Abstract
In this report we describe experiments to investigate a simple virulence model in which Pseudomonas aeruginosa PAO1 rapidly paralyzes and kills the nematode Caenorhabditis elegans. Our results imply that hydrogen cyanide is the sole or primary toxic factor produced by P. aeruginosa that is responsible for killing of the nematode. Four lines of evidence support this conclusion. First, a transposon insertion mutation in a gene encoding a subunit of hydrogen cyanide synthase (hcnC) eliminated nematode killing. Second, the 17 avirulent mutants examined all exhibited reduced cyanide synthesis, and the residual production levels correlated with killing efficiency. Third, exposure to exogenous cyanide alone at levels comparable to the level produced by PAO1 killed nematodes with kinetics similar to those observed with bacteria. The killing was not enhanced if hcnC mutant bacteria were present during cyanide exposure. And fourth, a nematode mutant (egl-9) resistant to P. aeruginosa was also resistant to killing by exogenous cyanide in the absence of bacteria. A model for nematode killing based on inhibition of mitochondrial cytochrome oxidase is presented. The action of cyanide helps account for the unusually broad host range of virulence of P. aeruginosa and may contribute to the pathogenesis in opportunistic human infections due to the bacterium.
Pseudomonas aeruginosa is a ubiquitous gram-negative bacterium that is virulent towards a wide range of organisms, including bacteria, plants, nematodes, insects, and mammals (5, 9, 17, 19, 35, 36, 41, 48, 49, 62). In humans, P. aeruginosa chronically infects the lungs of most cystic fibrosis patients, causes serious infections of burn wounds and eye lesions, and causes systemic infections of immunocompromised individuals (21, 29, 33, 39). The bacterium's pathogenic versatility is reflected in its large arsenal of secreted and surface-associated virulence factors and in the complexity of the regulatory circuitry with which it controls these factors. Among the specific virulence factors that it produces are adhesins, such as pili and filamentous hemagglutinin (14, 39); protein toxins, such as phospholipase, proteases, and ADP-ribosylating enzymes (39, 64); and small-molecule poisons, such as phenazines, rhamnolipid biosurfactant, and cyanide (4, 8, 44). Additionally, the genome of P. aeruginosa boasts the highest proportion of predicted regulatory genes of any of the bacterial genomes sequenced to date (61), which is indicative of the bacterium's remarkable ability to adapt and thrive in numerous pathogenic and nonpathogenic environments.
Several model systems for Pseudomonas pathogenesis have been developed recently, and numerous genes required for virulence towards model hosts are also required for virulence towards mammals. For example, mutants of P. aeruginosa PA-14 exhibiting reduced virulence towards Arabidopsis or Caenorhabditis elegans also exhibit reduced virulence in a burned-mouse infection model (49, 50, 62). In addition, a putative Pseudomonas signal transduction gene cluster required for full virulence towards Drosophila melanogaster also mediates mammalian epithelial cell injury (19, 37). Such examples help illustrate the value of using genetically tractable model organisms to identify P. aeruginosa virulence determinants (24, 25, 40).
We recently described a virulence model in which P. aeruginosa PAO1 rapidly paralyzes and kills the nematode C. elegans (17). This killing, termed paralytic killing, is mediated by a diffusible factor that is under control of both the LasR and RhlR quorum sensing regulators. This killing also requires a functional copy of the C. elegans gene egl-9. The EGL-9 protein, which is strongly expressed in the nematode body wall and pharyngeal muscles, has homologues in a wide range of organisms, including mammals and Drosophila (3, 22). Paralytic killing of nematodes by strain PAO1 may be distinct from two modes of nematode killing reported for strain PA-14 based on differences in gene and growth condition requirements (17, 41, 62).
In this report we describe experiments designed to identify bacterial factors that mediate paralytic killing of C. elegans by strain PAO1. Our results indicate that hydrogen cyanide is the primary toxic factor responsible for the phenomenon.
MATERIALS AND METHODS
Strains, plasmids, growth media, and culture conditions.
The P. aeruginosa strains used were PAO1 (34) from the laboratory of B. Iglewski, PAO-R1, a lasR mutant of PAO1 (26), two pvd strains carrying transposon insertions in the PA2401 and PA2424 genes (provided by D. D'Argenio), and the mTn5-Tc (20) insertion mutants listed in Table 1. The Escherichia coli strains used were DH5α (52) for plasmid construction and SM10λpir (55) for conjugal suicide plasmid delivery. The growth media used were brain heart infusion (BHI) agar (Difco), L agar (52), skim milk agar (57), King's B medium (38), and L broth. Plasmids were maintained in P. aeruginosa in media supplemented with 100 μg of carbenicillin per ml and in E. coli in media supplemented with 100 μg of ampicillin per ml or 40 μg of tetracycline per ml. To construct plasmids used for hcn complementation, an 8,968-bp XhoI fragment carrying the P. aeruginosa hcnABC operon was gel purified from an XhoI-BglII-ScaI digest of cosmid 011 (supplied by Matt Wolfgang and S. Lory), whose insert corresponds to nucleotides 2,396,530 to 2,441,543 in the PAO1 single contig sequence (www.pseudomonas.com). The XhoI fragment was cloned in both orientations into the SalI site of pUCP18 (53) to obtain pLG2 (Fig. 1) and pLG3. pLG3 was then digested with XbaI and religated to obtain pLG4 (Fig. 1). All constructs were confirmed by restriction analysis. For hcnC complementation assays, MP507 transformed with either pLG2, pLG4, or pUCP18 was tested in a standard worm killing assay after growth in individual chambers (see below) on BHI agar supplemented with 40 μg of tetracycline per ml and 100 μg of carbenicillin per ml. Standard molecular biology protocols were used throughout (52).
TABLE 1.
