Abstract
Subject
The Angiopoietin‐like 3 (ANGPTL3) gene has been reported to be associated with cardiovascular risk. This study is designed to compare the genetic variant (rs1748195) of the ANGPTL3 gene and the presence of a coronary artery occlusion of >50% in Iranian nation.
Method
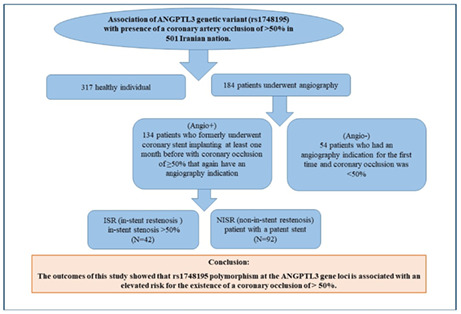
In this study, 184 patients underwent angiography and 317 healthy individuals were evaluated for polymorphism of rs1748195 the ANGPTL3 gene using Tetra‐ARMs PCR. Coronary patients who experience angiography were categorized into two groups: 54 patients who had an angiography indication for the first time and coronary occlusion was <50% (Angio−) and 134 patients who formerly underwent coronary stent implanting at least 1 month before with coronary occlusion of ≥50% that again have an angiography indication (Angio+).
In addition, individuals with angio+ are categorized in two groups: (1) non‐in‐stent restenosis (NISR); patient with a patent stent (N = 92). (2) in‐stent restenosis (ISR); in‐stent stenosis >50% (N = 42).
Result
The fundamental of characteristics of our study design population was categorized based on undergoing angiography or not. In the present study, we investigated that the CC genotype, and also the A allele corresponding to rs1748195 at the ANGPTL3 gene loci, was associated with negative angiogram and directly related to the risk of coronary occlusion >50%. In contrast, this result was not significant in genotypes of ANGPTL3 between non‐ISR and ISR groups.
Conclusion
The outcomes of this study showed that rs1748195 polymorphism at the ANGPTL3 gene loci is associated with an elevated risk for the existence of a coronary occlusion of >50%.
Keywords: angiography, angiopoietin‐like3 (ANGPTL3), atherosclerosis, cardiovascular diseases (CVD)
Summary of research, in this study, 184 patients underwent angiography and 317 healthy individuals were evaluated for polymorphism of rs1748195 the ANGPTL3 gene using Tetra‐ARMs PCR. Coronary patients who experience angiography were categorized into two groups: 54 patients who had an angiography indication for the first time and coronary occlusion was <50% (Angio–), 134 patients who formerly underwent coronary stent implanting at least one month before with coronary occlusion of ≥50% that again have an angiography indication (Angio+). In addition, individuals with angio+ categorized in two groups: (1) non‐in‐stent restenosis (NISR); patient with a patent stent (N = 92). (2) In‐stent restenosis (ISR); in‐stent stenosis >50% (N = 42). Finally, the outcomes of this study showed thatrs1748195 polymorphism at the ANGPTL3 gene loci is associated with an elevated risk for the existence of a coronary occlusion of > 50%.
1. INTRODUCTION
Cardiovascular disease (CVD), consisting of ischemic heart disease, heart failure, arterial disease and other heart problems, causes high mortality in the world (De Hert et al., 2018). Based on the research studies, CVD is one of the main reasons of reducing quality and duration of life (Mensah et al., 2019). Given to the World Health Organization (WHO) report in 2020, about 37% of premature non‐communicable deaths and approximately 50% of total annual deaths in Iran are caused by CVD (Roth et al., 2017) .
Atherosclerosis is a major cause of heart attack and stroke (Parsamanesh et al., 2021). Observations showed that cholesterol is an important component of arterial plaque that leads to atherosclerosis (Samadi et al., 2019). LDL cholesterol and Apo lipoprotein B (ApoB100) are directly related to the risk of atherosclerotic cardiovascular events (ASCVE) (Aghasizadeh, Bizhaem, et al., 2021; Levin, 2014). Angioprotein 3 is a secreted glycoprotein expressed in the liver (Estey et al., 2013) and contained 460 amino acid polypeptides containing a peptide signal, an N‐terminal domain, and a fibrinogen‐like C‐terminal domain (Zierk et al., 2013). This protein inhibits lipoprotein lipase, which has been one of the important enzymes associated with CVD risk. Besides controlling the activity of this enzyme is very important in preventing diseases. Studies show that this gene is a pathway for a therapeutic target for lipoprotein metabolism (Gomaraschi et al., 2011). Coronary angiography is a medical procedure that uses a special dye and x‐rays to visualize the lumen of blood vessel and the heart chambers (Parsamanesh et al., 2021). It has known as a serious diagnostic method of detecting atherosclerotic plaque. This medical technology can be used as a detection method of coronary heart and peripheral arterial diseases (PAD). Coronary angiography is often done along with catheterization (Aghasizadeh, Samadi, et al., 2021; Saberi‐Karimian et al., 2021). In recent dedicate, coronary angioplasty has made a considerable decrease in bypass operation in patients with atherosclerotic plaque, but however in 15%–40% of patients restenosis of arteries veins happen (Gimbrone Jr et al., 2000; Roth et al., 2017; Rubin et al., 2014) .
Restenosis is defined as stenosis in the coronary arteries >50% after percutaneous coronary intervention (PCI). This technique is the most important method of revascularization in the coronary heart disease therapy (Kivelä & Hartikainen, 2006; Oguri et al., 2007; Sadeghi et al., 2017). Repeated treatment of coronary angioplasty are cost high. Hence, to diminish this cost, the other method of treatment such as coronary bypass surgery, and implantation of drug‐eluting stents, is very critical to diminish the cost (Htay & Liu, 2005; Reiner et al., 2011; Teslovich et al., 2010). Although drug‐eluting stents implantation is one way to regenerate and prevent restenosis of the coronary arteries, stent thrombosis and in‐stent restenosis (ISR) demonstrate inflammatory response by increasing the diffusion of reactive oxygen species (ROS), and individual susceptibility play a critical role in their onset (Montone et al., 2013; Niccoli et al., 2010). This adverse effect has been shown in 15%–27% of implanted patients (Kathiresan et al., 2008; The 1000 Genomes Project Consortium, 2010; Willer et al., 2008).
