Abstract
The apolipoprotein E4 (APOE4) gene, a genetic risk factor for Alzheimer’s disease (AD), is abundantly expressed in both the brain and periphery. Here, we present evidence that peripheral apoE isoforms, separated from those in the brain by the blood-brain barrier (BBB), differentially impact AD pathogenesis and cognition. To evaluate the function of peripheral apoE, we developed conditional mouse models expressing human APOE3 or APOE4 in the liver with no detectable apoE in the brain. Liver-expressed apoE4 compromised synaptic plasticity and cognition by impairing cerebrovascular functions. Plasma proteome profiling revealed apoE isoform-dependent functional pathways highlighting cell adhesion, lipoprotein metabolism, and complement activation. ApoE3 plasma from young mice improved cognition and reduced vessel-associated gliosis when transfused into aged mice, whereas apoE4 compromised the beneficial effects of young plasma. A human iPSC-derived endothelial cell model recapitulated the plasma apoE isoform-specific effect on endothelial integrity, further supporting a vascular-related mechanism. Upon breeding with amyloid model mice, liver-expressed apoE4 exacerbated brain amyloid pathology, whereas apoE3 reduced it. Our findings demonstrate pathogenic effects of peripheral apoE4, providing a strong rationale for targeting peripheral apoE to treat AD.
INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disease and the leading cause of dementia in the elderly population. The accumulation and deposition of amyloid-β (Aβ) in the brain are central events in AD pathogenesis1. Epidemiological studies showed that vascular risk factors such as hypertension, diabetes, and atherosclerosis have significant impacts on AD risk2,3. Cerebrovascular dysfunction, including blood-brain barrier (BBB) leakage and impaired cerebral microcirculation, is tightly linked to neuronal dysfunction4-6. While genetic and environmental factors influence AD pathogenesis, the ε4 allele of the apolipoprotein E (APOE) gene dramatically increases the risk and lowers the age at onset of AD in a gene dose-dependent manner7. The APOE4 also accelerates aging-related cognitive decline in non-demented individuals8. Compared to the common APOE3 or the AD-protective APOE2 allele, APOE4 drives earlier and more abundant Aβ pathology, a major driver in AD pathogenesis9,10. APOE4 also regulates non-Aβ pathways such as vascular function, immune responses, and tau-mediated neurodegeneration11-13. A better understanding on how apoE isoforms impact brain functions and AD-related pathways should allow us to address why apoE4 increases AD risk and how we can target this protein for therapy14.
ApoE is produced primarily by astrocytes in the central nervous system (CNS) and serves as a major lipid carrier to neurons to support membrane homeostasis, synaptic integrity, and injury repair in the brain11. In the periphery, apoE is abundantly produced by the liver and macrophages, released into the blood to modulate lipid-related events. Importantly, peripheral apoE is separated from that in the CNS by the BBB, most evident in a liver transplantation study showing that apoE isoform status in the recipient’s plasma was changed to the donor’s genotype while it remained the same in the CSF15. Despite physical separation, several lines of evidence suggest that peripheral apoE might impact cerebrovascular and brain functions16,17. ApoE4 is a risk factor for hypercholesterolemia and atherosclerosis, conditions known to increase AD risk18. ApoE4 can also synergize with other risk factors including insulin resistance, dyslipidemia, and peripheral vascular diseases19, exerting confounding effects on AD. However, due to the presence of apoE in both the periphery and the brain, it has been difficult to assess the specific contribution of peripheral apoE isoforms to CNS functions and AD pathogenesis. To meet this challenge, we developed mouse models that conditionally express human APOE3 or APOE4 in the liver with no detectable apoE in the brain. Importantly, we found that liver-expressed apoE4 compromised synaptic plasticity and cognitive behaviors likely by impairing cerebrovascular functions. With single-cell RNA sequencing (scRNA-Seq), we showed that expression of apoE4 in the periphery promotes immune responses in endothelial cells along with an increase of vessel-associated gliosis, which may compromise barrier integrity and cerebrovascular functions. We also explored the therapeutic potential of peripheral treatment using young plasma with different APOE genotypes. We demonstrated that apoE3, but not apoE4 young plasma improved cognition and reduced vessel-associated gliosis in aged mice. In addition, liver-expressed apoE4 exacerbated brain amyloid pathology, whereas liver-expressed apoE3 led to beneficial effects on brain functions and ameliorated amyloid deposition. Our findings demonstrate differential effects of peripheral apoE isoforms on brain function and pathological outcomes.
RESULTS
Peripheral apoE isoforms differentially impact brain function
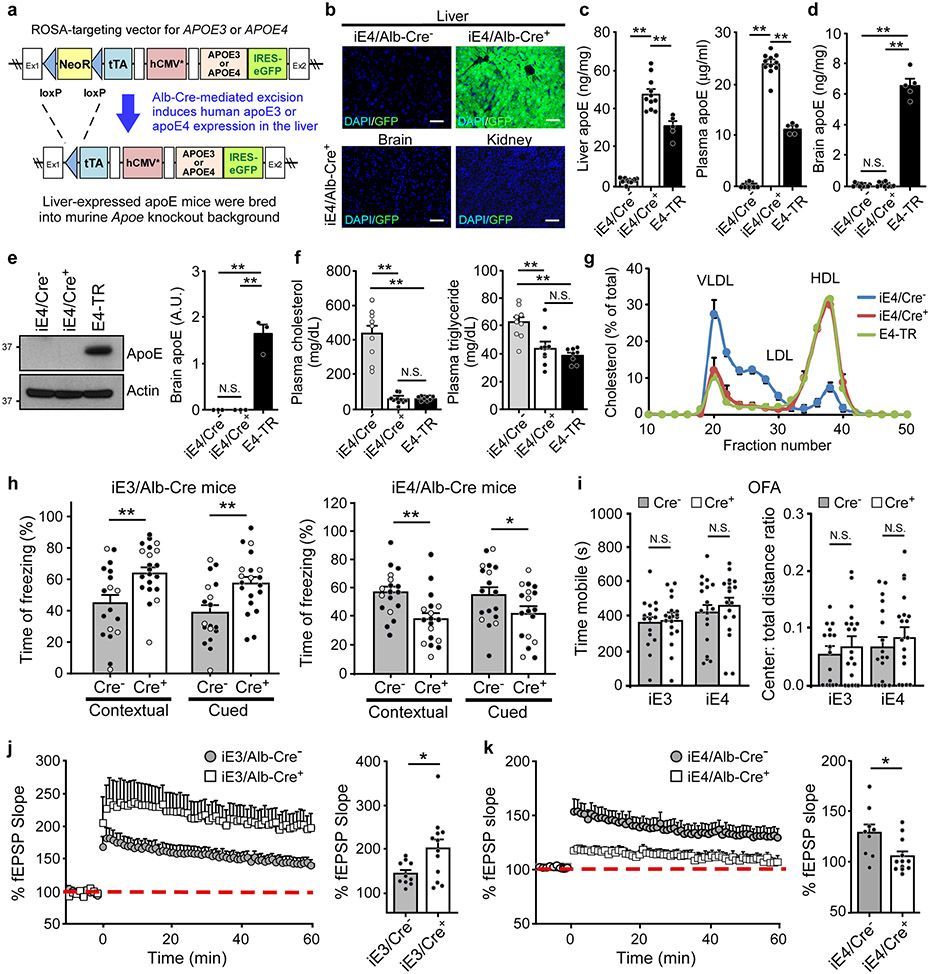
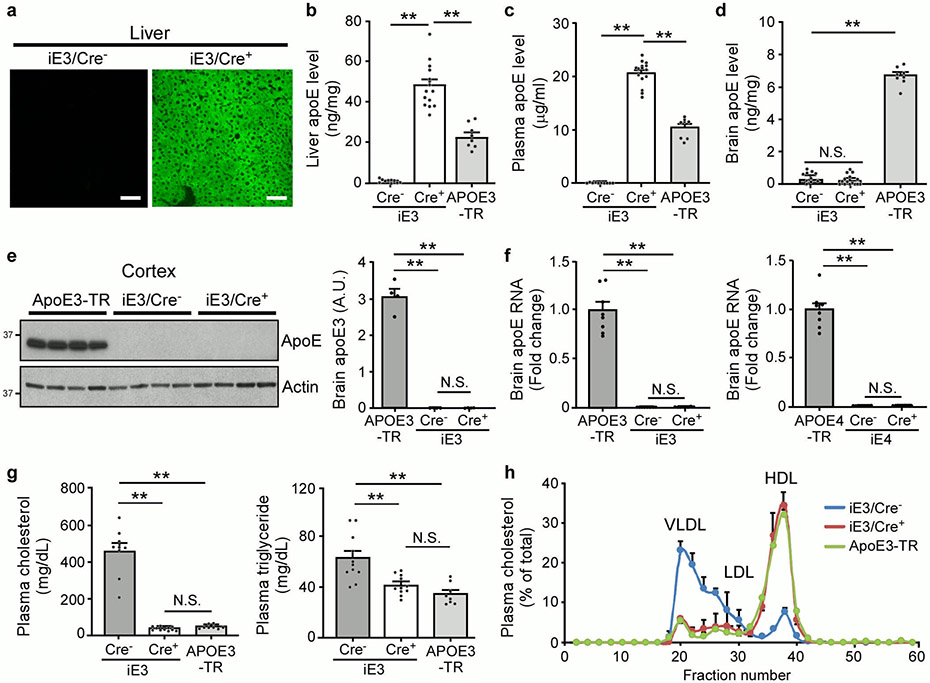
We developed animal models that allow for cell type-specific and inducible expression of apoE3 or apoE4 (termed iE3 and iE4 mice where “i” represents inducible) (Fig. 1a)20. After breeding with liver-specific albumin-Cre (Alb-Cre) mice, we generated mice expressing human apoE3 or apoE4 in the liver in an Apoe knockout (apoe-KO) background, allowing us to specifically address the effect of peripheral apoE on brain function. Further, comparison with apoe-KO littermate controls should provide clues as to whether liver-expressed apoE4 exhibits a gain-of-toxic function or a loss-of-protective function. As anticipated, a strong GFP fluorescence, a surrogate of apoE expression, was observed exclusively in the liver of iE3/Cre+ and iE4/Cre+ mice but not in other tissues or in Cre− mice (Fig. 1b and Extended Data Fig. 1a). Both apoE3 and apoE4 were expressed in the liver and secreted into the bloodstream in Alb-Cre+ mice with concentrations ~1.5-2 fold higher than those in apoE-targeted replacement (apoE-TR) (Fig. 1c and Extended Data Fig. 1b, c). Importantly, apoE was undetectable in the mouse brains (Fig. 1d, e and Extended Data Fig. 1d-f). The presence of apoE in the plasma of iE3/Cre+ or iE4/Cre+ mice rescued hypercholesterolemia, elevated triglyceride, and altered plasma lipoprotein profiles in Cre− (apoe-KO) mice to the extent comparable to those in apoE-TR mice (Fig. 1f, g and Extended Data Fig. 1g, h).
Fig. 1 ∣. Opposing effects of liver-expressed apoE3 and apoE4 on brain cognition and synaptic function.
a, The structure of the ROSA-targeting vector with the Tet-off regulatory cassette for APOE and eGFP expression. Breeding to Alb-Cre mice removed the loxP-flanked Neor gene and led to expression of human apoE3 or apoE4 in the liver. b, GFP was exclusively detected in the liver of iE4/Cre+ mice. Similar results were observed in at least three independent experiments. Scale bar, 50 μm. c, d, ApoE levels in the liver, plasma, or brain of iE4/Cre mice (Cre−, n=11; Cre+, n=11) and apoE4-TR mice (n=5) at 12-13 months of age were examined by ELISA. **, P<0.0001. e, No apoE was detected in the brains of iE4/Cre mice (n=3/group) at 12-13 months of age by Western blotting. **, P=0.0002. f, Total cholesterol and triglyceride levels in the plasma of iE4/Cre mice (Cre−, n=9; Cre+, n=10) and apoE4-TR mice (n=8) were examined. Cholesterol: **, P<0.0001; Triglyceride: **, P=0.003 (Cre− vs. Cre+); P=0.0004. g, Distribution of plasma lipoprotein cholesterol in iE4/Cre− or iE4/Cre+ mice (n=3/group). Pooled plasma from apoE4-TR mice (n=3) was included as controls. N.S., not significant. One-way analysis of variance (ANOVA) with Tukey’s post-hoc test. h, Memory performance of mice expressing apoE3 (Cre−, n=18; Cre+, n=21 for contextual test; **, P=0.003; Cre−, n=18; Cre+, n=20 for cued test; **, P=0.005) or apoE4 (Cre−, n=18; Cre+, n=18; **, P=0.003; *, P=0.048) in the liver examined by fear conditioning tests. The percentage of time spent showing freezing behavior in response to stimulus is shown. Black circle: male; Grey circle: female. i, Analyses of anxiety-related behaviors in iE3/Cre (Cre−, n=16; Cre+, n=19) and iE4/Cre mice (Cre−, n=18; Cre+, n=18) assessed by open field analysis (OFA). N.S., not significant. j, k, Normalized fEPSP responses to field stimulation were summarized during recordings from the CA1 region of hippocampal slices from iE3/Cre mice (Cre−, n=10; Cre+, n=12) or iE4/Cre mice (Cre−, n=9; Cre+, n=12). Averages of the last 5 min of fEPSP recording were quantified. Data represent mean ± s.e.m.; iE3: *, P=0.03; iE4: *, P=0.018, two-tailed Student's t-test.
To examine how peripheral expression of apoE3 or apoE4 impacts brain function, we performed behavioral and electrophysiology studies. Importantly, we found that apoE4 expression in the liver resulted in significant memory deficits in iE4/Cre+ mice compared to controls as assessed by contextual and cued fear conditioning tests, whereas apoE3 expression improved associative memory (Fig. 1h). The anxiety behaviors, locomotor activity, and motor coordination were not affected by apoE3 or apoE4 expression in the liver (Fig. 1i, and Supplementary Fig. 1). Consistently, peripheral apoE4 significantly suppressed synaptic plasticity measured by long-term potentiation (LTP); in contrast, peripheral apoE3 increased it (Fig. 1j, k). These results indicate that peripheral apoE3 has a beneficial effect, whereas peripheral apoE4 exhibits a gain-of-toxic effect, impairing brain function in the absence of brain apoE.
Peripheral apoE4 impairs cerebrovascular functions
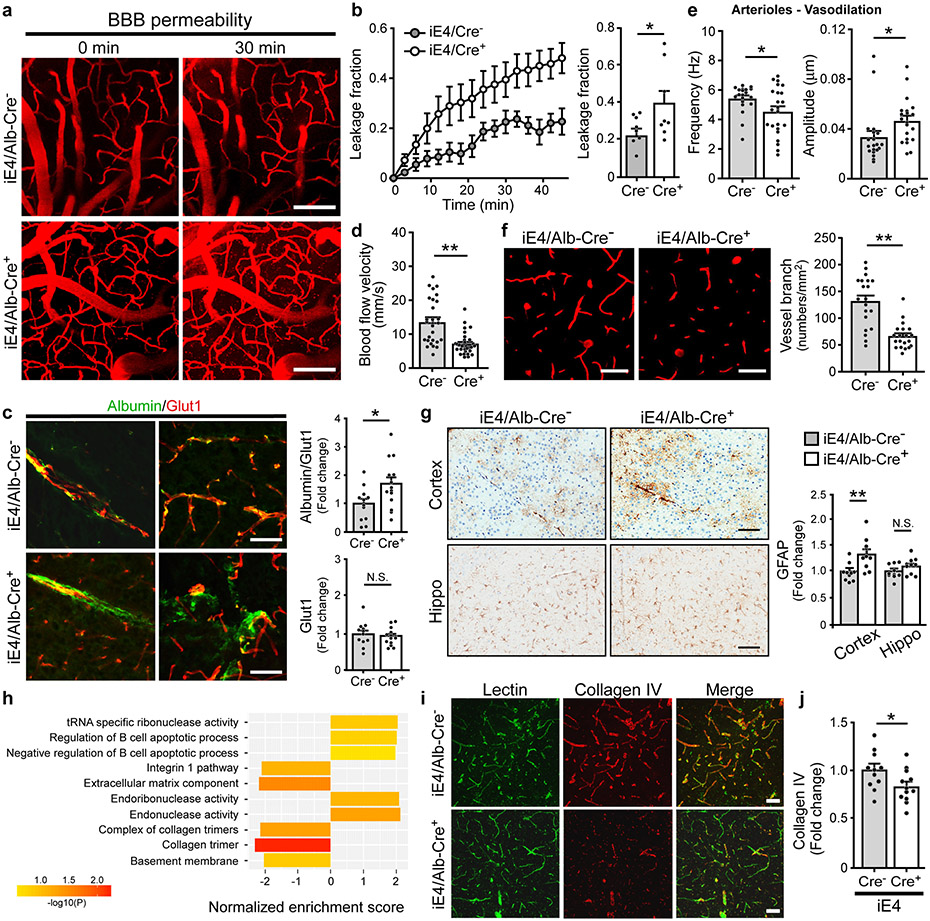
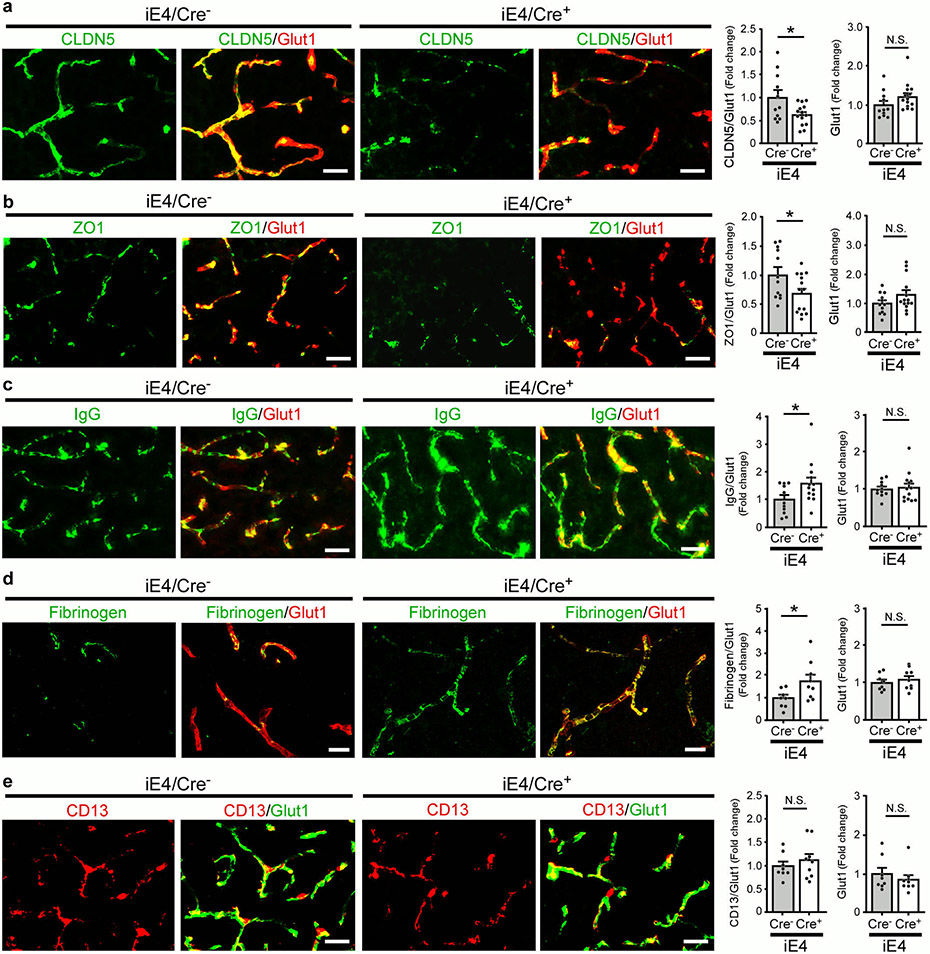
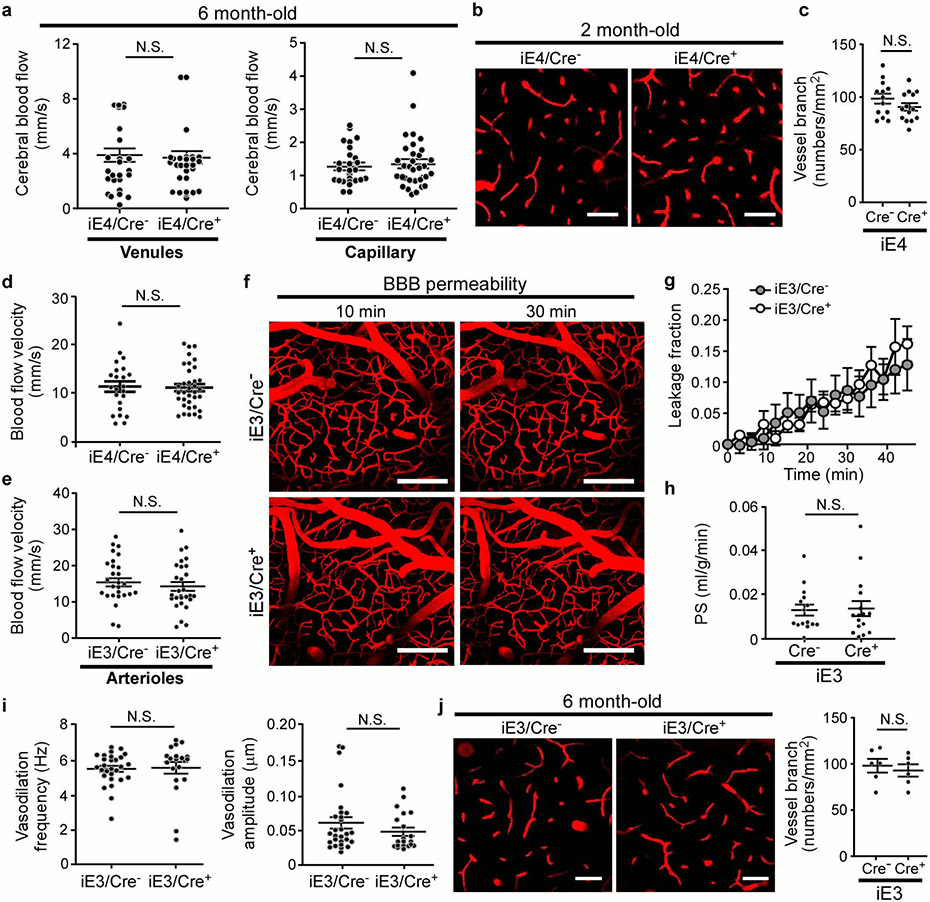
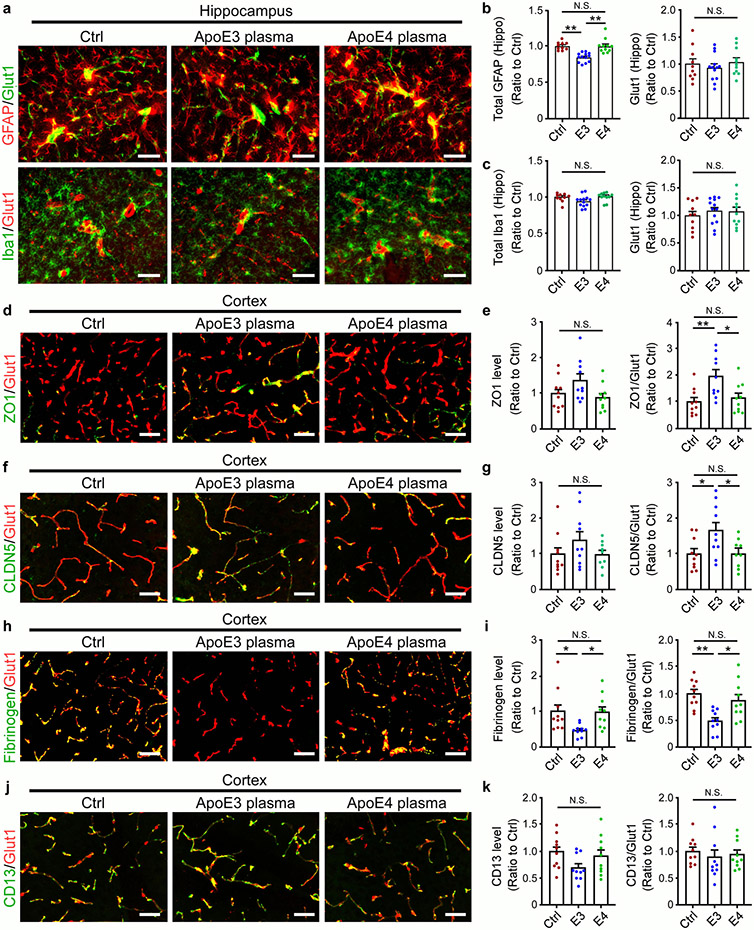
Cerebral hypoperfusion and BBB dysfunction have been implicated in AD pathogenesis5. To explore the potential pathological mechanisms by which liver-expressed apoE4 disrupts brain functions, we investigated the cerebrovascular permeability of mice at 6-7 months of age. Remarkably, an increase in BBB leakage in iE4/Cre+ mice was observed compared to iE4/Cre− mice measured by in vivo two-photon microscopy (Fig. 2a, b), consistent with previous finding in apoE4-TR mice12. We further examined the BBB integrity in mice at 12-13 months of age by analyzing the tight junction (TJ)-associated proteins claudin-5 (CLDN5), and scaffold protein zonula occludens 1 (ZO1) in endothelial cells. Immunostaining demonstrated that the levels of CLDN5 and ZO1 normalized by the vascular marker Glut1 were both reduced in iE4/Cre+ mice (Extended Data Fig. 2a, b). Upon BBB disruption, blood components and coagulation factors are shown to gain access to the brain12,21. To further assess if peripheral apoE4 impacts the brain vascular permeability, we examined the leakage of serum proteins to the brain by immunostaining for albumin (67 kDa), immunoglobulin G (IgG; 150 kDa) or fibrinogen (340 kDa), robust markers of impaired BBB integrity. An increased level of albumin in some perivascular areas and brain parenchymal regions near vessels was observed in iE4/Cre+ mice (Fig. 2c). In addition, an elevation of perivascular deposits of IgG was found in iE4/Cre+ mice (Extended Data Fig. 2c). The iE4/Cre+ mice also exhibited an increase of fibrinogen immunoreactivity although mostly confined to vessels (Extended Data Fig. 2d). These results suggest that the chronic BBB leakage might be selective according to the size of the molecules. As pericytes regulate the BBB integrity, we also assessed the CD13-positive pericyte coverage on the brain microvessels and found no differences between iE4/Cre+ and iE4/Cre− mice (Extended Data Fig. 2e).
