Abstract
Microsporidia are intracellular eukaryotes that infect many animals and cause opportunistic infections in AIDS patients. The disease is transmitted via environmentally resistant spores. Two spore wall constituents from the microsporidian Encephalitozoon intestinalis were characterized. Spore wall protein 1 (SWP1), a 50-kDa glycoprotein recognized by monoclonal antibody (MAb) 11B2, was detected in developing sporonts and at low levels on the surfaces of mature spores. In contrast, SWP2, a 150-kDa glycoprotein recognized by MAb 7G7, was detected on fully formed sporonts and was more abundant on mature spores than SWP1. Nevertheless, the SWPs appeared to be complexed on the surfaces of mature spores. SWP1 and SWP2 are similar at the DNA and protein levels and have 10 conserved cysteines in the N-terminal domain, suggesting similar secondary structures. The C-terminal domain of SWP2 has a unique region containing 50 repeating 12- or 15-amino-acid units that lacks homology to known protein motifs. Antibodies from mice infected with E. intestinalis recognized SWP1 and SWP2. The characterization of two immunogenic SWPs from E. intestinalis will allow the study of exospore structure and function and may lead to the development of useful tools in the diagnosis and treatment of microsporidiosis.
Microsporidia are obligate intracellular organisms that infect a wide variety of animals ranging from insects and fish to mammals, including humans. Of over 1,000 microsporidial species identified, at least 13 are known to infect humans (10). The species more commonly identified in humans are members of the families Enchephalitozoonidae and Enterocytozoonidae. In humans, microsporidiosis is found mostly in human immunodeficiency virus-infected and AIDS patients and commonly results in severe diarrhea and wasting (3, 20). However, microsporidiosis also occurs in immunocompetent individuals and common farm animals (21, 29, 32).
Microsporidia infect cells by a unique mechanism (reviewed in reference 31). Upon close association of a spore with a suitable host cell, a hollow polar filament is extruded from the spore into the host cell's cytoplasm. The infectious sporoplasm passes through the polar filament into the cell, initiating infection (18). Alternatively, the spore may be internalized by phagocytosis (33). The microsporidia then enter a stage of proliferative growth by nuclear fission called merogony, resulting in large and less structurally defined cells. In the family Encephalitozoonidae, the transition from meront to the next stage (sporont) is marked by aggregation of electron-dense material on the outer spore membrane. These immature cells are localized to the edge of the parasitophorous vacuole (PV). Then, in most cases, fully formed sporonts break away from the edge of the PV to reside internally (7, 12). Sporonts undergo continuous transition into sporoblasts, after which organelles organize and become more defined. At this stage, the cells form an electron-lucent material, the endospore, immediately inside the exospore region. The spore is considered mature when organelles are localized and fully formed.
Purified spores can survive heating to 56°C for 60 min, a pH of 9 or 4 for 24 h, or storage at 4°C for 2 years without losing infectivity (19, 28). The spore wall, comprising the exospore, endospore, and plasma membrane, provides structural rigidity and protects the mature spore from the environment. The endospore is composed of protein and chitin, and the exospore contains proteinaceous material (reviewed in reference 31). The proteins of the outer spore wall have been partially characterized in a few isolates. A glycine- and serine-rich protein was localized to the exospore of Encephalitozoon cuniculi (5). In Encephalitozoon intestinalis, a series of monoclonal antibodies (MAbs) reacted with proteins on the spore surface (22, 24); however, the reacting proteins were not characterized. In the present study, we characterized two spore wall proteins (SWPs) of E. intestinalis and the genes from which these proteins are derived. In addition, we determined the immunogenicities of these proteins in a mouse infection model.
MATERIALS AND METHODS
Parasite and host cell cultivation.
African green monkey kidney (Vero) cells were initially grown in Dulbecco's modified Eagle's medium (BioWhittaker, Walkersville, Md.) supplemented with l-glutamine (2 mM), penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin B (0.25 μg/ml), and 10% fetal bovine serum (FBS) (HyClone, Logan, Utah) in 5% CO2 at 37°C. For maintenance, 10% FBS was replaced with 2% FBS. Subconfluent host cell monolayers were infected with E. intestinalis spores. Following 12 to 15 days of infection, spores were harvested 2 to 3 times a week. The harvested spores were purified from host cell debris by washing it with 0.25% sodium dodecyl sulfate (SDS), followed by several washes with H2O. The washed spores were then mixed with an equal volume of Percoll (Sigma, St. Louis, Mo.) and centrifuged at 500 × g for 30 min. The pellet was washed and stored at 4°C in H2O.
Construction and screening of cDNA libraries.
The construction and screening methods for the subtracted cDNA library were described previously (15). In addition, a unidirectional full-length cDNA expression library was constructed from infected host cell mRNA using the Uni-ZAP XR Vector kit (Stratagene, La Jolla, Calif.). The library had a preamplification titer of 1.2 × 106 PFU/μg. After amplification, the library was screened with radiolabeled probes specific for clone 46, which had been isolated from the subtracted library. The expression library was also screened with MAbs, as previously described (22). All DNA probes were radiolabeled by random priming, and DNA sequencing was performed using the Dye Terminator cycle-sequencing kit (Beckman Coulter, Fullerton, Calif.) and the capillary array CEQ 2000 DNA analysis system (Beckman Coulter).
Southern blot analysis.
For Southern blotting, genomic DNA was isolated from infected host cells and digested with restriction endonucleases. After electrophoresis, the DNA was transferred to a nylon membrane by alkaline transfer as previously described (26). The blot was hybridized with randomly primed radiolabeled probes. The probe that hybridized with the genes for both SWP1 and SWP2 was a PCR fragment of common sequence representing the predicted amino acids 158 to 312 of SWP1. The swp1-specific probe was a PCR fragment that encoded the predicted amino acids 239 to 387 of SWP1. The SWP2-specific probe was a nested deletion clone that contained the 3′-terminal ≈500 bases of the swp2 open reading frame (ORF). The conditions for hybridization were similar to those used previously (15).
Inverse PCR and sequence analysis of swp1 and swp2.
The flanking regions of clone 46 (swp1) and clone 2.8 (swp2) were amplified by inverse PCR, cloned, and sequenced by primer walking. To sequence through the repeated motif of swp2, nested deletion clones were constructed using the Erase-A-Base system (Promega, Madison, Wis.). Based on the sizes of the deletion clones and overlapping sequence, the sequence of the repeated region was determined to be complete. The sequence was confirmed by priming from either end of a modified transposon element randomly inserted (EZ::TN <TET-1> insertion kit; Epicentre Technologies, Madison, Wis.) into the plasmid insert as described by the manufacturer.
Comparative analysis of the predicted amino acid sequences of SWP1 and SWP2 was performed using the Clustal W alignment program with an open gap penalty of 10.0 and an extended gap penalty of 0.05. The protein sequence motifs of swp1 and swp2 were analyzed using the protein subsequence analysis tools of the MacVector Sequence Analysis program (Genetics Computer Group, Madison, Wis.).
