Abstract
Introduction
Classical Philadelphia-negative myeloproliferative neoplasm (MPN) includes Essential Thrombocythemia (ET), Polycythemia Vera (PV) and Primary Myelofibrosis (PMF). The JAK2V617F mutation is part of the major criteria for diagnosis of MPN. WT1 is reported to be highly overexpressed in most hematological malignancy. Our aim was to explore the combination value of JAK2V617F allele burden and WT1 expression in distinguishing the subtype of MPN patients.
Methods
Allele specific real-time quantitative fluorescence PCR (AS-qPCR) was conducted to detect JAK2V617F allele burden. WT1 expression was assessed by RQ-PCR. Our study is a retrospective study.
Results
JAK2V617F allele burden and WT1 expression were different in MPN subgroups. The expression of WT1 in PMF and PV is higher than in ET. JAK2V617F allele burden in PMF and PV is also higher than in ET. ROC analysis indicated that combination of JAK2V617F allele burden and WT1 expression to discriminate ET and PV, ET and PMF, PV and PMF is 0.956, 0.871, 0.737 respectively. Furthermore, their ability to distinguish ET patients with high Hb levels from PV patients with high platelet counts is 0.891.
Conclusions
Our data revealed that combination of JAK2V617F allele burden and WT1 expression is useful in distinguishing the subtype of MPN patients.
Keywords: JAK2V617, WT1, MPN, ET, PV, PMF
Introduction
Myeloproliferative neoplasm (MPN) is a group of clonal hematopoietic stem cell disorders characterized by clonal proliferation of one or more hematopoietic progenitors in the bone marrow, and may progress to acute leukemia.1 Classical Philadelphia-negative (Ph-negative) MPN can be divided into three main sets: essential thrombocythemia (ET), Polycythemia vera (PV) and primary myelofibrosis (PMF).2 Currently, bone marrow (BM) histology, including megakaryocyte morphology and degree of myeloproliferation, is essential in the distinguish of ET from PV and PMF.3 However, when BM fibrosis was absent and the morphology changes of megakaryocyte are atypical, it is difficult to distinguish ET and Pre-primary myelofibrosis (Pre-PMF).4 In a large multicenter study, that included 1104 patients with ET and pre-PMF, significant differences in the occurrence of bleeding, rate of death, progression to overt myelofibrosis and transformation to leukemia were found.5 Since the different prognosis and treatment for MPN patients, it is urgently needed to find new tools to correctly distinguish the specific type of MPNs.
Human Janus kinase 2 (JAK2) gene is mapped to chromosome region 9p24.1.6 In normal hematopoietic cells, JAK2 consists two homologous kinases: one is an enzymatically active kinase domain and the other is a catalytically inactive pseudokinase domain (JH2).7 The most common mutation of JAK2 is the substitution of valine with phenylalanine at position 617 (JAK2-V617F). JAK2V617F mutations mainly occurred in MPN patients and a small part of AML patients.8 The mutation frequency of JAK2V617F in PV patients was more than 95.0% and 55.0-60.0% in ET and PMF patients.9 The finding that JAK2V617F was mutated in patients with myeloproliferative disorders has changed the diagnostic landscape of MPNs greatly.10 JAK2V617F mutation is essential in the screening of MPN. However, For JAK2V617F-positive patients, the patient can only be diagnosed as MPN, we could not distinguish their specific subtypes.
Wilms’ tumor 1 gene (WT1) is known for its function in organ development and cell differentiation.11 It is a transcription factor that guides the development of several organs and tissues.12 WT1 is essential in the maintenance of tissue homeostasis and can act as either a tumor suppressor or an oncogene in several solid tumors and leukemia.13 In a normal human bone marrow, WT1 is expressed in a small percentage and confined to CD34+ cells.14 However, existing studies suggested that WT1 tend to be highly overexpressed in most de novo acute myeloid leukemias (AML),15,16 myelodysplastic syndromes17,18 and chronic myelogenous leukemia (CML).19 Recent years, increasing number of studies have focused on evaluating the correlation between WT1 and MPNs. Guglielmelli et al.20 using an approach of transcriptomic micro array showed that WT1 level was higher in PMF-derived CD34+ cells than in PV and patients who had higher WT1 expression levels tend to be disease active. In 2016, Gallo et al.21 reported a positive correlation between WT1 transcript levels and IPSS score at diagnosis in PMF patients and they also found that WT1 expression can be a marker of response to therapy. Recently, Cottin and his colleagues22 validated WT1 expression was significantly increased during the myelofibrotic transformation of ET or PV. In their study, they also reported that WT1 transcription levels was related to age over 65, splenomegaly and thrombocytopenia. Based on the above findings, we assume that the combination of WT1 expression and JAK2V617F allele burden may be helpful in distinguishing the subtype of MPN patients.
