Abstract
Organoids recapitulate many aspects of the complex three-dimensional (3D) organization found within native tissues and even display tissue and organ-level functionality. Traditional approaches to organoid culture have largely employed a top-down tissue engineering strategy, whereby cells are encapsulated in a 3D matrix, such as Matrigel, alongside well-defined biochemical cues that direct morphogenesis. However, the lack of spatiotemporal control over niche properties renders cellular processes largely stochastic. Therefore, bottom-up tissue engineering approaches have evolved to address some of these limitations and focus on strategies to assemble tissue building blocks with defined multi-scale spatial organization. However, bottom-up design reduces the capacity for self-organization that underpins organoid morphogenesis. Here, we introduce an emerging framework, which we term middle-out strategies, that relies on existing design principles and combines top-down design of defined synthetic matrices that support proliferation and self-organization with bottom-up modular engineered intervention to limit the degrees of freedom in the dynamic process of organoid morphogenesis. We posit that this strategy will provide key advances to guide the growth of organoids with precise geometries, structures and function, thereby facilitating an unprecedented level of biomimicry to accelerate the utility of organoids to more translationally relevant applications.
Subject terms: Tissue engineering, Biomaterials, Regenerative medicine, Stem-cell biotechnology, Stem-cell niche
Organoids recapitulate many aspects of native tissues and even display tissue and organ-level functionality, although with limited control over morphogenesis. This Review describes an emerging framework, termed middle-out tissue engineering, that facilitates spatiotemporal control of tissue-specific cell niches to enable deterministic organoid self-organization and build more advanced in vitro tissue models.
Key points
Organoids have emerged as powerful in vitro models for a variety of organs and applications; however, their utility is currently limited by a lack of spatiotemporal control over the tissue-specific cell niches in which they reside.
Tissue engineering approaches facilitate the generation of more physiologically relevant in vitro cell niches to improve methods for organoid culture.
Top-down, scaffold-based tissue engineering approaches allow for macroscale control over organoid geometry and are amenable to cell-based remodelling and self-organization but suffer from a lack of spatiotemporal control of niche properties.
Bottom-up, modular tissue engineering approaches allow for precise control over cellular and extracellular tissue building blocks for precision engineering but at the cost of minimizing the capacity for cellular self-organization.
Middle-out, interventional tissue engineering approaches combine aspects of top-down and bottom-up tissue engineering methods to enable precise spatiotemporal control of engineered cell niches, thereby enabling deterministic control of cellular self-organization.
Introduction
Organoids are complex three-dimensional (3D) structures that arise through cell-intrinsic genetic programmes activated, in large part, by carefully selected soluble biochemical niche cues in suspension culture or a 3D extracellular matrix (ECM)-based culture platform that facilitates self-organization and morphogenesis. Organoids closely match the histoarchitecture of their in vivo counterparts and, importantly, they also contain many tissue-resident cell types and display some tissue-level and organ-level functionality1. Traditional methods for cell culture, often employing single cell-type populations with no 3D organization, cannot reach the in vitro complexity afforded by organoids. This key limitation has motivated the widespread use of organoids in fundamental biology, clinical and biomedical engineering laboratories. With their increased utility, the field has rapidly expanded and advanced since its modern conception by Eiraku et al., who cultured 3D, self-organizing cortical tissues from embryonic stem cells (ESCs)2, and soon thereafter by the work of Sato et al., who specified the modern definition of organoids by generating self-organizing murine intestinal 3D multicellular constructs (in this case, organoids) from single intestinal stem cells (ISCs) positive for leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5)3. Organoids are useful in fundamental biological studies as models of organ development and disease progression as well as for applied research related to drug screening and generation of transplantable tissues4–10. Protocols have been established for organoids derived from multipotent adult stem cells (ASCs) and pluripotent stem cells (PSCs) for numerous tissues and organs spanning all three germ layers, including the intestine11, brain12, heart13 and liver14. ASC-derived organoids are well suited to model tissue homeostasis, regeneration and adult-onset disease, whereas PSC-derived organoids accurately recapitulate developmental processes and can be used to study developmental disorders15. Organoids derived from model organisms, most notably from mice, have been transformative in developing protocols for organoid growth3, differentiation3,16 and cell-type enrichment17 and to understand the fundamental regulatory mechanisms underpinning symmetry breaking18,19, which is a crucial initial step for the establishment of complexity and 3D architecture. Building further upon these benefits, human organoids bridge the gap between non-human animal models, which have a notoriously low translational success rate, and more simplistic in vitro human cell models20. Moreover, patient-specific organoids can be generated from tissue biopsies or from PSCs, and are useful as clinical models for genetic disorders, including cystic fibrosis21 and microcephaly12, as well as for infections such SARS-CoV-2 (ref. 22). Finally, organoid models are promising for precision and/or personalized medicine and could improve the success rate of translational therapies.
Despite being powerful in vitro models, organoids are not without limitations. For example, they have low scalability, limited lifespan, difficulty in imposing the proper extracellular forces and incomplete matching of organ-level functionality15,23–25. Here, we focus on other limitations of organoids related to the lack of control of morphogenesis, composition and maturation, and identify methods to better assess key regulators of these processes. We focus on intestinal organoids as their biology has been well characterized and numerous publications have highlighted methods for their engineering.
Organoid cultures are established through a process of minimally controlled self-organization, most often by flooding cells cultured in suspension or 3D ECM-based matrices with biochemical cues to drive differentiation and symmetry breaking. The lack of spatiotemporal control of this self-organized morphogenesis leads to highly heterogeneous organoids with varying composition, shape and size. Protocols have been developed to better control or enrich for certain cell populations by refining the cocktail of biochemical cues17 and to induce morphogenesis by timed exposure or withdrawal of certain cues16,26. For example, initial culture of murine ASC-derived intestinal organoids in media maintaining a high stem population (containing EGF, Noggin, R-spondin, CHIR and valproic acid) facilitates uniform growth of spherical colonies compared with differentiation media. By contrast, withdrawing factors activating WNT and NOTCH signalling (for example, CHIR and valproic acid, respectively) leads to the differentiation of multiple intestinal cell types, including Paneth cells, enterocytes and goblet cells, while also inducing the formation of crypt buds reminiscent of the histoarchitecture of the native intestine16. However, these globally administered cues do not match the spatiotemporal signalling and dynamic boundary conditions of in vivo organogenesis, which involves the generation of local signalling gradients to establish, for example, the crypt–villus axis of the intestine27,28. Cells can interpret their spatial location based on their position along the gradient, making these cues crucial in establishing proper tissue organization29. Importantly, organoid protocols do not optimize for the effects of local, spatially defined soluble signalling that is known to be important in differentiation and in the shaping of developing tissues, but rather allow for cell differentiation and spatial organization resulting from global, user-defined cues to guide growth. With standard organoid culture methods using flood exposure to biochemical cues, local gradients arise in the presence of an already complex mixture of cues, making it difficult to tease out the roles of local, cell-secreted cues and globally provided ones.
In addition, controlled cell–cell interactions, particularly those with supporting mesenchymal cell populations that contribute to geometric changes through direct contact or by imparting mechanical forces, are understudied because many organoid cultures lack these cell populations as they are difficult to co-culture in vitro. Specifically, mesenchymal and/or stromal cells have a key role in shaping intestinal villus evolution through cell–cell interactions and compressive force generation mechanisms30. Cell–cell interactions are also essential in PSC-derived organoid maturation. The current state-of-the-art maturation protocols involve implantation into immunocompromised mice, which leads to maturation yet without knowledge of how this is achieved31,32. In implanted organoids, a perfusable vasculature is established, suggesting that cell–cell interactions between tissue-specific cell types and endothelial cells or paracrine signalling from endothelial cells might have a substantial role in maturation. Lymphatics have also been identified as a signalling hub in the intestinal crypt niche33. To establish better in vitro organoid models, control of the inputs, including the cell types present, will be crucial to obtain the desired outputs such as vascularization and innervation of organoids.
Cell–ECM interactions are also important in relaying outside-in signalling of the composition and mechanics of the local microenvironment. However, the role of the ECM is difficult to gauge because the organoid field primarily relies on Matrigel as a 3D matrix for in vitro experiments. Matrigel is a poorly characterized proteinaceous material that is generated from reconstituted Engelbreth–Holm–Swarm mouse sarcoma that contains thousands of different ECM components34. Although Matrigel is a versatile system for the culture of many types of organoids and is permissive to self-organization and the identification of soluble niche cues guiding cell fate specification, its complex bioactivity, numerous sites for integrin engagement and several mechanisms for cell-based remodelling render interpretation of cell–matrix interactions more difficult. The complex composition of Matrigel contributes to its versatility but introduces substantial heterogeneity between organoids. Moreover, it suffers from batch-to-batch variability in composition and it is difficult to tailor its mechanical properties, thereby limiting its utility for mechanistic studies related to ECM composition and mechanics35. Finally, because Matrigel is generated from a reconstituted mouse sarcoma, it comes from a non-human, non-tissue-specific source; therefore, there is a contextual mismatch between its composition and that of the ECM of the organoid of interest. Finally, it is a non-good manufacturing practice material, which limits its use for translational applications34.
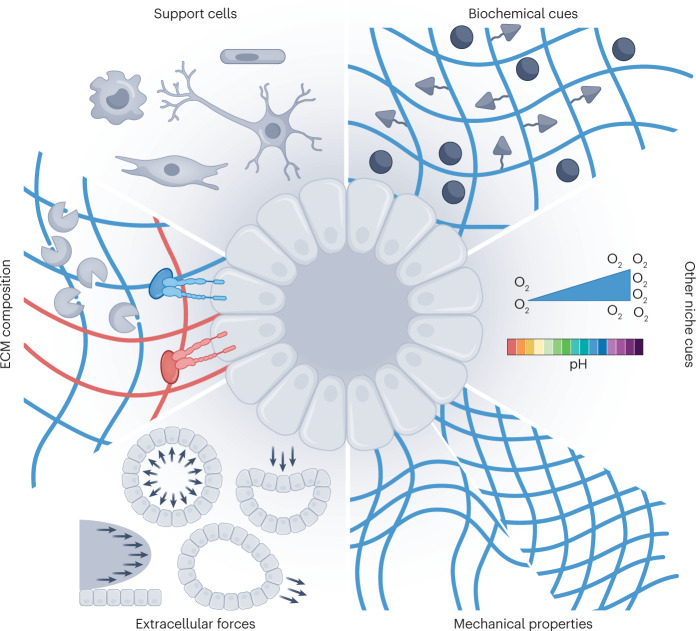
In the simplest terms, the limitations that persist with organoid culture arise because the tissue-specific stem cell niche is not adequately matched. Ultimately, this is a two-pronged problem: first, protocols suffer from a limited understanding of the evolving niche and tissue morphogenesis, and second, artificial niches are limited by the technical abilities to design platforms that accurately mimic key niche dynamic properties. Niche properties include soluble signalling cues, cell–cell interactions, cell–ECM interactions, oxygen levels, pH, mechanical properties and extracellular mechanical forces, including compression, strain and shear stress (Fig. 1). Importantly, these cues are not static, but change dynamically in space and time, in accordance with rapidly growing and differentiating organoids, introducing spatiotemporal regulation as an additional dimension in niche mimicry.
Fig. 1. The organoid niche.
The organoid niche is composed of: support cells, including endothelial cells, mesenchymal/stromal cells, neurons and immune cells; soluble and membrane-bound biochemical cues, which can exist as local gradients; local pH and oxygen concentration; mechanical properties, including stiffness and stress relaxation; extracellular forces such as shear stress, compression, external strain and osmotic swelling; and the extracellular matrix (ECM) composition, including degradability (for example, by cell-produced matrix metalloproteinases), integrin-binding specificity and the presence of niche-specific ECM proteins such as collagen IV or laminin.
Specifically, in this Review, we survey the use of tissue engineering principles and highlight how these advanced methods can be used to control the presentation of niche factors and, ultimately, control intestinal organoid morphogenesis. These strategies, which we term middle-out tissue engineering, combine the advantages of traditional top-down biomaterial scaffold-based approaches with modular bottom-up approaches by spatiotemporally controlling the properties of tissue-specific niches (Box 1). In other words, middle-out tissue engineering combines bottom-up techniques for the controlled assembly of cells and scaffold components, built on the knowledge of those building blocks, while keeping the tissue architecture in mind to top-down process the engineered organoid-laden constructs using patterning, 3D printing or other methods to achieve the targeted properties. Middle-out (in this case, interventional) approaches start in the middle (for example, with an already growing organoid) and combine both strategies to integrate control over organoid morphogenesis in space and time.