Fig. 2 ∣. Compromised cerebrovascular integrity and astrogliosis associated with liver-specific expression of apoE4.
a-e, Two-photon imaging was used to assess vascular functions in male iE4/Cre mice (Cre−, n=5; Cre+, n=5) at 6-7 months of age. b, d-f, In two-photon analyses, each data point represents blood vessel-related observation with varying number of measurements in each animal from 5 mice per genotype. a, The representative images at 0 and 30 mins after dextran injection are shown. Scale bar, 100 μm. b, The leakage of dextran signals were monitored over time (left panel). The fraction of BBB leakage at 30 min after injection of dextran was quantified (right panel). *, P=0.045. c, Brain sections from iE4/Cre mice (Cre−, n=12; Cre+, n=14) at 12-13 months of age were immunostained with albumin and Glut1 antibodies. The total Glut1 (red) and the albumin (green) normalized against Glut1 were quantified. Scale bar, 50 μm. *, P=0.024. d, The cerebral blood flow (CBF) in the cerebral arterioles of iE4/Cre mice at 6-7 months of age was measured. **, P<0.0001. e, The frequency and amplitude of arteriolar oscillation in iE4/Cre mice were measured. The data points represent the measurements for frequency (*, P=0.042) or amplitude (*, P=0.045) of arteriolar oscillation in each vessel from 5 mice/genotype. f, The vessel branch densities in the cerebral arterioles of iE4/Cre mice were examined. **, P<0.0001. Scale bar, 50 μm. g, Brain sections from iE4/Cre mice (Cre−, n=10; Cre+, n=10) at 12-13 months of age were immunostained with a GFAP antibody and quantified. **, P=0.005. Scale bar, 50 μm. h, Brain tissues from iE4/Cre mice (Cre−, n=8; Cre+, n=8) were subjected to RNA Sequencing. Top gene ontology and canonical pathways enriched for differential expression signals between Cre− and Cre+ mice were identified by GSEA. Minus sign for down-regulation; Positive sign for up-regulation. i, Collagen IV-positive (Red) basement membrane and lectin-positive endothelium (Green) in the cortex of iE4/Cre mice (Cre−, n=3; Cre+, n=4). Scale bar, 50 μm. j, Collagen IV levels in iE4/Cre mice (Cre−, n=11; Cre+, n=12) examined by ELISA. *, P=0.045. Data represent mean ± s.e.m. N.S., not significant, two-tailed Student's t-test.
To examine how apoE isoforms affect vascular functions, we next measured the cerebral blood flow (CBF) velocity in the cortical arterioles, capillaries, and venules in mice at 6-7 months of age by two-photon imaging (Supplementary Fig. 2). Interestingly, the expression of apoE4 in the liver dramatically suppressed CBF in the arterioles but not in the venules or capillaries (Fig. 2d; Extended Data Fig. 3a). Vascular tone dysregulation has been shown to cause reduced cerebrovascular reactivity and regional hypoperfusion, which can further promote brain Aβ accumulation5. We thus measured the oscillation of cerebral vessel tone by two-photon imaging. Importantly, we found that iE4/Cre+ mice exhibited a lower frequency but higher amplitude in vasodilation in the cerebral arterioles (Fig. 2e), which may contribute to reduced arterial CBF and impaired autoregulation. In addition, a significant reduction in the number of vessel branches was observed in iE4/Cre+ mice (Fig. 2f). No alteration was observed in vessel branch density and CBF of iE4/Cre+ mice at 2 months of age (Extended Data Fig. 3b-d), indicating that such change was not due to developmental defects. In contrast, we did not observe differences in the BBB permeability, CBF, or arteriolar oscillation in iE3/Cre+ mice (Extended Data Fig. 3e-j). Together, these results demonstrate that apoE4 expression in the periphery negatively impacts brain function likely by impairing cerebrovascular integrity and functions.
We next examined the impacts of peripheral apoE expression on synaptic integrity and neuroinflammation. No significant changes in the PSD-95 and synaptophysin levels were detected in the brains of experimental mice (Supplementary Fig. 3a). The GFAP-positive astrogliosis was significantly increased in the iE4/Cre+ mice, whereas it was decreased in iE3/Cre+ mice (Fig. 2g; Supplementary Fig. 3a, b). Interestingly, Iba1-positive microgliosis was significantly reduced in iE3/Cre+ but not in iE4/Cre+ mice (Supplementary Fig. 3c, d). Furthermore, the increased gliosis in iE4/Cre+ mice was most prominent in regions proximal to cerebral vessels shown by co-immunostaining for astrocytes (anti-GFAP), microglia (anti-Iba1) or perivascular macrophages (anti-CD206) (Supplementary Fig. 4). These results suggest that expression of apoE4 in the periphery promotes BBB dysfunction and vessel-associated gliosis.
Effects of peripheral apoE isoforms on brain transcriptomic signatures
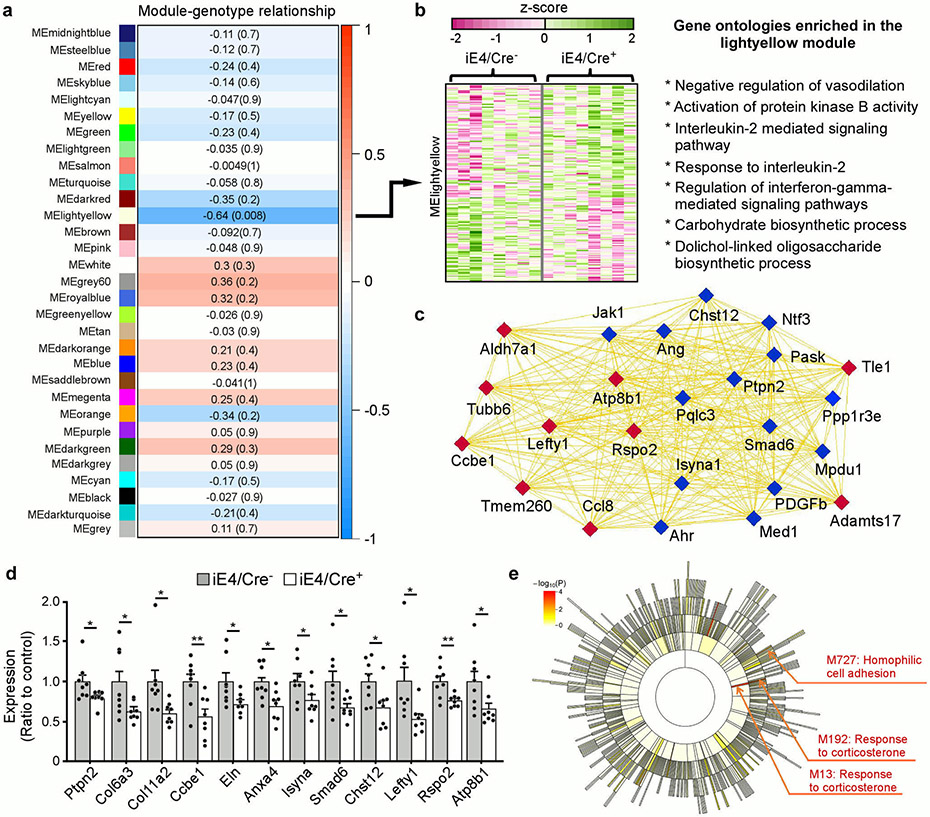
To further address the molecular mechanisms by which liver-expressed apoE affects brain functions, we performed RNA-Seq analysis to profile the transcriptional changes in mouse brains. Weighted gene correlation network analysis (WGCNA) showed that the cyan module has a significant positive correlation with peripheral apoE3 expression in which multiple pathways including WAVE/SCAR actin nucleation complex, cell-cell adhesion, and galactose metabolism were enriched (Supplementary Fig. 5a, b). Specifically, peripheral apoE3 expression was associated with the regulation of Pcdha gene clusters (Supplementary Fig. 5c), encoding a family of cell surface proteins that play pivotal roles in synaptic connections, learning, and memory22. More importantly, WGCNA revealed a critical association of peripheral apoE4 expression with the regulation of vasodilation, cell survival, and energy homeostasis (Extended Data Fig. 4a, b). The light yellow module (MElightyellow) which contained multiple genes important for vascular endothelial cell functions and BBB integrity (e.g., Ang, Ptpn-2, Pdgfb and Ahr)23-25 was found to be down-regulated in iE4/Cre+ mice (Extended Data Fig. 4c, d). Notably, Atp8b1, a hub gene in the module, encodes a BBB microvessel-enriched protein that mediates lipid metabolism. These results suggest that peripheral expression of apoE4 may impact brain vascular functions by affecting endothelial integrity. Interestingly, MElightyellow also contained genes linked to carbohydrate biosynthetic and metabolic processes (e.g., Mpdu1 Chst12, Isyna1, Pask, Aldh7a1), implying that dysregulation of cerebrovascular functions may disrupt energy homeostasis in the brain. To further identify coordinated changes at the pathway and network levels, we performed Gene Set Enrichment Analysis (GSEA)26 to screen for “enriched” gene ontology (GO) terms. The B-cell apoptotic process and endonuclease activity, which may be related to the regulation of immune responses and tissue/vascular remodeling, were up-regulated, whereas extracellular matrix component (ECM), basement membranes (BMs), collagens and integrin pathways were down-regulated in the brain of iE4/Cre+ mice (Fig. 2h). In particular, the expression of collagen family members (e.g., Col6a3, Col11a2) and elastin, the dominant ECM components in the arterial wall regulating blood flow, were down-regulated in iE4/Cre+ mice (Extended Data Fig. 4d). Moreover, we observed that collagen IV, a major component of the BMs, was more fragmented and reduced in iE4/Cre+ mice (Fig. 2i, j). Since endothelial and parenchymal cells secrete ECM proteins to generate and maintain the BMs, which directly regulate barrier integrity27, these observations together support that peripheral apoE4 negatively impacts brain functions by impairing BBB integrity.
Furthermore, we employed multiscale embedded gene co-expression network analysis (MEGENA)28 to explicitly build up gene co-expression networks from the RNA-Seq data to analyze how components of different pathways interact. Importantly, we found that the MEGENA modules related to hemophilic cell adhesion and responses to corticosterone were significantly up-regulated in iE4/Cre+ mice (Extended Data Fig. 4e), suggesting that increased neuroinflammation by peripheral apoE4 might contribute to vascular remodeling, conditioning the brain to be more susceptible to stress29,30.
Peripheral apoE4 promotes immune responses in the endothelial cells and increases vessel-associated gliosis
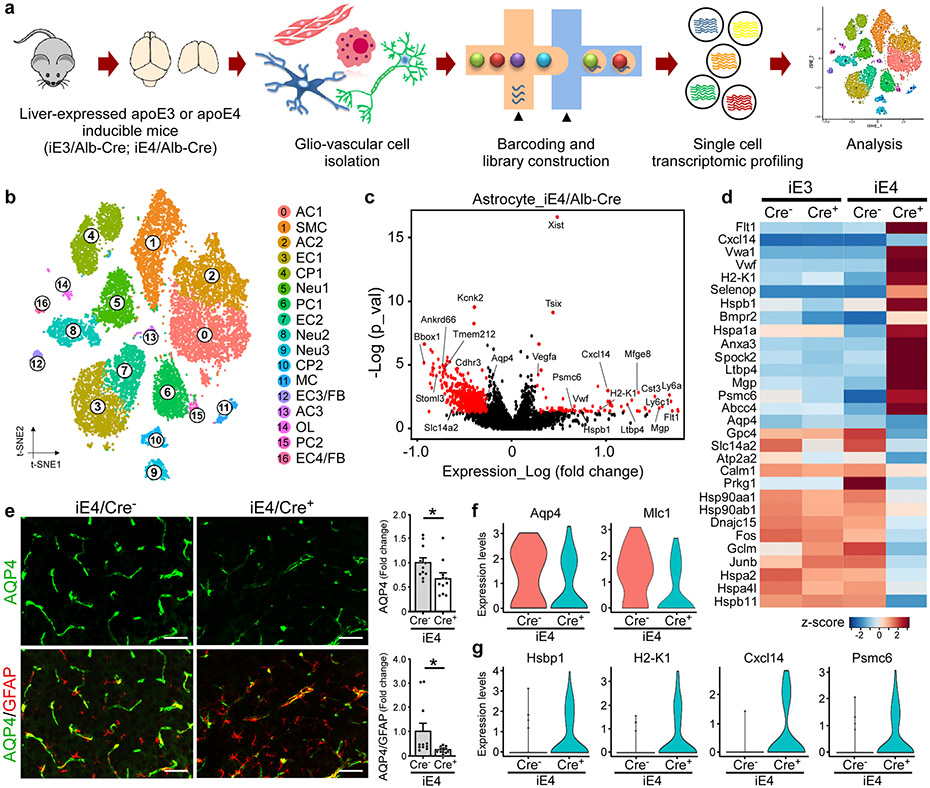
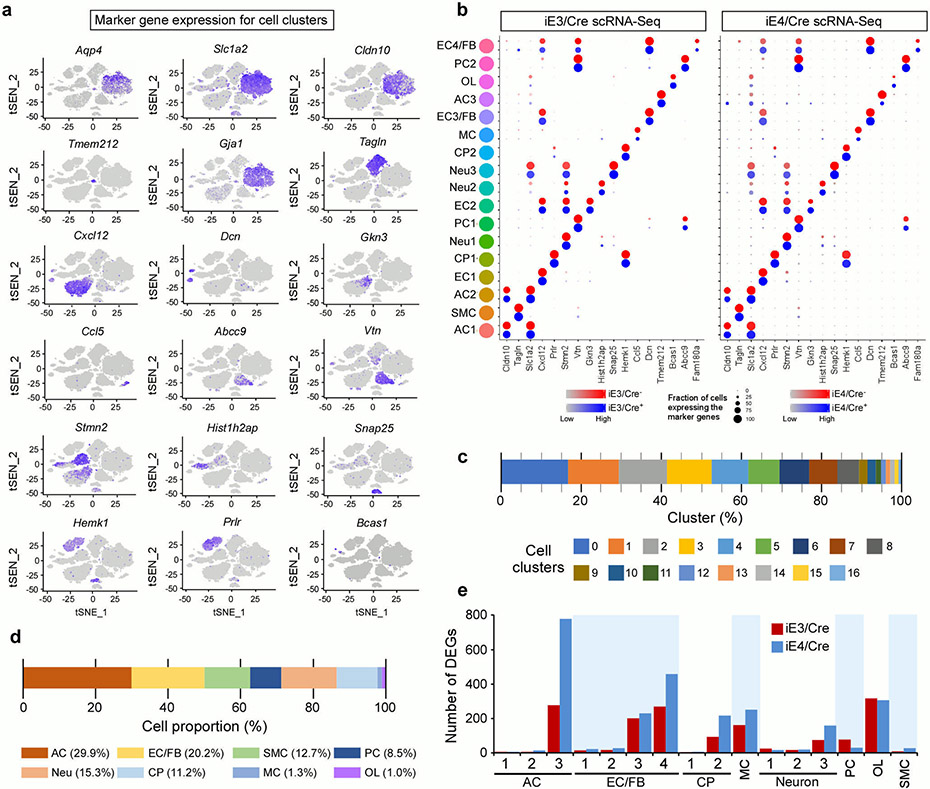
To further examine how liver-expressed apoE isoforms impact the transcriptomic changes within the glio-vascular unit, we performed single cell RNA-sequencing (scRNA-seq) on vascular cell-enriched cell population isolated from the brains of iE3/Cre and iE4/Cre mice (Fig. 3a). We first performed canonical correlation analysis of all four groups (i.e., iE3/Cre−, iE3/Cre+, iE4/Cre− and iE4/Cre+ mice) to identify common sources of variation, followed by integration analysis to identify cell clusters and conserved cell type markers (Extended Data Fig. 5a)32-34. The same cell clusters were identified in all four groups in which conserved cell markers were observed (Extended Data Fig. 5b). In total, seventeen cell clusters were identified, including astrocytes, endothelial cells, smooth muscle cells, pericytes, choroid plexus cells, neurons, oligodendrocytes, and myeloid cells (Fig. 3b; Extended Data Fig. 5c, d). We found that astrocyte, endothelial cell, and oligodendrocyte clusters had the most differentially expressed genes (DEGs), in particular the astrocyte cluster 3 (AC3) and endothelial cell clusters 3 and 4 (EC3 and EC4) (Extended Data Fig. 5e). EC3 and EC4, which are responsive to the peripheral apoE isoform expression, contained genes such as Icam1, Vcam1, Nr2f2, Cgnl1, Mgp, and decorin (Dcn), the latter is an ECM proteoglycan produced by endothelial cells in inflammation-associated angiogenesis32. As Dcn is expressed by stressed vascular endothelial cells as well as fibroblasts (FB), these two cell clusters were termed EC3/FB and EC4/FB.