Base on genome wide association studies (GWAS), single nucleotide polymorphism (SNP) is one of the major risk factor for cardiovascular disease (Kathiresan & Srivastava, 2012; Teslovich et al., 2010). Angiopoietin‐like 3 (ANGPTL3) gene has critical role in regulation of lipid profile and lipoprotein concentration therefore (Kharazmi‐Khorassani et al., 2019), this gene has been shown to be associated with CVD risk (Bea et al., 2021; Kathiresan & Srivastava, 2012).
The ANGPTL3 gene encodes Angiopoietin‐Like3 protein as a secretory protein that contributed in angiogenesis (Kersten, 2017; Willer et al., 2008). This gene is a liver derived circulating factor with a C‐terminal fibrinogen chain and a N‐terminal coiled‐coil domain that prohibits the enzyme lipoprotein lipase (LPL) (Kersten, 2017; Mattijssen & Kersten, 2012). LPL is a hydrolysis enzyme that break down triglycerides into free fatty acids (FFA) and monoacyl‐glycerols and then this product transported by very low density lipoproteins (VLDL) and chylomicrons from different organs to the bloodstream (Goldberg, 1996; Kobayashi et al., 2002; Zechner et al., 2000). Hence, LPL plays a critical role in lipid metabolism. Lipoprotein lipase deficiency is an infrequent inherited disease define by chylomicronaemia and severe hypertriglyceridemia, recurrent acute pancreatitis and increased risk of atherosclerosis or other complications (Gaudet et al., 2012; Nordestgaard et al., 1997). Higher level of LPL decrease bloodstream TG level and thus protecting individual from CVD risk (Rip et al., 2006). In 2017, genetic research in CVD in GWAS showed that single nucleotide polymorphisms (SNPs) can affect blood lipid levels and contributing factors that cause CVD (Li et al., 2018). Base on the GWAS study this rs1748195 is located at the enhancer histone signature site of the ANGPTL3 that effects gene transcription (Oldoni et al., 2016). This SNP is located on the forward (F) strand of the chromosome 1 that is encoding a protein involved in angiogenesis (Li et al., 2018).
The purpose of this study was to compare the genetic variant (rs1748195) of ANGPTL3 gene in angiographic patients with healthy ones.
2. METHODS
2.1. Study population
In this case‐control study, 501 subjects were recruited that 317 were healthy individuals and 184 underwent angiography. The following patients were categorized into two group:
1. Having a symptom to undergo angiography that defines as a subject who has an angiography indication for the first time and coronary occlusion was <50% (N = 54).
2. Having a symptom to underwent angiography that defines as angio+ group who formerly experienced coronary stent implanting and at least one month ago with coronary occlusion of ≥50% that again have an angiography indication (N = 134).
Also, classify individuals with angio+ in two groups include patients in‐stent restenosis (ISR); stent stenosis >50% (N = 42), and non‐in‐stent restenosis (NISR); patient with patent stent (N = 92).
2.2. Genotyping
DNA extracted from the whole blood of these individuals by salting out method was done, detailed in our pervious study (Valizadeh et al., 2021) and DNA samples was stored at −20°C. In this study, Tetra amplification‐refractory mutation system (ARMS) PCR reaction was used to determine the genotypes of rs1748195 polymorphism. Tetra ARMS PCR reaction was performed in a total volume of 20 ml containing DNA template‐dNTP‐primers and Mgcl2 buffer. To determine the rs1748195 genotype, a sample containing 10 μl of Master Mix (Pars Tous company), 0.5 μl of FO, RO and 1 μl of primer FI, RI and 2 μl of sample DNA and 5 μl values deionized is used. Primer sequences was “ACCTACTATACAAATTGTTGGCTTCA” for FO, “AGTTGTTTTACCAGTTGTGTTGATG” for RO, and “ACTTCTGGAGTAATAACAGGAATACTCTC “for FI, and “TATCTAATCAGCTGCTCTGATTATCC” for RI. After the reaction, the product was electrophoresed in agarose powder 2%. To end (Figure 1), the genotypes were established by sanger sequencing as shown in Figure 2 and all sequenced examples were analyzed by Finch TV (version 1.4.0).
FIGURE 1.
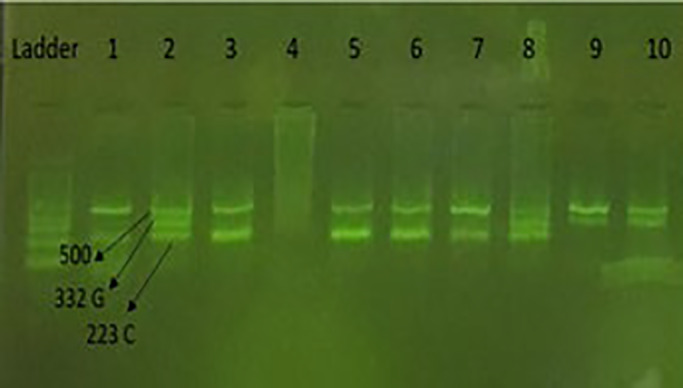
Agarose gel for rs1748195 polymorphism (C > G). Number 1, CC genotype. Number 2, CG genotype. Number 9, GG genotype.
FIGURE 2.
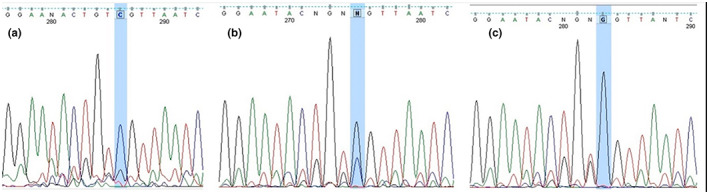
The results of DNA sequencing of rs1748195, ANGPTL3 gene; (a) CC genotype, (b) CG genotype, (c) GG genotype.
2.3. Biochemical measurements
HDL‐C, TG and TC concentration was determined by Pishtaz Teb kit and auto‐analyzer. Furthermore, Biosystem company kit was used to determined level of Hs‐CRP by autoanalyzer model BT3000.
2.4. Statistical analysis
All analyses were performed by SPSS software. To determine variables with normal distribution performed t‐test. Used square‐Chi test to determine variables without a normal. Estimated the risk ratio at a 95% confidence interval to determined logistic regression of the correlation between genotype and clinical information/patient pathology. The level of statistical significance was considered <0.05.