Materials and Methods
Sample and Collection
A total of 90 (38 men and 52 women, median age 62,22–85) bone marrow from new diagnosed MPN patients with JAK2V617F mutation positive were collected from January 2019 and October 2021 at Zhongnan Hospital of Wuhan University. Of all the 90 patients, 45 patients were ET, 26 patients were PV, and 19 patients were PMF. All patient details are de-identified in our study. The diagnosis was made according to the 2016 revision to WHO myeloid neoplasms and acute leukemia.23 All patients included in our study had no any other hematological malignancy. All MPN patients were analyzed at diagnosis before any treatment, and informed consent was obtained from all individuals involved in this study. Our study is a retrospective study.
DNA and RNA Extraction
Total genomic DNA was extracted from EDTA-anticoagulated bone marrow samples by using DNA Isolation Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s instructions. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) as described by the manufacturer. After extraction, DNA and RNA concentration was measured using Nanodrop 2000 spectrophotometer (Thermo Scientific Inc., Waltham, MA, USA).
JAK2V617F Allele Burden Detection and WT1 Expression Detection
Allele specific real-time quantitative fluorescence PCR (AS-qPCR) was conducted to detect JAK2V617F allele burden by using a quantitative fluorescent DNA detection kit (Yuanqi, Shanghai, China). JAK2 wild-type and mutation type were both amplified with respective primers, and JAK2V617F mutant allele burden was relatively quantified (JAK2V617F%) by determining the percentage of JAK2 mutation type quantity in the total JAK2 quantity. WT1 expression was assessed by RQ-PCR using WT1 ProfileQuant Kit (Yuanqi, Shanghai, China). Calibration curves with plasmids containing ABL1 and WT1 target sequences were used. The WT1 transcript values measured by RQ-PCR were normalized with respect to the number of ABL1 transcript and expressed per 104 ABL1 copies. The WT1 expression was transformed into a logarithm to ensure that values follow a normal distribution for statistical analysis. All PCR analyses were performed in duplicate. RQ-PCR was performed on ABI Prism 7500 system (Applied Biosystems).
Statistical Analysis
Patients’ characteristics were summarized as numbers (range) for qualitative variables. Shapiro-Wilk test was performed to check the normality of the distribution. For normally distributed data, student’s t test was used to compare 2 groups of continuous variables and one-way ANOVA was used for the comparison among multiple groups, while non-normally distributed data was analyzed by Kruskal-Wallis variance analysis. To evaluate the diagnostic value of JAK2 and WT1, receiver operating characteristic (ROC) curves were done, and the area under the ROC curves was determined. SPSS 17.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 5 software (GraphPad software, La Jolla, CA, USA) were used to analyze the data. All statistical tests were two-sided and P < .05 was considered statistically significant.
Results
Characteristics of the Studied Participants
The clinical and laboratory features of enrolled MPN patients are summarized in Table 1. A total of 90 patients were studied, 38 (42.2%) were male and 52 (57.8%) were female. Patients were classified into three groups: 45 patients with ET (18 men and 27 women, median age 58 years, 22–83), 26 patients with PV (10 men and 16 women, median age 66,41–77), and 19 patients with PMF(10 men and 9 women, median age 62,49–85). Of all the 90 patients, the regular laboratory examination is as follows: Hb 147.15 (range 58–242), platelet 486 (range 18–2291), LDH 312.5 (range 140–904), IL-6 4.11 (range 1.50–370.08), CRP 1.6 (range 0.2–207.20), D-dimer 795 (range 1–8203). There were no differences in gender, age, D-dimer whereas a significant difference in WBC, RBC, Hb, PLT, LDH, IL-6, CRP, JAK2V617F mutation burden and WT1 expression among the groups were found.
Table 1.