Fig. 3 ∣. Vascular enriched scRNA-Seq reveals reduced astrocytic endfeet and increased immune responses associated with liver-specific expression of apoE4.
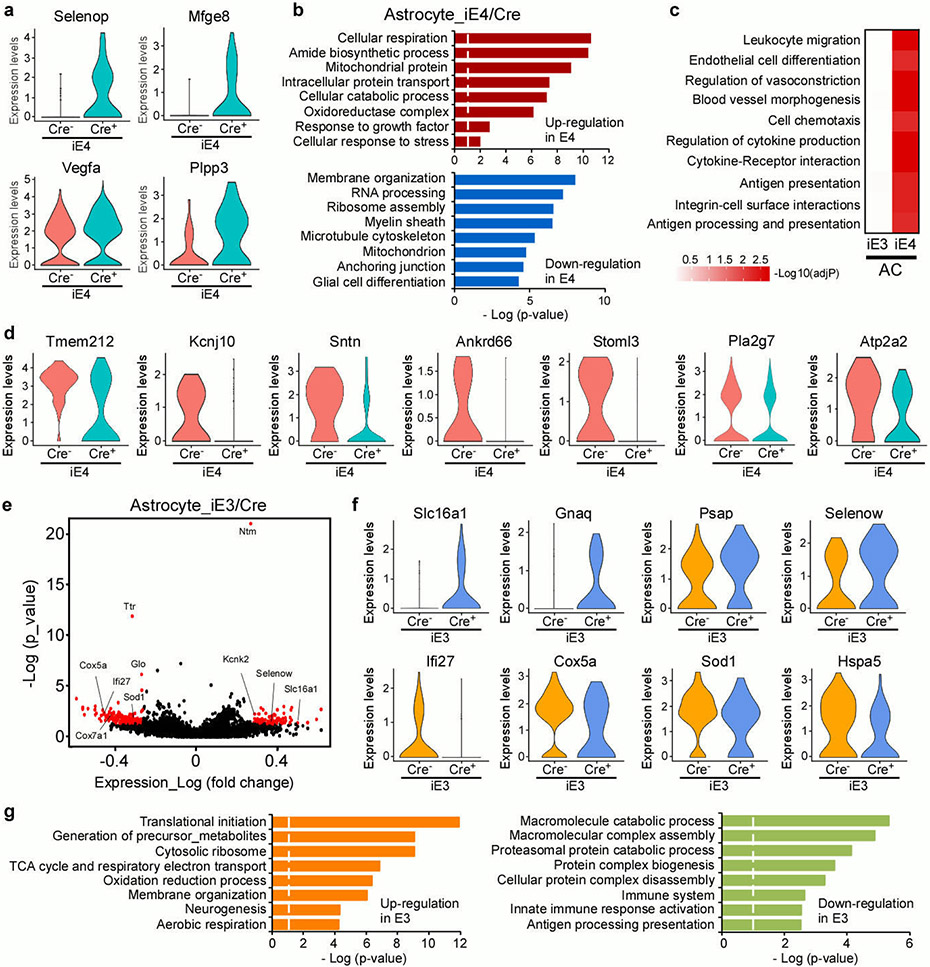
Brain cortical tissues from iE3/Cre and iE4/Cre mice (n=4/genotype) at 12-13 months of age were subjected to vascular and glial cell-enriched single cell RNA-Sequencing (scRNA-Seq). a, Diagram showing an overview of the scRNA-seq experiment. b, The t-SNE plot showing the clusters of single cell events captured in scRNA-seq. c, Volcano plot depicting up- and down-regulated genes in astrocytic cell populations (AC1, AC2, AC3) in iE4/Cre+ mice compared to iE4/Cre− control mice. Genes significant at the P value ≤ 0.05 and fold change ≥ 1.2 are denoted red in color. d, Heatmap revealing the scaled expression of differentially expressed genes in astrocytes from iE3/Cre and iE4/Cre mice. e, Brain sections from iE4/Cre− or iE4/Cre+ mice (Cre−, n=11; Cre+, n=11) at 12-13 months of age were immunostained with anti-AQP4 (green) and anti-GFAP (red) antibodies. The AQP4 immunoreactivity with or without normalization to GFAP signal was quantified. Scale bar, 100 μm. Data represent mean ± s.e.m. AQP4: *, P = 0.036; AQP4/GFAP: *, P = 0.038, two-tailed Student's t-test. f, g, Violin plots showing the mean and variance differences between iE4/Cre− and iE4/Cre+ astrocytes for genes regulating astrocyte endfeet (Aqp4 and Mlc1), innate immune response (H2-K1, Cxcl14 and Psmc6), and hypoxia/stress responses (Hsbp1).
In the astrocyte clusters from iE4/Cre+ mice, we observed an up-regulation of genes involved in innate immunity (Cxcl14, Cst3, Ly6c1, Ly6a), antigen-processing (H2-K1, Psmc6), VEGF signaling (Vegfα, Flt1), matrix remodeling and migration (Vwa1, Mgp, Spock2), oxidative stress (Hspb1, Selenop), and TGF-β signaling (Bmpr2, Ltbp4) (Fig. 3c, d, f, g; Extended Data Fig. 6a). Genes involved in synapse modifying function (Gpc4), energy homeostasis (Atp2a2, Slc1a4), and astrocyte endfeet (Aqp4) were down-regulated (Fig. 3f). GO term enrichment and GSEA analyses revealed common perturbed pathways in iE4 astrocyte clusters, including anchoring junction, regulation of vasoconstriction, cytokine-receptor interaction, and leukocyte migration (Extended Data Fig. 6b, c). Importantly, multiple genes previously identified in astrocyte endfeet transcriptomes (e.g., Aqp4, Tmem212, Mlc1, Kcnj10, Stoml3, Ankrd66, and Sntn) were down-regulated in the iE4/Cre+ mice (Fig. 3f; Extended Data Fig. 6d). As a robust association between astrocytic endfeet and the vasculature is vital for the BBB integrity, we performed immunostaining for marker of astrocyte endfeet, AQP4, and confirmed the reduction of AQP4 in iE4/Cre+ mice (Fig. 3e). Together, these findings indicate that peripheral apoE4 expression leads to transcriptional changes of genes involved in perivascular astrocyte endfeet and inflammatory responses within glio-vascular unit. Interestingly, peripheral apoE3 expression regulated transcriptomic alterations in energy homeostasis, cellular metabolism, and brain immune response-related genes in astrocytes (Extended Data Fig. 6e-g).
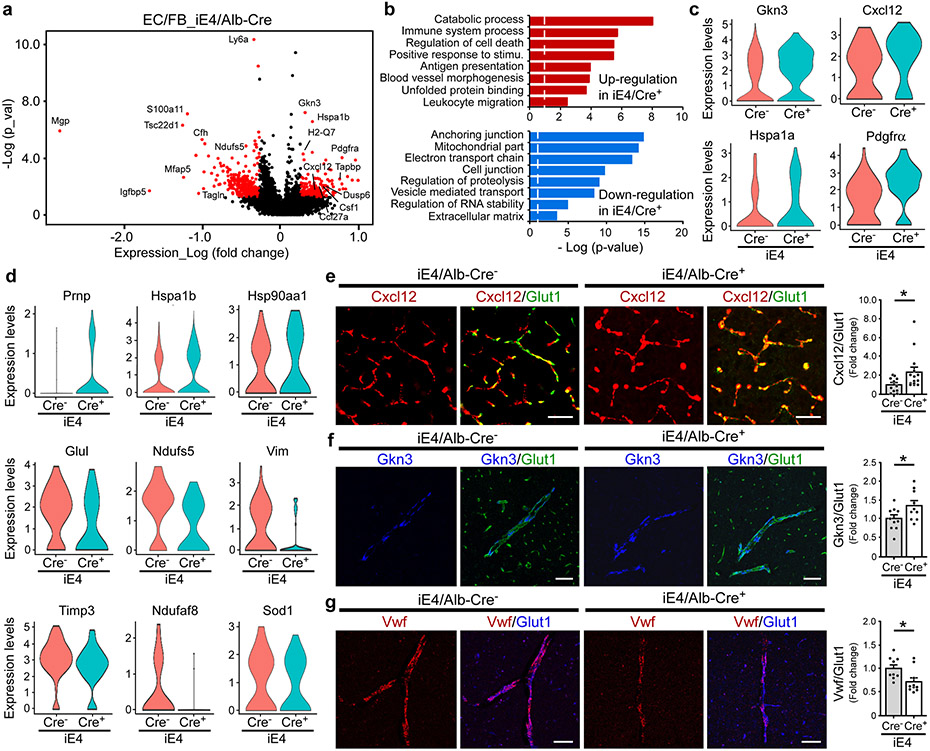
A recent study showed that brain endothelial cells can sense and relay signals between the blood and the brain34. The scRNA-Seq analysis showed that genes involved in IGF signaling (Csnk2a1, Ptpn11), tight junction formation (Vcl, Tcf7l2, Prkacb), actin cytoskeleton signaling (Arpc1b, Rhob), and maintenance of endothelial barrier function (Fgfbp1, Vasp) were up-regulated in the endothelial cells of iE3/Cre+ mice (Supplementary Fig. 6a, b). Major pathways modulated by apoE3 expression included cell junction, basement membrane, translational initiation, immune system processes, and cell death. (Supplementary Fig. 6c). Notably, genes mediating immune response (Ccl27a, Dusp1, Gkn3, Csf-1, Cxcl12), antigen presentation (H2-Q7, H2-T23, Tapbp), cell proliferation (Pdgfrα), and hypoxia/stress response (Hspa1a, Hspa1b, Hsp90aa1) were up-regulated in the endothelial cells of iE4/Cre+ mice (Fig. 4a, c, d). GO term enrichment analysis indicated that pathways involved in immune processes, antigen processing and presentation, unfolded protein binding, and leukocyte migration pathways were up-regulated, whereas pathways associated with anchoring junction, mitochondrial function, and extracellular matrix were down-regulated in the endothelial cells of iE4/Cre+ mice (Fig. 4b). Cxcl12 is a critical chemoattractant induced by hypoxia and contributes to the regulation of immune responses in the CNS35. Vwf was shown to play an important role in regulating BBB permeability in response to peripheral inflammatory stimuli36. Using immunostaining, we confirmed that Cxcl12 was up-regulated in the vessels of iE4/Cre+ mice (Fig. 4e). In addition, we examined the expression of Gkn3 and Vwf by in situ hybridization along with immunostaining for Glut1. We validated that Gkn3 was up-regulated, whereas Vwf was down-regulated in the vessel of iE4/Cre+ mice (Fig. 4f, g). Together, our data suggest that apoE4 in the periphery promotes immune responses in the brain endothelial cells along with an increase of vessel-associated gliosis, which potentially compromises barrier integrity and cerebrovascular functions.
Fig. 4 ∣. Transcriptional signatures in brain endothelial cells associated with liver-specific expression of apoE4.
a, Volcano plots illustrating up- or down-regulated genes in the endothelial cell populations (EC1, EC2, EC3/FB, EC4/FB) from iE4/Cre mice (Cre+ vs. Cre−). Genes significant at the P value ≤ 0.05 and fold change ≥ 1.2 are denoted red in color. b, Gene ontology analyses for up- or down-regulated genes in the endothelial cell populations of the iE4/Cre mice c, d, Violin plots showing the mean and variance difference between iE4/Cre− and iE4/Cre+ endothelial cells for genes regulating inflammatory response (Cxcl12 and Gkn3), cell proliferation (Pdgfrα), lactate homeostasis (Glu1), mitochondrial function (Ndufs5 and Ndufaf8), matrix assembly (Vim), oxidative stress (Prnp), regulators of MMP family (Timp3), and hypoxia/stress response (Sod1, Hspa1a, Hspa1b, and Hsp90aa1). e, Brain sections from iE4/Cre− or iE4/Cre+ mice (Cre−, n=11; Cre+, n=14) at 12-13 months of age were subjected to co-immunostaining for Cxcl12 (red) and Glut1 (green), and the Cxcl12/Glut1 signals were quantified. *, P = 0.036, two-tailed Student's t-test. Scale bar, 50 μm. f, Brain sections from the iE4/Cre− or iE4/Cre+ mice (Cre−, n=10; Cre+, n=10) at 12-13 months of age were subjected to RNAScope assay for Gkn3 (blue) and co-immunostaining for Glut1 (green). The Gkn3/Glut1 signal was quantified. *, P = 0.039. Scale bar, 50 μm. g, Brain sections from iE4/Cre− or iE4/Cre+ mice (Cre−, n=9; Cre+, n=10) at 12-13 months of age were subjected to RNAScope analysis for Vwf (red) and co-immunostaining for Glut1 (blue). The Vwf/Glut1 signal was quantified. *, P = 0.024. Scale bar, 50 μm. Data represent mean ± s.e.m.
Peripheral expression of apoE isoforms alters plasma proteomes
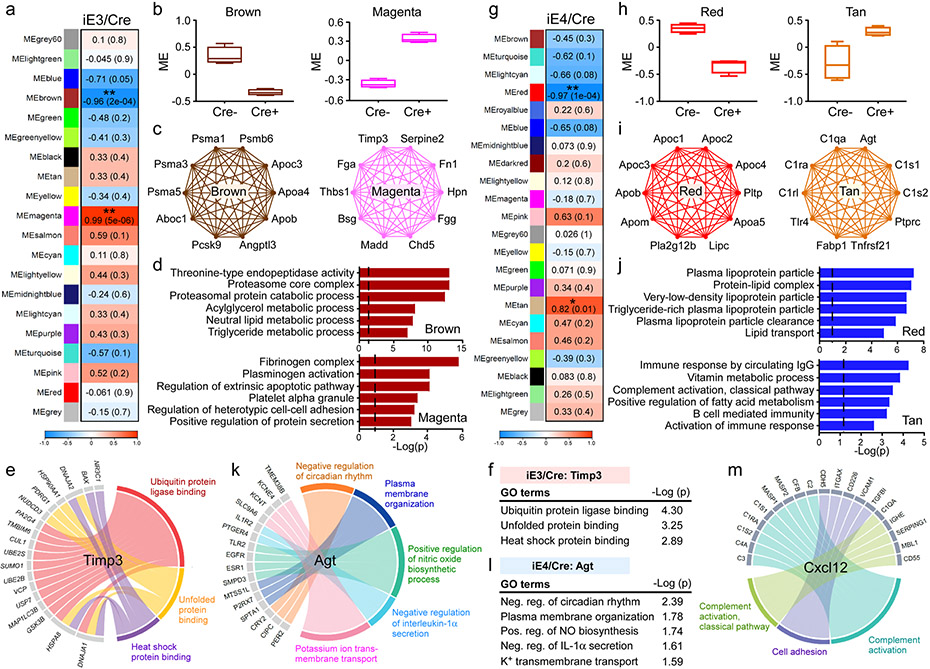
To identify potential blood factors mediating the apoE isoform effects on cerebrovasculature and brain function, we performed unbiased proteomic analysis of the plasma from iE3/Cre and iE4/Cre mice (Supplementary Table 1). With WGCNA analysis, we identified 20 co-expression modules, among which two were significantly associated with apoE3 plasma (Fig. 5a). Specifically, the magenta module was significantly up-regulated, whereas the brown module was down-regulated in iE3/Cre+ mice compared to iE3/Cre− mice (Fig. 5b). Functional annotation showed that proteins in the brown module (e.g., Psma1, Psma3, Aboc1, Apob) were associated with the proteasomal protein catabolic and lipid metabolic processes (Fig. 5c, d). The magenta module containing hub proteins such as Timp3, Bsg, Thbs1, Serpine2, Hpn, and Tgfb1 was enriched for fibrinogen complex, regulation of protein secretion, and cell-cell adhesion pathways (Fig. 5c, d). Timp3 is an ECM-binding protein that modulates the turnover of ECM through inhibiting the activity of metalloproteinases (MMPs)37,38. Thus, the increased Timp3 might be a mechanism contributing to improved vascular health and brain function of iE3/Cre+ mice39. Interestingly, the regulatory network analysis found a negative association between plasma Timp3 and ubiquitin protein ligase binding, unfolded protein, and heat shock protein binding in the endothelial cells of iE3/Cre+ mice (Fig. 5e, f), implying a down-regulation of stress responses.
Fig. 5 ∣. ApoE isoform-specific changes in plasma protein network and the association with vascular related pathway.
Plasma from iE3/Cre (a-f) mice (Cre−, n=4; Cre+, n=4), and iE4/Cre (g-m) mice (Cre−, n=4; Cre+, n=4) were subjected to proteomics analysis. The protein modules (MEs) associated with APOE3 or APOE4 genotype were identified by WGCNA analysis. a, g, Module-genotype correlation. Numbers in the table indicate the correlations of the corresponding module eigengenes and genotype, with the P values shown in parentheses. The red color represents positive correlation and blue represents negative correlation to APOE genotype. b, h, Modules that are significantly correlated with APOE3 (b) or APOE4 (h) genotype. The box in the plots displays 25th and 75th percentile values. The center line represents the median and the whiskers show minimum and maximum values. c, i, Network plots of the proteins with high intramodular connectivity in the brown and magenta modules in the iE3/Cre group (c) and red and tan modules in the iE4/Cre group (i). d, j, Pathway analysis using gene ontology (GO) showing the pathways enriched in the protein-sets of the brown and magenta modules in the iE3/Cre group (d), as well as red and tan modules in the iE4/Cre group (j). Limma’s moderate t-test statistics were used. e, Circos plot showing the associated genes and pathways in the endothelial cell clusters negatively correlated with the changes of plasma Timp3 in the iE3/Cre group. f, Timp3-regulated pathways in the endothelial cell clusters of the iE3/Cre group. k, Circos plot showing the associated genes and pathways in the endothelial cell clusters negatively correlated with the changes of plasma Agt in the iE4/Cre group. l, Agt-regulated pathways in the endothelial cell clusters of the iE4/Cre group. Limma’s moderate t-test statistics were used. m, Circos plot showing the associated proteins and pathways in the plasma correlated with the changes of Cxcl12 in endothelial cell clusters in the iE4/Cre group.
Among the co-expression modules associated with apoE4, the red module was down-regulated, whereas the tan module was up-regulated in iE4/Cre+ mice compared to iE4/Cre− mice (Fig. 5g, h). Hub genes in the red module included Apob, Apoc1, Apoc3, Apom, and Lipc which were involved in lipoprotein metabolism (Fig. 5i). Pathway analysis showed that this module was enriched for plasma lipoprotein particles, lipid transport, and clearance (Fig. 5j). Furthermore, the tan module, enriched for immune response, complement activation (e.g., C1qa, C1ra, C1s1, and C1rl), and fatty acid metabolism (e.g., Agt and Fabp1), was up-regulated in iE4/Cre+ mice (Fig. 5i, j). The regulatory network analysis found a negative association between plasma angiotensinogen (Agt), known to regulate blood pressure, and pathways related to the regulations of interleukin-1α (IL-1α) secretion, circadian rhythm, nitric oxide biosynthesis, and plasma membrane organization in the endothelial cells of iE4/Cre+ mice (Fig. 5k, l). Furthermore, a positive regulation of complement activation in the plasma was correlated with Cxcl12 which was increased in the endothelial cells of iE4/Cre+ mice (Fig. 5m).
APOE genotype-specific effects of young plasma on brain function
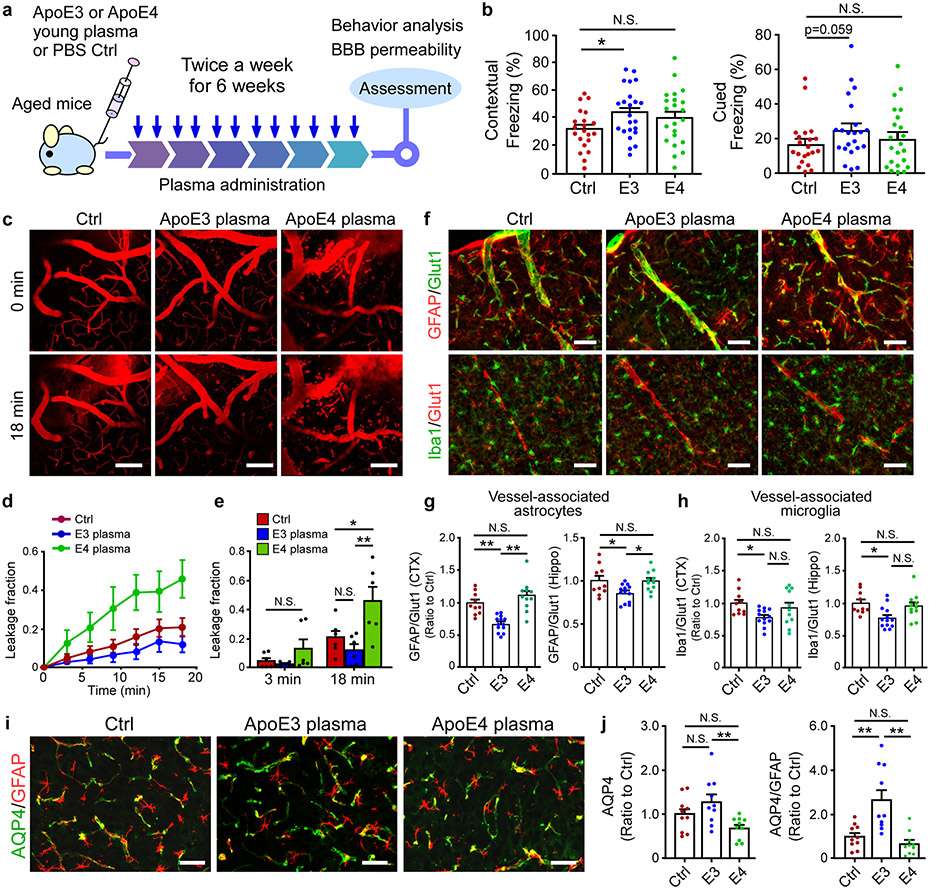
Recent studies showed that exposure to young blood counteracts age-related cognitive decline with the brain vasculature capable of sensing and transmitting the circulatory cues to the brain34,40,41. To examine how young plasma with different human apoE isoforms affects age-associated cognitive functions, plasma from young apoE3 or apoE4 mice was intravenously administrated into aged wild-type mice (Fig. 6a). Intriguingly, apoE3, but not apoE4 young plasma, improved associative memory of the aged animals assessed by contextual fear conditioning test compared to control treatment (Fig. 6b). It is possible that the negative effects of apoE4 offset the beneficial effects of young plasma. We next investigated whether BBB integrity was differentially modulated by peripheral young plasma treatment. As expected, aged animals treated with PBS exhibited apparent BBB impairment (Fig. 6c-e). Remarkably, exposure to young apoE4 plasma exacerbated BBB leakage, whereas young plasma from apoE3 mice trended to improve BBB integrity (Fig. 6c-e). The vessel-associated astrocytes and microglia were reduced in mice treated with apoE3 plasma, in particular in the cortex (Fig. 6f-h, Extended Data Fig. 7a-c). Intriguingly, apoE3 young plasma treatment increased the level of AQP4, whereas apoE4 young plasma did not have an effect (Fig. 6i, j). The levels of TJ proteins ZO1 and CLDN5 were increased in aged mice treated with apoE3, but not apoE4, young plasma (Extended Data Fig. 7d-g). The vessel-associated fibrinogen deposition was decreased in apoE3, but not apoE4, young plasma-treated mice (Extended Data Fig. 7h, i). Finally, young plasma treatment did not impact CD13-positive pericyte coverage in aged animals regardless of apoE isoform (Extended Data Fig. 7j, k). In summary, our results indicated that young plasma affects brain cognition and BBB functions in an apoE isoform-dependent manner.
