Abstract
Induction of ferroptosis is an emerging strategy to suppress melanoma progression. Strategies to enhance the sensitivity to ferroptosis induction would be a major advance in melanoma therapy. Here, we used a drug synergy screen that combined a ferroptosis inducer, RSL3, with 240 anti-tumor drugs from the FDA-approved drug library and identified lorlatinib to synergize with RSL3 in melanoma cells. We further demonstrated that lorlatinib sensitized melanoma to ferroptosis through inhibiting PI3K/AKT/mTOR signaling axis and its downstream SCD expression. Moreover, we found that lorlatinib's target IGF1R, but not ALK or ROS1, was the major mediator of lorlatinib-mediated sensitivity to ferroptosis through targeting PI3K/AKT/mTOR signaling axis. Finally, lorlatinib treatment sensitized melanoma to GPX4 inhibition in preclinical animal models, and melanoma patients with low GPX4 and IGF1R expression in their tumors survived for longer period. Altogether, lorlatinib sensitizes melanoma to ferroptosis by targeting IGF1R-mediated PI3K/AKT/mTOR signaling axis, suggesting that combination with lorlatinib could greatly expand the utility of GPX4 inhibition to melanoma patients with IGF1R-proficient expression.
Keywords: Melanoma, Ferroptosis, Lorlatinib, Synergy, IGF1R
1. Introduction
Ferroptosis is an iron-dependent and non-apoptotic form of programmed cell death characterized by lethal accumulation of lipid peroxides [1,2]. It has been well documented that therapy-resistant tumor cells, particularly those of the mesenchymal-like state and prone to metastasis, are highly susceptible to ferroptosis [3,4]. These works highlighted induction of ferroptosis by the inhibition of glutathione peroxidase 4 (GPX4) as a promising strategy for cancer treatment [2,5]. However, the sensitivity of ferroptosis varies greatly among cancer cells, and melanoma is relatively insensitive to erastin-induced ferroptosis, compared with other tumors, especially diffuse large B cell lymphoma and renal cell carcinomas [2,6]. Therefore, there is mounting interest in explore the mechanisms that underpin the sensitivity of melanoma cells to ferroptosis.
Several excellent studies have clarified multiple, complex and inter-related signaling pathway to regulate the susceptibility of ferroptosis to melanoma. For example, some microRNAs such as miR-9 and miR-137 affect the sensitivity of ferroptosis by regulating glutamine catabolism [7,8]. Moreover, some proteins regulated by ferroptosis induction, including aldo-keto-reductase-1C (AKR1C) 1/2/3, neuronal precursor cell-expressed developmentally downregulated 4 (NEDD4) and calcium/calmodulin dependent protein kinase 2 (CAMKK2), render melanoma cells not responsive any longer to ferroptosis through degrading the 12/15-LOX-generated lipid peroxides [9], down-regulating voltage dependent anion channel 2/3 (VADC2/3) expression [10], or activating the AMP-activated protein kinase (AMPK)/nuclear factor erythroid 2-related factor 2 (NRF2) pathway [11], respectively. Additionally, sterol regulatory element binding transcription factor 2 (SREBP2) or oleic acid protect circulating melanoma cells or melanoma cells in lymph from ferroptosis by inducing transferrin expression or reducing the amount/density of polyunsaturated fatty acids (PUFA) available for oxidation in membranes [12,13]. However, drugs targeting these ferroptosis suppressors are far from being used in the clinic, highlighting the significance of screening the FDA-approved drug library to synergize with GPX4 inhibition in melanoma cells.
Lorlatinib is an FDA-approved, third-generation, ATP-competitive small-molecule tyrosine kinase inhibitor for the treatment of non-small cell lung cancer caused by an abnormal anaplastic lymphoma kinase (ALK) gene [14]. However, the role of lorlatinib in melanoma and whether it is involved in the regulation of ferroptosis sensitivity are completely unknown. Here, we identified lorlatinib to synergize with GPX4 inhibition in melanoma cells from 240 FDA-approved anti-tumor drugs. Mechanistically, lorlatinib sensitizes melanoma to ferroptosis through targeting insulin like growth factor 1 receptor (IGF1R)-mediated PI3K/AKT/mTOR signaling axis and its downstream stearoyl-CoA desaturase 1 (SCD) expression. Consistently, melanoma patients with low GPX4 and IGF1R expression in their tumors survive for longer period. Thus, lorlatinib-mediated IGF1R inhibition plays a critical role in promoting melanoma ferroptosis, indicating that combination with lorlatinib could greatly expand the utility of GPX4 inhibition to melanoma patients with IGF1R-proficient expression.
2. Results
2.1. Identification of lorlatinib to synergize with GPX4 inhibition in melanoma
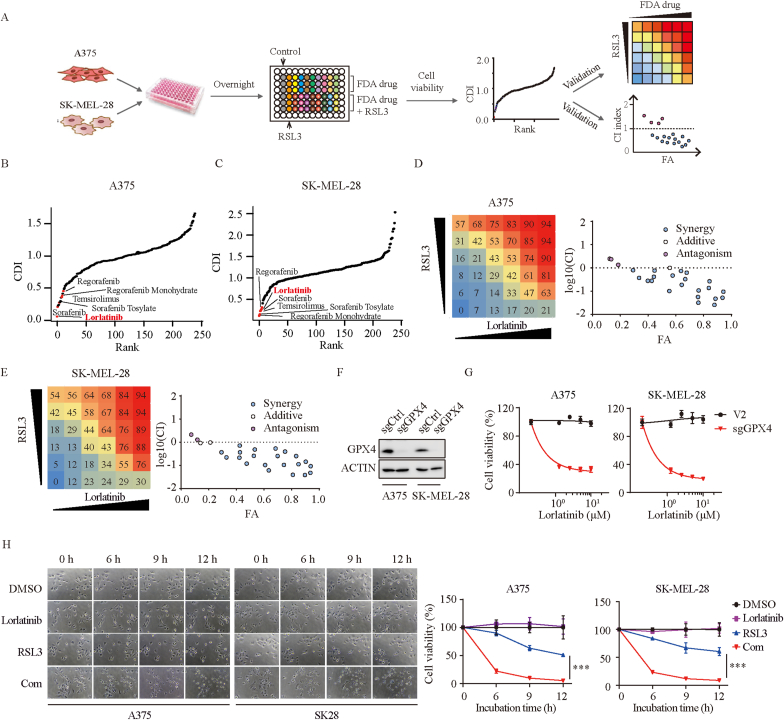
The sensitivity of ferroptosis varies greatly among cancer cells (Fig. S1A). To uncover clinically applicable drugs that synergize with GPX4 inhibition in melanoma, we performed a screening of 240 anti-tumor drugs identified from the FDA-approved drug library combined with ferroptosis inducer - RSL3 using in-vitro drug combination assay (Fig. 1A). The coefficient of drug interaction (CDI) was applied to evaluate the effect of combined medication [15]. Lorlatinib was identified as one of the most potential drugs in both A375 and SK-MEL-28 cells, in addition to several drugs including regorafenib and its monohydrate [16], temsirolimus [17], sorafenib and sorafenib tosylate [16], which have been reported to synergize with ferroptosis inducers in cancer cells (Fig. 1B–C). Consistent with previous findings, our data also demonstrated that sorafenib synergized with RSL3 in melanoma (Figs. S1B–C), thus supporting the validity of our screens. To further clarify whether lorlatinib synergizes with RSL3, we conducted a series of 6 × 6 screening experiments in both A375 and SK-MEL-28 cells, indicating a strong synergy (Fig. 1D–E). RSL3 functions primarily through binding and inactivation of peroxidase activity of GPX4 [2]. To further support our findings, we constructed GPX4 knockout melanoma cell lines (Fig. 1F), and found that GPX4 deficient melanoma cells was vulnerable to lorlatinib in a dose-dependent manner (Fig. 1G), suggesting that lorlatinib sensitizes melanoma to GPX4 inhibition. To visualize the morphological features after drugs treatment, we performed live cell time-lapse imaging. We observed that lorlatinib had minimal toxic effects on melanoma cells. However, after 6 h, melanoma cells cotreated with lorlatinib and RSL3 experienced more cell death which shared morphological characteristics of necrosis, including cell rounding, swelling and plasma membrane rupture (Fig. 1H).
Fig. 1.
Identification of lorlatinib to synergize with GXP4 inhibition in melanoma.
