Abstract
The brain µ-opioid receptor (MOR) is critical for the analgesic, rewarding, and addictive effects of opioid drugs. However, in rat models of opioid-related behaviors, the circuit mechanisms of MOR-expressing cells are less known because of a lack of genetic tools to selectively manipulate them. We introduce a CRISPR-based Oprm1-Cre knock-in transgenic rat that provides cell type-specific genetic access to MOR-expressing cells. After performing anatomic and behavioral validation experiments, we used the Oprm1-Cre knock-in rats to study the involvement of NAc MOR-expressing cells in heroin self-administration in male and female rats. Using RNAscope, autoradiography, and FISH chain reaction (HCR-FISH), we found no differences in Oprm1 expression in NAc, dorsal striatum, and dorsal hippocampus, or MOR receptor density (except dorsal striatum) or function between Oprm1-Cre knock-in rats and wildtype littermates. HCR-FISH assay showed that iCre is highly coexpressed with Oprm1 (95%-98%). There were no genotype differences in pain responses, morphine analgesia and tolerance, heroin self-administration, and relapse-related behaviors. We used the Cre-dependent vector AAV1-EF1a-Flex-taCasp3-TEVP to lesion NAc MOR-expressing cells. We found that the lesions decreased acquisition of heroin self-administration in male Oprm1-Cre rats and had a stronger inhibitory effect on the effort to self-administer heroin in female Oprm1-Cre rats. The validation of an Oprm1-Cre knock-in rat enables new strategies for understanding the role of MOR-expressing cells in rat models of opioid addiction, pain-related behaviors, and other opioid-mediated functions. Our initial mechanistic study indicates that lesioning NAc MOR-expressing cells had different effects on heroin self-administration in male and female rats.
SIGNIFICANCE STATEMENT The brain µ-opioid receptor (MOR) is critical for the analgesic, rewarding, and addictive effects of opioid drugs. However, in rat models of opioid-related behaviors, the circuit mechanisms of MOR-expressing cells are less known because of a lack of genetic tools to selectively manipulate them. We introduce a CRISPR-based Oprm1-Cre knock-in transgenic rat that provides cell type-specific genetic access to brain MOR-expressing cells. After performing anatomical and behavioral validation experiments, we used the Oprm1-Cre knock-in rats to show that lesioning NAc MOR-expressing cells had different effects on heroin self-administration in males and females. The new Oprm1-Cre rats can be used to study the role of brain MOR-expressing cells in animal models of opioid addiction, pain-related behaviors, and other opioid-mediated functions.
Keywords: mu opioid receptor, knockin, CRISPR, heroin self-administration, pain, caspase 3 lesion
Introduction
The µ-opioid receptor (MOR) is expressed in many brain areas (Akil et al., 1984; Mansour et al., 1995a; Emery and Akil, 2020). Activation of MORs mediates diverse effects of opioid agonists, such as heroin, morphine, and fentanyl, including analgesia, tolerance, and self-administration in mice, rats, monkeys, and humans, as well as addiction liability of opioid drugs in humans (Jaffe, 1990; Darcq and Kieffer, 2018).
During the last decade, investigators have developed transgenic mouse models that allow for the investigation of circuit mechanisms of MOR-expressing cells in different brain regions in the behavioral and physiological effects of opioid drugs. These include a knock-in MOR-mCherry mouse line to map MOR protein expression throughout the brain (Gardon et al., 2014) and a mouse line with a floxed Oprm1 gene (the gene encoding MOR) that allows for selective deletion of the receptor (Weibel et al., 2013; Charbogne et al., 2017). More recently, Bailly et al. (2020) introduced an Oprm1-Cre knock-in mouse line that allows for in vivo manipulation of activity of MOR-expressing cells in the brain to study their causal role in the behavioral and physiological effects of opioid drugs. In this Oprm1-Cre mouse line, a cDNA encoding a T2A cleavable peptide and Cre recombinase was fused to EGFP, and the genetic construct was inserted downstream of the Oprm1 coding sequence.
To date, the Cre line technology has not been applied to study the role of MOR-expressing cells and projections in opioid analgesia and self-administration in the rat. Mice provide a good model organism to study circuit mechanisms of unconditioned and simple conditioned behaviors related to opioid analgesia and reinforcement. However, it has been difficult to reliably study circuit mechanisms of opioid self-administration and relapse-related behaviors in this species because of technical limitations (small veins and difficulties in maintaining catheter patency) and limited repertoire of sophisticated learned behaviors. For example, established rat behavioral phenomena, such as incubation of drug craving (time-dependent increase in drug seeking during abstinence) and drug priming-induced reinstatement after extinction (Shaham et al., 2003; Wolf, 2016), are not readily observed in mouse models (Highfield et al., 2002; Terrier et al., 2016). Additionally, behavioral phenomena, such as context-induced relapse after extinction or punishment that are reliable in rats, have not yet been demonstrated in mice (Marchant et al., 2019).
Based on these considerations, we have created and characterized a knock-in rat (Oprm1-Cre) that coexpresses the MOR protein and an improved Cre recombinase from the endogenous MOR locus (Oprm1). The presence of the Cre transgene did not appear to change Oprm1 expression in nucleus accumbens (NAc), dorsal striatum (DS), and dorsal hippocampus (dHipp) or MOR receptor density (except DS) or function in these regions. Similarly, the presence of Cre did not change MOR-related behaviors in pain-related models or in heroin self-administration and relapse models.
Next, we used the knock-in rats to study the involvement of NAc MOR-expressing cells in heroin self-administration in male and female rats. We focused on NAc MOR-expressing cells because an early pharmacological study in male rats showed that local NAc injections of the preferential MOR antagonist methyl naloxonium chloride (a lipophobic quaternary derivative of naloxone) decreased the reinforcing effects of self-administered heroin (Vaccarino et al., 1985). Additionally, in Oprm1 KO mice, rescue of Oprm1 expression using a Pdyn-MOR transgene restored remifentanil self-administration (Cui et al., 2014). We injected the Cre-dependent vector AAV1-EF1a-Flex-taCasp3-TEVP (AAV-DIO-Casp3) (Takahashi et al., 2017) into Oprm1-Cre rats and their wildtype littermates, and selectively lesioned NAc MOR-expressing cells in only Oprm1-Cre rats. Beyond identifying potential sex differences in the mechanisms of heroin self-administration, our study serves as a proof of concept for the value of this Oprm1-Cre rat model in refining our understanding of the functions of endogenous opioid receptor systems.
Materials and Methods
Subjects
We performed the experiments in accordance with the National Institutes of Health's Guide for the care and use of laboratory animals (Ed 8), under protocols approved by the Animal Care and Use Committees of National Institute on Drug Abuse (NIDA) Intramural Research Program or the University of Michigan.
NIDA
We used 89 Oprm1-Cre heterozygotes (41 males and 48 females) and 92 wildtype littermates (45 males and 47 females) for our molecular and behavioral experiments. Before virus or intravenous surgery, the approximate weight range of the rats we used in the behavioral experiments was 350-550 g (males) or 175-300 g (females). We maintained the rats under a reverse 12:12 h light/dark cycle (lights off at 8:00 A.M.) with food and water freely available in the home cage. We housed the rats 2 or 3 per cage for all experiments, except those requiring intravenous surgery, which were housed 2 or 3 per cage before surgery and individually after surgery.
University of Michigan
We used 25 Oprm1-Cre heterozygotes (17 males and 8 females) and 25 wildtype littermates (17 males and 8 females) for our molecular and behavioral experiments. We excluded 1 rat because of health problems. The approximate weight range of the rats we used in the behavioral experiments was 350-550 g (males) or 175-300 g (females). We housed the rats 2 per cage (except 1 rat whose cage mate was excluded because of health problems) in a temperature- and humidity-controlled animal facility, on either a 12 h (lights on at 07:00; males) or a 14 h (lights on at 05:00; females) light cycle, with food and water freely available.
We excluded 1 female Oprm1-Cre rat from the proof-of-concept AAV-DIO-Casp3 experiment because of misplaced injection, 1 male (Experiment 2) and 2 female (Experiments 3B and 3C, 1/experiment) Oprm1-Cre rats because of poor health, 2 male rats (Experiment 4, 1/genotype) because of failure to acquire heroin self-administration, and 1 wildtype female rat (Experiment 5) because of failure to acquire food self-administration. For Experiments 4 and 5, we tested catheters' patency after the within-dose heroin maintenance phase. We found loss of patency in 3 Oprm1-Cre rats (Experiment 4, 2 males, 1 female) and 2 wildtype rats (Experiment 5, 1/sex) and only included their data in the food and heroin acquisition phase and food self-administration (Experiment 5, rats).
Rat Oprm1 iCre recombinase knock-in
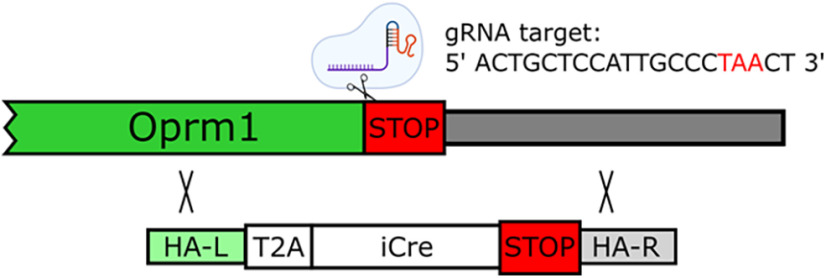
We used CRISPR/Cas9 technology to introduce the iCre recombinase coding sequence (Shimshek et al., 2002) before the termination codon of the rat Oprm1 gene. The knock-in approach followed the Easi-CRISPR method (Quadros et al., 2017). The canonical rat Oprm1 gene codes a 2306 nt mRNA (1196 coding bp) from four exons that translates a 398 aa protein (NCBI Ref RNA sequence NM_013071.2). We used the CRISPOR algorithm (www.CRISPR.tefor.net) (Concordet and Haeussler, 2018) to select a single guide RNA (sgRNA) that was predicted to cut between the second and third base pair of the stop codon located in Oprm1 exon 4. The guide sequence was 5′-ACTGCTCCATTGCCCTAACT-3′ (PAM = GGG) (see Fig. 1). This sgRNA has a high specificity score (CFD = 91) (Doench et al., 2016).
Figure 1.
CRISPR-mediated knock-in of T2A-iCre downstream of the rat Oprm1 coding sequence. Schematic of the target gene (rat Oprm1) with annotation for the location and sequence of the SpCas9 sgRNA that cleaves within the stop codon. The donor template encoding homologous arms and the T2A-iCre transgene are also shown.
The chemically modified sgRNA (Basila et al., 2017) was synthesized by Millipore Sigma. We tested and verified the sgRNA, which induced Cas9-mediated chromosome breaks, in primary rat embryonic fibroblasts. Briefly, ribonucleoprotein complexes (RNP) were formed by combining 4.85 µg sgRNA with 21.6 µg of enhanced specificity Cas9 protein (eSpCas9, www.sigmaaldrich.com) (Slaymaker et al., 2016) in 20 µl. We added RNP to a 0.2 cm cuvette containing 5 µg of a PGKpuro drug resistance plasmid (kind gift of Michael McBurney) (McBurney et al., 1994) and 750 million cells suspended in D-PBS. A Bio-Rad Gene Pulser was set to deliver a square wave pulse at 250 V, 2 ms pulse width for one pulse with unipolar polarity. We plated cells onto one 6 cm dish in culture medium (high glucose DMEM with the addition of 10% FBS, 4 mm glutamine, and penicillin-streptomycin at 10,000 U/ml). The next day, we changed the electroporation media. On days 2 and 3, we used media containing 2 µg/µl puromycin to eliminate cells that did not receive the PGKpuro plasmid. On days 4 and 5, we used media without puromycin to feed the cells. On the following day, we collected surviving cells for DNA extraction.
We used PCR primers to amplify a 776 bp genomic DNA fragment that included the sgRNA target (Oprm1 activity forward primer: 5′-ATGGAAGATGGAGCAAGAGAAAGAATTT-3', Oprm1 activity reverse primer: 5′-ATGTATCACTACAGTGAATTTAACAAGTGAC-3′). We submitted amplicons for Sanger sequencing. Chromatograms revealed superimposed peaks, typical of indels formed by nonhomologous endjoining repair of Cas9-induced chromosome breaks (Brinkman et al., 2014).
We used the sgRNA to produce iCre knock-in rats because of its high activity. The use of an sgRNA with high specificity in combination with eSpCas9 dramatically reduces the likelihood of Cas9 off target hits in generation zero (G0) founder animals (Anderson et al., 2018). The DNA donor was obtained as a long single-stranded DNA Megamer synthesis of 1312 nucleotides (Table 1; www.IDTDNA.com). We designed the DNA donor to include a Gly-Ser-Gly linker and T2A self-cleaving peptide after Oprm1 codon 398. In this way, Oprm1 and iCre will be expressed from a single bi-cistronic locus so that physiological levels of MOR protein will be present in cells in addition to the iCre protein (Kim et al., 2011). We performed pronuclear microinjection of 30 mouse zygotes with the RNP and DNA donor to demonstrate that the reagents did not interfere with mouse zygote development in vitro, with the expectation that such reagents would not then interfere with rat zygote development in utero. We obtained mouse zygotes for pronuclear microinjection from B6SJLF1 mice (The Jackson Laboratory stock #100012).
Table 1.
Sequence of donor DNA for generation of Oprm1-Cre knock-in rat
| 5′ arm of homology (nucleotides 63-98 are oprm1 exon 4) | CATCAATGTGCTCTCTAATGAGACCCCAGAACTCACTATCTTCACTCTTTCTCTTCTTTCAGCTAGAAAATCTGGAGGCAGAAACTGCTCCATTGCCC |
| Gly-Ser-Gly linker and T2A self-cleaving peptide sequence (lowercase) | GGTTCTGGCgagggcagaggaagtcttctaacatgcggtgacgtggaggagaatcccggccct |
| iCre recombinase coding sequence | GTGCCCAAGAAGAAGAGGAAAGTCTCCAACCTGCTGACTGTGCACCAAAACCTGCCTGCCCTCCCTGTGGATGCCACCTCTGATGAAGTCAG GAAGAACCTGATGGACATGTTCAGGGACAGGCAGGCCTTCTCTGAACACACCTGGAAGATGCTCCTGTCTGTGTGCAGATCCTGGGCTG CCTGGTGCAAGCTGAACAACAGGAAATGGTTCCCTGCTGAACCTGAGGATGTGAGGGACTACCTCCTGTACCTGCAAGCCAGAGGCCT GGCTGTGAAGACCATCCAACAGCACCTGGGCCAGCTCAACATGCTGCACAGGAGATCTGGCCTGCCTCGCCCTTCTGACTCCAATGCTG TGTCCCTGGTGATGAGGAGAATCAGAAAGGAGAATGTGGATGCTGGGGAGAGAGCCAAGCAGGCCCTGGCCTTTGAACGCACTGACT TTGACCAAGTCAGATCCCTGATGGAGAACTCTGACAGATGCCAGGACATCAGGAACCTGGCCTTCCTGGGCATTGCCTACAACACCCTG CTGCGCATTGCCGAAATTGCCAGAATCAGAGTGAAGGACATCTCCCGCACCGATGGTGGGAGAATGCTGATCCACATTGGCAGGACCA AGACCCTGGTGTCCACAGCTGGTGTGGAGAAGGCCCTGTCCCTGGGGGTTACCAAGCTGGTGGAGAGATGGATCTCTGTGTCTGGTGT GGCTGATGACCCCAACAACTACCTGTTCTGCCGGGTCAGAAAGAATGGTGTGGCTGCCCCTTCTGCCACCTCCCAACTGTCCACCCGGG CCCTGGAAGGGATCTTTGAGGCCACCCACCGCCTGATCTATGGTGCCAAGGATGACTCTGGGCAGAGATACCTGGCCTGGTCTGGCCA CTCTGCCAGAGTGGGTGCTGCCAGGGACATGGCCAGGGCTGGTGTGTCCATCCCTGAAATCATGCAGGCTGGTGGCTGGACCAATGT GAACATTGTGATGAACTACATCAGAAACCTGGACTCTGAGACTGGGGCCATGGTGAGGCTGCTCGAGGATGGGGAC |
| Oprm1 termination codon and 3′ arm of homology from oprm1 exon 4 | TAACTGGGTCTCACACCATCCAGACCCTCGCTAAGCTTAGAGGCCGCCATCTACGTGGAATCAGGTTGCTGTCAGGGTGTGTGGGAGGCTCTGGTTTCCTG |
To produce the rat Oprm1 iCre recombinase knock-in, we obtained rat zygotes from Sprague Dawley rats (Charles River Laboratory Strain Code 001). We performed pronuclear microinjection with a mixture containing RNP (30 ng/µl sgRNA mixed with 50 ng/µl eSpCas9 protein) and 5 ng/µl of the DNA donor as described (Filipiak and Saunders, 2006; Filipiak et al., 2019). Of 125 microinjected rat zygotes, 115 survived microinjection and were surgically transferred to pseudopregnant SAS Sprague Dawley rats (Charles River Strain Code 400). After 39 possible G0 founder rats were born, we extracted DNA from tail tip biopsies and amplified with primers specific for the iCre coding sequence (internal iCre forward primer 5′-AGAAGAAGAGGAAAGTCTCCAACCTGCT-3′ and internal iCre reverse primer: 5′-TTTCTGATTCTCCTCATCACCAGGGACA-3′; expected DNA fragment size: 379 bp). After screening 39 potential founder pups, 14 were positive for iCre recombinase.
