Kattenberg et al., 2020; Koepfli et al., 2017; Koepfli et al., 2015). The impact on parasite population structure and transmission dynamics after the rollout of LLINs, however, remains unresolved. We sought to characterise the impact of reduced prevalence on the population structure of sympatric P. falciparum and P. vivax populations. Microsatellite haplotypes were generated from P. falciparum and P. vivax samples collected in multiple cross sectional surveys from 2010-14 after two rounds of mass LLIN distribution and compared to published data from isolates collected before the intensified malaria control program (Jennison et al., 2015; Schultz et al., 2010). The results show the impact of declining prevalence on PNG parasite populations and identify the critical parameters for monitoring these changes using microsatellite markers.
MATERIALS AND METHODS
Study sites and design
The studies were conducted in two Provinces on the highly endemic north coast region of Papua New Guinea (PNG) (Figure 1). In the WHO Western Pacific Region (WPR), the malaria mortality rate declined by 58% over the period 2010–2015, however infection prevalence in Papua New Guinea (PNG) remains the highest in this region (and outside the African continent), contributing 81% of malaria cases and 86% of malaria deaths in 2017 in the region (WHO, 2017, 2018) primarily due to P. falciparum and P. vivax infections (Kattenberg et al., 2020; Koepfli et al., 2017; WHO, 2018). In 2003, a new national malaria control campaign was launched to achieve high levels of LLIN ownership and usage in PNG (Hetzel et al., 2014; Hetzel et al., 2012). Coverage with LLIN was low in most parts of the country before nationwide free distribution took place (2004-8 and 2009-2012) (Betuela et al., 2012; Genton et al., 1994; Hii et al., 2001). This resulted in a significant increase in ownership of bed nets across the country by 2010 (any type 80%; LLINs 65%) (Hetzel et al., 2014; Hetzel et al., 2012) and the average malaria incidence rate in sentinel sites dropped from 13/1,000 population to 2/1,000 (range 0.6-3.3/1000 post-LLIN) (Hetzel et al., 2016).
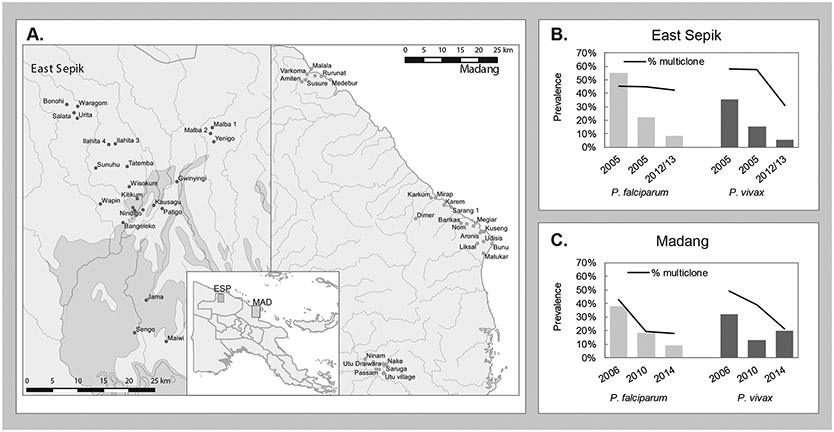
Figure 1. Map of the study areas and infection prevalence from 2005-2016.
(A) Map of East Sepik and Madang study area villages and locations on the north coast of Papua New Guinea (inset) The graphs show the pre-LLIN (2005/6) and post-LLIN (2010-2014) molecular prevalence for (B) East Sepik and (C) Madang for both P. falciparum (light grey) and P. vivax (dark grey) and proportion of multiclonal infections (black line) (Arnott et al., 2013; Barry et al., 2013; Kattenberg et al., 2020; Koepfli et al., 2017; Koepfli et al., 2015; Mueller et al., 2009; Senn et al., 2012).
On the hyperendemic north coast, P. falciparum PCR prevalence dropped from 38% to 12% and P. vivax prevalence decreased from 28% to 13% (Kattenberg et al., 2020; Koepfli et al., 2017) (Figure 1). Prevalence reductions were more substantial for P. falciparum than for P. vivax, as has been seen in many co-endemic areas (Feachem et al., 2010). In East Sepik Province, malaria decreased from a very high burden (73% of surveyed individuals infected in 2005 as measured by molecular detection (LDR-FMA (Mueller et al., 2009)) to heterogeneous transmission (prevalence in villages ranging from 1% to 61%, median 6%, as measured by qPCR (Kattenberg et al., 2020)) after two rounds of LLIN distribution. An initial round of LLIN distribution was conducted between 2004 and 2009, followed by additional distributions in 2011/2012 and subsequently in 2014/15. In Madang province, however, malaria prevalence decreased from 63% to 28% by qPCR after the first round of LLIN distributions (Koepfli et al., 2017; Koepfli et al., 2015; Schultz et al., 2010). After the second LLIN distribution (2010-2014), P. falciparum continued to drop, however an increase in P. vivax prevalence was observed (from 13% to 20% by qPCR (Koepfli et al., 2017)). In Madang province, malaria prevalence was less heterogenous in the sampled villages than in East Sepik Province (Kattenberg et al., 2020; Koepfli et al., 2017; Koepfli et al., 2015).
Whole blood samples were collected from participants in cross-sectional studies conducted between 2005 and 2014 along the North Coast of PNG (Figure 1) (Arnott et al., 2013; Kattenberg et al., 2020; Koepfli et al., 2017; Koepfli et al., 2015; Mueller et al., 2009; Schultz et al., 2010). In Madang Province (MAD), the same three catchment areas were studied in 2006 (Schultz et al., 2010; Senn et al., 2012), 2010 (Koepfli et al., 2015) and 2014 (Koepfli et al., 2017). The study area included a selection of villages along a coastal stretch of 70km (Mugil and Malala regions), surrounded by coconut and cocoa plantations and subsistence gardens, and one area approximately 50 km inland (Utu). Here, the climate is tropical with a rainy season from December to April. In East Sepik Province (ESP), participants in the Wosera Catchment (ESP1) including fourteen villages were sampled in 2005 during the dry-season (August-September) (Jennison et al., 2015; Senn et al., 2012). A broader survey (ESP2) was conducted in April-May 2005 including five catchment areas that were re-visited in 2012-13 (Kattenberg et al., 2020; Mueller et al., 2009). The study areas in ESP consist of an area of over 160 km2 with low hills and riverine plains with a wet, tropical climate (Genton et al., 1995). The natural vegetation is lowland hill forest that has mostly been replaced by re-growth following cultivation and wide grasslands on the plains near the Sepik River.
In all surveys, demographic and clinical information was collected, blood slides examined by expert microscopists and a blood sample collected in EDTA tubes for extraction of DNA. In the 2005 ESP studies, Plasmodium species were detected by Light Detection Reaction- Fluorescent Microspere Assay (LDR-FMA) (McNamara et al., 2006; Mueller et al., 2009), whereas in all other studies quantitative PCR (qPCR) detection by TaqMan™ assay was used (Rosanas-Urgell et al., 2010). To determine multiplicity of infection (MOI), P. falciparum positive samples were genotyped for Pfmsp2 and P. vivax positive samples were genotyped with Pvmsp1f3 and MS16 (ESP1 and MAD 2006) or Pvmsp1f3 and MS2 (MAD 2010 & 2014 and ESP2 2005 & ESP 2012-13) ), as previously described (Arnott et al., 2013; Kattenberg et al., 2020; Koepfli et al., 2017; Koepfli et al., 2015; Mueller et al., 2009; Schultz et al., 2010). The sample selection and genotyping procedures of the ESP1 2005 (Wosera) and MAD 2006 were as previously described (Arnott et al., 2013; Jennison et al., 2015; Schultz et al., 2010). For the other studies, samples with MOI of 1 were selected for further genotyping with the neutral microsatellite panels as described below. For the studies conducted after the large scale LLIN distribution (>2006) all monoclonal isolates (MOI=1) were included, but for the 2005 ESP2 population, a selection of samples was made for the analysis with the microsatellite panel (Table S1).
Ethical approval
Written informed consent was obtained from all study participants or their parents or legal guardians. The study was approved by the PNG IMR Institutional Review Board (IRB#11/16) and the PNG Medical Research Advisory Committee (MRAC 11/21), National Institutes of Health, Division of Microbiology and Infectious Diseases (DMID Protocol #10-0035) and Walter and Eliza Hall Institute Human Research Ethics Committee (HREC #12/10).
Genotyping procedures
For both species, a panel of 9-10 neutral microsatellite markers were amplified in the selected samples (Table S1) using a multiplex primary PCR followed by individual nested PCRs as previously described (Anderson et al., 2000; Jennison et al., 2015; Koepfli et al., 2013; Schultz et al., 2010). For P. falciparum, samples were genotyped at nine previously validated and commonly used, putatively neutral, microsatellite loci including TA1, TAA60, Polya, ARA2, Pfg377, TAA87, PfPK2, TAA81 and 2490 (Anderson et al., 2000; Schultz et al., 2010). For P. vivax, 10 putatively neutral microsatellites were genotyped as previously described: MS1, MS2, MS5, MS6, MS7, MS9, MS10, MS12, MS15, and MS20 (Jennison et al., 2015; Koepfli et al., 2013). All PCR products were sent to a commercial facility for fragment analysis on an ABI3730xl platform (Applied Biosystems) using the size standard LIZ500. Primers used were the same for all datasets (Jennison et al., 2015; Schultz et al., 2010).
