Abstract
The endosymbiotic theory posits that ancient eukaryotic cells engulfed O2-consuming prokaryotes, which protected them against O2 toxicity. Previous studies have shown that cells lacking cytochrome c oxidase (COX), required for respiration, have increased DNA damage and reduced proliferation, which could be improved by reducing O2 exposure. With recently developed fluorescence lifetime microscopy–based probes demonstrating that the mitochondrion has lower [O2] than the cytosol, we hypothesized that the perinuclear distribution of mitochondria in cells may create a barrier for O2 to access the nuclear core, potentially affecting cellular physiology and maintaining genomic integrity. To test this hypothesis, we utilized myoglobin-mCherry fluorescence lifetime microscopy O2 sensors without subcellular targeting (“cytosol”) or with targeting to the mitochondrion or nucleus for measuring their localized O2 homeostasis. Our results showed that, similar to the mitochondria, the nuclear [O2] was reduced by ∼20 to 40% compared with the cytosol under imposed O2 levels of ∼0.5 to 18.6%. Pharmacologically inhibiting respiration increased nuclear O2 levels, and reconstituting O2 consumption by COX reversed this increase. Similarly, genetic disruption of respiration by deleting SCO2, a gene essential for COX assembly, or restoring COX activity in SCO2−/− cells by transducing with SCO2 cDNA replicated these changes in nuclear O2 levels. The results were further supported by the expression of genes known to be affected by cellular O2 availability. Our study reveals the potential for dynamic regulation of nuclear O2 levels by mitochondrial respiratory activity, which in turn could affect oxidative stress and cellular processes such as neurodegeneration and aging.
Keywords: respiration, mitochondria, hypoxia, oxygen, nucleus, classification: cell biology, metabolism
Abbreviations: AA, antimycin A; Asc, ascorbate; cDNA, complementary DNA; COX, cytochrome c oxidase; DAPI, 4′,6-diamidino-2-phenylindole; Ex/Em, excitation/emission; FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; FLIM, fluorescence lifetime microscopy; HEK293T, human embryonic kidney 293T cell line; HIF-1α, hypoxia-inducible factor 1-alpha; mAb, monoclonal antibody; MB, myoglobin; Myxo, myxothiazol; ROS, reactive oxygen species; Rot, rotenone; Tg, transgene; TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine
Molecular oxygen (O2) has a dual nature. Its oxidative potential permits efficient aerobic metabolism for beneficial cellular energy production, whereas its unstable nature can result in the generation of toxic reactive oxygen species (ROS) that cause oxidative damage and serve as the basis for the free radical theory on aging (1, 2, 3). In developing a mechanistic explanation for this original theory of aging, mitochondria were proposed to be the major source of ROS responsible for oxidative damage, although ROS can also serve important signaling functions (4, 5). This theory, however, has been beset with some inconsistent results in models ranging from yeast to human, and the concept of the mitochondrion as the major source of ROS has been questioned (6, 7, 8).
Mitochondria have multiple sites from which ROS may be generated, but its production in vivo is dependent on a number of important factors, such as the substrate, mitochondrial membrane potential, matrix pH, and intracellular O2 availability (9). These factors, which may be interdependent in vivo, are difficult to control under experimental conditions to permit an accurate assessment of in vivo mitochondrial ROS production. Even the increase in ROS production associated with hypoxia has been suggested to be an experimental phenomenon (9). From another perspective, the mitochondria could have net antioxidant effects by consuming O2, the essential substrate for ROS production, and thereby preventing its genotoxicity (10). We previously showed that disrupting SCO2 (SCO2−/−), a gene regulated by tumor suppressor p53 and essential for respiration, results in higher intracellular levels of O2, increased oxidative DNA damage, and decreased proliferation under normal ambient O2 levels (∼20% O2) (11). Exposing these nonrespiring SCO2−/− cells to lower O2 levels (≤5%) decreased DNA damage and improved their growth; but other factors such as their propensity for regenerating NAD+ as also contributing to this improvement cannot be ruled out (11). Cellular O2 is mostly consumed by mitochondria; therefore, their dysfunction could result in elevated tissue O2 levels. In accord with this notion, more recent studies have shown increased brain tissue O2 levels in a mouse model of mitochondrial disease, and the neurodegeneration and shortened life span of these mice were ameliorated by decreasing ambient O2 exposure (12, 13).
The development of various probes and techniques for measuring O2 levels in cultured cells and tissues have contributed to understanding O2 homeostasis under normal and abnormal conditions (14, 15, 16, 17). Previous O2 homeostasis studies suggested the existence of substantial intracellular gradients under some O2-limited conditions (18). Consistent with the view of mitochondria as O2 sinks capable of creating intracellular O2 gradients (19, 20), recent advances in fluorescence lifetime microscopy (FLIM) probes have permitted the measurement of O2 concentration ([O2]) within the mitochondria and confirmed that it is lower relative to the rest of the cytosol (21). Notably, this phenomenon was observed to be dependent on active respiration. Because mitochondria are observed to have a perinuclear localization, we hypothesized that the nucleus would also have low [O2] relative to the cytosol, secondary to the hypoxic microenvironment of the surrounding mitochondria. In the current study, we report that the subcellular compartment of the nucleus is indeed capable of being maintained at lower O2 levels by mitochondrial respiration. This dependence of nuclear [O2] on the functional state of the mitochondria may have important implications, such as adding another dimension to the metabolic control of epigenetics or genomic stability in cancer.
