Summary
Background
Post-acute COVID-19 syndrome (PACS) is linked to severe organ damage. The identification and stratification of at-risk SARS-CoV-2 infected individuals is vital to providing appropriate care. This exploratory study looks for a potential liquid biopsy signal for PACS using both manual and machine learning approaches.
Methods
Using a high definition single cell assay (HDSCA) workflow for liquid biopsy, we analysed 100 Post-COVID patients and 19 pre-pandemic normal donor (ND) controls. Within our patient cohort, 73 had received at least 1 dose of vaccination prior to SARS-CoV-2 infection. We stratified the COVID patients into 25 asymptomatic, 22 symptomatic COVID-19 but not suspected for PACS and 53 PACS suspected. All COVID-19 patients investigated in this study were diagnosed between April 2020 and January 2022 with a median 243 days (range 16–669) from diagnosis to their blood draw. We did a histopathological examination of rare events in the peripheral blood and used a machine learning model to evaluate predictors of PACS.
Findings
The manual classification found rare cellular and acellular events consistent with features of endothelial cells and platelet structures in the PACS-suspected cohort. The three categories encompassing the hypothesised events were observed at a significantly higher incidence in the PACS-suspected cohort compared to the ND (p-value < 0.05). The machine learning classifier performed well when separating the NDs from Post-COVID with an accuracy of 90.1%, but poorly when separating the patients suspected and not suspected of PACS with an accuracy of 58.7%.
Interpretation
Both the manual and the machine learning model found differences in the Post-COVID cohort and the NDs, suggesting the existence of a liquid biopsy signal after active SARS-CoV-2 infection. More research is needed to stratify PACS and its subsyndromes.
Funding
This work was funded in whole or in part by Fulgent Genetics, Kathy and Richard Leventhal and Vassiliadis Research Fund. This work was also supported by the National Cancer InstituteU54CA260591.
Keywords: SARS-CoV-2, COVID-19, Post-acute COVID-19 syndrome (PACS), Post-COVID sequelae, Long COVID, Liquid biopsy
Research in context.
Evidence before this study
We searched PubMed for publications on post-acute COVID-19 syndrome, long COVID diagnosis criteria, post-COVID biomarkers and the use of liquid biopsy in COVID-19 and post-COVID-19 syndromes from April 2020 to May 2021. We retrieved studies using liquid biopsy in actively infected SARS-CoV-2 patients, finding elevated levels of endothelial cells, as well as studies documenting post-COVID sequelae for hospitalised COVID-19 patients up to 6 months. To this day, post-COVID-19 syndrome or long COVID is still an umbrella term for a wide variety of different symptoms. The currently available guidelines recommend a comprehensive, rule-out assessment with blood tests as conditional to the patient's symptoms.
Added value of this study
In this exploratory study, we aimed to explore a potential liquid biopsy biomarker for post-COVID-19 syndrome patients in comparison to a pre-pandemic control, using both manual and machine learning methods. This study motivates the potential of minimally invasive liquid biopsy to evaluate persons post-infection with SARS-CoV-2 and/or persons with post-COVID-19 syndrome.
Implications of all the available evidence
Currently, post-COVID-19 syndrome has many theories, such as persistent vasculature damage, neurological damage, and immune shifts. There are well over 200 symptoms ascribed to post-COVID sequelae, ranging from cardiovascular to psychological. As the world reaches half a billion confirmed SARS-CoV-2 infections, the need to determine physiological injury becomes more important to provide appropriate care. Liquid biopsy is a minimally invasive blood test that has been used to identify biomarkers for myocardial infarction and cancer. Using both manual data reduction of rare events and a machine learning patient classifier model, we have been able to find differences in our post-COVID cohort and the pre-pandemic controls. This suggests that there is a liquid biopsy signal in the peripheral blood that could be used to diagnose and stratify post-COVID conditions. A larger cohort is needed to validate our findings.
Introduction
Early in the COVID-19 pandemic, post-infection sequelae were considered the consequence of severe disease and hospitalisation.1 A large follow-up study in China showed the more severe the disease, the higher the risk of lung diffusion impairment and abnormal chest imaging with 49% of the 1279 hospitalised patients still reporting at least one symptom after 1 year.2 However, various studies, such as the one using health records from the U.S. Department of Veterans Affairs, found that there is an increased risk of post-acute sequelae for non-hospitalised COVID-19 survivors as well.3,4 Further research is needed to determine the risk profile for asymptomatic SARS-CoV-2 infection.5,6 To date, the global pandemic has resulted in over half a billion confirmed cases of SARS-CoV-2 infection,7,8 and the estimate of post-acute COVID-19 syndrome (PACS) ranges anywhere from 4 to 35%, depending on the demographic and disease severity of the study population.6,9, 10, 11, 12
Although the definition of PACS has evolved, it is most consistently defined as persistent and/or delayed symptoms after the SARS-CoV-2 infection that cannot be explained by an alternative diagnosis.13,14 Currently, the diagnostic criteria for PACS is all encompassing, ranging from cardiovascular to respiratory to neurological symptoms.14 Some common symptoms documented are fatigue, muscle weakness, cough, and shortness of breath, while neurological and psychological symptoms include headache, depression, cognitive impairment and brain fog.3,14,15
Now, the healthcare system must determine the best care for a syndrome spanning several organ systems.14,16 The interplay between physical and mental symptoms makes the ability to differentially diagnose the cause of and care for patients with PACS subsyndromes critical.5 The healthcare resources to care for patients with physiological injury vs purely psychological conditions are different, and the earlier a patient receives the appropriate care, the more likely for better outcomes. Targeted care will decrease burden to the healthcare system.
In this discovery study, we explore whether a comprehensive liquid biopsy can identify a disease specific profile, including injury specific biomarkers for PACS. Previously, we have used the 3rd generation high-definition single cell assay (HDSCA3.0) workflow to detect circulating tumour cells and circulating endothelial cells.17,18 We also explore whether a patient level classification model using liquid biopsy analytes can be used to separate individuals with a prior SARS-CoV-2 infection from those without.
