Abstract
A total of 35 homologs of the iron-sulfur flavoprotein (Isf) from Methanosarcina thermophila were identified in databases. All three domains were represented, and multiple homologs were present in several species. An unusually compact cysteine motif ligating the 4Fe-4S cluster in Isf is conserved in all of the homologs except two, in which either an aspartate or a histidine has replaced the second cysteine in the motif. A phylogenetic analysis of Isf homologs identified four subgroups, two of which were supported by bootstrap data. Three homologs from metabolically and phylogenetically diverse species in the Bacteria and Archaea domains (Af3 from Archaeoglobus fulgidus, Cd1 from Clostridium difficile, and Mj2 from Methanococcus jannaschii) were overproduced in Escherichia coli. Each homolog purified as a homodimer, and the UV-visible absorption spectra were nearly identical to that of Isf. After reconstitution with iron, sulfide, and flavin mononucleotide (FMN) the homologs contained six to eight nonheme iron atoms and 1.6 to 1.7 FMN molecules per dimer, suggesting that two 4Fe-4S or 3Fe-4S clusters and two FMN cofactors were bound to each dimer, which is consistent with Isf data. Homologs Af3 and Mj2 were reduced by CO in reactions catalyzed by cell extract of acetate-grown M. thermophila, but Cd1 was not. Homologs Af3 and Mj2 were reduced by CO in reactions catalyzed by A. fulgidus and M. jannaschii cell extracts. Cell extract of Clostridium thermoaceticum catalyzed CO reduction of Cd1. Our database sequence analyses and biochemical characterizations indicate that Isf is the prototype of a family of iron-sulfur flavoproteins that occur in members of all three domains.
Methane produced by the acetate fermentation pathway accounts for two-thirds of all biologically produced methane. In Methanosarcina thermophila, acetate is cleaved into carbonyl and methyl groups by the CO dehydrogenase (CODH)–acetyl coenzyme A (acetyl-CoA) synthase (ACS) enzyme complex. The methyl group is subsequently reduced to methane by electrons derived from oxidation of the carbonyl group to CO2 by the CODH-ACS complex (7). An iron-sulfur flavoprotein (Isf) from M. thermophila has been overproduced in Escherichia coli, purified, and characterized. Isf is purified as a homodimer which binds two 4Fe-4S clusters and two flavin mononucleotide (FMN) cofactors (2, 14). Ferredoxin, the electron acceptor of the CODH-ACS complex, is the physiological electron donor for Isf. Additional results support the hypothesis that Isf plays a role in electron transport during fermentation of acetate to methane (14). The deduced amino acid sequence of Isf contains an unusually compact cysteine motif (CX2CX2CX5C) that has been shown, on the basis of site-directed mutagenesis and electron paramagnetic resonance studies, to ligate the 4Fe-4S cluster (15). The midpoint potential values are -394 mV for the 4Fe-4S cluster and -277 mV for the FMN in Isf, suggesting that electrons derived from oxidation of the carbonyl group of acetate flow from ferredoxin to the 4Fe-4S cluster of Isf and then to the FMN (2). The downstream electron acceptor of Isf has not been identified yet.
When Isf was initially discovered, no sequences which exhibited significant overall identity were found in the databases (14). Subsequently, homologs were identified in the genomes of methanogenic and nonmethanogenic members of the Archaea domain (2) and several organisms belonging to the Bacteria domain (15). Here we describe an analysis of these homologs and 24 additional homologs and also describe biochemical characterization of three homologs from diverse species. Our findings indicate that Isf is the prototype of a family of iron-sulfur flavoproteins that are more widely distributed in nature than previously recognized.
MATERIALS AND METHODS
Materials.
Archaeoglobus fulgidus genomic DNA was a gift from Michael Adams, University of Georgia. A. fulgidus cells grown on lactate were a gift from Robert Kelly, North Carolina State University. Clostridium difficile genomic DNA was a gift from TechLab, Inc., Blacksburg, Va. Clostridium thermoaceticum cells grown on glucose and CO2 as previously described (1) was a gift from Steve Ragsdale, University of Nebraska. Methanococcus jannaschii clone AMJAI79 was purchased from the American Type Culture Collection. Autotrophically grown cells of M. jannaschii JAL-1 were a gift from Biswarup Mukhopadhyay, University of Illinois. Acetate-grown M. thermophila cells were a gift from Birthe Borup. pSJS1240 was a gift from Rosalind Kim, University of California, Berkeley. Restriction endonucleases, T4 DNA ligase, and Vent DNA polymerase were purchased from New England Biolabs, Inc. Oligonucleotide primers were synthesized at Integrated DNA Technologies, Inc. All E. coli strains were obtained from Novagen, Inc. Phenyl Sepharose high-performance matrix, HiTrapQ, HiTrap SP, HiTrap Desalting, PD-10, Superose-12, and Mono-Q columns were obtained from Pharmacia. Amicon stirred cell 8200 and YM-10 membranes were obtained from Millipore. Tryptone and yeast extract were obtained from Difco. All other chemicals were obtained from Sigma.
Sequence analysis.
BLAST searches were performed at http://www.ncbi.nlm.nih.gov and http://www.jgi.doe.gov/JGI_microbial/html/index.html. BLAST searches were also performed with preliminary genome data for Methanosarcina mazei from the Göttingen Genomic Laboratory at http://www.g21.bio.uni-goettingen.de/. Sequences were aligned by using Clustal X, version 1.64b. A phylogenetic analysis of protein sequences was performed with the MEGA program (13), using a neighbor-joining algorithm with gamma distance estimation (α = 2). A phylogenetic tree was constructed based on pairwise estimates of the expected number of amino acid replacements per site. A total of 1,500 bootstrap cycles were performed, and the values reported below are percentages of times that proteins grouped together on a branch.
Amplification, cloning, and expression of genes encoding Af3, Cd1, and Mj2.