Results
Respiring mitochondria surround the nucleus and decrease its O2 exposure
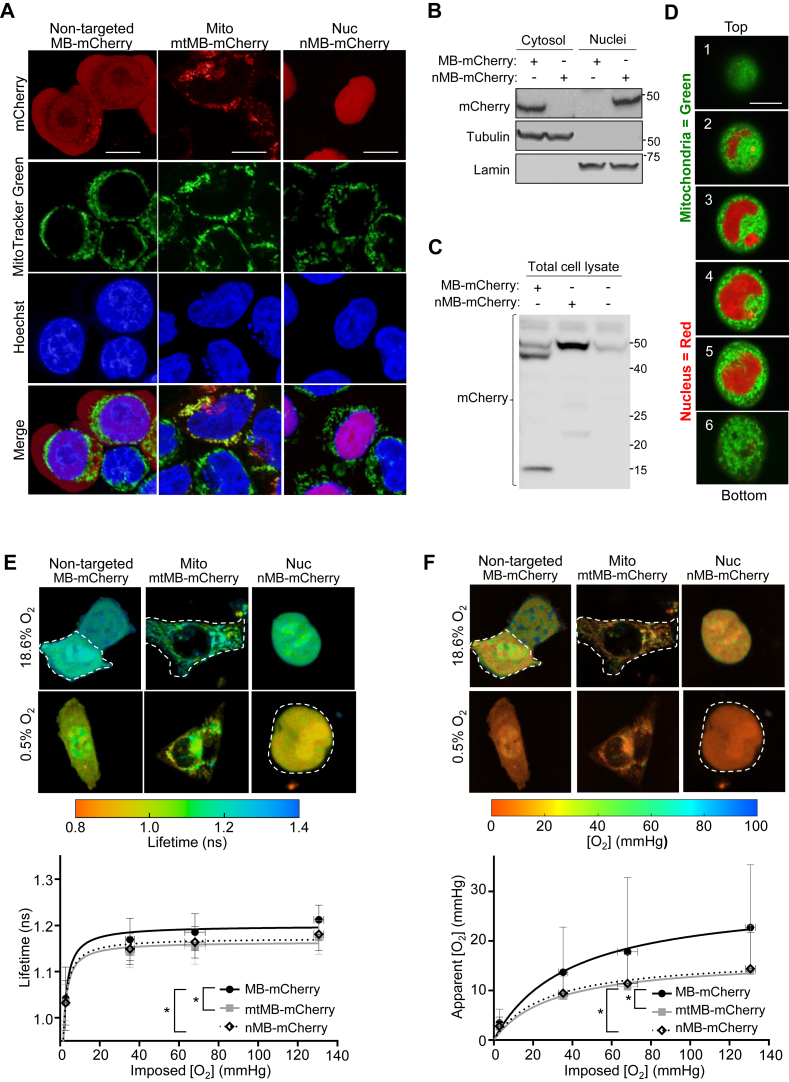
The red fluorescent protein mCherry was fused to the O2-binding protein myoglobin (MB) to create the O2 sensor MB-mCherry as previously described (21). The FRET-induced changes in the lifetime of MB-mCherry have been shown to be dependent on the O2-bound or O2-free (oxy/deoxy) state of MB and carefully validated as reflecting intracellular O2 levels (Fig. 1) (21, 22, 23). To test the idea that the perinuclear localization of mitochondria may deplete nuclear O2 levels, we transiently transfected human HCT116 cells with MB-mCherry constructs targeted to the different subcellular compartments: cytosol, nontargeted MB-mCherry; mitochondrion, mtMB-mCherry; or nucleus, nMB-mCherry. Generally, these three constructs revealed mCherry red fluorescence signals in their respective subcellular compartments by confocal fluorescence microscopy (Fig. 2A).
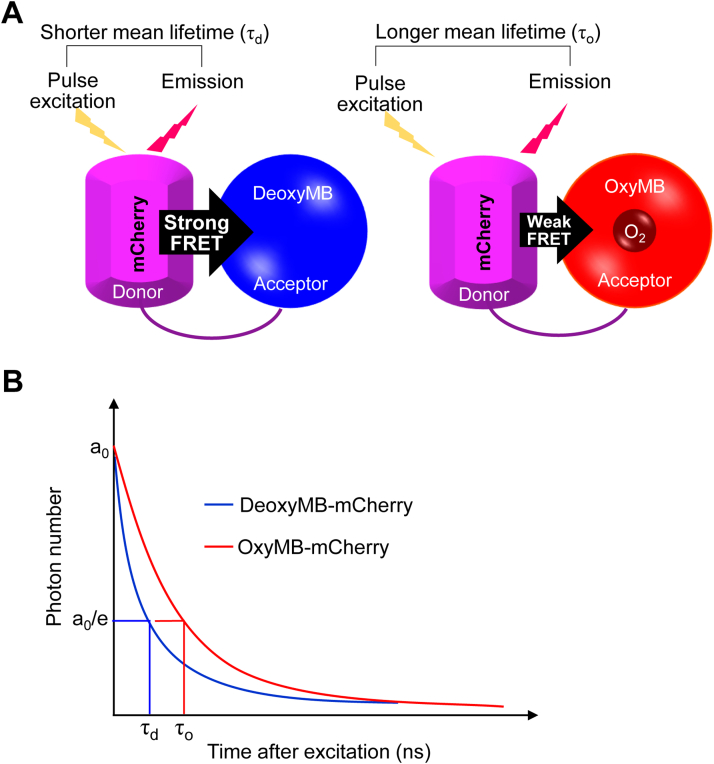
Figure 1.
Schematic outline of fluorescence lifetime imaging (FLIM) for intracellular O2mapping.A, diagram of O2-binding myoglobin (MB) fused to fluorescent protein mCherry to create an intracellular O2 sensor (MB-mCherry). Strong or weak FRET between mCherry and MB, depending on the O2-free or O2-bound state of MB, respectively, changes the duration of fluorescence lifetime as shown (21). B, amplitude of components (ao); lifetime of MB-mCherry in deoxy- (τd) and oxy- (τo) MB form.
Figure 2.