Methods
Ethics
The study was conducted with the approval of Cedars-Sinai Office of Research Compliance and Quality Improvement (Los Angeles, CA; reference IRB STUDY00001316, approval date 2nd March 2021) and University of Southern California Institutional Review Boards (Los Angeles, CA; reference IRB HS-21-00556, approval date 7th December 2021). Each patient has provided written informed consent.
Study design
SeroNet-CORALE is a prospective repeated-measurement cohort study established on November 3, 2020.19,20 Participants are recruited from a large healthcare system in the Los Angeles catchment area. The Cedars Sinai Health System is located in the diverse metropolis of Los Angeles, serving a catchment area over 5.7 million (57.6% of the total Los Angeles County population).21 Adult (≥18 years) patients who had previously been infected with SARS-CoV-2 were approached from April 2020 and January 2022 for enrollment into the Post-COVID cohort. From this cohort, 75 individuals were previously symptomatic for COVID-19 or are currently being evaluated for PACS (Post-SYMP COVID). Within the Post-SYMP COVID group, 53 patients were being evaluated for PACS (PACS Suspected), and 22 were not suspected to have PACS (PACS Not Suspected). Patients were categorised as being evaluated for PACS if they self-reported symptoms after their SARS-CoV-2 infection through community enrollment or they were being evaluated and treated at the long COVID clinic or outpatient pulmonary rehabilitation at Cedars-Sinai Medical Center. The remaining 25 had no notable symptoms during or after their SARS-CoV-2 infection (Post-ASYMP COVID). All clinical and demographic data were taken from electronic health records and self-completed surveys. Patients underwent a single-draw phlebotomy of 8 mL of peripheral blood (PB) into Streck tubes between June 2021 and February 2022.
For a comparative analysis, a total of 19 PB samples were collected and analysed from normal donors (NDs) collected between March 2017 and February 2020. These are individuals with no known disease. The dates of collection suggest these individuals have not had a prior COVID infection. Samples were procured from the Scripps Normal Blood Donor Service.
Sample processing
PB samples were collected in 10 mL blood collection tubes (Cell-free DNA, Streck, La Vista, NE, USA) and processed by the Convergent Science Institute in Cancer (CSI-Cancer) at the University of Southern California within 48 h from the time of collection as previously described.22
Immunofluorescence staining and imaging
For analysis, each test consists of a minimum of 2 slides with the exception of 1 patient in which the low cellularity of the received sample generated only 1 slide. Slides are stained with immunofluorescent (IF) markers at room temperature using the IntelliPATH FLX™ autostainer (Biocare Medical LLC, Irvine, CA, USA) with negative and positive controls as previously described and validated (Supplemental Fig. S1).23,24 Briefly, samples were stained with the Landscape assay using 2.5 μg/mL of a mouse IgG1 anti-human CD31:Alexa Fluor® 647 mAb (clone: WM59, MCA1738A647, BioRad, Hercules, CA, USA, RRID:AB_322463), permeabilized with cold methanol, followed by an antibody cocktail consisting of mouse IgG1/Ig2a anti-human cytokeratins (CKs) 1, 4, 5, 6, 8, 10, 13, 18, and 19 (clones: C-11, PCK-26, CY-90, KS-1A3, M20, A53-B/A2, C2562, Sigma, St. Louis, MO, USA, RRID:AB_476839), mouse IgG1 anti-human CK 19 (clone: RCK108, GA61561-2, Dako, Carpinteria, CA, USA), mouse anti-human CD45:Alexa Fluor® 647 (clone: F10-89-4, MCA87A647, AbD Serotec, Raleigh, NC, USA, RRID:AB_324730) and rabbit IgG anti-human vimentin (Vim) (clone: D21H3, 9854BC, Cell Signaling, Danvers, MA, USA, RRID:AB_10829352). Lastly, slides were incubated with Alexa Fluor® 555 goat anti-mouse IgG1 antibody (A21127, Invitrogen, Carlsbad, CA, USA. RRID:AB_2535769) and 4′,6-diamidino-2-phenylindole (DAPI; D1306, ThermoFisher). Samples were mounted with a glycerol-based aqueous mounting media and imaged using automated high-throughput fluorescence scanning microscopy at 100× magnification generating 2304 frames per slide.
To determine whether the elongated acellular events consist of platelets, we stained an additional sample slide (Patient ID PACS-C0005) with a platelet specific marker, CD41. A single patient slide was stained with an alternative custom assay using pan-cytokeratin (CK) and CD45 antibodies with DAPI as described above with the addition of a polyclonal CD41 antibody (0.25 mg/mL, PA5-22307, ThermoFisher, Waltham, MA, USA, RRID:AB_11155042; Alexa Fluor® 488 goat anti-mouse IgG2b secondary antibody, 1:500, A21141, Invitrogen, Carlsbad, CA, RRID:AB_2535778). The sample was chosen due to the presence of rare acellular events when using the Landscape staining protocol. The CD41 stained sample had rare acellular events that were CD41 positive, CD45 negative and morphologically similar to the acellular events in the original stain (Supplemental Fig. S2). This is preliminary confirmation that a subgroup of acellular events is composed of platelets which we will refer to as a platelet structure. When comparing the samples, we manually enumerated 86.5 acellular CD45/CD31 positive events/mL and 120.7 acellular CD41 positive, CD45 negative events/mL from the Landscape stain and the CD41 stain, respectively.
Image processing and event identification
From the IF imaging, each event was segmented using the “EBImage” R package (EBImage_4.12.2), 761 morphometrics were obtained for cellular events (DAPI positive) and 749 for acellular events (DAPI negative). Rare cell candidates were detected using a custom computational methodology termed OCULAR (Outlier Clustering Unsupervised Learning Automated Report), as previously reported.23,24
Manual rare event classification and enumeration
We manually classified liquid biopsy samples on the molecular level. The rare cell and acellular event candidates are presented to trained analysts for further manual data reduction. The rare cells and acellular events are then classified according to their IF marker expression, termed the channel-type classification. For the rare cells, which are positive in the DAPI channel, there are 8 channel-type classifications stemming from the three remaining IF channels. For the acellular events, which are negative in the DAPI channel, there are 7 channel-type classifications.
