Abstract
Porphyromonas gingivalis (P. gingivalis) is considered to be one of the main periodontal pathogens. The goal of this work was to confirm the ability of P. gingivalis to invade host cells. We detected P. gingivalis inside KB cells by confocal microscopy and analyzed the various aspects of the adherence and internalization process. Lysates of P. gingivalis-infected KB cells were also examined using anaerobic growth techniques. The results showed the viability and ability to replicate, inside the host cells, of the internalized pathogen. The production of vesicles was also tracked for the first time. Confocal microscopy revealed P. gingivalis in a perinuclear position.
The literature is unanimous in assigning Porphyromonas gingivalis (P. gingivalis) a role as a major periodontal pathogen (17, 39). While it occasionally expresses its virulence on its own, P. gingivalis more commonly acts in cooperation with other microorganisms (21, 23). P. gingivalis is an opportunistic pathogen (22, 24) which can express an outstanding arsenal of virulence factors (6). As with many enteropathogens (5, 7, 26, 32), invasion of host cells seems to be an important strategy used by P. gingivalis to protect itself against the host immune system and to advance through tissues (36). A possible connection has been made between the incidence of periodontal infections in pregnant women and an increased risk of their having preterm low-birth-weight babies (31). P. gingivalis has also been cited as a potential etiological factor in myocardial infarction and atherosclerosis (2, 27). The internalization was first quantified, and the ability of this pathogen to multiply within the eukaryotic cells was assessed. The survival conditions in the host cell were then determined.
Epithelial cell growth.
KB cells (ATCC CL17) derived from an epidermoid cancer of the oral cavity were used. The cells were inoculated into 24-well macroplates at a concentration of 105 cells/ml. The growth medium was replaced every day to maintain confluent cultures. For confocal microscopy, the cells were inoculated at the same concentration in glass culture dishes coated with 1% collagen I.
Bacterial growth.
P. gingivalis ATCC 33277 was maintained on blood agar plates in an anaerobic chamber at 37°C. Todd-Hewitt broth cultures were inoculated 48 h prior to each experiment.
Bacterial contamination.
The epithelial cells were washed twice with unsupplemented RPMI 1640 and covered with 500 μl of a bacterial suspension for 1 h at 37°C. The bacterial concentration was adjusted by dilution in phosphate-buffered saline (PBS) containing 1 mM β2-mercaptoethanol to a ratio of 100 bacteria per epithelial cell as determined by optical density. RPMI alone and bacteria alone served as negative controls.
Cell lysate culturing.
The epithelial cells were washed and incubated with 500 μl of metronidazole (100 μg/ml) for 3 h (25). The cells were lysed in 1 ml of sterile distilled water for 15 min. The lysates were serially diluted, and 200 μl of each dilution was spread on a blood agar plate. The plates were incubated in an anaerobic chamber at 37°C to quantify the level of bacterial invasion by counting the number of CFU. The CFU were determined on days 0, 1, 2, 3, 4, and 5. The results were analyzed by repeated measures of one-way analysis of variance using Statview 5.5.
Locating P. gingivalis using confocal microscopy.
For all microscopic protocols, the samples were fixed in 1% paraformaldehyde in PBS for 30 min. After washing, the samples were sealed between a slide and coverslip using Mounting Medium (Sigma).
SYBR and propidium iodide treatment.
The bacteria were centrifuged, washed with PBS, and incubated in an SYBR II solution (10−4) for 15 min. After the contamination and fixation procedures, the epithelial cells were incubated for 5 min with 2 μM propidium iodide prepared in PBS plus 0.2% Tween 20 and washed with PBT (PBS–0.2% bovine serum albumin–0.1% Tween 20).
Phalloidin-FITC and anti-P. gingivalis rhodamine antibody.
The cytoskeleton was visualized using phalloidin-fluorescein isothiocyanate (FITC). The samples were saturated with PBS containing 1% bovine serum albumin and 0.1% Tween 20 for 20 min with agitation. After removing the supernatant, rabbit anti-P. gingivalis antiserum was added and the plates were incubated for 20 min. The negative control wells contained PBT. After washing with PBT, 250 μl of the secondary rhodamine-coupled antibody was added together with 250 μl of phalloidin-FITC. The plates were incubated with agitation then rinsed.
