Abstract
Encystment in Azotobacter vinelandii is induced by n-butanol or β-hydroxybutyrate (BHB). We identified a gene, encoding an aldehyde dehydrogenase, that was named aldA. An aldA mutation impaired bacterial growth on n-butanol, ethanol, or hexanol as the sole carbon source. Expression of aldA increased in cells shifted from sucrose to n-butanol and was shown to be dependent on the alternative ς54 factor. A mutation in rpoN encoding the ς54 factor also impaired growth on alcohols. Encystment on n-butanol, but not on BHB, was impaired in aldA or rpoN mutants, indicating that n-butanol is not an inducer of encystment by itself but must be catabolized in order to induce encystment.
Azotobacter vinelandii is a soil nitrogen-fixing bacterium which produces the intracellular polyester poly-β-hydroxybutyrate (poly-BHB) and undergoes differentiation to form desiccation-resistant cysts. Encystment can be induced by removing sucrose from exponential-phase cells and replacing it with BHB or n-butanol (7, 20, 27): BHB, which is a natural metabolite in A. vinelandii (7), is believed to induce encystment because its addition results in conditions approximating poly-BHB degradation and because induction by n-butanol has been proposed to result from its conversion to BHB (20). Encystment induced by BHB or n-butanol or that occurring in late-stationary-phase glucose cultures is accompanied by the formation of a family of 5-n-alkylresorcinols and 6-n-alkylpirones that are lipids present only in encysting cells. They replace the membrane phospholipids and are a major component of the exine (16–18). Thus, encystment seems to be induced by a metabolic shift from carbohydrate metabolism to lipid metabolism, and this shift seems to be promoted in the presence of either n-butanol or BHB.
Aldehyde dehydrogenases participate in metabolic processes, such as the catabolism of alcohols (15); they catalyze the oxidation of aldehydes to their corresponding acid form. Some bacterial aldehyde dehydrogenases, such as Pseudomonas aeruginosa malonic semialdehyde dehydrogenase, have high specificity (12), while others such as acetaldehyde dehydrogenase II (AcDH-II) from Ralstonia eutropha function on a broad spectrum of substrates. AcDH-II is involved in the catabolism of acetoin as well as of ethanol (15).
We identified aldA, a gene encoding an aldehyde dehydrogenase, whose expression was increased in cells shifted from sucrose to n-butanol. We found that AldA activity is essential for catabolism of n-butanol and other alcohols and that aldA expression is controlled by the alternative ς54 factor. We also show that n-butanol has to be catabolized to induce encystment.
MATERIALS AND METHODS
Microbiological procedures.
The bacterial strains and plasmids used in this study are listed in Table 1. A. vinelandii was grown at 30°C on Burk's medium (5), supplemented with 2% sucrose, 0.2% n-butanol, or 0.2% BHB. A. vinelandii transformation was carried out as reported by Bali et al. (2). Encystment was determined by measuring resistance to desiccation as previously described (3). Bacterial strains were grown on liquid Burk's sucrose medium for 24 h, washed with 10 mM magnesium sulfate, and induced for encystment by incubation on plates of Burk's medium supplemented with n-butanol or BHB. β-Galactosidase activity was measured as reported by Miller (10); 1 U corresponds to 1 nmol of o-nitrophenyl-β-D-galactoside hydrolyzed per min per μg of protein. Protein was determined by the Lowry method (8). All measurements were done in triplicate.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains (A. vinelandii) | ||
| ATCC 9046 | Highly mucoid | Our collection |
| JG8 | alg8::Tc derivative of ATCC 9046 | 9 |
| SG9 | aldA::mini-Tn5 derivative of JG8 | This work |
| SG9A | aldA::mini-Tn5 derivative of ATCC 9046 | This work |
| SG9A::pSM9-Gm | derivative of SG9A with pSM9-Gm integrated in the chromosome | This work |
| CN10 | rpoN::Gm derivative of ATCC 9046 | This work |
| Plasmids | ||
| pBluescript SK(+) | Plasmid used for subcloning DNA, Apr | Stratagene |
| pUT-mini-Tn5–lacZ | Suicide vector for mutagenesis with mini-Tn5–lacZ | 4 |
| pBSL141 | 1 | |
| pSM888 | Derivative of pCP13 carrying A. vinelandii aldA region | This work |
| pSG9 | Derivative of pBluescript SK(+) carrying the aldA::Tn5 mutation from SG9 | This work |
| pSM9 | Derivative of pBluescript SK(+) carrying the aldA wild-type gene | This work |
| pSM9-Gm | Derivative of pSM9 with a Gm resistance gene | This work |
| pLV72 | Cosmid clone with the A. vinelandii rpoN gene | 26 |
| pSM6 | Derivative of pBluescript SK(+) carrying the A. vinelandii rpoN gene in a 2.3-kb EcoRI-ClaI fragment | This work |
| pCN10 | pSM6 derivative carrying a rpoN::Gm mutation | This work |
Transposon mutagenesis.
