Abstract
The immunosuppressive non-classical human leukocyte antigen-G (HLA-G) can elicits pro-viral activities by down-modulating immune responses. We analysed soluble forms of HLA-G, IL-6 and IL-10 as well as on immune effector cell expression of HLA-G and its cognate ILT-2 receptor in peripheral blood obtained from hospitalised and convalescent COVID-19 patients. Compared with convalescents (N = 202), circulating soluble HLA-G levels (total and vesicular-bound molecules) were significantly increased in hospitalised patients (N = 93) irrespective of the disease severity. During COVID-19, IL-6 and IL-10 levels were also elevated. Regarding the immune checkpoint expression of HLA-G/ILT-2 on peripheral immune effector cells, the frequencies of membrane-bound HLA-G on CD3+ and CD14+ cells were almost identical in patients during and post COVID-19, while the frequency of ILT-2 receptor on CD3+ and CD14+ cells was increased during acute infection. A multi-parametric correlation analysis of soluble HLA-G forms with IL-6, IL-10, activation markers CD25 and CD154, HLA-G, and ILT-2 expression on immune cells revealed a strong positive correlation of soluble HLA-G forms with membrane-bound HLA-G molecules on CD3+/CD14+ cells only in convalescents. During COVID-19, only vesicular-bound HLA-G were positively correlated with the activation marker CD25 on T cells. Thus, our data suggest that the elevated levels of soluble HLA-G in COVID-19 are due to increased expression in organ tissues other than circulating immune effector cells. The concomitant increased expression of soluble HLA-G and ILT-2 receptor frequencies supports the concept that the immune checkpoint HLA-G/ILT-2 plays a role in the immune-pathogenesis of COVID-19.
Keywords: HLA-G, Immune checkpoint, Extracellular vesicles, COVID-19, ILT-2
1. Introduction
Non-classical class I Human Leukocyte Antigen (HLA)-G is an important inhibitory immune checkpoint molecule capable of modulating innate and adaptive immune responses [1], [2], [3], [4]. HLA-G is predominantly expressed in immune-privileged tissues and induces immunological tolerance that prevents tissue damage from inflammatory responses [5]. In contrast to classical HLA class I antigens, HLA-G is less polymorphic and exists only in rather few different isoforms. So far, seven isoforms of HLA-G have been described, including four membrane-bound antigens (HLA-G1, -G2, -G3 and -G4) and three soluble molecules (sHLA-G5, -G6, and -G7) [6], [7], [8]. Soluble HLA-G can be released as free soluble molecules (sHLA-G) or within extracellular vesicles (EV) [9], [10], [11], [12], [13]. Under physiological conditions, the cell surface expression of HLA-G is restricted to maternal-fetal interfaces and immune-privileged adult tissues, where it mediates immune-inhibitory pathways, supporting immunologic tolerance [14], [15], [16]. Several immunosuppressive mechanisms on CD8+ cytotoxic T lymphocytes (CTL), natural killer (NK) cells, B cells, and dendritic cells mediated by HLA-G molecules have been uncovered [17], [18], [19], [20], [21], [22]. Fundamental to these immunomodulatory effects of HLA-G is the interaction with its specific inhibitory receptors, in particular the immunoglobulin-like transcript (ILT)-2, ILT-4, and inhibitory killer receptor (KIR)-2DL4, which are expressed on various immune cells [23], [24], [25]. By binding directly to these immune cell inhibitor receptors, HLA-G and its soluble forms can exert several immunosuppressive functions leading to an impairment of immune cells proliferation and diminished effector functions such as cytotoxicity, chemotaxis, and immunoglobulin production, while the release of anti-inflammatory cytokines is increased [13], [22], [26], [27], [28], [29], [30].
HLA-G has been associated with a variety of non-physiological conditions such as transplantation malignancies, and infectious diseases [27], [31], [32], [33], [34], [35], [36], [37], [38], [39]. Viruses have evolved multiple subversive strategies to evade recognition and elimination by the immune system. In fact, altered HLA-G expression is observed during infections with influenza A virus, various human herpes viruses, BK polyomavirus, rabies virus, human immunodeficiency virus, hepatitis C virus, hepatitis B viruses, and, as recently described, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [32], [40], [41], [42], [43], [44]. In addition, various cytokines as well as certain stress stimuli such as interferon (IFN)-γ, interleukin (IL)-10, transforming growth factor (TGF)-β, hypoxia, and heat stress increase HLA-G levels. In viral infections, two hypotheses have been proposed to explain the role of HLA-G in viral immune-pathogenesis [45]: by its immunosuppressive properties, HLA-G might either promote viral escape from immune surveillance, or the expression of HLA-G may reflect a beneficial response to prevent over-shooting immune responses to limit overt tissue damage, which can occur during viral infections [46].
Starting in December 2019, the novel coronavirus SARS-CoV-2 caused a global pandemic, resulting in hundreds of millions of infection and millions of deaths [47], [48]. The clinical severity of COVID-19 (“coronavirus disease 2019″) is highly variable, ranging from asymptomatic infections to mild flu-like courses to life-threatening systemic inflammation with acute respiratory distress syndrome and high mortality [49], [50]. The variability of COVID-19 courses complicates clinical assessment, especially when it comes to early identification of patients at risk for severe disease [51]. A large number of potential risk factors for a severe COVID-19 course were identified over the past three years [52], [53], [54], [55], [56], [57], [58]. In the pathogenesis of COVID-19, the interplay between direct virus-mediated damage and associated host responses seems to be crucial. Effective innate and adaptive host antiviral responses to SARS-CoV-2 include production of various pro-inflammatory cytokines [59]. This leads to the activation of CD4+ and CD8+ T cells, which is essential for controlling viral replication and spread [55], [57], [59]. However, SARS-CoV-2 infection often leads to a dysregulated and exaggerated immune response that manifests itself in enhanced cytokine production. This can cause a fulminant hyper-inflammatory syndrome and thus mediate more severe disease [60], [61]. In the context of COVID-19 and the role of HLA-G in the immunopathology of the disease, it has been suggested that upon the influence of IL-10, IL-6 and IL-8 in early inflammatory stage of the infection, HLA-G expression might be enhanced to prevent injuries. Thus, the expression or secretion of HLA-G might reflect a negative feedback response to inflammatory processes during viral infections. In line with this, high membrane-bound HLA-G expression during the early phase of SARS-CoV-2 infection and significantly higher serum levels of sHLA-G in patients with COVID-19 have been reported [62], [63], [64], [65], [66], [67]. These HLA-G levels appear to depend on the severity of COVID-19 infection [68]. Recently, an induction of HLA-G expression in damaged lung tissue and a negative correlation between HLA-G expression and COVID-19 course has been described [59].