(A) Schematic of the identification of clinically applicable drug from the FDA-approved drug library that sensitize melanoma to RSL3. (B-C) Summary scatter plot of CDI in A375 (B) and SK-MEL-28 (C) cells indicating lorlatinib as one of the most potential drugs that synergize with RSL3. Indicated was the drugs that have been reported to synergize with RSL3. (D-E) Percentage of inhibition rate was presented in a series of 6 × 6 screening experiments in A375 (D) and SK-MEL-28 (E) cells. Synergy was evaluated by Chou-Talalay combination index (CI) for lorlatinib and RSL3 across the indicated cell lines. The x axis of CI plots represents fraction affected. (F) GPX4 protein levels were quantified by western blotting in control (sgCtrl) and GPX4 deficient (sgGPX4) cells. (G) Cell viability of GPX4 deficient cells treated with different concentrations of lorlatinib for 12 h. (H) Cell morphological features at different time point after the indicated treatment. Lorlatinib, 5 μM; RSL3, 2.5 μM. Images were taken at 200X magnification. P values were calculated using two-way ANOVA analysis. ***, P < 0.001.
2.2. Combination of lorlatinib and GPX4 inhibition drives melanoma ferroptosis
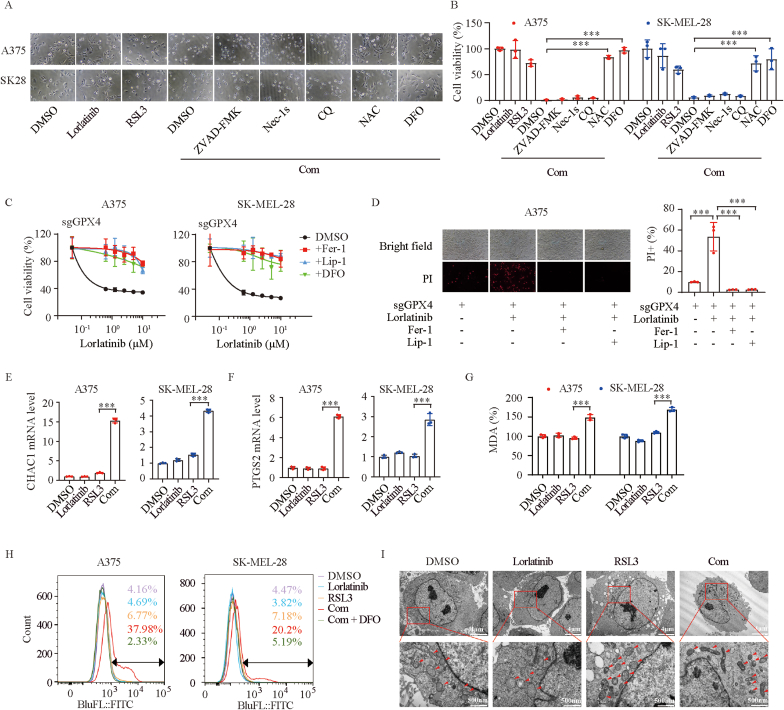
To further determine the type of cell death driven by lorlatinib and RSL3, we cotreated the cells with various cell death inhibitors. We found that the toxic effect of the combination therapy could be completely negated by the anti-oxidant N-acetyl-cysteine (NAC) and the iron chelator deferoxamine (DFO), but not by inhibitors of apoptosis (Z-VAD-FMK), necroptosis (Nec-1s), or autophagy (CQ) (Fig. 2A–B), suggesting that combination of lorlatinib and RSL3 drives melanoma ferroptosis. To further support the notion, we used four different approaches to assess whether ferroptosis was induced by lorlatinib and RSL3 co-treatment. First, more ferroptosis inhibitors including ferrostatin-1, liproxstatin-1 and DFO could reverse the cell death in GPX4-deficient melanoma cells cotreated with lorlatinib (Fig. 2C–D). Second, mRNA levels of CHAC1 and PTGS2, markers for assessment of ferroptosis, were notably increased after lorlatinib and RSL3 co-treatment (Fig. 2E–F). Third, malondialdehyde (MDA), aldehyde secondary products of lipid peroxidation, and lipid peroxidation, the hallmark of ferroptosis, were markedly increased in combination group, compared with other groups (Fig. 1G–H). Fourth, transmission electron microscopy analysis displayed a striking ferroptosis-associated morphologic change in melanoma cells with combination therapy, characterized by shrunken mitochondria with increased membrane density and reduced numbers of mitochondrial cristae (Fig. 1I). Collectively, these findings suggest that co-treatment of lorlatinib and RSL3 leads to melanoma ferroptosis.
Fig. 2.
Combination of lorlatinib and GPX4 inhibition drives melanoma ferroptosis.
(A-B) Indicated melanoma cells were treated with lorlatinib (5 μM), RSL3 (2.5 μM), or a combination of both drugs with or without cell death inhibitors (ZVAD-FMK, 5 μM; Necrostatin-1s, 10 μM; CQ, 10 μM; NAC, 1 mM; DFO, 100 μM) for 6h, and cell viability was assessed. (C) GPX4 deficient cells treated with different concentrations of lorlatinib for 12 h in the absence or presence of Fer-1 (2 μM), Lip-1 (10 μM) or DFO (100 μM). (D) Cell death of GPX4 deficient cells induced by the indicated treatment were shown by microscope and quantified by PI-staining coupled with flow cytometry. Lorlatinib, 5 μM; RSL3, 2.5 μM. (E-F) Real-time PCR analysis of CHAC1 (E) and PTGS2 (F) expression in A375 and SK-MEL-28 cells after the indicated treatment for 6 h. Lorlatinib, 5 μM; RSL3, 2.5 μM. (G) Analysis of MDA in A375 and SK-MEL-28 cells after the indicated treatment for 6 h. Lorlatinib, 5 μM; RSL3, 2.5 μM. (H) Lipid ROS were quantified with BODIPY-C11 using flow cytometry. Cells were treated as indicated for 6 h. Lorlatinib, 5 μM; RSL3, 2.5 μM. (I) Transmission electron microscopy images of A375 cells after the indicated treatment for 6 h. Lorlatinib, 5 μM; RSL3, 2.5 μM. Scale bar, upper, 2 μm; lower, 500 nm. One-way ANOVA analysis was performed in B, D, E, F, G. ***, P < 0.001.
2.3. Lorlatinib sensitizes melanoma to ferroptosis through SCD
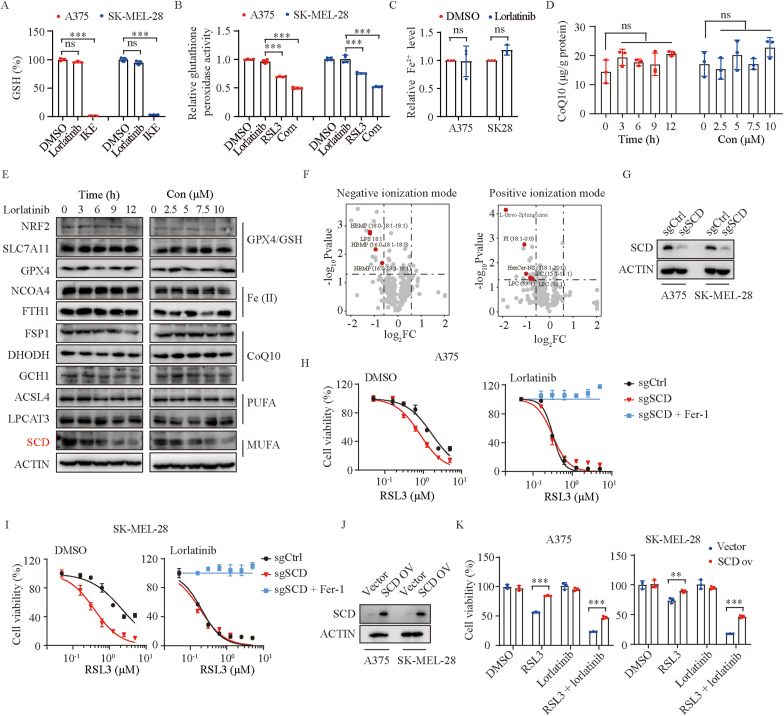
To illuminate the underlying mechanism by which lorlatinib enhance RSL3-mediated ferroptosis, we pretreated melanoma cells with lorlatinib overnight and then replaced with new medium only containing RSL3, finding that the effect of promoting ferroptosis still existed (Fig. S1D). This result suggested that pretreatment with lorlatinib puts melanoma cells in a state that is sensitized to ferroptosis. Several pathways have been reported to be associated with the sensitivity of ferroptosis [3,4]. We firstly check the effect of lorlatinib on the SLC7A11/GPX4 axis. As expected, the levels of GSH were dramatically reduced under imidazole ketone erastin treatment and GPX4 activity was significantly abrogated by RSL3 treatment [1,2]. However, lorlatinib treatment did not affect the levels of GSH and GPX4 activity (Fig. 3A–B). Moreover, lorlatinib treatment failed to affect the intracellular levels of iron (Fig. 3C) and the mRNA expression of genes for iron metabolism (Figs. S2A–B), as well as coenzyme Q10 (Fig. 3D), a main downstream of the newly discovered ferroptosis suppressor - FSP1 [18,19]. Furthermore, we generated DHODH and GCH1 deficient melanoma cells (Figs. S2C–D), and found that lorlatinib could still sensitize these cells to ferroptosis (Figs. S2E–F), indicating that lorlatinib-mediated sensitivity to ferroptosis is independent on the expression of DHODH or GCH1, another two newly discovered ferroptosis suppressors [20,21]. Consistent with these findings, lorlatinib treatment had no obvious effects on the protein expression of key regulators in GPX4/GSH axis, iron metabolism, or these ferroptosis suppressors (Fig. 3E).