Box 1 Tissue engineering definitions.
Top-down tissue engineering: combining one or more cell types of the targeted tissue with a biodegradable scaffold mimicking the macroscale architecture and the biophysical and biochemical cues of the organotypic niche of interest, which is permissive to cellular self-organization, proliferation, remodelling and morphogenesis. Exogenous spatial control is limited after initial cell seeding. Although stringent initial conditions, including well-characterized cell populations and controllable biophysical properties as well as macroscale boundary conditions, can be set, the local niche properties often rely on cell-mediated changes that cannot be controlled through top-down engineering methods. Ultimately, this limitation leads to uncontrolled cellular self-organization during organoid formation or to cells populating scaffolds to reach pre-defined boundary conditions without proceeding through intrinsic self-organization processes.
Bottom-up tissue engineering: engineered construction of tissue building blocks and their assembly in a modular approach. Well-defined cells or aggregates of cells are combined with other components of the tissue-specific niche in a spatially controlled manner. Using this method, morphogenesis is user-designed through defined spatial positioning and/or control of cell–cell and cell–matrix interactions as well as initial spatial control of other niche properties. Here, initial conditions and boundary conditions are set but endogenous remodelling and ECM dynamics are limited, thereby reducing the capacity for cellular self-organization.
Middle-out (interventional) tissue engineering: combining principles of top-down and bottom-up approaches to establish in vitro methods permissive to cellular self-organization and integrating the capacity to continuously and spatiotemporally adapt boundary conditions to match the developmental or regenerative extracellular matrix and exploit the instructive cellular machinery that can promote deterministic organ morphogenesis. Starting with growing tissue constructs generated with either top-down or bottom-up methods, an engineered cell and niche intervention is used to mimic dynamic cues presented at specific points in space and time at various stages throughout development or regeneration using a combination of engineering approaches.
Tissue-engineered organoids
At its inception in the early 1990s, the field of tissue engineering focused on generating functional tissue replacements by combining principles of biology and engineering36. The goals of the field have further evolved to establish biomimetic tissue models to understand regenerative and developmental biology as well as human disease. With this broadened scope, organoids represent a transformative achievement because they harness the benefits of cellular self-organization to form multicellular organoids that are typically difficult to achieve with strictly engineering principles.
Initial strategies to engineer tissues took a top-down, scaffold-based approach, where tissue-specific cells were cultured in biodegradable scaffolds with geometries that matched the tissue of interest at the macroscale37. As the field progressed with technological and engineering advancements, bottom-up approaches emerged to complement the top-down strategies, enabling the spatial organization of tissue building blocks composed of specific cell types and ECM components at multiple length scales38,39. At the time these techniques were emerging, the ability to culture PSCs did not yet exist40,41 and modern protocols for stem cell-derived organoids had not yet been established2,3. With the advances in tissue construction from a biological perspective, the definitions of these two approaches (in this case, bottom-up and top-down) have become blurred and transcend one another. We have thus redefined these two approaches (Box 1), with a particular focus on their use for organoid culture. Specifically, the ability for cells to self-organize in vitro is a concept that does not neatly fit into previously established tissue engineering methods. Material-based self-assembly is, by definition, a bottom-up process but, because cells can proliferate and differentiate as they self-organize, this process is not necessarily consistent with the classical definition of bottom-up tissue engineering. Namely, the building blocks are not assembled by engineering methods but rather produced and organized through intrinsic cellular processes that can be minimally controlled with static macroscale boundary conditions imparted by classical top-down tissue engineering approaches. Therefore, we consider cellular self-organization, in which cells are not assembled using engineering methods at the cellular scale, an aspect of top-down tissue engineering. Notably, classical definitions for top-down approaches allowed for proliferation and cell-based remodelling of the scaffold based on biodegradability and cell-produced ECM components, both of which are involved in organoid morphogenesis42,43. Considering all these factors, our refined definition categorizes most organoid protocols as top-down tissue engineering processes. Another important note here is that, in top-down methods guided by macroscale scaffold geometries and with the additional spatial control afforded by modular bottom-up approaches, the ability to harness the inherent capacity of cells to engineer themselves into functional tissues is minimized. Therefore, these methods are severely limited by the user’s understanding of morphogenesis and tissue organization. Finally, despite top-down approaches allowing for dynamic cell-based matrix remodelling or pre-programmed degradation, and both methods enabling temporal changes to the bulk biochemical make-up by staged changes to media conditions, neither method allows for spatiotemporal control of niche properties after initial cell seeding. With top-down tissue engineering, this limitation can lead to highly heterogeneous organoids generated by stochastic growth because they lack local changes to boundary conditions that are necessary to accommodate controlled self-organization as the organoid size changes over time; therefore, top-down tissue engineering cannot match the dynamic conditions present during native tissue morphogenesis. To combine the benefits of both methods, we define a new category, middle-out tissue engineering, where self-organization is engineered through interventions to the local boundary conditions at precise points in space and time to guide controlled and deterministic organoid growth with engineered niche dynamics.
Intestinal organoids as a model system
To provide the proper context for the development of engineering methods, the underlying biology must be well understood. Therefore, we focus here on the intestinal organoid as a model system, which was among the first to be developed, thereby establishing the definition of the modern organoid44, and has been well characterized through a series of in-depth studies3,11,16,18,45,46. Because the intestinal epithelium has among the highest turnover rates of any tissue47, ASC-derived intestinal organoids can be rapidly generated from a single LGR5+ ISC3. The ease of culture and rapid generation of intestinal organoids are attractive to engineers seeking to implement tissue engineering principles to establish more homogeneous and functionally relevant tissue models. These approaches can be adapted to other types of organoids, with optimization based on a biological understanding of tissue-specific morphogenesis. To best understand the factors that can be manipulated with engineering methods, we first describe the intestinal architectural organization and the biological mechanisms guiding murine ASC-derived intestinal organoids as well as human ASC-derived and induced PSC (iPSC)-derived organoids.
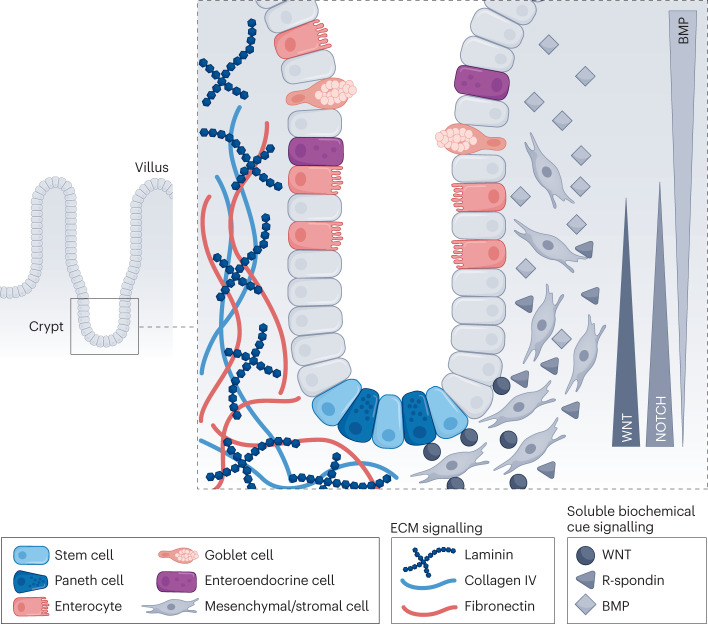
The cells of the small intestinal epithelium are organized into a crypt–villus structure, where ISCs populate the bottom of the crypt. ISCs can differentiate into all intestinal-resident cell types. Paneth cells, terminally differentiated progeny of ISCs, and ISCs form an intercalated pattern that helps maintain crypt homeostasis through cell–cell signalling. ISCs proliferate to produce bipotent progenitors that, before exiting the crypt, undergo fate specification into a variety of differentiated post-mitotic cell types, including absorptive enterocytes and secretory cells such as goblet and enteroendocrine cells48. Well-defined signalling pathways control homeostasis, including gradients of WNT, NOTCH, EGF and bone morphogenetic protein (BMP) that arise through epithelial and mesenchymal cell crosstalk28,49 (Fig. 2).
Fig. 2. The intestinal niche and the current state of niche mimicry for intestinal organoids.
The intestine is organized into a crypt–villus architecture. The intestinal crypt contains crypt-base cell populations (such as intestinal stem cells and Paneth cells) and other differentiated cell types, including enterocytes, goblet cells and enteroendocrine cells. The niche is regulated by epithelial interactions with the surrounding mesenchymal/stromal cells and gradients of extracellular matrix (ECM) composition and soluble biochemical cues. BMP, bone morphogenetic protein.
Furthermore, the composition and stiffness of the ECM surrounding intestinal cells also have an essential role in guiding cell fate specification and organization. The composition of the ECM is dynamic, with the rate of deposition of specific ECM proteins occurring in concert with intestinal morphogenetic events. For example, the highest concentration of laminin is found in the crypt and laminin synthesis increases as crypts bud50–53. These changes in ECM composition are associated with changes in the availability of integrin binding, with integrin α6 expression localized in the crypt and integrin β1 driving ISC proliferation and homeostasis48,52. In addition to laminin, the ISC niche is also comprised of fibronectin, collagens and glycosaminoglycans. The cells making up the intestinal niche, including epithelial cells, contribute to ECM deposition and remodelling54. Furthermore, the role of the mechanosensitive Yes-associated protein (YAP) has been established in symmetry-breaking events guiding crypt formation, cell fate and proliferation in the intestine18,55,56. Specifically, YAP contributes to intestinal regeneration through elevated integrin β1 signalling and upregulation of focal adhesion kinase (FAK) and SRC signalling associated with alterations in the local matrix environment, including upregulation of ECM proteins such as collagen I56,57. Taken together, these findings highlight the dynamic nature of the ECM in the intestinal microenvironment and reveal that outside-in signalling, through integrins and YAP, is crucial for homeostatic regeneration and in determining the fate of ISCs.
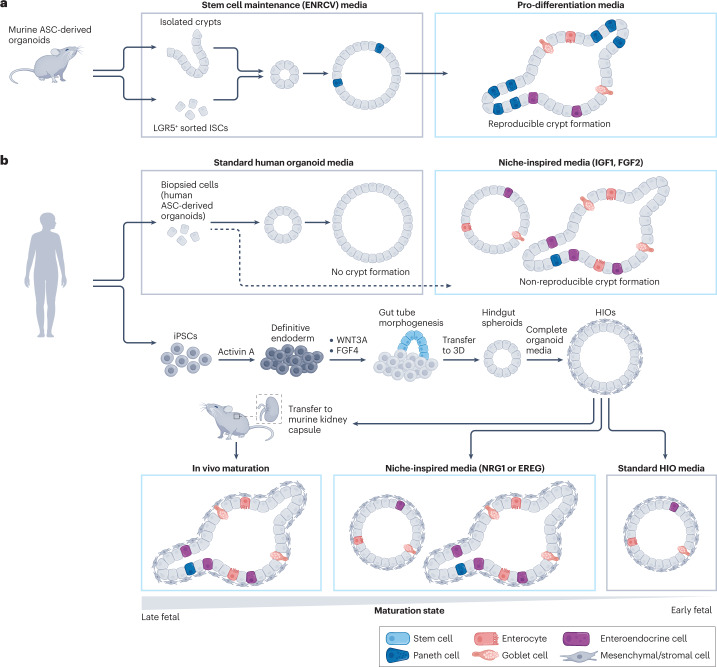
Murine ASC-derived intestinal organoids can be generated from a single LGR5+ ISC when embedded and cultured in 3D matrices such as Matrigel, and defined media conditions have been identified that activate key signalling pathways relevant to the ISC niche3,16 (Fig. 3a). ISCs proliferate and form spherical colonies with a polarized epithelium, which subsequently bud to form a crypt–villus structure reminiscent of in vivo organization. Budding occurs following a change to the media growth factor cocktail (for example, by withdrawal of CHIR and valproic acid) favouring cell differentiation. Changes in cell shape and phenotype occur concurrent to structural changes to the ECM, including matrix degradation, to yield highly biomimetic 3D organoids42. Although the generation of murine intestinal organoids is efficient and reproducible, suggesting that many key niche factors are presented to these organoids, the resulting organoids are highly heterogeneous, with uncontrollable growth arising from stochastic symmetry-breaking events. This stochasticity results in random budding and variable crypt dimensions, which highlights that the niche is not completely matched with spatiotemporal precision. To understand this randomness, transient cell–cell heterogeneity in YAP signalling has been observed to trigger Paneth cell differentiation where bud formation is initiated18. Additionally, Paneth cells at the site of crypt formation express higher levels of integrin β4 compared to other cell types, which allows them to preferentially bind to laminin and contribute to crypt formation58. Notably, changes to the ECM and YAP signalling are necessary to alter cellular identity. Therefore, activating YAP through differential ECM binding is of fundamental importance to understanding epithelial regenerative homeostasis. Changes to the ECM associated with diseases such as fibrosis can also alter these signalling pathways to reduce the regenerative potential of the epithelium and perpetuate intestinal dysfunction59. Furthermore, cell and niche mechanical forces have also been implicated in organoid morphogenesis60–62.