The genes encoding Af3 and Cd1 were amplified by performing PCR with A. fulgidus and C. difficile genomic DNAs, respectively. The gene encoding Mj2 was amplified by performing PCR with M. jannaschii clone AMJAI79 DNA. PCR primers were designed to introduce an NdeI site at the start codon and a BamHI site (or an EcoRI site for the gene encoding Cd1) downstream of the stop codon of each gene. The sequences of the primers used are as follows: (restriction sites are underlined): 5′-CGCATCGCATTCCATATGAAACTGCTGGCAATA-3′ (5′ primer) and 5′-GGTGAAGGATCCGGTGGTTTAGGACTCTC-3′ (3′ primer) for the gene encoding Af3, 5′-CATCGACCATATGGCAAAGATATTAGGT-3′ (5′ primer) and 5′-CCATCGAGAATTCGCAATAATCTATTACATCC-3′ (3′ primer) for the gene encoding Cd1, and 5′-CCCCCCGGGTTCCATATGAAAGTTATAGGGATAAGTGG-3′ (5′ primer) and 5′-CCGTAGGATCCAAAGAGAAGGTAAAAA-3′ (3′ primer) for the gene encoding Mj2. The PCR products were purified from agarose gels and digested with NdeI and BamHI (or NdeI and EcoRI for the gene encoding Cd1). The resulting DNA fragments were cloned into the NdeI and BamHI sites (or NdeI and EcoRI sites for the gene encoding Cd1) of pT7-7 to obtain recombinant plasmids pTZ993 (the gene encoding Af3), pFC991 (the gene encoding Cd1), and pTZ983 (the gene encoding Mj2). To confirm the gene sequences in the plasmids, DNA sequencing was done by the automated dideoxy method at the nucleic acid facility of the Pennsylvania State University Biotechnology Institute. To express the genes encoding Cd1 and Mj2, pFC991 and pTZ982, respectively, were used to transform E. coli BL21(DE3), and the transformed E. coli cells were grown at 37°C in Luria-Bertani broth containing 100 mg of ampicillin per liter. To express the gene encoding Af3, both pTZ993 and pSJS1240 (a plasmid expressing some rare E. coli tRNAs with spectinomycin resistance) were transformed into E. coli BL21(DE3), and the transformed E. coli cells were grown at 37°C in Luria-Bertani broth containing 100 mg of ampicillin per liter and 50 mg of spectinomycin per liter. The transformed E. coli cells were grown to an absorbance at 600 nm (A600) of 0.6 to 0.8; then the cultures were cooled (for approximately 10 min) to approximately 25°C and induced by adding 1% (wt/vol) (final concentration) lactose. Ferric ammonium citrate (1 g/liter) was also added at the same time to supply iron to the cultures. After 6 h of growth at 25°C, the cells were harvested by centrifugation at 5,322 × g for 10 min at 4°C. Cell pellets were stored at −80°C.
Purification of Af3, Cd1, and Mj2 overproduced in E. coli.
All steps in the purification procedure were performed anaerobically. Where appropriate, a COY anaerobic chamber (Coy Laboratory Products, Ann Arbor, Mich.) was employed. All steps were performed at 21°C except as indicated below. Cells (approximately 15 g [wet weight]) were resuspended in 90 ml of buffer (50 mM HEPES [pH 7.0] for Af3, 50 mM morpholineethanesulfonic acid [MES] [pH 6.2] for Cd1, and 50 mM Tris [pH 7.6] for Mj2) and lysed by two passages through a French press at 20,000 lb/in2 (138 MPa). The lysates were centrifuged at 74,400 × g for 20 min at 4°C. For Af3 and Mj2, the supernatants were heated at 65°C for 30 min anaerobically. For Cd1, streptomycin sulfate (final concentration, 1% [wt/vol]) was added to the supernatant. The treated supernatants were centrifuged as described above. The supernatants resulting from the second centrifugation were filtered (pore size, 0.45 μm) and loaded on 5-ml HiTrap columns (HiTrap SP for Af3 and Cd1 and HiTrap Q for Mj2). A 50-ml linear 0 to 1.0 M NaCl gradient was applied at a rate of 5 ml/min. Fractions containing the Isf homologs (as determined by a yellow-brown color and sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] [see Fig. 4]) were pooled. The pooled fractions were then adjusted so that the (NH4)2SO4 concentration in the resuspension buffer was 0.6 M by addition of 3.0 M (NH4)2SO4. The solutions were then passed over a custom-made 9-ml phenyl Sepharose high-performance column that had been preequilibrated with 0.6 M (NH4)2SO4 in the resuspension buffer. The flowthrough fractions containing Isf homologs were diluted 1:10 with 50 mM Tris (pH 7.6 for Mj2 and pH 8.6 for Af3 and Cd1), filtered (pore size, 0.2 μm), and loaded onto a Mono-Q 10/10 column that had been preequilibrated with the dilution buffer. A 50-ml 0 to 1.0 M NaCl gradient was applied at a rate of 2 ml/min. The fractions containing the desired proteins were pooled, and their purity was analyzed by SDS-PAGE and Coomassie blue staining.
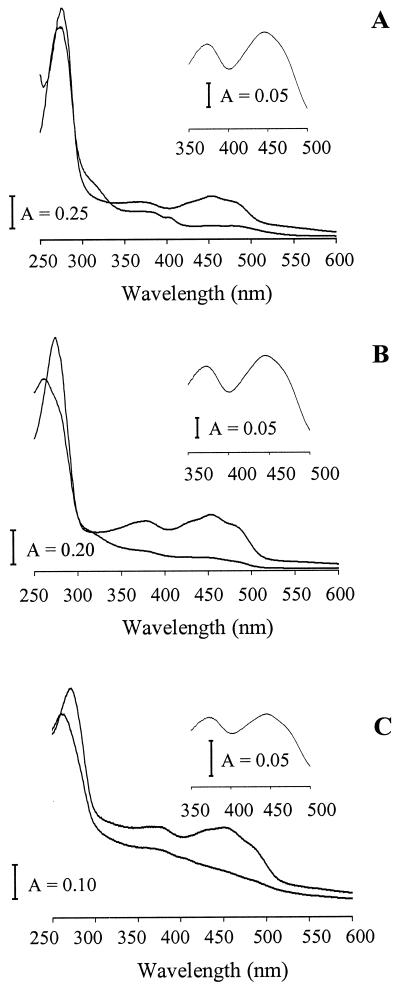
FIG. 4.
UV-visible spectra of reconstituted homologs Af3 (A), Cd1 (B), and Mj2 (C). In each panel, the upper and lower traces are the traces obtained before and after reduction by dithionite. The insets show the spectra obtained after mild acidification. The proteins were in 50 mM Tris-Cl (pH 7.6 for Mj2 and pH 8.6 for Af3 and Cd1). The spectra were recorded at room temperature.
Reconstitution of Af3, Cd1, and Mj2.
The reconstitution method used was a modified version of the method described previously (16). To 100 ml of anaerobic 50 mM Tris buffer (pH 7.6 for Mj2 and pH 8.6 for Af3 and Cd1), 800 μl of β-mercaptoethanol was added dropwise while the preparation was gently stirred. After 10 min, 2.5 to 10 mg of pure protein (eluted from the Mono-Q column) was added. Then 300 μl of 60 mM ferric chloride, 300 μl of 60 mM sodium sulfide, and 300 μl of 15 mM FMN (all in 50 mM Tris) were added stepwise at 10-min intervals. The mixtures were incubated anaerobically at 4°C overnight and then concentrated by using an ultrafiltration apparatus (Amicon stirred cell 8200 and YM-10 membrane) under an N2 atmosphere at 4°C. The excess Fe, S, and FMN were removed by several ultrafiltration cycles and/or by passage through a desalting column.
Analyses.
