Abstract
The wild relatives of rice hold unexplored genetic diversity that can be employed to feed an estimated population of 10 billion by 2050. The Oryza Map Alignment Project (OMAP) initiated in 2003 has provided comprehensive genomic resources for comparative, evolutionary, and functional characterization of the wild relatives of rice, facilitating the cloning of >600 rice genes, including those for grain width (GW5) and submergence tolerance (SUB1A). Following in the footsteps of the original project, the goal of ‘IOMAP: the Americas’ is to investigate the present and historic genetic diversity of wild Oryza species endemic to the Americas through the sequencing of herbaria and in situ specimens. The generation of a large diversity panel describing past and current genetic status and potential erosion of genetic variation in the populations will provide useful knowledge for the conservation of the biodiversity in these species. The wild relatives of rice in the Americas present a wide range of resistance traits useful for crop improvement and neodomestication approaches. In the race against time for a sustainable food future, the neodomestication of the first cereal species recently accomplished in O. alta opens the door to the potential neodomestication of the other wild Oryza species in Americas.
Keywords: Biodiversity conservation, genetic diversity, herbaria specimens, in situ specimens, IOMAP, neodomestication, population genetics, resistance traits, wild relatives of rice in the Americas
We review the relevance and major outcomes of IOMAP to the wild Oryzaspecies in the Americas, focusing on biodiversity conservation, and discuss achievements and future implications of the neodomestication of Oryza alta.
Introduction
Rice is one of the major cereal grains and a staple food for more than half of the world’s population (Bhandari, 2019). Rice is a significant food source representing 19% of per capita calorie consumption across the globe (Maclean et al., 2013), and its by-products (e.g. straw and husks) can be used to produce biofuels, paper, or fertilizers (Goodman, 2020). Asian rice (Oryza sativa) is the most consumed rice species, with China, India, Thailand, Vietnam, and Pakistan accounting for 74% of the global export (Bhandari, 2019). According to the United States Department of Agriculture (USDA), the world rice production in 2020/2021 was 509.3 Mt, with China and India generating 53.5% of the global production (https://apps.fas.usda.gov/). The domestication of Asian rice began ~9000 years ago in East Asia, possibly in the Yangtze valley in China, where O. sativa was domesticated from O. rufipogon (Molina et al., 2011). In contrast, African rice (O. glaberrima) is a minor crop grown in limited areas of West Africa and Suriname (South America). The domestication of O. glaberrima started ~3000 years ago in Africa with the cultivation of O. barthii along the delta of the Niger River and its later development into the modern African rice (Wang et al., 2014). In addition to the domesticated rice species, the Oryza genus contains 25 wild species distributed in the pan-tropics, which exhibit tremendous diversity in morphology, agronomic traits, and adaptations to different biotic and abiotic stress (Sanchez et al., 2013). The genus spans 15 million years (MYs) of evolutionary history, and includes species with different ploidy levels and genome architectures. Morphological, cytological, and molecular analyses permitted classification of the Oryza species into 11 genome types, six diploid (i.e. AA, BB, CC, EE, FF, and GG, 2n=2x=24 chromosomes), and five tetraploid (i.e. BBCC, CCDD, HHJJ, HHKK, and KKLL, 2n=4x=48 chromosomes). Based on their phylogenetic relationships and crossability with cultivated rice, Oryza species are further classified into three gene pools (Harlan and de Wet, 1971; Vaughan, 1989). The primary gene pool consists of eight species belonging to the AA genome type; the two cultivated species and six wild species (i.e. O. barthii, O. longistaminata, O. meridionalis, O. glumipatula, O. nivara, and O. rufipogon; however, the latter three species are often considered as synonyms). The AA genome species are also regarded as the O. sativa complex, one of the four Oryza complexes defined by the taxonomic studies of Tateoka in the 1960s (Tateoka, 1963). Species belonging to the O. sativa complex are characterized by high crossability, with wild species commonly used for trait introgression into rice cultivars via traditional breeding methods. The rate of successful hybridization between cultivated rice and species belonging to the secondary and tertiary gene pools is progressively lower given higher reproductive barriers and cross incompatibilities. The secondary gene pool comprises O. brachyantha (FF) and the O. officinalis complex, which contains 11 wild species and five genome types, namely BB (O. punctata); CC (O. eichingeri, O. officinalis, and O. rhizomatis); EE (O. australiensis); BBCC (O. malampuzhaensis, O. minuta, and O. schweinfurthiana); and CCDD (O. alta, O. grandiglumis, and O. latifolia). The tertiary gene pool includes the O. meyeriana complex (comprising the GG type O. meyeriana and O. granulata), the O. ridleyi complex (comprising the HHJJ type O. ridleyi and O. longiglumis), and the unclassified species O. schlecteri (HHKK) and O. coarctata (KKLL) (Vaughan, 1989).
The wild relatives of rice harbor large (and often unexplored) genetic variation that can potentially be used for crop improvement, and ultimately food security (Dempewolf et al., 2017; Brar and Khush, 2018). Through millions of years of evolution and genetic adaptation to different environments, the wild Oryza species have accumulated allelic diversity in genes useful for crop improvement, such as biotic stress resistance (e.g. insects, fungi, and bacteria), abiotic stress tolerance (e.g. wounding, salinity, cold, and flooding), and nutritional- and yield-related traits (Song et al., 2005). However, the utility of wild relatives for rice improvement depends on the availability of these genetic resources: in situ and ex situ conservation strategies are therefore crucial to preserve the wild relatives from extinction and to counteract the anthropogenic activity on their natural habitats (Medeiros et al., 2021).
The Oryza Map Alignment Project (OMAP) and the Oryza Genome Evolution Project (OGEP) were established almost two decades ago to provide comprehensive genomic resources for comparative, evolutionary, and functional characterization of the wild relatives of rice. This platform has allowed the discovery of novel genetic diversity in wild species to generate new rice genotypes with improved traits (Wing et al., 2007). The project was further extended to several international research alliances under the frame of the International Oryza Map Alignment Project (IOMAP; Jacquemin et al., 2013). The resources provided by IOMAP have allowed the generation of many advanced mapping populations through conventional trait introgression and molecular breeding techniques (Gutierrez et al., 2010; Ram et al., 2010; Bocco et al., 2012). With the development of genome editing and CRISPR/Cas [clustered regularly interspaced palindromic repeats(CRISPR)/CRISPR-associated protein] technology (Jinek et al., 2012, 2013; Doudna and Charpentier, 2014), an innovative approach called neodomestication (Gutaker et al., 2022) arose as a promising alternative to conventional crop improvement (Nooryazdan et al., 2011; Zsögön et al., 2017; Li et al., 2018). The recent publication of a high-quality genome reference of O. alta and a road-map for the rapid domestication of this species represents the first description of a neodomesticated polyploid cereal (Yu et al., 2021).
Whereas several studies subsequent to the IOMAP gave insights into Asian and African Oryza species’ evolutionary history (Ammiraju et al., 2007; Chen et al., 2013; Stein et al., 2018), a description of the genetic diversity of the wild Oryza species in the Americas is lagging behind. The project ‘IOMAP: the Americas’ was funded to investigate the genetic diversity of the wild relatives of rice endemic to the Americas. The project aims to provide a comprehensive description of the status of modern wild Oryza species by generating an extensive collection of resequenced specimens collected in North, Central, and South America. The project’s perspective is to inform on both the historical and current biodiversity status of the wild rice species endemic to the Americas, and identify putative candidates for neodomestication (Fornasiero et al., 2022).
Here, we review the primary outcomes of IOMAP and present major methods and goals of the project ‘IOMAP: the Americas’, giving a background on the evolutionary origin of the wild Oryza in the Americas, their biotic and abiotic resistance traits, their conservation status, and the recent neodomestication of O. alta.