Fig. 3. Murine and human intestinal organoids.
a, Murine adult stem cell (ASC)-derived intestinal organoids can be generated from isolated crypts or by sorting isolated crypts for leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5+) intestinal stem cells (ISCs). Symmetry breaking occurs with the differentiation of Paneth cells within spherical organoids. Following a change to media promoting differentiation, intestinal crypts form and other intestine-specific cells differentiate. Mouse intestinal organoids more closely match their in vivo counterpart than human organoids in terms of architecture and cell composition, with reproducible crypt formation and the ability to enrich for numerous cell types, based on the timing of exposure to various organoid media compositions. b, Human intestinal organoids can be generated from adult tissue using biopsy or from pluripotent stem cells (PSCs). In standard human organoid media, stem cell-rich organoids are generated without reproducible crypt formation. Using niche-inspired conditions, with the addition insulin-like growth factor 1 (IGF1) and fibroblast growth factor 2 (FGF2), increased cell-type diversity is achieved with some organoids containing crypts. Human induced PSCs (iPSC)-derived organoids (HIOs) are generated by differentiating iPSCs to definitive endoderm, followed by the formation of three-dimensional (3D) gut tubes emerging from the 2D culture surface. These structures are collected and embedded into 3D as hindgut spheroids. Culture in HIO media then results in HIOs, which contain a supporting mesenchymal population. Using standard HIO media, immature organoids are produced, which contain some differentiated cell types but no crypt structures. Using niche-inspired media conditions (either by the inclusion of neuregulin 1 (NRG1) or epiregulin (EREG) in the cell culture media at precise time points) leads to the emergence of more mature organoids, some containing crypt-like structures. Implantation of HIOs into the mouse kidney capsule results in the most mature phenotype, representing a later stage in development. To more closely match later stages of development or even generate post-natal (for example, paediatric, adult) organoids, additional niche cues will need to be presented with spatiotemporal precision. For organoids from all cell sources, more closely matching the niche will result in organoids that more accurately represent native tissue, providing potential for translational applications.
Many of the signalling pathways and regulatory mechanisms guiding morphogenesis are conserved between murine and human development, but important differences exist that have led to additional limitations with human intestinal organoid culture. Interestingly, it is more difficult to reproducibly obtain crypt structures in human ASC-derived and PSC-derived organoids compared to murine-derived ones, likely owing to a niche that is not optimized to match that of the human intestine (Fig. 3b). Efforts to more closely match the niche with the addition of supplementary soluble cues, such as fibroblast growth factor 2 (FGF2) and insulin-like growth factor 1 (IGF1), have led to increased cell-type diversity in human ASC-derived organoids but remain limited in matching the biomimetic spatial organization achieved with mouse organoids63. Knowing how to match the developmental niche to induce fate specification and maturation of iPSC-derived organoids is more difficult because of limitations in access to fetal tissue. However, new data sets have emerged, containing single-cell RNA sequencing and phenotypic characterization, that have enabled refinement of the composition of media cocktails and their timed exposure, specifically with exposure to neuregulin 1 (NRG1), to improve the maturation of iPSC-derived organoids and facilitate symmetry breaking26. For human fetal-derived organoids, exposure to epiregulin (EREG) can lead to similar effects, resulting in organoids with enhanced spatial organization64. In vivo organoid implantation can also enhance maturation by allowing increased cell–cell interactions and facilitating organoid vascularization, although the mechanistic details underlying in vivo maturation are not well understood31. Finally, imposing mechanical strain on implanted organoids with nitinol springs can also improve their maturation, highlighting that an externally applied force can be used as a means to advance organoid engineering65.
Taken together, knowledge of key biochemical factors of the intestinal niche has refined media conditions and the timing of their exposure to advance the maturation and morphogenesis of murine and human organoids. Other niche factors, which can be tuned with tissue engineering methods, have begun to be investigated but remain underexplored in the organoid field, and will thus likely address some limitations to current methods.
Top-down organoid tissue engineering
Top-down, scaffold-based tissue engineering approaches are common in the organoid field (Fig. 4). These methods combine tissue-specific cells, soluble factors and an engineered scaffold amenable to cell proliferation and self-organization, including those using Matrigel or synthetic, engineered ECM alternatives3,42,66. Because these methods harness uncontrolled cellular self-organization resulting in heterogeneous organoids, more homogeneous structures can be obtained by specifically controlling the initial conditions, including the starting cell populations, the biochemical cue composition and the initial scaffold properties. Intestinal organoids can be generated from single stem cells or from small clusters of cells sheared directly from intestinal crypts. In the case of a population started from a single cell, cells are sorted for one or more markers (for example, LGR5+ for intestinal organoids), providing a highly characterized initial cell condition. Controlled morphogenesis is more difficult to achieve from clusters of cells because their initial state is more difficult to define. For organoids that cannot be generated from single stem cells, bottom-up approaches that rely on highly regulated multicellular assembly are preferred to establish more refined initial conditions (see below). These refined cell populations can then be used as the starting point for top-down assembly and allowed to self-organize rather than be used as building blocks for bottom-up assembly19. From an engineering perspective, defining the biochemical cue composition is relatively simple with top-down approaches because flood exposure is the most common method. The composition of the media cocktail is, of course, tissue dependent and informed by knowledge of key signalling pathways regulating development and regeneration. These recipes are continually updated as new data are published and can be altered over time.
Fig. 4. Tissue engineering of organoids.
a, In top-down tissue engineering, cells (either whole organoids or single stem cell populations) are combined with defined biochemical media components and a scaffold with defined initial properties to impart biophysical cues to organoids. Over time, and depending on user-defined media conditions, the extracellular matrix undergoes remodelling (either cell imposed or pre-programmed) and cellular morphogenesis proceeds through proliferation, differentiation and architectural changes to generate heterogeneous organoids in scaffolds without controlled cell-scale boundary conditions (for example, Matrigel or monolithic synthetic hydrogels) or to populate scaffolds along macroscale boundary conditions designed to match the macro-architectural features of the tissue of interest. b, Bottom-up tissue engineering consists of constructing and assembling cellular building blocks, which can be generated by forced aggregation using the hanging drop method or commercially available Aggrewells. Organoid arrays can be used to generate homogeneous organoids at precise three-dimensional (3D) locations. Microfabrication can be used to generate organoids with different diameters and aspect ratios. Droplet microfluidics can be used to generate single organoid microenvironments with defined dimensions. Engineered assembly can be performed at the single-cell level by functionalizing cell membranes with complementary reactive groups. Microgels can be assembled using complementary linkages to form 3D structures. Finally, 3D printing can be used to place cells or organoids at precise locations in 3D.
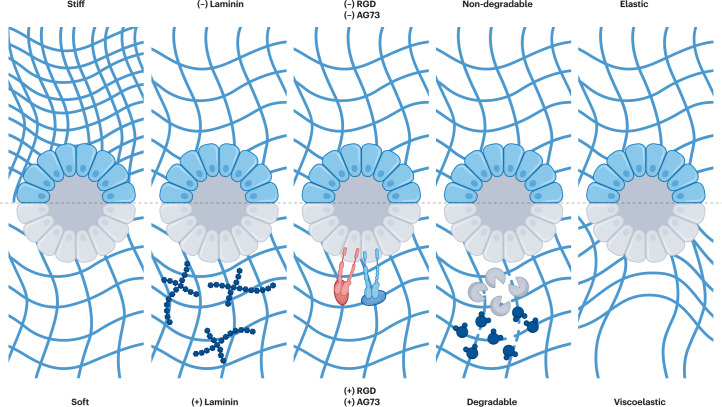
Additional niche factors can be regulated by modulating the initial scaffold properties. Altering the concentration of Matrigel can tune its properties albeit across a relatively small property range compared to tunable synthetic hydrogels and without allowing isolation of the role of each individual property in guiding cell behaviour. These properties, including ECM composition42,67, integrin binding42,68–72, degradation42,73, stiffness42,74 and viscoelasticity (stress relaxation)75–78, can all regulate cell signalling pathways through outside-in signalling to control organoid growth, cell differentiation and morphogenesis (Fig. 5). The use of so-called ‘blank slate’ hydrogels (for example, poly(ethylene glycol) (PEG), hyaluronic acid and alginate) as reductionist ECM mimics can address these limitations to understand the role of each factor in organoid morphogenesis and therefore build more reproducible organoids. Initial studies developed an effective blueprint for the design of these hydrogels by manipulating initial matrix stiffness (for example, by changing the concentration of hydrogel precursors), type and concentration of integrin-binding peptides (such as arginylglycylaspartic acid (RGD) and AG73) or supplemental ECM components (such as fibronectin, laminin 111 and collagen IV), and degradability (for example, by matrix metalloproteinase (MMP)-degradable crosslinkers or by pre-programmed hydrolytic degradation)42. Notably, the latter study on degradability demonstrated the temporal dependence of these niche properties, in other words, the properties optimized at the start of cell culture did not match those required for crypt morphogenesis. An initially stiff matrix (stiffer than Matrigel, ~1.3 kPa) and cell–RGD binding are needed to activate YAP signalling, whereas a programmed matrix softening in the presence of full-length laminin 111 (not laminin-derived peptides) is needed for crypt formation. Furthermore, the mode and timing of matrix softening are key, with cell-derived MMP-mediated rapid matrix degradation leading to organoids that are depolarized and disorganized, adopting an inflammation-like state. Shifting the timing of matrix degradation later in organoid culture facilitates the formation of crypts. Similar experiments have supported this notion by temporally photosoftening synthetic matrices through controlled light doses79. Importantly, both of these studies highlighted the need for laminin in crypt formation. It was later identified that properties need to be further optimized based on species (for example, mouse versus human) as well as for ASC-derived versus PSC-derived organoids42,69,71. Others have sought to replace Matrigel with matrices free from any animal-derived components80–82. Furthermore, ECM-mimetic materials with controllable viscoelasticity and stress relaxation have been shown to influence organoid growth81 and crypt formation77,78 by regulating symmetry breaking at early stages through differential YAP localization77. Notably, crypts formed using alginate–Matrigel interpenetrating networks with controllable stress relaxation much more closely match the properties of native tissue and of Matrigel compared to the synthetic dynamic hydrogels77. Understanding the role of stress relaxation in the absence of ECM components is an area of potential future interest to understand the regulators of early symmetry breaking.
Fig. 5. Tunable hydrogel scaffold properties.
Initial conditions of engineered extracellular matrices (ECMs) can be tuned to adjust matrix stiffness, ECM composition (with or without full-length proteins such as laminin), integrin-binding motifs, degradability (matrix metalloproteinase degradable or hydrolytically degradable versus non-degradable) and stress relaxation. Each of these properties can be easily tuned with engineered ECMs to better understand their role in organoid growth, differentiation and morphogenesis. RGD, arginylglycylaspartic acid.