We screened the 14 iCre-positive rats for correct genomic targeting with PCR primers in genomic DNA and in iCre to amplify the 5′ and 3′ junctions of the iCre insertion site. The 5′ junction primers were 5′ junction forward primer: 5′-AAGACAATGTTCAGTACAGTTCTCATACC-3′ and 5′ junction reverse primer ATTCTCCTTTCTGATTCTCCTCATCAC-3′; expected DNA fragment size from iCre coding sequence insertion: 595 bp. The 3′ junction primers were 3′ junction forward primer: 5′-GATGAACTACATCAGAAACCTGGACTC-3′ and 3′ junction reverse primer TTCAAGGTGAAAGTTTTAAGTTGGAAATG-3′; expected DNA fragment size from iCre coding sequence insertion: 571 bp. PCR amplicons showed that 1 of the 14 iCre-positive rats was positive for both 5′ and 3′ junctions. Sanger sequencing of the amplicons demonstrated iCre was inserted in the desired location. We also used spanning primers placed in genomic DNA to produce amplicons across the iCre insertion site. Spanning primers were spanning forward primer 5′-CAGAGGAAGTCTTCTAACATGCGGTGAC-3′, spanning reverse primer 5′-TCTGGATGGTGTGAGACCCAGTTAGTC-3′; expected DNA fragment size: 1123 bp. We gel-purified the PCR amplicons and subjected them to TOPO TA cloning and Sanger sequencing to confirm correct iCre insertion into the Oprm1 gene and that the iCre coding sequence was intact.
We mated the confirmed G0 rat with wildtype Sprague Dawley rats and obtained germline transmission from the G0 founder. Sequencing of DNA isolated from 14 obligate heterozygote G1 pups showed that they inherited the correctly targeted Oprm1 iCre recombinase knockin. We used G2 rats descended from two (G1) founders for further colony expansion.
Breeding and genotyping
NIDA and University of Michigan
We set up male and female Oprm1-Cre heterozygotes in breeding with wildtype mates (CD (Sprague Dawley) IGS, Charles River Labs, strain code #400) for at least four generations. At NIDA, we used at least 8 different breeding pairs at each generation after the 3rd generation and used heterozygote rats and wildtype littermates from the 4th and 5th generation. At University of Michigan, we used heterozygote rats and wildtype littermates from four genetically diverse litters from the 7th generation. Tail genotyping was performed by Transnetyx at NIDA and in-house PCR at University of Michigan. The rats bred at NIDA were registered with the Rat Genome Database (RGD#155641245) and deposited at the Rat Resource and Research Center (RRRC#975).
FISH chain reaction (HCR FISH) (University of Michigan)
We designed the Split-initiator DNA probes (version 2.0; Tables 2 and 3) (Choi et al., 2018; Kumar et al., 2021), and the probes were synthesized by Integrated DNA Technologies. We purchased DNA hairpins conjugated with B3 AlexaFluor-546 (AF-546) and B2 AF-647 from Molecular Instruments. We sectioned fresh-frozen rat brains at 30 µm in a cryostat: sections were from NAc, DS, and dHipp using AP coordinates from bregma of 1.2-2.0 mm for NAc and DS and −2.4 to −3.0 mm for dHipp. We optimized the HCR FISH method as described previously (Choi et al., 2018; Kumar et al., 2021).
Table 2.
Oprm1 probe: Seq ID NM_013071.2; HCR amplifier: B2-AlexaFluor647
| Odd | 1st half of initiator I1 + spacer (AA) + probe sequence | Even | Probe sequence + spacer (TA) + 2nd half of initiator I1 |
|---|---|---|---|
| 1 | CCTCgTAAATCCTCATCAAAGACACTCTGAAAGGGCAGTGTACTG | 2 | GAAGGGCCATGTTCCCATCAGGTAGAAATCATCCAgTAAACCgCC |
| 3 | CCTCgTAAATCCTCATCAAATCACGATCTTGCAGAGGATGGTTCC | 4 | TGAACATGTTGTAGTAATCTATTGAAAATCATCCAgTAAACCgCC |
| 5 | CCTCgTAAATCCTCATCAAAATGGTGCAGAGGGTGAATATGCTGG | 6 | CAGACAGCAATGTAGCGGTCCACGCAAATCATCCAgTAAACCgCC |
| 7 | CCTCgTAAATCCTCATCAAAAAGAGAGGATCCAGTTGCAGACGTT | 8 | TGAACATTACAGGCAGACCGATGGCAAATCATCCAgTAAACCgCC |
| 9 | CCTCgTAAATCCTCATCAAACCCTGCCTGTATTTTGTGGTTGCCA | 10 | GAGAACGTGAGGGTGCAATCTATGGAAATCATCCAgTAAACCgCC |
| 11 | CCTCgTAAATCCTCATCAAAGTTCTCCCAGTACCAGGTTGGGTGG | 12 | GAAGATAAAGACACAGATTTTGAGCAAATCATCCAgTAAACCgCC |
| 13 | CCTCgTAAATCCTCATCAAATGATGAGGACCGGCATGATGAAAGC | 14 | AGATCATCAGGCCGTAACACACAGTAAATCATCCAgTAAACCgCC |
| 15 | CCTCgTAAATCCTCATCAAAAGCATGCGAACGCTCTTGAGTCGTA | 16 | TTCCTGTCCTTTTCTTTGGAGCCCGAAATCATCCAgTAAACCgCC |
| 17 | CCTCgTAAATCCTCATCAAACACCATCCGGGTGATCCTGCGCAGA | 18 | GACGATAAATACAGCCACGACCACCAAATCATCCAgTAAACCgCC |
| 19 | CCTCgTAAATCCTCATCAAAGAAACGGTCTGAAATGTGGTTTCTG | 20 | TAACCCAAAGCAATGCAGAAGTGCCAAATCATCCAgTAAACCgCC |
| 21 | CCTCgTAAATCCTCATCAAAAACTGGATTCAGGCAGCTGTTCGTG | 22 | GAAGTTTTCATCCAGGAAGGCGTAAAAATCATCCAgTAAACCgCC |
| 23 | CCTCgTAAATCCTCATCAAATGCAGAACTCTCTGAAGCATCGCTT | 24 | GCTGTTCGATCGTGGACGAGGTTGGAAATCATCCAgTAAACCgCC |
| 25 | CCTCgTAAATCCTCATCAAATTCTGACGGACTCGAGTGGAGTTTT | 26 | TTAGCCGTGGAGGGATGTTCCCTAGAAATCATCCAgTAAACCgCC |
Table 3.
iCre. probe: Seq ID AY056050.1; HCR amplifier: B3-AlexaFluor546
| Odd | 1st half of initiator I1 + spacer (TT) + probe sequence | Even | Probe sequence + spacer (TT) + 2nd half of initiator I1 |
|---|---|---|---|
| 1 | gTCCCTgCCTCTATATCTTTCTTGGGCACCATGGTGGACAAGCTT | 2 | CAGCAGGTTGGAGACTTTCCTCTTCTTCCACTCAACTTTAACCCg |
| 3 | gTCCCTgCCTCTATATCTTTGGGCAGGCAGGTTTTGGTGCACAGT | 4 | CTTCATCAGAGGTGGCATCCACAGGTTCCACTCAACTTTAACCCg |
| 5 | gTCCCTgCCTCTATATCTTTAACATGTCCATCAGGTTCTTCCTGA | 6 | TGTTCAGAGAAGGCCTGCCTGTCCCTTCCACTCAACTTTAACCCg |
| 7 | gTCCCTgCCTCTATATCTTTCACAGACAGGAGCATCTTCCAGGTG | 8 | CTTGCACCAGGCAGCCCAGGATCTGTTCCACTCAACTTTAACCCg |
| 9 | gTCCCTgCCTCTATATCTTTCAGGGAACCATTTCCTGTTGTTCAG | 10 | GGTAGTCCCTCACATCCTCAGGTTCTTCCACTCAACTTTAACCCg |
| 11 | gTCCCTgCCTCTATATCTTTAGGCCTCTGGCTTGCAGGTACAGGA | 12 | AGGTGCTGTTGGATGGTCTTCACAGTTCCACTCAACTTTAACCCg |
| 13 | gTCCCTgCCTCTATATCTTTCCTGTGCAGCATGTTGAGCTGGCCC | 14 | GTCAGAAGGGCGAGGCAGGCCAGATTTCCACTCAACTTTAACCCg |
| 15 | gTCCCTgCCTCTATATCTTTTCATCACCAGGGACACAGCATTGGA | 16 | CATCCACATTCTCCTTTCTGATTCTTTCCACTCAACTTTAACCCg |
| 17 | gTCCCTgCCTCTATATCTTTAGGGCCTGCTTGGCTCTCTCCCCAG | 18 | TGGTCAAAGTCAGTGCGTTCAAAGGTTCCACTCAACTTTAACCCg |
| 19 | gTCCCTgCCTCTATATCTTTAGAGTTCTCCATCAGGGATCTGACT | 20 | CAGGTTCCTGATGTCCTGGCATCTGTTCCACTCAACTTTAACCCg |
| 21 | gTCCCTgCCTCTATATCTTTTGTTGTAGGCAATGCCCAGGAAGGC | 22 | TGGCAATTTCGGCAATGCGCAGCAGTTCCACTCAACTTTAACCCg |
| 23 | gTCCCTgCCTCTATATCTTTCGGGAGATGTCCTTCACTCTGATTC | 24 | TGGATCAGCATTCTCCCACCATCGGTTCCACTCAACTTTAACCCg |
| 25 | gTCCCTgCCTCTATATCTTTCACCAGGGTCTTGGTCCTGCCAATG | 26 | CAGGGCCTTCTCCACACCAGCTGTGTTCCACTCAACTTTAACCCg |
| 27 | gTCCCTgCCTCTATATCTTTCCACCAGCTTGGTAACCCCCAGGGA | 28 | CCACACCAGACACAGAGATCCATCTTTCCACTCAACTTTAACCCg |
| 29 | gTCCCTgCCTCTATATCTTTCATAGATCAGGCGGTGGGTGGCCTC | 30 | ATCTCTGCCCAGAGTCATCCTTGGCTTCCACTCAACTTTAACCCg |
| 31 | gTCCCTgCCTCTATATCTTTGCAGAGTGGCCAGACCAGGCCAGGT | 32 | GCCATGTCCCTGGCAGCACCCACTCTTCCACTCAACTTTAACCCg |
| 33 | gTCCCTgCCTCTATATCTTTTTCAGGGATGGACACACCAGCCCTG | 35 | ATTGGTCCAGCCACCAGCCTGCATGTTCCACTCAACTTTAACCCg |
| 35 | gTCCCTgCCTCTATATCTTTTGATGTAGTTCATCACTATGTTCAC | 36 | TGGCCCCAGTCTCAGAGTCCAGGTTTTCCACTCAACTTTAACCCg |
We fixed the sections in 4% PFA, washed with 5× sodium chloride/sodium citrate/0.01% Tween-20 (SSCTw) buffer for 3 times (5 min each), and then acetylated in 0.1 m triethanolamine, pH 8.0, with 0.25% v/v acetic anhydride solution for 10 min. After rinsing with ddH2O, we delipidated the sections in −20°C chilled acetone: methanol (1:1) for 5 min, washed with 5× SSCTw, and equilibrated in hybridization buffer (30% deionized formamide, 5× SSC, 9 mm citric acid, pH 6.0, 0.5 mg/ml yeast tRNA, 1× Denhardt's solution, 10% dextran sulfate, 0.1% Tween 20) for 60 min, and then incubated the sections in hybridization buffer containing 10 nm initiator-labeled probes at 37°C for 16 h.
After hybridization, we washed the sections at 37°C with probe wash buffer (30% formamide, 5× SSC, 0.1% Tween 20) 3 times and twice with 5× SSCTw for 15 min each. We equilibrated the sections in amplification buffer for 60 min (5× SSC, 10% dextran sulfate, 0.1% Tween 20). We diluted fluorophore-labeled hairpins separately from 3 μm stock to a 2.25 μm final concentration in 20× SSC, heated at 90°C for 90 s, and then snap-cooled to room temperature for 30 min in the dark. We further diluted snap-cooled hairpins to 60 nm final concentration in amplification buffer. We incubated the sections in amplification buffer with hairpins for 16 h at room temperature. Finally, we washed the sections in 5× SSCTw twice for 30 min and mounted and coverslipped slides with Vectashield antifade mounting medium (catalog #H-1000, Vector Labs).
Confocal microscopy (University of Michigan)
We acquired image stacks using an Olympus Fluoview-3000 confocal microscope that consisted of three channels: DAPI, Cre, and Oprm1. For quantitative colocalization analysis, we used 10× magnification objective lens (Olympus UPLSAPO10X2, NA 0.4/working distance 2.2 mm) to acquire image-stacks (xy dimension 1.59 µm/pixel × 1.59 µm/pixel and z step of 4.5 µm; 4-6 z slices per stack). For high-magnification representative images, we used 40× (silicone oil immersion, Olympus UPLSAPO40XS, NA 1.25/working distance 0.3 mm) objectives. We selected the image acquisition settings (mainly the PMT Voltage and the laser transmissivity) for optimal pixel saturation to avoid excessive or weak signal and kept these settings constant for all sections.
Image processing and analysis (University of Michigan)
We used open-source ImageJ/Fiji software (Schindelin et al., 2012; Schneider et al., 2012) for processing and quantitation, and Amira (Fisher Scientific) for visualization representation. We processed and quantified images of 2 or 3 sections per rat (n = 5 males/per genotype) per brain area. ROIs were drawn following the coronal diagrams and Nissl stain plates (Paxinos and Watson, 2007) and saved in the region of interest (ROI) manager for quantification. 3D image stacks were processed globally, first using the subtract background (rolling = 50 stack) tool and then filtered using nonlocal means denoising method (auto estimate σ), an adaptive-manifold-based approach which naturally preserves most features of objects and reduces background noise. Next, Gaussian blur (σ = 1) was used to concentrate the signal toward the wcenter of each cell and then segmented using auto local threshold (method = Phansalkar radius = 15 parameter_1 = 0 parameter_2 = 0). After a watershed split, we estimated iCre and Oprm1 cell numbers in each channel using analyze particle tool (size = 15-400, circularity = 0.3-1.0). Subsequently, the generated masks of each channel (iCre and Oprm1) were used to quantify the number of colocalized cell bodies through Image Calculator 'AND' and Analyze 'Particle' tools. We estimated area of ROIs using maximum thresholding values, which then were used to quantify neurons per mm2 (density number) and percent of colocalized cells and performed statistical analyses on both density and percent data.
RNAScope ISH and immunohistochemistry (NIDA)
We deeply anesthetized the rats with isoflurane and rapidly decapitated them. We extracted and flash-froze the brains in isopentane solution on dry ice. We sectioned the brains at 20 µm in a cryostat (−13°C to 15°C), air-dried them on the slides at −20°C, and stored them at −80°C. For the detection of Oprm1 mRNA, we used the RNAscope 2.5 HD Assay-RED kit (322360; Advanced Cell Diagnostics). We fixed the sections for 20 min in neutral buffered 10% formalin, followed by ethanol dehydration series on 50%, 70%, 95%, and 100%, 5 min each. We stored the sections overnight at −20°C in 100% ethanol.
Before hybridization, we incubated the sections in H2O2 followed by protease IV (322340, Advanced Cell Diagnostics). We hybridized the sections with the Oprm1 mRNA probe (catalog #410691, Advanced Cell Diagnostics) for 2 h at 40°C and amplified the probe using RNAScope amplifiers as directed by the manufacturer. For brightfield, we detected the Oprm1 probe with the chromophore Fast Red. We counterstained the sections with methylene blue, removed the excess dye, washed the sections 3 times for 1 min in ddH20, dried the sections in the oven at 60°C for 15 min, cooled the slides to room temperature, dipped them in Citrosolv (catalog #04-355-121, Fisher Scientific), and coverslipped the slides with Permount (catalog #SP15-100, Fisher Scientific).
Image processing and analysis (NIDA)
For brightfield microscopy, we imaged Oprm1 mRNA signal that was labeled Fast Red and nuclei that were labeled with methylene blue. To quantify Capase-3-induced lesions using Fiji ImageJ, we drew the ROI for NAc shell and core on both hemispheres, segmented fast red labeling from methylene blue staining using Giemsa color deconvolution, made thresholds for red grains, and segmented from channel 2 of the color deconvolution. We divided the pixels covered by grains by the pixels within the ROI to determine the percent of area covered by the Oprm1 mRNA signal (% area covered by red grains).
[35S]GTPγS autoradiography
We cut frozen brain sections at 20 µm using a cryostat and thaw-mounted the sections onto glass slides. We pipetted preincubation buffer onto each slide and incubated for 20 min at room temperature (50 mm Tris-HCl, 1 mm EDTA, 5 mm MgCl2, and 100 mm NaCl). We removed the buffer by aspiration and incubated the sections for 60 min in preincubation buffer containing 2 mm GDP and 1 μm DPCPX. We removed the GDP buffer and pipetted [35S]GTPγS cocktail (GDP buffer, 1 mm DTT, 0.625 nm [35S]GTPγS) with DAMGO (10 μm), without DAMGO (basal condition), or with a saturated concentration of nonradioactive GTP (for nonspecific binding) onto each slide and incubated for 90 min. We washed (2 × 5 min, 50 mm Tris-HCl, pH 7.4, 5 mm MgCl2), rinsed (30 s in ice water), and air-dried the slides. Along with radioactive standards (nCi/g), apposed them to a BAS-SR2040 phosphor screen (Fujifilm) for 3 d and imaged the slides using a PhosphorImager (Typhoon FLA 7000; GE Healthcare). We drew ROIs onto the sections with standards using Multigauge software (GE Healthcare) and expressed values as % basal.
[3H]DAMGO autoradiography
We cut frozen brain sections at 20 µm using a cryostat and thaw mounted the sections onto SuperPlus glass slides (Avantor). We preincubated slides (10 min, room temperature) in incubation buffer (50 mm Tris-HCl, pH 7.4), then incubated (60 min, room temperature) in incubation buffer containing [3H]DAMGO (5 nm). We determined nonspecific binding in the presence of 10 μm naloxone. After incubation, we washed (2 × 30 s, incubation buffer), rinsed (30 s in ice water), and air-dried the slides and, along with radioactive standards (nCi/g), apposed them to a BAS-TR2025 Phosphor Screen (Fujifilm) for 10 d and imaged using a PhosphorImager (Typhoon FLA 7000). We drew ROIs onto the sections with standards using Multigauge software (GE Healthcare) and expressed and calibrated values as nCi/g.
