Abstract
Herein we present the exploration of the utility of DNA demethylase enzymes for targeted protein degradation. Novel benzylguanine substrates are characterized for their ability to control protein degradation in cells. Our data demonstrate the utility of this approach to degrade fusion proteins in different localizations within living cells.
Keywords: adamantyl, protein degron, SNAP tag
Manipulating protein expression levels by chemical control of their degradation is an emergent technology for studying protein function in normal cellular function and disease.[1] Several approaches have been developed that rely on selective binding of small molecules to specific designed protein domains to either stabilize or encourage degradation of a protein of interest (POI), thereby offering control of protein concentration.[2–4] Until recently, majority of research in this area has been on de-stabilizing proteins,[3] recent work highlighting PROTACs for protein-specific degradation has been a major focus.
These technologies rely on chimeric small molecules consisting of a POI targeting unit coupled to a unit that recruits the proteosome. This second unit functions by either mimicking the exposure of a hydrophobic core, resulting in proteasomal recognition and degradation, or by direct recruitment of ubiquitin ligases to encourage ubiquitin transfer and subsequent degradation (Figure 1a). For the former, the adamantyl functional group has been demonstrated to work efficiently for protein degradation.[5,6] For the latter, derivatives of thalidomide are often synthesized to encourage cereblon (CRBN) ubiquitin ligase recruitment to a protein of interest.[7,8] These efforts, while valuable, are obviously protein specific and require the POI to have a unique known interaction with a small molecule.
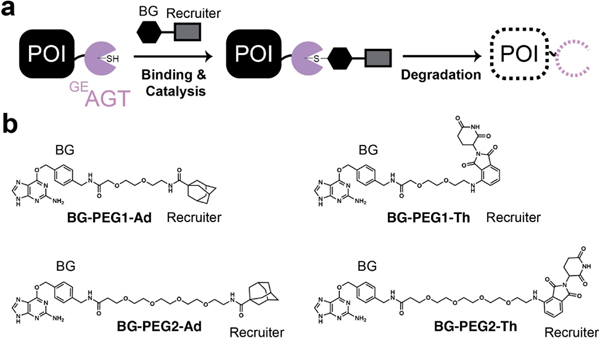
Figure 1.
Utilizing GEAGT for ligand-induced protein degradation. a. Schematic of benzylguanine ligand binding to GEAGT and inducing protein degradation. b. Chemical structures of ligands used in this study. POI=protein of interest. BG=benzylguanine. Ad=adamantyl. Th=thalidomide. PEG=polyethylene glycol.
The possibility of studying many POIs without developing individual targeting molecules is currently limited to HaloPROTACS and the dTAG system.[6,9–11] The HaloPROTAC system uses a protein fusion between HaloTag and the POI. Crews et al. demonstrated that selective degradation of the POI could be induced by using a halo ligand that was coupled to a degradation machinery-recruiting small molecule.[9,10] When HaloTag2 is used it is possible to employ an adamantyl as the recruitment unit, leading to robust protein degradation. However, when the more stable HaloTag7 fusion is used it is necessary for the halo unit to be coupled to a ligand specific for a particular E3 ligase (VHL). The dTAG system uses novel degrader of FKBP12F36V in-frame with a protein of interest, relying on cereblon-mediated protein decay. Such examples, which rely on specific protein complex recruitment may not always be optimal for POI degradation.[11,12] Additionally, having a single technology for controlled degradation of a wide variety of proteins means that multiplex experiments where multiple POIs are degraded simultaneously without protein specific ligands are not currently possible. Therefore, additional systems for controlled protein degradation are necessary to better understand the role of gene function at the protein level.
In an effort to advance the goal of multiplex experiments, we describe the utility of novel ligand-protein complexes that take advantage of the specificity and robust nature of the DNA repair protein O6-alkylguanine-DNA-alkyltransferase (hAGT).[13,14] Evolved, highly stable, derivatives of hAGT known as SNAP-tag[13,15,16] are routinely fused to POIs for both labelling and purification with O6-benzylguanine (BG) based ligands.[17] The wide-spread use and versatility of SNAP-tag technology and its known orthogonality with HaloTag suggested it as a starting point for the development of a non-POI specific protein degradation technology. We therefore designed, synthesized, and employed SNAP ligands meant to mimic hydrophobic protein unfolding. We anticipated that this would lead to nonspecific ubiquitin ligase recruitment and subsequent controlled SNAP-fusion protein degradation. Using these ligands, we demonstrated robust degradation of POIs localized to different cellular compartments.
