Abstract
Conventional wet laboratory testing, validations, and synthetic procedures are costly and time-consuming for drug discovery. Advancements in artificial intelligence (AI) techniques have revolutionized their applications to drug discovery. Combined with accessible data resources, AI techniques are changing the landscape of drug discovery. In the past decades, a series of AI-based models have been developed for various steps of drug discovery. These models have been used as complements of conventional experiments and have accelerated the drug discovery process. In this review, we first introduced the widely used data resources in drug discovery, such as ChEMBL and DrugBank, followed by the molecular representation schemes that convert data into computer-readable formats. Meanwhile, we summarized the algorithms used to develop AI-based models for drug discovery. Subsequently, we discussed the applications of AI techniques in pharmaceutical analysis including predicting drug toxicity, drug bioactivity, and drug physicochemical property. Furthermore, we introduced the AI-based models for de novo drug design, drug-target structure prediction, drug-target interaction, and binding affinity prediction. Moreover, we also highlighted the advanced applications of AI in drug synergism/antagonism prediction and nanomedicine design. Finally, we discussed the challenges and future perspectives on the applications of AI to drug discovery.
Keywords: MT: Bioinformatics, artificial intelligence, drug discovery and development, bioinformatics, data resources, molecular descriptors
Graphical abstract
Chen and colleagues reviewed the applications of AI in drug discovery from the following aspects including data resources, molecular descriptors, and representative applications of AI in drug discovery. The challenges and perspectives for the applications of AI in drug discovery and development were discussed as well.
Introduction
Drug discovery is a process through which new medications against diseases are discovered. It involves the use of a wide variety of technologies and expertise. In general, discovering and developing a drug takes US$2.8 billion and 15 years on average.1 The low-efficacy and high-cost characteristics of conventional methods have become the hurdles of drug discovery. Therefore, developing new methods to deal with such a time-consuming and expensive task is necessary.2
The revolution in high-performance computer hardware and the availability of multi-omics data have enabled artificial intelligence (AI) techniques to transcend from theoretical studies to real applications in multiple disciplines. The successful application of AI techniques, particularly to biological data analysis, has attracted the attention of the pharmaceutical industry. Thus far, AI techniques have been implemented in drug discovery processes, such as drug-target prediction,3 bioavailability prediction,4 and de novo drug design.5 Some major pharmaceutical companies, such as Bayer, Roche, and Pfizer, have also begun to collaborate with information technology (IT) companies to develop AI technique-based methods for drug design.6 Recently, with the help of AI, the Insilico Medicine company discovered the drug treating idiopathic pulmonary fibrosis, which exhibits positive results in Phase I trials (https://clinicaltrials.gov/ct2/show/NCT05154240). Hence, drawing the conclusion that AI techniques have modernized the field of drug discovery and development is reasonable.
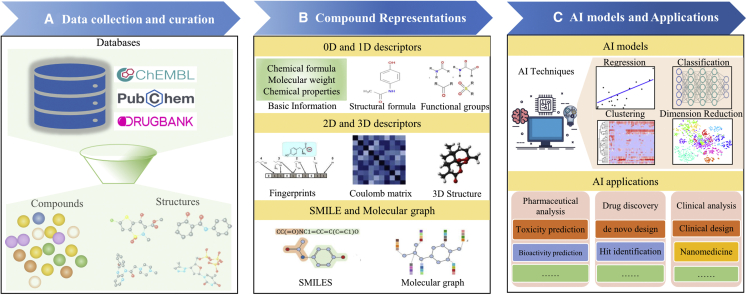
The basic schematics of applying AI techniques to drug discovery and evaluation are summarized in Figure 1. The major procedures include data collection and curation (Figure 1A), compound representation (Figure 1B), and AI methods and their applications in drug discovery (Figure 1C). To provide researchers with a catching-up view of the development in this field, we first introduced representative data resources, molecular representations and descriptors, and AI techniques in drug discovery. Then, we introduced the successful applications of AI to different stages of drug discovery. Finally, we discussed the challenges and future perspectives on applying AI to drug discovery.
Figure 1.
Framework of AI technique application to drug discovery and evaluation
Major procedures include (A) data collection and curation; (B) compound representations by using molecular descriptors; and (C) AI methods and their applications.
Resources and methods for AI-based drug discovery
As indicated in Figure 1, data resources, data representation schemes, and AI methods are the three key components of applying AI to drug discovery and evaluation. Accordingly, they will be introduced briefly in this section.
Data resources
A high-quality dataset is the key to applying AI to drug discovery. Advances in high-throughput sequencing and IT have boosted the generation of a series of free and open-access databases for drug discovery. These databases enable drug discovery to transit into the big data era and accelerate the drug discovery process. Representative databases, along with their web links, brief descriptions, and references, are listed in Table 1. Their applications are not reviewed in the present work due to the limited space.
Table 1.
Representative databases for drug discovery
| Database | Website URL | Description | Reference |
|---|---|---|---|
| ChEMBL | https://www.ebi.ac.uk/chembl/ | A manually curated database of bioactive molecules with drug-like properties. It gathers chemical, bioactivity, and genomic data to aid the translation of genomic information into effective new drugs. | Mendez et al.7 |
| ChemDB | http://cdb.ics.uci.edu | A chemical database that contains nearly 5 million commercially available small molecules, along with their predicted or experimentally determined physicochemical properties. | Chen et al.8 |
| COCONUT | https://coconut.naturalproducts.net/ | A database that contains 407,270 unique natural products, along with information about their molecular properties and molecular descriptors. | Sorokina et al.9 |
| DGIdb | http://www.dgidb.org | A database that provides information on DTI and druggable genomes from over 30 trusted sources. | Freshour et al.10 |
| DrugBank | http://www.drugbank.ca | A database of drugs, their targets, 3D structures, and other useful information. | Wishart et al.11 |
| DTC | http://drugtargetcommons.fimm.fi/ | A crowd-sourcing platform that provides drug-target bioactivity data and classification of targets. | Tang et al.12 |
| INPUT | http://cbcb.cdutcm.edu.cn/INPUT/ | A network pharmacology platform for traditional Chinese medicine. It contains 29,812 compounds isolated from 4,716 Chinese herbs. | Li et al.13 |
| PubChem | https://pubchem.ncbi.nlm.nih.gov/ | An open chemistry database that provides information about molecules, such as chemical structures, identifiers, chemical and physical properties, and biological activities. | Kim et al.14 |
| SIDER | http://sideeffects.embl.de | A database that provides information on marketed medicines and their recorded adverse reactions. | Campillos et al.15 |
| STITCH | http://stitch.embl.de/ | A database of known and predicted interactions between chemicals and proteins, including 9,643,763 proteins from 2,031 organisms. | Szklarczyk et al.16 |
ChEMBL is a manually curated database that currently contains more than 2 million compounds that exhibit drug-like properties.7 ChEMBL gathers information regarding the action mechanisms, molecular properties, absorption, distribution, metabolism, excretion, toxicity, therapeutic indications, and target interactions of the deposited compounds.
ChemDB is a freely accessible database that contains nearly 5 million commercially available small molecules and their physicochemical properties, such as molecular weight, solubility, and rotatable bonds.8 In addition, a series of cheminformatics tools, such as Smi2Depict, MOLpro, AquaSol, and Reaction Predictor, are embedded into ChemDB, making this database user-friendly for drug discovery.
The Collection of Open Natural Products (COCONUT) is one of the best annotated databases of natural products.9 It aggregates 407,270 elucidated and predicted natural products collected from a large number of chemical data sources. As a free database, COCONUT can be searched in multiple ways, such as molecule names, molecular structures, and structural properties. COCONUT also provides molecular properties and descriptors for each natural product. Moreover, all the data in COCONUT are available for download and can be queried programmatically via an application programming interface (API).
