Abstract
The COVID-19 pandemic has caused millions of deaths and has resulted in disastrous societal and economic impacts worldwide. During SARS-CoV-2 infection, abnormal levels of pro-inflammatory cytokines have been observed and were associated to the severity of the disease. Type I (-α/β) and Type III (IFN-λ) interferons are family members of cytokines that play an important role in fighting viral replication during the early phases of infection. The location and timing of the IFNs production have been shown to be decisive for the COVID-19 outcome. Despite the effectiveness of COVID-19 vaccines and with the emergence of new SARS-CoV-2 variants, a better understanding of the involvement of IFNs as players in antiviral immunity in the COVID-19 pathophysiology is necessary to implement additional potent prophylactic and/or therapeutic approaches. In this study, we investigated the role of type I and III IFN in COVID-19 pathophysiology. We first analyzed the IFN-α, IFN-β and IFN- λ mRNA expression in nasopharyngeal swabs and blood samples from Moroccan patients infected with SARS-CoV-2 and secondly correlated these IFNs expressions with COVID-19 clinical and biological parameters. Our results showed that in the upper airways of patients with mild, non-severe, or severe COVID-19 manifestations, the IFN- α, - β and - λ are expressed in the same manner as in controls. However, in blood samples their expression was downregulated in all groups. Univariate linear models with interferons as predictors to evaluate clinical-biological parameters highlighted that the main clinical-biological relations were found when testing: FiO2, Lymphocyte values and virus load. Furthermore, the multivariate models confirmed that quantifications of interferons during COVID-19 are good biological markers for tracking COVID-19 pathophysiology.
Keywords: COVID-19, Type I interferon, Type III interferon, Immunity, Pathophysiology
1. Introduction
In late 2019 in Wuhan, a new corona virus appeared, leading to hundreds of thousands of deaths worldwide in only few months, and causing a major global crisis [1]. The new corona virus known as SARS-Cov-2 is a positive-sense single-stranded (ss) RNA virus belonging to the large group of Beta coronavirus in the Coronaviruses family. It shares major structural and molecular characteristics with other coronaviruses, including the presence of multitude of proteins essential to its formation and stability [2].
The immune system has a multitude of weapons and well-planned strategies against any invaders, through receptors that can sense danger, and recognizes microorganisms either on the membrane or in the endosomal/cytosolic compartments. These receptors or sensors are called Pattern-Recognition Receptors (PRRs). One of the most studied families of PRRs is the Toll Like Receptor family (TLR) which are expressed in a variety of cells including the immune cells to detect pathogen-associated molecular patterns (PAMPs) from microorganisms. TLR7/8 can recognize single stranded RNA viruses through endosomal nucleic acid sensing [3]. As it has been shown previously, an increase of the TLR3 transcription levels via TRIF occurs in the 2nd day post infections with coronavirus, which contributes to a protective response against this infection [4]. After virus recognition by the TLRs, the MyD88 pathway is activated and triggers some transcription factors such as IRF7 and NF-κβ which are responsible for the production of IFN-I and pro-inflammatory cytokines [5]. Another important PRR family involved in the antiviral response is the RLR family, especially RIG-5 and MDA5 which can recognize cytosolic RNAs. These receptors can activate antiviral signaling pathways through MAVS, which results in the activation of the IRF3, IRF7, and NF-κB transcription factors leading to the production of IFN-I and IFN-III [6].
Because people infected by SARS-Cov-2 show different degrees of severities, it has been clear that a deep understanding of how our immune system works in this infection is crucial to deliver the appropriate treatment. Especially with the emergence of new variants, and the recent data showing that the virus hijacks the immune system, it is important to understand the role of the first barrier, the innate immune system, which acts fast against CoV infection and within hours, can determine the disease outcome. During SARS-CoV-2 infection, abnormal levels of pro-inflammatory cytokines can be observed, and is called either a “cytokine storm”, or a “cytokine release syndrome”. Several studies have underlined the correlation between this exacerbated inflammation and the severity and the disease outcome [7]. Interferons (IFN) are a family of cytokines that plays an important role in the innate and adaptive immune response. This family is composed of three types of interferons: Type I (Interferon α/β), Type II (Interferon γ), and Type III (Interferon λ). Even though it is one of the most powerful innate immune responses to fight viral replication during the early phases of infection. It is mostly known to increase the autolytic activity of macrophages and NK against viruses and restrict coronaviruses infection in humans and mice [8], [5]. The study of Trouillet Assant and colleagues highlighted the importance of IFN-I in COVID-19. They showed that COVID-19 patients with a defective or delayed IFN- α production presented poorer outcomes as the viral load tended to be higher in these patients [9]. This disruption or reduction of IFN-I in infected cells with viruses such as SARS-CoV, SARS-CoV-2, and MERS-CoV, has been shown to be caused by some encoded viral molecules, such as ORF-3b, ORF5, ORF6, nsp1, nsp3, nsp16, ORF3b, and M and N proteins, in the case of SARS-CoV-2. These viral proteins seem to disrupt the RLR/MAVS-mediated antiviral signaling pathways [10].
It is important to highlight that recent studies reported the importance of the location and timing of the IFNs production, as these parameters have been shown to be decisive for the disease outcome. Low levels of IFN type I and III proteins and higher levels of other pro-inflammatory cytokines and chemokines such as IL-6 and IL-1RA in the serum of COVID-19 patients resulted in severe pathology [5]. In COVID-19 patients, several mutations were identified and have been associated to decreased IFN I expression [11]. Low levels of IFN at the beginning of the infection are critical as they can determine the severity of the disease progression. Hadjadj et al. have shown a distinction of phenotypes between severe and critical patients that consisted of a highly impaired IFN type I response, which is characterized by no production of IFN-β and low levels of IFN-α. This dysregulation was associated with a persistent blood viral load and an exacerbated inflammatory response with a substantial increased production of IL-6 and TNF-α [12]. The excessive IFN production in the late stages of the disease was associated with the development of cytokine storm [13].
Although COVID-19 vaccines have proven to be safe, effective against severity, and with the emergence of new SARS-CoV-2 variants, a better understanding of the involvement of IFNs as players in antiviral immunity in the COVID-19 pathophysiology is necessary to implement additional potent prophylactic and/or therapeutic approaches. The aim of this study is to investigate the role of type I and III interferons in COVID-19 pathophysiology by i) analyzing the level of IFN I and IFN III expression in the upper airways and blood samples of COVID-19 Moroccan patients and ii) correlating these cytokines expression with the different biological and clinical parameters of COVID-19 disease.
2. Methods
2.1. Ethics statement
This research was conducted according to the principles set out in the Declaration of Helsinki and in local ethical guidelines. This study was approved by the local Ethics Committee of Cheikh Zaid Hospital, Rabat, Morocco (Project: CEFCZ/PR/2020-PR04). Written informed consent was obtained from all patients and healthy subjects used as controls in this study.
2.2. Patients
24 individuals suspected of SARS-CoV-2 infection (travelers, closeness to infected patients, and presence of symptoms) were recruited with informed consent at Cheikh Zaïd Hospital in Rabat Morocco from December 7th 2020 to March 8th 2021. 7 healthy individuals were also recruited and included in the study as controls. Patients were diagnosed as COVID-19-positive by nucleic acid test. Patients were tested for COVID-19-positivity at 5 time-points: On admission (Day 0), Day 6, Day 13, Day 20, and Day 30 +. Patients were analyzed after a median duration of 6 days (interquartile range, 5 to 7 days) following disease onset. Nasopharyngeal and Blood samples were taken from each subject (n = 31) and were stored at −80 °C until use. The tissue sampling procedures, as well as the method of sample handling, were identical for the nasopharyngeal and blood samples.
2.3. Extraction of total RNA and semiquantitative RT-PCR
Total RNA was extracted from exfoliated cells from the nasal cavity and purified leukocytes from blood using MagPurix Extraction system (Repository: Zinexts Life Science Corp. New Taipei, Taiwan) (https://www.zinexts.com/). A one-step real-time PCR kit (Tib-Molbiol, Berlin) for the detection of 2019-nCoV RdRp and E genes was purchased from Qiagen. The PCR reactions were performed using the Rotor-Gene Q thermocycler (Hilden, Germany). The PCR program consisted of 50 °C for 15 min, 95 °C for 5 min, 45 cycles of 95 °C for 10 sec, 55 °C for 45 sec, and was terminated by dissociation. Synthetic RNA for 2019-nCoV E gene and RdRp assay were used as positive controls (ACAGGTACGTTAATAGTTAATAGCGT, and GTGARATGGTCATGTGTGGCGG for E and RdRp genes, respectively).
For RT-PCR cytokines expression analysis, 1 μg of total RNA was reverse transcribed using reverse transcriptase (Qiagen, COURTABOEUF, France).
The cDNA was amplified for 40 cycles (annealing temperature 60 °C) using standard conditions. IFN- α, IFN- β, and IFN- λ and β -actin quantification was performed using qPCR (Bio-Rad CFX96, Marnes-la-Coquette, France), and a Brilliant II SYBR Green QPCR Master Mix (Bio-Rad, Marnes-la-Coquette, France). The primers used are listed in (Table 1 ). The conditions for the reactions were as follows: an initial 3-min denaturation step at 95 °C, followed by 40-s cycles at 95 °C, 60 °C, and 72 °C. The results were calculated using the relative quantification method normalized to β -actin.
Table 1.
Primer sequences for the RT-PCR analysis.
| Primers | Forward | Reverse |
|---|---|---|
| IFN- α IFN- β IFN- λ β-Actin |
3′-GCC-TCG-CCC-TTT-GCT-TTA-CT-5′ 3′-GCA-CAA-CAG-GTA-GTA-GGC-GA-5′ 3′-GGT-GAC-TTT-GGT-GCT-AGG-CT-5′ 3′-GAA CGG TGA AGG TGA CA-5′ |
3′-CTG-TGG-GTC-TCA-GGG-AGA-TCA-5′ 3′-TGG-AAA-GAG-CTG-TCG-TGG-AG-5′ 3′-GGC-CTT-CTT-GAA-GCT-CGC-TA-5′ 3′-TAG AGA GAA GTG GGG TGG-5′ |
2.4. Statistical analysis
The cytokines expression results were analyzed using the Mann-Whitney U test for nonparametric data. A p < 0.05 was considered statistically significant. Expression analyses were performed using GraphPad Prism software and version 3.4.3 of the R environment.