Respiring mitochondria surround the nucleus and decrease nuclear O2levels. The O2 sensor myoglobin (MB)-mCherry was expressed in different subcellular compartments of the HCT116 cell line by transfecting with nontargeted MB-mCherry or MB-mCherry fused to either the mitochondrial targeting sequence of TFAM (transcription factor A, mitochondrial; mtMB-mCherry, Mito) or SV40 nuclear localization signal (nMB-mCherry, Nuc). A, HCT116 cells were transfected with the three different MB-mCherry (red) constructs and stained with the mitochondrial dye MitoTracker Green (green) and nuclear DNA dye DAPI (blue) to confirm their specific subcellular localization. Note that the diffuse red fluorescence in the nuclei of nontargeted MB-mCherry-transfected cells does not result in significant purple color compared with that of nuclear nMB-mCherry when merged with the blue of nuclear Hoechst. Scale bar reprsents 5 μm. B, HCT116 cells were transfected with plasmids containing the indicated probes for 24 h, subcellularly fractionated to isolate the cytosolic and nuclear compartments, and the resulting samples were immunoblotted using anti-mCherry antibody. Tubulin and lamin serve as subcellular compartment markers for the cytosol and nucleus, respectively; all molecular weight markers indicated in kilodalton. C, the total cell lysates of HCT116 cells transfected with the indicated plasmids were immunoblotted using mCherry antibody. The predicted size of the nontargeted MB-mCherry construct is ∼44 kDa, but a smaller immunoreactive fragment at ∼15 kDa is evident. Note the presence of a nonspecific band at ∼48 kDa present in nontransfected control cells. All molecular weight markers are indicated in kilodalton. D, Z-stack images of a single cell transfected with nMB-mCherry and costained with MitoTracker Green from top (height ∼25 μm) to bottom (height ∼5 μm) of the well slide reveals a 3-dimensional shell of mitochondria around the nucleus. Scale bar represents 5 μm. E, representative pseudocolor FLIM images of HCT116 cells transfected with nontargeted MB-mCherry, mitochondrial mtMB-mCherry, and nuclear nMB-mCherry. The cells were incubated under different O2 concentrations ranging from 0.5% (∼2.8 mm Hg) to 18.6% (∼130 mm Hg) for lifetime measurements. Shorter lifetime values indicate lower [O2] (red), whereas longer lifetime indicates higher [O2] (green–blue). Note that for E and F, a threshold was introduced during the data analyses to remove the background noise. Dashed white lines have been placed around cells to indicate those with higher levels of probe expression and excluded background debris. Also, note that the FLIM SPCImage software does not provide scale bars. F, corresponding pseudo-color images of apparent cytosolic, mitochondrial, and nuclear [O2]. The lifetime value for each pixel in FLIM images shown in C and the calibration curve (obtained for nonrespiring rotenone/antimycin treated or SCO2−/− cells) were used to estimate apparent compartmental [O2]. The summary of data used in E and F is provided in Tables S1 and S2. In the color bars, red indicates lower values, whereas green–blue indicates higher values. Except at the lowest imposed [O2] (∼2.8 mm Hg), Mann–Whitney tests showed a significant difference between [O2] obtained for nontargeted versus mitochondria- or nuclear-localized probes. The differences between the mitochondrial and nuclear [O2] were not statistically significant (n = 33–53). Statistical difference by two-way ANOVA with Tukey’s post-test. Comparison was performed between mean values of the different compartments. Values are mean ± SD. ∗p < 0.01. DAPI, 4′,6-diamidino-2-phenylindole; FLIM, fluorescence lifetime microscopy.
The nontargeted MB-mCherry showed a diffuse pattern of red fluorescence throughout the cell, which contrasted with the more prominent perinuclear appearance of the mitochondrial targeted mtMB-mCherry construct nontargeted versus mito (Fig. 2A). The mtMB-mCherry red colocalized with the mitochondrial-specific dye MitoTracker Green to produce yellow merge signals, whereas the nuclear nMB-mCherry colocalized with the blue Hoechst DNA dye to result in purple merge color (mito versus nuc, Fig. 2A). Notably, the nontargeted MB-mCherry-transfected cells displayed some red mCherry fluorescence in their nuclei although immunoblotting of their subcellular fractions using anti-mCherry antibody showed nontargeted probe only in the cytosolic fraction but not nuclear fraction (Fig. 2, A and B). On the other hand, nMB-mCherry-transfected cells showed mCherry immunoreactivity only in the nuclear fraction confirming the specific targeting of this probe (Fig. 2B). Further investigation of nontargeted MB-mCherry-transfected cells revealed the presence of two mCherry immunoreactive bands in their whole cell lysates, a ∼44 kDa band corresponding to the predicted size of the fusion protein and a smaller fragment at ∼15 kDa (Fig. 2C). The mCherry sequence containing fragment derived from the nontargeted MB-mCherry construct may be able to diffuse into the nuclear compartment of intact cells but wash out during subcellular fractionation, potentially explaining our current observation. However, the mCherry fragment is too small to contain MB; therefore, it would not result in O2-sensitive FLIM measurements.
Two-dimensional fluorescence images of HCT116 cells transfected with nMB-mCherry and stained with MitoTracker Green revealed that the mitochondria are localized around the nucleus in a perinuclear pattern. Confocal fluorescence imaging using Z-stack technique in a single cell further revealed that the mitochondria form a 3-dimensional network encasing the nucleus, similar to a shell. This configuration may serve as a protective barrier for the nucleus by preventing exposure to high levels of oxygen (Fig. 2D).
FLIM measurements of cells transfected with these three MB-mCherry O2 probes were performed under different imposed O2 levels to demonstrate the dependence of O2 on its availability and consumption by mitochondria. The lifetime values of MB-mCherry were examined for any refractive index (or possible pH) changes in the intracellular environment by transfecting with control sensors: nontargeted, mitochondrial-targeted, or nuclear-targeted mCherry (alone, without an O2-responsive MB component) (Fig. S1). Lifetime measurements of all three MB-containing probes showed the characteristic hyperbolic O2-binding curve of MB when the transfected cells were exposed to controlled ambient O2 levels in tissue culture slide wells from ∼0.5% to 18.6% (taking into account water vapor and 5% CO2), corresponding to media [O2] of ∼2.8 mm Hg to 130 mm Hg (Fig. 2E). The shorter lifetimes observed for the mitochondrial- and nuclear-targeted probes across the range of imposed [O2] were consistent with “cytosolic” [O2] being higher compared with that of the mitochondrial and nuclear compartments. The overlapping FLIM values for the mtMB-mCherry and nMB-mCherry probes indicated that the mitochondrion and nucleus have similarly low [O2] levels and supported an association between these two compartments, as initially hypothesized (Fig. 2E). The subcellular FLIM measurements were converted to apparent compartmental [O2] (mm Hg) by using OxyLite Pro 1 point measurements of [O2] in the chamber culture medium exposed to 5% CO2 and appropriate N2 to decrease O2 levels as previously described (21). Likely because of diffusion limitations of O2 above the cell layer, even within low confluency cell culture conditions (24), the apparent [O2] levels of all three subcellular compartments were lower than that of the culture medium determined by the imposed ambient O2 level (Fig. 2F). To ensure that these findings were not limited to the HCT116 cell line, FLIM measurements were repeated in human embryonic kidney 293T (HEK293T) cells transfected with the three O2 sensor constructs, and similar results were observed (Fig. S2A). Taken together, these results supported the hypothesis that mitochondria act as O2 sinks within the cell, and that their perinuclear localization may secondarily result in the reduction of nuclear [O2].