The Drug-Gene Interaction Database (DGIdb) provides information on drug-gene interactions and genes or gene products that can interact with drugs.10 To date, DGIdb contains more than 40,000 genes and 10,000 drugs involved in over 100,000 drug-gene interactions. These data are mined from multiple diverse sources by performing expert curation and text mining. All the deposited genes in DGIdb are clustered into 43 categories. Users can either browse the genes in each category or enter a list of genes or drugs to retrieve drug-gene interactions in the search module. In addition, DGIdb can be accessed programmatically by API through the web-based interface.
DrugBank is a free-to-access reference drug database.11 It currently contains 14,746 drugs, along with comprehensive information about drug-drug interactions, drug-target associations, drug classifications, and drug reactions. Users can search, browse, and extract text, images, and structural data in DrugBank by using the embedded tools. DrugBank has become the world’s most widely used resource for drug screening, design, and metabolism prediction.
Drug Target Commons (DTC) is a freely accessible online resource that provides annotated and unannotated drug-target interaction (DTI) data.12 For its recent release, DTC includes clinical trial information and disease-gene associations, facilitating the chemical biology and drug-repurposing applications of compounds. As an open resource, DTC not only supports database dump but also API to access its deposited data.
The Intelligent Network Pharmacology Platform Unique for Traditional Chinese Medicine (INPUT) is an online analytical platform that is uniquely for traditional Chinese medicine.13 At present, INPUT contains 4,716 herbs, 29,812 herbal compounds, and 9,847 diseases collected from public databases and the literature. The herbs, compounds, and diseases are cross-linked through the herb-compound-gene-disease network in INPUT, which facilitates the discovery of herb-oriented drugs and the scientific interpretation of traditional Chinese medicine.
PubChem is a freely accessible chemical information resource that contains the biological, physical, chemical, and toxic information of chemical molecules.14 All these data are collected from more than 850 sources. Users can search for chemicals in PubChem by inputting molecular formula, structure, and other identifiers as keywords. At present, PubChem has become one of the foremost data sources for computational drug discovery and design.
The Side Effect Resource (SIDER) is a database that focuses on drugs and their side effects.15 The current release of SIDER includes 1,430 drugs, 5,880 side effects, and 140,064 drug-side effect pairs. These data can be browsed through either drugs or side effects. They have been used in many aspects, such as predicting drug indications, mining side effects, and identifying metabolic dysregulation.
The Search Tool for Interacting Chemicals (STITCH) is a database that contains known and predicted interactions between chemicals and proteins.16 These interactions encompass 9,643,763 proteins from 2,031 organisms, which were collected from computational prediction, knowledge transfer between organisms, and other databases. Users can query STITCH in multiple ways, such as through the names of chemicals and proteins, chemical structures, and protein sequences. For large-scale analyses, the data in STITCH can be obtained either via bulk download or accessed programmatically with API.
Molecular descriptors and structure representations
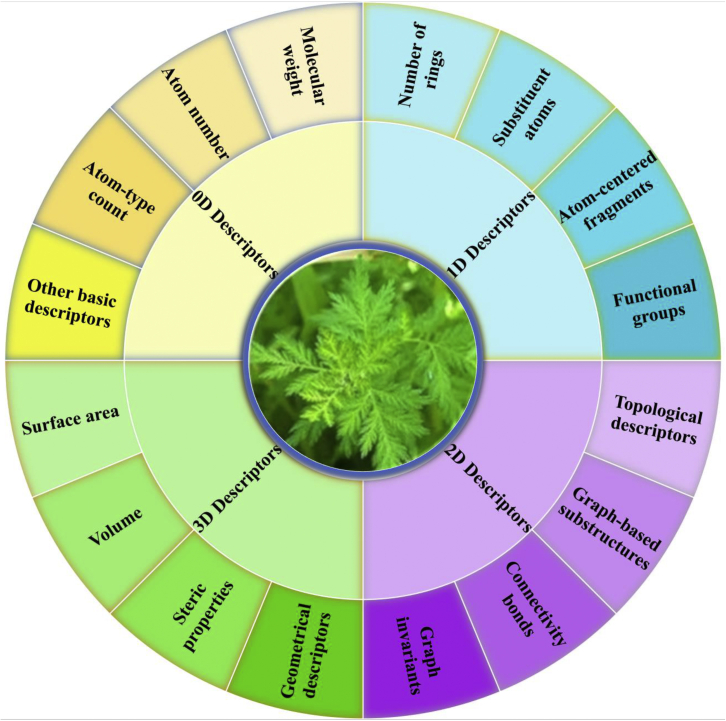
With the explosive growth of natural products, another key point in AI-based drug discovery and analysis is the transfer of molecules into computer-readable format, while keeping their intrinsic physicochemical properties.17 Various types of descriptors have been proposed to represent drugs; these descriptors can be classified into four categories in accordance with their dimensionality (Figure 2). To accelerate the drug discovery process, a series of open-source toolkits has been proposed for calculating molecular descriptors and structure representations, such as OpenBabel18 and ChemmineR.19
Figure 2.
Summary of molecular and structural representation schemes
The zero-dimensional (0D) descriptor is the simplest molecular representation; it is obtained in accordance with the chemical formula of drugs.20 The 0D descriptor typically includes molecular weight, atom number, atom-type count, and other basic descriptors (e.g., number of heavy atoms). The 0D descriptor is extremely simple, and it can only extract shallow information.
The one-dimensional (1D) descriptor encodes drugs in accordance with their substructures, such as the number of rings, functional groups, substituent atoms, and atom-centered fragments.20 The elements of the 1D descriptor are typically binary (e.g., 1/0 indicates the presence/absence of a substituent atom) or the occurrence frequencies of some substructures. Apart from the property-based 1D descriptor, the simplified molecular-input line-entry system (SMILES)21 is another type of 1D descriptor. SMILES represents drugs with a string of characters. SMILES depends on atom order, and thus, a drug will have several SMILES representations, and the normalization algorithm should be performed to obtain canonical SMILES.
The two-dimensional (2D) descriptor provides additional information to the 1D descriptor by considering adjacency, connectivity, and other types of topological features of the atoms. Therefore, 2D descriptors are typically derived by representing a drug as a graph, wherein the nodes indicate atoms and edges indicate bonds. Property-based 2D descriptors frequently include graph invariants, connectivity bonds, graph-based substructures, and topological descriptors. To extract more information, the molecular fingerprint (FP) was proposed for encoding molecules in binary form.22 FP indicates the presence/absence of particular substructures through a string with a given length and marked by 1/0. The commonly used 2D FPs are the molecular access system fingerprints,23 daylight-like fingerprint,18 and extended-connectivity fingerprints.24
The three-dimensional (3D) descriptor depicts a molecule in 3D space,25 and each atom of a molecule is spatially characterized by the x, y, and z coordinates. The 3D descriptor includes spatial and geometrical configuration information; it has high information content. Thus, information about surface area, volume, and steric properties can be obtained by using 3D descriptors. Non-property-based 3D descriptors, such as geometrical fingerprint26 and pharmacophore fingerprint,27 are also available. They can represent complex physicochemical properties of drugs and are widely used in drug discovery and virtual screening.