Random transposon mutagenesis of strain JG8 was carried out with a pUT derivative containing the mini-Tn5–lacZ2 transposon as described previously (4). We isolated 3,200 kanamycin derivatives. Unexpectedly, all 3,200 mutants showed blue on plates containing Burk's sucrose medium with sucrose and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal). The parental strain JG8, used as a control, showed white. It is unlikely that, in all kanamycin-resistant mutants, the lacZ gene was fused to an A. vinelandii promoter, thus suggesting that the lacZ gene is transcribed in A. vinelandii. However, about 10% of the mutants were bluer, suggesting that, in these strains, expression of lacZ was also initiated from A. vinelandii promoters.
Construction of strains SG9A and CN10.
Plasmid pSG9 (Fig. 1), which is unable to replicate in A. vinelandii, was introduced by transformation into strain ATCC 9046. Strain SG9A is a kanamycin-resistant derivative. The replacement of the aldA wild-type gene by the aldA–mini-Tn5 mutation on the chromosome of the SG9A was confirmed by Southern blotting using labeled pSG9 as a probe (data not shown).
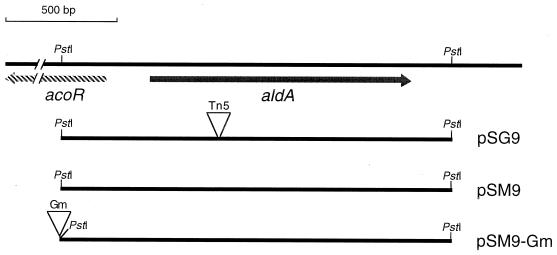
FIG. 1.
Physical map of the A. vinelandii aldA-acoR chromosomal region and plasmids carrying this region.
A 2.3-kb ClaI-EcoRI restriction fragment from cosmid pLV72 (26) containing the rpoN gene from A. vinelandii was cloned into plasmid pBluescript KS, producing plasmid pSM6. A 0.8-kb KpnI fragment containing a gentamicin (Gm) resistance gene from plasmid pBSL141 (1) was inserted in the KpnI site within the rpoN gene present in pSM6 to create a rpoN::Gm mutation. The resultant plasmid pCN10, which is unable to replicate in A. vinelandii, was introduced by transformation into strain ATCC 9046. One gentamicin-resistant transformant strain named CN10 was chosen and confirmed by Southern analysis to carry the rpoN::Gm mutation (data not shown).
Recombinant DNA techniques.
All DNA manipulations were performed by standard procedures (21).
Primer extension analysis.
Primer extension reactions were performed with a primer extension system (Amersham) as instructed by the manufacturer. Oligonucleotide primer 5′CACTGCCAGGATGGGCATAC was labeled with [γ-32P]dATP (Amersham) at the 5′ end by using polynucleotide kinase and was hybridized to 50 μg of total RNA. After extension with reverse transcriptase, cDNA products were examined by electrophoresis in an 8% polyacrylamide gel. To map transcriptional start points, sequencing reactions were performed on pSG9 DNA by the dideoxy chain method (22), using [γ-32P]dATP and a sequencing kit with the same primer employed for the primer extension reactions.