Fig. 3.
Lorlatinib sensitizes melanoma to ferroptosis through SCD.
(A) The relative levels of GSH were assayed in A375 and SK-MEL-28 cells treated with DMSO, lorlatinib (5 μM), or IKE (2.5 μM) for 12 h. (B) Relative glutathione peroxidase activity were quantified in A375 and SK-MEL-28 cells with the indicated treatment for 6 h. Lorlatinib, 5 μM; RSL3, 2.5 μM. (C) Relative Fe2+ levels in A375 and SK-MEL-28 cells following treatment with DMSO or lorlatinib (5 μM) for 12 h. (D) CoQ10 levels at different time point in A375 cells after treatment with 5 μM lorlatinib, or in A375 cells treated with different concentrations of lorlatinib for 12 h. (E) Western blotting analysis of proteins at different time point in A375 cells after treatment with lorlatinib (5 μM), or proteins in A375 cells treated with different concentrations of lorlatinib for 12 h. (F) Fold change of lipid species in A375 cells treated with 5 μM lorlatinib for 12 h in negative and positive ionization modes. (G) SCD protein levels were quantified by western blotting in control (sgCtrl) and SCD deficient (sgSCD) cells. (H-I) Dose response of RSL3-induced death of sgCtrl and sgSCD cells in the presence of DMSO or lorlatinib (5 μM) for 6 h. (J) SCD protein levels were quantified by western blotting in cells with control (vector) or SCD overexpression (SCD ov). (K) Viability of the indicated cells with control or SCD overexpression after treatment with RSL3 (2.5 μM), lorlatinib (2.5 μM), or RSL3 + lorlatinib. P values were determined using one-way ANOVA analysis in A, B, D. Two-tailed unpaired Student's t-test was performed in C. Two-way ANOVA analysis was performed in K. **, P < 0.01; ***, P < 0.001; ns, no significance.
Lipid metabolic processes has been reported to impinge on cells susceptibility toward ferroptosis [22]. We further evaluated the protein expression of genes regulating lipid metabolism, finding that SCD, but not ACSL4 or LPCAT3, was down-regulated by lorlatinib in a time- and dose-dependent manner (Fig. 3E). SCD converts saturated fatty acids to monounsaturated fatty acids (MUFA) and promotes ferroptosis resistance, providing a mechanistic explanation to our observation [23]. In line with this finding, lipidomics showed that lorlatinib could decrease the abundance of some lipids, especially some MUFA - phospholipids (MUFA-PLs) which tend to confer resistance to ferroptosis, while polyunsaturated fatty acyl-PLs (PUFA-PLs) were comparable with or without lorlatinib treatment (Fig. 3F, Fig. S3). Furthermore, we constructed SCD deficient cells (Fig. 3G), and observed that SCD knockout had a strong sensitization effect on RSL3-induced ferroptosis in the absence of lorlatinib, but almost no sensitization effect in the presence of lorlatinib (Fig. 3H–I). Conversely, SCD overexpression rendered melanoma cells resistant to the combination of lorlatinib with RSL3 (Fig. 3J–K). These data indicated that lorlatinib sensitized melanoma cells to RSL3-mediated ferroptosis through inhibiting the expression of SCD.
2.4. Lorlatinib inhibits the expression of SCD via PI3K/AKT/mTOR signaling
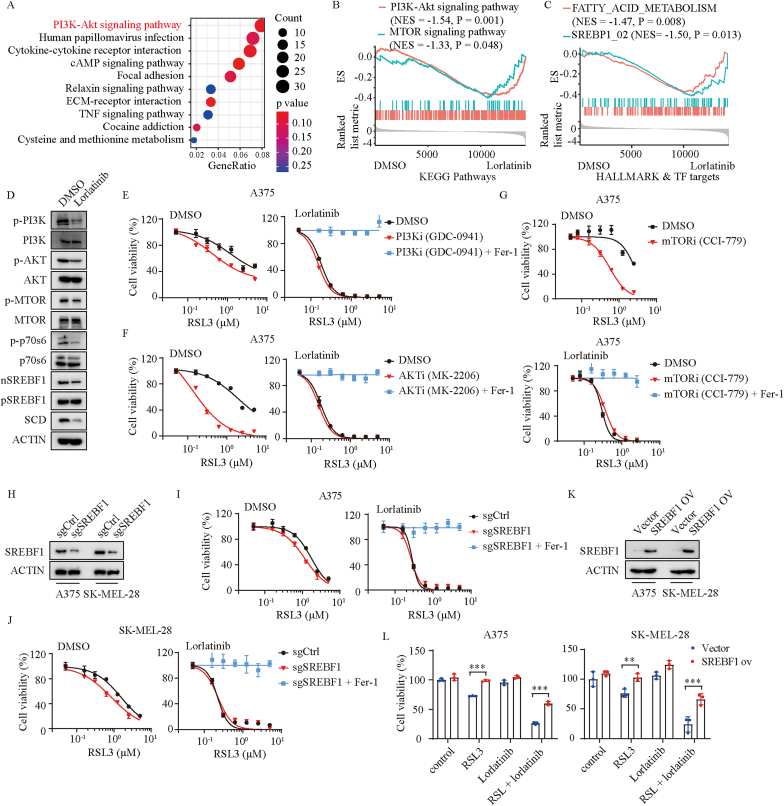
To elaborate how lorlatinib regulates the expression of SCD, we performed KEGG enrichment analysis based on RNA-seq data (Fig. S4A). Strikingly, as one of the pathways that are affected most by lorlatinib, PI3K/AKT signaling pathway was suggested to play a significant role (Fig. 4A). Gene Set Enrichment Analysis (GSEA) further demonstrated that lorlatinib could significantly inhibit PI3K/AKT/mTOR pathways (Fig. 4B, Fig. S4B). Previous studies have proved that inhibition of PI3K/AKT/mTOR signaling potentiates the cancer therapeutic effect of ferroptosis inducer via SREBP1/SCD-mediated lipogenesis [17]. SREBP1 is a transcription factor and particularly related to fatty acid metabolism [24]. We wondered whether lorlatinib regulated the expression of SCD through a similar mechanism. Interestingly, gene signatures associated with SREBP1 activity and fatty acid metabolism were markedly abrogated by the treatment of lorlatinib (Fig. 4C). In agreement with these findings, lorlatinib treatment caused a decrease of p-PI3K, p-AKT, p-mTOR, p-p70s6, and the level of the mature form of SREBP1 (SREBP1m), which could translocate into the nucleus to regulate its downstream transcriptional targets including SCD, FASN, ACLY and ACACA (Fig. 4D, Figs. S4C–F). Moreover, inhibition of PI3K/AKT/mTOR pathway with PI3K inhibitor (GDC-0941), AKT inhibitor (MK-2206), and mTOR inhibitor (CCI-779) strongly sensitized melanoma to RSL3-induced ferroptosis, but did not further enhance RSL3-induced ferroptosis in the presence of lorlatinib (Fig. 4E–G, Figs. S5A–C). Additionally, a marked degree of sensitization to RSL3 was observed after knockout of SREBP1 in control cells, but not in cells treated with lorlatinib (Fig. 4H–J). Conversely, SREBP1 overexpression protected melanoma cells from ferroptosis induced by the combination of RSL3 with lorlatinib (Fig. 4K-L). These findings suggested that lorlatinib inhibits the expression of SCD via PI3K/AKT/mTOR/SREBP1 signaling.