Statistical analyses were performed in R software environment version 4.0.2. Each cytokine quantification for COVID19 patients was taken as a univariate predictor in a linear model with a Gaussian family parameter to test relations with a different clinical and biological parameter. Unsupervised principal component analysis based on quantification of Interferons α, β, and λ was processed in FactoMineR R- package: p-value of group discrimination was obtained after the projection of each parameter on principal component axes.
3. Results
3.1. Patients screening and characterization
Our cohort included 31 individuals with 24 patients and 7 controls, whose clinical characteristics are detailed in (Table 2 ). Out of these 24 patients, 70.84% were male and 29.16% were female. The ages of our patients ranged from 19 to 86 years, with a mean age of 40 years (4.24 SD) and an average weight of 71 kg (8.48 SD). Patients who were identified as COVID-19 positive based on the presence of 2019-nCoV RdRp and E genes by RT-PCR analysis showed fever, cough, dyspnea, fatigue, headache, chest pain, and pharyngalgia as clinical signs. We enrolled 7 controls with 57.14% male and 42.86% female. Their ages ranged from 28 to 76 years, with a mean age of 51 years (16.15 SD) and an average weight of 70.85 (11.81 SD). They were declared negative by both RT-PCR amplification of 2019-nCoVRp and E genes and chest CT scans.
Table 2.
Clinical profile of patients included in the study.
| Patients | Sexe | Age (years) | Weight (Kg) | SARS-CoV-2 RNA |
|---|---|---|---|---|
| Control | F | 28 | 64 | – |
| Control | M | 76 | 60 | – |
| Control | F | 58 | 64 | – |
| Control | M | 55 | 68 | – |
| Control | F | 58 | 93 | – |
| Control | M | 34 | 66 | – |
| Control | M | 48 | 81 | – |
| Patient 1 | F | 43 | 77 | P |
| Patient 2 | F | 86 | 53 | P |
| Patient 3 | M | 77 | 94 | P |
| Patient 4 | M | 77 | 81 | P |
| Patient 5 | F | 19 | 83 | P |
| Patient 6 | M | 28 | 77 | P |
| Patient 7 | M | 22 | 95 | P |
| Patient 8 | M | 39 | 87 | P |
| Patient 9 | M | 50 | 67 | P |
| Patient 10 | M | 47 | 79 | P |
| Patient 11 | M | 65 | 75 | P |
| Patient 12 | M | 30 | 92 | P |
| Patient 13 | M | 69 | 86 | P |
| Patient 14 | M | 74 | 97 | P |
| Patient 15 | F | 80 | 70 | P |
| Patient 16 | F | 64 | 63 | P |
| Patient 17 | F | 22 | 81 | P |
| Patient 18 | M | 83 | 59 | P |
| Patient 19 | M | 28 | 88 | P |
| Patient 20 | M | 49 | 96 | P |
| Patient 21 | M | 65 | 79 | P |
| Patient 22 | M | 59 | 87 | P |
| Patient 23 | M | 54 | 92 | P |
| Patient 24 | F | 37 | 65 | P |
P: SarsCov-2 RNA positif.
3.2. Type I (-α, β) and III (λ) Interferon’s analysis expression in nasopharyngeal and blood samples of COVID-19 patients
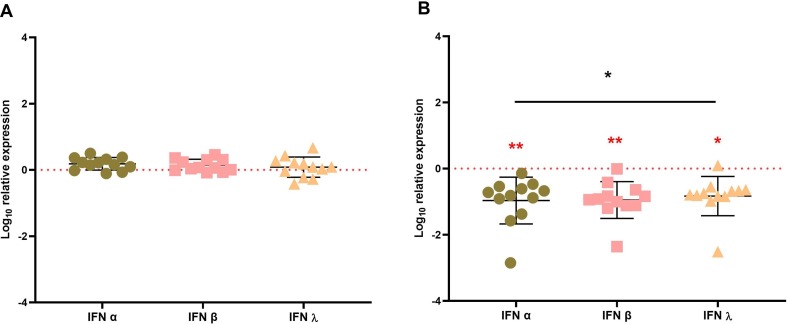
To investigate the involvement of interferons in early stages of COVID-19 pathogenesis, the expression of particularly IFN- α, IFN- β, and IFN- λ was monitored by qRT-PCR in nasopharyngeal and blood samples of SARS-CoV-2 patients. Our results showed no statistically significant difference in these cytokines expression in nasopharyngeal samples from SARS Cov-2 infected patients compared to controls (Fig. 1 A). On the other hand, cytokine levels in SARS-CoV-2 patients' blood samples were significantly lower (P < 0.05, p < 0.01) than in controls (Fig. 1 B).
Fig. 1.
IFN-α, IFN-β and IFN-λ mRNA expression in SARS Cov-2 infected patients. (A) The relative IFN-α, IFN-β and IFN- λ genes expression was analyzed by RT-qPCR in the upper airways; nasopharyngeal swabs and (B) in blood of SARS Cov-2-infected patients. IFN-α, IFN-β and IFN-λ mRNA expression levels relative to the β-actin gene are indicated on the Y axis. The 24 infected patients were normalized to the 7 healthy subjects (controls) as indicated by the dashed lines. The results are expressed with standard deviations. The statistical analyses were performed using the Mann-Whitney and ANOVA test for non-parametric data. ns, no significant, *p < 0.05, **p < 0.01.
3.3. The relationship between type I (-α, β) and III (λ) interferons and COVID-19 outcome
In order to understand the involvement of type I (-α, β) and III (λ) in the outcome of SARS-CoV-2 infections, the patients were stratified into COVID-19 pathological stages based on their clinical parameters (Table 3 ). Patients declared positive by the nucleic acid test were subsequently divided into mild, non-severe, and severe COVID-19 groups on the basis of clinical criteria using the American Thoracic Society guidelines for community-acquired pneumonia [14]. Patients with mild disease manifestations were discharged from the emergency room without being hospitalized (home confinement), while non-severe patients were admitted to the hospital (hospitalized), and severe patients (critically ill patients) were admitted to the Intensive Care Unit (ICU). The severity of the disease was established, and routine blood parameters were monitored at the time of admission for mild (n = 8), non-severe (n = 8), and severe cases of COVID-19 (n = 8).
Table 3.
Blood parameters and Pathological stages of the COVID-19 patients.
| Patients | Platelet Value | Lymphocyte Value | PLR | ALT Value | AST Value | LDH Value | D-dimers | CRP value | FiO2 | Hospitalization Time | PaO2 / FiO2 | Fibrinogen | ABO group |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 326 | 3,9 | 83,58974359 | 24,1 | 14,3 | 206 | 0,24 | 5,77 | NA | NA | NA | 187 | AB |
| Control | 189 | 0,4 | 472,5 | 25,7 | 25 | 513 | 1,09 | 14,52 | NA | NA | NA | 376 | A |
| Control | 241 | 2,2 | 109,5454545 | 14,2 | 14,5 | 189 | 0,12 | 11,8 | NA | NA | NA | 198 | O |
| Control | 120 | 0,96 | 125 | 49,7 | 60,1 | 742 | 1,77 | 26,47 | NA | 13 | NA | NA | AB |
| Control | 197 | 1,34 | 147,0149254 | 23,8 | 29,8 | 355 | 0,41 | 16,48 | NA | 14 | NA | NA | AB |
| Control | 85 | 3,74 | 22,72727273 | 67,8 | 46,5 | 806 | 0,35 | 40,82 | NA | 20 | NA | NA | AB |
| Control | 97 | 3,06 | 31,69934641 | 58,9 | 61,2 | 963 | 1,15 | 12,63 | NA | 35 | NA | NA | O |
| Mild | 242 | 1,6 | 151,25 | 15,3 | 26,8 | 211 | 0,3 | 18,42 | 60 | 4 | 150 | 183 | O |
| Mild | 196 | 1,9 | 103,1578947 | 37,7 | 36 | 543 | 0,14 | 2,98 | 40 | 6 | 150 | 249 | B |
| Mild | 287 | 0,9 | 318,8888889 | 22,6 | 24,8 | 282 | 0,77 | 25,47 | 60 | 11 | 150 | 174 | AB |
| Mild | 204 | 4,2 | 48,57142857 | 28,7 | 26,5 | 796 | 0,19 | 14,68 | 30 | 2 | 150 | 312 | O |
| Mild | 94 | 0,39 | 241,025641 | 51,2 | 35,6 | 468 | 1,05 | 36,74 | NA | 27 | NA | NA | O |
| Mild | 168 | 0,57 | 294,7368421 | 23,4 | 46,7 | 977 | 0,9 | 14,99 | NA | 32 | NA | NA | B |
| Mild | 186 | 1,73 | 107,5144509 | 21,7 | 19,4 | 1164 | 1,32 | 13,34 | NA | 32 | NA | NA | AB |
| Mild | 83 | 1,14 | 72,80701754 | 20 | 38,1 | 956 | 2,45 | 52,89 | NA | 3 | NA | NA | O |
| Non-Severe | 78 | 0,5 | 156 | 48,7 | 61,3 | 889 | 1,72 | 23,91 | NA | 8 | NA | 347 | B |
| Non-Severe | 259 | 1,7 | 152,3529412 | 26,4 | 22,1 | 179 | 0,26 | 17,01 | NA | 7 | NA | 225 | A |
| Non-Severe | 262 | 1,2 | 218,3333333 | 22,4 | 17,6 | 246 | 1,34 | 3,79 | NA | 14 | NA | 401 | AB |
| Non-Severe | 89 | 0,7 | 127,1428571 | 47,9 | 46,5 | 845 | 0,71 | 29,41 | NA | 8 | NA | 192 | A |
| Non-Severe | 78 | 1,13 | 69,02654867 | 41,5 | 34,7 | 798 | 0,61 | 6,31 | NA | 19 | NA | NA | B |
| Non-Severe | 134 | 0,8 | 167,5 | 42,1 | 55,3 | 1053 | 0,97 | 50,03 | NA | 35 | NA | NA | A |
| Non-Severe | 271 | 1,05 | 258,0952381 | 25,9 | 45 | 416 | 2,03 | 22,41 | NA | 15 | NA | NA | AB |
| Non-Severe | 142 | 0,28 | 507,1428571 | 19,6 | 13,2 | 954 | 0,89 | 4,91 | NA | 23 | NA | NA | A |
| Severe | 116 | 1,5 | 77,33333333 | 63,2 | 42,8 | 635 | 1,37 | 24,57 | 60 | 14 | 150 | 375 | A |
| Severe | 120 | 0,8 | 150 | 41,2 | 22,6 | 283 | 0,47 | 4,5 | 80 | 7 | 150 | 399 | AB |
| Severe | 91 | 0,9 | 101,1111111 | 45,8 | 39,9 | 940 | 2,08 | 12,39 | 60 | 13 | 150 | 538 | AB |
| Severe | 317 | 2,4 | 132,0833333 | 47 | 42,5 | 398 | 0,96 | 3,72 | 60 | 6 | 150 | 560 | B |
| Severe | 129 | 0,98 | 131,6326531 | 30,2 | 37,2 | 429 | 0,08 | 14,2 | 80 | 15 | 150 | NA | A |
| Severe | 341 | 2,02 | 168,8118812 | 36,1 | 45,2 | 318 | 0,95 | 5,76 | 80 | 21 | 150 | NA | AB |
| Severe | 158 | 1,01 | 156,4356436 | 48,9 | 55,4 | 586 | 1,15 | 51,87 | 100 | 14 | 150 | NA | AB |
| Severe | 187 | 0,14 | 1335,714286 | 33 | 41,5 | 490 | 0,17 | 9,27 | 60 | 13 | 150 | NA | B |
NA: not applicable.