Mutants defective in paralytic killing
| Strain | Insertion sitea | Geneb | Function | % Killingc |
|---|---|---|---|---|
| PAO1 | 99 (±0) | |||
| Class I (strongly avirulent strains) | ||||
| MP503 | 3,571,648 | eda | 2-Keto-3-deoxy-6-phosphogluconate aldolase; Entner-Doudoroff pathway | 4 (±2) |
| MP505 | 435,468 | proC | Δ-1-Pyrroline-5-carboxylate reductase; proline biosynthesis | 4 (±3) |
| MP506 | 435,230 | proC | Δ-1-Pyrroline-5-carboxylate reductase; proline biosynthesis | 0 (±0) |
| MP504 | 812,969 | PA0745d | Probably enoyl-coenzyme A hydratase; fatty acid degradation | 13 (±10) |
| MP507 | 2,415,450 | hcnC | Hydrogen cyanide synthase | 3 (±2) |
| MP508 | 45,846 | PA0041d | Homologue of fhaB, Bordetella filamentous hemagglutinin | 0 (±0) |
| MP501 | 4,423,808 | PA3946d | Homologue of bvgS, Bordetella two-component sensor kinase virulence gene regulator | 0 (±0) |
| MP502 | 1,015,249 | gacS | Homologue of Pseudomonas syringae two-component sensor kinase controlling disease lesion formation | 9 (±5) |
| MP511 | Unsequencede | 0 (±0) | ||
| Class II (moderately avirulent strains) | ||||
| MP554 | 3,572,897 | zwf | Glucose-6-phosphate dehydrogenase; Entner-Doudoroff pathway | 41 (±19) |
| MP555 | 6,098,814 | soxA | Sarcosine oxidase | 38 (±20) |
| MP556 | 6,100,002 | soxA | Sarcosine oxidase | 39 (±19) |
| MP557 | 1,032,886 | purM | Phosphoribosylaminoimldazole synthetase; purine biosynthesis | 92 (±3) |
| MP558 | 4,216,505 | purL | Phosphoribosylformylglycinamidine synthase; purine biosynthesis | 90 (±5) |
| MP559 | 874,405 | prpB | Carboxyphosphonoenolpyruvate phosphonomutase; fatty acid and phospholipid metabolism | 83 (±15) |
| MP560 | 873,168 | prpC | Citrate synthase 2 | 72 (±28) |
| MP561 | 1,758,910 | gpdA | Glycerol-3-phosphate dehydrogenase; specific to fatty acid and phospholipid metabolism | 57 (±23) |
| MP562 | 2,927,500 | PA2587 | Putative salicylate hydroxylase; quinolone signal synthesis | 27 (±16) |
| MP571 | 6,193,910 | znuB | Permease of ABC zinc transporter | 39 (±20) |
| MP573 | 4,290,026 | phpA | Aminopeptidase; protein modification, alginate biosynthesis | 28 (±13) |
| MP574 | 5,993,742 | algC | Lipopolysaccharide and alginate biosynthesis | 75 (±15) |
| MP572 | 5,100,129 | pilW | Type 4 pili | 56 (±10) |
| MP551 | 1,086,674 | PA1003d | Putative transcriptional regulator with LysR family signature | 31 (±22) |
| MP552 | 5,304,505 | PA4725d | Putative amino acid permease fused to putative two-component sensor histidine kinase | 42 (±15) |
| MP553 | 5,304,930 | PA4725d | Putative amino acid permease fused to putative two-component sensor histidine kinase | 76 (±16) |
The transposon insertion site corresponds to the chromosomal location in the PAO1 single contig sequence (www.pseudomonas.com).
Boldface type indicates known P. aeruginosa genes; lightface type indicates close homologues of known genes. PA numbers are designations assigned by the web site (www.pseudomonas.com).
Percentages of killing are averages based on at least three independent killing assays for each strain. The numbers in parentheses are standard errors of the means.
Gene not experimentally characterized in studies of pseudomonads.
Repeated attempts to sequence were unsuccessful.
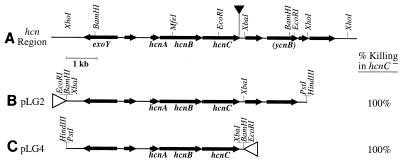
FIG. 1.
Complementation of the killing defect in hcnC mutant MP507. (A) Restriction map of the hcnABC region, showing the locations and orientations of known genes hcnA, hcnB, hcnC, and exoY and of putative genes (unlabeled arrows), including a homologue of the conserved hypothetical E. coli protein gene ycnB. The solid triangle indicates the location of the mTn5-Tc transposon insertion in the hcnC mutant MP507. (B and C) Maps of the insertion regions in recombinant plasmids carrying the hcnABC region. The results of nematode killing assays for hcnC mutant MP507 carrying these plasmids are also shown. The open triangles indicate the orientations of the Plac promoter in the pUCP18 vector. The killing percentages are averages based on three separate assays. MP507 carrying only the vector plasmid pUCP18 exhibited less than 1% killing.
The C. elegans strains used were wild-type Bristol strain N2 and JT330, an egl-9 mutant (17). Nematodes were grown at 22°C and were handled by using standard techniques (6, 72).
Nematode paralytic killing assay.
Unless indicated otherwise, all paralytic killing assays were carried out by spreading 150 μl of a 2- to 7-day-old P. aeruginosa colony suspended in BHI broth at an optical density at 660 nm (OD660) of ∼0.1 onto a 3.5-cm-diameter BHI agar plate containing 4 ml of BHI agar. After the plate was incubated for 24 h at 37°C, N2 nematodes from stock plates were collected in M9 buffer, and a 50-μl aliquot (containing 20 to 200 adult animals) was spotted onto the P. aeruginosa lawn. The plate was then incubated for 4 h at room temperature with the lid on, and paralytic nematode killing was scored with a dissecting microscope. As described previously (17), worms were considered dead if they did not move spontaneously and did not respond detectably to tapping of the assay plate against the microscope stage. For experiments in which individual chambers were used (see below), each 3.5-cm-diameter plate was enclosed in a 10-cm-diameter petri plate, which was then either sealed with Parafilm (sealed chamber) or left unsealed (unsealed chamber).
Transposon mutagenesis of PAO1.
Most transposon insertion mutants were generated by using transposon mTn5-Tc (20). MP501 and MP551 were generated by using ISphoA/hah-Tc, a transposon Tn5 derivative that will be described elsewhere (unpublished data), and MP508 was generated by using Tn5 (18). For transposon mutagenesis, a 37°C overnight aerated culture of E. coli SM10 λpir/pUT-mTn5-Tc (20) or SM10 λpir/pUT-IsphoA/hah-Tc grown in L broth supplemented with 100 μg of ampicillin per ml was diluted 1:10 into fresh L broth containing ampicillin and grown with aeration for 45 min at 37°C. A 0.5-ml aliquot of this culture was mixed with 0.5 ml of a 42°C nonaerated overnight L broth culture of PAO1. The mixture was filtered with a Nalgene analytical test filter (pore size, 0.45 μm) and washed with 1 ml of 10 mM Mg2SO4. The filter was then removed from the apparatus, transferred to an L agar plate, incubated at 37°C for 1 h to allow conjugation and transposition to occur, and then transferred to a test tube containing 1 ml of L broth, and the cells were washed from the filter by vortexing. Cells were plated onto L agar containing 10 μg of chloramphenicol per ml to counterselect for E. coli and 60 μg of tetracycline per ml to select for growth of P. aeruginosa cells carrying transposon insertions. Individual colonies appeared after 1 to 2 days of incubation at 37°C.