Western blot analysis.
Purified spores were processed for SDS-polyacrylamide gel electrophoresis (PAGE) in Laemmli sample buffer (Bio-Rad, Hercules, Calif.), and 30 μl (≈5 × 105 spores) was subjected to SDS-PAGE on a 4 to 20% Tris-glycine polyacrylamide gel (Invitrogen, Carlsbad, Calif.). Electrophoresis, transfer to nitrocellulose, and blocking were performed under standard conditions (26). Either MAb 11B2 or 7G7 (1:1,000 dilution of ascites) (22) was used as the primary antibody. The secondary antibody, a goat anti-mouse immunoglobulin linked to alkaline phosphatase (Southern Biotechnologies, Birmingham, Ala.), was detected using the Western Blue reagent (Promega).
For immunoprecipitation assays, an infected host cell monolayer from a 75-cm2 flask was lysed in 10 ml of lysis buffer containing 5 mM EDTA, 250 mM NaCl, 25 mM Tris (pH 7.5), 1% Triton X-100, and protease inhibitor cocktail (Roche, Indianapolis, Ind.). The lysates were centrifuged to remove cell debris, and MAbs 11B2 and 7G7 (1:500 dilution) were added to the cell lysate on ice for 1 h. Fifty microliters of protein A-agarose beads (Life Technologies, Rockville, Md.) was added to the MAb-lysate mixture and incubated on ice for 1 h. The beads were then washed with phosphate-buffered saline (PBS), resuspended in 70 μl of Laemmli sample buffer (Bio-Rad) with 2-mercaptoethanol, boiled for 5 min, and electrophoresed into a 4 to 20% Tris-glycine polyacrylamide gel.
To determine if SWP1 and SWP2 are glycosylated, 50 μl of concanavalin A (ConA)-agarose or wheat germ agglutinin (WGA)-agarose (Vector, Burlingame, Calif.) was reacted with 125 μl of infected cell lysate on ice for 1 h. For inhibition, either methyl-α-mannopyranoside (Sigma) or chitin hydrolysate (Vector) was added to the lysate at final concentrations of 0.2 M and 1:8, respectively. The beads were processed as described above. Following SDS-PAGE, the proteins were transferred to nitrocellulose and processed for Western blotting.
Immunoelectron microscopy (IEM).
Host cells, grown on Thermanox coverslips (Nunc, Naperville, Ill.) in 12-well plates, were infected with E. intestinalis spores. The coverslips were removed 5 to 7 days postinfection and rinsed with Hanks balanced salt solution. Then they were reacted for 2 h in fixative containing 3 parts solution A (0.1 M lysine–HCl–NaPO4), 1 part solution B (8% paraformaldehyde, 21.3 mg of sodium periodate, and 100 μl of 25% glutaraldehyde), and an additional 0.1% glutaraldehyde. The coverslips were then rinsed in PBS and permeabilized with either 0.05 or 0.025% saponin in PBS for 5 min at room temperature. For immunostaining, MAb (11B2 or 7G7) diluted 1:500 in a 3% globulin-free bovine serum albumin (BSA)-PBS (Sigma) solution was added at room temperature for 1 h. After PBS-BSA washes, the fluoronanogold anti-mouse immunoglobulin G Fab antibody (Nanoprobes, Yaphank, N.Y.), diluted 1:30 in PBS-BSA with either 0.05 or 0.025% saponin, was added for 1 h at room temperature. The coverslips were washed five times in PBS and stored at 4°C in postfixative (2.5% glutaraldehyde, 4% paraformaldehyde) until they were used. The coverslips were washed in H2O and reacted for 4 min in the dark with a solution of HQ silver reagents (Nanoprobes) at an equal ratio of red-blue-white. The coverslips were then washed three times in H2O and one time in 1% aqueous tannic acid for 5 min, followed by an H2O rinse. Next, the coverslips were reacted with a solution of reduced K4(FeCN)6 and 1% osmium tetroxide for 15 min, followed by two rinses in H2O. They were then subjected to a 5-min graded alcohol dehydration series of 50, 80, 95, and 100%, infiltrated with Spurr's resin, and polymerized at 60°C. The samples were then sectioned and examined using a Hitachi H7500 electron microscope equipped with a Hamamatsu digital camera (Advanced Microscopy Techniques Corp., Danvers, Mass.). The resulting images were digitally recorded.
Immunofluorescence and confocal microscopy.
Host cells, grown on glass coverslips in 12-well plates, were infected with E. intestinalis spores. When a majority of cells were infected, the coverslips were removed, fixed with acetone-methanol, and blocked with 1% FBS in PBS for 1 h at room temperature. MAbs (11B2 or 7G7) were diluted 1:500 in blocking solution (1% FBS; HyClone). After the coverslips were washed in PBS, fluorescein-conjugated goat anti-mouse immunoglobulin (1:500; Cappel, West Chester, Pa.) was added. The coverslips were mounted on glass slides with Vectashield (Vector) and viewed with either a Zeiss Axioplan fluorescence microscope or a Leica TCS-NT/SP confocal microscope. Controls included omission of primary antibody and staining of uninfected cells. The confocal images were magnified to ×100 with a zoom value of 2.7. Differential interference contrast images were collected at the same time as fluorescence images using the transmitted-light detector. The images were processed using Leica TCS-NT/SP software (version 1.6.551) and Photoshop version 3.0 (Adobe Systems).
Synchronized infection and RT-PCR.
Twelve-well tissue culture plates were seeded with 105 host cells. After 24 h, 6 × 107 spores were added per well for 3 h. The plates were extensively washed, and fresh medium was added. Infected cells were harvested at 12, 24, and 72 h postinfection. Three wells per time point were used in total RNA isolation (RNA STAT-60; Tel-Test, Inc., Friendswood, Tex.) following the manufacturer's protocol. The RNA was treated with DNase I, and reverse transcriptase (RT) PCR (Life Technologies) was performed following the manufacturer's description. The primers used for amplification were as follows: E. intestinalis beta-tubulin, 5′-GTTGACTGCAAGCTTCCTAAG and 5′-CAGAGTCGAGTGACTGCTTG (amplicon, 397 bp); swp1, 5′-GTTCCTTCTGTACCCTCATC and 5′-TCAGGATTCAACCCAGTCTTC (amplicon, 692 bp); and swp2, 5-AGTGACCGCTGTAGAAATCA and 5-TCAGGATTCAACCCAGTCTTC (amplicon, 371 bp). Controls included PCR amplification without prior RT elongation.
Mouse infection model.
