Abstract
Bacillus subtilis RB14 is a producer of the antifungal lipopeptide iturin A. Using a transposon, we identified and cloned the iturin A synthetase operon of RB14, and the sequence of this operon was also determined. The iturin A operon spans a region that is more than 38 kb long and is composed of four open reading frames, ituD, ituA, ituB, and ituC. The ituD gene encodes a putative malonyl coenzyme A transacylase, whose disruption results in a specific deficiency in iturin A production. The second gene, ituA, encodes a 449-kDa protein that has three functional modules homologous to fatty acid synthetase, amino acid transferase, and peptide synthetase. The third gene, ituB, and the fourth gene, ituC, encode 609- and 297-kDa peptide synthetases that harbor four and two amino acid modules, respectively. Mycosubtilin, which is produced by B. subtilis ATCC 6633, has almost the same structure as iturin A, but the amino acids at positions 6 and 7 in the mycosubtilin sequence are d-Ser→l-Asn, while in iturin A these amino acids are inverted (i.e., d-Asn→l-Ser). Comparison of the amino acid sequences encoded by the iturin A operon and the mycosubtilin operon revealed that ituD, ituA, and ituB have high levels of homology to the counterpart genes fenF (79%), mycA (79%), and mycB (79%), respectively. Although the overall level of homology of the amino acid sequences encoded by ituC and mycC, the counterpart of ituC, is relatively low (64%), which indicates that there is a difference in the amino acid sequences of the two lipopeptides, the levels of homology between the putative serine adenylation domains and between the asparagine adenylation domains in the two synthetases are high (79 and 80%, respectively), implying that there is an intragenic domain change in the synthetases. The fact that the flanking sequence of the iturin A synthetase coding region was highly homologous to the flanking sequence that of xynD of B. subtilis 168 and the fact that the promoter of the iturin A operon which we identified was also conserved in an upstream sequence of xynD imply that horizontal transfer of this operon occurred. When the promoter was replaced by the repU promoter of the plasmid pUB110 replication protein, production of iturin A increased threefold.
Many Bacillus subtilis strains produce a small peptide(s) with a long fatty moiety, the so-called lipopeptide antibiotics. The peptide portions of these compounds contain α-amino acids with a d configuration and are produced nonribosomally with templates of the multifunctional peptide synthetases. As in the synthesis of the peptide antibiotic gramicidin S, the peptide chain grows in a defined sequence by moving on the template of the multifunctional peptide synthetase (18, 35, 44). On the basis of the structural relationships, the lipopeptides that have been identified in B. subtilis are generally classified into three groups: the surfactin group (27), the plipastatin-fengycin group (16, 41, 42), and the iturin group (17). The members of the surfactin and plipastatin-fengycin groups are composed of one β-hydroxy fatty acid and 7 and 10 α-amino acids, respectively, while the members of the iturin group consist of one β-amino fatty acid and 7 α-amino acids. The presence of the β-amino fatty acid is the most striking characteristic of the iturin A group and distinguishes this group from the other two groups. The operons that encode surfactin (3), plipastatin-fengycin (16, 37, 38, 40), and mycosubtilin (4), which is a member of the iturin A group, have been sequenced and characterized. In particular, a study of the mycosubtilin operon of B. subtilis ATCC 6633 (4) showed that MycA, a novel template enzyme which has functional domain homology to β-ketoacyl synthetase and amino transferase and amino adenylation, was present, which implied that MycA is responsible for incorporation of the β-amino fatty acid.
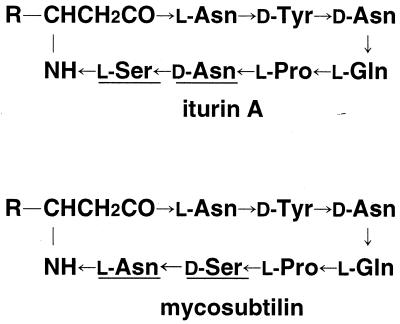
Recently, biological control agents for plant diseases have received considerable attention as alternatives to chemical pesticides (32). B. subtilis RB14, which has a suppressive effect against several phytopathogens, is expected to be used as a biocontrol agent (1, 5). We previously demonstrated that the biocontrol activity of RB14 can be attributed mainly to production of iturin A (Fig. 1) (10, 26). Iturin A, as well as mycosubtilin, is a member of the iturin group. The amino acid compositions of iturin A and mycosubtilin are almost identical, except that the sixth and seventh amino acids are inverted, as shown in Fig. 1. In our previous study, we cloned a gene, lpa-14, that encodes the 4′-phosphopantheteinyl transferase required for maturation of the template enzyme of iturin A (6, 8, 15). For further investigation of the details of the synthesis steps and because of the interesting evolutionary relationship between iturin A and mycosubtilin, cloning and sequencing of the complete iturin A synthetase operon are essential.
FIG. 1.
Structures of iturin A and mycosubtilin. R indicates an alkyl moiety (generally C14 to C17). The arrows represent peptide bonds in the -CO-NH- direction. The differences between the two lipopeptides are indicated by underlining.
In this study, we examined the features of the iturin A synthetase operon based on the nucleotide sequence and gene disruption. By comparing the iturin A operon with the mycosubtilin operon, we found that the difference between the two operons may be a result of intragenic swapping of amino acid adenylation domains. We also obtained genetic evidence of probable horizontal transfer of the iturin A operon. In addition, a promoter replacement experiment whose goal was construction of a iturin A hyperproducer is also described below.
MATERIALS AND METHODS
Strains and media.
