Abstract
Skin, the largest organ in the body, provides a passive physical barrier against infection and contains elements of the innate and adaptive immune systems. Skin consists of various cells, including keratinocytes, fibroblasts, endothelial cells and immune cells. This diversity of cell types could be important to gene therapies because DNA transfection could elicit different responses in different cell types. Previously, we observed the upregulation and activation of cytosolic DNA sensing pathways in several non-tumor and tumor cell types as well in tumors after the electroporation (electrotransfer) of plasmid DNA (pDNA). Based on this research and the innate immunogenicity of skin, we correlated the effects of pDNA electrotransfer to fibroblasts and keratinocytes to mouse skin using reverse transcription real-time PCR (RT-qPCR) and several types of protein quantification. After pDNA electrotransfer, the mRNAs of the putative DNA sensors DEAD (AspGlu-Ala-Asp) box polypeptide 60 (Ddx60), absent in melanoma 2 (Aim2), Z-DNA binding protein 1 (Zbp1), interferon activated gene 202 (Ifi202), and interferon-inducible protein 204 (Ifi204) were upregulated in keratinocytes, while Ddx60, Zbp1 and Ifi204 were upregulated in fibroblasts. Increased levels of the mRNAs and proteins of several cytokines and chemokines were detected and varied based on cell type. Mouse skin experiments in vivo confirmed our in vitro results with increased expression of putative DNA sensor mRNAs and of the mRNAs and proteins of several cytokines and chemokines. Finally, with immunofluorescent staining, we demonstrated that skin keratinocytes, fibroblasts and macrophages contribute to the immune response observed after pDNA electrotransfer.
Keywords: DNA sensors, Cytokines, DNA electrotransfer, Keratinocytes, Fibroblasts, Skin
1. Introduction
Genetically engineered vectors encoding specific DNA sequences can be used for gene therapies or vaccination. Viral and non-viral vectors have been developed to introduce DNA into cells [1]. Due to the potential for immunogenicity and cytotoxicity with viral vectors, the use of non-viral vectors is increasing [2]. However, delivery systems are essential to facilitate cellular internalization of plasmid DNA (pDNA) due to its lower cellular uptake.
Gene electrotransfer (GET) is a safe and efficient non-viral gene delivery method based on electroporation. The application of defined electric pulses to cells or tissues leads to increased cell membrane permeability to facilitate the transport of nucleic acids into cells [3]. This method is also used to deliver DNA or RNA to different tissues, including skin [4,5]. Skin is an attractive target for GET due to its easy accessibility, large treatment area and the presence of antigen-presenting cells, which are critical for eliciting an effective immune response in therapies where an immune response is desirable. GET to the skin has been used in many medical applications, including vaccination [6–8], wound healing [9,10] and cancer treatment [4]. Although immune cell infiltration is observed after pDNA GET to skin [11], providing an adjuvant effect, the response of skin to pDNA GET at the molecular level has yet to be elucidated.
Currently, potential applications of DNA vaccines such as cancer therapy [12], infectious disease [13], and autoimmune disease [14] are being investigated. GET of pDNA vaccines improves immunogenicity and increases gene expression [11,15–18]. The basic working principle of pDNA vaccines involves the use of pDNA encoding antigens of targeted pathogens or tumors [19].
Encoded protein antigen(s) can be expressed in stromal and dendritic cells. These antigens are processed and presented to naïve CD4 + or CD8 + T cells either directly or by cross-presentation, respectively [20]. The transfected pDNA can also interact with DNA-specific pattern recognition receptors known as DNA sensors to activate the production of Type I interferons (IFN) and pro-inflammatory cytokines and chemokines or cause cell death [21–23]. These pro-inflammatory proteins may induce innate immune responses that potentiate adaptive immune responses.
Skin is an ideal target for pDNA vaccine administration. Cutaneous GET is a safe and efficient delivery technique highly applicable to the clinical setting. The aim of our study was to evaluate if several putative cytosolic DNA sensors (Table 1) and their signaling pathways are upregulated in non-immune cell types in vitro, assess these pathways and the innate immune response in mouse skin in vivo, and determine which cell types in skin in vivo respond to entry of pDNA after electrotransfer.
Table 1.
Putative DNA sensors assayed in this study.
| Putative DNA Sensor | Reference |
|---|---|
| Endosomal | |
| TLR9 (Toll-like receptor 9) | Hemmi, 2000; [24] |
| Cytosolic/nuclear | |
| ZBP1 (Z-DNA binding protein 1) | Takaoka 2007, [25] |
| AIM2 (Absent in melanoma 2) | Burckstummer, 2009, [26] |
| RIG-I (Retinoic acid inducible gene I) | Ablasser, 2009; [27] |
| p202 (Interferon activated gene 202) | Roberts, 2009, [28] |
| IFI16/p204 (Interferon-inducible protein 204) | Yan, 2008; [29] |
| DHX9 (DEAH (AspGlu-Ala-His) box helicase 9) | Kim, 2010, [30] |
| DHX36 (DEAH (Asp-Glu-Ala-His) box helicase 36) | Kim, 2010, [30] |
| LRRFIP1 (Leucine-rich repeat flightless interacting protein 1) | Yang, 2010, [31] |
| DDX41 (DEAD (Asp-GluAla-Asp) box polypeptide 41) | Zhang, 2011, [32] |
| DDX60 (DEAD (AspGlu-Ala-Asp) box polypeptide 60) | Miyashita, 2011, [33] |
| Ku70 (Lupus Ku autoantigen protein p70) | Mimori, 1986; [34] |
| MRE11 (Meiotic recombination 11 homolog, double- | |
| strand break repair nuclease) | Kondo, 2013, [35] |
| cGAS (Cyclic guanosine monophosphate adenosine monophosphate synthase) | Sun, 2013,[36] |
| SOX2 (SRY (sex-determining region Y)-box 2) | Xia, 2015, [37] |
2. Material and methods
2.1. Cell lines
Mouse Kera-308 keratinocytes (CLS Cell Lines Service, Eppelheim, Germany) and L929 fibroblasts (American Type Culture Collection, Manassas, VA, USA) were cultured in Advanced Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 5% fetal bovine serum (FBS, Gibco), 10 mM L-glutamine (Gibco), 50 μg/ml Penicillin-Streptomycin (Merck, Germany) at 37 ◦C in a humidified incubator with 5% CO2. Cells were regularly tested for the presence of Mycoplasma spp. with the MycoAlert PLUS Mycoplasma Detection kit (Lonza, Basel, Switzerland) and found negative.
2.2. Animals
For immunofluorescent tissue staining, 12-week-old female BALB/cAnNCrl mice (Charles River L. s.r.l., Calco, Italy) were used. For protein expression analysis, 7–8-week-old female BALB/cJ mice (Jackson Laboratories, Bar Harbor, ME, USA) were used. All procedures were approved by the Administration of the Republic of Slovenia for Food Safety, Veterinary Sector and Plant Protection or by the University of South Florida Institutional Animal Care and Use Committee.
2.3. Plasmid DNA (pDNA)
The plasmid pEGFP-N1 (BD Biosciences, San Jose, CA, USA), encoding green fluorescent protein, was used to establish transfection efficiency. It was isolated after amplification in a competent Escherichia coli (TOP10; Thermo Fisher Scientific) and purified using Maxi-Endo Free Plasmid Kits (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration of isolated plasmid was measured with an Epoch Microplate Spectrophotometer (BioTek Instruments, Winooski, VT, USA). The plasmid vector gWizBlank (pDNA, purified by Aldevron, Fargo, ND, USA) was used for all other experiments. Unless otherwise noted, all plasmids were suspended at 2 μg/μl in physiological saline.
2.4. In vitro electrotransfer of pDNA
In vitro pDNA electrotransfer was carried out using either an Electro Cell B10 electroporator (LEROY biotech, Saint-Orens-de-Gameville, France) or an ECM 830 Square Wave Electroporation System (BTX, Holliston, MA, USA). One million cells were prepared in 40 μl of electroporation buffer. Ten μl of pDNA was added, and 50 μl of this suspension containing 1 × 106 cells was electroporated using stainless steel electrodes with a 2 mm gap. Eight square-waved pulses with a voltage-to-distance ratio of 600 V/cm, a pulse duration of 5 ms and a frequency of 1 Hz (EP1) or six square-waved pulses with a voltage-to-distance ratio of 1300 V/cm, pulse duration 100 μs, and a frequency of 4 Hz (EP2) were used [23].
2.5. Transfection efficiency in vitro
Two days after electrotransfer of 10 μg pEGFP-N1 per 1 × 106 cells, the cells were imaged by fluorescence microscopy (Olympus IX-70, Hamburg, Germany) then trypsinized and resuspended in 400 μl of PBS for flow cytometry analysis (FACSCanto II flow cytometer, BD Biosciences, Franklin Lakes, NJ, USA). A 488-nm laser (air-cooled, 20 mW solid-state) and 530/30-nm band-pass filter were used for the excitation and emission to detect green fluorescent protein fluorescence. A total of 20,000 events were measured. The percentage of transfected cells represented transfection efficiency, while the fluorescence intensity was determined as an indirect measure of protein expression and therefore the amount of pDNA that was introduced into the cells.
2.6. Cell survival measured by metabolic viability assay in vitro
After electrotransfer of 1, 2, and 3.5 mg/ml of pDNA, 1 × 103 cells were cultured in 100 μl of medium in 96-well plates and incubated for 72 h. Presto Blue™ viability reagent was used to measure cell metabolism, an indirect indicator of cell viability. Fluorescence was quantified using a Cytation 1 Imaging Multi-Mode Reader with a 560/590 nm (excitation/emission) filter (Biotek, Santa Clara, CA, USA).
2.7. Cell death mechanism in vitro
Cell death mechanisms were determined twenty hours after pDNA electrotransfer using a FITC Annexin V Apoptosis Detection Kit with 7-AAD (7-aminoactinomycin D) (BioLegend, San Diego, CA, USA) according to manufacturer’s instructions. Flow cytometry (FACSCanto II, BD Biosciences, Franklin Lakes, New Jersey, USA.). Data analysis was performed with FACSDiva V6.1.2 and FlowJo software (BD Biosciences). A total of 20,000 events were measured. Apoptosis was evaluated by phosphatidylserine detection in the outer plasma membrane leaflet using Annexin V. Necrotic cells were detected with 7-AAD, which has a high DNA-binding constant and can pass into the nucleus and bind to DNA in necrotic cells. Viable cells were Annexin V and 7-AAD negative.
Pyroptosis was determined with caspase activation using Caspase-Glo® 1 Inflammasome Assay (Promega, Madison, WI, USA) per manufacturer’s instructions and quantified using the Cytation 1 Imaging Multi-Mode Reader.
2.8. In vivo electrotransfer of pDNA
Twenty μl of 2 μg/μl pDNA was injected intradermally and pulses immediately applied to the injection area. For the RNA and histological studies, EP1 pulses were applied with a stainless steel 5 mm gap plate electrode using an Electro Cell B10 electroporator (LEROY biotech). For protein studies, two perpendicular sets of four EP1 pulses were delivered using a 4-plate electrode with 6 mm distance between the electrodes [38] with an ECM 830 Square Wave Electroporator (BTX, Holliston, MA, USA). Each mouse was monitored continuously until recovered from anesthesia as indicated by their ability to maintain sternal recumbency and exhibit purposeful movement.