Gamma interferon receptor null (IFN-γR−) mice (129-Ifngrtm1) and wild-type mice (129S3/SvImJ) (Jackson Laboratory, Bar Harbor, Maine) were infected orally with 2 × 108 E. intestinalis spores in H2O as previously described (13). Pooled infected or control sera were collected from each mouse on days 15, 29, 45, and 60 postinfection and used at 1:500 in Western blot analysis.
Nucleotide sequence accession numbers.
The sequence data for swp1 and swp2 have been submitted to the DDJ, EMBL, and GenBank databases under the accession numbers AF355749 and AF355750.
RESULTS
Isolation of two closely related cysteine-rich genes.
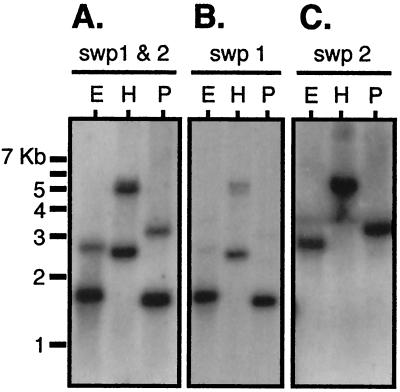
To study the molecular aspects of microsporidial infection and propagation, a previously constructed subtracted cDNA library was screened for parasite-specific genes (15). Clone 46 was isolated repeatedly in independent screenings of the subtracted cDNA library. Southern blot analysis of genomic DNA from infected cultures using three different enzymes and a clone 46 fragment as a probe unexpectedly showed two bands, suggesting a second related gene (Fig. 1A). The second gene (clone 2.8) was isolated from a conventional cDNA library using a fragment of clone 46 as a probe. Using DNA probes unique to either clone 46 or clone 2.8 in Southern blot analysis, the two hybridizing bands in Fig. 1A were accounted for (Fig. 1B and C).
FIG. 1.
Southern blot analysis of in vitro-infected host cell genomic DNA using probes that are either common or specific for swp1 or swp2. DNA probed with a purified PCR fragment common to both swp1 and swp2 (A); probed with a PCR fragment specific for swp1 (B); and probed with a PCR fragment specific for swp2 (C). Genomic DNA was restriction digested with either EcoRI (lanes E), HindIII (lanes H), or PstI (lanes P) in triplicate. Uninfected host cell genomic DNA did not hybridize with any of these probes (data not shown).
Localization of the protein gene products of clones 46 and 2.8 in infected host cells.
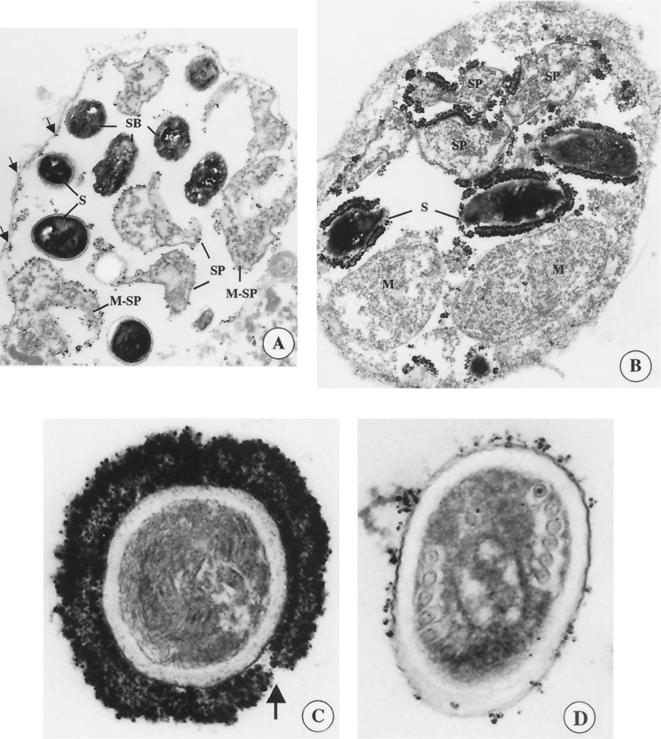
IEM was employed to determine the stage specificity and cellular location of the proteins encoded by clones 46 and 2.8. The insert of clone 46 was reisolated from the conventional cDNA expression library and, along with clone 2.8, was used to screen a battery of MAbs reactive with E. intestinalis and Encephalitozoon hellem (22). MAb 11B2 reacted specifically with clone 46 (SWP1), while MAb 7G7 reacted specifically with clone 2.8 (SWP2). MAb 11B2 localized swp1 to the thickened membranes of cells in transition from meronts to sporonts (Fig. 2A). Binding of MAb 11B2 to the cell surface diminished as the parasites developed, but some staining was evident on the surfaces of mature spores. Staining seen on the inside of the PV may represent residual protein from developing meronts that were attached to the PV but had since migrated to the lumen. In mature spores that were released from the PV, SWP1 was clearly located in the exospore region of the spore wall and not the endospore or plasma membrane (Fig. 2D).
FIG. 2.
IEM of E. intestinalis-infected host cells at different developmental stages using either MAb 11B2 or 7G7 followed by a fluoronanogold anti-mouse antibody and silver-staining enhancement. (A) PV reacted with MAb 11B2. The arrows indicate residual staining along the inside of the PV lining. (B) PV reacted with MAb 7G7. (C) Cross sections of mature spores that were released from the PV reacted with MAb 7G7. The arrow indicates a gap in the exospore staining. (D) Cross sections of mature spores that have been released from the PV reacted with MAb 11B2. PVs contain cells at different stages of development: meronts (M), sporoblasts (SB), sporonts (SP), and mature spores (S). Also shown are cells that do not have a completely defined dense membrane and are considered to be in transition from meronts into sporonts (M-SP).
In contrast to the reactivity of MAb 11B2, the 7G7 MAb did not react with developing sporonts (Fig. 2B). However, well-defined sporonts, which had a contiguous dense membrane and were located in the vacuolar lumen, showed heavy staining along the outer membrane. Unlike SWP1, whose expression diminished with spore development, SWP2 was expressed in mature spores inside the PV. The staining appeared to be in the “clear zone” that occurs between the sporoblast thickened spore membrane and the fibrillar matrix that is unique to E. intestinalis (7). In addition, individual spores released from the PV of an infected cell showed the same intense staining in the exospore region of the spore wall (Fig. 2C). Moreover, as with SWP1, SWP2 was not located in the endospore or plasma membrane. In spores that were released from the PV, MAb 7G7 staining was not always uniformly intense around the spore. A gap in the staining was occasionally observed (Fig. 2C) and may represent the area near the anchoring disk and polar filament.