Acridine orange and crystal violet.
The bacteria were stained with 0.01% acridine orange in PBS for 45 s as per the protocol of Miliotis (28). The cells were contaminated as described previously. After a wash, the KB cells were incubated with 0.01% crystal violet in PBS for 45 s to quench extracellular fluorescence and then washed.
DiO and anti-P. gingivalis rhodamine-conjugated antibody.
Marcia's (14) protocol was used. Prior to contamination, KB cells were incubated with DiO (3,3′-dioctadecyloxacarbocyanine perchlorate), a lipophilic membrane marker. It was diluted in culture medium to obtain a final concentration of 170 μg/ml. The KB cells were incubated at 37°C for 90 min and then washed in RPMI. Immunodetection of P. gingivalis was carried out as described previously.
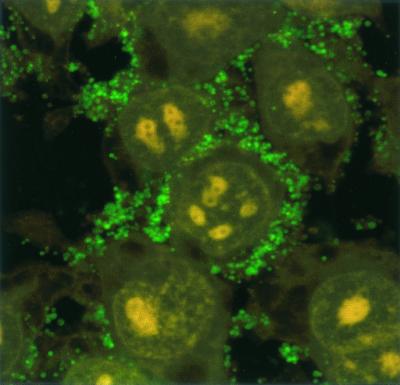
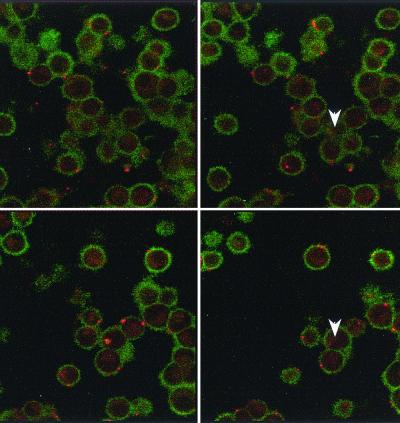
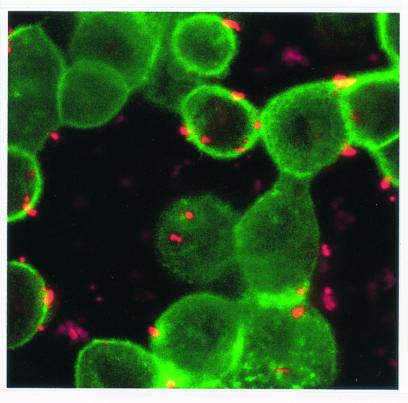
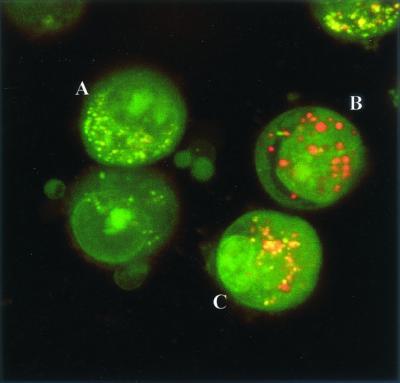
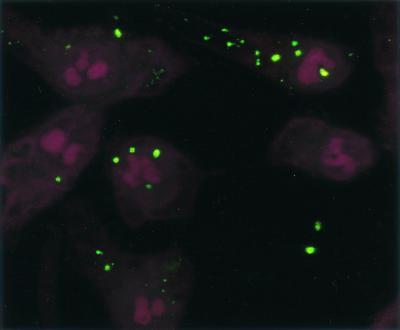
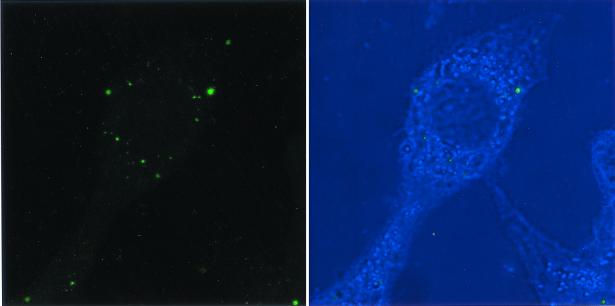
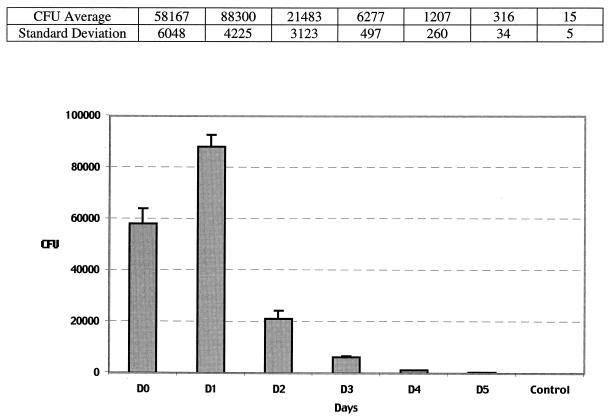
Confocal microscopy showed numerous SYBR II-stained bacteria adhering to the cell surface (Fig. 1). While a few bacteria could be seen at some distance from the epithelial cells, most appeared tightly attached to the cell membrane. The intracytoplasmic location of the bacteria was visualized by superimposing phase contrast and tetramethyl rhodamine isocyanate images of serial sections (Fig. 2). Phalloidin-FITC was used to reveal the condensation of membrane actin where the bacteria are attached to the epithelial cell (Fig. 3). The membrane modifications associated with the internalization process were confirmed by using DiO in combination with bacterial immunolabeling. The membrane invagination enveloping the bacteria being internalized was clearly visible. The bacteria appeared attached to the cell surface. The membrane was curved inwards and was thickened at the point of contact. So, a large number of bacteria were clearly visible inside the epithelial cell. While some bacterial cells were dispersed throughout the cytoplasm, they were located mainly in the perinuclear space. Once inside the cell, the bacteria appeared to either float freely in the cytoplasm or persist in the endocytotic vacuole. The vacuoles were marked with DiO and appeared green. The