Clinical and biologic characteristics of MPN patients.
| Characteristics | Overall | ET | PV | PMF | P |
|---|---|---|---|---|---|
| N = 90 | N = 45 | N = 26 | N = 19 | ||
| Sex | 0.581a | ||||
| Male | 38 | 18 | 10 | 10 | |
| Female | 52 | 27 | 16 | 9 | |
| Age [M (R)] | 0.110b | ||||
| Median | 62 | 58 | 66 | 62 | |
| Range | 22–85 | 22–83 | 41–77 | 49–85 | |
| WBC (×109/L) | 0.007b | ||||
| Median | 12.3 | 9.76 | 14.02 | 15.5 | |
| Range | 1.70–67.05 | 1.70–67.05 | 6.75–25.46 | 2.8–54.3 | |
| RBC (×1012/L) | <0.001b | ||||
| Median | 5.01 | 4.84 | 7.71 | 4.57 | |
| Range | 1.25–8.35 | 1.25–7.91 | 4.67–8.35 | 2.41–6.46 | |
| Hb (g/L) | <0.001b | ||||
| Median | 147.15 | 139.5 | 198.1 | 121.5 | |
| Range | 58–242 | 58–185 | 146–242 | 68–171 | |
| PLT (×109/L) | <0.001b | ||||
| Median | 486 | 673 | 470 | 280 | |
| Range | 18–2291 | 469–2291 | 144–1484 | 18–1175 | |
| LDH (U/L) | <0.001b | ||||
| Median | 312.5 | 245 | 343 | 519.5 | |
| Range | 140–904 | 140–471 | 228–650 | 280–904 | |
| IL-6 (pg/mL) | 0.001b | ||||
| Median | 4.11 | 3.77 | 3.73 | 8.165 | |
| Range | 1.50–370.08 | 1.50–370.08 | 1.99–42.78 | 1.5–77.4 | |
| CRP (mg/L) | 0.028b | ||||
| Median | 1.6 | 2.5 | 1.3 | 1.75 | |
| Range | 0.2–207.20 | 0.20–207.20 | 0.30–3.20 | 0.60–134.00 | |
| D-dimer (ng/ml) | 0.215b | ||||
| Median | 795 | 96 | 65 | 100 | |
| Range | 1–8203 | 3–8203 | 11–3642 | 1–2149 | |
| JAK2V617F mutation burden% | <0.001b | ||||
| Median | 46.325 | 25.94 | 75.11 | 82.09 | |
| Range | 2.78–97.47 | 2.78–80.56 | 34.49–91.8 | 11.48–97.47 | |
| WT1 expression (WT1/104copes ABL1) | <0.001b | ||||
| Median | 0.225 | 0.14 | 0.41 | 0.840 | |
| Range | 0.00–30.82 | 0.00–.99 | 0.00–2.51 | 0.01–30.82 |
Data are shown as Median and range. aChi-square test; bKruskal-Wallis. Abbreviation: LDH, lactate dehydrogenase.
JAK2V617F Allele Burden in MPN
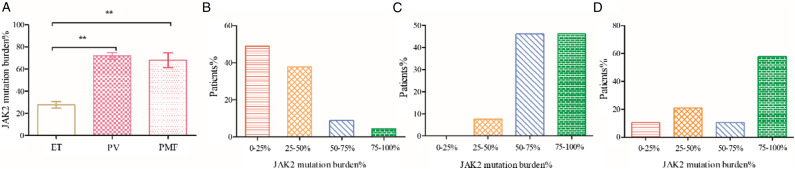
Allele specific real-time quantitative fluorescence PCR (AS-qPCR) was conducted to detect JAK2V617F allele burden. Our results revealed that JAK2V617F allele burden in PMF (68.09 ± 6.60 vs 27.8 ± 2.98 P < 0.001) and PV(72.03 ± 2.89 vs 27.8 ± 2.98 P < 0.001) is higher than in ET. No significant difference was found between PV and PMF (72.03 ± 2.89 vs 68.09 ± 6.60 P = 0.551). We further explored the JAK2V617F allele burden distribution in the MPN subgroup. Patients were grouped by quantities of JAK2V617F allele burden. PV patients were more often distributed in the 50-75% or 75-100% groups, PMF patients were more often distributed in the 75-100% groups, whereas ET patients more often classified into 0%–25% and 25-50% groups (Figure 1).