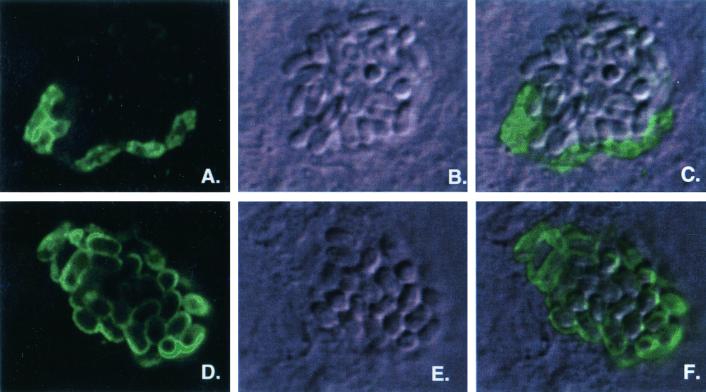
These data show that SWP1 was expressed in the transition stage between merogamy and sporogamy and that SWP2 was expressed in clearly defined sporonts and spores, suggesting a difference in expression of SWP1 and SWP2. To confirm differential expression, immunofluorescence assays (IFA) were performed on in vitro-infected host cells using the anti-SWP1 and anti-SWP2 MAbs (11B2 and 7G7, respectively). Five to 7 days postinfection, 11B2 (anti-SWP1) stained the immature cells lining the PV much more intensely than the well-formed, mature spores that reside in the lumen (Fig. 3A to C). The location, elongated shape, and lack of structural definition suggest that these cells are multinucleated immature cells that are developing a uniformly dense thick membrane (transitioning sporonts). In contrast, 7G7 (anti-SWP2 IFA showed that SWP2 was found on structurally well-defined, ovoid spores representing later developmental stages (Fig. 3D to F). These data confirm that SWP1 is expressed in an earlier developmental stage than SWP2.
FIG. 3.
Immunofluorescence and confocal imagery of in vitro-infected host cells using MAbs specific for SWP1 (11B2) and SWP2 (7G7). (A, B, and C) Localization of SWP1. (A) Immunofluorescent staining using the SWP1-specific MAb 11B2. (B) Differential interference contrast image (Nomarski) of the same microscopic field. (C) Layering of images in panels A and B. ( D, E, and F) Localization of SWP2. (D) Immunofluorescent staining using the SWP2 MAb 7G7. (E) Differential interference contrast image (Nomarski) of the same microscopic field. (F) Layering of images in panels D and E. All images are about 16 μm wide.
mRNA expression of SWP1 and SWP2 in a timed infection.
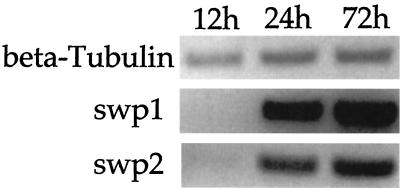
The IEM and IFA data suggested that SWP1 may be expressed earlier in spore development than SWP2. To correlate protein expression with mRNA expression, RT-PCR was performed on mRNA from “synchronized” infected host cells (Fig. 4). Although infection of host cells was performed so that those cells infected would be infected at the same time, Encephalitozoon species develop in an asynchronized fashion (7). This results in several developmental stages existing within a single PV and complicates the determination of stage-specific expression, but by 48 h postinfection, mature spores are formed (23). RT-PCR was performed using RNA purified from synchronized infected host cells at 12, 24, and 72 h postinfection. Transcripts for both swp1 and swp2 were first detected 24 h postinfection and increased with time; however, the level of swp1 mRNA was higher than that of swp2 mRNA at 24 h. This contrasted with the expression of beta-tubulin, which was first detected 12 h postinfection and increased slightly over time. While differences between the RNA stabilities of swp1 and swp2 could not be ruled out, these data suggest that the swp1 gene is transcribed at a higher level than the swp2 gene early in infection.
FIG. 4.
Examination of differentially expressed swp1 and swp2 transcripts by RT-PCR. RT-PCR was performed using mRNA isolated 12, 24, and 72 h postinfection and primers specific for E. intestinalis beta-tubulin, swp1, or swp2. The data are presented as an inverse image of an ethidium bromide-stained gel following equal-volume loading and electrophoresis of the products. Control PCR without RT yielded no products.
Sequence analysis of swp1 and swp2.
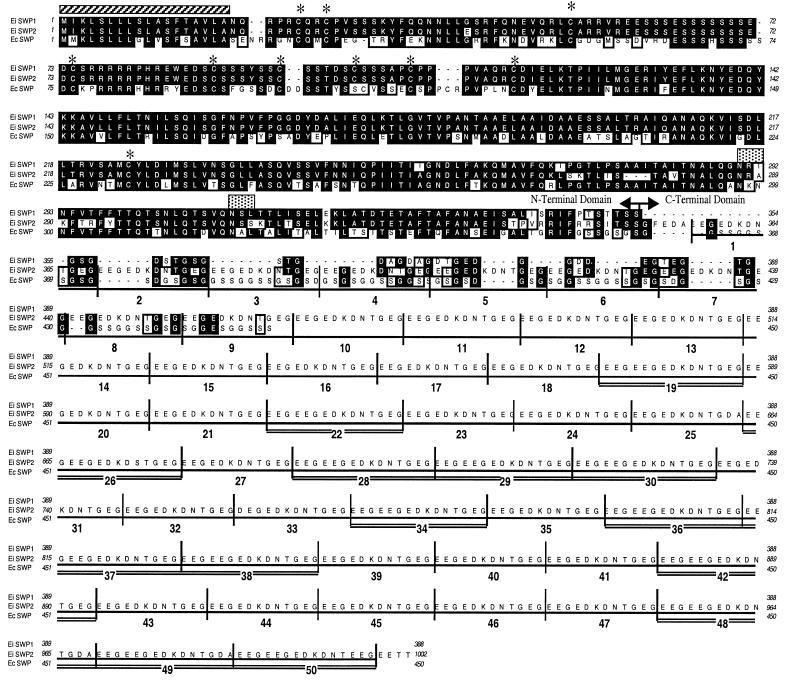
Inverse PCR and genomic sequence analyses were used to obtain the complete coding ORFs and flanking sequences of the swp1 and swp2 genes. These analyses showed that swp1 and swp2 are related genes that encode proteins of 388 and 1,002 amino acids, respectively (Fig. 5), and that both proteins have a predicted 18-amino-acid signal sequence at the amino (N) terminus. No transmembrane domains were found, suggesting that these proteins may be secreted. When the predicted amino acid sequences were aligned, two domains were identified based on sequence identity and length. The N-terminal domains of SWP1 (positions 1 to 354) and SWP2 (1 to 351) are 92% identical at the amino acid level. Comparison of the SWP1 and SWP2 N-terminal domains with that of the previously identified E. cuniculi SWP showed that the E. cuniculi SWP is 65 and 61% identical to E. intestinalis SWP1 and SWP2, respectively (5). In addition, 10 cysteine residues in this domain are conserved, suggesting similar secondary structures. Tyrosine phosphorylation sites are also conserved in these domains (positions 136 to 142); however, studies were inconclusive as to whether these sites are phosphorylated. SWP1 and SWP2 have N-linked glycosylation sites, but they are in slightly different locations (Fig. 5).