The strains and plasmids used in this study are listed in Table 1. Luria-Bertani (LB) medium was used for cultivation of Escherichia coli and B. subtilis (30). When necessary, antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 5 μg/ml; erythromycin, 10 μg/ml; tetracycline, 20 μg/ml; and neomycin, 20 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristica | Reference |
|---|---|---|
| E. coli JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 Δ(lac-proAB) /F′[traD36 proAB+lacIqlacZΔM15] | 30 |
| B. subtilis strains | ||
| RB14 | Wild type, IT+ SF+ PL+ | 5 |
| 1006 | RB14 ituA::Tn10, IT− SF+ PL+ | This study |
| RΔIA1 | RB14 ituD::neo, IT− SF+ PL+ | This study |
| R-PM1 | RB14 PrepU-neo-ituD, IT+ SF+ PL+ | This study |
| MI113 | Marburg 168 derivative, arg-15 trpC2 hsmM hsrM | 40 |
| E. coli plasmids | ||
| pUC19 | Cloning vector, Apr | 30 |
| pBEST502 | PrepU-neo cassette, Apr Nmr | 11 |
| pSC24Pst | pUC19 carrying the 2-kb Sau3AI-PstI fragment containing ituD, Apr | This study |
| pSC24PstNm | Nmr gene cassette inserted into the internal SalI-NsiI region of ituD in pSC24Pst, Apr Nmr | This study |
| pSC53EcoT | pUC19 carrying the 1.2-kb TthHB8I-EcoT14I fragment containing PituD, Apr | This study |
| pKODP4P5 | pUC19 carrying the 0.8-kb PCR product containing 5′-terminal region of ituD as well as the putative ribosome binding site, Apr | This study |
| pP4P5Nm | PreuU-neo inserted into the XbaI site located upstream of the putative ribosome binding site of ituD in pKODP4P5, Apr Nmr | This study |
| pUCIPNm | pP4P5Nm carrying the 0.8-kb NdeI-TthHB8I fragment containing 3′-terminal region of yxjF, Apr Nmr | This study |
| B. subtilis plasmids | ||
| pHV1249 | Mini-Tn10 Apr Emr Cmr | 25 |
| pTB522 | Cloning vector, Tcr | 9 |
| pE194 | Temperature sensitive for replication, Emr | 40 |
| pE24PstNmα | Disruption plasmid, pE194 carrying the 2.0-kb insert from pSC24PstNm, ituD::neo gene cassette, Emr Nmr | This study |
| pEIPNm2 | pE194 carrying the 3.0-kb insert from pUCIPNm, PituD-neo-ituD, Emr Nmr | This study |
IT, iturin A production; SF, surfactin production; PL, plipastatin-like antifungal agent production; Apr, ampicillin resistant; Emr, erythromycin resistant; Nmr, neomycin resistant; Tcr, tetracycline resistant.
Iturin A production in vitro was indicated by the formation of a clear inhibitory zone on LB agar (LB medium with 1.5% agar) containing a spore suspension of a phytopathogenic fungus, Fusarium oxysporum f. sp. lycopersici race J1 SUF119, as described previously (5). Number 3 medium was used for lipopeptide production in liquid cultures and contained (per liter) 10 g of Polypepton (Nihon Pharmaceutical Co., Tokyo, Japan), 10 g of glucose, 1 g of KH2PO4, and 0.5 g of MgSO4 · 7H2O (pH 6.8). Number 3S medium contained 10 g of Polypepton S (Nihon Pharmaceutical Co.) per liter instead of Polypepton because iturin A productivity was enhanced by Polypepton S.
Transformation and DNA manipulation.
B. subtilis RB14 was transformed by electroporation as described previously (19, 39). B. subtilis MI113 was transformed by a competent cell method as described previously (39). Chromosomal DNA of B. subtilis strains were prepared by using a method developed by Itaya and Tanaka (12). Routine DNA manipulation and E. coli transformation were performed as described previously (39). Plaque and Southern hybridizations were performed by a digoxigenin enzyme-linked immunosorbent assay using a DNA labeling and detection kit (Roche) as recommended in the instruction manual.
Transposon technique.
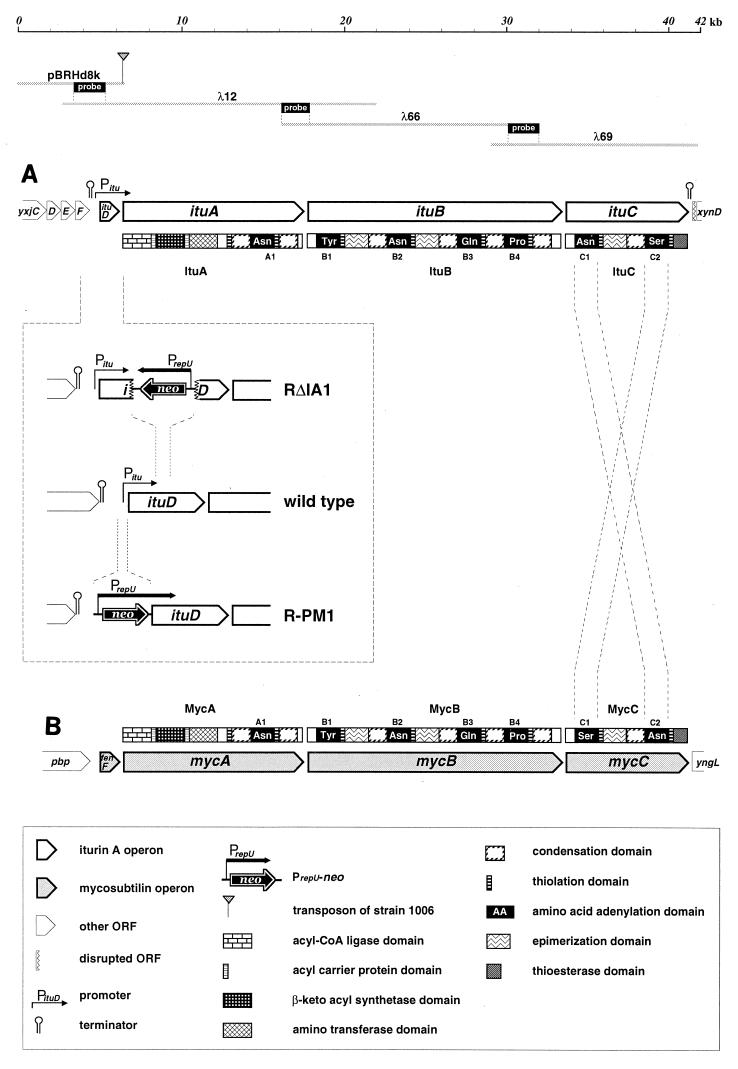
Transposon-harboring plasmid pHV1249 (25) was introduced into RB14 by electroporation, and random mutagenesis of mini-Tn10 was performed by a previously described method (25). Cloning of the mini-Tn10-disrupted gene was carried out as follows. To construct a B. subtilis MI113 plasmid library of the RB14 chromosome, chromosomal DNA of the transposon-containing mutant was digested with HindIII, ligated at the HindIII site of plasmid pTB522 which could be replicated in B. subtilis (9), and then transformed into B. subtilis MI113 competent cells. The plasmid of the chloramphenicol-resistant transformant resulting from cloning of a chromosomal fragment containing mini-Tn10 was obtained from the library and designated pTB1006. The HindIII insert of pTB1006 was transplanted into the HindIII site of pBR322 by transformation of E. coli JM109, resulting in pBRHd8k (Fig. 2).