UV-visible absorption spectra and absorbance were determined with a Beckman DU 600 spectrophotometer. The molecular masses of Af3, Cd1, and Mj2 subunits were estimated by SDS–15% PAGE, using a low-molecular-weight protein standard obtained from Sigma (14). The molecular masses of the native proteins were based on elution from a Superose-12 gel filtration column as described previously (14). The following proteins with known molecular masses were used as standards: bovine serum albumin (66 kDa), ovalbumin (45 kDa), carbonic anhydrase (29 kDa), and chymotrypsinogen (25 kDa) or horse heart cytochrome c (12.4 kDa). The flavins were extracted from the purified Af3, Cd1, and Mj2 (without reconstitution) by adding trichloroacetic acid (final concentration, 5% [vol/vol]) to the protein solutions. The precipitated proteins were removed by centrifugation at 4°C and 13,500 × g for 15 min. The supernatants containing the flavins were adjusted to pH 6.0 with ammonium acetate, and the flavins were identified as FMN by high-performance liquid chromatography as described previously (14). The quantity of extracted FMN was determined by measuring the A450 and using a standard curve generated from pure FMN. The iron content was determined as described previously (14). The biuret method was used to determine the protein concentration; bovine serum albumin was used as the standard. N-terminal sequences were determined with a model 477A protein sequencer (PE Biosystems, Inc., Foster City, Calif.) at the Macro Core Facility of the Pennsylvania State University Hershey Medical Center.
Reduction of the homologs and enzyme activity assays.
Cell extracts were made anaerobically as follows. Cells were resuspended in 50 mM Tris (pH 7.6) and then lysed by one passage through a French press at 20,000 lb/in2. The lysates were centrifuged at 30,000 to 40,000 × g for 30 min at 4°C. The supernatants (cell extracts) were divided into aliquots and stored in liquid nitrogen. The reduction experiments were performed at 21°C. In each reduction experiment, 180 μg of cell extract protein (or 250 μg of protein in the case of the M. jannaschii cell extract) in 50 mM Tris (pH 7.6) was equilibrated at 21°C for 10 min in a stoppered 1.0-ml cuvette. Each reduction reaction was initiated by adding 0.2 mg of Isf or an Isf homolog protein (or 0.125 mg of protein for reduction of Mj2 by M. jannaschii cell extract). The total volume of the reduction mixture was 0.5 ml. The UV-visible spectra during reduction of the proteins were recorded every 20 s with a Hewlett-Packard 8452A diode array spectrophotometer. The A480 was used to monitor the amount of protein reduced after the absorbance was corrected for turbidity. Assays to determine the CODH activities of the cell extracts were conducted at 21°C and pH 7.6 by using a Hewlett-Packard 8452A diode array spectrophotometer as described previously (22). Hydrogenase assays were conducted in the same way as the CODH assays, except that CO was replaced by H2.
RESULTS
Analysis of sequences from diverse prokaryotes with identity to Isf.
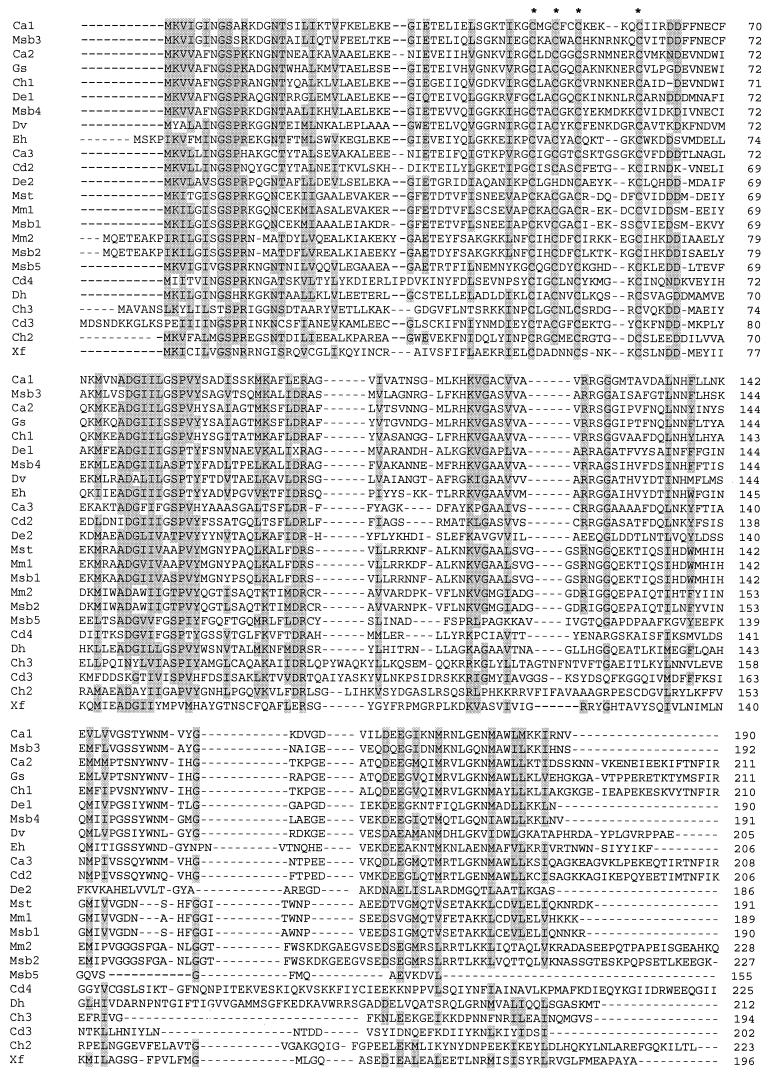
Previous database searches identified 11 homologs of Isf (2, 15). A recent search (15 July 2001) of all available databases with BLASTP and TBLASTN, which included finished and unfinished genomes, was conducted with the entire 191-amino-acid sequence of Isf as the query. Less biased BLAST searches with the Isf sequence minus the conserved region containing the cysteine motif (residues 41 to 61) yielded the same results. The searches identified 24 additional homologs (Fig. 1) that exhibited at least 20% overall identity to Isf and also contained the unique cysteine motif (CX2CX2CX4–7C) previously shown to ligate the 4Fe-4S cluster in Isf (15). This motif is conserved in all of the homologs except De3 and Xf, in which either an aspartate or a histidine has replaced the second cysteine in the motif. For all of the homologs the number of residues (155 to 228 residues) and the predicted subunit molecular masses (16.9 to 25.4 kDa) are within narrow ranges. Inspection of the predicted protein sequence immediately upstream of the annotated start of Mbt3 (21) revealed 16 residues with high levels of identity to the conserved N-terminal residues in all other homologs shown in Fig. 1, suggesting that the annotated start of Mbt3 was not correct. In general, the C-terminal halves of the homologs are less similar than the N-terminal halves. The homologs shown in Fig. 1 and those described previously are homologs from physiologically and phylogenetically distinct microbes representing all three domains. The species represented include methane-producing and sulfate-reducing members of the Archaea domain. The Bacteria domain is represented by gram-positive, green nonsulfur, and purple nonsulfur species. All of the species belonging to the Bacteria and Archaea domains are anaerobes except for Xylella fastidiosa. The only Isf homolog identified in a member of the Eucarya domain is a homolog from the intestinal anaerobic pathogen Entamoeba histolytica. The previously described homologs Mj1-2 and Af1-3 are from the hyperthermophiles M. jannaschii and A. fulgidus, which have optimum growth temperatures of 85 and 83°C, respectively (10). Multiple Isf homologs were found in many species (Fig. 1 and 2).