SNAP-tag was evolved in a stepwise manner from human hAGT, which functions through recognition of alkylated DNA and subsequent transfer of the undesired methyl group to a reactive cysteine residue.[18,19] Once alkylated, hAGT undergoes an allosteric conformational change that reveals lysine residues for ubiquitination and subsequent proteasomal degradation (t1/2 ~2.5 h).[20,21] The robust nature of the SNAP tag technology (t1/2~42.5 h) is the result of directed evolution efforts to create a highly stable protein complex that is specific for the recognition of BG derivatives for protein alkylation, without concomitant degradation.[16] To both enable selective, efficient alkylation and induce degradation, we reasoned that a version of hAGT that retained the selective BG binding ability of SNAP tag while being unstable post-binding, termed GEAGT,[16] would be the best starting point for testing degradation of fused proteins.
In designing the chimeric molecules, we therefore needed an O6-benzylguanine on one end for GEAGT binding and either a hydrophobic moiety such as adamantyl or a specific E3 ligase recruitment ligand (i.e.: thalidomide) on the other. Additionally, due to its chimeric nature the optimal linker for connecting the POI-recruitment and the degradation-induction units is often not obvious, suggesting the need to test linker lengths to identify suitable substrates.[22] Toward this end, we synthesized 4 potential substrates, all containing a BG (GEAGT-recruiting) and either adamantyl or thalidomide unit. We also utilized O6-benzylguanine as a baseline control because it has been demonstrated to encourage degradation of GEAGT upon binding and catalysis (Figure 1b). For detailed synthetic procedures and characterization data for all new compounds see the Supporting Information.
We first determined if the synthesized compounds were capable of binding to GEAGT in cellulo by a competitive binding in-gel fluorescence assay (Figure S1) utilizing the commercial cell-permeable fluorescent SNAP substrate TMR-Star. We transfected HEK293T cells with a plasmid expressing GEAGT, generating stable cell lines. The cells were then introduced to each ligand at 20uM for 1 hour. Cells were subsequently washed and TMR-Star was added at 1uM for 30 minutes. As shown in Figure S1, all ligands were able to prevent TMR-Star from binding when compared with the negative control. These results strongly suggest that all of our ligands are able to bind to active site in GEAGT.
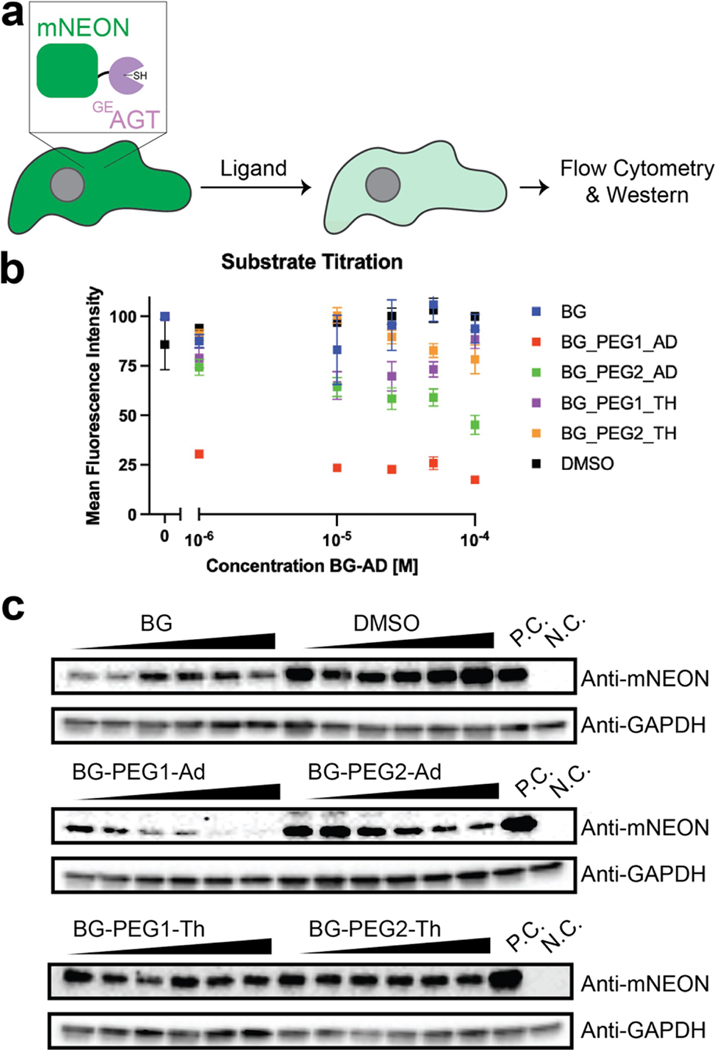
