Abstract
In primary Sjögren syndrome (pSS), chronic inflammation of exocrine glands results in tissue destruction and sicca symptoms, primarily of the mouth and eyes. Fatigue, arthralgia and myalgia are also common symptoms, whereas extraglandular manifestations that involve the respiratory, nervous and vascular systems occur in a subset of patients. The disease predominantly affects women, with an estimated female to male ratio of 14 to 1. The aetiology of pSS, however, remains incompletely understood, and effective treatment is lacking. Large-scale genetic and epigenetic investigations have revealed associations between pSS and genes in both innate and adaptive immune pathways. The genetic variants mediate context-dependent effects, and both sex and environmental factors can influence the outcome. As such, genetic and epigenetic studies can provide insight into the dysregulated molecular mechanisms, which in turn might reveal new therapeutic possibilities. This Review discusses the genetic and epigenetic features that have been robustly connected with pSS, putting them into the context of cellular function, carrier sex and environmental challenges. In all, the observations point to several novel opportunities for early detection, treatment development and the pathway towards personalized medicine.
Subject terms: Sjögren's disease, Disease genetics, Rheumatic diseases
This Review summarizes the genetic and epigenetic basis of primary Sjögren syndrome, including genetic interactions with factors such as sex and environment. Understanding these processes provides insight into the molecular basis of this disease and might reveal new treatment targets.
Key points
Advances in the past few years provide new insight into the genetic and epigenetic basis of primary Sjögren syndrome (pSS), and how environmental factors and carrier sex might interact with risk-associated loci.
Immunomodulatory treatments that affect disease progression and activity have not been identified for pSS and remain unmet clinical needs.
Data from genetic and epigenetic studies point to Toll-like receptor and interferon signalling, lymphocyte regulation, antigen presentation and target tissue maintenance as the most relevant processes for therapeutic targeting.
Of the implicated pathogenic processes, Toll-like receptor and interferon signalling, as well as lymphocyte regulation, contain targets of various treatments in development or in clinical trials.
Novel treatment approaches for pSS could include modification of epigenetic signatures, interference with antigen presentation pathways and antigen-specific immunotherapy.
Introduction
Primary Sjögren syndrome (pSS) is a common autoimmune disease that predominantly affects women1. The disease is characterized by inflammation and tissue destruction of the salivary and lacrimal glands, resulting in oral and ocular dryness (denoted as sicca symptoms). Fatigue and arthralgia affect most patients, but presentation of the condition is heterogeneous, and extraglandular manifestations such as synovitis, cutaneous vasculitis, polyneuropathy, interstitial lung disease and tubulointerstitial nephritis develop in 30–40% of patients2,3 (Fig. 1). For a diagnosis of pSS, current internationally accepted classification criteria require objective measures of decreased production of tears or saliva, as well as immunological aberrances verified either through detection of anti-SSA/Ro autoantibodies, or through histological findings of focal lymphocytic sialadenitis in labial salivary glands4. Patient-reported symptom onset often precedes diagnosis by many years, which might relate to an insidious onset of pSS and the fact that sicca symptoms are common in the general population5. Therefore, facilitating early diagnosis and identification of disease-modifying treatments that can rescue organ function are central objectives in current research on pSS.
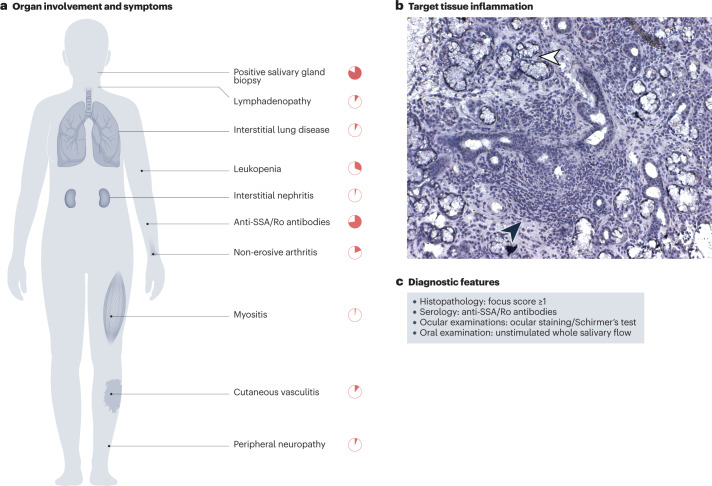
Fig. 1. Clinical features of Sjögren syndrome.
The clinical presentation of primary Sjögren syndrome (pSS) is heterogeneous, and patients can present with various symptoms and organ involvement. a, Lymphocyte infiltration and tissue destruction of the salivary glands, as well as the presence of anti-SSA/Ro autoantibodies are hallmarks of the disease, but other features such as leukopenia and non-erosive arthritis are relatively common. The frequency of the best-described organ manifestations in patients with pSS are represented3,32. b, Lymphocytic infiltrate in a haematoxylin-stained minor salivary gland biopsy section analysed as part of the diagnostic procedure for Sjögren syndrome. The infiltrate (filled black arrowhead) surrounds dilated secretory ducts and penetrates into glandular tissue. Some remaining acini are indicated by a white arrowhead. c, Diagnostic features required for classification as Sjögren syndrome include histological findings of at least one focus of lymphocytic sialadenitis in a 4 mm2 labial salivary gland biopsy (focus score >1), the presence of anti-SSA/Ro autoantibodies and objective measures of decreased production of tears (ocular staining score or Schirmer’s test) or saliva (unstimulated whole salivary flow rate).
Comorbidities are important contributors to morbidity and mortality in rheumatic disease, and in pSS include an increased risk of cardiovascular disease, especially in patients with autoantibodies6,7, and a much-increased incidence of lymphoma, with a lifetime risk of around 5%8,9. Moreover, in pregnant women, anti-SSA/Ro and anti-SSB/La autoantibodies transfer across the placenta and might cause neonatal lupus with a congenital atrioventricular block in the developing child10. Current treatment recommendations for pSS focus on the management of sicca symptoms and stimulation of residual glandular function with muscarinic agonists, but also suggest the use of various conventional synthetic DMARDs (such as hydroxychloroquine) and biologics (such as rituximab) for the treatment of specific organ manifestations11. Drugs with an effect on disease progression have not been successfully identified12, which relates both to the disease heterogeneity and challenges in adequately assessing disease activity in pSS.
pSS is thought to develop when individuals with genetic susceptibility traits are exposed to disease-triggering environmental risk factors, leading to activation of both innate and adaptive immunity. Important factors in pSS immune pathology include augmented activity of the interferon system and B cell hyperactivity, resulting in hypergammaglobulinaemia, disturbed B cell subpopulation frequencies and the production of autoantibodies13,14. Further, T cells and B cells infiltrate the affected glands, and can form autoantibody-producing ectopic germinal centres15,16. A more precise understanding of the underlying genetics, epigenetics and environmental risk factors is imperative to enable more timely diagnosis and treatment to rescue organ function and improve prognosis.
In this Review, we provide an overview of advances in the understanding of genetic and epigenetic factors in pSS, focusing on well-substantiated observations with genome-wide significance. We discuss how factors such as sex and environment might interact with genetics and influence epigenetic regulation and disease susceptibility and outline how such knowledge points towards specific dysregulated molecular mechanisms, which in turn can instruct the development of highly needed novel treatments.
Genetic studies in pSS
Genetics refers to the study of DNA sequences and variation therein such as single nucleotide polymorphisms (SNPs) or mutations. Most of the genetic factors associated with complex diseases are common single nucleotide variants (often defined as >1% carriers) in non-coding segments of our DNA, posing a challenge for understanding how genetic features translate into pathogenic mechanisms. Furthermore, concordance rates of autoimmune diagnoses in monozygotic twins are typically below 30%17,18, providing evidence that non-genetic factors are also of major importance in these diseases. Such extrinsic and intrinsic factors can, however, interact with genetic features19, and modulation of the epigenome represents one way in which these interactions can translate into phenotypic outcomes. Important approaches for achieving a better understanding of the underlying genetics of complex diseases include case–control association studies and the less commonly performed family linkage studies. Association studies typically either focus on a specific genetic region in candidate gene studies or use an unbiased approach through large-scale or genome-wide association studies (GWAS).
Several lines of evidence confirm the role of genetic predisposition in the development of pSS. For example, compared with the general population, the occurrence of pSS is higher among siblings of patients with pSS, the familial incidence of other autoimmune conditions is also higher20 and a few existing reports indicate increased co-occurrence of pSS in twins21–24, although estimates of twin concordance rates are still lacking. Knowledge on the genetics of pSS is building, but the number of publications from genetic studies remains low compared with other autoimmune diseases (such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA) and multiple sclerosis) (Fig. 2). Studies in pSS have identified associations in the HLA locus as well as in >20 non-HLA loci that reach genome-wide significance (P < 5 × 10−8) (Table 1). Associations that do not reach genome-wide significance might also be of interest to explore, as reviewed elsewhere25,26, although data should be interpreted with care. In this Review, we focus on those associations with genetic regions that reach genome-wide significance.
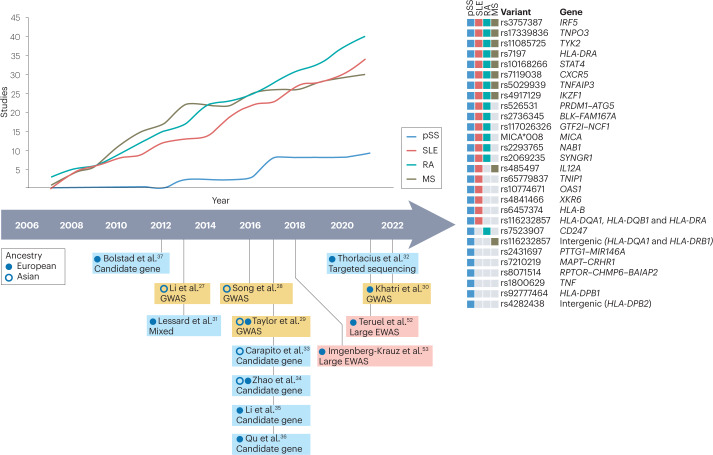
Fig. 2. Studies identifying genetic risk loci of genome-wide significance.
An increasing number of studies on primary Sjögren syndrome (pSS) and selected other autoimmune diseases are registered in the genome-wide association studies (GWAS) catalogue205. However, the number of studies that have identified genetic variants of genome-wide significance in pSS is still low compared with other autoimmune diseases. In the timeline, key papers in pSS that identify novel genetic loci at genome-wide significance are indicated, as well as large-scale epigenome-wide association studies (EWAS) that included ≥100 patients with pSS. Some genetic risk loci identified for pSS are also associated with other autoimmune disease. Many of the non-HLA variants associated with pSS are also associated with systemic lupus erythematosus (SLE), whereas fewer variants overlap with rheumatoid arthritis (RA) or multiple sclerosis (MS). Some variants seem to specifically increase the risk of pSS30,205.
Table 1.
Genetic loci associated with primary Sjögren syndrome
| Variant | Position (Chr:bp) | Gene | Function | Odds ratio | P value | Ref. |
|---|---|---|---|---|---|---|
| rs7523907 | 1:167427247 | CD247 | T cell receptor signalling | 0.85 | 9.33 × 10−9 a | 30 |
| rs10168266 | 2:191071078 | STAT4 | Intracellular signalling | 1.44 | 2 × 10−17 | 27 |
| rs2293765 | 2:191520845 | NAB1 | Transcriptional repression | 1.24 | 5.53 × 10−14 | 30 |
| rs485497 | 3:160001345 | IL12A | The 35-kDa subunit of IL-12 | 1.3 | 1 × 10−10 | 31 |
| rs6579837 | 5:151055333 | TNIP1 | Inhibits NF-κB activation | 1.43 | 3 × 10−8 | 31 |
| rs2431697 | 5:159879978 | PTTG1 | Anaphase-promoting complex substrate | 0.83 | 3.33 × 10−9 | 30 |
| MIR146A | Post-transcriptional regulation of gene expression | |||||
| rs6457374 | 6:31272261 | HLA-B | Presents peptides derived from the endoplasmic reticulum lumen | 3.27 | 3.52 × 10−27 | 33 |
| rs2523607 | 6:31322790 | HLA-B | Presents peptides derived from the endoplasmic reticulum lumen | 5.27 | 5.30 × 10−58 | 32 |
| MICA*008 | 6:31371356 | MICA | Stress-induced ligand for the natural killer cell-activating protein NKG2D | 2.24 | 2.61 × 10−35 | 33 |
| rs1800629 | 6:31543031 | TNF | Multifunctional pro-inflammatory cytokine | 2 | 2.48 × 10−10 | 37 |
| rs7197 | 6:32412580 | HLA-DRA | Presents peptides derived from extracellular proteins | 1.56 | 2.6 × 10−25 | 32 |
| rs116232857 | 6:32597064 | Intergenic (HLA-DQA1 and HLA-DRB1) | Presents peptides derived from extracellular proteins | 2.42 | 1.14 × 10−67 | 31 |
| Presents peptides derived from extracellular proteins | ||||||
| rs115575857 | 6:32659645 | HLA-DQA1 | Presents peptides derived from extracellular proteins | 3.65 | 3.7 × 10−90 | 31 |
| HLA-DQB1 | ||||||
| HLA-DRA | ||||||
| rs9277464 | 6:33053352 | HLA-DPB1 | Presents peptides derived from extracellular proteins | 1.65 | 3 × 10−7 | 29 |
| rs4282438 | 6:33072172 | Intergenic (HLA-DPB2) | Presents peptides derived from extracellular proteins | 1.58 | 8.77 × 10−25 | 27 |
| rs5029939 | 6:137874586 | TNFAIP3 | Inhibits NF-κB activation | 1.67 | 8 × 10−9 | 27 |
| rs526531 | 6:138243700 | PRDM1 | Repressor of IFNβ expression | 1.13 | 4.86 × 10−8 a | 30 |
| ATG5 | Involved in autophagic vesicle formation | |||||
| rs4917129 | 7:50323074 | IKZF1 | Transcription factor that regulates lymphocyte differentiation | 0.7 | 4.24 × 10−8 | 36 |
| rs117026326 | 7:74711703 | GTF2I | Phosphoprotein with roles in transcription and signal transduction | 2.2 | 1 × 10−53 | 27 |
| NCF1 | Subunit of neutrophil enzyme NADPH oxidase | |||||
| rs3757387 | 7:128936032 | IRF5 | Transcription factor with diverse roles | 1.44 | 3 × 10−19 | 31 |
| rs17339836 | 7:129041008 | TNPO3 | Nuclear import receptor | 1.58 | 2 × 10−16 | 31 |
| rs4841466 | 8:10829159 | XKR6 | Unknown | 1.17 | 3.77 × 10−8 | 30 |
| rs2736345 | 8:11494976 | BLK | Tyrosine kinase involved in B cell development | 1.3 | 5 × 10−10 | 31 |
| FAM167A | Unknown | |||||
| rs7119038 | 11:118867572 | CXCR5 | Receptor for the chemokine CXCL13 | 1.35 | 1 × 10−8 | 31 |
| rs10774671 | 12:113356943 | OAS1 | Interferon-induced antiviral enzyme | 0.75 | 2.59 × 10−9 | 35 |
| rs8071514 | 17:10462513 | RPTOR | Controls mTORC1 activity involved in cell growth and survival | 0.84 | 1.64 × 10−8 | 30 |
| CHMP6 | Probably involved in the biosynthesis of endosomes | |||||
| BAIAP2 | Adapter protein that links membrane-bound small G proteins to cytoplasmic effector proteins. | |||||
| rs7210219 | 17:78964083 | MAPT | Promotes microtubule assembly and stability | 0.78 | 2.4 × 10−10 | 30 |
| CRHR1 | A receptor that binds to corticotropin-releasing hormones | |||||
| rs11085725 | 19:39747780 | TYK2 | Involved in interferon and cytokine signalling | 0.78 | 7.17 × 10−13 | 30 |
| rs2069235 | 22:39747530 | SYNGR1 | Unknown | 1.21 | 5.06 × 10−10 | 30 |
bp, base pair; Chr, chromosome; mTORC1, mechanistic target of rapamycin complex 1; NF-κB, nuclear factor-κB. aP value from a meta-analysis combining more than one study or cohort.