Figure 1.
JAK2V617F allele burden in MPN and distribution in the subgroups. JAK2V617F allele burden were calculated by the following formula JAK2V617F Mutation/(Mutation + Wild Type). (A) JAK2V617F allele burden in ET, PV and PMF; (B) JAK2V617F allele burden distribution in ET; (C) JAK2V617F allele burden distribution in PV; (D) JAK2V617F allele burden in PMF. ET: essential thrombocythemia; PV: polycythemia vera; PMF: primary myelofibrosis, *P < .01, **P < .001.
WT1 Expression in MPN
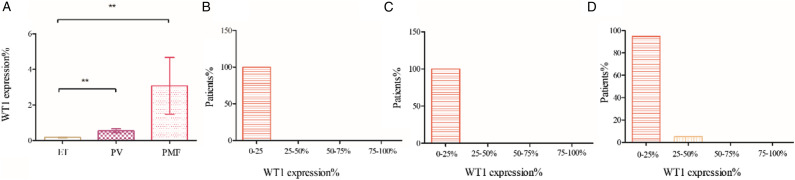
To investigate whether WT1 expression was different among ET, PV and PMF patients, one-way ANOVA test was performed. The results showed the expression of WT1 in PMF is higher than in ET (3.08 ± 1.59 vs 0.18 ± 0.029 P < .01). Besides, WT1 expression in PV is higher than in ET (0.55 ± 0.11 vs 0.18 ± 0.029 P < .001). When compared WT1 expression in PV and PMF, we found WT1 expression is higher in PMF than in PV, but the statics was not significant (3.08 ± 1.59 vs 0.55 ± 0.119 P = 0.069). To further investigate the relationship between WT1 expression and MPN patients, patients were grouped by quantities of WT1 expression. And we found that the patients were imbalanced distributed. All the ET and PV patients were populated in the 0-25%, as to the PMF patients, although most patients were classified into 0%–25%, there are a portion distributed in 25-50% groups (Figure 2).
Figure 2.
WT1 expression in MPN and distribution in the subgroups. WT1 levels were expressed as a ratio to ABL1 quantification. (A) WT1 expression in ET, PV and PMF; (B) WT1 expression distribution in ET; (C) WT1 expression distribution in PV; (D) WT1 expression distribution in PMF. ET: essential thrombocythemia; PV: polycythemia vera; PMF: primary myelofibrosis, *P < 0 .01, **P < 0 .001.
Diagnostic Value of JAK2V617F Allele Burden and WT1 Expression
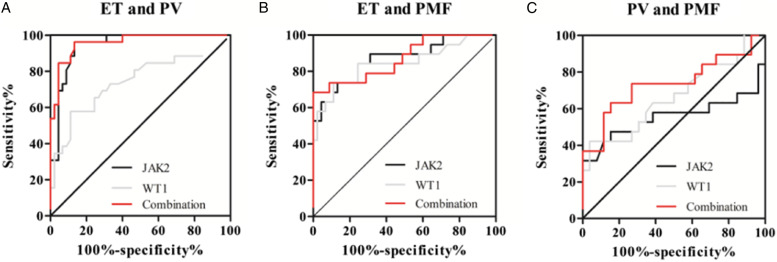
To assess whether JAK2V617F allele burden and WT1 expression could be used as potential diagnostic markers to distinguish the specific type of MPNs, ROC was carried out using 3 models: ET vs PV, ET vs PMF, PV vs PMF (Table 2). Results revealed that areas under the ROC curves of JAK2V617F allele burden and WT1 expression to discriminate ET from PV was 0 .9432 and 0.7436 respectively (Figure 3(a)); their value to distinguish ET from PMF is 0.869 and 0.8444 (Figure 3(b)). Although using JAK2V617F allele burden or WT1 expression alone to discriminate PV from PMF was not obvious, combination of JAK2V617F allele burden and WT1 expression possessed a moderate ability to discriminate PV from PMF with an area under the ROC curve of 0.7368 (Figure 3(c)).
Table 2.