3. RESULTS
3.1. Study population characteristics
The clinical characteristics of the three healthy, Angio− and Angio+ groups studied are summarized in Table 1. The mean age of healthy, Angio− and Angio+ individuals was 48.73 ± 7.69 and 54.68 ± 9.22 and 59.95 ± 8.65, respectively (p < 0.001). Based on the results, there was a significant difference in terms of gender between the study groups (p < 0.001). According to the analyses, the mean levels of HDL, LDL and TC in the healthy group were significantly higher compared to Angio− and Angio+ individuals (p < 0.001). Also, in comparison between the two groups of Angio− and Angio+, it was found that individuals with a history of hypertension, diabetes and dyslipidemia were more present in the Angio+ group (p < 0.001). The mean of other factors studied was different between the study groups, but these differences were not statistically significant (p > 0.05).
TABLE 1.
Baseline characteristics of healthy individuals, Angio− and Angio+
| Healthy | Angio− | Angio+ | p | |
|---|---|---|---|---|
| Age | 48.73 ± 7.69 | 54.68 ± 9.22 | 59.95 ± 8.65 | <0.001* |
| Female | 185 (59.1%) | 29 (58%) | 35 (26.7%) | <0.001* |
| HDL | 44.74 ± 12.95 | 36.21 ± 8.79 | 35.35 ± 8.87 | <0.001* |
| LDL | 114.65 ± 37.90 | 95.48 ± 32.20 | 93.80 ± 33.46 | <0.001* |
| TG | 136.11 ± 92.55 | 138.72 ± 84.53 | 130.15 ± 62.13 | 0.762 |
| TC | 192.13 ± 40.50 | 160.77 ± 42.62 | 154.30 ± 38.91 | <0.001* |
| SBP | 121.43 ± 19.64 | 121.16 ± 12.99 | 124.66 ± 14.20 | 0.257 |
| DBP | 79.39 ± 11.80 | 78.86 ± 7.84 | 78.59 ± 9.81 | 0.801 |
| BMI | 27.75 ± 4.77 | 27.33 ± 4.04 | 28.77 ± 20.96 | 0.654 |
| CRPnew | 5.37 ± 10.87 | 4.43 ± 4.22 | 3.55 ± 3.36 | 0.181 |
| PAB | 93.37 ± 46.30 | 1.04 ± 0.29 | 1.08 ± 0.30 | <0.001* |
| Hypertention | 96 (30.7%) | 23 (46%) | 76 (58%) | <0.001* |
| Diabetes | 51 (16.6%) | 20 (40%) | 50 (38.5%) | <0.001* |
| Dyslipidemia | 256 (81.8%) | 28 (56%) | 71 (54.2%) | <0.001* |
Note: Values are expressed as mean ± standard deviation or median (interquartile range 25th–75th) for normal and non‐normal distribution data respectively. *p ≤ 0.05 is indicates as significant.
3.2. Association of rs1748195 genotypes of ANGPTL3 gene with angiographic events
The frequency of AA, AC and CC, polymorphism rs1748195 genotypes in healthy, Angio− and Angio+ groups is shown in Table 2. According to univariate regression analysis, it was found that among the three studied genotypes, only the CC genotype of rs1748195 polymorphism has a significant relationship with the risk of negative angiogram in individuals (OR = 0.38, 95%CI = 0.15–0.97, p= 0.043). Also, based on dominant and additive genetic models, after univariate regression analysis, it was found that AC + CC and CC genotypes, respectively, increase the risk of negative angiogram compared to other genotypes (p < 0.05), while after adjusting the parameters of age, sex, BMI, Diabetes, HDL, LDL and TC in the multiple regression model this relationship did not remain significant (p > 0.05). The data showed that allele A of rs1748195 polymorphism was directly related to the risk of coronary occlusion <50% in individuals. In addition, the C allele frequency of this genetic variant was higher in the control group (p = 0.44).
TABLE 2.
Distribution of genotypes and allele frequencies and their association with Angio− and Angio+ groups
| Frequency | Univariate regression (odds ratio 95% CI) p‐value | Multivariate regression (odds ratio 95% CI) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Healthy | Angio− | Angio+ | p | Healthy | Angio− | Angio+ | Healthy | Angio− | Angio+ | |
| Rs1748195 | 0.101 | |||||||||
| AA | 102 (32.6) | 25 (50) | 42 (33.9) | Ref | Ref | Ref | Ref | Ref | Ref | |
| AC | 146 (46.6) | 19 (38) | 63 (50.8) | Ref | 0.53 (0.28, 1.01) 0.055 | 1.05 (0.66, 1.67) 0.844 | Ref | 0.50 (0.17, 1.43) 0.196 | 0.68 (0.28, 1.61) 0.378 | |
| CC | 65 (20.8) | 6 (12) | 19 (15.3) | Ref | 0.38 (0.15, 0.97) 0.043* | 0.71 (0.38, 1.33) 0.282 | Ref | 0.49 (0.22, 1.11) 0.089 | 0.92 (0.47, 1.79) 0.808 | |
| Dominant | 0.054 | |||||||||
| AA | 102 (32.6) | 25 (50) | 42 (33.9) | Ref | Ref | Ref | Ref | Ref | Ref | |
| AC + CC | 211 (67.4) | 25 (50) | 82 (66.1) | Ref | 0.48 (0.26, 0.88) 0.018* | 0.94 (0.61, 1.47) 0.797 | Ref | 0.49 (0.23, 1.04) 0.064 | 0.85 (0.45, 1.59) 0.603 | |
| Recessive | 0.192 | |||||||||
| AA + AC | 248 (79.2) | 44 (88) | 105 (84.7) | Ref | Ref | Ref | Ref | Ref | Ref | |
| CC | 65 (20.8) | 6 (12) | 19 (15.3) | Ref | 0.52 (0.21, 1.27) 0.153 | 0.69 (0.39, 1.21) 0.195 | Ref | 0.73 (0.27, 1.94) 0.529 | 0.71 (0.33, 1.54) 0.384 | |
| Additive | 0.088 | |||||||||
| AA | 102 (61.1) | 25 (80.6) | 42 (68.9) | Ref | Ref | Ref | Ref | Ref | Ref | |
| CC | 65 (38.9) | 6 (19.4) | 19 (31.1) | Ref | 0.38 (0.15, 0.97) 0.043* | 0.71 (0.38, 1.33) 0.282 | Ref | 0.43 (0.14, 1.33) 0.144 | 0.78 (0.31, 1.93) 0.587 | |
| Codominant | 0.308 | |||||||||
| AA + CC | 167 (53.4) | 31 (62) | 61 (49.2) | Ref | Ref | Ref | Ref | Ref | ||
| AC | 146 (46.6) | 19 (38) | 63 (50.8) | 0.70 (0.38, 1.29) 0.256 | 1.18 (0.78, 1.79) 0.433 | Ref | 0.63 (0.3, 1.32) 0.222 | 1.07 (0.59, 1.93) 0.824 | ||
| Allele | 0.044* | |||||||||
| A | 350 (55.9) | 69 (69) | 147 (59.3) | Ref | Ref | Ref | Ref | Ref | ||
| C | 276 (44.1) | 31 (31) | 101 (40.7) | 0.57 (0.36, 0.89) 0.015* | 0.87 (0.65, 1.17) 0.366 | Ref | 0.62 (0.39, 1.004) 0.052 | 0.91 (0.62, 1.34) 0.641 | ||
Note: Multivariate regression model adjusted with age, sex, BMI, Diabetes, HDL, TC, LD. Values are expressed as mean ± standard deviation or median (interquartile range 25th–75th) for normal and non‐normal distribution data respectively. *p ≤ 0.05 is indicates as significant.