Analysis
The electropherograms were analysed with Genemapper V4.0 (Applied Biosystems) with the same peak calling strategy as described previously (Jennison et al., 2015; Schultz et al., 2010). To avoid artefacts, precautions were taken to ensure allele calling was consistent (Jennison et al., 2015), and carefully reconstructing dominant haplotypes as per previously described methods (Anderson, Su, Bockarie, Lagog, & Day, 1999; Jennison et al., 2015; Schultz et al., 2010). Briefly, this involved setting the minimum fluorescence to 500 Random Fluorescence Units (RFU) for all colours except the size standard. Stutter window was set to 3.5 for 3bp repeats and 4.5 for 4bp repeats. The stutter ratio was set to 0.4 for all markers. The stutter detection was only applied to shorter alleles, with longer alleles within the stutter window subject to the standard 30% cut-off threshold. Samples with low fluorescence were manually reanalysed with a minimum fluorescence of 100 RFU. For the Madang 2005 and Wosera 2006 P. falciparum data, previously published cleaned and rounded microsatellite allele repeat numbers for P. falciparum single clone infections (Schultz et al., 2010) were converted back to allele sizes using the known number of nucleotides/repeat, whereas for P. vivax the raw data (allele calls) was available (Jennison et al., 2015). These data were combined with the newly generated MS data from the other studies before binning the alleles using the TANDEM software (Matschiner & Salzburger, 2009). Allele frequencies of the entire dataset (incl. previously genotyped datasets) were investigated and outlying alleles (most likely caused by PCR artefacts) were removed. Samples with missing data at six (60%) or more MS loci were excluded from further analysis. We attempted to calibrate the P. falciparum data from pre-LLIN Madang 2006 and Wosera (ESP1 2005) by converting rounded repeat numbers back to allele sizes, binning together with the newly generated data and removing outliers. However, there was strong population structure when compared to the new dataset, indicating experimental differences despite the use of the same protocols. Thus, we excluded direct comparisons between old and new datasets for P. falciparum.
To conduct the population genetic analyses, allele frequencies and input files for the various population genetics programs were created using CONVERT version 1.31 (Glaubitz) and PGD Spider version 2.1.0.1 (Lischer & Excoffier, 2012). Genetic diversity was measured by calculating the number of alleles (A), Nei’s gene diversity (expected heterozygosity (He) (Nei, 1987)) and allelic richness (Rs) (Hurlbert, 1971) that corrects for sample size, using FSTAT version 2.9.3.2 (Goudet, 1995). Pairwise genetic differentiation was measured by calculating pairwise Jost’s D (Jost, 2008) and Weir and Cockerhams FST (Weir & Cockerham, 1984) values and 95% confidence intervals were estimated with 1000 bootstraps using the diveRsity package in R (Keenan, McGinnity, Cross, Crozier, & Prodöhl, 2013). In contrast to some earlier studies (Schultz et al., 2010), where haploid genotypes were coded as diploid genotypes, but homozygote at each locus, the data in this study was analysed using haploid datasets (as in Jennison et al. (Jennison et al., 2015)). As a measure of inbreeding in each population, multilocus linkage disequilibrium (mLD) was calculated using LIAN version 3.7, applying a Monte Carlo test with 100,000 re-sampling steps (Haubold & Hudson, 2000). In the LIAN analysis only samples with complete haplotypes were included. For both species the dominant and single haplotypes were compared within catchments to identify any significant linkage disequilibrium (mLD) using LIAN (Haubold & Hudson, 2000) and FST using FSTAT version 2.9.3.2 (Goudet, 1995).
Bottleneck (Piry, Luikart, & Cornuet) was used to test for an excess of heterozygosity brought about by the loss of rare alleles following a population bottleneck. A two-phase mutational (TPM) model of 70% stepwise and 30% non-stepwise mutations and run 1000 iterations was used. In addition, the allele frequency distribution for all loci was examined for a ‘mode shift’ in the distribution (Luikart, Allendorf, Cornuet, & Sherwin, 1998). To investigate parasite population genetic structure, the Bayesian clustering software, STRUCTURE version 2.3.4 (Pritchard, Stephens, & Donnelly, 2000) was used to investigate whether haplotypes for each species clustered according to geographical origin and/or within time periods. The analysis was run 20 times for K = 1 to 15 for 100,000 Monte Carlo Markov Chain (MCMC) iterations after a burn-in period of 10,000 using the admixture model and correlated allele frequencies. The second order rate of change of LnP[D], ΔK was calculated according to the method of Evanno et al. (Evanno, Regnaut, & Goudet, 2005) to determine the most likely K (most likely number of populations). CLUMPP version 1.1.2 (Jakobsson & Rosenberg, 2007) was used to facilitate the interpretation of population genetic results using files generated with STRUCTURE HARVESTER Web v0.6.94 (Earl & vonHoldt, 2011) and Distruct 1.1 (Rosenberg, 2004) was used to visualize the structure plots with the data generated with CLUMPP. Statistical analysis of molecular epidemiological and population genetic parameters was done using non-parametric methods as indicated in the results-section using STATA v12.1 (StataCorp, USA). QGIS 2.18.24 (OpenSource Geospatial Foundation) was used to map the villages and households and maps were constructed with spatial layers from DIVA-GIS ("Diva GIS country level data,").
RESULTS
Multiplicity of Infection
Multiplicity of infection (MOI), determined by genotyping highly polymorphic markers and counting the numbers of alleles in each infection, is a proxy measure of the intensity of transmission. MOI in all areas was lower in all areas post-LLIN distribution decreasing from 1.8 to 1.3 (p=0.0463 Mann Whitney U test) and 2.0 to 1.4 (p=0.0495 Mann Whitney U test), respectively for P. falciparum and P. vivax. This corresponds with a lower proportion of multiclonal infections, except for P. falciparum in ESP in 2012-13 (Figure 1 and Table S1). Despite an increase in PCR prevalence of P. vivax in Madang Province in 2014, few multiclonal infections were detected (Figure 1 and Table S1).
Microsatellite haplotypes
We then genotyped parasite isolates at microsatellite loci to generate multilocus haplotypes for population genetic analyses. Multilocus haplotypes with at least five loci successfully genotyped (out of 9 for P. falciparum and 10 for P. vivax) were constructed for 860 P. falciparum samples (300 previously published) and 755 P. vivax samples (202 previously published) (Jennison et al., 2015; Schultz et al., 2010) (Table S1). Despite having genotyped the samples that were identified as MOI=1 by pfmsp2, pvmsp1f3 and pvMS2/MS16 genotyping, 31% of P. falciparum samples and 49% of P. vivax samples had more than one allele for at least one microsatellite locus, suggesting multiple clone infection and the increased resolution of the microsatellite panel. From these we created dominant haplotypes (Schultz et al., 2010). No significant changes in multilocus Linkage Disequilibrium (mLD) were found when comparing single vs. all haplotypes combined within each study (Table S2). Low genetic differentiation was found between single and dominant haplotypes for P. falciparum in MAD2014 (FST=0.063, p = 0.58), however, this can be explained by small sample size (n=9 dominant haplotypes). For P. vivax, low differentiation between single and dominant haplotypes in ESP 2012 (FST = 0.041, p = 0.33), was explained by a cluster of closely related haplotypes, all reconstructed from dominant alleles), which are described in more detail below. The fact that these related haplotypes were independently constructed from dominant haplotypes provides additional confidence in the allele-calling strategy. All other comparisons (within each province for at each time point) showed negligible genetic differentiation between single and dominant haplotypes. Therefore, the haplotypes were combined for further analysis.
Reduction in P. falciparum but not P. vivax genetic diversity post-LLIN
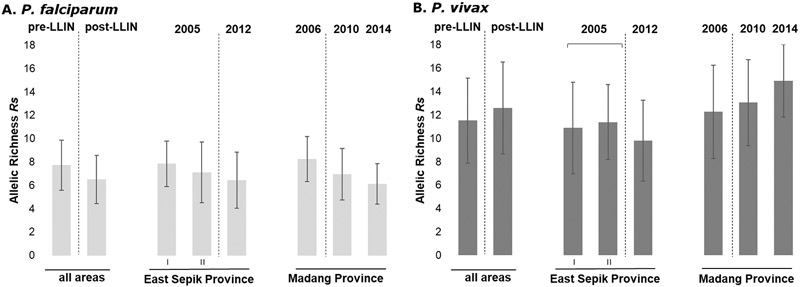
Based on the microsatellite haplotypes (n=860), the genetic diversity of P. falciparum populations was modestly but significantly lower post-LLIN compared to the earlier time points for pre-LLIN populations (ESP1 and 2 2005 and MAD 2006). Mean heterozygosity for P. falciparum over all areas combined decreased significantly from 0.76±0.1 to 0.71±0.1 (Mann-Whitney U test p =0.036; Table S1) and allelic richness from 7.7±2.2 to 6.5±2.1 (Mann-Whitney U test p=0.014; Figure 2). These parameters also showed a small but insignificant decline for the provinces analysed individually (Figure 2, p>0.05). For P. vivax, overall genetic diversity remained high (post-LLIN Rs=12.5; He=0.85, Table S1), but slightly different results were seen in each province (Figure 2, Table S1). In Madang Province after the distribution of LLINs, He values slightly increased from 0.85±0.07 in 2006 to 0.88±0.04 in 2014 (p=0.3, Table S1), with high allelic richness (pre-LLIN Rs 12.2±4.0 vs post-LLIN 14.0±3.4; p=0.2). Whereas in East Sepik Province, P. vivax genetic diversity decreased but not significantly with He values of 0.83±0.09 to He 0.80±0.08 (2-sample t-test, p=0.48) and Rs values of 11.1±3.5 vs post-LLIN Rs 9.8±3.5 (2-sample t-test, p=0.33) (Table S1, Figure 2). No significant correlation was found between prevalence (by PCR) and heterozygosity, allelic richness, mean MOI, or proportion of multiple clone infections, for either species (data not shown).