Aldehyde dehydrogenase assay.
A. vinelandii cells were grown to early stationary phase on Burk's sucrose medium at 30°C. The cultures were centrifuged, washed, and resuspended in Burk's n-butanol medium. After 20 h of induction in this medium, the cells were collected by centrifugation, resuspended in 1/25 volume of phosphate buffer, pH 7.4, and sonicated. The sonicated cell suspensions were centrifuged at 14,000 × g for 5 min, and the supernatants were centrifuged for 1.5 h at 192,000 × g. Ammonium sulfate was added to give 50% saturation. The precipitate was collected by centrifugation at 14,000 × g, resuspended in phosphate buffer, pH 7.4, and dialyzed against the same buffer. The activity was assayed spectrophotometrically as reported previously (13) by measuring the reduction of NAD+ at 340 nm. The reaction was initiated by addition of butyraldehyde or acetaldehyde. The rate of endogenous NAD+ reduction was recorded in the absence of butyraldehyde or acetaldehyde in the assay. One unit of activity is defined as the amount of enzyme reducing 1 nmol of NAD+ per min per mg of protein.
Cloning of aldA gene.
A cosmid clone pSM888 from an A. vinelandii gene library that harbored a 2.2-kb PstI fragment containing the aldA gene was identified by hybridization with a 0.5-kb SalI fragment from pSG9. The 2.2-kb PstI fragment was subcloned into plasmid pKS to give plasmid pSM9 (Fig. 1). A Gm resistance gene was cloned into the polylinker of pSM9 to give plasmid pSM9-Gm (Fig. 1).
Nucleotide sequence accession number.
The A. vinelandii aldA gene sequence reported here has been assigned GenBank accession no. AF277380.
RESULTS
Identification of A. vinelandii gene induced in n-butanol.
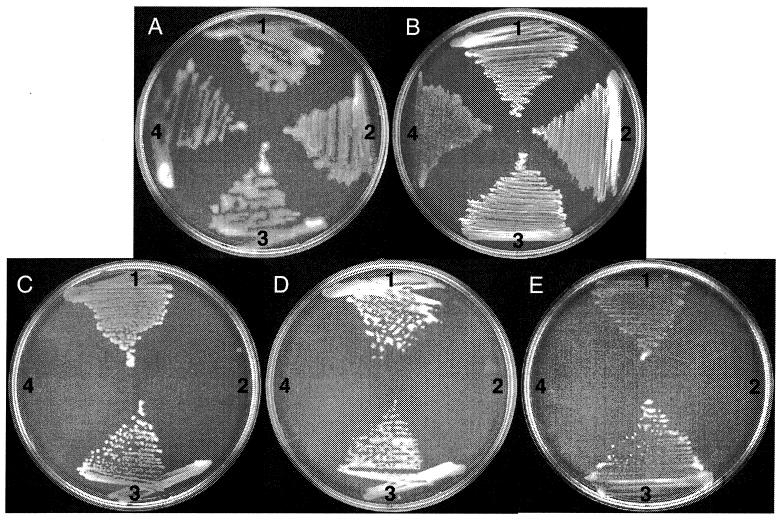
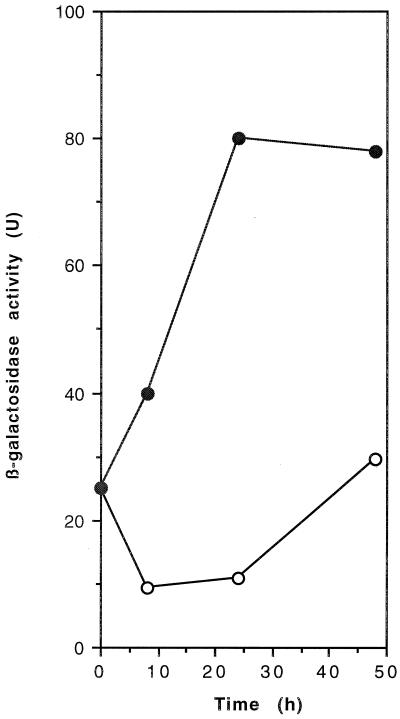
In A. vinelandii, encystment can be induced by transferring vegetative cells into plates of Burk's salt medium containing sublethal levels (0.2%) of n-butanol as the carbon source. To identify genes expressed during the induction of the encystment process, we isolated 3,200 mutants carrying mini-Tn5–lacZ insertions (see Materials and Methods). When mini-Tn5–lacZ is inserted downstream, a promoter in the correct orientation creates a gene fusion. We transferred the 3,200 mini-Tn5–lacZ mutants to plates of Burk's medium containing n-butanol as the only carbon source and X-Gal and to plates with Burk's medium with 2.0% sucrose and X-Gal. A mutant named SG9, that, although unable to grow on Burk's butanol, showed a bluer phenotype on Burk's butanol than on Burk's sucrose, was identified (data not shown). A 7.0-kb PstI fragment containing the mini-Tn5–lacZ insertion from SG9 was cloned into pBluescript KS. The resultant plasmid pSG9 (Fig. 1) was transformed to wild-type ATCC 9046. Strain SG9A, a transformant that was unable to grow on Burk's butanol (Fig. 