Relevance and major outcomes of the IOMAP
The first digital platform of the Oryza genus generated in the 2000s by the OMAP and OGEP consortia (Wing et al., 2007; Ammiraju et al., 2010a) included the release of BAC-end sequence/fingerprint-based physical maps of 17 species spanning all eight AA-genome species and one of each of the other nine extant genome types (i.e. BB, CC, BBCC, CCDD, EE, FF, GG, KKLL, and HHJJ). Alignment of the genome sequences against the International Rice Genome Sequencing Project (IRGSP) reference genome sequence allowed the characterization of transposable element (TE) abundance, distribution, and evolutionary dynamics (Piegu et al., 2006; Ammiraju et al., 2007, 2010b; Roulin et al., 2008), the study of the evolution of gene families (Ammiraju et al., 2008; Lu et al., 2009), and the characterizations of mutations and genome-wide rearrangements (Hurwitz et al., 2010). Ammiraju et al. (2008) sequenced a set of orthologous loci harboring the alcohol-dehydrogenase 1 and 2 genes (Adh-1 and Adh-2, two established loci for comparative genomic studies across plant species) in the six extant diploid genome types, and compared them with the Adh1 locus from the IRGSP reference sequence (OsAdh1RefSeq). Comparative analysis showed that the locus underwent recent rearrangements leading to rapid and lineage-specific diversification of the Oryza species analyzed, with gain and loss of gene family members and general synteny disruption. The analysis of TE insertion events at the Adh1 region revealed that long terminal repeat retrotransposable elements (LTR-RTs) are not conserved in the array of Oryza species and appeared to have inserted independently in the different genome types within the last 5 MYs after speciation (Ammiraju et al., 2008).
The goal of IOMAP was to produce reference genome sequences and transcriptome data for the 17 original species and generate a genome-scale and genus-wide platform available to the rice scientific community to address major questions related to comparative genomics. The primary foci were to: (i) gain new knowledge at the macro-evolutionary level (i.e. understand the dynamics of the evolution of genomes and polyploidy in the genus Oryza, unravel the history and impact of rice domestication, and explore the diversity and evolution of gene families useful for rice improvement); (ii) develop advanced mapping populations for functional and breeding purposes; and (iii) carry out population genomics and ecological studies for biodiversity conservation purposes (Jacquemin et al., 2013).
The vast array of genomic tools and data produced by IOMAP set the basis for generating chromosome-level reference assemblies of many Oryza species (Yu et al., 2002; International Rice Genome Sequencing Project and Sasaki, 2005; Chen et al., 2013; Zhang et al., 2015a). The comparative analysis of 13 Oryza reference genomes, including the AA genome species, O. punctata, O. brachyantha, and Leersia perrieri used as the outgroup, revealed important features of the evolution of the Oryza genomes (Stein et al., 2018). Using >6000 single-copy orthologs, Stein and colleagues inferred a phylogeny of 13 single-individual species in which the AA clade was estimated to diverge 2.5 MYs ago and the diversification occurred at a rate of ~0.50 net new species per MY. Analysis of TEs revealed lineage-specific amplification and rapid turnover of LTR-RTs in the genomes under study. Comparative annotation and phylogenetic analysis of LTR-RTs showed that the genomes underwent recent and lineage-specific bursts of transposition. By tracking 75 orthologous LTR-RT loci across the eight most closely related AA species, Stein et al. estimated an average loss rate of 3.62 kb per MY per LTR-RT. At the genome scale, this implies that a quarter of an AA genome would be eliminated within 3–4 MYs in the absence of new bursts of transposition. Comparative analysis of gene annotations showed that lineage-specific gene families in the Oryzeae clade exhibited features of newly emerging genes, such as shorter coding sequences, lower expression, more rapid evolution rates (i.e. relaxed purifying selection), and evolutionary instability compared with gene families emerged in the common ancestors of grasses (Poaceae). However, Oryzeae-specific loci showed a high degree of conserved synteny even among distantly related species in the clade (Stein et al., 2018).
Finally, the generation and release of high-quality reference genomes of cultivated and non-cultivated rice species by IOMAP has allowed the cloning of >600 rice genes, many of which are not present in the IRGSP reference itself, including those for grain width (GW5), and submergence tolerance (SUB1A; Wing, 2015).
IOMAP: the Americas
One of the key goals of IOMAP is the establishment of diversity panels of wild Oryza species for evolutionary studies and conservation efforts. Following in the footsteps of IOMAP, ‘the Americas project’ was funded to investigate the present and historic genetic diversity found in wild relatives of rice endemic to North, Central, and South America. There are four wild Oryza species endemic to the Americas: one AA genome type—O. glumipatula Steud, and three CCDD types—O. alta Swallen, O. grandiglumis (Doell) Prod, and O. latifolia Desv (Vaughan, 1989). Insights into the distribution, diversification, and genetic diversity will inform: (i) the identification of candidate accessions as suitable targets for the neodomestication and/or the genetic improvement of rice crops to respond to environmental challenges due to climate change; and (ii) the establishment of a valuable resource to be used to promote conservation of the wild Oryza endemic to the Americas. To accomplish these objectives, we will generate a collection of both contemporary specimens collected in the Americas (with established collaborations with the Universidad de Costa Rica, the Universidade Federal de Pelotas in Brazil, the Universidad de las Americas in Ecuador, and CIAT in Colombia), and herbarium specimens from the Royal Botanical Gardens, Kew (RBG Kew, London, UK), the Natural History Museum (NHM) in London, the New York Botanical Garden (NYBG, New York, USA), the Smithsonian National Museum of Natural History (Washington DC, USA), and the Muséum National d’Histoire Naturelle (MNHN) in Paris. Whole-genome sequencing of historical and contemporary collections, and analysis of single nucleotide polymorphisms (SNPs), will be the starting point to study evolutionary processes to estimate loss or gain of genetic diversity over time, detect hybridization and admixture events between populations/species, and identify candidate regions involved in adaptation to local conditions (Fig. 1). To generate the SNP panel, we will take advantage of the assembled and annotated Platinum Standard Reference Sequences (PSRefSeqs) of the four species previously generated in the Wing lab and deposited in NIH GenBank (Benson et al., 2013; https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA48429). The genetic information gained will be useful to evaluate the condition, threat of genetic erosion, and conservation status of wild populations to inform on their long-term viability status and, possibly, the need for conservation/protection strategies. Detection of adaptive variation can be harnessed for future crop improvement and neodomestication efforts (Godfray et al., 2010).
Fig. 1.
Anthropogenic activities, such as agriculture, and natural events affect the interaction between the wild species and its native environment, altering the connections with other species, and causing genetic threats (i.e. drift, inbreeding, and hybridization). Human intervention can have a high impact on the demography and structure of the populations and can lead to (from left to right): new contacts among species/populations; expansion to new habitats; habitat fragmentation; population expansion; hybridization; and introgression of new alleles from crops or non-indigenous species.