Fig. 4.
Lorlatinib inhibits the expression of SCD via PI3K/AKT/mTOR signaling.
(A) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of the differentially expressed genes between DMSO and lorlatinib treated A375 cells. (B) GSEA showing that PI3K-Akt and MTOR signaling pathways were down regulated in lorlatinib treatment group. (C) GSEA showing that FATTY_ACID_METABOLISM and SREBP1 signaling were down regulated in lorlatinib treatment group. (D) Western blotting analysis of the indicated proteins in A375 cells treated with DMSO or lorlatinib (5 μM) for 12 h. (E) Dose response of RSL3-induced death of DMSO or PI3Ki (GDC-0941) treated- A375 cells in the absence or presence of lorlatinib for 6 h. (F) Dose response of RSL3-induced death of DMSO or AKTi (MK-2206) treated- A375 cells in the absence or presence of lorlatinib for 6 h. (G) Dose response of RSL3-induced death of DMSO or mTORi (CCI-779) treated- A375 cells in the absence or presence of lorlatinib for 6 h. (H) SREBP1 protein levels were quantified by western blotting in control (sgCtrl) and SREBP1 deficient (sgSREBF1) cells. (I-J) Dose response of RSL3-induced death of sgCtrl and sgSREBF1 A375 (I) or SK-MEL-28 (J) cells in the presence of DMSO or lorlatinib (5 μM) for 6 h. (K) SREBP1 protein levels were quantified by western blotting in cells with control (vector) or SREBF1 overexpression (SREBF1 ov). (L) Viability of the indicated cells with control or SREBF1 overexpression after treatment with RSL3 (2.5 μM), lorlatinib (2.5 μM), or RSL3 + lorlatinib. P values were calculated using two-way ANOVA analysis in L. **, P < 0.01; ***, P < 0.001.
2.5. ALK and ROS1 are not the major mediators of lorlatinib-mediated sensitivity to ferroptosis
Lorlatinib is well known as an FDA-approved, third-generation, ATP-competitive small-molecule tyrosine kinase inhibitor of ALK/ROS1 [25]. We next sought to determine whether lorlatinib-mediated sensitivity to ferroptosis is dependent on the expression of ALK and ROS1. However, ALK or ROS1 knockdown by shRNA failed to sensitize melanoma cells to RSL3-induced ferroptosis (Figs. S6A–D). Consistently, ALK or ROS1 knockdown by siRNA or knockout by sgRNA still could not sensitize melanoma cells to RSL3-induced ferroptosis (Figs. S6E–I). To further rule out the possibility of ALK and ROS1 as the major mediator of lorlatinib-mediated sensitivity to ferroptosis, we performed a 6 × 6 screening experiments. We observed that ALK inhibitor (alectinib or alectinib hydrochloride) or inhibitors targeting both ALK and ROS1 (entrectinib or brigatinib) failed to synergize with RSL3 in melanoma cells (Figs. S6J–K). Furthermore, ferroptosis induction could still be potently sensitized by lorlatinib even after ALK or ROS1 knockdown. ALK or ROS1 overexpression also failed to reverse the inhibitory effect caused by lorlatinib and RSL3 treatment (Fig. S6L-M). These results demonstrated that lorlatinib-mediated sensitivity to ferroptosis is independent of the expression ALK and ROS1.
2.6. Lorlatinib regulates melanoma susceptibility to ferroptosis and PI3K/AKT/mTOR pathway through IGF1R
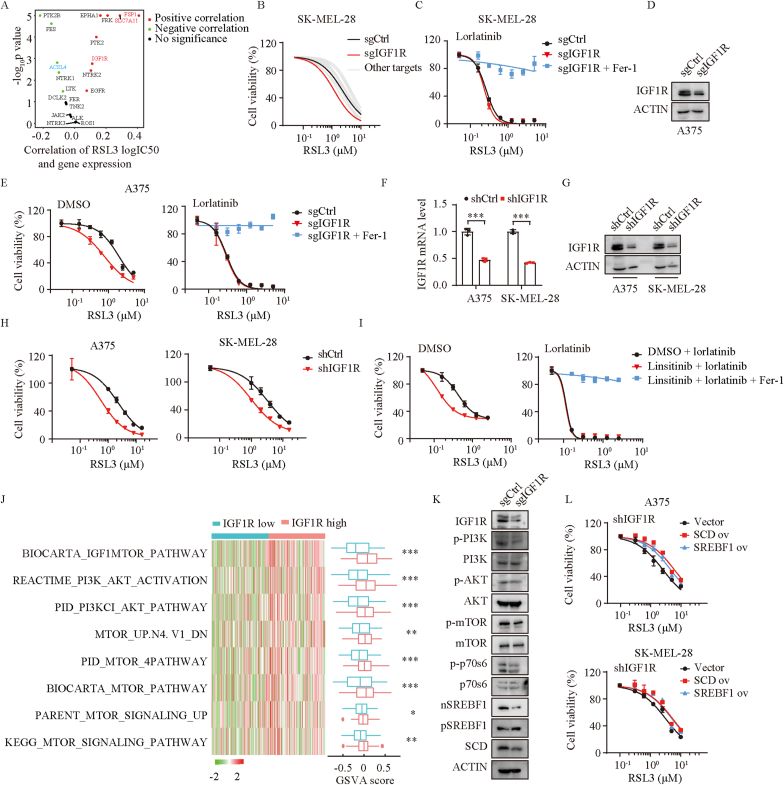
Lorlatinib is also reported to potently inhibit other tyrosine kinases, including LTK, FER, FES, PTK2B, TNK2, PTK2, NTRK1/2/3, FRK, EGFR, IGF1R, TSSK2, EPHA1, JAK2 and INSR [14]. By analyzing the associations between GPX4 inhibitors and the expression of lorlatinib targets using the Cancer Therapeutics Response Portal, we found that IGF1R is one of these kinases positively correlated with resistance to ferroptosis inducers including RSL3, erastin, ML162, and ML120. As expected, FSP1 and SLC7A11 were positively while ACSL4 were negatively associated with the logIC50 of these ferroptosis inducers (Fig. 5A, Figs. S7A–C). To further evaluate the association between lorlatinib targets and the sensitivity of ferroptosis, we generated these kinases knockout cells using CRISPR/Cas9 technology, respectively (Fig. S7D), finding that IGF1R knockout sensitized melanoma to ferroptosis mostly, but failed to further enhance ferroptosis in the presence of lorlatinib in SK-MEL-28 cells (Fig. 5B–C). This result was further validated in A375 cells (Fig. 5D–E). Consistently, knockdown of IGF1R using shRNA also sensitized melanoma cells to ferroptosis (Fig. 5F–H). Pharmacologically, IGF1R selective inhibitor linsitinib sensitized the effect of RSL3 on the induction of ferroptosis (Fig. S7E), but failed to further enhance RSL3-induced ferroptosis in the presence of lorlatinib (Fig. 5I). Collectively, these finding indicated that IGF1R is the major mediator of lorlatinib-mediated sensitivity to ferroptosis.
Fig. 5.
Lorlatinib regulates melanoma susceptibility to ferroptosis and PI3K/AKT/mTOR pathway through IGF1R.