The schematic diagrams illustrating the representations of compounds by using 0D-3D descriptors are shown in Figure 1B. In addition to these encoding schemes, graph-based methods have also been proposed recently to encode molecules. Examples of the graph-based schemes include the spectral and spatial graph convolutional network. For more details about graph-based molecular representation methods, readers can refer to a recent review.28
Commonly used AI techniques
To be a fit-for-purpose approach, the selection and application of AI techniques are problem-oriented. Two common types of AI techniques, namely, supervised and unsupervised learning, are used in the field of drug discovery.29 A supervised learning technique uses input-labeled data to train models that are capable of classifying or predicting outcomes of new data. By contrast, an unsupervised learning technique deals with unlabeled data and aims to develop models that are capable of identifying recurring patterns and clustering of the input data in a manner without prior knowledge.30 Supervised learning techniques can be further classified into classification and regression algorithms, and unsupervised learning techniques include clustering and dimensionality reduction algorithms. To facilitate users in applying these AI techniques, a series of open-source packages and frameworks, such as Scikit-learn,31 PyTorch,32 and Keras (https://github.com/fchollet/keras), have been developed for practicing the aforementioned algorithms. Widely used AI techniques in drug discovery are listed in Table 2 and briefly discussed below.
Table 2.
Widely used AI techniques in drug discovery
| Category | Task | Method | Representative application | Reference |
|---|---|---|---|---|
| Supervised learning | Regression analysis | MLR | DTI | Talevi et al.29 |
| DT | Adverse drug reactions | Hammann et al.33 | ||
| LR | Drug-drug interaction | Schober and Vetter34 | ||
| Classification | SVM | Compound classification | Maltarollo et al.35 | |
| CNN | Bioactivity prediction | El-Attar et al.36 | ||
| RNN | De novo drug design | Gupta et al.37 | ||
| GAN | Molecule discovery | Blanchard et al.38 | ||
| Unsupervised learning | Clustering | k-means | Drug candidate selection | Shen et al.39 |
| Hierarchical | Molecular scaffold analysis | Manelfi et al.40 | ||
| Dimension reduction | PCA | QSAR | Yoo and Shahlaei41 | |
| t-SNE | Chemical space mapping | Karlov et al.42 |
CNN, convolution neural network; DT, drug target; GAN, generative adversarial network; LR, logistic regression; MLR, multiple linear regression; PCA, principal-component analysis; RNN, recurrent neural network; SVM, support vector machine; t-SNE, T-distributed stochastic neighbor embedding.
Regression analysis technique
Multiple linear regression (MLR) is a modeling technique that aims to estimate the relationship between independent variables and the dependent variable by fitting a linear equation into observed data.29 The ordinary least squares method is used to find the best-fit line by reducing the sum of squared errors, which are the differences between the observed value and the fitted value given by the model.
A decision tree (DT) is a nonlinear supervised learning technique that can be used in classification and regression tasks.33 The primary components of a DT model are nodes (including root nodes, internal nodes, and leaf nodes) and branches. The algorithm starts at the root node and selects a branch according to the decision rule of the root node. Subsequently, the algorithm reaches the internal nodes and further makes decisions on the basis of this node. Finally, the algorithm will reach leaf nodes that represent possible outcomes within the dataset.
Logistic regression (LR) is a supervised learning technique that can be used to estimate the probability of occurrence of an event on the basis of log odds ratio.34 LR can be classified into three categories, namely, binary, nominal, and ordinal LR, in accordance with the categories of response variables.
Classification technique
Support vector machine (SVM) is a classical supervised learning technique that is widely used in drug discovery.35 The basic idea of SVM is to cast data into higher-dimensional feature space by using kernel functions and find the optimal separating hyperplane that maximizes the margin of training data.
Convolution neural network (CNN) is a deep learning technique with feedforward neural network architecture.36 The CNN model includes three types of layers: the convolutional, pooling, and fully connected layers. The convolutional layer aims to learn feature representations of the input. The pooling layer is used to reduce the number of trainable parameters. The fully connected layer aims to produce classification scores and perform reasoning. Compared with conventional machine learning methods, the advantages of CNN include automatically extracting non-handcrafter features from raw input.
Recurrent neural network (RNN) is a feedforward artificial neural network (ANN) that specializes in dealing with sequential data.37 RNN consists of numerous successive recurrent layers, and its information cycles through a loop. These features make RNN distinct from the traditional neural network. Hence, RNN has the ability to capture contextual content from input data. RNN has also been used in drug design and discovery given its great promise in handling sequential data.43
Generative adversarial network (GAN) is a deep learning framework with two components: the generator and the discriminator.38 The former is used to generate new data with the same characteristics as the training data. The latter is used to distinguish actual samples from the generated fake ones. Compared with conventional machine learning methods and other deep learning techniques, GAN is good at solving problems with a small sample size.
Clustering technique
k-means clustering is one of the most important and popular clustering algorithms.39 It aims to group similar data into clusters, such that samples in the same cluster are more similar to each other than to those in other clusters. This algorithm iteratively identifies a certain number of centroids (i.e., the arithmetic mean of all data points assigned to a particular cluster) within a dataset and allocates every datum to the nearest cluster. These procedures are repeated until cluster assignments stop changing.
Hierarchical clustering is another type of clustering algorithm that is used to group data into clusters on the basis of similarity measures.40 Distinct from k-means clustering, hierarchical clustering initially regards each datum as an individual cluster and then identifies the two closest clusters and merges them together. These procedures are iterated until all the clusters are merged together. The final result is presented in a dendrogram.
Dimension reduction
Principal-component analysis (PCA) is a linear dimensionality reduction technique that can transform a large dataset into a smaller one while maintaining most of the original information.41 The basic idea of PCA is to find principal components that explain a large portion of the variation in a dataset. The procedures for conducting PCA include standardizing data, computing the covariance matrix, computing the eigenvalues and eigenvectors, identifying the principal components, and remodeling the data.
T-distributed stochastic neighbor embedding (t-SNE) is a nonlinear dimensionality reduction technique that is capable of visualizing high-dimensional data in 2D or 3D space.42 The t-SNE algorithm first converts similarities between data points into joint probabilities. Then, it minimizes the Kullback-Leibler divergence between the joint probabilities of high-dimensional data and low-dimensional embedding.
Application of AI to pharmaceutical analysis
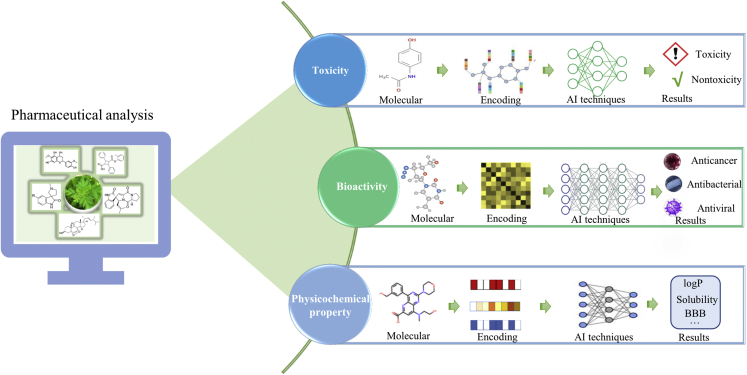
Pharmaceutical analysis involves the processes of identification, determination, quantification, and purification of pharmaceutical raw materials; it is an essential part of drug discovery. Qualitative and quantitative analyses are the two major types of experimental methods in pharmaceutical analysis. Although these techniques exhibit high accuracy, their cost for screening novel drug candidates from a huge amount of natural products is still expensive. Compared with experimental techniques, the costs required by computational methods are negligible. Hence, AI techniques have been used in pharmaceutical analysis to complement experimental techniques. The representative applications of AI techniques in pharmaceutical analysis are summarized in Figure 3.
Figure 3.