The AUC of JAK2V617F allele burden and WT1 expression for MPN subgroups.
| Group | Gene | AUC (95% Confidence Intervals) | P | Se (%) | Sp (%) | Cut-Off Value |
|---|---|---|---|---|---|---|
| ET vs PV | JAK2V617F | 0.943 (.891–.995) | <0.01 | 96.15 | 86.67 | 47.09% |
| WT1 | 0.744 (.615–.873) | <0.01 | 57.69 | 88.89 | 0.34 | |
| Combination of JAK2V617F and WT1 | 0.956 (.916–1.000) | <0.01 | 96.15 | 86.67 | ||
| ET vs PMF | JAK2V617F | 0.869 (.766–.973) | <0.01 | 73.68 | 86.67 | 46.33% |
| WT1 | 0.844 (.724–.965) | <0.01 | 73.68 | 88.89 | 0.33 | |
| Combination of JAK2V617F and WT1 | 0.871 (.768–.975) | <0.01 | 68.42 | 100 | ||
| PV vs PMF | JAK2V617F | 0.555 (.356–.754) | 0.54 | 47.37 | 84.62 | 86.84% |
| WT1 | 0.665 (.498–.832) | 0.06 | 42.11 | 96.15 | 1.335 | |
| Combination of JAK2V617F and WT1 | 0.737 (.578–.897) | <0.01 | 63.16 | 84.62 |
Abbreviation: Se: Sensitivity; Sp: Specificity; AUC: area under the curve.
Figure 3.
Receiver operating characteristic (ROC) curves of JAK2V617F allele burden and WT1 expression for MPN subgroups. (A) ET vs PV; (B) ET vs PMF; (C) PV vs PMF.
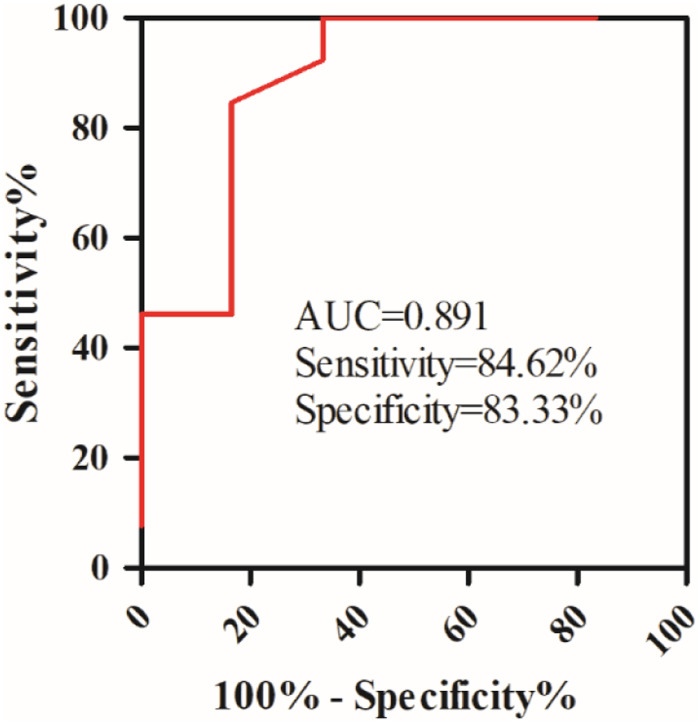
Clinically, there are some ET patients with high Hb levels and PV patients with high platelet counts, for these patients, it is difficult to make a precise diagnosis. In our study, 6 ET patients with high Hb levels and 13 PV patients with high platelet counts were included, we conduct an ROC analysis, and found the combination of JAK2V617F allele burden and WT1 expression has a high ability to distinguish ET patients with high Hb levels from PV patients with high platelet counts with an AUC of 0.891 (Figure 4).
Figure 4.
Receiver operating characteristic (ROC) curves of Combination of JAK2V617F and WT1 for ET patients with high Hb levels and PV patients with high platelet counts.
Discussion
Studies have revealed that JAK2V617F mutation allele burden may be useful in evaluating the response to therapy.24 Furthermore, researches have also reported JAK2V617F mutation allele burden can be used as a follow-up tool during the treatment of JAK2V617F positive patients.25 Another perspective value of the allele burden is its predict value of both vascular events and fibrotic transformation.26 However, the specific utility of the JAK2V617F allele burden for the differential diagnosis of MPN yet to be established. Our study for the first time explored the diagnostic value of JAK2V617F mutation allele burden to distinguish MPN subtypes.