FIG. 2.
ORF organization of iturin A (A) and mycosubtilin (B) operons. The positions of the plasmid and phages sequenced are shown above the iturin A operon. The sequences of derivative strains RΔIA1 and R-PM1 associated with ituD are shown in a box. The intersecting dotted lines indicate the difference in the amino acid adenylation domain arrangement between the two operons. The mycosubtilin operon was drawn by referring to reference 4.
Phage library construction and screening.
The RB14 chromosome was digested partially with EcoRI and then was separated by electrophoresis to obtain homogeneous fragments that were 10 to 20 kb long. The fragments obtained were used for lambda DASH II library construction with a Lambda DASH II/EcoRI vector kit (Stratagene) as recommended in the instruction manual. First, screening by plaque hybridization in which the 1.9-kb NdeI-PstI fragment of pBRHd8k was used as a probe identified a positive phage with a 19.2-kb insert, designated λ12 (Fig. 2). By using the internal 1.7-kb EcoRI-HindIII fragment of λ12 as a probe, λ66, harboring a 16.0-kb insert, was obtained (Fig. 2). The end of the 1.9-kb EcoRI fragment of λ66 was employed in the next screening, and λ69 with a 12.4-kb insert was obtained (Fig. 2).
Disruption of ituD by the plasmid pop in-pop out method.
A 2-kb Sau3AI-PstI fragment from pBRHd8k, which harbored the entire ituD gene, was inserted at the BamHI-PstI site of pUC19, generating pSC24Pst. The SalI-NsiI fragment of pSC24Pst was removed, and the SalI-PstI fragment of the neomycin resistance gene cassette (neo) from pBEST502 was inserted (11), resulting in pSC24PstNm. The HindIII-KpnI fragment of pSC24PstNm, which harbored the disrupted ituD gene, was inserted at the PstI site of pE194 by blunt-end ligation. The ligated mixture was transformed into MI113. We selected a neomycin-resistant colony and thus obtained pE24PstNmα. The ituD coding region in RB14 was disrupted by using the thermosensitive replication origin of pE194, as described previously (40). First, pE24PstNmα was transformed into RB14 by electroporation. Strain RB14(pE24PstNmα) was plated onto LB agar containing neomycin and erythromycin and then incubated at 48°C. The resulting strain, RB14::pE24PstNmα, was cultivated in LB medium without selective pressure at 30°C for 10 generations. The culture was diluted and plated onto LB agar to obtain single colonies. The neomycin resistance and erythromycin resistance of the resulting colonies were assayed. Finally, the disrupted mutant, RΔIA1, was isolated by screening for neomycin-resistant and erythromycin-sensitive colonies.
Primer extension analysis.
Total mRNA was prepared from a stationary-phase culture of RB14 cultivated in Number 3 medium for 17 h. Four milliliters of the culture was centrifuged and washed with STE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.1 M NaCl). The pellet was then resuspended in 50 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and 50 μl of TE buffer containing lysozyme (20 mg/ml) was added. The suspension was incubated at room temperature for 10 min. Further purification was performed with an RNeasy mini kit (Qiagen). For primer extension analysis, IRD41-labeled primer ITU-PREX (5′-ATCGCATCGCTCGCTTCTTCAAAC-3′) (Fig. 3), which was complementary to the sequence from nucleotide 96 to nucleotide 119 downstream of the putative start codon of ituD, was purchased from NisSHINBO Co. (Tokyo, Japan). Total RNA was ethanol precipitated and dissolved in 20 μl of hybridization buffer [80 mM piperazine-N, N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 6.4), 2 mM EDTA, 800 mM NaCl, 50 % formamide]. The solution was supplemented with 1.8 μl of primer ITU-PREX (1 pmol/μl), denatured at 80°C for 15 min, and then cooled to 30°C with gentle shaking. After ethanol precipitation, the pellet was dissolved in extension buffer (4 μl of Moloney murine leukemia virus reverse transcriptase [Toyobo Inc., Osaka, Japan], 8 μl of 5× buffer for Moloney murine leukemia virus reverse transcriptase, 16 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 2.5 mM, 10 μl of H2O, 2 μl of RNase inhibitor) and then incubated at 42°C for 1 h. The resulting cDNA was subjected to Li-cor dNA automated DNA sequencing.
FIG. 3.
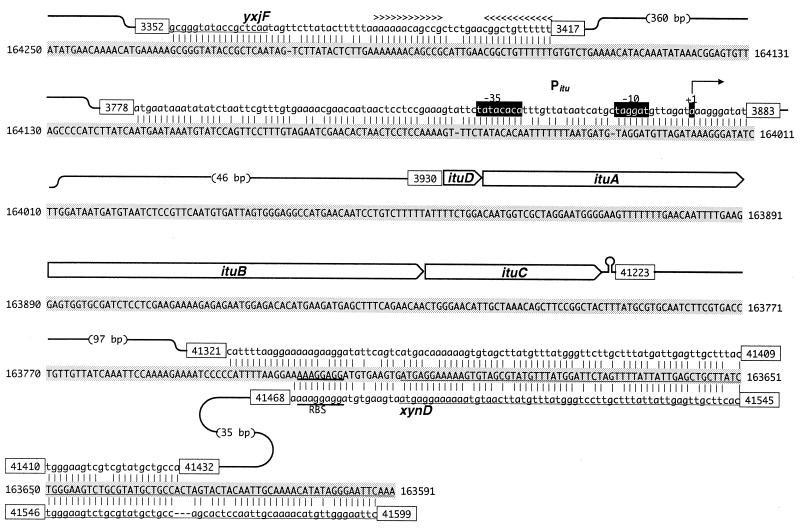
Determination of the transcription initiation site of ituD by primer extension analysis. (A) The position of the band corresponding to the ituD-specific primer extension product is indicated by arrows. (B) Nucleotide sequence of the ituD promoter region (PituD). The positions of the ituD transcription initiation site (highlighted, +1), putative −10 and −35 regions (highlighted), and putative ribosome binding site (RBS) (underlined) are indicated. Putative protein sequences of YxjF and ItuD are shown below the DNA sequence (shaded). The sequence complementary to the oligonucleotide used for primer extension analysis (ITU-PREX) is double underlined. The putative ρ-independent terminator downstream of yxjF is indicated by > and < above the sequence.
Promoter exchange.