Mutant screening.
To screen for non-nematode-killing mutants, individual transposon insertion mutants were suspended in BHI broth at a density sufficient to make the broth visibly turbid. Then 150 μl of each suspension was plated onto a 3.5-cm-diameter BHI agar plate, and after 24 h of incubation at 37°C worm killing was assayed. Strains which exhibited at least a 10% reduction in killing compared to the wild type were saved and retested. Strains that arose from 37 independent mutagenesis events were screened.
DNA sequencing.
The chromosomal DNA flanking the transposon insertions was sequenced after semirandom PCR amplification or cloning. For semirandom PCR, a variation of a protocol described by Chun et al. (12) was used. One microliter of a 50-μl boiled single-colony suspension in distilled H2O was used as the template DNA in a 20-μl PCR mixture containing primer MTN5I.1 (5′-CGAGGGCTTTACTAAGCTG-3′) and either primer CEKG 2A (5′-GGCCACGCGTCGACTAGTACN10AGAG-3′), CEKG 2B (5′-GGCCACGCGTCGACTAGTACN10ACGCC-3′), or CEKG 2C (5′-GGCCACGCGTCGACTAGTACN10GATAT-3′); 1 μl of a 1:5 dilution of this reaction mixture was used as the template DNA for a second PCR performed with primers MTN5O.1 (5′-ATTCGTCGACAAGCTTCGG-3′) and CEKG 4 (5′-GGCCACGCGTCGACTAGTAC-3′). For the first reaction, the thermocycler conditions were 94°C for 2 min, followed by six cycles of 94°C for 30 s, 42°C for 30 s (with the temperature reduced 1°C per cycle), and 72°C for 3 min and then 25 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min; for the second reaction, the thermocycler conditions were 30 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min. Samples that produced distinct bands on an agarose gel after the second reaction were cleaned with a PCR purification kit (Qiagen) and sequenced by using primer MNT5S.1 (5′-GACAAGCTTCGGCCGCCT-3′). For cloning, chromosomal DNA digested with PstI was ligated into PstI-digested pUC18 (73). The ligation mixture was electroporated into DH5α, and transformants were selected with tetracycline. The chromosomal locations of the insertions were determined by BLAST analysis of the transposon-adjacent chromosomal DNA sequences compared with the complete PAO1 genome (www.pseudomonas.com).
Exoproduct assays.
To measure cyanide production, we used a protocol modified from a protocol generously supplied by D. Haas and based on the method of Gewitz et al. (27). Strains were grown on 3.5-cm-diameter BHI agar plates in individual unsealed chambers for 24 h at 37°C and then enclosed without lids in individual sealed chambers which also contained a 1-ml reservoir of 4 M NaOH (in an inverted 3.5-cm-diameter plate lid). After 4 h of incubation at 30°C, the NaOH was collected and diluted to 0.09 M with double-distilled H2O. If necessary, the sample was further diluted with 0.09 M NaOH to bring the cyanide concentration to within the linear range of the detection procedure (0 to 10 μM). The cyanide in the sample was quantified by comparison with standards of KCN dissolved in 0.09 M NaOH: 105-μl aliquots of the samples were mixed with 350-μl aliquots of a fresh 1:1 mixture of 0.1 M o-dinitrobenzene (Sigma) in ethylene-glycol monomethyl ether (Sigma) and 0.2 M p-nitrobenzaldehyde (Sigma) in ethylene-glycol monomethyl ether. After exactly 30 min of incubation at the ambient temperature (22°C), the OD578 was measured.
Pyocyanin production was assayed by the method of Essar et al. (23): 24-h plate cultures were grown as described above for the nematode killing assay in unsealed individual chambers. The lawn-bearing agar from each plate was diced and extracted for 3 h with 4 ml of chloroform. The chloroform was then extracted with one-seventh volume of 0.2 M HCl, and the pyocyanin in the aqueous phase was quantified by measuring the OD520.
Pyoverdine production was assayed by previously described methods (15, 60) by measuring the OD404, relative to that of pvd strains (generously supplied by D. D'Argenio), of cell-free supernatants from saturated overnight 37°C aerated cultures grown in King's B medium and adjusted for culture density (38). Exoprotease production was assessed by spotting 5-μl aliquots of cultures at an OD660 of ∼0.1 onto skim milk agar plates, incubating the plates at 37°C overnight, and measuring zones of clearance from the edges of the growth spots.
Treatment of nematodes with exogenous cyanide.
For direct exposure to exogenous cyanide, nematodes were placed on a 3.5-cm-diameter BHI agar plate without a lid, and this plate was then sealed in a 10-cm-diameter petri plate containing an inverted 3.5-cm-diameter lid. The inverted lid contained separated 0.25-ml aliquots of 0.18 M HCl and a defined amount of KCN dissolved in 0.09 M NaOH. After the 10-cm-diameter plate was sealed, the aliquots were mixed by tipping the plate, thus acidifying the cyanide solution and releasing HCN gas. For experiments in which cyanide exposure in the presence of bacteria was examined (see Fig. 3), worms were placed on a standard 24-h pregrown lawn of bacteria rather than in an empty BHI agar plate.
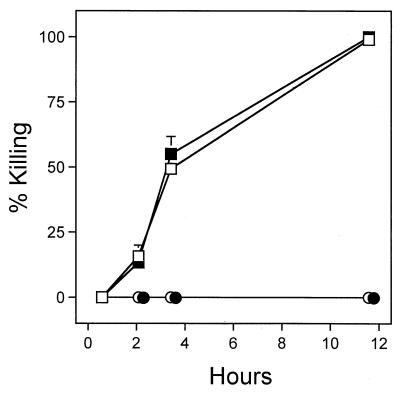
FIG. 3.