Apparatus (food and drug self-administration)
We trained and tested the rats in standard Med Associates self-administration chambers. Each chamber had two retractable levers located 7.5-8 cm above the grid floor on the right wall with a food receptacle between them, and an inactive nonretractable lever on the left side. A tone cue is located above one of the levers and a light cue is located above the other lever. Lever presses on the retractable levers activated either the infusion pump or a pellet dispenser. Lever presses on the inactive lever had no programmed consequences. In Experiment 2, the self-administration and extinction contexts differed in their auditory, visual, and tactile cues, as in our previous studies (Adhikary et al., 2017; Bossert et al., 2019). We refer to the contexts as A and B, where A is the context of self-administration training and reacquisition, and B is the context of extinction. We counterbalanced the physical environments of Contexts A and B.
Drugs
NIDA
We received heroin hydrochloride (HCl) and morphine sulfate from the NIDA pharmacy and dissolved them in sterile saline. The heroin unit doses of Experiment 2 are based on our previous work (Bossert et al., 2004, 2016, 2022) and the heroin unit doses in Experiments 4 and 5 are based on Stewart et al. (1996). The morphine (1 ml/kg, s.c.) doses and lactic acid (catalog #L1250, Sigma Aldrich, dissolved in sterile water, 1 ml/kg, i.p.) concentrations in Experiment 3 are based on Baldwin et al. (2022) and Reiner et al. (2021). We injected morphine 30 min before behavioral testing or 20 min before lactic acid injections; we injected lactic acid 10 min before behavioral testing.
University of Michigan
We prepared morphine sulfate from a pharmaceutical liquid stock (Mitigo USP preservative free, 25 mg/ml, Piramal Critical Care) that we diluted with sterile saline (Fresenius Kabi). We dissolved naloxone HCl (Tocris Bioscience) in sterile saline. We injected both drugs (i.p.) in a volume of 1 ml/kg. We injected naloxone 30 min before morphine, which we injected 60 min before behavioral testing.
Surgery
Intracranial surgery for viral delivery
In our proof-of-concept experiment, we injected AAV1-EF1a-Flex-taCasp3-TEVP (NIDA Genetics & Engineering Viral Vectors Core [GEVVC], lot #AAV-2015-11-10-B, titer: 5.16E + 11 vg/ml) unilaterally into the right hemisphere of NAc shell and PBS into the left hemisphere; injections were 500 nl/side. In Experiment 4, we injected AAV1-EF1a-Flex-taCasp3-TEVP (NIDA GEVVC, lot #AAV-2015-11-10-B, titer: 5.16E + 11 vg/ml) bilaterally into NAc shell; injections were 1000 nl/side. In Experiment 5, we injected AAV1-EF1a-DIO-EYFP (NIDA GEVVC, lot #AAV-2015-02-17-B, titer: 2.73E + 12 vg/ml) bilaterally into NAc shell; injections were 1000 nl/side. We used the following coordinates from bregma: AP, 1.6 mm; ML, 2.5 mm (10° angle); DV, −7.5 mm (males) and −7.3 mm (females). These coordinates are based on a previous study (Marchant et al., 2016). We delivered the AAVs using Nanofil syringes (WPI, 33 gauge) at a rate of 100 nl/min. After each injection, we left the injection needle in place for 3 min to allow for diffusion. After the injections, we filled the drilled holes with bone wax and closed the wounds using autoclips (Texas Scientific Instruments).
Intravenous surgery
We anesthetized the rats with isoflurane (5% induction; 2%-3% maintenance, Covetrus). We attached Silastic catheters to a modified 22-gauge cannula cemented to polypropylene mesh (Industrial Netting), inserted the catheter into the jugular vein, and fixed the mesh to the mid-scapular region of the rat (Caprioli et al., 2015; Fredriksson et al., 2020). We injected the rats with ketoprofen (2.5 mg/kg, s.c., Covetrus) during surgery and on the following day to relieve pain and decrease inflammation. We also injected Enrofloxacin (2.27% diluted 1:9 in sterile saline, s.c., Covetrus) during surgery, 4-5 d after surgery, and if we observed an infection during the experiment. The rats recovered for 6-8 d before heroin self-administration training. During all experimental phases, we flushed the catheters daily with gentamicin in sterile saline (4.25 mg/ml, 0.1 ml, Fresenius Kabi).
Behavioral experiments
Experiment 1: food self-administration
The goal of Experiment 1 was to determine whether there are differences between Oprm1-Cre rats and their wildtype littermates in operant learning and performance for a nondrug reward. For this purpose, we used 45 mg high carbohydrate food pellets (TestDiet, catalog #1811155) that food-sated rats strongly prefer over heroin, fentanyl, and methamphetamine (Caprioli et al., 2015; Venniro et al., 2017; Reiner et al., 2020). We trained and tested the rats (8 males, 12 females) ∼5 h after the onset of the dark cycle (8:00 A.M.). The experiment consisted of two phases: (1) acquisition of food self-administration for 7 d for 1 h/d under a fixed-ratio 1 (FR1) 20 s timeout reinforcement schedule, and (2) tests for food self-administration after increasing the response requirements from FR1 to FR8 in the following sequence: FR1 (3 d), FR2 (1 d), FR4 (1 d), FR6 (1 d), and FR8 (1 d).
Each session began with the illumination of the houselight and the insertion of the food-paired active lever 10 s later. During the first 7 daily acquisition sessions, we mildly food-restricted the rats (removed their food between 8:00 A.M. and 9:00 A.M.) and gave them 1 h magazine-training sessions before the operant training during which 1 pellet was delivered noncontingently every 2 min. Lever presses led to the delivery of one 45 mg pellet and each pellet delivery was paired with a 20 s white-light cue.
Experiment 2: heroin self-administration and relapse-related behaviors
The goal of Experiment 2 was to determine whether there are differences between Oprm1-Cre rats and their wildtype littermates in heroin self-administration and commonly used relapse-related behaviors: extinction responding, context-induced reinstatement, and reacquisition (Bossert et al., 2013; Venniro et al., 2016; Khoo et al., 2017). We used a variation of the ABA context-induced reinstatement (renewal) procedure in which rats are trained to self-administration heroin in Context A, are tested for extinction of heroin-reinforced responding in Context B, and then tested for context-induced reinstatement of heroin seeking and reacquisition in Context A (Bossert et al., 2020, 2022).
Training in Context A (12 d)
We trained the rats (12 males, 16 females) to self-administer heroin HCl in Context A for 6 h/day (six 1 h sessions separated by 10 min) for 12 d. Each session began with the illumination of the houselight that remained on for the entire session; the active lever was inserted into the chamber 10 s after the houselight was illuminated. During training, the rats earned heroin infusions by pressing on the active lever; infusions were paired with a compound tone–light cue for 3.5 s under an FR1 20 s timeout reinforcement schedule. Heroin was infused at a volume of 100 µl over 3.5 s at a dose of 100 µg/kg/infusion (first 6 sessions) and then 50 µg/kg/infusion (last 6 sessions). Lever presses on the active lever during the timeout period were recorded but did not result in heroin infusions. Presses on the inactive lever were recorded but had no programmed consequences. At the end of each 1 h session, the houselight turned off and the active lever was retracted. If we suspected catheter failure during training, we tested patency with Diprivan (propofol, NIDA pharmacy, 10 mg/ml, 0.1-0.2 ml injection volume, i.v.).
Extinction responding in Context B (7 d)
We ran the rats under extinction conditions in Context B for 6 h per day (six 1 h sessions separated by 10 min) for 7 d. During this phase, presses on the previously active lever led to presentation of the discrete tone-light cue but not heroin infusions.
Context-induced reinstatement in contexts A and B (2 d)
We tested the rats under extinction conditions (see above) for 6 h per day for 2 d in Context A and Context B in a counterbalanced order.
Reacquisition of heroin self-administration in Context A (1 d)
We tested reacquisition of heroin self-administration during one 6 h session in Context A. During testing, lever presses were reinforced by heroin (50 µg/kg/infusion, FR1 20 s timeout reinforcement schedule) and the discrete tone-light cue. After the 6 h session, we tested catheter patency with propofol (NIDA pharmacy, 10 mg/ml, 0.1-0.2 ml injection volume, i.v.).
Experiment 3: evaluation of pain-related responses using von Frey test, tail flick test, and lactic acid-induced suppression of operant responding
The goal of Experiment 3 was to determine whether there are differences between Oprm1-Cre rats and their wildtype littermates in pain sensitivity and morphine analgesia using three different methods: von Frey test, tail flick test, and lactic acid-induced suppression of operant responding.
Experiment 3a: von Frey test
We performed all testing in a quiet, dimly lit room; and we gave the rats ∼30 min to habituate to the testing environment before testing began. We assessed sensitivity to mechanical stimulation using nylon von Frey filaments (BiosEB). We placed the rats (14 males, 6 females) on a stainless-steel 1 cm square mesh grid and applied von Frey filaments to the plantar surface of both hind paws using the sampling method described by Wang et al. (2005). We obtained paw withdrawal threshold scores for both hind paws and averaged them to produce a single composite withdrawal score for each rat. Following an initial test of baseline response, we injected rats with increasing doses of morphine (0.625, 1.25, and 2.5 mg/kg, i.p.) 60 min before test, 1 dose per day. After acute analgesia testing, we injected naloxone (1 mg/kg, i.p.) 30 min before morphine (2.5 mg/kg), which was injected 60 min before test. Following this test, we injected the rats daily for 21 d with 2.5 mg/kg morphine to induce analgesic tolerance, and then retested their analgesic response to this dose of morphine. We compared the analgesic response after tolerance development to the response to the same dose during the acute analgesic phase of the experiment.
Experiment 3b: tail flick test
We performed all testing in a quiet, dimly lit room; and we gave the rats ∼30 min to habituate to the testing environment before testing began. We used a commercially available tail flick apparatus (IITC Life Science) to assess latency to tail flick from a noxious thermal stimulus. We calibrated the testing intensity to provide reliable tail flick latencies of ∼4 s at baseline, and an automatic cutoff time of 15 s to prevent tissue damage. We gently restrained the rats (10 males, 10 females) and then applied the noxious thermal stimulus to the caudal one-third of the tail and automatically recorded latency to flick the tail away from the stimulus. We conducted 2 tests per rat at least 1 min apart at distinctive locations along the tail to prevent sensitization. We averaged the latencies to produce a single composite latency score for each rat. After an initial test of baseline response, we injected the rats with increasing doses of morphine (1.25, 2.5, 5, and 10 mg/kg, i.p.) 60 min before test, 1 dose per day. After acute analgesia testing, we injected naloxone (1 mg/kg, i.p.) 30 min before morphine (5 mg/kg), which was injected 60 min before test. Following this test, we injected the rats daily for 21 d with 5 mg/kg morphine to induce analgesic tolerance, and then retested their analgesic response to this dose of morphine. We compared the analgesic response after tolerance development to the response to the same dose during the acute analgesic phase of the experiment.
Experiment 3c: lactic acid-induced behavioral depression
Using the 45 mg pellets described in Experiment 1, we trained and tested the rats (8 males, 8 females) ∼5 h after the onset of the dark cycle (8:00 A.M.). The experiment consisted of four phases: (1) acquisition of food self-administration for 6 d for 1 h/d under an FR1 20 s timeout reinforcement schedule, (2) acute injections of lactic acid (0%, 0.9%, 1.3%, and 1.8%) 10 min before food self-administration sessions, (3) acute injections of morphine (0, 1, 3, and 10 mg/kg) 30 min before food self-administration sessions, and (4) acute injections of morphine (0, 1, and 3 mg/kg) 20 min before acute injections of lactic acid (1.8%) 10 min before food self-administration sessions. All injections were counterbalanced within each phase, and we ran baseline sessions (no injections) in between every test day during all phases. During the first three acquisition sessions, we mildly food-restricted the rats (removed their food between 8:00 A.M. and 9:00 A.M.) and gave them 1 h magazine-training sessions before the operant training as described above. Lever presses led to the delivery of one 45 mg pellet, and each pellet delivery was paired with a 20 s white-light cue.
Experiment 4: effect of cre-dependent AAV1-EF1a-Flex-taCasp3-TEVP (AAV-DIO-Casp3) NAc lesions on acquisition and maintenance of heroin self-administration
Striatal MORs have been implicated in heroin self-administration in rats and mice (Vaccarino et al., 1985; Wise, 1989; Koob, 1992; Cui et al., 2014). Based on this knowledge, the goal of Experiment 4 was to behaviorally validate the Oprm1-Cre knock-in rat by demonstrating that Cre-dependent lesions of NAc MOR-expressing neurons will decrease acquisition and maintenance of heroin self-administration in Oprm1-Cre rats.
Experiment 4 consisted of five phases: (1) concurrent acquisition of food (morning) and heroin (afternoon) self-administration (12 d, 3 h/d, 3 d/heroin dose), (2) within-session heroin dose–response (1 d, 2 h/dose), (3) within-session heroin FR response (1 d, 1 h/FR requirement), (4) extended access heroin session (1 d, 9 h), and (5) within-session food FR response (1 d, 1 h/FR requirement).
Acquisition: Food and heroin self-administration
We trained the rats (28 males, 28 females) to self-administer food and heroin with a 3 h food session (1 pellet per reward delivery) in the morning and a 3 h heroin session in the afternoon; food pellets were paired with a 20 s white-light cue on one lever, and heroin infusions were paired with a 5 s tone on a different lever. Heroin was infused at a volume of 100 µl over 3.5 s at a dose of 12.5 µg/kg/infusion (the first 3 d) and then 25, 50, and 100 µg/kg/infusion for each subsequent 3 d. This acquisition procedure is based on a previous study of Stewart et al. (1996).
Heroin maintenance: within-session dose–response
After the rats learned to self-administer heroin, we tested them using an ascending within-session dose–response curve procedure (Deroche et al., 1999; Fredriksson et al., 2017). We tested the ascending heroin doses of 12.5, 25, 50, and 100 µg/kg/infusion for 2 h per dose under an FR1 20 s timeout reinforcement schedule. We used two sets of stock solutions and manipulated the intended drug dose by using two different infusion times (1.75 s for the 12.5 and 50 µg/kg unit doses, and 3.5 s for the 25 and 100 µg/kg unit doses). We excluded 2 male rats (n = 1 per genotype) because they showed no evidence of acquiring heroin self-administration (three infusions per day or less during the 12 acquisition sessions) despite having patent catheters. We also excluded 3 Oprm1-Cre rats (2 males, 1 female) because of loss of catheter patency when tested at the end of the within-session dose–response. We also excluded these rats from the subsequent tests described below.
Heroin maintenance: ascending within-session fixed-ratio response
We tested the rats in 1 h consecutive sessions for heroin self-administration (50 µg/kg/infusion) under FR1, FR2, FR4, FR8, FR16, FR32, and FR64 reinforcement schedules. This procedure is based on a study by Chow et al. (2022).
Heroin maintenance: extended access
We tested the rats in a single 9 h heroin (50 µg/kg/infusion, FR1 20 s timeout reinforcement schedule) self-administration session. We retested 1 rat whose tubing was disconnected during the test session for determining heroin FR response.
Food self-administration: ascending within-session FR response
We tested some of the rats (15 males, 11 females) in 1 h consecutive sessions for food self-administration under FR1, FR2, FR4, FR8, FR16, FR32, and FR64 reinforcement schedules. We ran Experiment 4 in two cohorts, and only performed this test on the second cohort.
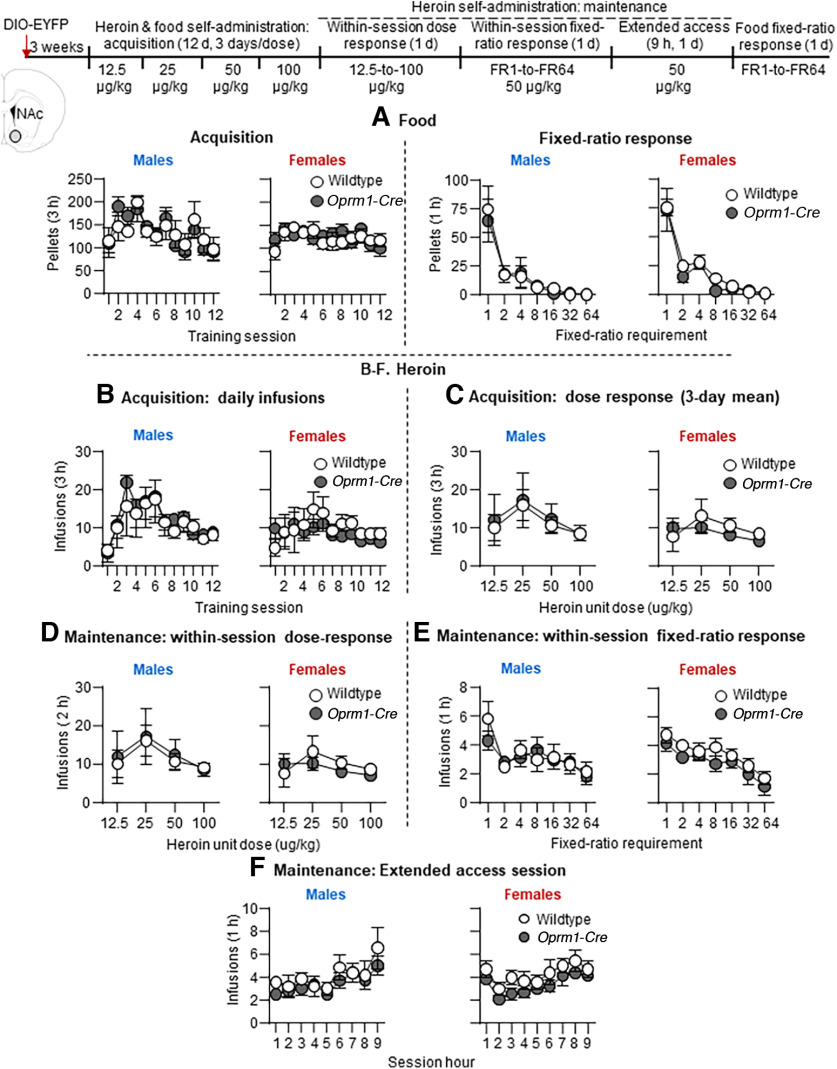
Experiment 5: effect of control virus AAV1-EF1a-DIO-EYFP (AAV-DIO-EYFP) into NAc on acquisition and maintenance of heroin self-administration
The goal of Experiment 5 was to demonstrate the specificity of the effect of AAV-DIO-Casp3 in Oprm1-Cre rats by using a control virus. For this purpose, we injected a virus that has the same Cre-dependent mechanism and same promoter as AAV-DIO-Casp3 but does contain the taCasp-TEVP component that activates the cells and induces apoptosis.