FIG. 1.
Amino acid sequence alignment for 24 Isf homologs from diverse microorganisms not previously reported (15). The sequence of M. thermophila Isf is included for reference. The sequences included exhibit more than 20% identity with the M. thermophila Isf sequence (191 residues) and bear the conserved cysteine motif. Abbreviations: Ca1, Ca2, and Ca3, ORFs from a C. acetobutylicum unfinished fragment of the complete genome; Cd2, Cd3, and Cd4, ORFs from C. difficile unfinished fragments of the complete genome; Ch1, Ch2, and Ch3, ORFs from Carboxydothermus hydrogenoformans unfinished fragments of the complete genome; De1 and De2, ORFs from two Dehalococcoides ethenogenes unfinished fragments of the complete genome; Dh, ORF from preliminary sequence data for the genome of Desulfitobacterium hafniense (http://www.jgi.doe.gov/JGI_microbial/html/index.html); Dv, ORF from a Desulfovibrio vulgaris unfinished fragment of the complete genome; Eh, ORF from E. histolytica unfinished fragment of the complete genome; Gs, ORF from a Geobacter sulfurreducens unfinished fragment of the complete genome; Mm1 and Mm2, ORFs from preliminary sequence data for the genome of M. mazei provided by the Göttingen Genomic Laboratory (http://www.g21.bio.uni-goettingen.de/); Msb1, Msb2, Msb3, Msb4, and Msb5, ORFs from preliminary sequence data for the genome of Methanosarcina barkeri: Mst, M. thermophila iron-sulfur flavoprotein (GenBank accession no. U50189); and Xf, X. fastidiosa ORF XF1919 (GenBank accession no. AE004011). Residues that are conserved in at least 7 of 10 sequences are shaded. The cysteines in the cysteine motif ligating the 4Fe-4S cluster are indicated by asterisks.
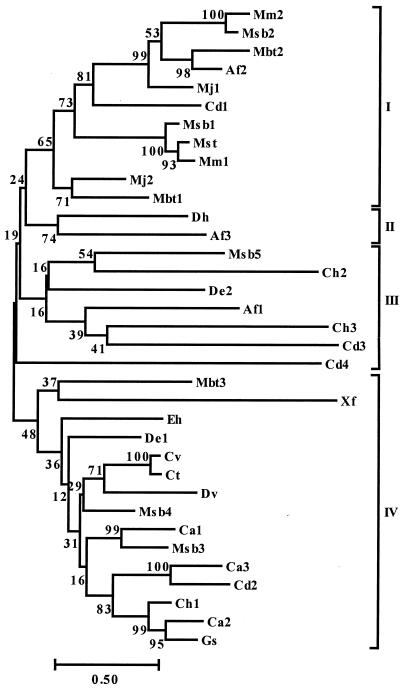
FIG. 2.
Phylogenetic tree for 35 Isf homologs. The following homologs were described previously (15) and are not included in Fig. 1: Af1, Af2, and Af3, A. fulgidus ORFs AF1436, AF1519, and AF1896 (GenBank accession no. AE001004, AE000997, and AE000972), respectively; Cd1, ORF from a C. difficile unfinished fragment of the complete genome; Ct, ORF from a Chlorobium tepidum unfinished fragment of the complete genome; Cv, ORF (EMBL accession no. Z83933) in a 15-kb genomic DNA fragment of Chlorobium vibrioforme; Mbt1, Mbt2, and Mbt3, M. thermoautotrophicum ORFs MTH135, MTH1473, and MTH1595 (GenBank accession no. AE000908 and AE000919), respectively; and Mj1 and Mj2, M. jannaschii ORFs MJ0731 and MJ1083 (GenBank accession no. C64391 and B64435), respectively. The tree was constructed based on pairwise distance estimates of the expected number of amino acid replacements per site. Bar = 0.50 amino acid replacement per site. The bootstrap values are indicated to the left of the branches. The four subgroups (subgroups I to IV) are indicated on the right.
A neighbor-joining phylogenetic tree for the previously described homologs and the homologs in Fig. 1 is shown in Fig. 2. Although the number of homologs is relatively small, there appear to be four subgroups. Bootstrap data support the grouping of subgroups I and II. Closely related sequences are present in physiologically and phylogenetically diverse species, and several strains have homologs belonging to different subgroups, suggesting that they have different functions in the cell. Subgroup I contains homologs that are mostly from the Archaea domain, and subgroups III and IV contain mainly homologs from the Bacteria domain.
Previously described Isf homologs accounted for the first nine pairwise alignments with the lowest E values produced by the BLASTP search. These homologs were immediately followed by three FMN-binding tryptophan repressor binding proteins, the Isf homolog Xf, two additional tryptophan repressor binding proteins, and then a flavodoxin from Clostridium acetobutylicum exhibiting 28% identity with Isf residues 42 to 154. The identity of Isf and the flavodoxin was reported previously (14). No other alignments with flavodoxins were obtained up to an E value of 8.2. The search also resulted in pairwise alignments with the FMN-containing enzymes 1,4-benzoquinone reductase (E = 0.002) and NADH dehydrogenase (E = 0.70).
Biochemical characterization of Af3, Cd1, and Mj2.