2.9. mRNA expression in vitro and in vivo
Four hours after pDNA electrotransfer, total RNA was isolated from 1 × 106 cells using a peqGOLD Total RNA Kit (VWR Life Science, Germany) or from skin tissue using TRIzol™ Reagent (Thermo Fischer Scientific, USA) and purified (peqGOLD Total RNA Kit, VWR Life Science, Germany). Complementary DNA was synthesized from 500 ng of RNA (SuperScript VILO, Thermo Fischer Scientific, USA) and diluted 1:10. Relative mRNA levels were determined with quantitative real time PCR (See Supplementary Table 1 for primer sequences) and PowerUp SYBR Green Master Mix (Thermofisher Scientific, USA) on a Quant-Studio™ 3 Real-Time PCR System (Thermo Fisher Scientific, USA). Messenger RNA expression was quantified using the ΔΔCt method [39] relative to the reference mRNAs β-actin and glyceraldehyde 3-phosphate dehydrogenase and normalized to the control group.
2.10. In vitro ELISAs
Mouse IFN-β (PBL Assay Science, Piscataway, NJ, USA), IL-1β (R&D Systems, Minneapolis, MN, USA), and IL-6 (R&D Systems, Minneapolis, MN, USA) secreted by 106 viable keratinocytes or fibroblasts into 1 ml medium were quantified by ELISA six hours after pDNA electrotransfer.
2.11. Protein analysis in vivo
Four hours after pDNA electrotransfer, mice were humanely euthanized and skin samples snap-frozen on dry ice. The samples were homogenized in NP-40 Lysis buffer (Alfa Aesar, Ward Hill, MA, USA) with the addition of Halt Protease Inhibitor cocktail EDTA-free (Thermo Fisher Scientific, Rockford, IL, USA) using a gentle MACS OctoDissociator (Miltenyi Biotec, Auburn, CA, USA). The total protein concentration was normalized, and 25 μg of each sample was analyzed using a Mouse Cytokine/Chemokine Magnetic Bead Panel (Millipore, Burlington, MA, USA) using a MAGPIX System (Luminex, Austin, TX, USA) per manufacturer’s instructions. Confirming ELISAs to quantify IFN-β (PBL Assay Science, Piscataway, NJ, USA), IL-6 (R&D Systems, Minneapolis, MN, USA), and CXCL1 (RayBiotech, Peachtree Corners, GA, USA) were performed on the same homogenates.
2.12. Immunofluorescence (IF) staining in vivo
Six hours after pDNA electrotransfer, excised skin samples were fixed in 4% paraformaldehyde for 17 h, then incubated 24 h in 30% sucrose for cryoprotection and frozen in OCT medium (VWR). Fourteenmicrometer thick frozen sections were cut using a Leica CM1850 cryostat and mounted onto the Superfrost plus glass slides (ThermoFisher Scientific). Slides were then stained with primary and secondary antibodies (Table 2). Because the anti-mouse Pan-cytokeratin antibody (ThermoFisher Scientific) had the same host as the skin samples, we used Fab fragments (Jackson Immunoresearch, West Grove, PA, USA) to block endogenous immunoglobulins in tissue sections or on cell surfaces. Nuclei were stained with Hoechst 33342 solution (3 μg/ml). Slides were mounted with ProLong™ Glass Antifade Mountant (Thermo Fisher Scientific). Samples were imaged with an LSM 800 confocal microscope (Carl Zeiss) with a 20x objective (NA 0.8). Hoechst 33342, Alexa Fluor 488, Cy3 and Alexa Fluor 647 were excited with lasers with excitation wavelengths of 405 nm, 488 nm, 561 nm and 640 nm, respectively. Emitted light was captured with Gallium Arsenide Phosphide (GaAsP) detector combined with a variable dichroic and filters at channel specific wavelengths: 410 – 545 nm (Hoechst 33342), 488 – 545 nm (Alexa Fluor 488), 565 – 620 nm (Cy3) and 645 – 700 nm (Alexa Fluor 647). The images were visualized and analyzed with Imaris software (Bitplane Image Analysis software, Switzerland) and FIJI-ImageJ software [40]. For quantitating IF data, we used FIJI-ImageJ software. Cells containing colocalizations were selected as regions of interest in which the relative levels of specific molecules were measured by mean fluorescent intensity of Cy3 [41].
Table 2.
Primary and Secondary antibodies used in mouse skin experiments.
| Primary antibody | Dilution | Host | Secondary antibody |
|---|---|---|---|
| Fibroblast marker MA1–40076 (ThermoFisher Scientific) | 1:100 | Rat | Alexa Fluor® 647 AffiniPure Donkey Anti-Rat IgG (H+L) (Jackson Immunoresearch) |
| Cytokeratin Pan Antibody Cocktail MA5–13203 (ThermoFisher Scientific) | 1:200 | Mouse | Donkey Anti-Mouse IgG H&L (Alexa Fluor® 488) (ab150105, (Abcam)) |
| Anti-Mo F4/80, 14–4801–82 (ThermoFisher Scientific) | 1:200 | Rat | Alexa Fluor® 488 AffiniPure Donkey Anti-Rat IgG (H+L) Jackson Immunoresearch) |
| IFN-β PA5 20390 (ThermoFisher Scientific) | 1:200 | Rabbit | Cy™3 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson Immunoresearch) |
| TNFα 6671 (Abcam) | 1:50 | Rabbit | Cy™3 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson Immunoresearch) |
| IL-1β 9722 (Abcam) | 1:200 | Rabbit | Cy™3 AffiniPure Donkey Anti-Rabbit IgG (H+L) (Jackson Immunoresearch) |
2.13. Statistical analysis
GraphPad Prism 9 (La Jolla, CA, USA) was used for statistical analysis and graph preparation. Data were tested for normality distribution and found to be normally distributed. Significance was determined by a two-tailed t-test or one-way ANOVA test followed by a Dunnett’s multiple comparisons test. A P < 0.05 was considered statistically significant.
3. Results
3.1. Transfection efficiency, cytotoxicity, and cell death mechanisms in vitro
Transfection efficiency, cell survival, and cell death mechanisms were quantified after electrotransfer of pEGFP-N1 into L929 fibroblasts and KERA 308 keratinocytes using two different pulse protocols. The transfection efficiency was pulse protocol-dependent in keratinocytes, with 4.5% fluorescent cells using the EP1 pulse protocol and 46.0% using the EP2 pulse protocol (Fig. 1a). In fibroblasts 5.2% of cells were transfected with the EP1 pulse protocol, and 10.1% with the EP2 pulse protocol. A statistically significant difference was detected in fibroblast transfection efficiency between the two pulse protocols (Fig. 1b). Although the transfection efficiency varied between the cell types, the median fluorescence intensity, indicating the level of protein expression, was higher using the EP1 pulse protocol in both cell types.
Fig. 1.
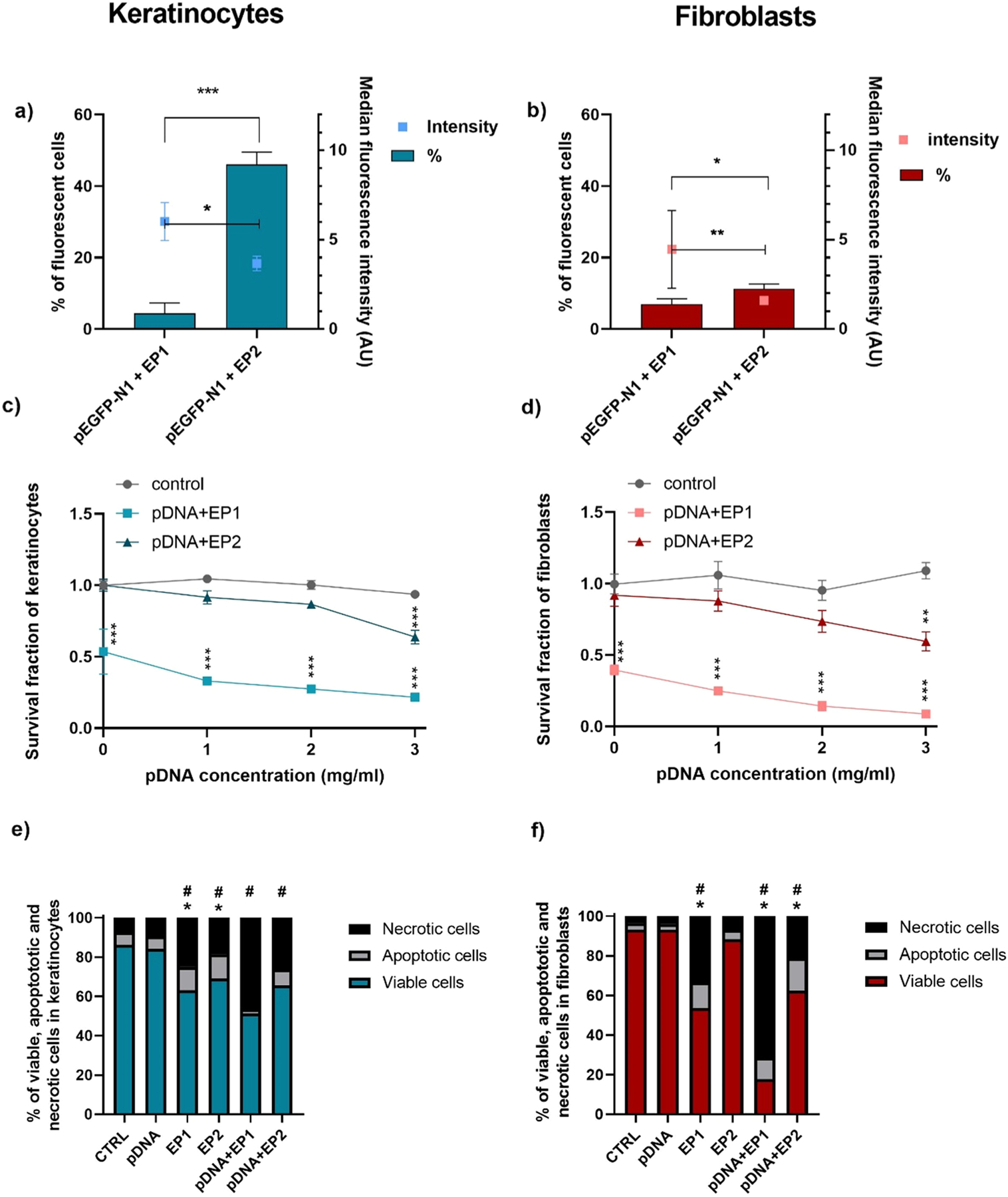
Transfection efficiency, cytotoxicity, and cell death mechanisms after electrotransfer of pDNA to keratinocytes and fibroblasts. Transfection efficiency was measured by flow cytometry in a) keratinocytes and b) fibroblasts (n = 3). Cell survival was measured by metabolic activity in c) keratinocytes and d) fibroblasts (n = 3–4) data were normalized to control. Apoptosis and necrosis were quantified by detecting Annexin V and 7-AAD staining using flow cytometry in e) keratinocytes and f) fibroblasts (n = 3) data were normalized to control. In a and b * **p < 0.001, * *p < 0.01, *p < 0.05 compared to different pulse protocols; in c and d * **p < 0.001, * *p < 0.01, *p < 0.05 compared to control; in e and f *p < 0.05 refers to apoptotic cells, #p < 0.05 refers to necrotic cells.
Cell cytotoxicity was evaluated 72 h after delivery of varying pDNA concentrations. Pulse protocol EP1 alone had a greater effect on cell viability than protocol EP2 in both keratinocytes and fibroblasts (p > 0.01). The addition of plasmid DNA further decreased viability with either pulse protocols in each cell type, indicating that entry of pDNA into the cell further induced cell death (Fig. 1c, d). Therefore, cell death mechanisms were investigated twenty hours after electrotransfer of pDNA. A high percentage of necrotic cells was detected in each cell lines after applying EP1 with or without pDNA (Fig. 1e, f). After electrotransfer of pDNA, 46.5% of the keratinocyte cells were necrotic and 2.2% were apoptotic using protocol EP1, while 26.1% were necrotic and 8.1% were apoptotic using protocol EP2 (Fig. 1e). In fibroblasts, 71.4% of the cells were necrotic and 10.8% were apoptotic using protocol EP1 to deliver pDNA, while 21.2% were necrotic and 16.3% were apoptotic using protocol EP2 (Fig. 1 f).