The wild Oryza species in the Americas
Oryza glumipatula is the only diploid wild rice species in the New World, and ranges from Central to South America and the Caribbean. It grows in flooded areas, swamps, rivers, and humid areas with clay soils. The life cycle depends on the geographical location of the populations, ranging from perennial to annual or biannual (Karasawa et al., 2007). The plant is characterized by a bushy growth, with fragile stems near the base that can detach and fluctuate on rising water, forming new populations downstream (Vaughan et al., 2003). Flowering occurs predominantly through self-pollination, although cross-pollination is also possible (Fuchs et al. 2016; Table 1). There are no clear morphological differences between O. glumipatula and AA genomes from Asia and Australia. The Plants of the World Online (POWO), an international collaborative program facilitated by the RBG Kew, considers O. glumipatula as a synonym for O. rufipogon (https://powo.science.kew.org/results?f=&q=Oryza%20glumipatula). However, some sterility barriers occur between O. glumipatula and Asian AA species (Juliano et al., 1998), suggesting speciation. Wambugu et al. (2015) used whole chloroplast sequences to clarify the phylogenetic relationships among the AA genomes. The phylogenetic tree defined two primary clades: a basal South American/African clade containing O. glumipatula and O. longistaminata, and a second clade containing all other Oryza AA genome species (Wambugu et al., 2015). A phylogenetic analysis of the 13 Oryza genome dataset by Stein et al. (2018) showed that O. glumipatula is more closely related to the African O. barthii than to the Asian O. rufipogon. Accordingly, clear signatures of introgressions were found in 10 out of 12 chromosomes between O. glumipatula and the African species, but not the Asian species (Stein et al., 2018). Reciprocal introgression lines between O. glumipatula and O. sativa have been developed, and facilitated the identification of genes such as Rhw, controlling hybrid weakness restoration, and S22, regulating pollen semi-sterility and hybrid sterility (Vaughan et al., 2003). Genetic diversity studies for this species have been carried out in Brazil and Costa Rica: in Brazil, two main groups were found in the Amazon basin and one in the Pantanal (Karasawa et al., 2007); for Costa Rica, two main groups were found—one in the north-east and one in the north-west side of the country (Fuchs et al., 2016). Being predominantly self-pollinated implies a tendency for O. glumipatula to lose genetic diversity and increase genetic differentiation among populations, with the risk of fragmentation and lower flexibility in response to environmental changes (Karasawa et al., 2007).
Table 1.
Brief description of the wild Oryza species endemic to the Americas
| Species | Karyotype | Genome type |
Genome Size (Mbp) | Morhology | Geographical Distribution | Natural Habitat | Traits of interest |
|---|---|---|---|---|---|---|---|
| Oryza glumipatula | 24 | AA | 464 | Bushy growth with fragile stem near the base that can detach and float on rising water | Brazil, Colombia, Costa Rica, French Guiana, Cuba, Suriname, and Venezuela | Flooded areas that become seasonally dry, swamps, rivers, and humid areas with clay soils; open and sunny areas | Resistance to blast, M. graminicola, and rice black streaked dwarf virus (RBSDV); source of cytoplasmic male sterility (CMS); submergence tolerance |
|
Oryza
alta |
48 | CCDD | 895 | Tall (up to 4 m), erect plant with broad leaves (generally >5 cm), spikelet length >7 mm | Belize, Brazil, Colombia, Guyana, Paraguay, and Venezuela | Found mainly in savannah, sometimes in woodland, in seasonally flooded habitats; open and sunny areas | Resistance to striped stem borer (SSB); salinity tolerance; high biomass production |
| Oryza grandiglumis | 48 | CCDD | 865 | Tall (up to 4 m), broad leaves (3–5 cm), pubescent ligule, sterile lemma | Argentina, Bolivia, Brazil, Colombia, Costa Rica, Ecuador, French Guiana, Paraguay, Peru, and Venezuela | Found in savannah or woodland, in seasonally flooded areas; open and shaded areas | Submergence tolerance; high biomass production |
| Oryza latifolia | 48 | CCDD | 1043 | Both short (usually <1 m) and tall (≥2 m) forms exist. Broad leaves, typically <5 cm; spikelet length <7 mm | Argentina, Belize, Bolivia, Brazil, Colombia, Costa Rica, Cuba, Dominican Republic, Ecuador, El Salvador, French Guiana, Guatemala, Guyana, Haiti, Honduras, Mexico, Nicaragua, Panama, Paraguay, Peru, Puerto Rico, Surinam, Trinidad, and Venezuela | Found in low forest, rainforest, open woodland, undulating savannah, pasture, cultivated fields, and open swamp. Often grows in or near water. In seasonally dry, open or semi-open, sunny areas | Resistance to bacterial blight (BB), brown planthopper (BPH), and white-backed planthopper (WBPH); salinity tolerance; high biomass production |
Oryza alta, O. grandiglumis, and O. latifolia are widely distributed in Central America, South America, and the Caribbean, and show similar morphological characteristics and distribution, with few distinctions. Oryza alta (considered a synonym for O. latifolia by POWO, https://powo.science.kew.org/results?f=&q=oryza%20alta) is a perennial type characterized by tall forms growing up to 400 cm in height, with broad leaves and a long spikelet length. It is found along river basins, streams, lake edges, and canals in deep water, usually in open and sunny areas. Oryza grandiglumis is a perennial tall plant with open panicles and a sterile lemma, and grows in savannas as well as woodlands with clay and alluvial soils. Its characteristic culm elongation, which may reach up to 760 cm in height, makes this species suitable to grow in areas subject to seasonal flooding. Oryza latifolia is a perennial plant found in aquatic ecosystems (such as swamps, savannas, grassland, and woodlands, and along hill slopes and the shores of rivers and coasts) and its growth depends on seasonal variations and water levels. Oryza latifolia shows both dwarf and tall forms, broad leaves, long panicles, high biomass, and a large number of spikelets (Table 1).
The origin of subgenome composition in the CCDD species is controversial. The three CCDD genomes are assumed to track back to a single polyploidization event (Vaughan et al., 2003), where the CC genome (O. officinalis or O. rhizomatis; Bao and Ge, 2004; Ammiraju et al., 2010b) contributed as the maternal donor. However, the origin of the DD subgenome remains unclear, and the progenitor genome is presumed to be extinct, or yet to be discovered (Vaughan et al., 2003; Shenton et al., 2020). Oryza australiensis (genome type EE) has been proposed as the potential donor of the DD subgenome in many studies (e.g. Bao and Ge, 2004; Hirsch et al., 2009). However, the study of Li et al. (2001) supported the original cytological definition of a distinct DD genome. Using multicolor genomic in situ hybridization (McGISH) in O. alta with probes from both the CC (O. eichingeri and O. officinalis) and EE genomes (O. australiensis), the authors found that the EE genome is closer to the CC than to the DD subgenome, and therefore rejected the hypothesis of the EE genome as the direct donor of the DD subgenome (Li et al., 2001).
A succession of phylogenetic studies of the Oryza species at different times [e.g. using restriction fragment length polymorphisms (RFLPs), amplified fragmernt length polymorphisms (AFLPs), chloroplast fragments, nuclear genes, and whole-genome SNPs] showed a very close proximity among the three CCDD genomes compared with the other species of the genus, with O. alta and O. grandiglumis more closely related to each other than to O. latifolia (Wang et al., 1992; Aggarwal et al., 1999; Bao and Ge, 2004; Shenton et al., 2020).
Although highly related, the CCDD genome species appear to have diversified in relation to different ecological conditions. For example, field reports from Shimamoto, Ohara, and Akimoto in the Rio Solimões (in the Amazon state) describe an atypical population of O. grandiglumis showing some morphological characteristics of O. alta. Oliveira et al. (1994) reported a population in the region of Iranduba (Amazon state) comprising both typical specimens of O. grandiglumis and O. alta, and specimens displaying intermediate forms. In the region of Santarem (Parà state), they found a population of O. alta showing morphological deviations towards O. grandiglumis (Oliveira et al., 1994). Such field observations highlight the importance of collecting genetic and ecological information on these species over a range of ecotypes to help in locating the number and size of populations for appropriate conservation strategies (Vaughan et al., 2003). This topic will be discussed further below.
Wide range of resistances to stress
Genomic and genetic research of rice cultivars and their wild relatives has greatly advanced the understanding of resistance traits for crop improvement. The wild Oryza species in the Americas possess many naturally occurring resistance/tolerance traits, such as resistance to biotic diseases caused by bacteria [e.g. bacterial blight (BB)], insects [e.g. brown planthopper (BPH)], fungi (e.g. blast), and nematodes (e.g. Meloidogyne graminicola), and tolerance to abiotic stresses (e.g. flooding and salinity), as shown in Table 1.