In our study, we found an obvious differences between JAK2V617F mutations burden and WT1 expression within three subtypes of MPN. Both our studies and previous researches revealed that JAK2V617F allele burden27,28 and WT1 expression29 will gradually increase with the progress of MPN. It is reported that JAK2V617F mutations mainly occurred in MPN patients and a small part of AML patients8 and elevated expression of WT1 often imply patients to have malignant tumor.30 In the present study, only new diagnosed MPN patients were included which will not be significantly affected factors other than subtypes.
Previous study has reported that WT1 transcript levels were higher in PMF than in ET and PV.22 Our data showed the expression of WT1 in PMF and PV is higher than in ET. When comparing WT1 expression in PV and PMF, although no difference was found (P = 0.069), we think this is due to small size of this study. Gallo et al. found that hyperexpression of the WT1 gene in PMF is associated with the IPSS prognostic score and WT1 expression increased during leukemic transformation of MPN.21 Our research focused on exploring combined value of WT1 expression and JAK2V617F allele burden in distinguishing the subtype of MPN patients. The combination of JAK2V617F allele burden and WT1 expression possessed a high ability to discriminate ET from PV and ET from PMF with area under the ROC curve of 0.9585 and 0.8713. And their combined value to discriminate PV from PMF is 0.7368. PV patients with thrombocytosis at disease presentation were frequently misclassified as JAK2V617F-positive ET patients.31,32 Our study found the combination of JAK2V617F allele burden and WT1 expression could distinguish ET patients with high Hb levels from PV patients with high platelet counts with an AUC of 0.891.
Limitations may exist in our study. First, in our retrospective study, although the diagnosis of MPN was made according to the WHO 2016 classification, but no pre-PMF patients were enrolled in our study, we were therefore unable to evaluated the value of WT1 expression and JAK2V617F allele burden to discriminate ET and pre-PMF that is difficult to differentiate by bone marrow biopsy; second although we only included patients that were firstly diagnosed before any treatment, there might be patients with initial diagnosis rather than initial onset; third the sample size selected for our study was not calculated and justified, and the statistical power of the study may be limited.
In conclusion, diagnosis of classical Ph-negative MPN should integrate several clinical, laboratory features, as well as morphology and immunophenotyping. When a patient is suspected to be MPN, JAK2 allele burden and WT1 expression can be a helpful additional diagnostic marker to distinguish the subtype of MPN patients. A prospective study with large sample is in need to confirm our findings. In our future study, we will explore whether the combination of JAK2V617F allele burden and WT1 expression in peripheral blood is helpful in distinguishing the subtype of MPN patients to overcome the obstacle due to the failure of the bone marrow aspirate in patients affected by myelofibrosis.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was supported by National Natural Science Foundation of China (81670125), Natural Science Foundation of Hubei Province (2018CFB243).
Ethics approval: The study protocol was approved by the Medical Ethical Committee of Zhongnan Hospital of Wuhan University (Ethical approval 2023022K, February 2023).