A 0.8-kb fragment of the 5′ region of ituD, which contained up to the ribosome binding site but not the promoter, was obtained by PCR. PCR amplification with primers ITUP4-F (5′-CCCCTGTTCTAGATGATCGGAGGAATCTC-3′; underlining indicates an XbaI site, and italics indicates substituted bases) and ITUP5-R (5′-TGCATCGATTCTGTCCATCTAACCGGCATC-3′; underlining indicates a ClaI site) was performed with the RB14 chromosome by using KOD DNA polymerase (Toyobo Inc.). The fragment obtained was double digested with XbaI and ClaI and then inserted between the XbaI and ClaI sites of pUC19. The resulting plasmid was confirmed to have the correct sequence and was designated pKODP4P5. PrepU accompanied by the neomycin resistance gene cassette (PrepU-neo) was excised from pBEST502 (11) by digestion with XbaI and then inserted into the XbaI site of pKODP4P5. A plasmid with PrepU-neo, whose direction of transcription is the same as that of ituD, was selected and designated pP4P5Nm. The 0.8-kb NdeI-TthHB8I fragment, which was upstream of the ituD transcription start site prepared from plasmid subclone pSC53EcoT of the phage library, was blunt ended and inserted into the SmaI site of pP4P5Nm, generating pUCIPNm. The HindIII-KpnI fragment of pUCIPNm, which was PrepU-neo accompanied by the Pitu flanking region, was inserted into the PstI site of pE194 by blunting and ligation and then transformed into MI113, which resulted in plasmid pEIPNm2. Replacement of Pitu of RB14 by PrepU-neo by the plasmid pop in-pop out method using pEIPNm2 was performed by the method described above for ituD disruption. Promoter replacement was confirmed by PCR (data not shown).
Quantitative analysis of iturin A and surfactin.
A culture of B. subtilis in 40 ml of Number 3 medium was acidified to pH 2.0 with 12 N HCl. Then the precipitate was collected by centrifugation and extracted with methanol. Iturin A and surfactin in the extracted solution were quantified by reversed-phase high-performance liquid chromatography (HPLC), as described previously (1, 40).
Nucleotide sequence analysis.
Double-stranded DNA cloned in pUC19 was sequenced with a Li-cor dNA 4000L DNA sequencer by using an IRD41 dye-labeled primer (Nisshinbo Co., Tokyo, Japan) and a Thermo Sequenase cycle sequencing kit (Amersham Pharmacia Biotech Co.). A partial sequence of the insert of the screened lambda DASH II phage was determined by Nippon Seifun Co., Atsugi, Japan. Motif retrieval for proteins was performed by using the PROSITE package. Harrplot analysis was performed with the Genetyx package (Software Development Co., Tokyo, Japan). Multiple-alignment analysis and phylogenetic analysis were performed by using Clustal W.
Nucleotide sequence accession number.
The nucleotide sequence of the iturin A operon of RB14 has been deposited in the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession number AB050629.
RESULTS
Cloning, sequencing, and analysis of iturin A operon.
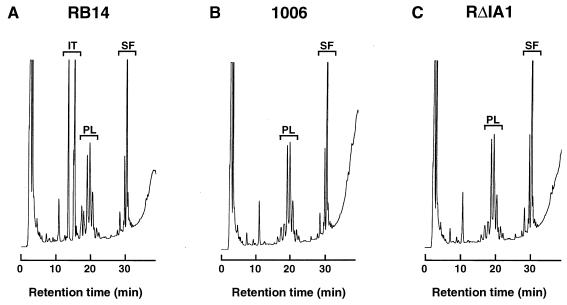
To identify the genes responsible for iturin A production, we performed transposon mutagenesis with mini-Tn10. About 5,000 transposon-containing colonies were replicated on an LB agar plate containing F. oxysporum to assay for iturin A production deficiency. Fifteen colonies exhibited no antifungal activity on the plate, and none of these colonies had the ability to produce iturin A, as determined by HPLC. Ten of the colonies were selected at random, and chromosomal DNA were prepared, digested by HindIII, and subjected to Southern hybridization. All 10 colonies contained mini-Tn10 in the same 8-kb HindIII fragment (data not shown). We selected one colony, designated strain 1006, for further study. No iturin A production by 1006 was observed (Fig. 4). The 8-kb HindIII fragment with a transposon was screened from the plasmid library of the strain 1006 chromosome by using the chloramphenicol resistance marker of mini-Tn10.
FIG. 4.
Qualitative HPLC analysis of the lipopeptides produced by RB14 (A), 1006 (B), and RΔIA1 (C). Peaks corresponding to iturin A (IT) and surfactin (SF) are indicated. Plipastatin-like peaks (not identified) (PL) are also indicated.
To clone a complete iturin A operon, an RB14 chromosome library was constructed by using the lambda DASH II phage, and four contiguous inserts that collectively encompassed a 42-kb region were obtained. Figure 2 shows that this region contains nine open reading frames (ORFs). All nine ORFs are oriented in the same direction. The four ORFs located upstream in the region sequenced have high levels of homology to the B. subtilis 168 genes yxjC (77% identical), yxjD (83%), yxjE (83%), and yxjF (77%). These genes are thought to encode 3-oxoadipate coenzyme A (CoA) transferase (yxjD and yxjE) and gluconate 5-dehydrogenase (yxjF) (14), but their actual functions have not been determined. There is a putative rho-independent terminator downstream of yxjF that appears to terminate transcription from both directions. The fifth ORF, designated ituD, is separated from yxjF by a 0.6-kb intercoding region. The 45-kDa ItuD protein has a high level of homology to FenF encoded by the mycosubtilin synthetase operon of B. subtilis ATCC 6633 (79%) (4) and FenF of B. subtilis F29-3 (89%) (2) and lower levels of homology to malonyl-CoA transacylase of E. coli (44%) (43) and B. subtilis 168 (37%) (23), suggesting that ituD encodes malonyl-CoA transacylase. The transposon-containing ORF, located 19 bp downstream of ituD, is designated ituA. The deduced amino acid sequence encoded by ituA corresponds to a 449-kDa protein that has a high level of homology to MycA (79%), which is the first subunit of mycosubtilin synthetase. The striking feature of MycA, the fact that three functional domains homologous to β-ketoacyl synthetase, amino transferase, and amino acid adenylation are combined, is conserved in ItuA. The gene 43 bp downstream of ituA is called ituB. ItuB is a 609-kDa peptide synthetase consisting of four amino acid adenylation domains, two of which are flanked by an epimerization domain. ItuB also exhibits 79% homology to MycB. The next gene is designated ituC, which encodes a peptide synthetase that has two adenylation domains, one epimerization domain, and a thioesterase domain that is probably responsible for peptide cyclization. Although ItuD, ItuA, and ItuB have high levels of homology to their counterparts in mycosubtilin synthetase, ItuC exhibits only 64% homology to MycC, which may reflect a structural difference between iturin A and mycosubtilin. There is a putative rho-independent terminator downstream of ituC. The final ORF exhibits homology to the xynD gene (29) of B. subtilis 168, which encodes endo-1,4-xylanase. As shown in Fig. 5, there is a sequence that has a high level of homology to the xynD gene and is located between ituC and xynD in a direct-repeat manner (see below).