To further elucidate the role of the HLA-G/ILT-2 ligand-receptor immune checkpoint axis in the context of COVID-19, we assessed (i) total soluble HLA-G (sHLA-Gtot), (ii) EV-bound HLA-G (sHLA-GEV), (iii) the cellular expression HLA-G and its receptor ILT-2 on peripheral immune cells, (iv) the cytokines IL-6 and IL-10, and (v) the activation markers CD25 and CD154 in SARS-CoV-2-infected and convalescent patients.
2. Methods
2.1. Study cohort
Recruitment of patients attending the Department of Infectious Diseases of the University Hospital Essen for this prospective study started in March 2020 and ended in April 2021. In total, we included 266 unvaccinated SARS-CoV-2-positive patients with at least one positive real-time reverse transcription polymerase chain reaction (RT-PCR) test result. Ninety-three EDTA blood samples were obtained from patients hospitalised for moderate or severe COVID-19 and 202 blood samples were procured post-recovery from individuals enrolled in our outpatient clinic. Peripheral blood samples for immunological analysis on peripheral blood lymphocytes were collected from 58 patients.
Hospitalized patients were grouped into categories according to their worst disease manifestation at the time point of blood sampling, by the ECDC (European Center of Disease Prevention and Control, 2021) criteria: 69 non-severe hospitalized (ECDC 0); 15 severe hospitalized admitted to an intensive care unit or became dependent on mechanical ventilation (ECDC 1) and 9 patients with COVID-19-related deaths either during the hospital stay or within a follow-up of 30 days (ECDC 2).
For further statistical analyses, demographic data, medical history, and laboratory parameters at the time of hospitalization were documented for each patient. The clinical characteristics of the patient cohort were comprehensively recorded and are summarized in Table 1 .
Table 1.
Demographic and clinical characteristics of the study cohort.
| Parameter | Total (n = 266) |
|
|---|---|---|
| n | % | |
| Age (≥60) | 83 | 31.2 |
| Male sex | 135 | 50.8 |
| Obesity (BMI ≥ 30) | 100 | 37.6 |
| Diabetes* | 31 | 11.8 |
| Cardiovascular disease* | 44 | 16.7 |
| Hypertension* | 76 | 28.9 |
| Lung disease* | 44 | 16.7 |
| Immunosuppression** | 31 | 11.7 |
*Information not available for three patients, **information not available for one patient.
The study was approved by the local ethics committee (approval no. 20-9374-BO and 20-9529-BO) and performed in accordance with ethics standards noted in the 1964 Declaration of Helsinki and its later amendments or comparable ethics standards. All patients provided informed consent for participation in the study.
2.2. Blood sampling
After blood collection, the samples were centrifuged at 1500 g for 10 min. The corresponding supernatants were stored at −80 °C until use.
Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll density gradient centrifugation. Thereafter, PBMC were stored in freezing media (RPMI 1640, Thermo Fisher Scientific, Darmstadt, Germany) with 10% (v/v) fetal bovine serum (PAN-Biotech GmbH, Aidenbach, Germany) and 0.556 µg DNAse (Roche, Mannheim, Germany) in liquid nitrogen until usage.
2.3. Quantification of soluble HLA-G plasma levels
Soluble HLA-G was quantified as previously described, using anti-HLA-G MEM-G/09 (Exbio, Praha, Czech Republic) as capture antibody and anti-β2-microglobulin antibody (Dako, Denmark) as secondary antibody conjugated with peroxidase [7], [35].
2.4. Isolation and characterization of extracellular vesicle
The preparation of EV was performed from patient plasma as described previously [10], [12] by precipitation using ExoQuick (System Biosciences, CA, USA) according to manufacturer's instructions. After precipitation, the pellets were resuspended in PBS and the EVs fractions were stored at −20 °C until further processing.
2.5. Quantification of IL-6 and IL-10 levels
Quantification of IL-6 and IL-10 levels in undiluted plasma samples was performed using commercially available kit (ThermoFisher, San Diego, CA, USA) according to manufacturer’s protocols.
2.6. Flow cytometry analysis
Frozen PBMC from healthy donors were thawed in RPMI-1640 and 0.556 μg DNAse (Roche). Subsequently, the surface expression on peripheral blood mononuclear cells (PBMC) was analysed with an antibody cocktail containing fluorochrome-conjugated mononuclear antibodies recognizing CD3 (BV421), CD25 (BV650), ILT-2 (FITC), CD14 (BV510), or HLA-G (APC). In addition, a second antibody cocktail containing anti-human CD154 (FITC) and anti-human CD3 (ECD) was used.
All antibodies were provided by BioLegend (Koblenz, Germany) with the exception of ECD-conjugated CD3 (Beckman Coulter, Krefeld Germany). Isotype-matched antibodies served as negative controls (Beckman Coulter or BioLegend). Samples were subjected to multi-colour flow cytometry using a CytoFlexS cytometer (Beckman Coulter). Data acquisition of at least 200.000 events was performed with CytExpert Version 2.1 software (Beckman Coulter) and analysed using Kaluza Analysis 2.1 software. The general gating strategy for flow cytometric analysis is shown in Supplementary Fig. S1.
2.7. Statistical and graphical analysis
Statistical analyses were performed using SPSS 25.0 (SPSS Inc.), GraphPad Prism 9.0 (GraphPad Software), or R. Data are presented as mean ± standard deviation (SD). After testing for Gaussian distribution, the differences between two or more groups were assessed by the Mann-Whitney test or the Kruskal-Wallis test. Correlation was performed with the R package corrplot (version 0.4.0; https://CRAN.R-project.or/package = corrplot). Statistical significance was defined as p ≤ 0.05. The graphs were created using GraphPad Prism 9 or R.
3. Results
3.1. Total amounts of sHLA-G and HLA-G-bearing EVs are elevated during COVID-19 and are associated with COVID-19 disease severity
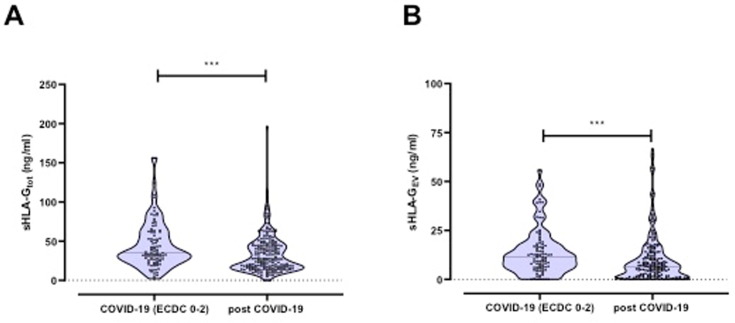
To determine whether the SARS-CoV-2 infection stimulates the induction of sHLA-G in peripheral blood, we quantified plasma levels of sHLA-Gtot and sHLA-GEV in acute COVID-19 patients. SARS-CoV-2 convalescents served as controls. The comparison of sHLA-Gtot and HLA-GEV revealed significantly higher levels during acute COVID-19 in comparison to convalescents (sHLA-Gtot: acute COVID-19 patients ECDC 0–2: 45.2 ± 30.0 ng/ml vs. post COVID-19: 33.5 ± 24.3 ng/ml; p = 0.0003; sHLA-GEV: acute COVID-19 patients ECDC 0–2: 13.9 ± 11.8 ng/ml vs. post COVID-19: 10.3 ± 12.6 ng/ml; p = 0.0045, Fig. 1 A and B).