Figure 2. Changing diversity of P. falciparum and P. vivax populations over an intensifying period of malaria control (2005-2014).
Allelic Richness (Rs) in P. falciparum (A) (n= 860) and P. vivax (B) (n=755) populations pre- (≤2006) and post-LLIN (≥2010) mass-distributions. Error bars indicate standard deviations.
Significant multilocus linkage disequilibrium (mLD) for both P. falciparum and P. vivax post-LLIN
For P. falciparum, matching haplotypes (allowing missing loci) were seen in all post-LLIN datasets and the pre-LLIN ESP2 2005 dataset. However, for P. vivax, matching haplotypes (allowing missing loci) were rarely seen and only in post-LLIN data sets. Among the 332 complete P. falciparum multilocus haplotypes (9-loci successfully genotyped) from all study sites, 16 repeated haplotypes were found, with 11 haplotypes represented two times, three represented three times, and two represented four times. Clonal haplotypes were always found within the same year and province, and in all cases except one in the same catchment area, but not always in the same village (7/16 haplotypes found in neighbouring villages). In ESP2 2005, one clonal haplotype was found in two villages (Yenigo and Sengo, Figure 1) from different catchment areas, roughly 40km apart. Among the 303 complete P. vivax multilocus haplotypes (10-loci), two haplotypes were repeated, with one haplotype represented two times (in two different villages in ESP 2012), and one represented four times (three in one village, one in a neighbouring village in MAD 2010).
To investigate whether inbreeding was present in these populations (Smith, Smith, O’Rourke, & Spratt, 1993), non-random associations among the microsatellite loci (mLD) were calculated for all complete and unique haplotypes. Whilst mLD was absent from most pre-LLIN populations, significant mLD was observed in P. falciparum and P. vivax infections post-LLIN (Table 1). In ESP 2012-13, mLD was high with unique infections indicating the circulation of closely related haplotypes in the population, suggesting near clonal transmission (low recombination between diverse clones and high levels of inbreeding) or the presence of high proportions of meiotic siblings among isolates (Bright et al., 2014; Smith et al., 1993), as observed in the village of Sunuhu (Table S3). Low, but significant mLD was found for P. falciparum in Madang in 2006 (Table 1), however this population was structured (Schultz et al., 2010), resulting in a phenomenon called the Wahlund effect, confirmed by the fact that linkage equilibrium was restored when mLD was analysed separately for subpopulations (Jennison et al., 2015; Wahlund, 1928) (Table S4). In post-LLIN Madang, the observed mLD for P. falciparum is not the result of subpopulation structure, as significant mLD remained in the subpopulations (Table S4). In the Mugil area in 2014, significant mLD for P. falciparum remains due to the circulation of a few very closely related haplotypes in the Megiar village (Table S4, Dataset 1).
Table 1. Estimates of multi-locus linkage disequilibrium (mLD) for Plasmodium populations pre- and post-LLIN mass-distribution.
IAS is the standardized index of association; n = number of complete haplotypes. Single clone infection mLD is shown in Table S2.
| All haplotypes |
Unique haplotypes |
||||||
|---|---|---|---|---|---|---|---|
| P. falciparum | n | I A S | p-value | n | I A S | p-value | |
| all pre-LLIN studies | 199 | 0.0051 | 0.036 | 196 | 0.0048 | 0.046 | |
| all post-LLIN studies | 136 | 0.0075 | 0.020 | 116 | −0.003 | 0.787 | |
| East Sepik I | 2005 | 15 | −0.0038 | 0.566 | 15 | −0.0038 | 0.566 |
| East Sepik II | 2005 | 103 | 0.0004 | 0.446 | 100 | −0.0003 | 0.517 |
| East Sepik | 2012-13 | 21 | 0.0454 | 0.002 | 19 | 0.0202 | 0.088 |
| Madang | 2006 | 81 | 0.0108 | 0.011 | 81 | 0.0108 | 0.011 |
| Madang | 2010 | 82 | 0.0113 | 0.021 | 72 | −0.0046 | 0.802 |
| Madang | 2014 | 33 | 0.0725 | <0.00001 | 25 | 0.0198 | 0.059 |
| All studies | 335 | 0.0044 | 0.021 | 312 | 0.0027 | 0.103 | |
| P. vivax | |||||||
| all pre-LLIN studies | 179 | −0.0002 | 0.540 | 179 | −0.0002 | 0.539 | |
| all post-LLIN studies | 125 | 0.0126 | <0.00001 | 120 | 0.007 | 0.001 | |
| East Sepik I | 2005 | 48 | 0.0066 | 0.099 | 48 | 0.0066 | 0.099 |
| East Sepik II | 2005 | 37 | 0.0064 | 0.149 | 37 | 0.0064 | 0.149 |
| East Sepik | 2012-13 | 20 | 0.2154 | <0.00001 | 19 | 0.1981 | <0.00001 |
| Madang | 2006 | 94 | −0.0022 | 0.796 | 94 | −0.0022 | 0.796 |
| Madang | 2010 | 80 | 0.0113 | 0.0005 | 77 | 0.0004 | 0.436 |
| Madang | 2014 | 24 | 0.0073 | 0.209 | 24 | 0.0073 | 0.209 |
| All studies | 303 | 0.0025 | 0.013 | 299 | 0.0017 | 0.057 | |
Population Bottleneck for P. falciparum but not P. vivax post-LLIN
Bottleneck analysis was performed using a two-phase mutational model (TPM) and testing for heterozygosity excess with a 2-tailed Wilcoxon sign rank test (see Materials and Methods). Significant heterozygosity excess was observed for P. falciparum in ESPII 2005 (but not in ESPI 2005), MAD 2006 and 2010 populations (p=0.020, p=0.049 and p= 0.010, respectively). However, for P. vivax significant heterozygosity excess was only observed in a single pre-LLIN population, Wosera 2005 (p=0.042, these samples were collected in the relatively dry season) and not in any of the other P. vivax populations. Despite finding significant heterozygosity excess, a mode-shifted distribution of allele frequencies (as is frequently observed in bottlenecked populations) was not observed in any of the time points and provinces.
Contrasting and dynamic patterns of population structure for both P. falciparum and P. vivax
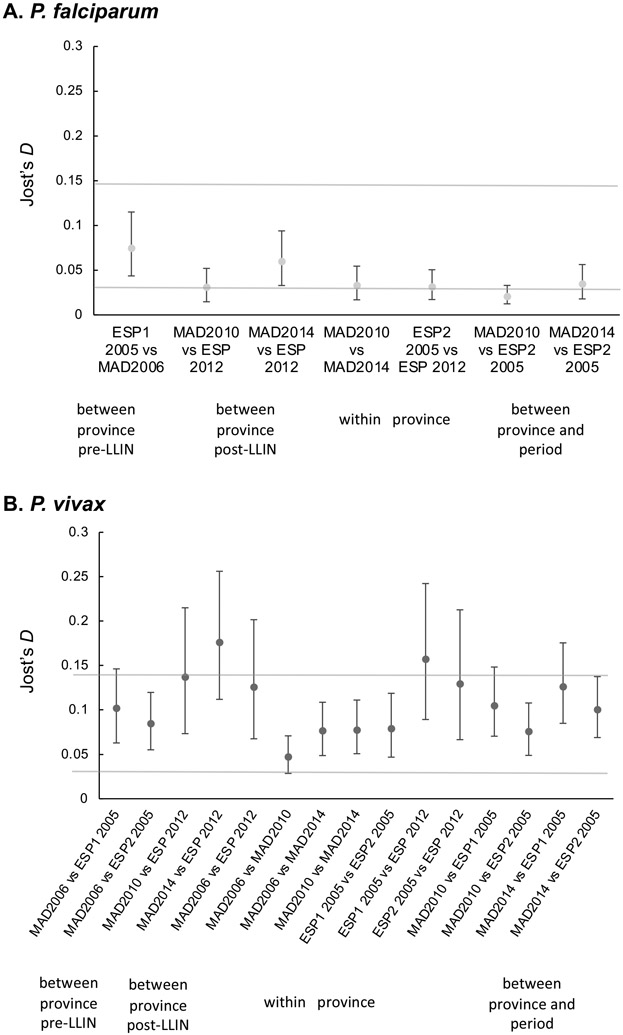
As previously described, for P. falciparum, low to moderate genetic differentiation was seen between the Wosera 2005 (ESP1) and Madang 2006 studies (Jennison et al., 2015; Schultz et al., 2010) (Figure 3, S1). Comparisons were not done between Pf 2005/6 and other Pf populations as it was not possible to calibrate data through combining allele calls before binning (see Materials and Methods). Post-LLIN, there remains low to moderate genetic differentiation between ESP (2012-2013) and Madang (2014) (Figure 3, S1). However, there was little genetic differentiation of East Sepik P. falciparum populations pre-LLIN (ESP2 2005) compared to post-LLIN (2012-13), nor between Madang 2010 and 2014 populations (Figure 3, S1). For P. vivax, a different pattern can be seen, with low genetic differentiation between provinces pre-LLIN, which increases in the post LLIN-studies (Figure 3, S1). Similar to P. falciparum, within province genetic differentiation between the different time points does not increase post-LLIN.
Figure 3. Genetic differentiation estimates among P. falciparum and P. vivax populations pre- and post-LLIN mass-distributions.
Pairwise Jost’s D values for (A) P. falciparum and (B) P. vivax. Pairwise Jost’s D values and 95% confidence intervals were estimated with 1000 bootstraps using the diveRsity package in R. Pairwise FST values (Weir and Cockerham) are shown in figure S1.