Experiment 5 consisted of the same five phases as Experiment 4 and was run in the same sequence (13 males, 15 females).
Statistical analysis
We analyzed datasets without any missing values with GLM procedure of SPSS (version 27). We analyzed datasets with missing values with linear mixed effects modeling (Gelman and Hill, 2006) in JMP 16. Specifically, for the von Frey and tail flick tests, we analyzed the dose–response data with Genotype (nominal) as a fixed between-subjects factor, Dose (nominal) as a fixed within-subjects factor, and Subject as a random factor. For the Morphine + Naloxone and Morphine + Tolerance data, we analyzed the data with Genotype (nominal) as a fixed between-subjects factor, Treatment Condition (nominal) as a fixed within-subjects factor, and Subject as a random factor. We also used Linear mixed effect modeling in JMP 16 to analyze the heroin self-administration data because some rats were disconnected from the tubing line (9 events of 660 events for acquisition and 3 events of 450 events for extended access). For acquisition, we analyzed the data with Genotype and Sex (both nominal) as fixed between-subjects factors, Dose (nominal) as a fixed within-subjects factor, and Subject as a random factor. For extended access, we analyzed the data with Genotype and Sex (both nominal) as fixed between-subjects factors, Hour (nominal) as a fixed within-subjects factor, and Subject as a random factor.
In the figures, we indicate post hoc (Fisher PLSD test) genotype differences between each sex and within each sex after significant main effects or interactions (see Results). Because our ANOVAs yielded multiple main and interaction effects, we only report statistical effects that are critical for data interpretation in Results. We used Sex as an experimental factor in Experiment 4 because it is the only experiment that was statistically powered to detect sex differences. For complete statistical results, see Table 4.
Table 4.
| Figure | Factor name | F | p | Partial Eta2 |
|---|---|---|---|---|
| Figure 2: males only | ||||
| 2A: HCR FISH analysis: Oprm1 mRNA NAc | Between-subjects: Genotype | F(1,8) = 3.0 | 0.123 | 0.270 |
| 2A: HCR FISH analysis: Oprm1 mRNA DS | Between-subjects: Genotype | F(1,8) = 3.9 | 0.083 | 0.330 |
| 2A: HCR FISH analysis: Oprm1 mRNA dHipp | Between-subjects: Genotype | F(1,8) = 0.5 | 0.480 | 0.064 |
| 2B: HCR FISH analysis: Cre ± NAc | Within-subjects: Cre ± | F(1,4) = 356.1 | <0.001* | 0.989 |
| 2B: HCR FISH analysis: Cre ± DS | Within-subjects: Cre ± | F(1,4) = 652.4 | <0.001* | 0.994 |
| 2B: HCR FISH analysis: Cre ± dHipp | Within-subjects: Cre ± | F(1,4) = 172.3 | <0.001* | 0.977 |
| Figure 3: males and females | ||||
| 3A: NAc [35S]GTPγS activity | Between-subjects: Genotype | F(1,10) = 2.1 | 0.178 | 0.173 |
| 3A: DS [35S]GTPγS activity | Between-subjects: Genotype | F(1,10) = 0.2 | 0.636 | 0.023 |
| 3B: NAc [3H]DAMGO binding | Between-subjects: Genotype | F(1,10) = 0.05 | 0.820 | 0.005 |
| 3B: DS [3H]DAMGO binding | Between-subjects: Genotype | F(1,10) = 6.7 | 0.027* | 0.400 |
| 3C: NAc [3H]DAMGO binding: NAc shell | Between-subjects: Genotype | F(1,9) = 0.2 | 0.666 | 0.022 |
| Within-subjects: Lesion (Vehicle, Caspase3) | F(1,9) = 11.3 | 0.008* | 0.556 | |
| Genotype × Lesion | F(1,9) = 8.4 | 0.018* | 0.482 | |
| 3C: NAc [35S]GTPγS activity: NAc shell | Between-subjects: Genotype | F(1,9) = 1.7 | 0.224 | 0.159 |
| Within-subjects: Lesion (Vehicle, Caspase3) | F(1,9) = 3.2 | 0.106 | 0.264 | |
| Genotype × Lesion | F(1,9) = 4.8 | 0.056 | 0.348 | |
| 3D: RNAscope: NAc shell | Between-subjects: Genotype | F(1,9) = 3.6 | 0.089 | 0.288 |
| Within-subjects: Lesion (Vehicle, Caspase3) | F(1,9) = 12.5 | 0.006* | 0.581 | |
| Genotype × Lesion | F(1,9) = 9.9 | 0.012* | 0.525 | |
| 3D: RNAscope: NAc core | Between-subjects: Genotype | F(1,9) = 1.7 | 0.230 | 0.156 |
| Within-subjects: Lesion (Vehicle, Caspase3) | F(1,9) = 5.9 | 0.038* | 0.396 | |
| Genotype × Lesion | F(1,9) = 1.6 | 0.236 | 0.152 | |
| Figure 4: males and females | ||||
| 4A: Acquisition: Pellets | Between-subjects: Genotype | F(1,18) = 0.04 | 0.839 | 0.002 |
| Within-subjects: Session | F(6108) = 24.1 | <0.001* | 0.573 | |
| Genotype × Session | F(6108) = 0.5 | 0.833 | 0.025 | |
| 4A: Acquisition: Lever presses | Between-subjects: Genotype | F(1,18) = 0.1 | 0.711 | 0.008 |
| Within-subjects: Session | F(6108) = 7.8 | <0.001* | 0.303 | |
| Genotype × Session | F(6108) = 0.3 | 0.923 | 0.018 | |
| 4A: FR response: Pellets | Between-subjects: Genotype | F(1,18) = 0.0 | 0.987 | 0.000 |
| Within-subjects: FR requirement | F(4,72) = 34.4 | <0.001* | 0.656 | |
| Genotype × FR requirement | F(4,72) = 1.4 | 0.248 | 0.071 | |
| 4A: FR response: Lever presses | Between-subjects: Genotype | F(1,18) = 0.03 | 0.874 | 0.001 |
| Within-subjects: FR requirement | F(4,72) = 12.3 | <0.001* | 0.407 | |
| Genotype × FR requirement | F(4,72) = 1.2 | 0.311 | 0.063 | |
| 4B: Heroin SA: infusions | Between-subjects: Genotype | F(1,25) = 0.1 | 0.767 | 0.004 |
| Within-subjects 1: Heroin Dose | F(1,25) = 62.2 | <0.001* | 0.713 | |
| Within-subjects 2: Session | F(5125) = 5.5 | <0.001* | 0.181 | |
| Genotype × Session | F(5125) = 0.9 | 0.515 | 0.033 | |
| Heroin Dose × Genotype | F(1,25) = 0.9 | 0.365 | 0.033 | |
| Heroin Dose × Session | F(5125) = 10.0 | <0.001* | 0.286 | |
| Genotype × Heroin Dose × Session | F(5125) = 0.5 | 0.768 | 0.020 | |
| 4B: Heroin: Extinction responding | Between-subjects: Genotype | F(1,25) = 1.3 | 0.273 | 0.048 |
| Within-subjects: Session (1-7) | F(6150) = 49.9 | <0.001* | 0.666 | |
| Genotype × Session | F(6150) = 0.1 | 0.994 | 0.005 | |
| 4B: Heroin: Context-induced reinstatement | Between-subjects: Genotype | F(1,25) = 0.00 | 0.963 | 0.000 |
| Within-subjects: Context (A, B) | F(1,25) = 65.4 | <0.001* | 0.723 | |
| Genotype × Context | F(1,25) = 0.1 | 0.742 | 0.004 | |
| 4B: Heroin: Reacquisition | Between-subjects: Genotype | F(1,25) = 0.5 | 0.474 | 0.021 |
| Within-subjects: Session Hour (1-6) | F(5125) = 6.0 | <0.001* | 0.194 | |
| Genotype × Session Hour | F(5125) = 1.6 | 0.166 | 0.060 | |
| Figure 5: males and females | ||||
| 5A: von Frey: Dose–response | Between-subjects: Genotype | F(1,22.35) = 0.0 | 0.931 | |
| Within-subjects: Dose | F(3,47.72) = 73.0 | <0.001* | ||
| Genotype × Dose | F(3,47.72) = 0.1 | 0.931 | ||
| 5A: von Frey: morphine + naloxone | Between-subjects: Genotype | F(1,17.45) = 0.0 | 0.917 | |
| Within-subjects: Condition | F(2,17.45) = 65.4 | <0.001* | ||
| Genotype × Condition | F(2,17.45) = 0.1 | 0.924 | ||
| 5A: von Frey: morphine + tolerance | Between-subjects: Genotype | F(1,30.54) = 0.0 | 0.897 | |
| Within-subjects: Condition | F(2,30.54) = 87.4 | <0.001* | ||
| Genotype × Condition | F(2,30.54) = 0.1 | 0.905 | ||
| 5B: Tail flick: Dose–response | Between-subjects: Genotype | F(1,19.4) = 0.0 | 0.979 | |
| Within-subjects: Dose | F(4,61.58) = 58.1 | <0.001* | ||
| Genotype × Dose | F(4,61.58) = 0.6 | 0.700 | ||
| 5B: Tail flick: morphine + naloxone | Between-subjects: Genotype | F(1,19.75) = 0.1 | 0.723 | |
| Within-subjects: Condition | F(2,23.01) = 22.8 | <0.001* | ||
| Genotype × Condition | F(2,23.01) = 0.1 | 0.908 | ||
| 5B: Tail flick: morphine + tolerance | Between-subjects: Genotype | F(1,18.93) = 0.2 | 0.658 | |
| Within-subjects: Condition | F(2,33.63) = 20.8 | <0.001* | ||
| Genotype × Condition | F(2,33.63) = 0.1 | 0.885 | ||
| 5C: Lactic acid dose–response: change baseline | Between-subjects: Genotype | F(1,13) = 0.02 | 0.889 | 0.002 |
| Within-subjects: Dose | F(3,39) = 17.2 | <0.001* | 0.569 | |
| Genotype × Dose | F(3,39) = 0.9 | 0.471 | 0.062 | |
| 5C: Morphine dose–response: change baseline | Between-subjects: Genotype | F(1,13) = 0.4 | 0.557 | 0.027 |
| Within-subjects: Dose | F(3,39) = 58.1 | <0.001* | 0.817 | |
| Genotype × Dose | F(3,39) = 0.05 | 0.984 | 0.004 | |
| 5C: Morphine + lactic acid: change baseline | Between-subjects: Genotype | F(1,13) = 1.7 | 0.217 | 0.115 |
| Within-subjects: Dose | F(2,26) = 25.0 | <0.001* | 0.658 | |
| Genotype × Dose | F(2,26) = 0.2 | 0.828 | 0.014 | |
| Figure 6: males and females | ||||
| 6A: Food Acquisition: Pellets | Between-subjects 1: Genotype | F(1,51) = 0.1 | 0.753 | 0.002 |
| Between-subjects 2: Sex | F(1,51) = 1.2 | 0.27 | 0.024 | |
| Genotype × Sex | F(1,51) = 0.3 | 0.605 | 0.005 | |
| Within-subjects: Session | F(11,561) = 24.2 | <0.001* | 0.322 | |
| Genotype × Session | F(11,561) = 1.7 | 0.072 | 0.032 | |
| Sex × Session | F(11,561) = 1.6 | 0.109 | 0.030 | |
| Genotype × Sex × Session | F(11,561) = 0.7 | 0.727 | 0.014 | |
| 6A: Food FR requirement: Pellets | Between-subjects 1: Genotype | F(1,22) = 1.0 | 0.339 | 0.042 |
| Between-subjects 2: Sex | F(1,22) = 0.001 | 0.981 | 0.000 | |
| Genotype × Sex | F(1,22) = 0.2 | 0.665 | 0.009 | |
| Within-subjects: FR requirement | F(6132) = 49.8 | <0.001* | 0.693 | |
| Genotype × FR requirement | F(6132) = 0.6 | 0.761 | 0.025 | |
| Sex × FR requirement | F(6132) = 1.7 | 0.133 | 0.071 | |
| Genotype × Sex × FR requirement | F(6132) = 0.5 | 0.773 | 0.024 | |
| 6B: Heroin Acquisition: Dose–response | Between-subjects 1: Genotype | F(1,49) = 5.1 | 0.029* | |
| Between-subjects 2: Sex | F(1,49) = 4.1 | 0.049* | ||
| Genotype × Sex | F(1,49) = 1.9 | 0.169 | ||
| Within-subjects 1: Dose | F(3147) = 0.7 | 0.545 | ||
| Dose × Sex | F(3147) = 1.5 | 0.225 | ||
| Dose × Genotype | F(3147) = 0.9 | 0.437 | ||
| Dose × Sex × Genotype | F(3147) = 2.1 | 0.109 | ||
| 6C: Heroin Acquisition 3 d mean: Dose–response | Between-subjects 1: Genotype | F(1,49) = 5.1 | 0.029* | 0.094 |
| Between-subjects 2: Sex | F(1,49) = 4.1 | 0.049* | 0.077 | |
| Genotype × Sex | F(1,49) = 2.0 | 0.167 | 0.039 | |
| Within-subjects: Dose | F(3147) = 0.7 | 0.547 | 0.014 | |
| Sex × Dose | F(3147) = 1.5 | 0.224 | 0.029 | |
| Genotype × Dose | F(3147) = 0.9 | 0.437 | 0.018 | |
| Sex × Genotype × Dose | F(3147) = 2.0 | 0.111 | 0.040 | |
| 6D: Heroin Maintenance: Within-session dose–response | Between-subjects 1: Genotype | F(1,46) = 4.7 | 0.035* | 0.093 |
| Between-subjects 2: Sex | F(1,46) = 3.0 | 0.088 | 0.062 | |
| Genotype × Sex | F(1,46) = 0.1 | 0.714 | 0.003 | |
| Within-subjects: Dose | F(3138) = 88.2 | <0.001* | 0.657 | |
| Dose × Genotype | F(3138) = 3.1 | 0.029* | 0.063 | |
| Dose × Sex | F(3138) = 0.8 | 0.502 | 0.017 | |
| Dose × Genotype × Dose | F(3138) = 1.2 | 0.301 | 0.026 | |
| 6E: Heroin Maintenance: Within-session FR response | Between-subjects 1: Genotype | F(1,46) = 14.0 | <0.001 | 0.234 |
| Between-subjects 2: Sex | F(1,46) = 3.0 | 0.088 | 0.062 | |
| Genotype × Sex | F(1,46) = 0.8 | 0.387 | 0.016 | |
| Within-subjects: FR requirement | F(6276) = 54.6 | <0.001* | 0.543 | |
| FR requirement × Genotype | F(6276) = 1.7 | 0.130 | 0.035 | |
| FR requirement × Sex | F(6276) = 0.6 | 0.741 | 0.013 | |
| FR requirement × Genotype × Sex | F(6276) = 1.0 | 0.395 | 0.022 | |
| 6F: Heroin Maintenance: Extended access | Between-subjects 1: Genotype | F(1,46) = 5.4 | 0.025* | |
| Between-subjects 2: Sex | F(1,46) = 8.0 | 0.007* | ||
| Genotype × Sex | F(1,46) = 0.03 | 0.870 | ||
| Within-subjects: Hour | F(8365) = 5.1 | <0.001* | ||
| Hour × Genotype | F(8365) = 2.0 | 0.041* | ||