3.2. Cytokine mRNA upregulation in keratinocytes and fibroblasts
The mRNA levels of interferon beta (Ifn-β1), tumor necrosis factor alpha (Tnfα), interleukin 1 beta (Il-1β), interleukin-6 (Il-6) and interleukin 18 (Il-18) 4 h after electrotransfer of pDNA were next evaluated. A 3- to 7-fold increase in Ifn-β1 mRNA levels in response to either pulse protocol in combination with pDNA was induced in keratinocytes (Fig. 2a, Supplementary figure 1). In addition, Il-6 and Tnfα mRNAs were 2-fold higher in keratinocytes after pDNA electrotransfer using the EP1 protocol. These cells responded to pDNA electrotransfer with a 3.5-fold increase in Il-1β mRNA using EP1 and a 1.7-fold increase in Il-1β mRNA using EP2. IL-18 mRNA levels did not change. Fibroblasts (Fig. 2b, Supplementary figure 2) responded to pDNA electrotransfer by either method with an approximately 1000-fold increase in Ifn-β1 mRNA levels. These cells responded to EP1 pulses alone with an approximately 95-fold increase in Tnfα and a 13-fold increase in Il-6 mRNA levels; for each cytokine, these levels increased an additional 2.5-fold in the presence of DNA. EP2 pulses alone did not induce Il-6 mRNA upregulation, although the combination with pDNA induced an upregulation of nearly 18-fold. Tnfα mRNA was upregulated 95-fold using the EP1 protocol and combination with pDNA induced a 244-fold upregulation. In these cells, no increase in Il-1β and Il-18 mRNA level was detected using either protocol.
Fig. 2.
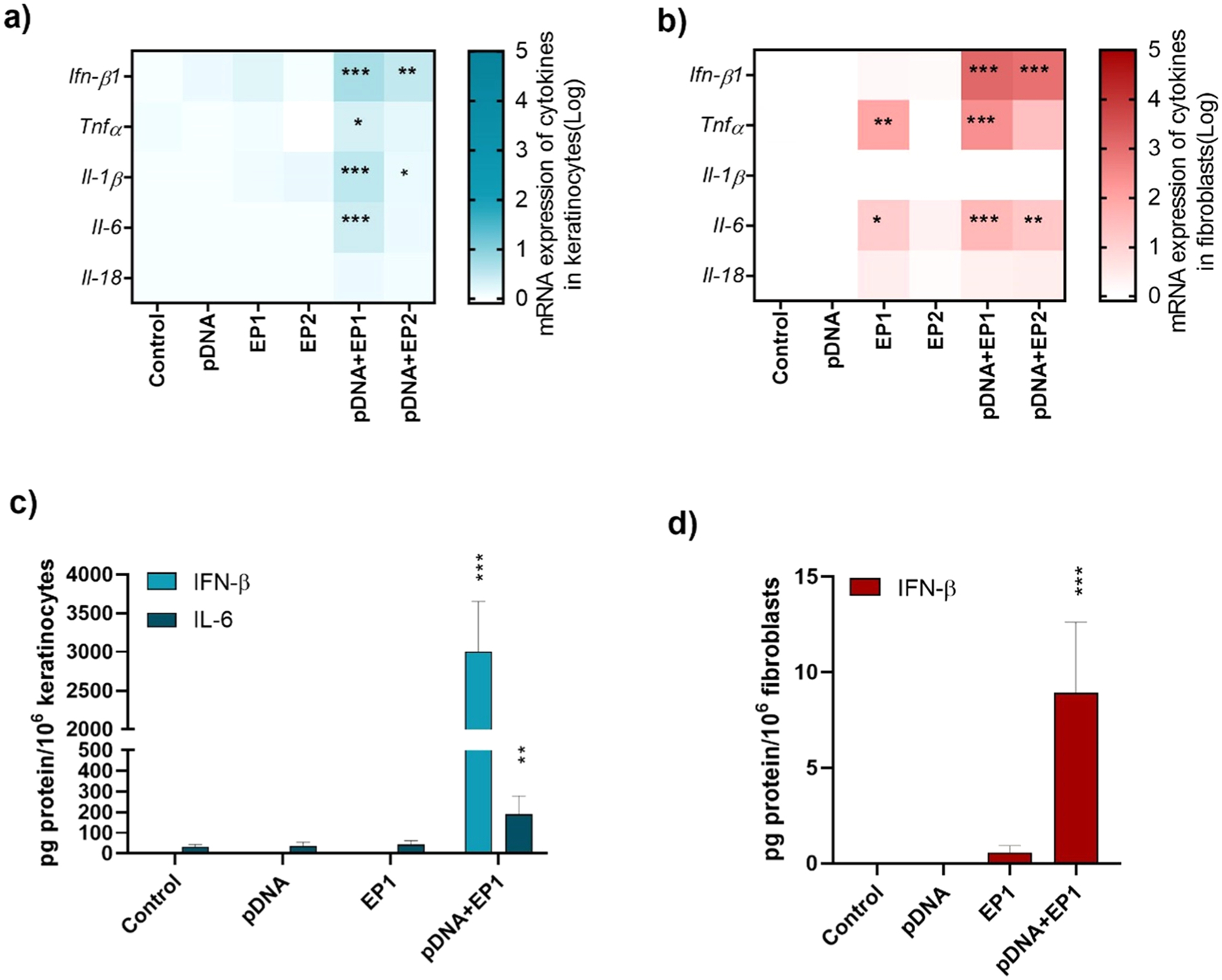
Normalized expression of cytokine mRNA in a) keratinocytes and b) fibroblasts (n = 3–6). Protein detected in the medium by ELISA in c) keratinocytes and d) fibroblasts (n = 4), * **p < 0.001, * *p < 0.01, * p < 0.05 compared to control. Fold expression is shown in a logarithmic scale.
Due to the higher expression of cytokines and DNA sensor mRNAs after delivery with EP1, this pulse protocol was used for protein analysis. While both cell lines secreted significant levels of IFN-β after pDNA electrotransfer, keratinocytes were much more responsive (nearly 300-fold) than fibroblasts (Fig. 2c, d). Keratinocytes also secreted IL-6 (Fig. 2c), while fibroblasts did not secrete detectable levels. In keratinocytes, the levels of IL-1β were below the levels of detection by ELISA (data not shown).
3.3. Increased expression of DNA sensor mRNAs in keratinocytes and fibroblasts
The levels of the mRNAs encoding several putative DNA sensors described in the literature (Table 1) were evaluated in keratinocytes and fibroblasts (Fig. 3, Supplementary figure 3, 4). The mRNAs encoding Tlr9, Ddx60, Dhx9, Dhx36, Aim2, Cgas, Zbp1, Ddx41, LrrfipI, Ifi202, Ifi204, Mre11, Ku70 and the adapter protein Sting (stimulator of interferon genes) were expressed in both cell lines. Ddx58 mRNA was not detected in either cell line, while Sox2 mRNA was not detected in fibroblasts.
Fig. 3.
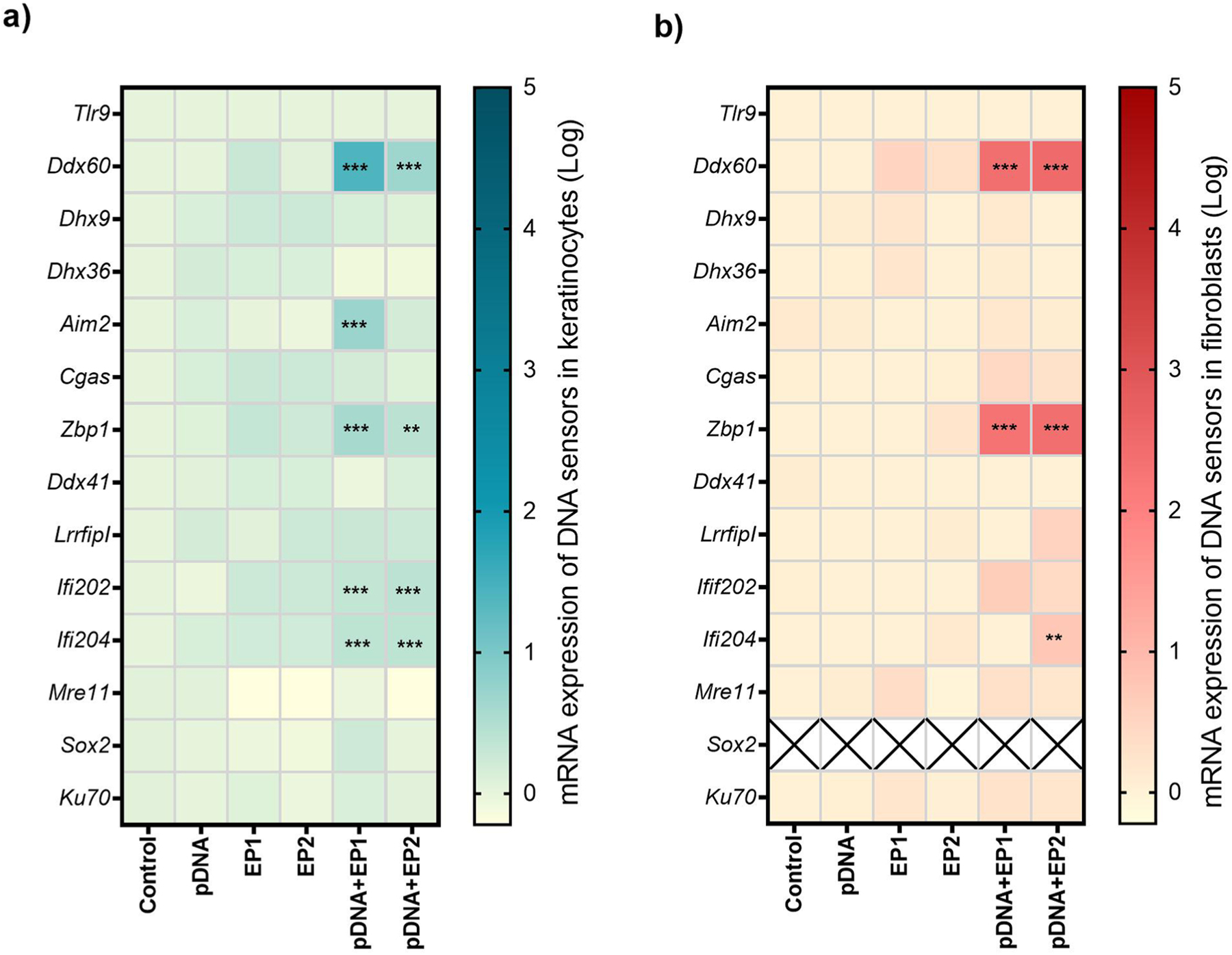
Normalized expression of putative DNA sensor mRNA in a) keratinocytes and b) fibroblasts (n = 3–6). * **p < 0.001, * *p < 0.01, *p < 0.05 compared to control. Fold expression is shown in a logarithmic scale.