3.3.1. Type I (-α, β) and III (λ) Interferon’s analysis expression in different COVID-19 stages
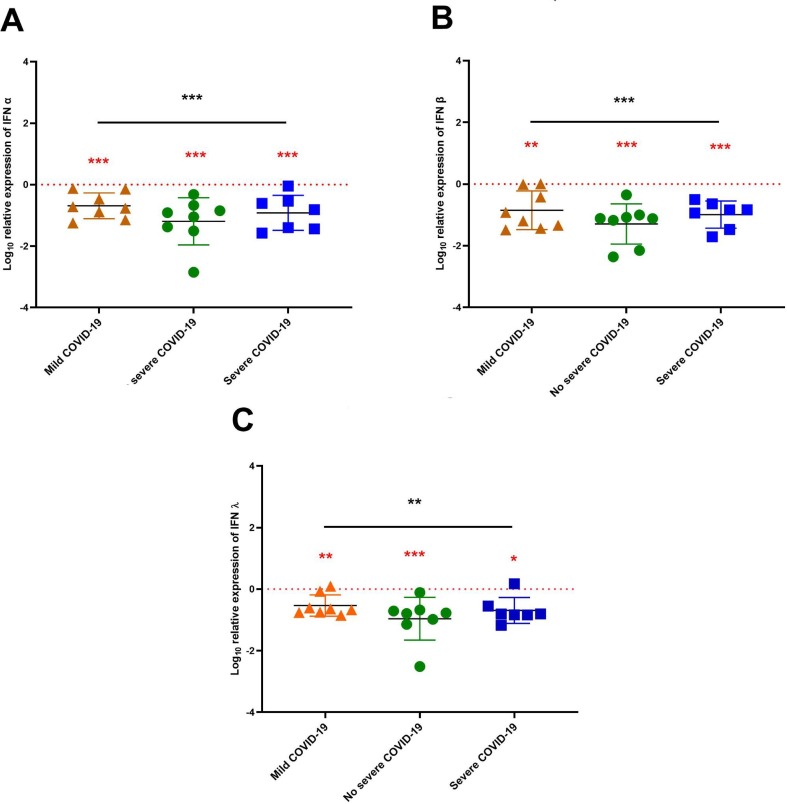
In order to investigate whether type I and type III IFN correlated with the severity of SARS-CoV-2 infection, we evaluated the levels of IFN- α, IFN- β, and IFN- λ mRNA expression in blood samples of the three groups of COVID-19 patients (mild, non-severe and severe COVID19) by RT-qPCR. We found significant (p < 0.001) lower levels of IFN-α (Fig. 2 A), IFN- β (Fig. 2B), and IFN- λ (Fig. 2C) in COVID-19 patients compared to healthy controls. This decrease was observed in all SARS-CoV-2-stages, including non-severe, mild, and severe (Fig. 2).
Fig. 2.
Interferon’s mRNA expression levels according to COVID-19 severity stages. The patients were stratified into three stages of severity based on the clinical parameters. IFN-α, IFN-β and IFN- λ mRNA expression levels in blood according to stages of severity was normalized to the 7 controls as indicated by the red dotted lines. The results are expressed as means ± standard deviations. The statistical analyses were performed using the Mann-Whitney and ANOVA test for non-parametric data. *p < 0.5, **p < 0.01, ***p < 0.001.
3.3.2. Type I (α, β) and III (λ) interferons are associated with COVID-19 clinical-biological parameters.
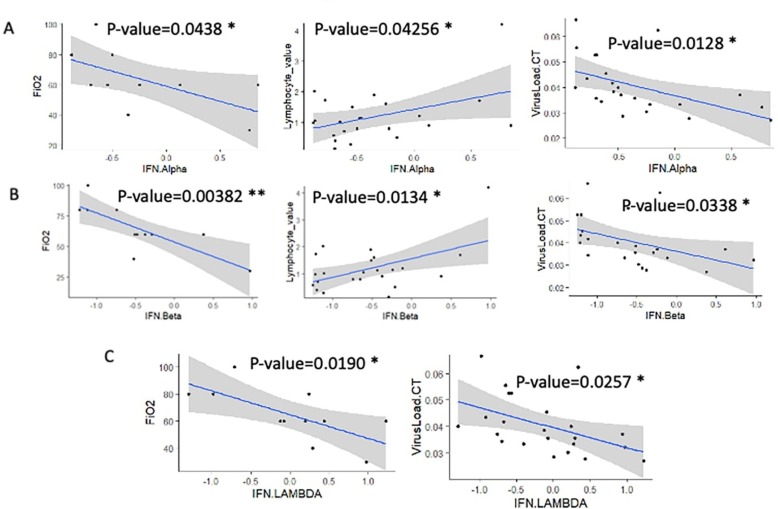
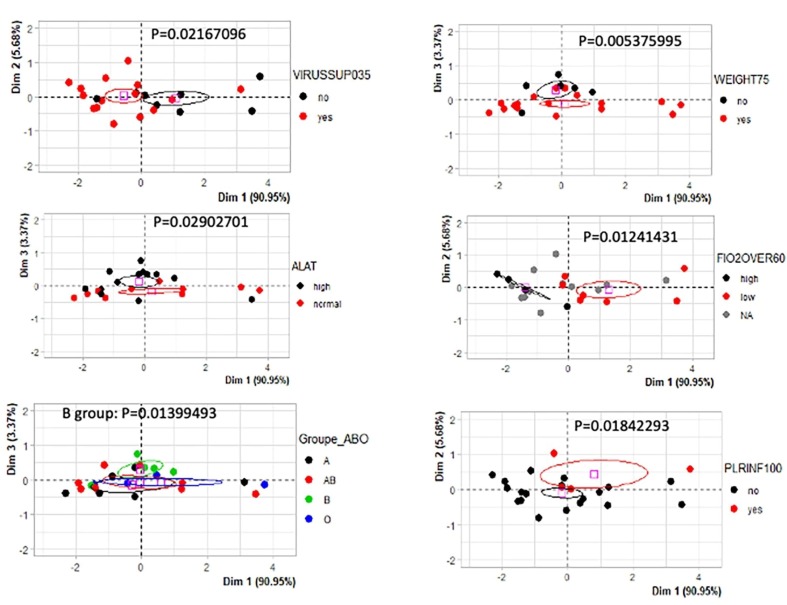
During the follow-up of the cohort, the quantification of interferons α, β, and λ was investigated. Univariate linear models to test relations between interferon showed that interferons were found highly associated between them. A significant association was found between interferons (Table 4 ). Univariate linear models with interferons as predictors to evaluate clinical-biological parameters (Table 4) highlighted that the main clinical-biological relations were found when testing: FiO2, Lymphocyte values, and virus load (Fig. 3 A-C). Furthermore, unlike α, and β interferons being correlated with three distinct clinical criteria, λ interferon correlated only with two clinical criteria. This finding suggests that α and β interferons would be more efficient than λ and that interferons quantifications might be useful markers for monitoring COVID-19 pathophysiology. For these reasons, we decided to build an unsupervised multivariate model-based principal component analysis based on the three interferon quantifications. This model confirmed a significant stratification of the patients with distinct parameters like: virus load (threshold = 0.35, p-value = 0.021, Fig. 4 ), also stratified B blood group from the others (p-value = 0.013, Fig. 4), also stratified weight of the patients (threshold = 75 kg, p-value = 0.0054, Fig. 4), also stratified ALAT value (p-value = 0.029, Fig. 4), stratified patients FiO2 values (threshold over 60, p-value = 0.012, Fig. 4) and stratified PLR patient values (threshold over 100, p-value = 0.018, Fig. 4). This multivariate model confirmed that quantifications of Interferons during COVID-19 are good markers to monitor COVID-19 pathophysiology.