3.3. Association of polymorphism rs1748195 genotypes with non‐ISR and ISR
The frequencies of AA, AC and CC genotypes associated rs1748195 in the non‐ISR group were 33.3%, 50.6% and 16.1%, and in the ISR group were 37.5%, 50% and 12.5%, respectively. According to the obtain results, no significant differences were observed between rs1748195 polymorphism genotypes and non‐in‐stent restenosis and in‐stent restenosis>50% in dominant, recessive, additive and codominant genetic models (p > 0.05) (Table 3).
TABLE 3.
Genotype distribution and frequency of alleles and its relationship with non‐ISR and ISR
| Non‐ISR | ISR | p | Univariate regression (odds ratio 95% CI) | Multivariate regression (odds ratio 95% CI) | |||||
|---|---|---|---|---|---|---|---|---|---|
| N (%) | N (%) | Non‐ISR | ISR | p | Non‐ISR | ISR | p | ||
| Rs1748195 | 0.829 | ||||||||
| AA | 29 (33.3) | 15 (37.5) | Ref | Ref | Ref | Ref | |||
| AC | 44 (50.6) | 20 (50) | Ref | 0.88 (0.39, 1.99) | 0.757 | Ref | 0.56 (0.20, 1.54) | 0.262 | |
| CC | 14 (16.1) | 5 (12.5) | Ref | 0.69 (0.21, 2.28) | 0.544 | Ref | 0.42 (0.09, 1.97) | 0.272 | |
| Dominant | 0.647 | ||||||||
| AA | 29 (33.3) | 15 (37.5) | Ref | Ref | Ref | Ref | |||
| AC + CC | 58 (66.7) | 25 (62.5) | Ref | 0.83 (0.38, 1.82) | 0.647 | Ref | 0.53 (0.20, 1.39) | 0.195 | |
| Recessive | 0.598 | ||||||||
| AA + AC | 73 (83.9) | 35 (87.5) | Ref | Ref | Ref | Ref | |||
| CC | 14 (16.1) | 5 (12.5) | Ref | 0.74 (0.25, 2.23) | 0.599 | Ref | 0.59 (0.14, 2.44) | 0.466 | |
| Additive | 0.543 | ||||||||
| AA | 29 (67.4) | 15 (75) | Ref | Ref | Ref | Ref | |||
| CC | 14 (32.6) | 5 (25) | Ref | 0.69 (0.21, 2.28) | 0.544 | Ref | 0.40 (0.08, 2.02) | 0.270 | |
| Codominant | 0.952 | ||||||||
| AA + CC | 43 (49.4) | 20 (50) | Ref | Ref | Ref | Ref | |||
| CC | 44 (50.6) | 20 (50) | Ref | 0.98 (0.46, 2.07) | 0.952 | Ref | 0.71 (0.28, 1.79) | 0.464 | |
3.4. Association between genotypes of genetic variants rs1748195 and lipid profile
According to the data in Table 4 which was analyzed with Pearson χ 2 tests, there was a significant difference between lipid profiles (HDL and TC) and rs1748195 in ANGPTL3 gene with AA, AC and CC genotypes in all groups (p < 0.05). Analysis have revealed that the level of PAB in the group of healthy individuals in all studied genotypes is higher than the other two groups (p < 0.05). Evaluation of some disorders related to CVD events such as hypertension, diabetes and dyslipidemia showed that these disorders are more common in individuals with negative angiogram and carriers of AC genotype compared to Angio+ group (p < 0.001). In contrast, the investigation of TG, SBP, DBP, BMI and CRP factors in three groups of healthy individuals, Angio− and Angio+ did not show a significant relationship in any of the rs1748195 polymorphism genotypes (p > 0.05).
TABLE 4.