Exposure to pDNA or pulses alone did not change mRNA levels. However, after pDNA delivery, several mRNAs were upregulated. This upregulation varied with cell type and with the pulse protocol used to deliver the DNA. In keratinocytes (Fig. 3a, Supplementary figure 3), Ddx60 mRNA levels increased nearly 26-fold after DNA delivery with EP1 and nearly 5-fold after delivery with EP2. Zpb1 mRNA regulation was independent of the pulse protocol used, but increased 3- to 4-fold. Ifi202 and Ifi204 mRNA levels increased significantly in keratinocytes. Interestingly, Aim2 mRNA levels were significantly upregulated after pDNA delivery with EP1 but not EP2. Fibroblasts (Fig. 3b, Supplementary figure 4) responded more intensely to intracellular pDNA but with a more limited repertoire of DNA sensor mRNA regulation. In these cells, the expression of Ddx60 mRNA increased to the similar level after delivery of pDNA with either of the pulse protocol (~250-fold after delivery of pDNA with EP1, ~ 300-fold after delivery with EP2). Zbp1 mRNA increased approximately 200-fold regardless of the pulse protocol used. Finally, Ifi204 mRNA increased modestly but significantly over 5-fold after delivery with EP2, but not after EP1 delivery.
3.4. Caspase 1 and IL-1β detection in keratinocytes
AIM2 initiates the inflammasome assembly, an innate immune complex that leads to the activation of inflammatory caspases [42]. Increased expression of the mRNAs of the cytokine Il-1β and DNA sensor Aim2 were detected in keratinocytes (Fig. 2a, 3 a), which implicated the activity of caspase 1. This caspase proteolytically cleaves the IL-1β precursor, allowing the release of the mature protein [42]. IL-1β protein secretion was not detected by keratinocytes after electrotransfer of pDNA (data not shown). Finally, activation of caspase 1 using Caspase-Glo® 1 Inflammasome Assay, indicating pyroptosis, was not detected (data not shown).
3.5. Expression of DNA sensor and cytokine mRNA in mouse skin after pDNA electrotransfer
After the in vitro studies, we evaluated the expression of DNA sensor and cytokine mRNAs in mouse skin four hours after pDNA electrotransfer with the EP1 pulse protocol. In the in vitro experiments, pDNA delivery with this protocol induced more potent responses to DNA sensing. We quantified the mRNAs of putative DNA sensors Ddx60, Zbp1, Aim2, Ifi202, Ifi204 and the cytokines Ifn-β1, Tnfα, Il-1β, Il-6 since these were upregulated in keratinocytes and fibroblasts. The cytokines were significantly upregulated with a 20-fold increase of Tnfα, a 100- fold increase of Il-1β and a 160- fold increase of Il-6 after electrotransfer of pDNA. The fold expression of Ifn-β1 increased more than 2000-fold (Fig. 4a, Supplementary figure 5). Exposure to pDNA or pulses alone did not change mRNA levels; however, after electrotransfer of pDNA, all the assayed DNA sensor mRNAs were upregulated (Fig. 4b, Supplementary figure 6). In skin, a 44- fold increase in Ddx60, a 70-fold increase in Zbp1, a 14-fold increase in Aim2, a 32-fold increase in Ifi202and a 23.5-fold increase in Ifi204 mRNA levels were detected (Fig. 4b).
Fig. 4.
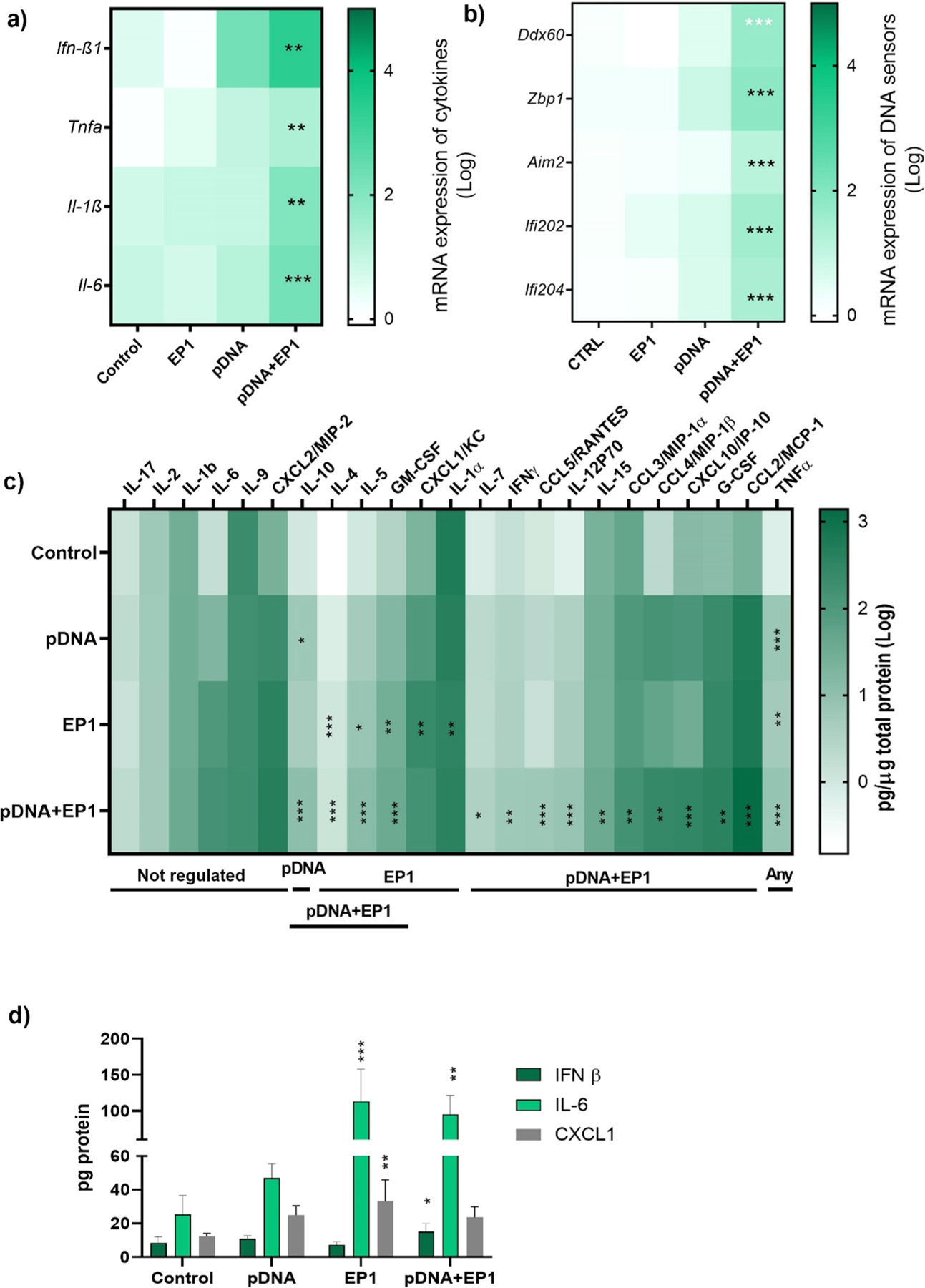
Normalized mRNA and protein expression in mouse skin 4 h after pDNA delivery. a) Expression of pro-inflammatory cytokine mRNA (Ifn-β1, Tnfα, Il1β, Il-6, n = 3) b) Expression of putative DNA sensor mRNAs (Ddx60, Zbp1, Aim2, Ifi202, Ifi204, n = 3) Cytokine and chemokine protein expression in skin 4 h after electrotransfer of pDNA determined by c) bead array or d) ELISA. * **p < 0.001, * *p < 0.01, * p < 0.05 compared to control. Interleukin 17 (IL-17); interleukin 2 (IL-2); interleukin 1 beta (IL-1 β); interleukin 6 (IL-6); interleukin 9 (IL-9); chemokine (C-X-C motif) ligand/macrophage inflammatory protein 2 (CXCL2/MIP-2); interleukin 10 (IL-10); interleukin 4 (IL-4); interleukin 5 (IL-5); granulocyte-macrophage colony-stimulating factor (GM-CSF); chemokine (C-X-C motif) ligand 1 (CXCL1/KC); interleukin 1 alpha (IL-1α); interleukin 7 (IL-7); interferon gamma (IFNγ); chemokine (C-C motif) ligand 5/regulated on activation, Normal T Expressed and Secreted (CCL5/RANTES); interleukin 12P70 (IL-12P70); interleukin 15 (IL-15); chemokine (C-C motif) ligand 3/macrophage inflammatory protein 1-alpha (CCL3/MIP-1α); Chemokine (C-C motif) ligand 4/ macrophage inflammatory protein 1 beta (CCL4/MIP-1β); C-X-C motif chemokine ligand 10/interferon gamma-induced protein 10 (CXCL10/IP-10); granulocyte-colony stimulating factor (G-CSF); chemokine (C-C motif) ligand 2/monocyte chemoattractant protein-1 (CCL2/MCP-1). Fold expression is showed on a logarithmic scale.
3.6. Cytokine and chemokine protein expression in mouse skin after electrotransfer of pDNA
A bead array analysis of cytokines and chemokines in mouse skin homogenates was performed 4 h after pDNA electrotransfer and demonstrated the production of several of these signaling proteins (Fig. 4c). IL-1β, IL-2, IL-6, IL-9, IL-17, and the chemokine C-X-C motif chemokine ligand 2 (CXCL2) were expressed but not significantly upregulated. TNFα was upregulated approximately 8- to 12-fold by any manipulation (p < 0.001). IL-1α protein was downregulated minimally but significantly after application of the EP1 protocol alone; this downregulation was alleviated by the addition of pDNA. IL-10 was upregulated approximately 6-fold by pDNA injection only; this increased to approximately 11-fold with the addition of pulses. IL-4, IL-5, and GM-CSF were upregulated equivalently by pulse application alone or in the presence of pDNA. We confirmed the secretion of CXCL1 detected by ELISA after pulses alone, but this effect was ameliorated by the addition of pDNA electrotransfer (Fig. 4d). Finally, the levels of nearly half the chemokine and cytokine proteins assayed were upregulated only by the combination of pDNA and pulses to varying levels. IL-15 was minimally but significantly upregulated 1.5-fold, C-C motif chemokine ligand 3 (CCL3), IFNγ, IL-7, and CCL5 were upregulated 3- to 7- fold, IL-12p70, CCL2, and CXCL10 were upregulated 13- to 22-fold, while CCL4 was upregulated 122-fold. The bead array confirmed the upregulation of IL-6 mRNA levels. However, the IL-6 ELISA demonstrated that only groups receiving pulses were significantly upregulated, whether alone or with pDNA. Finally, by ELISA, we observed that IFN-β was significantly secreted in the skin 4 h after pDNA electrotransfer (Fig. 4d).
3.7. Effect of electrotransfer of pDNA on DNA sensing pathways in mouse skin and local immune cells in vivo
We used immunofluorescent staining on frozen skin sections to identify the specific cells that expressed IFNβ, TNFα, and IL-1β six hours after electrotransfer of pDNA and to quantify each protein. Each section was stained with a marker for a specific cell type (keratinocytes, macrophages or fibroblasts), the nucleus and the specific cytokine. In each cell type, we detected increase and equivalent expression of TNFα and IFN-β (Figs. 5, 6, 7). After pDNA electrotransfer, increased expression of Il-1β was detected in keratinocytes and particularly in macrophages but not in fibroblasts (Figs. 7,8).
Fig. 5.