The discovery of pSS-associated loci
In pSS, four GWAS27–30, one large-scale study employing the ImmunoChip (a custom SNP array) and other large-scale arrays31, a targeted sequencing study32 and five candidate gene studies33–37 have identified various loci of genome-wide significance in either European populations, Asian populations or both (Fig. 2).
More than a decade ago in 2012, the first association at genome-wide significance was reported in a candidate gene study of the HLA class III locus by Bolstad et al.37. The study identified an association between a SNP in the promoter region of TNF and pSS; however, given the high linkage disequilibrium of the whole HLA region, whether this observation represents an independent signal is unclear. In 2013, Lessard et al.31 studied patients of European ancestry using several different large-scale arrays and reported genome-wide significant associations in six non-HLA loci (IRF5–TNPO3, STAT4, IL12A, FAM167A–BLK, CXCR5 and TNIP1), as well as confirming previously reported HLA associations. Results from a GWAS performed by Li et al.27 in a Han Chinese population were published in the same issue of Nature Genetics. The study established associations between pSS and GTF2IRD1–GTF2I and TNFAIP3 (although the association with TNFAIP3 had already been indicated in a previous candidate gene study38) and confirmed the association of STAT4 and the HLA region with pSS in Han Chinese patients. The GTF2IRD1–GTF2I association was later confirmed by Song et al.28 in a 2016 GWAS of Han Chinese female patients, and further characterized by Zhao et al.34, who identified a missense variant in NCF1 located 62 kb from the associated SNP rs117026326.
In 2017, Taylor et al.29 analysed patients of both European and Asian ancestry, as part of the Sjögren’s International Collaborative Clinical Alliance (SICCA) initiative, and revealed pronounced ancestry-specific differences in genetic associations with pSS, including in the HLA locus, while confirming associations at IRF5 and STAT4. In cohorts of patients from France and the UK, another 2017 study by Carapito et al.33 reported an association between pSS and an allele of the non-conventional MHC-encoded class I gene (MICA). The association was later confirmed as being independent of pSS-associated MHC class II signals32.
In an alternative approach that involved cis-expression quantitative trait locus (eQTL) analyses of type I interferon-inducible transcripts (Supplementary Box 1), Li et al.35 found multiple associations between pSS and a SNP in OAS1 that resulted in shifted splicing of the gene, and in a following candidate gene investigation, a meta-analysis of cohorts of patients of European ancestry identified OAS1 as a susceptibility locus. Also employing a candidate gene approach, Qu et al.36 identified a genome-wide significant association with IKZF1 in a Han Chinese population. Using targeted sequencing, Thorlacius et al.32 confirmed and refined the signal from the HLA locus, revealing three independent signals in patients with pSS and anti-SSA/Ro and/or anti-SSB/La autoantibodies. This study also observed the highest odds ratio so far at 6.1 for a genetic association with anti-SSA/Ro autoantibody- and/or anti-SSB/La autoantibody-positive pSS (Fig. 3).
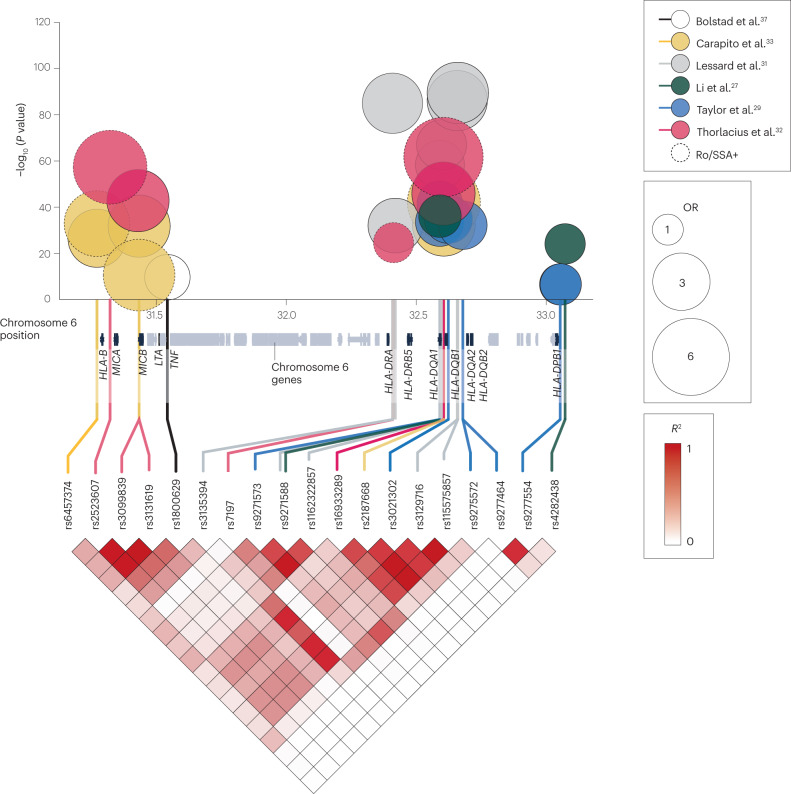
Fig. 3. Associations between variants within the HLA locus and primary Sjögren syndrome.
A large number of variants that are associated with primary Sjögren syndrome (pSS) are located within the HLA locus on chromosome 6. The graph depicts chromosome position (x axis), odds ratio (OR; bubble size) and significance level (y axis) of these disease-associated genetic variants, which are coloured by study. Dotted lines indicate data generated from cohorts that included only patients with pSS who are positive for anti-SSA/Ro and/or anti-SSB/La autoantibodies. Linkage disequilibrium (expressed in terms of the squared correlation (R2)) between the depicted variants in Utah residents of northern and western European ancestry (a population of the 1000 Genomes Project, Code CEU) is indicated in the bottom triangle (data taken from LDlink206). Genes near the associated variants are annotated (using data from the UCSC Genome Browser on Human (GRCh37/hg19)207).
Most recently, a GWAS in patients of European ancestry by Khatri et al.30 identified seven novel loci (NAB1, PTTG1–MIR146A, XKR6, MAPT–CRHR1, RPTOR–CHMP6–BAIAP2, TYK2 and SYNGR1), and a meta-analysis performed with additional ImmunoChip-derived data revealed three additional loci (CD247, PRDM1–ATG5 and TNFAIP3) that are associated with pSS at genome-wide significance. In this study, polygenic risk scores for pSS were calculated for the first time, reaching similar predictive values to those that had previously been indicated for SLE and RA30.
Associations with subphenotypes and comorbidities of pSS
pSS is a heterogeneous condition with several subphenotypes, and it is often accompanied by comorbidities, both features of which are probably dependent on discrete genetic variations. Several genetic associations of genome-wide significance are specific to patients positive for anti-SSA/Ro and/or anti-SSB/La autoantibodies32. This group of patients are more prone to systemic disease with extraglandular manifestations than other patients with pSS32, and although it has not yet been demonstrated at a genome-wide significant level, it is possible that genetic variants associated with anti-SSA/Ro and/or anti-SSB/La autoantibodies associate with discrete systemic engagement or comorbidities in pSS.
Studies of genetic variations in relation to other pSS-related features such as specific symptoms or comorbidities are few, but in 2021, a meta-analysis of patients with pSS from Norwegian and Swedish cohorts identified a genome-wide significant association between variants in the RTP4–MASP1 locus and fatigue in patients with pSS39. No genome-wide significant associations have yet been identified for the pSS-associated comorbidity lymphoma, but several studies imply a role for variants in TNFAIP3 in this comorbidity40–42. This gene, together with other genetic loci implicated in lymphoma development in pSS but for which the reported associations do not reach genome-wide significance, have been reviewed elsewhere43,44.
Epigenetic studies in pSS
Epigenetics refers to processes that influence gene expression without affecting the actual DNA sequence. These processes include histone modifications, non-coding RNA activity and DNA methylation, the last of which is the most thoroughly studied epigenetic mechanism in pSS. The addition of methyl (CH3) groups at cytosine guanine dinucleotide (CpG) sites in gene promoter regions (hypermethylation) generally results in repression of gene expression45. Conversely, hypomethylation of DNA is associated with augmented gene expression levels45.
In keeping with this association, many of the genes identified as hypomethylated or hypermethylated in studies of pSS are differentially expressed at the mRNA level (Table 2).
Table 2.
Top sites identified as differentially methylated in primary Sjögren syndrome
| CpG site | Position (Chr:bp) | Gene | P value | Methylation status (hypermethylated (+) or hypomethylated (−)) | Tissue | Interferon induced?a | Refs. |
|---|---|---|---|---|---|---|---|
| cg07515989 | 1:43250763 | NA | 1.39 × 10−8 | + | Salivary glands | NA | 53 |
| cg03607951 | 1:79085586 | IFI44L | 9.9 × 10−67 | − | Blood and B cells | Yes | 52,53 |
| cg04268125 | 1:154579384 | ADAR | 1.83 × 10−11 | − | Blood | Yes | 52 |
| cg10549986 | 2:7018153 | RSAD2 | 1.4 × 10−17 | − | B cells | Yes | 53 |
| cg09858955 | 2:58135951 | VRK2 | 2.14 × 10−14 | − | Blood | Yes | 52 |
| cg22930808 | 3:122281881 | PARP9–DTX3L | 2.4 × 10−55 | − | Blood and B cells | Yes | 52,53 |
| cg06981309 | 3:146260954 | PLSCR1 | 4.1 × 10−51 | − | Blood and B cells | Yes | 52,53 |
| cg24678928 | 4:169240829 | DDX60 | 2.84 × 10−16 | − | Blood | Yes | 52 |
| cg17608381 | 6:29911550 | HLA-A | 3.4 × 10−15 | − | Blood | Yes | 53 |
| cg24399349 | 7:55517045 | NA | 2.38 × 10−8 | + | Salivary glands | NA | 53 |
| cg02680398 | 8:4629524 | CSMD1 | 1.22 × 10−8 | − | Salivary glands | No | 53 |
| cg01232121 | 8:58177661 | LOC286177 | 1.59 × 10−8 | − | Salivary glands | No | 53 |
| cg14864167 | 8:66751182 | PDE7A | 2.55 × 10−10 | − | Blood | Yes | 52 |
| cg06188083 | 10:91093005 | IFIT3 | 8.85 × 10−18 | − | Blood | Yes | 52 |
| cg05552874 | 10:91153143 | IFIT1 | 2.7 × 10−56 | − | Blood and B cells | Yes | 52,53 |
| cg06999856 | 10:135260170 | NA | 2.79 × 10−8 | + | Salivary glands | NA | 53 |
| cg23570810 | 11:315102 | IFITM1 | 6.1 × 10−38 | − | Blood and B cells | Yes | 52,53 |
| cg23198815 | 11:87184120 | NA | 3.73 × 10−8 | − | Salivary glands | NA | 53 |
| cg14943355 | 12:3980704 | PARP11 | 2.85 × 10−16 | − | Blood | Yes | 52 |
| cg04740359 | 12:5603542 | NTF3 | 1.39 × 10−9 | − | Salivary glands | Yes | 53 |
| cg20870559 | 12:113416518 | OAS2 | 1.02 × 10−9 | − | Salivary glands | Yes | 53 |
| cg12439472 | 13:43565399 | EPSTI1 | 2.32 × 10−17 | − | Blood | Yes | 52 |
| cg18329187 | 14:103989711 | CKB | 2.8 × 10−8 | + | Salivary glands | Yes | 53 |
| cg03425812 | 15:45005363 | B2M | 6.11 × 10−11 | − | Blood | Yes | 52 |
| cg07839457 | 16:57023022 | NLRC5 | 7.59 × 10−20 | − | Blood | Yes | 52 |
| cg25330422 | 17:40467382 | STAT3 | 3.2 × 10−13 | + | B cells | Yes | 53 |
| cg00871371 | 17:75371476 | SEPTIN9 | 3.15 × 10−8 | + | Salivary glands | No | 53 |
| cg16638092 | 17:78800774 | RPTOR | 1.72 × 10−8 | + | Salivary glands | No | 53 |
| cg05825244 | 20:2730488 | EBF4 | 2.4 × 10−11 | + | Blood | No | 53 |
| cg22862003 | 21:42797588 | MX1 | 7.9 × 10−63 | − | Blood and B cells | Yes | 52,53 |
| cg14293575 | 22:18635460 | USP18 | 9.7 × 10−14 | − | B cells | Yes | 53 |
bp, base pair; Chr, chromosome; CpG, cytosine guanine dinucleotide; NA, not available. aDefined as absolute fold change of 2.0 in the interferome database204.
Researchers initially discovered that type I interferon-regulated genes were expressed at high levels in the blood46,47 and salivary glands48,49 of patients with pSS and later found DNA hypomethylation at these genes. Methylation levels are influenced by environmental factors, including drugs, and different cells and tissues have distinct methylation patterns (known as methylomes), which are linked to cell-specific activation of particular signalling pathways and transcription factors50. Initial studies in pSS, which will not be reviewed here, assessed global DNA methylation levels and specific CpG sites in candidate genes51. Since 2014, the involvement of DNA methylation has been studied in several epigenome-wide association studies (EWAS)52–59 to identify genetic regions where methylation levels differ between patients with pSS and healthy individuals.