Distribution of genotypes and clinical characteristics of individuals and their relationship with angio
| AA | AC | CC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Healthy | Angio− | Angio+ | p | Healthy | Angio− | Angio+ | p | Healthy | Angio− | Angio+ | p | |
| Age | 48.04 ± 7.77 | 56.80 ± 8.60 | 60.98 ± 8.09 | <0.001* | 49.08 ± 7.74 | 54.79 ± 8.83 | 59.48 ± 9.37 | <0.001* | 49 ± 7.5 | 45.50 ± 8.55 | 60.26 ± 8.01 | <0.001* |
| Female | 52 (51) | 17 (68) | 15 (35.7) | 0.035* | 94 (64.4) | 9 (47.4) | 13 (20.6) | <0.001* | 39 (60) | 3 (50) | 6 (31.6) | 0.091 |
| HDL | 44.09 ± 12.38 | 38.21 ± 9.07 | 35.12 ± 7.62 | <0.001* | 44.98 ± 13.37 | 35.47 ± 6.90 | 36.34 ± 9.12 | <0.001* | 45.23 ± 13.04 | 30.33 ± 10.86 | 34.12 ± 11.48 | 0.001* |
| LDL | 109.82 ± 34.41 | 93.97 ± 26 | 88.97 ± 33.58 | 0.002* | 112.73 ± 35.99 | 92.99 ± 36.25 | 96.86 ± 31.03 | 0.003* | 126.45 ± 44.81 | 108.60 ± 44.60 | 96.92 ± 45.20 | 0.50 |
| TG | 130.71 ± 79.28 | 130.64 ± 74.98 | 138.02 ± 67 | 0.868 | 143.36 ± 102.33 | 129.76 ± 75.68 | 124.91 ± 60.39 | 0.398 | 128.28 ± 88.80 | 197.83 ± 130.64 | 134.41 ± 58.19 | 0.179 |
| TC | 187.01 ± 40.24 | 160.84 ± 37.90 | 151.70 ± 40.34 | <0.001* | 192.88 ± 36.70 | 154.41 ± 44.53 | 157.07 ± 35.92 | <0.001* | 198.51 ± 47.99 | 178.5 ± 57.62 | 157.29 ± 50.83 | 0.010* |
| SBP | 122.24 ± 19.05 | 118.75 ± 10.86 | 125.29 ± 15.07 | 0.364 | 120.89 ± 18.98 | 122.14 ± 16.61 | 124.33 ± 14.02 | 0.475 | 121.38 ± 22.18 | 130 ± 7.91 | 124.06 ± 14.52 | 0.622 |
| DBP | 79.14 ± 10.96 | 76.46 ± 6.67 | 79.29 ± 8.01 | 0.461 | 78.95 ± 11.60 | 81.07 ± 9.44 | 78.87 ± 11.42 | 0.794 | 80.78 ± 13.52 | 83.33 ± 5.16 | 76.25 ± 8.47 | 0.346 |
| BMI | 27.81 ± 4.98 | 28.18 ± 4.92 | 26.24 ± 3.38 | 0.156 | 27.64 ± 4.80 | 26.56 ± 3.14 | 30.03 ± 27.39 | 0.516 | 27.93 ± 4.41 | 26.57 ± 2.61 | 31.75 ± 24.71 | 0.429 |
| CRPnew | 6.78 ± 11.68 | 4.09 ± 3.59 | 4.12 ± 3.85 | 0.227 | 4.78 ± 10.19 | 5.54 ± 5.30 | 3.30 ± 3.03 | 0.468 | 4.47 ± 10.98 | 2.73 ± 2.66 | 2.82 ± 2.09 | 0.815 |
| PAB | 77.79 ± 34.37 | 1.06 ± 0.31 | 1.05 ± 0.25 | <0.001* | 100.70 ± 46.05 | 1.04 ± 0.29 | 1.13 ± 0.35 | <0.001* | 80.72 ± 56.52 | 0.98 ± 0.13 | 0.99 ± 0.11 | 0.005* |
| Hypertention | 27 (26.5) | 9 (36) | 19 (45.2) | 0.085 | 49 (33.6) | 10 (52.6) | 39 (61.9) | <0.001* | 20 (30.8) | 4 (66.7) | 14 (73.7) | 0.002* |
| Diabetes | 10 (10.1) | 12 (48) | 11 (26.8) | <0.001* | 26 (17.9) | 6 (31.6) | 28 (44.4) | <0.001* | 15 (23.8) | 2 (33.3) | 7 (36.8) | 0.504 |
| Hyperlipidemia | 76 (74.5) | 16 (64) | 26 (61.9) | 0.257 | 124 (84.9) | 8 (42.1) | 33 (52.4) | <0.001* | 56 (86.2) | 4 (66.7) | 8 (42.1) | |
Note: Values are expressed as mean ± standard deviation or median (interquartile range 25th–75th) for normal and non‐normal distribution data respectively. *p ≤ 0.05 is indicates as significant.
4. DISCUSSION
Statistical analyzes in the present study demonstrated that lipid profiles (including LDL, HDL, and TC), history of hypertension, diabetes, and dyslipidemia were associated with angiography. According to the results, we found that the CC genotype of the rs1748195 polymorphism at the ANGPTL3 gene loci has a significant association with the negative angiogram. Whereas, the frequency of C allele of this SNP was higher in the group of healthy individuals. In contrast, we did not find any significant differences between the genotypes of this genetic variant and the NISR and in‐stent restenosis>50% groups in any of the studied genetic models. Also, the clinical characteristics of individuals, such as PAB and some disorders related to CVD events such as blood pressure, showed a significant and effective difference in the types of genotypes.
In the reviews, we found that, Cardiovascular disease is the leading cause of death worldwide and involves a range of circulatory disorders (Francula‐Zaninovic & Nola, 2018). One of the most common causes of CVD is an inflammatory disease called atherosclerosis, in which the accumulation of LDL particles containing cholesterol blocks the coronary arteries of the heart (Lnsis, 2000). ANGPTL3 belongs to the family of ANGPTLs and is expressed almost exclusively in stages of embryonic development and in adults in the liver (Lupo & Ferri, 2018). ANGPTLs are regulators of plasma lipids in animals and humans (Su & Peng, 2018). This protein inhibits LPL and EL and thus affects the level of plasma TGs (Olkkonen et al., 2018). Dewey et al. reported that one type of loss function in ANGPTL3 was associated with a 41% reduction in CAD risk. It was also found that the anti‐ANGPTL3 monoclonal antibody, called evinacumab can reduce plasma TG and LDL‐C levels by up to 76% and 23%, respectively (Dewey et al., 2017). Several GWAS have shown that many single nucleotide polymorphisms, including the genetic variant rs1748195 of the ANGPTL3 gene, have a significant relationship with serum TGA levels. In this way, it plays an effective role in cardiovascular disease. (Jeemon et al., 2011). Willer et al. (2008) in the study of 18,243 samples stated that this polymorphism with p = 1.7*10–10 is related to plasma triglyceride levels. Also, by studying Murray et al. on 1155 eligible Italians, after determining the effect of this SNP on the TG level, it was determined that the C allele of this polymorphism with frequency = 0.675 is the TG risk‐increasing allele (Murray et al., 2009). In addition, the study of Chung et al. which was performed on 3060 staff of Korean medical clinics (Chung et al., 2014) and 5537 individuals from the cohort study, confirmed the association of serum TG levels with genetic variants rs10889353 and rs1748195 of ANGPTL3 gene (Cho et al., 2009). In 2021, a study by Aghasizadeh et al. on 1002 samples reported a strong association between CVD events and different genotypes of ANGPTL3 gene polymorphisms, including rs1748195 polymorphism. In contrast, no significant association was found between this SNP and HTN, dyslipidemia and metabolic syndrome (MetS) (Aghasizadeh, Zare‐Feyzabadi, et al., 2021). Based on the results of a study by Li et al. in 2018 on 568 patients with CAD and 539 patients with ischemic stroke, it was found that the genetic variant rs1748195 would increase the risk of atherosclerosis as well as CAD (Li et al., 2018).