Fig. 1.
A) Total plasma levels of soluble HLA-G (sHLA-G) as well as B) level of HLA-G bearing extracellular vesicles (EVs) in COVID-19 patients and in convalescents.
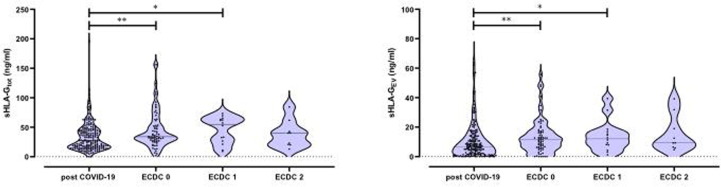
Next, we analysed sHLA-G levels in relation to the severity of COVID-19 disease. The highest levels of sHLA-Gtot were found in patients with severe COVID-19 (ECDC 1; see Fig. 1A). Of note, none-severe and severe hospitalized COVID-19 patients (ECDC 0 and ECDC 1; sHLA-Gtot 45.7 ± 32.5 ng/ml and 45.7 ± 22.5 ng/ml) had increased levels of sHLA-G compared to convalescent post-COVID-19 patients (sHLA-Gtot: 33.5 ± 24.3 ng/ml; p = 0.0024 and p = 0.036). It should be emphasised that in critically ill patients with fatal outcome (ECDC 2; 38.5 ± 23.0 ng/ml), the plasma levels of sHLA-Gtot appeared lower compared to the other COVID-19 severity levels (ECDC 0 or ECDC 1). Although sHLA-Gtot levels were higher in patients with fatal outcome than in patients after COVID-19, no significant difference was observed between these two groups, most likely due to the small number of patients with fatal outcome. Similar results were obtained for sHLA-GEV (see Fig. 2 B).
Fig. 2.
A) Total plasma levels of soluble HLA-G (sHLA-G) as well as B) level of HLA-G bearing extracellular vesicles (EVs) in convalescents and patients with different COVID-19 severity.
3.2. IL-6 and IL-10 plasma levels significantly increase during COVID-19
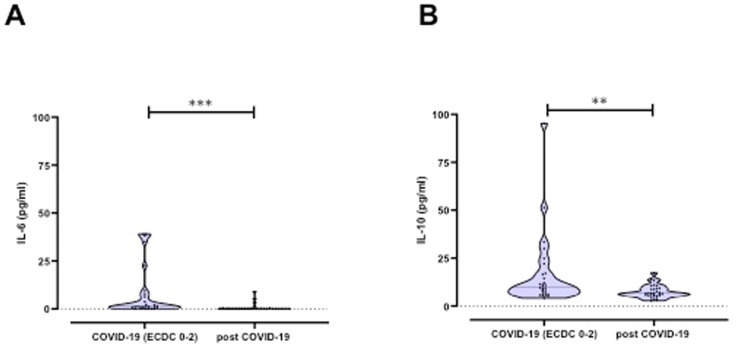
Plasma levels of the acute phase protein IL-6 known to be associated with severe COVID-19, as well as the immunosuppressive cytokine IL-10 were significantly enhanced in 28 acute COVID-19 patients (IL-6: 7.7 ± 13.3 pg/ml and IL-10: 16.3 ± 18.7 pg/ml) compared to 30 convalescents (IL-6: 0.6 ± 1.9 pg/ml and IL-10: 7.4 ± 3.3 pg/ml; IL-6p < 0.0001 and IL-10p = 0.004) (Fig. 3 A and B).
Fig. 3.
A) Plasma levels of IL-6 as well as B) IL-10 in convalescents and patients with different COVID-19 severity.
3.3. The frequency of ILT-2-positive cells is increased during acute COVID-19 compared to convalescence
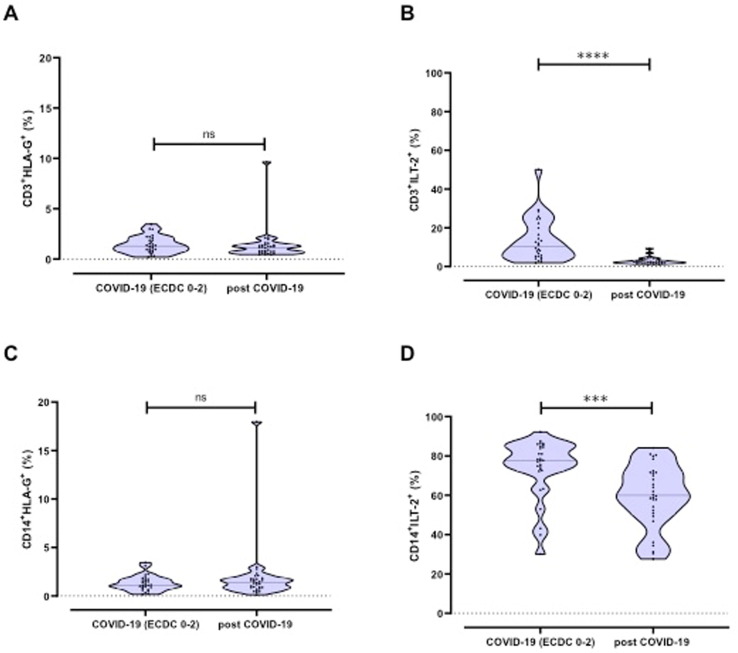
To determine whether the increased sHLA-G expression (both total and EV-resident) during COVID-19 merely reflects the HLA-G expression on PBMC, frequencies of HLA-G-positive CD3+ and CD14+ immune effector cells were determined in 58 patients by flow cytometry (Fig. 4 ). Since the immunomodulatory effect of HLA-G is mainly mediated by its interaction with the ILT-2 receptor, we additionally determined the expression of the ILT-2 receptor on these cells. Expression frequencies of HLA-G and ILT-2 on immune effector cells from COVID-19 patients (N = 28) were compared with those from convalescents (N = 30). As shown in Fig. 4, HLA-G frequencies on CD3+ and CD14+ cells in patients with acute COVID-19 did not significantly differ from convalescent patients post COVID-19. When comparing receptor frequencies on CD3+ and CD14+ immune cell populations, the proportion of ILT-2-positive cells was significantly higher in COVID-19 patients compared to convalescents (CD3+: 13.6 ± 11.1 % vs. 2.8 ± 1.7 %, p < 0.0001; CD14+: 73.7 ± 15.3 % vs. 59.5 ± 15.9 % p = 0.0002).