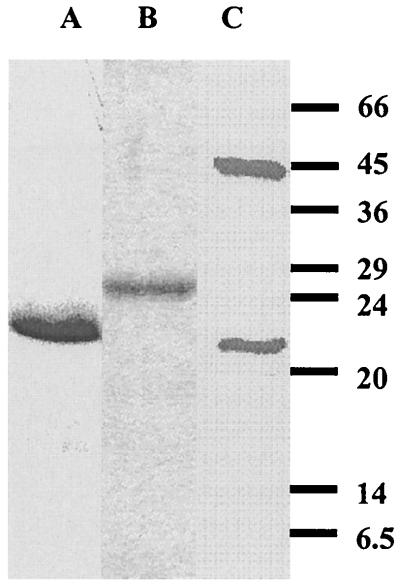
Isf homologs from physiologically and phylogenetically diverse species in the Archaea and Bacteria domains were selected for production in E. coli to determine if they have biochemical properties characteristic of Isf. The homologs chosen were Af3 from the sulfate-reducing archaeon A. fulgidus, Mj2 from the CO2-reducing hyperthermophilic methanoarchaeon M. jannaschii, and Cd1 from C. difficile, which is an intestinal pathogen classified in the Bacteria domain. Cd1 is also one of the sequences in Fig. 1 with the largest number of residues. Sequencing of the expression plasmids containing the genes encoding Af3, Cd1, and Mj2 confirmed that the correct sequences were present. Af3 was produced at low levels in E. coli; however, when plasmid pSJS1240 was cotransformed with the expression plasmid, the level of production of Af3 increased more than 10-fold (data not shown). Plasmid pSJS1240 harbors and expresses argU and ileX, genes that encode the rare E. coli tRNAarg (AGA/AGG) and tRNAile (AUA) codons. All 11 arginines and 5 of 16 isoleucines in Af3 are encoded by the rare codons. Heating E. coli cell extracts containing Af3 or Mj2 precipitated most E. coli proteins, whereas the recombinant proteins from the hyperthermophiles remained soluble, which facilitated purification of Af3 and Mj2. The proteins produced in E. coli were purified to apparent homogeneity, as determined by SDS-PAGE (Fig. 3). Two bands, at 46 and 20 kDa, were observed for Mj2. N-terminal analysis revealed that the first five residues of both bands were the same as the residues deduced from the DNA sequence, suggesting that the two bands correspond to the dimer and monomer forms of Mj2. Without boiling, the amount of the 46-kDa band increased significantly, whereas the amount of the 20-kDa band decreased (data not shown), findings which further support the identity of the bands as dimer and monomer forms of Mj2. The dimer band persisted even when Mj2 was boiled for 20 min in the presence of dithiothreitol, which indicated that Mj2 dimerizes strongly at high temperatures, as would be expected for the hyperthermophilic organism M. jannaschii. N-terminal sequencing confirmed the identities of Af3 and Cd1 produced in E. coli. The experimental subunit molecular masses agreed with the predicted values deduced from the gene sequences (Table 1). The experimental native molecular masses suggest that each Isf homolog was purified as an α2 dimer, although each value was slightly higher than the calculated value (Table 1). Each of the proteins purified from E. coli contained FMN, as shown by high-performance liquid chromatography analysis of the extracted flavins (data not shown). The UV-visible spectra of the proteins purified from E. coli were similar to those of Isf (data not shown), suggesting that iron-sulfur centers were present in addition to FMN. However, the intensity of absorbance centered at 430 nm varied among preparations, indicating that there was incomplete incorporation of the iron-sulfur center. Prior to quantification of iron and FMN, each purified protein was reconstituted with iron, sulfur, and FMN by a procedure similar to that previously described for apo-Isf (15). The reconstituted proteins had UV-visible spectra nearly identical to that of Isf, with absorbance maxima at approximately 484, 452, 430, 378, and 280 nm (Fig. 4). Exposure to air did not alter the spectra, indicating that the reconstituted proteins were in their oxidized states (data not shown). Mild acidification of the reconstituted proteins to destroy the iron-sulfur clusters produced visible spectra with absorbance maxima at approximately 445, 374, and 269 nm typical of flavins (Fig. 4, insets). Analyses of the reconstituted proteins showed that 1.6 to 1.7 mol of FMN and 5.6 to 7.5 mol of nonheme iron were bound to 1 mol of dimer (Table 1), indicating that two FMN molecules and two 4Fe-4S or 3Fe-4S clusters were bound to each dimer. Addition of dithionite decreased the absorbance at 350 to 550 nm (Fig. 4), whereas exposure to air resulted in recovery of the initial spectra, which indicated that Af3, Cd1, and Mj2 are redox-active proteins. In summary, Af3, Cd1, and Mj2 have the distinguishing biochemical characteristics of Isf.
FIG. 3.
SDS-PAGE of purified homologs Af3 (lane A), Cd1 (lane B), and Mj2 (lane C). The positions of molecular mass markers (in kilodaltons) are indicated on the right.
TABLE 1.
Properties of Isf homologs
| Homolog | Predicted isoelectric point | Predicted molecular mass (kDa) | Subunit molecular mass (kDa) | Native molecular mass (kDa) | Amt of FMN (mol/mol of dimers)a | Amt of iron (mol/mol of dimers)a |
|---|---|---|---|---|---|---|
| Af3 | 8.9 | 22 | 21 | 51 | 1.6 ± 0.1 | 6.6 ± 0.2 |
| Cd1 | 6.9 | 24 | 26 | 58 | 1.7 ± 0.1 | 7.5 ± 0.1 |
| Mj2 | 6.3 | 21 | 20 | 51 | 1.6 ± 0.1 | 5.6 ± 02 |
Means ± standard deviations based on three measurements.
Reconstituted Af3 and Mj2 were reduced by CO in reactions catalyzed by cell extract of acetate-grown M. thermophila, but Cd1 was not (Table 2). Reconstituted Af3 and Mj2 were reduced by CO in reactions catalyzed by A. fulgidus and M. jannaschii cell extracts, respectively (Table 3). C. difficile cell extract was not available; however, C. thermoaceticum cell extract catalyzed CO reduction of Cd1 (Table 3). When H2 replaced CO as the electron donor, only Isf and Mj2 showed significant reduction (Table 3). Most cell extracts had significant CODH and hydrogenase activities (Table 4); the only exception was A. fulgidus cell extract, which had no detectable hydrogenase activity, which is consistent with previous reports (25).
TABLE 2.
Rates of CO-dependent reduction of Isf and Isf homologs catalyzed by M. thermophila cell extract
| Protein | Rate of reduction (nmol/min/mg of cell extract protein)a | Relative rate (%)b |
|---|---|---|
| Isf | 20 ± 2 (0.37 ± 0.03) | 100 |
| Af3 | 7 ± 1 (0.1 ± 0.02) | 35 |
| Cd1 | NDc | NAd |
| Mj2 | 9.7 ± 0.8 (0.12 ± 0.01) | 49 |
Each rate was determined from the slope of the linear portion of the time course. The values in parentheses are the changes in A480 per minute per milligram of cell extract protein. The values are means ± standard deviations based on three determinations.
Relative rates were normalized to the rate of reduction of Isf.
ND, not detected (below the level of detection, 0.1 nmol/min/mg of cell extract protein).
NA, not applicable.
TABLE 3.
Rates of CO- or H2-dependent reduction of Isf and Isf homologs catalyzed by cell extracts
| Protein | Cell extract | Rates of reduction with the following electron donors (nmol/min/mg of cell extract protein)a
|
|
|---|---|---|---|
| CO | H2 | ||
| Isf | M. thermophila | 20 ± 2 (0.37 ± 0.03) | 2.7 ± 0.5 (0.05 ± 0.01) |
| Af3 | A. fulgidus | 12 ± 2 (0.18 ± 0.03) | NDb |
| Cd1 | C. thermoaceticum | 6.2 ± 0.6 (0.11 ± 0.01) | ND |
| Mj2 | M. jannaschii | 1.2 ± 0.2 (0.024 ± 0.003) | 0.4 ± 0.1 (0.007 ± 0.001) |
Each value was determined from the slope of the linear portion of the time course. The values in parentheses are the changes in A480 per minute per milligram of cell extract protein. The values are means ± standard deviations based on three determinations.