bacteria were immunolabeled with rhodamine and appeared red. Figure 4 shows a mixture of cells with and without internalized bacteria. The serial sections (0.75 μm) enable adherent bacteria to be differentiated from internalized bacteria. The internalized bacteria appeared red when free in the cytoplasm and orange (fluorochrome codetection) when in endocytotic vacuoles. The bacterial viability was assessed using the protocol of Miliotis (28). Dead bacteria stain red because their DNA is denatured. However, as can be seen in Fig. 5, the bacteria all appeared green, indicating that they remained viable. Confocal microscopic examinations of specimens where both the bacteria and the contents of the epithelial cells were marked showed the presence of internalized bacteria over a span of 5 days (D0 to D5). Twenty-four hours after the contamination, a large number of bacteria could already be seen in the cytoplasm, mainly in the perinuclear space, with no apparent modification of the cell (Fig. 6). After 48 h, there appeared to be fewer bacteria, while the epithelial cells were beginning to show signs of distress, with significant vacuolization of the cytoplasm. At high magnification, however, it could be seen that these cytoplasmic vacuoles, which seemed to occupy most of the cytosol, did not intrude into the spaces occupied by the small vesicles, whose staining indicated that they were of bacterial origin (Fig. 7). The persistence of viable bacteria within the epithelial cells was assessed by CFU counts obtained by inoculating lysates of contaminated cells on blood agar plates. The average number of internalized viable bacteria for all experiments was 58,167 ± 6,046 CFU (0.6% of the initial inoculum). When blood agar plates were inoculated with the contents of the wells that contained only bacteria, no CFU were detected, indicating the effectiveness of a 100-μg/ml dose of metronidazole and revealing no bacterial survival without KB cells. Twenty-four hours after contamination, the number of CFU was 52% higher with respect to day 0 (P < 0.05), indicating that the bacteria were able to multiply inside the KB cells. Between days 2 and 5 (P < 0.05), there was a gradual drop in the number of CFU, indicating a drop in bacterial viability. The survival rate dropped from 0.4% on day 3 to 0.005% on day 5 (Fig. 8).