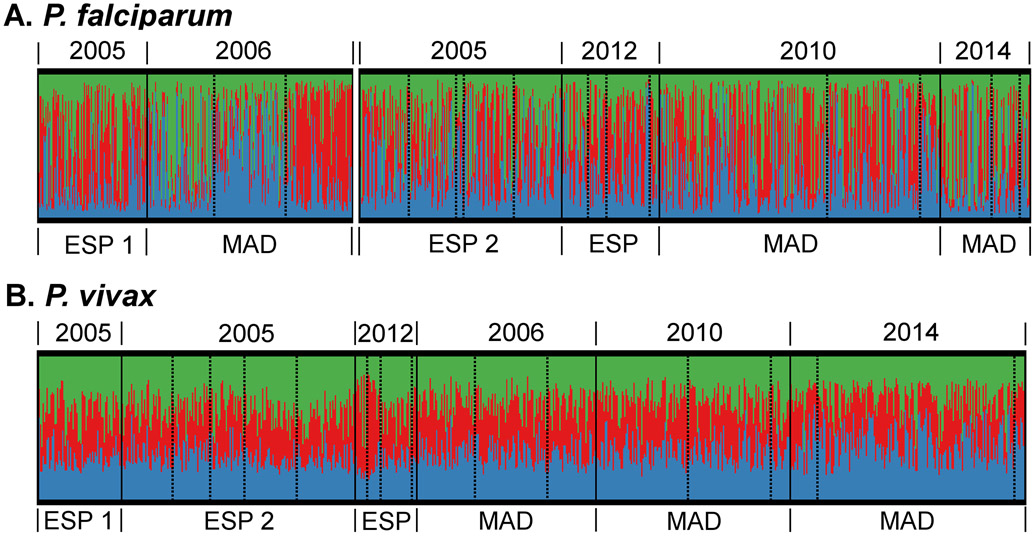
Population genetic structure was further investigated by Bayesian cluster analysis using STRUCTURE (Pritchard et al., 2000). Haplotypes or populations with ancestry in more than one cluster are considered admixed and indicates that substantial gene flow occurs between defined geographic areas. Our analysis identified three P. falciparum and three P. vivax clusters (Figure 4, S2). The clustering patterns show that the P. falciparum populations in later years are more mixed than the populations of 2005/2006, where populations clustered according to geographical locations including amongst the three catchment areas within Madang Province (Figure 4, S2) (Schultz et al., 2010). On the contrary, P. vivax populations were very diverse and displayed little population structure in all time points, despite the increase in differentiation between ESP and MAD post-LLIN populations (Figures 4 and 5).
Figure 4. Bayesian cluster analysis of P. falciparum (A) and P. vivax (B) of pre- (2005/6) and post-LLIN studies (2010-2014).
Individual samples are sorted by province and year (solid lines), catchment area (dashed line) and cluster membership (colour). Madang catchments are organised as Malala, Mugil, Utu and ESP2 and 2012-13 as Brukham, Burui, Ilahita, Ulupu, and Wombisa (no infections in 2012). As identified in the genetic differentiation analysis (Jost’s D, Figure 3), there was moderate differentiation for P. falciparum between the ESP1 (Wosera area) 2005 and Madang 2006 versus the other studies (only one province included pre-LLIN), which is believed to be for a large part caused by experimental- and data analysis differences. Therefore, these P. falciparum studies were grouped separately for the population structure analysis, in order to avoid artificial changes in ancestry over time.
Figure 5. Unrooted neighbour-joining tree of P. vivax isolates in East Sepik province in 2012-13.
Relatedness among haplotypes was defined by calculating the pairwise distance and neighbour-joining tree using the APE package in R and the tree was visualized using Phylocanvas through microreact.org (Argimon et al., 2016; "Microreact,"). Colours and shapes indicate the village where the isolates were collected. Very closely related isolates are observed in the village of Sunuhu (dark grey circles).
For P. vivax, as we observed significant mLD post-LLIN in ESP, local phylogenetic analysis was conducted. This supports focal transmission as shown by the clustering of haplotypes from the same village: Sunuhu (Figure 5). The STRUCTURE analysis also shows some evidence of this inbred cluster (Figure 4B). Interestingly, this village had the highest prevalence in the region in the 2012-13 survey (36% infected with P. vivax by qPCR compared to 0.5-9% in other villages (Kattenberg et al., 2020). In Sunuhu, clonal and closely related haplotypes (≤2 unmatched alleles) were observed in 48% (11/23) of the haplotypes from that village (see supplementary file 1). The 11 closely related haplotypes were observed throughout the village, were not clustered in neighbouring households, and were not associated with participant characteristics (p>0.05), such as age and sex (Table S3).
DISCUSSION
Parasite genetic diversity has been proposed as a key indicator of malaria transmission dynamics to track control and elimination progress (Dalmat et al., 2019). This demands an evaluation of whether genotyping provides insights about changes in transmission following intensified malaria control efforts. Here we have monitored the population genetics of P. falciparum and P. vivax over an eight-year period of LLIN-induced transmission decline in PNG, an area of high year round transmission. Despite large reductions in parasite prevalence and multiple infections from very high to low/moderate levels (Kattenberg et al., 2020; Koepfli et al., 2017; Koepfli et al., 2015), we show that population genetic changes were minimal with populations remaining diverse and unstructured. P. falciparum diversity decreased somewhat, though remained high relative to other malaria endemic areas outside Africa (Anderson et al., 2000; Branch et al., 2011; Chenet et al., 2012; Chenet et al., 2015; dalla Martha, Tada, Ferreira, da Silva, & Wunderlich, 2007; Noviyanti et al., 2015; Orjuela-Sanchez et al., 2009; Orjuela-Sanchez et al., 2013; Pava et al., 2017; Susomboon et al., 2008), whereas P. vivax diversity was unchanged throughout the study period. Surprisingly, P. falciparum populations that were structured pre-LLIN (2005, 2006) (Jennison et al., 2015; Schultz et al., 2010), were unstructured post-LLIN (2010, 2014), although a reduction in multiple infections and an increase in multilocus LD due to clonal haplotypes were detected. For P. vivax, there was also no evidence of population structure after LLIN, however increasing pairwise genetic differentiation within and between provinces was observed and clonal transmission and inbreeding had emerged in at least one village. These results demonstrate that declining transmission does not result in an immediate decrease in overall population diversity nor an increase in population structure. Sustained low transmission may be needed to observe these changes using these small panels of microsatellite markers. However, changes in multiple infection prevalence and multilocus LD indicate increasing heterogeneity in transmission within populations. These results have implications with respect to monitoring changing transmission for any pathogen using population genetic approaches.
Nkhoma and colleagues previously reported a limited impact on P. falciparum diversity following a decrease from moderate to low transmission on the Thai-Myanmar border over 10 years. However, as we have observed in PNG, there was an increase in multilocus LD which and decreasing proportions of multiclonal infections (Nkhoma et al., 2013). Daniels et al. have also reported decreasing multiclonal infections, increasing proportions of repeated genotypes and multilocus LD, and long-term persistence of particular clones in Senegal (Daniels et al., 2013). These studies utilised 96 and 24 biallelic SNP markers respectively, compared to our study using a small number of microsatellite markers. Similar panels of SNPs may reveal additional insights in the PNG context. As for P. vivax, Gunawardena and colleagues also reported sustained high P. vivax microsatellite diversity and multiclonal infections during a five-year period as the country progressed towards malaria elimination (Gunawardena et al., 2014). Population genetic signals of declining transmission might take longer to emerge for P. vivax due to new blood-stage infections from the hypnozoite reservoir and could explain why we only observed mLD in one village of East Sepik.
In PNG, the limited decline in P. falciparum diversity and loss of parasite population structure after LLIN distribution indicates that there may be increased gene flow between the sampled parasite populations, which was unexpected. Population structure prior to intensified control was thought to be the result of limited historical human migration due to the rugged terrain and lack of direct transport connections (Mueller, Bockarie, Alpers, & Smith, 2003; Riley, 1983) or population bottlenecks due to past control efforts or emergence of drug resistance (Anderson et al., 2000; Jennison et al., 2015; Schultz et al., 2010; Tessema et al., 2015). Changes in first-line treatment policies, for example the introduction of sulphadoxine/pyrimethamine (SP) in the early 2000’s and artemether-lumefantrine in 2008, might have played a role in shaping parasite population structure (Mu et al., 2005). Chloroquine (CQ) and/or SP resistance (near fixation of resistant pfmdr1 and pfdhps resistant alleles were observed in the same areas (Barnadas et al., 2015; Koleala et al., 2015; Mita et al., 2018)) and may have contributed to the observed bottleneck and mLD in pre-LLIN P. falciparum populations, with consequent reductions in effective population size, while outcrossing due to high transmission preserved within-population genetic diversity as the resistance mutation spread (Mita et al., 2018). From 2000 to 2011 the PNG population increased by over two million people (National Census Report, 2011), and local observations suggest that large numbers of migrants from East Sepik have moved into Madang in the last decade seeking employment opportunities. As a result, post-LLIN, greater connectivity between human populations may have broken down P. falciparum population structure and maintained high gene flow between P. vivax populations (Fola et al., 2018).
For the post-LLIN East Sepik P. vivax population where prevalence dropped to below 5%, significant mLD was observed resulting from very closely related parasite strains circulating in a residual pocket of relatively high transmission within a single village. This suggests considerable inbreeding of parasites in that village, in a background of high genetic diversity resulting in a lack of evidence of a bottleneck at the population level. This paradoxical signature of significant mLD with high diversity and a considerable proportion of multiple clone infections of P. vivax appears to be a hallmark of lower transmission areas (Barry et al., 2015; Batista et al., 2015; Chenet et al., 2012; Delgado-Ratto et al., 2016; Delgado-Ratto et al., 2014; Ferreira et al., 2007; Hong et al., 2016; Noviyanti et al., 2015; Orjuela-Sanchez et al., 2013). Similar to P. falciparum populations though, there was a correlation between mLD and prevalence of infection for P. vivax. This shows that reductions in multiclonal infections and mLD is an earlier indicator of reduced transmission than genetic diversity and population structure (for both species).