Table 4.
Univariate analysis of each cohort parameter as compared to the three interferon quantifications. This table includes the p-values obtained by univariate linear model for each tested variable (blue cell: tendency, yellow cell: significant p-value < 0.05).
| Parameters | Subgroups | IFN- α | IFN- β | IFN- λ |
|---|---|---|---|---|
| Age | 0.582 | 0.4005 | 0.567 | |
| ALT_value | 0.768 | 0.651 | 0.674 | |
| AST_value | 0.241 | 0.375 | 0.331 | |
| C Reactive Protein | 0.685 | 0.5867 | 0.238 | |
| D. dimers | 0.940 | 0.9766 | 0.529 | |
| Fibrinogen | 0.400 | 0.416 | 0.318 | |
| FiO2 | 0.0438 * | 0.00382 ** | 0.0190 * | |
| Groupe_ABO (ref = O) | Groupe_ABO[A] | 0.551 | 0.297 | 0.616 |
| Groupe_ABO[B] | 0.397 | 0.486 | 0.948 | |
| Groupe_ABO[AB] | 0.612 | 0.347 | 0.830 | |
| Hospitalization_Time | 0.0535 | 0.00437 ** | 0.00831 ** | |
| LDH_value | 0.896 | 0.572 | 0.437 | |
| Lymphocyte_value | 0.04256 * | 0.0134 * | 0.1543 | |
| Platelet_value | 0.726 | 0.5540 | 0.854 | |
| PLR | 0.3197 | 0.80238 | 0.727 | |
| Severity | 0.591 | 0.800 | 0.822 | |
| Sexe | 0.17052 | 0.53004 | 0.641 | |
| State (ref = non severe) | State[T.Mild] | 0.956 | 0.8183 | 0.800 |
| State[T.Severe] | 0.489 | 0.5586 | 0.561 | |
| Virus Load.CT | 0.0128 * | 0.0338 * | 0.0257 * | |
| Weight | 0.661 | 0.709 | 0.486 | |
| _ | ||||
| IFN- α | NA | 0.0000000502 *** | 0.0000000161 *** | |
| IFN- β | 0.0000000502 *** | NA | 0.000000942 *** | |
| IFN-λ | 0.0000000161 *** | 0.000000942 *** | NA |
Fig. 3.
Univariate analysis of type I and III Interferon quantifications during COVID-19 infection: (A) univariate linear models 2 for significant parameters against quantification of IFN- α (p-value obtained by linear model). (B) univariate linear models for significant parameters against quantification of IFN- β (p-value obtained by linear model). (C) univariate linear models for significant parameters against quantification of IFN- λ (p-value obtained by linear model).
Fig. 4.
Univariate analysis of type I and III Interferon quantifications during COVID-19 infection: Principal component analyses based on Interferon quantifications (α, β, λ) and testing the distinct clinico-biological parameters, p-values were obtained by group stratifications on principal component axes.
4. Discussion
COVID-19 pandemic has caused millions of deaths and has had disastrous societal and economic impacts worldwide. A better understanding of the immune responses related to clinical manifestations of COVID-19 can guide the identification of potential pharmacological targets.
During viral infection, IFN-I α, β and other members of the extended IFN-I family are produced rapidly and exhibit important antiviral properties within infected cells, by limiting virus proliferation and spreading. Furthermore, IFN-I enhances both the innate and adaptive immune responses to viral infections [15], [16]. These properties of IFN-I are shared by IFN-III (IFN-λ), despite the differential expression of their respective receptors. IFN-I and IFN-III are powerful antiviral cytokines via the induction of IFN-stimulated genes (ISGs), and the possibility of using clinical grade recombinant IFN-I or IFN-III as treatments have raised much interest [17]. However, to date, conflicting findings have hampered our view of the played role by IFN-I and IFN-III families’ members during SARS-CoV-2 infection. As previously mentioned, the timing of the response appears to be critical, with early, transitory IFN production being associated with milder disease. Another layer of complexity linked not only to the timing, but also to the response localization, with efficient induction of IFNs in the upper airways being protective, while sustained production of IFNs in the lung or in the blood driving detrimental effects [18].
Despite the effectiveness of COVID-19 vaccines, a better understanding of the role of innate antiviral immunity players in the progression of the disease in SARS-COV-2 infected patients is imperative to implement effective additional prophylactic and/or therapeutic strategies. In this study, we investigated the role of type I and III IFN in COVID-19 pathophysiology. We first analyzed the IFN- α, IFN- β, and IFN- λ mRNA in nasopharyngeal swabs and blood samples from Moroccan patients infected with SARS-COV-2 during the early phases of infection and secondly correlated these IFNs expressions with clinical and biological parameters of COVID-19. We found that in the upper airways of patients with mild, non-severe, or severe COVID-19 manifestations, the IFN- α, - β and - λ mRNA are expressed in the same manner as in controls. However, in blood samples these interferons expression was downregulated in all groups; whether it is mild, non-severe, or severe COVID-19 groups. This is the first study carried out on a Moroccan cohort and our results confirm previous cross-sectional studies that found decreased blood IFN-I and IFN-III levels in COVID-19 patients compared to controls.
Hadjadj et al. have also reported reduced IFN responses in the peripheral blood of COVID-19 patients. Furthermore, Blanco-Melo et al. have demonstrated, by comparing the transcriptional responses of several respiratory viruses, that the host response to SARS-CoV-2 is unable to activate an IFN-I and -III robust response [19]. Since the first SARS outbreak in Asia in February 2003, several studies have shown that SARS-CoV and MERS-CoV use distinct mechanisms to escape and suppress type I IFN-mediated immune responses. Indeed, it has been shown in SARS-CoV infection that a subdued IFN-I response reduces antigen presentation and the antiviral adaptive Th-1 immune response [20]. In SARS-CoV-2 infection, several recent studies have confirmed that SARS-CoV-2 inhibits type I and III induction. In fact, it has been shown that SARS-CoV-2 infection prevents the release of mRNA from transcription sites and/or initiates transcript degradation in the nucleus, which reduces type I and III IFN production at post-transcriptional levels. Furthermore, several proteins that are encoded by SARS-CoV-2 interfere with RLR sensing pathways as well as IFN induction, signaling, or effector functions. Through interfering with RIG-I and MDA5 pathways, SAR-CoV-2 proteins’ ORF9b, N, and M inhibit IFN-β and pro-inflammatory cytokines expression. Furthermore, ORF3b inhibits IFN-I induction, while ORF6 and ORF8 abolish IFN-β expression and ISGs activation. Additionally, the SARS-CoV-2 N protein prevents viral RNA from aggregating with MAVS to impede IFN pathway induction [21]. In addition to the decreased IFN response triggered by SARS-CoV-2, recent investigations have shown that genetic predisposition or the formation of autoantibodies that neutralize IFN can cause further restrictions on IFN function [22]. By using a candidate gene approach Q. Zhang et al. identified patients with severe COVID-19 who have mutations in genes related to the type I and III IFN immunity regulation. They discovered an enrichment of these genes in patients, suggesting that genetics may have a role in the infection's clinical outcome. In about 10% of patients with severe COVID-19 pneumonia, Bastard et al. identified individuals with high titers of neutralizing autoantibodies against type I IFN which were found only in COVID-19 severe patients [22]. According to another study, at least 3.5% of patients with life-threatening COVID-19 pneumonia had genetic mutations at potential loci known to be involved in TLR3- and IRF7-dependent IFN-I production and amplification [23].
Furthermore, our data showed that the IFNs expression was linked to the different biological and clinical parameters associated with SARS-COV-2 infection. Based on the quantification of the interferons, a principal component analysis model was constructed in order to discriminate the samples. During the projection of the clinical-biological information of the samples on the graphic representation of this multivariate model, it was possible to discriminate the criteria: “virus load” for a threshold +/- 0.35 (p-value = 0.022), ALAT (p-value = 0.029), blood group B of other blood groups (p-value = 0.014), patient's weight with a threshold +/- 75 kg (p-value = 0.0054), FiO2 for a threshold of +/- − 60 (p-value = 0.012), PLR for a threshold of +/- 100 (p-value = 0.018). Although this model could not be directly related to the severity of the disease (data not shown), these results suggest that this multivariate model based on the combined quantification of the three interferons is discriminatory for the study of clinical-biological information of patients with COVID-19. Our findings reconcile a large portion of the literature on IFNs and emphasize the importance of these interferons, notably IFN- α and - β, as good indicators for monitoring COVID-19 progression.
The presence of interferon deficiency in COVID-19 patients suggests that type I and III IFN modulation could be used as a treatment option in these patients. IFN-α has recently been proposed as a potential therapeutic strategy for COVID-19 disease, owing to the fact that the innate immune system releases IFN-α quickly as the first line of defense against viral infections. On the other hand, it might have immunoregulatory effects, which can lead to pathogenic damage and uncontrolled inflammatory responses [24]. Due to the absence of a reliable animal model and the availability of clinically approved IFN-I treatments, multiple clinical trials have been done administering different subtypes of IFN-Is via different modes of administration [20]. Another type I IFN member, IFN-β, has shown promise in clinical trials. The inhaled interferon beta-1a was assessed in patients with COVID-19, in a randomized, double-blind phase 2 pilot trial in the United Kingdom. Patients who received nebulized IFN-β-1a had a lower risk of developing severe disease and showed significant clinical improvement when compared to the placebo group, suggesting that IFN-β-1a therapy may help COVID-19 patients recover faster [25]. Future studies are warranted to explore the effectiveness of this therapeutic approach in COVID-19, to better understand the association between distinct IFN-subtypes and antiviral effects, and the decreased immunoregulatory responses elicited by various IFN subtypes. These shreds of evidence would help select the optimal drug candidate for effective COVID-19 treatment.
