Abstract
Motivation
IntLIM uncovers phenotype-dependent linear associations between two types of analytes (e.g. genes and metabolites) in a multi-omic dataset, which may reflect chemically or biologically relevant relationships.
Results
The new IntLIM R package includes newly added support for generalized data types, covariate correction, continuous phenotypic measurements, model validation and unit testing. IntLIM analysis uncovered biologically relevant gene–metabolite associations in two separate datasets, and the run time is improved over baseline R functions by multiple orders of magnitude.
Availability and implementation
IntLIM is available as an R package with a detailed vignette (https://github.com/ncats/IntLIM) and as an R Shiny app (see Supplementary Figs S1–S6) (https://intlim.ncats.io/).
Supplementary information
Supplementary data are available at Bioinformatics Advances online.
1 Introduction
In recent years, many biomedical fields have begun to explore multi-omic mechanisms of disease, clinical outcomes and other phenotypic traits. However, integrating and interpreting multi-omic data to discover latent interdependencies remains a challenge (Eicher et al., 2020). We introduce IntLIM 2.0, an R package that uncovers phenotype-dependent linear associations between two types of analytes (e.g. genes and metabolites). IntLIM 2.0 extends IntLIM 1.0 (Siddiqui et al., 2018) to support generalized analyte measurement data types, continuous phenotypic measurement, covariate correction, model validation and unit testing. Several other tools support global multi-omic correlations or phenotype-dependent correlation analysis for either discrete or continuous phenotypic measurements (Fukushima, 2013; Langfelder and Horvath, 2008; Ma et al., 2019; Shi et al., 2019; Siska et al., 2016). IntLIM 2.0 is unique as it supports continuous and discrete phenotypic measurements, and is based on linear models, which allow for adjustment of independent effects (e.g. clinical variables and technical effects).
2 IntLIM functionality
For each pair of analytes in a dataset, IntLIM 2.0 solves Equation (1) in a streamlined manner, where ai and aj are measurements for two separate analytes, p is the phenotypic measurement, C = {c1,…c|C|} is a set of continuous or discrete clinical covariates (potential model confounders as determined by data analyst), and β0–β3+ |C| is a set of coefficients corresponding to the model intercept, aj, p, the interaction between aj and p, and additional model covariates. Notably, the order of ai and aj may affect the outcome and should be motivated by biology (e.g. effect of gene expression level ai on metabolite abundance aj).
| (1) |
The input to IntLIM 2.0 is described in the Supplementary Files. Users have the option to filter individual analytes by percentage of missing values, mean measurement and coefficient of variation, but it is expected that the sample metadata and analyte measurements have already been otherwise filtered, normalized and/or imputed prior to analysis.
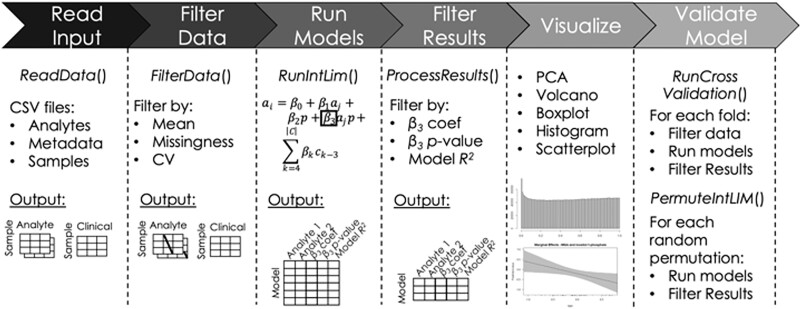
Analyte pairs with significant phenotype-dependent associations are filtered and returned using one or more of the following criteria: Benjamini–Hochberg false discovery rate adjusted P-value, percentile and coefficient of determination (R2) value (Supplementary Files). To ensure the validity of the P-value as a measure of significance, it is expected that aj follows a Gaussian distribution. We note that multiple visualization functions are also included in IntLIM 2.0, including a trendline and residual plot, a newly developed figure of analyte measurements marginalized over phenotype and a plot of sorted β3 values (Supplementary Files). An overview of the functionality of IntLIM is illustrated in Figure 1.
Fig. 1.