2C) and showed a bluer phenotype on Burk's butanol was isolated, confirming that these phenotypes were caused by the mini-Tn5 insertion. We also confirmed induction of transcription under encystment conditions of the identified gene by measuring β-galactosidase activity. Cultures of strain SG9A were grown for 24 h in liquid Burk's medium supplemented with 2.0% sucrose. Cells from these cultures were transferred to agar plates containing Burk's medium supplemented with n-butanol or sucrose. As expected, an increase in β-galactosidase activity was observed during incubation on n-butanol (Fig. 3).
FIG. 2.
Growth phenotypes of A. vinelandii ATCC 9046 (1), SG9A (2), SG9A::pSM9-Gm (3), and CN10 (4) at 30°C on Burk's medium supplemented with 10 mM NH4Cl as the nitrogen source and with 2.0% sucrose (A), 0.2% BHB (B), n-butanol (C), ethanol (D), or hexanol (E) as the sole carbon source.
FIG. 3.
β-Galactosidase activity of strain SG9A, during incubation in solid Burk's medium with sucrose (open symbols) or n-butanol (closed symbols).
Cloning and DNA sequence.
Plasmid pSG9 (Fig. 1) was used to determine 2.3 kb of the A. vinelandii DNA sequence surrounding the transposon. Analysis of this sequence revealed an open reading frame encoding a polypeptide of 506 amino acid residues with a calculated molecular mass of 55,061 Da. The location of the mini-Tn5–lacZ mutation was determined by sequencing across the transposon insertion junction and was found to lie within codons 135 to 137. Comparison of the deduced amino acid sequence with those in the databases revealed a high degree of similarity with aldehyde dehydrogenases from different sources. Thus, we named this A. vinelandii gene aldA. The deduced protein product shares 89% identity with the P. aeruginosa putative aldehyde dehydrogenase PA4022 (24) (accession no. F83142), 88% with P. aeruginosa ExaC (23) (accession no. AF068264), and 69% with R. eutropha AcDH-II (P46368), an enzyme responsible for the oxidation of acetaldehyde to acetate during growth on ethanol or acetoin (15). In the sequenced DNA fragment, 303 nucleotides upstream of aldA, an incomplete open reading frame comprised of 207 nucleotides and divergently oriented is present (Fig. 1). A Blast P GenBank search for the deduced protein revealed similarity to the acetoin catabolism regulatory protein AcoR (SWISS PROT P28614), the putative activator for acetoin catabolism genes of R. eutropha H16 (6). It is a member of the NifA family of transcriptional activators known as enhancer binding proteins, required for transcription from ς54-dependent genes (11). The highest identity of this A. vinelandii AcoR homologue (89%) was found with the P. aeruginosa PAO1 putative transcriptional activator PA4021, which is located upstream and in opposite orientation to that of the putative aldehyde dehydrogenase PA4022 (24).