5. Conclusion
In conclusion, our results support the accumulating data that COVID-19 severity is associated with IFNs down-regulation and extend this effect to the fact that SARS-CoV-2 infection is linked to a lack of host interferon responses. We further showed that the early expression of IFN type I and III genes was linked to various biological and clinical parameters associated with SARS-CoV-2 infection, suggesting that IFNs could be useful markers for tracking COVID-19 pathogenesis. In order to better anticipate disease severity and optimize patient management, a more expanded study with a larger size of patients, and a deeper understanding of the relationship between IFN deficiency, and viral pathogenesis, as well as the different immune responses triggered in COVID-19 are required.
6. Authors’ contributions
KA contributed to the conception and design of the experiments. RE and AO Performed experimental work. DD, CD, AO, IH and KA prepared all figures, analyzed the results and wrote the manuscript draft. FG contributed to review and editing of the manuscript with YZ, FH, ML participation. All authors read and approved the final version of the manuscript.
Funding
This research has been partially funded by COVID-19 program of Hassan II University of Casablanca.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Dr. Damien ARNOULT from INSERM, UMR_S 1197, Hôpital Paul Brousse, Villejuif, France, and Ayyoub KIHEL from Ain Chock Faculty of Sciences, Casablanca, Morocco for their technical assistance.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cyto.2023.156172.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.About the virus, (2022). https://www.euro.who.int/en/health-topics/health-emergencies/coronavirus-covid-19/novel-coronavirus-2019-ncov (accessed January 6, 2022).
- 2.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2019;9(2020):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan X., Sun L., Chen J., Chen Z.J. Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol. 2018;72:447–478. doi: 10.1146/ANNUREV-MICRO-102215-095605. [DOI] [PubMed] [Google Scholar]
- 4.Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-Like Receptor 3 Signaling via TRIF Contributes to a Protective Innate Immune Response to Severe Acute Respiratory Syndrome Coronavirus Infection. MBio. 2015;6:1–14. doi: 10.1128/MBIO.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S.J., Channappanavar R., Kanneganti T.D. Coronaviruses: Innate Immunity, Inflammasome Activation, Inflammatory Cell Death, and Cytokines. Trends Immunol. 2020;41:1083–1099. doi: 10.1016/J.IT.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ablasser A., Hur S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 2020;21:17–29. doi: 10.1038/S41590-019-0556-1. [DOI] [PubMed] [Google Scholar]
- 7.Darif D., Hammi I., Kihel A., El Idrissi Saik I., Guessous F., Akarid K. The pro-inflammatory cytokines in COVID-19 pathogenesis: What goes wrong? Microb. Pathog. 2021;153 doi: 10.1016/J.MICPATH.2021.104799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhary S., Sharma K., Silakari O. The interplay between inflammatory pathways and COVID-19: A critical review on pathogenesis and therapeutic options. Microb. Pathog. 2021;150 doi: 10.1016/J.MICPATH.2020.104673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trouillet-Assant S., Viel S., Gaymard A., Pons S., Richard J.C., Perret M., Villard M., Brengel-Pesce K., Lina B., Mezidi M., Bitker L., Belot A., Mouton W., Oriol G., Compagnon C., Generenaz L., Cheynet V., Ader F., Becker A., Benech N., Chauvelot P., Chidiac C., Conrad A., Ferry T., Miailhes P., Perpoint T., Perry M., Pouderoux C., Roux S., Triffault-Fillit C., Valour F., Hodane Y., Chauvelot L., Chabert P., Provoost J., David G., Folliet L., Lecam P., Billaud G., Bouscambert M., Escuret V., Frobert E., Bal A., Destras G., Josset L., Morfin F., Munier C., Valette M., Venet F., Garnier L., Pescarmona R., Lombard C., Walzer T. Type I IFN immunoprofiling in COVID-19 patients. J. Allergy Clin. Immunol. 2020;146:206. doi: 10.1016/J.JACI.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birra D., Benucci M., Landolfi L., Merchionda A., Loi G., Amato P., Licata G., Quartuccio L., Triggiani M., Moscato P. COVID 19: a clue from innate immunity. Immunol. Res. 2020;68:161–168. doi: 10.1007/S12026-020-09137-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourdon M., Manet C. Xavier Montagutelli, Host genetic susceptibility to viral infections: the role of type I interferon induction. Genes Immun. 2020;21:365–379. doi: 10.1038/s41435-020-00116-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pène F., Marin N., Roche N., Szwebel T.A., Merkling S.H., Treluyer J.M., Veyer D., Mouthon L., Blanc C., Tharaux P.L., Rozenberg F., Fischer A., Duffy D., Rieux-Laucat F., Kernéis S., Terrier B. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/SCIENCE.ABC6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onomoto K., Onoguchi K., Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell. Mol. Immunol. 2021;183(18):539–555. doi: 10.1038/s41423-020-00602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J.P. Metlay, G.W. Waterer, A.C. Long, A. Anzueto, J. Brozek, K. Crothers, L.A. Cooley, N.C. Dean, M.J. Fine, S.A. Flanders, M.R. Griffin, M.L. Metersky, D.M. Musher, M.I. Restrepo, C.G. Whitney, Diagnosis and Treatment of Adults with Community-acquired Pneumonia. An Official Clinical Practice Guideline of the American Thoracic Society and Infectious Diseases Society of America, Am. J. Respir. Crit. Care Med. 200 (2019) E45–E67. 10.1164/RCCM.201908-1581ST. [DOI] [PMC free article] [PubMed]
- 15.Park A., Iwasaki A. Type I and Type III Interferons – Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe. 2020;27:870. doi: 10.1016/J.CHOM.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazear H.M., Schoggins J.W., Diamond M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity. 2019;50:907–923. doi: 10.1016/J.IMMUNI.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.L. Prokunina-Olsson, N. Alphonse, R.E. Dickenson, J.E. Durbin, J.S. Glenn, R. Hartmann, S. V Kotenko, H.M. Lazear, T.R. O’brien, C. Odendall, O.O. Onabajo, H. Piontkivska, D.M. Santer, N.C. Reich, A. Wack, I. Zanoni, COVID-19 and emerging viral infections: The case for interferon lambda, (2020). 10.1084/jem.20200653. [DOI] [PMC free article] [PubMed]
- 18.Zanoni I. Interfering with SARS-CoV-2: are interferons friends or foes in COVID-19? Curr. Opin. Virol. 2021;50:119. doi: 10.1016/J.COVIRO.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eskandarian Boroujeni M., Sekrecka A., Antonczyk A., Hassani S., Sekrecki M., Nowicka H., Lopacinska N., Olya A., Kluzek K., Wesoly J., Bluyssen H.A.R. Dysregulated Interferon Response and Immune Hyperactivation in Severe COVID-19: Targeting STATs as a Novel Therapeutic Strategy. Front. Immunol. 2022;13:2153. doi: 10.3389/FIMMU.2022.888897/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schreiber G. The Role of Type I Interferons in the Pathogenesis and Treatment of COVID-19. Front. Immunol. 2020;11 doi: 10.3389/FIMMU.2020.595739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diamond M.S., Kanneganti T.D. Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol. 2022;232(23):165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.P. Bastard, L.B. Rosen, Q. Zhang, E. Michailidis, H.H. Hoffmann, Y. Zhang, K. Dorgham, Q. Philippot, J. Rosain, V. Béziat, J. Manry, E. Shaw, L. Haljasmägi, P. Peterson, L. Lorenzo, L. Bizien, S. Trouillet-Assant, K. Dobbs, A.A. de Jesus, A. Belot, A. Kallaste, E. Catherinot, Y. Tandjaoui-Lambiotte, J. Le Pen, G. Kerner, B. Bigio, Y. Seeleuthner, R. Yang, A. Bolze, A.N. Spaan, O.M. Delmonte, M.S. Abers, A. Aiuti, G. Casari, V. Lampasona, L. Piemonti, F. Ciceri, K. Bilguvar, R.P. Lifton, M. Vasse, D.M. Smadja, M. Migaud, J. Hadjadj, B. Terrier, D. Duffy, L. Quintana-Murci, D. van de Beek, L. Roussel, D.C. Vinh, S. Tangye, F. Haerynck, D. Dalmau, J. Martinez-Picado, P. Brodin, M.C. Nussenzweig, S. Boisson-Dupuis, C. Rodríguez-Gallego, G. Vogt, T.H. Mogensen, A.J. Oler, J. Gu, P.D. Burbelo, J.I. Cohen, A. Biondi, L.R. Bettini, M. DÁngio, P. Bonfanti, P. Rossignol, J. Mayaux, F. Rieux-Laucat, E.S. Husebye, F. Fusco, M.V. Ursini, L. Imberti, A. Sottini, S. Paghera, E. Quiros-Roldan, C. Rossi, R. Castagnoli, D. Montagna, A. Licari, G.L. Marseglia, X. Duval, J. Ghosn, J.S. Tsang, R. Goldbach-Mansky, K. Kisand, M.S. Lionakis, A. Puel, S.Y. Zhang, S.M. Holland, G. Gorochov, E. Jouanguy, C.M. Rice, A. Cobat, L.D. Notarangelo, L. Abel, H.C. Su, J.L. Casanova, A.A. Arias, B. Boisson, S. Boucherit, J. Bustamante, M. Chbihi, J. Chen, M. Chrabieh, T. Kochetkov, T. Le Voyer, D. Liu, Y. Nemirovskaya, M. Ogishi, D. Papandrea, C. Patissier, F. Rapaport, M. Roynard, N. Vladikine, M. Woollett, P. Zhang, A. Kashyap, L. Ding, M. Bosticardo, Q. Wang, S. Ochoa, H. Liu, S.D. Chauvin, M. Stack, G. Koroleva, N. Bansal, C.L. Dalgard, A.L. Snow, J. Abad, S. Aguilera-Albesa, O.M. Akcan, I.A. Darazam, J.C. Aldave, M.A. Ramos, S.A. Nadji, G. Alkan, J. Allardet-Servent, L.M. Allende, L. Alsina, M.A. Alyanakian, B. Amador-Borrero, Z. Amoura, A. Antolí, S. Arslan, S. Assant, T. Auguet, A. Azot, F. Bajolle, A. Baldolli, M. Ballester, H.B. Feldman, B. Barrou, A. Beurton, A. Bilbao, G. Blanchard-Rohner, I. Blanco, A. Blandinières, D. Blazquez-Gamero, M. Bloomfield, M. Bolivar-Prados, R. Borie, A.A. Bousfiha, C. Bouvattier, O. Boyarchuk, M.R.P. Bueno, J. Bustamante, J.J.C. Agra, S. Calimli, R. Capra, M. Carrabba, C. Casasnovas, M. Caseris, M. Castelle, F. Castelli, M.C. de Vera, M. V. Castro, E. Catherinot, M. Chalumeau, B. Charbit, M.P. Cheng, P. Clavé, B. Clotet, A. Codina, F. Colkesen, F. Colkesen, R. Colobran, C. Comarmond, A.G. Corsico, D. Dalmau, D.R. Darley, N. Dauby, S. Dauger, L. de Pontual, A. Dehban, G. Delplancq, A. Demoule, A. Di Sabatino, J.L. Diehl, S. Dobbelaere, S. Durand, W. Eldars, M. Elgamal, M.H. Elnagdy, M. Emiroglu, E.H. Erdeniz, S.E. Aytekin, R. Euvrard, R. Evcen, G. Fabio, L. Faivre, A. Falck, M. Fartoukh, M. Faure, M.F. Arquero, C. Flores, B. Francois, V. Fumadó, F. Fusco, B.G. Solis, P. Gaussem, J. Gil-Herrera, L. Gilardin, M.G. Alarcon, M. Girona-Alarcón, J.C. Goffard, F. Gok, R. González-Montelongo, A. Guerder, Y. Gul, S.N. Guner, M. Gut, J. Hadjadj, F. Haerynck, R. Halwani, L. Hammarström, N. Hatipoglu, E. Hernandez-Brito, M.S. Holanda-Peña, J.P. Horcajada, S. Hraiech, L. Humbert, A.D. Iglesias, A. Íñigo-Campos, M. Jamme, M.J. Arranz, I. Jordan, F. Kanat, H. Kapakli, I. Kara, A. Karbuz, K.K. Yasar, S. Keles, Y.K. Demirkol, A. Klocperk, Z.J. Król, P. Kuentz, Y.W.M. Kwan, J.C. Lagier, Y.L. Lau, F. Le Bourgeois, Y.S. Leo, R.L. Lopez, D. Leung, M. Levin, M. Levy, R. Lévy, Z. Li, A. Linglart, J.M. Lorenzo-Salazar, C. Louapre, C. Lubetzki, C.E. Luyt, D.C. Lye, D. Mansouri, M. Marjani, J.M. Pereira, A. Martin, D.M. Pueyo, J. Martinez-Picado, I. Marzana, A. Mathian, L.R.B. Matos, G. V. Matthews, J. Mayaux, J.L. Mège, I. Melki, J.F. Meritet, O. Metin, I. Meyts, M. Mezidi, I. Migeotte, M. Millereux, T. Mirault, C. Mircher, M. Mirsaeidi, A.M. Melián, A.M. Martinez, P. Morange, C. Mordacq, G. Morelle, S. Mouly, A. Muñoz-Barrera, C. Nafati, J.F. Neves, L.F.P. Ng, Y.N. Medina, E.N. Cuadros, J. Gonzalo Ocejo-Vinyals, Z. Orbak, M. Oualha, T. Özçelik, Q.P. Hammarström, C. Parizot, T. Pascreau, E. Paz-Artal, R.P. de Diego, A. Philippe, Q. Philippota, L. Planas-Serra, D. Ploin, J. Poissy, G. Poncelet, M. Pouletty, P. Quentric, D. Raoult, A.S. Rebillat, I. Reisli, P. Ricart, J.C. Richard, N. Rivet, J.G. Rivière, G.R. Blanch, C. Rodrigo, C. Rodriguez-Gallego, A. Rodríguez-Palmero, C.S. Romero, A. Rothenbuhler, F. Rozenberg, M.Y.R. del Prado, J.S. Riera, O. Sanchez, S. Sánchez-Ramón, A. Schluter, M. Schmidt, C.E. Schweitzer, F. Scolari, A. Sediva, L.M. Seijo, D. Sene, S. Senoglu, M.R.J. Seppänen, A.S. Ilovich, M. Shahrooei, D. Smadja, A. Sobh, X.S. Moreno, J. Solé-Violán, C. Soler, P. Soler-Palacín, Y. Stepanovskiy, A. Stoclin, F. Taccone, Y. Tandjaoui-Lambiottea, J.L. Taupin, S.J. Tavernier, B. Terrier, C. Thumerelle, G. Tomasoni, J. Toubiana, J.T. Alvarez, S. Trouillet-Assanta, J. Troya, A. Tucci, M.V. Ursini, Y. Uzunhan, P. Vabres, J. Valencia-Ramos, A.M. van Den Rym, I. Vandernoot, H. Vatansev, V. Vélez-Santamaria, S. Viel, C. Vilain, M.E. Vilaire, A. Vincent, G. Voiriot, F. Vuotto, A. Yosunkaya, B.E. Young, F. Yucel, F. Zannad, M. Zatz, A. Belota, G. Foti, G. Bellani, G. Citerio, E. Contro, A. Pesci, M.G. Valsecchi, M. Cazzaniga, C. Bole-Feysot, S. Lyonnet, C. Masson, P. Nitschke, A. Pouliet, Y. Schmitt, F. Tores, M. Zarhrate, L. Abela, C. Andrejak, F. Angoulvant, D. Bachelet, R. Basmaci, S. Behillil, M. Beluze, D. Benkerrou, K. Bhavsar, F. Bompart, L. Bouadma, M. Bouscambert, M. Caralp, M. Cervantes-Gonzalez, A. Chair, A. Coelho, C. Couffignal, S. Couffin-Cadiergues, E. D’ortenzio, C. da Silveira, M.P. Debray, D. Deplanque, D. Descamps, M. Desvallées, A. Diallo, A. Diouf, C. Dorival, F. Dubos, X. Duval, P. Eloy, V.V.E. Enouf, H. Esperou, M. Esposito-Farese, M. Etienne, N. Ettalhaoui, N. Gault, A. Gaymard, J. Ghosn, T. Gigante, I. Gorenne, J. Guedj, A. Hoctin, I. Hoffmann, S. Jaafoura, O. Kafif, F. Kaguelidou, S. Kali, A. Khalil, C. Khan, C. Laouénan, S. Laribi, M. Le, Q. Le Hingrat, S. Le Mestre, H. Le Nagard, F.X. Lescure, Y. Lévy, C. Levy-Marchal, B. Lina, G. Lingas, J.C. Lucet, D. Malvy, M. Mambert, F. Mentré, N. Mercier, A. Meziane, H. Mouquet, J. Mullaert, N. Neant, M. Noret, J. Pages, A. Papadopoulos, C. Paul, N. Peiffer-Smadja, V. Petrov-Sanchez, G. Peytavin, O. Picone, O. Puéchal, M. Rosa-Calatrava, B. Rossignol, P. Rossignol, C. Roy, M. Schneider, C. Semaille, N.S. Mohammed, L. Tagherset, C. Tardivon, M.C. Tellier, F. Téoulé, O. Terrier, J.F. Timsit, T. Treoux, C. Tual, S. Tubiana, S. van der Werf, N. Vanel, A. Veislinger, B. Visseaux, A. Wiedemann, Y. Yazdanpanah, L. Abelc, A. Alcover, H. Aschard, K. Astrom, P. Bousso, P. Bruhns, A. Cumano, C. Demangel, L. Deriano, J. Di Santo, F. Dromer, G. Eberl, J. Enninga, J. Fellay, I. Gomperts-Boneca, M. Hasan, S. Hercberg, O. Lantz, H. Mouquet, E. Patin, S. Pellegrini, S. Pol, A. Rausell, L. Rogge, A. Sakuntabhai, O. Schwartz, B. Schwikowski, S. Shorte, F. Tangy, A. Toubert, M. Touvier, M.N. Ungeheuer, M.L. Albert, D. Duffy, L. Quintana-Murci, L. Alavoine, K.K.A. Amat, S. Behillil, J. Bielicki, P. Bruijning, C. Burdet, E. Caumes, C. Charpentier, B. Coignard, Y. Costa, S. Couffin-Cadiergues, F. Damond, A. Dechanet, C. Delmas, D. Descamps, X. Duval, J.L. Ecobichon, V. Enouf, H. Espérou, W. Frezouls, N. Houhou, E. Ilic-Habensus, O. Kafif, J. Kikoine, Q. Le Hingrat, D. Lebeaux, A. Leclercq, J. Lehacaut, S. Letrou, B. Lina, J.C. Lucet, D. Malvy, P. Manchon, M. Mandic, M. Meghadecha, J. Motiejunaite, M. Nouroudine, V. Piquard, A. Postolache, C. Quintin, J. Rexach, L. Roufai, Z. Terzian, M. Thy, S. Tubiana, S. van der Werf, V. Vignali, B. Visseaux, Y. Yazdanpanah, M. van Agtmael, A.G. Algera, F. van Baarle, D. Bax, M. Beudel, H.J. Bogaard, M. Bomers, L. Bos, M. Botta, J. de Brabander, G. Bree, M.C. Brouwer, S. de Bruin, M. Bugiani, E. Bulle, O. Chouchane, A. Cloherty, P. Elbers, L. Fleuren, S. Geerlings, B. Geerts, T. Geijtenbeek, A. Girbes, B. Goorhuis, M.P. Grobusch, F. Hafkamp, L. Hagens, J. Hamann, V. Harris, R. Hemke, S.M. Hermans, L. Heunks, M.W. Hollmann, J. Horn, J.W. Hovius, M.D. de Jong, R. Koning, N. van Mourik, J. Nellen, F. Paulus, E. Peters, T. van der Poll, B. Preckel, J.M. Prins, J. Raasveld, T. Reijnders, M. Schinkel, M.J. Schultz, A. Schuurman, K. Sigaloff, M. Smit, C.S. Stijnis, W. Stilma, C. Teunissen, P. Thoral, A. Tsonas, M. van der Valk, D. Veelo, A.P.J. Vlaar, H. de Vries, M. van Vugt, W. Joost Wiersinga, D. Wouters, A.H. Zwinderman, D. van de Beek, L. Abelb, F. Iuti, S. Al Muhsen, F. Al-Mulla, M.S. Anderson, A.A. Arias, H.B. Feldman, D. Bogunovic, A. Bolze, A. Bondarenko, A.A. Bousfiha, P. Brodin, Y. Bryceson, C.D. Bustamante, M. Butte, S. Chakravorty, J. Christodoulou, E. Cirulli, A. Condino-Neto, M.A. Cooper, C.L. Dalgard, J.L. DeRisi, M. Desai, B.A. Drolet, S. Espinosa, J. Fellay, C. Flores, J.L. Franco, P.K. Gregersen, F. Haerynck, D. Hagin, J. Heath, S.E. Henrickson, E. Hsieh, K. Imai, Y. Itan, T. Karamitros, K. Kisanda, C.L. Ku, Y.L. Lau, Y. Ling, C.L. Lucas, T. Maniatis, D. Mansouri, L. Marodi, J.D. Milner, K. Mironska, T. Morio, L.D. Notarangeloa, G. Novelli, A. Novelli, C. O’Farrelly, S. Okada, T. Ozcelik, R.P. de Diego, A.M. Planas, C. Prando, A. Pujol, L. Quintana-Murci, L. Renia, A. Renieri, C. Rodríguez-Gallego, V. Sancho-Shimizu, V. Sankaran, K.S. Barrett, M. Shahrooei, A. Snow, P. Soler-Palacín, A.N. Spaan, S. Turvey, F. Uddin, M.J. Uddin, D. van de Beek, S.E. Vazquez, D.C. Vinh, H. von Bernuth, N. Washington, P. Zawadzki, H.C. Sua, J.L. Casanovaa, Autoantibodies against type I IFNs in patients with life-threatening COVID-19, Science. 370 (2020). 10.1126/SCIENCE.ABD4585.