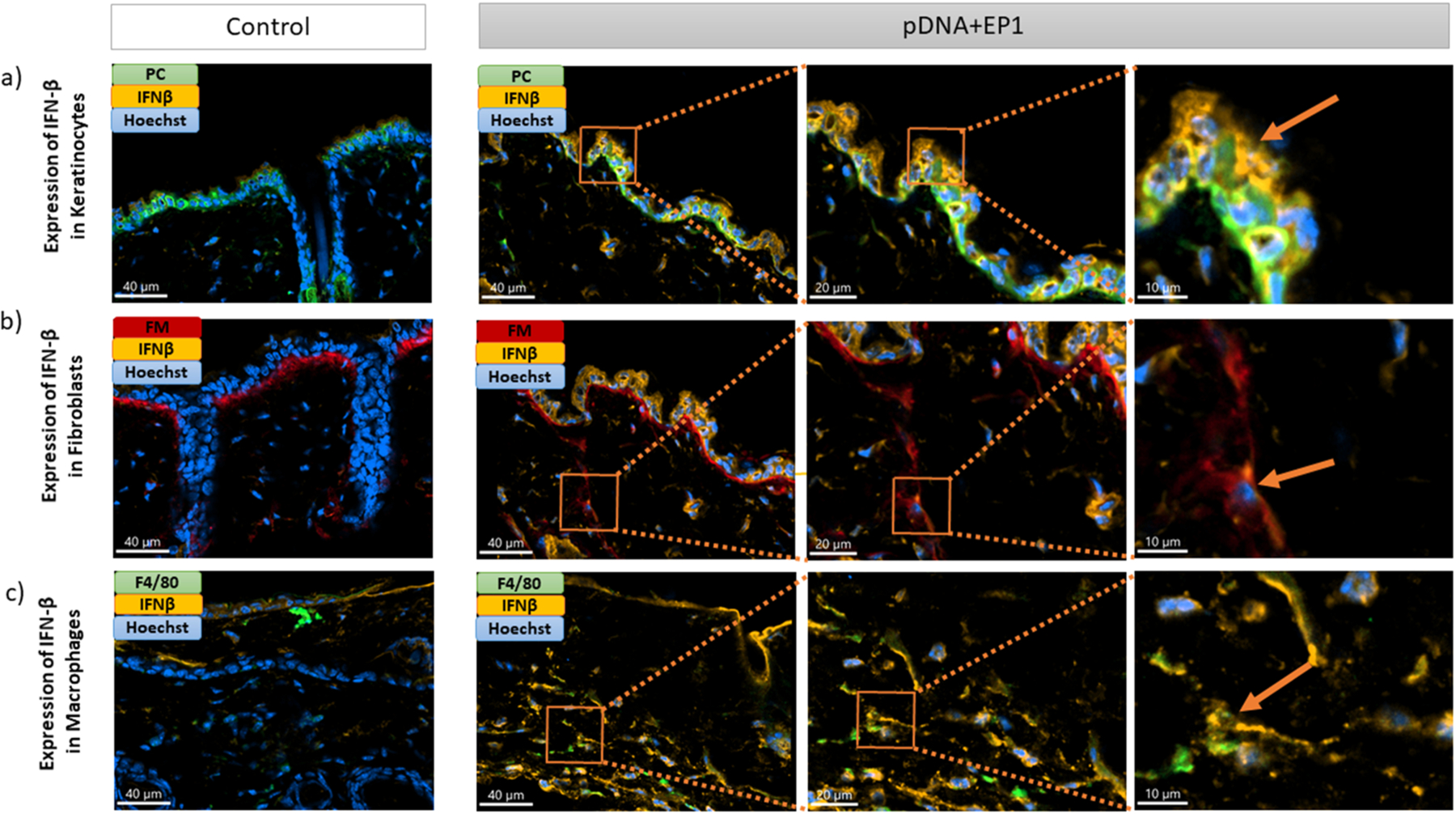
Detection of IFN-β (IFN-β -orange) in a) keratinocytes (Pan-cytokeratin -green), b) fibroblasts (Fibroblast marker-red) and c) macrophages (F4/80–green) on frozen mouse skin Section 6 hours after pDNA electrotransfer. Three different images on the right side represent the zoom into the region of interest (ROI). Arrows show detection of IFN-β in different cells. Scale bar: 40 μm for the control column and first column of pDNA+EP1 (right side), 20 μm for the second column of pDNA+EP1, 10 μm for the third column of pDNA+EP1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
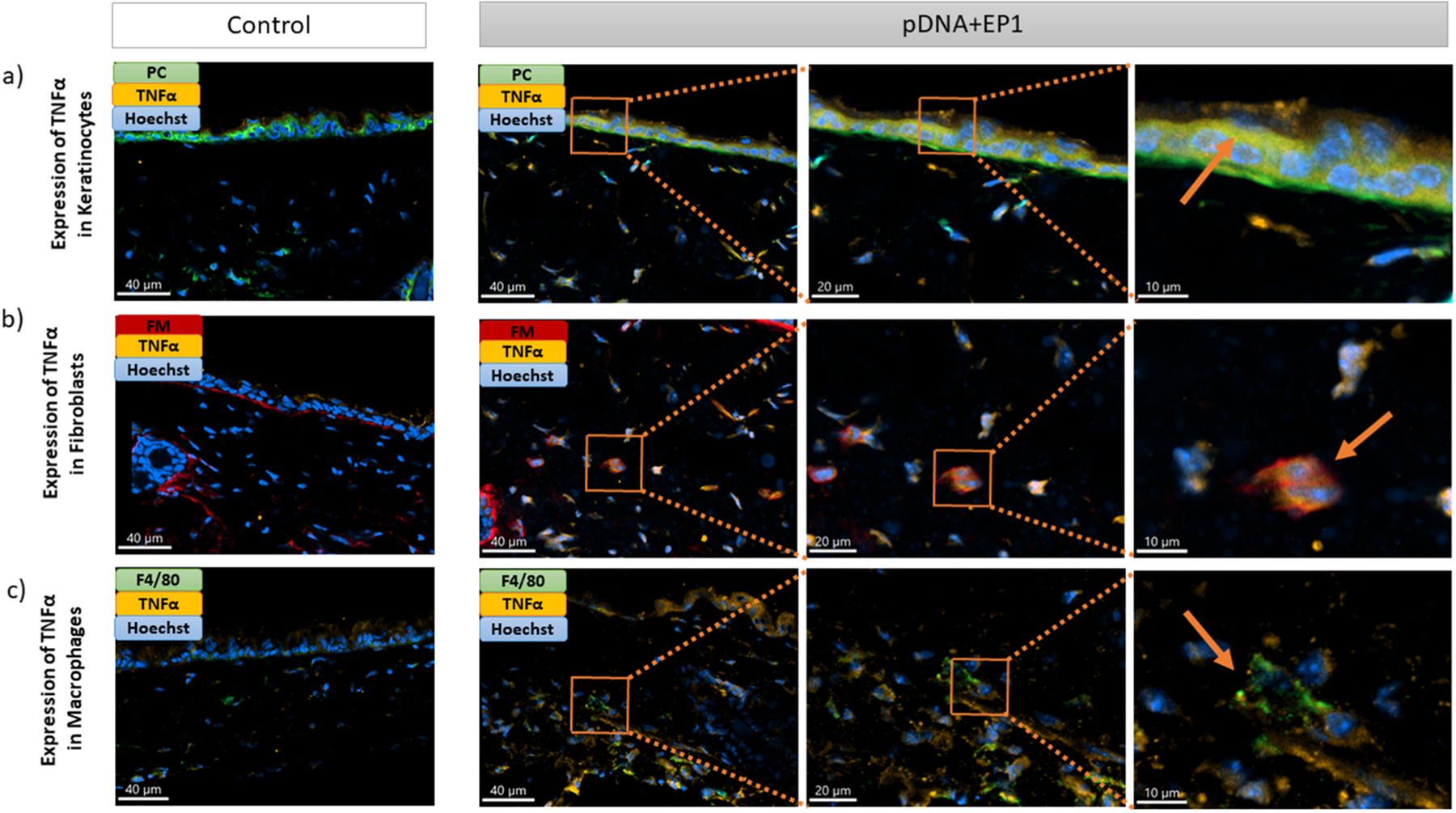
Detection of TNFα (TNFα -orange) in a) keratinocytes (Pan cytokeratin-green), b) fibroblasts (Fibroblast marker-red) and c) macrophages (F4/80-green) on frozen mouse skin Section 6 hours after pDNA electrotransfer. Three different images on the right side represent the zoom into the ROI. Arrows show detection of TNFα in different cells. Scale bar: 40 μm for the control column and first column of pDNA+EP1 (right side), 20 μm for the second column of pDNA+EP1, 10 μm for the third column of pDNA+EP1.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
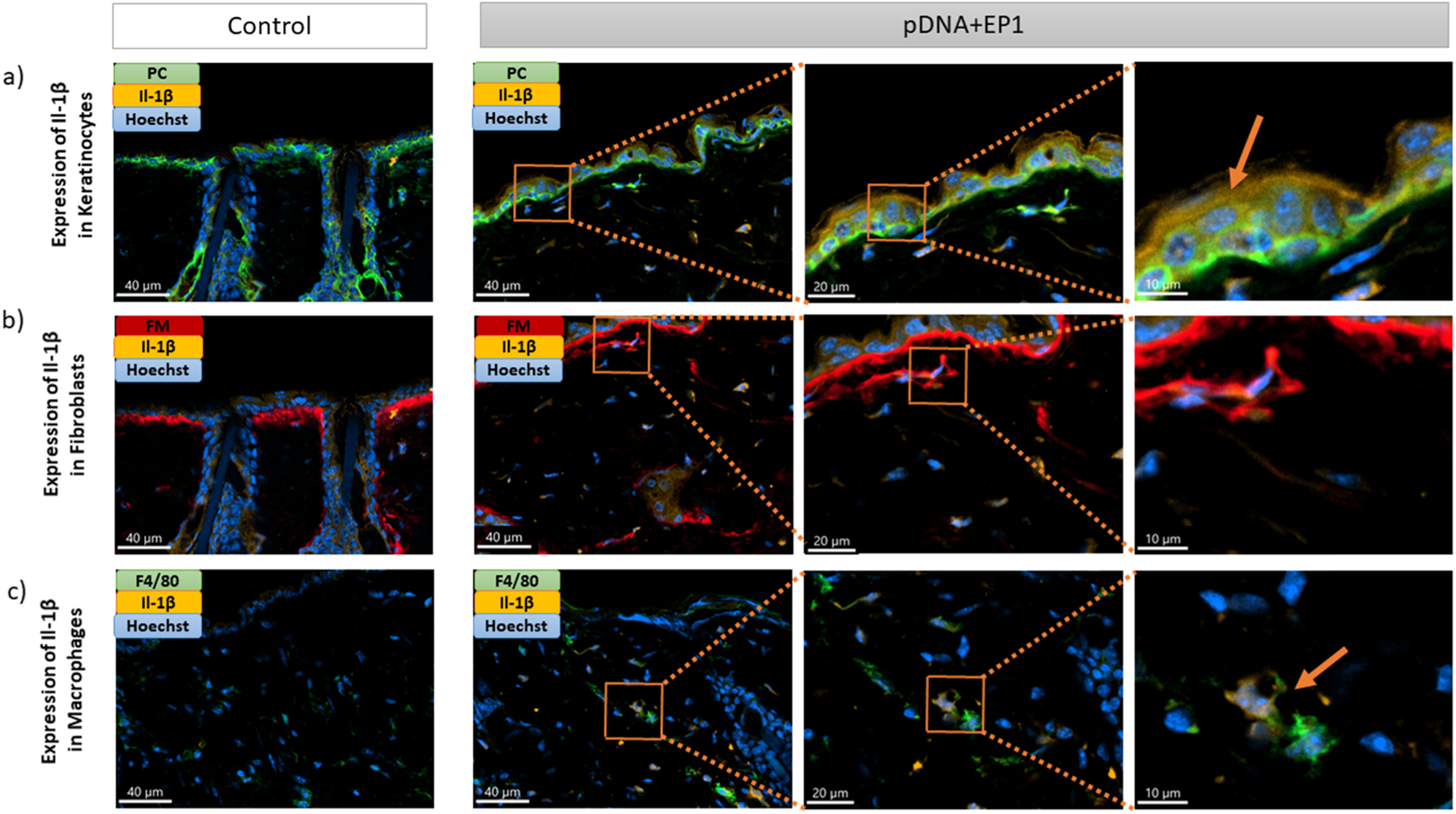
Expression of Il-1β (Il-1β -orange) in a) keratinocytes (Pan cytokeratin-green), b) fibroblasts (Fibroblast marker-red) and c) macrophages (F4/80-green), on frozen mouse skin Section 6 hours after pDNA electrotransfer. Three different images on the right side represent the zoom into the ROI. Arrows show detection of Il-1β in different cells. Scale bar: 40 μm for the control column and first column of pDNA+EP1 (right side), 20 μm for the second column of pDNA+EP1, 10 μm for the third column of pDNA+EP1.(For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
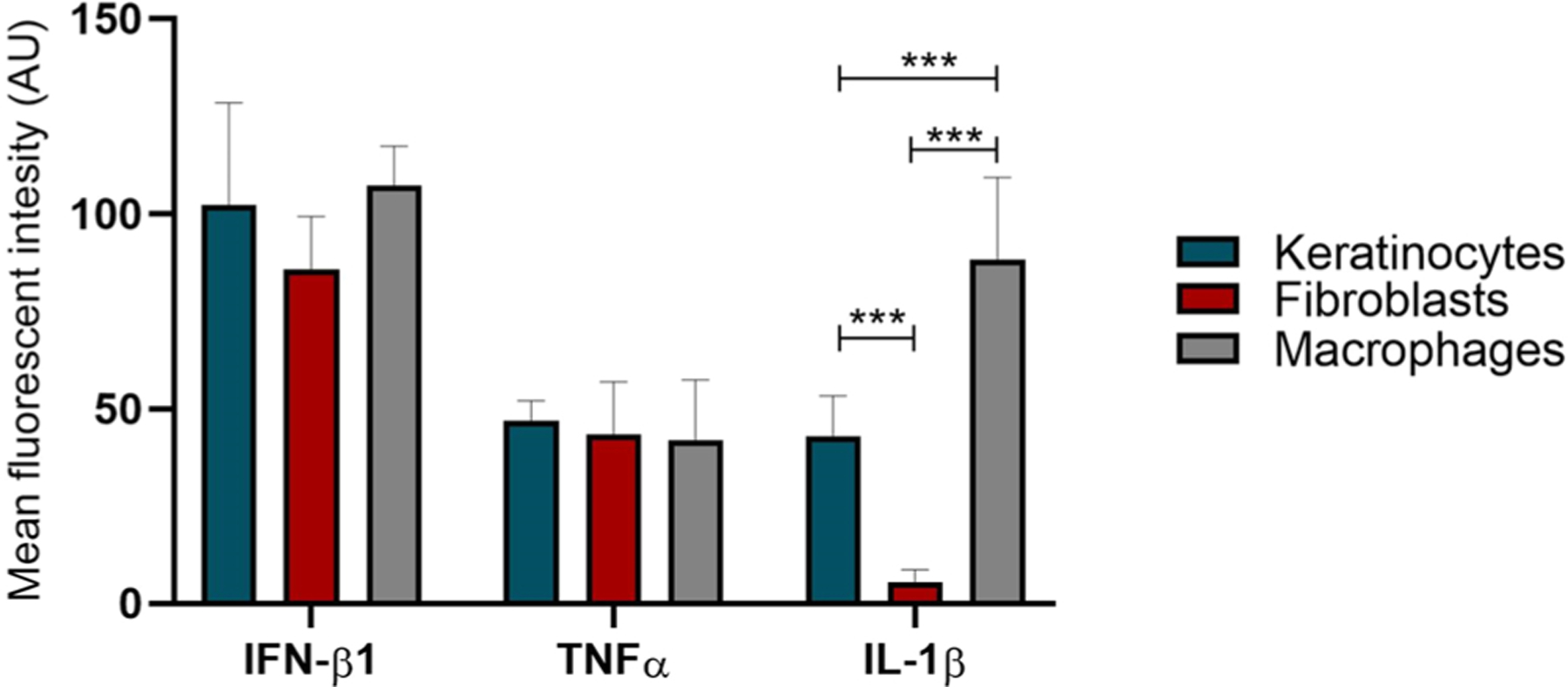
Quantification of cytokine induction in keratinocytes, fibroblasts and macrophages after electrotransfer of pDNA in mouse skin based on fluorescence intensity. * **p < 0.001 statistically significant difference between cell types.
4. Discussion
We previously demonstrated that transfection with exogenous nucleic acids induces the production of pro-inflammatory cytokines and chemokines and the upregulation of DNA sensor mRNAs and proteins in several cell types [22,23,43]. Using mouse myoblasts, we demonstrated that several DNA sensors physically bind pDNA after transfection [43]. We also observed antitumor effects in B16F10 melanoma tumors [44–46]. This study correlates the responses of skin cells in vitro to those of skin in vivo, an inherently immunogenic tissue, to pDNA electrotransfer. To the best of our knowledge, this is the first study evaluating this response in skin. We used two standard electrotransfer pulse protocols to deliver backbone plasmid DNA to keratinocytes and skin fibroblasts in vitro or to skin in vivo. We found that these cells responded to DNA transfection with DNA-dose dependent cell death in vitro, by the production of pro-inflammatory cytokines and chemokines and with the upregulation of DNA sensors mRNAs and protein in vitro and in vivo, implicating DNA sensor signaling.
In vitro, we first measured the transfection efficiency and cell death types. After delivery of a plasmid encoding EGFP, we found that keratinocytes and fibroblasts were transfected to similar levels with the EP1 pulse protocol but that keratinocytes were transfected to a higher level than fibroblasts with EP2. In each case, the median fluorescence, indicating the level of plasmid expression, was higher using the EP1 pulse protocol. Cell death, a potential outcome of DNA sensor activation, was significantly higher after EP1 pulse application alone and increased in a direct relationship with the plasmid concentration. We quantified relative levels of apoptosis and necrosis but did not detect pyroptosis [47]. Fibroblasts underwent higher levels of necrotic cell death than keratinocytes (p > 0.05). At least four DNA sensors, AIM2, cGAS, IFI204, and ZBP1, are associated with cell death pathways [48–50]. AIM2 initiates the inflammasome assembly, an innate immune complex that leads to the activation of inflammatory caspases leading to both apoptotic and pyroptotic cell death [26,51,52]. The presence and increased expression of Aim2 and Il-1β mRNAs implicated activation of the AIM2 signaling pathway in keratinocytes after electrotransfer of pDNA, but neither caspase 1 activity nor increased IL-1β protein levels were detected. The mRNAs for cGas and STING are expressed in both keratinocytes and fibroblasts. STING agonists can induce apoptosis, implicating cGAS activation, and cGAS-STING pathway activation can also induce necrosis [53]. We observed both apoptotic and necrotic cell death.