BB, caused by the bacterium Xanthomonas oryzae pv. oryzae (Xoo), is widespread in almost all rice-growing countries and can cause yield loss of up to 50% depending on the variety, growth stage, geographic location, and environmental conditions of the rice crop (Jiang et al., 2020). According to Jiang et al. (2020), >40 resistance genes conferring host resistance to various strains of Xoo have been identified in rice [e.g. receptor-like kinase (RLK) genes, sugar will eventually be exported transporter (SWEET) genes, and executor genes]. The RLK gene Xa21, which originated from O. longistaminata, was the first cloned resistance gene in rice (Song et al., 1995). Subsequently, interspecific crosses between an elite breeding line of O. sativa and O. latifolia resulted in new lines showing resistance to BPH and BB (Multani et al., 2003).
BPH (Nilaparvata lugens Stål) has become a severe constraint in rice production. Between 2005 and 2008, it caused a combined production loss of 2.7 Mt of rice in China (Brar et al., 2009). This migratory herbivore is a vector for plant viruses. An outbreak of BPH in Vietnam in 2005 caused the transmission of ragged stunt virus (RRSV) and rice grassy stunt virus (RGSV), resulting in the loss of 828 000 t of rice (Cabauatan et al., 2009). The most adopted method to control BPH is the application of pesticides, whose intensive use has led to environmental pollution, the death of pests’ natural enemies, and the development of insecticide-resistant BPH populations. Host plant resistance is therefore a more desirable, sustainable, and economic strategy for the control or management of BPH (Hu et al., 2016). Qiu et al. (2012) constructed pyramid lines containing the partially dominant BPH12 gene from O. latifolia, and the BPH6 gene. The combination of the two BPH resistance genes resulted in an additive effect and overall increase in resistance compared with single-gene-resistant lines (Qiu et al., 2012).
Magnaporthe oryzae, the aetiological agent of blast, is a devastating fungal disease that infects rice plants from the vegetative (leaf blast) to the reproductive stage (neck blast), and can cause up to 30% of crop loss annually (Devi et al., 2020). This pathogen evolves rapidly and presents species- and cultivar-specific races. According to Li et al. (2019), ~100 resistance genes (Pi genes) and 500 quantitative trait loci (QTLs) associated with blast resistance have been identified in rice. Due to the high variability of the fungus and the fast breaking down of resistant lines, breeders prefer varieties that contain multiple resistance genes and have broad-spectrum blast resistance (Li et al., 2019). Devi et al. (2020) characterized a major effect QTL from O. glumipatula, qBL3, and identified a putative candidate gene, Pi68(t), conferring field resistance to leaf and neck blast (Devi et al., 2020).
Root-knot nematodes (RKNs) are damaging parasites of the genus Meloidogyne infecting roots of many plants, including rice. RKN M. graminicola is widespread in subtropical and tropical regions, and is responsible for yield losses ranging from 11% to 80% in irrigated rice systems in Asia and the Americas (Mattos et al., 2019). Given that resistance to M. graminicola reported in O. sativa is limited, Mattos and colleagues evaluated resistance and response mechanisms to this RKN in the American wild rice species. Oryza glumipatula showed high levels of resistance to infection of M. graminicola, suggesting it as a potential source of resistance to this nematode in breeding programs, whereas O. alta and O. grandiglumis were found to be moderately resistant and susceptible, respectively (Mattos et al., 2019).
Both O. glumipatula and O. grandiglumis show submergence tolerance traits (Hattori et al., 2009; Okishio et al., 2014; Dos Santos et al., 2017). There are two known coping mechanisms to flooding in rice: an escape response to prolonged submergence at the mature stage, where internode elongation allows the plant to progressively emerge and float on the water; and a quiescent response, where the shoot growth is reduced during flash floods in completely submerged seedlings and resumes to normal growth after the flood recedes (Gumi and Aliero, 2018). Floating ability during the escape response in rice is driven by two ethylene response factors (ERFs), namely SNORKEL1 (SK1) and SNORKEL2 (SK2), whose expression induces rapid internodal elongation in response to ethylene in O. sativa (Hattori et al., 2009). Although SK genes were not found in O. grandiglumis, this species showed enhanced elongation of internodes upon rising water levels, suggesting an SK-independent internode elongation mechanism to prolonged flooding (Okishio et al., 2014). Oryza glumipatula responds to deep water conditions and was shown to possess SK2 and SK2-like genes, but not SK1 (Hattori et al., 2009), indicating the importance of SK2 in prolonged submergence response. A different accession of O. glumipatula possessing both SK1 and SK2 homolog sequences was used by Sasayama et al. to investigate submergence response induced by hypoxia. The authors found that hypoxia-induced internodal elongation in this accession occurred even when ethylene biosynthesis was inhibited, and suggested hypoxia as the first signal of elongation growth in this species (Sasayama et al., 2018). The submergence tolerance through a quiescent response is associated with the SUB1 locus (Xu and Mackill, 1996), which is conserved on chromosome 9 in many Oryza species including the AA, BB, and FF genome types (Dos Santos et al., 2017). Xu et al. (2006) sequenced this region in the tolerant cultivar FR13A and found three ERF-encoding genes, namely SUB1A, SUB1B, and SUB1C. Overexpression of the allele SUB1A-1 in flooding-susceptible O. sativa varieties showed that this allele is sufficient to induce a flooding survival response. Accessions carrying the SUB1A-2 allele are sensitive to this stress (Xu et al., 2006). Although O. grandiglumis survives complete submergence, Okishio and colleagues could not find homologs of the SUB1A gene in O. grandiglumis (Okishio et al., 2014), and found that submergence tolerance response in this species is not regulated by ethylene (Okishio et al., 2015). Interestingly, SUB1B- and SUB1C-like genes were found in O. glumipatula, whereas a SUB1A-like gene sequence was not detected (Dos Santos et al., 2017).
Prusty et al. (2018) evaluated salt stress tolerance in wild Oryza species, comparing them with cultivated salt-tolerant and sensitive lines. The evaluation was based on many factors, including a visual salt injury score, number of days of seedling survival, shoot length and biomass, chlorophyll content, leaf and root ion content, and level of expression of salt tolerance genes (i.e. OsHKT1;4, OsHKT1;5, and OsSOS1) under stress conditions. All CCDD genomes exhibited salt stress tolerance properties, with O. alta and O. latifolia showing similar levels of tolerance to O. coarctata (a well-known halophyte in the genus), followed by O. grandiglumis and other wild species (Prusty et al., 2018).
Biodiversity conservation
One of the main foci of IOMAP is the identification of populations of wild Oryza species for diversity analysis and conservation efforts (Jacquemin et al., 2013). Rice crop improvement via crop wild relatives (CWRs) is contingent on the effective availability and conservation of these genetic resources (Thomas et al., 2017). Conservation of1 diversity for CWRs can be achieved through: (i) ex situ conservation strategies (i.e. conservation in seed banks, herbaria collections, or artificial environments reproducing conditions of the original environment) and (ii) in situ conservation strategies (i.e. conservation of CWRs within their original species range) (Vincent et al., 2019).