(A) The correlation between logIC50 of RSL3 and gene expression of lorlatinib targets. Red dots, significant positive correlation; blue dots, significant negative correlation; black dots, no significance. (B) Dose response of RSL3-induced death of SK-MEL-28 cells transfected with control sgRNA or sgRNA of lorlatinib targets. SK-MEL-28 cells was sensitive to RSL3 the most when IGF1R was deficient. (C) Dose response of RSL3-induced death of sgCtrl and sgIGF1R SK-MEL-28 cells in the presence of lorlatinib for 6 h. (D) IGF1R protein levels were quantified by western blotting in control (sgCtrl) and IGF1R deficient (sgIGF1R) cells. (E) Dose response of RSL3-induced death of sgCtrl and sgIGF1R A375 cells in the presence of DMSO or lorlatinib (5 μM) for 6 h. (F) IGF1R knock down efficiency in A375 and SK-MEIL-28 cells assessed by real-time PCR (F) and western blotting (G). (H) Dose response of RSL3-induced death of shCtrl and shIGF1R cells. (I) Dose response of RSL3-induced death of DMSO or IGF1R inhibitor (linsitinib) treated- A375 cells in the absence or presence of lorlatinib for 6 h. (J) Gene set variation analysis (GSVA) of TCGA-SKCM segregated by IGF1R expression. High IGF1R group demonstrated significant activation of PI3K/AKT/mTOR related pathways. (K) Western blotting analysis of the indicated proteins in sgCtrl and sgIGF1R A375 cells. (L) Dose response of RSL3-induced death of shIGF1R A375 (upper) and SK-MEL-28 (lower) cells overexpressed vector, SCD or SREBF1. P values were calculated using two-tailed unpaired Student's t-test in F, J. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
IGF1R activates a 110-kDa lipid kinase PI3K subgroup Iα (p110α) and causes the phosphorylation and activation of AKT [26,27]. To support the notion, we performed Gene Set Variation Analysis (GSVA) based on the TCGA-SKCM datasets, finding that PI3K/AKT/mTOR signaling pathways were significantly enriched in the IGF1R proficient group (Fig. 5J). Conversely, the expressions of IGF1R, p-PI3K, p-AKT, p-MTOR, p-p70s6, nSREBF1 and SCD were significantly reduced by the knockout of IGF1R (Fig. 5K). Furthermore, SCD or SREBF1 overexpression reversed melanoma cells sensitivity to ferroptosis induced by IGF1R knockdown (Fig. 5L). Taken together, these results unveiled that lorlatinib sensitized melanoma to ferroptosis by targeting IGF1R-mediated PI3K/AKT/mTOR signaling axis.
2.7. Combination of lorlatinib with GPX4 inhibition causes melanoma repression in vivo
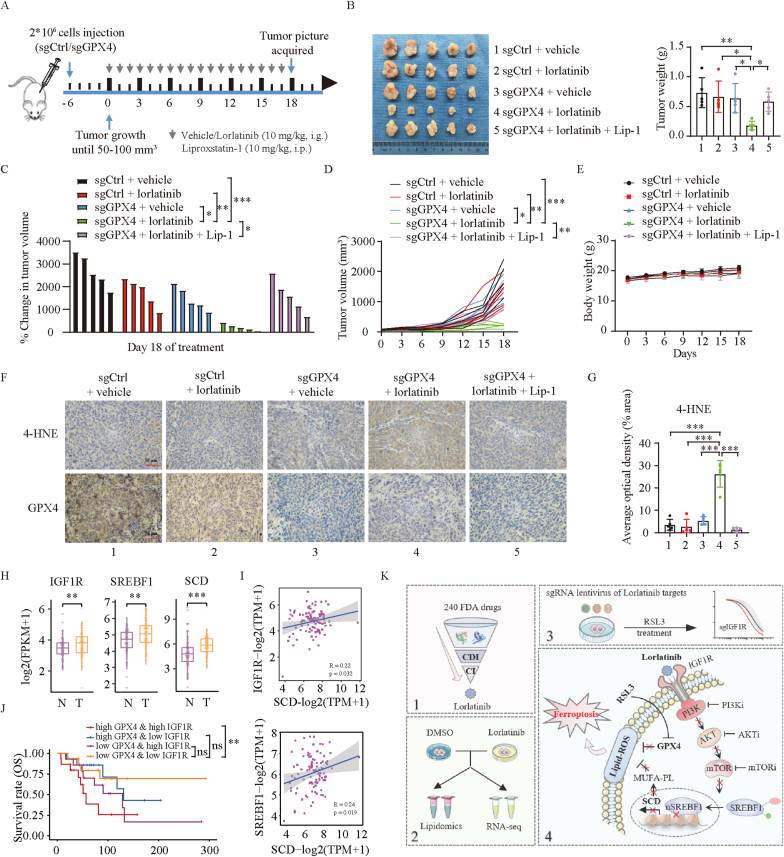
To evaluate the melanoma therapeutic potential of combining lorlatinib with ferroptosis induction, GPX4 knockout or control A375 cells were inoculated into the right flank of nude mice to generate subcutaneous xenograft models (Fig. 6A). When the tumor size reached 50–100 mm3, tumor-bearing mice were randomly allocated into groups and treated with vehicle (2% DMSO+30% PEG300, per day by orally) or lorlatinib (10 mg/kg, per day by orally). Liproxstatin-1 (10 mg/kg) was administrated through intraperitoneal injection per day. As expected, GPX4 knockout had minimal effect on A375 cell viability and tumor progression in vivo [28]. However, only the combination of GPX4 knockout with lorlatinib treatment, but not either alone, repressed melanoma growth (Fig. 6B–D), without significant changes in body weight (Fig. 6E). The melanoma regression was almost completely abolished by treatment with ferroptosis inhibitor liproxtatin-1 (Fig. 6B–D). Immunohistochemical analysis of 4-HNE, a major product of lipid peroxidation, supported such synergistic effect of the combining inhibition of GPX4 and lorlatinib in inducing tumor ferroptosis in vivo (Fig. 6F–G).
Fig. 6.
Combination of lorlatinib with GPX4 inhibition causes melanoma repression in vivo.
(A) Treatment schedule of tumor-bearing mice for drug administration. Nude mice were injected with sgCtrl or sgGPX4 (2 × 106) and treated with lorlatinib (10 mg/kg orally, every day) and vehicle at day 6 when the tumor size reached 50–100 mm [3]. Lip-1 was given 10 mg/kg intraperitoneally every day. Tumor volume was calculated every three days. (B-E) Tumor weight (B), percentage of change in tumor volume (C), tumor volume (D), and body weight (E) in the indicated groups. (F) IHC staining with antibodies against 4-HNE and GPX4 in the indicated group. Magnification, 400 × . Scale bar = 50 μm. (G) Quantification by Image J of 4-HNE in IHC staining. (H) Gene expression of IGF1R, SREBF1 and SCD within normal skin and melanoma patients in Xiangya cohorts. Num (N) = 77, num (T) = 99. N, normal skin; T, tumor. (I) Pearson correlation assay between IGF1R, SREBF1 and SCD gene expression in Xiangya melanoma cohorts. (J) Kaplan-Meier survival analysis with log-rank test of GPX4 and IGF1R gene expression in Xiangya melanoma cohorts. Num (high GPX4 & high IGF1R) = 15, Num (high GPX4 & low IGF1R) = 14, Num (low GPX4 & high IGF1R) = 14, Num (low GPX4 & low IGF1R) = 16. (K) Schematic summary for the findings in the present study. One-way ANOVA analysis was performed in B, C, D, G, J. Two-tailed unpaired Student's t-test was performed in H. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To further investigate the clinical significance of our findings, we carried out bioinformatics analyses using Xiangya melanoma datasets based on previous reports [29]. IGF1R, SREBF1 and SCD were all significantly increased in melanoma tissue compared with adjacent tissues (Fig. 6H, Fig. S8A). Pearson correlation assay showed that IGF1R and SREBF1 were positively correlated with SCD expression (Fig. 6I). An overall survival assay further revealed that the low expression of GPX4 combined with the low expression of IGF1R decreased the mortality of melanoma patients (Fig. 6J). These results were consistent in TCGA-SKCM datasets (Figs. S8B–F).
In conclusion, the present study identified a clinically applicable drug, lorlatinib, to synergize with GPX4 inhibition in melanoma through a large-scale screening. By using RNA seq, lipidomics and CRISPR/Cas9 based-lorlatinib's targets screening, we unveiled that lorlatinib sensitizes melanoma to ferroptosis by targeting IGF1R-mediated PI3K/AKT/mTOR signaling axis (Fig. 6K). These findings demonstrate that the combination of IGF1R inhibition with ferroptosis induction is a promising therapeutic approach for the treatment of melanoma with the IGF1R-proficient expression.