FIG. 5.
Predictive amino acid sequence alignment of SWPs from E. intestinalis and E. cuniculi. The predicted amino acid sequences of E. intestinalis (EI) swp1 and swp2 and the E. cuniculi (EC) SWP are compared. Identical amino acids are indicated by reverse shading, while similarities are boxed. Dashes represent gaps introduced to maximize homology. The arrows denote separation of the SWPs into two domains. The hatched box indicates the predicted signal sequence. The asterisks denote 10 conserved cysteine residues. N-glycosylation sites for swp1 (positions 290 to 292) and swp2 (positions 308 to 310) are indicated by stippled boxes. The C-terminal domain of swp2 contains a 12- or 15-amino-acid repeat that is bracketed and numbered. The repeats containing 15 amino acids are double underlined. The predicted amino acid sequence for E. cuniculi was obtained from GenBank (accession number AJ133745).
The two SWPs have divergent C-terminal domains that have several distinct features (Fig. 5). For example, SWP1 has a glycosaminoglycan attachment site at position 356 but lacks the required acidic residue immediately upstream, which may render the site nonfunctional (6). An unusual feature of the SWP2 C-terminal domain is a repeat region where a 12- or 15-amino-acid motif is duplicated 50 times. The amino acid sequence is conserved in most of the repeats, except where a missense mutation occurs (repeats 25, 26, 33, 48, 49, and 50).
The C termini of both proteins consist mainly of amino acids with either uncharged polar side chains or very acidic or basic polar side chains. These relatively hydrophilic amino acids are usually positioned externally in proteins, suggesting that the C-terminal domains of these proteins are externally exposed on the molecule. This is more apparent in swp2, where fully two-thirds of the 651 amino acids in this domain are considered structurally external. These repeated sequences may represent a unique external structural repeating motif that requires further functional analysis.
The flanking regions of the swp1 and swp2 genes were amplified by inverse PCR, and their sequences were analyzed (Fig. 6). The 5′ flanking regions of swp1 and swp2 (−1 to −61) are 75% AT rich and completely identical, suggesting that these genes are transcriptionally regulated in the same fashion. The transcription start site of swp1, mapped by 5′ rapid amplification of cDNA ends, was variable, with initiation at the −2, −3, −4, or −6 position relative to the translational start codon. Nevertheless, it remained within the AT-rich patch immediately upstream of the ATG, a common transcriptional feature among protozoan genes (27). In addition, an apparent TATA box was identified approximately 25 bases upstream from the transcriptional start site, as is typical for most eukaryotic genes. Further computational sequence analysis of this region did not reveal any motifs similar to known transcriptional regulatory elements. Sequence analysis of the 3′ flanking regions revealed putative nonconsensus polyadenylation sites for both swp1 and swp2. The site for swp1 differed from the AATAAA consensus by 1 bp, while the putative swp2 polyadenylation site contained the rare alternative hexanucleotide sequence known to be active in eukaryotes (25). Further studies are required to determine if these sites are utilized in vivo.
FIG. 6.
Nucleotide sequence comparison of the 5′ and 3′ flanking regions of swp1 and swp2 ORFs. (A) Comparison of the 5′ flanking sequences of swp1 (top) and swp2 (bottom). The nucleotide numbering is relative to the translational start site (boxed). The boldface lettering indicates the putative eukaryotic TATA box promoter. The putative transcriptional start sites for swp1 are indicated by overlines. (B) Comparison of the 3′ flanking sequences of swp1 (top) and swp2 (bottom). The nucleotide numbering of the 3′ flanking regions begins at the termination codon (boxed). The boldface letters indicate predicted polyadenylation sites. The asterisks denote sequence identity.
SWP1 and SWP2 are glycosylated and complexed.
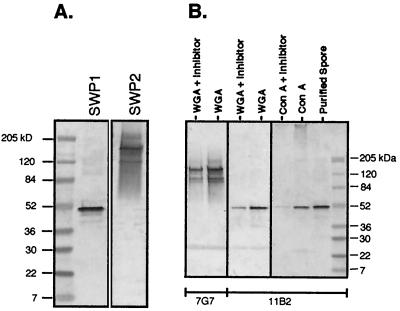
To characterize the SWP1 and SWP2 proteins, Western blot analyses of in vitro-purified spore proteins were performed (Fig. 7A). Western blot analysis using MAb 11B2 (anti-SWP1) detected a protein of about 50 kDa, which differs greatly from the 41 kDa estimated from the swp1 ORF. In addition, Western blot analysis using MAb 7G7 (anti-SWP2) identified a protein of 150 kDa, which is also larger than the estimated size of SWP2 (107 kDa). To determine if the proteins are glycosylated at the sites identified by computational sequence analysis, lysates from host cells infected with E. intestinalis were reacted with immobilized lectins (ConA and WGA). Western blot analysis of the bound proteins indicated that SWP1 and SWP2 were glycosylated and that they contain at least the core sugar residues of an N-linked oligosaccharide (Fig. 7B).
FIG. 7.
Western blot analyses of reduced E. intestinalis spore protein or infected cell lysate reacted with agarose-bound lectins and detected with either MAb 11B2 or 7G7. (A) Purified spores were reduced with 2-mercaptoethanol, run on an SDS-PAGE gel, and detected with MAbs to SWP1 (11B2) and SWP2 (7G7). (B) Cultured infected cell lysate was reacted with the agarose-bound lectin ConA or WGA with or without the inhibiting sugar (methyl-α-mannopyranoside for ConA; chitin hydrolysate for WGA).
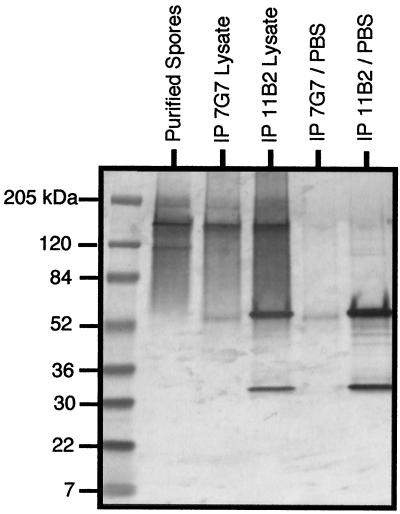
Further studies indicated that SWP1 and SWP2 form a protein complex in the spore wall. Western blots of denatured, nonreduced spore proteins detected with either MAb 11B2 (anti-SWP1) or MAb 7G7 (anti-SWP2) showed no product, suggesting that the protein(s) may be complexed and too large for migration into a polyacrylamide gel. However, when MAb 11B2 or 7G7 immunoprecipitation products were reduced and detected by Western blot analysis with MAb 7G7 (anti-SWP2), a 150-kDa band (SWP2) was detected, indicating that SWP1 and SWP2 are part of a protein complex (Fig. 8).