Fig. 6 ∣. Differential effects of apoE3 and apoE4 young plasma on cerebrovascular integrity and cognitive function in aged mice.
a, Schematic illustration of the experimental procedures for young plasma treatment. b, Memory performance of mice treated with Ctrl, or young plasma from apoE3-TR or apoE4-TR mice examined by fear conditioning contextual test (Ctrl: n=21; E3: n=23; E4: n=22), and cued test (Ctrl: n=21; E3: n=22; E4: n=22). The percentage of time spent showing freezing behavior in response to stimulus in contextual or cued test is shown. *, P=0.046, Wilcoxon rank sum test. c-e, Two-photon imaging was used to assess BBB integrity in 28-month-old mice (n=3/group) treated with Ctrl, apoE3 or apoE4 young plasma. Mice were intravenously injected with Texas Red conjugated dextran (40 kDa), and the BBB permeability was examined. Scale bar, 100 μm. d, The fractions of BBB leakage over time are shown. e, The fractions of BBB leakage at 3 or 18 min after injection of dextran were quantified. *, P = 0.01; **, P = 0.0006. f, g, Vessel-associated astrocytes or microglia in the cortex were examined by double labeling with anti-GFAP (astrocyte) or anti-Iba1 (for microglia) together with endothelial marker Glut1 antibody (for visualizing vasculature). Representative images of cortex from experimental mice are shown. Scale bar, 50 μm. Data from individual animals are shown. g, Fluorescence intensity for GFAP signals surrounding vasculature in the cortex (Ctrl vs. E3: **, P = 0.0009; E3 vs. E4: **, P<0.0001) or hippocampus (Ctrl vs. E3: *, P=0.04; E3 vs. E4: *, P=0.039) of mice (Ctrl: n=10; E3: n=13; E4: n=11) was quantified. h, Fluorescence intensity for Iba1 signals surrounding vasculature in the cortex (*, P=0.045) or hippocampus (*, P=0.017) of mice (Ctrl: n=10; E3: n=13; E4: n=11) was quantified. i, j, Brain sections from young plasma-treated mice (10 mice/group) were immunostained with anti-AQP4 (green) and anti-GFAP (red) antibodies. The AQP4 immunoreactivity (**, P=0.008) and AQP4/GFAP signals (Ctrl vs. E3: **, P=0.002; E3 vs. E4: **, P=0.0002) was quantified. Scale bar, 50 μm. Data represent mean ± s.e.m. One-way ANOVA with a Tukey’s post-hoc test.
ApoE4 diminished the beneficial effect of young plasma on the integrity of human brain endothelial cells
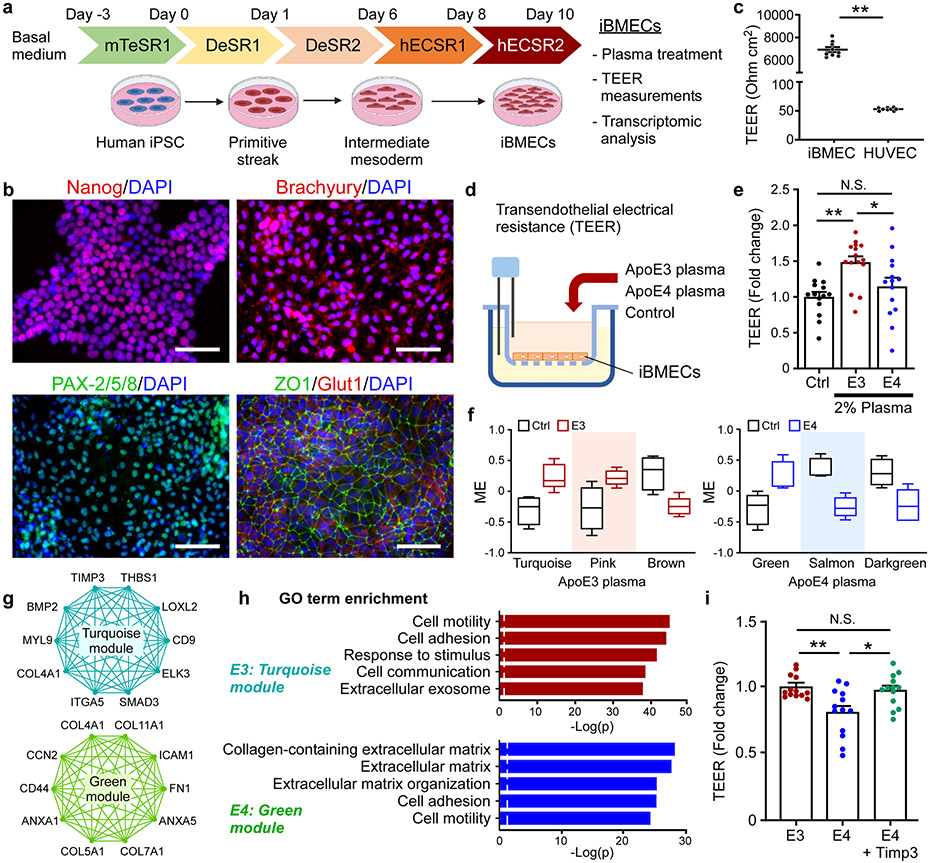
To explore how brain endothelial cells respond to the apoE-regulated blood factors, we established a model of brain microvascular endothelial cells (BMECs) differentiated from human induced pluripotent stem cells (iPSCs) (Fig. 7a). The markers for the pluripotency of human iPSCs, mesodermal differentiation, and endothelial cells (ZO1 and Glut1) were validated (Fig. 7a, b). Furthermore, human iBMECs showed significantly higher trans-endothelial electrical resistance (TEER) values compared to human umbilical vein endothelial cells (HUVECs), indicating the formation of robust endothelial junctions (Fig. 7c). To investigate the effect of young plasma with different apoE isoforms on the endothelial barrier integrity, iBMECs were treated with control (PBS-citrate), apoE3, or apoE4 young plasma for 24 hours (Fig. 7d). Remarkably, compared to the control, we observed a significant increase of the TEER values in the apoE3, but not in the apoE4, young plasma-treated group (Fig. 7e). This result indicates that apoE3 young plasma increased the barrier integrity of iBMECs, while the apoE4 young plasma failed to do so. To further investigate the associated molecular changes, plasma-treated human iBMECs were subjected to RNA-seq, followed by WGCNA analysis. Two modules (turquoise and pink) were significantly up-regulated, and two modules (brown and purple) were down-regulated in apoE3-treated cells (Supplementary Fig. 7a; Fig. 7f). Three modules were correlated with apoE4, including one up-regulated (green) and two down-regulated modules (darkgreen and salmon) (Supplementary Fig. 7d; Fig. 7f). To further evaluate the functional changes of iBMECs associated with apoE3 or apoE4 plasma treatment, we performed GO term enrichment analysis on these modules. The modules associated apoE3 plasma were enriched for various cellular responses, such as endosome organization, cell division, and double-strand break repair (Supplementary Fig. 7b, c). Importantly, genes in the turquoise module including TIMP3, THBS1, CD9, ITGA5, and ELK3, were enriched for cell adhesion, cell communication, and extracellular exosome pathways (Fig. 7g, h). The modules associated with apoE4 plasma treatment were enriched for lipid biosynthetic and metabolic processes, and ribosome biogenesis (Supplementary Fig. 7e, f). Additionally, genes in green module associated with apoE4 plasma treatment were enriched for collagen-containing ECM, and cell mobility, with hub genes including COL4A1, COL11A1, ICAM1, ANXA1, CD44 and FN1 (Fig. 7g, h), suggesting that peripheral apoE4 may affect vascular function through regulating ECM and endothelial junction integrity. Given that Timp3 was up-regulated in the plasma of iE3/Cre+ mice (Fig. 5d), we next examined whether addition of Timp3 can modulate apoE4-mediated endothelial barrier integrity. Interestingly, co-treatment of Timp3 and apoE4 young plasma significantly increased the TEER compared to apoE4 plasma treatment alone (Fig. 7i). This result implies that apoE4 young plasma might have a compromised ability to support the endothelial barrier integrity via influencing Timp3-mediated pathway and/or associated ECM organization.
Fig. 7 ∣. ApoE isoform-specific effects of young plasma on human endothelial barrier integrity and transcriptomic signature.
a, Schematic diagram of human brain endothelial cell (iBMEC) differentiation from iPSCs and the experimental design. b, Characterization of brain endothelial cell differentiation at different time points. Similar results were observed in at least three independent experiments. The iPSC pluripotency and mesoderm lineage differentiation were confirmed by Nanog, Brachyury (a primitive streak marker), or PAX2/5/8 (an intermediate mesoderm marker) staining, respectively. The iPSC-derived iBMECs were stained for Glut1 (an endothelial marker) and ZO1 (a tight junction protein) at Day 12. Scale bars: 100 μm. c, The transendothelial electrical resistance (TEER) in iBMECs and human umbilical vein endothelial cells (HUVECs) was measured. **, P < 0.0001. d, The transwell setup in which the iBMECs were treated with control or young plasma, followed by the assessment of the endothelial cell integrity. e, Human iBMECs cultured in the transwell system were treated with PBS-citrate (control), apoE3 young plasma (2%) or apoE4 young plasma (2%) for 24 hr. The TEER values were measured and compared to the control. Data represent mean ± s.e.m. from three independent experiments. **, P=0.002; *, P=0.034; N.S., not significant. One-way ANOVA with a Tukey’s post-hoc test. f, Modules significantly correlated with apoE3 or apoE4 plasma treatment compared to control identified from RNA-Seq. The box in the plots displays 25th and 75th percentile values. The center line represents the median and the whiskers show minimum and maximum values. g, Network plots of the proteins with high intramodular connectivity in the turquoise module of apoE3 group, and in the green module of apoE4 group. h, Pathway analysis using gene ontology showing the pathways enriched in the gene-sets of the turquoise module and green module. i, The human iBMECs were treated with apoE3 or apoE4 young plasma, or apoE4 plasma together with Timp3 (0.5 μg/ml) for 24 hr, and the TEER values were measured. Data represent mean ± s.e.m. from three independent experiments. **, P=0.003; *, P=0.012; N.S., not significant. One-way ANOVA with a Tukey’s post-hoc test was used to determine the significance between groups.
Differential effects of peripheral apoE isoforms on brain amyloid
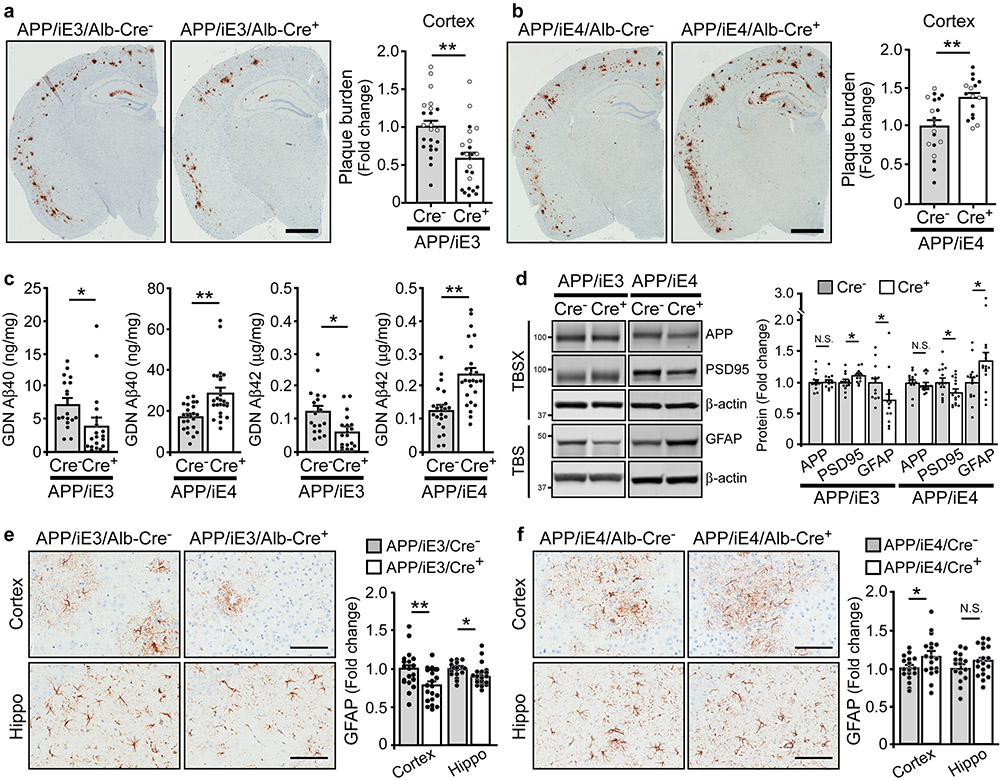
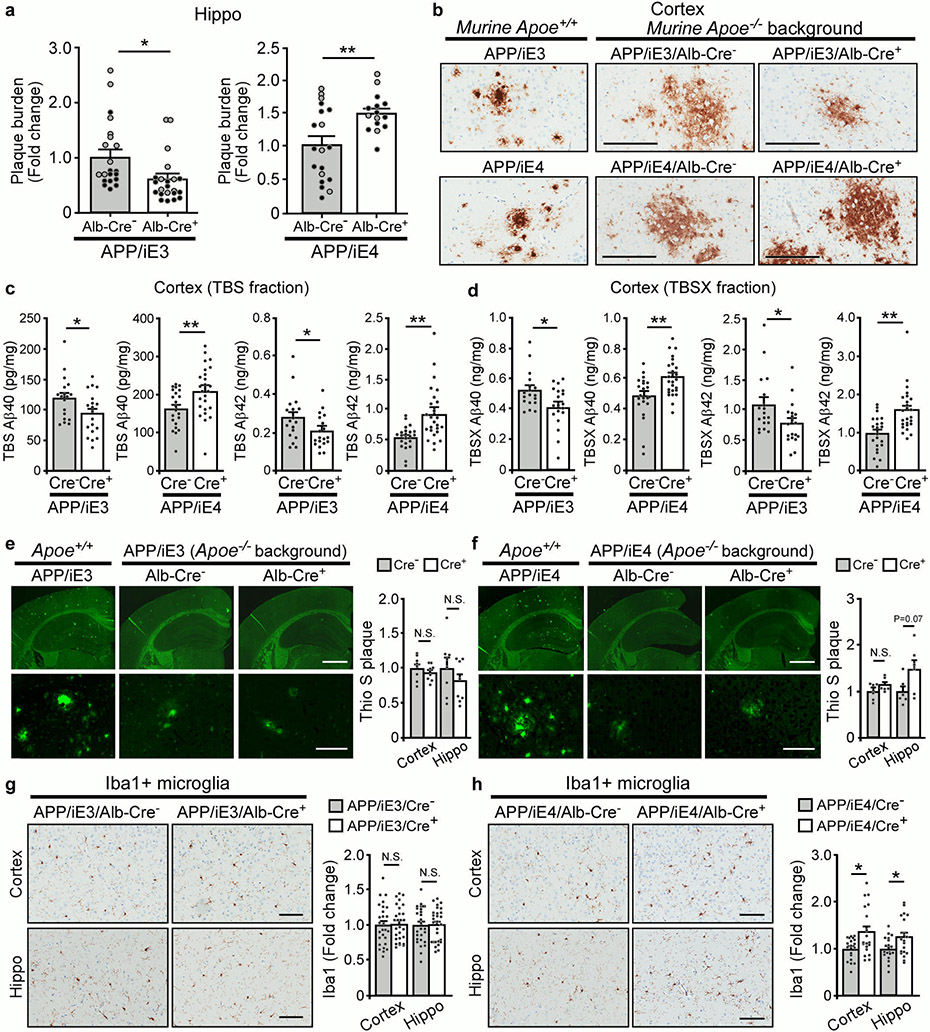
Studies show that brain amyloid deposition is apoE isoform-dependent (E4>E3≥E2)9. However, whether peripheral apoE isoforms affect amyloid pathogenesis and AD-related pathways in vivo remains unclear. We thus bred our liver-specific apoE3 and apoE4 mice with APP/PS1 mice42 in the apoe-KO background to examine their effects on amyloid pathology. Interestingly, we found that expression of apoE3 in the liver suppressed, whereas apoE4 enhanced amyloid pathology (Fig. 8a, b; Extended Data Fig. 8a, b). To analyze how liver-expressed apoE affects the dynamic pools of Aβ, we fractionated brain lysates into TBS-soluble, detergent-soluble (TBSX), and insoluble (guanidine-HCl, GDN) fractions, and quantified Aβ by ELISA43. Consistently, we found that soluble and insoluble Aβ40 and Aβ42 levels were decreased in APP/iE3/Cre+ mice but increased in APP/iE4/Cre+ mice at 9 months of age with total APP levels unchanged (Fig. 8c, d; Extended Data Fig. 8c, d). It is possible that peripheral apoE4 impairs the basement membrane proteins and endothelial functions in cerebral blood vessels which in turn disrupt the efficiency of perivascular drainage of Aβ from the brain44. The absence of apoE in the brain of peripheral apoE models led to minimal fibrillary plaques that were not different among the experimental groups examined by Thioflavin S (Thio S) staining (Extended Data Fig. 8e, f)45,46. In addition, the level of PSD-95 was slightly increased in APP/iE3/Cre+ mice but reduced in APP/iE4/Cre+ mice (Fig. 8d), suggesting their differential impacts on Aβ-mediated synaptic impairment. Also, we found a profound decrease of astrogliosis in APP/iE3/Cre+ mice, whereas an opposite effect was seen in APP/iE4/Cre+ mice (Fig. 8d-f)20,47. Microgliosis was not changed in APP/iE3/Cre+ mice, whereas a significant increase was observed in APP/iE4/Cre+ mice compared (Extended Data Fig. 8g, h). These results demonstrate that liver-expressed apoE isoforms differentially affect amyloid pathology and related gliosis.
Fig. 8 ∣. Opposing effects of liver-expressed of apoE3 and apoE4 on Aβ plaque deposition and associated neuroinflammation.
a, b, Brain sections from 9-month-old APP/iE3/Alb-Cre mice (Cre−, n=23; Cre+, n=24) and APP/iE4/Alb-Cre mice (Cre−, n=19; Cre+, n=16) were immunostained with a pan-Aβ antibody. Representative images of Aβ staining are shown, and the plaque burden in the cortex was quantified. Scale bar, 1 mm. Black circles are males; grey circles are females. Data represent mean ± s.e.m. E3: **, P=0.005; E4: **, P=0.002, two-tailed Student's t-test. c, Insoluble Aβ40 and Aβ42 levels in the cortex of 9-month-old APP/iE3/Alb-Cre mice (Cre−, n=18; Cre+, n=19) and APP/iE4/Alb-Cre mice (Cre−, n=22; Cre+, n=25) were examined by specific Aβ ELISA. E3: Aβ40 (*, P=0.03); Aβ42 (**, P=0.005). E4: Aβ40 (**, P=0.0004); Aβ42 (**, P<0.0001), two-tailed Student's t-test. d, The levels of APP, postsynaptic marker PSD-95, and GFAP in the cortex of APP/iE3/Alb-Cre mice (Cre−, n=14; Cre+, n=14) and APP/iE4/Alb-Cre mice (Cre−, n=15; Cre+, n=16) at 9 months of age were examined by Western blotting and quantified. E3: PSD-95 (*, P=0.026); GFAP (*, P=0.04). E4: PSD-95 (*, P=0.037); GFAP (*, P=0.037). e, f, Brain sections from APP/iE3/Alb-Cre mice (Cre−, n=19; Cre+, n=21) and APP/iE4/Alb-Cre mice (Cre−, n=17; Cre+, n=18) at 9 months of age were immunostained with GFAP antibody and quantified. Scale bar, 100 μm. Data represent mean ± s.e.m. E3: Cortex (**, P=0.008); Hippo (*, P=0.005). E4: Cortex (*, P=0.046). N.S., not significant, two-tailed Student's t-test.