Nuclear O2 level is dependent on mitochondrial respiration
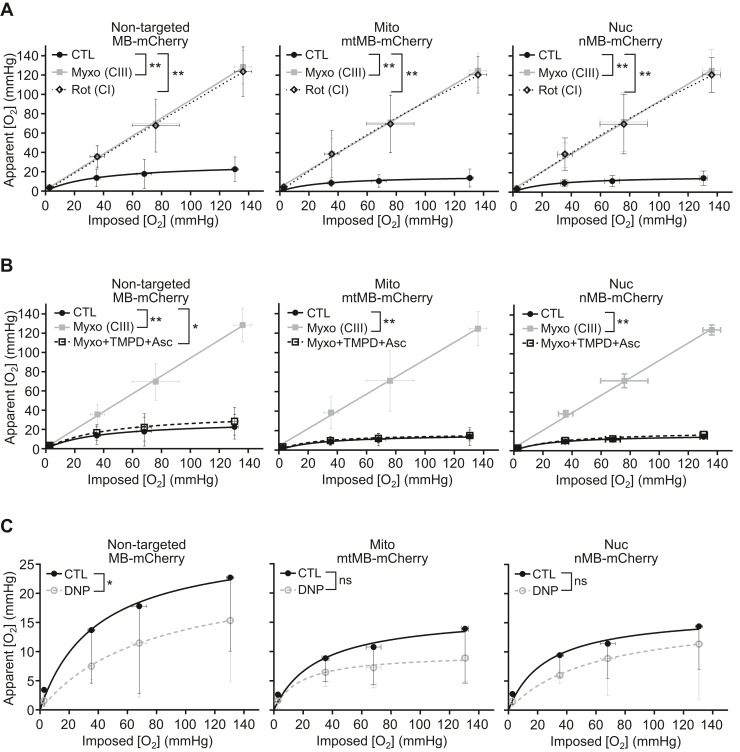
We next tested whether mitochondrial respiratory activity can directly affect nuclear [O2] as previously shown for mitochondria (21). The pharmacologic inhibition of mitochondrial respiratory complex I by rotenone (Rot) or complex III by myxothiazol (Myxo) resulted in increased subcellular [O2] both in the mitochondrial and nuclear compartments of HCT116 and HEK293T cells (Figs. 3A and S2B). Notably, there was also a pattern of significantly increased [O2] even in the cytosol, consistent with a previous study utilizing a nontargeted phosphorescent O2 probe that showed intracellular [O2] dependence on respiration (Fig. 3A) (11).
Figure 3.
Pharmacologic modulation of mitochondrial respiration affects nuclear O2levels. HCT116 cells were transfected with the indicated O2 sensor MB-mCherry constructs (nontargeted MB-mCherry; mitochondrial [Mito] mtMB-mCherry; nuclear [Nuc] nMB-mCherry) and apparent [O2] measurements obtained by fluorescence lifetime microscopy [FLIM] at the indicated imposed [O2]. A, apparent [O2] measurements of untreated control (CTL) cells or those treated with inhibitors of mitochondrial respiratory complex III (CIII) myxothiazol (Myxo) or complex I (CI) rotenone (Rot). Except at the lowest imposed [O2], there was a statistically significant difference between the apparent [O2] values obtained for the respiring cells and those treated with Rot/antimycin. B, apparent [O2] measurements of cells with no treatment (CTL), inhibition of respiration by Myxo, or rescue of Myxo inhibition of respiration by metabolic bypass using redox mediator TMPD and ascorbate (Asc). C, apparent [O2] measurements of cells treated with mitochondrial uncoupler 2,4-dinitrophenol (DNP) for maximal O2 consumption (n = 12–53). Statistical difference by two-way ANOVA with Tukey’s post-test. Comparison was performed between mean values of the different treatments. Values are mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ns (nonsignificant). MB, myoglobin; TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine.
Because cytochrome c oxidase (COX) (complex IV) is the major site of O2 consumption, we predicted that reconstituting its activity even with inhibition of upstream electron transfer would prevent the rise in mitochondrial and nuclear O2 levels. Indeed, in the presence of Myxo, directly feeding electrons to COX by using the reductant ascorbate (Asc) and redox mediator N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) resulted in decreased mitochondrial and nuclear O2 levels (Fig. 3B). In contrast to the observation with respiratory inhibitors, stimulating mitochondrial O2 consumption with the uncoupling agent 2,4-dinitrophenol resulted in a further lowering of [O2] in all three subcellular compartments as expected in our proposed model (Fig. 3C).
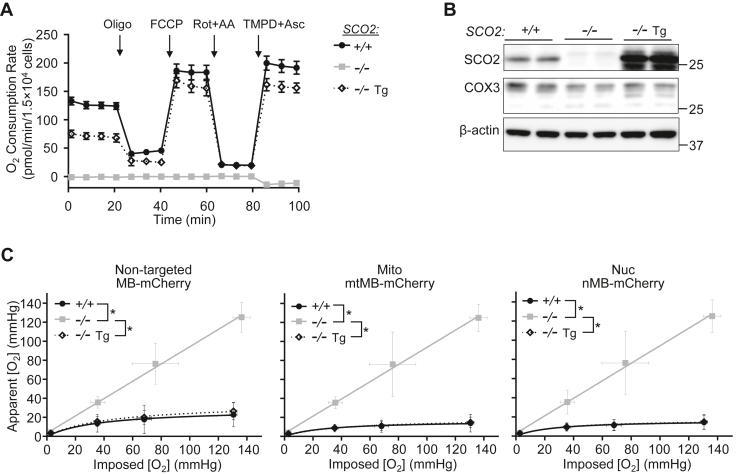
To further confirm the effects of these pharmacologic agents on mitochondrial and nuclear [O2], we utilized a nonrespiring HCT116 cell line that had been created previously by homozygous disruption of SCO2 (SCO2−/−), a metallochaperone gene involved in cellular copper homeostasis and essential for COX assembly (25, 26, 27). Measurements of O2 consumption using mitochondrial inhibitors and uncoupler carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) revealed the complete absence of respiration in SCO2−/− cells in comparison with isogenic wildtype cells (Fig. 4A). Furthermore, the absence of O2 consumption upon treatment with TMPD and Asc was consistent with the essential role that SCO2 plays in the assembly and stabilization of the COX complex, whereas the reintroduction of SCO2 complementary DNA (cDNA) (transgene, Tg) into SCO2−/− cells (SCO2−/− Tg) rescued COX3 subunit protein levels, respiratory activity, and the effects of TMPD/Asc on O2 consumption (Fig. 4, A and B). Importantly, the genetic disruption of respiration in SCO2−/− cells increased [O2] levels in both the mitochondrial and nuclear compartments, whereas the respiration-rescued SCO2−/− Tg cells showed reversal of these changes (Fig. 4C). These observations confirmed the specificity of the pharmacologic inhibitors of mitochondrial respiration used in our current and previous FLIM O2 studies (21).