In addition to the ‘blank slate’ approach with hydrogel matrices, other strategies have been pursued to more closely match the tissue-specific ECM context, including the use of decellularized intestinal tissue as a scaffold83,84. These materials, along with the use of collagen I-based hydrogels85,86, can capture other aspects of the fibrillar structure of ECM proteins absent in most amorphous synthetic hydrogels. However, tuning their properties often involves altering protein concentrations; therefore, it is more difficult to decouple the role of mechanics from the chemistry, ligand density and other niche properties. Nonetheless, some tunability is afforded using collagen I, especially regarding fibre self-assembly and dimensions (diameter and density)87. New methods for synthetic fibre generation will help elucidate the role of this understudied niche cue in future organoid studies88.
Although most of the approaches previously discussed have exploited monolithic hydrogels of cylindrical or dome shape, top-down approaches allow for the production of macroscale architectures that can be generated with microfabrication, laser ablation or 3D scaffold biomaterial printing. However, another important clarification of terminology is needed here. In the tissue engineering context, 3D printing has been referred to as both a top-down89 and a bottom-up approach23,25. Indeed, 3D bioprinting of cell-based bio-inks90 constitutes a bottom-up approach because cells are placed at precise points in space as building blocks of an engineered tissue. However, 3D printing of cell-free biomaterial inks followed by cell seeding within printed scaffolds is a top-down approach, fitting well with both the classical and our refined definitions. Therefore, scaffolds generated by 3D printing that match the dimensions and architecture of the intestine that are later seeded with organoid-generating cells are top-down approaches. Culture platforms with macroscale geometries that set initial boundary conditions matching the in vivo intestinal architecture can be generated by microfabrication19 or laser ablation91 and then seeded with cells. An important advantage of these systems is the ability to access the lumen and subject the luminal compartment to peristaltic fluid flow. Micropatterning can also be used to control the initial geometry of PSC-derived tissues and thereby guide self-organization of differentiating cells by spatially controlling cell–cell interactions and local gradients that can shape developing tissues92,93.
Overall, top-down approaches have revealed niche cues that guide self-organization and enable macroscale construction of tissue-engineered intestines. However, the lack of cell-scale spatial control and spatiotemporal control after initial cell seeding limits their utility to match the ever-changing intestinal cell niche.
Bottom-up organoid tissue engineering
Bottom-up approaches allow for precise construction of building blocks composed of cells and niche cues and assembly of those building blocks at precise points in space. The scale of building blocks used from bottom-up tissue engineering ranges from the single-cell level to tightly controlled whole organoids spanning hundreds of microns in size (Fig. 4b).
Methods for the generation of uniform multicellular spheroids using forced aggregation can be extended to the generation of uniform organoids of defined cell density. These methods, including hanging drop94 or commercially available microwells (such as Aggrewells)95, induce cell–cell contact, resulting in spatially defined organoids. For some cell types, such as intestinal organoids, ECM components can also be included to improve growth96. Moreover, microfabrication by soft lithography is a versatile method to generate organoids at pre-determined x–y coordinates on the same z-plane, which simplifies imaging and enables high throughput arrays of homogeneous organoids for drug screening96. Furthermore, arrays of wells of defined diameter and aspect ratio allow the culture of organoids with defined initial geometries19. These defined building blocks can be combined with top-down scaffolds to understand the role of organoid geometry in driving symmetry breaking19. For example, cell packing (in this case internuclear distance) and cell shape, as controlled by engineered well dimensions, were identified as key regulators of spatial heterogeneities in YAP that contribute to symmetry breaking.
Despite not yet being applied in intestinal organoid culture, droplet microfluidics can be used to generate iPSC-derived islet organoids with highly uniform morphometrics97. In addition to precise control over organoid size, these methods can also generate tunable, cell-laden microgels98,99, the properties of which can be modulated to alter single-cell fate or as designer single organoid microenvironments. Microgels can be jammed to generate bulk granular hydrogels that contain individual cell-laden building blocks or to assemble mixed population microgels. Microgels can also be annealed through a variety of surface functionalization methods, including complementary DNA assembly100, guest–host interactions101 and a host of commonly used covalent crosslinking chemistries102,103. Assembly can also be mediated by light, magnetism and acoustic waves104–106, where intricate patterns of building blocks are generated in 3D space. These methods are in a nascent stage for organoid assembly, but one can envisage many applications in the coming years, for example, combining spatially organized organoids with supporting mesenchymal, vascular or immune cells to enhance their complexity and mimic the organoid niche.
In addition to precisely defining the spatial localization of cell-laden microgels, these materials, or similarly functionalized microparticles or microbeads, can also be loaded with biochemical cues and spatially oriented to study the role of local signalling cues or designed for controlled release over time103. For example, WNT-loaded microbeads spatially restrict WNT signalling and direct fate at the single-cell level depending on their relative distance from the source of WNT107. Spatially organized patterns of bound biochemical cues or integrin-binding motifs have also been generated by light-mediated reactions to guide cell behaviour in 3D and would provide interesting new avenues of inquiry for guided organoid growth108–111.
Besides annealing of cell-laden microgels, several other methods are being explored to assemble cellular building blocks in 3D. Indeed, 3D bioprinting is growing as a popular method to establish spatially defined 3D cellular organization and has rapidly advanced to printing with cell-based bio-inks, including microgels112. The benefits of using 3D printing methods for organoid culture have been demonstrated using extrusion-based printing methods for several types of organoids, including cortical113, kidney114 and intestinal organoids115. In the case of the intestine, other supporting cell populations, such as branching, lumenized endothelial cells and mesenchymal cells, have been spatially deposited to generate more complex, multicellular constructs92,115. The generation of 3D-printed cortical organoids also represents a merging of several technologies, including one-pot multi-lineage differentiation with inducible transcription factor overexpression to generate vascularized and spatially patterned cortical organoids113. Although not yet used for intestinal organoid assembly, acoustic116,117, magnetic118,119, optical120,121 and electrical assembly122 have all been used for other cell types and could theoretically be used with intestinal organoids. Finally, cell membranes can be chemically functionalized for synthetic cellular assembly. For example, liposomes have been used to modify cell membranes with bio-orthogonal lipids that are subsequently crosslinked to other cells through bio-click chemistry123. In a complementary way, DNA has been used as a synthetic cell adhesion molecule to create bottom-up 3D tissues with programmable initial size, shape, cellular composition and spatial organization124,125. Synthetic biology methods have allowed the generation of engineered cell surface ligands and receptors based on synthetic NOTCH signalling to generate programmable multicellular structures126.
In sum, bottom-up approaches address some of the limitations of top-down approaches by enabling tight control over cellular spatial organization from the single-cell to the tissue-level scale. These technologies enable exquisite control over cell number, construct size and construct shape to establish reproducible tissue building blocks. Numerous assembly methods can spatially organize these building blocks into complex, biomimetic architectures. However, they are highly dependent on a priori knowledge of the cell composition and spatial organization of the desired end product and will require continual adaptation as knowledge is gained in this area. Furthermore, these methods are often restrictive of cellular self-organization, thereby likely eliminating steps in the natural trajectory that leads to the final tissue form, which could be important for the generation of fully functional tissues.
Middle-out organoid tissue engineering
The tissue-specific niche contains a multitude of cues that can guide morphogenesis at multiple length scales, from the cellular to the whole organoid level, and the optimal cue combination for each step in morphogenesis changes over space and time. These spatiotemporal changes are difficult to control with the available methods; therefore, advances are needed in biomaterial chemistry, processing methods and 4D cell culture (that is, cell culture that is controllable in space and time) to accurately impart these changes.
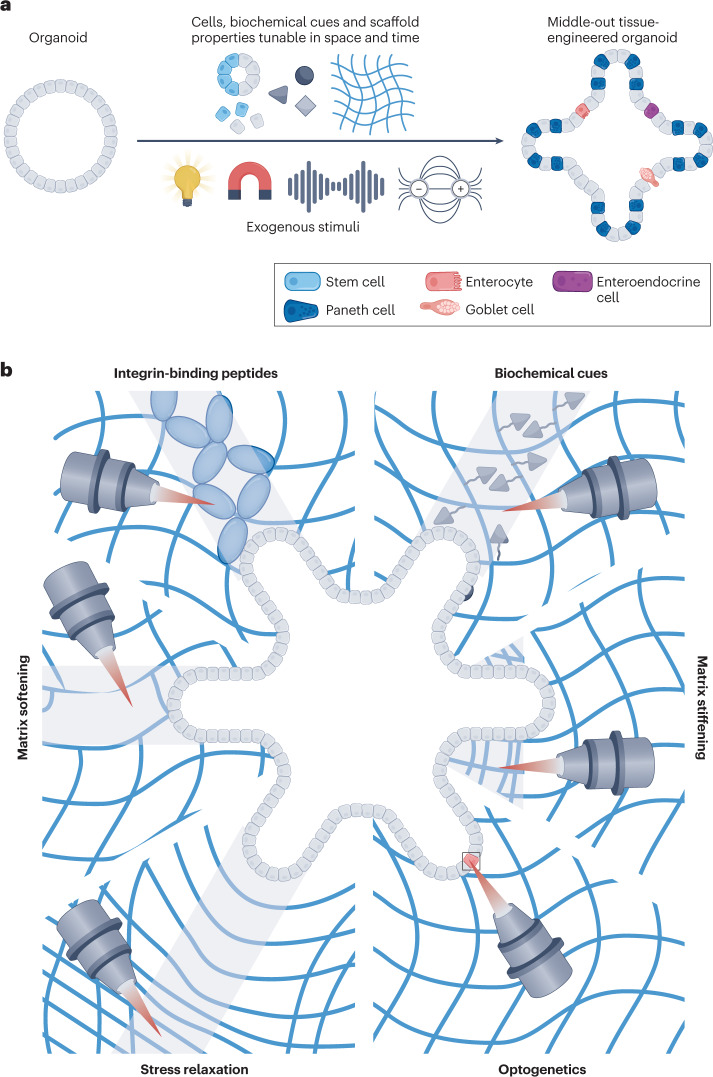
Bottom-up strategies can precisely control initial cellular composition and spatial positioning and can be combined with top-down biomaterials amenable to self-organization to maximize biomimicry115 and generate more homogeneous organoids to study the impact of spatial organization on symmetry breaking19. However, the lack of controllable dynamics with even these powerful combinatorial methods limits the control of morphogenesis. Here, we suggest several nascent and prospective strategies for middle-out tissue engineering of organoids, which we define for the first time in this article, using sequential or reversible reactions or exogenously activatable stimuli to manipulate the spatiotemporal composition of engineered niches to guide organoid morphogenesis (Fig. 6a).
Fig. 6. Middle-out tissue engineering of organoids.
a, Organoids can be manipulated with stimuli-mediated changes through light, magnetism, ultrasound or the application of an electric field to directly modulate cell behaviour or gene expression, localization of biochemical cues, and scaffold properties at any point in space or time to result in deterministic, middle-out tissue-engineered organoids. b, At precise points in time, modular extracellular matrix properties can be induced by external stimuli to make changes to material or cell properties, dynamically tuning the boundary conditions and guiding self-organization. Middle-out tissue engineering with focused laser light can pattern integrin-binding peptides, biochemical cues, matrix stiffening, optogenetic changes, stress relaxation or matrix softening to deterministically control symmetry breaking and crypt formation in intestinal organoids.
To engineer niche properties in space and time, the cells or materials must be sensitive to the cytocompatible stimuli of change, including controlled light doses, magnetic fields, ultrasound or electric fields. Light-based changes have been studied in most detail because these chemistries allow accurate and real-time manipulation of spatiotemporal matrix properties (Fig. 6b). Originally, fibroblasts were encapsulated in proteolytically degradable synthetic hydrogels composed of photocrosslinkable PEG diacrylate macromers, followed by acrylate-functionalized RGD peptide integration into the existing hydrogel with a photoinitiator and controlled light exposure. A two-photon laser scanning confocal microscope was used to visualize cells and precisely pattern regions of RGD attached to the pre-existing hydrogel. These changes facilitated directed cell migration along the region of patterned RGD127. Different chemistries can be used, such as copper-free click reactions to enable initial cell encapsulation, followed by bio-orthogonal thiol-ene photocoupling of RGD at precise points in space and time to sequentially pattern peptides and guide cell migration128. For example, reversible and repeatable peptide and protein exchange in 3D using allyl sulfide addition–fragmentation chain transfer allows PEG hydrogel functionalization to sequentially tether three consecutive peptides and provide additional control over the local niche129. Because these changes can theoretically occur at any point in time and space after initial cell encapsulation, it would be compelling to perform similar experiments with organoids to study the timing and spatial localization of these modifications and understand the role of differential integrin binding in organoid cell fate67,68,70 and morphogenesis72. In essence, the initial hydrogel used for encapsulation could be optimized using the blueprint laid out in the top-down approaches with any of the numerous cytocompatible photochemistries130, and any cell adhesive peptides131 or even full-length proteins132 could be spatiotemporally presented to single cells or geometrically defined regions surrounding an organoid.