FIG. 1.
Confocal photomicrograph. Adherence of P. gingivalis to KB epithelial cells is shown. The bacteria are stained with SYBR II (green), and the cell nuclei are stained with propidium iodide (red).
FIG. 2.
Confocal photomicrograph: the serial sections (0.75 μm) show the presence of P. gingivalis in the cytoplasm.
FIG. 3.
The internalization of P. gingivalis is accompanied by actin condensation (phalloidin-FITC) at the point of invagination.
FIG. 4.
(A) A cell with no internalized bacteria. The endocytic vesicles are green. (B) The serial sections allow the adherent red-stained bacteria to be visualized. (C) The internalized bacteria are either stained red (free in cytoplasm) or orange (in vesicle).
FIG. 5.
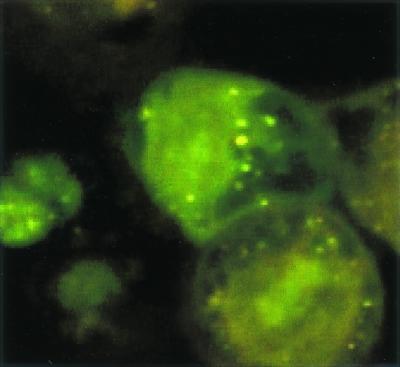
Viability of the internalized bacteria as shown by acridine orange staining. The fluorescence of the adhering bacteria is quenched by using crystal violet.
FIG. 6.
Confocal photomicrograph. Immunolabeling of P. gingivalis reveals the presence of numerous bacteria in each cell on day 2.
FIG. 7.
On day 3, the epithelial cell shows signs of significant vacuole formation, while the presence of bacteria and bacterial vesicles is revealed by immunolabeling.
FIG. 8.
Intracellular viability of P. gingivalis over time (measured as the number of CFU). Control, for each time, the number of CFU recovered from the bacterial suspension without KB cells (negligible CFU number).
The adherence mechanism is mediated by a certain number of factors grouped together under the general heading of adhesins. These include external membrane lipopolysaccharides, hemagglutinins, and various proteases, as well as fimbriae, which are also considered as a virulence factor (1, 9, 20, 40). Dorn et al. (8) recently reported no relationship between the ability to adhere and the ability to invade. We used confocal microscopy to demonstrate that P. gingivalis first adhered to and then entered the KB cells. The technique involved (i) the use of antibody specifically directed against P. gingivalis to detect the internalized bacteria and (ii) the parallel use of cell component markers to help locate the precise position of P. gingivalis in the cytoplasm. Confocal microscopy, unlike the transmission electron microscopy techniques used by most authors (10, 21, 25, 38), allows entire cells to be examined and the integrity of the cell membrane to be ascertained. Finlay (11) described the great advantage of using confocal microscopy for studying host-parasite interactions because of the three-dimensional reconstructions that allow molecular events during infection processes to be analyzed. The fact that a larger number of adherent bacteria was observed than typically seen in transmission electron microscopy images was most likely due to the gentler protocol we used. The results presented here corroborate those of Huard-Delcourt et al. (15). The bacterial invasion of cells that are not professional phagocytes involves certain events common to many pathogens, notably (i) the recognition of specific receptors on the eukaryotic cell surface, (ii) the transduction of molecular signals from the membrane to various intracellular locations in the host cell, and (iii) the rearrangement of the cytoskeleton to allow the entry of the bacteria via endocytotic vacuoles (12). In the case of P. gingivalis, internalization takes place according to the following schema: adherence of the bacterium to the epithelial cell is induced by bacterium-host cell molecular signals, formation of a membrane invagination that surrounds the bacterium, and rapid internalization of the bacterium (30). The formation of endocytotic vacuoles by the cell membrane involves a rearrangement of the cytoskeleton, which is an active phenomenon requiring the participation of the eukaryotic cell and a viable bacterium. Sandros et al. (37) suggests that the internalization process is more likely mediated by microtubules than microfilaments, whose condensation is induced by fimbriae (30). The specific receptors of these adhesins are well known for professional eukaryotic phagocytes, such as macrophages (41): the β2 integrin (CD11/CD18). On