Two large-scale EWAS that each included ≥100 patients have been performed52,53, and the top associated loci are listed in Table 2. In one of the EWAS, Teruel et al.52 assessed differentially methylated positions and regions in whole blood from 189 patients with pSS and 220 healthy individuals. The most robustly associated differentially methylated positions and regions were located within type I interferon-regulated genes, but differentially methylated positions were also enriched in pathways related to the metabolism of collagen and the organization of the extracellular matrix. Notably, the detected epigenetic signatures were observed only for anti-SSA/Ro antibody-positive patients and were not present in autoantibody-negative patients. Similarly, the other EWAS by Imgenberg-Kreuz et al.53 found extensive hypomethylation at regulatory enhancer and promoter regions of interferon-regulated genes in whole-blood analysis of 100 patients and 400 healthy individuals, and this signature was more pronounced in autoantibody-positive patients. Subset analyses of CD19+ B cells and minor salivary gland biopsy samples also revealed hypomethylation at interferon-regulated genes, which correlated with increased gene expression levels in B cells. In whole blood, the most prominent finding in both of these large-scale studies was hypomethylation at the type I interferon-regulated gene IFI44L.
Several smaller EWAS have also assessed epigenome-wide DNA methylation in minor salivary gland biopsy samples53,56,59, CD4+ T cells54,55, CD19+ B cells53,55 and/or cultured minor salivary gland epithelial cells57 from patients with pSS. Altogether, the findings suggest that differential DNA methylation is more pronounced in B cells than in T cells55, and most studies detect prominent hypomethylation at interferon-regulated genes. A 2021 study of minor salivary gland biopsy samples, which included 64 patients with pSS and 67 individuals with sicca symptoms but not pSS, found that close to half of the detected differentially methylated regions were located in the HLA locus, with corresponding methylation quantitative trait loci (meQTLs) in the regions encompassing the HLA-DQA1, HLA-DQB1 and HLA-DQA2 loci59. Conversely, relatively few non-HLA loci genetically associated with pSS have been identified as differentially methylated in EWAS.
Implicated pathogenic processes
Important features of pSS immunopathogenesis include augmented B cell activity, crosstalk between B cells and T cells and the resulting formation of ectopic lymphoid tissue, disruption of salivary gland tissue associated with increased apoptosis and exposure of autoantigens, persistently elevated levels of pro-inflammatory cytokines and high expression of type I interferon-regulated genes13,60. Interestingly, most of the identified genetic variants and epigenetic signals associated with pSS can be assigned to genes involved in these processes, including Toll-like receptor (TLR) and interferon pathways, lymphocyte regulation, antigen presentation and target tissue maintenance (Fig. 4), as discussed in the next section.
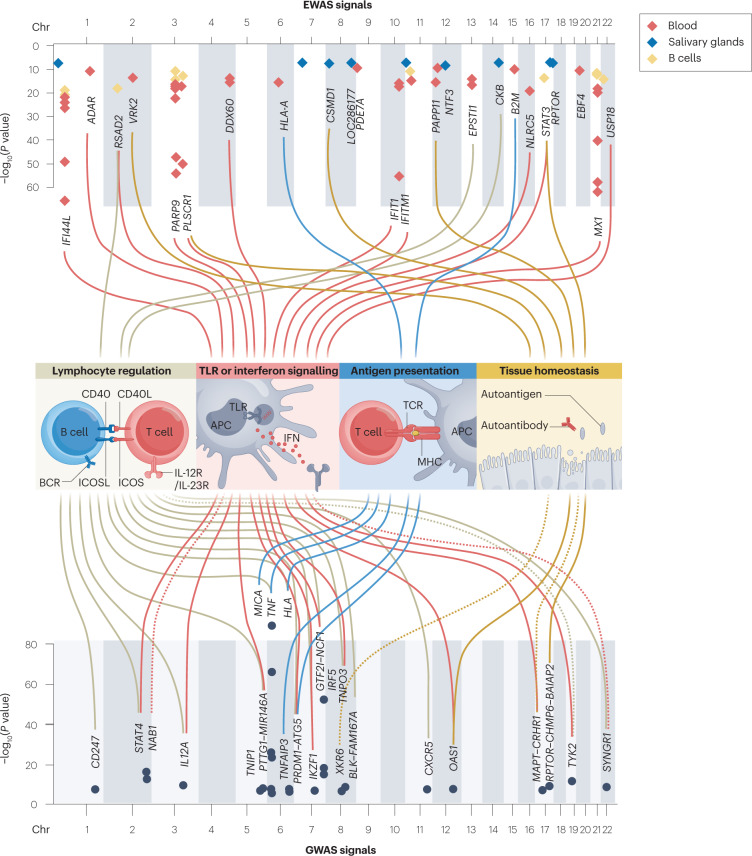
Fig. 4. Pathogenic processes implicated by genetic and epigenetic studies.
Genetic and epigenetic studies implicate various genes in the development of primary Sjögren syndrome (pSS). Most of these genes have functions in lymphocyte regulation, Toll-like receptor (TLR) or interferon (IFN) signalling, antigen presentation and tissue homeostasis, pointing to a role for these processes in the pathogenesis of pSS. The position and significance level of methylation signals associated with pSS for various tissues are shown in the upper Manhattan plot, including associations found in blood (red dots), salivary glands (blue dots) and B cells (yellow dots). Genetic variants associated with pSS at genome-wide significance and their annotated genes are shown in the lower Manhattan plot. The genes are linked by coloured cords to the four main pathogenic processes, according to gene function; solid lines indicate a direct function, whereas dotted lines indicate an indirect connection to these processes. Data taken from the studies listed in Tables 1 and 2. APC, antigen-presenting cell; BCR, B cell receptor; Chr, chromosome; EWAS, epigenome-wide association studies; GWAS, genome-wide association studies; ICOS, inducible T cell co-stimulator; ICOSL, inducible T cell co-stimulator ligand; TCR, T cell receptor.
TLR signalling and interferon pathways
The type I and type II interferon systems are highly activated in the peripheral blood and target tissue of most patients with pSS47,48,61,62. This feature of increased type I and type II interferon activity is also evident in DNA methylation studies, in which interferon-regulated genes are hypomethylated and the expression of the corresponding genes is increased in pSS52,53. Activity of the type I interferon system can be assessed by measuring hypomethylation at type I interferon-regulated genes63, a method that has been employed to define patient subsets in pSS64. Beyond DNA methylation studies, several disease-associated loci of genome-wide significance encode genes related to type I and/or type II interferon signalling, including IRF5, STAT4, TYK2, IL12A and OAS1 (Fig. 4).
The expression of the transcription factor interferon regulatory factor 5 (IRF5) is induced by TLR and type I interferon signalling, and IRF5 in turn regulates the expression of type I interferon and other pro-inflammatory cytokines65. The genetic association of IRF5 with disease has been replicated across ancestries in pSS, and also in several other autoimmune diseases (Fig. 2). Interestingly, variants in IRF5 and STAT4 have additive effects on the risk of pSS66. STAT4 encodes a transcription factor that functions downstream of IL-12 and type I interferon receptors, thus functionally connecting this gene to other pSS-associated loci. STAT1 is a neighbour of STAT4 on chromosome 2, and the mRNA levels of STAT1 differ considerably depending on the risk variants in the STAT1–STAT4 locus in B cell lines67 and monocyte-derived macrophages68, implicating signal transducer and activator of transcription 1 (STAT1) in systemic autoimmune diseases. However, the STAT4 risk allele rs7574865[T] is not associated with differential protein levels of STAT1 or STAT4 in resting peripheral blood mononuclear cells from patients with SLE69. Also, no eQTLs were identified for risk variants in the STAT1–STAT4 locus in peripheral blood from patients with pSS and healthy individuals31. However, as STAT1 is directly implicated in pSS pathology, the effect of genetic variants in this region on STAT1 expression and function requires further investigation.
The tyrosine kinase TYK2 transmits signals downstream of multiple cytokine receptors, including the type I interferon and IL-12 receptors, and is therefore central in both interferon signalling and lymphocyte activation70. Genetic variants in TYK2 associated with pSS are protective, possibly owing to a reduction in signalling downstream of pro-inflammatory cytokine receptors71. IL12A encodes the p35 subunit of IL-12, which signals through the Janus kinase (JAK)–STAT pathway (including TYK2 and STAT4) and has several functions but is especially important for the differentiation of naive T cells into T helper type 1 (TH1) cells72. Activation of the IL-12 receptor leads to the production of interferon-γ (IFNγ) by T cells and natural killer cells73. Finally, OAS1, as a type I interferon-induced gene, has an important role in antiviral responses. The pSS-associated SNP rs10774671 in OAS1 results in alternative splicing of the gene and generates an isoform of 2′–5′-oligoadenylate synthase 1 (OAS1) with a reduced function35; in addition to being connected with pSS, this SNP is also associated with increased risk of severe coronavirus disease 2019 (COVID-19)74, as well as susceptibility to West Nile virus and hepatitis C virus infection75,76.
Regarding the most recently discovered associations, variants in NAB1 have previously been associated with seropositive systemic autoimmunity in a GWAS meta-analysis of systemic sclerosis, SLE, RA and idiopathic inflammatory myopathies77. The encoded protein NGFI-A binding protein 1 (NAB1) functions as a transcriptional repressor78, and, in mice, downregulates the expression of Ifngr1 (ref. 79). Interestingly, a variant in TNFAIP3 is an eQTL for IFNGR1 in the salivary gland, further tying type II interferon responses to potential pathogenic processes30. The microRNA (miRNA) encoded by MIR146A regulates inflammation through a negative feedback loop downstream of TLR and cytokine receptors80,81, and genetic variation in this gene is associated with numerous inflammatory diseases82. High expression of miR-146a has been reported in peripheral blood mononuclear cells from patients with pSS83–87, primarily in CD4+ T cells88.
Lymphocyte regulation
Features of B cell hyperactivity in pSS include hypergammaglobulinaemia, high levels of B cell-activating factor (BAFF), disturbances in B cell subset proportions with high relative numbers of naive B cells and low frequencies of CD27+ memory B cells89, and the development of B cell lymphomas8. T cells are present at sites of autoimmune inflammation and stimulate the autoreactive B cells90. In keeping with these processes, many genetic regions associated with pSS contain genes involved in the regulation of lymphocytes30.
Disease-associated variants in the FAM167A–BLK locus were first described in a candidate gene study in Swedish and Norwegian patients91. A later study described a prominent disease-associated variant in the shared promoter region of the two genes, which are transcribed in opposite directions, along with notable eQTLs for both genes31. BLK encodes the Src family tyrosine kinase BLK, which is an important signalling molecule downstream of the B cell receptor, whereas FAM167A encodes the protein FAM167A (also known as DIORA1), which is highly expressed in B cells and in the lung92,93. Variants near CXCR5 implicate the importance of the CXCR5–CXCL13 axis in pSS immunopathology, a signalling pathway that is crucially involved in the migration of B cells and T cells as well as the formation of germinal centres94. A risk variant in this locus is an eQTL for CXCR5 in CD19+ B cells94. Notably, mice lacking Cxcr5 fail to develop several types of peripheral lymph node and form few and abnormal Peyer’s patches95. Furthermore, dysregulation of the CXCR5–CXCL13 axis is amply documented in pSS96. DDX6 is a neighbour of CXCR5 and a SNP in the promoter/enhancer region of DDX6 and CXCR5 is an eQTL of DDX6 in T cells97, which is of interest not least because DDX6 is implicated in the suppression of interferon-stimulated genes98.
Other pSS-associated loci also contain several genes involved in lymphocyte regulation. For instance, CD247 encodes the T cell receptor-ζ, which is part of the T cell receptor–CD3 complex and is thus important for signal transduction upon antigen stimulation. Notably, CD247 is hypomethylated in CD4+ T cells in pSS54, further supporting the relevance of the association of the CD247 locus with the disease. Another relevant gene is PRDM1, which encodes B lymphocyte-induced maturation protein 1 (BLIMP1), an IRF5-regulated transcription factor crucial for lymphocyte development and the regulation of B cell differentiation99. Finally, a pSS-associated region contains IKZF1, which encodes a transcription factor that is also involved in lymphocyte differentiation and proliferation and is regulated by various IRFs, including IRF5 (ref. 100).
Antigen presentation
The HLA locus harbours by far the strongest genetic associations with pSS30. This gene-dense region is difficult to dissect owing to high linkage disequilibrium, but disease-associated signals in both the HLA class I and II regions are well established and contain eQTLs that often lead to increased expression of HLA molecules31 (Supplementary Box 1).
Data from several studies suggest that the HLA associations occur predominantly in patients with anti-SSA/Ro and/or anti-SSB/La autoantibodies, with the most relevant being HLA class II genes in the HLA-DR and HLA-DQ loci32. Specifically, DRB1*03:01, DQA1*05:01 and DQB1*02:01 have the best established associations101, although researchers have also identified associations with variants in the region containing HLA-B, MICA and HCP5 (refs. 32,33). Emerging data suggest that blood levels of IFNα in pSS are associated with polymorphisms in the HLA-DQ locus, indicating crosstalk between these two pathways102.
The proteins encoded by the HLA genes present antigens to T cells and have a central role in activation and regulation of the immune system. Notably, a pSS-associated genomic region contains ATG5, which encodes a protein required for proper phagocytosis, processing and presentation of peptides on HLA class II103.
Target tissue maintenance
Ductal cells and salivary gland epithelial cells (SGECs) in affected glands are thought to actively participate in the pathogenesis of pSS through the secretion of chemoattractants and the expression of HLA class I molecules and adhesion molecules104. Defective apoptotic mechanisms in SGECs are implicated in pSS105, and indeed several genes in loci associated with pSS have a role in these processes (Fig. 4).
Interestingly, a disease-associated variant in the PRDM1–ATG5 locus is an eQTL for ATG5 in salivary gland tissue30. ATG5 is involved in autophagy and apoptosis, among several other immune processes106 and is expressed at high levels in large and organized salivary gland infiltrates in pSS107. Regarding the disease-associated locus MIR146A–PTTG1, securin (encoded by PTTG1) is a regulatory protein involved in chromosome stability, cell cycle regulation and apoptosis108. However, miR-146a (encoded by MIR146A) is perhaps the most likely candidate gene to have a functional impact on pSS disease processes, given its properties in modulating immune responses. Within the disease-associated region RPTOR–CHMP6–BAIAP2, RPTOR encodes a scaffold protein that is a component of the mechanistic target of rapamycin complex 1 (mTORC1), which has an importance for both cell growth and apoptosis109, but this complex has not yet been studied in pSS. Finally, two disease-associated regions contain TNFAIP3 and TNIP1, encoding A20 and an A20-binding protein, which function to suppress nuclear factor-κB (NF-κB) signalling as well as TNF-induced apoptosis110. In addition to pSS, genetic variants in the TNFAIP3 locus are associated with RA, SLE, multiple sclerosis and inflammatory bowel disease, among other diseases111. The expression of TNFAIP3 is downregulated in B cells in pSS, and a missense polymorphism in the gene is associated with lymphoma development41.
EWAS have revealed dysregulated methylomes in the minor salivary glands of patients with pSS that probably affect target tissue maintenance53,56,59. For example, an analysis of minor salivary gland biopsy samples found that global methylation levels are reduced in pSS and are associated with glandular lymphocyte infiltration and the presence of anti-SSB/La autoantibodies, but are not associated with EULAR Sjögren’s Syndrome Disease Activity Index (ESSDAI) scores112. Furthermore, a separate study found that the expression of DNA methylation mediators (including DNA methyltransferases 1 and 3B) in the minor salivary glands is higher in patients with pSS than in individuals with non-pSS sicca symptoms, which in turn is positively associated with the expression of LINE-1 retroelements113; this is of interest as LINE-1 retroelements might function as endogenous triggers of immune activation in the context of interferon-driven autoimmune diseases114. Notably, dysregulated methylation levels have also been reported in saliva from patients with pSS115.