Application of AI techniques to pharmaceutical analysis
Drug toxicity prediction
Toxicity is a measure of the unwanted or adverse effects of chemicals.44 Toxicity evaluation is one of the fundamental steps in drug discovery, and it aims to identify substances that have harmful effects on humans.45 However, the in vivo test requires animal tests and thus increases the costs of drug discovery. Computational methods exhibit the advantages of being able to predict a chemical’s toxicity with low cost and high efficiency.46 Accordingly, a series of AI technique-based methods have been developed to predict the toxicity of chemicals.47,48 To assess the performance of different computational methods for predicting the toxicity of chemicals, the scientific community proposed the “Toxicology in the 21st Century (Tox21)” challenge.46
DeepTox is an ensemble model for predicting the toxicity of chemicals, and its fundamental framework is based on a three-layer deep neural network (DNN).49 After performing data cleaning and quality control, the remaining chemicals are encoded by using the aforementioned 0D to 3D molecular descriptors, which are used as input of DNN. The DeepTox pipeline is obtained by tuning and optimizing a set of hyperparameters, such as number of hidden units, learning rate, and dropout rate. Comparative results based on the Tox21 dataset demonstrate that DeepTox outperforms its counterparts in toxicity prediction.49
Drug bioactivity prediction
In reality, a large number of drugs derived from natural products are ineffective due to the lack of bioactivity. Hence, drug bioactivity assessment has become an active area in drug discovery. Although in vitro and in vivo experiments can mimic the functions of molecules in the human body, they are still time-consuming and expensive. Given their cost-effectiveness and time economy, AI techniques have been effectively applied to predicting drug bioactivities, such as anticancer, antiviral, and antibacterial activities.50,51,52
For example, Stokes et al. proposed a directed message passing neural network that is capable of predicting antibacterial activity.53 For each molecule, they first constructed a molecular graph in accordance with its SMILES and then obtained the feature vector based on atomic features (e.g., number of bonds for each atom and atomic number) and bond features (e.g., bond type and stereochemistry).53 By applying the message passing operation multiple times, the optimized feature vector was fed into the feedforward neural network that outputted the antibacterial probability of a molecule.53 This model is available at http://chemprop.csail.mit.edu/, and it can facilitate the discovery of antibacterial molecules.
Drug physicochemical property prediction
Physicochemical properties are intrinsic characteristics of drugs. Knowledge about physicochemical properties is required for understanding and modeling the action of drugs. Among the numerous types of physicochemical properties, solubility is important because it affects the pharmacokinetic properties and formulations of drugs.54,55 However, laborious and costly experimental techniques have precluded rapid solubility prediction; hence, considerable effort has been devoted to develop AI-based solubility prediction models.
Panapitiya et al. assessed different deep learning methods (i.e., fully connected neural networks, RNNs, graph neural networks, and SchNet) and molecular representation approaches (i.e., molecular descriptors, SMILES, molecular graphs, and 3D atomic coordinates) for solubility prediction.54 Based on the same test dataset, the authors found that the fully connected neural network achieved the best performance for solubility prediction by leveraging molecular descriptors. In addition, the authors analyzed the importance of different features for prediction and found that 2D molecular descriptors made the greatest contributions. To facilitate further research on solubility prediction, an open-source code was provided at https://github.com/pnnl/solubility-prediction-paper.
AI in natural product-inspired drug discovery
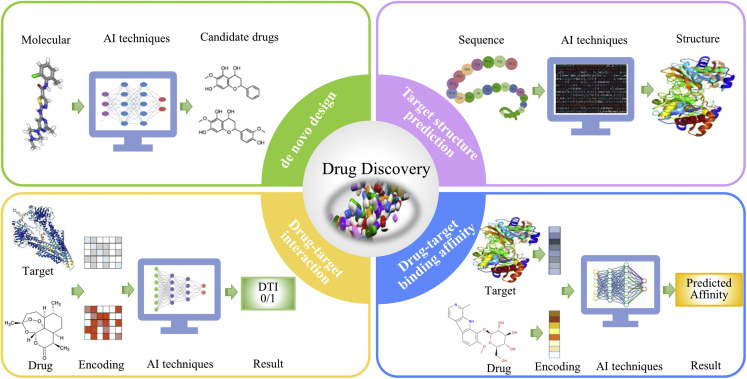
Drug discovery is a process of identifying active compounds with therapeutic effects on the intended diseases. Although a high-throughput screening technique can scan thousands of different compounds one at a time, it is still time-consuming and costly.56 To address these challenges, AI techniques have been applied to nearly all aspects of drug discovery. The applications of AI to natural product-inspired drug discovery, such as de novo drug design, target structure prediction, DTI prediction, and drug-target binding affinity prediction, are illustrated in Figure 4.
Figure 4.
AI techniques for natural product-inspired drug discovery
De novo drug design
De novo drug design refers to the process of generating novel drug-like compounds without a starting template. Although conventional structure-based and ligand-based drug design methods have enhanced the discovery of small-molecule drug candidates, they respectively rely on knowledge about the active site of a biological target or the pharmacophores of a known active binder,57 hindering their applications to modern drug discovery. The boom of AI techniques has offered new opportunities to de novo drug design and accelerated the drug discovery process.
In recent years, various deep learning-based models have been proposed for de novo drug design, such as the reinforcement learning-based model ReLeaSE,58 the encoder-decoder-based model ChemVAE,59 the GAN-based model GraphINVENT,60 and the RNN-based model MolRNN.61 Another key point of de novo drug design is molecular representation. SMILES, fingerprint, molecular graph, and 3D geometry have been used as input of deep learning algorithms. The fundamental framework of deep learning-based de novo drug design methods is shown in the left upper corner of Figure 4. Detailed information about deep learning-based de novo drug design models is provided recent reviews.57,62
Target structure prediction
Most drug targets are proteins that play important roles in enzymatic activities, cell signaling, and cell-cell transduction. The functions of proteins are determined by their structures. Although conventional experimental techniques, such as X-ray crystallography, cryogenic electron microscopy, and nuclear magnetic resonance spectroscopy, have been proposed to determine protein structures, they are still time-consuming and costly.63 As reported, experimental techniques have only deciphered the structures of 100,000 unique proteins, which account for only a small part of known proteins.64 Therefore, developing novel methods to fill the gap between the number of protein sequences and known protein structures is an urgent need.65
With the rapid growth of computational power and the breakthroughs of AI techniques, many computational approaches have been proposed for protein structure prediction. The basic schematics of computational protein structure prediction models are presented in the right upper corner of Figure 4. The neural network-based AlphaFold method developed by DeepMind is the best-performing method, and it is able to predict the 3D structures of proteins from their amino acid sequences and achieve accuracies competitive with experiments.64 The descriptions of the algorithm and architecture of AlphaFold are provided in Senior et al.66 The source code of AlphaFold is available at https://github.com/deepmind/alphafold.
DTI prediction
DTI prediction refers to the interaction between chemical compounds and protein targets in living organisms.67 DTI prediction is an essential process for drug discovery. Hence, experimental methods have been used to determine DTI, such as co-immunoprecipitation,68 phage display technology,69 and yeast two-hybrid.70 However, these wet laboratory techniques are time-consuming when they are used to predict DTI. Recently, the ever-increasing biological data have paved the way for the in silico prediction of DTI. Therefore, computational methods are being increasingly used in DTI prediction. These methods, which were summarized in a recent review,71 can be classified into the following categories: ligand-based methods, docking simulations, gene ontology-based methods, text mining-based methods, and network-based methods.