Killing of C. elegans by hydrogen cyanide gas with and without exposure to bacteria. Wild-type (squares) or egl-9 (circles) nematodes placed on plates containing either no bacteria (open symbols) or hcnC mutant MP507 (solid symbols) were exposed to 1 μmol of hydrogen cyanide in sealed 10-cm-diameter petri plates. Each datum point represents the average based on triplicate experiments.
RESULTS
P. aeruginosa mutants with impaired ability to kill C. elegans.
To help identify the substance or substances produced by P. aeruginosa that are toxic to C. elegans, we screened chromosomal mTn5-Tc transposon insertion mutants for reduced nematode killing. Of approximately 3,000 mutants screened, 25 strains with significant defects were recovered (Table 1). Slowly growing mutants that formed small colonies on nutrient agar were not included in the analysis. The mutants could be grouped into two classes based on the strength and reproducibility of their killing defects. Nine of the mutants (class I) killed ≤13% of the nematodes, whereas 16 of the mutants (class II) killed 27 to 92% of the nematodes. The class I mutants were quantitatively more reproducible than the class II mutants in terms of the defects in killing observed in different trials. Three of the mutant strains listed in Table 1 (MP508, MP552, and MP553) exhibited abundant papillation of secondary colonies upon prolonged incubation (several days) on rich media, suggesting that there was a reduction in the viability of the parent strains with outgrowth of fitter variants (data not shown).
We identified the transposon insertion sites for 24 of the 25 mutants by PCR amplification of the genomic sequence flanking each transposon, DNA sequencing, and comparison with the PAO1 genome sequence. Twenty-one genes were represented in the mutant set. These genes included regulatory genes, genes encoding metabolic enzymes, a gene for a probable metal transporter, and five other genes with known or postulated virulence functions (Table 1). Four of the 21 genes that were interrupted (PA0041, PA0745, PA3946, and PA4725) have not been identified previously except as part of the PAO1 genome sequence.
One mutant (MP507) carried an insertion in the hcnC gene, which encodes a subunit of hydrogen cyanide synthase (47). This finding suggested that hydrogen cyanide contributed to nematode killing. In addition, we discovered previously that worm killing was inefficient if the petri plate lid was removed during the 4-h killing assay, suggesting that a volatile factor (such as hydrogen cyanide) contributed to the killing (data not shown).
Cyanide production strongly correlates with nematode killing.
To verify that the killing defect in strain MP507 was due to inactivation of the hcnC gene rather than to polar effects of the transposon insertion, we complemented the HCN synthase defect by introducing the hcnABC gene cluster (lacking downstream open reading frames) in trans. The nematode killing phenotype was fully restored in the complemented mutant (Fig. 1), implying that paralytic killing truly depends on the hcn genes.
To determine whether other killing-defective mutants exhibited reduced cyanide production, we measured the level of cyanide generated by each strain under growth conditions mimicking those used to assay nematode killing (see above). As shown in Table 2, there was an excellent correlation between decreased cyanide production and reduced killing for both class I and class II mutants. These data implicate cyanide as a primary component of PAO1 virulence towards C. elegans.
TABLE 2.
Exoproduct production by P. aeruginosa mutants
| Strain | Mutant gene | % Killinga | Amt of cyanide (nmol)b | Production of:
|
||
|---|---|---|---|---|---|---|
| Pyocyaninc | Pyoverdinec | Proteased | ||||
| PAO1 | None | 100 (±1) | 300 (±56) | 1.00 (±0.047) | 1.00 (±0.079) | ++ |
| PAO-R1 | lasR | 0 (±0) | <10 (±0) | 0.011 (±0.000) | 1.54 (±0.013) | +/− |
| Class I (strongly avirulent strains) | ||||||
| MP503 | eda | 7 (±1) | <15 (±8) | 0.038 (±0.015) | 0.083 (±0.099) | + |
| MP505 | proC | 9 (±6) | 31 (±3) | ND | ND | ND |
| MP506 | proC | 0 | 44 (±34) | 0.608 (±0.057) | 0.303 (±0.006) | ++ |
| MP504 | PA0745 (fad-1) | 19 (±21) | <11 (±2) | 0.036 (±0.006) | 0.379 (±0.018) | ++ |
| MP507 | hcnC | 13 | <10 (±0) | 2.35 (±0.040) | 1.54 (±0.009) | ++ |
| MP508 | PA0041 (fhaB) | 0 | <10 (±0) | 1.71 (±0.059) | 0.134 (±0.035) | +++ |
| MP501 | PA3946 (bvgS) | 0 (±0) | <17 (±6) | 0.521 (±0.025) | 1.58 (±0.002) | ++ |
| MP502 | gacS | 11 | 21 (±12) | 0.127 (±0.002) | 0.064 (±0.070) | ++ |
| MP511 | Unknown | 0 | 36 (±37) | 0.034 (±0.006) | 0.033 (±0.026) | +/− |
| Class II (moderately avirulent strains) | ||||||
| MP554 | zwf | 74 (±25) | 200 (±86) | 0.254 (±0.032) | 0.447 (±0.169) | ++ |
| MP555 | soxA | 3 | 64 (±7) | 1.00 (±0.085) | 0.353 (±0.009) | ++ |
| MP556 | soxA | 17 | 55 (±15) | ND | ND | ND |
| MP562 | PA2587 | 0 (±0) | 36 (±31) | 0.013 (±0.006) | 1.03 (±0.283) | ++ |
| MP571 | znuB | 4 | 96 | 0.767 (±0.042) | 0.597 (±0.476) | ++ |
| MP573 | phpA | 14 | 150 | 1.11 (±0.021) | 0.879 (±0.046) | ++ |
| MP572 | pilW | 36 | 84 | ND | ND | ND |
| MP551 | PA1003 | 0 (±0) | <23 (±23) | 0.008 (±0.000) | 1.54 (±0.039) | ++ |
Worm killing was assayed in trials parallel to two of the three cyanide collection trials by using plates inoculated and incubated exactly like the plates used for cyanide collection. The numbers in parentheses are standard errors of the means determined when two assays were conducted.
The values are averages based on one to three independent assays. The numbers in parentheses are standard errors of the means determined when multiple assays were conducted.
The values are expressed in arbitrary units normalized to the amount in PAO1 and are averages based on two independent assays. The numbers in parentheses are standard errors of the means. ND, not done.