Sequential addition of integrin-binding peptides (or proteins) can mimic protein deposition that could be essential in organoid growth. These binding peptides can also be removed with spatiotemporal precision to mimic ECM remodelling and study the role of dynamic (subtractive) ECM cues. For example, photocleavable RGD or full-length proteins can be incorporated into PEG-based hydrogels and, upon controlled light exposure, be removed at precise points in space and time. Removal of RGD enhances ECM production by encapsulated mesenchymal stromal cells and favours chondrogenic fate specification133. Removing integrin-binding peptides and assessing their role in organoid cell fate and subsequent morphogenesis is an interesting line of inquiry applying middle-out principles and co-opting old techniques to answer new questions in organoid biology.
These principles can be used to spatiotemporally define other niche cues that are commonly presented as soluble factors in organoid culture media. Photocaging techniques are widely employed to spatiotemporally expose active groups for chemical photopatterning. Coumarin-caged thiols were initially used in agarose hydrogels, where two-photon irradiation at geometrically defined 3D coordinates resulted in uncaging of thiols, which could react with free diffusing thiol-reactive maleimide-functionalized barnase134. This process can be repeated at different 3D coordinates with maleimide-functionalized streptavidin. Fusion proteins (barstar functionalized and biotin functionalized) can then be diffused throughout the hydrogel to react specifically with photopatterned regions, enabling simultaneous patterning of multiple factors in the presence of cells134. The initial cell-laden hydrogel have been further advanced from agarose to more controllable and cell-compatible PEG-based hydrogels135 and even to naturally derived hydrogels136, which could prove especially useful for organoid culture. Patterning has also progressed from initially requiring a multi-step process with multiple washes to enabling more streamlined, one-step patterning reactions137, as well as the capacity for repeatable and/or reversible patterning with photolabile linkages138, multiple wavelengths with orthogonal chemistries132,138–141 or addition–fragmentation chain transfer142 and the preservation of bioactivity of a range of proteins with sortase-mediated transpeptidation to enable spatiotemporally defined conjugation to biomaterials143.
Additionally, gradients of growth factors144–147 and other niche cues, such as oxygen148–151, can be generated using microfluidics152 or photochemistry using the controlled motion of photomasks140. For example, diffusion-based engineered gradients of WNT3A and/or R-SPONDIN1 in a microfluidic device compartmentalize proliferative cells and crypt formation to the location along the gradient with the higher concentration of these two morphogens, which exist at higher concentrations in the crypt compartment in vivo152. These tools can directly manipulate one or more factors and their spatiotemporal presentation to isolate the effects of morphogen gradients in organoid morphogenesis, which have been previously difficult to manipulate. Such experiments would allow researchers to better understand the role of the spatial distances over which gradients act and the magnitude of the concentration gradient on cell behaviour.
Furthermore, niche-resident cells constantly remodel their surroundings through ECM deposition and degradation, thereby changing the mechanical properties of their microenvironment. These parameters (in this case, stiffness and stress relaxation) can also be tuned with spatiotemporal precision and decoupled from other niche properties using photoresponsive chemistries such as photolabile degradation of crosslinkers133,153 or photostiffening-induced crosslinking154,155. Specifically, photocleavable ortho-nitrobenzyl derivatives have evolved to enable their use as crosslinkers in different types of hydrogels. The use of this photolabile crosslinker has advanced from its initial utility to photodegrade channels to allow for mesenchymal stromal cell migration133, to facilitating matrix modification at the single-cell level156, allowing study of the role of spatial subcellular patterns of matrix stiffness on cell fate decisions157, and guiding neural network growth into eroded channels158. Phototunable matrix softening through photocleavage of ortho-nitrobenzyl crosslinkers has also been used to guide deterministic crypt growth of intestinal organoids19. For example, matrix mechanics can first be tuned to facilitate organoid colony growth by varying the initial matrix stiffness and quantifying colony forming efficiency; photosoftening of spatially defined regions adjacent to live organoids with a laser scanning confocal microscope can then be optimized in terms of the degree of softening, dimensions and density of softened regions to facilitate crypt formation. This controlled matrix softening mimics the ECM softening needed for crypt bud initiation42 and results in crypt formation in the photopatterned regions, which can be quantified by measuring the crypt-forming efficiency and characterized by investigating the localization of crypt-resident cell types, including LGR5+ stem cell and Paneth cells. This type of location-specific matrix manipulation is a particularly powerful tool to study the role of mechanosensing in intestinal organoids because cues regulated by ECM stiffness are crucial in symmetry breaking (for example, YAP18) and can be precisely tuned in space and time. Other hydrogels, including those with allyl sulfide crosslinkers79, could also be used to spatially pattern matrix softening. Because a single photon can initiate multiple events through the allyl sulfide addition–fragmentation chain transfer process, the photodegradation is amplified, resulting in rapid degradation on the order of seconds that could be useful to increase the throughput of middle-out applications or to study and control mechanotransduction, which can occur on the timescale of seconds or less159,160. Spatially restricted photostiffening can be similarly used, for example, by conjugating coumarin-derivatives (7-hydroxycoumarin-3-carboxylate (HCC) or 7-carboxymethoxy-4-methylcoumarin) to a multi-arm PEG backbone or to gelatin to generate photocrosslinkable hydrogels. When mixed with Matrigel, photocrosslinking of HCC–gelatin is initiated after organoid encapsulation to restrict intestinal organoid growth to defined regions161, and HCC–PEG can be used to pattern stiffened regions adjacent to organotypic epithelial lung bud cultures and direct branching162. This so-called ‘gel-in-gel’ printing strategy is useful as a middle-out approach to study the role of local matrix stiffening on symmetry-breaking events because cells adjacent to bud formation have increased cell–ECM interactions58. Similarly, local matrix accumulation can drive branching in other epithelial organoids163, possibly because of increased matrix stiffness in those regions. Stress relaxation has also been suggested to be a key player in symmetry breaking by facilitating heterogeneities in YAP localization. Therefore, it would be beneficial to spatiotemporally regulate stress relaxation at defined regions adjacent to growing organoids, like the methods described for modulating stiffness. Such photo-induced stress relaxation is possible by covalent adaptability with allyl sulfide-based crosslinks that can undergo rearrangement rather than degradation when exposed to the proper conditions164.
Although gradients of soluble cues have well-established roles in development and organoid morphogenesis, the effects of stiffness gradients are less understood. Stiffness gradients, which can be self-generated, have been suggested to have an important role in development and tissue homeostasis165. Although they have not been studied in organoid growth, stiffness gradients can be generated by tuning hydrogel depth166, using a moving photomask to expose different regions to different light doses167 or maskless lithography to generate submicron gradients with any input grey-scale image168.
Light can also be used to specifically and spatiotemporally impact gene expression, rather than changes to the surrounding niche, through optogenetics169. This technique has been applied to an organoid model of neural tube development to locally activate Sonic Hedgehog signalling170. For patterning morphogenesis, OptoShroom3 has been used to control apical constriction with spatiotemporal precision in neural organoids171, and optoYAP172,173 could be similarly used as an initiator of symmetry breaking in intestinal or other organoid models.
Other stimuli, including magnetic fields, electric fields, ultrasound and acoustic waves, compression and fluid flow, can be applied to manipulate various engineered organoids174,175. For example, when subjected to a magnetic field, magnetically labelled cells within neural organoids induce mechanical tissue changes and drive asymmetric growth, thereby enhancing patterning through an external stimulus176. To study the effects of biomimetic peristaltic flow, intestinal organ-on-a-chip technologies have been optimized for organoid-derived cell culture177,178. In future studies, methods to control organoid growth using top-down strategies of geometrically confined self-organization91 could reveal the impact of peristaltic fluid flow on engineered intestinal tissue. Furthermore, the ability to impart local forces at the cellular scale will be useful to advance our understanding of local forces on cell fate and to exogenously control morphogenesis using middle-out methods.
Other emerging techniques use combinatorial approaches, including facilitating exogenous user-defined control using light-responsive chemistries coupled with logic-gate controlled endogenous cell-guided biomaterial remodelling, where unique control over morphogenesis can be achieved179. This technique can spatiotemporally control at multiple scales and pattern multiple cell types independently within one construct. Finally, emerging strategies, including the use of shape-morphing hydrogels, take a unique approach to guiding cellular assembly based on patterns of cell contractility180. These methods could be used to construct organoids with a predictable and controllable shape.
Outlook
Tissue engineering approaches have improved control over organoid culture and have uncovered specific roles for numerous niche cues that could not be studied using standard Matrigel. Moving forward, we posit that applying middle-out approaches to organoids will continue to advance the reproducibility of organoid culture while providing fundamental insights into key regulators of morphogenesis to scale up and improve their translational potential25. Engineering organoid self-organization will likely involve combining strategies to manipulate cells from the subcellular to the tissue-level scale and tuning different niche dynamics independently and simultaneously. Biomaterial strategies are advancing and will be specifically developed for organoid cultures as our understanding of morphogenesis progress. It is likely that added cellular complexity, including mesenchymal/stromal, vascular, nerve and immune cells, will be necessary to build fully functional tissues and to achieve adult-like maturation states for organoids cultured from PSCs. Many tissue-engineered models that include multiple cell types do not account for their tissue-specific nature (for example, tissue-specific vasculature181) or include poorly defined mesenchymal/stromal populations, whose heterogeneity is becoming more appreciated with emerging data sets from cell atlases46. These cell atlases46,182–184 have also proven to be a valuable resource to improve niche-inspired organoid protocols by identifying new factors that have been added to organoid culture media and, importantly, the optimal duration and timing for these factors to improve differentiation and maturation26,64. Technical advancements in multiplexed antibody-based imaging185, spatial sequencing33,186, individual organoid sequencing187 and multimodal phenotyping188 will continue to advance architectural and compositional biomimicry and provide opportunities to mine existing data sets for other regulators of cellular processes. All this information should prove beneficial for labs with different interests (for example, development biology, stem cell biologists, biomaterial scientists, tissue engineers) by providing new directions for the design of tissue-specific culture niches with spatiotemporal precision. User-selected subsets of cells subjected to specific spatiotemporally induced cues could also be collected using photodegradation139 for, theoretically, any type of analysis. Furthermore, advances in real-time microscopy and characterization with large imaging data sets and deep learning have enabled the detection of features, such as shape and cell composition, and quantitative classification of organoid phenotypes45,189–191. These advances, coupled with the ability to generate multi-colour reporter organoids192,193 and to precisely control the spatial location and size of organoids with bottom-up techniques and high throughput arrays96, will likely continue to advance our understanding of key cell players and their localization throughout organoid morphogenesis by generating heat maps with overlays of multiple organoids to improve statistical power compared with traditional imaging methods. Finally, methods for expansion microscopy have been developed to image organoids cultured in 3D matrices and enable super-resolution imaging194.
Complementary approaches can also be used to characterize the dynamic ECM secreted by cells in organoids, including metabolic labelling with non-canonical amino acids coupled with mass spectrometry to understand how cellular remodelling contributes to organoid growth and morphogenesis195. Furthermore, methods to measure local rheological properties196 and forces197 could be used to characterize niche cues and inform the design of new tissue engineering strategies to construct organoids.