- 23.Q. Zhang, Z. Liu, M. Moncada-Velez, J. Chen, M. Ogishi, B. Bigio, R. Yang, A.A. Arias, Q. Zhou, J.E. Han, A.C. Ugurbil, P. Zhang, F. Rapaport, J. Li, A.N. Spaan, B. Boisson, S. Boisson-Dupuis, J. Bustamante, A. Puel, M.J. Ciancanelli, S.Y. Zhang, V. Béziat, E. Jouanguy, L. Abel, A. Cobat, J.L. Casanova, P. Bastard, C. Korol, J. Rosain, Q. Philippot, M. Chbihi, L. Lorenzo, L. Bizien, A.L. Neehus, G. Kerner, Y. Seeleuthner, J. Manry, T. Le Voyer, B. Boisson, S. Boisson-Dupuis, J. Bustamante, A. Puel, S.Y. Zhang, V. Béziat, E. Jouanguy, L. Abel, A. Cobat, P. Bastard, J. Rosain, Q. Philippot, M. Chbihi, L. Lorenzo, L. Bizien, A.L. Neehus, G. Kerner, Y. Seeleuthner, J. Manry, T. Le Voyer, B. Boisson, S. Boisson-Dupuis, J. Bustamante, A. Puel, S.Y. Zhang, V. Béziat, E. Jouanguy, L. Abel, A. Cobat, J. Le Pen, W.M. Schneider, B.S. Razooky, H.H. Hoffmann, E. Michailidis, C.M. Rice, I.K.D. Sabli, S. Hodeib, V. Sancho-Shimizu, K. Bilguvar, J. Ye, T. Maniatis, A. Bolze, A.A. Arias, A.A. Arias, Y. Zhang, L.D. Notarangelo, H.C. Su, Y. Zhang, L.D. Notarangelo, H.C. Su, F. Onodi, S. Korniotis, L. Karpf, V. Soumelis, L. Bonnet-Madin, A. Amara, K. Dorgham, G. Gorochov, N. Smith, D. Duffy, L. Moens, I. Meyts, P. Meade, A. García-Sastre, F. Krammer, A. Corneau, C. Masson, Y. Schmitt, A. Schlüter, A. Pujol, T. Khan, N. Marr, J. Fellay, J. Fellay, J. Fellay, L. Roussel, D.C. Vinh, M. Shahrooei, M. Shahrooei, M.F. Alosaimi, F. Alsohime, R. Hasanato, D. Mansouri, H. Al-Saud, F. Almourfi, F. Al-Mulla, S.Z. Al-Muhsen, S. Al Turki, S. Al Turki, D. van de Beek, A. Biondi, L.R. Bettini, M. D’Angio, P. Bonfanti, L. Imberti, A. Sottini, S. Paghera, E. Quiros-Roldan, C. Rossi, A.J. Oler, M.F. Tompkins, C. Alba, C.L. Dalgard, I. Vandernoot, G. Smits, J.C. Goffard, I. Migeotte, F. Haerynck, P. Soler-Palacin, A. Martin-Nalda, R. Colobran, P.E. Morange, S. Keles, F. Çölkesen, T. Ozcelik, K.K. Yasar, S. Senoglu, Ş.N. Karabela, C. Rodríguez-Gallego, C. Rodríguez-Gallego, G. Novelli, S. Hraiech, Y. Tandjaoui-Lambiotte, Y. Tandjaoui-Lambiotte, X. Duval, C. Laouénan, X. Duval, C. Laouénan, C. Laouénan, A.L. Snow, C.L. Dalgard, J.D. Milner, T.H. Mogensen, A.N. Spaan, J. Bustamante, M.J. Ciancanelli, T. Maniatis, V. Soumelis, M. Nussenzweig, A. García-Sastre, A. García-Sastre, A. García-Sastre, R.P. Lifton, R.P. Lifton, R.P. Lifton, G. Foti, G. Bellani, G. Citerio, E. Contro, A. Pesci, M.G. Valsecchi, M. Cazzaniga, J. Abad, I. Blanco, C. Rodrigo, S. Aguilera-Albesa, O.M. Akcan, I.A. Darazam, J.C. Aldave, M.A. Ramos, S.A. Nadji, G. Alkan, J. Allardet-Servent, L.M. Allende, L. Alsina, M.A. Alyanakian, B. Amador-Borrero, S. Mouly, D. Sene, Z. Amoura, A. Mathian, A. Antolí, G.R. Blanch, J.S. Riera, X.S. Moreno, S. Arslan, S. Assant, T. Auguet, A. Azot, F. Bajolle, J. Bustamante, R. Lévy, M. Oualha, A. Baldolli, M. Ballester, H.B. Feldman, B. Barrou, A. Beurton, A. Bilbao, G. Blanchard-Rohner, A. Blandinières, N. Rivet, D. Blazquez-Gamero, M. Bloomfield, M. Bolivar-Prados, P. Clavé, R. Borie, C. Bosteels, B.N. Lambrecht, E. van Braeckel, A.A. Bousfiha, C. Bouvattier, A. Vincent, O. Boyarchuk, M.R.P. Bueno, M. V. Castro, L.R.B. Matos, M. Zatz, J.J.C. Agra, S. Calimli, R. Capra, M. Carrabba, G. Fabio, C. Casasnovas, V. Vélez-Santamaria, M. Caseris, A. Falck, G. Poncelet, M. Castelle, F. Castelli, M.C. de Vera, E. Catherinot, M. Chalumeau, J. Toubiana, B. Charbit, Z. Li, S. Pellegrini, M.P. Cheng, B. Clotet, A. Codina, F. Colkesen, F. Çölkesen, R. Colobran, C. Comarmond, D. Dalmau, D. Dalmau, D.R. Darley, N. Dauby, S. Dauger, F. Le Bourgeois, M. Levy, L. de Pontual, A. Dehban, G. Delplancq, A. Demoule, J.L. Diehl, S. Dobbelaere, S. Durand, C. Mircher, A.S. Rebillat, M.E. Vilaire, W. Eldars, M. Elgamal, M.H. Elnagdy, M. Emiroglu, E.H. Erdeniz, S.E. Aytekin, R. Euvrard, R. Evcen, L. Faivre, M. Fartoukh, Q. Philippot, M. Faure, M.F. Arquero, C. Flores, C. Flores, C. Flores, C. Flores, B. Francois, V. Fumadó, V. Fumadó, V. Fumadó, F. Fusco, M.V. Ursini, B.G. Solis, R.P. de Diego, A.M. van Den Rym, P. Gaussem, J. Gil-Herrera, L. Gilardin, M.G. Alarcon, M. Girona-Alarcón, J.C. Goffard, F. Gok, A. Yosunkaya, R. González-Montelongo, A. Íñigo-Campos, J.M. Lorenzo-Salazar, A. Muñoz-Barrera, A. Guerder, Y. Gul, S.N. Guner, M. Gut, J. Hadjadj, F. Haerynck, R. Halwani, L. Hammarström, N. Hatipoglu, E. Hernandez-Brito, C. Heijmans, M.S. Holanda-Peña, J.P. Horcajada, L. Hoste, E. Hoste, S. Hraiech, L. Humbert, C. Mordacq, C. Thumerelle, F. Vuotto, A.D. Iglesias, M. Jamme, M.J. Arranz, I. Jordan, P. Jorens, F. Kanat, H. Kapakli, I. Kara, A. Karbuz, K.K. Yasar, S. Senoglu, S. Keles, Y.K. Demirkol, A. Klocperk, Z.J. Król, P. Kuentz, Y.W.M. Kwan, J.C. Lagier, Y.L. Lau, D. Leung, Y.S. Leo, B.E. Young, R.L. Lopez, M. Levin, A. Linglart, B. Loeys, C. Louapre, C. Lubetzki, C.E. Luyt, D.C. Lye, M. Marjani, J.M. Pereira, A. Martin, P. Soler-Palacín, D.M. Pueyo, J. Martinez-Picado, I. Marzana, G. V. Matthews, J. Mayaux, C. Parizot, P. Quentric, J.L. Mège, D. Raoult, I. Melki, J.F. Meritet, O. Metin, M. Mezidi, I. Migeotte, F. Taccone, M. Millereux, T. Mirault, M. Mirsaeidi, A.M. Melián, A.M. Martinez, P. Morange, G. Morelle, L. Naesens, C. Nafati, J.F. Neves, L.F.P. Ng, Y.N. Medina, E.N. Cuadros, J. Gonzalo Ocejo-Vinyals, Z. Orbak, T. Özçelik, Q. Pan-Hammarström, T. Pascreau, E. Paz-Artal, A. Philippe, L. Planas-Serra, A. Schluter, D. Ploin, S. Viel, J. Poissy, M. Pouletty, I. Reisli, P. Ricart, J.C. Richard, J.G. Rivière, C. Rodriguez-Gallego, C. Rodriguez-Gallego, A. Rodríguez-Palmero, C.S. Romero, A. Rothenbuhler, F. Rozenberg, M.Y.R. del