Figure 4.
Genetic modulation of mitochondrial respiration affects nuclear O2levels.A, characterization of mitochondrial respiratory activity in wildtype respiring SCO2+/+, nonrespiring SCO2−/−, and respiration-rescued SCO2−/− transgene (Tg) (re-expressing SCO2) HCT116 cells using the Seahorse XFe96 Analyzer (n = 10). B, immunoblots of SCO2 and cytochrome c oxidase subunit 3 (COX3) in SCO2+/+, SCO2−/−, and SCO2−/− Tg cells. β-actin serves as loading control; all molecular weight markers are indicated in kilodalton. C, SCO2+/+, SCO2−/−, and SCO2−/− Tg cells were transfected with the indicated myoglobin (MB)-mCherry constructs (nontargeted MB-mCherry; mitochondrial [Mito] mtMB-mCherry; nuclear [Nuc] nMB-mCherry) and apparent O2 measurements obtained by FLIM at the indicated imposed [O2] (n = 27–53). Statistical difference by ANOVA with Tukey’s post-test. Comparison was performed between mean values of the different SCO2 genotypes. Values are mean ± SD. ∗p < 0.01. FLIM, fluorescence lifetime microscopy.
Mitochondrial respiration modulates the expression of genes sensitive to intracellular O2 levels
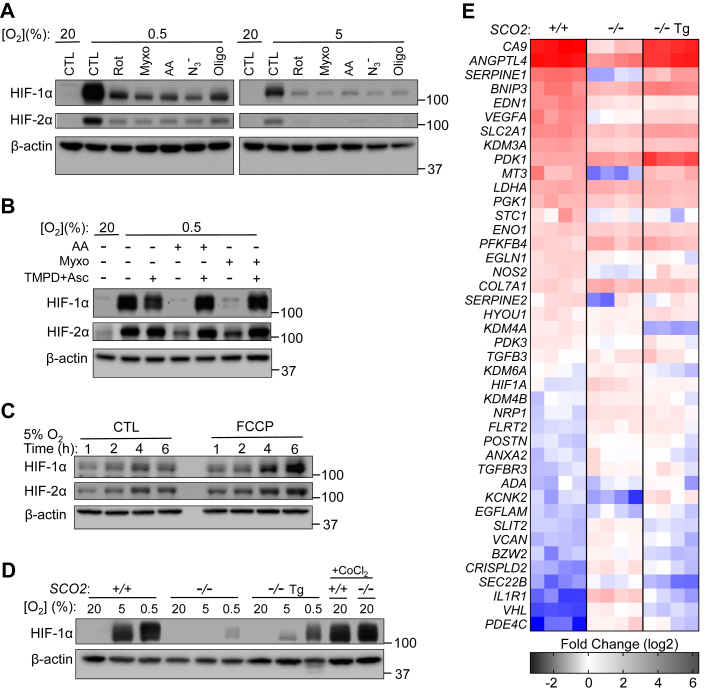
The protein levels of nuclear transcription factor hypoxia-inducible factor 1-alpha (HIF-1α) are sensitive to intracellular [O2] as its hydroxylation by HIF prolyl hydroxylase for subsequent degradation requires O2 as substrate. Because inhibiting respiration increases intracellular O2 levels, we examined whether the levels of HIF-1α could serve as a sensitive biologic read out of changes in [O2]. Treating wildtype HCT116 cells with mitochondrial inhibitors targeting the different respiratory complexes all prevented the stabilization of HIF-1α under both 5% and 0.5% O2, which normally increase its levels, suggesting a state of increased intracellular O2 availability confirmed by our FLIM measurements (Fig. 5A). Bypassing the electron transfer blockade at complex III (Myxo or antimycin A) using TMPD/Asc to resume O2 consumption resulted in recovery of HIF-1α and HIF-2α stabilization, whereas stimulating O2 consumption with the uncoupling agent FCCP further increased their levels compared with controls (Fig. 5, B and C). Thus, these observations demonstrated that mitochondrial O2 consumption may contribute to the stabilization of HIF-1α by decreasing intracellular [O2]. Consistent with this mechanism, there was mild stabilization of HIF-1α only at the most hypoxic imposed [O2] (0.5% O2) in nonrespiring SCO2−/− cells, further supporting an important role for mitochondrial respiration in O2 homeostasis (Fig. 5, C and D). We also confirmed that the diminished stabilization of HIF-1α in SCO2−/− cells by hypoxia was not because of a defect in HIF or its intrinsic regulation because treatment with CoCl2, a direct inactivator of HIF prolyl hydroxylase, stabilized HIF-1α to levels identical to that in wildtype cells (Fig. 5D).
Figure 5.