Understanding if cell-based remodelling can override engineered material properties or if material properties can override intrinsic genetic programmes will be important for understanding how specific niche cues regulate organoid growth. The competition and/or synergy between biochemical and biophysical cues remains unknown. Middle-out approaches hold great potential to deconvolute these complex pathways. Moreover, finding the proper middle ground for controlled self-organization to enable genetically encoded programmes to proceed with exogenously controlled boundary conditions remains a challenge and will likely require comparisons with in vivo data to ensure biomimetic tissue models are achieved. It is yet unknown if the pathway to the formation of the final tissue construct matters towards its ultimate function. Differentiating multiple cell types separately and then combing them post-differentiation show variations from methods for co-emergence of different cell types using ‘one-pot’ approaches, indicating that the trajectory from multipotency to terminally differentiated cell type might be important198. Finally, achieving the cellular diversity, maturation and full functionality of organoids will require a better understanding of niche dynamics and engineering strategies to perturb or control these dynamics.
Acknowledgements
We thank Peter Dempsey, P. Sean McGrath, F. Max Yavitt, Ella Hushka and Mark Young for useful discussions related to the topics discussed in this manuscript.
Author contributions
M.R.B. and K.S.A. conceptualized, wrote, and edited the manuscript.
Peer review
Peer review information
Nature Reviews Bioengineering thanks Nuria Montserrat, Virgile Viasnoff, who co-reviewed with Geetika Sahni, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 2.Eiraku M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 4.Hsia GSP, Esposito J, da Rocha LA, Ramos SLG, Okamoto OK. Clinical application of human induced pluripotent stem cell-derived organoids as an alternative to organ transplantation. Stem Cell Int. 2021;2021:6632160. doi: 10.1155/2021/6632160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calandrini C, Drost J. Normal and tumor-derived organoids as a drug screening platform for tumor-specific drug vulnerabilities. STAR Protoc. 2022;3:101079. doi: 10.1016/j.xpro.2021.101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim J, Koo B-K, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 2020;21:571–584. doi: 10.1038/s41580-020-0259-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lancaster MA, Huch M. Disease modelling in human organoids. Dis. Model. Mech. 2019;12:dmm039347. doi: 10.1242/dmm.039347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu JL, Todhunter ME, LaBarge MA, Gartner ZJ. Opportunities for organoids as new models of aging. J. Cell Biol. 2017;217:39–50. doi: 10.1083/jcb.201709054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–955. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe S, et al. Transplantation of intestinal organoids into a mouse model of colitis. Nat. Protoc. 2022;17:649–671. doi: 10.1038/s41596-021-00658-3. [DOI] [PubMed] [Google Scholar]
- 11.Spence JR, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lancaster MA, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang P, Wang J, Conklin BR, Healy KE, Ma Z. Generation of spatial-patterned early-developing cardiac organoids using human pluripotent stem cells. Nat. Protoc. 2018;13:723–737. doi: 10.1038/nprot.2018.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebe T, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 15.Hofer M, Lutolf MP. Engineering organoids. Nat. Rev. Mater. 2021;6:402–420. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin X, et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boonekamp KE, Dayton TL, Clevers H. Intestinal organoids as tools for enriching and studying specific and rare cell types: advances and future directions. J. Mol. Cell Biol. 2020;12:562–568. doi: 10.1093/jmcb/mjaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serra D, et al. Self-organization and symmetry breaking in intestinal organoid development. Nature. 2019;569:66–72. doi: 10.1038/s41586-019-1146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gjorevski N, et al. Tissue geometry drives deterministic organoid patterning. Science. 2022;375:eaaw9021. doi: 10.1126/science.aaw9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FitzGerald G, et al. The future of humans as model organisms. Science. 2018;361:552–553. doi: 10.1126/science.aau7779. [DOI] [PubMed] [Google Scholar]
- 21.Dekkers JF, et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 2013;19:939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- 22.Monteil V, et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913.e7. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marti-Figueroa CR, Ashton RS. The case for applying tissue engineering methodologies to instruct human organoid morphogenesis. Acta Biomater. 2017;54:35–44. doi: 10.1016/j.actbio.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurent J, et al. Convergence of microengineering and cellular self-organization towards functional tissue manufacturing. Nat. Biomed. Eng. 2017;1:939–956. doi: 10.1038/s41551-017-0166-x. [DOI] [PubMed] [Google Scholar]
- 25.Brassard JA, Lutolf MP. Engineering stem cell self-organization to build better organoids. Cell Stem Cell. 2019;24:860–876. doi: 10.1016/j.stem.2019.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Kilik U, et al. Maturation of human intestinal epithelium from pluripotency in vitro. bioRxiv. 2021 doi: 10.1101/2021.09.24.460132. [DOI] [Google Scholar]
- 27.Farin HF, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 28.Shyer AE, Huycke TR, Lee C, Mahadevan L, Tabin CJ. Bending gradients: how the intestinal stem cell gets its home. Cell. 2015;161:569–580. doi: 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolpert L. Positional information and the spatial pattern of cellular differentiation. J. Theor. Biol. 1969;25:1–47. doi: 10.1016/S0022-5193(69)80016-0. [DOI] [PubMed] [Google Scholar]
- 30.Shyer Amy E, et al. Villification: how the gut gets its villi. Science. 2013;342:212–218. doi: 10.1126/science.1238842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watson CL, et al. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cakir B, et al. Engineering of human brain organoids with a functional vascular-like system. Nat. Methods. 2019;16:1169–1175. doi: 10.1038/s41592-019-0586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niec RE, et al. Lymphatics act as a signaling hub to regulate intestinal stem cell activity. Cell Stem Cell. 2022 doi: 10.1016/j.stem.2022.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–1890. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 35.Aisenbrey EA, Murphy WL. Synthetic alternatives to Matrigel. Nat. Rev. Mater. 2020;5:539–551. doi: 10.1038/s41578-020-0199-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langer R, Vacanti Joseph P. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 37.Cao Y, Vacanti JP, Paige KT, Upton J, Vacanti CA. Transplantation of chondrocytes utilizing a polymer-cell construct to produce tissue-engineered cartilage in the shape of a human ear. Plast. Reconstr. Surg. 1997;100:297–302. doi: 10.1097/00006534-199708000-00001. [DOI] [PubMed] [Google Scholar]
- 38.Elbert DL. Bottom-up tissue engineering. Curr. Opin. Biotechnol. 2011;22:674–680. doi: 10.1016/j.copbio.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichol JW, Khademhosseini A. Modular tissue engineering: engineering biological tissues from the bottom up. Soft Matter. 2009;5:1312–1319. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 42.Gjorevski N, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–564. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 43.Jowett GM, et al. ILC1 drive intestinal epithelial and matrix remodelling. Nat. Mater. 2021;20:250–259. doi: 10.1038/s41563-020-0783-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corrò C, Novellasdemunt L, Li VSW. A brief history of organoids. Am. J. Physiol. Cell Physiology. 2020;319:C151–C165. doi: 10.1152/ajpcell.00120.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukonin I, et al. Phenotypic landscape of intestinal organoid regeneration. Nature. 2020;586:275–280. doi: 10.1038/s41586-020-2776-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu Q, et al. Charting human development using a multi-endodermal organ atlas and organoid models. Cell. 2021;184:3281–3298.e22. doi: 10.1016/j.cell.2021.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simons BD, Clevers H. Stem cell self-renewal in intestinal crypt. Exp. Cell Res. 2011;317:2719–2724. doi: 10.1016/j.yexcr.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 48.Meran L, Baulies A, Li VSW. Intestinal stem cell niche: the extracellular matrix and cellular components. Stem Cell Int. 2017;2017:11. doi: 10.1155/2017/7970385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beumer J, Clevers H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 2020 doi: 10.1038/s41580-020-0278-0. [DOI] [PubMed] [Google Scholar]
- 50.Scarpa S, et al. Expression and synthesis of fibronectin and laminin by an intestinal epithelial cell line. Tissue Cell. 1988;20:305–312. doi: 10.1016/0040-8166(88)90066-3. [DOI] [PubMed] [Google Scholar]
- 51.Simo P, et al. Changes in the expression of laminin during intestinal development. Development. 1991;112:477. doi: 10.1242/dev.112.2.477. [DOI] [PubMed] [Google Scholar]
- 52.Simon-Assmann P, et al. Differential expression of laminin isoforms and α6-β4 integrin subunits in the developing human and mouse intestine. Dev. Dyn. 1994;201:71–85. doi: 10.1002/aja.1002010108. [DOI] [PubMed] [Google Scholar]
- 53.Teller IC, et al. Laminins in the developing and adult human small intestine: relation with the functional absorptive unit. Dev. Dyn. 2007;236:1980–1990. doi: 10.1002/dvdy.21186. [DOI] [PubMed] [Google Scholar]
- 54.Quaroni A, Isselbacher KJ, Ruoslahti E. Fibronectin synthesis by epithelial crypt cells of rat small intestine. Proc. Natl Acad. Sci. USA. 1978;75:5548. doi: 10.1073/pnas.75.11.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moya IM, Halder G. Hippo–YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019;20:211–226. doi: 10.1038/s41580-018-0086-y. [DOI] [PubMed] [Google Scholar]
- 56.Yui S, et al. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell. 2018;22:35–49.e7. doi: 10.1016/j.stem.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huels DJ, Medema JP. Think about the environment: cellular reprogramming by the extracellular matrix. Cell Stem Cell. 2018;22:7–9. doi: 10.1016/j.stem.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 58.Langlands AJ, et al. Paneth cell-rich regions separated by a cluster of Lgr5+ cells initiate crypt fission in the intestinal stem cell niche. PLoS Biol. 2016;14:e1002491. doi: 10.1371/journal.pbio.1002491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okamoto R. Epithelial regeneration in inflammatory bowel diseases. Inflamm. Regen. 2011;31:275–281. doi: 10.2492/inflammregen.31.275. [DOI] [Google Scholar]
- 60.Pérez-González C, et al. Mechanical compartmentalization of the intestinal organoid enables crypt folding and collective cell migration. Nat. Cell Biol. 2021;23:745–757. doi: 10.1038/s41556-021-00699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, et al. Volumetric compression induces intracellular crowding to control intestinal organoid growth via Wnt/β-catenin signaling. Cell Stem Cell. 2021;28:63–78.e7. doi: 10.1016/j.stem.2020.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Q, et al. Cell fate coordinates mechano-osmotic forces in intestinal crypt formation. Nat. Cell Biol. 2021;23:733–744. doi: 10.1038/s41556-021-00700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujii M, et al. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell. 2018;23:787–793.e6. doi: 10.1016/j.stem.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 64.Childs CJ, et al. A development-inspired niche for homeostatic human mini-intestines. bioRxiv. 2022 doi: 10.1101/2022.06.12.495827. [DOI] [Google Scholar]