Keratinocytes constitute more than 90% of epidermal cells. Their primary function is defense against pathogens and the formation of the skin’s dead superficial layer (keratinized layer). These superficial keratinized cells continuously desquamate from the surface and are replaced by cells in the lowest layer (basal layer). Pathogens invading the upper layers of the epidermis can cause keratinocytes to produce pro-inflammatory mediators, particularly chemokines such as CXCL10 and CCL2, which attract monocytes, natural killer cells, T cells, and DCs to the site of pathogen invasion [54]. Due to the skin’s immunoprotective role, several studies of DNA sensing pathways in keratinocytes have been performed. Abundant cytosolic DNA and increased AIM2 expression in keratinocytes were detected in psoriatic lesions. In cultured keratinocytes, cytosolic DNA triggered the release of IL-1β via the AIM2 inflammasome [55]. Human papillomavirus genomic DNA in keratinocytes led to AIM2 inflammasome activation and IL-1β release [56]. The DNA sensors cGAS and IFI16 are required to fully activate the innate immune response to exogenous DNA and DNA viruses in human keratinocytes [57]. In our study, we found that the mRNAs of cytokines Ifn-β1, Tnfα, Il-1β, Il-6 and DNA sensors Ddx60, Zbp1, Aim2, Ifi202, Ifi204 were upregulated in keratinocytes after pDNA electrotransfer. Coordinate upregulation of the IFN-β and IL-6 proteins was confirmed.
Fibroblasts hold an essential role in wound healing and structural support and act as immune sentinels. These cells can produce inflammatory mediators including cytokines, chemokines, and growth factors, as well as act in response to infections [58,59]. The first cytosolic DNA sensor described, ZBP1, was discovered in fibroblasts [25]. Another study confirmed the activation of ZBP1 after infection with human cytomegalovirus [60]. In human dermal fibroblasts, the increased levels of IFI16 protein were associated with the induction of IFN-β [61]. In our study, the mRNAs of the cytokines Ifn-β1, Tnfα, Il-1β, Il-6, Il-18 and DNA sensors Ddx60, Zbp1 and Ifi204 were upregulated in fibroblasts after pDNA electrotransfer. Interestingly, while both transfection efficiency and IFN-β secretion were higher in keratinocytes, the DNA sensor mRNAs in fibroblasts were much more highly upregulated. Ifn-β1 mRNA upregulation and protein secretion by both fibroblasts and keratinocytes implicated the activation of DNA sensor signaling.
The skin contains multiple cell types, including non-immune cells such as fibroblasts and keratinocytes and immune cells such as Langerhans cells, which are likely to respond to DNA delivery. Cells in an in vivo environment may respond differently than cells in culture. In skin, the same as in vitro in both cell types, we observed upregulation of the mRNAs of the pro-inflammatory cytokines Ifn-β1, Tnfα, Il-1β and Il-6. This confirmed the observation by Roos, et al. that Il-1β mRNA was regulated in skin 24 h after pDNA electrotransfer [62]. However, protein regulation differed from the RNA regulation at the time points tested. TNFα protein was upregulated by any manipulation four hours after electrotransfer of pDNA. While many cytokine and chemokine proteins were upregulated, IL-1β was not, which confirmed the in vitro results.
The fold expression of Ifn-β1 after pDNA electrotransfer to skin increased more than 2000-fold, implicating the activation of several DNA sensors [25,33,63–65]. The mRNAs of the five DNA sensors upregulated in keratinocytes or fibroblasts, Ddx60, Zbp1, Aim2, Ifi202, and Ifi204, were upregulated in the skin. Sensing of endogenous nucleic acids by ZBP1 induces skin inflammation [50]. In an early study in pigs, pulse application increased the efficacy of DNA vaccines targeted to the skin [66]. These authors hypothesized that inflammatory cytokines induced by electroporation and subsequent cellular infiltration were responsible for this effect.
Many aspects of this study confirm previous studies performed to B16F10 melanoma tumors, specifically increased expression of the Il-1β, CCL3, CCL4, IL6, and IL10 [67] and IFN-β [68] proteins after pDNA electrotransfer. Similarly, Zbp1, Ddx0 and Ifi204 mRNAs were upregulated [68]. Our data also supported the early results of a study of DNA vaccine delivery by electroporation to skin [18]. In this study, we confirmed the increase in IL-15 protein observed in the dermis at day 3. The cytokine expression observed in our study after pulses alone tended to correlate with the findings of Todorova et al. in macaque skin biopsy supernatants where GM-CSF and TNFα levels were significantly higher in the electroporated skin than in non-electroporated skin [17].