Effect of aldA mutation on aldehyde dehydrogenase activity.
The inability of SG9A to grow on the carbon source n-butanol and the high similarity of aldA with aldehyde dehydrogenases suggested that this gene encodes an enzyme with butyraldehyde dehydrogenase activity. We assayed this activity in extracts of the wild-type strain ATCC 9046 and SG9A.
After induction on n-butanol, coenzyme A-independent butyraldehyde dehydrogenase activity was detected in the wild type and a significant reduction of this activity was found in the SG9A mutant (Table 2). The aldA mutation did not completely abrogate butyraldehyde activity, suggesting that another enzyme is the source of the background activity. AldA activity on acetaldehyde as a substrate was also tested. As shown in Table 2, AldA also displays acetaldehyde dehydrogenase activity. In agreement with this result, strain SG9A was unable to grow with ethanol or hexanol as the carbon source (Fig. 2D and E). These data indicate that the AldA activity is not specific for n-butanol and participates in the catabolism of several alcohols.
TABLE 2.
Aldehyde dehydrogenase activity in A. vinelandii aldA mutanta
| Strain | Genotype | Activity of aldehyde dehydrogenase (nmol min−1 mg−1)b found with:
|
|
|---|---|---|---|
| Butyraldehyde | Acetaldehyde | ||
| ATCC 9046 | Wild type | 75.0 ± 3.0 | 104.1 ± 3.0 |
| SG9A | aldA | 20.4 ± 1.4 | 23.6 ± 2.6 |
| CN10 | rpoN | 22.4 ± 1.7 | 21.3 ± 0.6 |
Cells were induced on Burk's medium with n-butanol as the sole carbon source.
Values are the mean of three determinations ± standard deviations.
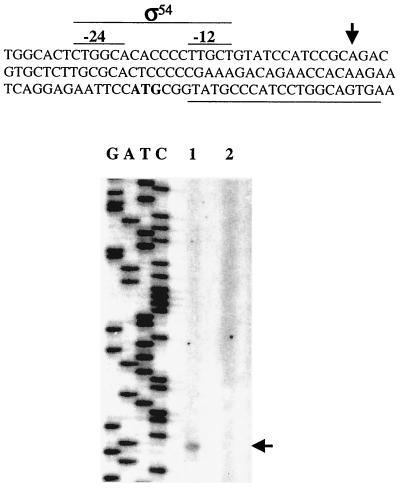
Identification of an aldA ς54 promoter.
Primer extension experiments were performed to determine the transcriptional start site of aldA. Figure 4 shows the primer extension products obtained with RNA isolated from strain ATCC 9046 grown in Burk's medium supplemented with n-butanol. A transcriptional start site corresponding to an A residue located 57 nucleotides upstream of the proposed aldA start codon was identified (Fig. 4). Examination of the 5′ upstream region revealed the presence of putative −24 (CTGGCA) and −12 (TTGCT) promoter sequences recognized by ς54. We therefore carried out primer extension analysis of aldA in strain CN10, an ATCC 9046 derivative carrying a mutation in rpoN encoding ς54. As shown in Fig. 4, no primer extension products were detected with RNA from strain CN10.
FIG. 4.
Primer extension analysis of aldA transcription in strains ATCC 9046 and CN10. (A) DNA sequence of the 5′ end of aldA. The arrow indicates the start site of aldA transcription; the ς54 promoter is also indicated at the top. The ATG initiation codon is shown in boldface. The complementary sequence where the oligonucleotide used for primer extension was generated is underlined. (B) Primer extension of the aldA gene in strain ATCC 9046 (lane 1) and in strain CN10 (lane 2) incubated in Burk's medium plus ammonia with sucrose. The aldA sequence ladder (GATC) was produced with the oligonucleotide used for primer extension.
Effect of rpoN mutation on aldehyde dehydrogenase activity and on growth on alcohols.