Genes with unknown or other functions
Various pSS susceptibility loci encompass genes with no immediately obvious link to pathways implicated in the disease, either because little is known about the gene function or because the function is difficult to connect with pSS immunopathology. Examples of such genes include XKR6, TNPO3, CHMP6, MAPT, CRHR1, SYNGR1 and BAIAP2 (refs. 30,31) (Table 1). Further investigation into the function of these genes and a better understanding of the pathogenic process of pSS should reveal whether the genes are indeed important for disease development.
Interaction of genetic susceptibility with other risk factors
Although tremendous progress has been made in understanding the genetic prerequisites for pSS, disease-associated loci generally convey only a modestly increased risk of pSS, with odds ratios between 1 and 2 (Table 1) when estimated without regard to other intrinsic or extrinsic factors. Thus, genetic factors seem to explain only a limited proportion of pSS development. Studies of other rheumatic diseases show that genetic susceptibility and other risk factors can interact. The contribution of environmental factors in the pathogenesis of pSS is, however, understudied, and replication is needed for the few published findings19. In this section, we discuss the available studies and how identified risk factors might interact with genetic traits.
Environmental factors
Environmental risk factors that have been suggested for pSS include infection, vitamin D deficiency, stress, smoking and silicone breast implants, as reviewed elsewhere19. The most evidence has been presented for infection, and several epidemiological studies provide evidence of an association between prior infection and subsequent development of pSS5,116. These studies imply that both bacterial and viral infection can increase the risk of pSS, but experimental investigations have focused mostly on viruses. Specifically, some data, such as the expression of viral antigens in the minor salivary glands of patients with pSS117–119, implicate a role for Epstein–Barr virus (EBV) infection in driving local chronic inflammation. Intriguingly, a substantial number of pSS risk loci are potentially bound by the EBV protein EBNA2 (ref. 30), substantiating a potential gene–environment link in the context of EBV infection. Regarding other environmental factors implicated in autoimmunity, smoking is an established risk factor for seropositive RA120. By contrast, smoking is not thought to increase the risk of pSS development121–124. Instead, in the years preceding a diagnosis, individuals who later develop pSS are more prone to stop smoking than the general population121, and no discernible interactions have been detected between smoking and risk-related HLA alleles121.
Female sex
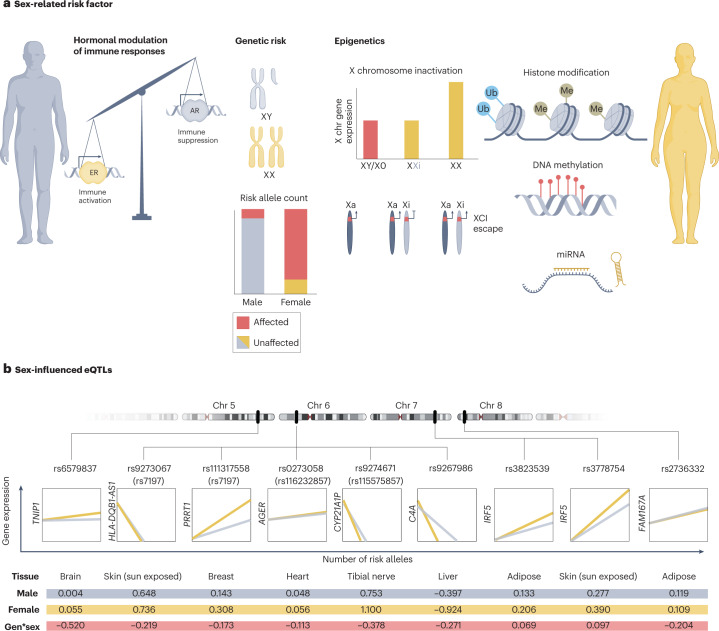
The strongest known risk factor for developing pSS is female sex. The sex disparity in pSS is among the most pronounced of all autoimmune diseases, with an average 14:1 female to male ratio across geolocations and ethnicities1,125. The factors that underlie this difference are, however, poorly explored, and both general differences in immune regulation between women and men, as well as overlaid pSS-specific features, probably contribute126. The differences in immune responses between women and men in general have long been recognized127, and they became particularly evident during the COVID-19 pandemic, with more profound antibody responses and fewer cases of severe disease occurring in women than in men128,129. Increasing data from large-scale analyses of gene expression in tissues52,130,131 and cells stimulated in vitro with different classes of pathogen132 confirm that immune responses differ substantially between the sexes. Sex hormones, sex chromosome dosage and sex-influenced regulation of disease-associated genetics have all been suggested as relevant genetic or genetically related factors for sex differences in human complex traits133 (Fig. 5). In this section, we summarize the current understanding of these factors and their role in pSS.
Fig. 5. Potential mechanisms connecting biological sex with risk of primary Sjögren syndrome.
Sex hormones affect biological functions directly and indirectly. The figure illustrates how genetic risk factors can result in discordant effects depending on the sex of the individual. a, Suggested hypotheses to explain this discordance include differential hormonal modulation of immune responses, men requiring a higher number of genetic risk loci to develop disease and differences in X chromosome (X chr) dosage. Genes located on the X chromosome, including important immune genes such as TLR7, can escape X chromosome inactivation (XCI), resulting in higher expression of the protein in women than in men. Other epigenetic modifications can also differ between men and women, including microRNA (miRNA) expression, histone modification and DNA methylation patterns. b, Some single nucleotide polymorphisms (SNPs) associated with primary Sjögren syndrome (pSS) affect the expression of genes in a sex-dependent fashion (known as sex-influenced expression quantitative trait loci (eQTL)). Variants associated with pSS with discordant effects on gene expression between men and women that reach a particular significance threshold (P < 0.05) for sex–genotype interactions are included (data taken from the Genotype-Tissue Expression (GTEx) database). All variants in high linkage disequilibrium with the Sjögren syndrome-associated variants were considered. Variants reported as associated with pSS (Table 1) are included in parenthesis if a sex-eQTL effect was identified for a linked variant. The table summarizes the tissue the effect was identified in, the effect size (expressed as a regression coefficient) in men and in women, and the genotype-by-sex (gen*sex) interaction effect size. AR, androgen receptor; ER, oestrogen receptor; Me, methyl group; Ub, ubiquitin.
Sex hormones
Differential immunomodulatory effects elicited by sex hormones have obvious potential to contribute to the sex bias in pSS134; however, studies on sex hormone levels in patients with pSS are remarkably limited, and the results are inconsistent135–138. Thus, sex hormones might contribute non-specifically to pSS as part of a general enhancement of immune responses that is more pronounced in women than in men. Indeed, oestrogen receptors are potent modulators of immune responses, including the type I interferon responses often seen in patients with pSS (reviewed elsewhere139).
X chromosome gene dosage
Another aspect in the context of female risk of pSS is the disparity in X chromosome number between men and women, which could provide a genetic explanation for the sex bias. Indeed, many important immune genes including TLR7, TLR8, BTK, FOXP3, IL2RG and CXCR3 are located on the X chromosome. Although no SNP associations with pSS have yet been identified on this chromosome30, the importance of the X chromosome in pSS is supported by studies of aneuploidy140. The frequency of aneuploidy in patients with pSS is low, but several studies support an X chromosome dosage effect on risk of pSS. One study found that the frequency of men with Klinefelter syndrome (47, XXY) is higher among patients with pSS than in the general population141: four men with the karyotype 47, XXY were identified in a cohort of 136 men with pSS, as compared with the 1:500 reported population prevalence of this condition. In another study, women with an extra X chromosome (47, XXX) were present at a higher-than-expected rate in a cohort of patients with pSS142. As these women have the same levels of sex hormones as women without an extra X chromosome (46, XX), the X chromosome load rather than hormonal factors was suggested to increase the risk of pSS.
In carriers of multiple X chromosomes, the paternal or maternal X chromosome is randomly silenced in each cell during embryogenesis in an epigenetic process denoted X chromosome inactivation (XCI). Skewed XCI or escape from XCI can result in increased expression of X chromosome genes, including immune genes. Indeed, as much as 15–23% of X chromosome genes are reported to escape XCI143. Although data for pSS specifically is currently lacking, XCI escape of TLR7 (ref. 144) and TASL145 is known to occur in immune cells of men with Klinefelter syndrome and in women. Both genes are implicated in SLE146,147 and are functionally connected; indeed, TASL functions as an immune adaptor for TLR7, TLR8 and TLR9 signalling and mediates activation of IRF5 (ref. 148). Escape from XCI is thus a potentially important genetic mechanism for sex-dependent development of autoimmunity dependent on type I interferon pathways.
Sex-influenced eQTLs
Sex differences at the DNA sequence level are restricted to the X and Y chromosomes. However, all genetic variants so far associated with pSS at genome-wide significance are located on autosomal chromosomes and thus have the same frequencies in men and women in the general population149. However, female carriers of these polymorphisms are more prone to develop pSS than male carriers150, indicating that the context of ‘female sex’ influences the effect of these polymorphisms on promoting disease development. Disease-associated variants can have discordant effects on the expression of nearby genes in women versus men151, known as sex-influenced eQTLs (Supplementary Box 1).
A role for sex-influenced eQTLs in complex diseases that differ by sex in terms of disease prevalence, course or severity was suggested more than a decade ago151, but only after GWAS grew common did these large-scale studies provide the substantial data needed to support the hypothesis152–154. Indeed, many genetic variants associated with pSS have genotype-dependent effects on gene expression that relate to the biological sex of the carrier. For example, CD74 (TNIP1 locus), PXK, CTSB (FAM167A–BLK locus) and ARCN1 (CXCR5 locus) are differentially expressed in B cells depending on the genotype and sex of the carrier150. Similarly, disease-associated variants in the HLA locus differentially affect the expression of HLA-DRB5 in whole blood in a sex-dependent manner152.
Large-scale efforts such as the Genotype-Tissue Expression (GTEx) consortium (see Related links) have made possible the examination of sex-influenced eQTLs in many different tissues155. Of the 27 independent pSS-associated genetic signals, six had notable sex-specific effects on the expression of several genes, including TNIP1, IRF5 and C4A in five tissues (Fig. 5). Sex-specific differences have been further studied during immune activation, in which cells from men and women were exposed to lipopolysaccharide to identify sex-specific responses to microbial stimuli156. Interestingly, seven genes in pSS-associated loci showed significant sex by treatment interactions, including four HLA genes, IRF5, OAS1 and IKZF1 (ref. 156). Overall, eight of the 27 loci independently associated with pSS contain sex-influenced eQTLs, clearly demonstrating that the biological sex of an individual can influence the effect of a risk allele on gene expression and subsequent outcomes. Notably, in 2020, researchers confirmed the interaction between sex and pSS-associated genetic variants of the C4 locus157, firmly supporting the relevance of sex-influenced eQTLs not only for gene expression levels, but also for disease development.
Sex influence on epigenetic regulation
An emerging area of investigation is the involvement of epigenetic mechanisms in sex-influenced eQTLs. The X chromosome is rich in miRNA genes, whereas the Y chromosome contains relatively few miRNAs158, suggesting that miRNAs or other epigenetic factors have a role in mediating sex differences in pSS. Sex is known to influence autosomal DNA methylation159, and evidence suggests that oestrogen can regulate both methylation and miRNA expression (reviewed in160). Sex-influenced QTLs that regulate the expression of miRNA (known as miRNA eQTLs) have been identified and implicated in autoimmune diseases161. Various miRNAs, including miR-146s, are already implicated in the phenotypes commonly seen in pSS such as parotid swelling and dry eyes85, and whether such epigenetic mechanisms differ by sex is worth exploring.
Genetically inferred therapeutic targets
Genetic, epigenetic and mechanistic studies have greatly expanded our understanding of the immunopathogenic processes that underlie pSS. Nevertheless, no immunomodulatory treatments that can affect the activity and/or progression of the disease are yet available12. Ongoing and completed randomized controlled trials (RCTs) generally use targeted approaches aimed at depleting specific cell populations, inhibiting co-stimulation, blocking cytokine–receptor interactions or targeting kinases important for intracellular signalling. Therapies evaluated in major RCTs for pSS (trials with >100 patients) include infliximab (a TNF inhibitor)162, rituximab (an anti-CD20, B cell-depleting antibody)163,164, abatacept (a fusion protein that blocks CD28–CD80/CD86 co-stimulation)165, tocilizumab (an IL-6 inhibitor)166 and ianalumab (a BAFF inhibitor and B cell-depleting antibody)167 (Fig. 6). Of these trials, only ianalumab met the primary end point of the study167. Numerous smaller RCTs and open label studies have also been carried out, with varying degrees of success (summarized elsewhere12). The failure of clinical trials in pSS could be due to the selection of therapeutic target, factors such as disease heterogeneity, choice of clinical end point, difficulties in distinguishing between organ damage and disease activity and/or power limitations. Novel responder indices and end point measures have been developed for pSS, which could benefit future studies168,169.
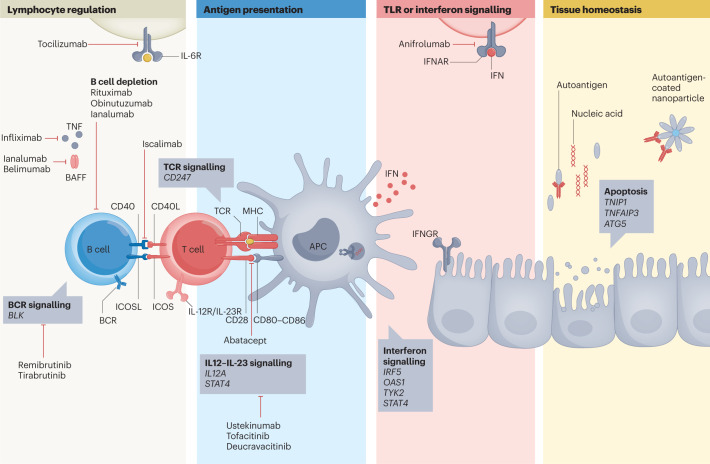
Fig. 6. Treatment targets implicated by genetic studies in primary Sjögren syndrome.
The processes implicated by genetic studies in the pathogenesis of primary Sjögren syndrome (pSS) include lymphocyte regulation, antigen presentation, Toll-like receptor (TLR) or interferon (IFN) signalling and tissue homeostasis (including apoptosis and autophagy). Selected pSS risk genes involved in these processes are indicated in boxes. These processes (particularly lymphocyte regulation and TLR or interferon signalling pathways) include targets of current and emerging therapies. APC, antigen-presenting cell; BAFF, B cell-activating factor; BCR, B cell receptor; TCR, T cell receptor.