the other hand, no receptors have been identified for cells that are not professional phagocytes. While the membrane receptor of KB cells for P. gingivalis fimbriae has not yet been identified, Park and Lamont (33) have reported the neosynthesis of class III proteins as soon as a P. gingivalis bacterium attaches to the epithelial cell. These enable internalized bacteria to act directly on the metabolism of the cell being invaded. The internalization of P. gingivalis is also correlated with the tyrosine phosphorylation of a eukaryotic cell protein (37) corresponding to the mitogen-activated protein kinase involved in the internalization processes of other pathogens (4, 35). It has also been shown that P. gingivalis induces an increase in intracellular calcium flux. Calcium ions are also involved in numerous intercellular interactions (16). Contamination times vary greatly, ranging from a few minutes to 1 or 2 h (3, 25). Since the speed of P. gingivalis internalization did not enter into our study, we settled on a contamination time of 1 h. Adherence and internalization are more efficient with epithelial cells from primary gingival tissue cultures, as shown by Belton (3), who obtained a 90% contamination rate after 12 min. However, we decided to use the established KB cell line, whose proliferation rate and viability are constant, thus providing more reproducible results. While many articles have been published on the adherence and internalization mechanisms of P. gingivalis, the fate of the bacterium once inside the eukaryotic cell has not been totally elucidated. Most recent studies report that P. gingivalis survives and proliferates inside epithelial cells in vitro (25), but the life span of the endocytotic vacuole has not been established. The bacteria remain viable after internalization, as shown by the acridine orange staining, and the production of intracellular vesicles is additional proof of their viability. The presence of the pathogen inside the vacuoles was confirmed using a dextran-Texas red conjugate. There are several explanations for this behavior: P. gingivalis acts to prevent lysosomal fusion, inhibits proton pumps, thus preventing acidification of the vacuoles, or possesses mechanisms that enable it to survive in a low-pH environment. More work will be required to shed additional light on the survival strategies used by this anaerobic bacterium. Our study revealed for the first time the intracellular production of bacterial vesicles. These vesicles contain large quantities of proteolytic material (27) because they are formed from extrusions of the external membrane of the bacterium. When released into the extracellular environment, they act both as decoys that thwart the host immune system and as remote weapons that project the virulence of the bacterium through their enzymatic contents. As with previous studies (3, 25), significant cytoplasmic vacuole formation was noted on day 2. This sign of distress could be a reflection of damage to the epithelial cell caused by the P. gingivalis vesicles. Our study has confirmed that the pathogen is found mainly in the perinuclear space (3). According to Belton, this may be due to interactions between the bacterium and cell organites or the presence of the pathogen in late endosomes, which are located in the perinuclear space. Confocal microscopic observations subsequent to day 0 showed that the viability of the pathogen within the eucaryotic cell was closely related to the life span of the endocytotic vacuoles. Lamont and Jenkinson (20) have reported that the vacuoles are lysed very soon after the entry of the bacterium into the cell. With Shigella flexneri, this event is correlated with the capacity to replicate and express a motility phenotype linked to the production of a surface protein (IcsA). IcsA directs the polymerization of actin filaments that act as flagella (34). On the other hand, Njoroge et al. (30) have reported that the vacuoles are not immediately lysed. It has been suggested that cell invasion not only protects the bacterium against the host immune system but also allows transcytosis, leading to the invasion of underlying