ND, not detected (below the limit of detection, 0.1 nmol/min/mg of cell extract protein).
TABLE 4.
Enzyme activities in cell extracts
| Cell extract | Enzyme activities (μmol of methyl viologen reduced/min/mg of cell extract protein)a
|
|
|---|---|---|
| CODH | Hydrogenase | |
| M. thermophila | 2.29 ± 0.07 | 0.46 ± 0.02 |
| A. fulgidus | 0.136 ± 0.003 | NDb |
| C. thermoaceticum | 1.31 ± 0.04 | 0.96 ± 0.02 |
| M. jannaschii | 0.072 ± 0.005 | 1.06 ± 0.03 |
Each rate was determined from the slope of the linear portion of the time course. The values are means ± standard deviations based on three measurements. The assays were performed at 21°C.
ND, not detected (below the limit of detection, 0.01 μmol of methyl viologen reduced/min/mg of cell extract protein).
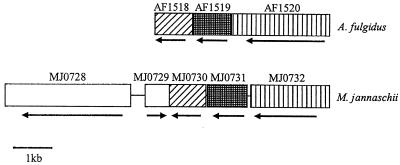
Open reading frames (ORFs) near the genes encoding the Isf homologs in all of the completed genomes (A. fulgidus, M. jannaschii, Methanobacterium thermoautotrophicum, and X. fastidiosa) were examined to determine if the ORFs encode redox proteins that could partner with the homologs. An ORF annotated as the shikimate 5-dehydrogenase ORF (aroE) is directly adjacent to and downstream of the gene encoding the M. jannaschii Mj2 homolog and is transcribed in the same direction (4). Also, an ORF annotated as the ORF encoding a heterodisulfide reductase-related protein (MTH139) is located four ORFs downstream of the gene encoding the M. thermoautotrophicum homolog Mbt1, and the two regions are transcribed in the same direction (21). The genes encoding the A. fulgidus Af2 and M. jannaschii Mj1 homologs are flanked by two ORFs annotated as ORFs encoding flavoproteins (AF1518, AF1520, MJ0730, and MJ0732) (Fig. 5) (12, 21). The deduced sequences of the putative proteins encoded by AF1518 and MJ0730 are very similar (50% identity and 73% similarity), as are the sequences of the flavoproteins encoded by AF1520 and MJ0732 (46% identity and 65% similarity). The deduced sequences of Af2 and Mj1 are 56% identical and 74% similar. Thus, it appears that the A. fulgidus and M. jannaschii genomes have the same flavoprotein-Isf homolog-flavoprotein gene arrangement. An ORF (MJ0728) encoding the CODH catalytic subunit (CooS) in M. jannaschii and an unidentified ORF (MJ0729) are located upstream of MJ0730 (Fig. 5); however, the two ORFs immediately upstream of A. fulgidus AF1518 exhibit little similarity with MJ0729 and MJ0728, and the homologous CooS protein in A. fulgidus is encoded by a remote ORF (AF1849) (12). No ORFs encoding redox proteins were found adjacent to the ORF encoding homolog Xf in X. fastidiosa.
FIG. 5.
Flavoprotein-Isf homolog-flavoprotein gene arrangements in the genomes of A. fulgidus and M. jannaschii. ORFs that encode homologous proteins are aligned and are indicated by the same pattern. The transcription direction for each ORF is indicated by the arrow below the ORF. AF1519 and MJ0731 are the ORFs that encode Af2 and Mj1, respectively. AF1518, AF1520, MJ0730, and MJ0732 encode flavoproteins. MJ0728 encodes the catalytic subunit (CooS) of CODH. MJ0729 is an undefined ORF.
DISCUSSION
The results presented here indicate that physiologically and phylogenetically diverse organisms belonging to all three domains contain homologs of Isf from M. thermophila, suggesting that this protein is distributed more broadly than previously recognized. Although the databases contained data for aerobes and facultative anaerobes, the BLAST searches primarily identified Isf homologs in organisms capable of anaerobic growth and metabolism. The only exception was the aerobe X. fastidiosa. However, these results should not be interpreted to indicate that Isf homologs are found in all anaerobes or are not also widespread in aerobes.
Our results also indicate that the homologs belongs to a new family of flavoproteins. In addition to the overall sequence identity that separates Isf from other flavoproteins, a distinguishing characteristic is the unique cysteine motif which ligates the 4Fe-4S center (15). This motif is conserved in all of the Isf homologs, suggesting that all of these homologs have an iron-sulfur center. The iron contents and UV-visible spectra of homologs Af3, Cd1, and Mj1 support this interpretation, although it cannot be determined from the results presented here if the clusters are of the 4Fe-4S or 3Fe-4S type. Histidine and aspartate replace the second cysteine in the motif of De3 and Xf, suggesting that these residues are ligands for the iron-sulfur cluster in these homologs. A role for histidine and aspartate in ligation of iron-sulfur centers is uncommon but not without precedent (3, 5, 8, 9, 24, 26). Following the previously described Isf homologs, the BLASTP alignments with the next lowest E values were the alignments with several FMN-binding tryptophan repressor binding proteins that do not contain iron-sulfur clusters. Of the two other flavoproteins whose alignments had E values less than 1.0, only NADH dehydrogenase contains iron-sulfur clusters; however, this enzyme has low overall identity with Isf (E = 0.70) and does not contain the unique cysteine motif characteristic of Isf homologs. The BLAST searches also produced a pairwise alignment with a flavodoxin, as previously reported (14), which exhibited significant identity with selected regions of Isf; however, no other flavodoxin alignments were obtained, and members of the flavodoxin family do not contain iron-sulfur clusters. Indeed, one of the defining features of the flavodoxin family is the ability to replace ferredoxin under iron-limiting growth conditions when the synthesis of iron-sulfur proteins is compromised. Nonetheless, the pairwise alignments of Isf with flavoproteins belonging to diverse families identify residues in Isf with the potential to bind FMN. Finally, less biased BLAST searches with only the Isf sequences that predominantly aligned with the sequences of flavoproteins (complete Isf sequence minus residues spanning the cysteine motif) produced the same alignments of flavoproteins as searches with the complete Isf sequence produced. These results support the notion that Isf is the prototype of a new family of flavoproteins.
The Isf homologs are relatively small (155 to 228 residues) compared to members of other families of flavoproteins (17, 23). The sizes of most of the Isf homologs are close to the sizes of flavodoxins (ca. 175 residues) that contain no iron-sulfur clusters and one FMN per subunit. Thus, Isf homologs are unusually compact considering that two iron-sulfur clusters and two FMN are bound per dimer. Only the N-terminal regions of the homologs Eh, Mbt1, Msb2, Mm2, and Cd3 deviate from the highly conserved N-terminal region by having an extension consisting of 4 to 10 residues, the function of which is unknown.