Multilocus LD in post-LLIN P. vivax populations was explained by both focal clonal transmission and inbreeding, as similarly observed in other studies in Peru, Vietnam, and Papua Indonesia (Delgado-Ratto et al., 2014; Hong et al., 2016; Noviyanti et al., 2015). The explanation for this observation will most likely lie in unique P. vivax characteristics related to hypnozoites, relapses and transmission dynamics (Abdullah et al., 2013; Bright et al., 2014; Chen, Auliff, Rieckmann, Gatton, & Cheng, 2007; Delgado-Ratto et al., 2014; Ferreira et al., 2007; Fola et al., 2018; Iwagami et al., 2012; White, 2011). At high transmission (e.g. pre-LLIN) with high prevalence and high multiplicity of infection, these clusters of similar haplotypes might also occur, but could be obscured due to sampling limitations and the large number of different haplogroups circulating at the same time. As transmission declines, infections have fewer clones and the diversity of the hypnozoite reservoir decreases, resulting in fewer circulating haplogroups, lower levels of recombination between distinct genomes and more frequent clonal transmission and inbreeding, resulting in increased mLD as in this and other studies (Barry et al., 2015; Batista et al., 2015; Chenet et al., 2012; Delgado-Ratto et al., 2016; Delgado-Ratto et al., 2014; Ferreira et al., 2007; Hong et al., 2016; Noviyanti et al., 2015; Orjuela-Sanchez et al., 2013). These signatures are more likely to be seen in a population with more sustained low transmission such as was the case for the East Sepik Province. In this region, malaria transmission is heterogeneous between villages. Besides the national malaria control program, other initiatives were also distributing LLINs in East Sepik Province prior to the nationwide distribution (Hetzel et al., 2014; Hetzel et al., 2012; Hetzel et al., 2016) suggesting that longer term sustained control efforts have been effective.
Considerable variance in the impact of the LLIN program was observed in the two provinces. In Madang, whilst P. falciparum rates steadily declined over the entire study period, there was a resurgence of submicroscopic P. vivax infections in 2014 (Koepfli et al., 2017). Although multiplicity of infection remained low, the lack of mLD and population structure suggests that this is not due to an outbreak, but more likely the residual P. vivax population was able to gain a foothold once again despite the intensive control efforts. In addition, an increase in the prevalence of pvdhfr triple and quadruple mutants, related with SP resistance, were observed in Madang province in 2010 (Barnadas et al., 2015), and a high proportion of resistant parasites could be a possible explanation for the higher infection prevalence by 2014. Different studies have shown that selective pressure of drugs such as CQ and/or SP have had an impact on shaping worldwide P. vivax populations in recent history (Hupalo et al., 2016; Pearson et al., 2016). However, a population bottleneck as seen in P. falciparum populations (Mita et al., 2018) did not occur in P. vivax populations of PNG. Malaria control had a greater impact on P. vivax prevalence in East Sepik and population structure was observed in one village post-LLIN.
This study has some limitations related to sampling and the genotyping approach used. The samples were collected in serial cross-sectional surveys over a period of malaria control initiated at different times in the two provinces. Fewer years of sustained control pressure compared to East Sepik might explain why, despite substantial prevalence decline in Madang Province by 2010, we did not observe any signature of changing population structure. Whilst, in East Sepik 2012, a cluster of closely related parasites was observed in one village suggesting more focal transmission than previous years. The microsatellite panels were selected as these have been the gold standard genotyping tool for large-scale malaria population genetic studies for many years (Anderson et al., 1999; Imwong et al., 2006; Karunaweera et al., 2006). However, they are limited in number (less than one per chromosome), rapidly evolving and prone to error. Although these markers are extremely useful for measuring parasite population structure on local and global scales (Auburn & Barry, 2017; Barry et al., 2015; Koepfli & Mueller, 2017; Sutton, 2013), they are not ideal for cross-study comparisons due to the difficultly in calibrating alleles from different data sources. The lack of raw data from the previously published P. falciparum study (Schultz et al., 2010), prevented the direct comparison of haplotypes and thus the analysis of population structure between the earlier P. falciparum time points for the East Sepik II (Wosera) and Madang populations. Furthermore, dominant haplotypes derived from multiple clone infections can be reconstructed incorrectly, thus inflating diversity values (Messerli, Hofmann, Beck, & Felger, 2017). However, only haplotype-based analyses such as multilocus LD and phylogenetic analysis were vulnerable to these possible artefacts, and would result in an overestimate of diversity and underestimate of LD. All other analyses were conducted using mean values across markers or allele frequencies and thus limit the impact of such errors. Moreover, the dataset included a large proportion of single infection haplotypes in all populations, and the detected clones included dominant haplotypes suggesting that allele calling was reliable. Finally, the highly polymorphic and rapidly evolving nature of microsatellite markers (Ellegren, 2004) may limit the resolution of the population genetic parameters such as population level diversity and population structure in high transmission areas (Branch et al., 2011). This may both lead to false assignment of unrelated parasites (e.g. from different provinces) as related and reduce the detection of truly related parasites (identical by descent), both of which would result in a lack of population structure. Other more stable markers, such as SNPs (Baniecki et al., 2015; R. Daniels et al., 2008) or whole genome sequencing (Hupalo et al., 2016; Miotto et al., 2013; Mu et al., 2005; Pearson et al., 2016; Volkman et al., 2007) may be more sensitive and specific to detect changes in parasite population structure.
According to the data presented, in two high transmission provinces of PNG, recent reductions in malaria transmission result in limited changes in parasite population diversity and structure as determined by microsatellite markers. The high parasite diversity and lack of population structure are consistent with both species maintaining a large and evolutionarily fit population immediately after prevalence decline suggesting the gains made are fragile. However, fewer multiclonal infections, and the emergence of significant mLD for both species indicates there is focally interrupted transmission and suggests that these parameters may be useful markers for measuring control impact and early changes in parasite population structure with intervention. The results were in contrast to our expectations of decreasing diversity and increasing population structure (Jennison et al., 2015) and show that long term sustained control efforts may need to be maintained to observe significant change in population structure, at least as measured by the microsatellite panels used in this study. The PNG national malaria control program has made an impact on the malaria burden through the substantial reductions in infections circulating in the community (Hetzel et al., 2016; Kattenberg et al., 2020; Koepfli et al., 2017), however it appears that this has not been sustained long enough for the underlying parasite population to fragment. Finally, this study demonstrates how parasite population genetics can inform whether malaria intervention has reduced and fragmented the parasite population or if significantly more control effort will be required to do so.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the people who participated in the study, and for the support and collaboration of the participating communities. We appreciate the efforts of the laboratory, field team members and support staff from the Papua New Guinea Institute of Medical Research for their involvement in the included studies. This research was supported by National Health and Medical Research Council (NHMRC) of Australia Project Grants (1010069 and 1027108), the International Centres of Excellence in Malaria Research (ICEMR) for the South West Pacific, NIH U19 AI089686 and a Bill & Melinda Gates Foundation Grant (TransEpi Consortium). IM and LR are supported by NHMRC Research Fellowships.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.37pvmcvfh
Data accessibility
Dataset 1. Microsatellite genotypes were deposited in Dryad: https://doi.org/10.5061/dryad.37pvmcvfh.