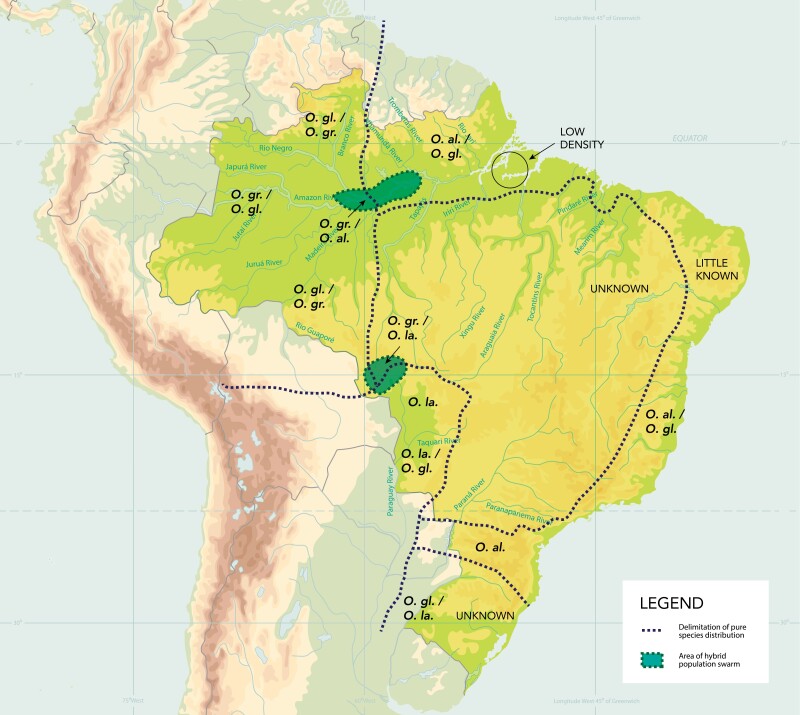
Germplasm collections of wild and cultivated rice species started in the 1950s. During the Green Revolution in India and the Philippines in the late 1960s, crop improvement programs boosted the collections of landrace rice germplasm by the international scientific community (Vaughan et al., 2003). Since the 1960s, the International Rice Research Institute (IRRI) in the Philippines has carried out programs for the genetic improvement of rice, has maintained an international collection of rice genetic resources (IRGC) for germplasm preservation, and has provided seeds freely for scientific research purposes (Evenson and Douglas, 1997). The Germplasm Resource Information Network (GRIN) database indicates that IRRI currently maintains seed resources for 13 accessions of O. alta, 52 of O. glumipatula, 9 of O. grandiglumis, and 77 of O. latifolia (https://gringlobal.irri.org/gringlobal/search). A program for botanical exploration and collection of wild crop relatives, rice included, was initiated in Brazil after the establishment of the Brazilian Agricultural Research Corporation (Embrapa) in 1973. CWRs of rice have been conserved in the Embrapa’s Rice Genebank since 1992, with 177 accessions of O. alta, O. glumipatula, O. grandiglumis, and O. latifolia present in 2015 (Medeiros et al., 2021). In 2002, Morishima collected the reports made by the Japanese National Institute of Genetics between 1957 and 1997, describing field trips and observations of wild and cultivated rice species around the world, including the Americas (Fig. 2). In the preface of his book, he claims that habitats and populations of rice species described therein changed or were lost since the time of the initial expeditions (Morishima, 2002).
Fig. 2.
The map depicts the geographical distribution of the wild Oryza species in Brazil as described by Giancarlo C.X. Oliveira after his field trips to the Amazon in 1988, 1992, and 1993 and a survey of Brazilian herbaria. O. gl., O. glumipatula; O. gr., O. grandiglumis; O. al, O. alta; O. la, O. latifolia. Limits of species distribution are based on Oliveira GCX, Morishima H, Martins PS. 1994. Investigations of plant genetic resources in the Amazon basin with the emphasis on the genus Oryza: Report of 1992/93 Amazon Project—funded by FAPESP (São Paulo State Foundation for the Support of Research) and the Ministry of Education of Japan, with permission.
The characterization of ecological and genetic aspects of a species is necessary to efficiently prioritize populations for conservation. An extended definition of conservation biology includes the application of genomics to conservation (Allendorf et al., 2010). Through increasingly accessible genomics technologies, the genotyping of hundreds of thousands of loci in large sample sizes improves the estimation of the parameters widely used in conservation genetics, such as population diversity, structure, effective size, and demographic history (i.e. the variation of population size through time). This information is crucial to define units of conservation for diversity protection and management plans (Allendorf et al., 2010).
Medeiros et al. (2021) described the conservation status of four native species related to crops in Brazil, which included rice. They used 130 germplasm accessions of the four wild Oryza species conserved ex situ and 112 herbarium records to evaluate the status of ex situ conservation, and collected 47 new accessions in five sites in the Pantanal and the Amazon to evaluate the status of in situ conservation. This study was carried out within a global initiative supported by the Government of Norway, and managed by the Global Crop Diversity Trust and the Millennium Seed Bank at the RBG Kew, whose goal is the establishment of national programs for the collection and ex situ preservation of prioritized germplasm. The authors estimated gaps of representativeness of the four Oryza species in ex situ collections, and highlighted the need to develop complementary in situ conservation strategies: one proposed action is the expansion and consolidation of protected areas in the Cerrado region, which currently show a narrow geographical range and are under threat of high anthropogenic impact (Vincent et al., 2019; Medeiros et al., 2021).
Anthropogenic activities such as conversion of land to agriculture, and, in general, human intervention in natural patterns and processes are associated with threats to population viability and biodiversity conservation (e.g. changes in the environment, disturbance of the interaction with other species, and genetic threats). These threats can lead to different outcomes, such as habitat fragmentation, or gene flow from non-indigenous or crop species (Fig. 1). Habitat fragmentation implies the shattering of a continuous habitat with a progressive isolation of populations, and generally leads to a decrease in population size and decrease in migrations among populations. This results in reduced genetic diversity and lower adaptive capability and overall fitness. Between-species hybridization can lead to introgression and reduced genetic integrity of the native species, especially in small populations which tend to behave as sinks (Oostermeijer, 2003). Using AFLPs, Fuchs et al. found indirect evidence that O. glumipatula individuals were admixed with cultivated O. sativa varieties. The risk of losing local genetic diversity via introgression in the largest population of O. glumipatula in Costa Rica led the authors to highlight the importance of developing in situ and ex situ conservation strategies for this rice wild relative (Fuchs et al., 2016). The effect of climate change on ecosystems can alter the geographic connectivity of species and populations, and affect the amount of gene flow among them (Huang and Schaal, 2012). Thomas et al. (2017) assessed the genetic diversity, population structure, and natural distribution range of the wild relatives of rice in Colombia. They estimated the spatiotemporal overlap between geographic areas suitable for the growth of wild and cultivated rice under current and predicted future climate conditions, and assessed the potential scale of gene flow from the latter to the former. They found that all the wild relatives, except O. latifolia, are predicted to experience range expansion under climate change. On the other hand, most habitats suitable for the four wild rice species are located outside protected areas in Colombia, hampering an effective in situ conservation program in areas with high risk of alien introgression from cultivated rice. The authors suggested that effective strategies for ex situ and in situ conservation in Colombia should consider the prioritization of populations showing the highest level of genetic diversity, complementarity in terms of genetic differentiation (i.e. population units), and adaptive traits (Thomas et al., 2017).
The neodomestication of Oryza alta
Neodomestication has revolutionized the concept of crop improvement by shifting the focus of incremental gains in domesticated species to giant leaps forward in CWRs. The core of the technique resides in the editing of domestication-related genes in a CWR to obtain novel genotypes with the desired agronomic characteristics while keeping the natural genetic diversity and adaptive traits of the CWR (Zhu and Zhu, 2021). Recently, a successful neodomestication of a wild rice tetraploid was accomplished in O. alta (Yu et al., 2021). Yu and colleagues identified gene homologs of domestication-related genes in O. alta that regulate important domestication (e.g. seed shattering, awn length, hull and pericarp color, panicle shape, and grain width), and agronomic traits in rice cultivars (e.g. genes regulating yield, grain quality, efficient use of nutrients, and resistance to biotic and abiotic stresses) as targets for gene editing. The essential tools required to accomplish this task were: (i) a high-quality annotated reference genome to enable the identification of target genes, and (ii) a genotype that is suitable for regeneration and genome editing. Yu et al. developed a protocol for tissue culture and genetic transformation of O. alta, and implemented a genome editing system that uses CRISPR/Cas9. Subsequently, Yu et al. improved yield in O. alta by inducing a mutation in an ortholog of O. sativa GS3, a gene that regulates grain length in diploid rice. Spontaneous seed shattering was inhibited by targeting a QTL associated with abscission layer formation (i.e. qSH1), an essential and ancient trait found in almost all domesticated cereals. The Green Revolution gene SD1, which controls plant height with shorter culms and consequential resistance to lodging, was also edited. Combining improved domestication- and agricultural-related traits with natural adaptation to native environments can be obtained at an unprecedented speed compared with traditional techniques involving breeding and introgression. Through their work, Yu and colleagues showed a route for the generation of a new crop in a wild tetraploid Oryza species that opens the door to the exploitation of other wild polyploid genomes to support global food security.