We next sought to understand if increased protein degradation occurred post ligand binding. Toward this end, we generated stable cell lines with a nuclear localized GEAGT-mNeonGreen fusion construct and incubated cells with a range of concentrations for 24 hours. mNeonGreen fluorescence was then quantified using flow cytometry (Figure 2a). As shown in Figure 2b, we observed that the adamantyl chimeric compound with the shorter PEG linker (BG-PEG1-Ad) resulted in the most noticeable reduction of mNeonGreen fluorescence. Fluorescence was ~20% of the control signal even at the lowest tested concentration (1 μM). We surprisingly observed that the thalidomide-containing compounds only had limited degradation of the GEAGT-mNeonGreen fusion construct. Corroboration of these results by mNeonGreen Western blot also demonstrated that incubation with BG-PEG1-Ad resulted in the most protein degradation (Figure 2c). Time-course experiments demonstrated that the majority of protein was degraded in 24 hours (Figure S2).
Figure 2.
Validation of various BG ligands for protein degradation. a. Schematic of BG ligand binding to GEAGT and inducing degradation of both the protein and fluorescent reporter. b. Normalized flow cytometry data based on the mean fluorescent shift of the mNeonGreen reporter. Cells were treated for 24 h and ligand concentration is noted in the x-axis. N=3. FACS results were gated against fluorescence and differences in cell populations above selected gates were used to calculate mean fluorescence intensity. c. Western blot of various BG ligands. Cells were treated for 24 h and the concentration of ligands increased from 0.5 uM to 50 uM. P.C.=positive control; plasmid transfection of the mNeonGreen reporter without vehicle or small molecule exposure. N.C.=negative control; no plasmid transfection of the mNeonGreen reporter.
To further demonstrate the selectivity, we treated cells with proteasome inhibitors MG-132 and Bortezomib.[23] Incubation of the inhibitors resulted in increased fluorescence, suggesting that inhibition of the proteosome directly contributes to the majority of degradation – this is consistent with hydrophobicity-induced degradation from the adamantly functional group.[6] Increased concentration of Lenalidomide, which is known to inhibit E3-ligase dependent degradation of phthalidomide PROTACs[24] did not have a dramatic effect on the degradation, further suggesting that our AGT construct does not seem to be amenable to cereblon (CRBN) ubiquitin ligation and degradation (Figure S3).
We also determined the cell toxicity in this assay by measuring the BG-PEG1-Ad at varying concentrations over 24 hours. As shown in Figure S4, we did not observe significant amounts of cell death with any ligand, suggesting that our compounds are not detrimental to cell survival even at higher concentrations or over longer periods of time. These results overall suggest that BG-PEG1-Ad is a reasonable ligand to induce degradation of GEAGT-fused proteins and does so with kinetics that are on par with other chimeric degrons that have been developed. These results prompted us to continue to better understand degradation induced by BG-PEG1-Ad.