FIG. 8.
Immunoprecipitation analysis of protein lysates from infected host cells. Infected cell lysates were immunoprecipitated with MAb to SWP1 and protein A (IP 11B2 Lysate) or with MAb to SWP2 and protein A (IP 7G7 Lysate). Following SDS-PAGE, Western blotting detection was conducted using MAb 7G7 (anti-SWP2). Negative controls included immunoprecipitations using PBS instead of protein lysate (IP 7G7/PBS and IP 11B2/PBS). The detected bands in these lanes are the reduced antibody proteins. As a positive control for Western blotting with 7G7, purified spores were included.
SWP1 and SWP2 are immunogenic in a mouse model infection.
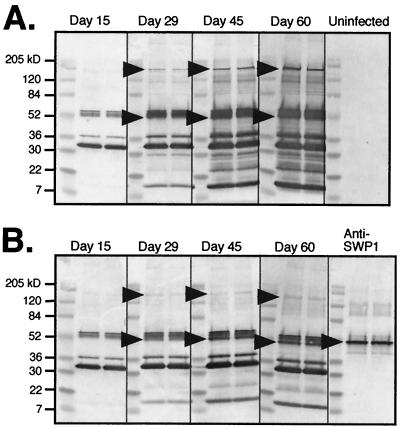
Because SWP1 and SWP2 are localized to the exospore region of the spore wall, the proteins are probably exposed to the host environment. Since it is well known that most immunocompetent individuals mount an immune response to microsporidia yet have no overt signs of disease (reviewed in reference 9), it was of interest to determine if the SWPs were involved in this process. Toward this end, a well-established mouse infection model system (1, 13) was employed to determine if the SWPs are immunogenic. IFN-γR− mice and wild-type control mice were infected in vivo with in vitro-isolated spores. Sera, collected from the mice at 15, 29, 45, and 60 days postinfection, were used in Western blot analysis of proteins from purified spores (Fig. 9). Both groups of mice mounted an antibody response to E. intestinalis within 15 days of infection. However, as the infection progressed, the IFN-γR− mice showed more intense reactions, as evidenced by the increase in the number of spore proteins recognized by their sera and the increased intensity of the banding pattern. Furthermore, IFN-γR− mice showed a more pronounced immune response to proteins of the same molecular mass as SWP1 and SWP2 (Fig. 9). Antibodies to proteins that were the same size as the SWPs were evident on day 29 postinfection, and the intensity of this response strengthened through day 60 of the infection. In addition, pooled sera from IFN-γR− mice (day 60) readily detected SWP1 and SWP2 from a lambda phage expression library (data not shown). These data indicate that SWP1 and SWP2 are immunogenic and suggest that the immune response toward these proteins may be, in part, responsible for the clearance of the organisms in immunocompetent individuals.
FIG. 9.
Western blot analysis of reduced E. intestinalis spore proteins detected with sera from in vivo-infected IFN-γR− mice and wild-type control mice. (A) Sera from six infected IFN-γR− mice were collected on days 15, 29, 45, and 60 postinfection, pooled at each time point, and reacted with reduced E. intestinalis spore proteins in duplicate. (B) Sera from six infected wild-type control mice were collected on days 15, 29, 45, and 60 postinfection, pooled at each time point, and reacted with reduced E. intestinalis spore proteins in duplicate. The arrowheads at ≈50 kDa indicate the expected size of SWP1, and the arrowheads at ≈150 kDa indicate the expected size of SWP2. SWP1 is the bottom band of the triple banding pattern seen at about 52 kDa. Controls included sera from uninfected mice and the anti-SWP1 MAb 11B2.
DISCUSSION
Two closely related cysteine-rich genes were isolated from E. intestinalis cDNA libraries. These genes encode proteins (SWP1 and SWP2) that localize to the exospores of mature spores. The remarkable sequence similarity between SWP1 and SWP2 is, for the most part, limited to the N-terminal region and is responsible for the two-band hybridizing pattern in Southern blot analysis. Although phylogenetic analysis of rRNA shows that E. intestinalis, E. hellem, and E. cuniculi are closely related (2, 17), Southern blot analyses of E. hellem and E. cuniculi show a single hybridizing band (5), suggesting that these species have only one SWP. Thus, despite E. intestinalis recently being reclassified in the genus Encephalitozoon (14), the identification of two SWPs in E. intestinalis supports the morphologic finding (7) that E. intestinalis is unique within the genus.
At the amino acid level, E. intestinalis SWP1 and SWP2 are considerably similar to the SWP of E. cuniculi (5). Most notable is the conservation of cysteine residues among these SWPs. While the E. cuniculi SWP has 11 cysteine residues and SWP1 and SWP2 have 10 each, the spacing is conserved, suggesting that these proteins have similar secondary structures and functions. Interestingly, immunoprecipitations show that E. intestinalis SWP1 and SWP2 form a protein complex. The fact that E. cuniculi has only one SWP with cysteine residues in similar locations suggests that other, unidentified proteins may bind to the SWPs of both species. Because the extent of the disulfide linkage between SWP1 and SWP2 is unknown, other proteins may participate in the E. intestinalis SWP complex.
The localization of SWP1 and SWP2 to the exospore region of mature spores suggests that they may be part of the matrix that confers rigidity and/or environmental protection. However, computational sequence analyses of SWP1 and SWP2 failed to reveal homology to any sequences in the databases. The 12- or 15-amino-acid repeated motif in the C-terminal domain of SWP2 is a novel element that may be important in protein interactions, but BLAST searches using the repeated sequence do not show any similarity to known protein binding motifs. Furthermore, little similarity exists between the repeated motifs of SWP2 and those of E. cuniculi SWP, except for repeat length (12 or 15 amino acids versus ≈17 amino acids, respectively) and glycine and aspartic acid in each of the repeats. The amino acids within the SWP2 repeated regions are hydrophilic residues, indicating that the C terminus of SWP2 may be externally exposed. Computational analyses of protein secondary structure for the SWP2 C-terminal domain were ambiguous, with one program predicting alpha-helical structure while another predicted a repeating turn. Nonetheless, the hydrophilicity and repeating nature of the SWP2 C terminus suggest that it contains a unique external repeating structure. Further analyses are required to determine the function(s) of this region.