REFERENCES
- Abdullah NR, Barber BE, William T, Norahmad NA, Satsu UR, Muniandy PK, … Auburn S (2013). Plasmodium vivax population structure and transmission dynamics in Sabah Malaysia. PLoS One, 8(12), e82553. doi: 10.1371/journal.pone.0082553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TJ, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, … Day KP (2000). Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol, 17(10), 1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247 [DOI] [PubMed] [Google Scholar]
- Anderson TJ, Su XZ, Bockarie M, Lagog M, & Day KP (1999). Twelve microsatellite markers for characterization of Plasmodium falciparum from finger-prick blood samples. Parasitology, 119 (Pt 2), 113–125. doi: 10.1017/s0031182099004552 [DOI] [PubMed] [Google Scholar]
- Argimon S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, … Aanensen DM (2016). Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom, 2(11), e000093. doi: 10.1099/mgen.0.000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott A, Barnadas C, Senn N, Siba P, Mueller I, Reeder JC, & Barry AE (2013). High genetic diversity of Plasmodium vivax on the north coast of Papua New Guinea. Am J Trop Med Hyg, 89(1), 188–194. doi: 10.4269/ajtmh.12-0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott A, Barry AE, & Reeder JC (2012). Understanding the population genetics of Plasmodium vivax is essential for malaria control and elimination. Malar J, 11, 14. doi: 10.1186/1475-2875-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auburn S, & Barry AE (2017). Dissecting malaria biology and epidemiology using population genetics and genomics. Int J Parasitol, 47(2-3), 77–85. doi: 10.1016/j.ijpara.2016.08.006 [DOI] [PubMed] [Google Scholar]
- Babiker HA, Ranford-Cartwright LC, Currie D, Charlwood JD, Billingsley P, Teuscher T, & Walliker D (1994). Random mating in a natural population of the malaria parasite Plasmodium falciparum. Parasitology, 109 ( Pt 4), 413–421. doi: 10.1017/s0031182000080665 [DOI] [PubMed] [Google Scholar]
- Baniecki ML, Faust AL, Schaffner SF, Park DJ, Galinsky K, Daniels RF, … Sabeti PC (2015). Development of a single nucleotide polymorphism barcode to genotype Plasmodium vivax infections. PLoS Negl Trop Dis, 9(3), e0003539. doi: 10.1371/journal.pntd.0003539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnadas C, Timinao L, Javati S, Iga J, Malau E, Koepfli C, … Mueller I (2015). Significant geographical differences in prevalence of mutations associated with Plasmodium falciparum and Plasmodium vivax drug resistance in two regions from Papua New Guinea. Malar J, 14, 399. doi: 10.1186/s12936-015-0879-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Schultz L, Senn N, Nale J, Kiniboro B, Siba PM, … Reeder JC (2013). High levels of genetic diversity of Plasmodium falciparum populations in Papua New Guinea despite variable infection prevalence. Am J Trop Med Hyg, 88(4), 718–725. doi: 10.4269/ajtmh.12-0056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Waltmann A, Koepfli C, Barnadas C, & Mueller I (2015). Uncovering the transmission dynamics of Plasmodium vivax using population genetics. Pathog Glob Health, 109(3), 142–152. doi: 10.1179/2047773215Y.0000000012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista CL, Barbosa S, Da Silva Bastos M, Viana SA, & Ferreira MU (2015). Genetic diversity of Plasmodium vivax over time and space: a community-based study in rural Amazonia. Parasitology, 142(2), 374–384. doi: 10.1017/S0031182014001176 [DOI] [PubMed] [Google Scholar]
- Bei AK, Niang M, Deme AB, Daniels RF, Sarr FD, Sokhna C, … Toure-Balde A (2018). Dramatic Changes in Malaria Population Genetic Complexity in Dielmo and Ndiop, Senegal, Revealed Using Genomic Surveillance. J Infect Dis, 217(4), 622–627. doi: 10.1093/infdis/jix580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betuela I, Maraga S, Hetzel MW, Tandrapah T, Sie A, Yala S, … Mueller I (2012). Epidemiology of malaria in the Papua New Guinean highlands. Trop Med Int Health, 17(10), 1181–1191. doi: 10.1111/j.1365-3156.2012.03062.x [DOI] [PubMed] [Google Scholar]
- Branch OH, Sutton PL, Barnes C, Castro JC, Hussin J, Awadalla P, & Hijar G (2011). Plasmodium falciparum genetic diversity maintained and amplified over 5 years of a low transmission endemic in the Peruvian Amazon. Mol Biol Evol, 28(7), 1973–1986. doi: 10.1093/molbev/msq311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright AT, Manary MJ, Tewhey R, Arango EM, Wang T, Schork NJ, … Winzeler EA (2014). A high resolution case study of a patient with recurrent Plasmodium vivax infections shows that relapses were caused by meiotic siblings. PLoS Negl Trop Dis, 8(6), e2882. doi: 10.1371/journal.pntd.0002882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Auliff A, Rieckmann K, Gatton M, & Cheng Q (2007). Relapses of Plasmodium vivax infection result from clonal hypnozoites activated at predetermined intervals. J Infect Dis, 195(7), 934–941. doi: 10.1086/512242 [DOI] [PubMed] [Google Scholar]
- Chenet SM, Schneider KA, Villegas L, & Escalante AA (2012). Local population structure of Plasmodium: impact on malaria control and elimination. Malar J, 11, 412. doi: 10.1186/1475-2875-11-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenet SM, Taylor JE, Blair S, Zuluaga L, & Escalante AA (2015). Longitudinal analysis of Plasmodium falciparum genetic variation in Turbo, Colombia: implications for malaria control and elimination. Malar J, 14, 363. doi: 10.1186/s12936-015-0887-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, & Escalante AA (2006). The origin and age of Plasmodium vivax. Trends Parasitol, 22(12), 558–563. doi: 10.1016/j.pt.2006.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- dalla Martha RC, Tada MS, Ferreira RG, da Silva LH, & Wunderlich G (2007). Microsatellite characterization of Plasmodium falciparum from symptomatic and non-symptomatic infections from the Western Amazon reveals the existence of non-symptomatic infection-associated genotypes. Mem Inst Oswaldo Cruz, 102(3), 293–298. doi: 10.1590/s0074-02762007005000044 [DOI] [PubMed] [Google Scholar]
- Dalmat R, Naughton B, Kwan-Gett TS, Slyker J, & Stuckey EM (2019). Use cases for genetic epidemiology in malaria elimination. Malar J, 18(1), 163. doi: 10.1186/s12936-019-2784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Volkman SK, Milner DA, Mahesh N, Neafsey DE, Park DJ, … Wiegand RC (2008). A general SNP-based molecular barcode for Plasmodium falciparum identification and tracking. Malar J, 7, 223. doi: 10.1186/1475-2875-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RF, Schaffner SF, Wenger EA, Proctor JL, Chang HH, Wong W, … Wirth DF (2015). Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci U S A, 112(22), 7067–7072. doi: 10.1073/pnas.1505691112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Ratto C, Gamboa D, Soto-Calle VE, Van den Eede P, Torres E, Sanchez-Martinez L, … D'Alessandro U (2016). Population Genetics of Plasmodium vivax in the Peruvian Amazon. PLoS Negl Trop Dis, 10(1), e0004376. doi: 10.1371/journal.pntd.0004376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Ratto C, Soto-Calle VE, Van den Eede P, Gamboa D, Rosas A, Abatih EN, … D'Alessandro U (2014). Population structure and spatio-temporal transmission dynamics of Plasmodium vivax after radical cure treatment in a rural village of the Peruvian Amazon. Malar J, 13, 8. doi: 10.1186/1475-2875-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diva GIS country level data. Retrieved from http://www.diva-gis.org/gdata
- Earl DA, & vonHoldt BM (2011). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361. doi: 10.1007/s12686-011-9548-7 [DOI] [Google Scholar]
- Ellegren H (2004). Microsatellites: simple sequences with complex evolution. Nat Rev Genet, 5(6), 435–445. doi: 10.1038/nrg1348 [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, & Goudet J (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol, 14(8), 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- Feachem RG, Phillips AA, Hwang J, Cotter C, Wielgosz B, Greenwood BM, … Snow RW (2010). Shrinking the malaria map: progress and prospects. Lancet, 376(9752), 1566–1578. doi: 10.1016/S0140-6736(10)61270-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MU, Karunaweera ND, da Silva-Nunes M, da Silva NS, Wirth DF, & Hartl DL (2007). Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis, 195(8), 1218–1226. doi: 10.1086/512685 [DOI] [PubMed] [Google Scholar]
- Fola AA, Harrison GLA, Hazairin MH, Barnadas C, Hetzel MW, Iga J, … Barry AE (2017). Higher Complexity of Infection and Genetic Diversity of Plasmodium vivax Than Plasmodium falciparum Across All Malaria Transmission Zones of Papua New Guinea. Am J Trop Med Hyg, 96(3), 630–641. doi: 10.4269/ajtmh.16-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fola AA, Nate E, Abby Harrison GL, Barnadas C, Hetzel MW, Iga J, … Barry AE (2018). Nationwide genetic surveillance of Plasmodium vivax in Papua New Guinea reveals heterogeneous transmission dynamics and routes of migration amongst subdivided populations. Infect Genet Evol, 58, 83–95. doi: 10.1016/j.meegid.2017.11.028 [DOI] [PubMed] [Google Scholar]
- Gatei W, Kariuki S, Hawley W, ter Kuile F, Terlouw D, Phillips-Howard P, … Shi YP (2010). Effects of transmission reduction by insecticide-treated bed nets (ITNs) on parasite genetics population structure: I. The genetic diversity of Plasmodium falciparum parasites by microsatellite markers in western Kenya. Malar J, 9, 353. doi: 10.1186/1475-2875-9-353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genton B, al-Yaman F, Beck HP, Hii J, Mellor S, Narara A, … Alpers MP (1995). The epidemiology of malaria in the Wosera area, East Sepik Province, Papua New Guinea, in preparation for vaccine trials. I. Malariometric indices and immunity. Ann Trop Med Parasitol, 89(4), 359–376. doi: 10.1080/00034983.1995.11812965 [DOI] [PubMed] [Google Scholar]
- Genton B, Hii J, al-Yaman F, Paru R, Beck HP, Ginny M, … Alpers MP (1994). The use of untreated bednets and malaria infection, morbidity and immunity. Ann Trop Med Parasitol, 88(3), 263–270. doi: 10.1080/00034983.1994.11812866 [DOI] [PubMed] [Google Scholar]