Compared with other types of methods, deep learning-based methods frequently exhibit better performance in DTI prediction.72 The common workflow of the deep learning-based DTI prediction method is illustrated at the left bottom corner of Figure 4. First, compounds and proteins are encoded by using their corresponding features. Then, the feature embedding of the compounds and proteins is used as the input of deep learning methods. In accordance with this strategy, models based on deep belief neural network,73 CNN,72 and multiple layer perceptron74 have been proposed for drug-protein interaction prediction, considerably facilitating drug discovery.
In real life, many diseases lack well-defined targets. Hence, finding drugs for these diseases is impossible by using the aforementioned methods. Zhu et al. recently proposed a deep learning-based efficacy prediction system (DLEPS) that can identify drug candidates in accordance with the changes in gene expression profiles rather than specific targets.75 First, compounds were encoded using SMILES and used as input of CNN to fit gene expression changes. Subsequently, the potential efficacy of compounds against diseases was evaluated on the basis of gene signatures specific to certain diseases and sorted using a method similar to gene set enrichment analysis. DLEPS provides novel insights into identifying new drugs for complex diseases.
Drug-target binding affinity prediction
In most cases, DTI prediction is regarded as a binary classification problem, but binding affinity between a drug and its target is disregarded.67 Binding affinity reflects the strength of drug-target pair interactions, and it is considerably informative for drug discovery. Although binding affinity can be experimentally determined by measuring dissociation and inhibition constants, the time cost and financial expenses of these procedures are extremely high. Therefore, developing computational methods for predicting binding affinity is necessary.
In 2018, Öztürk et al. proposed the first deep learning model, called DeepDTA, for predicting binding affinity between drugs and their targets.76 In DeepDTA, the drug and the target were encoded using SMILES and amino acid letters, respectively, which were then used as input for CNN. The basic framework of DeepDTA is shown at the right bottom corner of Figure 4. The comparative results demonstrated that DeepDTA suppressed KronRLS77 and SimBoost78 for drug-target binding affinity prediction. Inspired by DeepDTA, a series of deep learning-based models has been sequentially proposed, such as WideDTA76 and DeepAffinity,79 which have become useful tools in drug discovery.
Advanced applications of AI in drug design
AI in drug synergism/antagonism prediction
Synergism and antagonism are the two categories of drug combination effects. The former can overcome primary and secondary drug resistance, and it is effective for the treatment of cancers,80 AIDS,81 and bacterial infections,82 whereas the latter reduces the effectiveness of drugs. With the ever-increasing number of drugs, their possible combinations are astronomical. Thus, experimentally investigating drug combination effect is costly and time-consuming. The advancements of AI techniques have made them applicable to exploring possible drug combinations at lower cost and with more efficiency.
In 2015, Li et al. proposed a Bayesian network model for exploring and analyzing drug combinations.83 In the same year, Wildenhain et al. developed a random forest-based model for predicting compound synergism from chemical-genetic interactions.84 Recently, Preuer et al. proposed DeepSynergy,85 a deep learning-based model for predicting the synergism of anticancer drugs. The inputs of DeepSynergy included the chemical information of drugs and the genomic information of diseases, which were then propagated through the network to the output unit. The comparative results from a publicly available synergy dataset demonstrated that DeepSynergy outperformed its counterparts in predicting drug synergism. The web server and source code of DeepSynergy are provided at www.bioinf.jku.at/software/DeepSynergy and https://github.com/KristinaPreuer/DeepSynergy, respectively.
AI in nanomedicine design
Nanotechnology has been applied to design nanomedicines by using nanometric-scale materials in the clinical setting.86 Nanomedicines are developed by materials at the nanometric scale, and, thus, they can penetrate the barriers to interact with targets in the body. At present, some nanomedicines have already been approved by the U.S. Food and Drug Administration, and they have exhibited better performance in the treatment of cancers87 and HIV-1 infection.88 However, the lack of quantitative and qualitative understanding of nanomaterial properties and biological responses precluded the wide application of nanomedicines.
A combination of nanotechnology and AI provides novel solutions to deal with this dilemma. For example, Li et al. proposed an ANN for the task of nanomedicine composition optimization.89 Muñiz Castro et al. developed a 3D printing nanomaterial formulation pipeline that can predict the extrusion temperature, filament mechanical characteristics, and dissolution time of nanomaterials.90 In addition, the effectiveness of a nanomedicine is affected by its cellular uptake. Hence, a cellular uptake prediction model will considerably help researchers in predicting nanomedicine effectiveness. On the basis of an ANN, Alafeef et al. developed a platform for predicting nanoparticle cellular internalization in different cell types.91 Other applications of AI to nanomedicine design and their principles were summarized in a recent comprehensive review.80
AI in oligonucleotide design
Besides the drugs derived from natural products, oligonucleotide therapeutics composed of short strands of DNA or RNA have become a novel class of drugs.92 Antisense oligonucleotides (ASO), small interfering RNA (siRNA), and CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated protein) are the main oligonucleotide therapeutics systems that enable the precise treatment of diverse diseases. Since experimental designing these oligonucleotides will cost enormous resources, the AI approaches have also been used to help researchers to identify and design the oligonucleotide-based drugs. For example, Chiba et al. proposed a machine learning-based model, eSkip-Finder, to identify effective exon skipping ASOs.93 Dar et al. developed SMEpred to predict the efficacy of siRNAs.94
Concluding remarks and prospects
Over the past few years, we have witnessed the wide applications of AI techniques to various steps of drug discovery and development. The boom of AI techniques has made substantial contributions to the acceleration of drug discovery. The application of Chat Generative Pre-Trained Transformer (ChatGPT) is also a promising topic in drug discovery and development. Since it can provide methods to identify potential targets, design new drugs, and optimize the pharmacodynamics of drug candidates, ChatGPT has the potential to speed up drug development process. However, AI techniques are not versatile tools for drug discovery due to the following challenges.
The first key point is the availability of high-quality data that can be used to train AI technique-based models. Although the amount of biological and chemical data is increasing, the issue of poor data quality hinders the full use of these data. To solve this issue, data curation can be performed to organize and manage raw data. For this objective, academic institutions and pharmaceutical companies should cooperate to develop data standards and frameworks that will be helpful in data collection and clearance. Data quantity is also important for the applications of AI techniques. In real cases, the number of positive samples is smaller than that of negative ones. The sample imbalance problem will directly affect the performance of the models. Thus, oversampling and undersampling methods are suggested to be used to balance samples in the datasets.
Another typical issue of AI technique-based models for drug discovery is the lack of interpretability. A model’s interpretability is the degree to which humans can understand the processes it uses to arrive at its outcomes. In most cases, the proposed models fall short in interpreting their biological and pharmaceutical meanings. Hence, trusting the predictive results obtained by AI techniques is difficult for experimental scientists.95 In addition, the lack of interpretability also makes models unable to troubleshoot these approaches when their performance is poor on the test data. To deal with this issue, post hoc explanation techniques are suggested to be used when building models.96 Popular techniques for post hoc interpretations include text explanation, visualization explanation, and attention mechanism explanation. Text explanation techniques can provide qualitative interpretations by presenting human-understandable verbal words. Visualization explanation techniques, such as t-SNE, can visualize the learned latent high-dimensional features in 2D space.96 Attention mechanism explanation techniques can automatically learn and calculate the contribution of input to output, making the model interpretable.97
The availability and accessibility of the proposed models are also challenges in drug discovery. Although many AI technique-based models have been developed, neither related freely accessible web servers nor source codes are provided for most of these models. Even though some smart tools have been designed, they are only commercially available. These issues preclude their applications to drug discovery and development. Hence, developing open-source tools or packages, which will become invaluable sources in the near future, is necessary.