In the development of mature spores, the transition from meront to sporont, sporogamy, involves the accumulation of an electron-dense material forming the thick membrane (7). IEM and IFA studies show that SWP1 is located on the surfaces of cells undergoing sporogamy. The localization of SWP1 to the surfaces of spores in the PV diminished as they entered the final stages of maturation, when the organelles become organized and the endospore develops; however, Western blot analysis showed that SWP1 was abundant in mature spores. It is possible that as spores mature, the SWP1 epitope recognized by the antibody becomes inaccessible. Furthermore, the fully formed sporont in the PV lumen was the earliest parasite stage in which SWP2 was detected. With the appearance of SWP2 on the sporont surface, SWP1 was no longer detectable, suggesting that expression of SWP2 blocks the 11B2 SWP1 epitope. This theory is supported by immunoprecipitation of infected cell lysates, which show that SWP1 and SWP2 form a protein complex. Thus, these data indicate that SWP1 is expressed on the surfaces of developing sporonts, and as SWP2 is expressed on the surfaces of fully formed sporonts, a protein complex forms that becomes part of the exospore, which could contribute to the rigidity and environmental protection exhibited by mature spores.
Ultrastructural studies of E. hellem show that three layers form the exospore region of the spore wall: an outer electron-dense layer, an electron-lucent intermediate layer, and an inner fibrous layer (4). Freeze-fracture analysis of mature spores reveals that the outer layer of the exospore consists of closely packed protruding spikes. Although similar studies have not been performed with E. intestinalis, the dense coat of material labeled in our IEM studies using anti-SWP2 MAb may represent this outer layer, and SWP2 may be a major component of the spiny layer of the exospore in E. intestinalis. Because the C-terminal domain of SWP2 may contain an external repeating structure, the extensive structural repeat motif of SWP2 may be part of these spiny projections. Moreover, if SWP1 is complexed with SWP2 in this spiny outer layer, the epitopes recognized by MAbs to SWP1 may be buried, as suggested by our studies.
It is well documented that microsporidiosis is problematic for immunocompromised animals and that immunocompetent animals elicit antibody responses that confer resistance to infection (8, 11, 16). Since SWP1 and SWP2 are expressed in the spore wall exospore, they should be exposed to the host cell environment. Therefore, antibody responses to SWP1 and SWP2 were addressed in an established in vivo mouse model using IFN-γR− and wild-type control mice (1). Interestingly, immune sera from both control and IFN-γR− mice recognized a series of E. intestinalis proteins. This differs from the findings of El Fakhry et al. (13), which showed an increase in immunoglobulin G levels in IFN-γR− mice but no increase in antibody levels in controls. In our study, by day 29 postinfection, serum antibody levels in IFN-γR− mice were sufficient to detect several immunogenic proteins, including SWP1 and SWP2. Control mice also mounted a substantial immune response to E. intestinalis, but the levels of reactivity, judged by the banding pattern and intensity, were less than those of IFN-γR− mice. While the natures of the other immunogenic proteins await analysis, SWP1 and SWP2 were identified as immunogenic in both IFN-γR− and control mice.
In summary, the two SWPs, SWP1 and SWP2, of the microsporidian E. intestinalis are differentially expressed and localized to the exospores of developing and mature spores. SWP1 and SWP2 show remarkable similarity at both the DNA and protein levels, but SWP2 has a unique, extensive repeat region in its C terminus that may be important for the structural formation of highly environmentally resistant spores. Additional studies will determine the structures and functions of these repeated motifs, as well as those of the SWP1 and SWP2 proteins themselves. Immunoprecipitation and Western blot analyses indicate that SWP1 and SWP2 are components of a protein complex and that they are immunogenic in mouse infections, suggesting that they could be, in part, responsible for the immune resistance of immunocompetent individuals. Not only will the characterization of differentially expressed SWPs be of benefit in delineating microsporidial developmental stages, but having specific MAbs for SWPs may allow development of antibody-based immunodiagnostics. Thus, the identification of two SWPs in E. intestinalis will not only improve our understanding of the biological and immunological interactions involved in microsporidiosis but may also advance the diagnosis and epidemiological study of microsporidiosis caused by E. intestinalis.
ACKNOWLEDGMENTS
We thank Owen Schwartz for assistance in the confocal and differential interference contrast imaging. We are also grateful to Sara Davis-Hayman for critical review of the manuscript.
REFERENCES
- 1.Achbarou A, Ombrouck C, Gneragbe T, Charlotte F, Renia L, Desportes-Livage I, Mazier D. Experimental model for human intestinal microsporidiosis in interferon gamma receptor knockout mice infected by Encephalitozoon intestinalis. Parasite Immunol. 1996;18:387–392. doi: 10.1046/j.1365-3024.1996.d01-128.x. [DOI] [PubMed] [Google Scholar]
- 2.Baker M D, Vossbrinck C R, Didier E S, Maddox J V, Shadduck J A. Small subunit ribosomal DNA phylogeny of various microsporidia with emphasis on AIDS related forms. J Eukaryot Microbiol. 1995;42:564–570. doi: 10.1111/j.1550-7408.1995.tb05906.x. [DOI] [PubMed] [Google Scholar]
- 3.Beaugerie L, Carbonnel F, Carrat F, Rached A A, Maslo C, Gendre J P, Rozenbaum W, Cosnes J. Factors of weight loss in patients with HIV and chronic diarrhea. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:34–39. doi: 10.1097/00042560-199809010-00005. [DOI] [PubMed] [Google Scholar]
- 4.Bigliardi E, Selmi M G, Lupetti P, Corona S, Gatti S, Scaglia M, Sacchi L. Microsporidian spore wall: ultrastructural findings on Encephalitozoon hellem exospore. J Eukaryot Microbiol. 1996;43:181–186. doi: 10.1111/j.1550-7408.1996.tb01388.x. [DOI] [PubMed] [Google Scholar]