Hypoxia sensing is impaired by pharmacologic or genetic disruption of mitochondrial respiration.A, HIF-1α and HIF-2α immunoblots of wildtype HCT116 SCO2+/+ cells exposed to hypoxia (0.5% or 5% O2) and the indicated mitochondrial complex inhibitor rotenone (Rot, CI), myxothiazol (Myxo, CIII), antimycin A (AA, CIII), azide (N3−, CIV), or oligomycin (Oligo, CV) for 3 h. No inhibitor treatment control (CTL). β-actin serves as protein loading control; all molecular weight markers are indicated in kilodalton. B, HIF-1α and HIF-2α immunoblots of SCO2+/+ cells exposed to hypoxia (0.5% O2) and the indicated complex III inhibitor (Myxo or AA) or bypass of this inhibition with redox mediator TMPD and ascorbate (Asc) for 3 h. C, immunoblot of HIF-1α and HIF-2α stabilization in SCO2+/+ cells upon 5% O2 hypoxia exposure and treatment with FCCP (uncoupler) to increase O2 consumption for the indicated period. D, HIF-1α immunoblot of SCO2+/+, SCO2−/−, and SCO2−/− Tg cells after exposure to 0.5% or 5% O2 for 3 h. Treatment with 100 μM CoCl2 serves as positive control for HIF-1α stabilization. E, fold change in the expression of a panel of genes known or reported to be O2 sensitive in SCO2+/+, SCO2−/−, and SCO2−/− Tg cells exposed to 0.5% O2 for 72 h compared with control 20% O2 condition. The mRNA levels of these O2-sensitive genes were assessed by real-time RT–PCR. FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; HIF, hypoxia-inducible factor; TMPD, N,N,N′,N′-tetramethyl-p-phenylenediamine.
Changes in intracellular O2 levels may impact gene expression regulated not only by HIF transcription factors but also by chromatin modifications such as histone methylations that induce or silence genes. Among the post-translational modifications of histones, lysine (K) methylation in particular has been shown to be sensitive to the cellular levels of O2 (28). Methylation of histone H3 at amino acid residues K4, K9, and K27 has been shown to be increased under hypoxia, possibly because of limited O2, a substrate required for demethylation by histone lysine demethylases (29, 30, 31). Therefore, as a biologic read out of intracellular O2 levels to complement the FLIM probe measurements and a simple demonstration of respiration-driven alterations in [O2], we examined the expression of genes known to be sensitive to O2 levels via epigenetic regulation or HIF-1α transactivation. We exposed HCT116 cells of the three different SCO2 genotypes to hypoxia (0.5% O2) and performed RT–quantitative PCR of a panel of hypoxia-sensitive genes known to be regulated by HIF1 and histone H3 lysine methylation, specifically trimethylated H3K9 as previously reported (29). Notably, unlike in respiring wildtype cells, the regulation of various O2-sensitive genes was blunted in SCO2−/− cells, whereas there was recovery of this regulation with the rescue of respiration in isogenic SCO2−/− Tg cells (Fig. 5E). This effect mirrored the diminished stabilization of HIF-1α in nonrespiring SCO2−/− cells when exposed to 0.5% O2 (Fig. 5D), underscoring again that mitochondrial respiration indeed significantly contributes to intracellular O2 homeostasis.
Discussion
Using FLIM O2 sensors targeted to the nucleus, we report that direct measurements of the nuclear compartment reveal relatively low O2 levels compared with the cytosol. Previous work using O2 probes targeted to the mitochondria had revealed a hypoxic microenvironment (21), so the perinuclear localization of mitochondria suggested the possibility of a secondary hypoxic nuclear core, which we have now demonstrated. We also show that both pharmacologic and genetic alterations of mitochondrial respiration consistently associate O2 consumption with the maintenance of low [O2] in the nucleus. The expression patterns of O2-sensitive genes as a read out of intracellular hypoxia revealed biological sequelae consistent with the FLIM [O2] results. We should note here that diffusion limitation of O2-causing gradients in cultured cells has been reviewed extensively and should be considered here (24); however, the apparent differences in [O2] levels of all three subcellular compartments that consistently depend on respiration, whether pharmacologically or genetically altered, can still be interpreted as being significant.
It is tempting to speculate that the mitochondria serving to maintain low levels of O2 in the nucleus may represent a principle of the endosymbiotic theory of the mitochondrion, which holds that O2-consuming prokaryotes hosted by primordial anaerobic eukaryotes provided protection against genotoxic O2 (10, 32). More mechanistically, the concept of the mitochondrion as an O2 sink had been proposed even before direct measurements of mitochondrial [O2] were feasible (20). Previous works have also demonstrated respiration-dependent changes in intracellular [O2], including by a phosphorescent probe that accumulates in the nucleus (33, 34). In this study, we have specifically targeted the nuclear, mitochondrial, and “cytosolic” subcellular compartments using FLIM O2 probes and have performed parallel measurements of their intracellular O2 levels as a function of respiration.
Our finding of relative hypoxia in the coupled mitochondrial and nuclear compartments is also consistent with observations from disparate fields. As reflected by our FLIM measurements under typical physiologic tissue [O2] (≤50 μM or pO2 ∼40 mm Hg) (17), the [O2] necessary to decrease O2 consumption rate in respiring mammalian mitochondria has been reported to be less than 2 μM, which falls within the range of our mitochondrial mtMB-mCherry measurements (Fig. 2F) (35). Furthermore, the reported submicromolar Km for O2 as substrate of the COX complex indicates that intracellular [O2] within the microenvironment of the mitochondria may indeed be low. Our current work suggests that the perinuclear localization of O2-consuming mitochondria results in the low [O2] measured using our nuclear nMB-mCherry probe. It should be noted that at least two independent studies have previously associated the perinuclear localization of mitochondria with HIF-1α stabilization, proposed to be ROS mediated, but O2 depletion by respiration was not examined (36, 37). Although ROS in certain circumstances may be involved in HIF-1α stabilization (38), recent emerging studies suggest a direct role of decreased O2 availability (39, 40). It is also notable that HIF prolyl-4-hyroxylase 2 has been reported to be present in the nucleus and to interact with HIF-1α in the nucleus, suggesting nuclear O2 homeostasis may play a role in HIF-1α stabilization (41). In summary, our observation of dynamic subcellular [O2] regulation by respiration links mitochondrial metabolism to nuclear transcriptional and epigenetic regulatory processes via O2 homeostasis, raising new questions in fields of study ranging from evolutionary biology to genomic instability.
Experimental procedures
Cells and tissue culture
Wildtype HCT116 (SCO2+/+) and HEK293T cell lines were obtained from American Type Culture Collection and cultured in McCoy’s 5A and Dulbecco's modified Eagle's medium medium, respectively, supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin. The generation of isogenic nonrespiring SCO2−/− and rescued SCO2−/− Tg (re-expressing SCO2) cell lines has previously been described (11, 26). The nonrespiring SCO2−/− cells were cultured in the McCoy’s 5A medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin, and their media were replaced every 2 days for improved growth. To decrease the potential effects of O2 toxicity, SCO2−/− were normally maintained at lower ambient O2 (5% O2) for longer term cultures but were exposed to identical O2 conditions as respiring cells for cDNA transfection and FLIM studies.