Statistics
The sample size for this study was not predetermined, as it is exploratory in nature and there is no established standard. The Wilcoxon rank sum test was used to compare channel classification enumerations across cohorts.25 The Wilcoxon rank-sum test was chosen as it constitutes a non-parametric, rank test. Namely, no assumptions were made for the underlying distribution of the data. Furthermore, the rank-based nature of the test allows for robustness of the results with respect to outliers.
Patient classifier model
We built a machine learning model to classify liquid biopsy samples on the patient level. The input data used is the output of the OCULAR algorithm,23,24 which constitutes a collection of rare cells identified in the target slides combined with all the detected acellular events. We iteratively group these cellular and acellular events into clusters by mapping them onto a previously defined, morphometric based space and applying a hierarchical clustering algorithm. The number of events per mL of blood in each cluster is the input for a random forest classification model, which is trained to separate the samples based on clinical classification labels. A detailed description of the classifier model is provided in the Supplemental.
Classifier model training and evaluation
For each classification model iteration, the samples were split into training and test sets with a random stratified 80/20 split for train and test, respectively. Subsequently, class imbalance was addressed in the training samples by undersampling the majority class, such that the two classes have equal number of samples. The excess samples of the majority class were added to the test set. Undersampling the majority class yields limited and balanced training sets but maximises the amount of samples in the test set, providing a rigorous assessment of model generalisability. The model's performance on the test set was quantified by calculating the accuracy, sensitivity and specificity. The test set was withheld during the training process and was used only after the final model was built. The model hyperparameters were chosen algorithmically to fit the training set with no human intervention. We repeat these steps 10 times to evaluate the model, allowing for obtaining the average and confidence intervals for the classifier performance metrics.
Cohort pairs for classifier model
The classifier model was applied on seven cohort pairs. They consisted of comparisons of the Post-COVID cohort and its sub cohorts with the ND cohort: Post-COVID, Post-SYMP COVID, Post-ASYMP COVID, PACS Suspected, PACS Not Suspected vs ND. In addition, they included two comparisons within the sub cohorts of the Post-COVID cohort: Post-ASYMP COVID vs Post-SYMP COVID and PACS Not Suspected vs PACS Suspected.
Role of the funders
This work was funded in whole or in part by Fulgent Genetics (PK), Kathy and Richard Leventhal (PK), Vassiliadis Research Fund (PK) and the National Cancer Institute U54CA260591 (NN, AM, JCF). None of the sponsors and funders had any role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Results
Clinical demographics
A total of 100 patients investigated in this study were diagnosed with COVID-19 between April 2020 and January 2022 with PB collection occurring between June 2021 and March 2022. The median time from COVID-19 diagnosis to the blood draw was 232 days (range 16–669 days, IQR 202.3). The median age of the patients was 52 years (range 21–87 years, IQR 27.2) and 59% were female. The demographic and clinical characteristics of these patients are summarised in Table 1.
Table 1.
Baseline demographic and clinical characteristics of the post-COVID cohort (n = 100).
| Post-COVID (n = 100) | Post-symptomatic (n = 75) |
Post-asymptomatic (n = 25) | ||
|---|---|---|---|---|
| PACS suspected (n = 53) | PACS not suspected (n = 22) | |||
| Age, years | ||||
| Median (IQR) | 52 (27.2) | 56 (24) | 45 (28.3) | 50 (25.7) |
| Race, n (%) | ||||
| White | 58 (57) | 31 (58.5) | 13 (59.1) | 14 (56) |
| Black | 5 (5) | 3 (5.7) | 0 (0) | 2 (8) |
| Asian | 10 (10) | 4 (7.5) | 1 (4.5) | 5 (20) |
| Other | 21 (21) | 10 (18.9) | 7 (31.8) | 4 (16) |
| Not available | 6 (6) | 5 (9.4) | 1 (4.5) | 0 (0) |
| Ethnicity, n (%) | ||||
| Hispanic/Latino | 49 (49) | 27 (50.9) | 12 (54.5) | 10 (40) |
| Sex, n (%) | ||||
| Male | 39 (39) | 21 (39.6) | 9 (40.9) | 9 (36) |
| Female | 59 (59) | 30 (56.6) | 13 (59.1) | 16 (64) |
| Not available | 2 (2) | 2 (3.8) | 0 (0) | 0 (0) |
| BMI | ||||
| Median (IQR) | 26.9 (7.38) | 28.2 (9.07) | 26.3 (5.29) | 24.4 (4.99) |
| Time between COVID diagnosis to blood draw, days | ||||
| Median (IQR) | 232 (202.3) | 234 (108) | 278 (174.8) | 98 (297) |
| Infected after fully vaccinateda, n (%) | 27 (27) | 10 (18.9) | 2 (9.1) | 15 (60) |
| Hospitalised, n (%) | ||||
| Yes | 42 (42) | 36 (67.9) | 6 (27.3) | 0 (0) |
| Vaccinated, n (%) | ||||
| None | 11 (11) | 5 (11.3) | 4 (18.2) | 2 (8) |
| Fully vaccinated | 89 (89) | 48 (90.6) | 18 (81.8) | 23 (88) |
| Booster shot | 57 (57) | 30 (56.6) | 11 (50) | 16 (64) |
| Comorbidities, n (%) | ||||
| Respiratory | 3 (3) | 2 (3.8) | 0 (0) | 1 (4) |
| Cardiovascular | 12 (12) | 10 (18.9) | 2 (9.0) | 0 (0) |
| Psychological | 7 (7) | 6 (11.3) | 0 (0) | 1 (4) |
| Other | 13 (13) | 8 (15.1) | 4 (18.2) | 1 (4) |
| Not available | 65 (65) | 31 (58.5) | 13 (59.1) | 12 (48) |
| Cancer, n (%) | 45 (45) | 19 (35.8) | 15 (68.1) | 11 (44) |
| Haematological malignancy | 31 (31) | 10 (18.9) | 10 (45.4) | 11 (44) |
| Solid malignancy | 12 (12) | 7 (13.2) | 5 (22.7) | 0 (0) |
| Unknownb | 2 (2) | 2 (3.8) | 0 (0) | 0 (0) |
Fully vaccinated does not include the booster shot.