tissue. This invasion model, which has been accepted for a number of internalizing bacterial species (13, 34), would also seem to apply to P. gingivalis. Recent studies have suggested that P. gingivalis can invade the lower layers of connective tissue via a paracellular pathway by degrading cell junction complexes (18). Some 0.6% of the initial CFU were recovered after lysis of the contaminated KB cells, which is very close to the 0.65% obtained by Sandros et al. (38). Njoroge et al. (30) and Duncan et al. (10), however, reported lower levels of internalization, i.e., 0.02% and 0.07%, respectively. Only Madianos et al. (25) reported extremely high levels of bacterial internalization (60%) on day 0. Most studies used essentially the same ratio, i.e., 100 bacteria per cell (10, 30, 38), while Madianos et al. (25) used a very low ratio of 2 bacteria per cell. This suggests that epithelial cells that are subjected to less aggression internalize more bacteria. P. gingivalis did not survive without cells. The bacterial suspension, without KB cells, did not lead to any CFU during the entire experiments. These results are in agreement with the findings of previous studies (38), which revealed no bacterial survival in the absence of epithelial cells. Moreover, Madianos et al. (25) demonstrated that the supernatant counts remained low and never exceeded 5% of the intracellular counts. All these results demonstrated that CFU recovered from infected KB cells represent only the intracellular P. gingivalis. Bacterial multiplication occurred between days 0 and 2. Several authors have described the intracellular replication of a number of pathogens, including Salmonella enterica serovar Typhimurium (19) and Vibrio hollisae (29). We observed an increase in the number of bacteria on day 1, after which the number of CFU dropped. This early multiplication of the internalized bacteria may thwart the antibacterial mechanisms of the contaminated cell and thus allow a sufficient number of pathogens to survive to maintain a recurring infection. The production of toxic antibacterial substances by the invaded cell may also, however, explain the gradual reduction in the number of viable internalized bacteria. Another hypothesis is that the pathogen may induce a message leading to cell death. However, Madianos et al. (25) have described a bacterial stress that would explain why intracellular multiplication begins only after day 2. On the other hand, they also reported a gradual drop in numbers until day 8. This difference with respect to our results could be due to the small bacterial inoculum they used. Furthermore, the in vitro model does not reproduce all in vivo events, and pathogen-host cell interactions may thus be modified. This work confirms that P. gingivalis adheres to and enters KB cells. The pathogen is able to survive and multiply either free in the cytoplasm or within endocytotic vacuoles. This mechanism allows P. gingivalis to avoid host defenses and penetrate the epithelium, thus causing infections in deeper tissue layers. The signs of cell suffering observed confirm the cytotoxicity of the pathogen and/or its vesicles, which provoke superficial periodontitis by lysing keratinocytes.
Acknowledgments
We thank Roselyne Primault for help with the confocal microscopy and Gene Bourgeau and Céline Allaire for editorial assistance.
This study was supported by the Fondation Langlois, the Institut Français de Recherche Odontologique, and the Conseil Régional de Bretagne.
REFERENCES
- 1.Amano A, Shizukuishi S, Horie H, Kimura S, Morisaki I, Hamada S. Binding of Porphyromonas gingivalis fimbriae to proline-rich glycoproteins in parotid saliva via a domain shared by major salivary components. Infect Immun. 1998;66:2072–2077. doi: 10.1128/iai.66.5.2072-2077.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck J, Garcia R, Heiss G, Vokonas P S, Offenbacher S. Periodontal disease and cardiovascular disease. J Periodontol. 1996;67:1123–1137. doi: 10.1902/jop.1996.67.10s.1123. [DOI] [PubMed] [Google Scholar]
- 3.Belton C M. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. 1999;1:215–223. doi: 10.1046/j.1462-5822.1999.00022.x. [DOI] [PubMed] [Google Scholar]