- Gilabert A, Otto TD, Rutledge GG, Franzon B, Ollomo B, Arnathau C, … Rougeron V (2018). Plasmodium vivax-like genome sequences shed new insights into Plasmodium vivax biology and evolution. PLoS Biol, 16(8), e2006035. doi: 10.1371/journal.pbio.2006035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaubitz JC convert: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. [Google Scholar]
- Goudet J (1995). FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. Journal of Heredity, 86(6), 485–486. [Google Scholar]
- Grad YH, & Lipsitch M (2014). Epidemiologic data and pathogen genome sequences: a powerful synergy for public health. Genome Biology, 15(11), 538. doi: 10.1186/s13059-014-0538-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S, Ferreira MU, Kapilananda GM, Wirth DF, & Karunaweera ND (2014). The Sri Lankan paradox: high genetic diversity in Plasmodium vivax populations despite decreasing levels of malaria transmission. Parasitology, 141(7), 880–890. doi: 10.1017/S0031182013002278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubold B, & Hudson RR (2000). LIAN 3.0: detecting linkage disequilibrium in multilocus data. Linkage Analysis. Bioinformatics, 16(9), 847–848. doi: 10.1093/bioinformatics/16.9.847 [DOI] [PubMed] [Google Scholar]
- Hedtke SM, Kuesel AC, Crawford KE, Graves PM, Boussinesq M, Lau CL, … Grant WN (2019). Genomic Epidemiology in Filarial Nematodes: Transforming the Basis for Elimination Program Decisions. Front Genet, 10, 1282. doi: 10.3389/fgene.2019.01282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel MW, Choudhury AA, Pulford J, Ura Y, Whittaker M, Siba PM, & Mueller I (2014). Progress in mosquito net coverage in Papua New Guinea. Malar J, 13, 242. doi: 10.1186/1475-2875-13-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel MW, Gideon G, Lote N, Makita L, Siba PM, & Mueller I (2012). Ownership and usage of mosquito nets after four years of large-scale free distribution in Papua New Guinea. Malar J, 11(1), 192. doi: 10.1186/1475-2875-11-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzel MW, Reimer LJ, Gideon G, Koimbu G, Barnadas C, Makita L, … Mueller I (2016). Changes in malaria burden and transmission in sentinel sites after the roll-out of long-lasting insecticidal nets in Papua New Guinea. Parasit Vectors, 9(1), 340. doi: 10.1186/s13071-016-1635-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hii JL, Smith T, Vounatsou P, Alexander N, Mai A, Ibam E, & Alpers MP (2001). Area effects of bednet use in a malaria-endemic area in Papua New Guinea. Trans R Soc Trop Med Hyg, 95(1), 7–13. doi: 10.1016/s0035-9203(01)90315-3 [DOI] [PubMed] [Google Scholar]
- Hofmann NE, Karl S, Wampfler R, Kiniboro B, Teliki A, Iga J, … Mueller I (2017). The complex relationship of exposure to new Plasmodium infections and incidence of clinical malaria in Papua New Guinea. Elife, 6. doi: 10.7554/eLife.23708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong NV, Delgado-Ratto C, Thanh PV, Van den Eede P, Guetens P, Binh NT, … Rosanas-Urgell A (2016). Population Genetics of Plasmodium vivax in Four Rural Communities in Central Vietnam. PLoS Negl Trop Dis, 10(2), e0004434. doi: 10.1371/journal.pntd.0004434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupalo DN, Luo Z, Melnikov A, Sutton PL, Rogov P, Escalante A, … Carlton JM (2016). Population genomics studies identify signatures of global dispersal and drug resistance in Plasmodium vivax. Nat Genet, 48(8), 953–958. doi: 10.1038/ng.3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert SH (1971). The Nonconcept of Species Diversity: A Critique and Alternative Parameters. Ecology, 52(4), 577–586. doi: 10.2307/1934145 [DOI] [PubMed] [Google Scholar]
- Imwong M, Nair S, Pukrittayakamee S, Sudimack D, Williams JT, Mayxay M, … Anderson TJ (2007). Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol, 37(8-9), 1013–1022. doi: 10.1016/j.ijpara.2007.02.010 [DOI] [PubMed] [Google Scholar]
- Imwong M, Sudimack D, Pukrittayakamee S, Osorio L, Carlton JM, Day NP, … Anderson TJ (2006). Microsatellite variation, repeat array length, and population history of Plasmodium vivax. Mol Biol Evol, 23(5), 1016–1018. doi: 10.1093/molbev/msj116 [DOI] [PubMed] [Google Scholar]
- Iwagami M, Fukumoto M, Hwang SY, Kim SH, Kho WG, & Kano S (2012). Population structure and transmission dynamics of Plasmodium vivax in the Republic of Korea based on microsatellite DNA analysis. PLoS Negl Trop Dis, 6(4), e1592. doi: 10.1371/journal.pntd.0001592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, & Rosenberg NA (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics, 23(14), 1801–1806. doi: 10.1093/bioinformatics/btm233 [DOI] [PubMed] [Google Scholar]
- Jennison C, Arnott A, Tessier N, Tavul L, Koepfli C, Felger I, … Barry AE (2015). Plasmodium vivax populations are more genetically diverse and less structured than sympatric Plasmodium falciparum populations. PLoS Negl Trop Dis, 9(4), e0003634. doi: 10.1371/journal.pntd.0003634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L (2008). G(ST) and its relatives do not measure differentiation. Mol Ecol, 17(18), 4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x [DOI] [PubMed] [Google Scholar]
- Karunaweera ND, Ferreira MU, Hartl DL, & Wirth DF (2006). Fourteen polymorphic microsatellite DNA markers for the human malaria parasite Plasmodium vivax. Molecular Ecology Notes, 7(1), 172–175. doi: 10.1111/j.1471-8286.2006.01534.x [DOI] [Google Scholar]
- Kattenberg JH, Gumal DL, Ome-Kaius M, Kiniboro B, Philip M, Jally S, … Robinson LJ (2020). The epidemiology of Plasmodium falciparum and Plasmodium vivax in East Sepik Province, Papua New Guinea, pre- and post-implementation of national malaria control efforts Accepted for publication in Malaria Journal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan K, McGinnity P, Cross TF, Crozier WW, & Prodöhl PA (2013). diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods in Ecology and Evolution, 4(8), 782–788. doi: 10.1111/2041-210X.12067 [DOI] [Google Scholar]
- Koepfli C, & Mueller I (2017). Malaria Epidemiology at the Clone Level. Trends Parasitol, 33(12), 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli C, Ome-Kaius M, Jally S, Malau E, Maripal S, Ginny J, … Robinson LJ (2017). Sustained malaria control over an eight-year period in Papua New Guinea: the challenge of low-density asymptomatic infections. J Infect Dis. doi: 10.1093/infdis/jix507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli C, Robinson LJ, Rarau P, Salib M, Sambale N, Wampfler R, … Mueller I (2015). Blood-Stage Parasitaemia and Age Determine Plasmodium falciparum and P. vivax Gametocytaemia in Papua New Guinea. PLoS One, 10(5), e0126747. doi: 10.1371/journal.pone.0126747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepfli C, Timinao L, Antao T, Barry AE, Siba P, Mueller I, & Felger I (2013). A Large Plasmodium vivax Reservoir and Little Population Structure in the South Pacific. PLoS One, 8(6), e66041. doi: 10.1371/journal.pone.0066041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleala T, Karl S, Laman M, Moore BR, Benjamin J, Barnadas C, … Davis TM (2015). Temporal changes in Plasmodium falciparum anti-malarial drug sensitivity in vitro and resistance-associated genetic mutations in isolates from Papua New Guinea. Malar J, 14, 37. doi: 10.1186/s12936-015-0560-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin E, Kiniboro B, Gray L, Dobbie S, Robinson L, Laumaea A, … Mueller I (2010). Differential patterns of infection and disease with P. falciparum and P. vivax in young Papua New Guinean children. PLoS One, 5(2), e9047. doi: 10.1371/journal.pone.0009047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischer HEL, & Excoffier L (2012). PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics, 28(2), 298–299. doi: 10.1093/bioinformatics/btr642 [DOI] [PubMed] [Google Scholar]
- Liu W, Li Y, Shaw KS, Learn GH, Plenderleith LJ, Malenke JA, … Sharp PM (2014). African origin of the malaria parasite Plasmodium vivax. Nat Commun, 5, 3346. doi: 10.1038/ncomms4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy DE, Liu W, Li Y, Learn GH, Plenderleith LJ, Sundararaman SA, … Hahn BH (2017). Out of Africa: origins and evolution of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. International Journal for Parasitology, 47(2), 87–97. doi: 10.1016/j.ijpara.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, Allendorf FW, Cornuet JM, & Sherwin WB (1998). Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered, 89(3), 238–247. [DOI] [PubMed] [Google Scholar]
- malEra Consultative Group on Monitoring, E., & Surveillance. (2011). A research agenda for malaria eradication: monitoring, evaluation, and surveillance. PLoS Med, 8(1), e1000400. doi: 10.1371/journal.pmed.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M, & Salzburger W (2009). TANDEM: integrating automated allele binning into genetics and genomics workflows. Bioinformatics, 25(15), 1982–1983. doi: 10.1093/bioinformatics/btp303 [DOI] [PubMed] [Google Scholar]
- McNamara DT, Kasehagen LJ, Grimberg BT, Cole-Tobian J, Collins WE, & Zimmerman PA (2006). Diagnosing infection levels of four human malaria parasite species by a polymerase chain reaction/ligase detection reaction fluorescent microsphere-based assay. Am J Trop Med Hyg, 74(3), 413–421. [PMC free article] [PubMed] [Google Scholar]