Cancer diagnosis documented but unknown type.
Within the Post-SYMP COVID cohort, the PACS Suspected cohort had a median age of 56 years (range 21–80, IQR 24) and 30 (56.6%) were female. There were also 36 patients (67.9%) who were hospitalised during their SARS-CoV-2 infection and, to date, 48 (90.6%) are fully vaccinated. Ten (18.9%) of the patients were infected after being fully vaccinated with an average of 128 days after their last dose. Of the PACS Suspected cohort, 19 (35.8%) patients had or currently have cancer with 10 (18.9%) haematological malignancies, 7 (13.2%) solid malignancies and 2 (3.8%) unknown cancer types.
The PACS Not Suspected cohort had a median age of 45 years (range 25–87, IQR 28.3) with 6 (27.3%) of the 22 patients having been hospitalised. Two (9.1%) of the patients were infected after being fully vaccinated with an average of 118 days after their last dose. Of these, 15 (68.1%) patients had or currently have cancer with 10 (45.4%) haematological malignancies and 5 (22.7%) solid malignancies.
For the Post-ASYMP COVID cohort, the COVID-19 diagnosis dates range from April 2020 to January 2022. The median time from COVID-19 diagnosis to blood draw was 98 days (range 25–669 days, IQR 297). Of these 25, 23 (88%) were fully vaccinated, and 15 (60%) of the patients were infected after being fully vaccinated with an average of 269 days after their last dose. 11 (44%) patients had or currently have cancer with all being haematological malignancies.
Manual classification of rare events
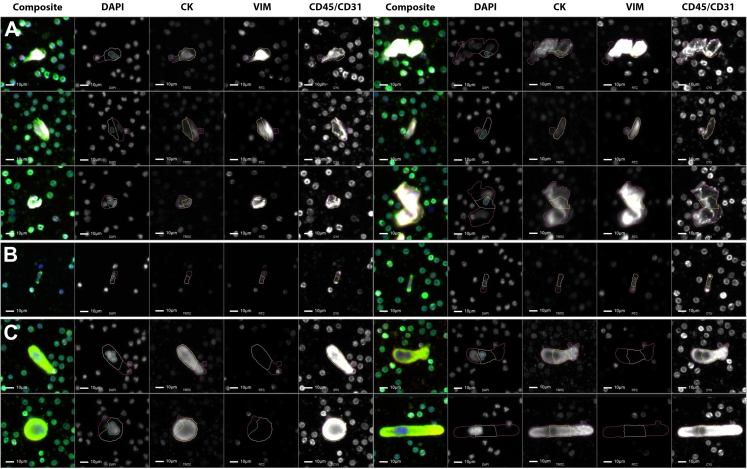
Five patients from the PACS Suspected cohort were manually curated by a trained analyst. These patients were experiencing respiratory symptoms (i.e., shortness of breath while lying down or exercising). From these patients, there were a number of Vim positive and CD45/CD31 positive cells with variable CK expression that were larger than surrounding WBCs and had an elongated cellular body with a punctated CD45/CD31 signal (Fig. 1A and B). These cells were observed both individually, as well as in clusters in the liquid biopsy. The morphology and channel expression are consistent with what is known of CECs. Another cellular phenotype that was observed unique to the PACS Suspected cohort was CK positive and CD45/CD31 positive cells that were larger than surrounding WBCs, with a multilobular nucleus and variable cellular shape and size (Fig. 1C). These have characteristics consistent with circulating megakaryocytes.
Fig. 1.
Gallery of rare cellular events found in the PB of Post-COVID patients. (A) DAPI positive, Vim positive, and CD45/CD31 positive with varying morphology; (B) Elongated cells with CD45/CD31 foci at both ends; (C) large DAPI positive and CD45/CD31 positive events. Blue: DAPI, Red: CK, White: VIM, Green: CD45/CD31. Images taken at 100× magnification. Scale bar = 10 μm.
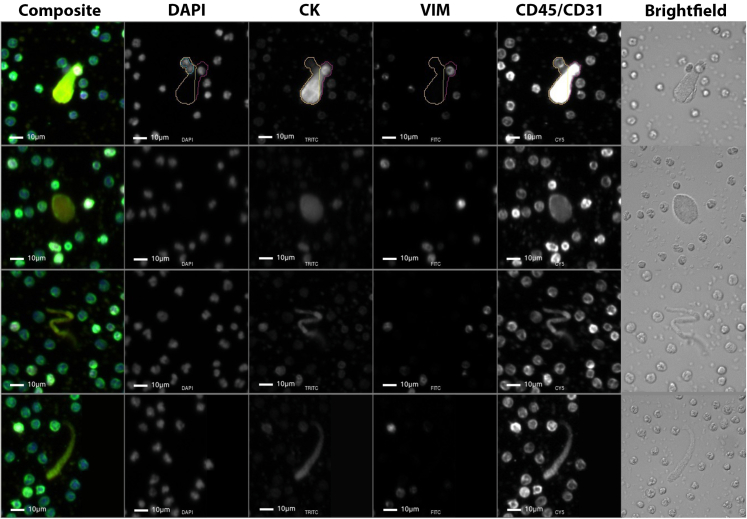
In the Post-COVID cohort, we observed the presence of acellular structures with predominantly CD45/CD31 expression. These events are morphologically larger and/or more elongated compared to the surrounding WBCs, demonstrating a predominantly diffuse CD45/CD31 signal pattern with occasional punctate signals at the event surface (Fig. 2). To better understand the event structure, images were acquired in brightfield. These acellular events appear semi-transparent with highly refractive regions similar to the surrounding cells. These events seem to have a slightly different diffraction and scattering pattern at the edges compared to the cells. The morphology and channel positivity is consistent with platelet structures.
Fig. 2.
Gallery of rare acellular events found in the PB of Post-COVID patients. Blue: DAPI, Red: CK, White: VIM, Green: CD45/CD31. Images taken at 100× magnification. Scale bar = 10 μm.