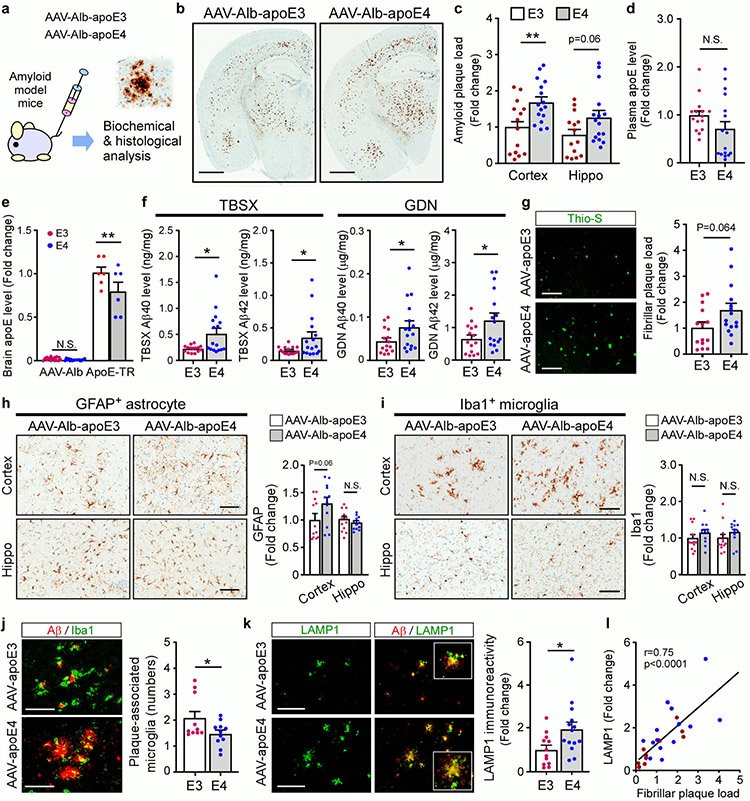
To further validate our findings in a model where brain apoE is present, we examined the effects of liver-expressed apoE isoforms on amyloid pathology in 5xFAD mice transduced with AAV-Alb-apoE3 or AAV-Alb-apoE4 viruses (Extended Data Fig. 9a). We found that AAV-Alb-apoE4 mice had increased amyloid plaque load in the cortex compared with AAV-Alb-apoE3 mice (Extended Data Fig. 9b, c). The human apoE levels in the plasma were not statistically different between mice expressing apoE3 and apoE4 though it was trending lower in apoE4 mice, while the human apoE was not detectable in the brain (Extended Data Fig. 9d, e). Both detergent-soluble and -insoluble Aβ40 and Aβ42 were higher in mice transduced with AAV-Alb-apoE4 compared to those transduced with AAV-Alb-apoE3 (Extended Data Fig. 9f). A trend of increased fibrillar plaque deposition was observed in mice expressing apoE4 in the liver (Extended Data Fig. 9g). In addition, AAV-Alb-apoE3 mice had a greater number of plaque-associated microglia than AAV-Alb-apoE4 mice, whereas the total immunoreactivity for microglia or astrocytes was not significantly different (Extended Data Fig. 9h-j). Finally, AAV-Alb-apoE4 mice had increased LAMP1-immunoreactive neuritic dystrophy that was positively correlated with fibrillar plaque load (Extended Data Fig. 9k, l). To further evaluate the peripheral apoE isoform effect, we conducted the experiments using the same viruses along with a control AAV-Alb-GFP virus (Supplementary Fig. 8a). Consistently, AAV-Alb-apoE4 mice exhibited an elevated amount of Aβ plaques compared to AAV-Alb-apoE3 mice, whereas their plaque loads were not significantly different from the control cohort (Supplementary Fig. 8b, c). The levels of astrogliosis and microgliosis in AAV-Alb-apoE4 mice were higher than those in AAV-Alb-apoE3 or control mice (Supplementary Fig. 8d-f). Together, our results indicate that peripheral apoE isoforms differentially impact brain amyloidosis, plaque-associated microglial responses, and neuritic dystrophy.
Discussion
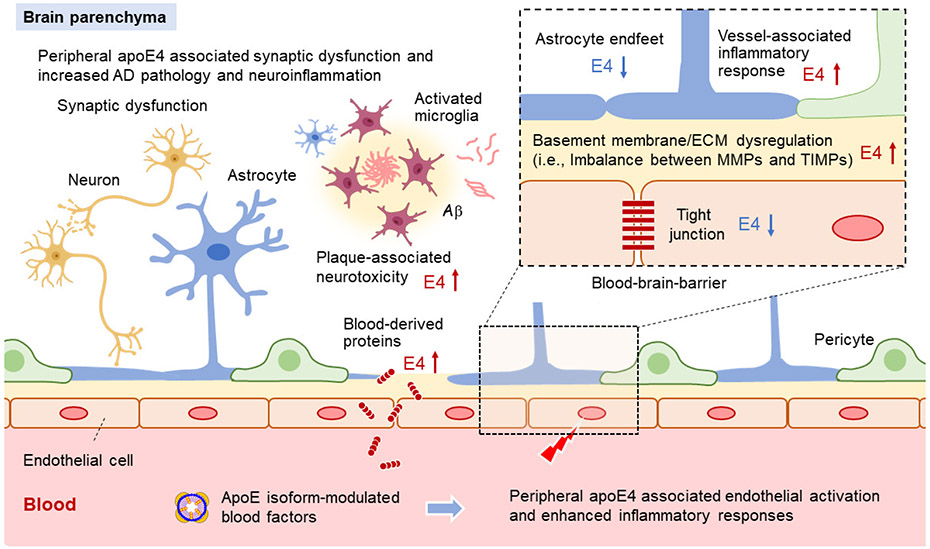
APOE4 increases AD risk by multiple pathways. Our current study shows that peripheral apoE4 is sufficient to impair brain functions and exacerbate amyloid pathogenesis. Mice with peripheral apoE4 expression have compromised cerebrovascular integrity and function, and elevated astrogliosis which may collectively contribute to behavioral impairments and accelerated AD pathogenesis. Transcriptomic profiling indicates that apoE4 expression in the liver may lead to an impairment of ECM and basement membranes contributing to endothelial dysfunction. The scRNA-Seq analysis suggests that peripheral apoE isoforms differentially affect immune responses within the glio-vascular units, which in turn can impact vascular function and BBB integrity. Conversely, peripheral expression of apoE3 improves synaptic function and memory, and ameliorates the amyloid pathology. Finally, peripheral administration of young blood plasma with different apoE isoforms shows differential effects on cognitive functions, BBB integrity, and vessel-associated inflammatory responses in aged mice. As drug targeting of apoE for AD therapy will likely involve regulating apoE both in the brain and the periphery, our studies provide mechanistic insights as to how apoE isoforms should be targeted in AD through individualized therapy.
Emerging evidence indicates that peripheral systemic factors, such as blood components, microbial metabolites, and immune cells, can modulate brain homeostasis and functions with age and in diseases48,49. Young blood infusion was found to promote memory function, hippocampal neurogenesis, and vascular remodeling in the aged brain41. A recent AMBAR clinical trial assessing the effects of plasma protein replacement therapy also showed a positive impact in reducing the cognitive decline in mild and moderate AD patients50. These studies support the therapeutic potential of modulating peripheral factors to treat age-related brain dysfunction and AD. Plasma apoE isoform levels have also been shown to correlate with regional brain volume, cerebral glucose metabolism, and possibly cognitive performance51. Our findings show that apoE3 young plasma improves cognitive function and reduces vessel-associated gliosis in aged animals. However, exposure to young apoE4 plasma impairs BBB integrity without deteriorating cognition, likely attributed to the overall beneficial effects of young plasma. It is possible that apoE isoforms influence the systemic factors in the periphery which differentially impact brain functions. Using iBMECs, we show that apoE3, but not apoE4 young plasma is able to increase endothelial junction integrity, whereas addition of exogenous Timp3, one of the blood factors up-regulated in apoE3 plasma, enhanced tight junctions in the presence of apoE4. Loss of Timp3 increases inflammation and aggravates atherosclerosis in apoe-KO mice52. Timp3 levels are decreased in cardiovascular diseases, while replenishment of Timp3 can ameliorate the disease through inhibiting excess MMP activity and inflammation37, supporting a critical role of Timp3 in regulating vascular function. A tight regulation in the levels of Timp family and MMP proteins is critical for vascular ECM remodelling37. It is possible that peripheral apoE3 expression may regulate blood factors, such as Timp3, which improves endothelial integrity and vascular health. Interestingly, LRP1, an apoE receptor, was shown to bind to Timp3 and mediate its endocytosis and degradation53. Additionally, plasma proteomic profiling reveals an up-regulation of immune response and complement upon peripheral apoE4 expression. Complement activation, which regulates the release of cytokines, vascular permeability, and leukocyte extravasation54, may contribute to the vascular dysfunction associated with apoE4. Future studies toward elucidating apoE isoform-dependent differences in plasma proteome, lipidome, and metabolome in preclinical models and plasma samples from healthy individuals and AD patients with different APOE genotypes can facilitate the identification of effective systematic peripheral factors.
Vascular dysfunction and BBB breakdown are implicated in normal aging and AD pathogenesis55. Capillary brain endothelial cells are sensitive to blood circulatory factors and are capable of transducing aging signals to the brain34. We found that peripheral apoE3 expression regulates pathways associated with energy homeostasis, cellular metabolism, cell junction, and brain immune response in glio-vascular units which may contribute to the improvement of cognitive and synaptic functions (Extended Data Fig. 10). Peripheral apoE4 expression leads to up-regulation of immune-related and stress-related pathways, and down-regulation of anchoring junction, mitochondrial function, and ECM pathways in brain endothelial cells. This suggests that peripheral expression of apoE4 may promote vascular inflammation, which transduces inflammatory cues to the brain along with activation of astrocytes and microglia. In addition, liver-expressed apoE4 results in transcriptomic changes in astrocyte endfeet, inflammatory responses, and immune cell infiltration within the glio-vascular unit (Extended Data Fig. 10). It is possible that the aberrant activation of astrocytes in iE4/Cre+ mice compromises the barrier function of perivascular astrocyte endfeet and neurotrophic functions of astrocytes. Finally, an increased accumulation of blood-derived proteins (e.g., albumin, IgG and fibrinogen) in the vessels, perivascular regions, and/or brain parenchyma around the vessels is evident in the iE4/Cre+ mice, likely due to BBB impairment. BBB dysfunction is associated with normal aging and various neurodegenerative diseases56. Fibrinogen leakage upon BBB disruption has been shown to trigger perivascular microglial clustering, microglia-dependent release of neurotoxic reactive oxygen species (ROS), synapse loss, and cognitive impairment21,57. Knockout of fibrinogen in AD model mice reduces dystrophic neurites, amyloid plaque load, and cerebral amyloid angiopathy (CAA) while improving cognition58. Taken together, the vascular related detrimental outcomes may synergistically propagate peripheral apoE4-related effects to the CNS translating to cognitive deficits and exacerbated pathology. Whether microglia and their responses play a vital role in peripheral apoE-mediated brain function and AD pathogenesis requires further investigation.
An early study suggests that greater than 90% of the apoE in the plasma is synthesized by the liver15. However, apoE secreted from macrophages, vascular mural cells, or other organs in the periphery may also affect brain functions. As such, additional studies are required to further define the cell type-specific effects of apoE isoforms on brain functions and AD pathogenesis. In addition, we cannot rule out the possibility that minute amounts of apoE may leak into the brain and contribute to a confounding effect on brain functions. Furthermore, Apoe deficiency in mice leads to vascular deficits, neuronal dysfunction, and memory impairments12,59. As such, we cannot rule out the possibility that apoe-KO background might increase brain susceptibility to peripheral apoE4. This concern is partially alleviated by our complementary approaches using apoE isoform-specific young plasma or AAV-apoE delivery in the wild-type background with similar findings. As astrocytic apoE4 expression is sufficient to drive amyloid pathology20,60, it is reasonable to assume that both CNS and peripheral apoE4 exhibit gain-of-toxic effects on brain functions and AD-related pathology. Supporting this, apoE immunotherapy administered via a peripheral route improves cognitive performance and enhances Aβ clearance in amyloid mouse models61,62. Interestingly, a recent study shows that deletion of apoE in hepatocytes of APP/PS1 mice, which reduces plasma apoE but not brain apoE levels, does not affect the amount of amyloid plaques63. Whether there is a synergistic interaction between peripheral and CNS apoE on amyloid pathogenesis requires further investigation. Importantly, our current findings shed light on potential apoE isoform-dependent mechanisms on cerebrovascular functions. Whether multiple pathways synergistically influence cerebrovascular integrity and brain functions warrants further examination. Future studies should also explore the therapeutic potential of human young plasma with defined APOE genotype such as APOE2/3 and APOE3/3 on cognitive functions in apoE4 model mice. Our findings offer a strong rationale that peripheral apoE is a viable and individualized target alternative to brain apoE for AD therapy.
Methods
Generation of mice expressing apoE isoforms in the liver
The cell type-specific and inducible apoE mouse models were generated by a knock-in strategy targeting the ROSA-26 locus with a vector system containing a floxed STOP cassette and tetracycline-regulatory elements64. The construct for apoE3 or apoE4 includes: 1) ROSA short and long arms at either ends for targeted integration of ROSA26 locus; 2) a loxP-flanked STOP cassette that harbors a Neor gene for selection of integrated clones; 3) a Tet-off cassette that includes tetracycline-controlled transactivator (tTA) (expressed after Cre-mediated excision of the Neor cassette) and a CMV promoter with a tetracycline-responsive element (TRE); 4) human APOE3 or APOE4 cDNA; and 5) an IRES-eGFP cassette. The constructs are termed inducible APOE3-eGFP and inducible APOE4-eGFP. The resulting inducible apoE3 or apoE4 mice were named in short as iE3 and iE4 mice20. To examine the effects of liver-expressed apoE isoforms on brain functions, iE3 and iE4 mice were crossed with Albumin (Alb)-driven Cre recombinase mice (Jackson Lab) and further bred into the murine Apoe knockout background; thus, apoE was exclusively expressed in the liver and not in the brain. The iE3/Alb-Cre mice (named in short as iE3/Cre− and iE3/Cre+) and iE4/Alb-Cre (named in short as iE4/Cre− and iE4/Cre+) mice (including both male and female mice) were used. The iE3/Cre and iE4/Cre mice in the Apoe null background were further bred with APPSWE/PS1ΔE9 (hereafter referred to as APP/PS1) amyloid mouse model42. Mice expressing apoE3 (APP/iE3/Cre− or APP/iE3/Cre+) or apoE4 (APP/iE4/Cre− or APP/iE4/Cre+) in the liver (including both male and female mice) were used to examine the effects of liver-expressed apoE isoforms on amyloid pathology. To examine the effects of liver-expressed human apoE3 and apoE4 in the presence of murine Apoe, 5xFAD amyloid mice (Jackson Lab) were transduced with AAV8-Alb-GFP, AAV8-Alb-apoE3 or AAV8-Alb-apoE4 virus (titer ~1013 vg/ml) at 1-1.5 month of age via intravenous injection. The animals were harvested at 4 months of age for the experiments. The animal housing conditions follows the Guide for the Care and Use of Laboratory Animals (Humidity: 30-70% relative humidity; Temperature: 68-79°F). All animal procedures were approved by the Mayo Clinic Institutional Animal Care and Use Committee (IACUC) and in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals.
Preparation of brain homogenates
Brain tissues were dissected and kept frozen at −80°C until further analysis. Some brain tissues were fixed in 10% neutralized formalin for histological analysis. Mouse brain tissues for biochemical analysis were processed through sequential extraction43. Briefly, frozen brain tissues were homogenized with TBS buffer, supplemented with Protease Inhibitor Cocktail (Sigma) and PhosSTOP (Sigma), and centrifuged at 100,000g for 60 min at 4°C with supernatant defined as TBS-soluble fraction. Pellets were re-suspended in TBS buffer containing 1% Triton X-100 (TBSX) and mixed gently by rotation at 4°C for 30 min, followed by a second centrifugation at 100,000g for 60 min with supernatant defined as TBSX-soluble fraction. The TBSX-insoluble pellets were re-suspended with 5M guanidine, mixed by rotation at room temperature overnight, and centrifuged at 16,000g for 30 min with supernatant defined as insoluble fraction (GDN-soluble fraction).
Immunohistochemical staining
Paraffin-embedded sections were immunostained with pan-Aβ (Aβ 33.1.1; human Aβ 1-16 specific), anti-GFAP (BioGenex; PU020-UP), and anti-Iba-1 (Wako; 019-19741) antibodies65,66. Immunohistochemically stained sections were captured using the ImageScope AT2 image scanner (Aperio Technologies) and analyzed using the ImageScope software. The immunoreactivities of GFAP and Iba-1 staining in the hippocampus were calculated using the Positive Pixel Count Algorithm available with the ImageScope software (Aperio Technologies). Fibrillar Aβ was stained with Thioflavin S. The images were captured by Aperio Fluorescent Scanner and the stained areas were quantified by Image J. To examine the expression of GFP reporter, frozen slides were mounted and observed under confocal microscopy. For brain Collagen IV staining, frozen brain sections (40 μm thickness) were fixed with acetone/ethanol, and then blocked with 5% normal donkey serum. Sections were incubated in Collagen IV antibody (Rabbit, Millipore, AB756P, 1:300) at 4°C overnight. Sections were then incubated in Alexa Fluor 568 secondary antibody diluted in 1:300. Finally, sections were incubated in fluorescein-labeled Lectin (Vector Laboratories, FL-1171, 1:100) at 4°C overnight to visualize the endothelial cells. Z-stack images of the stained sections were acquired using confocal microscope (model LSM510 Invert, Carl Zeiss), and converted into maximum z-stack projection images. For Cxcl12 and Glut1 co-staining, frozen brain sections were incubated with anti-Cxcl12 (R&D Systems; MAB350; 1:100) and anti-Glut1 (Abcam; ab15309; 1:300) at 4°C overnight. The brain sections were subsequently subjected to incubation with secondary antibodies (goat anti-mouse alexa-568 and goat anti-rabbit alexa-488). Images were acquired using Keyence BZ-X800 fluorescence microscope. The area of Cxcl12 signal relative to Glut1 was quantified using ImageJ. Quantification of immunostaining was performed in a blinded manner.
In vivo two-photon imaging and data analyses
For cranial window preparation, the experimental male mice were anesthetized by the inhalation of isoflurane (4% for induction; 2% for surgery, and 0.5 to 1% for imaging) and immobilized on a custom-made stereotactic apparatus. Approximately 5-10 min before the surgery, dexamethasone (2 mg/kg) and buprenorphine (0.2 mg/kg) were subcutaneously administered to reduce inflammation and pain. Body temperature was monitored by a rectal probe and maintained at 37°C by a heating blanket (Homeothermic blanket systems, Harvard Apparatus). After the removal of the scalp, a surgical drill was used to make a 3-4 mm-diameter craniotomy over the somatosensory and motor cortex, and a 5 mm cover slip was added onto the exposed cortex for protection. Experiments were performed only if physiological variables remained within normal limits.
Dextran conjugated with Texas Red (40 kDa, 50 mg/kg, Sigma) was systemically administered to visualize the vasculature and blood flow in the cerebral cortex. Time-lapse imaging of small cortical sub-volumes (30-40 image planes with 1-2 μm axial spacing) was performed for at least 40 min to track the permeability in the cerebral cortex. The interval duration between stack sequences was 2-3 min; PMT settings (including gain and offset) and laser excitation power were kept constant during time-lapse imaging. In addition, a three-dimension stack image (step size: 1 μm) was captured to track and visualize 3D morphology of vasculature. Arterioles, veins, and capillaries (diameter < 5 μm) were discriminated by the following three criteria: blood flow direction from the pial surface, the patterns of line scan, and vessel diameter (see Supplementary Fig. 2). Line scan was performed along the central axis of the single vessel and perpendicular to a single vessel to measure blood flow velocity and vessel diameter, respectively (see Supplementary Fig. 2).
The upright laser scanning microscope (BX61WI, Olympus) attached to a Ti:sapphire pulsed laser system (80 MHz repetition rate, <100 fs pulse width, Spectra Physics) and the Prairie view 5.3 software (Bruker) was used for two-photon fluorescence imaging. The 20x (NA, 1.00; WD, 2 mm, Olympus) and 40x water-immersion objectives (NA 0.80; WD, 3.3 mm, Olympus) were used for fluorescent imaging in vivo. An excited wavelength of 830 nm was applied to excite the fluorescence of Texas Red-conjugated dextran, and emission light was differentiated and detected with 615/50 filters. The average laser power for imaging was less than 50 mW.
For data analysis, images were processed using open-source software Fiji (NIH) and commercial software Matlab (Version 8.5.0 R2015a, Mathworks). Registration had been taken to perform the intensity-based alignment of images at the different time points with imregister function in Matlab and StackReg plugin in Fiji. To measure the BBB permeability in the cerebrovasculature, regions of interest (ROIs) were manually circumscribed within the brain parenchyma and a corresponding vessel (5-6 ROIs), and the mean fluorescence intensity (MFI) of each ROI was calculated minute by minute. The relative intensity of fluorescence signals (leakage fraction) in the brain parenchyma was defined as ΔF= (Fn–F0)/F0 (where Fn and F0 were fluorescence intensity at any given time point and the initial time point respectively). The BBB permeability surface (PS) area product was calculated by the following equation12: PS=(1-Hct) 1/lv x Vx dlt/dt. Hct is the hematocrit (45%); lv is the initial fluorescence intensity of ROI within the vessel. lt is the intensity of corresponding ROI within the brain at time t. V is the vessel volume, assuming 1 gram of brain is equal to 50 cm2. Our current permeability data indicated that the fluorescence intensity increased linearly with the time, so the PS product was simulated and calculated with the linear regression model of dlt/dt. In addition, Radon transform algorithm had been used to measure blood flow velocity in the cerebral cortex67. Maximum intensity projection was used to calculate vessel diameter and diameter change, and full width at half maximum intensity was defined as vessel diameter.