To determine which specific skin cell type responded to pDNA electrotransfer, we performed immunofluorescent staining on frozen sections using different antibodies for keratinocytes, fibroblasts and macrophages. Cytoplasmic DNA activates a transcriptional response leading to the activation of nuclear factor-κB and interferon regulatory factor 3 and the subsequent production of pro-inflammatory cytokines and type I interferons [69]. Therefore, the response to pDNA electrotransfer was measured by staining for the co-association IFN-β1, TNFα, and IL-1β with specific cell types on frozen skin sections. These immune modulators are known to be expressed in skin fibroblasts, keratinocytes and macrophages after infection, skin injury, in response to ultraviolet B and in inflammatory conditions [70–74]. In our study, we also show the expression of these cytokines after pDNA electrotransfer. IFN-β1 was detected in fibroblasts, keratinocytes and macrophages in skin which correlated with our observed gene and protein expression in vitro and in vivo. TNFα was detected in all skin cell types tested, which also confirmed our detection of the mRNAs in vitro and the mRNAs and proteins in vivo. Il-1β was detected in keratinocytes and macrophages but not in skin fibroblasts after pDNA electrotransfer. This supported our observation that Il-1β mRNA levels increased in keratinocytes but not in fibroblasts. However, we did not detect Il-1β protein secretion by keratinocytes in vitro, which may have been due to the time point tested or to the sensitivity of the assay. We also demonstrated increased accumulation of macrophages after electrotransfer of pDNA in skin. The recruitment of macrophages after electrotransfer of pDNA was also detected in tumors. [75,76]. We previously demonstrated reporter gene expression in the epidermis and dermis after pDNA electrotransfer [77]. Our results confirmed this localization since we demonstrated a response in keratinocytes in the epidermis and in fibroblasts and macrophages in the dermis.
5. Conclusions
The skin constitutes a complex environment, with constant interactions of immune and stromal cell types which are capable of performing immune functions. Our research shows that pDNA electrotransfer drives signaling pathways that lead to the production of many cytokines and chemokines in keratinocytes and fibroblasts in vitro and in skin. Our results confirm the essential immune functions of both immune and non-immune cells in the skin which are essential for vaccination and gene therapy.
Supplementary Material
Acknowledgments
We would like to thank Mira Lavric and colleagues from the Cytopathology Department for their help with cell cultures, experiments in vivo and flow cytometry measurements (Institute of Oncology Ljubljana, Ljubljana, Slovenia). The graphical abstract was created with Bio-Render.com.
Funding
This work was supported in part by the National Cancer Institute of the National Institutes of Health under award number R01CA196796 and by the Slovenian Research Agency (programs no. P3–0003). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Footnotes
CRediT authorship contribution statement
Bošnjak Masa: Conceptualization, Methodology, Validation, Investigation, Writing – original draft, Writing – review & editing, Visualization, Znidar Katarina: Conceptualization, Methodology, Validation, Investigation, Writing – original draft, Writing – review & editing, Visualization. Sales Conniff Amanda: Visualization, Writing – review & editing, Jesenko Tanja: Methodology, Validation, Investigation, Writing – review & editing, Visualization, Markelc Bostjan: Methodology, Validation, Investigation, Writing – review & editing, Visualization, Semenova Nina: Methodology, Validation, Investigation, Formal analysys Tur Jared: Investigation, Writing – review & editing, Kohena Kris: Investigation, Kranjc Simona: Methodology, Investigation, Review & editing, Heller Loree: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition, Cemazar Maja: Conceptualization, Methodology, Resources, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2022.113088.
References
- [1].Lou D, Saltzman WM, Synthetic DNA delivery systems, Nat. Biotechnol 18 (2000) 33–37, 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- [2].Ramamoorth M, Narvekar A, Non viral vectors in gene therapy - An overview, J. Clin. Diagn. Res. 9 (2015) GE01–GE06, 10.7860/JCDR/2015/10443.5394. [DOI] [PMC free article] [PubMed]
- [3].Yarmush ML, Golberg A, Serša G, Kotnik T, Miklavčič D, Electroporation-based technologies for medicine: principles, applications, and challenges, Annu. Rev. Biomed. Eng 16 (2014) 295–320, 10.1146/annurev-bioeng-071813-104622. [DOI] [PubMed] [Google Scholar]
- [4].Heller R, Schultz J, Lucas ML, Jaroszeski MJ, Heller LC, Gilbert RA, Moelling K, Nicolau C, Intradermal delivery of interleukin-12 plasmid DNA by in vivo electroporation, DNA Cell Biol 20 (2001) 21–26, 10.1089/10445490150504666. [DOI] [PubMed] [Google Scholar]
- [5].Pavšelj N, Préat V, DNA electrotransfer into the skin using a combination of one high- and one low-voltage pulse, J. Control. Release 106 (2005) 407–415, 10.1016/j.jconrel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- [6].Medi BM, Singh J, Skin targeted DNA vaccine delivery using electroporation in rabbits II, Saf., Int. J. Pharm 308 (2006) 61–68, 10.1016/j.ijpharm.2005.10.035. [DOI] [PubMed] [Google Scholar]
- [7].Dobaño C, Widera G, Rabussay D, Doolan DL, Enhancement of antibody and cellular immune responses to malaria DNA vaccines by in vivo electroporation, Vaccine 25 (2007) 6635–6645, 10.1016/j.vaccine.2007.06.036. [DOI] [PubMed] [Google Scholar]
- [8].Roos A-K, Eriksson F, Timmons JA, Gerhardt J, Nyman U, Gudmundsdotter L, Bråve A, Wahren B, Pisa P, Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment, PLoS One 4 (2009), e7226, 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ferraro B, Cruz YL, Coppola D, Heller R, Intradermal delivery of plasmid VEGF165 by electroporation promotes wound healing, Mol. Ther 17 (2009) 651–657, 10.1038/mt.2009.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Steinstraesser L, Lam MC, Jacobsen F, Porporato PE, Chereddy KK, Becerikli M, Stricker I, Hancock RE, Lehnhardt M, Sonveaux P, Préat V, Vandermeulen G, Skin electroporation of a plasmid encoding hCAP-18/LL-37 host defense peptide promotes wound healing, Mol. Ther 22 (2014) 734–742, 10.1038/mt.2013.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Babiuk S, Baca-Estrada ME, Foldvari M, Baizer L, Stout R, Storms M, Rabussay D, Widera G, Babiuk L, Needle-free topical electroporation improves gene expression from plasmids administered in porcine skin, Mol. Ther 8 (2003) 992–998, 10.1016/j.ymthe.2003.09.008. [DOI] [PubMed] [Google Scholar]
- [12].Fioretti D, Iurescia S, Fazio VM, Rinaldi M, DNA vaccines: developing new strategies against cancer, J. Biomed. Biotechnol 2010 (2010), 10.1155/2010/174378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Maslow JN, Zika vaccine development—current progress and challenges for the future, Trop. Med. Infect. Dis 4 (2019) 104, 10.3390/tropicalmed4030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zhang N, Nandakumar KS, Recent advances in the development of vaccines for chronic inflammatory autoimmune diseases, Vaccine 36 (2018) 3208–3220, 10.1016/j.vaccine.2018.04.062. [DOI] [PubMed] [Google Scholar]
- [15].Bråve A, Nyström S, Roos A, Applequist SE, Plasmid DNA vaccination using skin electroporation promotes poly-functional CD4 T-cell responses, Immunol. Cell Biol 89 (2011) 492–496, 10.1038/icb.2010.109. [DOI] [PubMed] [Google Scholar]
- [16].Cashman KA, Wilkinson ER, Shaia CI, Facemire PR, Bell TM, Bearss JJ, Shamblin JD, Wollen SE, Broderick KE, Sardesai NY, Schmaljohn CS, A DNA vaccine delivered by dermal electroporation fully protects cynomolgus macaques against Lassa fever, Hum. Vaccin. Immunother 13 (2017) 2902–2911, 10.1080/21645515.2017.1356500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Todorova B, Adam L, Culina S, Boisgard R, Martinon F, Cosma A, Ustav M, Kortulewski T, Le Grand R, Chapon C, Electroporation as a vaccine delivery system and a natural adjuvant to intradermal administration of plasmid DNA in macaques, Sci. Rep 7 (2017) 1–11, 10.1038/s41598-017-04547-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Adam L, Tchitchek N, Todorova B, Rosenbaum P, Joly C, Poux C, Chapon C, Spetz A-L, Ustav M, Le Grand R, Martinon F, Innate molecular and cellular signature in the skin preceding long-lasting T cell responses after electroporated DNA vaccination, J. Immunol 204 (2020) 3375–3388, 10.4049/jimmunol.1900517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Qin F, Xia F, Chen H, Cui B, Feng Y, Zhang P, Chen J, Luo M, A guide to nucleic acid vaccines in the prevention and treatment of infectious diseases and cancers: from basic principles to current applications, Front. Cell Dev. Biol 9 (2021) 1–13, 10.3389/fcell.2021.633776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hobernik D, Bros M, DNA vaccines—how far from clinical use? Int. J. Mol. Sci 19 (2018) 3605, 10.3390/ijms19113605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Coban C, Kobiyama K, Jounai N, Tozuka M, Ishii KJ, DNA vaccines, Hum. Vaccin. Immunother 9 (2013) 2216–2221, 10.4161/hv.25893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Znidar K, Bosnjak M, Cemazar M, Heller LC, Cytosolic DNA sensor upregulation accompanies DNA electrotransfer in B16.F10 melanoma cells, Mol. Ther. Nucleic Acids 5 (2016), e322, 10.1038/mtna.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Znidar K, Bosnjak M, Semenova N, Pakhomova O, Heller L, Cemazar M, Tumor cell death after electrotransfer of plasmid DNA is associated with cytosolic DNA sensor upregulation, Oncotarget 9 (2018) 18665–18681, 10.18632/oncotarget.24816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S, A Toll-like receptor recognizes bacterial DNA, Nature 408 (2000) 740–745, 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- [25].Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T, DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response, Nature 448 (2007) 501–505, 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- [26].Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G, An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome, Nat. Immunol 10 (2009) 266–272, 10.1038/NI.1702. [DOI] [PubMed] [Google Scholar]
- [27].Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V, RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate, Nat. Immunol 10 (2009) 1065–1072, 10.1038/NI.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ, HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA, Science 323 (2009) 1057–1060, 10.1126/SCIENCE.1169841. [DOI] [PubMed] [Google Scholar]
- [29].Yan H, Dalal K, Hon BK, Youkharibache P, Lau D, Pio F, RPA nucleic acid-binding properties of IFI16-HIN200, Biochim. Biophys. Acta 2008. (1784) 1087–1097, 10.1016/J.BBAPAP.2008.04.004. [DOI] [PubMed] [Google Scholar]