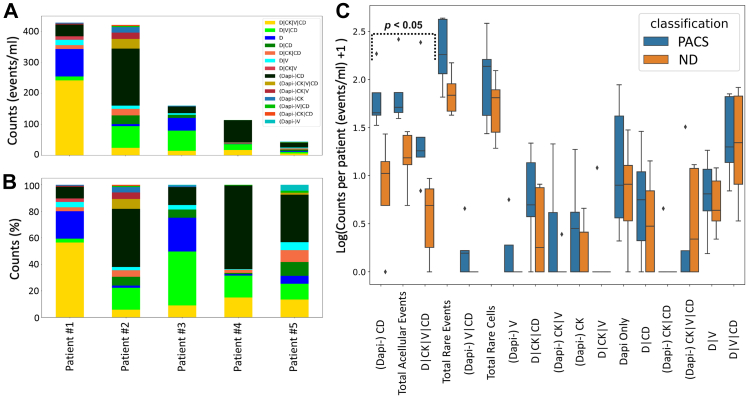
Five PACS Suspected patient samples were compared to the manual analysis of 5 ND samples (Fig. 3). An average of 242.8 (median 180.6, range 64.8–433.3) total events/mL were detected in the PACS Suspected cohort, while the ND presented with 78.6 (median 67.6 range 41.7–148.2) total events/mL. Three categories of events were observed at a significantly higher incidence in the PACS Suspected cohort compared to the ND: acellular CD45/CD31 positive events (p-value = 0.0090, Wilcoxon rank sum test), total acellular events (p-value = 0.0090, Wilcoxon rank sum test), and DAPI positive, CK positive, Vim positive and CD45/CD31 positive events (p-value = 0.028, Wilcoxon rank sum test). The acellular CD45/CD31 positive events were detected at 74.9 (median 44.2, range 32.4–184.3) events/mL in PACS Suspected and 10.5 (median 9.5, range 0.0–26.2) events/mL in ND. We hypothesise these events to be platelet structures. Total acellular event detection for the PACS Suspected patient samples had an average of 93.3 events/mL (median 94.5 range 38.4–260.8) while the ND samples had an average of 16.7 events/mL (median 14.3, range 3.9–27.8). The CK positive, Vim positive and CD45/CD31 positive rare cell category represents a morphologically heterogeneous population of cells. In the PACS Suspected patient samples, these cells were detected at 60.8 (median 17.1, range 6.0–242) cells/mL. In the ND samples, we observed an average of 3.9 (median 3.9, range 0.0–8.4) cells/mL. We hypothesise that rare cell populations expressing CK, Vim and CD45/CD31 include circulating endothelial cells (CECs). All other categories of circulating rare events were not significantly different between cohorts.
Fig. 3.
Rare event detection using HDSCA3.0 in PB samples collected from PACS Suspected (n = 5) and ND (n = 5). (A) Enumeration and (B) frequency of each rare event by channel-type specification for PACS Suspected patient samples. (C) Box plot of the channel-type rare events/mL between PACS Suspected and ND samples ordered by degree of statistical significance. The median value is depicted by the midline of the boxplot, the third and first quartiles are the upper and lower bounds of the box, and the whiskers cover 1.5 times the IQR. Channel-type specifications that were statistically significant across the two classifications are highlighted (p < 0.05).
Patient level classification model
To conduct an automated analysis of the circulating rare events in the Post-COVID cohort and determine if the liquid biopsy has potential in stratifying the patient populations, the classifier model was trained to separate ND from Post-COVID samples and clinical subdivisions thereof. The performance of the classifier on the test set is shown in Table 2. When comparing the Post-COVID cohort and its sub cohorts with NDs, the training set consisted of 15 samples for each class. For Post-ASYMP COVID vs Post-SYMP COVID the training set consisted of 20 samples and for PACS Not Suspected vs PACS Suspected the training set consisted of 18 samples for each class.
Table 2.
Classification model performance for post-COVID, NDs and different cohort subdivisions.
| Cohort 1 | Cohort 2 | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Normal donors | Post-COVID | 90.1% ± 1.1% | 90.6% ± 1.1% | 80.0% ± 3.3% |
| Normal donors | Post-SYMP COVID | 92.5% ± 1.0% | 92.8% ± 1.0% | 87.5% ± 5.6% |
| Normal donors | Post-ASYMP COVID | 90.7% ± 2.1% | 97% ± 1.5% | 75.0% ± 6.5% |
| Normal donors | PACS suspected | 87.1% ± 1.5% | 86.6% ± 1.7% | 92.5% ± 3.8% |
| Normal donors | PACS not suspected | 90.0% ± 3.2% | 94.3% ± 3.2% | 82.5% ± 6.5% |
| Post-ASYMP COVID | Post-SYMP COVID | 70.3% ± 1.8% | 69.6% ± 1.8% | 78.0% ± 8.1% |
| PACS not suspected | PACS suspected | 58.7% ± 3.7% | 59.4% ± 4.2% | 52.5% ± 9.5% |
Averages and standard errors are reported for model accuracy, sensitivity, and specificity.
We now investigate model performance between Post-COVID cohort subdivisions and NDs. The classifier performed well when separating the NDs with Post-COVID, Post-SYMP COVID, Post-ASYMP COVID, PACS Suspected, and PACS Not Suspected with average accuracies of 90.1%, 92.5%, 90.7%, 87.1%, and 90.0%, respectively. Due to the imbalance in the test set, the model sensitivity reflects the accuracy, with all values being greater than 90% except for NDs vs PACS Suspected with a sensitivity of 86.6%. Furthermore, the specificity was greater than or equal to 80% for all cases, except ND vs Post-ASYMP COVID, with a specificity of 75%. Finally, the classifier retained fair performance after excluding cancer patients from the analysis (Supplemental Table S1).
Next, we investigate model performance in differentiating two clinical subdivisions of the Post-COVID cohort. When training the model to separate Post-ASYMP COVID vs Post-SYMP COVID, the overall test set accuracy is fair, with a value of 70.3% with a sensitivity of 69.6% and specificity of 78%. Furthermore, PACS Not Suspected vs PACS Suspected, demonstrated a poor model performance with a low accuracy of 58.7%.