Although there exist the above-mentioned challenges, AI techniques have been incorporated into drug discovery and development industry. It is believable that AI techniques will bring revolutionary changes for this field.
Acknowledgments
This study was supported by Natural Science Foundation of Sichuan (No. 2023NSFSC0683) and Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (No: ZYYCXTD-D-202209).
Author contributions
S.C. and W.C. conceived the work, X.L and S.Z. participated in reviewing and revising the manuscript. S.C. and W.C. wrote and revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Wei Chen, Email: greatchen@ncst.edu.cn.
Shilin Chen, Email: slchen@icmm.ac.cn.
References
- 1.Fleming N. How artificial intelligence is changing drug discovery. Nature. 2018;557:S55–S57. doi: 10.1038/d41586-018-05267-x. [DOI] [PubMed] [Google Scholar]
- 2.Chen S., Li Z., Zhang S., Zhou Y., Xiao X., Cui P., Xu B., Zhao Q., Kong S., Dai Y. Emerging biotechnology applications in natural product and synthetic pharmaceutical analyses. Acta Pharm. Sin. B. 2022;12:4075–4097. doi: 10.1016/j.apsb.2022.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You Y., Lai X., Pan Y., Zheng H., Vera J., Liu S., Deng S., Zhang L. Artificial intelligence in cancer target identification and drug discovery. Signal Transduct. Targeted Ther. 2022;7:156. doi: 10.1038/s41392-022-00994-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei M., Zhang X., Pan X., Wang B., Ji C., Qi Y., Zhang J.Z.H. HobPre: accurate prediction of human oral bioavailability for small molecules. J. Cheminf. 2022;14:1. doi: 10.1186/s13321-021-00580-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaschke T., Arús-Pous J., Chen H., Margreitter C., Tyrchan C., Engkvist O., Papadopoulos K., Patronov A. Reinvent 2.0: an AI tool for de novo drug design. J. Chem. Inf. Model. 2020;60:5918–5922. doi: 10.1021/acs.jcim.0c00915. [DOI] [PubMed] [Google Scholar]
- 6.Mak K.K., Pichika M.R. Artificial intelligence in drug development: present status and future prospects. Drug Discov. Today. 2019;24:773–780. doi: 10.1016/j.drudis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M.P., Mosquera J.F., Mutowo P., Nowotka M., et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019;47:D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Swamidass S.J., Dou Y., Bruand J., Baldi P. ChemDB: a public database of small molecules and related chemoinformatics resources. Bioinformatics. 2005;21:4133–4139. doi: 10.1093/bioinformatics/bti683. [DOI] [PubMed] [Google Scholar]
- 9.Sorokina M., Merseburger P., Rajan K., Yirik M.A., Steinbeck C. COCONUT online: collection of open natural products database. J. Cheminf. 2021;13:2. doi: 10.1186/s13321-020-00478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freshour S.L., Kiwala S., Cotto K.C., Coffman A.C., McMichael J.F., Song J.J., Griffith M., Griffith O.L., Wagner A.H. Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49:D1144–D1151. doi: 10.1093/nar/gkaa1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang J., Tanoli Z.U.R., Ravikumar B., Alam Z., Rebane A., Vähä-Koskela M., Peddinti G., van Adrichem A.J., Wakkinen J., Jaiswal A., et al. Drug target commons: a community effort to build a consensus knowledge base for drug-target interactions. Cell Chem. Biol. 2018;25:224–229.e2. doi: 10.1016/j.chembiol.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X., Tang Q., Meng F., Du P., Chen W. INPUT: an intelligent network pharmacology platform unique for traditional Chinese medicine. Comput. Struct. Biotechnol. J. 2022;20:1345–1351. doi: 10.1016/j.csbj.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., et al. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021;49:D1388–D1395. doi: 10.1093/nar/gkaa971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campillos M., Kuhn M., Gavin A.C., Jensen L.J., Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- 16.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., Doncheva N.T., Legeay M., Fang T., Bork P., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David L., Thakkar A., Mercado R., Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. J. Cheminf. 2020;12:56. doi: 10.1186/s13321-020-00460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: an open chemical toolbox. J. Cheminf. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y., Charisi A., Cheng L.C., Jiang T., Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grisoni F., Ballabio D., Todeschini R., Consonni V. Molecular descriptors for structure-activity applications: a hands-on approach. Methods Mol. Biol. 2018;1800:3–53. doi: 10.1007/978-1-4939-7899-1_1. [DOI] [PubMed] [Google Scholar]
- 21.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Model. 1988;28:31–36. [Google Scholar]
- 22.Capecchi A., Probst D., Reymond J.L. One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J. Cheminf. 2020;12:43. doi: 10.1186/s13321-020-00445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo M., Shin H.K., Myung Y., Hwang S., No K.T. Development of natural compound molecular fingerprint (NC-MFP) with the dictionary of natural products (DNP) for natural product-based drug development. J. Cheminf. 2020;12:6. doi: 10.1186/s13321-020-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 25.Matter H., Pötter T. Comparing 3D pharmacophore triplets and 2D fingerprints for selecting diverse compound subsets. J. Chem. Inf. Comput. Sci. 1999;39:1211–1225. [Google Scholar]
- 26.Yin S., Proctor E.A., Lugovskoy A.A., Dokholyan N.V. Fast screening of protein surfaces using geometric invariant fingerprints. Proc. Natl. Acad. Sci. USA. 2009;106:16622–16626. doi: 10.1073/pnas.0906146106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood D.J., de Vlieg J., Wagener M., Ritschel T. Pharmacophore fingerprint-based approach to binding site subpocket similarity and its application to bioisostere replacement. J. Chem. Inf. Model. 2012;52:2031–2043. doi: 10.1021/ci3000776. [DOI] [PubMed] [Google Scholar]
- 28.Li Z., Jiang M., Wang S., Zhang S. Deep learning methods for molecular representation and property prediction. Drug Discov. Today. 2022;27:103373. doi: 10.1016/j.drudis.2022.103373. [DOI] [PubMed] [Google Scholar]
- 29.Talevi A., Morales J.F., Hather G., Podichetty J.T., Kim S., Bloomingdale P.C., Kim S., Burton J., Brown J.D., Winterstein A.G., et al. Machine learning in drug discovery and development Part 1: a primer. CPT Pharmacometrics Syst. Pharmacol. 2020;9:129–142. doi: 10.1002/psp4.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dara S., Dhamercherla S., Jadav S.S., Babu C.M., Ahsan M.J. Machine learning in drug discovery: a review. Artif. Intell. Rev. 2022;55:1947–1999. doi: 10.1007/s10462-021-10058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 32.Paszke A., Gross S., Massa F., Lerer A., Bradbury J., Chanan G., Killeen T., Lin Z., Gimelshein N., Antiga L., Desmaison A. Vol. 32. Curran Associates, Inc.; 2019. PyTorch: an imperative style, high-performance deep learning library; pp. 8024–8035. (Advances in Neural Information Processing Systems). [Google Scholar]
- 33.Hammann F., Gutmann H., Vogt N., Helma C., Drewe J. Prediction of adverse drug reactions using decision tree modeling. Clin. Pharmacol. Ther. 2010;88:52–59. doi: 10.1038/clpt.2009.248. [DOI] [PubMed] [Google Scholar]