The CO-dependent reduction of homologs Af3 and Cd1 by extracts of A. fulgidus and C. thermoaceticum, respectively, indicates that Isf homologs could potentially have a role in energy-yielding pathways in these nonmethanogenic anaerobes analogous to the role proposed for Isf during acetotrophic growth of M. thermophila (2, 14). When the sulfate-reducing organism A. fulgidus grows on lactate, a CODH-ACS complex cleaves acetyl-CoA, oxidizes the carbonyl group, and donates electrons to ferredoxin (6); thus, it is hypothesized that Af3 or another Isf homolog in A. fulgidus accepts electrons from the ferredoxin, thereby participating in the electron transport chain for reduction of sulfate to sulfide. The strictly anaerobic homoacetogen C. thermoaceticum belonging to the Bacteria domain utilizes the energy-yielding Wood-Ljungdahl pathway. Central to this pathway is CODH-ACS, which synthesizes acetyl-CoA from a methyl group, CoA, and CO obtained by reduction of CO2 catalyzed by CODH-ACS. Analogous to the situation in M. thermophila and A. fulgidus, ferredoxin is the proposed electron donor for CODH-ACS in C. thermoaceticum (18, 19). Thus, it is hypothesized that an Isf homolog similar to Cd1 in C. difficile is present in C. thermoaceticum and that this homolog donates electrons to the ferredoxin. Contrary to this hypothesis, extracts of C. thermoaceticum were unable to catalyze H2-dependent reduction of the Isf homolog Cd1, although ferredoxins are electron acceptors for the hydrogenase of C. thermoaceticum. Clearly, additional experiments are necessary to test the hypotheses described above.
BLAST searches of the unfinished C. difficile genome revealed two ORFs which encode proteins that exhibit 45 and 37% identity to the α subunit (contig 939, nucleotides 49244 to 51367) and β subunit (contig 939, nucleotides 35452 to 37368), respectively, of the CODH-ACS complex of C. thermoaceticum, in addition to two ORFs that encode proteins homologous to C. thermoaceticum ferredoxin I (data not shown). These results are consistent with a role for Cd1 in electron transport coupled to CODH-ACS in C. difficle, as shown previously for M. thermophila.
The CO-dependent reduction of Mj2 catalyzed by extracts of M. jannaschii suggests that the Isf homologs are components of electron transport chains coupled to CODH in this methanoarchaeon. Indeed, the annotated genome of M. jannaschii contains the CODH homologs CdhA from M. thermophila and CooS from Rhodospirillum rubrum (4). CdhA from M. thermophila catalyzes reversible oxidation of CO to CO2 and is a subunit of the CODH-ACS complex. M. jannaschii oxidizes H2 and reduces CO2 to methane for growth and, therefore, does not utilize the CODH-ACS complex for energy generation; however, the CODH-ACS complex is required by this autotroph for synthesis of acetyl-CoA from 2CO2 for cell carbon (20). The ability of M. jannaschii extracts to link CO oxidation and H2 oxidation to reduction of Mj2 is consistent with a role for Isf homologs in an electron transport chain required for H2-dependent reduction of CO2 to CO for incorporation into acetyl-CoA. Thus, it is hypothesized that Mj2 or another Isf homolog in M. jannaschii reduces a ferredoxin that then donates electrons to the CdhA homolog for reduction of CO2 to CO, analogous to the reverse role of Isf in M. thermophila during growth with acetate. The Isf homologs identified in the autotrophic CO2-reducing methanoarchaeon M. thermoautotrophicum may also have a role analogous to the role postulated for acetyl-CoA synthesis in M. jannaschii. During growth of R. rubrum in the dark, CooS oxidizes CO, which is coupled to the reduction of protons to H2 and generation of ATP (11). The role of the CooS homolog in M. jannaschii is unknown; however, the low level of overall identity between CooS and CdhA in M. jannaschii (less than 7%) suggests that these proteins have different functions. The close proximity of the genes encoding Mj1 and CooS in M. jannaschii is consistent with a role for Mj1 in an electron transport chain coupled to CooS that may involve the flavoproteins flanking Mj1. Further research is needed to test the hypotheses described above.
Although the results presented here lead to hypothesized roles in electron transport for the Isf homologs, the electron donors and acceptors are unknown, except that ferredoxin is the electron donor for Isf in M. thermophila. The genes encoding putative flavoproteins flank the genes encoding Mj1 and Af2, which is consistent with the hypothesis that these Isf homologs interact with the flavoproteins. The same flavoprotein-Isf homolog-flavoprotein gene arrangement, together with the high level of identity between the proteins encoded by these genes in A. fulgidus and M. jannaschii, is consistent with this hypothesis. The Isf homologs in Fig. 1 exhibit less identity in their C-terminal halves, which may reflect adaptations for interactions with different redox partners or cofactors. The results obtained for Cd1 of C. difficile are consistent with this interpretation. Although C. thermoaceticum extract catalyzed robust CO-dependent reduction of Cd1, no CO-dependent reduction of Cd1 by M. thermophila cell extract was detected. Cd1 is one of the largest Isf homologs identified and has an extended C-terminal region compared to the C-terminal regions of Isf, Af3, and Mj1. The most straightforward interpretation of these results is that Cd1 is unable to interact with the ferredoxin that donates electrons to Isf in M. thermophila (14). Most species in which Isf homologs were found contain more than one homolog, which not only reflects the importance of this iron-sulfur flavoprotein family in physiology but also underscores the potential for diverse functions. Finally, the levels of identity for some Isf homologs from the same organism are lower than the levels of identity for homologs from different organisms; for example, Af2 exhibits 76% identity with Mbt2 but only 29 and 30% identity with Af3 and Af1, respectively, which indicates that homologs in the same organism may have different functions.
The same flavoprotein-Isf homolog-flavoprotein gene arrangement and the high level of deduced sequence identity of the flavoproteins of A. fulgidus and M. jannaschii are consistent with lateral gene transfer. On the other hand, the G+C content of each gene in an arrangement is similar to the total G+C content of the genome to which it belongs (49 to 50% G+C for the three genes from A. fulgidus and 33 to 38% G+C for the genes from M. jannaschii), suggesting that if lateral gene transfer occurred, the event was not recent.
ACKNOWLEDGMENTS
We thank Nancy L. Scott and Juliette T. Lecomte of the Chemistry Department of the Pennsylvania State University for their kindness and help with the high-performance liquid chromatography analysis of the flavins extracted from Af3, Cd1, and Mj2. We also especially thank Rebecca D. Miles for critically reading the manuscript. We thank the Göttingen Genomic Laboratory for providing the preliminary genome data for M. mazei.