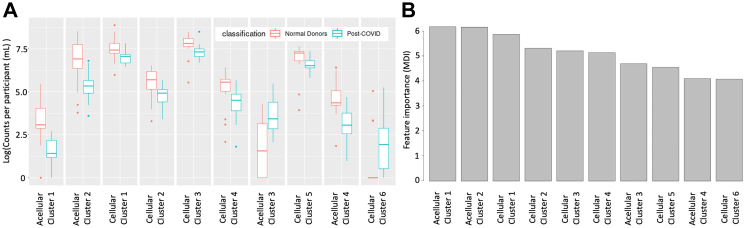
The classifier is based on event enumerations. Thus, one can investigate the cellular and acellular distributions that are contributing to the separation. Herein, we visualise the top ten cellular and acellular cluster occupancy profiles that drive the separation in a model iteration on the most populated cohorts, ND vs Post-COVID. The event enumerations and corresponding feature importance metrics for the top 10 event clusters are depicted in Fig. 4A and B respectively. As shown, 8 out of 10 clusters have increased median counts in NDs compared to Post-COVID cohort individuals. Furthermore, 4 out of 10 clusters involve acellular events.
Fig. 4.
Enumeration profile and feature importance of the ten top clusters out of a total of 306 for the classifier built on the ND (N = 19) and Post-COVID (N = 100) cohorts. (A) Box plot showing the cellular and acellular cluster enumeration profile of the ND and Post-COVID cohorts. The median value is depicted by the midline of the boxplot, the third and first quartiles are the upper and lower bounds of the box, and the whiskers cover 1.5 times the IQR. (B) Feature importance of top ten clusters quantified by the mean decrease in impurity obtained from the Random Forest component of the classifier model.
Discussion
Herein, we investigate whether individuals after SARS-CoV-2 infection carry a liquid biopsy signature in their PB using an assay configuration that was previously validated in carcinoma and myocardial infarction patients. To this end, we utilise the liquid biopsy assay on PB samples drawn from individuals with a prior SARS-CoV-2 infection and ND PB samples collected pre-pandemic. In PACS Suspected samples, via manual data reduction of rare events, we detected an increased count of hypothesised endothelial cells and platelet structures. Using a machine learning classifier approach, based on the rare event profile, we were able to separate controls from COVID-19 recovered individuals and sub cohorts thereof. Furthermore, the classifier exhibited a fair performance when separating Post-SYMP COVID and Post-ASYMP COVID cohorts. Our findings suggest that rare event analysis from the liquid biopsy should be further investigated as an inclusion in Post-COVID patient management.
Circulating endothelial cells have been used to identify cardiovascular disease and COVID-19 related cardiovascular damage.26, 27, 28 In a previous study, we determined that our liquid biopsy pipeline was able to identify circulating endothelial cells in myocardial infarction patients.18 Many studies have described the vascular dysfunction during active SARS-CoV-2 infection,29 and some suggest that COVID-19 related capillary damage may contribute to long term symptoms relating to tissue hypoxia.30 Consistent with previous findings, we were able to detect endothelial-like events in the Post-COVID cohort. The channel-type classification of rare cells associated with endothelial-like cells was identified at a statistically higher incidence in the PACS Suspected compared to ND. Direct evidence of cardiovascular damage through a liquid biopsy analysis may allow for healthcare providers to focus treatment and target resources appropriately. Further molecular characterisation of the hypothesised endothelial cells is needed to confirm their origin.
Another well documented event in SARS-CoV-2 infection is the presence of microclots or platelet aggregates.31,32 One autopsy study found microclots to be 9 times more prevalent in the pulmonary capillaries of deceased COVID-19 patients than those who died of influenzae.33 Preliminary studies have also suggested the presence of persistent microclots in the PB of PACS patients.34 In our patient samples, through manual data reduction, we detected a statistically significant increase of acellular events compared to that of the NDs. These acellular events match the morphology and biomarker expression profile of platelet structures. We hypothesise that these events are associated with the formation of microclots in the capillaries due to vascular damage. Microclots may be the cause of capillary blockage and reduced oxygen exchange in tissues, leading to fatigue and muscle weakness which are common symptoms of PACS.35 While molecular characterisation and clinical correlation analysis is still needed, platelet aggregates or microclots may be a therapeutic target for PACS treatments.
The cohort level classifier was successful in separating ND samples from the Post-COVID cohort and its clinical subdivisions. This result indicates differences in the frequency of liquid biopsy analytes within the PB of Post-COVID cohort and NDs. When separating Post-COVID sub cohorts with NDs, the classifier sensitivity was above or equal to 90% for all COVID-19 sub cohorts except PACS Suspected, with a value of 87.1%. Furthermore, the specificity was greater than or equal to 80% for all cases, except ND vs Post-ASYMP COVID, with a specificity of 75%. The lower performance on separating Post-ASYMP COVID from ND is anticipated as asymptomatic COVID-19 patients are expected to have less physiological damage. The separation suggests post-infection changes in the circulatory system, and, with a median time between COVID-19 diagnosis and blood draw of over 200 days for the Post-COVID cohort, these changes seem to be long lasting.
When separating major clinical subdivisions of the Post-COVID cohort, the classifier demonstrated a mixed performance. In the case of Post-SYMP COVID vs Post-ASYMP COVID, the model exhibited fair performance with sensitivity of 69.6%, and specificity of 78.0%, suggesting that liquid biopsy analytes can reflect on the severity of symptoms during infection. In the case of PACS Suspected vs PACS Not Suspected the model demonstrated poor performance, with sensitivity of 59.4% and specificity of 52.5%. In contrast to the symptomatic identifier, which has a clear clinical definition, PACS remains ill defined. Furthermore, PACS can be diagnosed due to symptoms in a variety of organ systems and combinations thereof, explaining poor model performance. Taken together, these attributes may explain the poor performance of the classifier in this setting.