Fig. 4.
A-D HLA-G and ILT-2 frequency on immune effector cells of COVID-19 patients and convalescent patients post COVID; A) HLA-G on CD3+ cells; B) ILT-2 on CD3+ cells; C) HLA-G on CD14+ cells; D) ILT-2 on CD14+ cells.
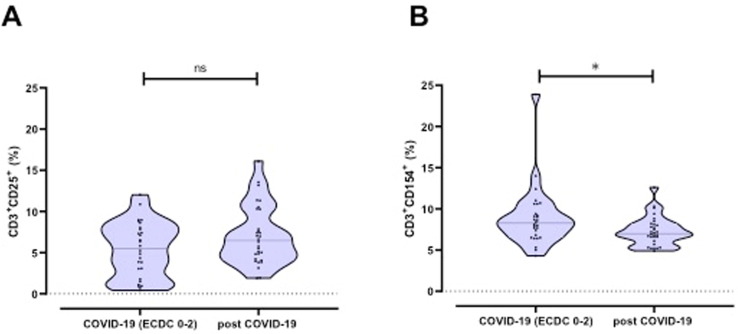
3.4. The frequency of CD3+ CD154+ cells is increased in COVID-19 patients
In patients with acute COVID-19, we found an increase in frequency of CD3+ CD154+ cells (p = 0.03), while there was no difference in frequency of CD3+ CD25+ cells when comparing patients with COVID-19 with post COVID-19 individuals (Fig. 5 A and B).
Fig. 5.
A-B: Frequencies of the activation markers CD25 and CD154 on CD3+ cells.
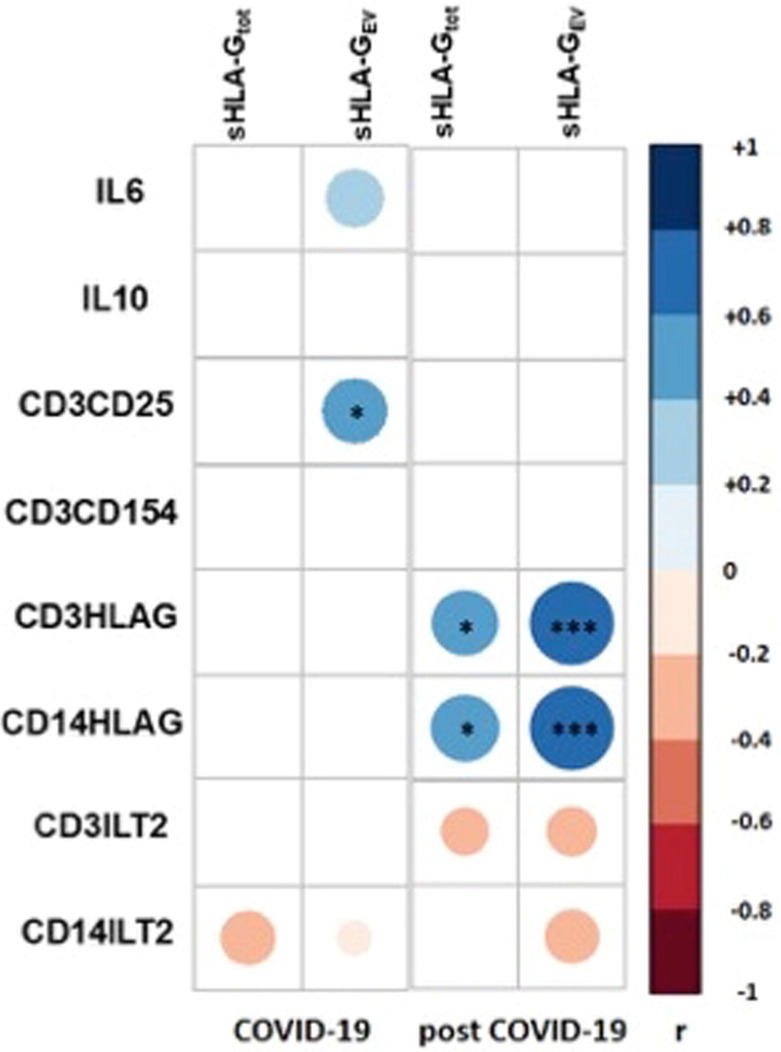
3.5. Exclusively correlation of sHLA-G with HLA-G expression on CD3+ and CD14+ immune effector cells in convalescence
In individuals who recovered from COVID-19, there was a strong positive correlation between soluble (total and EV-resident) HLA-G levels and membrane-bound HLA-G expression on CD3+ and CD14+ immune effector cells (Fig. 6 ). Conversely, during COVID-19, no correlation was found between sHLA-G levels and the HLA-G expression on CD3+ and CD14+ cells. Furthermore, a correlation analysis showed a significant association between sHLA-GEV molecules and the frequency of CD3+ CD25+ cells during COVID-19. A negative yet non-significant trend towards an association between sHLA-G levels and ILT-2 frequency on immune effector cells was found.
Fig. 6.
Correlation map - Pearson correlation map with association to functional markers for patients with COVID-19 and convalescents: Pearson correlation coefficients are represented by different colours defined in the scale on the right side of the correlation map. Significant correlations are highlighted by *: <0.05 and **: <0.01.
4. Discussion
Numerous studies reporting clinical and immunological findings regarding the immune-pathological features of COVID-19 have been published. Nevertheless, the molecular mechanisms involved in the dysregulation of cellular immune responses against SARS-CoV-2 infection are far from being fully understood. Defining parameters that may contribute to the high clinical variability of COVID-19 is critical in predicting disease outcomes and for the development of effective therapeutic strategies. To enable efficient replication, SARS-CoV-2 evolved various strategies to escape from antiviral immune responses of the host that may also affect the disease progression. Early in the pandemic, high membrane and epithelial expression of HLA-G was described in a patient infected with SARS-CoV2 [69]. This led to the hypothesis that HLA-G might modulate the hyper-inflammation induced by SARS-CoV2 [45], [46]. The hypothesis was supported by results of a large genome-wide association study that - among others - identified a HLA-G single nucleotide polymorphism within the 3′untranslated region (rs9380142) as a genetic marker strongly associated with COVID-19 severity [70].