Conclusions
The generation of a vast array of publicly available genomic resources by (I)OMAP starting two decades ago helped the rice scientific community to address complex biological questions on a whole-genome scale, such as the dynamics of the evolution of the genus, the impacts of domestication, the diversity and evolution of gene families useful for crop improvement (Wing et al., 2007; Jacquemin et al., 2013), etc. The main objective of ‘IOMAP: the Americas’ is thus to provide a valuable genomic asset describing the genetic diversity, population structure and demographic history, gene flow, and potential hybridization of populations of wild rice species endemic to the Americas. Putative candidate regions for adaptive traits, such as resistance to pathogens and tolerance to abiotic stress, will be identified and explored. The genomic characterization of these species will help the identification of accessions as potential candidates for future neodomestication efforts and the genetic improvement of stress-sensitive rice crops. The effect of climate change is predicted to modify the distribution ranges of the wild relatives of rice in the Americas (Thomas et al., 2017), and to alter inundation patterns in western Amazonian rivers with significant consequences for lowland agriculture, including cultivation of rice (Coomes et al., 2016). The adaptive capacity of the wild Oryza species in the Americas may be exploited to cope with effects of climate change on the environment. For example, neodomesticated submergence-resistant O. glumipatula and O. grandiglumis could be cultivated in their native aquatic and flooded habitats in the Amazon basin to replace non-adapted rice cultivars; neodomesticated salt-tolerant O. latifolia and O. alta could be cultivated in areas where the rising sea level and reduced freshwater flow from upper catchments is causing severe saltwater intrusion, soil salinity increase, and damage to rice crop production, similar to that reported for the Mekong Delta River in Vietnam and the Satkhira River in Bangladesh (Radanielson et al., 2018; Paik et al., 2020). The generation of a vast diversity panel spanning a large geographic area and a temporal range of decades will inform the potential erosion of genetic variation in the populations/species due to climate change, human activity, and hybridization events, focusing conservation efforts where they are needed most, and facilitating future efforts of comparative evolutionary genomics studies. Eventually, we trust that the knowledge of the genetic diversity and health status of the wild Oryza species endemic to the Americas will help identify the most appropriate accessions to include in ex situ core collections and the development of guidelines for conservation of biodiversity in situ that are sustainable. Rather than establishing protected reserves that preserve species’ presence only, future research on the wild relatives of rice should be applied to establish reserves based on genetic diversity, as proposed by Vincent et al. (2019), with suitable management plans aimed at the maximization and maintenance (or even enhancement) of the genetic diversity of priority rice specimens.
Acknowledgements
We thank João D. Santos for his contribution in the writing of the grant proposal for the project ‘IOMAP: the Americas’, Giancarlo Conde Xavier Oliveira for the discussion on his investigation of the rice genetic resources in the Amazon basin, and Robyn Palescandolo for designing the figures.
Glossary
Abbreviations
- BB
bacterial blight
- BPH
brown planthopper
- CWR
crop wild relative
- IOMAP
International Oryza Map Alignment Project
- IRGSP
International Rice Genome Sequencing Project
- LTR-RT
long terminal repeat retrotransposable element
- MY
million years.
Contributor Information
Aseel Alsantely, Center for Desert Agriculture, Biological and Environmental Sciences & Engineering Division (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia.
Rafal Gutaker, Royal Botanic Gardens, Kew, Kew Green, Richmond, Surrey TW9 3AE, UK.
María E Navarrete Rodríguez, Center for Desert Agriculture, Biological and Environmental Sciences & Engineering Division (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia.
Griselda Arrieta-Espinoza, Centro de Investigación en Biología Celular y Molecular, Universidad de Costa Rica, Ciudad de la Investigación-C.P., San José 11501-2050, Costa Rica.
Eric J Fuchs, Escuela de Biología, Universidad de Costa Rica, San José 11501-2060, Costa Rica.
Antonio Costa de Oliveira, Plant Genomics and Breeding Center, Eliseu Maciel School of Agronomy, Federal University of Pelotas, Pelotas-RS, Brazil.
Joe Tohme, International Center for Tropical Agriculture (CIAT), Cali 763537, Colombia.
Andrea Zuccolo, Center for Desert Agriculture, Biological and Environmental Sciences & Engineering Division (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia; Crop Science Research Center, Sant’Anna School of Advanced Studies, Pisa 56127, Italy.
Rod A Wing, Center for Desert Agriculture, Biological and Environmental Sciences & Engineering Division (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia; Arizona Genomics Institute, School of Plant Sciences, University of Arizona, Tucson, AZ 85721, USA.
Alice Fornasiero, Center for Desert Agriculture, Biological and Environmental Sciences & Engineering Division (BESE), King Abdullah University of Science and Technology (KAUST), Thuwal 23955-6900, Saudi Arabia.
Fabrizio Costa, University of Trento, Italy.
Author contributions
AA and AF: writing—original draft preparation; RG, GA-E, EJF, ACDO, JT, AZ, RAW, and AF: writing—review and editing; MENR and AF: visualization; RG, RAW, and AF: conceptualization; RAW and AF: supervision.
Conflict of interest
No conflict of interest declared.
Funding
This work was supported by King Abdullah University of Science and Technology (KAUST) under Award No. ORA-CRG10-2021-4734.
References
- Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS.. 1999. Phylogenetic relationships among Oryza species revealed by AFLP markers. Theoretical and Applied Genetics 98, 1320–1328. [Google Scholar]
- Allendorf FW, Hohenlohe PA, Luikart G.. 2010. Genomics and the future of conservation genetics. Nature Reviews. Genetics 11, 697–709. [DOI] [PubMed] [Google Scholar]
- Ammiraju JSS, Fan C, Yu Y, et al. 2010b. Spatio-temporal patterns of genome evolution in allotetraploid species of the genus Oryza. The Plant Journal 63, 430–442. [DOI] [PubMed] [Google Scholar]
- Ammiraju JSS, Lu F, Sanyal A, et al. 2008. Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. The Plant Cell 20, 3191–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammiraju JSS, Song X, Luo M, et al. 2010a. The Oryza BAC resource: a genus-wide and genome scale tool for exploring rice genome evolution and leveraging useful genetic diversity from wild relatives. Breeding Science 60, 536–543. [Google Scholar]
- Ammiraju JSS, Zuccolo A, Yu Y, et al. 2007. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. The Plant Journal 52, 342–351. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D.. 2010. Submergence tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice 3, 138–147. [Google Scholar]
- Bao Y, Ge S.. 2004. Origin and phylogeny of Oryza species with the CD genome based on multiple-gene sequence data. Plant Systematics and Evolution 249, 55–66. [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW.. 2013. GenBank. Nucleic Acids Research 41, D36–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari H. 2019. Global rice production, consumption and trade: trends and future directions. In: Proceedings of the Korean Society of Crop Science Conference, 5-5.