In recent years, genome‐related studies have provided new insights into genetics and its association with cardiovascular disease. Although many studies have been performed to find an association between the ANGPTL3 gene and CVD on this gene and its polymorphisms, there are still many unknowns. Therefore, it is suggested that more studies be done in this regard so that the resulting information can have more effective clinical applications for us.
5. CONCLUSION
In the present study, we investigated that the CC genotype, and also the A allele corresponding to rs1748195 at the ANGPTL3 gene loci was associated with negative angiogram and directly related to the risk of coronary occlusion >50%. In contrast, this result was not significant between ISR and non‐ISR groups. Furthermore, statistical analysis demonstrates that the fundamental characteristics of this study population were related to the existence of a coronary occlusion of >50%. Therefore, it is suggested that more studies should be done to have more effective clinical applications for us.
AUTHOR CONTRIBUTIONS
We declare that we contributed significantly towards the research study; MA, NA, NS, and SGH designed the experiments and revised the manuscript. MA and NA performed the experiments. MA, SS, MR and GF wrote the manuscript. ARE, KK and HE carried out the data analysis. RAD and MGM were supervisor of the study. All authors reviewed, considered, and approved the manuscript.
AVAILABILITY OF DATA AND MATERIALS
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Informed consent was obtained from all subjects using protocols approved by the Ethics Committee of the Mashhad University of Medical Science.
ACKNOWLEDGMENTS
We gratefully acknowledge the contributions of the data collection team and the individuals who participated in this study.
Afkhami, N. , Aghasizadeh, M. , Ghiasi Hafezi, S. , Zare‐Feyzabadi, R. , Saffar Soflaei, S. , Rashidmayvan, M. , Rastegarmoghadam–Ebrahimian, A. , Khanizadeh, K. , Safari, N. , Ferns, G. A. , Esmaily, H. , Darban, R. A. , & Ghayour‐Mobarhan, M. (2023). Evaluation of rs1748195 ANGPTL3 gene polymorphism in patients with angiographic coronary artery disease compared to healthy individuals. Molecular Genetics & Genomic Medicine, 11, e2105. 10.1002/mgg3.2105
Nafise Afkhami, Malihe Aghasizadeh and Somayeh Ghiasi Hafezi co‐first authors.
Contributor Information
Reza Assaran Darban, Email: assaran@mshdiau.ac.ir.
Majid Ghayour‐Mobarhan, Email: ghayourm@mums.ac.ir.
REFERENCES
- Aghasizadeh, M. , Bizhaem, S. K. , Baniasadi, M. , Khazdair, M. R. , & Kazemi, T. (2021). Evaluation of LDL goal achievement in statin consumption, south east of Iran. Scientific Reports, 11(1), 10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghasizadeh, M. , Samadi, S. , Sahebkar, A. , Miri‐Moghaddam, E. , Esmaily, H. , Souktanloo, M. , Avan, A. , Mansoori, A. , Ferns, G. A. , Kazemi, T. , & Ghayour‐Mobarhan, M. (2021). Serum HDL cholesterol uptake capacity in subjects from the MASHAD cohort study: Its value in determining the risk of cardiovascular endpoints. Journal of Clinical Laboratory Analysis, 35(6), e23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghasizadeh, M. , Zare‐Feyzabadi, R. , Kazemi, T. , Avan, A. , Ferns, G. A. , Esmaily, H. , Miri‐Moghaddam, E. , & Ghayour‐Mobarhan, M. (2021). A haplotype of the ANGPTL3 gene is associated with CVD risk, diabetes mellitus, hypertension, obesity, metabolic syndrome, and dyslipidemia. Gene, 782, 145525. [DOI] [PubMed] [Google Scholar]
- Bea, A. , Franco‐Marin, E. , Marco‐Benedí, V. , Jarauta, E. , Gracia‐Rubio, I. , Cenarro, A. , Civeira, F. , & Lamiquiz‐Moneo, I. (2021). ANGPTL3 gene variants in subjects with familial combined hyperlipidemia. Scientific Reports, 11(1), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. S. , Go, M. J. , Kim, Y. J. , Heo, J. Y. , Oh, J. H. , Ban, H.‐J. , Yoon, D. , Lee, M. H. , Kim, D. J. , Park, M. , Cha, S. H. , Kim, J. W. , Han, B. G. , Min, H. , Ahn, Y. , Park, M. S. , Han, H. R. , Jang, H. Y. , Cho, E. Y. , … Kim, H. L. (2009). A large‐scale genome‐wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nature Genetics, 41(5), 527–534. [DOI] [PubMed] [Google Scholar]
- Chung, S.‐K. , Yu, H. , Park, A. Y. , Kim, J. Y. , & Cha, S. (2014). Genetic loci associated with changes in lipid levels leading to constitution‐based discrepancy in Koreans. BMC Complementary and Alternative Medicine, 14(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hert, M. , Detraux, J. , & Vancampfort, D. (2018). The intriguing relationship between coronary heart disease and mental disorders. Dialogues in Clinical Neuroscience, 20(1), 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey, F. E. , Gusarova, V. , Dunbar, R. L. , O'Dushlaine, C. , Schurmann, C. , Gottesman, O. , McCarthy, S. , van Hout, C. , Bruse, S. , Dansky, H. M. , Leader, J. B. , Murray, M. F. , Ritchie, M. D. , Kirchner, H. L. , Habegger, L. , Lopez, A. , Penn, J. , Zhao, A. , Shao, W. , … Baras, A. (2017). Genetic and pharmacologic inactivation of ANGPTL3 and cardiovascular disease. New England Journal of Medicine, 377(3), 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estey, M. P. , Cohen, A. H. , Colantonio, D. A. , Chan, M. K. , Marvasti, T. B. , Randell, E. , Delvin, E. , Cousineau, J. , Grey, V. , Greenway, D. , Meng, Q. H. , Jung, B. , Bhuiyan, J. , Seccombe, D. , & Adeli, K. (2013). CLSI‐based transference of the CALIPER database of pediatric reference intervals from Abbott to Beckman, Ortho, Roche and Siemens clinical chemistry assays: Direct validation using reference samples from the CALIPER cohort. Clinical Biochemistry, 46(13–14), 1197–1219. [DOI] [PubMed] [Google Scholar]