As transcription of aldA seems to be impaired by the rpoN mutation, growth on alcohols and aldehyde dehydrogenase activity in the rpoN mutant were expected to be similar to those of the aldA mutant. The results shown in Table 2 and Fig. 2 indicate that this is indeed the case and confirm the dependence of the aldA promoter on the ς54 factor. Together these data confirm that aldA is transcribed from a single ς54 promoter and that its product AldA is essential for the utilization of several alcohols as carbon sources.
Effect of aldA and rpoN mutations on encystment.
Induction of encystment by n-butanol is proposed to be due to its conversion to BHB (20). If this hypothesis is correct, AldA and ς54 are expected to be essential for encystment induced by n-butanol. We tested encystment of strains SG9A and CN10 in either n-butanol or BHB. As expected, strain SG9A was unable to produce desiccation-resistant cells when induced with n-butanol but not with BHB (Table 3). Growth of strain SG9A on BHB is similar to that of the wild-type strain (Fig. 2). However, it produced cysts resistant to desiccation with a reduced frequency when BHB rather than n-butanol was the sole carbon source (Table 3). This result supports the hypothesis that n-butanol induces encystment because it is converted to BHB; however, it does not rule out the possibility that the lack of encystment of SG9A is caused by its inability to grow on n-butanol. We added 0.1% sucrose to the Burk's n-butanol encysting medium to promote growth of strain SG9A during encystment; however, no mature cysts were detected under this condition, either in the wild type or the aldA mutant (data not shown). This result is in agreement with the abortion of encystment that was observed when glucose was added to encysting A. vinelandii cultures (7).
TABLE 3.
Encystment in A. vinelandii aldA and rpoN mutants
| Strain | Resistance to desiccation (%)a with:
|
|
|---|---|---|
| n-Butanol | BHB | |
| ATCC 9046 | 17.9 ± 4.0 | 11.6 ± 2.4 |
| SG9A | <10−4 | 1.3 ± 0.03 |
| SG9A::pSM9-Gm | 13 ± 3.4 | ND |
| CN10 | <10−4 | 10.0 ± 2.8 |
Resistance to desiccation of cells induced on Burk's medium with n-butanol or BHB as the sole carbon source. Values are the mean of three determinations. Values are given ± standard deviations. ND, not determined.
We tested encystment of strain CN10. Similar to strain SG9A, strain CN10 formed mature cysts when induced with BHB but not with n-butanol (Table 3). As the rpoN mutation impairs diazotrophic growth (26), this result implies that encystment by strain CN10 in Burk's medium supplemented with BHB in the absence of ammonia occurred under nongrowing conditions. These data further suggest that the inability to catabolize n-butanol imposed by the aldA or the rpoN mutation causes its cyst-defective phenotype and support the hypothesis that catabolism of n-butanol is essential to induce encystment.
SG9A complementation by aldA wild-type gene.
Plasmid pSM9-Gm (Fig. 1), which is unable to replicate in A. vinelandii and carries the aldA gene flanked by 500 nucleotides upstream of the ATG start codon and 200 nucleotides downstream of the stop codon, was transformed into SG9A for integration into the chromosome. Strain SG9A::pSM9-Gm, a gentamicin-resistant transformant, was selected. The presence of a wild-type aldA gene was confirmed by PCR analysis using SG9A::pSM9-Gm chromosomal DNA as a template as well as oligonucleotides aldA1 5′- and aldA2 3′- (data not shown). Strain SG9A::pSM9-Gm grew in n-butanol, ethanol, or hexanol (Fig. 2) and produced cells resistant to desiccation after induction with n-butanol (Table 3). We do not know whether the aldA mutation has a polar effect on downstream genes; however, integration of pSM9-Gm downstream of the mini-Tn5–lacZ insertion (in strain SG9A::pSM9-Gm) should restore the activity of these genes. Therefore, this result confirms that, either polar or nonpolar, the aldA mutation causes an inability to grow on alcohols and to encyst.