What can we learn from genetic and epigenetic studies regarding treatment approaches? As summarized in the section on ‘Implicated pathogenic processes’, results from these studies implicate four major pathogenic processes in pSS: TLR and interferon signalling; lymphocyte regulation; antigen presentation; and target tissue maintenance (Figs. 4 and 6). In this section, we discuss current and emerging therapeutic strategies that target these processes. Although the first two processes contain main targets of current treatment approaches, the latter two remain underexplored.
TLR signalling and interferon pathways
Genetic associations as well as the frequent detection of hypomethylation at interferon-regulated genes underscore the importance of TLR and interferon signalling in pSS. Activity of the type I interferon system is high in the blood and target tissue of most patients with pSS13. As such, several approaches have been developed to target these pathways. For example, the JAK inhibitor tofacitinib inhibits both type I and type II interferon, and a randomized trial in pSS is currently recruiting170. Notably, in ATG5-deficient 3D-acini (a relevant model given the previously identified genetic associations at ATG5 (ref. 30)), tofacitinib could suppress IL-6 production caused by deficient autophagy171. Another JAK inhibitor, baricitinib, indicated possible efficacy in a small pilot study involving 11 patients with pSS172.
Regarding the genetic association at TYK2, selective inhibition of TYK2 in pSS is a potential therapeutic avenue173, and this strategy is currently being tested in SLE using deucravacitinib174. Blockade of IL-12 and IL-23, which results in inhibition of the type II interferon pathway, can be achieved using the monoclonal antibody ustekinumab and is currently being evaluated in a pilot trial in pSS175. Targeting of the BDCA2 antigen on plasmacytoid dendritic cells (a cell source of type I interferon) using monoclonal antibodies has been evaluated in a phase I trial in patients with cutaneous lupus erythematosus, with preliminary results suggesting some efficacy176, highlighting another possible approach for the treatment of pSS.
Finally, the monoclonal antibody anifrolumab, which targets the type I interferon receptor IFNAR1, restored gene expression-based type I interferon scores to normal levels after 12 weeks of treatment in patients with SLE177, and was approved in 2021 for the treatment of moderate to severe SLE178. A phase IIa proof-of-concept trial of anifrolumab for evaluation as a potential treatment of pSS is in recruitment phase179. From an epigenetic perspective, one question of interest is whether treatment with anifrolumab influences hypomethylation at interferon-regulated genes. Altogether, targeting TLR and type I interferon signalling in pSS constitutes an attractive approach and can be achieved in many ways.
Lymphocyte regulation
Most proteins, as well as the miRNA miR-146a, that are encoded by pSS genetic risk loci are involved in lymphocyte regulation (Fig. 4). Additionally, variants at genes involved in TLR and interferon signalling might contribute to augmented activation of B cells and T cells, and polymorphisms in FAM167A–BLK, PRDM1 and IKZF1 might have direct effects on B cell lineages, supporting the potential of B cell-targeted therapies.
Many small studies as well as two large RCTs have evaluated the monoclonal anti-CD20 B cell-depleting antibody rituximab in patients with pSS12. Although saliva flow rates improved, the large RCTs did not meet their primary end points163,164. Nevertheless, in clinical practice rituximab is considered effective for treating certain manifestations including cryoglobulinaemia-associated vasculitis11. Second-generation anti-CD20 drugs such as obinutuzumab, which mediate more efficient B cell depletion than rituximab, have been developed but have not yet been tested in pSS.
An alternative approach to B cell targeting is to block B cell activation. BAFF is an important cytokine in pSS pathogenesis owing to its many B cell-promoting properties, and the anti-BAFF monoclonal antibody belimumab is an approved treatment for SLE180. In one open label study of belimumab in 30 patients with pSS, the primary end point was reached in 60% of patients181. In a randomized phase II study evaluating co-administration of belimumab with rituximab, depletion of CD20+ B cells in minor salivary gland biopsies and CD19+ B cells in the periphery was greater compared with monotherapy with rituximab or belimumab alone, and reduction in ESSDAI scores was numerically greater in the combination group, highlighting the potential benefit of combined therapy182. Another approach has been to block transmembrane activator and cyclophilin ligand interactor (TACI), which is a receptor for both BAFF and a proliferation-inducing ligand (APRIL), and a phase II trial of telitacicept (a TACI–Fc fusion protein) has been completed, but the results have not yet been published183,184. In 2022, a phase IIb dose-finding trial of ianalumab (an anti-BAFF receptor monoclonal antibody) was the first large RCT in pSS to reach its primary end point (reduction in ESSDAI at 24 weeks)167. Future approaches could involve specific targeting of pathogenic B cells (as discussed in the next section), but means to accomplish such target specificity is still needed. In terms of other potential B cell targets, FAM167A is expressed in peripheral B cells and the encoded protein is present in plasma cells in the minor salivary glands of patients with pSS92, yet the function of this intracellular protein remains unknown, and should therefore be further studied and assessed as a potential therapeutic target.
Several genetic signals such as those from CXCR5, CD247 and the HLA locus point to the importance of B cell–T cell interactions, which can be targeted in several ways. In pSS, two phase III RCTs assessed blockade of the CD28–CD80/86 axis using the CTLA4–immunoglobulin fusion protein abatacept, but both studies failed to reach the clinical end points165,185. Another approach has been to block CD40–CD40L interactions using iscalimab (an anti-CD40 antibody), an approach that showed preliminary success in terms of efficacy in a proof-of-concept study186, and further trials targeting this interaction are underway187. A third approach has been to block the interaction of inducible T cell co-stimulator (ICOS) with its ligand, ICOSL, using a monoclonal antibody (MEDI5872); however, this approach did not result in improvement in disease activity in a phase IIb study in pSS188. Interestingly, the promoter region of CXCR5 is hypomethylated in B cells of patients with pSS189, and a pSS-associated variant near CXCR5 has an eQTL effect in B cells and seems to influence the cell surface expression of CXCR5; therefore, targeting the CXCR5–CXCL13 chemokine axis is another potential therapeutic strategy that remains to be explored94.
pSS associations at TYK2 and BLK suggest that intracellular kinases are important in pSS-related disease pathways, which could be targeted with small-molecule inhibitors. For example, Bruton’s tyrosine kinase (BTK) has an important role in signalling downstream of the B cell receptor190. Mutations in BTK cause the immunodeficiency disease X-linked agammaglobulinaemia191, and overexpression of BTK in mice causes an autoimmune phenotype192. A phase II trial of the BTK inhibitor remibrutinib in pSS showed that this drug has a favourable safety profile and potential efficacy (as assessed by reduction in ESSDAI score and improvement in salivary flow rates at 24 weeks)193. By contrast, a phase II trial that included assessment of tirabrutinib, another BTK inhibitor, found no indication of efficacy in pSS194. Other approaches for blocking pathways crucial for lymphocyte activation include inhibition of one of the signalling molecules spleen tyrosine kinase (SYK) or phosphatidylinositol 3-kinase (PI3K); however, inhibitors of these kinases have thus far shown no obvious efficacy in small trials187,194.
Antigen presentation
Although the HLA locus has long been characterized as the major genetic risk locus in pSS, antigen presentation remains a challenge to target therapeutically. Future approaches to treat autoimmunity include antigen-specific immunotherapy, which aims to specifically target the pathogenic immune cells. This concept involves specifically targeting autoreactive B cells and T cells through the administration of tolerogenic peptides, the use of tolerizing antigen-specific vaccines, administration of tolerogenic dendritic cells, clearance of pathogenic autoantibodies and/or the use of chimeric antigen receptor (CAR)-transduced T cell therapy195. For example, in various mouse models of autoimmunity, administration of nanoparticles coated with autoimmune-associated antigens bound to MHC class II peptides could shift disease-primed cells into regulatory cells196. Antigen-specific immunotherapy relies on adequate selection of autoantigens, and whether the known autoepitopes in pSS, such as those of the Ro52 and Ro60 proteins197, would enable disease modification remains to be explored.
Target tissue maintenance and multiple target strategies
Dysregulated apoptosis of salivary gland epithelial cells occurs in pSS198. Whether this process is an important initial event that leads to development of the autoimmunity and disease is unclear, but exploring this process further might uncover treatment options for the very early phase of disease, with the prospect of preventing tissue destruction.
Given the polygenic nature of disease risk in pSS, targeting a single cell type or signalling pathway might be insufficient, and targeting multiple processes could be necessary. Hence, various trials have tested drugs that modulate several disease pathways. For instance, the tetravalent bispecific antibody tibulizumab targets both IL-17A and BAFF199. Although targeting multiple pathways might prove to be more efficient than targeting a single pathway, a higher risk of adverse events could be expected.
Targeting DNA methylation
Modification of DNA methylation is a potential therapeutic intervention for various diseases, as exemplified by successful employment of such drugs in the field of immuno-oncology200. Available drugs that modify DNA methylation are employed for the treatment of various cancers200. However, these drugs, including azacytidine and decitabine, are DNA methyltransferase inhibitors, and they function in a nonspecific manner to reduce genomic DNA methylation, which could be counterproductive for the treatment of pSS, given that pSS-associated genes tend to be hypomethylated. Indeed, the drugs hydralazine and procainamide, which inhibit DNA methylation201, have long been known to confer a high risk of triggering drug-induced SLE202. To date, epigenetic modifiers have not been tested in pSS but constitute an interesting future treatment approach.
Conclusions
Emerging genetic and epigenetic data in pSS highlight the polygenic nature of the disease, and the influence of environmental triggers as well as female sex on pSS development. The identified genetic and epigenetic signals hold promise for addressing the many unmet clinical needs in pSS, including possibilities for earlier diagnosis, novel tools for patient subphenotyping, prognostic markers for comorbidities such as cardiovascular disease and lymphoma, and, ultimately, efficacious treatment to ameliorate disease activity, stop disease progression and rescue organ function. As outlined in this Review, the genetic and epigenetic findings point towards four interconnected immune processes of central importance to disease development and pathology. Within these processes, some therapeutic prospects have already been identified and attempted, but many potential targets remain to be explored. Interestingly, calculating genetic or epigenetic risk scores already holds potential for better stratification of disease subtypes and could be considered in future clinical drug trials30.
Notably, only a few epigenetic signals overlap with identified genetic risk loci in pSS. This lack of overlap suggests that many genes that increase the risk of pSS function upstream of cellular events and lead to epigenetic changes and differences in gene expression but are not epigenetically modified themselves and could thus constitute potential molecular targets. Strong evidence highlights the involvement of TLR and interferon signalling, including disease associations with genetic variants of genes in these pathways, as well as the extensive hypomethylation of interferon-regulated genes. Both genetic and epigenetic findings also support the strong association of disease with the HLA locus, which has been recognized for decades. Although many drugs currently in the pipeline are aimed at TLR and interferon pathways, treatments specifically targeting antigen presentation or aimed at tolerance induction have not been successfully developed for pSS and constitute future potential areas for development.
Epigenetic alterations differ between tissues and cell types, but a pattern in pSS is emerging, involving extensive alterations of DNA methylation in both B cells and target tissue. A future challenge will be to identify drugs that can specifically reverse these epigenetic changes, unlike the currently available drugs, which function in a largely nonspecific manner. Other epigenetic treatment prospects in immune cells and target tissue include modulation of non-coding RNAs and alteration of histone acetylation patterns as well as nucleosome positioning. Also, the emerging field of epitranscriptomics, which refers to post-transcriptional modifications of RNA203, deserves attention but has not yet been studied in pSS.
Beyond indicating which processes could be of relevance to target, knowledge of the underlying genetics and epigenetics can facilitate earlier diagnosis through risk scores and enable subphenotyping of patients. Future studies performed in larger cohorts that include sequencing of RNA and DNA as well as use of other novel technologies and application of sophisticated analytical approaches to combine data obtained by these high-throughput technologies is expected to greatly enhance our understanding of pSS and open novel possibilities to address the unmet needs of the disease in the upcoming years.
Supplementary information
Acknowledgements
The work of the authors is funded by the Swedish Research Council, the Swedish Heart–Lung Foundation, the Swedish Rheumatism Foundation, the Stockholm County Council, the FOREUM Foundation for Research in Rheumatology, the NECESSITY grant of EU Innovative Medicines Initiative 2, the Norwegian Research Council grant 316120, the King Gustaf V 80-year Foundation, the Freemason Children Foundation Stockholm, the Torsten Söderberg Foundation, the Tore Nilsson Foundation, the Magnus Bergvall Foundation, the Rheumatology Foundations at Karolinska Institutet, the Nanna Svartz Foundation, the Åke Wiberg Foundation, the Lars Hierta Foundation, and the Swedish Women and Health Foundation.
Author contributions
G.E.T., A.B. and M.W.-H. researched data for this article, provided substantial contributions to discussions of content, wrote the article and reviewed and/or edited the final manuscript before submission.
Peer review
Peer review information
Nature Reviews Rheumatology thanks A. Tzioufas, who co-reviewed with A. Goules, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Competing interests
M.W.-H. has received research grants from Merck KGaA and Janssen Pharmaceutica NV during the past 5 years. G.E.T. and A.B. declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Genotype-Tissue Expression (GTEx): https://gtexportal.org/home/
UCSC Genome Browser: http://genome.ucsc.edu
These authors contributed equally: Gudny Ella Thorlacius, Albin Björk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41584-023-00932-6.