Mitochondrial modulator treatment
Cells were treated with the following final concentrations of mitochondrial modulators for FLIM, O2 consumption (Seahorse), and HIF stabilization studies (all Sigma–Aldrich): Rot, 1 μM; antimycin A, 1 μM; Myxo, 2 μM; NaN3, 2 mM; oligomycin A, 1.5 μM; 2,4-dinitrophenol, 40 μM; FCCP, 0.5 μM; TMPD, 100 μM; and sodium Asc, 2 mM.
Subcellular fractionation
Nuclear and cytoplasmic subcellular fractionation was performed as previously described (42). Briefly, the cells were homogenized in fractionation buffer (250 mM sucrose, 50 mM Tris–HCl [pH 7.4], and 5 mM MgCl2) using a tissue grinder and centrifuged at 800g for 15 min. The resulting pellet and supernatant were used to isolate the nuclear and cytosolic fractions, respectively. The pellet was resuspended in nuclear buffer (20 mM Hepes [pH 7.9], 1.5 mM MgCl2, 0.5 M NaCl, 0.2 mM EDTA, 20% glycerol, and 1% Triton X-100) and centrifuged at 9000g for 30 min to yield the soluble nuclear fraction within the resulting supernatant. The initial supernatant was centrifuged at 11,000g for 10 min, and the resulting supernatant yielded the cytosolic fraction.
Antibodies
The following antibodies were utilized for immunoblotting: α-tubulin mouse monoclonal antibody (mAb) (T5168), β-actin mouse mAb (A5441) (Sigma–Aldrich); HIF-1α rabbit polyclonal antibody (pAb) (catalog no.: 10006421) (Cayman Chemical); HIF-2α rabbit pAb (7096) (Cell Signaling Technology, Inc); COX3 (MT-CO3) mouse mAb (catalog no.: ab110259), COX4 mouse mAb (catalog no.: ab33985) (Abcam PLC); COX17 rabbit pAb (catalog no.: 11464-1-AP) (Proteintech Group, Inc); and mCherry mouse mAb (catalog no.: MA5-32977), SCO2 rabbit pAb (catalog no.: PA5-76209) (Thermo Fisher Scientific, Inc).
Immunoblotting
Protein samples were solubilized in cold radioimmunoprecipitation assay buffer supplemented with protease/phosphatase inhibitors (Roche) and centrifuged at 160,00g for 15 min. The supernatant was mixed with SDS protein sample buffer, resolved by Tris–glycine SDS-PAGE, and transferred to Immobilon-P membrane (Millipore) for standard ECL immunoblotting.
FLIM probes
Plasmids containing the following constructs were made: nontargeted MB-mCherry (“cytosolic” MB-mCherry); MB-mCherry fused to the TFAM (transcription factor A, mitochondrial) mitochondrial targeting sequence (mitochondrial mtMB-mCherry) (21); and MB-mCherry fused to three repeats of the nuclear localization signal of SV40 T antigen (nuclear nMB-mCherry). Nontargeted mCherry with no MB (mCherry alone) or its targeted forms with either nuclear localization signal (n-mCherry) or mitochondrial targeting sequence (mt-mCherry) served as O2-insensitive controls for FLIM measurements. The MB sequence was deleted from the nMB-mCherry construct by PCR amplification using In-Fusion HD Cloning Plus (Takara Bio USA) with the primer set listed in the supporting information section. n-mCherry was sequenced, and nuclear localization was confirmed by 4′,6-diamidino-2-phenylindole (DAPI) colocalization after transfection. Plasmids were amplified in BL21 Star Escherichia coli (Thermo Fisher Scientific) and purified using NucleoSpin Plasmid Kit (Macherey–Nagel, Takara Bio USA, Inc).
Plasmid transfection and cell culture for FLIM
Cells were plated at a subconfluent density in 6- or 24-well plates (∼3.25 × 105 or 6.5 × 104 cells/well, respectively), allowed to attach for 24 h, and then transfected with Lipofectamine 2000 or FuGene HD per manufacturer’s instruction in Opti-MEM I Reduced Serum Medium (Gibco) in a standard room air with 5% CO2 tissue culture incubator. Cells were allowed to recover for 24 h after transfection, trypsinized, counted, replated at a density of 1.0 × 104 ∼ 1.0 × 105 cells/cm2 in μ-Slide 4 Well or 8 Well (ibidi), and allowed to attach for an additional 24 h prior to FLIM analysis.
Confocal fluorescence microscopy
For the subcellular colocalization imaging studies, cells were plated at 1.1 × 104 cells/cm2 (1.0 × 105 cells/dish) in 35 mm poly-d-lysine–coated glass bottom dishes (MatTek Co), allowed to attach for 24 h, transfected, and incubated another 24 h prior to analysis. The transfected cells were washed, incubated in 100 nM MitoTracker Green FM 100 (Invitrogen) for 30 min and 10 μg/ml DAPI (or Hoechst) at 37 °C for 5 min, washed and imaged in prewarmed Hank’s balanced salt solut supplemented with Ca2+/Mg2+ (1.26 and 0.9 mM, respectively), and d-glucose (5.6 mM). Images of the cells were acquired using a Zeiss LSM 780 confocal microscope (Carl Zeiss) equipped with a 63× oil immersion objective. Filter parameters were excitation/emission (Ex/Em) 355/410 to 481 nm for DAPI; Ex/Em 488/499 to 534 nm for MitoTracker Green (FITC); and Ex/Em 594/597 to 633 nm for MB-mCherry (Rhodamine). Images were processed using AxioVision 4.8 software.