There were several limitations to our study. First, we primarily relied on patient self-reported symptoms during recruitment and only collected a single blood draw from each patient. Future trial designs will use a stricter inclusion criteria with physician evaluated PACS and a narrower range of symptoms as well as incorporate multiple blood draws for longitudinal observation. Furthermore, the current study has a limited sample size and the generalisability of the results can be tested in a larger cohort. Another limitation is the use of only five biomarkers in the current four channel immunofluorescence analysis. However, using the HDSCA3.0 workflow, we can conduct downstream molecular characterisation of single events, using single cell genomics and targeted multiplexed proteomics.17,36, 37, 38, 39, 40 Proteomic analysis may confirm our hypotheses on CECs and platelet structures presented here, while also describing the immune cell profile of Post-COVID patients. Since the Post-COVID cohort did include cancer patients, genomic analysis will be important to differentiate the liquid biopsy signal for cancer vs Post-COVID or PACS. We also recognise that additional comorbidities may also affect the outcome of this study and will be further investigated in larger cohorts. A final limitation was the number of ND samples analysed as controls were limited due to the difficulty in acquiring PB draws collected prior to the beginning of the pandemic.
Conclusion
With the declining mortality and increased infectivity of SARS-CoV-2, finding a specific analyte or a collection of analytes will be important to help determine the cause of unexplained symptoms for the growing population of COVID-19 survivors. If we are able to correlate biological analytes with symptoms, we can further refine the definition of PACS or even provide an objective measure to differentiate PACS subsyndromes. This will help the healthcare providers to understand the cause of PACS symptoms and target resources to best care for the patient.
Contributors
EQ, GC, JM, JCF, SNS and PK contributed to the study concept and design. EQ, GC and JM conducted statistical analyses on the data. GC, JM, KLR and EK were involved in data curation. EQ, GC, EL, JM, SNS and PK interpreted the data. EQ, GC, JM and SNS verified the data and conducted the formal analysis. EQ, GC, KLR, JM and SNS were involved in the investigation. EQ, GC, JM, SNS and PK were responsible for the methodology. NMM, AM and JCF were involved in funding acquisition. EH, NN, MN, SG, KLR, NMM, AM and JCF were responsible for patient accrual and clinical resources. EQ and GC wrote the original draft. All authors agreed to the content of the manuscript, reviewed manuscript drafts, and approved the final version. All authors take final responsibility for the decision to submit for publication.
Data sharing statement
All data discussed in this manuscript is included in the main manuscript text and Supplementary Material. The images of the single cells and event clusters from the patient classification model are available through the BloodPAC Data Commons Accession ID BPDC000128 (https://data.bloodpac.org/discovery/BPDC000128).
Declaration of interests
NN reports grants from NCI (1U54CA260591) during the conduct of the study. SG reports IBSA pharmaceutical grant and pending patent during the conduct of the study. KLR reports honoraria from AstraZeneca, Janssen and Lilly. AM reports grants from NCI (1U54CA260591) during the conduct of the study. JCF reports grants from NCI (1U54CA260591) during the conduct of the study. PK reports gift donations from Fulgent Genetics, Kathy and Richard Leventhal and Vassiliadis Research Fund. No other disclosures were reported by the other authors.
Acknowledgments
We would like to thank all the patients who participated in this study. We would also like to thank all the clinical research staff who contributed to the study. We also thank all the past and current technical staff at CSI-Cancer for processing the samples.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104519.
Appendix A. Supplementary data
References
- 1.Huang C., Huang L., Wang Y., et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397(10270):220–232. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang L., Yao Q., Gu X., et al. 1-Year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. Lancet. 2021;398(10302):747–758. doi: 10.1016/S0140-6736(21)01755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Aly Z., Xie Y., Bowe B. High-dimensional characterization of post-acute sequelae of COVID-19. Nature. 2021;594(7862):259–264. doi: 10.1038/s41586-021-03553-9. [DOI] [PubMed] [Google Scholar]
- 4.Augustin M., Schommers P., Stecher M., et al. Post-COVID syndrome in non-hospitalised patients with COVID-19: a longitudinal prospective cohort study. Lancet Reg Health Eur. 2021;6 doi: 10.1016/j.lanepe.2021.100122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzunova G., Pallanti S., Hollander E. Presentation and management of anxiety in individuals with acute symptomatic or asymptomatic COVID-19 infection, and in the post-COVID-19 recovery phase. Int J Psychiatry Clin Pract. 2021;25(2):115–131. doi: 10.1080/13651501.2021.1887264. [DOI] [PubMed] [Google Scholar]
- 6.Malkova A., Kudryavtsev I., Starshinova A., et al. Post COVID-19 syndrome in patients with asymptomatic/mild form. Pathogens. 2021;10(11):1408. doi: 10.3390/pathogens10111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevention CfDCa COVID data tracker 2022. https://covid.cdc.gov/covid-data-tracker/#datatracker-home Available from:
- 8.Organization WH WHO coronavirus (COVID-19) dashboard 2022. https://covid19.who.int/ Available from:
- 9.Moreno-Pérez O., Merino E., Leon-Ramirez J.M., et al. Post-acute COVID-19 syndrome. Incidence and risk factors: a Mediterranean cohort study. J Infect. 2021;82(3):378–383. doi: 10.1016/j.jinf.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Kessel S.A.M., Olde Hartman T.C., Lucassen P., van Jaarsveld C.H.M. Post-acute and long-COVID-19 symptoms in patients with mild diseases: a systematic review. Fam Pract. 2022;39(1):159–167. doi: 10.1093/fampra/cmab076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alkodaymi M.S., Omrani O.A., Fawzy N.A., et al. Prevalence of post-acute COVID-19 syndrome symptoms at different follow-up periods: a systematic review and meta-analysis. Clin Microbiol Infect. 2022;28(5):657–666. doi: 10.1016/j.cmi.2022.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lara Bull-Otterson P., Baca S., Sharon Saydah P., et al. 2022. Post–COVID conditions among adult COVID-19 survivors aged 18–64 and ≥65 years — United States, March 2020–November 2021.https://www.cdc.gov/mmwr/volumes/71/wr/mm7121e1.htm#suggestedcitation [updated May 26, 2022. 71]. Available from: [Google Scholar]