| Hour × Sex | F(8365) = 1.7 | 0.100 | ||
| Hour × Genotype × Sex | F(8365) = 1.6 | 0.112 | ||
| 6G: NAc [3H]DAMGO binding | Between-subjects: Genotype | F(1,41) = 20.7 | <0.001* | 0.335 |
| Between-subjects: Sex | F(1,41) = 9.5 | 0.004* | 0.189 | |
| Genotype × Sex | F(1,41) = 1.4 | 0.240 | 0.033 | |
| Figure 6: males only | ||||
| 6A: Food Acquisition: Pellets | Between-subjects 1: Genotype | F(1,26) = 0.4 | 0.528 | 0.016 |
| Within-subjects: Session | F(11,286) = 14.3 | <0.001* | 0.355 | |
| Session × Genotype | F(11,286) = 1.4 | 0.182 | 0.050 | |
| 6A: Food FR requirement: Pellets | Between-subjects: Genotype | F(1,13) = 1.2 | 0.298 | 0.894 |
| Within-subjects: FR requirement | F(6,78) = 21.8 | <0.001* | 0.627 | |
| Genotype × FR requirement | F(6,78) = 0.6 | 0.712 | 0.046 | |
| 6B: Heroin Acquisition: Dose–response | Between-subjects: Genotype | F(1,24) = 8.0 | 0.009* | |
| Within-subjects 1: Dose | F(3,72) = 1.0 | 0.418 | ||
| Dose × Genotype | F(3,72) = 1.5 | 0.209 | ||
| 6C: Heroin Acquisition 3 d mean: Dose–response | Between-subjects: Genotype | F(1,24) = 8.0 | 0.009* | 0.250 |
| Within-subjects: Dose | F(3,72) = 1.0 | 0.420 | 0.038 | |
| Dose × Genotype | F(3,72) = 1.5 | 0.214 | 0.060 | |
| 6D: Heroin Maintenance: Within-session dose–response | Between-subjects: Genotype | F(1,22) = 4.2 | 0.053 | 0.160 |
| Within-subjects: Dose | F(3,66) = 49.4 | <0.001* | 0.692 | |
| Dose × Genotype | F(3.66) = 4.6 | 0.006* | 0.172 | |
| 6E: Heroin Maintenance: Within-session FR response | Between-subjects: Genotype | F(1,22) = 5.2 | 0.033* | 0.191 |
| Within-subjects: FR requirement | F(6132) = 40.6 | <0.001* | 0.649 | |
| FR requirement × Genotype | F(6132) = 0.4 | 0.854 | 0.019 | |
| 6F: Heroin Maintenance: Extended access | Between-subjects: Genotype | F(1,22.03) = 2.6 | 0.122 | |
| Within-subjects: Hour | F(8174.1) = 2.0 | 0.053 | ||
| Hour × Genotype | F(8174.1) = 0.6 | 0.739 | ||
| 6G: NAc [3H]DAMGO binding | Between-subjects: Genotype | F(1,20) = 5.7 | 0.027* | 0.221 |
| Figure 6: females only | ||||
| 6A: Food Acquisition: Pellets | Between-subjects 1: Genotype | F(1,25) = 0.02 | 0.894 | 0.001 |
| Within-subjects: Session | F(11,275) = 11.3 | <0.001* | 0.312 | |
| Session × Genotype | F(11,275) = 1.0 | 0.443 | 0.039 | |
| 6A: Food FR requirement: Pellets | Between-subjects: Genotype | F(1,9) = 0.1 | 0.729 | 0.014 |
| Within-subjects: FR requirement | F(6,54) = 30.9 | <0.001* | 0.775 | |
| Genotype × FR requirement | F(6,54) = 0.5 | 0.785 | 0.055 | |
| 6B: Heroin Acquisition: Dose–response | Between-subjects: Genotype | F(1,25) = 0.3 | 0.578 | |
| Within-subjects 1: Dose | F(3,75) = 1.2 | 0.324 | ||
| Dose × Genotype | F(3,75) = 1.5 | 0.228 | ||
| 6C: Heroin Acquisition 3 d mean: Dose–response | Between-subjects: Genotype | F(1,25) = 0.3 | 0.579 | 0.013 |
| Within-subjects: Dose | F(3,75) = 1.2 | 0.323 | 0.045 | |
| Dose × Genotype | F(3,75) = 1.5 | 0.230 | 0.056 | |
| 6D: Heroin Maintenance: Within-session dose–response | Between-subjects: Genotype | F(1,25) = 2.0 | 0.168 | 0.075 |
| Within-subjects: Dose | F(3,75) = 40.0 | <0.001* | 0.616 | |
| Dose × Genotype | F(3,75) = 0.4 | 0.719 | 0.018 | |
| 6E: Heroin Maintenance: Within-session FR response | Between-subjects: Genotype | F(1,24) = 9.1 | 0.006* | 0.276 |
| Within-subjects: FR requirement | F(6144) = 20.5 | <0.001* | 0.461 | |
| FR requirement × Genotype | F(6144) = 2.0 | 0.073 | 0.076 | |
| 6F: Heroin Maintenance: Extended access | Between-subjects: Genotype | F(1,24.02) = 2.8 | 0.105 | |
| Within-subjects: Hour | F(8191) = 4.4 | <0.001* | ||
| Hour × Genotype | F(8191) = 2.6 | 0.010* | ||
| 6G: NAc [3H]DAMGO binding | Between-subjects: Genotype | F(1,21) = 16.4 | <0.001* | 0.439 |
| Figure 7: males and females | ||||
| 7A: Food Acquisition: Pellets | Between-subjects 1: Genotype | F(1,26) = 0.01 | 0.913 | 0.000 |
| Within-subjects: Session | F(11,286) = 6.8 | <0.001* | 0.208 | |
| Session × Genotype | F(11,286) = 0.7 | 0.729 | 0.027 | |
| 7A: Food FR requirement: Pellets | Between-subjects: Genotype | F(1,26) = 0.7 | 0.420 | 0.025 |
| Within-subjects: FR requirement | F(6156) = 50.7 | <0.001* | 0.661 | |
| Genotype × FR requirement | F(6156) = 0.2 | 0.980 | 0.007 | |
| 7B: Heroin Acquisition: Dose–response | Between-subjects: Genotype | F(1,26) = 2.4 | 0.132 | 0.085 |
| Within-subjects 1: Dose | F(3,78) = 4.7 | 0.005* | 0.153 | |
| Within-subjects 2: Session | F(2,52) = 4.6 | 0.014* | 0.152 | |
| Dose × Genotype | F(3,78) = 1.7 | 0.168 | 0.062 | |
| Dose × Session | F(6156) = 2.5 | 0.023* | 0.089 | |
| Session × Genotype | F(2,52) = 1.1 | 0.335 | 0.041 | |
| Dose × Genotype × Session | F(6156) = 1.1 | 0.380 | 0.040 | |
| 7C: Heroin Acquisition: 3 d mean dose–response | Between-subjects: Genotype | F(1,26) = 0.001 | 0.972 | 0.000 |
| Within-subjects 1: Dose | F(3,78) = 7.0 | <0.001* | 0.212 | |
| Dose × Genotype | F(3,78) = 0.7 | 0.576 | 0.025 | |
| 7D: Heroin Maintenance: Within-session dose–response | Between-subjects: Genotype | F(1,24) = 0.6 | 0.449 | 0.024 |
| Within-subjects: Dose | F(3,72) = 49.1 | <0.001* | 0.672 | |
| Dose × Genotype | F(3,72) = 0.3 | 0.816 | 0.013 | |
| 7E: Heroin Maintenance: Within-session FR response | Between-subjects 1: Genotype | F(1,24) = 1.0 | 0.325 | 0.040 |
| Within-subjects: FR requirement | F(6144) = 16.4 | <0.001* | 0.405 | |
| FR requirement × Genotype | F(6144) = 0.3 | 0.922 | 0.013 | |
| 7F: Heroin Maintenance: Extended access | Between-subjects 1: Genotype | F(1,24) = 1.9 | 0.182 | 0.073 |
| Within-subjects: Hour | F(8192) = 10.6 | <0.001* | 0.306 | |
| Hour × Genotype | F(8192) = 0.4 | 0.939 | 0.015 |
aPartial Eta2 = proportion of explained variance. FR, fixed ratio; SA, self-administration.*
Results
Anatomical and cellular validation of the CRISPR-mediated knock-in of T2A-iCre
Figure 1 shows a schematic of the target gene (rat Oprm1) with annotation for the location and sequence of the SpCas9 gRNA that cleaves immediately before the stop codon.
HCR FISH assay
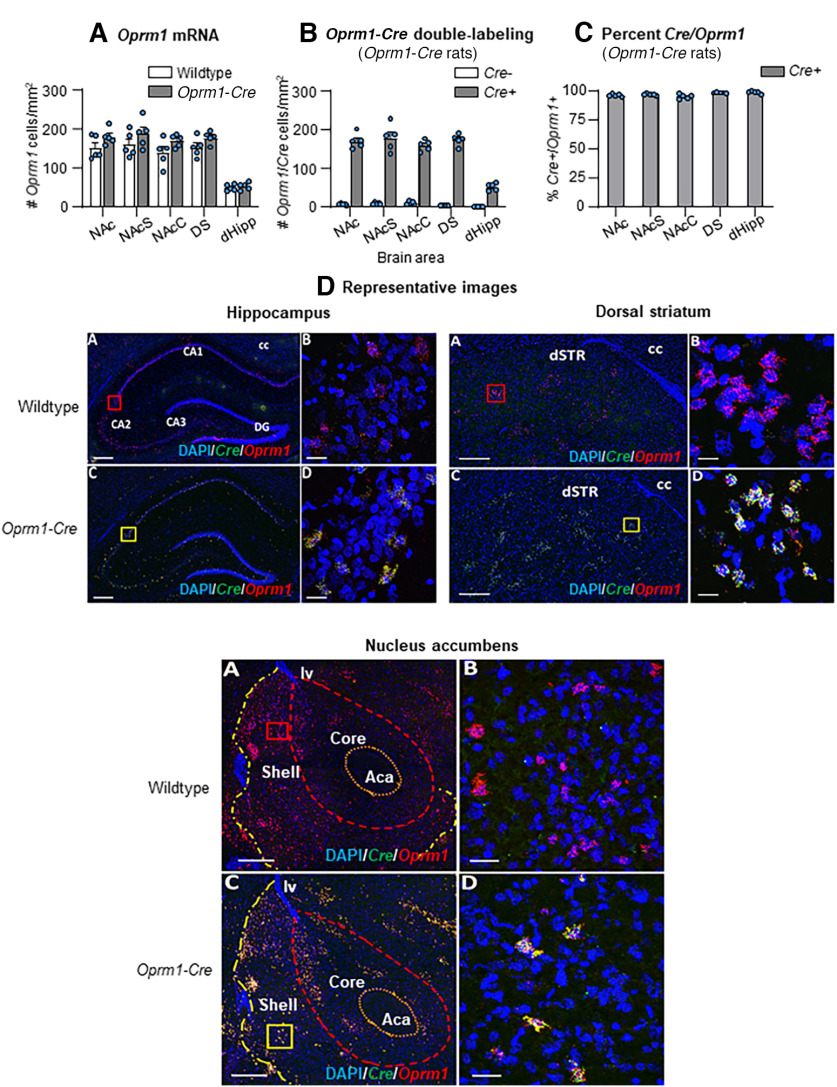
We used HCR FISH to label Oprm1, iCre, and Oprm1 + iCre mRNA double-labeled cells in Oprm1-Cre male rats and their wildtype littermates. We found no genotype differences in Oprm1+ cells per mm2 in NAc, DS, or dHipp (Fig. 2A). We found a significant number of double-labeled Oprm1+/Cre+ cells (compared with Oprm1+Cre– cells) in Oprm1-Cre rats in all brain areas: NAc: F(1,4) = 356.1, p < 0.001, DS: F(1,4) = 652.4, p < 0.001, dHipp: F(1,4) = 172.3, p < 0.001 (Fig. 2B). The percent of Cre+/Oprm1+ cells in the different brain areas was 95%-98% (Fig. 2C), and Cre was not detected in wildtype littermates.
Figure 2.
iCre mRNA and Oprm1 mRNA in NAc, DS, and dHipp. A, Oprm1+ cells per mm2 for Oprm1 mRNA (wildtype and Oprm1-Cre, n = 5/genotype; males only). B, Oprm1+/Cre+ double-labeled cells per mm2 (Oprm1-Cre rats only). C, Percent Cre+/Oprm1+ (Oprm1-Cre rats only). D, Representative confocal photomicrographs of Oprm1-Cre rat brains showing colocalization (yellow) between Oprm1+ neurons (red) and Cre+ neurons (green) in dHipp, DS, and NAc compared with wildtype rats which only showed Oprm1 expression (red). Objective lens magnification: A, C, 10×; B, D, 40×. Scale bars: A, C, 300 µm; B, D, 25 µm. Aca, Anterior commissure; CA1, CA2, CA3, hippocampal subfields; cc, corpus callosum; DG, dentate gyrus; lv, left ventricle.
[35S]GTPγS autoradiography and [3H]DAMGO autoradiography assays
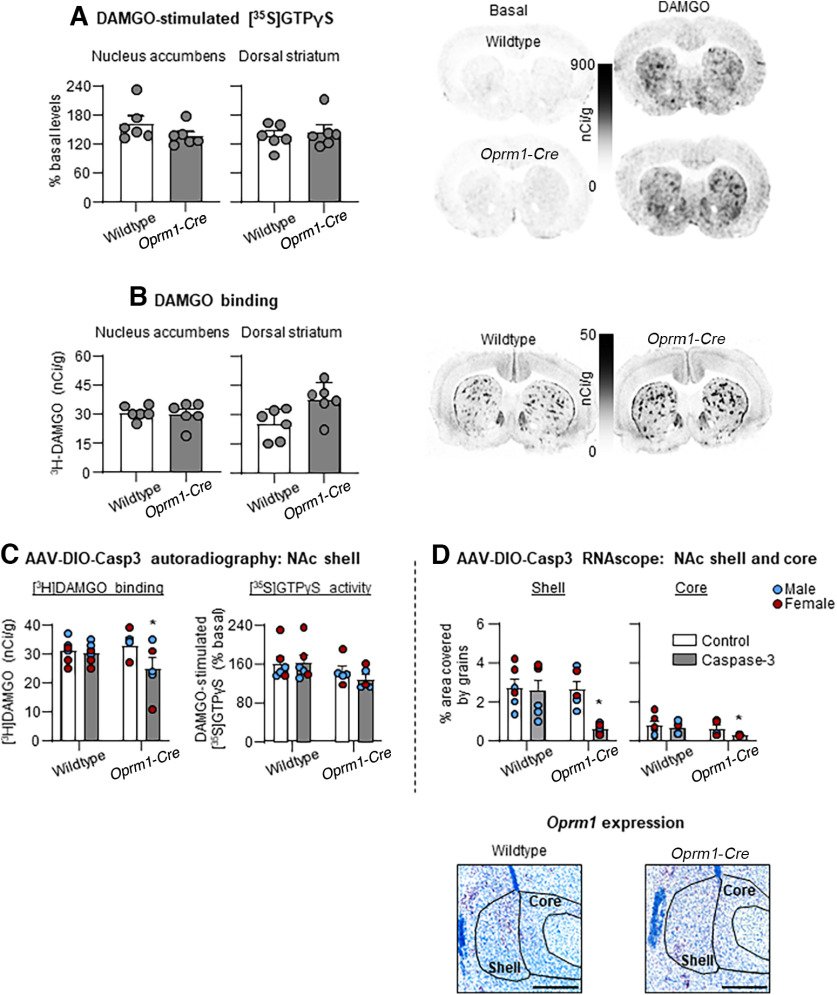
We used autoradiography to measure MOR activity (via [35S]GTPγS) and binding (via [3H]DAMGO) in Oprm1-Cre male and female rats and their wildtype littermates. We found no genotype differences in DAMGO-stimulated [35S]GTPγS recruitment in NAc and DS (p > 0.05) (Fig. 3A). We also found no genotype differences in [3H]DAMGO binding in NAc (p > 0.05). However, [3H]DAMGO binding was higher binding in DS in Oprm1-Cre rats than in wildtype littermates (F(1,10) = 6.7, p = 0.027) (Fig. 3B).
Figure 3.
Autoradiography in Oprm1-Cre rats and wildtype littermates. A, DAMGO-stimulated [35S]GTPyS in NAc and DS in wildtype and Oprm1-Cre rats (n = 6/genotype & sex). Values are calibrated and expressed as % basal. B, [3H]DAMGO binding in NAc and DS in wildtype and Oprm1-Cre rats (n = 6/genotype & sex). Values are calibrated and expressed as nCi/g. AAV-DIO-Casp3 lesion in NAc: autoradiography and RNAscope. We injected AAV1-EF1a-Flex-taCasp3-TEVP unilaterally into the right hemisphere of NAc shell and PBS into the left hemisphere; injections were 500 nl/side. C, [3H]DAMGO binding (left) and DAMGO-stimulated [35S]GTPyS (right) in NAc of wildtype and Oprm1-Cre rats (n = 5 or 6/sex & genotype). Values are calibrated and expressed as nCi/g and % basal, respectively. D, Mean ± SEM Oprm1+ cells expressed as red grains per area (% area covered by red grains) in NAc shell and core in wildtype and Oprm1-Cre rats (n = 5 or 6/genotype & sex). C, D, Individual data points are depicted for males (blue) and females (red). Scale bar, 500 µm. *p < 0.05; different from the control hemisphere.
AAV-DIO-Casp3 lesion in NAc: [3H]DAMGO autoradiography and RNAscope
We injected AAV-DIO-Casp3 unilaterally (500 nl/side) into NAc shell and measured Oprm1 mRNA expression and MOR activity and binding in Oprm1-Cre male and female rats and their wildtype littermates. The analysis of DAMGO binding and DAMGO-stimulated [35S]GTPγS recruitment in NAc, which included the between-subjects factor of Genotype (wildtype, Oprm1-Cre) and the within-subjects factor of Lesion (Vehicle, AAV-DIO-Casp3), showed significant Genotype × Lesion interaction for binding (F(1,9) = 8.4, p = 0.018) and approaching significant interaction for DAMGO-stimulated [35S]GTPγS activity (F(1,9) = 4.8, p = 0.056) (Fig. 3C). The analysis of % area covered by red grains in NAc shell and core, which included the between-subjects factor of Genotype and the within-subjects factor of Lesion, showed significant effects of Genotype × Lesion interaction in NAc shell (F(1,9) = 9.9, p = 0.012), but not in NAc core (Fig. 3D).
Together, these results indicate that AAV-DIO-Casp3 NAc shell injections selectively decreased Oprm1 mRNA and MOR binding activity in the injected hemisphere of Oprm1-Cre male and female rats, but not in the vehicle-injected hemisphere or in wildtype littermates.
Experiment 1: food self-administration
There were no genotype differences in acquisition of food self-administration and subsequent responding under the different FR requirements (Fig. 4A). The analysis of acquisition data, which included the between-subjects factor of Genotype (wildtype, Oprm1-Cre) and the within-subjects factor of Session (1-7) showed a significant effect of Session for both the number of pellets and number of active lever presses (F(6108) = 24.1, p < 0.001; F(6108) = 7.8, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1). The analysis of the FR response data, which included the between-subjects factor of Genotype and the within-subjects factor of FR requirement (FR1-FR8), showed a significant effect of FR requirement for both the number of pellets and number of active lever presses (F(4,72) = 34.4, p < 0.001; F(4,72) = 12.3, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1).
Figure 4.