- 4.Cossart P, Boquet P, Normark S, Rappuoli R. Cellular microbiology emerging. Science. 1996;271:315–316. doi: 10.1126/science.271.5247.315. [DOI] [PubMed] [Google Scholar]
- 5.Danielova V, Holubova J, Schramlova J, Sobotkova J. Interaction of virulent and non-virulent Yersinia enterocolitica strains and an epithelial and a phagocytic cell lines. Cent Eur J Public Health. 1995;3:149–153. [PubMed] [Google Scholar]
- 6.Deshpande R G, Khan M, Genco C A. Invasion strategies of the oral pathogen Porphyromonas gingivalis: implications for cardiovascular disease. Invasion Metastatis. 1998;18:57–69. doi: 10.1159/000024499. [DOI] [PubMed] [Google Scholar]
- 7.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 8.Dorn B R, Leung K L, Progulske-Fox A. Invasion of human oral epithelial cells by Prevotella intermedia. Infect Immun. 1998;66:6054–6057. doi: 10.1128/iai.66.12.6054-6057.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du L, Pellen-Mussi P, Chandad F, Mouton C, Bonnaure-Mallet M. Fimbriae and the hemagglutinating adhesin HA-Ag2 mediate adhesion of Porphyromonas gingivalis to epithelial cells. Infect Immun. 1997;65:3875–3881. doi: 10.1128/iai.65.9.3875-3881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan M J, Nakao S, Skobe Z, Xie H. Interactions of Porphyromonas gingivalis with epithelial cells. Infect Immun. 1993;61:2260–2265. doi: 10.1128/iai.61.5.2260-2265.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finlay B B. Fluorescence and confocal microscopy reveal host-microbe interactions. ASM News. 2000;66:601–608. [Google Scholar]
- 12.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galan J E. Interactions of bacteria with non-phagocytic cells. Curr Opin Immunol. 1994;6:590–595. doi: 10.1016/0952-7915(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 14.Honig M G, Hume R I. Fluorescent carbocyanine dyes allow living neurons of identified origin to be studied in long-term cultures. J Cell Biol. 1986;103:171–187. doi: 10.1083/jcb.103.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huard-Delcourt A, Menard C, Du L, Pellen-Mussi P, Tricot-Doleux S, Bonnaure-Mallet M. Adherence of Porphyromonas gingivalis to epithelial cells: analysis by flow cytometry. Eur J Oral Sci. 1998;106:938–944. doi: 10.1046/j.0909-8836.1998.eos106506.x. . (Erratum, 106:1051.) [DOI] [PubMed] [Google Scholar]
- 16.Izutsu K T, Belton C M, Chan A, Fatherazi S, Kanter J P, Park Y, Lamont R J. Involvement of calcium in interactions between gingival epithelial cells and Porphyromonas gingivalis. FEMS Microbiol Lett. 1996;144:145–150. doi: 10.1111/j.1574-6968.1996.tb08521.x. [DOI] [PubMed] [Google Scholar]
- 17.Joucla A, Suzuki J B. Les maladies parodontales se ressemblent-elles? J Parodontol Implantol Orale. 1994;13:111–124. [Google Scholar]
- 18.Katz J, Sambandam V, Wu J H, Michalek S M, Balkovetz D F. Characterization of Porphyromonas gingivalis-induced degradation of epithelial cell junctional complexes. Infect Immun. 2000;68:1441–1449. doi: 10.1128/iai.68.3.1441-1449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lajarin F, Rubio G, Galvez J, Garcia-Penarrubia P. Adhesion, invasion and intracellular replication of Salmonella typhimurium in a murine hepatocyte cell line. Effect of cytokines and LPS on antibacterial activity of hepatocytes. Microb Pathog. 1996;21:319–329. doi: 10.1006/mpat.1996.0065. [DOI] [PubMed] [Google Scholar]
- 20.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamont R J, Oda D, Persson R E, Persson G R. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol Immunol. 1992;7:364–367. doi: 10.1111/j.1399-302x.1992.tb00637.x. [DOI] [PubMed] [Google Scholar]
- 22.Lantz M S. New insights into mechanisms of bacterial pathogenesis in periodontitis. Curr Opin Periodontol. 1996;3:10–18. [PubMed] [Google Scholar]
- 23.Li J, Ellen R P, Hoover C I, Felton J R. Association of proteases of Porphyromonas (Bacteroides) gingivalis with its adhesion to Actinomyces viscosus. J Dent Res. 1991;70:82–86. doi: 10.1177/00220345910700021501. [DOI] [PubMed] [Google Scholar]