- 65.Poling HM, et al. Mechanically induced development and maturation of human intestinal organoids in vivo. Nat. Biomed. Eng. 2018;2:429–442. doi: 10.1038/s41551-018-0243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kratochvil MJ, et al. Engineered materials for organoid systems. Nat. Rev. Mater. 2019;4:606–622. doi: 10.1038/s41578-019-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanton AE, Tong X, Yang F. Extracellular matrix type modulates mechanotransduction of stem cells. Acta Biomater. 2019;96:310–320. doi: 10.1016/j.actbio.2019.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keselowsky BG, Collard DM, García AJ. Integrin binding specificity regulates biomaterial surface chemistry effects on cell differentiation. Proc. Natl Acad. Sci. USA. 2005;102:5953–5957. doi: 10.1073/pnas.0407356102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cruz-Acuña R, et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017;19:1326–1335. doi: 10.1038/ncb3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Below CR, et al. A microenvironment-inspired synthetic three-dimensional model for pancreatic ductal adenocarcinoma organoids. Nat. Mater. 2022;21:110–119. doi: 10.1038/s41563-021-01085-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hernandez-Gordillo V, et al. Fully synthetic matrices for in vitro culture of primary human intestinal enteroids and endometrial organoids. Biomaterials. 2020;254:120125. doi: 10.1016/j.biomaterials.2020.120125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li S, et al. Hydrogels with precisely controlled integrin activation dictate vascular patterning and permeability. Nat. Mater. 2017;16:953–961. doi: 10.1038/nmat4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Madl CM, et al. Maintenance of neural progenitor cell stemness in 3D hydrogels requires matrix remodelling. Nat. Mater. 2017;16:1233–1242. doi: 10.1038/nmat5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 75.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat. Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cameron AR, Frith JE, Cooper-White JJ. The influence of substrate creep on mesenchymal stem cell behaviour and phenotype. Biomaterials. 2011;32:5979–5993. doi: 10.1016/j.biomaterials.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 77.Chrisnandy A, Blondel D, Rezakhani S, Broguiere N, Lutolf MP. Synthetic dynamic hydrogels promote degradation-independent in vitro organogenesis. Nat. Mater. 2022;21:479–487. doi: 10.1038/s41563-021-01136-7. [DOI] [PubMed] [Google Scholar]
- 78.Elosegui-Artola A, et al. Matrix viscoelasticity controls spatiotemporal tissue organization. Nat. Mater. 2022 doi: 10.1038/s41563-022-01400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hushka EA, Yavitt FM, Brown TE, Dempsey PJ, Anseth KS. Relaxation of extracellular matrix forces directs crypt formation and architecture in intestinal organoids. Adv. Healthc. Mater. 2020;9:1901214. doi: 10.1002/adhm.201901214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rezakhani S, Gjorevski N, Lutolf MP. Low-defect thiol-michael addition hydrogels as matrigel substitutes for epithelial organoid derivation. Adv. Funct. Mater. 2020;30:2000761. doi: 10.1002/adfm.202000761. [DOI] [Google Scholar]
- 81.Hunt DR, et al. Engineered matrices enable the culture of human patient-derived intestinal organoids. Adv. Sci. 2021;8:2004705. doi: 10.1002/advs.202004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yavitt FM, et al. The effect of thiol structure on allyl sulfide photodegradable hydrogels and their application as a degradable scaffold for organoid passaging. Adv. Mater. 2020;32:1905366. doi: 10.1002/adma.201905366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim S, et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat. Commun. 2022;13:1692. doi: 10.1038/s41467-022-29279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim S, et al. Intestinal extracellular matrix hydrogels to generate intestinal organoids for translational applications. J. Ind. Eng. Chem. 2022;107:155–164. doi: 10.1016/j.jiec.2021.11.044. [DOI] [Google Scholar]
- 85.Sachs N, Tsukamoto Y, Kujala P, Peters PJ, Clevers H. Intestinal epithelial organoids fuse to form self-organizing tubes in floating collagen gels. Development. 2017;144:1107–1112. doi: 10.1242/dev.143933. [DOI] [PubMed] [Google Scholar]
- 86.Curvello R, Alves D, Abud HE, Garnier G. A thermo-responsive collagen-nanocellulose hydrogel for the growth of intestinal organoids. Mater. Sci. Eng. C. 2021;124:112051. doi: 10.1016/j.msec.2021.112051. [DOI] [PubMed] [Google Scholar]
- 87.Nguyen-Ngoc KV, Ewald AJ. Mammary ductal elongation and myoepithelial migration are regulated by the composition of the extracellular matrix. J. Microsc. 2013;251:212–223. doi: 10.1111/jmi.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Davidson MD, et al. Programmable and contractile materials through cell encapsulation in fibrous hydrogel assemblies. Sci. Adv. 2021;7:eabi8157. doi: 10.1126/sciadv.abi8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Song HHG, Rumma RT, Ozaki CK, Edelman ER, Chen CS. Vascular tissue engineering: progress, challenges, and clinical promise. Cell Stem Cell. 2018;22:340–354. doi: 10.1016/j.stem.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Groll J, et al. A definition of bioinks and their distinction from biomaterial inks. Biofabrication. 2018;11:013001. doi: 10.1088/1758-5090/aaec52. [DOI] [PubMed] [Google Scholar]
- 91.Nikolaev M, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–578. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 92.Sahni G, et al. A micropatterned human-specific neuroepithelial tissue for modeling gene and drug-induced neurodevelopmental defects. Adv. Sci. 2021;8:2001100. doi: 10.1002/advs.202001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uchida H, et al. A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells. JCI Insight. 2017 doi: 10.1172/jci.insight.86492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Panek M, Grabacka M, Pierzchalska M. The formation of intestinal organoids in a hanging drop culture. Cytotechnology. 2018;70:1085–1095. doi: 10.1007/s10616-018-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yoon S-J, et al. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brandenberg N, et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 2020;4:863–874. doi: 10.1038/s41551-020-0565-2. [DOI] [PubMed] [Google Scholar]
- 97.Liu H, et al. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. 2020;7:1903739. doi: 10.1002/advs.201903739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan HF, et al. Rapid formation of multicellular spheroids in double-emulsion droplets with controllable microenvironment. Sci. Rep. 2013;3:3462. doi: 10.1038/srep03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mao AS, et al. Deterministic encapsulation of single cells in thin tunable microgels for niche modelling and therapeutic delivery. Nat. Mater. 2017;16:236–243. doi: 10.1038/nmat4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li CY, Wood DK, Hsu CM, Bhatia SN. DNA-templated assembly of droplet-derived PEG microtissues. Lab Chip. 2011;11:2967–2975. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Harada A, Kobayashi R, Takashima Y, Hashidzume A, Yamaguchi H. Macroscopic self-assembly through molecular recognition. Nat. Chem. 2011;3:34–37. doi: 10.1038/nchem.893. [DOI] [PubMed] [Google Scholar]
- 102.Caldwell AS, Aguado BA, Anseth KS. Designing microgels for cell culture and controlled assembly of tissue microenvironments. Adv. Funct. Mater. 2020;30:1907670. doi: 10.1002/adfm.201907670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daly AC, Riley L, Segura T, Burdick JA. Hydrogel microparticles for biomedical applications. Nat. Rev. Mater. 2020;5:20–43. doi: 10.1038/s41578-019-0148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheikhi A, et al. Microfluidic-enabled bottom-up hydrogels from annealable naturally-derived protein microbeads. Biomaterials. 2019;192:560–568. doi: 10.1016/j.biomaterials.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ouyang L, Armstrong JPK, Salmeron-Sanchez M, Stevens MM. Assembling living building blocks to engineer complex tissues. Adv. Funct. Mater. 2020;30:1909009. doi: 10.1002/adfm.201909009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Armstrong JPK, Stevens MM. Using remote fields for complex tissue engineering. Trends Biotechnol. 2020;38:254–263. doi: 10.1016/j.tibtech.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Habib SJ, et al. A localized wnt signal orients asymmetric stem cell division in vitro. Science. 2013;339:1445–1448. doi: 10.1126/science.1231077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luo Y, Shoichet MS. A photolabile hydrogel for guided three-dimensional cell growth and migration. Nat. Mater. 2004;3:249–253. doi: 10.1038/nmat1092. [DOI] [PubMed] [Google Scholar]
- 109.Aizawa Y, Wylie R, Shoichet M. Endothelial cell guidance in 3D patterned scaffolds. Adv. Mater. 2010;22:4831–4835. doi: 10.1002/adma.201001855. [DOI] [PubMed] [Google Scholar]
- 110.Hahn MS, Miller JS, West JL. Three-dimensional biochemical and biomechanical patterning of hydrogels for guiding cell behavior. Adv. Mater. 2006;18:2679–2684. doi: 10.1002/adma.200600647. [DOI] [Google Scholar]
- 111.Tam RY, Smith LJ, Shoichet MS. Engineering cellular microenvironments with photo- and enzymatically responsive hydrogels: toward biomimetic 3D cell culture models. Acc. Chem. Res. 2017;50:703–713. doi: 10.1021/acs.accounts.6b00543. [DOI] [PubMed] [Google Scholar]
- 112.Hull SM, Brunel LG, Heilshorn SC. 3D Bioprinting of cell-laden hydrogels for improved biological functionality. Adv. Mater. 2022;34:2103691. doi: 10.1002/adma.202103691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Skylar-Scott MA, et al. Orthogonally induced differentiation of stem cells for the programmatic patterning of vascularized organoids and bioprinted tissues. Nat. Biomed. Eng. 2022;6:449–462. doi: 10.1038/s41551-022-00856-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lawlor KT, et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater. 2021;20:260–271. doi: 10.1038/s41563-020-00853-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Brassard JA, Nikolaev M, Hübscher T, Hofer M, Lutolf MP. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 2021;20:22–29. doi: 10.1038/s41563-020-00803-5. [DOI] [PubMed] [Google Scholar]
- 116.Zhu, Y. et al. In Organoids: Stem Cells, Structure, and Function (ed Kursad Turksen) 301–312 (Springer New York, 2019).
- 117.Chen P, Güven S, Usta OB, Yarmush ML, Demirci U. Biotunable acoustic node assembly of organoids. Adv. Healthc. Mater. 2015;4:1937–1943. doi: 10.1002/adhm.201500279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Parfenov VA, et al. Magnetic levitational bioassembly of 3D tissue construct in space. Sci. Adv. 2020;6:eaba4174. doi: 10.1126/sciadv.aba4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ferreira JN, et al. A magnetic three-dimensional levitated primary cell culture system for the development of secretory salivary gland-like organoids. J. Tissue Eng. Regen. Med. 2019;13:495–508. doi: 10.1002/term.2809. [DOI] [PubMed] [Google Scholar]
- 120.Kirkham GR, et al. Precision assembly of complex cellular microenvironments using holographic optical tweezers. Sci. Rep. 2015;5:8577. doi: 10.1038/srep08577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Au - Yamazaki T, et al. Manipulating living cells to construct stable 3D cellular assembly without artificial scaffold. J. Vis. Exp. 2018 doi: 10.3791/57815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sebastian A, Buckle A-M, Markx GH. Tissue engineering with electric fields: immobilization of mammalian cells in multilayer aggregates using dielectrophoresis. Biotechnol. Bioeng. 2007;98:694–700. doi: 10.1002/bit.21416. [DOI] [PubMed] [Google Scholar]
- 123.O’Brien PJ, Luo W, Rogozhnikov D, Chen J, Yousaf MN. Spheroid and tissue assembly via click chemistry in microfluidic flow. Bioconjug. Chem. 2015;26:1939–1949. doi: 10.1021/acs.bioconjchem.5b00376. [DOI] [PubMed] [Google Scholar]
- 124.Gartner GJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl Acad. Sci. USA. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Todhunter ME, et al. Programmed synthesis of three-dimensional tissues. Nat. Methods. 2015;12:975–981. doi: 10.1038/nmeth.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA. Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science. 2018;361:156–162. doi: 10.1126/science.aat0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee S-H, Moon JJ, West JL. Three-dimensional micropatterning of bioactive hydrogels via two-photon laser scanning photolithography for guided 3D cell migration. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.DeForest CA, Polizzotti BD, Anseth KS. Sequential click reactions for synthesizing and patterning three-dimensional cell microenvironments. Nat. Mater. 2009;8:659–664. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gandavarapu NR, Azagarsamy MA, Anseth KS. Photo-click living strategy for controlled, reversible exchange of biochemical ligands. Adv. Mater. 2014;26:2521–2526. doi: 10.1002/adma.201304847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brown TE, Anseth KS. Spatiotemporal hydrogel biomaterials for regenerative medicine. Chem. Soc. Rev. 2017;46:6532–6552. doi: 10.1039/C7CS00445A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huettner N, Dargaville TR, Forget A. Discovering cell-adhesion peptides in tissue engineering: beyond RGD. Trends Biotechnol. 2018;36:372–383. doi: 10.1016/j.tibtech.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 132.DeForest CA, Tirrell DA. A photoreversible protein-patterning approach for guiding stem cell fate in three-dimensional gels. Nat. Mater. 2015;14:523–531. doi: 10.1038/nmat4219. [DOI] [PubMed] [Google Scholar]
- 133.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wylie RG, et al. Spatially controlled simultaneous patterning of multiple growth factors in three-dimensional hydrogels. Nat. Mater. 2011;10:799–806. doi: 10.1038/nmat3101. [DOI] [PubMed] [Google Scholar]
- 135.Mosiewicz KA, et al. In situ cell manipulation through enzymatic hydrogel photopatterning. Nat. Mater. 2013;12:1072–1078. doi: 10.1038/nmat3766. [DOI] [PubMed] [Google Scholar]