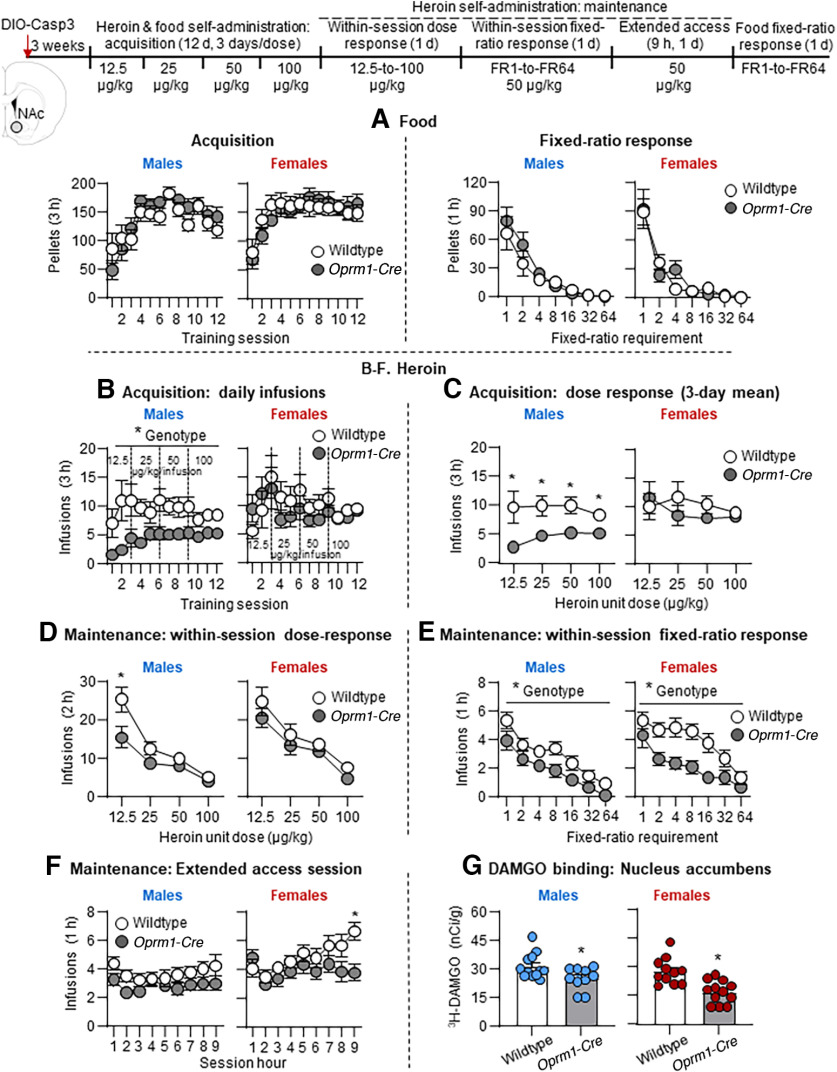
Food self-administration, heroin self-administration, and heroin relapse-related behaviors in Oprm1-Cre rats and wildtype littermates. A, Food self-administration. Acquisition (left) and fixed-ratio response (right). Mean ± SEM number of pellets consumed (left) and active lever presses (right). Wildtype (3 males, 6 females), Oprm1-Cre (4 males, 6 females); data were combined for males and females. B, Heroin self-administration. Mean ± SEM number of heroin infusions during heroin self-administration training (days 1-6, 0.1 mg/kg/infusion; days 7-12, 0.05 mg/kg/infusion). Extinction responding. Mean ± SEM number of active lever presses during the seven 6 h extinction sessions. Active lever presses led to contingent presentations of the tone-light cue, but not heroin. Context-induced reinstatement. Mean ± SEM number of active lever presses during the 6 h reinstatement tests in Contexts B and A. Active lever presses led to contingent presentations of the tone-light cue, but not heroin. Individual data points are depicted for males (blue) and females (red). Reacquisition. Mean ± SEM number of heroin infusions (0.05 mg/kg/infusion) per hour during reacquisition. Active lever presses led to the delivery of heroin infusions and the tone-light cue. Wildtype (6 males, 8 females), Oprm1-Cre (5 males, 8 females); data were combined for males and females.
Together, the results of Experiment 1 indicate that the knock-in manipulation had no effect on acquisition of palatable food self-administration in mildly food-restricted rats or the effort to self-administer food pellets in food-sated rats.
Experiment 2: heroin self-administration and relapse-related behaviors
Heroin self-administration (Context A)
There were no genotype differences in acquisition of heroin self-administration (Fig. 4B, far left). The statistical analysis of number of infusions, which included the between-subjects factor of Genotype, and the within-subjects factors of Training session (1-7) and Training dose (50, 100 µg/kg/infusion), showed significant effects of Training session × Training dose (F(5125) = 10.0, p < 0.001). There were no significant effects of Genotype or interactions with this factor (p > 0.1).
Extinction responding (Context B)
There were no genotype differences in extinction of heroin self-administration (Fig. 4B, mid left). The statistical analysis of number of active lever presses, which included the between-subjects factor of Genotype and the within-subjects factor of Extinction session, showed a significant effect of Extinction session (F(6150) = 49.9, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1).
Context-induced reinstatement (Contexts A and B)
There were no genotype differences in context-induced reinstatement of heroin seeking (Fig. 4B, far right). The statistical analysis of number of active lever presses, which included the between-subjects factor of Genotype and the within-subjects factor of Context (A, B), showed a significant effect of Context (F(1,25) = 65.4, p < 0.001) but no significant effects of Genotype and interaction between the two factors (p > 0.1).
Reacquisition (Context B)
There were no genotype differences in reacquisition of heroin self-administration (Fig. 4B, far right). The statistical analysis of number of infusions, which included the between-subjects factor of Genotype and the within-subjects factor of Session hour (1-6), showed significant effects of Session hour (F(5125) = 6.0, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1).
Together, the results of Experiment 2 indicate that the knock-in manipulation had no effect on heroin self-administration, extinction responding, context-induced reinstatement, and reacquisition of heroin self-administration.
Experiment 3: evaluation of pain-related responses using von Frey test, tail flick test, and lactic acid-induced suppression of operant responding
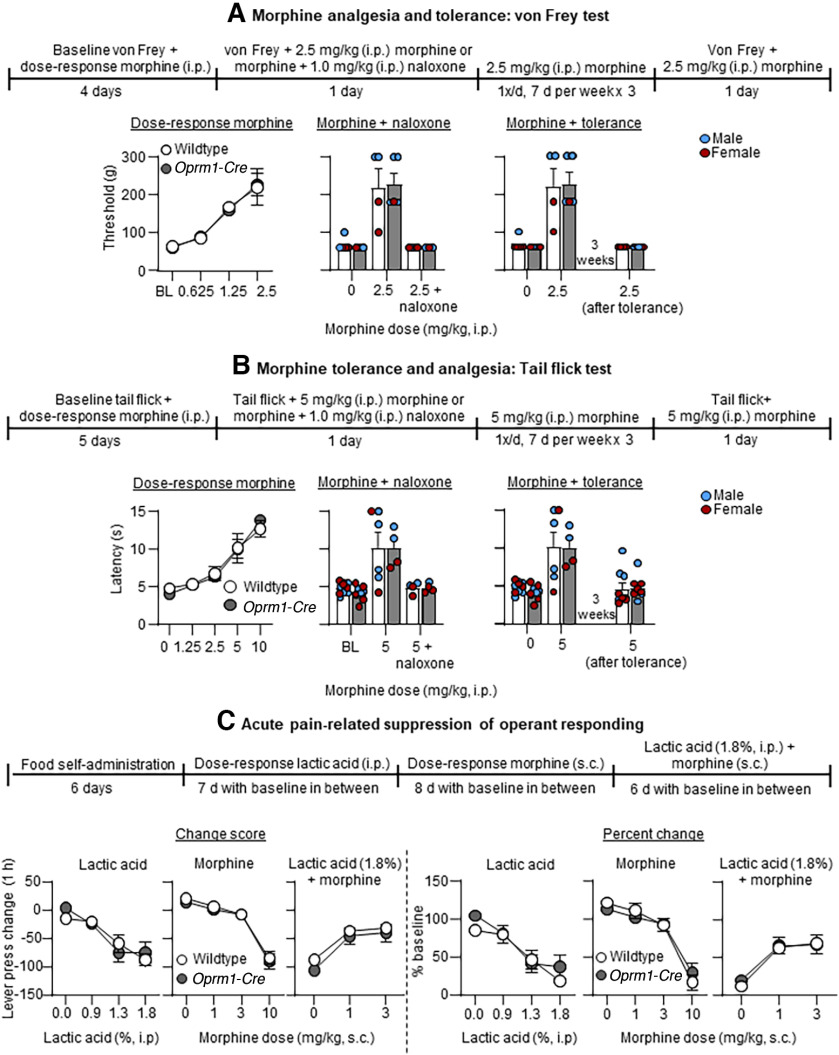
Experiment 3a: von Frey test
There were no genotype differences in the morphine dose–response of paw withdrawal thresholds (Fig. 5A, left). The statistical analysis of threshold, which included the between-subjects factor of Genotype and the within-subjects factor of Dose (0, 0.625, 1.25, 2.5 mg/kg), showed significant effects of Dose (F(3,47.72) = 73.0, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1). There were also no genotype differences in response to naloxone or to the analgesic tolerance to morphine (Fig. 5A, middle and right). The statistical analysis of threshold, which included the between-subjects factor of Genotype and the within-subjects factor of Condition (Morphine + Naloxone, or Morphine + Tolerance), showed significant effects of Condition for response to naloxone (F(2,17.45) = 65.4, p < 0.001) and for analgesic tolerance (F(2,30.54) = 87.4, p < 0.001), but no significant effects of Genotype or interaction between the two factors (p > 0.1).
Figure 5.
Morphine analgesia, tolerance, and pain-related suppression of operant responding in Oprm1-Cre rats and wildtype littermates. A, von Frey test and timeline of experiment for morphine analgesia and tolerance. Left, Baseline and von Frey thresholds (g) after ascending doses of morphine (0.625, 1.25, and 2.5 mg/kg, i.p.). Middle, von Frey thresholds (g) after vehicle, 2.5 mg/kg morphine, or 2.5 mg/kg morphine + naloxone (1.0 mg/kg, i.p.). Right, von Frey thresholds (g) after vehicle, 2.5 mg/kg morphine, or 2.5 mg/kg morphine after 21 d of chronic morphine (2.5 mg/kg/day, i.p.). Wildtype (7 males, 3 females), Oprm1-Cre (7 males, 3 females). Individual data points are depicted for males (blue) and females (red). B, Tail flick test and timeline of experiment for morphine analgesia and tolerance. Left, Latency (s) measured after ascending doses of vehicle and morphine (1.25, 2.5, 5, and 10 mg/kg, i.p.). Middle, Latency (s) after vehicle, 5 mg/kg morphine, or 5 mg/kg morphine + naloxone (1.0 mg/kg, i.p.). Right, Latency (s) after vehicle, 5 mg/kg morphine, or 5 mg/kg morphine after 21 d of chronic morphine (5 mg/kg/day, i.p.). Wildtype (5 males, 5 females), Oprm1-Cre (5 males, 4 females). Individual data points are depicted for males (blue) and females (red). C, Acute lactic acid-induced suppression of operant responding for food pellets. Left panels, Mean ± SEM pellet intake change score from baseline after injections of lactic acid (0%, 0.9%, 1.35%, and 1.8%, i.p.), morphine (0, 1, 3, and 10 mg/kg, s.c.), and lactic acid (1.8%) plus morphine (0, 1, and 3 mg/kg, s.c). Right panels, Mean ± SEM percent change from baseline of the data presented on the left panels. Wildtype (4 males, 4 females) and Oprm1-Cre (4 males, 3 females) rats.
Experiment 3b: tail flick test
There were no genotype differences in morphine dose–response tail flick latencies (Fig. 5B, left). The statistical analysis of latency, which included the between-subjects factor of Genotype and the within-subjects factor of Dose (0, 1.25, 2.5, 5, 10 mg/kg) showed significant effects of Dose (F(4,61.58) = 58.1, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1). There were also no genotype differences in response to naloxone or to the analgesic tolerance to morphine (Fig. 5B, middle and right). The statistical analysis of latency, which included the between-subjects factor of Genotype and the within-subjects factor of Condition (Morphine + Naloxone, or Morphine + Tolerance) showed significant effects of Condition for response to naloxone (F(2,23.01) = 22.8, p < 0.001) and for analgesic tolerance (F(2,33.63) = 20.8, p < 0.001) but no significant effects of Genotype or interaction between the two factors (p > 0.1).
Experiment 3c: lactic acid-induced suppression of operant responding for food
There were no genotype differences in lactic acid concentration response, morphine dose–response, and reversal of lactic acid-induced suppression of food responding by morphine (Fig. 5C, left panels). Unlike in Experiment 1, in Experiments 4 and 5, there were baseline differences in pellet intake between Oprm1-Cre rats and their wildtype littermates (mean ± SEM number of pellets per session during the 3 baseline days before lactic acid injections was 106 ± 8 for wildtype and 133 ± 6 for Oprm1-Cre rats). Thus, we calculated change scores from baseline pellet intake for data presentation and statistical analyses. We also show in Figure 5C (right panels), the percent change score from baseline.
The statistical analysis of lever press change score for lactic acid, which included the between-subjects factor of Genotype and the within-subjects factor of Dose (0%, 0.9%, 1.3%, 1.8%), showed significant effects of Dose (F(3,39) = 17.2, p < 0.001). The statistical analysis of lever press change score for morphine, which included the between-subjects factor of Genotype and the within-subjects factor of Morphine Dose (0, 1, 3, 10 mg/kg), showed significant effects of Dose (F(3,39) = 58.1, p < 0.001). The statistical analysis of pellet intake change score for lactic acid (1.8%) + morphine, which included the between-subjects factor of Genotype and the within-subjects factor of Morphine Dose (0, 1, 3 mg/kg), showed significant effects of F(2,26) = 25.0, p > 0.001). In all analyses, there were no significant effects of Genotype or interaction between the other factors (p > 0.1).
Together, the results of Experiment 3 indicate that the knock-in manipulation had no effect on pain sensitivity to mechanical stimulation, to a noxious thermal stimulus, to acute morphine analgesia, to morphine analgesic tolerance, to suppression of operant responding by lactic acid or morphine, or to reversal of the suppression effect of lactic acid on operant responding by morphine.
Experiment 4: Effect of Cre-dependent AAV-DIO-Casp3 NAc lesions on acquisition and maintenance of heroin self-administration
Food self-administration
Acquisition
Bilateral AAV-DIO-Casp3 lesions had no effect on acquisition of food self-administration in Oprm1-Cre male and female rats or their wildtype littermates (Fig. 6A, left panels). The statistical analysis of number of pellets, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of Session (1-12), showed significant effects of Session (F(11,561) = 24.2, p < 0.001), but no other significant main or interaction effects (p > 0.1).
Figure 6.
Effect of AAV-DIO-Casp3 NAc lesions on acquisition and maintenance of food and heroin self-administration in heterozygote Oprm1-Cre rats and wildtype littermates. A, Food. Left, Acquisition. Mean ± SEM number of pellets consumed during the 3 h sessions. Wildtype (14 males, 12 females), Oprm1-Cre (14 males, 15 females). Right, Within-session fixed-ratio response. Mean ± SEM number of pellets consumed during the 1 h sessions. Wildtype (8 males, 5 females), Oprm1-Cre (8 males, 6 females). B–F, Heroin. B, Acquisition: daily infusions. Mean ± SEM number of heroin infusions during the 3 h heroin self-administration sessions. C, Acquisition: dose–response (3 d mean). Mean ± SEM of heroin infusions at each unit dose. Wildtype (13 males, 12 females), Oprm1-Cre (13 males, 15 females) for B, C. D, Maintenance: within-session dose–response. Mean ± SEM number of heroin infusions at each unit dose. E, Maintenance: within-session fixed-ratio response. Mean ± SEM number of heroin infusions at each hour of heroin self-administration for each fixed-ratio requirement. F, Maintenance: extended access. Mean ± SEM number of heroin infusions during the 9 h heroin self-administration session. Wildtype (13 males, 12 females), Oprm1-Cre (11 males, 14 females) for D–F. G, [3H]DAMGO binding in NAc in wildtype (12 males, 11 females) and Oprm1-Cre (10 males, 12 females) rats. Values are calibrated and expressed as nCi/g. Individual data points are depicted for males (blue) and females (red). *p < 0.05; different from the control hemisphere. *p < 0.05; different from the Oprm1-Cre group.
Fixed-ratio response
AAV-DIO-Casp3 had no effect on food self-administration under the different FR requirements in Oprm1-Cre male and female rats or their wildtype littermates (Fig. 6A, right panels). The statistical analysis of number of pellets, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of FR requirement (1-64), showed significant effects of FR requirement (F(6142) = 49.8, p < 0.001), but no other significant main or interaction effects (p > 0.1).
Heroin self-administration
Acquisition
AAV-DIO-Casp3 lesions decreased acquisition of heroin self-administration in Oprm1-Cre male, but not female, rats (Fig. 6B). The statistical analysis of number of daily infusions, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of Heroin Dose (12.5, 25, 50, 100 µg/kg), showed significant effects of Genotype (F(1,49) = 5.1, p = 0.029) and Sex (F(1,49) = 4.1, p = 0.049) but no other significant main or interaction effects (p > 0.1). For males, a follow-up mixed ANOVA, which included the between-subjects factor of Genotype and the within-subjects factor of Heroin Dose, showed significant effects of Genotype (F(1,24) = 8.0, p = 0.009) but no significant main effect of Dose or interaction (p > 0.1). The same analysis for females did not show any significant effects.
Finally, a mixed ANOVA of the 3 d mean infusions within each Heroin Dose (Fig. 6C), which included the between-subjects factors of Genotype and Sex and the within-subjects factor of Heroin Dose, showed significant effects of Genotype (F(1,49) = 5.1, p = 0.029) and Sex (F(1,49) = 4.1, p = 0.049) but no significant effects of Genotype or interactions between the different factors (p > 0.1). Follow-up ANOVA within each sex showed a significant effect of Genotype for males (F(1,24) = 8.0, p = 0.009) but not females (p > 0.1).