3. Discussion
Ferroptosis is a novel type of regulated cell death driven by excessive accumulation of iron-dependent lipid peroxidation, which is tightly associated with melanoma progression. Primary tumor-derived melanoma cells are characterized by four different differentiation status, which is inversely correlated with ferroptosis sensitivity [30]. Moreover, metastasizing melanoma cells avert ferroptosis through oleic acid in lymph or SREBP2-mediated transferrin expression in blood [12,13]. More interestingly, melanoma cells in therapy-resistant state are highly vulnerable to ferroptosis inducers [5,28], implying that ferroptosis induction could be a promising therapy strategy in melanoma.
To further improve the sensitivity of melanoma to ferroptosis, a drug screening was performed and lorlatinib was identified as a clinically applicable sensitizer of ferroptosis induction. SLC7A11/GSH/GPX4 axis represents the mainstay in ferroptosis control [1,2]. Iron metabolism is necessary for ferroptosis induction [31]. FSP1, GCH1 and DHODH were the main ferroptosis suppressors identified recently [[18], [19], [20], [21]]. However, lorlatinib did not affect these signaling through detecting the downstream or genetic modulation. Instead, we found that SCD expression was notably decreased after lorlatinib treatment, in line with the findings in lipidomics. Therefore, exploring the role of lorlatinib in SCD regulation could provide a mechanistic explanation to our observation.
SCD catalyzes the rate-limiting step in monounsaturated fatty acid synthesis, including oleic acid (18:1) and palmitoleic acid (16:1) [32], which could be transcriptionally regulated by several transcription factors including peroxisome proliferator activated receptor α (PPARα), liver X receptor (LXR), CCAAT/enhancer binding protein α (C/EBP-α), nuclear transcription factor Y (NF-Y), neurofibromin 1 (NF-1), specificity protein 1 (SP1) and SREBP1 [33,34]. RNA seq demonstrated that PI3K/AKT/mTOR signaling was significantly inhibited after lorlatinib treatment. mTOR has been reported to regulate ferroptosis sensitivity through three different mechanisms: (1) promoting GPX4 protein synthesis; (2) upregulating the SREBP1/SCD axis; (3) inhibiting autophagy [35]. Unchanged GPX4 expression after lorlatinib treatment and lorlatinib-promoting ferroptosis in the presence of autophagy inhibitor - CQ suggested that lorlatinib sensitized ferroptosis and regulated SREBP1/SCD axis through PI3K/AKT/mTOR signaling. The subsequent results validated the hypothesis.
Lorlatinib is an inhibitor of ALK and ROS1 developed for the treatment of non-small cell lung cancer [25]. Some cases reports demonstrated that those melanomas expressing oncogenic ALK or ROS1 fusion also had a good response to ALK or ROS1 inhibitors [36,37]. In our study, we found that lorlatinib alone shows little effect on the viability of A375 cells or SK-MEL-28 melanoma cells, mostly because both of them do not include abnormal ALK or ROS1. We further demonstrated that IGF1R, but not ALK or ROS1, is the major mediator of lorlatinib-mediated ferroptosis sensitivity. IGF1R is overexpressed in malignant melanoma and promotes melanoma progression [38]. Several studies further demonstrated that PI3K/AKT activation induced by persistent IGF1R signaling enabled V600E mutant melanomas resistant to BRAF inhibitors [39]. Activation of the IGF1R/MEK5/ERK5 signaling leads to acquired resistance to MAPK inhibitors in melanoma [40]. These results indicated that targeting IGF1R could be a promising treatment in melanoma. Here, we further showed that IGF1R inhibition sensitized melanoma to ferroptosis induction through PI3K/AKT/mTOR signaling, providing a potential combination therapy between IGF1R inhibition and ferroptosis induction.
It's worth to mention that the effect of lorlatinib on ferroptosis sensitivity under GPX4 inhibition condition might not be perfectly explained by IGF1R/PI3K/AKT/mTOR axis. Because some other kinases targets of lorlatinib such as EGFR and JAK2 has also been reported to be associated with the susceptibility to ferroptosis. For example, chlorogenic acid alleviates ferroptosis via the inhibition of the IL-6/JAK2/STAT3 signaling pathway [41]. O-glycosylation of EGFR activates mTOR activity to suppress ferroptosis [42]. Besides, some unidentified non-kinase targets of lorlatinib might also contribute to ferroptosis sensitivity. Even so, among these kinases' targets, IGF1R knockout sensitized melanoma to ferroptosis mostly and melanoma patients with low GPX4 and IGF1R expression in their tumors survive for longer period, highlighting its critical role in lorlatinib-mediated ferroptosis sensitivity.
In summary, we identified an FDA-approved drug, lorlatinib that synergized with GPX4 inhibition in melanoma. Mechanistically, lorlatinib sensitized melanoma to ferroptosis through targeting IGF1R-mediated PI3K/AKT/mTOR signaling axis and its downstream SCD expression. These insights are potentially translatable toward novel therapies for melanoma and other diseases involving ferroptosis.
4. Materials and methods
4.1. Cell culture
SK-MEL-28, A375, WM35, SK-MEL-5, 786-O, Caki-1, MDA-231, and HEK293T cells were obtained from American Type Culture Collection (ATCC). All cells were incubated at 37 °C in humid air with 5% CO2 and maintained in Dulbecco's Modified Eagle's Medium (Biological Industries) supplemented with 10% fetal bovine serum (Biological Industries) and 1% penicillin-streptomycin solution (Beyotime Biotechnology).
4.2. Lentiviral transduction and RNA interference
Stable cell lines were generated as described previously. In Brief, the plasmid used for knockdown, knockout or overexpression of target gene were co-transfected with lentivirus package plasmid (Addgene) into HEK293T cells for two days using TurboFect (Thermo Fisher Scientific, R0531) according to the manufacturer's instructions. Then the viral-containing supernatants were added to culture cells for additional 48 h and selected in the presence of puromycin (Thermo Fisher Scientific) at 2 μg/mL for three days. Knockdown of IGF1R with siRNA was performed by transfection with siALK ot siROS1 (GenePharma) using Turbofect according to the manufacturer's protocol. Knockdown efficiency was quantified by real-time PCR and western blotting. The sequence of sgRNAs were shown in Table S1.
4.3. RNA extraction and real-time PCR
Total RNA was extracted using Magzol reagent (Magen, R4801) according to the manufacturer's instruction. cDNA was synthesized from total RNA using HiScript Q RT SuperMix kit (Vazyme, R223-01). Real-time PCR was performed with SYBR Green Master Mix (bimake, B21703) in Applied Biosystems QuantStudio™ 3 Real-Time PCR System (Thermo Fisher Scientific). GAPDH was used as internal control. Primers are summarized in Table S2.
4.4. Western blotting
Total protein was extracted from cells using NP-40 buffer (Beyotime Biotechnology). Cytoplasmic and nuclear fractions were extracted from cells using the Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology, P0027) according to the manufacturer's protocol. The cell lysates were separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membrane. The membranes were then blocked with 5% skim milk and incubated with primary antibodies. Signals were detected with secondary antibodies (ABclonal) and visualized with Western ECL Blotting Substrates. The primary antibodies were: GPX4 (CST, #52455), NCOA4 (CST, #66849), NRF2 (CST, #12721), FTH1 (CST, #4393), xCT/SLC7A11 (CST, #12691), AIFM2/FSP1 (CST, #24972), DHODH (CST, #26381), LPCAT3 (Abcam, ab232958), GCH1 (Santa Cruz, sc-271482), ACSL4 (Santa Cruz, sc-365230), SCD (Santa Cruz, sc-81776), Actin (Santa Cruz, sc-8432), Phospho-PI3 Kinase p85 Tyr458/p55 Tyr199 (CST, #17366), PI3K p110 (Santa Cruz, sc-8010), Phospho-Akt Ser473 (CST, #4060), Akt (CST, #2920), Phospho-mTOR Ser2448 (CST, #5536), mTOR (CST, #2983), Phospho-p70 S6 Kinase Thr389 (CST, #2920), p70 S6 Kinase (CST, #34475), ALK (CST, #3633), ROS1 (CST, #3287), LTK (Santa Cruz, sc-393465), Fer (Santa Cruz, sc-390484), Fes (Santa Cruz, sc-377179), PTK2B (Santa Cruz, sc-393181), TNK2 (Santa Cruz, sc-28336), PTK2 (Santa Cruz, sc-271126), FRK (Santa Cruz, sc-166478), EGFR (Santa Cruz, sc-373746), IGF1R (Santa Cruz, sc-462), TSSK2 (Santa Cruz, sc-100437), JAK2 (Santa Cruz, sc-390539), EPHA1 (Santa Cruz, sc-65993).