Lipid and lipoprotein analysis
Plasma cholesterol and triglyceride levels were determined by Amplex Red Cholesterol Assay Kit (ThermoFisher Scientific) and Triglyceride Assay Kit (Cayman) according to manufacturer's instructions, respectively. To determine plasma lipoprotein profiles, the plasma from iE3/Cre (Cre− or Cre+) and iE4/Cre (Cre− or Cre+) mice (n=3/group) at 12 months of age was fractionated by size exclusion chromatography using fast protein liquid chromatography (FPLC) (GE Healthcare) with tandem Superose 6 columns (GE Healthcare) in PBS containing 50 mM sodium phosphate, PH 7.4, with 150 mM NaCl, 1 mM EDTA, and 0.02% sodium azide at a flow rate of 0.4 ml/min68. As controls, pooled plasma from three apoE3-TR or three apoE4-TR mice was included. The total cholesterol in the fractions was measured by Amplex Red Cholesterol Assay (ThermoFisher Scientific) and the percentage of cholesterol was shown. Human high density lipoproteins (HDL), low density lipoproteins (LDL) and very-low density lipoproteins (VLDL) (Athens Research & Technology) were included as controls.
Behavioral studies
Open field assay, elevated plus maze (for anxiety), fear conditioning (for associative learning and memory), and rotarod (for motor learning and coordination) were performed as previously described69. For open field analysis, the animals were placed in the open field chamber (40 x 40 x 30 cm) for 15 min in standard room-lighting conditions. Activity in the open field was monitored by an overhead camera to track movement with AnyMaze software (Stoelting Co.). Mice were analyzed for multiple measures, including total distance traveled, average speed, and the time spent in the center of the chamber (digitally designated by an 8 × 8 cm region) compared to the perimeter. For elevated plus maze, the apparatus consisted of two opposing open arms (50 x 10 cm) and two opposing closed arms with roofless gray walls (40 cm) connected by a central square platform and positioned 50 cm above the ground. Mice were placed in the open arms facing an open arm, and their behavior was tracked for 5 min with an overhead camera and AnyMaze software. The fear conditioning test was conducted in a sound attenuated chamber with a grid floor capable of delivering an electric shock. Freezing was measured with an overhead camera and FreezeFrame software (Actimetrics). Mice were initially placed into the chamber and undisturbed for 2 min, during which time baseline freezing behavior was recorded. An 80-dB white noise served as the conditioned stimulus (CS) and was presented for 30 sec. During the final 2 sec of this noise, mice received a mild foot shock (0.5 mA), which served as the unconditioned stimulus (US). After 1 min, another CS-US pair was presented. The mouse was removed 30 sec after the second CS-US pair. To assess contextual learning, the animals were placed back into the training context 24 hr post-training and scored for freezing for 5 min. To assess cued learning, the animals were then placed in a different context (novel odor, cage floor, and visual cues) for 3 min and then the auditory CS was presented and freezing was recorded for another 3 min. Baseline freezing behavior obtained during training was subtracted from the context or cued tests to control for animal variability. For the rotarod test, mice were placed on the rotating rod with a linear increase in rotation speed from 4 to 40 rpm and ability to maintain balance on a rotating cylinder was measured. Mice were subjected to 4 trials per day for 2 consecutive days, and the latency to remain on the rotarod was averaged for each trial.
Extracellular field recording from hippocampal slices
To examine the LTP in the hippocampus of experimental mice, extracellular recordings were performed as described with modification70. Mice were sacrificed and transverse slices were prepared for electrophysiological experimental paradigms in ice-cold cutting solution containing 110 mM sucrose, 60 mM NaCl, 3 mM KCl, 1.25 mM NaH2PO4, 28 mM NaHCO3, 0.6 mM sodium ascorbate, 5 mM glucose, 7 mM MgCl2 and 0.5 mM CaCl2. Field excitatory post-synaptic potentials (fEPSPs) were obtained from area CA1 stratum radiatum with the use of a glass microelectrodes (2–4 mΩ) filled with artificial cerebrospinal fluid (aCSF) containing 125 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 25 mM NaHCO3, 25 mM glucose, 1 mM MgCl2 and 2 mM CaCl2. The fEPSPs were evoked through stimulation of the Schaffer collaterals using a 0.1 msec biphasic pulse delivered every 20 sec. After a consistent response to a voltage stimulus was established, threshold voltage for evoking fEPSPs was determined and the voltage was increased incrementally every 0.5-1 mV until the maximum amplitude of the fEPSP was reached (I/O curve). All other stimulation paradigms were induced at the same voltage, defined as 50-60% of the stimulus voltage used to produce the maximum fEPSP amplitude, for each individual slice. Paired-pulse facilitation (PPF) was induced with two paired-pulses given with an initial delay of 20 msec and the time to the second pulse incrementally increased 20 msec until a final delay of 300 msec was reached. A fEPSP baseline response was then recorded for 20 min. The tetanus used to evoke LTP was a theta-burst stimulation (TBS) protocol consisting of five trains of four pulse bursts at 200 Hz separated by 200 msec, repeated six times with an inter-train interval of 10 sec. Following TBS, fEPSPs were recorded for 60 min. Potentiation was measured as the increase of the mean fEPSP descending slope following TBS normalized to the mean fEPSP descending slope of baseline recordings.
RNA isolation and real-time PCR analysis
Total RNA was isolated by using Trizol (Invitrogen) followed by RNeasy Mini kit (QIAGEN). For real-time PCR analysis, cDNA was synthesized from total RNA by SuperScript III reverse transcriptase (Invitrogen). The primer sequences were as follows: Human apoE, TGTCTGAGCAGGTGCAGGAG; TCCAGTTCCGATTTGTAGG. Mouse β-actin, AGTGTGACGTTGACATCCGTA; GCCAGAGCAGTAATCTCCTTC. Mouse PTPN-2, GTGATCCATTGCAGTGCG; TTCCATCAGAACAAGACAGGTAT. Mouse Smad6, ATCACCTCCTGCCCCTGT; CTGGGGTGGTGTCTCTGG. Mouse Chst12, GCCCCCAGCAAGCCAGT-CTTG; AGGCTGGCGTTGGCACAGAAGT. Mouse Isyna, CCTTGGTGCTCCATAATACCT-G; AGTCTGTGCAGAAGCTCACG. Mouse Lefty1, TGTGTGTGCTCTTTGCTTCC; GGGGATTCTGTCCTTGGTTT. Mouse Rspo2, ATAGAGGCCGCTGCTTTG; TGCCGTGTT-CTGGTTTCC. Col11a2 (Mm.PT.58.6983762), Ccbe1 (Mm.PT.58.6423596), and Anxa4 (Mm.PT.58.9558059) predesigned primers were ordered from IDT-DNA. Mouse Col6a3, and Eln primers were purchased from Qiagen. Reactions were prepared using a 20-μl mixture containing universal SYBR Green Supermix (Bio-Rad). Real-time quantification was performed on CFX96 real-time PCR detection system (Bio-Rad). Relative mRNA levels were calculated by ΔΔCt method with β-actin used as a reference.
Western blotting
The detailed procedures were carried out as previously described20. Briefly, after the membranes were blocked, proteins were detected with primary antibody. Membrane was probed with LI-COR IRDye secondary antibodies and detected using the Odyssey infrared imaging system (LI-COR). To examine whether apoE was detectable in the brain, blots were incubated with horseradish peroxidase-conjugated secondary antibody, detected by SuperSignal West Femto Chemiluminescent Substrate (Pierce). The following antibodies were used in this study: anti-GFAP (Millipore; MAB360), anti-apoE (WUE4; NB110-60531) anti-PSD-95 (Cell Signaling; #3450), anti-synaptophysin (Millipore; MAB5258), and anti-β-actin (Sigma; A2228) antibodies.
ELISA quantification
ApoE was measured by ELISA as previously described68. Aβ levels in the brain lysates were determined by ELISA with end-specific mAb 2.1.3 (human Aβx-42 specific) and mAb 13.1.1 (human Aβx-40 specific) for capture and HRP-conjugated mAb Ab5 (human Aβ1-16 specific) for detection as described71. The ELISAs were developed using Super Slow ELISA TMB (Sigma). The level of brain Collagen IV was examined by mouse Collagen IV ELISA (LifeSpan BioSciences) according to manufacturer's instructions. Colorimetric quantification was performed on a Synergy HT plate reader (BioTek).
RNA sequencing, quality control, and normalization
RNAs were extracted via the Trizol/chloroform method, followed by DNase and Cleanup using RNase-Free DNase Set and RNeasy Mini Kit (QIAGEN). The quantity and quality of all RNA samples were determined by the Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano Chip (Agilent Technologies, CA). A total of 32 samples (8 samples/ genotype) with RNA Integrity Number (RIN) ≥ 9.0 were used for RNA sequencing (RNAseq) at Mayo Clinic Core facility using Illumina HiSeq 4000. Reads were mapped to the mouse genome mm10. Raw gene read counts, along with sequencing quality control, were generated using the Mayo Clinic RNA-Seq analytic pipeline: MAP-RSeq Version 2.1.172. Conditional Quantile Normalization (CQN) was performed on the raw gene counts to correct for GC bias and gene length differences, and to obtain similar quantile-by-quantile distributions of gene expression levels across samples73. Based on the bi-modal distribution of the CQN normalized and log2-transformed reads per kb per million (RPKM) gene expression values, genes with average log2 RPKM >= 0 in at least one genotype group were considered expressed above detection threshold. Using this selection threshold, 18,841 genes were included in the downstream analysis. One iE3 sample was determined as an outlier by Principal Component Analysis (PCA) and excluded from further analyses. It is noted that the mouse Apoe mRNA was detected in RNA-Seq analysis likely due to the gene inactivation approach initially used to generate this apoe-KO mice31. Nonetheless, we confirmed that there is no mouse apoe protein detected in the brain.
Weighted correlation network analysis
To identify groups of genes that are correlated with the APOE genotype, we performed weighted correlation network analysis (WGCNA)74 using residual expression values calculated from adjusting for tissue harvest batch, which contributed significantly to the variation of gene expression based on source of variation analysis. Separate WGCNA analyses were performed for the iE3 and iE4 datasets. Unsigned hybrid co-expression networks were built for both WGCNA analyses. Based on the relationships between power and scale independence, the power of 12 was chosen for building scale-free topology for the iE3 dataset, and the power of 8 was chosen for the iE4 dataset. We used hybrid dynamic tree cutting, a minimum module size of 60 genes, and a minimum height for merging modules at 0.4. Each module was summarized by the first principal component of the scaled (standardized) module expression profiles (module eigengene). Each module was assigned a unique color identifier, and genes that did not fulfil these criteria for any of the modules were assigned to the gray module. To assess the correlation of modules to genotype, we defined the iE3/Cre− genotype as 0, and iE3/Cre+ as 1 for the iE3 dataset, and iE4/Cre− genotype as 0, and iE4/Cre+ as 1 for the iE4 dataset. Modules significantly associated with genotype were annotated using WGCNA R function GOenrichmentAnalysis. Genes with high connectivity in the respective modules were considered hub genes. Intramodular gene-gene connection was visualized using VisANT.
Gene Set Enrichment Analysis and co-expression network analysis
Gene functional enrichment analysis was executed in a cutoff-free manner with Broad Institute’s Gene Set Enrichment Analysis (GSEA) to screen for “enriched” MSigDB gene ontology (GO) and canonical pathway annotation gene sets26. In GSEA, limma’s moderate t-test statistics were used to rank the transcriptome-wide gene-phenotype association.
To further interrogate the interactions of genes, we applied MEGENA75, a multiscale approach that allows for identification of overlapping gene modules as well as individual gene-gene interactions. MEGENA complements WGCNA in its capability of simplifying densely connected co-expression networks and identifying multiscale modular structures. Briefly, Pearson correlation coefficients (PCCs) were firstly computed for all gene pairs. As the sample size in each genotype group is relatively small (7~8 mice per group), we pooled the samples from each strain (E4 or E3) together to boost the sample size to derive a more robust correlation relationship among transcriptome-wide genes. The gene pairs with significant correlation (adjusted P value less than 0.05) were ranked and iteratively tested for planarity to grow a Planar Filtered Network (PFN). Multiscale clustering analysis was conducted with the resulting PFN to identify co-expression modules at different network scale topology under the default parameter setting of the MEGENA package (version 1.3.4-1). Modules were annotated for enrichment of MSigDB GO/pathway gene sets using the hypergeometric test. In addition, GSEA was applied to test for modules enriched for gene differential expression signals.
Single cell RNA sequencing for brain vascular-enriched cell populations
Brain vascular-enriched cell populations were prepared from cortical tissues of iE3/Cre−, iE3/Cre+, iE4/Cre−, iE4/Cre+ mice (n=4/group; male:female=1:1) as previously described76,77. Brain endothelial cells, vascular mural cells, glial cells, and other vascular-related cells were isolated as described33 with modifications. Cortical tissues were enzymatically digested using the Neural Tissue Dissociation kit (P) (Cat. #130-092-628, Miltenyi Biotec), and myelin was removed using magnetic bead separation (Myelin Removal Beads II, Cat. #130-096-733, Miltenyi Biotec). After myelin removal and red blood cell removal (Cat.# 130-094-183, Miltenyi Biotec), microglia and debris were removed by magnetic separation using anti-CD11b+ microbeads (Cat. #130-049-601, Miltenyi Biotec), followed by a dead cell removal kit (Cat. #130-090-101, Miltenyi Biotec) according to the manufacturer’s instruction. The final cell pellet was suspended in Ca/Mg-free PBS supplemented with 0.5% FBS. In all samples, more than 90% of the cells were viable based on trypan blue examination. More than 10,000 single cell events derived from cortical tissues of mice (2 male and 2 female/genotype) were captured. Approximately 3,000 glio-vascular cells from one animal were loaded into each sample well on a chip and combined with Gel Beads containing barcoded oligonucleotides using a 10x Chromium Controller (10x Genomics). Single cell libraries were constructed according to the manufacturer’s instructions and were sequenced by an Illumina HiSeq 4000 Sequencing Systems at an average depth of 50,000 reads per cell.
The single cell sequence preprocessing was performed using the standard 10x Genomics Cell Ranger Single Cell Software Suite (v3.0.0). Briefly, raw sequencing data were demultiplexed, aligned to the mouse genome mm10, and the reads aligned to each gene were counted. Cell filtration, normalization, clustering, and differential expression analyses were performed using standard Seurat package procedures (v.2.3.4). For quality assurance, cells that had unique gene counts over 4,000 or less than 200, and >25% mitochondrial content were removed. Canonical correlation analysis (CCA) was performed to identify common sources of variation between cases and controls in the iE3 or iE4 dataset. Based on the measurements of correlation strength, the first 20 canonical correlation vectors were used for dimension reduction and clustering analysis. Clusters were visualized using t-distributed stochastic nearest neighbor embedding. Function “FindConservedMarkers” was used to identify cluster-specific genes that were conserved across conditions. Cell clusters were manually annotated as specific cell types based on literature and the datasets for murine cell taxonomy. Differentially expressed genes (DEGs) in each pairwise comparison of cell clusters were identified by function “FindMarkers” with default parameters (p < 0.05 and ∣Fold change∣ >=1.2). Gene ontology (GO) and pathway analysis of DEGs from each cluster was performed using both the hypergeometric test (equivalent to Fisher’s exact test, FET) and the gene set enrichment analysis (GSEA) [Pubmed 16199517]. GO and pathway annotations were derived from the MSigDB v6.0 (Pubmed 26771021) after converting human genes to mouse orthologs. Relevant terms were selected and shown in the figures.
Regulatory network construction
To understand a regulatory network of Timp3, we constructed regulatory networks with ARACNe using data obtained from vascular cell-enriched scRNA-seq. To construct the regulatory network of Timp3, we extracted endothelial cell clusters from iE3/Cre mice and performed preprocessing (e.g., filtered genes with no expression are removed), and counts-per-million (CPM) normalized using the Seurat R package. The normalized data were summed and log10 transformed to generate in-silico sample-level transcriptomic data. ARACNe was run with 100 bootstrap iterations using in-silico single-cell RNA-seq data. We used a set of regulators provided by ARACNe, including 651 mouse ligands, 1,465 transcriptional factors (TFs), 840 transcription cofactors (coTFs), 3,233 surface proteins. We also added Timp3 as a regulator. Parameters were set to zero DPI (Data Processing Inequality) tolerance and MI (Mutual Information) p-value threshold of 10−8.
We also generated co-expression subnetworks for Agt and Cxcl12 in endothelial cell clusters of iE4/Cre mice. The Agt subnetwork was generated by correlating its protein abundance from the plasma proteomics data with genes from endothelial cell clusters (as in-silico bulk transcriptomics). Conversely, the Cxcl12 subnetwork was produced by correlating its expression level with plasma proteins. The top correlated genes and proteins were used to construct co-expression networks and subsequent enrichment analysis.
Plasma administration
Aged male C57BL/6 mice were obtained from the National Institute on Aging (NIA). Human apoE-TR mice expressing human apoE3 or apoE4 under the control of the mouse Apoe promoter on a pure C57BL/6J background were purchased from Taconic. Mice were bred and aged in-house and lived under a 12-hour light-dark cycle in pathogenic-free conditions, in accordance with the Guide for Care and Use of Laboratory Animals of the National Institutes of Health and the Mayo Clinic Institutional Animal Care and Use Committee. Blood was collected from the femoral vein of young (2–3-month-old) male human apoE-TR mice with 3.2% sodium citrate (12-8480-10, Medicago, Sweden) as anticoagulant, and centrifuged at 1000g for 10 min at 4°C with a break set to 5 or less. Plasma was collected and immediately snap frozen on dry ice and stored at −80°C until further processing. Mice at 26 months of age were subjected to blood plasma injection. Briefly, mice were treated with plasma from young apoE3-TR or apoE4-TR or phosphate-buffered saline (PBS)/sodium citrate via intravenous injections (100 μL/injection) into the tail vein twice a week for 6 weeks.
Viral vector construction and production
The AAV8-Alb-apoE3, AAV8-Alb-apoE4, and AAV8-Alb-GFP plasmid construction and viral production were performed by the Hope Center Viral Vectors Core at the Washington University School of Medicine. Briefly, the Alb promoter was obtained from Addgene as ALB-GFP (a gift from Snorri Thorgeirsson (Addgene plasmid # 55759; RRID: Addgene_55759)78 and was sub-cloned into the AAV8 vector. The AAV8 viruses were produced as previously described79.
Immunostaining and quantification for markers of tight junction, blood-derived protein, astrocyte endfeet, and brain vessel-associated gliosis
Brains were sectioned coronally at 40 μm with a cryomicrotome (Leica Camera) and stored in cryoprotective medium. For immunostaining, sections were permeabilized in 0.25% Triton X-100 and blocked with blocking buffer containing 4% normal donkey serum, 2% BSA, and 1 M glycine in PBS. Sections were then incubated with primary antibodies in blocking buffer for 48 hr at 4°C. Primary antibodies used were: anti-Glut1 (Abcam; ab40084; 1:500), anti-Glut1 (Abcam; ab15309; 1:500), anti-ZO1 (Invitrogen; 33-9100; 1:500), anti-AQP4 (Alomone labs; AQP-004; 1:100), anti-CLDN5 (Thermo fisher scientific; # 35-2500; 1:300), anti-Fibrinogen (DAKO; A008002-2; 1:100), anti-CD13 (R&D; AF2335; 1:300), anti-GFAP (Millipore; MAB360; 1:500), anti-CD206 (Bio-Rad; MCA2235; 1:500), anti-Iba1 (Wako; 019-19741; 1:500), and albumin antibody (Bethyl Laboratories; A90-134; 1:300). After washing three times with PBS, samples were incubated with fluorescently conjugated secondary antibodies (Alexa Fluor 488 or 594 conjugates, Invitrogen, 1:500) overnight at 4°C and washed three times with PBS before mounting with a glass coverslip using Vectashield (Vector Labs). Polyclonal goat anti-mouse IgG antibody conjugated with Alexa 488 (Invitrogen; A-11001; 1:300) was used to detect mouse IgG. Z-stack images of the stained sections were acquired by Keyence fluorescence microscopy (model BZ-X, Keyence) and converted into maximum z-stack projection images.
At least two different brain slides per mouse were used for image acquisition. The intensity of GFAP or Iba-1 surrounding the selected vessels was quantified and normalized to the Glut1 signals. The immunoreactivities were quantified by ImageJ software (NIH, ImageJ bundled with 64-bit Java 1.8.0_112). For the quantification of brain vessel associated GFAP or Iba1 fluorescence intensity, continuous vessels with clear Glut1 immunoreactivity (>100 μm) were randomly selected as regions of interest (ROIs). The image acquisition and staining quantification were performed by investigators who were blind to the experimental conditions.