ORCID iD
Xuelan Zuo https://orcid.org/0000-0002-8577-347X
References
- 1.Thomas S, Krishnan A. Platelet Heterogeneity in Myeloproliferative Neoplasms. Arterioscler Thromb Vasc Biol. 2021;41:2661-2670. doi: 10.1161/atvbaha.121.316373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tefferi A. Primary myelofibrosis: 2021 update on diagnosis, risk-stratification and management. Am J Hematol. 2021;96(1):145-162. doi: 10.1002/ajh.26050 [DOI] [PubMed] [Google Scholar]
- 3.Kvasnicka HM, Thiele J. Prodromal myeloproliferative neoplasms: the 2008 WHO classification. Am J Hematol. 2010;85(1):62-69. doi: 10.1002/ajh.21543 [DOI] [PubMed] [Google Scholar]
- 4.Guglielmelli P, Pacilli A, Rotunno G, Rumi E, Rosti V, Delaini F, et al. Presentation and outcome of patients with 2016 WHO diagnosis of prefibrotic and overt primary myelofibrosis. Blood 2017;129(24):3227-3236. doi: 10.1182/blood-2017-01-761999 [DOI] [PubMed] [Google Scholar]
- 5.Barbui T, Thiele J, Passamonti F, Rumi E, Boveri E, Ruggeri M, et al. Survival and disease progression in essential thrombocythemia are significantly influenced by accurate morphologic diagnosis: an international study. J Clin Oncol 2011;29(23):3179-3184. doi: 10.1200/jco.2010.34.5298 [DOI] [PubMed] [Google Scholar]
- 6.Tang G, Sydney Sir Philip JK, Weinberg O, Tam W, Sadigh S, Lake JI. et al. Hematopoietic neoplasms with 9p24/JAK2 rearrangement: a multicenter study. Mod Pathol 2019;32(4):490-498. doi: 10.1038/s41379-018-0165-9 [DOI] [PubMed] [Google Scholar]
- 7.Azevedo AP, Silva SN, Reichert A, Lima F, Júnior E, Rueff J, et al. Prevalence of the Janus kinase 2 V617F mutation in Philadelphia-negative myeloproliferative neoplasms in a Portuguese population. Biomed Rep 2017;7(4):370-376. doi: 10.3892/br.2017.977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook AM, Li L, Ho Y, Lin A, Li L, Stein A, et al. Role of altered growth factor receptor-mediated JAK2 signaling in growth and maintenance of human acute myeloid leukemia stem cells. Blood 2014;123(18):2826-2837. doi: 10.1182/blood-2013-05-505735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langabeer SE, Andrikovics H, Asp J, Bellosillo B, Carillo S, Haslam K, et al. Molecular diagnostics of myeloproliferative neoplasms. Eur J Haematol 2015;95(4):270-279. doi: 10.1111/ejh.12578 [DOI] [PubMed] [Google Scholar]
- 10.Kralovics R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005;352(17):1779-1790. doi: 10.1056/NEJMoa051113 [DOI] [PubMed] [Google Scholar]
- 11.Huff V. Wilms’ tumours: about tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer. 2011;11(2):111-121. doi: 10.1038/nrc3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toska E, Roberts SGE. Mechanisms of transcriptional regulation by WT1 (Wilms’ tumour 1). Biochem J. 2014;461(1):15-32. doi: 10.1042/bj20131587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Artibani M, Sims AH, Slight J, Aitken S, Thornburn A, Muir M, et al. WT1 expression in breast cancer disrupts the epithelial/mesenchymal balance of tumour cells and correlates with the metabolic response to docetaxel. Sci Rep 2017;7:45255. doi: 10.1038/srep45255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosen N, Sonoda Y, Oji Y, Kimura T, Minamiguchi H, Tamaki H, et al. Very low frequencies of human normal CD34+ haematopoietic progenitor cells express the Wilms’ tumour gene WT1 at levels similar to those in leukaemia cells. Br J Haematol 2002;116(2):409-420. [DOI] [PubMed] [Google Scholar]
- 15.Adnan-Awad S, Meligui YME, Salem SE, Salaheldin O, Ayoub MA, Kamel MM, et al. Prognostic Impact of WT-1 and survivin gene expression in acute myeloid leukemia patients. Clin Lab 2019;65(4). doi: 10.7754/Clin.Lab.2018.180329 [DOI] [PubMed] [Google Scholar]
- 16.Zhu YM, Wang PP, Huang JY, Chen YS, Chen B, Dai YJ, et al. Gene mutational pattern and expression level in 560 acute myeloid leukemia patients and their clinical relevance. J Transl Med 2017;15(1):178. doi: 10.1186/s12967-017-1279-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mashima K, Ikeda T, Toda Y, Ito S, Umino K, Minakata D, et al. Associations between the peripheral blood Wilms tumor gene 1 level and both bone marrow blast cells and the prognosis in patients with myelodysplastic syndrome. Leuk Lymphoma 2019;60(3):703-710. doi: 10.1080/10428194.2018.1504940 [DOI] [PubMed] [Google Scholar]