4.5. Cell death and viability assays
As previously described, cell death was identified by propidium iodide (Beyotime Biotechnology, C2015 M) staining and cell viability was calculated by the Cell Counting Kit-8 (CCK-8) assay (Bimake, B34302) [43].
4.6. In-vitro drug combination screen
Cells were seeded in 96-well plates at a density of 9000 cells per well. The next day, cells were treated with DMSO, RSL3 (2.5uM), 240 FDA-approved anti-tumor drugs (Selleck Chemicals, Table S3) (5uM) or both drugs for 12h. Subsequently, cell viability was calculated by the CCK-8 assay. To test for drug synergy, the coefficient of drug interaction (CDI) was calculated using the following formula: CDI = (2 drug combination divided by the control)/(drug 1 divided by the control) x (drug 2 divided by the control) [15]. CDI less than 1 indicates synergy; CDI equal to 1 indicates additivity; and CDI greater than 1 indicates antagonism. The combination index (CI) was calculated using the CompuSyn software based on the Chou-Talalay methodology and less than 1 indicates synergy.
4.7. Lipid peroxidation assay
Cells were seeded in 6-well plates and treated as indicated. After treatment, the cells were harvested and resuspended with PBS containing 5 μM 581/591 C-11 BODIPY dye (Thermo Fisher Scientific, D3861), then incubated at 37 °C for 30 min. Lipid peroxidation was evaluated by flow cytometer.
4.8. GSH assay
GSH levels were determined using GSH and GSSG assay (Beyotime Biotechnology, S0053) according to the manufacturer's instruction. Briefly, after the indicated treatment, the harvested cells were lysed and centrifuged at 10,000 g for 10 min. The supernatant was loaded into a 96-well plate and mixed with glutathione reductase, DTNB and assay buffer. After incubation for 5 min at room temperature, NADPH was added and the plate was read at 412 nm.
4.9. GPX4 activity measurement
GPX4 activity was measured with Glutathione Peroxidase Assay Kit (Abcam, ab102530). The indicated cells were harvested and homogenized in 200 μl assay buffer. After centrifuging 15 min at 4 °C at 10,000g, the supernatants were collected and mixed well with reaction mix, followed by a 15 min incubation at room temperature. Cumene hydroperoxide solution was added to the sample well and the initial output was measured on a microplate reader at 340 nm. After incubation for 5 min at room temperature, the second output was recorded at 340 nm.
4.10. CoQ10
CoQ10 measurements were performed using CoQ10 ELISA Kit (CUSABIO, CSB-E14081h) according to manufacturer's instruction. After the indicated treatment, the collected cells were homogenized in PBS and centrifuged at 10,000 g for 10 min. The supernatants were loaded into assay plate and mixed well with HRP-conjugate, then incubated for 40 min at 37 °C. After five washes with wash buffer, TMB Substrate was added to each well and incubate for 20 min at 37 °C in the dark. Finally, the stop solution was added and the plate was read at 450 nm.
4.11. MDA assay
MDA levels were evaluated by Lipid Peroxidation Assay Kit (Merck, MAK085). Cells were homogenized on ice in MDA Lysis Buffer containing BHT. The samples were centrifuged at 13,000 g for 10 min and the supernatants were placed into a microcentrifuge tube. TBA solution was then added to form the MDA-TBA adduct. The reaction mixture was then incubated for 60 min at 95 °C, and cooled to room temperature. 200 μL of reaction mixture was then placed to 96-well plate and read at 532 nm.
4.12. Iron assay
The relative Fe2+ concentration in cells was assessed using Iron Assay Kit (Abcam, ab83366). Briefly, cells were homogenized in 4–10 vol of iron assay buffer and centrifuged at 16,000 g for 10 min to remove insoluble materials. The supernatants were added to 96-well plate and adjusted to the volume of 100 μl with assay buffer. After incubation at 37 °C for 30 min, 100 μl iron probe was added and the reaction was incubated at 37 °C for 60 min. The absorbance at 593 nm was measured on a colorimetric microplate reader.
4.13. Immunohistochemistry
Tumor tissues were fixed in formalin and embedded in paraffin. Embedded tissues were sectioned and stained as previously [43]. The primary antibodies, 4-HNE (Abcam, ab46545) and GPX4 (CST, #52455), were incubated overnight at 4 °C. Images were taken at 400× magnification using microscope and quantified by Image J.
4.14. Transmission electron microscopy
Cells were fixed in 2.5% glutaraldehyde solution with Millonig's phosphate buffer (pH = 7.3). After washing three times with Millonig's phosphate buffer, cells were incubated for 1 h in 1% osmium tetroxide, and then washed three times with Millonig's phosphate buffer. The samples were dehydrated in increasing concentrations of acetone, embedded in 1:1 mix of acetone:resin for 12 h, and polymerized in 100% resin overnight at 37 °C and then 12 h at 60 °C. Ultrathin sections were cut in a Leica Ultracut microtome (Leica EM UC7). After staining with uranyl acetate and lead nitrate, the specimens were examined in a Hitachi HT-7700 electron microscope.
4.15. RNA-seq
A375 cells were treated with DMSO or 5 μM lorlatinib for 12 h. Total RNA were extracted with Magzol and used for RNA-seq analysis. Library construction was validated on the Agilent Technologies 2100 bioanalyzer for quality control. Libraries were sequenced on a BGISEQ-500RS. Only genes with at least 1 read in each of six samples and at least 50 reads in total among all samples were retained for following analyses [44]. Differential expression genes were defined with |log2 fold-change| > 1 and Q value < 0.05. Data were presented in Table S4. RNA seq data of melanoma patients were from the research files at Xiangya Hospital of Central South University, as previous reported [33]. Ethical approval for the study was obtained from the Ethics Committee of Central South University and written informed consent was obtained from each patient.
4.16. Lipidomics
Cells were homogenized in 750 μl methanol and transferred into a glass tube with a Teflon lined cap for extraction. The extracted lipids were processed and analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). UHPLC-MS/MS analyses were performed using a Vanquish UHPLC system (Thermo Fisher Scientific) coupled with an Orbitrap Q ExactiveTM HF mass spectrometer (Thermo Fisher Scientific) in Novogene Co., Ltd. (Beijing, China). Liquid chromatography was performed on a Thermo Accucore C30 column (150 × 2.1 mm, 2.6 μm) using a 20-min linear gradient at a flow rate of 0.35 mL/min. Mobile phase buffer A consisted of acetonitrile/water (6/4) and buffer B consisted of acetonitrile/isopropanol (1/9) both containing 10 mM ammonium acetate and 0.1% formic acid. The gradient was as follows: 30% B, initial; 30% B, 2min; 43% B, 5 min; 55% B, 5.1 min; 70% B, 11 min; 99%B, 16 min; 30%B, 18.1min. The column temperature was set at 40 °C. The raw data generated by UHPLC-MS/MS were processed using the Compound Discoverer 3.01 (CD3.1, Thermo Fisher). Statistical analyses were performed using the statistical software R (R version R-3.4.3), Python (Python 2.7.6 version) and CentOS (CentOS release 6.6). Data were presented in Table S5.
4.17. Animal study
All animal experiments were approved by the Ethical Review of Experimental Animals at Central South University. To generate subcutaneous tumors, 2 × 106 control A375 cells or GPX4 KO cells were suspended in 100 μl PBS and injected subcutaneously into nude mice (Shanghai SLAC). Tumor-bearing mice were randomly allocated into groups and treated with vehicle (2% DMSO+30% PEG300, per day by orally) or lorlatinib (10 mg/kg, per day by orally). Liproxstatin-1 (10 mg/kg) was administrated through intraperitoneal injection per day. Tumors were weighted and photographed on day 18 after treatment. Tumor size were recorded every three days and calculated as [(length × width × width)/2].
4.18. Statistical analyses
Data were presented as mean ± SD and analyzed with GraphPad Prism 8. Two-tailed unpaired student's t-test was used for comparison between two groups. ANOVA analysis with Tukey's multiple comparison test were employed for comparison among the different groups. A P value of <0.05 was considered statistically significant.
Contribution
F. Z., G.D., L.Y, and Q.Z. carried out the experiments and analyzed the data. G.D., Y.Z., H.L. and X.C. supervised the project, designed experiments. F. Z., and G.D. wrote the manuscript. Y.H. and G.D. conducted the bioinformatics.