FIG. 5.
Alignment of iturin A operon of RB14 and xynD regions of strain 168. Shaded uppercase letters indicate the sequence of the xynD region of strain 168 (based on the complementary sequence from positions 164, 250 to 163, 691 of the accession no. Z99113 sequence). Lowercase letters indicate the iturin A operon sequence (accession no. AB050629) that has homology to the strain 168 sequence. Sequences encoding iturin A synthetase are indicated by arrow-shaped boxes. Identical nucleotides in the two sequences are indicated by vertical lines. The numbers in the boxes are the positions in the deposited sequence. The lines indicate linkage between discrete homologous segments of the iturin A operon. Underlining indicates coding regions of yxjF and xynD. −35 and −10 are promoter sites of ituD, and +1 is a transcription start site of ituD. The alignment was constructed based on the results of a BLAST search. RBS, ribosome binding site.
Disruption of the ituD gene to derive the iturin A-deficient phenotype.
To confirm that ituD is responsible for iturin A synthesis, ituD was disrupted (Fig. 2). The resulting disruptant, RΔIA1, was inoculated into Number 3S medium and cultivated at 30°C for 60 h to assay for iturin A production. As shown in Fig. 4, analytical HPLC of an RΔIA1 culture extract resulted in no iturin A peak, while the production of the lipopeptide surfactin by RΔIA1 was the same as the production by the wild type, indicating that ituD disruption only resulted in an iturin A deficiency. Based on these results, we concluded that ituD is essential specifically for iturin A synthesis.
Promoter analysis of the ituD gene.
The transcription start site of ituD was determined by the primer extension method. Total RNA of RB14 was extracted from a 17-h culture in Number 3 medium. The total RNA obtained was hybridized with oligoDNA by using a fluorescent probe that was designed to hybridize with nucleotides 96 to 119 of ituD from the translation initiation codon. cDNA was synthesized with reverse transcriptase, and the resulting cDNA was then sequenced. As shown in Fig. 3, the transcription start site of ituD was found to be an A residue 56 bp upstream of the first residue of the ituD initiation codon. Upstream of this start site, we found a TATACACA-16 bp-TAGGAT sequence that exhibited low levels of homology to the consensus −10 and −35 (TTGACA-17 bp-TATAAT) sequences of ςA (Fig. 3). This promoter was designated Pitu. We also searched for other transcription start sites up to the putative rho-independent terminator of yxjF but did not find any other such sites.
Promoter replacement of the iturin A operon for high iturin A production.
Because the mutant with disrupted ituD had an iturin A deficient phenotype and there is no coding region between ituD and ituC, which implies that there is a promoter, it is possible that Pitu governs expression of the iturin A operon. To confirm this, we replaced Pitu with a constitutively expressed promoter by using the neomycin resistance gene cassette, PrepU-neo, of pBEST502 (10). PrepU-neo is the chimera of the promoter of the replication protein gene (PrepU) and the neomycin resistance gene (neo) of plasmid pUB110 (20). Since PrepU of PrepU-neo lacks autoregulation of transcription by RepU (24), constitutive expression of PrepU is expected. Moreover, PrepU is sufficiently strong that neomycin resistance can be conferred by a single copy of neo in the chromosome, and there is no terminator to stop transcription starting from PrepU. The presence of the neomycin resistance gene cassette in a transformant, therefore, indicates that PrepU has been introduced and implies that constitutive and enforced expression of genes located downstream of PrepU-neo occurs in a polycistronic manner. The neomycin resistance gene cassette was ligated with a PCR-amplified fragment of the 5′ portion of ituD at the ribosome binding site of ituD and then attached to the upstream fragment of Pitu in front of the neomycin resistance gene. The resulting three-fragment array was ligated to the thermosensitive plasmid pE194 and transformed by electroporation, and then the promoter was replaced by the pop in-pop out method, as described above for ituA disruption. The resulting strain, R-PM1, contained the inserted PrepU-neo instead of 250 bp of the Pitu sequence (Fig. 2).
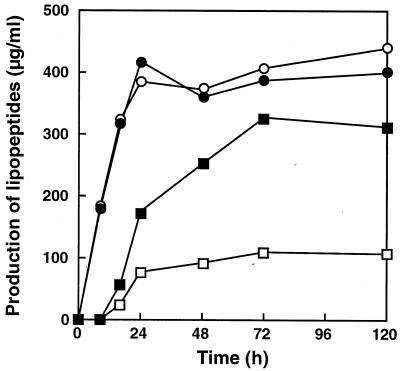
The strain generated and the wild type were inoculated into Number 3 medium and cultured at 30°C for 120 h to monitor iturin A production. The changes in the growth of strain R-PM1 and the pH of Number 3 medium over time were the same as those observed with the wild type (data not shown). However, as shown in Fig. 6, the rate of iturin A production by R-PM1 was significantly greater than that by the wild type. The concentration of iturin A for R-PM1 after 72 h of cultivation was 330 μg/ml, whereas the concentration for the wild type was 110 μg/ml, indicating that production was threefold greater in R-PM1. Since the two strains did not differ significantly in terms of surfactin production (Fig. 6), we concluded that the promoter exchange affected only iturin A production.
FIG. 6.
Time courses for production of two lipopeptides, iturin A produced by RB14 (□) and R-PM1 (■) and surfactin produced by RB14 (○) and R-PM1 (●).