Total secreted protease production was assessed by determining the relative zones of clearing around same-size overnight bacterial patches on skim milk agar as follows: +/−, no clearing beyond edge of patch; +, 1 to 2 mm clearing; ++, 4 to 5 mm clearing; +++, >6 mm clearing; ND, not done.
Cyanide alone is sufficient to kill C. elegans.
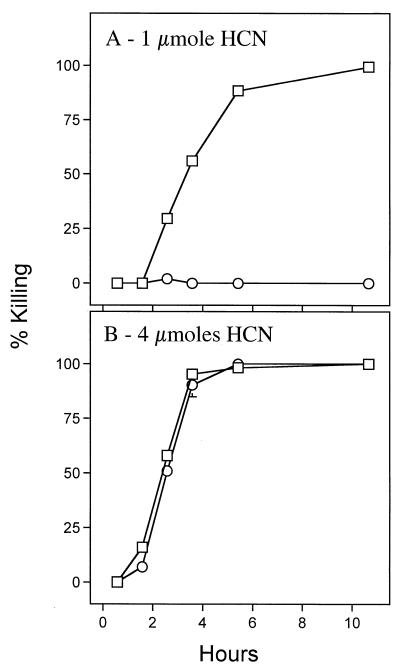
We next examined the response of C. elegans to hydrogen cyanide alone at concentrations comparable to those produced by bacteria. When exposed to 1 μmol of cyanide gas (HCN) in a sealed chamber (see above), wild-type worms exhibited a gradual slowing of movement, and more than 85% of the worms became fully immobile and unresponsive to touch by 5 h after exposure began (Fig. 2A). By 10 h all of the worms were immobile and unresponsive. In contrast, although mutant egl-9 worms exhibited a sluggishness similar to that of wild-type worms soon after cyanide exposure began, they recovered completely within a few hours and remained fully viable. Cyanide gas thus killed C. elegans with kinetics and genetic dependency similar to the kinetics and genetic dependency of P. aeruginosa-induced paralytic killing, in which complete killing of wild-type worms but not egl-9 worms occurs after 4 h of exposure to bacteria (17). The 1 μmol of HCN used in this protocol approximates the ∼300 nmol recovered from wild-type bacteria grown under standard worm-killing conditions (Table 2).
FIG. 2.
Direct exposure of C. elegans to cyanide gas. Wild-type (□) or egl-9 (○) nematodes were exposed to 1 μmol (A) or 4 μmol (B) of cyanide gas in sealed 10-cm-diameter petri plates. Worms were considered dead if they did not respond detectably when the assay plate was tapped repeatedly against the microscope stage. Each datum point represents the average level of killing based on three separate assays.
Exposure to an increased amount of cyanide (4 μmol) killed both egl-9 and wild-type worms with indistinguishable kinetics (Fig. 2B). This result suggests that either an additional mechanism of killing operates at the higher cyanide level or inactivation of egl-9 simply raises the threshold of sensitivity to cyanide.
Cyanide as the sole toxic component.
Although the results described above indicated that the amount of cyanide normally produced by PAO1 should be enough to kill C. elegans (Table 2 and Fig. 2A), we wondered whether additional factors produced by the bacteria contribute to the killing. To address this possibility, we compared the kinetics of killing by cyanide gas when the worms were placed on a lawn of hcnC mutant bacteria and on agar lacking bacteria. As shown in Fig. 3, the effect of the hcnC mutant bacteria on the kinetics of the response to cyanide was negligible for both wild-type and egl-9 nematodes. The bacteria thus did not augment the toxicity of the cyanide added, suggesting that hydrogen cyanide alone can kill nematodes.
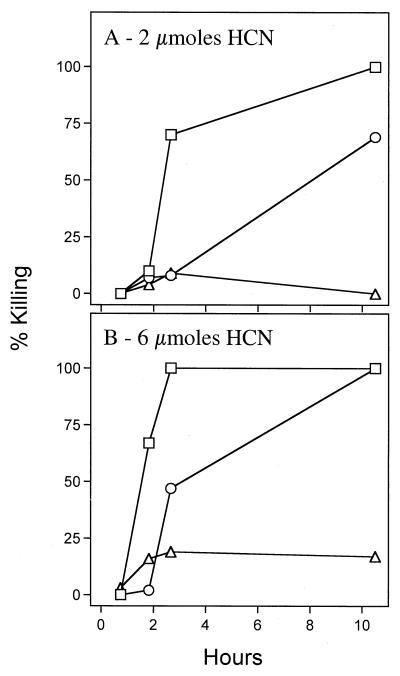
In similar augmentation experiments performed with the other class I mutants, we discovered that the two proC mutants (MP505 and MP506) were unique in that they strongly protected the worms from cyanide-induced killing. Exposure even to 8 μmol of cyanide failed to kill worms placed on a lawn of either mutant (data not shown). We examined whether the proC mutant bacteria need to be in direct contact with the nematodes to provide their protective effect by enclosing worms in a sealed chamber together with a second plate on which bacteria had been grown and then generating cyanide gas in the chamber. As shown in Fig. 4, the cyanide gas killed the worms when either no bacteria or hcnC mutant bacteria were present on the second plate (athough the hcnC bacteria conferred some protection early in the experiment). However, when the proC mutant was present on the second plate, generation of even 6 μmol of cyanide did not kill the worms. Hence, the proC mutant lawn protected the nematodes from the cyanide, even when it was not in contact with them. This protection presumably occurred by inactivation or sequestering of the cyanide by the proC mutant bacteria. One possibility is that the proC mutant bacteria secrete the proline biosynthetic intermediate glutamic-5-semialdehyde, which reacts with cyanide to form a cyanohydrin.
FIG. 4.
Killing of C. elegans by hydrogen cyanide gas when bacteria were present but not in contact with the nematodes. Wild-type nematodes were exposed to 2 μmol (A) or 6 μmol (B) of hydrogen cyanide in sealed 10-cm-diameter petri plates when plates containing either no bacteria (□), a 24-h lawn of hcnC mutant MP507 (○), or a 24-h lawn of proC mutant MP506 (▵) were also present in the chamber but were not in contact with the nematodes.
Phenazine production does not correlate with nematode killing.