- Bocco R, Lorieux M, Seck PA, Futakuchi K, Manneh B, Baimey H, Ndjiondjop MN.. 2012. Agro-morphological characterization of a population of introgression lines derived from crosses between IR 64 (Oryza sativa indica) and TOG 5681 (Oryza glaberrima) for drought tolerance. Plant Science 183, 65–76. [DOI] [PubMed] [Google Scholar]
- Brar DS, Khush GS.. 2018. Wild relatives of rice: a valuable genetic resource for genomics and breeding research. In: Mondal TK, Henry RJ, eds. Compendium of plant genomes. The wild Oryza genomes. Cham: Springer International Publishing, 1–25. [Google Scholar]
- Brar DS, Virk PS, Jena KK, Khush GS.. 2009. Breeding for resistance to planthoppers in rice. In: Heong KL, Hardy B, eds. Planthopper: new threats to the sustainability of intensive rice production systems in Asia. Manilla: International Rice Research Institute, 401–427 [Google Scholar]
- Cabauatan PQ, Cabunagan RC, Choi IR.. 2009. Rice viruses transmitted by the brown planthopper Nilaparvata lugens Stål .In: Heong KL, Hardy B, eds. Planthopper: new threats to the sustainability of intensive rice production systems in Asia. Manila: International Rice Research Institute, 357–368. [Google Scholar]
- Chen J, Huang Q, Gao D, et al. 2013. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nature Communications 4, 1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coomes OT, Lapointe M, Templeton M, List G.. 2016. Amazon River flow regime and flood recessional agriculture: flood stage reversals and risk of annual crop loss. Journal of Hydrology 539, 214–222. [Google Scholar]
- Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L.. 2017. Past and future use of wild relatives in crop breeding. Crop Science 57, 1070–1082. [Google Scholar]
- Devi SJSR, Singh K, Umakanth B, et al. 2020. Identification and characterization of a large effect QTL from Oryza glumaepatula revealed Pi68(t) as putative candidate gene for rice blast resistance. Rice 13, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos RS, Farias D da R, Pegoraro C, Rombaldi CV, Fukao T, Wing RA, de Oliveira AC.. 2017. Evolutionary analysis of the SUB1 locus across the Oryza genomes. Rice 10, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E.. 2014. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346, 1258096. [DOI] [PubMed] [Google Scholar]
- Evenson RE, Gollin D.. 1997. Genetic resources, international organizations, and improvement in rice varieties. Economic Development and Cultural Change 45, 471–500. [Google Scholar]
- Fornasiero A, Wing RA, Ronald P.. 2022. Rice domestication. Current Biology 32, R20–R24. [DOI] [PubMed] [Google Scholar]
- Fuchs EJ, Martínez AM, Calvo A, Muñoz M, Arrieta-Espinoza G.. 2016. Genetic diversity in Oryza glumaepatula wild rice populations in Costa Rica and possible gene flow from O. sativa. PeerJ 4, e1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray H, Charles J, Beddington John R, et al. 2010. Food security: the challenge of feeding 9 billion people. Science 327, 812–818. [DOI] [PubMed] [Google Scholar]
- Goodman BA. 2020. Utilization of waste straw and husks from rice production: a review. Journal of Bioresources and Bioproducts 5, 143–162. [Google Scholar]
- Gumi AM, Aliero AA.. 2018. Oryza grandiglumis (Doell) Prod. In: Mondal TK, Henry RJ, eds. Compendium of plant genomes. The wild Oryza genomes. Cham: Springer International Publishing, 137–143. [Google Scholar]
- Gutaker RM, Chater CCC, Brinton J, Castillo-Lorenzo E, Breman E, Pironon S.. 2022. Scaling up neodomestication for climate-ready crops. Current Opinion in Plant Biology 66, 102169. [DOI] [PubMed] [Google Scholar]
- Gutiérrez AG, Carabalí SJ, Giraldo OX, Martínez CP, Correa F, Prado G, Tohme J, Lorieux M.. 2010. Identification of a Rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines. BMC Plant Biology 10, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR, de Wet JMJ.. 1971. Toward a rational classification of cultivated plants. Taxon 20, 509–517. [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, et al. 2009. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. [DOI] [PubMed] [Google Scholar]
- Hirsch CD, Wu Y, Yan H, Jiang J.. 2009. Lineage-specific adaptive evolution of the centromeric protein CENH3 in diploid and allotetraploid Oryza species. Molecular Biology and Evolution 26, 2877–2885. [DOI] [PubMed] [Google Scholar]
- Hu J, Xiao C, He Y.. 2016. Recent progress on the genetics and molecular breeding of brown planthopper resistance in rice. Rice 9, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Schaal BA.. 2012. Association between the geographic distribution during the last glacial maximum of Asian wild rice, Oryza rufipogon (Poaceae), and its current genetic variation. American Journal of Botany 99, 1866–1874. [DOI] [PubMed] [Google Scholar]
- Hurwitz BL, Kudrna D, Yu Y, Sebastian A, Zuccolo A, Jackson SA, Ware D, Wing RA, Stein L.. 2010. Rice structural variation: a comparative analysis of structural variation between rice and three of its closest relatives in the genus Oryza. The Plant Journal 63, 990–1003. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project, Sasaki T.2005. The map-based sequence of the rice genome. Nature 436, 793–800. [DOI] [PubMed] [Google Scholar]
- Jacquemin J, Bhatia D, Singh K, Wing RA.. 2013. The International Oryza Map Alignment Project: development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Current Opinion in Plant Biology 16, 147–156. [DOI] [PubMed] [Google Scholar]
- Jiang N, Yan J, Liang Y, Shi Y, He Z, Wu Y, Zeng Q, Liu X, Peng J.. 2020. Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)—an updated review. Rice 13, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E.. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J.. 2013. RNA-programmed genome editing in human cells. eLife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano AB, Naredo MEB, Jackson MT.. 1998. Taxonomic status of Oryza glumaepatula Steud. I. Comparative morphological studies of New World diploids and Asian AA genome species. Genetic Resources and Crop Evolution 45, 197–203. [Google Scholar]
- Karasawa MMG, Vencovsky R, Silva CM, Zucchi MI, Oliveira GCX, Veasey EA.. 2007. Mating system of Brazilian Oryza glumaepatula populations studied with microsatellite markers. Annals of Botany 99, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C-B, Zhang D-M, Ge S, Lu B-R, Hong D-Y.. 2001. Differentiation and inter-genomic relationships among C, E and D genomes in the Oryza officinalis complex (Poaceae) as revealed by multicolor genomic in situ hybridization. Theoretical and Applied Genetics 103, 197–203. [Google Scholar]
- Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, Dong W, Gao C, Xu C.. 2018. Domestication of wild tomato is accelerated by genome editing. Nature Biotechnology 36, 1160–1163. [DOI] [PubMed] [Google Scholar]
- Li W, Chern M, Yin J, Wang J, Chen X.. 2019. Recent advances in broad-spectrum resistance to the rice blast disease. Current Opinion in Plant Biology 50, 114–120. [DOI] [PubMed] [Google Scholar]
- Lu F, Ammiraju JSS, Sanyal A, et al. 2009. Comparative sequence analysis of MONOCULM1-orthologous regions in 14 Oryza genomes. Proceedings of the National Academy of Sciences, USA 106, 2071–2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean J, Hardy B, Hettel G.. 2013. Rice Almanac: source book for one of the most economic activities on Earth. Manila: International Rice Research Institute. [Google Scholar]
- Mattos VS, Leite RR, Cares JE, Gomes ACMM, Moita AW, Lobo VLS, Carneiro RMDG.. 2019. Oryza glumaepatula, a new source of resistance to Meloidogyne graminicola and histological characterization of its defense mechanisms. Phytopathology 109, 1941–1948. [DOI] [PubMed] [Google Scholar]
- Medeiros MB, Valls JFM, Abreu AG, Heiden G, Ribeiro-Silva S, José SCBR, Santos IRI, Passos AMA, Burle ML.. 2021. Status of the ex situ and in situ conservation of Brazilian crop wild relatives of rice, potato, sweet potato, and finger millet: filling the gaps of germplasm collections. Agronomy 11, 638. [Google Scholar]