- 136.Batalov I, Stevens KR, DeForest CA. Photopatterned biomolecule immobilization to guide three-dimensional cell fate in natural protein-based hydrogels. Proc. Natl Acad. Sci. USA. 2021;118:e2014194118. doi: 10.1073/pnas.2014194118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Broguiere N, et al. Morphogenesis guided by 3D patterning of growth factors in biological matrices. Adv. Mater. 2020;32:1908299. doi: 10.1002/adma.201908299. [DOI] [PubMed] [Google Scholar]
- 138.Azagarsamy MA, Anseth KS. Wavelength-controlled photocleavage for the orthogonal and sequential release of multiple proteins. Angew. Chem. Int. Ed. 2013;52:13803–13807. doi: 10.1002/anie.201308174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.DeForest CA, Anseth KS. Cytocompatible click-based hydrogels with dynamically tunable properties through orthogonal photoconjugation and photocleavage reactions. Nat. Chem. 2011;3:925–931. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.DeForest CA, Anseth KS. Photoreversible patterning of biomolecules within click-based hydrogels. Angew. Chem. Int. Ed. 2012;51:1816–1819. doi: 10.1002/anie.201106463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin C-C, Ki CS, Shih H. Thiol-norbornene photo-click hydrogels for tissue engineering applications. J. Appl. Polym. Sci. 2015;132:41563. doi: 10.1002/app.41563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Grim JC, et al. A reversible and repeatable thiol–ene bioconjugation for dynamic patterning of signaling proteins in hydrogels. ACS Cent. Sci. 2018;4:909–916. doi: 10.1021/acscentsci.8b00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shadish JA, Benuska GM, DeForest CA. Bioactive site-specifically modified proteins for 4D patterning of gel biomaterials. Nat. Mater. 2019;18:1005–1014. doi: 10.1038/s41563-019-0367-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Uzel SGM, et al. Simultaneous or sequential orthogonal gradient formation in a 3D cell culture microfluidic platform. Small. 2016;12:612–622. doi: 10.1002/smll.201501905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Manfrin A, et al. Engineered signaling centers for the spatially controlled patterning of human pluripotent stem cells. Nat. Methods. 2019;16:640–648. doi: 10.1038/s41592-019-0455-2. [DOI] [PubMed] [Google Scholar]
- 146.Cederquist GY, et al. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Demers CJ, et al. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development. 2016;143:1884–1892. doi: 10.1242/dev.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lewis DM, Blatchley MR, Park KM, Gerecht S. O2-controllable hydrogels for studying cellular responses to hypoxic gradients in three dimensions in vitro and in vivo. Nat. Protoc. 2017;12:1620–1638. doi: 10.1038/nprot.2017.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Blatchley MR, et al. Discretizing three-dimensional oxygen gradients to modulate and investigate cellular processes. Adv. Sci. 2021;8:2100190. doi: 10.1002/advs.202100190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rodenhizer D, et al. A three-dimensional engineered tumour for spatial snapshot analysis of cell metabolism and phenotype in hypoxic gradients. Nat. Mater. 2016;15:227–234. doi: 10.1038/nmat4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Koens R, et al. Microfluidic platform for three-dimensional cell culture under spatiotemporal heterogeneity of oxygen tension. APL Bioeng. 2020;4:016106. doi: 10.1063/1.5127069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Attayek PJ, et al. In vitro polarization of colonoids to create an intestinal stem cell compartment. PLoS One. 2016;11:e0153795. doi: 10.1371/journal.pone.0153795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rosales AM, Anseth KS. The design of reversible hydrogels to capture extracellular matrix dynamics. Nat. Rev. Mater. 2016;1:15012. doi: 10.1038/natrevmats.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Guvendiren M, Burdick JA. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun. 2012;3:792. doi: 10.1038/ncomms1792. [DOI] [PubMed] [Google Scholar]
- 155.Stowers RS, Allen CS, Suggs LJ. Dynamic phototuning of 3D hydrogel stiffness. Proc. Natl Acad. Sci. USA. 2015;112:1953–1958. doi: 10.1073/pnas.1421897112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tibbitt MW, Kloxin AM, Dyamenahalli KU, Anseth KS. Controlled two-photon photodegradation of PEG hydrogels to study and manipulate subcellular interactions on soft materials. Soft Matter. 2010;6:5100–5108. doi: 10.1039/c0sm00174k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Yang C, et al. Spatially patterned matrix elasticity directs stem cell fate. Proc. Natl Acad. Sci. USA. 2016;113:E4439–E4445. doi: 10.1073/pnas.1609731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.McKinnon DD, Brown TE, Kyburz KA, Kiyotake E, Anseth KS. Design and characterization of a synthetically accessible, photodegradable hydrogel for user-directed formation of neural networks. Biomacromolecules. 2014;15:2808–2816. doi: 10.1021/bm500731b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Brown TE, Marozas IA, Anseth KS. Amplified photodegradation of cell-laden hydrogels via an addition–fragmentation chain transfer reaction. Adv. Mater. 2017;29:1605001. doi: 10.1002/adma.201605001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Urciuolo A, et al. Intravital three-dimensional bioprinting. Nat. Biomed. Eng. 2020;4:901–915. doi: 10.1038/s41551-020-0568-z. [DOI] [PubMed] [Google Scholar]
- 162.Elvassore N, et al. Four-dimensional hydrogel-in-hydrogel bioprinting for the spatiotemporal control of organoid and organotypic cultures. Research Square. 2021 doi: 10.21203/rs.3.rs-952864/v1. [DOI] [Google Scholar]
- 163.Nerger BA, et al. Local accumulation of extracellular matrix regulates global morphogenetic patterning in the developing mammary gland. Curr. Biol. 2021;31:1903–1917.e6. doi: 10.1016/j.cub.2021.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marozas IA, Cooper-White JJ, Anseth KS. Photo-induced viscoelasticity in cytocompatible hydrogel substrates. N. J. Phys. 2019;21:045004. doi: 10.1088/1367-2630/ab1309. [DOI] [Google Scholar]
- 165.Shellard A, Mayor R. Collective durotaxis along a self-generated stiffness gradient in vivo. Nature. 2021;600:690–694. doi: 10.1038/s41586-021-04210-x. [DOI] [PubMed] [Google Scholar]
- 166.Kloxin AM, Tibbitt MW, Kasko AM, Fairbairn JA, Anseth KS. Tunable hydrogels for external manipulation of cellular microenvironments through controlled photodegradation. Adv. Mater. 2010;22:61–66. doi: 10.1002/adma.200900917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kloxin AM, Benton JA, Anseth KS. In situ elasticity modulation with dynamic substrates to direct cell phenotype. Biomaterials. 2010;31:1–8. doi: 10.1016/j.biomaterials.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Norris SCP, Tseng P, Kasko AM. Direct gradient photolithography of photodegradable hydrogels with patterned stiffness control with submicrometer resolution. ACS Biomater. Sci. Eng. 2016;2:1309–1318. doi: 10.1021/acsbiomaterials.6b00237. [DOI] [PubMed] [Google Scholar]
- 169.Guo Q, et al. Light activated cell migration in synthetic extracellular matrices. Biomaterials. 2012;33:8040–8046. doi: 10.1016/j.biomaterials.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Legnini I, et al. Spatio-temporal, optogenetic control of gene expression in organoids. bioRxiv. 2022 doi: 10.1101/2021.09.26.461850. [DOI] [Google Scholar]
- 171.Martínez-Ara G, et al. Optogenetic control of apical constriction induces synthetic morphogenesis in mammalian tissues. Nat. Commun. 2022;13:5400. doi: 10.1038/s41467-022-33115-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Illes B, et al. Spatio-selective activation of nuclear translocation of YAP with light directs invasion of cancer cell spheroids. iScience. 2021;24:102185. doi: 10.1016/j.isci.2021.102185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Toh PJY, et al. Optogenetic control of YAP cellular localisation and function. EMBO Rep. 2022;23:e54401. doi: 10.15252/embr.202154401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gelmi A, Schutt CE. Stimuli-responsive biomaterials: scaffolds for stem cell control. Adv. Healthc. Mater. 2021;10:2001125. doi: 10.1002/adhm.202001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Park SE, Georgescu A, Huh D. Organoids-on-a-chip. Science. 2019;364:960–965. doi: 10.1126/science.aaw7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Abdel Fattah AR, Kolaitis N, Van Daele K, Rustandi AG, Ranga A. Local actuation of organoids by magnetic nanoparticles. bioRxiv. 2022 doi: 10.1101/2022.03.17.484696. [DOI] [Google Scholar]
- 177.Kasendra M, et al. Development of a primary human small intestine-on-a-chip using biopsy-derived organoids. Sci. Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Workman MJ, et al. Enhanced utilization of induced pluripotent stem cell–derived human intestinal organoids using microengineered chips. Cell. Mol. Gastroenterol. Hepatol. 2018;5:669–677.e2. doi: 10.1016/j.jcmgh.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Badeau BA, Comerford MP, Arakawa CK, Shadish JA, DeForest CA. Engineered modular biomaterial logic gates for environmentally triggered therapeutic delivery. Nat. Chem. 2018;10:251–258. doi: 10.1038/nchem.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Viola JM, et al. Guiding cell network assembly using shape-morphing hydrogels. Adv. Mater. 2020;32:2002195. doi: 10.1002/adma.202002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Nguyen J, Lin Y-Y, Gerecht S. The next generation of endothelial differentiation: tissue-specific ECs. Cell Stem Cell. 2021;28:1188–1204. doi: 10.1016/j.stem.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 182.Holloway EM, et al. Mapping development of the human intestinal niche at single-cell resolution. Cell Stem Cell. 2021;28:568–580.e4. doi: 10.1016/j.stem.2020.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bock C, et al. The organoid cell atlas. Nat. Biotechnol. 2021;39:13–17. doi: 10.1038/s41587-020-00762-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Regev A, et al. The human cell atlas. eLife. 2017;6:e27041. doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Hickey JW, et al. Spatial mapping of protein composition and tissue organization: a primer for multiplexed antibody-based imaging. Nat. Methods. 2022;19:284–295. doi: 10.1038/s41592-021-01316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.He Z, et al. Lineage recording in human cerebral organoids. Nat. Methods. 2022;19:90–99. doi: 10.1038/s41592-021-01344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bues J, et al. Deterministic scRNA-seq captures variation in intestinal crypt and organoid composition. Nat. Methods. 2022;19:323–330. doi: 10.1038/s41592-021-01391-1. [DOI] [PubMed] [Google Scholar]
- 188.Wahle P, et al. Multimodal spatiotemporal phenotyping of human organoid development. bioRxiv. 2022 doi: 10.1101/2022.03.16.484396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.de Medeiros G, et al. Multiscale light-sheet organoid imaging framework. Nat. Commun. 2022;13:4864. doi: 10.1038/s41467-022-32465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Beck LE, et al. Systematically quantifying morphological features reveals constraints on organoid phenotypes. Cell Syst. 2022 doi: 10.1016/j.cels.2022.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Beghin A, et al. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods. 2022 doi: 10.1038/s41592-022-01508-0. [DOI] [PubMed] [Google Scholar]
- 192.Artegiani B, et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR–Cas9 precision genome editing. Nat. Cell Biol. 2020;22:321–331. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 193.Martyn I, Kanno TY, Ruzo A, Siggia ED, Brivanlou AH. Self-organization of a human organizer by combined Wnt and Nodal signalling. Nature. 2018;558:132–135. doi: 10.1038/s41586-018-0150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Blatchley MR, et al. In situ super-resolution imaging of organoids and extracellular matrix interactions via phototransfer by allyl sulfide exchange-expansion microscopy (PhASE-ExM) Adv. Mater. 2022;34:2109252. doi: 10.1002/adma.202109252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Loebel C, et al. Metabolic labeling of secreted matrix to investigate cell–material interactions in tissue engineering and mechanobiology. Nat. Protoc. 2022;17:618–648. doi: 10.1038/s41596-021-00652-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Xia Q, Xiao H, Pan Y, Wang L. Microrheology, advances in methods and insights. Adv. Colloid Interface Sci. 2018;257:71–85. doi: 10.1016/j.cis.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 197.Roca-Cusachs P, Conte V, Trepat X. Quantifying forces in cell biology. Nat. Cell Biol. 2017;19:742–751. doi: 10.1038/ncb3564. [DOI] [PubMed] [Google Scholar]
- 198.Matthys OB, Hookway TA, McDevitt TC. Design principles for engineering of tissues from human pluripotent stem cells. Curr. Stem Cell Rep. 2016;2:43–51. doi: 10.1007/s40778-016-0030-z. [DOI] [PMC free article] [PubMed] [Google Scholar]