DISCUSSION
In an effort to understand the nature of genes involved in the induction of the encystment and the molecular mechanisms that control this process in A. vinelandii, we have identified, through mini-Tn5–lacZ mutagenesis and subsequent screening for lacZ gene fusions whose expression increased when shifted from sucrose to n-butanol, a gene encoding a protein with high similarity to aldehyde dehydrogenases. The gene was named aldA. Inactivation of aldA by mini-Tn5–lacZ led to a significant reduction in aldehyde dehydrogenase activity, demonstrating that this gene indeed encodes an aldehyde dehydrogenase. Inactivation of aldA also led to the inability to grow on n-butanol, ethanol, or hexanol as the sole carbon source. Thus, AldA is a wide substrate specificity enzyme that is responsible for the oxidation of aldehydes during growth on alcohols.
An interesting finding was the presence upstream from and in opposite orientation to aldA of an open reading frame whose deduced amino acid sequence is highly similar to that of the activator AcoR. The highest similarity of both AldA and this AcoR homologue was found with the not-yet-characterized P. aeruginosa PA4022 and PA4021 gene products, respectively. The physical organization of these two genes was also similar in both bacteria. It would be interesting to find out whether these P. aeruginosa genes are involved in alcohol catabolism.
This study demonstrates that expression of aldA is under ς54 control. ς54 controls diverse and unrelated functions in different groups of bacteria (for a recent review, see reference 25). Some of these functions include the catabolism of acetoin and ethanol in Alcaligenes eutrophus (13, 14). Our finding that aldA is transcribed from a ς54-dependent promoter and the inability of rpoN and aldA mutants to grow on ethanol, n-butanol, or hexanol add the catabolism of these alcohols to the processes controlled by rpoN.
Transcription from ς54-dependent genes requires additional transcriptional factors known as enhancer binding proteins (11); this implies the requirement of an activator protein for aldA transcription. In fact the presence upstream of aldA of an acoR homologue, encoding a member of the NifA family of transcriptional activators, suggests that this AcoR might be the activator of the aldA promoter. In R. eutropha, AcoR is the putative activator for acetoin and AcDH-II genes (6).
The inability of rpoN and aldA mutants to form mature cysts on n-butanol but not in BHB supports the hypothesis that n-butanol is not by itself an inductor but that it induces encystment because it is converted to BHB or other related metabolites. The lack of encystment by SG9A and CN10 on n-butanol could be caused by their inability to grow under these conditions. However, we showed that encystment can occur under nongrowing conditions, as was the case for strain CN10 when it was induced in BHB without ammonia. In addition, encystment is initiated by the rounding up of the vegetative cell and the termination of cell division (19).
It is important that lack of growth of strain CN10 on BHB in the absence of ammonia is due to the negative effect of the rpoN mutation on nitrogen fixation and that this process has been shown to be arrested upon initiation of encystment (19, 20). In contrast, lack of growth on n-butanol of either SG9A or CN10 strain is caused by each strain's inability to catabolize alcohols. Thus, a source of carbon (able to induce a shift from glucose to BHB metabolism) seems to be essential for the encystment process even under nongrowing conditions.
In conclusion, we identified an aldehyde dehydrogenase AldA gene regulated by ς54 that is essential for the catabolism of alcohols and provided evidence that the catabolism of n-butanol by AldA activity is required to induce encystment.
ACKNOWLEDGMENTS
This work was supported by grants 27767-N from CONACyT and IN209399 from DGAPA/PAPIIT UNAM to G.E.
We thank G. Soberón and M. Villanueva for reviewing the manuscript.