In our study, we showed to our knowledge for the first time that in patients hospitalized for mild or severe but not fatal SARS-CoV-2 infection, the total amount of sHLA-G as well as HLA-GEV in the circulation is increased, whereas the frequency of HLA-G-positive CD3+ and CD14+ cells was similar in acute and post COVID-19 patients. However, the frequency of ILT-2 on CD3+ and CD14+ cells was increased in acute COVID-19. While the level of soluble HLA-G was positively correlated with membrane-bound frequency on T-cells and monocytes in convalescent individuals, this was not the case in patients acutely infected with SARS-CoV-2, implying that the high amount of soluble HLA-G is independent of the expression on immune effector cells during COVID-19. Thus, it is very likely that the increase of soluble HLA-G during SARS-CoV-2 infection might originate from additional organ systems. In this context it is interestingly that the induction of HLA-G expression in damaged lung epithelial tissue was recently described in COVID-19 and was higher than in lung epithelia of influenza-infected patients [71]. This suggests that although HLA-G is upregulated in response to various infections, it may be differentially regulated depending on the infectious pathogen. In this recently published study, increased immune cell infiltration was observed in the lungs of SARS-CoV-2 infected patients. Other studies have found that soluble HLA-G is significantly increased in patients with COVID-19 and is related to disease severity, which is consistent with our data [62], [63], [64], [65], [66], [67], [68]. In our study, the lack of longitudinal sampling and the small number of patients with fatal outcome are limitations of the study that allow limited conclusions about HLA-G levels in critically ill patients. In addition, control patients with other respiratory viral infections are lacking to clarify whether or not the effects observed in our study are specific to SARS-CoV-2.
Previous studies have described a vast increase in CD25 expression, alpha chain of the IL-2 receptor, on T cells in both comorbid and severe COVID-19 cases and have linked this finding to the ability of SARS-CoV-2 to trigger massive activation of immunological responses in some infected individuals [72], [73], [74], [75]. Interestingly, we were able to demonstrate that the release of sHLA-GEV during COVID-19 was positively correlated with the expression of the activation marker CD25 on T cells, which may reflect the negative feedback on inflammatory processes during severe COVID-19. Moreover, increased expression of CD25 was found in murine models under the influence of extracellular vesicles [76], [77].
COVID-19 has been described to be associated with a dysregulated immune state and an IL-6-driven hyper-inflammation, as well as T and B cell lymphopenia [78], [79]. Similar to other studies, we also confirmed the finding of elevated IL-6 and IL-10 levels during COVID-19 in our cohort [80]. However, in our cohort, we could not demonstrate a significant correlation of IL-6 or IL-10 levels with soluble HLA-G forms. Of note, HLA-G expression is influenced by several immunomodulatory molecules [81], and IL-10 is one of the cytokines known to increase the expression of HLA-G [82], [83], [84]. Therefore, our data suggest the existence of additional mechanisms beyond increased IL-10 secretion contributing to increased HLA-G levels during COVID-19.
To assess the role of the HLA-G/ILT-2 ligand-receptor axis as an important immune checkpoint in the periphery, we additionally analysed the frequency of ILT-2 receptors on T-cells and monocytes. Indeed, an increase of ILT-2 frequency was observed in our study cohort during acute SARS-CoV-2 infection compared to the post COVID-19 status. The up-regulation of inhibitory receptors on immune cells may enhance their immune suppressive properties. HLA-G interacts with several receptors that can induce immunomodulatory effects and originate from different receptor families. The ILT-2 and ILT-4 receptors belong to the leukocyte immunoglobulin-like receptor (LILR) family, and the inhibitory killer receptor (KIR)-2DL4 belongs to the killer cell Ig-like receptors (KIR) [23], [24], [25]. The binding of HLA-G to its receptors is influenced by multiple mechanisms [85]. In particular, the ILT-2 receptor binds ß2-microglobulin-associated HLA-G isoforms (being the HLA-G1 and HLA-G5 isoforms). Since previous studies have shown that the monoclonal HLA-G specific MEM-G/9 antibody recognises HLA-G molecules only in the presence of ß2-microglobulin, we focused exclusively on the ILT-2 receptor in this study [86]. We have previously shown that soluble HLA-G can induce phenotypic changes in immune cells, including the up-regulation of ILT-2 [9], [25]. In addition, the transcriptional up-regulation has been observed in the presence of soluble HLA-G [87]. However, in the present study, we did not find a positive correlation between soluble HLA-G and ILT-2 during acute SARS-CoV-2 infection. This indicates that the enhanced ILT-2 expression on T cells and monocytes is independent of soluble HLA-G expression, but is rather attributable to the viral infectious status. Of note, ILT-2 expression is regulated by a number of polymorphisms in the gene promoter region that may influence the individual expression frequency in addition to HLA-G levels [88]. Moreover, for NK cells and CD56+ T-cells, it was shown that ILT-2 expression is up-regulated under the influence of IL-2 and IL-15 [89].
Nevertheless, the concomitant increased expression of soluble HLA-G and ILT-2 frequencies detected in our study supports the concept that the HLA-G/ILT-2 immune checkpoint plays a role in the immune-pathogenesis of COVID-19. This would qualify HLA-G and ILT-2 not only as prognostic markers but also as potential therapeutic targets. It still needs to be discussed how this therapeutic approach should look like for viral infections such as SARS-CoV-2 as recently described in tumour biology. The HLA-G-specific CAR-T-cells recently described are certainly less suitable, as they would target all HLA-G-expressing cells [90]. In COVID-19, where HLA-G expression has been described in the lung, this approach would increase cytotoxic and hyper-inflammatory responses in lung tissue [71] and may result in lung failure. In contrast to tumor biology, COVID-19 is characterized by opposing immunological processes in its two disease phases [45]. In the early phase of infection functionally blocking antibodies targeting the HLA-G/ILT-2 axis would be of interest, to impede HLA-G mediated viral escape from immune surveillance. In this context, it is interesting to note that novel therapeutic anti-ILT-2 antibody and anti-HLA-G antibody have been described and are currently under evaluation in tumour immunology [91], [92]. In later phases during hyperinflammation agonistic HLA-G/ILT-2 immune checkpoint antibodies or soluble HLA-G molecules could be promising immunotherapeutic approaches to attenuate inflammation and prevent further tissue damage. Here, it would be of great interest to study the amount of soluble HLA-G in COVID-19 convalescent plasma being used for the treatment of COVID-19 patients and to relate the results to therapeutic outcome.