RNAscope fluorescent in situ hybridization
Deparaffinized sections were subjected to fluorescence in situ hybridization (FISH) using RNAscope® Multiplex Fluorescent Reagent Manual Assay Kit V2 (#323100, Advanced Cell Diagnostics [ACD], Hayward, CA) according to the manufacturer’s protocol. In brief, following rehydration of slides through a series of xylene and ethanol incubations, slides were boiled in 1X Target Retrieval Reagent for 15 min and immediately washed for 15 sec in water. Slides were then incubated in 100% ethanol for 3 min and subsequently allowed to air dry before drawing a hydrophobic barrier around the tissue and placing the slides in a desiccator at room temperature to dry overnight. The following day, slides were incubated with Protease Plus digestion reagent for 30 min at 40°C in a HybEZ Oven and moist Humidity Control Tray and then washed twice with water using gentle agitation. Then the tissue was incubated for 2 hr at 40°C with ACD probes for mouse Vwf (499111 Lot 21266B; target NM_011708.4, region 2 - 4933bp), mouse Gkn3 (512061-C2 Lot 21181B; target NM_026860.1, region 2-778bp), DapB (negative control 310043 Lot 2011629; target EF191515, region 414-862bp) or mouse Ppib (positive control 313911 Lot 21140A; target NM_011149.2, region 98-856bp), then washed two times in 1X wash buffer. Amplification and detection steps were performed following the manufacturer’s protocol using detection probes from AKOYA biosciences fluorophore Opal 650 or Opal 570, diluted in TSA buffer (ACD). Prior to the counterstain, sections were blocked with 10% normal goat serum in TBS with 0.1% BSA and 0.3% Triton X-100 for 1 hr at room temperature. Next, the slides were incubated with rabbit anti-Glut1 (Abcam; ab15309; 1:300) diluted in blocking buffer at 4°C overnight. The following day, slides were washed three times with TBST (1X TBS-10% Tween-20) and incubated with goat anti-rabbit Alexa Fluor 488 or Alexa Fluor 647 (1:500) for 2 hr at room temperature. Slides were subsequently counterstained with DAPI (ACD) for 30 sec and then mounted using Vectashield or PermaFluor (Epredia). Images were acquired using a Zeiss LSM 800 confocal microscope and quantification was performed in a blinded manner. The area of Vwf and Gkn3 signal relative to Glut1 was quantified using ImageJ.
Proteomic analysis for mouse plasma samples
The plasma specimens were profiled based on a previously reported protocol80,81 with modifications. The 16 mouse plasma samples from iE3/Cre−, iE3/Cre+, iE4/Cre−, iE4/Cre+ mice (n=4/group) were precipitated by 80% cold acetone at −20°C for 2 hr and resuspended in fresh lysis buffer [50 mM HEPES, pH 8.5, 8 M urea, 0.5% sodium deoxycholate and phosphatase inhibitor cocktail (PhosSTOP, Sigma-Aldrich)]. The protein concentration was estimated by Coomassie-stained short SDS gel with BSA as a standard82. Each protein sample was digested and TMT labeled using our previously optimized protocol83,84. Briefly, the quantified protein (~200 μg per TMT channel) was digested in the lysis buffer with Lys-C (Wako; 1:100 w/w) at 21°C for 3 hr, then diluted with 50 mM HEPES (pH 8.5) to lower urea concentration to 2 M, and further digested by trypsin (Promega; 1:50 w/w) at 21°C overnight. The digested peptides were reduced by dithiothreitol (DTT, 1 mM) for 30 min, alkylated by iodoacetamide (IAA, 10 mM) at dark for 30 min, and further quenched with DTT (30 mM) for 30 min. The peptides were acidified by the addition of 1% trifluoroacetic acid (TFA), then centrifuged at 21,000 × g for 10 min. The supernatant was de-salted with a C18 Micro SpinColumn (Harvard apparatus) and dried by speedvac. The samples were re-dissolved in 50 mM HEPES, pH 8.5 (~2 μg/μL), reacted with 400 μg of TMTpro reagents for 30 min, then quenched with 0.5% hydroxylamine for 15 min. The labeled samples were pooled equally, and desalted again before LC/LC–MS/MS.
The pooled TMT labeled peptides were fractionated by an offline basic pH RPLC with two tandem XBridge C18 columns (3.5 μm particle size, 4.6 mm × 25 cm, Waters; buffer A: 10 mM ammonium formate, pH 8.0; buffer B: 90% AcN, 10 mM ammonium formate, pH 8.0) and a 140 min gradient of 15-50% buffer B. Each fraction was collected with 0.33 min, ending with a total of 424 fractions. The peptide concentration in each fraction was measured by BCA assay, then the collected fractions were divided into 5 categories according to the peptide amount in each fraction (<1.25 μg, 1.25~2.5 μg, 2.5~5 μg, 5~10 μg, >10 μg). The fractions within each category were concatenated by combining early, middle, and late LC fractions, generating 80 concatenated fractions. Each fraction was analyzed on a self-packed column (75 μm × 25 cm with 1.9 μm C18 resin) coupled with a Q Exactive HF Orbitrap MS (Thermo Fisher Scientific). A 120 min LC gradient of 15-45% buffer B (buffer A: 0.2% formic acid, 5% DMSO; buffer B: buffer A plus 65% AcN) was used to resolve peptides. MS settings included MS1 scans (60,000 resolution, 460-1600 m/z scan range, 1 x 106 AGC, and 50 ms maximal ion time) and 20 data-dependent MS2 scans (60,000 resolution, starting from 120 m/z, 1 x 105 AGC, 150 ms maximal ion time, 1.0 m/z isolation window with 0.2 m/z offset, HCD, 32% specified normalized collision energy (NCE), and 20 s dynamic exclusion).
The protein identification and quantification were performed by the JUMP search engine85. The mouse protein database was generated by combining downloaded Swiss-Prot, TrEMBL, and UCSC databases and removing redundancy (59,423 entries). The target-decoy database was used to estimate the false discovery rate (FDR)86. Major database search parameters included precursor and product ion mass tolerance (± 15 ppm), full trypticity, two maximal missed cleavages, static modification for TMTpro tags (+304.20715 on Lys and N-termini), and carbamidomethylation on (+57.02146 on Cys), dynamic modification for oxidation (+15.99491 on Met). The resulting PSMs were filtered by mass accuracy and then grouped by precursor ion charge state followed by the cutoffs of JUMP-based matching scores (Jscore and ΔJn) to reduce FDR below 1% for proteins. When the same peptide was derived from numerous homologous proteins, the peptide was matched to the protein with the top PSM number, according to the rule of parsimony. The protein quantification was performed using the TMT reporter ion intensities as previously described87.
Human iPSC-derived brain endothelial cell (iBMEC) model and treatment with young plasma
Human iBMECs were differentiated as previously described with modifications88. In brief, human iPSCs from normal individual were disassociated to single cells using Accutase and reseeded at 1.6 × 104/cm2 on Matrigel-coated plates in mTeSR1 supplemented with 10 μM Y27632 (Stem Cell Technologies). Cells were cultured in mTeSR1 for 3 days prior to differentiation. To initiate the differentiation, medium was changed to DeSR1 medium with 6 μM CHIR99021 (Selleckchem) at day 0 [DeSR1 medium: DMEM/Ham’s F12 (Thermo Fisher Scientific), 1X MEM-NEAA (Thermo Fisher Scientific), 0.5X GlutaMAX (Thermo Fisher Scientific), and 0.1 mM β-mercaptoethanol (Sigma)]. After 24 hr, the medium was removed and cells were transitioned to DeSR2 medium [DeSR1 medium with 1X B27 (Thermo Fisher Scientific)]. DeSR2 medium was changed every day for 5 days. Cells were switched to hECSR1 medium at day 6 for 2 days [hESFM (Thermo Fisher Scientific) supplemented with bFGF (20 ng/ml), 10 μM RA, and 1× B27]. At day 8, cells in hECSR1 were reseeded at 2.0x104 cells/cm2 onto Matrigel-coated 48-well tissue culture plates or Transwell inserts (0.4 μm pore size). After reseeding, iBMECs were maintained in hECSR2 (hECSR1 lacking RA and bFGF) for further marker staining and TEER measurement.
For immunostaining, cells were fixed in 4% paraformaldehyde for 30 min then washed with PBS three times. After fixation, sections were permeabilized in 0.25% Triton X-100 and blocked with blocking buffer (Protein Block Serum-Free Ready-to-use; Dako Agilent Technologies, Santa Clara, USA). Sections were then incubated with primary antibodies in background reducing dilution buffer (Dako Agilent Technologies) overnight at 4°C. The information of primary antibodies and their dilutions used in this study are as follows: Nanog (Cell Signaling; 4903; 1:300), Brachyury (R&D; AF2085; 1:300), PAX2/5/8 (Santa Cruz Biotechnology; sc-377181; 1:300). After washing three times with PBS, samples were incubated with fluorescently conjugated secondary antibodies (Alexa Fluor 488 and 594 conjugates, Invitrogen; 1:500) for 2 hr at room temperature and washed three times with PBS before mounting with a glass coverslip using Vectashield. Fluorescent signals were detected by Keyence fluorescence microscopy (model BZ-X, Keyence).
For treatment with young plasma, iBMECs were seeded on the upper side of the Matrigel (Corning)-coated polyester membrane of a Transwell insert (0.4 μm pore size; Cat. #3470, Corning) at a density of 2.0×104 cells/cm2, and plating day of iBMECs on membrane was defined as day 0. Young plasma collected from 2-3-month-old young apoE3-TR or apoE4-TR mice were added to the upper chamber on day 2 at a concentration of 2% in hECSR2 medium and incubated for 24 hr. In a rescue treatment, iBMECs were treated with apoE3 or apoE4 plasma (2%), or apoE4 young plasma together with Timp3 (0.5 μg/ml). To evaluate the integrity of iBMECs, the transendothelial electrical resistance (TEER) was measured using an EVOM3 Epithelial Voltammeter (World Precision Instrument). The TEER of Matrigel-coated cell-free filters was subtracted from the measured TEERs and shown as ohm·cm2.
Statistical analysis
All quantified data represents an average of samples. Statistical analyses were performed with GraphPad Prism software. Comparisons between two groups were performed with two-tailed Student’s t tests, and those among more than two groups were performed with ANOVA with a Tukey’s post-hoc comparisons. A P value of < 0.05 was considered statistically significant. No statistical methods were used to predetermine sample sizes, but our samples sizes are comparable to similar studies41,46,79.
Extended Data
Extended Data Fig. 1. Liver-expressed apoE3 restores peripheral lipid profiles.
a, EGFP, a surrogate of apoE expression, was detected in liver of iE3/Cre+ but not iE3/Cre− mice. Similar results were observed in at least three independent experiments. Scale bar, 50 μm. b-d, ApoE levels in the liver, plasma, and brain of iE3/Cre−, iE3/Cre+ (Cre−, n=10; Cre+, n=15), and apoE3-TR (n=8) mice were examined by ELISA. **, P<0.0001. e, The apoE was not detected by Western blotting in the brains of iE3/Cre− or iE3/Cre+ mice compared to apoE3-TR mice (n=4/group). **, P<0.0001. f, The RNA levels for apoE in the brains of iE3/Cre, apoE3-TR, iE4/Cre, and apoE4-TR mice (n=8/group) were examined by real-time PCR. **, P<0.0001. g, Total cholesterol and triglyceride levels in the plasma of iE3/Cre− or iE3/Cre+ mice (Cre−, n=9; Cre+, n=11) and apoE3-TR mice (n=8) were examined. Cholesterol: **, P<0.0001. Triglyceride: Cre− vs. Cre+: **, P=0.002; Cre− vs. E3: **, P=0.0002. h, Distribution of plasma lipoprotein cholesterol in iE3/Cre− or iE3/Cre+ mice (n=3/group) at 12 months of age. Pooled plasma from apoE3-TR mice (n=3/group) was included as control. Data represent mean ± s.e.m. One-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used for statistical analyses. N.S., not significant.
Extended Data Fig. 2. Expression of apoE4 in the liver leads to compromised tight junction integrity in the brain.
a, Brain sections from iE4/Cre− or iE4/Cre+ mice (Cre−, n=11; Cre+, n=14) at 12-13 months of age were immunostained with anti-claudin-5 (CLDN5; green) and anti-Glut1 (red) antibodies. The total Glut1 signals and the CLDN5 signals normalized against Glut1 were quantified. *, P=0.024. Scale bar, 25 μm. b, Brain sections from the iE4/Cre− or the iE4/Cre+ mice (Cre−, n=12; Cre+, n=14) at 12-13 months of age were immunostained with anti-ZO1 (green) and anti-Glut1 (red) antibodies. The total Glut1 signals and the ZO1 signals normalized against Glut1 were quantified. *, P=0.038. Scale bar, 25 μm. c, Brain sections from iE4/Cre− or iE4/Cre+ mice (Cre−, n=11; Cre+, n=13) at 12-13 months of age were immunostained with anti-IgG (green) and anti-Glut1 (red) antibodies. The total Glut1 signals and the IgG signals normalized against Glut1 were quantified. *, P=0.047. Scale bar, 10 μm. Data represent mean ± s.e.m. d, Brain sections from iE4/Cre− or iE4/Cre+ mice (Cre−, n=8; Cre+, n=9) at 12-13 months of age were immunostained with anti-fibrinogen (green) and anti-Glut1 (red) antibodies. The total Glut1 signals and the fibrinogen signals normalized against Glut1 were quantified. *, P=0.034. Scale bar, 10 μm. Data represent mean ± s.e.m. e, Brain sections from the iE4/Cre− or iE4/Cre+ mice (Cre−, n=8; Cre+, n=9) at 12-13 months of age were immunostained with anti-CD13 (red) for pericytes and anti-Glut1 (green) antibodies. The total Glut1 signals and the CD13 signals normalized against Glut1 were quantified. Scale bar, 25 μm. Data represent mean ± s.e.m. N.S., not significant, two-tailed Student's t-test.
Extended Data Fig. 3. Cerebral vascular function in mice expressing apoE3 or apoE4 in the liver by two-photon imaging.
a-d, The BBB integrity, CBF, and vasomotion in the arterioles of iE4/Cre− and iE4/Cre+ mice were assessed by two-photon imaging. a, CBF in the cerebral veins and capillaries of iE4/Cre− or iE4/Cre+ mice at 6-7 months of age were measured. Each data point represents blood vessel-related measurement with various number of measurements in each animal from 5 animals per genotype. b, c, Vessel branch densities in the cerebral arterioles of iE4/Cre− or iE4/Cre+ mice (Cre−, n=3; Cre+, n=3) at 1.5~2 months of age were assessed. Scale bar, 50 μm. d, CBF in the cerebral arterioles of iE4/Cre− or iE4/Cre+ mice at young age (1.5~2-month-old) was examined. e-j, BBB permeability, CBF, and vasomotion in the arterioles of iE3/Cre− and iE3/Cre+ mice (Cre−, n=3; Cre+, n=3) at 6-7 months of age were assessed by two-photon imaging. Each data point represents blood vessel-related measurement with various number of measurements in each animal from 3 animals per genotype. e, CBF in the cerebral arterioles of iE3/Cre− and iE3/Cre+ mice was examined. f, BBB integrity was examined after intravenous injection of Texas Red conjugated dextran (40 kDa) for 30 mins. Scale bar, 100 μm. g, Fractions of BBB leakage over time are shown. h, BBB leakage, calculated as BBB permeability surface (PS) area product for Texas Red-conjugated dextran, in the cortex of iE3/Cre mice was quantified. i, The frequency and amplitude of arteriolar oscillation in iE3/Cre− and iE3/Cre+ mice were measured. j, Vessel branch densities in the cerebral arterioles of iE3/Cre− and iE3/Cre+ mice were examined. Scale bar, 50 μm. Data expressed as mean ± s.e.m. N.S., not significant, two-tailed Student's t-test.
Extended Data Fig. 4. Peripheral apoE4 expression is associated with gene expression profiles consistent with reduced vascular functions and compromised energy homeostasis.
Brain cortical tissues from iE4/Cre mice (Cre−, n=8; Cre+, n=8) at 12-13 month of age were subjected to RNA-Sequencing. a, Module-genotype correlation. Each rectangle represents a module and selected modules are shown. The number in the front of each module is the correlation coefficient (r) between the module eigengene to genotype; the correlation p-value is in the parentheses. Red represents positive correlation to iE4/Cre+ and blue represents negative correlation to iE4/Cre+. b, Heatmaps of genes within the lightyellow module and selected gene ontologies enriched in the module. c, Interaction of genes involved in the gene ontologies (blue nodes) and hub genes (red nodes) in the lightyellow module. The thickness of the lines represents the strength of gene-gene connection. d, Expression of collagen family members (i.e., Col6a3, Col11a2), collagen and calcium binding EGF domain-containing protein 1 (Ccbe1), elastin (Eln), annexin A4 (Anxa4), and genes involved in carbohydrate biosynthetic process as well as the hub genes (Lefty1, Rspo2 and Atp8b1) in the lightyellow module shown in (c) were validated in iE4/Cre mice (Cre−, n=8; Cre+, n=8) by real-time PCR analysis. Data expressed as mean ± s.e.m. Ptpn2 (*, P=0.042); Col6a3 (*, P=0.036); Col1a2 (*, P=0.027); Ccbe1 (**, P=0.006); Eln (*, P=0.04); Anxa4 (*, P=0.014); lsyna (*, P=0.047); Smad6 (*, P=0.041); Chst12 (*, P=0.028); Lefty1 (*, P=0.021); Rspo2 (**, P=0.006); Atp8b1 (*, P=0.031), two-tailed Student's t-test. e, Sunburst plot showing the module hierarchical structure relationship. Each rectangular block denotes a module. A total of 1002 modules were identified in the E4 network, with module size ranging from 10 to 960. The color intensity denotes the FDR adjusted P value significance of GSEA based enrichment for differential expression signals. Functional annotations are highlighted for modules overrepresented with GO/pathway genes (red text for positive GSEA enrichment score with regard to differential expression signal).
Extended Data Fig. 5. Evaluation of glio-vascular-enriched single cell transcriptomics in mice with peripheral expression of apoE isoforms.
Brain cortical tissues from iE3/Cre and iE4/Cre mice (n=4/genotype) at 12-13 months of age were subjected to vascular and glial cell-enriched single cell RNA-sequencing (scRNA-seq). a, Feature plot of canonical markers defining major cell types. b, Split dot plot depicting marker genes for each cell population in iE3/Cre and iE4/Cre scRNA-seq datasets. Marker genes were identified in an unbiased fashion blind to known cell type markers. The expression level (color intensity) and the percentage of cells in a cluster expressing a given gene (size) are reflected in circles (Cre−, red; Cre+, blue). c, The proportions of cells in each cluster. d, The numbers for each major cell type identified are as follow: Astrocyte (5881), endothelial cell (3970), smooth muscle cell (2495), pericyte (1667), neuron (3014), choroid plexus (2212), microglia (252), and oligodendrocyte (206). The proportions of cell types are shown. e, The number of differentially-expressed genes (DEGs) in glio-vascular unit upon peripheral expression of apoE3 (red) or apoE4 (blue). AC, astrocyte; EC, endothelial cell; FB, fibroblast; SMC, smooth muscle cell; PC, pericyte; MC, myeloid cell; Neu, neuron; CP, choroid plexus.
Extended Data Fig. 6. Brain transcriptional changes in astrocytes influenced by peripheral apoE isoform expression.
a, Violin plots showing the mean and variance differences between iE4/Cre− and iE4/Cre+ astrocytes for genes regulating oxidative stress, lipid metabolism, and hypoxia/stress responses (Selenop, Mfge8, Plpp3, and Vegfa). b, Gene ontology (GO) enrichment analysis for genes upregulated (red) or down-regulated (blue) in astrocytes from iE4/Cre+ mice compared to iE4/Cre− mice. c, Top gene ontology and canonical pathways enriched for DEGs in the astrocytes from iE3/Cre and iE4/Cre mice were identified by gene set enrichment analysis (GSEA). d, Violin plots showing the mean and variance differences between iE4/Cre− and iE4/Cre+ astrocytes for genes regulating astrocyte endfeet (Tmem212, Kcnj10, Sntn, Ankrd66, Stoml3, Pla2g7, Atp2a2). e, Volcano plot depicting up- and down-regulated genes in astrocytic cell populations (AC1, AC2, AC3) in iE3/Cre+ mice compared to iE3/Cre− control mice. Genes significant at the P value ≤ 0.05 and fold change ≥ 1.2 are denoted in red. f, Violin plots showing the mean and variance differences between iE3/Cre− and iE3/Cre+ astrocytes for genes regulating lactate homeostasis (Slc16a1), cellular protection (Psap and Selenow), immune response (Ifi27, Cox5a), and cellular senescence/stress response (Sod1 and Hspa5). g, Gene ontology enrichment analysis for genes up-regulated (orange) or down-regulated (green) in astrocytes from iE3/Cre+ mice compared to iE3/Cre− control mice. Limma’s moderate t-test statistics were used to rank the transcriptome-wide gene-phenotype association.
Extended Data Fig. 7. Reduced vessel-associated astrocytes and microglia in aged mice treated with apoE3 young plasma.
Brain tissues from aged mice treated with Ctrl (PBS/sodium citrate), apoE3, or apoE4 young plasma were subjected to co-immunostaining for GFAP (astrocyte) or Iba1 (for microglia) together with Glut1 (endothelial marker). a, Representative images for the hippocampus of treated experimental mice are shown. Scale bar, 50 μm. b, Quantification of total GFAP and Glut1 (Ctrl: 10 mice; E3: 13 mice; E4: 11 mice) in the hippocampus. **, P<0.0001. c, Quantification of total Iba1 and Glut1 (Ctrl: 10 mice; E3: 13 mice; E4: 11 mice) in the hippocampus. d, e, Cortical brain tissues from young plasma-treated mice were co-stained for tight junction (TJ) protein ZO1 (green) and an endothelial marker Glut1 (red). Scale bar, 50 μm. e, The total ZO1 and TJ coverage (ZO1 against Glut1) was quantified (Ctrl: 10 mice; E3: 10 mice; E4: 10 mice). *, P=0.02; **, P=0.007. f, g, Cortical brain tissues from young plasma-treated mice were co-stained for TJ protein CLDN5 (green) and an endothelial marker Glut1 (red). Scale bar, 50 μm. g, The total CLDN5 and TJ coverage (CLDN5 against Glut1) was quantified (Ctrl: 10 mice; E3: 10 mice; E4: 9 mice). Ctrl vs. E3 (*, P=0.034); E3 vs. E4 (*, P=0.044). h, i, Cortical brain tissues from young plasma-treated mice were co-stained for blood protein fibrinogen (green) and an endothelial marker Glut1 (red). Scale bar, 50 μm. i, The total fibrinogen and the fibrinogen/Glut1 signal was quantified (Ctrl: 10 mice; E3: 10 mice; E4: 10 mice). Fibrinogen: Ctrl vs. E3 (*, P=0.04); E3 vs. E4 (*, P=0.045). Fibrinogen/Glut1: Ctrl vs. E3 (**, P=0.001); E3 vs. E4 (*, P=0.015). j, k, Cortical brain tissues from young plasma-treated mice were co-stained for a pericyte marker CD13 (green) and an endothelial marker Glut1 (red). Scale bar, 50 μm. k, The total CD13 signal and pericyte coverage (CD13 against Glut1) was quantified (Ctrl: 10 mice; E3: 10 mice; E4: 10 mice). b-k, Data represent mean ± s.e.m. N.S., not significant, one-way ANOVA with a Tukey’s post-hoc test.