Declaration of competing interest
All the authors declare no competing interests.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (no. 82272849, 82102803 to G.D., and no. 82103183 to F.Z., and no. 31800979 to H.L.). the Natural Science Foundation of China for outstanding Young Scholars (no. 82022060 to H.L.), the fellowship of China postdoctoral Science Foundation (no. 2022T150744 to F.Z., no. 2021T140748 to G.D.), the Natural Science Foundation of Hunan Province of China (no. 2021JJ40976 to G.D., no. 2022JJ40767 to F.Z.)
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102653.
Contributor Information
Guangtong Deng, Email: dengguangtong@outlook.com.
Xiang Chen, Email: chenxiangck@126.com.
Hong Liu, Email: hongliu1014@csu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- 1.Dixon S.J., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang W.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X., Kang R., Kroemer G., Tang D. Broadening horizons: the role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021;18:280–296. doi: 10.1038/s41571-020-00462-0. [DOI] [PubMed] [Google Scholar]
- 4.Lei G., Zhuang L., Gan B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer. 2022 doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viswanathan V.S., et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453–457. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown C.W., et al. Prominin2 drives ferroptosis resistance by stimulating iron export. Dev. Cell. 2019;51:575–586 e574. doi: 10.1016/j.devcel.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo M., et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ. 2018;25:1457–1472. doi: 10.1038/s41418-017-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang K., et al. miR-9 regulates ferroptosis by targeting glutamic-oxaloacetic transaminase GOT1 in melanoma. Mol. Carcinog. 2018;57:1566–1576. doi: 10.1002/mc.22878. [DOI] [PubMed] [Google Scholar]
- 9.Gagliardi M., et al. Aldo-keto reductases protect metastatic melanoma from ER stress-independent ferroptosis. Cell Death Dis. 2019;10:902. doi: 10.1038/s41419-019-2143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., et al. Nedd4 ubiquitylates VDAC2/3 to suppress erastin-induced ferroptosis in melanoma. Nat. Commun. 2020;11:433. doi: 10.1038/s41467-020-14324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S., et al. CAMKK2 defines ferroptosis sensitivity of melanoma cells by regulating AMPKNRF2 pathway. J. Invest. Dermatol. 2022;142:189–200 e188. doi: 10.1016/j.jid.2021.05.025. [DOI] [PubMed] [Google Scholar]
- 12.Hong X., et al. The lipogenic regulator SREBP2 induces transferrin in circulating melanoma cells and suppresses ferroptosis. Cancer Discov. 2021;11:678–695. doi: 10.1158/2159-8290.CD-19-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ubellacker J.M., et al. Lymph protects metastasizing melanoma cells from ferroptosis. Nature. 2020;585:113–118. doi: 10.1038/s41586-020-2623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson T.W., et al. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(m etheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J. Med. Chem. 2014;57:4720–4744. doi: 10.1021/jm500261q. [DOI] [PubMed] [Google Scholar]
- 15.Simpkins F., et al. Dual src and MEK inhibition decreases ovarian cancer growth and targets tumor initiating stem-like cells. Clin. Cancer Res. 2018;24:4874–4886. doi: 10.1158/1078-0432.CCR-17-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan B., et al. Membrane damage during ferroptosis is caused by oxidation of phospholipids catalyzed by the oxidoreductases POR and CYB5R1. Mol Cell. 2021;81:355–369 e310. doi: 10.1016/j.molcel.2020.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Yi J., Zhu J., Wu J., Thompson C.B., Jiang X. Oncogenic activation of PI3K-AKT-mTOR signaling suppresses ferroptosis via SREBP-mediated lipogenesis. Proc. Natl. Acad. Sci. U. S. A. 2020;117:31189–31197. doi: 10.1073/pnas.2017152117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bersuker K., et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doll S., et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 20.Kraft V.A.N., et al. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 2020;6:41–53. doi: 10.1021/acscentsci.9b01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao C., et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J., Conrad M. The metabolic underpinnings of ferroptosis. Cell Metabol. 2020;32:920–937. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 23.Tesfay L., et al. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 2019;79:5355–5366. doi: 10.1158/0008-5472.CAN-19-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Audet-Walsh E., et al. SREBF1 activity is regulated by an AR/mTOR nuclear Axis in prostate cancer. Mol. Cancer Res. 2018;16:1396–1405. doi: 10.1158/1541-7786.MCR-17-0410. [DOI] [PubMed] [Google Scholar]
- 25.Zou H.Y., et al. PF-06463922 is a potent and selective next-generation ROS1/ALK inhibitor capable of blocking crizotinib-resistant ROS1 mutations. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3493–3498. doi: 10.1073/pnas.1420785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X., et al. A cathelicidin-related antimicrobial peptide suppresses cardiac hypertrophy induced by pressure overload by regulating IGFR1/PI3K/AKT and TLR9/AMPKalpha. Cell Death Dis. 2020;11:96. doi: 10.1038/s41419-020-2296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao H., Han X., Han X. The cardioprotection of the insulin-mediated PI3K/Akt/mTOR signaling pathway. Am. J. Cardiovasc. Drugs. 2014;14:433–442. doi: 10.1007/s40256-014-0089-9. [DOI] [PubMed] [Google Scholar]
- 28.Hangauer M.J., et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature. 2017;551:247–250. doi: 10.1038/nature24297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farshidfar F., et al. Integrative molecular and clinical profiling of acral melanoma links focal amplification of 22q11.21 to metastasis. Nat. Commun. 2022;13:898. doi: 10.1038/s41467-022-28566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsoi J., et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell. 2018;33:890–904 e895. doi: 10.1016/j.ccell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Yu C., Kang R., Tang D. Iron metabolism in ferroptosis. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.590226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H., et al. Crystal structure of human stearoyl-coenzyme A desaturase in complex with substrate. Nat. Struct. Mol. Biol. 2015;22:581–585. doi: 10.1038/nsmb.3049. [DOI] [PubMed] [Google Scholar]
- 33.Duvel K., et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tracz-Gaszewska Z., Dobrzyn P. Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers. 2019;11 doi: 10.3390/cancers11070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lei G., Zhuang L., Gan B. mTORC1 and ferroptosis: regulatory mechanisms and therapeutic potential. Bioessays. 2021;43 doi: 10.1002/bies.202100093. [DOI] [PubMed] [Google Scholar]
- 36.Couts K.L., et al. ALK inhibitor response in melanomas expressing EML4-ALK fusions and alternate ALK isoforms. Mol. Cancer Therapeut. 2018;17:222–231. doi: 10.1158/1535-7163.MCT-17-0472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couts K.L., et al. Acral lentiginous melanoma harboring a ROS1 gene fusion with clinical response to entrectinib. JCO Precis Oncol. 2017;1:1–7. doi: 10.1200/PO.16.00013. [DOI] [PubMed] [Google Scholar]
- 38.Damsky W., et al. mTORC1 activation blocks BrafV600E-induced growth arrest but is insufficient for melanoma formation. Cancer Cell. 2015;27:41–56. doi: 10.1016/j.ccell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villanueva J., et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benito-Jardon L., et al. Resistance to MAPK inhibitors in melanoma involves activation of the igf1r-MEK5-erk5 pathway. Cancer Res. 2019;79:2244–2256. doi: 10.1158/0008-5472.CAN-18-2762. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., et al. Chlorogenic acid alleviates chronic stress-induced duodenal ferroptosis via the inhibition of the IL-6/JAK2/STAT3 signaling pathway in rats. J. Agric. Food Chem. 2022;70:4353–4361. doi: 10.1021/acs.jafc.2c01196. [DOI] [PubMed] [Google Scholar]
- 42.Li H.W., et al. GALNT14 regulates ferroptosis and apoptosis of ovarian cancer through the EGFR/mTOR pathway. Future Oncol. 2022;18:149–161. doi: 10.2217/fon-2021-0883. [DOI] [PubMed] [Google Scholar]
- 43.Deng G., et al. EEF2K silencing inhibits tumour progression through repressing SPP1 and synergises with BET inhibitors in melanoma. Clin. Transl. Med. 2022;12:e722. doi: 10.1002/ctm2.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu H., et al. ADORA1 inhibition promotes tumor immune evasion by regulating the ATF3-PD-L1 Axis. Cancer Cell. 2020;37:324–339 e328. doi: 10.1016/j.ccell.2020.02.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.