Schematic of IntLIM 2.0 functionality. New features include filtering by coefficient of variation in FilterData(), addition of covariate terms in RunIntLim(), filtering by model R2 in ProcessResults(), and the RunCrossValidation() and PermuteIntLIM() functions
3 New features
IntLIM 2.0 includes several major expansions to IntLIM 1.0 (Patt et al., 2019; Siddiqui et al., 2018). First, in addition to the gene ∼ metabolite models supported by IntLIM 1.0, IntLIM 2.0 supports both inter- and intra-omics (e.g. metabolite ∼ metabolite) models and other types of omics data (e.g. microbial abundance, protein abundance, methylation level or mutation rate) (Do et al., 2015; Langfelder and Horvath, 2008; Van Der Knaap and Verrijzer, 2016). Second, IntLIM 2.0 supports correction for covariates (e.g. batch effects, demographic or clinical covariates). Relatedly, a new option to filter models by R2 value allows users to evaluate models by goodness of fit. Third, IntLIM 2.0 supports continuous phenotypic measurements (e.g. disease severity, drug response metrics, etc.) in addition to data with two phenotype categories of interest (e.g. case/control). Fourth, IntLIM 2.0 supports model validation using (i) cross-validation and (ii) random permutation models, along with accompanying visualizations described further in Supplementary Files. Finally, the introduction of unit tests using the testthat package (Wickham, 2011) makes IntLIM 2.0 more robust to programming errors than IntLIM 1.0.
4 Results
The utility of IntLIM 2.0 is illustrated using datasets with continuous and discrete phenotypic measurements. The NCI-60 dataset (continuous) includes 57 cell lines from NCI-60 (Shoemaker, 2006), each with 17 987 gene expression levels, 280 metabolite abundance levels (Su et al., 2011) and the average drug concentration that inhibits cell growth by 50% (IC50) over 48 h (drug score) (Reinhold et al., 2012). The BRCA dataset (discrete) includes 61 tumor and 47 adjacent normal breast tissue samples, each with 20 254 gene expression levels and 536 metabolite abundance levels (Terunuma et al., 2014). Patient age and race covariates were adjusted for this dataset. Detailed vignettes on running these models in the NCI-60 and BRCA vignettes are available at tinyurl.com/du5xv5pc and tinyurl.com/2p94sfde, respectively. Results are summarized in Table 1, Supplementary Files and vignettes.
Table 1.
Summary of IntLIM 2.0 results on NCI-60 and BRCA datasets
| Dataset | Filtered analyte count (gene/metabolite) | Significant paira count | Significant pairs with knowledge supportb |
|---|---|---|---|
| NCI-60 | 16 188/280 | 2517 | 26 |
| BRCA | 18 288/536 | 14 583 | 555 |
FDR-adjusted P-value < 0.1 for β3, |β3| percentile > 0.5, R2 > 0.2.
Shared pathway and/or reaction in RaMP-DB 2.0 (Braisted et al., 2022; Zhang et al., 2018).
Significant associations found in the NCI-60 data have been implicated in tumor progression and/or treatment: namely, cholesterol with HHAT (Callahan and Wang, 2015) and CDKN1A (Moon et al., 2019). These pairs and 19 pairs with knowledgebase support were insignificant in all 100 permutations of the data (Supplementary Fig. S7), supporting non-randomness. Further, 1778 of the 2517 pairs were significant in at least one leave-one-out cross-validation fold, and 734 were significant in more than half of the folds. Additionally, the BRCA analysis also uncovered associations previously reported using IntLIM 1.0 (Siddiqui et al., 2018); namely, ASNS and glutamine, SLC7A1 and glutamine, and GPT2 and 2-hydroxyglutarate. These pairs and 413 pairs with knowledgebase support were insignificant in all 100 permutations (Supplementary Fig. S8). 19 of the 14 583 pairs were significant in at least one leave-one-out cross-validation fold, which is likely attributable to the smaller sample size.
Runtime of IntLIM 2.0 was considerably faster than the linear mixed model function lm() in the stats R package when tested on all metabolite levels and the first 100 gene expression levels in the NCI-60 data (Table 2).
Table 2.