5. Conclusion
In summary, the data presented here support an important role of the immune checkpoint axis HLAG/ILT-2 in the immune-pathogenesis of COVID-19 and thus qualify HLA-G and ILT-2 not only as prognostic markers but also as potential therapeutic approaches.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the “Stiftung Universitätsmedizin” (approval no. 2020 4699 135), University of Duisburg-Essen and the “Rudolf Ackermann Stiftung”. Special thanks go to the patients kindly providing their samples. We are grateful for the technical support to the team of the Department of Infectious Diseases and the colleagues from the Institute for Transfusion Medicine, both University Hospital Essen.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humimm.2023.03.002.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Tronik-Le Roux D., et al. The HLA-G immune checkpoint: a new immuno-stimulatory role for the alpha1-domain-deleted isoform. Cell. Mol. Life Sci. 2022;79(6):310. doi: 10.1007/s00018-022-04359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carosella E.D. The tolerogenic molecule HLA-G. Immunol. Lett. 2011;138(1):22–24. doi: 10.1016/j.imlet.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Carosella E.D., Dausset J., Rouas-Freiss N. Immunotolerant functions of HLA-G. Cell. Mol. Life Sci. 1999;55(3):327–333. doi: 10.1007/s000180050295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amiot L., et al. Biology of HLA-G in cancer: a candidate molecule for therapeutic intervention? Cell. Mol. Life Sci. 2011;68(3):417–431. doi: 10.1007/s00018-010-0583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonzalez A., et al. The immunosuppressive molecule HLA-G and its clinical implications. Crit. Rev. Clin. Lab. Sci. 2012;49(3):63–84. doi: 10.3109/10408363.2012.677947. [DOI] [PubMed] [Google Scholar]
- 6.Martelli-Palomino G., et al. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PLoS One. 2013;8(10):e71742. doi: 10.1371/journal.pone.0071742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rebmann V., et al. Quantification and identification of soluble HLA-G isoforms. Tissue Antigens. 2007;69(Suppl 1):143–149. doi: 10.1111/j.1399-0039.2006.763_5.x. [DOI] [PubMed] [Google Scholar]
- 8.Amodio G., Gregori S. HLA-G genotype/expression/disease association studies: success, hurdles, and perspectives. Front. Immunol. 2020;11:1178. doi: 10.3389/fimmu.2020.01178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwich E., et al. Soluble HLA-G and HLA-G bearing extracellular vesicles affect ILT-2 positive and ILT-2 negative CD8 T cells complementary. Front. Immunol. 2020;11:2046. doi: 10.3389/fimmu.2020.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwich E., et al. Vesicular-bound HLA-G as a predictive marker for disease progression in epithelial ovarian cancer. Cancers (Basel) 2019;11(8) doi: 10.3390/cancers11081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alegre E., et al. In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur. J. Immunol. 2013;43(7):1933–1939. doi: 10.1002/eji.201343318. [DOI] [PubMed] [Google Scholar]
- 12.Konig L., et al. The prognostic impact of soluble and vesicular HLA-G and its relationship to circulating tumor cells in neoadjuvant treated breast cancer patients. Hum. Immunol. 2016;77(9):791–799. doi: 10.1016/j.humimm.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Nardi Fda S., et al. Soluble monomers, dimers and HLA-G-expressing extracellular vesicles: the three dimensions of structural complexity to use HLA-G as a clinical biomarker. HLA. 2016;88(3):77–86. doi: 10.1111/tan.12844. [DOI] [PubMed] [Google Scholar]
- 14.Alegre E., et al. Maternal antigen presenting cells are a source of plasmatic HLA-G during pregnancy: longitudinal study during pregnancy. Hum. Immunol. 2007;68(8):661–667. doi: 10.1016/j.humimm.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Carosella E.D., et al. The role of HLA-G in immunity and hematopoiesis. Cell. Mol. Life Sci. 2011;68(3):353–368. doi: 10.1007/s00018-010-0579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouas-Freiss N., et al. Role of the HLA-G immune checkpoint molecule in pregnancy. Hum. Immunol. 2021;82(5):353–361. doi: 10.1016/j.humimm.2021.01.003. [DOI] [PubMed] [Google Scholar]
- 17.Adrian Cabestre F., et al. HLA-G expression in human melanoma cells: protection from NK cytolysis. J. Reprod. Immunol. 1999;43(2):183–193. doi: 10.1016/s0165-0378(99)00037-6. [DOI] [PubMed] [Google Scholar]
- 18.Bahri R., et al. Soluble HLA-G inhibits cell cycle progression in human alloreactive T lymphocytes. J. Immunol. 2006;176(3):1331–1339. doi: 10.4049/jimmunol.176.3.1331. [DOI] [PubMed] [Google Scholar]
- 19.Bahri R., et al. Dendritic cells secrete the immunosuppressive HLA-G molecule upon CTLA4-Ig treatment: implication in human renal transplant acceptance. J. Immunol. 2009;183(11):7054–7062. doi: 10.4049/jimmunol.0803054. [DOI] [PubMed] [Google Scholar]
- 20.Carosella E.D., Gregori S., LeMaoult J. The tolerogenic interplay(s) among HLA-G, myeloid APCs, and regulatory cells. Blood. 2011;118(25):6499–6505. doi: 10.1182/blood-2011-07-370742. [DOI] [PubMed] [Google Scholar]
- 21.Wu C.L., et al. Inhibition of iNKT cells by the HLA-G-ILT2 checkpoint and poor stimulation by HLA-G-expressing tolerogenic DC. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.608614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzo R., et al. A functional role for soluble HLA-G antigens in immune modulation mediated by mesenchymal stromal cells. Cytotherapy. 2008;10(4):364–375. doi: 10.1080/14653240802105299. [DOI] [PubMed] [Google Scholar]
- 23.Carosella E.D., Gregori S., Tronik-Le Roux D. HLA-G/LILRBs: a cancer immunotherapy challenge. Trends Cancer. 2021;7(5):389–392. doi: 10.1016/j.trecan.2021.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Jacquier A., et al. Cytometry-based analysis of HLA-G functions according to ILT2 expression. Hum. Immunol. 2020;81(4):168–177. doi: 10.1016/j.humimm.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Rohn H., et al. Effect of HLA-G5 immune checkpoint molecule on the expression of ILT-2, CD27, and CD38 in splenic B cells. J. Immunol. Res. 2022;2022:4829227. doi: 10.1155/2022/4829227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carosella E.D., et al. HLA-G: an immune checkpoint molecule. Adv. Immunol. 2015;127:33–144. doi: 10.1016/bs.ai.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 27.Rebmann V., et al. HLA-G as a tolerogenic molecule in transplantation and pregnancy. J. Immunol. Res. 2014;2014 doi: 10.1155/2014/297073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebmann V., et al. The potential of HLA-G-bearing extracellular vesicles as a future element in HLA-G immune biology. Front. Immunol. 2016;7:173. doi: 10.3389/fimmu.2016.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouas-Freiss N., et al. HLA-G promotes immune tolerance. J. Biol. Regul. Homeost. Agents. 2000;14(2):93–98. [PubMed] [Google Scholar]