Recent studies of a form of killing (fast killing) of C. elegans by a different strain of P. aeruginosa (PA-14) showed that production of the blue phenazine pigment pyocyanin occurred in a subset of mutants defective in killing (41). Phenazines are redox-active compounds secreted by pseudomonads, and pyocyanin is the characteristic phenazine produced by P. aeruginosa (23, 31, 66). To examine the potential involvement of phenazines in nematode killing by strain PAO1, we measured the amount of pyocyanin produced by our mutants under the growth conditions used to assay killing. As shown in Table 2, only about one-half of the mutants were defective in production of pyocyanin. Curiously, the hcnC mutant (MP507) produced more than twice as much pyocyanin as its parent. These data show that there was not a strong correlation between reduced pyocyanin production and loss of virulence towards nematodes in the assay described here.
Defects in pyocyanin production in some of the non-worm-killing mutants may simply reflect pleiotropic effects of the mutations (51, 70). To further assess pleiotropy in the killing-defective strains, we assayed production of total secreted protease (7) and the secreted siderophore pyoverdine (13, 43). Of the 14 mutants examined, 7 showed reduced pyoverdine production and 2 showed reduced secreted protease production (Table 2). One of the nine class I mutants, the hcnC mutant, was exceptional in that reduced pyocyanin, pyoverdine, or secreted protease production was not evident.
DISCUSSION
In this report we describe experiments in which we investigated the mechanism by which P. aeruginosa PAO1 rapidly paralyzes and kills C. elegans (17). Our results imply that the poison hydrogen cyanide is the sole or primary bacterial factor responsible for killing of the nematode. That cyanide is necessary for the virulence is implied by the finding that 17 transposon insertion mutants impaired in worm killing, including 1 mutant in which inactivated hydrogen cyanide synthase itself was inactivated, all exhibited reduced cyanide production. That cyanide is sufficient for nematode killing is implied by the finding that exposure to exogenous cyanide at levels comparable to that produced by the bacteria kills nematodes with kinetics similar to those observed with bacteria. Furthermore, a nematode mutant (egl-9) resistant to P. aeruginosa killing was also resistant to killing by exogenous HCN.
Hydrogen cyanide is a typical pseudomonad secondary metabolite, a compound which is not required for growth, energy storage, or primary metabolism but which may provide some ecological advantage to the organism (67). In addition to cyanide, the pseudomonad secondary metabolites include siderophores, such as pyoverdine; redox-active compounds, such as phenazines; and polyketide antibiotics (10). Cyanide is produced in Pseudomonas strains by oxidative decarboxylation of glycine by the three-subunit membrane-bound flavoenzyme encoded by hcnABC (4). P. aeruginosa produces HCN maximally in the late exponential and early stationary phases under microaerophilic conditions (4), and transcription of the hcn genes appears to depend directly on the quorum sensor regulators LasR and RhlR, as well as the anaerobic regulator Anr (7, 47, 70). Additional components of the complex regulatory circuitry controlling the production of cyanide and other secondary metabolites have been identified (2, 11, 42, 46, 71).
The mutations that we identified which reduced cyanide production and virulence towards C. elegans affect a variety of regulatory and metabolic functions (Table 1). Two of the regulatory mutations affect quorum sensing indirectly; one is in gacS, which encodes a two-component sensor that influences autoinducer levels (51), and the other is in a locus (PA2587) needed for synthesis of a quinolone signal required for RhlI-RhlR function (E. Pesci, personal communication). Mutations in three additional putative regulators were also identified; one of these regulators (PA3946) is homologous to the Bordetella pertussis virulence regulator BvgS (1), another (PA1003) belongs to the LysR family (32), and the third (PA4725) resembles a two-component sensor fused to a membrane permease. A mutation affecting PA1003 was previously identified in a study to screen for mutations that reduce virulence towards Arabidopsis (50). The mutations affecting metabolic functions inactivate enzymes that participate in central carbon metabolism, fatty acid breakdown, and proline biosynthesis. With the notable exception of the mutation of the HCN synthase mutant, all of the strongest (class I) non-worm-killing mutations reduced the production of pyocyanin, pyoverdine, or secreted protease. This pleiotropy would have made identification of cyanide as the worm-killing poison difficult if the hcnC mutant had not been isolated. An unanticipated benefit of the genetic approach taken in this study is that it appears to have identified several new regulatory and metabolic components of the circuitry controlling the production of secondary metabolites.
Studies of a different strain of P. aeruginosa (PA-14) showed that about one-half of a collection of transposon insertion mutations that eliminated a fast-killing form of virulence towards C. elegans also reduced production of pyocyanin, as did a constructed deletion mutation (ΔphnAB) that decreased phenazine biosynthesis (41). The results were interpreted in terms of a model in which phenazines are one component of a multifactorial killing process. Phenazines are toxic to a variety of cell types and are thought to act by generating reactive oxygen species by redox cycling (59). For strain PAO1, we found no convincing indication that pyocyanin or any other phenazine plays a direct role in killing C. elegans. Although five of nine strongly avirulent mutants produced significantly less pyocyanin than the parent, the reduction in production is readily explained by the pleiotropy of the mutations (Table 2). Indeed, two of the mutations affect regulators (LasR and GacS) already known to be required for expression of multiple genes, and a third affects an enzyme of central carbon catabolism in Pseudomonas (Entner-Douderoff aldolase) whose loss might also be expected to be highly pleiotropic (51, 63, 70). Furthermore, since exposure of nematodes to HCN in the absence of bacteria reproduced the nematode paralytic killing phenomenon, no additional bacterial substances are required.
The classic cellular target of cyanide inhibition is cytochrome oxidase, although other metalloenzymes are also sensitive to the poison (58). Inhibition of mitochondrial respiration can easily account for the rapid and dramatic paralytic killing of nematodes by P. aeruginosa PAO1. Pseudomonads appear to protect themselves from cyanide poisoning by expressing an unusual cyanide-resistant cytochrome oxidase (16). Studies of human and animal cyanide poisoning indicate that the poison strongly affects neurological tissue (69), and it is possible that nematode killing also reflects hypersensitivity of neuromuscular tissues to the poison.