- Messerli C, Hofmann NE, Beck HP, & Felger I (2017). Critical Evaluation of Molecular Monitoring in Malaria Drug Efficacy Trials and Pitfalls of Length-Polymorphic Markers. Antimicrob Agents Chemother, 61(1). doi: 10.1128/AAC.01500-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O, Almagro-Garcia J, Manske M, Macinnis B, Campino S, Rockett KA, … Kwiatkowski DP (2013). Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet, 45(6), 648–655. doi: 10.1038/ng.2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita T, Hombhanje F, Takahashi N, Sekihara M, Yamauchi M, Tsukahara T, … Ohashi J (2018). Rapid selection of sulphadoxine-resistant Plasmodium falciparum and its effect on within-population genetic diversity in Papua New Guinea. Scientific Reports, 8(1), 5565. doi: 10.1038/s41598-018-23811-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Awadalla P, Duan J, McGee KM, Joy DA, McVean GA, & Su XZ (2005). Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol, 3(10), e335. doi: 10.1371/journal.pbio.0030335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller I, Bockarie M, Alpers M, & Smith T (2003). The epidemiology of malaria in Papua New Guinea. Trends Parasitol, 19(6), 253–259. [DOI] [PubMed] [Google Scholar]
- Mueller I, Widmer S, Michel D, Maraga S, McNamara DT, Kiniboro B, … Zimmerman PA (2009). High sensitivity detection of Plasmodium species reveals positive correlations between infections of different species, shifts in age distribution and reduced local variation in Papua New Guinea. Malar J, 8, 41. doi: 10.1186/1475-2875-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neafsey DE, Galinsky K, Jiang RH, Young L, Sykes SM, Saif S, … Carlton JM (2012). The malaria parasite Plasmodium vivax exhibits greater genetic diversity than Plasmodium falciparum. Nat Genet, 44(9), 1046–1050. doi: 10.1038/ng.2373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M (1987). Molecular Evolutionary Genetics: Columbia University Press. [Google Scholar]
- Nkhoma SC, Nair S, Al-Saai S, Ashley E, McGready R, Phyo AP, … Anderson TJ (2013). Population genetic correlates of declining transmission in a human pathogen. Mol Ecol, 22(2), 273–285. doi: 10.1111/mec.12099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noviyanti R, Coutrier F, Utami RA, Trimarsanto H, Tirta YK, Trianty L, … Auburn S (2015). Contrasting Transmission Dynamics of Co-endemic Plasmodium vivax and P. falciparum: Implications for Malaria Control and Elimination. PLoS Negl Trop Dis, 9(5), e0003739. doi: 10.1371/journal.pntd.0003739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira-Ferreira J, Lacerda MV, Brasil P, Ladislau JL, Tauil PL, & Daniel-Ribeiro CT (2010). Malaria in Brazil: an overview. Malar J, 9, 115. doi: 10.1186/1475-2875-9-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olliaro PL, Barnwell JW, Barry A, Mendis K, Mueller I, Reeder JC, … Wongsrichanalai C (2016). Implications of Plasmodium vivax Biology for Control, Elimination, and Research. Am J Trop Med Hyg, 95(6 Suppl), 4–14. doi: 10.4269/ajtmh.16-0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjuela-Sanchez P, Da Silva-Nunes M, Da Silva NS, Scopel KK, Goncalves RM, Malafronte RS, & Ferreira MU (2009). Population dynamics of genetically diverse Plasmodium falciparum lineages: community-based prospective study in rural Amazonia. Parasitology, 136(10), 1097–1105. doi: 10.1017/S0031182009990539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjuela-Sanchez P, Sa JM, Brandi MC, Rodrigues PT, Bastos MS, Amaratunga C, … Ferreira MU (2013). Higher microsatellite diversity in Plasmodium vivax than in sympatric Plasmodium falciparum populations in Pursat, Western Cambodia. Exp Parasitol, 134(3), 318–326. doi: 10.1016/j.exppara.2013.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul RE, Packer MJ, Walmsley M, Lagog M, Ranford-Cartwright LC, Paru R, & Day KP (1995). Mating patterns in malaria parasite populations of Papua New Guinea. Science, 269(5231), 1709–1711. [DOI] [PubMed] [Google Scholar]
- Pava Z, Noviyanti R, Handayuni I, Trimarsanto H, Trianty L, Burdam FH, … Auburn S (2017). Genetic micro-epidemiology of malaria in Papua Indonesia: Extensive P. vivax diversity and a distinct subpopulation of asymptomatic P. falciparum infections. PLoS One, 12(5), e0177445. doi: 10.1371/journal.pone.0177445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson RD, Amato R, Auburn S, Miotto O, Almagro-Garcia J, Amaratunga C, … Kwiatkowski DP (2016). Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nat Genet, 48(8), 959–964. doi: 10.1038/ng.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piry S, Luikart G, & Cornuet J BOTTLENECK : A program for detecting recent effective population size reductions from allele data frequencies. Retrieved from http://www1.montpellier.inra.fr/CBGP/software/Bottleneck/pub.html
- Pritchard JK, Stephens M, & Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics, 155(2), 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley ID (1983). Population change and distribution in Papua New Guinea: an epidemiological approach. Journal of Human Evolution, 12(1), 125–132. doi: 10.1016/S0047-2484(83)80017-7 [DOI] [Google Scholar]
- Robinson LJ, Wampfler R, Betuela I, Karl S, White MT, Li Wai Suen CS, … Mueller I (2015). Strategies for understanding and reducing the Plasmodium vivax and Plasmodium ovale hypnozoite reservoir in Papua New Guinean children: a randomised placebo-controlled trial and mathematical model. PLoS Med, 12(10), e1001891. doi: 10.1371/journal.pmed.1001891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosanas-Urgell A, Mueller D, Betuela I, Barnadas C, Iga J, Zimmerman PA, … Felger I (2010). Comparison of diagnostic methods for the detection and quantification of the four sympatric Plasmodium species in field samples from Papua New Guinea. Malar J, 9, 361. doi: 10.1186/1475-2875-9-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA (2004). distruct: a program for the graphical display of population structure. Molecular Ecology Notes, 4(1), 137–138. doi: 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- Salgueiro P, Vicente JL, Figueiredo RC, & Pinto J (2016). Genetic diversity and population structure of Plasmodium falciparum over space and time in an African archipelago. Infect Genet Evol, 43, 252–260. doi: 10.1016/j.meegid.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Schultz L, Wapling J, Mueller I, Ntsuke P, Senn N, Nale J, … Barry A (2010). Multilocus haplotypes reveal variable levels of diversity and population structure of Plasmodium falciparum in Papua New Guinea, a region of intense perennial transmission. Malaria Journal, 9(1), 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn N, Rarau P, Stanisic DI, Robinson L, Barnadas C, Manong D, … Mueller I (2012). Intermittent Preventive Treatment for Malaria in Papua New Guinean Infants Exposed to Plasmodium falciparum and P. vivax: A Randomized Controlled Trial. PLOS Medicine, 9(3), e1001195. doi: 10.1371/journal.pmed.1001195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O'Rourke M, & Spratt BG (1993). How clonal are bacteria? Proceedings of the National Academy of Sciences, 90(10), 4384–4388. doi: 10.1073/pnas.90.10.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susomboon P, Iwagami M, Tangpukdee N, Krusood S, Looareesuwan S, & Kano S (2008). Differences in genetic population structures of Plasmodium falciparum isolates from patients along Thai-Myanmar border with severe or uncomplicated malaria. Malar J, 7, 212. doi: 10.1186/1475-2875-7-212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton PL (2013). A call to arms: on refining Plasmodium vivax microsatellite marker panels for comparing global diversity. Malar J, 12, 447. doi: 10.1186/1475-2875-12-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessema SK, Monk SL, Schultz MB, Tavul L, Reeder JC, Siba PM, … Barry AE (2015). Phylogeography of var gene repertoires reveals fine-scale geospatial clustering of Plasmodium falciparum populations in a highly endemic area. Mol Ecol, 24(2), 484–497. doi: 10.1111/mec.13033 [DOI] [PubMed] [Google Scholar]
- Vardo-Zalik AM, Zhou G, Zhong D, Afrane YA, Githeko AK, & Yan G (2013). Alterations in Plasmodium falciparum genetic structure two years after increased malaria control efforts in western Kenya. Am J Trop Med Hyg, 88(1), 29–36. doi: 10.4269/ajtmh.2012.12-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman SK, Sabeti PC, DeCaprio D, Neafsey DE, Schaffner SF, Milner DA Jr., … Wirth DF (2007). A genome-wide map of diversity in Plasmodium falciparum. Nat Genet, 39(1), 113–119. doi: 10.1038/ng1930 [DOI] [PubMed] [Google Scholar]
- Wahlund S (1928). Zusammensetzung Von Populationen Und Korrelationserscheinungen Vom Standpunkt Der Vererbungslehre Aus Betrachtet. Hereditas, 11(1), 65–-106. doi: 10.1111/j.1601-5223.1928.tb02483.x [DOI] [Google Scholar]
- Waltmann A, Darcy AW, Harris I, Koepfli C, Lodo J, Vahi V, … Mueller I (2015). High Rates of Asymptomatic, Sub-microscopic Plasmodium vivax Infection and Disappearing Plasmodium falciparum Malaria in an Area of Low Transmission in Solomon Islands. PLoS Negl Trop Dis, 9(5), e0003758. doi: 10.1371/journal.pntd.0003758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltmann A, Koepfli C, Tessier N, Karl S, Fola A, Darcy AW, … Barry AE (2018). Increasingly inbred and fragmented populations of Plasmodium vivax associated with the eastward decline in malaria transmission across the Southwest Pacific. PLoS Negl Trop Dis, 12(1), e0006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, & Cockerham CC (1984). Estimating F-Statistics for the Analysis of Population Structure. Evolution, 38(6), 1358–1370. doi: 10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- White NJ (2011). Determinants of relapse periodicity in Plasmodium vivax malaria. Malar J, 10, 297. doi: 10.1186/1475-2875-10-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. (2017). World Malaria Report 2017. Retrieved from Geneva: [Google Scholar]
- WHO. (2018). World Malaria Report 2018. Retrieved from Geneva: http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/ [Google Scholar]
- WHO. (2019). World Malaria Report 2019. Retrieved from Geneva: [Google Scholar]
- Wlodarska M, Johnston JC, Gardy JL, & Tang P (2015). A microbiological revolution meets an ancient disease: improving the management of tuberculosis with genomics. Clinical Microbiology Reviews, 28(2), 523–539. doi: 10.1128/CMR.00124-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are openly available in Dryad at https://doi.org/10.5061/dryad.37pvmcvfh
Dataset 1. Microsatellite genotypes were deposited in Dryad: https://doi.org/10.5061/dryad.37pvmcvfh.