- Francula‐Zaninovic, S. , & Nola, I. A. (2018). Management of measurable variable cardiovascular disease'risk factors. Current Cardiology Reviews, 14(3), 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet, D. , Méthot, J. , & Kastelein, J. (2012). Gene therapy for lipoprotein lipase deficiency. Current Opinion in Lipidology, 23(4), 310–320. [DOI] [PubMed] [Google Scholar]
- Gimbrone, M. A., Jr. , Topper, J. N. , Nagel, T. , Anderson, K. R. , & Garcia‐Cardeña, G. (2000). Endothelial dysfunction, hemodynamic forces, and atherogenesis. Annals of the New York Academy of Sciences, 902(1), 230–240. [DOI] [PubMed] [Google Scholar]
- Goldberg, I. J. (1996). Lipoprotein lipase and lipolysis: Central roles in lipoprotein metabolism and atherogenesis. Journal of Lipid Research, 37(4), 693–707. [PubMed] [Google Scholar]
- Gomaraschi, M. , Obici, L. , Simonelli, S. , Gregorini, G. , Negrinelli, A. , Merlini, G. , Franceschini, G. , & Calabresi, L. (2011). Effect of the amyloidogenic L75P apolipoprotein AI variant on HDL subpopulations. Clinica Chimica Acta, 412(13–14), 1262–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htay, T. , & Liu, M. W. (2005). Drug‐eluting stent: A review and update. Vascular Health and Risk Management, 1(4), 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeemon, P. , Pettigrew, K. , Sainsbury, C. , Prabhakaran, D. , & Padmanabhan, S. (2011). Implications of discoveries from genome‐wide association studies in current cardiovascular practice. World Journal of Cardiology, 3(7), 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan, S. , Melander, O. , Guiducci, C. , Surti, A. , Burtt, N. P. , Rieder, M. J. , Cooper, G. M. , Roos, C. , Voight, B. F. , Havulinna, A. S. , Wahlstrand, B. , Hedner, T. , Corella, D. , Tai, E. S. , Ordovas, J. M. , Berglund, G. , Vartiainen, E. , Jousilahti, P. , Hedblad, B. , … Orho‐Melander, M. (2008). Six new loci associated with blood low‐density lipoprotein cholesterol, high‐density lipoprotein cholesterol or triglycerides in humans. Nature Genetics, 40(2), 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan, S. , & Srivastava, D. (2012). Genetics of human cardiovascular disease. Cell, 148(6), 1242–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten, S. (2017). Angiopoietin‐like 3 in lipoprotein metabolism. Nature Reviews Endocrinology, 13(12), 731–739. [DOI] [PubMed] [Google Scholar]
- Kharazmi‐Khorassani, S. , Kharazmi‐Khorassani, J. , Rastegar‐Moghadam, A. , Samadi, S. , Ghazizadeh, H. , Tayefi, M. , Ferns, G. A. , Ghayour‐Mobarhan, M. , Avan, A. , & Esmaily, H. (2019). Association of a genetic variant in the angiopoietin‐like protein 4 gene with metabolic syndrome. BMC Medical Genetics, 20(1), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivelä, A. , & Hartikainen, J. (2006). Restenosis related to percutaneous coronary intervention has been solved? Annals of Medicine, 38(3), 173–187. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y. , Nakajima, T. , & Inoue, I. (2002). Molecular modeling of the dimeric structure of human lipoprotein lipase and functional studies of the carboxyl‐terminal domain. European Journal of Biochemistry, 269(18), 4701–4710. [DOI] [PubMed] [Google Scholar]
- Levin, K. A. (2014). Urban–rural differences in adolescent eating behaviour: A multilevel cross‐sectional study of 15‐year‐olds in Scotland. Public health Nutrition, 17(8), 1776–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.‐J. , Yin, R.‐X. , Cao, X.‐L. , Chen, W.‐X. , Huang, F. , & Wu, J.‐Z. (2018). DOCK7‐ANGPTL3 SNPs and their haplotypes with serum lipid levels and the risk of coronary artery disease and ischemic stroke. Lipids in Health and Disease, 17(1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lnsis, A. J. (2000). Atherosclenrosis. Nature, 407, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo, M. G. , & Ferri, N. (2018). Angiopoietin‐like 3 (ANGPTL3) and atherosclerosis: Lipid and non‐lipid related effects. Journal of Cardiovascular Development and Disease, 5(3), 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattijssen, F. , & Kersten, S. (2012). Regulation of triglyceride metabolism by angiopoietin‐like proteins. Biochimica et Biophysica Acta (BBA)‐Molecular and Cell Biology of Lipids, 1821(5), 782–789. [DOI] [PubMed] [Google Scholar]
- Mensah, G. A. , Roth, G. A. , & Fuster, V. (2019). The global burden of cardiovascular diseases and risk factors: 2020 and beyond. American College of Cardiology Foundation. [DOI] [PubMed] [Google Scholar]
- Montone, R. A. , Sabato, V. , Sgueglia, G. A. , & Niccoli, G. (2013). Inflammatory mechanisms of adverse reactions to drug‐eluting stents. Current Vascular Pharmacology, 11(4), 392–398. [DOI] [PubMed] [Google Scholar]
- Murray, A. , Cluett, C. , Bandinelli, S. , Corsi, A. M. , Ferrucci, L. , Guralnik, J. , Singleton, A. , Frayling, T. , & Melzer, D. (2009). Common lipid‐altering gene variants are associated with therapeutic intervention thresholds of lipid levels in older people. European Heart Journal., 30(14), 1711–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccoli, G. , Montone, R. A. , Ferrante, G. , & Crea, F. (2010). The evolving role of inflammatory biomarkers in risk assessment after stent implantation. Journal of the American College of Cardiology, 56(22), 1783–1793. [DOI] [PubMed] [Google Scholar]
- Nordestgaard, B. G. , Abildgaard, S. , Wittrup, H. H. , Steffensen, R. , Jensen, G. , & Tybjærg‐Hansen, A. (1997). Heterozygous lipoprotein lipase deficiency: Frequency in the general population, effect on plasma lipid levels, and risk of ischemic heart disease. Circulation, 96(6), 1737–1744. [DOI] [PubMed] [Google Scholar]