- 34.Schober P., Vetter T.R. Logistic regression in medical research. Anesth. Analg. 2021;132:365–366. doi: 10.1213/ANE.0000000000005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maltarollo V.G., Kronenberger T., Espinoza G.Z., Oliveira P.R., Honorio K.M. Advances with support vector machines for novel drug discovery. Expet Opin. Drug Discov. 2019;14:23–33. doi: 10.1080/17460441.2019.1549033. [DOI] [PubMed] [Google Scholar]
- 36.El-Attar N.E., Hassan M.K., Alghamdi O.A., Awad W.A. Deep learning model for classification and bioactivity prediction of essential oil-producing plants from Egypt. Sci. Rep. 2020;10:21349. doi: 10.1038/s41598-020-78449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta A., Müller A.T., Huisman B.J.H., Fuchs J.A., Schneider P., Schneider G. Generative recurrent networks for de novo drug design. Mol. Inform. 2018;37:1880141. doi: 10.1002/minf.201700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanchard A.E., Stanley C., Bhowmik D. Using GANs with adaptive training data to search for new molecules. J. Cheminf. 2021;13:14. doi: 10.1186/s13321-021-00494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen M., Xiao Y., Golbraikh A., Gombar V.K., Tropsha A. Development and validation of k-nearest-neighbor QSPR models of metabolic stability of drug candidates. J. Med. Chem. 2003;46:3013–3020. doi: 10.1021/jm020491t. [DOI] [PubMed] [Google Scholar]
- 40.Manelfi C., Gemei M., Talarico C., Cerchia C., Fava A., Lunghini F., Beccari A.R. “Molecular Anatomy”: a new multi-dimensional hierarchical scaffold analysis tool. J. Cheminf. 2021;13:54. doi: 10.1186/s13321-021-00526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo C., Shahlaei M. The applications of PCA in QSAR studies: a case study on CCR5 antagonists. Chem. Biol. Drug Des. 2018;91:137–152. doi: 10.1111/cbdd.13064. [DOI] [PubMed] [Google Scholar]
- 42.Karlov D.S., Sosnin S., Tetko I.V., Fedorov M.V. Chemical space exploration guided by deep neural networks. RSC Adv. 2019;9:5151–5157. doi: 10.1039/c8ra10182e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yasonik J. Multiobjective de novo drug design with recurrent neural networks and nondominated sorting. J. Cheminf. 2020;12:14. doi: 10.1186/s13321-020-00419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guengerich F.P. Mechanisms of drug toxicity and relevance to pharmaceutical development. Drug Metabol. Pharmacokinet. 2011;26:3–14. doi: 10.2133/dmpk.dmpk-10-rv-062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basile A.O., Yahi A., Tatonetti N.P. Artificial intelligence for drug toxicity and safety. Trends Pharmacol. Sci. 2019;40:624–635. doi: 10.1016/j.tips.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raies A.B., Bajic V.B. In silico toxicology: computational methods for the prediction of chemical toxicity. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2016;6:147–172. doi: 10.1002/wcms.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rim K.T. In silico prediction of toxicity and its applications for chemicals at work. Toxicol. Environ. Health Sci. 2020;12:191–202. doi: 10.1007/s13530-020-00056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang H., Sun L., Li W., Liu G., Tang Y. In silico prediction of chemical toxicity for drug design using machine learning methods and structural alerts. Front. Chem. 2018;6:30. doi: 10.3389/fchem.2018.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mayr A., Klambauer G., Unterthiner T., Hochreiter S. DeepTox: toxicity prediction using deep learning citation. Front. Environ. Sci. 2016;3 [Google Scholar]
- 50.Aguero-Chapin G., Galpert-Canizares D., Dominguez-Perez D., Marrero-Ponce Y., Perez-Machado G., Teijeira M., Antunes A. Emerging computational approaches for antimicrobial peptide discovery. Antibiotics. 2022;11:936. doi: 10.3390/antibiotics11070936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Covell D.G., Huang R., Wallqvist A. Anticancer medicines in development: assessment of bioactivity profiles within the National Cancer Institute anticancer screening data. Mol. Cancer Therapeut. 2007;6:2261–2270. doi: 10.1158/1535-7163.MCT-06-0787. [DOI] [PubMed] [Google Scholar]
- 52.Huang R., Xu M., Zhu H., Chen C.Z., Zhu W., Lee E.M., He S., Zhang L., Zhao J., Shamim K., et al. Biological activity-based modeling identifies antiviral leads against SARS-CoV-2. Nat. Biotechnol. 2021;39:747–753. doi: 10.1038/s41587-021-00839-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stokes J.M., Yang K., Swanson K., Jin W., Cubillos-Ruiz A., Donghia N.M., MacNair C.R., French S., Carfrae L.A., Bloom-Ackermann Z., et al. A deep learning approach to antibiotic discovery. Cell. 2020;181:475–483. doi: 10.1016/j.cell.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Panapitiya G., Girard M., Hollas A., Sepulveda J., Murugesan V., Wang W., Saldanha E. Evaluation of deep learning architectures for aqueous solubility prediction. ACS Omega. 2022;7:15695–15710. doi: 10.1021/acsomega.2c00642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye Z., Ouyang D. Prediction of small-molecule compound solubility in organic solvents by machine learning algorithms. J. Cheminf. 2021;13:98. doi: 10.1186/s13321-021-00575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiMasi J.A., Grabowski H.G., Hansen R.W. Innovation in the pharmaceutical industry: new estimates of R&D costs. J. Health Econ. 2016;47:20–33. doi: 10.1016/j.jhealeco.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 57.Mouchlis V.D., Afantitis A., Serra A., Fratello M., Papadiamantis A.G., Aidinis V., Lynch I., Greco D., Melagraki G. Advances in de Novo drug design: from conventional to machine learning methods. Int. J. Mol. Sci. 2021;22:1676. doi: 10.3390/ijms22041676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popova M., Isayev O., Tropsha A. Deep reinforcement learning for de novo drug design. Sci. Adv. 2018;4:eaap7885. doi: 10.1126/sciadv.aap7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gómez-Bombarelli R., Wei J.N., Duvenaud D., Hernández-Lobato J.M., Sánchez-Lengeling B., Sheberla D., Aguilera-Iparraguirre J., Hirzel T.D., Adams R.P., Aspuru-Guzik A. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 2018;4:268–276. doi: 10.1021/acscentsci.7b00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mercado R., Rastemo T., Lindelöf E., Klambauer G., Engkvist O., Chen H., Jannik Bjerrum E. Graph networks for molecular design. Mach. Learn, Sci. Technol. 2021;2:025023. [Google Scholar]
- 61.Li Y., Zhang L., Liu Z. Multi-objective de novo drug design with conditional graph generative model. J. Cheminf. 2018;10:33. doi: 10.1186/s13321-018-0287-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang M., Wang Z., Sun H., Wang J., Shen C., Weng G., Chai X., Li H., Cao D., Hou T. Deep learning approaches for de novo drug design: an overview. Curr. Opin. Struct. Biol. 2022;72:135–144. doi: 10.1016/j.sbi.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Callaway E. ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures. Nature. 2020;588:203–204. doi: 10.1038/d41586-020-03348-4. [DOI] [PubMed] [Google Scholar]