Extended Data Fig. 8. Liver-specific expression of apoE3 decreases, whereas apoE4 increases Aβ plaque deposition.
a, Brain sections from 9-month-old APP/PS1 mice expressing apoE3 (Cre− , n=21; Cre+, n=22) or apoE4 (Cre−, n=19; Cre+, n=15) in the liver were immunostained with a pan-Aβ antibody. The Aβ plaque burden in the hippocampus was quantified. Black circle: male; Grey circle: female. E3: *, P=0.041; E4: **, P=0.009, two-tailed Student's t-test. b, Representative images of Aβ staining in the cortex of APP/Alb/iE3 or APP/Alb/iE4 mice (murine Apoe−/− background) are shown. Scale bar, 200 μm. Images from APP/iE mice (murine Apoe+/+ background) were included as visual representation. Note that only diffused plaques were observed in APP/Alb/iE3 or iE4 mice due to the absence of murine apoE in the brain. c, d, TBS- and TBSX-soluble Aβ40 and Aβ42 levels in the cortex of 9-month-old APP/iE3/Cre mice (Cre−, n=18; Cre+, n=19) or APP/iE4/Cre mice (Cre−, n=22; Cre+, n=25) were examined by specific Aβ ELISA. c, TBS-E3_Aβ40: *, P=0.036; Aβ42: *, P=0.045. TBS-E4_Aβ40: **, P=0.008; Aβ42: **, P=0.001. d, TBSX-E3_Aβ40: *, P=0.024; Aβ42: *, P=0.030. TBSX-E4_Aβ40: **, P=0.002; Aβ42: **, P=0.0003. e, f, Brain sections from APP/PS1 mice expressing apoE3 (Cre−, n=8; Cre+, n=8) or apoE4 (Cre−, n=6; Cre+, n=6) in the liver (murine Apoe−/− background) were labeled for fibrillar Aβ using Thioflavin S (Thio S). Scale bar (upper panels), 1 mm; Scale bar (bottom panels), 100 μm. Images from APP/iE mice (murine Apoe+/+ background) were included for comparison. The amount of fibrillar plaques in the APP/Alb/iE3 or APP/Alb/iE4 mice was minimal due to the absence of murine apoE in the brain. The percentage of area covered by Thio S-positive plaques in the cortex and hippocampus of experimental mice was quantified. g, h, Brain sections from 9-month-old APP/PS1 mice expressing apoE3 (Cre−, n=27; Cre+, n=28) or apoE4 (Cre−, n=19; Cre+, n=19) in the liver were immunostained with an Iba1 antibody. Scale bar, 100 μm. The immunoreactivity of Iba1 in cortex and hippocampus were quantified. Data expressed as mean ± s.e.m. Cortex: *, P = 0.011; Hippo: *, P = 0.030. N.S., not significant, two-tailed Student's t-test.
Extended Data Fig. 9. AAV-mediated liver expression of apoE4 enhances amyloid pathology and related toxicity.
a, Schematic illustration of the experimental paradigm. 5xFAD amyloid mice at 1-1.5 month of age were transduced with AAV-Alb-apoE3 or AAV-Alb-apoE4 virus via intravenous injection. b, c, The amyloid deposition in the brain of experimental mice at 4 months of age was examined by immunostaining for Aβ. Scale bar, 1 mm. The amyloid plaque burdens in the cortex and hippocampus (E3, n=14; E4, n=16) were quantified. **, P = 0.007. d, e, ApoE levels in the plasma and brain of experimental mice (E3, n=15; E4, n=16) were measured by ELISAs. **, P = 0.0098. f, TBSX-soluble and -insoluble (guanidine; GDN) Aβ40 and Aβ42 levels in the cortex of 4-month-old 5xFAD mice transduced with AAV-Alb-apoE3 (n=15) or AAV-Alb-apoE4 (n=16) were examined by specific Aβ ELISA. TBSX-Aβ40 (*, P = 0.012); TBSX-Aβ42 (*, P = 0.031). GDN-Aβ40 (*, P = 0.049); GDN-Aβ42 (*, P = 0.033). g, Thio S-positive plaques in the cortex of experimental mice were shown and quantified. Scale bar, 100 μm. h, i, Brain sections from experimental mice (n=12/genotype) were immunostained with GFAP or Iba1 antibody. Scale bar, 100 μm. The immunoreactivity of GFAP and Iba1 in cortex and hippocampus were quantified. j, Representative images of plaque-associated microglia in mice expressing apoE3 or apoE4 in the liver are shown. Scale bar, 50 μm. The number of Iba1-positive microglia (green) surrounding Aβ plaque (red) between 50-300 μm2 plaque sizes were quantified. Each dot represents the average value from an individual mouse (E3, n=10; E4, n=11). *, P = 0.049. k, Co-immunofluorescence staining of LAMP1 (green) and Aβ plaques (red) was used to examine plaque-associated neuritic dystrophy. Scale bar, 50 μm. The LAMP1 immunoreactivity was quantified. *, P = 0.027. l, LAMP1 immunoreactivity was positively correlated with Thio S-positive fibrillar plaques. c-l, Data represent mean ± s.e.m., two-tailed Student's t-test.
Extended Data Fig. 10. Potential mechanisms by which peripheral apoE4 impacts cerebrovascular integrity, brain function, and AD pathology.
Peripheral apoE4 may modulate plasma factors that promote inflammatory responses and hypoxia/stress in endothelial cells along with cells in the glio-vascular unit1. Tight junction markers (i.e., Claudin-5 and ZO1) and astrocyte end-feet marker (i.e., AQP4) are down-regulated, whereas the dysregulation of ECM and vessel-associated gliosis are exacerbated in mice expressing apoE4 in the liver. Compromised BBB integrity along with an increase of blood-derived proteins (e.g., albumin, IgG, fibrinogen) in the brain, some of which have been shown to increase microglia-mediated oxidative stress and disrupt synaptic function2, may contribute to cognitive deficits in apoE4 mice. Additionally, the impairment of cerebrovascular functions and altered microglial responses may directly or indirectly influence Aβ clearance and Aβ deposition, which together exacerbate amyloid pathology in apoE4 mice. Not depicted here, peripheral apoE3 expression may regulate blood factors that benefit the endothelial integrity and vascular health. For example, Timp3, previously shown to ameliorate the vascular diseases through inhibiting excess matrix metalloproteinase (MMP) activity and inflammation3,4, is elevated in apoE3 mice and increases apoE4 plasma-associated endothelial integrity in the human iPSC-derived cellular model.
Supplementary Material
Acknowledgments:
We are grateful to Dr. Dennis Dickson, Monica Castanedes Casey, Linda Rousseau, Virginia Phillips, and Ariston Librero at Mayo Clinic Histology Core for the immunohistochemical analyses. We thank Mike Heckman at Mayo Clinic for the consultation of the statistical analysis. We thank Drs. Mingjie Li and Joyce Snyder from the Hope Center Viral Vectors Core at the Washington University School of Medicine for producing the AAV viruses.
Funding:
This work was supported by NIH grants RF1AG046205, R37AG027924, RF1AG057181, U19AG069701 (to G.B.) and a grant from the Cure Alzheimer’s Fund (to G.B.), NIH grants R21AG057981 and R01AG62110 (to C.-C.L.), a grant from the BrightFocus Foundation (to C.-C.L.), and NIH grants U01AG046170, RF1AG054014 and U01AG052411 (to B.Z.).
Footnotes
Competing interests: G.B. consults for SciNeuro and Lexeo, is on the scientific advisory board for Kisbee, and has consulted for AbbVie, E-Scape, Eisai, and Vida Ventures. Other authors declare no competing interests.
Data availability
The datasets generated and/or analyzed in the current study are attached. The bulk RNA-Seq data and scRNA-Seq data that support the findings of this study were deposited in GEO repository (Accession number for bulk RNA-Seq: GSE125501; Accession number for scRNA-Seq: GSE206204).
References
- 1.Guo T, et al. Molecular and cellular mechanisms underlying the pathogenesis of Alzheimer's disease. Molecular neurodegeneration 15, 40 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de la Torre JC Alzheimer disease as a vascular disorder: nosological evidence. Stroke 33, 1152–1162 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Whitmer RA, Karter AJ, Yaffe K, Quesenberry CP Jr. & Selby JV Hypoglycemic episodes and risk of dementia in older patients with type 2 diabetes mellitus. JAMA 301, 1565–1572 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zlokovic BV Neurovascular pathways to neurodegeneration in Alzheimer's disease and other disorders. Nature reviews. Neuroscience 12, 723–738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Marco LY, Farkas E, Martin C, Venneri A & Frangi AF Is Vasomotion in Cerebral Arteries Impaired in Alzheimer's Disease? J Alzheimers Dis 46, 35–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen B, Bix G & Yao Y Basal lamina changes in neurodegenerative disorders. Molecular neurodegeneration 16, 81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrer LA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278, 1349–1356 (1997). [PubMed] [Google Scholar]
- 8.Hyman BT, et al. Apolipoprotein E and cognitive change in an elderly population. Ann Neurol 40, 55–66 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Castellano JM, et al. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med 3, 89ra57 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiman EM, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A 106, 6820–6825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CC, Kanekiyo T, Xu H & Bu G Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9, 106–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell RD, et al. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature 485, 512–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamazaki Y, Zhao N, Caulfield TR, Liu CC & Bu G Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat Rev Neurol 15, 501–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams T, Borchelt DR & Chakrabarty P Therapeutic approaches targeting Apolipoprotein E function in Alzheimer's disease. Molecular neurodegeneration 15, 8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linton MF, et al. Phenotypes of apolipoprotein B and apolipoprotein E after liver transplantation. J Clin Invest 88, 270–281 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lane-Donovan C, et al. Genetic Restoration of Plasma ApoE Improves Cognition and Partially Restores Synaptic Defects in ApoE-Deficient Mice. J Neurosci 36, 10141–10150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Morillo E, et al. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer's disease patients and controls. Acta Neuropathol 127, 633–643 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Tai LM, et al. The role of APOE in cerebrovascular dysfunction. Acta Neuropathol 131, 709–723 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haan MN, Shemanski L, Jagust WJ, Manolio TA & Kuller L The role of APOE epsilon4 in modulating effects of other risk factors for cognitive decline in elderly persons. JAMA 282, 40–46 (1999). [DOI] [PubMed] [Google Scholar]
- 20.Liu CC, et al. ApoE4 Accelerates Early Seeding of Amyloid Pathology. Neuron 96, 1024–1032 e1023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davalos D, et al. Fibrinogen-induced perivascular microglial clustering is required for the development of axonal damage in neuroinflammation. Nat Commun 3, 1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukuda E, et al. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. Eur J Neurosci 28, 1362–1376 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Siddiqui MR, Mayanil CS, Kim KS & Tomita T Angiopoietin-1 Regulates Brain Endothelial Permeability through PTPN-2 Mediated Tyrosine Dephosphorylation of Occludin. PLoS One 10, e0130857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bell RD, et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 68, 409–427 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheng J & Xu Z Three decades of research on angiogenin: a review and perspective. Acta Biochim Biophys Sin (Shanghai) 48, 399–410 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baeten KM & Akassoglou K Extracellular matrix and matrix receptors in blood-brain barrier formation and stroke. Dev Neurobiol 71, 1018–1039 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song WM & Zhang B Multiscale Embedded Gene Co-expression Network Analysis. PLoS Comput Biol 11, e1004574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson-Leary J, et al. Inflammation and vascular remodeling in the ventral hippocampus contributes to vulnerability to stress. Transl Psychiatry 7, e1160 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piskunov A, et al. Chronic combined stress induces selective and long-lasting inflammatory response evoked by changes in corticosterone accumulation and signaling in rat hippocampus. Metab Brain Dis 31, 445–454 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Piedrahita JA, Zhang SH, Hagaman JR, Oliver PM & Maeda N Generation of mice carrying a mutant apolipoprotein E gene inactivated by gene targeting in embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America 89, 4471–4475 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nelimarkka L, et al. Decorin is produced by capillary endothelial cells in inflammation-associated angiogenesis. Am J Pathol 158, 345–353 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanlandewijck M, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Chen MB, et al. Brain Endothelial Cells Are Exquisite Sensors of Age-Related Circulatory Cues. Cell Rep 30, 4418–4432 e4414 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceradini DJ, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med 10, 858–864 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Noubade R, et al. von-Willebrand factor influences blood brain barrier permeability and brain inflammation in experimental allergic encephalomyelitis. Am J Pathol 173, 892–900 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan D & Kassiri Z Biology of Tissue Inhibitor of Metalloproteinase 3 (TIMP3), and Its Therapeutic Implications in Cardiovascular Pathology. Front Physiol 11, 661 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dewing JM, Carare RO, Lotery AJ & Ratnayaka JA The Diverse Roles of TIMP-3: Insights into Degenerative Diseases of the Senescent Retina and Brain. Cells 9(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckhouse SR, et al. Local hydrogel release of recombinant TIMP-3 attenuates adverse left ventricular remodeling after experimental myocardial infarction. Sci Transl Med 6, 223ra221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Middeldorp J, et al. Preclinical Assessment of Young Blood Plasma for Alzheimer Disease. JAMA Neurol 73, 1325–1333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villeda SA, et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med 20, 659–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jankowsky JL, et al. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet 13, 159–170 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Liu CC, et al. Neuronal heparan sulfates promote amyloid pathology by modulating brain amyloid-beta clearance and aggregation in Alzheimer's disease. Science translational medicine 8, 332ra344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hawkes CA, et al. Disruption of arterial perivascular drainage of amyloid-beta from the brains of mice expressing the human APOE epsilon4 allele. PLoS One 7, e41636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holtzman DM, et al. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America 97, 2892–2897 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ulrich JD, et al. ApoE facilitates the microglial response to amyloid plaque pathology. J Exp Med 215, 1047–1058 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heneka MT, et al. Neuroinflammation in Alzheimer's disease. Lancet Neurol 14, 388–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulgart HR, Neczypor EW, Wold LE & Mackos AR Microbial involvement in Alzheimer disease development and progression. Molecular neurodegeneration 15, 42 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pluvinage JV & Wyss-Coray T Systemic factors as mediators of brain homeostasis, ageing and neurodegeneration. Nat Rev Neurosci 21, 93–102 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Boada M, et al. A randomized, controlled clinical trial of plasma exchange with albumin replacement for Alzheimer's disease: Primary results of the AMBAR Study. Alzheimers Dement 16, 1412–1425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen HM, et al. Peripheral apoE isoform levels in cognitively normal APOE epsilon3/epsilon4 individuals are associated with regional gray matter volume and cerebral glucose metabolism. Alzheimers Res Ther 9, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stohr R, et al. Loss of TIMP3 exacerbates atherosclerosis in ApoE null mice. Atherosclerosis 235, 438–443 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Thevenard J, et al. Low-density lipoprotein receptor-related protein-1 mediates endocytic clearance of tissue inhibitor of metalloproteinases-1 and promotes its cytokine-like activities. PLoS One 9, e103839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Markiewski MM & Lambris JD The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol 171, 715–727 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nation DA, et al. Blood-brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat Med 25, 270–276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hussain B, Fang C & Chang J Blood-Brain Barrier Breakdown: An Emerging Biomarker of Cognitive Impairment in Normal Aging and Dementia. Front Neurosci 15, 688090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Merlini M, et al. Fibrinogen Induces Microglia-Mediated Spine Elimination and Cognitive Impairment in an Alzheimer's Disease Model. Neuron 101, 1099–1108 e1096 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cortes-Canteli M, et al. Fibrinogen and beta-amyloid association alters thrombosis and fibrinolysis: a possible contributing factor to Alzheimer's disease. Neuron 66, 695–709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan Y, et al. 7,8-Dihydroxyflavone Ameliorates Cognitive Impairment by Inhibiting Expression of Tau Pathology in ApoE-Knockout Mice. Front Aging Neurosci 8, 287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hartman RE, et al. Behavioral phenotyping of GFAP-apoE3 and -apoE4 transgenic mice: apoE4 mice show profound working memory impairments in the absence of Alzheimer's-like neuropathology. Exp Neurol 170, 326–344 (2001). [DOI] [PubMed] [Google Scholar]
- 61.Kim J, et al. Anti-apoE immunotherapy inhibits amyloid accumulation in a transgenic mouse model of Abeta amyloidosis. J Exp Med 209, 2149–2156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao F, et al. Anti-ApoE antibody given after plaque onset decreases Abeta accumulation and improves brain function in a mouse model of Abeta amyloidosis. J Neurosci 34, 7281–7292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huynh TV, et al. Lack of hepatic apoE does not influence early Abeta deposition: observations from a new APOE knock-in model. Molecular neurodegeneration 14, 37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miyazaki S, Miyazaki T, Tashiro F, Yamato E & Miyazaki J Development of a single-cassette system for spatiotemporal gene regulation in mice. Biochem Biophys Res Commun 338, 1083–1088 (2005). [DOI] [PubMed] [Google Scholar]
- 65.Chakrabarty P, et al. IFN-gamma promotes complement expression and attenuates amyloid plaque deposition in amyloid beta precursor protein transgenic mice. J Immunol 184, 5333–5343 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu CC, et al. APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia. Sci Transl Med 13, eabc9375 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drew PJ, Blinder P, Cauwenberghs G, Shih AY & Kleinfeld D Rapid determination of particle velocity from space-time images using the Radon transform. J Comput Neurosci 29, 5–11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao J, et al. Retinoic acid isomers facilitate apolipoprotein E production and lipidation in astrocytes through the retinoid X receptor/retinoic acid receptor pathway. J Biol Chem 289, 11282–11292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu CC, et al. Deficiency in LRP6-mediated Wnt signaling contributes to synaptic abnormalities and amyloid pathology in Alzheimer's disease. Neuron 84, 63–77 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rogers JT, et al. Subacute ibuprofen treatment rescues the synaptic and cognitive deficits in advanced-aged mice. Neurobiol Aging 53, 112–121 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu CC, et al. Tau and apolipoprotein E modulate cerebrovascular tight junction integrity independent of cerebral amyloid angiopathy in Alzheimer's disease. Alzheimers Dement 16, 1372–1383 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kalari KR, et al. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics 15, 224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hansen KD, Irizarry RA & Wu Z Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13, 204–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Langfelder P & Horvath S WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song W-M & Zhang B Multiscale Embedded Gene Co-expression Network Analysis. PLoS Comput Biol 11, e1004574 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yamazaki A, et al. Preparation of single cell suspensions enriched in mouse brain vascular cells for single-cell RNA sequencing. STAR Protoc 2, 100715 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yamazaki Y, et al. Vascular ApoE4 Impairs Behavior by Modulating Gliovascular Function. Neuron 109, 438–447 e436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heo J, et al. Hepatic precursors derived from murine embryonic stem cells contribute to regeneration of injured liver. Hepatology 44, 1478–1486 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Hu J, et al. Opposing effects of viral mediated brain expression of apolipoprotein E2 (apoE2) and apoE4 on apoE lipidation and Abeta metabolism in apoE4-targeted replacement mice. Molecular neurodegeneration 10, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dey KK, et al. Deep undepleted human serum proteome profiling toward biomarker discovery for Alzheimer's disease. Clin Proteomics 16, 16–16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang H, et al. Integrated analysis of ultra-deep proteomes in cortex, cerebrospinal fluid and serum reveals a mitochondrial signature in Alzheimer’s disease. Molecular neurodegeneration 15, 43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu P, Duong DM & Peng JM Systematical Optimization of Reverse-Phase Chromatography for Shotgun Proteomics. J Proteome Res 8, 3944–3950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Z, et al. 27-Plex Tandem Mass Tag Mass Spectrometry for Profiling Brain Proteome in Alzheimer’s Disease. Analytical Chemistry 92, 7162–7170 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Z, et al. High-throughput and Deep-proteome Profiling by 16-plex Tandem Mass Tag Labeling Coupled with Two-dimensional Chromatography and Mass Spectrometry. JoVE, e61684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, et al. JUMP: a tag-based database search tool for peptide identification with high sensitivity and accuracy. Molecular & cellular proteomics : MCP 13, 3663–3673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Peng J, Elias JE, Thoreen CC, Licklider LJ & Gygi SP Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2, 43–50 (2003). [DOI] [PubMed] [Google Scholar]
- 87.Niu M, et al. Extensive Peptide Fractionation and y1 Ion-Based Interference Detection Method for Enabling Accurate Quantification by Isobaric Labeling and Mass Spectrometry. Anal Chem 89, 2956–2963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qian T, et al. Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells. Sci Adv 3, e1701679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in the current study are attached. The bulk RNA-Seq data and scRNA-Seq data that support the findings of this study were deposited in GEO repository (Accession number for bulk RNA-Seq: GSE125501; Accession number for scRNA-Seq: GSE206204).