Maintenance: within-session dose–response
AAV-DIO-Casp3 lesions decreased heroin self-administration in Oprm1-Cre male, but not female, rats; this effect was stronger at the lower heroin unit doses (Fig. 6D). The statistical analysis of number of infusions, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of Heroin Dose (12.5, 25, 50, 100 µg/kg), showed significant effects of Genotype (F(1,46) = 4.7, p = 0.035), Heroin Dose (F(3138) = 88.2, p < 0.001), and Genotype × Heroin dose (F(3138) = 3.1, p = 0.029) but no other significant main or interaction effects (p > 0.1). For males, a follow-up mixed ANOVA, which included the between-subjects factor of Genotype and the within-subjects factor of Heroin Dose, showed significant effects of Heroin Dose (F(3,66) = 49.4, p < 0.001) and Genotype × Heroin Dose (F(3,66) = 4.6, p = 0.006) but no other significant main or interaction effects (p > 0.1). The same analysis for females showed a significant effect of Heroin Dose (F(3,75) = 40.0, p < 0.001), but no significant effects of Genotype or interactions between the two factors (p > 0.1).
Maintenance: fixed-ratio response
AAV-DIO-Casp3 lesions decreased heroin self-administration in both Oprm1-Cre male and female rats; the lesion effect was stronger at the intermediate FR requirements and appeared stronger in females (Fig. 6E). The statistical analysis of number of infusions, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of FR requirement (FR1-FR64), showed significant effects of Genotype (F(1,46) = 14.0, p > 0.001) and FR requirement (F(6276) = 54.6, p < 0.001) but no other significant main or interaction effects (p > 0.1). For males, a follow-up mixed ANOVA, which included the between-subjects factor of Genotype and the within-subjects factor of FR requirement, showed significant effects of Genotype (F(1,22) = 5.2, p = 0.033) and FR requirements (F(6132) = 40.6, p > 0.001) but no significant interaction (p > 0.1). The same analysis for females showed a significant effect of Genotype (F(1,24) = 9.1, p = 0.006) and FR requirements (F(6144) = 20.5, p > 0.001) but no significant interaction (p > 0.1).
Maintenance: extended access session (9 h)
AAV-DIO-Casp3 lesions decreased extended access heroin self-administration in Oprm1-Cre female, but not male, rats (Fig. 6F). The statistical analysis of number of infusions, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of Hour (1-9), showed significant effects of Genotype (F(1,46) = 5.4, p = 0.025), Sex (F(1,46) = 8.0, p = 0.007), Hour (F(8365) = 5.1, p < 0.001), and Genotype × Hour (F(8365) = 2.0, p = 0.041) but no other significant main or interaction effects (p > 0.1). For males, a follow-up mixed ANOVA, which included the between-subjects factor of Genotype and the within-subjects factor of Hour, showed no significant effects (p > 0.05). The same analysis for females showed a significant effect of Hour (F(8191) = 4.4, p > 0.001) and Genotype × Hour (F(8191) = 2.6, p = 0.010).
[3H]DAMGO autoradiography
AAV-DIO-Casp3 lesions decreased [3H]DAMGO binding in Oprm1-Cre male and female rats, but not wildtype littermates (22 males, 23 females, n = 10-12 per genotype) (Fig. 6G). The statistical analysis of the [3H]DAMGO binding values, which included the between-subjects factors of Genotype and Sex, showed significant main effects of Genotype (F(1,41) = 20.7, p < 0.001) and Sex (F(1,41) = 9.5, p < 0.004), but no interaction (p > 0.05).
Together, the results of Experiment 4 indicate that AAV-DIO-Casp3 NAc lesions in Oprm1-Cre rats, but not wildtype littermates, (1) decreased acquisition of heroin self-administration in males but not females, (2) decreased heroin self-administration under an FR1 reinforcement schedule at low, but not higher, heroin unit doses in males but not females, (3) decreased heroin self-administration when the response requirement was increased in both males and females, (4) decreased heroin self-administration in females, but not males, when the daily session was increased to 9 h (extended access condition), and (5) decreased NAc [3H]DAMGO binding in Oprm1-Cre male and female rats. In contrast, NAc Caspase-3 lesions had no effect on acquisition and maintenance of food self-administration under different FR reinforcement schedules in Oprm1-Cre rats.
Experiment 5: effect of control virus AAV1-DIO-EYFP into NAc on acquisition and maintenance of heroin self-administration
Food self-administration
Acquisition
AAV-DIO-EYFP had no effect on acquisition of food self-administration in Oprm1-Cre rats or their wildtype littermates (Fig. 7A, left panels). The statistical analysis of number of pellets, which included the between-subjects factors of Genotype and Sex and the within-subjects factor of Session (1-12), showed significant effects of Session (F(11,286) = 6.8, p < 0.001), but no significant effects of Genotype or interaction (p > 0.1).
Figure 7.
Effect of NAc injections of AAV-DIO-EYFP on acquisition and maintenance of food and heroin self-administration in heterozygote Oprm1-Cre rats and wildtype littermates. A, Food. Left, Acquisition. Mean ± SEM number of pellets consumed during the 3 h sessions. Wildtype (7 males, 8 females), Oprm1-Cre (6 males, 7 females). Right, Within-session fixed-ratio response. Mean ± SEM number of pellets consumed during the 1 h sessions. Wildtype (7 males, 8 females), Oprm1-Cre (6 males, 7 females). B–F, Heroin. B, Acquisition: daily infusions. Mean ± SEM number of heroin infusions during the 3 h heroin self-administration sessions. C, Acquisition: dose–response (3 d mean). Mean ± SEM of heroin infusions for each unit dose. Wildtype (7 males, 8 females), Oprm1-Cre (6 males, 7 females) for B, C. D, Maintenance: within-session dose–response. Mean ± SEM number of heroin infusions for each unit dose. E, Maintenance: within-session fixed-ratio response. Mean ± SEM number of heroin infusions during each hour of heroin self-administration for each fixed-ratio requirement. F, Maintenance: extended access. Mean ± SEM number of heroin infusions during the 9 h heroin self-administration session. Wildtype (6 males, 7 females), Oprm1-Cre (6 males, 7 females) for D–F.
Fixed-ratio response
AAV-DIO-EYFP had no effect on food self-administration under different FR requirements in Oprm1-Cre rats or their wildtype littermates (Fig. 7A, right panels). The statistical analysis of number of pellets, which included the between-subjects factor of Genotype and the within-subjects factor of FR requirement (1-64), showed significant effects of FR requirement (F(6156) = 50.7, p < 0.001), but no significant effects of Genotype or interaction (p > 0.1).
Heroin self-administration
Acquisition
AAV-DIO-EYFP had no effect on acquisition of heroin self-administration in Oprm1-Cre rats or their wildtype littermates (Fig. 7B). The statistical analysis of number of daily infusions, which included the between-subjects factor of Genotype and the within-subjects factors of Session (1-3) and Heroin Dose (12.5, 25, 50, 100 µg/kg), showed a significant effect of Heroin Dose (F(3,78) = 4.7, p = 0.005), Session (F(2,52) = 4.6, p = 0.014), and Session × Heroin Dose (F(6156) = 2.5, p = 0.023) but no significant effects of Genotype or interactions between Genotype and the other factors (p > 0.1). A mixed ANOVA of the 3 d mean infusions within each Heroin Dose (Fig. 7C), which included the between-subjects factor of Genotype and the within-subjects factors of Heroin Dose, showed a significant effect of Heroin Dose (F(3,78) = 7.0, p < 0.001) but no significant effects of Genotype or interaction (p > 0.1).
Maintenance: within-session dose–response
AAV-DIO-EYFP had no effect on heroin self-administration in Oprm1-Cre rats or wildtype littermates (Fig. 7D). The statistical analysis of number of infusions, which included the between-subjects factor of Genotype and the within-subjects factor of Heroin Dose (12.5, 25, 50, 100 µg/kg), showed significant effects of Heroin Dose (F(3,72) = 49.1, p < 0.001) but no significant effect of Genotype or interaction (p > 0.1).
Maintenance: fixed-ratio response
AAV-DIO-EYFP had no effect on heroin self-administration in Oprm1-Cre rats or wildtype littermates (Fig. 7E). The statistical analysis of number of infusions, which included the between-subjects factor of Genotype and the within-subjects factor of FR requirement (FR1-FR64), showed significant effects of FR requirement (F(6144) = 16.4, p < 0.001) but no significant effect of Genotype or interaction (p > 0.1).
Maintenance: extended access session (9 h)
AAV-DIO-EYFP had no effect on extended access heroin self-administration in Oprm1-Cre rats or their wildtype littermates (Fig. 7F). The statistical analysis of number of infusions, which included the between-subjects factor of Genotype and Sex and the within-subjects factor of Hour (1-9), showed significant effects of Hour (F(8192) = 10.6, p < 0.001) but no significant effects of Genotype or interaction between Genotype and the other factors (p > 0.1).
Together, the results of Experiment 5 indicate that NAc DIO-EYFP had no effect on any of the heroin self-administration behaviors in either Oprm1-Cre rats or their wildtype littermates, suggesting that nonlesioning Cre-dependent manipulations do not affect MOR expression or opioid-mediated behaviors.
Discussion
We performed anatomic and behavioral characterization of a novel CRISPR-mediated knock-in rat that coexpresses Cre-recombinase and MOR under the endogenous Oprm1 gene promoter. We report four main findings. First, the knock-in manipulation had no effect on Oprm1 mRNA expression in dHipp, NAc, and DS, MOR function in NAc and DS, and MOR density in NAc; however, MOR density in DS was higher in Oprm1-Cre rats. Second, insertion of T2A-iCre resulted in >95% colocalization of Oprm1 and Cre in Oprm1-Cre rats in NAc, DS, and dHipp; additionally, Cre was not detected in wildtype littermates. Third, the knock-in of Cre into the Oprm1 gene had no effect on operant responding for food pellets or MOR-mediated behaviors, including pain sensitivity, morphine analgesia and tolerance, heroin self-administration, and heroin relapse-related behaviors. Fourth, lesions of NAc MOR-expressing cells using a Cre-dependent AAV-DIO-Casp3 virus decreased Oprm1 mRNA expression and MOR density in Oprm1-Cre rats but not wildtype littermates. Additionally, the lesions had different effects on acquisition and maintenance of heroin self-administration in male and female rats. Together, these results indicate that MOR expression and function are preserved in the novel Oprm1-Cre knock-in rat, and that we can selectively target and manipulate brain MOR-expressing cells to study MOR-mediated behaviors.
The Oprm1 knock-in manipulation had no effect on several behavioral measures and MOR expression and function
We behaviorally characterized the Oprm1 knock-in rats in three ways: (1) operant responding for food pellets, (2) pain sensitivity and morphine analgesia using three different methods to evaluate pain-related behaviors, and (3) heroin self-administration and relapse-related behaviors.
Oprm1-Cre rats did not differ from their wildtype littermates in acquisition of food self-administration and sensitivity to increasing the effort (response requirement) to obtain the food reinforcer. Oprm1-Cre rats also did not differ from their wildtype littermates in three different pain-related assays: tail flick test, von Frey test, and lactic acid inhibition of operant responding for food. Oprm1-Cre rats also did not differ from their wildtype littermates in sensitivity to morphine-induced analgesia, the development of morphine tolerance, and reversal of morphine's analgesic effects by naloxone. Finally, Oprm1-Cre rats also did not differ from their wildtype littermates in acquisition and maintenance of heroin self-administration, and lever responding in three commonly used relapse-related measures (Shalev et al., 2002; Reiner et al., 2019): extinction responding, context-induced reinstatement, and reacquisition after extinction. Together, our initial behavioral characterization indicates that the Oprm1 knock-in manipulation did not alter normal operant learning, sensitivity to food reward, and prototypical MOR-mediated behaviors, such as opioid analgesia, opioid tolerance, and opioid self-administration.
Methodological considerations
The insertion of 2A-Cre into the coding sequence of Oprm1 may negatively affect how MOR is expressed at both the RNA and protein level, leading to altered MOR functionality. Our anatomical (and behavioral) characterization suggests that the CRISPR-based insertion of T2A-iCre did not change Oprm1 expression or function. However, this conclusion should be interpreted with caution because of several technical issues. Specifically, in our HCR FISH experiment using an Oprm1-specific probe on heterozygous Oprm1-Cre rats, we did not detect statistically significant differences between Oprm1-Cre rats and wildtype littermates in number of Oprm1-postive cells in dHipp, NAc, or DS. However, our experiment did not address whether the heterozygous genotype results in transcriptional changes at the cellular level. Additionally, because we cannot differentiate the transgenic mRNA from the endogenous mRNA that is normally expressed in wildtype rats, we are unable to conclude that the transgene expression is restricted to cells that would otherwise express endogenous Oprm1. Because of this, we cannot rule out the possibility that the Cre insertion will affect Oprm1 expression in terms of spatial distribution, temporal dynamics, and response to external stimuli, such as opioid drugs and stress. Additionally, the reasons for the somewhat higher [3H]DAMGO binding but not activity in DS are unknown. Finally, while we report no differences in HCR FISH or [35S]GTPγS activity assays, and pharmacology/behavioral assays, it remains to be determined whether these “nonphenotypes” generalize to homozygote Oprm1-Cre rats.
Effect of lesions of MOR-expressing cells in NAc on heroin self-administration in males and females
Studies using systemic injections of selective or preferential (e.g., naloxone, naltrexone) antagonists of MOR and other opioid receptors indicate that activation of MOR is critical for the reinforcing effects of opioid agonists in the drug self-administration procedure in male rodents and monkeys (Goldberg et al., 1971; Ettenberg et al., 1982; Wise, 1989; Mello and Negus, 1996). There is also evidence from studies using systemic injections of opioid receptor antagonists in male rats that activation of MOR is critical for reinstatement of opioid seeking induced by drug priming, and drug cues and contexts (Shaham and Stewart, 1996; Bossert et al., 2019; Reiner et al., 2019).
Studies using site-specific injections of lipophobic preferential MOR antagonists (methyl naltrexone or methyl naloxonium) into different brain areas indicate that the critical site of action for the systemic effect of MOR antagonists on opioid self-administration in male rats is NAc; there is also evidence for a role of VTA and periaqueductal gray (Vaccarino and Corrigall, 1987; Corrigall and Vaccarino, 1988). Additionally, in Oprm1 KO mice, rescue of Oprm1 expression using a PDYN-MOR transgene in striatum restored remifentanil self-administration (Cui et al., 2014). On the other hand, deletion of Oprm1 mRNA from GABAergic forebrain neurons, which primarily target NAc and DS, increased heroin self-administration (Charbogne et al., 2017).
Based on the literature described above, we used the new Oprm1-Cre rat to determine the involvement of MOR-expressing cells in NAc in initiation and maintenance of heroin self-administration, using a within-subjects ascending heroin dose–response curve procedure (Stewart et al., 1996). We found that injections of Cre-dependent AAV1-EF1a-Flex-taCasp3-TEVP into NAc of Oprm1-Cre rats, which selectively lesion MOR-expressing cells, had different effects on heroin self-administration in male and female rats. During the acquisition phase, the lesions selectively decreased heroin self-administration at all four heroin doses in males but not females. During the maintenance phase, the lesions decreased responding for the lowest heroin dose in males but not females, decreased extended-access heroin self-administration in females but not males, and had a more pronounced inhibitory effect on the effort (response requirement) to self-administer heroin in females than in males.
Our results suggest mechanistic differences in the involvement of MOR-expressing cells in heroin seeking and taking. This notion is supported by results from our recent studies on the effects of TRV130 (a selective partial agonist of MOR) and BU08028 (a mixed partial agonist of MOR and nociceptin/orphanin/FQ peptide receptor) on context-induced reinstatement and reacquisition of heroin and oxycodone self-administration in male and female rats (Bossert et al., 2020, 2022). These MOR-targeted pharmacological manipulations had different effects on context-induced reinstatement and reacquisition in outbred Sprague Dawley male and female rats despite the lack of sex differences in opioid self-administration, extinction responding, context-induced reinstatement, and reacquisition. A question for future research is what the mechanism(s) of the different effects of the AAV-DIO-Casp3 lesions on heroin self-administration in male and female rats is/are. One possibility is sex-specific modulation of striatal MOR by local estrogen and progesterone receptors (Mansour et al., 1988; Tobiansky et al., 2018), which play a role in striatal-dependent (Kalivas and Stewart, 1991) locomotor sensitization to opioid drugs (Craft et al., 2006). Another possibility is that sex-specific differences in the brain immune response (e.g., glial activation) (Midavaine et al., 2021; Hankerd et al., 2022; Reiss et al., 2022) affected the outcome of the AAV-DIO-Casp3 manipulation and resulted in different effects on NAc neuronal activity that normally contributes to heroin self-administration.
Overall, our results with the new Oprm1-Cre rats extend results from previous pharmacological studies on the role of NAc MOR in heroin self-administration in male rats and suggest that MOR-expressing cells in this region play distinct roles in heroin reinforcement in males and females. However, from a statistical perspective, an interpretation caveat of our data is that our conclusions are based on post hoc analysis within each sex, without a statistically significant sex × genotype interaction in the initial full factorial ANOVAs.
In conclusion, we anatomically and behaviorally validated a CRISPR-based Oprm1-Cre knock-in rat that allows cell type-specific genetic access to measure and manipulate brain MOR-expressing cells. We used the Oprm1-Cre rats to show a potential sex-specific role of NAc MOR-expressing cells in heroin self-administration. The ability to specifically alter the neurons expressing MOR, and to target MOR itself and its downstream signaling, offers significant mechanistic advantages given the lack of receptor selectivity of many of the endogenous opioid ligands and of several classical opioid agonist and antagonist drugs (Heyman et al., 1986; Mansour et al., 1995b). The new Oprm1-Cre rats can be used to study both the general role and the sex-specific role of brain MOR-expressing cells in rat models of opioid addiction and pain-related behaviors, and other opioid-mediated behaviors.
Footnotes
The work was supported by Intramural Research Program of National Institute on Drug Abuse ZIA-DA000434-22 and ZIA-DA000069; and National Institute on Drug Abuse U01DA043098, ONR 00014-19-1-2149, and HDRF to H.A. and S.J.W. The rats bred at National Institute on Drug Abuse were registered with the Rat Genome Database (RGD #155641245) and deposited at the Rat Resource and Research Center (RRRC #975).
The authors declare no competing financial interests.