- Oguri, M. , Kato, K. , Hibino, T. , Yokoi, K. , Segawa, T. , Matsuo, H. , Watanabe, S. , Nozawa, Y. , Murohara, T. , & Yamada, Y. (2007). Genetic risk for restenosis after coronary stenting. Atherosclerosis, 194(2), e172–e178. [DOI] [PubMed] [Google Scholar]
- Oldoni, F. , Palmen, J. , Giambartolomei, C. , Howard, P. , Drenos, F. , Plagnol, V. , Humphries, S. E. , Talmud, P. J. , & Smith, A. J. P. (2016). Post‐GWAS methodologies for localisation of functional non‐coding variants: ANGPTL3. Atherosclerosis, 246, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen, V. M. , Sinisalo, J. , & Jauhiainen, M. J. (2018). New medications targeting triglyceride‐rich lipoproteins: Can inhibition of ANGPTL3 or apoC‐III reduce the residual cardiovascular risk? Atherosclerosis, 272, 27–32. [DOI] [PubMed] [Google Scholar]
- Parsamanesh, N. , Karami‐Zarandi, M. , Banach, M. , Penson, P. E. , & Sahebkar, A. (2021). Effects of statins on myocarditis: A review of underlying molecular mechanisms. Progress in Cardiovascular Diseases, 67, 53–64. [DOI] [PubMed] [Google Scholar]
- Reiner, Ž. , Catapano, A. , & De Backer, G. (2011). ESC/EAS Džepne smjernice. ESC/EAS smjernice za liječenje dislipidemija, prilagođeno prema ESC smjernicama za liječenje dislipidemija. European Heart Journal, 32, 1769–1818.21712404 [Google Scholar]
- Rip, J. , Nierman, M. C. , Ross, C. J. , Jukema, J. W. , Hayden, M. R. , Kastelein, J. J. , Stroes, E. S. , & Kuivenhoven, J. A. (2006). Lipoprotein lipase S447X: A naturally occurring gain‐of‐function mutation. Arteriosclerosis, Thrombosis, and Vascular Biology, 26(6), 1236–1245. [DOI] [PubMed] [Google Scholar]
- Roth, G. A. , Johnson, C. , Abajobir, A. , Abd‐Allah, F. , Abera, S. F. , Abyu, G. , Ahmed, M. , Aksut, B. , Alam, T. , Alam, K. , Alla, F. , Alvis‐Guzman, N. , Amrock, S. , Ansari, H. , Ärnlöv, J. , Asayesh, H. , Atey, T. M. , Avila‐Burgos, L. , Awasthi, A. , … Murray, C. (2017). Global, regional, and national burden of cardiovascular diseases for 10 causes, 1990 to 2015. Journal of the American College of Cardiology, 70(1), 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin, G. D. , Leipsic, J. , Schoepf, U. J. , Fleischmann, D. , & Napel, S. (2014). CT angiography after 20 years: A transformation in cardiovascular disease characterization continues to advance. Radiology, 271(3), 633–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi‐Karimian, M. , Safarian‐Bana, H. , Mohammadzadeh, E. , Kazemi, T. , Mansoori, A. , Ghazizadeh, H. , Samadi, S. , Nikbakht‐Jam, I. , Nosrati, M. , Ferns, G. A. , Esmaily, H. , Aghasizadeh, M. , & Ghayour‐Mobarhan, M. (2021). A pilot study of the effects of crocin on high‐density lipoprotein cholesterol uptake capacity in patients with metabolic syndrome: A randomized clinical trial. BioFactors, 47(6), 1032–1041. [DOI] [PubMed] [Google Scholar]
- Sadeghi, M. , Haghdoost, A. A. , Bahrampour, A. , & Dehghani, M. (2017). Modeling the burden of cardiovascular diseases in Iran from 2005 to 2025: The impact of demographic changes. Iranian Journal of Public Health, 46(4), 506–516. [PMC free article] [PubMed] [Google Scholar]
- Samadi, S. , Mehramiz, M. , Kelesidis, T. , Mobarhan, M. G. , Sahebkar, A. H. , Esmaily, H. , Moohebati, M. , Farjami, Z. , Ferns, G. A. , Mohammadpour, A. , & Avan, A. (2019). High‐density lipoprotein lipid peroxidation as a molecular signature of the risk for developing cardiovascular disease: Results from MASHAD cohort. Journal of Cellular Physiology, 234(9), 16168–16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, X. , & Peng, D. (2018). New insights into ANGPLT3 in controlling lipoprotein metabolism and risk of cardiovascular diseases. Lipids in Health and Disease, 17(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich, T. M. , Musunuru, K. , Smith, A. V. , Edmondson, A. C. , Stylianou, I. M. , Koseki, M. , Pirruccello, J. P. , Ripatti, S. , Chasman, D. I. , Willer, C. J. , Johansen, C. T. , Fouchier, S. W. , Isaacs, A. , Peloso, G. M. , Barbalic, M. , Ricketts, S. L. , Bis, J. C. , Aulchenko, Y. S. , Thorleifsson, G. , … Kathiresan, S. (2010). Biological, clinical and population relevance of 95 loci for blood lipids. Nature, 466(7307), 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium . (2010). A map of human genome variation from population scale sequencing. Nature, 467(7319), 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valizadeh, M. , Aghasizadeh, M. , Nemati, M. , Hashemi, M. , Aghaee‐Bakhtiari, S. H. , Zare‐Feyzabadi, R. , Esmaily, H. , Ghazizdaeh, H. , Sahebi, R. , Ahangari, N. , Ferns, G. A. , Pasdar, A. , & Ghayour‐Mobarhan, M. (2021). The association between a fatty acid binding protein 1 (FABP1) gene polymorphism and serum lipid abnormalities in the MASHAD cohort study. Prostaglandins, Leukotrienes and Essential Fatty Acids, 172, 102324. [DOI] [PubMed] [Google Scholar]
- Willer, C. J. , Sanna, S. , Jackson, A. U. , Scuteri, A. , Bonnycastle, L. L. , Clarke, R. , Heath, S. C. , Timpson, N. J. , Najjar, S. S. , Stringham, H. M. , Strait, J. , Duren, W. L. , Maschio, A. , Busonero, F. , Mulas, A. , Albai, G. , Swift, A. J. , Morken, M. A. , Narisu, N. , … Abecasis, G. R. (2008). Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nature Genetics, 40(2), 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner, R. , Strauss, J. , Frank, S. , Wagner, E. , Hofmann, W. , Kratky, D. , Hiden, M. , & Levak‐Frank, S. (2000). The role of lipoprotein lipase in adipose tissue development and metabolism. International Journal of Obesity, 24(4), S53–S56. [DOI] [PubMed] [Google Scholar]
- Zierk, J. , Arzideh, F. , Haeckel, R. , Rascher, W. , Rauh, M. , & Metzler, M. (2013). Indirect determination of pediatric blood count reference intervals. Clinical Chemistry and Laboratory Medicine, 51(4), 863–872. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