- 24.Loos B G, Dyer D W, Whittam T S, Selander R K. Genetic structure of populations of Porphyromonas gingivalis associated with periodontitis and other oral infections. Infect Immun. 1993;61:204–212. doi: 10.1128/iai.61.1.204-212.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madianos P N, Papapanou P N, Nannmark U, Dahlen G, Sandros J. Porphyromonas gingivalis FDC381 multiplies and persists within human oral epithelial cells in vitro. Infect Immun. 1996;64:660–664. doi: 10.1128/iai.64.2.660-664.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menard R, Dehio C, Sansonetti P J. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 27.Meyer D H, Fives-Taylor P M. Oral pathogens: from dental plaque to cardiac disease. Curr Opin Microbiol. 1998;1:88–95. doi: 10.1016/s1369-5274(98)80147-1. [DOI] [PubMed] [Google Scholar]
- 28.Miliotis M D. Acridine orange stain for determining intracellular enteropathogens in HeLa cells. J Clin Microbiol. 1991;29:830–831. doi: 10.1128/jcm.29.4.830-831.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miliotis M D, Tall B D, Gray R T. Adherence to and invasion of tissue culture cells by Vibrio hollisae. Infect Immun. 1995;63:4959–4963. doi: 10.1128/iai.63.12.4959-4963.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Njoroge T, Genco R J, Sojar H T, Hamada N, Genco C A. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect Immun. 1997;65:1980–1984. doi: 10.1128/iai.65.5.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Offenbacher S, Katz V, Fertik G, Collins J, Boyd D, Maynor G, McKaig R, Beck J. Periodontal infection as a possible risk factor for preterm low birth weight. J Periodontol. 1996;67:1103–1113. doi: 10.1902/jop.1996.67.10s.1103. [DOI] [PubMed] [Google Scholar]
- 32.Pace J, Hayman M J, Galan J E. Signal transduction and invasion of epithelial cells by S. typhimurium. Cell. 1993;72:505–514. doi: 10.1016/0092-8674(93)90070-7. [DOI] [PubMed] [Google Scholar]
- 33.Park Y, Lamont R J. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect Immun. 1998;66:4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Philpott D J, Yamaoka S, Israel A, Sansonetti P J. Invasive Shigella flexneri activates NF-kappa B through a lipopolysaccharide-dependent innate intracellular response and leads to IL-8 expression in epithelial cells. J Immunol. 2000;165:903–914. doi: 10.4049/jimmunol.165.2.903. [DOI] [PubMed] [Google Scholar]
- 35.Rosenshine I, Ruschkowski S, Foubister V, Finlay B B. Salmonella typhimurium invasion of epithelial cells: role of induced host cell tyrosine protein phosphorylation. Infect Immun. 1994;62:4969–4974. doi: 10.1128/iai.62.11.4969-4974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saglie F R, Smith C T, Newman M G, Carranza F A, Jr, Pertuiset J H, Cheng L, Auil E, Nisengard R J. The presence of bacteria in the oral epithelium in periodontal disease. II. Immunohistochemical identification of bacteria. J Periodontol. 1986;57:492–500. doi: 10.1902/jop.1986.57.8.492. [DOI] [PubMed] [Google Scholar]
- 37.Sandros J, Madianos P N, Papapanou P N. Cellular events concurrent with Porphyromonas gingivalis invasion of oral epithelium in vitro. Eur J Oral Sci. 1996;104:363–371. doi: 10.1111/j.1600-0722.1996.tb00093.x. [DOI] [PubMed] [Google Scholar]
- 38.Sandros J, Papapanou P N, Nannmark U, Dahlen G. Porphyromonas gingivalis invades human pocket epithelium in vitro. J Periodont Res. 1994;29:62–69. doi: 10.1111/j.1600-0765.1994.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 39.Socransky S S, Haffajee A D. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 40.Sojar H T, Hamada N, Genco R J. Isolation and characterization of fimbriae from a sparsely fimbriated strain of Porphyromonas gingivalis. Appl Environ Microbiol. 1997;63:2318–2323. doi: 10.1128/aem.63.6.2318-2323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeshita A, Murakami Y, Yamashita Y, Ishida M, Fujisawa S, Kitano S, Hanazawa S. Porphyromonas gingivalis fimbriae use β2 integrin (CD11/CD18) on mouse peritoneal macrophages as a cellular receptor, and the CD18 β chain plays a functional role in fimbrial signaling. Infect Immun. 1998;66:4056–4060. doi: 10.1128/iai.66.9.4056-4060.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]