References
- Adhikary S, Caprioli D, Venniro M, Kallenberger P, Shaham Y, Bossert JM (2017) Incubation of extinction responding and cue-induced reinstatement, but not context- or drug priming-induced reinstatement, after withdrawal from methamphetamine. Addict Biol 22:977–990. 10.1111/adb.12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akil H, Watson SJ, Young E, Lewis ME, Khachaturian H, Walker JM (1984) Endogenous opioids: biology and function. Annu Rev Neurosci 7:223–255. 10.1146/annurev.ne.07.030184.001255 [DOI] [PubMed] [Google Scholar]
- Anderson KR, et al. (2018) CRISPR off-target analysis in genetically engineered rats and mice. Nat Methods 15:512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly J, Rossi D, Runtz N, Li L, Park JJ, Scherrer D, Tanti G, Birling A, Darcq E, Kieffer BL (2020) Targeting morphine-responsive neurons: generation of a knock-in mouse line expressing Cre recombinase from the mu-opioid receptor gene locus. eNeuro 7:ENEURO.0433-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AN, Banks ML, Marsh SA, Townsend EA, Venniro M, Shaham Y, Negus SS (2022) Acute pain-related depression of operant responding maintained by social interaction or food in male and female rats. Psychopharmacology (Berl) 239:561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basila M, Kelley ML, Smith AV (2017) Minimal 2′-O-methyl phosphorothioate linkage modification pattern of synthetic guide RNAs for increased stability and efficient CRISPR-Cas9 gene editing avoiding cellular toxicity. PLoS One 12:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert J, Liu S, Lu L, Shaham Y (2004) A role of ventral tegmental area glutamate in contextual cue-induced relapse to heroin seeking. J Neurosci 24:10726–10730. 10.1523/JNEUROSCI.3207-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Marchant NJ, Calu DJ, Shaham Y (2013) The reinstatement model of drug relapse: recent neurobiological findings, emerging research topics, and translational research. Psychopharmacology (Berl) 229:453–476. 10.1007/s00213-013-3120-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Adhikary S, St Laurent R, Marchant NJ, Wang HL, Morales M, Shaham Y (2016) Role of projections from ventral subiculum to nucleus accumbens shell in context-induced reinstatement of heroin seeking in rats. Psychopharmacology (Berl) 233:1991–2004. 10.1007/s00213-015-4060-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Hoots JK, Fredriksson I, Adhikary S, Zhang M, Venniro M, Shaham Y (2019) Role of mu, but not delta or kappa, opioid receptors in context-induced reinstatement of oxycodone seeking. Eur J Neurosci 50:2075–2085. 10.1111/ejn.13955 [DOI] [PubMed] [Google Scholar]
- Bossert JM, Kiyatkin E, Korah H, Hoots JK, Afzal A, Perekopskiy D, Thomas S, Fredriksson I, Blough BE, Negus SS, Epstein DH, Shaham Y (2020) In a rat model of opioid maintenance, the G-protein-biased MOR agonist TRV130 decreases relapse to oxycodone seeking and taking, and prevents oxycodone-induced brain hypoxia. Biol Psychiatry 88:935–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossert JM, Townsend EA, Altidor LK, Fredriksson I, Shekara A, Husbands S, Sulima A, Rice KC, Banks ML, Shaham Y (2022) Sex differences in the effect of chronic delivery of the buprenorphine analogue BU08028 on heroin relapse and choice in a rat model of opioid maintenance. Br J Pharmacol 179:227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, van Steensel B (2014) Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli D, Venniro M, Zeric T, Li X, Adhikary S, Madangopal R, Marchant NJ, Lucantonio F, Schoenbaum G, Bossert JM, Shaham Y (2015) Effect of the novel positive allosteric modulator of metabotropic glutamate receptor 2 AZD8529 on incubation of methamphetamine craving after prolonged voluntary abstinence in a rat model. Biol Psychiatry 78:463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbogne P, et al. (2017) Mu opioid receptors in gamma-aminobutyric acidergic forebrain neurons moderate motivation for heroin and palatable food. Biol Psychiatry 81:778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HM, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA (2018) Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JJ, Beacher N, Chabot JM, Oke M, Venniro M, Lin DT, Shaham Y (2022) Characterization of operant social interaction in rats: effects of access duration, effort, peer familiarity, housing conditions, and choice between social interaction versus food or remifentanil. Psychopharmacology (Berl) 239:2093–2108. 10.1007/s00213-022-06064-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concordet JP, Haeussler M (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46:W242–W245. 10.1093/nar/gky354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigall WA, Vaccarino FJ (1988) Antagonist treatment in nucleus accumbens or periaqueductal grey affects heroin self-administration. Pharmacol Biochem Behav 30:443–450. 10.1016/0091-3057(88)90478-9 [DOI] [PubMed] [Google Scholar]
- Craft RM, Clark JL, Hart SP, Pinckney MK (2006) Sex differences in locomotor effects of morphine in the rat. Pharmacol Biochem Behav 85:850–858. 10.1016/j.pbb.2006.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Ostlund SB, James AS, Park CS, Ge W, Roberts KW, Mittal N, Murphy NP, Cepeda C, Kieffer BL, Levine MS, Jentsch JD, Walwyn WM, Sun YE, Evans CJ, Maidment NT, Yang XW (2014) Targeted expression of mu-opioid receptors in a subset of striatal direct-pathway neurons restores opiate reward. Nat Neurosci 17:254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darcq E, Kieffer BL (2018) Opioid receptors: drivers to addiction? Nat Rev Neurosci 19:499–514. [DOI] [PubMed] [Google Scholar]
- Deroche V, Le Moal M, Piazza PV (1999) Cocaine self-administration increases the incentive motivational properties of the drug in rats. Eur J Neurosci 11:2731–2736. 10.1046/j.1460-9568.1999.00696.x [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34:184–191. 10.1038/nbt.3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery MA, Akil H (2020) Endogenous opioids at the intersection of opioid addiction, pain, and depression: the search for a precision medicine approach. Annu Rev Neurosci 43:355–374. 10.1146/annurev-neuro-110719-095912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettenberg A, Pettit HO, Bloom FE, Koob GF (1982) Heroin and cocaine intravenous self-administration in rats: mediation by separate neural systems. Psychopharmacology (Berl) 78:204–209. 10.1007/BF00428151 [DOI] [PubMed] [Google Scholar]
- Filipiak WE, Saunders TL (2006) Advances in transgenic rat production. Transgenic Res 15:673–686. 10.1007/s11248-006-9002-x [DOI] [PubMed] [Google Scholar]
- Filipiak WE, Hughes ED, Gavrilina GB, LaForest AK, Saunders TL (2019) Next generation transgenic rat model production. Methods Mol Biol 2018:97–114. 10.1007/978-1-4939-9581-3_4 [DOI] [PubMed] [Google Scholar]
- Fredriksson I, Adhikary S, Steensland P, Vendruscolo LF, Bonci A, Shaham Y, Bossert JM (2017) Prior exposure to alcohol has no effect on cocaine self-administration and relapse in rats: evidence from a rat model that does not support the gateway hypothesis. Neuropsychopharmacology 42:1001–1011. 10.1038/npp.2016.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson I, Applebey SV, Minier-Toribio A, Shekara A, Bossert JM, Shaham Y (2020) Effect of the dopamine stabilizer (–)-OSU6162 on potentiated incubation of opioid craving after electric barrier-induced voluntary abstinence. Neuropsychopharmacology 45:770–779. 10.1038/s41386-020-0602-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardon O, Faget L, Chu Sin Chung P, Matifas A, Massotte D, Kieffer BL (2014) Expression of mu opioid receptor in dorsal diencephalic conduction system: new insights for the medial habenula. Neuroscience 277:595–609. 10.1016/j.neuroscience.2014.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Hill J (2006) Data analysis using regression and multilevel/hierarchical models. Cambridge: Cambridge UP. [Google Scholar]
- Goldberg SR, Woods JH, Schuster CR (1971) Nalorphine-induced changes in morphine self-administration in rhesus monkeys. J Pharmacol Exp Ther 176:464–471. [PubMed] [Google Scholar]
- Hankerd K, McDonough KE, Wang J, Tang SJ, Chung JM, La JH (2022) Postinjury stimulation triggers a transition to nociplastic pain in mice. Pain 163:461–473. 10.1097/j.pain.0000000000002366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman JS, Koslo RJ, Mosberg HI, Tallarida RJ, Porreca F (1986) Estimation of the affinity of naloxone at supraspinal and spinal opioid receptors in vivo: studies with receptor selective agonists. Life Sci 39:1795–1803. 10.1016/0024-3205(86)90099-8 [DOI] [PubMed] [Google Scholar]
- Highfield D, Mead A, Grimm J, Rocha B, Shaham Y (2002) Reinstatement of cocaine seeking in 129X1/SvJ mice: effects of cocaine priming, cocaine cues and food deprivation. Psychopharmacology (Berl) 161:417–424. 10.1007/s00213-002-1047-9 [DOI] [PubMed] [Google Scholar]
- Jaffe JH (1990) Drug addiction and drug abuse. In: Goodman and Gilman's the pharmacological basis of therapeutics, Ed 8 (Gilman AG, Rall TW, Nies AS, Taylor P, eds), pp 522–573. New York: Pergamon. [Google Scholar]
- Kalivas PW, Stewart J (1991) Dopamine transmission in the initiation and expression of drug- and stress-induced sensitization of motor activity. Brain Res Brain Res Rev 16:223–244. 10.1016/0165-0173(91)90007-u [DOI] [PubMed] [Google Scholar]
- Khoo SY, Gibson GD, Prasad AA, McNally GP (2017) How contexts promote and prevent relapse to drug seeking. Genes Brain Behav 16:185–204. 10.1111/gbb.12328 [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee SR, Li LH, Park HJ, Park JH, Lee KY, Kim MK, Shin BA, Choi SY (2011) High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:1–8. 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koob GF (1992) Neural mechanisms of drug reinforcement. Ann NY Acad Sci 654:171–191. 10.1111/j.1749-6632.1992.tb25966.x [DOI] [PubMed] [Google Scholar]
- Kumar V, Krolewski DM, Hebda-Bauer EK, Parsegian A, Martin B, Foltz M, Akil H, Watson SJ (2021) Optimization and evaluation of fluorescence in situ hybridization chain reaction in cleared fresh-frozen brain tissues. Brain Struct Funct 226:481–499. 10.1007/s00429-020-02194-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour A, Khachaturian H, Lewis ME, Akil H, Watson SJ (1988) Anatomy of CNS opioid receptors. Trends Neurosci 11:308–314. 10.1016/0166-2236(88)90093-8 [DOI] [PubMed] [Google Scholar]
- Mansour A, Fox CA, Akil H, Watson SJ (1995a) Opioid-receptor mRNA expression in the rat CNS: anatomical and functional implications. Trends Neurosci 18:22–29. 10.1016/0166-2236(95)93946-U [DOI] [PubMed] [Google Scholar]
- Mansour A, Hoversten MT, Taylor LP, Watson SJ, Akil H (1995b) The cloned mu, delta and kappa receptors and their endogenous ligands: evidence for two opioid peptide recognition cores. Brain Res 700:89–98. 10.1016/0006-8993(95)00928-J [DOI] [PubMed] [Google Scholar]
- Marchant NJ, Campbell EJ, Whitaker LR, Harvey BK, Kaganovsky K, Adhikary S, Hope BT, Heins RC, Prisinzano TE, Vardy E, Bonci A, Bossert JM, Shaham Y (2016) Role of ventral subiculum in context-induced relapse to alcohol seeking after punishment-imposed abstinence. J Neurosci 36:3281–3294. 10.1523/JNEUROSCI.4299-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant NJ, Campbell EJ, Pelloux Y, Bossert JM, Shaham Y (2019) Context-induced relapse after extinction versus punishment: similarities and differences. Psychopharmacology (Berl) 236:439–448. 10.1007/s00213-018-4929-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Fournier S, Jardine K, Sutherland L (1994) Intragenic regions of the murine Pgk-1 locus enhance integration of transfected DNAs into genomes of embryonal carcinoma cells. Somat Cell Mol Genet 20:515–528. 10.1007/BF02255842 [DOI] [PubMed] [Google Scholar]
- Mello NK, Negus SS (1996) Preclinical evaluation of pharmacotherapies for treatment of cocaine and opioid abuse using drug self-administration procedures. Neuropsychopharmacology 14:375–424. 10.1016/0893-133X(95)00274-H [DOI] [PubMed] [Google Scholar]
- Midavaine E, Cote J, Marchand S, Sarret P (2021) Glial and neuroimmune cell choreography in sexually dimorphic pain signaling. Neurosci Biobehav Rev 125:168–192. 10.1016/j.neubiorev.2021.01.023 [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C (2007) The rat brain in stereotaxic coordinates, Ed 6. San Diego: Academic. [DOI] [PubMed] [Google Scholar]
- Quadros RM, et al. (2017) Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18:1–15. 10.1186/s13059-017-1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Fredriksson I, Lofaro OM, Bossert JM, Shaham Y (2019) Relapse to opioid seeking in rat models: behavior, pharmacology and circuits. Neuropsychopharmacology 44:465–477. 10.1038/s41386-018-0234-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Lofaro OM, Applebey SV, Korah H, Venniro M, Cifani C, Bossert JM, Shaham Y (2020) Role of projections between piriform cortex and orbitofrontal cortex in relapse to fentanyl seeking after palatable food choice-induced voluntary abstinence. J Neurosci 40:2485–2497. 10.1523/JNEUROSCI.2693-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner DJ, Townsend EA, Orihuel J, Applebey SV, Claypool SM, Banks ML, Shaham Y, Negus SS (2021) Lack of effect of different pain-related manipulations on opioid self-administration, reinstatement of opioid seeking, and opioid choice in rats. Psychopharmacology (Berl) 238:1885–1897. 10.1007/s00213-021-05816-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss D, Maduna T, Maurin H, Audouard E, Gaveriaux-Ruff C (2022) Mu opioid receptor in microglia contributes to morphine analgesic tolerance, hyperalgesia, and withdrawal in mice. J Neurosci Res 100:203–219. 10.1002/jnr.24626 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham Y, Stewart J (1996) Effects of opioid and dopamine receptor antagonists on relapse induced by stress and re-exposure to heroin in rats. Psychopharmacology (Berl) 125:385–391. 10.1007/BF02246022 [DOI] [PubMed] [Google Scholar]
- Shaham Y, Shalev U, Lu L, De Wit H, Stewart J (2003) The reinstatement model of drug relapse: history, methodology and major findings. Psychopharmacology (Berl) 168:3–20. 10.1007/s00213-002-1224-x [DOI] [PubMed] [Google Scholar]
- Shalev U, Grimm J, Shaham Y (2002) Neurobiology of relapse to heroin and cocaine seeking: a review. Pharmacol Rev 54:1–42. 10.1124/pr.54.1.1 [DOI] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hubner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R (2002) Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis 32:19–26. 10.1002/gene.10023 [DOI] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351:84–88. 10.1126/science.aad5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J, Woodside B, Shaham Y (1996) Ovarian hormones do not affect the initiation and maintenance of intravenous self-administration of heroin in the female rat. Psychobiology 24:154–159. 10.3758/BF03331967 [DOI] [Google Scholar]
- Takahashi YK, Batchelor HM, Liu B, Khanna A, Morales M, Schoenbaum G (2017) Dopamine neurons respond to errors in the prediction of sensory features of expected rewards. Neuron 95:1395–1405.e3. 10.1016/j.neuron.2017.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrier J, Luscher C, Pascoli V (2016) Cell type-specific insertion of GluA2-lacking AMPARs with cocaine exposure leading to sensitization, cue-induced seeking, and incubation of craving. Neuropsychopharmacology 41:1779–1789. 10.1038/npp.2015.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobiansky DJ, Wallin-Miller KG, Floresco SB, Wood RI, Soma KK (2018) Androgen regulation of the mesocorticolimbic system and executive function. Front Endocrinol 9:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino FJ, Corrigall WA (1987) Effects of opiate antagonist treatment into either the periaqueductal grey or nucleus accumbens on heroin-induced locomotor activation. Brain Res Bull 19:545–549. 10.1016/0361-9230(87)90071-2 [DOI] [PubMed] [Google Scholar]
- Vaccarino FJ, Bloom FE, Koob GF (1985) Blockade of nucleus accumbens opiate receptors attenuates intravenous heroin reward in the rat. Psychopharmacology (Berl) 86:37–42. 10.1007/BF00431681 [DOI] [PubMed] [Google Scholar]
- Venniro M, Caprioli D, Shaham Y (2016) Animal models of drug relapse and craving: from drug priming-induced reinstatement to incubation of craving after voluntary abstinence. Prog Brain Res 224:25–52. 10.1016/bs.pbr.2015.08.004 [DOI] [PubMed] [Google Scholar]
- Venniro M, Zhang M, Shaham Y, Caprioli D (2017) Incubation of methamphetamine but not heroin craving after voluntary abstinence in male and female rats. Neuropsychopharmacology 42:1126–1135. 10.1038/npp.2016.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Lim G, Yang L, Zeng Q, Sung B, Jeevendra Martyn JA, Mao J (2005) A rat model of unilateral hindpaw burn injury: slowly developing rightwards shift of the morphine dose–response curve. Pain 116:87–95. 10.1016/j.pain.2005.03.044 [DOI] [PubMed] [Google Scholar]
- Weibel R, Reiss D, Karchewski L, Gardon O, Matifas A, Filliol D, Becker JA, Wood JN, Kieffer BL, Gaveriaux-Ruff C (2013) Mu opioid receptors on primary afferent nav1.8 neurons contribute to opiate-induced analgesia: insight from conditional knockout mice. PLoS One 8:e74706. 10.1371/journal.pone.0074706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RA (1989) Opiate reward: sites and substrates. Neurosci Biobehav Rev 13:129–133. 10.1016/s0149-7634(89)80021-1 [DOI] [PubMed] [Google Scholar]
- Wolf ME (2016) Synaptic mechanisms underlying persistent cocaine craving. Nat Rev Neurosci 17:351–365. 10.1038/nrn.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]