References
- 1.Kvarnström M, Ottosson V, Nordmark B, Wahren-Herlenius M. Incident cases of primary Sjögren’s syndrome during a 5-year period in Stockholm County: a descriptive study of the patients and their characteristics. Scand. J. Rheumatol. 2015;44:135–142. doi: 10.3109/03009742.2014.931457. [DOI] [PubMed] [Google Scholar]
- 2.Mariette X, Criswell LA. Primary Sjögren’s syndrome. N. Engl. J. Med. 2018;378:931–939. doi: 10.1056/NEJMcp1702514. [DOI] [PubMed] [Google Scholar]
- 3.Brito-Zerón P, et al. Epidemiological profile and north-south gradient driving baseline systemic involvement of primary Sjögren’s syndrome. Rheumatology. 2020;59:2350–2359. doi: 10.1093/rheumatology/kez578. [DOI] [PubMed] [Google Scholar]
- 4.Shiboski CH, et al. 2016 American College of Rheumatology/European League Against Rheumatism classification criteria for primary Sjögren’s syndrome: a consensus and data-driven methodology involving three international patient cohorts. Ann. Rheum. Dis. 2017;76:9–16. doi: 10.1136/annrheumdis-2016-210571. [DOI] [PubMed] [Google Scholar]
- 5.Mofors J, et al. Infections increase the risk of developing Sjögren’s syndrome. J. Intern. Med. 2019;285:670–680. doi: 10.1111/joim.12888. [DOI] [PubMed] [Google Scholar]
- 6.Mofors J, et al. Concomitant Ro/SSA and La/SSB antibodies are biomarkers for the risk of venous thromboembolism and cerebral infarction in primary Sjögren’s syndrome. J. Intern. Med. 2019;286:458–468. doi: 10.1111/joim.12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartoloni E, et al. Cardiovascular disease risk burden in primary Sjögren’s syndrome: results of a population-based multicentre cohort study. J. Intern. Med. 2015;278:185–192. doi: 10.1111/joim.12346. [DOI] [PubMed] [Google Scholar]
- 8.Nocturne G, Mariette X. Sjögren syndrome-associated lymphomas: an update on pathogenesis and management. Br. J. Haematol. 2015;168:317–327. doi: 10.1111/bjh.13192. [DOI] [PubMed] [Google Scholar]
- 9.Theander E, et al. Lymphoma and other malignancies in primary Sjögren’s syndrome: a cohort study on cancer incidence and lymphoma predictors. Ann. Rheum. Dis. 2006;65:796–803. doi: 10.1136/ard.2005.041186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrosi A, Sonesson SE, Wahren-Herlenius M. Molecular mechanisms of congenital heart block. Exp. Cell Res. 2014;325:2–9. doi: 10.1016/j.yexcr.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Ramos-Casals M, et al. EULAR recommendations for the management of Sjögren’s syndrome with topical and systemic therapies. Ann. Rheum. Dis. 2020;79:3–18. doi: 10.1136/annrheumdis-2019-216114. [DOI] [PubMed] [Google Scholar]
- 12.Seror R, Nocturne G, Mariette X. Current and future therapies for primary Sjögren syndrome. Nat. Rev. Rheumatol. 2021;17:475–486. doi: 10.1038/s41584-021-00634-x. [DOI] [PubMed] [Google Scholar]
- 13.Bodewes ILA, Björk A, Versnel MA, Wahren-Herlenius M. Innate immunity and interferons in the pathogenesis of Sjögren’s syndrome. Rheumatology. 2019 doi: 10.1093/rheumatology/key360. [DOI] [PubMed] [Google Scholar]
- 14.Nocturne G, Mariette X. B cells in the pathogenesis of primary Sjögren syndrome. Nat. Rev. Rheumatol. 2018;14:133–145. doi: 10.1038/nrrheum.2018.1. [DOI] [PubMed] [Google Scholar]
- 15.Salomonsson S, et al. Cellular basis of ectopic germinal center formation and autoantibody production in the target organ of patients with Sjögren’s syndrome. Arthritis Rheum. 2003;48:3187–3201. doi: 10.1002/art.11311. [DOI] [PubMed] [Google Scholar]
- 16.Bombardieri M, Lewis M, Pitzalis C. Ectopic lymphoid neogenesis in rheumatic autoimmune diseases. Nat. Rev. Rheumatol. 2017;13:141–154. doi: 10.1038/nrrheum.2016.217. [DOI] [PubMed] [Google Scholar]
- 17.Ulff-Møller CJ, Svendsen AJ, Viemose LN, Jacobsen S. Concordance of autoimmune disease in a nationwide Danish systemic lupus erythematosus twin cohort. Semin. Arthritis Rheum. 2018;47:538–544. doi: 10.1016/j.semarthrit.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Silman AJ, et al. Twin concordance rates for rheumatoid arthritis: results from a nationwide study. Br. J. Rheumatol. 1993;32:903–907. doi: 10.1093/rheumatology/32.10.903. [DOI] [PubMed] [Google Scholar]
- 19.Björk A, Mofors J, Wahren-Herlenius M. Environmental factors in the pathogenesis of primary Sjögren’s syndrome. J. Intern. Med. 2020;287:475–492. doi: 10.1111/joim.13032. [DOI] [PubMed] [Google Scholar]
- 20.Kuo CF, et al. Familial risk of Sjögren’s syndrome and co-aggregation of autoimmune diseases in affected families: a nationwide population study. Arthritis Rheumatol. 2015;67:1904–1912. doi: 10.1002/art.39127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WS, Yoo WH. Primary Sjögren’s syndrome in monozygotic twins. Int. J. Rheum. Dis. 2014;17:578–579. doi: 10.1111/1756-185X.12398. [DOI] [PubMed] [Google Scholar]
- 22.Bolstad AI, Haga HJ, Wassmuth R, Jonsson R. Monozygotic twins with primary Sjögren’s syndrome. J. Rheumatol. 2000;27:2264–2266. [PubMed] [Google Scholar]
- 23.Houghton KM, Cabral DA, Petty RE, Tucker LB. Primary Sjögren’s syndrome in dizygotic adolescent twins: one case with lymphocytic interstitial pneumonia. J. Rheumatol. 2005;32:1603–1606. [PubMed] [Google Scholar]
- 24.Scofield RH, Kurien BT, Reichlin M. Immunologically restricted and inhibitory anti-Ro/SSA in monozygotic twins. Lupus. 1997;6:395–398. doi: 10.1177/096120339700600409. [DOI] [PubMed] [Google Scholar]
- 25.Imgenberg-Kreuz J, Rasmussen A, Sivils K, Nordmark G. Genetics and epigenetics in primary Sjögren’s syndrome. Rheumatology. 2021;60:2085–2098. doi: 10.1093/rheumatology/key330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ice JA, et al. Genetics of Sjögren’s syndrome in the genome-wide association era. J. Autoimmun. 2012;39:57–63. doi: 10.1016/j.jaut.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjögren’s syndrome at 7q11.23. Nat. Genet. 2013;45:1361–1365. doi: 10.1038/ng.2779. [DOI] [PubMed] [Google Scholar]
- 28.Song IW, et al. Identification of susceptibility gene associated with female primary Sjögren’s syndrome in Han Chinese by genome-wide association study. Hum. Genet. 2016;135:1287–1294. doi: 10.1007/s00439-016-1716-0. [DOI] [PubMed] [Google Scholar]
- 29.Taylor KE, et al. Genome-wide association analysis reveals genetic heterogeneity of Sjögren’s syndrome according to ancestry. Arthritis Rheumatol. 2017;69:1294–1305. doi: 10.1002/art.40040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khatri B, et al. Genome-wide association study identifies Sjögren’s risk loci with functional implications in immune and glandular cells. Nat. Commun. 2022;13:4287. doi: 10.1038/s41467-022-30773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessard CJ, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjögren’s syndrome. Nat. Genet. 2013;45:1284–1292. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorlacius GE, et al. Genetic and clinical basis for two distinct subtypes of primary Sjögren’s syndrome. Rheumatology. 2021;60:837–848. doi: 10.1093/rheumatology/keaa367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carapito R, et al. A new MHC-linked susceptibility locus for primary Sjögren’s syndrome: MICA. Hum. Mol. Genet. 2017;26:2565–2576. doi: 10.1093/hmg/ddx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, et al. A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat. Genet. 2017;49:433–437. doi: 10.1038/ng.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. Identification of a Sjögren’s syndrome susceptibility locus at OAS1 that influences isoform switching, protein expression, and responsiveness to type I interferons. PLoS Genet. 2017;13:e1006820. doi: 10.1371/journal.pgen.1006820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu S, et al. Common variants near IKZF1 are associated with primary Sjögren’s syndrome in Han Chinese. PLoS ONE. 2017;12:e0177320. doi: 10.1371/journal.pone.0177320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolstad AI, et al. Association between genetic variants in the tumour necrosis factor/lymphotoxin α/lymphotoxin β locus and primary Sjogren’s syndrome in Scandinavian samples. Ann. Rheum. Dis. 2012;71:981–988. doi: 10.1136/annrheumdis-2011-200446. [DOI] [PubMed] [Google Scholar]
- 38.Musone SL, et al. Sequencing of TNFAIP3 and association of variants with multiple autoimmune diseases. Genes Immun. 2011;12:176–182. doi: 10.1038/gene.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norheim KB, et al. Genetic variants at the RTP4/MASP1 locus are associated with fatigue in Scandinavian patients with primary Sjögren’s syndrome. RMD Open. 2021;7:e001832. doi: 10.1136/rmdopen-2021-001832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nocturne G, et al. Germline and somatic genetic variations of TNFAIP3 in lymphoma complicating primary Sjogren’s syndrome. Blood. 2013;122:4068–4076. doi: 10.1182/blood-2013-05-503383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nocturne G, et al. Germline variation of TNFAIP3 in primary Sjögren’s syndrome-associated lymphoma. Ann. Rheum. Dis. 2016;75:780–783. doi: 10.1136/annrheumdis-2015-207731. [DOI] [PubMed] [Google Scholar]
- 42.Nezos A, et al. TNFAIP3 F127C coding variation in Greek primary Sjogren’s syndrome patients. J. Immunol. Res. 2018;2018:6923213. doi: 10.1155/2018/6923213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nezos A, Evangelopoulos ME, Mavragani CP. Genetic contributors and soluble mediators in prediction of autoimmune comorbidity. J. Autoimmun. 2019;104:102317. doi: 10.1016/j.jaut.2019.102317. [DOI] [PubMed] [Google Scholar]
- 44.Goules AV, Tzioufas AG. Lymphomagenesis in Sjögren’s syndrome: predictive biomarkers towards precision medicine. Autoimmun. Rev. 2019;18:137–143. doi: 10.1016/j.autrev.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 45.Auclair G, Weber M. Mechanisms of DNA methylation and demethylation in mammals. Biochimie. 2012;94:2202–2211. doi: 10.1016/j.biochi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 46.Emamian ES, et al. Peripheral blood gene expression profiling in Sjögren’s syndrome. Genes Immun. 2009;10:285–296. doi: 10.1038/gene.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wildenberg ME, van Helden-Meeuwsen CG, van de Merwe JP, Drexhage HA, Versnel MA. Systemic increase in type I interferon activity in Sjögren’s syndrome: a putative role for plasmacytoid dendritic cells. Eur. J. Immunol. 2008;38:2024–2033. doi: 10.1002/eji.200738008. [DOI] [PubMed] [Google Scholar]
- 48.Gottenberg JE, et al. Activation of IFN pathways and plasmacytoid dendritic cell recruitment in target organs of primary Sjögren’s syndrome. Proc. Natl Acad. Sci. USA. 2006;103:2770–2775. doi: 10.1073/pnas.0510837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hjelmervik TO, Petersen K, Jonassen I, Jonsson R, Bolstad AI. Gene expression profiling of minor salivary glands clearly distinguishes primary Sjögren’s syndrome patients from healthy control subjects. Arthritis Rheum. 2005;52:1534–1544. doi: 10.1002/art.21006. [DOI] [PubMed] [Google Scholar]
- 50.Sun B, et al. DNA methylation perspectives in the pathogenesis of autoimmune diseases. Clin. Immunol. 2016;164:21–27. doi: 10.1016/j.clim.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 51.Imgenberg-Kreuz J, Sandling JK, Nordmark G. Epigenetic alterations in primary Sjögren’s syndrome – an overview. Clin. Immunol. 2018;196:12–20. doi: 10.1016/j.clim.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 52.Teruel M, et al. Integrative epigenomics in Sjögren´s syndrome reveals novel pathways and a strong interaction between the HLA, autoantibodies and the interferon signature. Sci. Rep. 2021;11:23292. doi: 10.1038/s41598-021-01324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imgenberg-Kreuz J, et al. Genome-wide DNA methylation analysis in multiple tissues in primary Sjögren’s syndrome reveals regulatory effects at interferon-induced genes. Ann. Rheum. Dis. 2016;75:2029–2036. doi: 10.1136/annrheumdis-2015-208659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altorok N, et al. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjögren’s syndrome. Arthritis Rheumatol. 2014;66:731–739. doi: 10.1002/art.38264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miceli-Richard C, et al. Overlap between differentially methylated DNA regions in blood B lymphocytes and genetic at-risk loci in primary Sjögren’s syndrome. Ann. Rheum. Dis. 2016;75:933–940. doi: 10.1136/annrheumdis-2014-206998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cole MB, et al. Epigenetic signatures of salivary gland inflammation in Sjögren’s syndrome. Arthritis Rheumatol. 2016;68:2936–2944. doi: 10.1002/art.39792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charras A, et al. Cell-specific epigenome-wide DNA methylation profile in long-term cultured minor salivary gland epithelial cells from patients with Sjögren’s syndrome. Ann. Rheum. Dis. 2017;76:625–628. doi: 10.1136/annrheumdis-2016-210167. [DOI] [PubMed] [Google Scholar]
- 58.Brække Norheim K, et al. Epigenome-wide DNA methylation patterns associated with fatigue in primary Sjögren’s syndrome. Rheumatology. 2016;55:1074–1082. doi: 10.1093/rheumatology/kew008. [DOI] [PubMed] [Google Scholar]
- 59.Chi C, et al. Hypomethylation mediates genetic association with the major histocompatibility complex genes in Sjögren’s syndrome. PLoS ONE. 2021;16:e0248429. doi: 10.1371/journal.pone.0248429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jonsson R. Disease mechanisms in Sjögren’s syndrome: what do we know? Scand. J. Immunol. 2022;95:e13145. doi: 10.1111/sji.13145. [DOI] [PubMed] [Google Scholar]
- 61.Bodewes ILA, Versnel MA. Interferon activation in primary Sjögren’s syndrome: recent insights and future perspective as novel treatment target. Expert Rev. Clin. Immunol. 2018;14:817–829. doi: 10.1080/1744666X.2018.1519396. [DOI] [PubMed] [Google Scholar]
- 62.Brkic Z, et al. Prevalence of interferon type I signature in CD14 monocytes of patients with Sjogren’s syndrome and association with disease activity and BAFF gene expression. Ann. Rheum. Dis. 2013;72:728–735. doi: 10.1136/annrheumdis-2012-201381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Björk A, et al. Protein and DNA methylation-based scores as surrogate markers for interferon system activation in patients with primary Sjögren’s syndrome. RMD Open. 2020;6:e000995. doi: 10.1136/rmdopen-2019-000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Imgenberg-Kreuz J, et al. DNA methylation-based interferon scores associate with sub-phenotypes in primary Sjögren’s syndrome. Front. Immunol. 2021;12:702037. doi: 10.3389/fimmu.2021.702037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazzari E, Jefferies CA. IRF5-mediated signaling and implications for SLE. Clin. Immunol. 2014;153:343–352. doi: 10.1016/j.clim.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 66.Nordmark G, et al. Additive effects of the major risk alleles of IRF5 and STAT4 in primary Sjögren’s syndrome. Genes. Immun. 2009;10:68–76. doi: 10.1038/gene.2008.94. [DOI] [PubMed] [Google Scholar]