- 18.Rautenberg C, Germing U, Pechtel S, Lamers M, Fischermanns C, Jäger P, et al. Prognostic impact of peripheral blood WT1-mRNA expression in patients with MDS. Blood Cancer J 2019;9(11):86. doi: 10.1038/s41408-019-0248-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montano G, Vidovic K, Palladino C, Cesaro E, Sodaro G, Quintarelli C, et al. WT1-mediated repression of the proapoptotic transcription factor ZNF224 is triggered by the BCR-ABL oncogene. Oncotarget 2015;6(29):28223-28237. doi: 10.18632/oncotarget.4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guglielmelli P, Zini R, Bogani C, Salati S, Pancrazzi A, Bianchi E, et al. Molecular profiling of CD34+ cells in idiopathic myelofibrosis identifies a set of disease-associated genes and reveals the clinical significance of Wilms’ tumor gene 1 (WT1). Stem Cell 2007;25(1):165-173. doi: 10.1634/stemcells.2006-0351 [DOI] [PubMed] [Google Scholar]
- 21.Gallo D, Nicoli P, Calabrese C, Gaidano V, Petiti J, Rosso V, et al. The Wilms’ tumor (WT1) gene expression correlates with the International Prognostic Scoring System (IPSS) score in patients with myelofibrosis and it is a marker of response to therapy. Cancer Med 2016;5(7):1650-1653. doi: 10.1002/cam4.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cottin L, Riou J, Boyer F, Bouvier A, Zannetti A, Blouet A, et al. WT1 gene is overexpressed in myeloproliferative neoplasms, especially in myelofibrosis. Blood Cells Mol Dis 2019;75:35-40. doi: 10.1016/j.bcmd.2018.12.004 [DOI] [PubMed] [Google Scholar]
- 23.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127(20):2391-2405. doi: 10.1182/blood-2016-03-643544 [DOI] [PubMed] [Google Scholar]
- 24.Barbui T, Thiele J, Gisslinger H, Kvasnicka HM, Vannucchi AM, Guglielmelli P, et al. The 2016 WHO classification and diagnostic criteria for myeloproliferative neoplasms: document summary and in-depth discussion. Blood Cancer J 2018;8(2):15. doi: 10.1038/s41408-018-0054-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malysz J, Crisan D. Correlation of JAK2 V617F mutant allele quantitation with clinical presentation and type of chronic myeloproliferative neoplasm. Ann Clin Lab Sci. 2009;39(4):345-350. [PubMed] [Google Scholar]
- 26.Stuckey R, Gómez-Casares MT. Recent advances in the use of molecular analyses to inform the diagnosis and prognosis of patients with polycythaemia vera. Int J Mol Sci. 2021;22(9):5042. doi: 10.3390/ijms22095042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carobbio A, Finazzi G, Antonioli E, Guglielmelli P, Vannucchi AM, Dellacasa CM, et al. JAK2V617F allele burden and thrombosis: a direct comparison in essential thrombocythemia and polycythemia vera. Exp Hematol 2009;37(9):1016-1021. doi: 10.1016/j.exphem.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pedersen RK, Andersen M, Knudsen TA, Sajid Z, Gudmand-Hoeyer J, Dam MJB, et al. Data-driven analysis of JAK2V617F kinetics during interferon-alpha2 treatment of patients with polycythemia vera and related neoplasms. Cancer Med 2020;9(6):2039-2051. doi: 10.1002/cam4.2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Ye X, Luo S, Xu X, Wang S, Jin K, et al. Clinical features and next-generation sequencing landscape of essential thrombocythemia, prefibrotic primary myelofibrosis, and overt fibrotic primary myelofibrosis: a Chinese monocentric retrospective study. J Cancer Res Clin Oncol 2022. doi: 10.1007/s00432-022-04067-1 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Yan WT, Yang ZY, Li YL, Tan XN, Jiang J, et al. The role of WT1 in breast cancer: clinical implications, biological effects and molecular mechanism. Int J Biol Sci 2020;16(8):1474-1480. doi: 10.7150/ijbs.39958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barbui T, Thiele J, Carobbio A, Gisslinger H, Finazzi G, Rumi E, et al. Masked polycythemia vera diagnosed according to WHO and BCSH classification. Am J Hematol 2014;89(2):199-202. doi: 10.1002/ajh.23617 [DOI] [PubMed] [Google Scholar]
- 32.Barbui T, Thiele J, Carobbio A, Guglielmelli P, Rambaldi A, Vannucchi AM, et al. Discriminating between essential thrombocythemia and masked polycythemia vera in JAK2 mutated patients. Am J Hematol 2014;89(6):588-590. doi: 10.1002/ajh.23694 [DOI] [PubMed] [Google Scholar]