- [30].Kim T, Pazhoor S, Bao M, Zhang Z, Hanabuchi S, Facchinetti V, Bover L, Plumas J, Chaperot L, Qin J, Liu YJ, Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase a helicases sense microbial DNA in human plasmacytoid dendritic cells, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 15181–15186, 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang P, An H, Liu X, Wen M, Zheng Y, Rui Y, Cao X, The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type i interferon via a В-catenin-dependent pathway, Nat. Immunol 11 (2010) 487–494, 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- [32].Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ, The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells, Nat. Immunol 12 (2011) 959–965, 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Miyashita M, Oshiumi H, Matsumoto M, Seya T, DDX60, a DEXD/H Box Helicase, Is a Novel Antiviral Factor Promoting RIG-I-Like Receptor-Mediated Signaling, Mol. Cell. Biol 31 (2011) 3802–3819, 10.1128/mcb.01368-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mimori T, Ohosone Y, Hama N, Suwa A, Akizuki M, Homma M, Griffith AJ, Hardin JA, Isolation and characterization of cDNA encoding the 80-kDa subunit protein of the human autoantigen Ku (p70/p80) recognized by autoantibodies from patients with scleroderma-polymyositis overlap syndrome, Proc. Natl. Acad. Sci. U. S. A 87 (1990) 1777–1781, 10.1073/pnas.87.5.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T, DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 2969–2974, 10.1073/PNAS.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sun L, Wu J, Du F, Chen X, Chen ZJ, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway, Science 339 (2013) 786–791, 10.1126/SCIENCE.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xia P, Wang S, Gao P, Gao G, Fan Z, DNA sensor cGAS-mediated immune recognition, Protein Cell 7 (2016) 777–791, 10.1007/s13238-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Heller LC, Jaroszeski MJ, Coppola D, McCray AN, Hickey J, Heller R, Optimization of cutaneous electrically mediated plasmid DNA delivery using novel electrode, Gene Ther 14 (2007) 275–280, 10.1038/SJ.GT.3302867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Livak KJ, Schmittgen TD, Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method, Methods 25 (2001) 402–408, 10.1006/METH.2001.1262. [DOI] [PubMed] [Google Scholar]
- [40].Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, Fiji - an Open Source platform for biological image analysis, Nat. Methods 9 (2012) 676–682, 10.1038/NMETH.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Shihan MH, Novo SG, Le Marchand SJ, Wang Y, Duncan MK, A simple method for quantitating confocal fluorescent images, Biochem. Biophys. Rep 25 (2021), 100916, 10.1016/j.bbrep.2021.100916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lugrin J, Martinon F, The AIM2 inflammasome: sensor of pathogens and cellular perturbations, Immunol. Rev 281 (2018) 99–114, 10.1111/imr.12618. [DOI] [PubMed] [Google Scholar]
- [43].Semenova N, Bosnjak M, Markelc B, Znidar K, Cemazar M, Heller L, Multiple cytosolic DNA sensors bind plasmid DNA after transfection, Nucleic Acids Res 47 (2019) 10235–10246, 10.1093/nar/gkz768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Heller L, Coppola D, Electrically mediated delivery of vector plasmid DNA elicits an antitumor effect, Gene Ther 9 (2002) 1321–1325, 10.1038/sj.gt.3301802. [DOI] [PubMed] [Google Scholar]
- [45].Heller L, Todorovic V, Cemazar M, Electrotransfer of single-stranded or double-stranded DNA induces complete regression of palpable B16.F10 mouse melanomas, Cancer Gene Ther 20 (2013) 695–700, 10.1038/cgt.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bosnjak M, Jesenko T, Kamensek U, Sersa G, Lavrencak J, Heller L, Cemazar M, Electrotransfer of different control plasmids elicits different antitumor effectiveness in B16.F10 melanoma, Cancers 10 (2018) 1–14, 10.3390/cancers10020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen X, He WT, Hu L, Li J, Fang Y, Wang X, Xu X, Wang Z, Huang K, Han J, Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis, Cell Res 26 (2016) 1007–1020, 10.1038/cr.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Vanpouille-Box C, Alard A, Aryankalayil MJ, Sarfraz Y, Diamond JM, Schneider RJ, Inghirami G, Coleman CN, Formenti SC, Demaria S, DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity, Nat. Commun 8 (2017) 15618, 10.1038/ncomms15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Paludan SR, Reinert LS, Hornung V, DNA-stimulated cell death: implications for host defence, inflammatory diseases and cancer, Nat. Rev. Immunol 19 (2019) 141–153, 10.1038/s41577-018-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Devos M, Tanghe G, Gilbert B, Dierick E, Verheirstraeten M, Nemegeer J, de Reuver R, Lefebvre S, de Munck J, Rehwinkel J, Vandenabeele P, Declercq W, Maelfait J, Sensing of endogenous nucleic acids by zbp1 induces keratinocyte necroptosis and skin inflammation, J. Exp. Med 217 (2020), 10.1084/jem.20191913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Dombrowski Y, Koglin S, Schauber J, DNA-triggered AIM2 inflammasome activation in keratinocytes: Comment on Kopfnagel et al. Exp. Dermatol 20 (2011) 1027–1029. Exp Dermatol. 21 (2012) 474–476. 〈 10.1111/j.1600-0625.2012.01466.x〉. [DOI] [PubMed] [Google Scholar]
- [52].Sagulenko V, Thygesen SJ, Sester DP, Idris A, Cridland JA, Vajjhala PR, Roberts TL, Schroder K, Vince JE, Hill JM, Silke J, Stacey KJ, AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC, Cell Death Differ 2013 209 (20) (2013) 1149–1160, 10.1038/cdd.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liu S, Guan W, STING signaling promotes apoptosis, necrosis, and cell death: an overview and update, Mediat. Inflamm 2018 (2018), 10.1155/2018/1202797. [DOI] [PMC free article] [PubMed]
- [54].Wang JN, Li M, The immune function of keratinocytes in anti-pathogen infection in the skin, Int. J. Dermatol. Venereol 3 (2020) 231–238, 10.1097/JD9.0000000000000094. [DOI] [Google Scholar]
- [55].Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Göß C, Anz D, Simanski M, Gläser R, Harder J, Hornung V, Gallo RL, Ruzicka T, Besch R, Schauber J, Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions, Sci. Transl. Med 3 (2011), 10.1126/scitranslmed.3002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Reinholz M, Kawakami Y, Salzer S, Kreuter A, Dombrowski Y, Koglin S, Kresse S, Ruzicka T, Schauber J, HPV16 activates the AIM2 inflammasome in keratinocytes, Arch. Dermatol. Res 305 (2013) 723–732, 10.1007/s00403-013-1375-0. [DOI] [PubMed] [Google Scholar]
- [57].Almine JF, O’Hare CAJ, Dunphy G, Haga IR, Naik RJ, Atrih A, Connolly DJ, Taylor J, Kelsall IR, Bowie AG, Beard PM, Unterholzner L, IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes, Nat. Commun 8 (2017) 1–15, 10.1038/ncomms14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jordana M, Sarnstrand B, Sime P, Ramis I, Immune-inflammatory functions of fibroblasts, Eur. Respir. J 7 (1994). [DOI] [PubMed] [Google Scholar]
- [59].Bautista-Hernández LA, Gómez-Olivares JL, Buentello-Volante B, Bautista-de Lucio VM, Fibroblasts: the unknown sentinels eliciting immune responses against microorganisms, Eur. J. Microbiol. Immunol 7 (2017) 151–157, 10.1556/1886.2017.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Früh K, Human cytomegalovirus induces the interferon response via the DNA Sensor ZBP1, J. Virol 84 (2010) 585–598, 10.1128/jvi.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Duan X, Ponomareva L, Veeranki S, Choubey D, IFI16 induction by glucose restriction in human fibroblasts contributes to autophagy through activation of the ATM/AMPK/p53 pathway, PLoS One 6 (2011), e19532, 10.1371/journal.pone.0019532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Roos AK, Eriksson F, Timmons JA, Gerhardt J, Nyman U, Gudmundsdotter L, Bråve A, Wahren B, Pisa P, Skin electroporation: effects on transgene expression, DNA persistence and local tissue environment, PLoS One 4 (2009), 10.1371/journal.pone.0007226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG, IFI16 is an innate immune sensor for intracellular DNA, Nat. Immunol 11 (2010) 997–1004, 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Keating SE, Baran M, Bowie AG, Cytosolic DNA sensors regulating type I interferon induction, Trends Immunol (2011), 10.1016/j.it.2011.08.004. [DOI] [PubMed]
- [65].Goubau D, van der Veen AG, Chakravarty P, Lin R, Rogers N, Rehwinkel J, Deddouche S, Rosewell I, Hiscott J, Reis e Sousa C, Mouse superkiller-2-like helicase DDX60 is dispensable for type I IFN induction and immunity to multiple viruses, Eur. J. Immunol 45 (2015) 3386–3403, 10.1002/eji.201545794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Babiuk S, Baca-Estrada ME, Foldvari M, Storms M, Rabussay D, Widera G, Babiuk LA, Electroporation improves the efficacy of DNA vaccines in large animals, Vaccine 20 (2002) 3399–3408, 10.1016/S0264-410X(02)00269-4. [DOI] [PubMed] [Google Scholar]
- [67].Heller LC, Cruz YL, Ferraro B, Yang H, Heller R, Plasmid injection and application of electric pulses alter endogenous mRNA and protein expression in B16.F10 mouse melanomas, Cancer Gene Ther 17 (2010) 864–871, 10.1038/CGT.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Znidar K, Bosnjak M, Cemazar M, Heller LC, Cytosolic DNA sensor upregulation accompanies DNA electrotransfer in B16.F10 melanoma cells, Mol. Ther. - Nucleic Acids 5 (2016), e322, 10.1038/mtna.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hornung V, Latz E, Intracellular DNA recognition, Nat. Rev. Immunol 10 (2010) 123–130, 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- [70].Zhang LJ, Type1 interferons potential initiating factors linking skin wounds with psoriasis pathogenesis, Front. Immunol 10 (2019) 1–8, 10.3389/fimmu.2019.01440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS, Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages, Cell Host Microbe 11 (2012) 469–480, 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kurane I, Janus J, Ennis FA, Dengue virus infection of human skin fibroblasts in vitro production of IFN-β, IL-6 and GM-CSF, Arch. Virol 124 (1992) 21–30, 10.1007/BF01314622. [DOI] [PubMed] [Google Scholar]
- [73].Bhatt DL, Fox KA, Hacke W, Berger PB, Black HR, Boden WE, Cacoub P, Cohen EA, Creager MA, Easton JD, Flather MD, Haffner SM, Hamm CW, Hankey GJ, Johnston SC, Mak KH, Mas JL, Montalescot G, Pearson TA, Steg PG, Steinhubl SR, Weber MA, Brennan DM, Fabry-Ribaudo L, Booth J, Topol EJ, C. Investigators, Clopidogrel and aspirin versus aspirin alone for the prevention of atherothrombotic events, N. Engl. J. Med 354 (2006) 1706–1717, 10.1056/NEJMoa060989. [DOI] [PubMed] [Google Scholar]
- [74].Cheung KT, Yuen Sze DM, Chan KH, Mei Leung PH, Involvement of caspase-4 in IL-1 beta production and pyroptosis in human macrophages during dengue virus infection, Immunobiology 223 (2018) 356–364, 10.1016/j.imbio.2017.10.044. [DOI] [PubMed] [Google Scholar]
- [75].Komel T, Bosnjak M, Kranjc Brezar S, De Robertis M, Mastrodonato M, Scillitani G, Pesole G, Signori E, Sersa G, Cemazar M, Gene electrotransfer of IL-2 and IL-12 plasmids effectively eradicated murine B16.F10 melanoma, Bioelectrochemistry 141 (2021), 107843, 10.1016/j.bioelechem.2021.107843. [DOI] [PubMed] [Google Scholar]
- [76].Lampreht Tratar U, Loiacono L, Cemazar M, Kamensek U, Fazio VM, Sersa G, Signori E, Gene electrotransfer of plasmid-encoding IL-12 Recruits the M1 macrophages and antigen-presenting cells inducing the eradication of aggressive B16F10 murine melanoma, Mediat. Inflamm 2017 (2017), 10.1155/2017/5285890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kos S, Blagus T, Cemazar M, Lampreht Tratar U, Stimac M, Prosen L, Dolinsek T, Kamensek U, Kranjc S, Steinstraesser L, Vandermeulen G, Préat V, Sersa G, Electrotransfer parameters as a tool for controlled and targeted geneexpression in skin, Mol. Ther. - Nucleic Acids 5 (2016), e356, 10.1038/mtna.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