Runtime of IntLIM 2.0 and lm() on two separate machines for a reduced dataset
| R function | Runtime (Machine 1) (s) | Runtime (Machine 2) (s) |
|---|---|---|
| RunIntLim() | 12 | 11 |
| lm() | 1622 | 482 |
Machine 1 is a MacBook Pro laptop running macOS 12.3 with an Intel Core i7 CPU, and Machine 2 is a single compute node of a SLURM high-performance computing cluster running CentOS Linux 7 with 2 allocated Intel Gold 6140 CPU’s.
Supplementary Material
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). We thank Elizabeth Baskin for contributing to the R package, Ke Wang and John Braisted for server deployment and repository management, and the NIH Individual Graduate Partnership Program for training and mentoring.
Contributor Information
Tara Eicher, Division of Preclinical Innovation, National Center for Advancing Translational Sciences, NIH, Rockville, MD 20892, USA; Department of Computer Science and Engineering, The Ohio State University, Columbus, OH 43210, USA.
Kyle D Spencer, Division of Preclinical Innovation, National Center for Advancing Translational Sciences, NIH, Rockville, MD 20892, USA; Biomedical Sciences Graduate Program, The Ohio State University, Columbus, OH 43210, USA.
Jalal K Siddiqui, Comprehensive Cancer Center, The Ohio State University, Columbus, OH 43210, USA.
Raghu Machiraju, Department of Computer Science and Engineering, The Ohio State University, Columbus, OH 43210, USA; Biomedical Informatics Department, The Ohio State University, Columbus, OH 43210, USA; Department of Pathology, The Ohio State University, Columbus, OH 43210, USA.
Ewy A Mathé, Division of Preclinical Innovation, National Center for Advancing Translational Sciences, NIH, Rockville, MD 20892, USA.
Funding
This work was supported in part by the National Center for Advancing Translational Sciences (NCATS) Informatics Research Core [ZIC TR000410-03].
Conflict of Interest: none declared.
References
- Braisted J. et al. (2022) RaMP-DB 2.0: a renovated knowledgebase for deriving biological and chemical insight from metabolites, proteins, and genes. Bioinformatics, 39, btac726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.P., Wang C. (2015) Hedgehog cholesterolysis: specialized gatekeeper to oncogenic signaling. Cancers (Basel), 7, 2037–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do K.T. et al. (2015) Network-based approach for analyzing intra- and interfluid metabolite associations in human blood, urine, and saliva. J. Proteome Res., 14, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Eicher T. et al. (2020) Metabolomics and multi-omics integration: a survey of computational methods and resources. Metabolites, 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima A. (2013) DiffCorr: an R package to analyze and visualize differential correlations in biological networks. Gene, 518, 209–214. [DOI] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics, 9, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. (2019) Differential network enrichment analysis reveals novel lipid pathways in chronic kidney disease. Bioinformatics, 35, 3441–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S.H. et al. (2019) p53 represses the mevalonate pathway to mediate tumor suppression. Cell, 176, 564–580.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patt A. et al. (2019) Integration of metabolomics and transcriptomics to identify gene-metabolite relationships specific to phenotype. Methods Mol. Biol., 1928, 441–468. [DOI] [PubMed] [Google Scholar]
- Reinhold W.C. et al. (2012) CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res., 72, 3499–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W.J. et al. (2019) Unsupervised discovery of phenotype-specific multi-omics networks. Bioinformatics, 35, 4336–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker R.H. (2006) The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer, 6, 813–823. [DOI] [PubMed] [Google Scholar]
- Siddiqui J.K. et al. (2018) IntLIM: integration using linear models of metabolomics and gene expression data. BMC Bioinformatics, 19, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siska C. et al. (2016) The discordant method: a novel approach for differential correlation. Bioinformatics, 32, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su G. et al. (2011) Integrated metabolome and transcriptome analysis of the NCI60 dataset. BMC Bioinformatics, 12, S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terunuma A. et al. (2014) MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Invest., 124, 398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Knaap J.A., Verrijzer C.P. (2016) Undercover: gene control by metabolites and metabolic enzymes. Genes Dev., 30, 2345–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2011) testthat: get started with testing. R Journal, 3, 5–10. [Google Scholar]
- Zhang B. et al. (2018) RaMP: a comprehensive relational database of metabolomics pathways for pathway enrichment analysis of genes and metabolites. Metabolites, 8, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.