- 30.Seliger B. The non-classical antigens of HLA-G and HLA-E as diagnostic and prognostic biomarkers and as therapeutic targets in transplantation and tumors. Clin. Transpl. 2013:465–472. [PubMed] [Google Scholar]
- 31.Rebmann V., et al. Systematic evaluation of HLA-G 3'Untranslated region variants in locally advanced, non-metastatic breast cancer patients: UTR-1, 2 or UTR-4 are predictors for therapy and disease outcome. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.817132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rohn H., et al. HLA-G 3' untranslated region gene variants are promising prognostic factors for BK polyomavirus replication and acute rejection after living-donor kidney transplant. Hum. Immunol. 2020;81(4):141–146. doi: 10.1016/j.humimm.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 33.Schwich E., et al. HLA-G 3' untranslated region variants +3187G/G, +3196G/G and +3035T define diametrical clinical status and disease outcome in epithelial ovarian cancer. Sci. Rep. 2019;9(1):5407. doi: 10.1038/s41598-019-41900-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer G., et al. HLA-G is a potential tumor marker in malignant ascites. Clin. Cancer Res. 2003;9(12):4460–4464. [PubMed] [Google Scholar]
- 35.Guberina H., et al. Recipient HLA-G +3142 CC genotype and concentrations of soluble HLA-G impact on occurrence of CMV infection after living-donor kidney transplantation. Int. J. Mol. Sci. 2017;18(11) doi: 10.3390/ijms18112338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avokpaho E., et al. HLA-G expression during hookworm infection in pregnant women. Acta Trop. 2019;196:52–59. doi: 10.1016/j.actatropica.2019.04.026. [DOI] [PubMed] [Google Scholar]
- 37.Brugiere O., et al. Chronic lung allograft dysfunction is associated with an early increase of circulating cytotoxic CD4+CD57+ILT2+ T cells, selectively inhibited by the immune check-point HLA-G. J. Heart Lung Transpl. 2022;41(5):626–640. doi: 10.1016/j.healun.2022.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Tizaoui K., et al. The relationship of 3'UTR HLA-G14-bp insertion/deletion and +3142 C/G polymorphisms and soluble HLA-G expression with gynecological cancers: an updated meta-analysis. Immun. Inflamm. Dis. 2022;10(7):e645. doi: 10.1002/iid3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebmann V., Wagner S., Grosse-Wilde H. HLA-G expression in malignant melanoma. Semin. Cancer Biol. 2007;17(6):422–429. doi: 10.1016/j.semcancer.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Bertol B.C., et al. HLA-G liver expression and HLA-G extended haplotypes are associated with chronic hepatitis C in HIV-negative and HIV-coinfected patients. Clin. Immunol. 2020;217 doi: 10.1016/j.clim.2020.108482. [DOI] [PubMed] [Google Scholar]
- 41.Jasinski-Bergner S., et al. Role of HLA-G in viral infections. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.826074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laaribi A.B., et al. Increased levels of soluble HLA-G molecules in Tunisian patients with chronic hepatitis B infection. J. Viral Hepat. 2017;24(11):1016–1022. doi: 10.1111/jvh.12718. [DOI] [PubMed] [Google Scholar]
- 43.Laaribi A.B., et al. Association of an HLA-G 14-bp Insertion/Deletion polymorphism with high HBV replication in chronic hepatitis. J. Viral Hepat. 2015;22(10):835–841. doi: 10.1111/jvh.12395. [DOI] [PubMed] [Google Scholar]
- 44.Morandi F., et al. Recent advances in our understanding of HLA-G biology: lessons from a wide spectrum of human diseases. J. Immunol. Res. 2016;2016:4326495. doi: 10.1155/2016/4326495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zidi I. Puzzling out the COVID-19: therapy targeting HLA-G and HLA-E. Hum. Immunol. 2020;81(12):697–701. doi: 10.1016/j.humimm.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin A., Yan W.H. Perspective of HLA-G induced immunosuppression in SARS-CoV-2 infection. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.788769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu N., et al. A Novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seibert F.S., et al. Detection of SARS-CoV-2 pneumonia: two case reports. J. Med. Case Rep. 2020;14(1):242. doi: 10.1186/s13256-020-02551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guberina H., et al. A patient with severe respiratory failure caused by novel human coronavirus. Infection. 2014;42(1):203–206. doi: 10.1007/s15010-013-0509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thummler L., et al. Cellular and humoral immunity against different SARS-CoV-2 variants is detectable but reduced in vaccinated kidney transplant patients. Vaccines (Basel) 2022;10(8) doi: 10.3390/vaccines10081348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohlendick B., et al. The GNB3 c.825C>T (rs5443) polymorphism and protection against fatal outcome of corona virus disease 2019 (COVID-19) Front. Genet. 2022;13 doi: 10.3389/fgene.2022.960731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y., et al. Impacts of immunosuppression and immunodeficiency on COVID-19: a systematic review and meta-analysis. J. Infect. 2020;81(2):e93–e95. doi: 10.1016/j.jinf.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wunsch K., et al. COVID-19 in elderly, immunocompromised or diabetic patients-from immune monitoring to clinical management in the hospital. Viruses. 2022;14(4) doi: 10.3390/v14040746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Konik M., et al. Long-term SARS-CoV-2 specific immunity is affected by the severity of initial COVID-19 and patient age. J. Clin. Med. 2021;10(19) doi: 10.3390/jcm10194606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paniskaki K., et al. Superior humoral immunity in vaccinated SARS-CoV-2 convalescence as compared to SARS-COV-2 infection or vaccination. Front. Immunol. 2022;13:1031254. doi: 10.3389/fimmu.2022.1031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thummler L., et al. Long-term cellular immune response in immunocompromised unvaccinated COVID-19 patients undergoing monoclonal antibody treatment. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.980698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindemann M., et al. SARS-CoV-2-specific humoral and cellular immunity in two renal transplants and two hemodialysis patients treated with convalescent plasma. J. Med. Virol. 2021;93(5):3047–3054. doi: 10.1002/jmv.26840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paniskaki K., et al. Immune response in moderate to critical breakthrough COVID-19 infection after mRNA vaccination. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.816220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lucas C., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584(7821):463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20(6):355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang S., et al. Dynamics of peripheral immune cells and their HLA-G and receptor expressions in a patient suffering from critical COVID-19 pneumonia to convalescence. Clin. Transl. Immunol. 