DISCUSSION
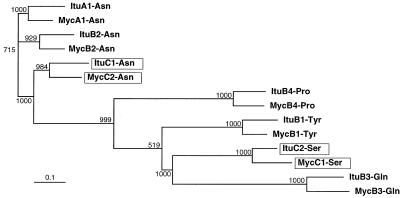
In this study, we identified the 42-kb region of B. subtilis RB14 that contains the complete iturin A synthetase operon, which is more than 38 kb long. The iturin A operon is composed of four ORFs, ituD, ituA, ituB, and ituC, in that order. The iturin A operon closely resembles the mycosubtilin operon. ItuD, as well as FenF of B. subtilis ATCC 6633 (4) and F29-3 (2), exhibits homology to the malonyl CoA-transacylase FabD, which participates in fatty acid synthesis in E. coli (43) and B. subtilis 168 (23). It is thought that in some Streptomyces spp. malonyl CoA-transacylase may be responsible for not only fatty acid synthesis but also type II polyketide antibiotic synthesis (28, 36). To clarify whether ItuD is involved in iturin A production and/or fatty acid synthesis, we disrupted ituD, which resulted in a specific iturin A deficiency. Since neither the growth of nor surfactin production by strain RΔIA1, a strain in which ituD was disrupted, was inhibited, we concluded that ituD is indispensable for iturin A production. This is the first evidence that a gene related to fenF participates in synthesis of an iturin group antibiotic. On the other hand, ItuA exhibits homology to β-ketoacyl synthetase, amino transferase, and peptide synthetase in one molecule and is probably responsible for synthesis of a β-amino fatty acid accompanied by ItuD dipeptide (β-amino fatty acid–Asn) formation, as has been proposed for the mycosubtilin synthetase MycA (4). These features are consistent with the report that cerulenin, which inhibits β-ketoacyl synthetase, is more active in iturin A β-amino fatty acid synthesis than in fatty acid synthesis, which implies that β-ketoacyl synthesis occurs during iturin A production independent of fatty acid synthesis (7). Other genes, ituB and ituC, encode large peptide synthetases that build peptide chains on the precursor from ItuA. The level of homology between ituC and mycC, the counterpart of ituC in the mycosubtilin operon, is relatively low (64%) compared to the levels of homology for other groups (79%), reflecting the difference in the amino acid arrangements in ItuC and MycC. As shown in Fig. 2, ItuC is predicted to be responsible for the d-Asn→l-Ser portion of iturin A, while MycC is thought to synthesize the d-Ser→l-Asn part of mycosubtilin. Therefore, it is not unreasonable to assume that swapping between two adenylation domains occurs. To examine the possibility that intragenic swapping occurs in the adenylation domain, the sequences of ituC and mycC were compared. The results of a dot matrix analysis of the nucleotide sequences of ituC and the other relevant synthetase genes (mycC, ituA, and ituB), including ituC itself, are shown in Fig. 7. Clearly, two regions of ituC exhibit discrete homology to mycC; the homology junction is roughly 10 amino acids upstream of L(TS)xEL (A1 motif sequence [18]) and the C terminus of GRxDxQVKIRGxRIELGEIE (A8 motif sequence [18]). Homologies are observed between asparagine adenylation domains (ItuC2 and MycC1) and between serine adenylation domains (ItuC1 and MycC2), implying that domain swapping occurs. Since there are three asparagine adenylation domains (ItuA1, ItuB2, and ItuC1) in iturin A synthetases (Fig. 2), it is likely that the asparagine adenylation domain of ItuC1 exhibits the highest level of homology to ItuA1 or Itu2. However, as shown in Fig. 8, phylogenetic analysis of 14 nucleotide sequences encoding the adenylation domain of iturin A and mycosubtilin revealed that the sequence most similar to the sequence encoding the asparagine adenylation domain of ItuC1 is the sequence encoding the asparagine adenylation domain of MycC2. All iturin A domains also show phylogenetic similarities to their counterparts in mycosubtilin domains. These results imply that iturin A and mycosubtilin have a common ancestor. Therefore, we propose a model in which ituC or mycC swapped nucleotide sequences encoding adenylation domains after a common ancestor became established.
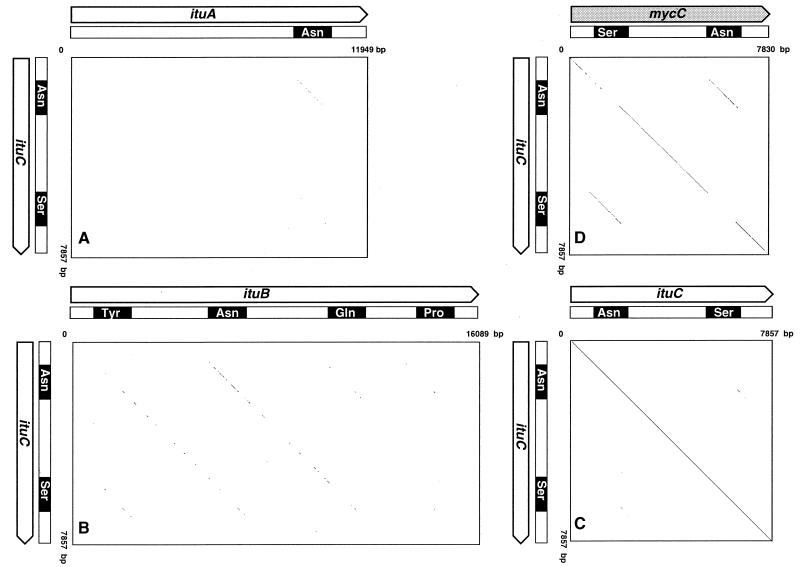
FIG. 7.
Dot matrices for ituC and ituA (A), ituB (B), ituC (C), and mycC (D). Dots were placed at locations with identical nucleotides when more than 45 of 60 nucleotides were identical. Amino acid adenylation domains of functional modules of synthetases are indicated by black boxes.
FIG. 8.
Phylogenetic tree constructed by the neighbor-joining method based on the sequences of the adenylation domains (from part of the A3 motif, PKG to the A6 motif, GELC[Y]) of iturin A synthetase and mycosubtilin synthetase. The numbers are bootstrap values based on 1,000 replicates.
Although the overall sequence of the iturin A operon is highly homologous to that of the mycosubtilin operon, the flanking region of the iturin A operon is significantly different from that of the mycosubtilin operon. It has been shown that the mycosubtilin operon of ATCC 6633, which is not a plipastatin producer, is between pbp and yngL (4), while the plipastatin operon (167° to 171°), instead of the mycosubtilin operon, is present in strain 168 (40). In the case of RB14, the iturin A operon lies between yxjF and xynD. Since strain RB14 is also a producer of a plipastatin-like compound (Fig. 4), it is reasonable to conclude that the iturin A operon of RB14 is in a region other than the plipastatin region. However, in the case of strain 168, yxjF (341°) is located on the side opposite xynD (166°) in the chromosome (14). This can be explained by the fact that the SfiI digestion pattern of the RB14 genome, as determined by pulsed-field gel electrophoresis, was significantly different from that of strain 168 (data not shown), suggesting that the entire genomic structure of RB14 has little similarity to that of strain 168, including the location of yxjF and/or xynD.