- Molina J, Sikora M, Garud N, et al. 2011. Molecular evidence for a single evolutionary origin of domesticated rice. Proceedings of the National Academy of Sciences, USA 108, 8351–8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishima H. 2002. Reports of the study-tours for investigation of wild and cultivated rice species. Conducted by the staff of National Institute of Genetics, Japan and coworkers during 1957–1997. Part I
- Multani DS, Khush GS, Delos Reyes BG, Brar DS.. 2003. Alien genes introgression and development of monosomic alien addition lines from Oryza latifolia Desv. to rice, Oryza sativa L. Theoretical and Applied Genetics 107, 395–405. [DOI] [PubMed] [Google Scholar]
- Nooryazdan H, Serieys H, David J, Bacilieri R, Bervillé AJ.. 2011. Construction of a crop–wild hybrid population for broadening genetic diversity in cultivated sunflower and first evaluation of its combining ability: the concept of neodomestication. Euphytica 178, 159–175. [Google Scholar]
- Okishio T, Sasayama D, Hirano T, Akimoto M, Itoh K, Azuma T.. 2014. Growth promotion and inhibition of the Amazonian wild rice species Oryza grandiglumis to survive flooding. Planta 240, 459–469. [DOI] [PubMed] [Google Scholar]
- Okishio T, Sasayama D, Hirano T, Akimoto M, Itoh K, Azuma T.. 2015. Ethylene is not involved in adaptive responses to flooding in the Amazonian wild rice species Oryza grandiglumis. Journal of Plant Physiology 174, 49–54. [DOI] [PubMed] [Google Scholar]
- Oliveira GCX, Morishima H, Martins PS.. 1994. Investigations of plant genetic resources in the Amazon basin with the emphasis on the genus Oryza: report of 1992/93 Amazon Project. Monbusho International Scientific Research Program/FAPESP, 10–15. [Google Scholar]
- Oostermeijer JGB. 2003. Threats to rare plant persistence. In: Brigham CA, Schwartz MW, eds. Ecological Studies. Population viability in plants: conservation, management, and modeling of rare plants. Berlin, Heidelberg: Springer, 17–58. [Google Scholar]
- Paik S, Le DTP, Nhu LT, Mills BF.. 2020. Salt-tolerant rice variety adoption in the Mekong River Delta: farmer adaptation to sea-level rise. PLoS One 15, e0229464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, et al. 2006. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research 16, 1262–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty MR, Kim S-R, Vinarao R, Entila F, Egdane J, Diaz MGQ, Jena KK.. 2018. Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Frontiers in Plant Science 9, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Guo J, Jing S, Zhu L, He G.. 2012. Development and characterization of japonica rice lines carrying the brown planthopper-resistance genes BPH12 and BPH6. Theoretical and Applied Genetics 124, 485–494. [DOI] [PubMed] [Google Scholar]
- Radanielson AM, Gaydon DS, Rahman Khan MM, Chaki AK, Rahman MA, Angeles O, Li T, Ismail A.. 2018. Varietal improvement options for higher rice productivity in salt affected areas using crop modelling. Field Crops Research 229, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram T, Deen R, Gautam SK, Kandu R, Rao YK, Brar DS.. 2010. Identification of new genes for brown planthopper resistance in rice introgressed from O. glaberrima and O. minuta. Rice Genetics Newsletter 25, 67–69. [Google Scholar]
- Ray S, Vijayan J, Chakraborti M, Sarkar S, Bose LK, Singh ON.. 2018. Oryza officinalis complex. In: Mondal TK, Henry RJ, eds. Compendium of plant genomes. The wild Oryza Genomes. Cham: Springer International Publishing, 239–258. [Google Scholar]
- Roulin A, Piegu B, Wing RA, Panaud O.. 2008. Evidence of multiple horizontal transfers of the long terminal repeat retrotransposon RIRE1 within the genus Oryza. The Plant Journal 53, 950–959. [DOI] [PubMed] [Google Scholar]
- Sanchez, PL, Wing, RA, Brar, DS.. 2013. The wild relative of rice: genomes and genomics. In: Zhang Q, Wing R, eds. Genetics and genomics of rice. Plant genetics and genomics: crops and models, vol 5. New York: Springer, 9–25. [Google Scholar]
- Sasayama D, Okishio T, Hirano T, Fukayama H, Hatanaka T, Akimoto M, Azuma T.. 2018. Internodal elongation under submergence in the Amazonian wild rice species Oryza glumaepatula: the growth response is induced by hypoxia but not by ethylene. Plant Growth Regulation 85, 123–132. [Google Scholar]
- Shenton M, Kobayashi M, Terashima S, et al. 2020. Evolution and diversity of the wild rice Oryza officinalis complex, across continents, genome types, and ploidy levels. Genome Biology and Evolution 12, 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W-Y, Wang G-L, Chen L-L, et al. 1995. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Song Z, Li B, Chen J, Lu B-R.. 2005. Genetic diversity and conservation of common wild rice (Oryza rufipogon) in China. Plant Species Biology 20, 83–92. [Google Scholar]
- Stein JC, Yu Y, Copetti D, et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nature Genetics 50, 285–296. [DOI] [PubMed] [Google Scholar]
- Tateoka T. 1963. Taxonomic studies of Oryza III. Key to the species and their enumeration. Botanical Magazine Tokyo 76, 165–173. [Google Scholar]
- Thomas E, Tovar E, Villafañe C, Bocanegra JL, Moreno R.. 2017. Distribution, genetic diversity and potential spatiotemporal scale of alien gene flow in crop wild relatives of rice (Oryza spp.) in Colombia. Rice 10, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan DA. 1989. Genus Oryza L. current status of taxonomy. IRRI Research Paper Series 138, 1–21. [Google Scholar]
- Vaughan DA. 1994. The wild relatives of rice: a genetic resources handbook. Manila: International Rice Research Institute. [Google Scholar]
- Vaughan DA, Morishima H, Kadowaki K.. 2003. Diversity in the Oryza genus. Current Opinion in Plant Biology 6, 139–146. [DOI] [PubMed] [Google Scholar]
- Vincent H, Amri A, Castañeda Álvarez NP, Dempewolf H, Dulloo E, Guarino L, Hole D, Mba C, Toledo A, Maxted N.. 2019. Modeling of crop wild relative species identifies areas globally for in situ conservation. Communications Biology 2, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ.. 2015. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Scientific Reports 5, 13957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yu Y, Haberer G, et al. 2014. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nature Genetics 46, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Second G, Tanksley SD.. 1992. Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theoretical and Applied Genetics 83, 565–581. [DOI] [PubMed] [Google Scholar]
- Wing RA. 2015. Harvesting rice’s dispensable genome. Genome Biology 16, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing RA, Kim HR, Goicoechea JL, et al. 2007. The Oryza Map Alignment Project (OMAP): a new resource for comparative genomics studies within Oryza. Rice functional genomics: challenges, progress and prospects. New York: Springer, 395–409. [Google Scholar]
- Xu K, Mackill DJ.. 1996. A major locus for submergence tolerance mapped on rice chromosome 9. Molecular Breeding 2, 219–224. [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ.. 2006. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708. [DOI] [PubMed] [Google Scholar]
- Yu H, Lin T, Meng X, et al. 2021. A route to de novo domestication of wild allotetraploid rice. Cell 184, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92. [DOI] [PubMed] [Google Scholar]
- Zamora A, Barboza C, Lobo J, Espinoza AM.. 2003. Diversity of native rice (Oryza Poaceae) species of Costa Rica. Genetic Resources and Crop Evolution 50, 855–870. [Google Scholar]
- Zhang Y, Zhang S, Liu H, et al. 2015a. Genome and comparative transcriptomics of African wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility. Molecular Plant 8, 1683–1686. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou J, Yang Y, Li J, Xu P, Deng X, Deng W, Wu Z, Tao D.. 2015b. A novel gene responsible for erect panicle from Oryza glumaepatula. Euphytica 205, 739–745. [Google Scholar]
- Zhu X-G, Zhu J-K.. 2021. Precision genome editing heralds rapid de novo domestication for new crops. Cell 184, 1133–1134. [DOI] [PubMed] [Google Scholar]
- Zsögön A, Cermak T, Voytas D, Peres LE.. 2017. Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: case study in tomato. Plant Science 256, 120–130. [DOI] [PubMed] [Google Scholar]