- 5.Bohne W, Ferguson D J, Kohler K, Gross U. Developmental expression of a tandemly repeated glycine- and serine-rich spore wall protein in the microsporidian pathogen Encephalitozoon cuniculi. Infect Immun. 2000;68:2268–2275. doi: 10.1128/iai.68.4.2268-2275.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourdon M A, Krusius T, Campbell S, Schwartz N B, Ruoslahti E. Identification and synthesis of a recognition signal for the attachment of glycosaminoglycans to proteins. Proc Natl Acad Sci USA. 1987;84:3194–3198. doi: 10.1073/pnas.84.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cali A, Kotler D P, Orenstein J M. Septata intestinalis N. G., N. Sp., an intestinal microsporidian associated with chronic diarrhea and dissemination in AIDS patients. J Eukaryot Microbiol. 1993;40:101–112. doi: 10.1111/j.1550-7408.1993.tb04889.x. [DOI] [PubMed] [Google Scholar]
- 8.Canning E U, Hollister W S. Enterocytozoon bieneusi (Microspora): prevalence and pathogenicity in AIDS patients. Trans R Soc Trop Med Hyg. 1990;84:181–186. doi: 10.1016/0035-9203(90)90247-c. [DOI] [PubMed] [Google Scholar]
- 9.Didier E, Bessinger G T. Host-parasite relationships in microsporidiosis: animal models and immunology. In: Wittner M, Weiss L M, editors. The microsporidia and microsporidiosis. Washington, D.C.: ASM Press; 1999. pp. 225–257. [Google Scholar]
- 10.Didier E S, Snowden K F, Shadduck J A. Biology of microsporidian species infecting mammals. Adv Parasitol. 1998;40:283–320. doi: 10.1016/s0065-308x(08)60125-6. [DOI] [PubMed] [Google Scholar]
- 11.Didier E S, Varner P W, Didier P J, Aldras A M, Millichamp M, Murphey-Corb N J, Bohm R, Shadduck J A. Experimental microsporidiosis in immunocompetent and immunodeficient mice and monkeys. Folia Parasitol. 1994;41:1–11. [PubMed] [Google Scholar]
- 12.Didier P J, Didier E S, Orenstein J M, Shadduck J A. Fine structure of a new human microsporidian, Encephalitozoon hellem, in culture. J Protozool. 1991;38:502–507. doi: 10.1111/j.1550-7408.1991.tb04824.x. [DOI] [PubMed] [Google Scholar]
- 13.El Fakhry Y, Achbarou A, Desportes-Livage I, Mazier D. Encephalitozoon intestinalis: humoral responses in interferon-gamma receptor knockout mice infected with a microsporidium pathogenic in AIDS patients. Exp Parasitol. 1998;89:113–121. doi: 10.1006/expr.1998.4267. [DOI] [PubMed] [Google Scholar]
- 14.Hartskeerl R A, Van Gool T, Schuitema A R, Didier E A, Terpstra W J. Genetic and immunological characterization of the microsporidian Septata intestinalis Cali, Kotler and Orenstein, 1993: reclassification to Encephalitozoon intestinalis. Parasitology. 1995;110:277–285. doi: 10.1017/s0031182000080860. [DOI] [PubMed] [Google Scholar]
- 15.Hayman J R, Nash T E. Isolating expressed microsporidial genes using a cDNA subtractive hybridization approach. J Eukaryot Microbiol. 1999;46:21S–24S. [PubMed] [Google Scholar]
- 16.Hermanek J, Koudela B, Kucerova Z, Ditrich O, Travnicek J. Prophylactic and therapeutic immune reconstitution of SCID mice infected with Encephalitozoon cuniculi. Folia Parasitol. 1993;40:287–291. [PubMed] [Google Scholar]
- 17.Keeling P J, Fast N M, McFadden G I. Evolutionary relationship between translation initiation factor eIF-2gamma and selenocysteine-specific elongation factor SELB: change of function in translation factors. J Mol Evol. 1998;47:649–655. doi: 10.1007/pl00006422. [DOI] [PubMed] [Google Scholar]
- 18.Keohane E M, Orr G A, Takvorian P M, Cali A, Tanowitz H B, Wittner M, Weiss L M. Purification and characterization of a microsporidian polar tube protein. Mol Biochem Parasitol. 1996;79:255–259. doi: 10.1016/0166-6851(96)02666-7. [DOI] [PubMed] [Google Scholar]
- 19.Koudela B, Kucerova S, Hudcovic T. Effect of low and high temperatures on infectivity of Encephalitozoon cuniculi spores suspended in water. Folia Parasitol. 1999;46:171–174. [PubMed] [Google Scholar]
- 20.Lambl B B, Federman M, Pleskow D, Wanke C A. Malabsorption and wasting in AIDS patients with microsporidia and pathogen-negative diarrhea. AIDS. 1996;10:739–744. doi: 10.1097/00002030-199606001-00007. [DOI] [PubMed] [Google Scholar]
- 21.Lopez-Velez R, Turrientes M C, Garron C, Montilla P, Navajas R, Fenoy S, del Aguila C. Microsporidiosis in travelers with diarrhea from the tropics. J Travel Med. 1999;6:223–227. doi: 10.1111/j.1708-8305.1999.tb00522.x. [DOI] [PubMed] [Google Scholar]
- 22.Luján H D, Conrad J T, Clark C G, Touz M C, Delbac F, Vivares C P, Nash T E. Detection of microsporidia spore-specific antigens by monoclonal antibodies. Hybridoma. 1998;17:237–243. doi: 10.1089/hyb.1998.17.237. [DOI] [PubMed] [Google Scholar]
- 23.Pakes S P, Shadduck J A, Cali A. Fine structure of Encephalitozoon cuniculi from rabbits, mice and hamsters. J Protozool. 1975;22:481–488. doi: 10.1111/j.1550-7408.1975.tb05213.x. [DOI] [PubMed] [Google Scholar]
- 24.Prigneau O, Achbarou A, Bouladoux N, Mazier D, Desportes-Livage I. Identification of proteins in Encephalitozoon intestinalis, a microsporidian pathogen of immunocompromised humans: an immunoblotting and immunocytochemical study. J Eukaryot Microbiol. 2000;47:48–56. doi: 10.1111/j.1550-7408.2000.tb00010.x. [DOI] [PubMed] [Google Scholar]
- 25.Proudfoot N J, Brownlee G G. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Seeber F. Consensus sequence of translational initiation sites from Toxoplasma gondii genes. Parasitol Res. 1997;83:309–311. doi: 10.1007/s004360050254. [DOI] [PubMed] [Google Scholar]
- 28.Shadduck J A, Polley M B. Some factors influencing the in vitro infectivity and replication of Encephalitozoon cuniculi. J Protozool. 1978;25:491–496. doi: 10.1111/j.1550-7408.1978.tb04174.x. [DOI] [PubMed] [Google Scholar]
- 29.Svenungsson B, Capraru T, Evengard B, Larsson R, Lebbad M. Intestinal microsporidiosis in a HIV-seronegative patient. Scand J Infect Dis. 1998;30:314–316. doi: 10.1080/00365549850161043. [DOI] [PubMed] [Google Scholar]
- 30.Undeen A H, Vander Meer R K. Microsporidian intrasporal sugars and their role in germination. J Invertebr Pathol. 1999;73:294–302. doi: 10.1006/jipa.1998.4834. [DOI] [PubMed] [Google Scholar]
- 31.Vavra J, Larsson J I R. Structure of the microsporidia. In: Wittner M, Weiss L M, editors. The microsporidia and microsporidiosis. Washington, D.C.: ASM Press; 1999. pp. 7–84. [Google Scholar]
- 32.Visvesvara G S, Belloso M, Moura H, Da Silva A J, Moura I N, Leitch G J, Schwartz D A, Chevez-Barrios P, Wallace S, Pieniazek N J, Goosey J D. Isolation of Nosema algerae from the cornea of an immunocompetent patient. J Eukaryot Microbiol. 1999;46:10S. [PubMed] [Google Scholar]
- 33.Weidner E. Interactions between Encephalitozoon cuniculi and macrophages. Parasitophorous vacuole growth and the absence of lysosomal fusion. Z Parasitenkd. 1975;47:1–9. doi: 10.1007/BF00418060. [DOI] [PubMed] [Google Scholar]