However, unexpectedly, as shown in Fig. 5, the promoter region of the iturin A operon has significant homology to the upstream sequence of xynD of strain 168. Since the downstream region of the iturin A operon has two tandem repeat DNA segments with significant homology to the xynD region of strain 168, it is highly probable that the coding region of the iturin A operon is transferred into the promoter of xynD and resides in this promoter, rather than that the promoter of xynD is simply transferred into the iturin A operon. The possibility that such a large antibiotic gene transfer occurs was also suggested for the tyrocidine operon, in which some peptide synthetase genes are integrated into the parental gramicidin S operon (21). In terms of horizontal transfer, it is possible that large DNA transfers occur frequently. The duplicated copy of xynD may indicate transient existence of a tandem repeat of early iturin A operons that were intermediates in the adenylation domain swapping mentioned above.
Our attempt to introduce a repU promoter instead of an intrinsic promoter into RB14 resulted in increased production of iturin A (Fig. 6), and the advantage of using the PrepU-neo cassette was identified. This method might be useful for generating bacteria that act as effective biological control agents.
At present, we are able to compare very similar operons, such as surfactin operons of B. subtilis (3) and two lichenysin operons of Bacillus licheniformis (13, 45), plipastatin (37, 38) and fengycin (16) operons, and iturin A and mycosubtilin (4) operons. These comparisons may provide good techniques for elucidating the boundary of the functional domain that directs engineered peptide synthesis (22, 31, 33, 34).
ACKNOWLEDGMENTS
We thank T. Ano and S. Inoue for helpful suggestions and assistance with the experiments.
REFERENCES
- 1.Asaka O, Shoda M. Biocontrol of Rhizoctonia solani damping-off of tomato with Bacillus subtilis RB14. Appl Environ Microbiol. 1996;62:4081–4085. doi: 10.1128/aem.62.11.4081-4085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C L, Chang L K, Chang Y S, Liu S T, Tschen J S M. Transposon mutagenesis and cloning of the genes encoding the enzymes of fengycin biosynthesis in Bacillus subtilis. Mol Gen Genet. 1995;248:121–125. doi: 10.1007/BF02190792. [DOI] [PubMed] [Google Scholar]
- 3.Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol. 1993;8:821–831. doi: 10.1111/j.1365-2958.1993.tb01629.x. [DOI] [PubMed] [Google Scholar]
- 4.Duitman E H, Hamoen L W, Rembold M, Venema G, Seitz H, Saenger W, Bernhard F, Reinhardt R, Schmidt M, Ullrich C, Stein T, Leenders F, Vater J. The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc Natl Acad Sci USA. 1999;96:13294–13299. doi: 10.1073/pnas.96.23.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiraoka H, Asaka O, Ano T, Shoda M. Characterization of Bacillus subtilis RB14, coproducer of peptide antibiotics iturin A and surfactin. J Gen Appl Microbiol. 1992;38:635–640. [Google Scholar]
- 6.Hiraoka H, Ano T, Shoda M. Molecular cloning of a gene responsible for the biosynthesis of the lipopeptide antibiotics iturin and surfactin. J Ferment Bioeng. 1992;74:323–326. [Google Scholar]
- 7.Hourdou M L, Besson F, Michel G. Specific inhibition of iturin biosynthesis by cerulenin. Can J Microbiol. 1989;36:164–168. doi: 10.1139/m90-029. [DOI] [PubMed] [Google Scholar]
- 8.Huang C C, Ano T, Shoda M. Nucleotide sequence and characteristics of the gene, lpa-14, responsible for biosynthesis of the lipopeptide antibiotics iturin A and surfactin from Bacillus subtilis RB14. J Ferment Bioeng. 1993;76:445–450. [Google Scholar]
- 9.Imanaka T, Himeno T, Aiba S. Effect of in vitro DNA rearrangement in the NH2-terminal region of the penicillinase gene from Bacillus licheniformis on the mode of expression in Bacillus subtilis. J Gen Microbiol. 1985;131:1753–1763. doi: 10.1099/00221287-131-7-1753. [DOI] [PubMed] [Google Scholar]
- 10.Isogai I, Takayama S, Murakoshi S, Suzuki A. Structures of β-amino acids in antibiotics iturin A. Tetrahedron Lett. 1982;23:3065–3068. [Google Scholar]
- 11.Itaya M, Kondo K, Tanaka T. A neomycin resistance gene cassette selectable in a single copy state in the Bacillus subtilis chromosome. Nucleic Acids Res. 1989;17:4410. doi: 10.1093/nar/17.11.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itaya M, Tanaka T. Complete physical map of the Bacillus subtilis 168 chromosome constructed by a gene-directed mutagenesis method. J Mol Biol. 1991;220:631–648. doi: 10.1016/0022-2836(91)90106-g. [DOI] [PubMed] [Google Scholar]
- 13.Konz D, Doekel S, Marahiel M A. Molecular and biochemical characterization of the protein template controlling biosynthesis of the lipopeptide lichenysin. J Bacteriol. 1999;181:133–140. doi: 10.1128/jb.181.1.133-140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunst F, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 15.Lambalot R H, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily—the phosphopantetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 16.Lin T S, Chen C L, Chang L K, Tschen J S, Liu S T. Functional and transcriptional analyses of a fengycin synthetase gene, fenC, from Bacillus subtilis. J Bacteriol. 1999;181:5060–5067. doi: 10.1128/jb.181.16.5060-5067.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maget-Dana R, Peypoux F. Iturins, a special class of pore-forming lipopeptides: biological and physicochemical properties. Toxicology. 1994;87:151–174. doi: 10.1016/0300-483x(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 18.Marahiel M A, Stachelhaus T, Mootz H D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 19.Matsuno Y, Ano T, Shoda M. High-efficiency transformation of Bacillus subtilis NB22, an antifungal antibiotic iturin producer, by electroporation. J Ferment Bioeng. 1992;73:261–264. [Google Scholar]