REFERENCES
- 1.Alexeyev M F, Shokolenko I, Croughan T P. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160:63–67. doi: 10.1016/0378-1119(95)00108-i. [DOI] [PubMed] [Google Scholar]
- 2.Bali A, Blanco G, Hill S, Kennedy C. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol. 1992;58:1711–1718. doi: 10.1128/aem.58.5.1711-1718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos M E, Martínez-Salazar J, Lloret L, Moreno S, Núñez C, Espín G, Soberón-Chávez G. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy C, Gamal R, Humphrey R, Ramos J, Brigle K, Dean D. The nifH, nifM and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1 gene banks. Mol Gen Genet. 1986;205:318–325. [Google Scholar]
- 6.Krüger N, Steinbüchel A. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J Bacteriol. 1992;174:4391–4400. doi: 10.1128/jb.174.13.4391-4400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin L P, Sadoff H L. Encystment and polymer production by Azotobacter vinelandii in the presence of β-hydroxybutyrate. J Bacteriol. 1968;98:1335–1341. doi: 10.1128/jb.95.6.2336-2343.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 9.Mejía-Ruíz, Guzmán H J, Moreno S, Soberón-Chávez G, Espín G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- 10.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 431–435. [Google Scholar]
- 11.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura K, Bernheim F. Studies on malonic semialdehyde dehydrogenase from Pseudomonas aeruginosa. Biochim Biophys Acta. 1961;50:147–152. doi: 10.1016/0006-3002(61)91071-x. [DOI] [PubMed] [Google Scholar]
- 13.Palosaari N R, Rogers P. Purification and properties of inducible coenzyme A-linked butyraldehyde dehydrogenase from Clostridium acetobutylicum. J Bacteriol. 1988;170:2971–2976. doi: 10.1128/jb.170.7.2971-2976.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priefert H, Hein S, Krüger N, Zeh K, Schmidt B, Steinbüchel A. Identification and molecular characterization of the Alcaligenes eutrophus H16 aco operon genes involved in acetoin catabolism. J Bacteriol. 1991;173:4056–4071. doi: 10.1128/jb.173.13.4056-4071.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priefert H, Krüger N, Jendrossek D, Schmidt B, Steinbüchel A. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J Bacteriol. 1992;174:899–907. doi: 10.1128/jb.174.3.899-907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reusch R N, Sadoff H L. 5-n-Alkylresorcinols from encysting Azotobacter vinelandii: isolation and characterization. J Bacteriol. 1979;139:448–453. doi: 10.1128/jb.139.2.448-453.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reusch R N, Sadoff H L. Lipid metabolism during encystment of Azotobacter vinelandii. J Bacteriol. 1981;145:889–895. doi: 10.1128/jb.145.2.889-895.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reusch R N, Sadoff H L. Novel lipid components of the Azotobacter vinelandii cyst membrane. Nature. 1983;302:268–270. doi: 10.1038/302268a0. [DOI] [PubMed] [Google Scholar]
- 19.Sadoff H L, Berke E, Loperfido B. Physiological studies of encystment in Azotobacter vinelandii. J Bacteriol. 1971;105:185–189. doi: 10.1128/jb.105.1.185-189.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadoff H L. Encystment and germination in Azotobacter vinelandii. Bacteriol Rev. 1975;39:516–539. doi: 10.1128/br.39.4.516-539.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schobert M, Görisch H. Cytochrome c550 is an essential component of the quinoprotein ethanol oxidation system in Pseudomonas aeruginosa: cloning and sequencing of the genes encoding cytochrome c550 and adjacent acetaldehyde dehydrogenase. Microbiology. 1999;145:471–481. doi: 10.1099/13500872-145-2-471. [DOI] [PubMed] [Google Scholar]
- 24.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S L, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrook-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R M, Smith K A, Spencer D H, Wong G K S, Wu Z, Paulsen I T, Reizer J, Saier M H, Hancock R E W, Lory S, Olson M V. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 25.Studholme D J, Buck M. The biology of enhancer-dependent transcriptional regulation in bacteria: insights from genome sequences. FEMS Microbiol Lett. 2000;186:1–9. doi: 10.1111/j.1574-6968.2000.tb09074.x. [DOI] [PubMed] [Google Scholar]
- 26.Toukdarian A, Kennedy C. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 1986;5:399–407. doi: 10.1002/j.1460-2075.1986.tb04225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyss O, Newmann M G, Sokolofsky M D. Development and germination of the Azotobacter cyst. J Biophys Biochem Cytol. 1961;10:555–565. doi: 10.1083/jcb.10.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]