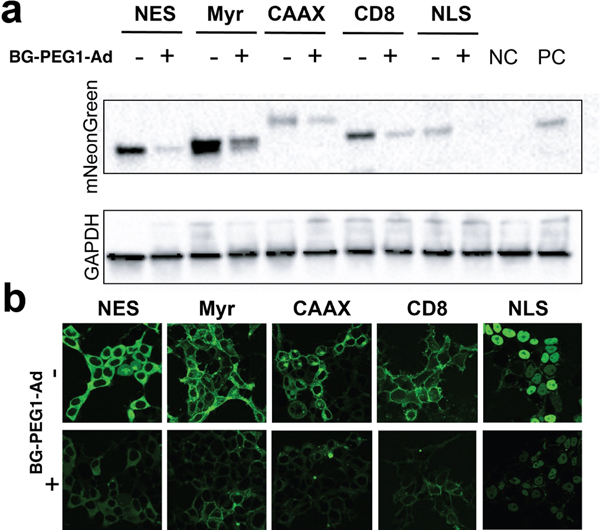
We next aimed to understand the scope of fusion GEAGT degradation with BG-PEG1-Ad in regard to cellular localization of POIs. To do this, we cloned fusion proteins representing the nucleus (NLS-fused), cytoplasm (NES-fused), membrane bound (N-terminal myristoylation (Myr), C-terminal farnesylation (CAAX)) and transmembrane (CD8) (using the CD8 TM region, mNeonGreen being on the extracellular-facing N-terminus and the GEAGT on the cytoplasmic-facing C-terminus). We first demonstrated that each fusion protein complex was able to bind BG-PEG1-Ad, utilizing the TMR-Star competition experiment described above (Figure S5). After stable transfection of each construct, we incubated the cells with 20uM BG-PEG1-Ad for 24 hours. We observed significant reduction in protein amount for each construct using Western blot analysis, consistent with the reduction in fluorescence observed via microscopy (Figure 3a). Gratifyingly, we observed a marked reduction of cellular fluorescence for all constructs. We then used live cell imaging to demonstrate that the target proteins both retained their desired localization and had reduced expression levels (Figure 3b). Overall, these data demonstrate the flexibility of our BG-PEG1-Ad- GEAGT pair for targeted protein degradation and the flexibility of fusing GEAGT proteins to POIs in different parts of the cell for efficient reduction in protein abundance.
Figure 3.
Degradation of fusion proteins at various localizations of the cell. a. visualization of protein degradation with western blot b. images of protein degradation before and after treatment with 20 uM of BG-PEG1-Ad for 24 hours. NC/=untransfected cells. PC=Cells Transfected with NLS-fusion protein.
Herein we have described the utility of a novel ligand-protein complex that can be deployed for targeted protein degradation. Comparison of the different ligand structures has demonstrated that a shorter linker paired with an adamantyl hydrophobic unit gave the best substrate-GEAGT pair. We demonstrated that our approach was able to induce substantial knockdown of protein expression, in line with PROTACS/Halo-PROTACS systems. Additionally, the fusion of GEAGT to proteins with subcellular localization results in significant reduction of protein expression levels in the expected locals. As such, we anticipate this approach could be applied to a wide variety of proteins. Future work is focused on testing this approach for the knockdown of endogenous proteins. Eventually we anticipate transitioning this technique to in vivo settings, since extensive studies on BG derivatives show them to have an exceptional safety profile, with demonstrated reactivity in several tissues.[25,26] This work will be reported in due course.