- 67.Patel ZH, et al. A plausibly causal functional lupus-associated risk variant in the STAT1–STAT4 locus. Hum. Mol. Genet. 2018;27:2392–2404. doi: 10.1093/hmg/ddy140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raj P, et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. Elife. 2016;5:e12089. doi: 10.7554/eLife.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hagberg N, et al. The STAT4 SLE risk allele rs7574865[T] is associated with increased IL-12-induced IFN-γ production in T cells from patients with SLE. Ann. Rheum. Dis. 2018;77:1070–1077. doi: 10.1136/annrheumdis-2017-212794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang Y, et al. Therapeutic potential of tyrosine kinase 2 in autoimmunity. Expert Opin. Ther. Targets. 2014;18:571–580. doi: 10.1517/14728222.2014.892925. [DOI] [PubMed] [Google Scholar]
- 71.Hagberg N, Lundtoft C, Rönnblom L. Immunogenetics in systemic lupus erythematosus: transitioning from genetic associations to cellular effects. Scand. J. Immunol. 2020;92:e12894. doi: 10.1111/sji.12894. [DOI] [PubMed] [Google Scholar]
- 72.Manetti R, et al. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J. Exp. Med. 1993;177:1199–1204. doi: 10.1084/jem.177.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.D’Andrea A, et al. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J. Exp. Med. 1993;178:1041–1048. doi: 10.1084/jem.178.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huffman JE, et al. Multi-ancestry fine mapping implicates OAS1 splicing in risk of severe COVID-19. Nat. Genet. 2022;54:125–127. doi: 10.1038/s41588-021-00996-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim JK, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhao Y, Kang H, Ji Y, Chen X. Evaluate the relationship between polymorphisms of OAS1 gene and susceptibility to chronic hepatitis C with high resolution melting analysis. Clin. Exp. Med. 2013;13:171–176. doi: 10.1007/s10238-012-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Acosta-Herrera M, et al. Genome-wide meta-analysis reveals shared new loci in systemic seropositive rheumatic diseases. Ann. Rheum. Dis. 2019;78:311–319. doi: 10.1136/annrheumdis-2018-214127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc. Natl Acad. Sci. USA. 1995;92:6873–6877. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kearney SJ, et al. Type I IFNs downregulate myeloid cell IFN-γ receptor by inducing recruitment of an early growth response 3/NGFI-A binding protein 1 complex that silences Ifngr1 transcription. J. Immunol. 2013;191:3384–3392. doi: 10.4049/jimmunol.1203510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Boldin MP, et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mortazavi-Jahromi SS, Aslani M, Mirshafiey A. A comprehensive review on miR-146a molecular mechanisms in a wide spectrum of immune and non-immune inflammatory diseases. Immunol. Lett. 2020;227:8–27. doi: 10.1016/j.imlet.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 83.Pauley KM, et al. Altered miR-146a expression in Sjögren’s syndrome and its functional role in innate immunity. Eur. J. Immunol. 2011;41:2029–2039. doi: 10.1002/eji.201040757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zilahi E, et al. Increased microRNA-146a/b, TRAF6 gene and decreased IRAK1 gene expressions in the peripheral mononuclear cells of patients with Sjögren’s syndrome. Immunol. Lett. 2012;141:165–168. doi: 10.1016/j.imlet.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 85.Shi H, Zheng LY, Zhang P, Yu CQ. miR-146a and miR-155 expression in PBMCs from patients with Sjögren’s syndrome. J. Oral. Pathol. Med. 2014;43:792–797. doi: 10.1111/jop.12187. [DOI] [PubMed] [Google Scholar]
- 86.Chen JQ, et al. MicroRNA expression profiles identify disease-specific alterations in systemic lupus erythematosus and primary Sjögren’s syndrome. PLoS ONE. 2017;12:e0174585. doi: 10.1371/journal.pone.0174585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang X, et al. MicroRNA-146a-5p enhances T helper 17 cell differentiation via decreasing a disintegrin and metalloprotease 17 level in primary sjögren’s syndrome. Bioengineered. 2021;12:310–324. doi: 10.1080/21655979.2020.1870321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang-Renault SF, et al. Deregulation of microRNA expression in purified T and B lymphocytes from patients with primary Sjögren’s syndrome. Ann. Rheum. Dis. 2018;77:133–140. doi: 10.1136/annrheumdis-2017-211417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szabó K, Papp G, Szántó A, Tarr T, Zeher M. A comprehensive investigation on the distribution of circulating follicular T helper cells and B cell subsets in primary Sjögren’s syndrome and systemic lupus erythematosus. Clin. Exp. Immunol. 2016;183:76–89. doi: 10.1111/cei.12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tengnér P, Halse AK, Haga HJ, Jonsson R, Wahren-Herlenius M. Detection of anti-Ro/SSA and anti-La/SSB autoantibody-producing cells in salivary glands from patients with Sjögren’s syndrome. Arthritis Rheum. 1998;41:2238–2248. doi: 10.1002/1529-0131(199812)41:12<2238::AID-ART20>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 91.Nordmark G, et al. Association of EBF1, FAM167A(C8orf13)–BLK and TNFSF4 gene variants with primary Sjögren’s syndrome. Genes Immun. 2011;12:100–109. doi: 10.1038/gene.2010.44. [DOI] [PubMed] [Google Scholar]
- 92.Aqrawi LA, et al. Clinical associations and expression pattern of the autoimmunity susceptibility factor DIORA-1 in patients with primary Sjögren’s syndrome. Ann. Rheum. Dis. 2018;77:1840–1842. doi: 10.1136/annrheumdis-2018-213634. [DOI] [PubMed] [Google Scholar]
- 93.Mentlein L, et al. The rheumatic disease-associated FAM167A–BLK locus encodes DIORA-1, a novel disordered protein expressed highly in bronchial epithelium and alveolar macrophages. Clin. Exp. Immunol. 2018;193:167–177. doi: 10.1111/cei.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Aqrawi LA, et al. Diminished CXCR5 expression in peripheral blood of patients with Sjögren’s syndrome may relate to both genotype and salivary gland homing. Clin. Exp. Immunol. 2018;192:259–270. doi: 10.1111/cei.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Förster R, et al. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–1047. doi: 10.1016/S0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- 96.Chen W, Yang F, Xu G, Ma J, Lin J. Follicular helper T cells and follicular regulatory T cells in the immunopathology of primary Sjögren’s syndrome. J. Leukoc. Biol. 2021;109:437–447. doi: 10.1002/JLB.5MR1020-057RR. [DOI] [PubMed] [Google Scholar]
- 97.Wiley MM, et al. POS0096. Sjögren’s disease and systemic lupus erythematosus DDX6-CXCR5 risk intervals reveal common SNPs with functional significance in immune and salivary gland cells. Ann. Rheum. Dis. 2022;81:269–270. doi: 10.1136/annrheumdis-2022-eular.2503. [DOI] [Google Scholar]
- 98.Lumb JH, et al. DDX6 represses aberrant activation of interferon-stimulated genes. Cell Rep. 2017;20:819–831. doi: 10.1016/j.celrep.2017.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lien C, et al. Critical role of IRF-5 in regulation of B-cell differentiation. Proc. Natl Acad. Sci. USA. 2010;107:4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang CM, et al. Unique contribution of IRF-5–Ikaros axis to the B-cell IgG2a response. Genes Immun. 2012;13:421–430. doi: 10.1038/gene.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cruz-Tapias P, Rojas-Villarraga A, Maier-Moore S, Anaya JM. HLA and Sjögren’s syndrome susceptibility. A meta-analysis of worldwide studies. Autoimmun. Rev. 2012;11:281–287. doi: 10.1016/j.autrev.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 102.Trutschel D, et al. Variability in primary Sjögren’s syndrome is driven by interferon alpha, and genetically associated with the class II HLA DQ locus. Arthritis Rheumatol. 2022;74:1991–2002. doi: 10.1002/art.42265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oh DS, Lee HK. Autophagy protein ATG5 regulates CD36 expression and anti-tumor MHC class II antigen presentation in dendritic cells. Autophagy. 2019;15:2091–2106. doi: 10.1080/15548627.2019.1596493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brito-Zerón P, et al. Sjögren syndrome. Nat. Rev. Dis. Prim. 2016;2:16047. doi: 10.1038/nrdp.2016.47. [DOI] [PubMed] [Google Scholar]
- 105.Tzioufas AG, Kapsogeorgou EK, Moutsopoulos HM. Pathogenesis of Sjögren’s syndrome: what we know and what we should learn. J. Autoimmun. 2012;39:4–8. doi: 10.1016/j.jaut.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 106.Ye X, Zhou XJ, Zhang H. Exploring the role of autophagy-related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front. Immunol. 2018;9:2334. doi: 10.3389/fimmu.2018.02334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Colafrancesco S, et al. Autophagy occurs in lymphocytes infiltrating Sjögren’s syndrome minor salivary glands and correlates with histological severity of salivary gland lesions. Arthritis Res. Ther. 2020;22:238. doi: 10.1186/s13075-020-02317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith VE, Franklyn JA, McCabe CJ. Pituitary tumor-transforming gene and its binding factor in endocrine cancer. Expert Rev. Mol. Med. 2010;12:e38. doi: 10.1017/S1462399410001699. [DOI] [PubMed] [Google Scholar]
- 109.Martin R, et al. Caspase-mediated cleavage of raptor participates in the inactivation of mTORC1 during cell death. Cell Death Discov. 2016;2:16024. doi: 10.1038/cddiscovery.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Priem D, et al. A20 protects cells from TNF-induced apoptosis through linear ubiquitin-dependent and -independent mechanisms. Cell Death Dis. 2019;10:692. doi: 10.1038/s41419-019-1937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu Y, He X, Huang N, Yu J, Shao B. A20: a master regulator of arthritis. Arthritis Res. Ther. 2020;22:220. doi: 10.1186/s13075-020-02281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Konsta OD, et al. Defective DNA methylation in salivary gland epithelial acini from patients with Sjögren’s syndrome is associated with SSB gene expression, anti-SSB/LA detection, and lymphocyte infiltration. J. Autoimmun. 2016;68:30–38. doi: 10.1016/j.jaut.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 113.Mavragani CP, et al. Defective regulation of L1 endogenous retroelements in primary Sjogren’s syndrome and systemic lupus erythematosus: role of methylating enzymes. J. Autoimmun. 2018;88:75–82. doi: 10.1016/j.jaut.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhao K, et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J. Autoimmun. 2018;90:105–115. doi: 10.1016/j.jaut.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 115.Karagianni P, Goules AV, Tzioufas AG. Epigenetic alterations in Sjögren’s syndrome patient saliva. Clin. Exp. Immunol. 2020;202:137–143. doi: 10.1111/cei.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nielsen PR, Kragstrup TW, Deleuran BW, Benros ME. Infections as risk factor for autoimmune diseases – a nationwide study. J. Autoimmun. 2016;74:176–181. doi: 10.1016/j.jaut.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 117.Maitland N, Flint S, Scully C, Crean SJ. Detection of cytomegalovirus and Epstein–Barr virus in labial salivary glands in Sjogren’s syndrome and non-specific sialadenitis. J. Oral. Pathol. Med. 1995;24:293–298. doi: 10.1111/j.1600-0714.1995.tb01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wen S, et al. Association of Epstein–Barr virus (EBV) with Sjögren’s syndrome: differential EBV expression between epithelial cells and lymphocytes in salivary glands. Am. J. Pathol. 1996;149:1511–1517. [PMC free article] [PubMed] [Google Scholar]
- 119.Croia C, et al. Implication of Epstein–Barr virus infection in disease-specific autoreactive B cell activation in ectopic lymphoid structures of Sjögren’s syndrome. Arthritis Rheumatol. 2014;66:2545–2557. doi: 10.1002/art.38726. [DOI] [PubMed] [Google Scholar]
- 120.Klareskog L, Rönnelid J, Saevarsdottir S, Padyukov L, Alfredsson L. The importance of differences; on environment and its interactions with genes and immunity in the causation of rheumatoid arthritis. J. Intern. Med. 2020;287:514–533. doi: 10.1111/joim.13058. [DOI] [PubMed] [Google Scholar]
- 121.Mofors J, et al. Cigarette smoking patterns preceding primary Sjögren’s syndrome. RMD Open. 2020;6:e001402. doi: 10.1136/rmdopen-2020-001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Olsson P, Turesson C, Mandl T, Jacobsson L, Theander E. Cigarette smoking and the risk of primary Sjögren’s syndrome: a nested case control study. Arthritis Res. Ther. 2017;19:50. doi: 10.1186/s13075-017-1255-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stone DU, et al. Effect of tobacco smoking on the clinical, histopathological, and serological manifestations of Sjögren’s syndrome. PLoS ONE. 2017;12:e0170249. doi: 10.1371/journal.pone.0170249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gebreegziabher EA, et al. Associations between smoking and primary Sjögren syndrome classification using the Sjögren’s international collaborative clinical alliance cohort. ACR Open Rheumatol. 2022;4:231–237. doi: 10.1002/acr2.11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brito-Zerón P, et al. Influence of geolocation and ethnicity on the phenotypic expression of primary Sjögren’s syndrome at diagnosis in 8310 patients: a cross-sectional study from the Big Data Sjögren Project Consortium. Ann. Rheum. Dis. 2017;76:1042–1050. doi: 10.1136/annrheumdis-2016-209952. [DOI] [PubMed] [Google Scholar]
- 126.Brandt JE, Priori R, Valesini G, Fairweather D. Sex differences in Sjögren’s syndrome: a comprehensive review of immune mechanisms. Biol. Sex. Differ. 2015;6:19. doi: 10.1186/s13293-015-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klein SL, Flanagan KL. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 128.Scully EP, Haverfield J, Ursin RL, Tannenbaum C, Klein SL. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Takahashi T, Iwasaki A. Sex differences in immune responses. Science. 2021;371:347–348. doi: 10.1126/science.abe7199. [DOI] [PubMed] [Google Scholar]
- 130.Jansen R, et al. Sex differences in the human peripheral blood transcriptome. BMC Genomics. 2014;15:33. doi: 10.1186/1471-2164-15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lopes-Ramos CM, et al. Sex differences in gene expression and regulatory networks across 29 human tissues. Cell Rep. 2020;31:107795. doi: 10.1016/j.celrep.2020.107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Piasecka B, et al. Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges. Proc. Natl Acad. Sci. USA. 2018;115:E488–E497. doi: 10.1073/pnas.1714765115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Khramtsova EA, Davis LK, Stranger BE. The role of sex in the genomics of human complex traits. Nat. Rev. Genet. 2019;20:173–190. doi: 10.1038/s41576-018-0083-1. [DOI] [PubMed] [Google Scholar]