- 20.McKenzie T, Hoshino T, Tanaka T, Sueoka N. The nucleotide sequencing of pUB110: some salient features in relation to replication and its regulation. Plasmid. 1986;15:93–103. doi: 10.1016/0147-619x(86)90046-6. [DOI] [PubMed] [Google Scholar]
- 21.Mootz H D, Marahiel M A. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mootz H D, Marahiel M A. Design and application of multimodular peptide synthetases. Curr Opin Biotechnol. 1999;10:341–348. doi: 10.1016/S0958-1669(99)80062-7. [DOI] [PubMed] [Google Scholar]
- 23.Morbidoni H R, de Mendoza D, Cronan J E., Jr Bacillus subtilis acyl carrier protein is encoded in a cluster of lipid biosynthesis genes. J Bacteriol. 1996;178:4794–4800. doi: 10.1128/jb.178.16.4794-4800.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller A K, Rojo F, Alonso J C. The level of the pUB110 replication initiator protein is autoregulated, which provides an additional control for plasmid copy number. Nucleic Acids Res. 1995;23:1894–1900. doi: 10.1093/nar/23.11.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petit M A, Bruand C, Janniere L, Ehrlich S D. Tn10-derived transposons active in Bacillus subtilis. J Bacteriol. 1990;172:6736–6740. doi: 10.1128/jb.172.12.6736-6740.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peypoux F, Guinand M, Michel G, Delcambe L, Das B C, Lederer E. Structure of iturin A, peptidolipid antibiotic from Bacillus subtilis. Biochemistry. 1978;17:3992–3996. doi: 10.1021/bi00612a018. [DOI] [PubMed] [Google Scholar]
- 27.Peypoux F, Bonmatin J M, Wallach J. Recent trends in the biochemistry of surfactin. Appl Microbiol Biotechnol. 1999;51:553–563. doi: 10.1007/s002530051432. [DOI] [PubMed] [Google Scholar]
- 28.Revill W P, Bibbe M J, Hopwood D A. Purification of a malonyltransferase from Streptomyces coelicolor A3(2) and analysis of its genetic determinant. J Bacteriol. 1995;177:3946–3952. doi: 10.1128/jb.177.14.3946-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose M, Entian K D. New genes in the 170 region of the Bacillus subtilis genome encode DNA gyrase subunits, a thioredoxin, a xylanase and an amino acid transporter. Microbiology. 1996;142:3097–3101. doi: 10.1099/13500872-142-11-3097. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schneider A, Stachelhaus T, Marahiel M A. Targeted alteration of the substrate specificity of peptide synthetases by rational module swapping. Mol Gen Genet. 1998;257:308–318. doi: 10.1007/s004380050652. [DOI] [PubMed] [Google Scholar]
- 32.Shoda M. Bacterial control of plant diseases. J Biosci Bioeng. 2000;89:515–521. doi: 10.1016/s1389-1723(00)80049-3. [DOI] [PubMed] [Google Scholar]
- 33.Stachelhaus T, Schneider A, Marahiel M A. Rational design of peptide antibiotics by targeted replacement of bacterial and fungal domains. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 34.Stachelhaus T, Schneider A, Marahiel M A. Engineered biosynthesis of peptide antibiotics. Biochem Pharmacol. 1996;52:177–186. doi: 10.1016/0006-2952(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 35.Stein T, Vater J, Kruft V, Otto A, Wittmann-Liebold B, Franke P, Panico M, McDowell R, Morris H R. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J Biol Chem. 1996;271:15428–15435. doi: 10.1074/jbc.271.26.15428. [DOI] [PubMed] [Google Scholar]
- 36.Summers R G, Ali A, Shen B, Wessel W A, Hutchinson C R. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: a possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34:9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 37.Tognoni A, Franchi E, Magistrelli C, Colombo E, Cosmina P, Grandi G. A putative new peptide synthase operon in Bacillus subtilis: partial characterization. Microbiology. 1995;141:645–648. doi: 10.1099/13500872-141-3-645. [DOI] [PubMed] [Google Scholar]
- 38.Tosato V, Albertini A M, Zotti M, Sonda S, Bruschi C V. Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis 168. Microbiology. 1997;143:3443–3450. doi: 10.1099/00221287-143-11-3443. [DOI] [PubMed] [Google Scholar]
- 39.Tsuge K, Ano T, Shoda M. Isolation of a gene essential for biosynthesis of the lipopeptide antibiotics plipastatin B1 and surfactin in Bacillus subtilis YB8. Arch Microbiol. 1996;165:243–251. doi: 10.1007/s002030050322. [DOI] [PubMed] [Google Scholar]
- 40.Tsuge K, Ano T, Hirai M, Nakamura Y, Shoda M. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob Agents Chemother. 1999;43:2183–2192. doi: 10.1128/aac.43.9.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umezawa H, Aoyagi T, Nishikiori T, Okuyama A, Yamagishi Y, Hamada M, Takeuchi T. Plipastatins: new inhibitors of phospholipase A2, produced by Bacillus cereus BMG202-fF67. I. Taxonomy, production, isolation and preliminary characterization. J Antibiot. 1986;39:737–744. doi: 10.7164/antibiotics.39.737. [DOI] [PubMed] [Google Scholar]
- 42.Vanittanakom N, Loeffler W, Koch U, Jung G. Fengycin—a novel antifungal lipopeptide antibiotic produced by Bacillus subtilis F-29-3. J Antibiot. 1986;39:888–901. doi: 10.7164/antibiotics.39.888. [DOI] [PubMed] [Google Scholar]
- 43.Verwoert I I G S, Verbree E C, van der Linden K H, Nijkamp H J J, Stuitje A R. Cloning, nucleotide sequence, and expression of the Escherichia coli fabD gene, encoding malonyl coenzyme A-acyl carrier protein transacylase. J Bacteriol. 1992;174:2851–2857. doi: 10.1128/jb.174.9.2851-2857.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.von Döhren H, Keller U, Vater J, Zocher R. Multifunctional peptide synthetases. Chem Rev. 1997;97:2675–2705. doi: 10.1021/cr9600262. [DOI] [PubMed] [Google Scholar]
- 45.Yakimov M M, Kröger A, Slepak T N, Giuliano L, Timmis K N, Golyshin P N. A putative lichenysin A synthetase operon in Bacillus licheniformis: initial characterization. Biochim Biophys Acta. 1998;1399:141–153. doi: 10.1016/s0167-4781(98)00096-7. [DOI] [PubMed] [Google Scholar]