REFERENCES
- 1.Andreesen J R, Schaupp A, Neurauter C, Brown A, Ljungdahl L G. Fermentation of glucose, fructose, and xylose by Clostridium thermoaceticum: effect of metals on growth yield, enzyme, and the synthesis of acetate from CO2. J Bacteriol. 1973;114:743–751. doi: 10.1128/jb.114.2.743-751.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker D F, Leartsakulpanich U, Surerus K K, Ferry J G, Ragsdale S W. Electrochemical and spectroscopic properties of the iron-sulfur flavoprotein from Methanosarcina thermophila. J Biol Chem. 1998;273:26462–26469. doi: 10.1074/jbc.273.41.26462. [DOI] [PubMed] [Google Scholar]
- 3.Britt R D, Sauer K, Klein M P, Knaff D B, Kriauciunas A, Yu C A, Yu L, Malkin R. Electron spin echo envelope modulation spectroscopy supports the suggested coordination of two histidine ligands to the Rieske Fe-S centers of the cytochrome b6f complex of spinach and the cytochrome bc1 complex of Rhodospirillum rubrum, Rhodobacter sphaeroides R-26, and bovine heart mitochondria. Biochemistry. 1991;30:1891–1901. doi: 10.1021/bi00221a023. [DOI] [PubMed] [Google Scholar]
- 4.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 5.Busch J L, Breton J L, Bartlett B M, Armstrong F A, James R, Thomson A J. [3Fe-4S]↔[4Fe-4S] cluster interconversion in Desulfovibrio africanus ferredoxin III: properties of an Asp14→Cys mutant. Biochem J. 1997;323:95–102. doi: 10.1042/bj3230095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai Y-R, Reed D W, Millstein J H, Hartzell P L, Grahame D A, DeMoll E. Acetyl-CoA decarbonylase/synthase complex from Archaeoglobus fulgidus. Arch Microbiol. 1998;169:525–529. doi: 10.1007/s002030050606. [DOI] [PubMed] [Google Scholar]
- 7.Ferry J G. Enzymology of the fermentation of acetate to methane by Methanosarcina thermophila. BioFactors. 1997;6:25–35. doi: 10.1002/biof.5520060104. [DOI] [PubMed] [Google Scholar]
- 8.Gurbiel R J, Batie C J, Sivaraja M, True A E, Fee J A, Hoffman B M, Ballou D P. Electron-nuclear double resonance spectroscopy of 15N-enriched phthalate dioxygenase from Pseudomonas cepacia proves that two histidines are coordinated to the [2Fe-2S] Rieske-type clusters. Biochemistry. 1989;28:4861–4871. doi: 10.1021/bi00437a051. [DOI] [PubMed] [Google Scholar]
- 9.Gurbiel R J, Ohnishi T, Robertson D E, Daldal F, Hoffman B M. Q-band ENDOR spectra of the Rieske protein from Rhodobacter capsulatus ubiquinol-cytochrome c oxidoreductase show two histidines coordinated to the [2Fe-2S] cluster. Biochemistry. 1991;30:11579–11584. doi: 10.1021/bi00113a013. [DOI] [PubMed] [Google Scholar]
- 10.Horikoshi K, Grant W D, editors. Extremophiles: microbial life in extreme environments. New York, N.Y: John Wiley & Sons, Inc.; 1998. [Google Scholar]
- 11.Kerby R L, Ludden P W, Roberts G P. Carbon monoxide-dependent growth of Rhodospirillum rubrum. J Bacteriol. 1995;177:2241–2244. doi: 10.1128/jb.177.8.2241-2244.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 13.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetic analysis. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 14.Latimer M T, Painter M H, Ferry J G. Characterization of an iron-sulfur flavoprotein from Methanosarcina thermophila. J Biol Chem. 1996;271:24023–24028. doi: 10.1074/jbc.271.39.24023. [DOI] [PubMed] [Google Scholar]
- 15.Leartsakulpanich U, Antonkine M L, Ferry J G. Site-specific mutational analysis of a novel cysteine motif proposed to ligate the 4Fe-4S cluster in the iron-sulfur flavoprotein of the thermophilic methanoarchaeon Methanosarcina thermophila. J Bacteriol. 2000;182:5309–5316. doi: 10.1128/jb.182.19.5309-5316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li N, Zhao J D, Warren P V, Warden J T, Bryant D A, Golbeck J H. PsaD is required for the stable binding of PsaC to the photosystem I core protein of Synechococcus sp. PCC 6301. Biochemistry. 1991;30:7863–7872. doi: 10.1021/bi00245a028. [DOI] [PubMed] [Google Scholar]
- 17.Lim L W, Shamala N, Mathews F S, Steenkamp D J, Hamlin R, Xuong N H. Three dimensional structure of the iron-sulfur flavoprotein trimethylamine dehydrogenase at 2.4-Å resolution. J Biol Chem. 1986;261:15140–15146. [PubMed] [Google Scholar]
- 18.Ragsdale S W, Clark J E, Ljungdahl L G, Lundie L L, Drake H L. Properties of purified carbon monoxide dehydrogenase from Clostridium thermoaceticum, a nickel, iron-sulfur protein. J Biol Chem. 1983;258:2364–2369. [PubMed] [Google Scholar]
- 19.Ragsdale S W, Ljungdahl L G, DerVartanian D V. 13C and 61Ni isotope substitutions confirm the presence of a nickel (III)-carbon species in acetogenic CO dehydrogenase. Biochem Biophys Res Commun. 1983;115:658–665. doi: 10.1016/s0006-291x(83)80195-8. [DOI] [PubMed] [Google Scholar]
- 20.Selkov E, Maltsev N, Olsen G J, Overbeek R, Whitman W B. A reconstruction of the metabolsim of Methanococcus jannaschii from sequence data. Gene. 1997;197:11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 21.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terlesky K C, Nelson M J K, Ferry J G. Isolation of an enzyme complex with carbon monoxide dehydrogenase activity containing corrinoid and nickel from acetate-grown Methanosarcina thermophila. J Bacteriol. 1986;168:1053–1058. doi: 10.1128/jb.168.3.1053-1058.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanoni M A, Curti B. Glutamate synthase: a complex iron-sulfur flavoprotein. Cell Mol Life Sci. 1999;55:617–638. doi: 10.1007/s000180050319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volbeda A, Charon M H, Piras C, Hatchikian E C, Frey M, Fontecilla-Camps J C. Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- 25.Vorholt J, Kunow J, Stetter K O, Thauer R K. Enzymes and coenzymes of the carbon monoxide dehydrogenase pathway for autotrophic CO2 fixation in Archaeoglobus lithotrophicus and the lack of carbon monoxide dehydrogenase in the heterotrophic A. profundus. Arch Microbiol. 1995;163:112–118. [Google Scholar]
- 26.Zhou Z H, Adams M W. Site-directed mutations of the 4Fe-ferredoxin from the hyperthermophilic archaeon Pyrococcus furiosus: role of the cluster-coordinating aspartate in physiological electron transfer reactions. Biochemistry. 1997;36:10892–10900. doi: 10.1021/bi9708141. [DOI] [PubMed] [Google Scholar]