Prado, O. Sanchez, S. Sánchez-Ramón, M. Schmidt, C.E. Schweitzer, F. Scolari, A. Sediva, L.M. Seijo, M.R.J. Seppänen, A.S. Ilovich, H. Slabbynck, D.M. Smadja, A. Sobh, J. Solé-Violán, C. Soler, Y. Stepanovskiy, A. Stoclin, Y. Tandjaoui-Lambiotte, J.L. Taupin, S.J. Tavernier, B. Terrier, G. Tomasoni, J.T. Alvarez, S. Trouillet-Assant, J. Troya, A. Tucci, Y. Uzunhan, P. Vabres, J. Valencia-Ramos, S. van de Velde, J. van Praet, I. Vandernoot, H. Vatansev, C. Vilain, G. Voiriot, F. Yucel, F. Zannad, A. Belot, C. Bole-Feysot, S. Lyonnet, C. Masson, P. Nitschke, A. Pouliet, Y. Schmitt, F. Tores, M. Zarhrate, M. Shahrooei, L. Abel, C. Andrejak, F. Angoulvant, D. Bachelet, K. Bhavsar, L. Bouadma, A. Chair, C. Couffignal, C. Da Silveira, M.P. Debray, X. Duval, P. Eloy, M. Esposito-Farese, N. Ettalhaoui, N. Gault, J. Ghosn, I. Gorenne, I. Hoffmann, O. Kafif, S. Kali, A. Khalil, C. Laouénan, S. Laribi, M. Le, Q. Le Hingrat, F.X. Lescure, J.C. Lucet, F. Mentré, J. Mullaert, N. Peiffer-Smadja, G. Peytavin, C. Roy, M. Schneider, N.S. Mohammed, L. Tagherset, C. Tardivon, M.C. Tellier, J.F. Timsit, T. Trioux, S. Tubiana, R. Basmaci, S. Behillil, M. Beluze, D. Benkerrou, C. Dorival, A. Meziane, F. Téoulé, F. Bompart, M. Bouscambert, A. Gaymard, B. Lina, M. Rosa-Calatrava, O. Terrier, M. Caralp, M. Cervantes-Gonzalez, E. D’Ortenzio, O. Puéchal, C. Semaille, A. Coelho, A. Diouf, A. Hoctin, M. Mambert, S. Couffin-Cadiergues, D. Deplanque, D. Descamps, B. Visseaux, M. Desvallées, C. Khan, A. Diallo, N. Mercier, C. Paul, V. Petrov-Sanchez, F. Dubos, V.V.E. Enouf, H. Mouquet, H. Esperou, S. Jaafoura, A. Papadopoulos, M. Etienne, T. Gigante, B. Rossignol, J. Guedj, H. Le Nagard, G. Lingas, N. Neant, F. Kaguelidou, Y. Lévy, A. Wiedemann, Y. Lévy, A. Wiedemann, C. Levy-Marchal, D. Malvy, M. Noret, J. Pages, O. Picone, P. Rossignol, C. Tual, A. Veislinger, S. van der Werf, N. Vanel, Y. Yazdanpanah, L. Alavoine, Y. Costa, X. Duval, J.L. Ecobichon, W. Frezouls, E. Ilic-Habensus, A. Leclercq, J. Lehacaut, S. Letrou, M. Mandic, M. Nouroudine, C. Quintin, J. Rexach, S. Tubiana, V. Vignali, K.K.A. Amat, S. Behillil, V. Enouf, S. van der Werf, J. Bielicki, P. Bruijning, C. Burdet, C. Burdet, E. Caumes, C. Charpentier, F. Damond, D. Descamps, Q. Le Hingrat, B. Visseaux, B. Coignard, S. Couffin-Cadiergues, C. Delmas, H. Espérou, L. Roufai, A. Dechanet, N. Houhou, O. Kafif, J. Kikoine, P. Manchon, V. Piquard, A. Postolache, Z. Terzian, D. Lebeaux, B. Lina, J.C. Lucet, D. Malvy, M. Meghadecha, J. Motiejunaite, M. Thy, M. van Agtmael, M. Bomers, O. Chouchane, S. Geerlings, B. Goorhuis, M.P. Grobusch, V. Harris, S.M. Hermans, J.W. Hovius, J. Nellen, E. Peters, T. van der Poll, J.M. Prins, T. Reijnders, M. Schinkel, K. Sigaloff, C.S. Stijnis, M. van der Valk, M. van Vugt, W. Joost Wiersinga, A.G. Algera, F. van Baarle, L. Bos, M. Botta, S. de Bruin, E. Bulle, P. Elbers, L. Fleuren, A. Girbes, L. Hagens, L. Heunks, J. Horn, N. van Mourik, F. Paulus, J. Raasveld, M.J. Schultz, M. Smit, W. Stilma, P. Thoral, A. Tsonas, H. de Vries, D. Bax, A. Cloherty, M. Beudel, M.C. Brouwer, R. Koning, D. van de Beek, H.J. Bogaard, J. de Brabander, G. de Bree, M. Bugiani, B. Geerts, M.W. Hollmann, B. Preckel, D. Veelo, T. Geijtenbeek, F. Hafkamp, J. Hamann, R. Hemke, M.D. de Jong, A. Schuurman, C. Teunissen, A.P.J. Vlaar, D. Wouters, A.H. Zwinderman, L. Abel, A. Aiuti, S. Al Muhsen, F. Al-Mulla, M.S. Anderson, A.A. Arias, H.B. Feldman, D. Bogunovic, Y. Itan, A. Bolze, E. Cirulli, K.S. Barrett, N. Washington, A. Bondarenko, A.A. Bousfiha, P. Brodin, Y. Bryceson, C.D. Bustamante, M. Butte, G. Casari, S. Chakravorty, J. Christodoulou, S. Le Mestre, A. Condino-Neto, M.A. Cooper, C.L. Dalgard, A. David, J.L. DeRisi, J.L. DeRisi, M. Desai, B.A. Drolet, S. Espinosa, J. Fellay, C. Flores, J.L. Franco, P.K. Gregersen, F. Haerynck, D. Hagin, J. Heath, S.E. Henrickson, E. Hsieh, K. Imai, T. Karamitros, K. Kisand, C.L. Ku, Y.L. Lau, Y. Ling, C.L. Lucas, T. Maniatis, D. Mansouri, L. Marodi, J. Milner, K. Mironska, T. Morio, L.D. Notarangelo, H.C. Su, A. Novelli, C. O’Farrelly, S. Okada, T. Ozcelik, R.P. de Diego, A.M. Planas, C. Prando, A. Pujol, L. Quintana-Murci, L. Renia, A. Renieri, C. Rodríguez-Gallego, V. Sancho-Shimizu, V. Sankaran, M. Shahrooei, A. Snow, P. Soler-Palacín, A.N. Spaan, S. Tangye, S. Turvey, F. Uddin, M.J. Uddin, M.J. Uddin, D. van de Beek, S.E. Vazquez, D.C. Vinh, H. von Bernuth, P. Zawadzki, H. Jing, W. Tung, K. Meguro, E. Shaw, H. Jing, S. Shafer, L. Zheng, Z. Zhang, S. Kubo, S.D. Chauvin, K. Meguro, E. Shaw, M. Lenardo, C.R. Luthers, B.M. Bauman, S. Shafer, L. Zheng, Z. Zhang, S. Kubo, S.D. Chauvin, M. Lenardo, J. Lack, E. Karlins, D.M. Hupalo, J. Rosenberger, G. Sukumar, M.D. Wilkerson, X. Zhang, Inborn errors of type I IFN immunity in patients with life-threatening COVID-19, Science. 370 (2020). 10.1126/SCIENCE.ABD4570.
- 24.Yang L., Wang J., Hui P., Yarovinsky T.O., Badeti S., Pham K., Liu C. Potential role of IFN-α in COVID-19 patients and its underlying treatment options. Appl. Microbiol. Biotechnol. 2021;105:4005–4015. doi: 10.1007/S00253-021-11319-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monk P.D., Marsden R.J., Tear V.J., Brookes J., Batten T.N., Mankowski M., Gabbay F.J., Davies D.E., Holgate S.T., Ho L.P., Clark T., Djukanovic R., Wilkinson T.M.A., Crooks M.G., Dosanjh D.P., Siddiqui S., Rahman N.M., Smith J.A., Horsley A., Harrison T.W., Saralaya D., McGarvey L., Watson A., Foster E., Fleet A., Singh D., Hemmings S., Aitken S., Dudley S., Beegan R., Thompson A., Rodrigues P.M. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. Respir. Med. 2021;9:196. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.