It is striking that loss-of-function mutations in a single nematode gene (egl-9) confer strong resistance to cyanide poisoning. The mechanism underlying this resistance is mysterious. Since HCN is predominantly uncharged at physiological pH (pK 9.3) and is expected to diffuse freely through membranes, it appears unlikely that a cyanide transporter is eliminated by the mutations. One possibility is that the egl-9 mutations constitutively activate an adaptive response to hypoxia (54), thus conferring some resistance to cytochrome oxidase inhibition by cyanide. Another possibility is that reactive oxygen species generated by cyanide inhibition activate an Egl-9-dependent pathway, such as a stress-dependent MAP (mitogen-activated protein) kinase pathway (65), leading to paralysis and death. Homologues of egl-9 exist in humans (3, 22) and may represent potential therapeutic targets for countering the toxic effects of cyanide.
Cyanide is a potent poison expected to be active against most eukaryotic species (4, 58). This compound thus could contribute profoundly to the broad pathogenic host range of P. aeruginosa (5, 9, 36, 40, 45). It is thought that cyanide inhibition of fungal growth helps account for the suppression of several plant root and leaf fungal diseases (30, 68). The activities of cyanide and other small-molecule poisons may also contribute to the pathogenesis accompanying the variety of opportunistic infections caused by P. aeruginosa (39). Although the role of cyanide in Pseudomonas pathogenesis in humans is largely unexplored, an early study of burn infections detected this poison (28). The recent finding that sputa of cystic fibrosis patients contain P. aeruginosa in the appropriate quorum-sensing physiological state to produce cyanide (56) suggests that the poison could also contribute to the tissue destruction that accompanies lung infections in this disease.
ACKNOWLEDGMENTS
We thank Jeannie Bailey, Chris Cosma, Dave D'Argenio, Creg Darby, Steve Lory, Maynard Olson, James Thomas, and Mike Vasil for helpful discussions; Christina Buchanan, Brian Buchwitz, Denise Gaunt, and Allyson McCormick for assisting with the mutant screening; Everett Pesci for providing unpublished data; and Dieter Haas for sharing the cyanide quantification protocol.
This research was supported by a grant from the Cystic Fibrosis Foundation.
REFERENCES
- 1.Akerley B J, Miller J F. Understanding signal transduction during bacterial infection. Trends Microbiol. 1996;4:141–146. doi: 10.1016/0966-842x(96)10024-x. [DOI] [PubMed] [Google Scholar]
- 2.Albus A M, Pesci E C, Runyen-Janecky L J, West S E, Iglewski B H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Koonin E V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. Res. Vol. 2. 2001. http://genomebiology.com/2001/2/3/research/0007/ [Online.] http://genomebiology.com/2001/2/3/research/0007/. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumer C, Haas D. Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch Microbiol. 2000;173:170–177. doi: 10.1007/s002039900127. [DOI] [PubMed] [Google Scholar]
- 5.Boman H G, Nilsson I, Rasmuson B. Inducible antibacterial defence system in Drosophila. Nature. 1972;237:232–235. doi: 10.1038/237232a0. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Britigan B E, Railsback M A, Cox C D. The Pseudomonas aeruginosa secretory product pyocyanin inactivates α1 protease inhibitor: implications for the pathogenesis of cystic fibrosis lung disease. Infect Immun. 1999;67:1207–1212. doi: 10.1128/iai.67.3.1207-1212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucher G E, Stephens J M. A disease of grasshoppers caused by the bacterium Pseudomonas aeruginosa (Schroeter) Migula. Can J Microbiol. 1957;3:611–625. doi: 10.1139/m57-067. [DOI] [PubMed] [Google Scholar]
- 10.Budzikiewicz H. Secondary metabolites from fluorescent pseudomonads. FEMS Microbiol Rev. 1993;10:209–228. doi: 10.1111/j.1574-6968.1993.tb05868.x. [DOI] [PubMed] [Google Scholar]
- 11.Chugani S A, Whiteley M, Lee K M, D'Argenio D, Manoil C, Greenberg E P. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98:2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun K T, Edenberg H J, Kelley M R, Goebl M G. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast. 1997;13:233–240. doi: 10.1002/(SICI)1097-0061(19970315)13:3<233::AID-YEA88>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 13.Cox C D, Adams P. Siderophore activity of pyoverdin for Pseudomonas aeruginosa. Infect Immun. 1985;48:130–138. doi: 10.1128/iai.48.1.130-138.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croft L, Beatson S A, Whitchurch C B, Huang B, Blakeley R L, Mattick J S. An interactive web-based Pseudomonas aeruginosa genome database: discovery of new genes, pathways and structures. Microbiology. 2000;146:2351–2364. doi: 10.1099/00221287-146-10-2351. [DOI] [PubMed] [Google Scholar]
- 15.Cunliffe H E, Merriman T R, Lamont I L. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham L, Pitt M, Williams H D. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol Microbiol. 1997;24:579–591. doi: 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- 17.Darby C, Cosma C L, Thomas J H, Manoil C. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:15202–15207. doi: 10.1073/pnas.96.26.15202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darby C B. Paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa: a genetically tractable model for bacterial pathogenesis. Ph.D. thesis. Seattle: University of Washington; 1998. [Google Scholar]
- 19.D'Argenio D A, Gallagher L A, Berg C A, Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J Bacteriol. 2000;183:1466–1471. doi: 10.1128/JB.183.4.1466-1471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doring G. Pseudomonas aeruginosa infection in cystic fibrosis patients. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 245–273. [Google Scholar]
- 22.Dupuy D, Aubert I, Duperat V G, Petit J, Taine L, Stef M, Bloch B, Arveiler B. Mapping, characterization, and expression analysis of the SM-20 human homologue, c1orf12, and identification of a novel related gene, SCAND2. Genomics. 2000;69:348–354. doi: 10.1006/geno.2000.6343. [DOI] [PubMed] [Google Scholar]
- 23.Essar D W, Eberly L, Hadero A, Crawford I P. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J Bacteriol. 1990;172:884–900. doi: 10.1128/jb.172.2.884-900.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finlay B B. Bacterial disease in diverse hosts. Cell. 1999;96:315–318. doi: 10.1016/s0092-8674(00)80544-9. [DOI] [PubMed] [Google Scholar]
- 25.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gewitz H S, Pistorius E K, Voss H H, Vennesland B. Cyanide formation in preparations from Chlorella vulgaris Beijerinch: effect of sonication and amygdalin addition. Planta. 1976;131:145–148. doi: 10.1007/BF00389986. [DOI] [PubMed] [Google Scholar]