2020;9(5):e1128. doi: 10.1002/cti2.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al-Bayatee N.T., Ad'hiah A.H. Soluble HLA-G is upregulated in serum of patients with severe COVID-19. Hum. Immunol. 2021;82(10):726–732. doi: 10.1016/j.humimm.2021.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamed R.M., Mahmood M.M., Ad'hiah A.H. Evaluation of serum soluble HLA-G levels post-recovery from COVID-19 and post-vaccination (Sinopharm and Pfizer-BioNTech) Hum. Immunol. 2022 doi: 10.1016/j.humimm.2022.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.da Silva-Neto P.V., et al. Matrix metalloproteinases on severe COVID-19 lung disease pathogenesis: cooperative actions of MMP-8/MMP-2 axis on immune response through HLA-G shedding and oxidative stress. Biomolecules. 2022;12(5) doi: 10.3390/biom12050604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramzannezhad S., et al. The association of decreased HLA-G(+) immune cell frequencies with critical COVID-19 patients. Microb Pathog. 2022;167 doi: 10.1016/j.micpath.2022.105550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bortolotti D., et al. Increased sHLA-G is associated with improved COVID-19 outcome and reduced neutrophil adhesion. Viruses. 2021;13(9) doi: 10.3390/v13091855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cordeiro J.F.C., et al. The severity of COVID-19 affects the plasma soluble levels of the immune checkpoint HLA-G molecule. Int. J. Mol. Sci. 2022;23(17) doi: 10.3390/ijms23179736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rizzo R., et al. SARS-CoV-2 nucleocapsid protein and ultrastructural modifications in small bowel of a 4-week-negative COVID-19 patient. Clin. Microbiol. Infect. 2021;27(6):936–937. doi: 10.1016/j.cmi.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pairo-Castineira E., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 71.Seliger B., et al. Induction of pulmonary HLA-G expression by SARS-CoV-2 infection. Cell. Mol. Life Sci. 2022;79(11):582. doi: 10.1007/s00018-022-04592-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.El-Badawy O., et al. COVID-19 infection in patients with comorbidities: clinical and immunological insight. Clin. Appl. Thromb.-Hemost. 2022:28. doi: 10.1177/10760296221107889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reddy M., et al. Comparative analysis of lymphocyte activation marker expression and cytokine secretion profile in stimulated human peripheral blood mononuclear cell cultures: an in vitro model to monitor cellular immune function. J. Immunol. Methods. 2004;293(1–2):127–142. doi: 10.1016/j.jim.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 74.Thevarajan I., et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26(4):p. 453-+. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biasi M. Covid-19 and labour law in Italy. Eur. Labour Law J. 2020;11(3):306–313. [Google Scholar]
- 76.Zhang B., et al. Mesenchymal stromal cell exosome-enhanced regulatory T-cell production through an antigen-presenting cell-mediated pathway. Cytotherapy. 2018;20(5):687–696. doi: 10.1016/j.jcyt.2018.02.372. [DOI] [PubMed] [Google Scholar]
- 77.Wang R., et al. Bone mesenchymal stem cell-derived exosome-enclosed miR-181a induces CD4(+)CD25(+)FOXP3(+) regulatory T cells via SIRT1/acetylation-mediated FOXP3 stabilization. J. Oncol. 2022;2022:8890434. doi: 10.1155/2022/8890434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giamarellos-Bourboulis E.J., et al. Complex immune dysregulation in COVID-19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992–1000 e3. doi: 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu B., Huang S., Yin L. The cytokine storm and COVID-19. J. Med. Virol. 2021;93(1):250–256. doi: 10.1002/jmv.26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Amodio G., et al. Combined plasma levels of IL-10 and testosterone, but not soluble HLA-G5, predict the risk of death in COVID-19 patients. Andrology. 2022 doi: 10.1111/andr.13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kordelas L., et al. Elevated soluble human leukocyte antigen G levels in patients after allogeneic stem cell transplantation are associated with less severe acute and chronic graft-versus-host disease. Bone Marrow Transpl. 2018;53(9):1149–1156. doi: 10.1038/s41409-018-0145-1. [DOI] [PubMed] [Google Scholar]
- 82.Hasanah N., et al. Interleukin 10 induces the expression of membrane-bound HLA- G and the production of soluble HLA-G on HeLa CCL-2 cells. Open Access Maced. J. Med. Sci. 2019;7(21):3554–3558. doi: 10.3889/oamjms.2019.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hviid T.V., et al. Polymorphism in the 5' upstream regulatory and 3' untranslated regions of the HLA-G gene in relation to soluble HLA-G and IL-10 expression. Hum. Immunol. 2006;67(1–2):53–62. doi: 10.1016/j.humimm.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 84.Garziera M., Scarabel L., Toffoli G. Hypoxic modulation of HLA-G expression through the metabolic sensor HIF-1 in human cancer cells. J. Immunol. Res. 2017;2017:4587520. doi: 10.1155/2017/4587520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Attia J.V.D., et al. The molecular and functional characteristics of HLA-G and the interaction with its receptors: where to intervene for cancer immunotherapy? Int. J. Mol. Sci. 2020;21(22) doi: 10.3390/ijms21228678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Menier C., et al. Characterization of monoclonal antibodies recognizing HLA-G or HLA-E: new tools to analyze the expression of nonclassical HLA class I molecules. Hum. Immunol. 2003;64(3):315–326. doi: 10.1016/s0198-8859(02)00821-2. [DOI] [PubMed] [Google Scholar]
- 87.Naji A., et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J. Immunol. 2014;192(4):1536–1546. doi: 10.4049/jimmunol.1300438. [DOI] [PubMed] [Google Scholar]
- 88.Oliveira M.L.G., et al. Genetic diversity of the LILRB1 and LILRB2 coding regions in an admixed Brazilian population sample. HLA. 2022;100(4):325–348. doi: 10.1111/tan.14725. [DOI] [PubMed] [Google Scholar]
- 89.Li N.L., et al. Modulation of the inhibitory receptor leukocyte Ig-like receptor 1 on human natural killer cells. Front. Immunol. 2011;2:46. doi: 10.3389/fimmu.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anna F., et al. First immunotherapeutic CAR-T cells against the immune checkpoint protein HLA-G. J. Immunother. Cancer. 2021;9(3) doi: 10.1136/jitc-2020-001998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mandel I., et al. BND-22, a first-in-class humanized ILT2-blocking antibody, promotes antitumor immunity and tumor regression. J. Immunother. Cancer. 2022;10(9) doi: 10.1136/jitc-2022-004859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilead Buys into Tizona's Anti-HLA-G Strategy, 2020. Cancer Discov. 10(10), 1433. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.