- 64.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pakhrin S.C., Shrestha B., Adhikari B., Kc D.B. Deep learning-based advances in protein structure prediction. Int. J. Mol. Sci. 2021;22:5553. doi: 10.3390/ijms22115553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., Qin C., Žídek A., Nelson A.W.R., Bridgland A., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 67.Nag S., Baidya A.T.K., Mandal A., Mathew A.T., Das B., Devi B., Kumar R. Deep learning tools for advancing drug discovery and development. 3 Biotech. 2022;12:110. doi: 10.1007/s13205-022-03165-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Husain A., Begum N.A., Kobayashi M., Honjo T. Native Co-immunoprecipitation assay to identify interacting partners of chromatin-associated proteins in mammalian cells. Bio. Protoc. 2020;10:e3837. doi: 10.21769/BioProtoc.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nixon A.E., Sexton D.J., Ladner R.C. Drugs derived from phage display: from candidate identification to clinical practice. mAbs. 2014;6:73–85. doi: 10.4161/mabs.27240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hamdi A., Colas P. Yeast two-hybrid methods and their applications in drug discovery. Trends Pharmacol. Sci. 2012;33:109–118. doi: 10.1016/j.tips.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 71.Bagherian M., Sabeti E., Wang K., Sartor M.A., Nikolovska-Coleska Z., Najarian K. Machine learning approaches and databases for prediction of drug-target interaction: a survey paper. Briefings Bioinf. 2021;22:247–269. doi: 10.1093/bib/bbz157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Öztürk H., Özgür A., Ozkirimli E. DeepDTA: deep drug-target binding affinity prediction. Bioinformatics. 2018;34:i821–i829. doi: 10.1093/bioinformatics/bty593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen M., Zhang Z., Niu S., Sha H., Yang R., Yun Y., Lu H. Deep-learning-based drug-target interaction prediction. J. Proteome Res. 2017;16:1401–1409. doi: 10.1021/acs.jproteome.6b00618. [DOI] [PubMed] [Google Scholar]
- 74.Lee I., Keum J., Nam H. DeepConv-DTI: prediction of drug-target interactions via deep learning with convolution on protein sequences. PLoS Comput. Biol. 2019;15:e1007129. doi: 10.1371/journal.pcbi.1007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu J., Wang J., Wang X., Gao M., Guo B., Gao M., Liu J., Yu Y., Wang L., Kong W., et al. Prediction of drug efficacy from transcriptional profiles with deep learning. Nat. Biotechnol. 2021;39:1444–1452. doi: 10.1038/s41587-021-00946-z. [DOI] [PubMed] [Google Scholar]
- 76.Öztürk H., Ozkirimli E., Arzucan Özgür. WideDTA: prediction of drugtarget binding affinity. arXiv. 2019 doi: 10.48550/arXiv.1902.04166. Preprint at. [DOI] [Google Scholar]
- 77.Pahikkala T., Airola A., Pietilä S., Shakyawar S., Szwajda A., Tang J., Aittokallio T. Toward more realistic drug-target interaction predictions. Briefings Bioinf. 2015;16:325–337. doi: 10.1093/bib/bbu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He T., Heidemeyer M., Ban F., Cherkasov A., Ester M. SimBoost: a read-across approach for predicting drug-target binding affinities using gradient boosting machines. J. Cheminf. 2017;9:24. doi: 10.1186/s13321-017-0209-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karimi M., Wu D., Wang Z., Shen Y. DeepAffinity: interpretable deep learning of compound-protein affinity through unified recurrent and convolutional neural networks. Bioinformatics. 2019;35:3329–3338. doi: 10.1093/bioinformatics/btz111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Al-Lazikani B., Banerji U., Workman P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012;30:679–692. doi: 10.1038/nbt.2284. [DOI] [PubMed] [Google Scholar]
- 81.Murphy E.M., Jimenez H.R., Smith S.M. Current clinical treatments of AIDS. Adv. Pharmacol. 2008;56:27–73. doi: 10.1016/S1054-3589(07)56002-3. [DOI] [PubMed] [Google Scholar]
- 82.Tamma P.D., Cosgrove S.E., Maragakis L.L. Combination therapy for treatment of infections with gram-negative bacteria. Clin. Microbiol. Rev. 2012;25:450–470. doi: 10.1128/CMR.05041-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li P., Huang C., Fu Y., Wang J., Wu Z., Ru J., Zheng C., Guo Z., Chen X., Zhou W., et al. Large-scale exploration and analysis of drug combinations. Bioinformatics. 2015;31:2007–2016. doi: 10.1093/bioinformatics/btv080. [DOI] [PubMed] [Google Scholar]
- 84.Wildenhain J., Spitzer M., Dolma S., Jarvik N., White R., Roy M., Griffiths E., Bellows D.S., Wright G.D., Tyers M. Prediction of synergism from chemical-genetic interactions by machine learning. Cell Syst. 2015;1:383–395. doi: 10.1016/j.cels.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Preuer K., Lewis R.P.I., Hochreiter S., Bender A., Bulusu K.C., Klambauer G. DeepSynergy: predicting anti-cancer drug synergy with Deep Learning. Bioinformatics. 2018;34:1538–1546. doi: 10.1093/bioinformatics/btx806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wagner V., Dullaart A., Bock A.K., Zweck A. The emerging nanomedicine landscape. Nat. Biotechnol. 2006;24:1211–1217. doi: 10.1038/nbt1006-1211. [DOI] [PubMed] [Google Scholar]
- 87.Shi J., Kantoff P.W., Wooster R., Farokhzad O.C. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Roy U., Rodríguez J., Barber P., das Neves J., Sarmento B., Nair M. The potential of HIV-1 nanotherapeutics: from in vitro studies to clinical trials. Nanomedicine. 2015;10:3597–3609. doi: 10.2217/nnm.15.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y., Abbaspour M.R., Grootendorst P.V., Rauth A.M., Wu X.Y. Optimization of controlled release nanoparticle formulation of verapamil hydrochloride using artificial neural networks with genetic algorithm and response surface methodology. Eur. J. Pharm. Biopharm. 2015;94:170–179. doi: 10.1016/j.ejpb.2015.04.028. [DOI] [PubMed] [Google Scholar]
- 90.Muñiz Castro B., Elbadawi M., Ong J.J., Pollard T., Song Z., Gaisford S., Pérez G., Basit A.W., Cabalar P., Goyanes A. Machine learning predicts 3D printing performance of over 900 drug delivery systems. J. Contr. Release. 2021;337:530–545. doi: 10.1016/j.jconrel.2021.07.046. [DOI] [PubMed] [Google Scholar]
- 91.Alafeef M., Srivastava I., Pan D. Machine learning for precision breast cancer diagnosis and prediction of the nanoparticle cellular internalization. ACS Sens. 2020;5:1689–1698. doi: 10.1021/acssensors.0c00329. [DOI] [PubMed] [Google Scholar]
- 92.Moumné L., Marie A.C., Crouvezier N. Oligonucleotide therapeutics: from discovery and development to patentability. Pharmaceutics. 2022;14:260. doi: 10.3390/pharmaceutics14020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiba S., Lim K.R.Q., Sheri N., Anwar S., Erkut E., Shah M.N.A., Aslesh T., Woo S., Sheikh O., Maruyama R., et al. eSkip-Finder: a machine learning-based web application and database to identify the optimal sequences of antisense oligonucleotides for exon skipping. Nucleic Acids Res. 2021;49:W193–W198. doi: 10.1093/nar/gkab442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dar S.A., Gupta A.K., Thakur A., Kumar M. SMEpred workbench: a web server for predicting efficacy of chemicallymodified siRNAs. RNA Biol. 2016;13:1144–1151. doi: 10.1080/15476286.2016.1229733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G., Li B., Madabhushi A., Shah P., Spitzer M., Zhao S. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 2019;18:463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen P., Dong W., Wang J., Lu X., Kaymak U., Huang Z. Interpretable clinical prediction via attention-based neural network. BMC Med. Inf. Decis. Making. 2020;20:131. doi: 10.1186/s12911-020-1110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang Q., Nie F., Zhao Q., Chen W. A merged molecular representation deep learning method for blood-brain barrier permeability prediction. Briefings Bioinf. 2022;23:bbac357. doi: 10.1093/bib/bbac357. [DOI] [PubMed] [Google Scholar]