- 134.Hughes GC, Choubey D. Modulation of autoimmune rheumatic diseases by oestrogen and progesterone. Nat. Rev. Rheumatol. 2014;10:740–751. doi: 10.1038/nrrheum.2014.144. [DOI] [PubMed] [Google Scholar]
- 135.Brennan MT, et al. Sex steroid hormones in primary Sjögren’s syndrome. J. Rheumatol. 2003;30:1267–1271. [PubMed] [Google Scholar]
- 136.McCoy SS, Sampene E, Baer AN. Association of Sjögren’s syndrome with reduced lifetime sex hormone exposure: a case-control study. Arthritis Care Res. 2020;72:1315–1322. doi: 10.1002/acr.24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Valtysdóttir ST, Wide L, Hällgren R. Low serum dehydroepiandrosterone sulfate in women with primary Sjögren’s syndrome as an isolated sign of impaired HPA axis function. J. Rheumatol. 2001;28:1259–1265. [PubMed] [Google Scholar]
- 138.Forsblad-d’Elia H, Carlsten H, Labrie F, Konttinen YT, Ohlsson C. Low serum levels of sex steroids are associated with disease characteristics in primary Sjogren’s syndrome; supplementation with dehydroepiandrosterone restores the concentrations. J. Clin. Endocrinol. Metab. 2009;94:2044–2051. doi: 10.1210/jc.2009-0106. [DOI] [PubMed] [Google Scholar]
- 139.Kovats S. Estrogen receptors regulate innate immune cells and signaling pathways. Cell Immunol. 2015;294:63–69. doi: 10.1016/j.cellimm.2015.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chatzis LG, Goules AV, Tzioufas AG. Searching for the “X factor” in Sjögren’s syndrome female predilection. Clin. Exp. Rheumatol. 2021;39:206–214. doi: 10.55563/clinexprheumatol/88dyrn. [DOI] [PubMed] [Google Scholar]
- 141.Harris VM, et al. Klinefelter’s syndrome (47,XXY) is in excess among men with Sjögren’s syndrome. Clin. Immunol. 2016;168:25–29. doi: 10.1016/j.clim.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu K, et al. X chromosome dose and sex bias in autoimmune diseases: increased prevalence of 47,XXX in systemic lupus erythematosus and Sjögren’s syndrome. Arthritis Rheumatol. 2016;68:1290–1300. doi: 10.1002/art.39560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Youness A, Miquel CH, Guéry JC. Escape from X chromosome inactivation and the female predominance in autoimmune diseases. Int. J. Mol. Sci. 2021;22:1114. doi: 10.3390/ijms22031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Souyris M, et al. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018;3:eaap8855. doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- 145.Odhams CA, et al. Interferon inducible X-linked gene CXorf21 may contribute to sexual dimorphism in systemic lupus erythematosus. Nat. Commun. 2019;10:2164. doi: 10.1038/s41467-019-10106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat. Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brown GJ, et al. TLR7 gain-of-function genetic variation causes human lupus. Nature. 2022;605:349–356. doi: 10.1038/s41586-022-04642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Heinz LX, et al. TASL is the SLC15A4-associated adaptor for IRF5 activation by TLR7–9. Nature. 2020;581:316–322. doi: 10.1038/s41586-020-2282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boraska V, et al. Genome-wide meta-analysis of common variant differences between men and women. Hum. Mol. Genet. 2012;21:4805–4815. doi: 10.1093/hmg/dds304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lindén M, et al. Sex influences eQTL effects of SLE and Sjögren’s syndrome-associated genetic polymorphisms. Biol. Sex. Differ. 2017;8:34. doi: 10.1186/s13293-017-0153-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Yao C, et al. Sex- and age-interacting eQTLs in human complex diseases. Hum. Mol. Genet. 2014;23:1947–1956. doi: 10.1093/hmg/ddt582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kukurba KR, et al. Impact of the X chromosome and sex on regulatory variation. Genome Res. 2016;26:768–777. doi: 10.1101/gr.197897.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Dimas AS, et al. Sex-biased genetic effects on gene regulation in humans. Genome Res. 2012;22:2368–2375. doi: 10.1101/gr.134981.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Oliva M, et al. The impact of sex on gene expression across human tissues. Science. 2020;369:eaba3066. doi: 10.1126/science.aba3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Stein MM, et al. Sex-specific differences in peripheral blood leukocyte transcriptional response to LPS are enriched for HLA region and X chromosome genes. Sci. Rep. 2021;11:1107. doi: 10.1038/s41598-020-80145-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kamitaki N, et al. Complement genes contribute sex-biased vulnerability in diverse disorders. Nature. 2020;582:577–581. doi: 10.1038/s41586-020-2277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ghorai A, Ghosh U. miRNA gene counts in chromosomes vary widely in a species and biogenesis of miRNA largely depends on transcription or post-transcriptional processing of coding genes. Front. Genet. 2014;5:100. doi: 10.3389/fgene.2014.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Singmann P, et al. Characterization of whole-genome autosomal differences of DNA methylation between men and women. Epigenetics Chromatin. 2015;8:43. doi: 10.1186/s13072-015-0035-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Khan D, Dai R, Ansar Ahmed S. Sex differences and estrogen regulation of miRNAs in lupus, a prototypical autoimmune disease. Cell Immunol. 2015;294:70–79. doi: 10.1016/j.cellimm.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 161.Shen JJ, Wang YF, Yang W. Sex-interacting mRNA- and miRNA-eQTLs and their implications in gene expression regulation and disease. Front. Genet. 2019;10:313. doi: 10.3389/fgene.2019.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mariette X, et al. Inefficacy of infliximab in primary Sjögren’s syndrome: results of the randomized, controlled Trial of Remicade in Primary Sjögren’s Syndrome (TRIPSS) Arthritis Rheum. 2004;50:1270–1276. doi: 10.1002/art.20146. [DOI] [PubMed] [Google Scholar]
- 163.Bowman SJ, et al. Randomized controlled trial of rituximab and cost-effectiveness analysis in treating fatigue and oral dryness in primary Sjögren’s syndrome. Arthritis Rheumatol. 2017;69:1440–1450. doi: 10.1002/art.40093. [DOI] [PubMed] [Google Scholar]
- 164.Devauchelle-Pensec V, et al. Treatment of primary Sjögren syndrome with rituximab: a randomized trial. Ann. Intern. Med. 2014;160:233–242. doi: 10.7326/M13-1085. [DOI] [PubMed] [Google Scholar]
- 165.Baer AN, et al. Efficacy and safety of abatacept in active primary Sjögren’s syndrome: results of a phase III, randomised, placebo-controlled trial. Ann. Rheum. Dis. 2020;80:339–348. doi: 10.1136/annrheumdis-2020-218599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Felten R, et al. Interleukin 6 receptor inhibition in primary Sjögren syndrome: a multicentre double-blind randomised placebo-controlled trial. Ann. Rheum. Dis. 2021;80:329–338. doi: 10.1136/annrheumdis-2020-218467. [DOI] [PubMed] [Google Scholar]
- 167.Bowman SJ, et al. Safety and efficacy of subcutaneous ianalumab (VAY736) in patients with primary Sjögren’s syndrome: a randomised, double-blind, placebo-controlled, phase 2b dose-finding trial. Lancet. 2022;399:161–171. doi: 10.1016/S0140-6736(21)02251-0. [DOI] [PubMed] [Google Scholar]
- 168.Seror R, et al. Development and preliminary validation of the Sjögren’s tool for assessing response (STAR): a consensual composite score for assessing treatment effect in primary Sjögren’s syndrome. Ann. Rheum. Dis. 2022;81:979–989. doi: 10.1136/annrheumdis-2021-222054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Arends S, et al. Composite of relevant endpoints for Sjögren’s syndrome (CRESS): development and validation of a novel outcome measure. Lancet Rheumatol. 2021;3:e553–e562. doi: 10.1016/S2665-9913(21)00122-3. [DOI] [PubMed] [Google Scholar]
- 170.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT04496960 (2023).
- 171.Barrera MJ, et al. Tofacitinib counteracts IL-6 overexpression induced by deficient autophagy: implications in Sjögren’s syndrome. Rheumatology. 2021;60:1951–1962. doi: 10.1093/rheumatology/keaa670. [DOI] [PubMed] [Google Scholar]
- 172.Bai W, et al. Pilot study of baricitinib for active Sjogren’s syndrome. Ann. Rheum. Dis. 2022;81:1050–1052. doi: 10.1136/annrheumdis-2021-222053. [DOI] [PubMed] [Google Scholar]
- 173.Gonciarz M, Pawlak-Buś K, Leszczyński P, Owczarek W. TYK2 as a therapeutic target in the treatment of autoimmune and inflammatory diseases. Immunotherapy. 2021;13:1135–1150. doi: 10.2217/imt-2021-0096. [DOI] [PubMed] [Google Scholar]
- 174.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT05617677 (2023).
- 175.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT04093531 (2022).
- 176.Furie R, et al. Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J. Clin. Invest. 2019;129:1359–1371. doi: 10.1172/JCI124466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Morand EF, et al. Trial of anifrolumab in active systemic lupus erythematosus. N. Engl. J. Med. 2020;382:211–221. doi: 10.1056/NEJMoa1912196. [DOI] [PubMed] [Google Scholar]
- 178.Deeks ED. Anifrolumab: first approval. Drugs. 2021;81:1795–1802. doi: 10.1007/s40265-021-01604-z. [DOI] [PubMed] [Google Scholar]
- 179.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT05383677 (2022).
- 180.Fanouriakis A, Tziolos N, Bertsias G, Boumpas DT. Update οn the diagnosis and management of systemic lupus erythematosus. Ann. Rheum. Dis. 2021;80:14–25. doi: 10.1136/annrheumdis-2020-218272. [DOI] [PubMed] [Google Scholar]
- 181.Mariette X, et al. Efficacy and safety of belimumab in primary Sjögren’s syndrome: results of the BELISS open-label phase II study. Ann. Rheum. Dis. 2015;74:526–531. doi: 10.1136/annrheumdis-2013-203991. [DOI] [PubMed] [Google Scholar]
- 182.Mariette X, et al. A randomized, phase II study of sequential belimumab and rituximab in primary Sjögren’s syndrome. JCI Insight. 2022;7:e163030. doi: 10.1172/jci.insight.163030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xu D, et al. Efficacy and safety of telitacicept in primary Sjögren’s syndrome: a randomized, double-blind, placebo-controlled, phase 2 trial. Arthritis Rheumatol. 2022;74(Suppl. 9):Abstr. 2031. doi: 10.1093/rheumatology/kead265. [DOI] [PubMed] [Google Scholar]
- 184.US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT05673993 (2023).
- 185.van Nimwegen JF, et al. Abatacept treatment for patients with early active primary Sjogren’s syndrome: a single-centre, randomised, double-blind, placebo-controlled, phase 3 trial (ASAP-III study) Lancet Rheumatol. 2020;2:E153–E163. doi: 10.1016/S2665-9913(19)30160-2. [DOI] [PubMed] [Google Scholar]
- 186.Fisher BA, et al. Assessment of the anti-CD40 antibody iscalimab in patients with primary Sjogren’s syndrome: a multicentre, randomised, double-blind, placebo-controlled, proof-of-concept study. Lancet Rheumatol. 2020;2:E142–E152. doi: 10.1016/S2665-9913(19)30135-3. [DOI] [PubMed] [Google Scholar]
- 187.Retamozo S, Sisó-Almirall A, Flores-Chávez A, Ramos-Casals M, Brito-Zerón P. An update of targeted therapeutic options for primary Sjögren syndrome: current status and future development. Expert Opin. Pharmacother. 2021;22:2359–2371. doi: 10.1080/14656566.2021.1951224. [DOI] [PubMed] [Google Scholar]
- 188.Mariette X BM, et al. A phase 2a study of MEDI5872 (AMG557), a fully human anti-ICOS ligand monoclonal antibody in patients with primary Sjögren’s syndrome. Arthritis Rheumatol. 2019;71(Suppl. 10):Abstr. 2417. [Google Scholar]
- 189.Imgenberg-Kreuz J, et al. Transcription profiling of peripheral B cells in antibody-positive primary Sjögren’s syndrome reveals upregulated expression of CX3CR1 and a type I and type II interferon signature. Scand. J. Immunol. 2018;87:e12662. doi: 10.1111/sji.12662. [DOI] [PubMed] [Google Scholar]
- 190.Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton’s tyrosine kinase in B cell malignancies. Nat. Rev. Cancer. 2014;14:219–232. doi: 10.1038/nrc3702. [DOI] [PubMed] [Google Scholar]
- 191.Griffin DD, Dolen WK. B cell disorders in children: part II. Curr. Allergy Asthma Rep. 2020;20:64. doi: 10.1007/s11882-020-00963-z. [DOI] [PubMed] [Google Scholar]
- 192.Kil LP, et al. Btk levels set the threshold for B-cell activation and negative selection of autoreactive B cells in mice. Blood. 2012;119:3744–3756. doi: 10.1182/blood-2011-12-397919. [DOI] [PubMed] [Google Scholar]
- 193.Dörner T, et al. Remibrutinib (LOU064) in Sjögren’s syndrome: safety and efficacy results from a 24-week placebo-controlled proof-of-concept study. Arthritis Rheumatol. 2022;74(Suppl. 9):Abstr. 1113. [Google Scholar]
- 194.Price E, et al. Safety and efficacy of filgotinib, lanraplenib, and tirabrutinib in Sjögren’s syndrome: randomised, phase 2, double-blind, placebo-controlled study. Rheumatology. 2022;61:4797–4808. doi: 10.1093/rheumatology/keac167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pozsgay J, Szekanecz Z, Sármay G. Antigen-specific immunotherapies in rheumatic diseases. Nat. Rev. Rheumatol. 2017;13:525–537. doi: 10.1038/nrrheum.2017.107. [DOI] [PubMed] [Google Scholar]
- 196.Clemente-Casares X, et al. Expanding antigen-specific regulatory networks to treat autoimmunity. Nature. 2016;530:434–440. doi: 10.1038/nature16962. [DOI] [PubMed] [Google Scholar]
- 197.Wahren-Herlenius M, Muller S, Isenberg D. Analysis of B-cell epitopes of the Ro/SS-A autoantigen. Immunol. Today. 1999;20:234–240. doi: 10.1016/S0167-5699(99)01458-9. [DOI] [PubMed] [Google Scholar]
- 198.Verstappen GM, Pringle S, Bootsma H, Kroese FGM. Epithelial-immune cell interplay in primary Sjögren syndrome salivary gland pathogenesis. Nat. Rev. Rheumatol. 2021;17:333–348. doi: 10.1038/s41584-021-00605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Benschop RJ, et al. Development of tibulizumab, a tetravalent bispecific antibody targeting BAFF and IL-17A for the treatment of autoimmune disease. MAbs. 2019;11:1175–1190. doi: 10.1080/19420862.2019.1624463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer. 2019;19:151–161. doi: 10.1038/s41568-019-0109-9. [DOI] [PubMed] [Google Scholar]
- 201.Cornacchia E, et al. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol. 1988;140:2197–2200. doi: 10.4049/jimmunol.140.7.2197. [DOI] [PubMed] [Google Scholar]
- 202.He Y, Sawalha AH. Drug-induced lupus erythematosus: an update on drugs and mechanisms. Curr. Opin. Rheumatol. 2018;30:490–497. doi: 10.1097/BOR.0000000000000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat. Rev. Genet. 2021;22:119–131. doi: 10.1038/s41576-020-00295-8. [DOI] [PubMed] [Google Scholar]
- 204.Rusinova I, et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013;41:D1040–D1046. doi: 10.1093/nar/gks1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Sollis E, et al. The NHGRI-EBI GWAS Catalog: knowledgebase and deposition resource. Nucleic Acids Res. 2023;51:D977–D985. doi: 10.1093/nar/gkac1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–2557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.