- 13.Soriano J.B., Murthy S., Marshall J.C., Relan P., Diaz J.V., WHO Clinical Case Definition Working Group on Post-COVID-19 Condition A clinical case definition of post-COVID-19 condition by a Delphi consensus. Lancet Infect Dis. 2022;22(4):e102–e107. doi: 10.1016/S1473-3099(21)00703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nalbandian A., Sehgal K., Gupta A., et al. Post-acute COVID-19 syndrome. Nat Med. 2021;27(4):601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceban F., Ling S., Lui L.M.W., et al. Fatigue and cognitive impairment in post-COVID-19 syndrome: a systematic review and meta-analysis. Brain Behav Immun. 2022;101:93–135. doi: 10.1016/j.bbi.2021.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacedonia D., Scioscia G., De Pace C.C., et al. How are we handling the post-COVID patients? The dance of uncertainties. Respiration. 2022;101(2):210–213. doi: 10.1159/000518330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chai S., Ruiz-Velasco C., Naghdloo A., et al. Identification of epithelial and mesenchymal circulating tumor cells in clonal lineage of an aggressive prostate cancer case. NPJ Precis Oncol. 2022;6(1):41. doi: 10.1038/s41698-022-00289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bethel K., Luttgen M.S., Damani S., et al. Fluid phase biopsy for detection and characterization of circulating endothelial cells in myocardial infarction. Phys Biol. 2014;11(1) doi: 10.1088/1478-3975/11/1/016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Figueiredo J.C., Merin N.M., Hamid O., et al. Longitudinal SARS-CoV-2 mRNA vaccine-induced humoral immune responses in patients with cancer. Cancer Res. 2021;81(24):6273–6280. doi: 10.1158/0008-5472.CAN-21-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figueiredo J.C., Ihenacho U., Merin N.M., et al. SARS-CoV-2 vaccine uptake, perspectives, and adverse reactions following vaccination in patients with cancer undergoing treatment. Ann Oncol. 2022;33(1):109–111. doi: 10.1016/j.annonc.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bureau USC . 2020. Redistricting file–PL 94-171. [Google Scholar]
- 22.Marrinucci D., Bethel K., Kolatkar A., et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9(1) doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chai S., Matsumoto N., Storgard R., et al. Platelet-coated circulating tumor cells are a predictive biomarker in patients with metastatic castrate-resistant prostate cancer. Mol Cancer Res. 2021;19(12):2036–2045. doi: 10.1158/1541-7786.MCR-21-0383. [DOI] [PubMed] [Google Scholar]
- 24.Shishido S.N., Sayeed S., Courcoubetis G., et al. Characterization of cellular and acellular analytes from pre-cystectomy liquid biopsies in patients newly diagnosed with primary bladder cancer. Cancers (Basel) 2022;14(3):758. doi: 10.3390/cancers14030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann H.B., Whitney D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann Math Stat. 1947;18(1):50–60. [Google Scholar]
- 26.Varga Z., Flammer A.J., Steiger P., et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nizzoli M.E., Merati G., Tenore A., et al. Circulating endothelial cells in COVID-19. Am J Hematol. 2020;95(8):E187–E188. doi: 10.1002/ajh.25881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guervilly C., Burtey S., Sabatier F., et al. Circulating endothelial cells as a marker of endothelial injury in severe COVID -19. J Infect Dis. 2020;222(11):1789–1793. doi: 10.1093/infdis/jiaa528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lei Y., Zhang J., Schiavon C.R., et al. SARS-CoV-2 spike protein impairs endothelial function via downregulation of ACE 2. Circ Res. 2021;128(9):1323–1326. doi: 10.1161/CIRCRESAHA.121.318902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Østergaard L. SARS CoV-2 related microvascular damage and symptoms during and after COVID-19: consequences of capillary transit-time changes, tissue hypoxia and inflammation. Physiol Rep. 2021;9(3) doi: 10.14814/phy2.14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishikawa M., Kanno H., Zhou Y., et al. Massive image-based single-cell profiling reveals high levels of circulating platelet aggregates in patients with COVID-19. Nat Commun. 2021;12(1):7135. doi: 10.1038/s41467-021-27378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venter C., Bezuidenhout J.A., Laubscher G.J., et al. Erythrocyte, platelet, serum ferritin, and P-selectin pathophysiology implicated in severe hypercoagulation and vascular complications in COVID-19. Int J Mol Sci. 2020;21(21):8234. doi: 10.3390/ijms21218234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ackermann M., Verleden S.E., Kuehnel M., et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383(2):120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pretorius E., Vlok M., Venter C., et al. Persistent clotting protein pathology in long COVID/post-acute sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc Diabetol. 2021;20(1):172. doi: 10.1186/s12933-021-01359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kell D.B., Laubscher G.J., Pretorius E. A central role for amyloid fibrin microclots in long COVID/PASC: origins and therapeutic implications. Biochem J. 2022;479(4):537–559. doi: 10.1042/BCJ20220016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shishido S.N., Welter L., Rodriguez-Lee M., et al. Preanalytical variables for the genomic assessment of the cellular and acellular fractions of the liquid biopsy in a cohort of breast cancer patients. J Mol Diagn. 2020;22(3):319–337. doi: 10.1016/j.jmoldx.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welter L., Xu L., McKinley D., et al. Treatment response and tumor evolution: lessons from an extended series of multianalyte liquid biopsies in a metastatic breast cancer patient. Cold Spring Harb Mol Case Stud. 2020;6(6) doi: 10.1101/mcs.a005819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerdtsson E., Pore M., Thiele J.A., et al. Multiplex protein detection on circulating tumor cells from liquid biopsies using imaging mass cytometry. Converg Sci Phys Oncol. 2018;4(1) doi: 10.1088/2057-1739/aaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malihi P.D., Morikado M., Welter L., et al. Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg Sci Phys Oncol. 2018;4(1) doi: 10.1088/2057-1739/aaa00b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerdtsson A.S., Setayesh S.M., Malihi P.D., et al. Large extracellular vesicle characterization and association with circulating tumor cells in metastatic castrate resistant prostate cancer. Cancers (Basel) 2021;13(5):1056. doi: 10.3390/cancers13051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.