- 28.Goldfarb W B, Margraf H. Cyanide production by Pseudomonas aeruginosa. Ann Surg. 1967;165:104–110. doi: 10.1097/00000658-196701000-00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govan J R, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas D, Blumer C, Keel C. Biocontrol ability of fluorescent pseudomonads genetically dissected: importance of positive feedback regulation. Curr Opin Biotechnol. 2000;11:290–297. doi: 10.1016/s0958-1669(00)00098-7. [DOI] [PubMed] [Google Scholar]
- 31.Hassan H M, Fridovich I. Mechanism of the antibiotic action of pyocyanine. J Bacteriol. 1980;141:156–163. doi: 10.1128/jb.141.1.156-163.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henikoff S, Haughn G W, Calvo J M, Wallace J C. A large family of bacterial activator proteins. Proc Natl Acad Sci USA. 1988;85:6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holder I A. Pseudomonas aeruginosa burn infections: pathogenesis and treatment. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 275–295. [Google Scholar]
- 34.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jander G, Rahme L G, Ausubel F M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol. 2000;182:3843–3845. doi: 10.1128/jb.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarrell K F, Kropinski A M. The virulence of protease and cell surface mutants of Pseudomonas aeruginosa for the larvae of Galleria mellonella. J Invertebr Pathol. 1982;39:395–400. doi: 10.1016/0022-2011(82)90065-9. [DOI] [PubMed] [Google Scholar]
- 37.Kang P J, Hauser A R, Apodaca G, Fleiszig S M, Wiener-Kronish J, Mostov K, Engel J N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol. 1997;24:1249–1262. doi: 10.1046/j.1365-2958.1997.4311793.x. [DOI] [PubMed] [Google Scholar]
- 38.King E O, Ward M K, Raney A B. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 39.Lyczak J B, Cannon C L, Pier G B. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 40.Mahajan-Miklos S, Rahme L G, Ausubel F M. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol Microbiol. 2000;37:981–988. doi: 10.1046/j.1365-2958.2000.02056.x. [DOI] [PubMed] [Google Scholar]
- 41.Mahajan-Miklos S, Tan M W, Rahme L G, Ausubel F M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 42.McKnight S L, Iglewski B H, Pesci E C. The Pseudomonas quinolone signal regulates rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McMorran B J, Merriman M E, Rombel I T, Lamont I L. Characterisation of the pvdE gene, which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene. 1996;176:55–59. doi: 10.1016/0378-1119(96)00209-0. [DOI] [PubMed] [Google Scholar]
- 44.Olvera C, Goldberg J B, Sanchez R, Soberon-Chavez G. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol Lett. 1999;179:85–90. doi: 10.1111/j.1574-6968.1999.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 45.Patty F A. The production of hydrocyanic acid by Bacillus pyocyaneus. J Infect Dis. 1921;29:73–77. [Google Scholar]
- 46.Pesci E C, Milbank J B, Pearson J P, McKnight S, Kende A S, Greenberg E P, Iglewski B H. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pessi G, Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J Bacteriol. 2000;182:6940–6949. doi: 10.1128/jb.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plotnikova J M, Rahme L G, Ausubel F M. Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 2000;124:1766–1774. doi: 10.1104/pp.124.4.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahme L G, Stevens E J, Wolfort S F, Shao J, Tompkins R G, Ausubel F M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 50.Rahme L G, Tan M W, Le L, Wong S M, Tompkins R G, Calderwood S B, Ausubel F M. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci USA. 1997;94:13245–13250. doi: 10.1073/pnas.94.24.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–121. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 54.Semenza G L. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 55.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 56.Singh P K, Schaefer A L, Parsek M R, Moninger T O, Welsh M J, Greenberg E P. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 57.Sokol P A, Ohman D E, Iglewski B H. A more sensitive plate assay for detection of protease production by Pseudomanas aeruginosa. J Clin Microbiol. 1979;9:538–540. doi: 10.1128/jcm.9.4.538-540.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Solomonson L P. Cyanide as a metabolic inhibitor. In: Vennesland B, Conn E E, Knowles C J, Westley J, Wissing F, editors. Cyanide in biology. London, United Kingdom: Academic Press; 1981. pp. 11–28. [Google Scholar]
- 59.Sorenson R, Joseph F. Phenazine pigments in Pseudomonas aeruginosa infection. In: Campa M, Bendinelli M, Friedman H, editors. Pseudomonas aeruginosa as an opportunistic pathogen. New York, N.Y: Plenum Press; 1993. pp. 43–57. [Google Scholar]
- 60.Stintzi A, Evans K, Meyer J M, Poole K. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol Lett. 1998;166:341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x. [DOI] [PubMed] [Google Scholar]
- 61.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 62.Tan M W, Mahajan-Miklos S, Ausubel F M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci USA. 1999;96:715–720. doi: 10.1073/pnas.96.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Temple L, Sage A, Schweizer H P, Phibbs P. Carbohydrate catabolism in Pseudomonas aeruginosa. In: Montie T, editor. Pseudomonas. New York, N.Y: Plenum Press; 1998. pp. 35–72. [Google Scholar]
- 64.Terada L S, Johansen K A, Nowbar S, Vasil A I, Vasil M L. Pseudomonas aeruginosa hemolytic phospholipase C suppresses neutrophil respiratory burst activity. Infect Immun. 1999;67:2371–2376. doi: 10.1128/iai.67.5.2371-2376.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tibbles L A, Woodgett J R. The stress-activated protein kinase pathways. Cell Mol Life Sci. 1999;55:1230–1254. doi: 10.1007/s000180050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Turner J M, Messenger A J. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/s0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 67.Vining L C. Functions of secondary metabolites. Annu Rev Microbiol. 1990;44:395–427. doi: 10.1146/annurev.mi.44.100190.002143. [DOI] [PubMed] [Google Scholar]
- 68.Voisard C, Keel C, Haas D, Défago G. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 1989;8:351–358. doi: 10.1002/j.1460-2075.1989.tb03384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Way J L. Cyanide intoxication and its mechanism of antagonism. Annu Rev Pharmacol Toxicol. 1984;24:451–481. doi: 10.1146/annurev.pa.24.040184.002315. [DOI] [PubMed] [Google Scholar]
- 70.Whiteley M, Lee K M, Greenberg E P. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whiteley M, Parsek M R, Greenberg E P. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wood W B. The nematode Caenorhabditis elegans. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 73.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]