Supplementary Material
Acknowledgements
R. C. S. is a Pew Biomedical Scholar.
Footnotes
Supporting information for this article is available on the WWW under https://doi.org/10.1002/cbic.202200053
Conflict of Interest
The authors declare no conflict of interest.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Schapira M, Calabrese MF, Bullock AN, Crews CM, Nat. Rev. Drug Discovery 2019, 18, 949–963. [DOI] [PubMed] [Google Scholar]
- [2].Banaszynski LA, Chen L-C, Maynard-Smith LA, Ooi AGL, Wandless TJ, Cell 2006, 126, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rakhit R, Navarro R, Wandless TJ, Chem. Biol 2014, 21, 1238–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stankunas K, Bayle JH, Gestwicki JE, Lin Y-M, Wandless TJ, Crabtree GR, Mol. Cell 2003, 12, 1615–1624. [DOI] [PubMed] [Google Scholar]
- [5].Gustafson JL, Neklesa TK, Cox CS, Roth AG, Buckley DL, Tae HS, Sundberg TB, Stagg DB, Hines J, McDonnell DP, Norris JD, Crews CM, Angew. Chem. Int. Ed 2015, 54, 9659–9662; Angew. Chem. 2015, 127, 9795–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Neklesa TK, Tae HS, Schneekloth AR, Stulberg MJ, Corson TW, Sundberg TB, Raina K, Holley SA, Crews CM, Nat. Chem. Biol 2011, 7, 538–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fang Y, He Q, Cao J, Curr. Med. Chem 2021.
- [8].Naito M, Ohoka N, Shibata N, Tsukumo Y, Front. Chem 2019, 7, 849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Buckley DL, Raina K, Darricarrere N, Hines J, Gustafson JL, Smith IE, Miah AH, Harling JD, Crews CM, ACS Chem. Biol 2015, 10, 1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Simpson LM, Macartney TJ, Nardin A, Fulcher LJ, Röth S, Testa A, Maniaci C, Ciulli A, Ganley IG, Sapkota GP, Cell Chem. Biol 2020, 27, 1164–1180.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Nomura DK, Dey M, Cell Chem. Biol 2021, 28, 887–888. [DOI] [PubMed] [Google Scholar]
- [12].Belcher BP, Ward CC, Nomura DK, Biochemistry 2021, DOI: 10.1021/acs.biochem.1c00464. [DOI] [PMC free article] [PubMed]
- [13].Kolberg K, Puettmann C, Pardo A, Fitting J, Barth S, Curr. Pharm. Des 2013, 19, 5406–5413. [DOI] [PubMed] [Google Scholar]
- [14].Liss V, Barlag B, Nietschke M, Hensel M, Sci. Rep 2015, 5, 17740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K, Chem. Biol 2003, 10, 313–317. [DOI] [PubMed] [Google Scholar]
- [16].Mollwitz B, Brunk E, Schmitt S, Pojer F, Bannwarth M, Schiltz M, Rothlisberger U, Johnsson K, Biochemistry 2012, 51, 986–994. [DOI] [PubMed] [Google Scholar]
- [17].Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K, Nat. Biotechnol 2003, 21, 86–89. [DOI] [PubMed] [Google Scholar]
- [18].Natarajan AT, Vermeulen S, Darroudi F, Valentine MB, Brent TP, Mitra S, Tano K, Mutagenesis 1992, 7, 83–85. [DOI] [PubMed] [Google Scholar]
- [19].Tano K, Shiota S, Collier J, Foote RS, Mitra S, Proc. Natl. Acad. Sci. USA 1990, 87, 686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Srivenugopal KS, Yuan XH, Friedman HS, Ali-Osman F, Biochemistry 1996, 35, 1328–1334. [DOI] [PubMed] [Google Scholar]
- [21].Kanugula S, Goodtzova K, Pegg AE, Biochem. J 1998, 329, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hughes SJ, Ciulli A, Essays Biochem. 2017, 61, 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goldberg AL, J. Cell Biol 2012, 199, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lopez-Girona A, Mendy D, Ito T, Miller K, Gandhi AK, Kang J, Karasawa S, Carmel G, Jackson P, Abbasian M, Mahmoudi A, Cathers B, Rychak E, Gaidarova S, Chen R, Schafer PH, Handa H, Daniel TO, Evans JF, Chopra R, Leukemia 2012, 26, 2326–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yang G, de Castro Reis F, Sundukova M, Pimpinella S, Asaro A, Castaldi L, Batti L, Bilbao D, Reymond L, Johnsson K, Heppenstall PA, Nat. Methods 2015, 12, 137–139. [DOI] [PubMed] [Google Scholar]
- [26].Bojkowska K, Santoni de Sio F, Barde I, Offner S, Verp S, Heinis C, Johnsson K, Trono D, Chem. Biol 2011, 18, 805–815. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.