FLIM set up
Two-photon FLIM was performed using a Leica SP5 confocal laser scanning microscope equipped with a tunable Chameleon Ti:Sapphire femtosecond laser (Coherent) with a wavelength set to 780 nm for the excitation of mCherry. The laser light was passed through a 685 nm LP dichroic mirror and directed to a Leica Plan-Apochromat 40×, 1.1 numerical aperture water immersion microscope objective (laser power ≤7 mW at the objective). A 647/57 nm bandpass filter (Semrock BrightLine) was used to filter mCherry signal. The electrical pulse output from the hybrid photomultiplier detectors (HyD; Leica Microsystems) was directed into an SPC-150 photon counting card (Becker & Hickl). Synchronization with the pixel, line, and frame clock from the scanning unit of the microscope was used for image construction in time-correlated single-photon counting mode. Single cells were imaged for 30 to 50 s (depending on the intensity), image size was set to 256 × 256 (pixels)2, and TCSPC histograms were collected with 256 channels in a 12.5 ns time window. A miniature incubation chamber with a gas mixing system (CO2 − O2−MI; Bioscience Tools) was mounted onto the microscope stage to keep the temperature at 37 °C and provide 5% CO2 and a stable % O2 (v/v) of 18.6%, 10%, 5%, or 0.5% during the imaging; ∼0.5% is the lowest percent of O2 attainable by our system (24). The media-imposed [O2] (pO2 in mm Hg) at each percent of O2 was monitored by a 250 μm diameter bare-fiber O2 sensor (NX-BF/O/E; Optronix Ltd) connected to an OxyLite, 1 Channel monitor (Optronix Lt). The [O2] values were measured at the bottom of the 4-well or 8-well chambers (ibidi) with or without live cells present in their respective culturing medium as previously described (21). The pO2 values measured by OxyLite Pro 1 were 130.6 ± 2.47, 68.1 ± 5.09, 35.3 ± 2.43, and 2.8 ± 0.61 mm Hg at 18.6, 10, 5, and 0.5% [O2], respectively.
FLIM analysis and intracellular mapping of pO2
FLIM images were processed as previously described (22). Briefly, the fluorescence lifetime decays of MB-mCherry in the cytosol, mitochondria, or nucleus were obtained by a double-exponential decay model in SPCImage (Becker & Hickl) at optimized goodness of fit (χ2). The mean lifetime was obtained for each single image (via amplitude weighting for each pixel) and averaged across multiple cells (n > 30). Then, the resulting lifetime values (τ(pO2)) were plotted against the media-imposed external pO2, and a hyperbolic curve was fit to the data using MATLAB R2020b (The MathWorks, Inc)
| (1) |
where τmax is the longest average lifetime for MB-mCherry at 18.6% O2 and K is a fitting parameter related to the affinity of MB for O2. The τ(pO2) values for SCO2−/− cells or those treated with Rot/antimycin (nonrespiring cells) were used as a reference for the lifetime of the probe at the environmental level of pO2 present in solution. Rearranging (Equation 1), fixing the K and τmax to the values obtained from the reference curve, the effective pO2 at each lifetime value was back calculated for the respiring cells. Then, pseudocolor mappings of pO2 in the cytosolic, mitochondrial, and nuclear environments were obtained using MATLAB R2020b (The MathWorks, Inc) equipped with the Image Processing Toolbox. More detailed description can be found elsewhere (22). The effects of intracellular refractive index on the lifetime values were corrected by using mCherry (alone) lifetime probes that were nontargeted or targeted to the mitochondria or nucleus.
Hypoxia
The indicated number of cells for each genotype (SCO2+/+, 1.0 × 106; SCO2−/−, 2.5 × 105; SCO2−/− Tg, 5.0 × 105) were plated in each well of 6-well tissue culture plates and allowed to attach in 20% O2 for 24 h prior to hypoxia exposure. Tissue culture plates were placed in modular incubator chambers (Billups-Rothenberg) with humidification, flushed for 5 min with gas containing 5% CO2 and appropriate mix of O2/N2, sealed, and placed in a 37 °C incubator for 3 h. As positive control for HIF stabilization, cells were treated in a standard incubator with 100 μM CoCl2 for 1 h. At the end of hypoxia treatment, the tissue culture plates were immediately transferred onto ice, media were removed, and washed with ice-cold and hypoxia mix–equilibrated Hank’s balanced salt solution. Then the cells were immediately lysed with 1× SDS protein sample buffer supplemented with 2% β-mercaptoethanol and protease and phosphatase inhibitors. Cells were sheared through a 28-gauge needle with eight strokes of a 1 ml syringe. Samples were heated at 85 °C for 10 min and stored at −80 °C.
RNA quantification by real-time RT–PCR
Total RNA was isolated from cells using RNeasy Plus Universal Kit (QIAGEN). cDNA was synthesized by using reverse transcription (High-Capacity cDNA Reverse Transcription Kit; Thermo Fisher), and RT–quantitative PCR was performed using a 7900HT Sequence Detection System (Applied Biosystems) as previously described (43). Relative gene expression levels were calculated from cycle threshold values normalized by the housekeeping genes reported to be best suited for HCT116 cells, phosphomannomutase 1 (PMM1), and ribosomal protein lateral stalk subunit P0 (RPLP0) (44). Primer sequences are listed in the supporting information section.
Mitochondrial respiration studies
For mitochondrial respiration studies using the Seahorse XF96e Analyzer, 1.5 × 104 HCT116 SCO2+/+, SCO2−/−, and SCO2−/− Tg cells from three different thawing batches were plated in Seahorse XF96 V3 PS Cell Culture Microplates and incubated for 16 h at standard conditions. XFe96 Sensor Cartridge was hydrated with ultrapure water overnight and, 45 min to 1 h prior to the commencement of the experiment, the cartridge was incubated with XF Calibrant solution at 37 °C in CO2-free humidified incubator. Cell-containing plate was washed with XF Assay medium, pH 7.4, supplemented with d-glucose 3 g/l, pyruvate 1 mM, and l-glutamine 1 mM at 37 °C according to the manufacturer’s recommended protocol. Injection ports were loaded with 10× concentrations of the stock agents listed in the aforementioned “Mitochondrial modulator treatment” section. Appropriates volumes of 10× stock were added to a starting medium volume of 180 μl containing the cells.
Statistical analysis
For each condition, FLIM was recorded in at least 30 cells. ANOVA with Tukey’s post-test or Mann–Whitney U tests were used to evaluate whether the values in the independent groups are significantly different from each other. Analyses were carried out using SPSS 14.0 (a subsidiary of IBM) software, and statistical significance was defined at p < 0.05 (95% confidence level).
Data availability
All data described are located in this article.
Supporting information
This article contains supporting information with primer sequences obtained from PrimerBank (45).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank current and past members of our laboratory, especially Ho Joong Sung, whose previous work inspired this current study. We also acknowledge the National Heart, Lung, and Blood Institute Light Microscopy Core for the use of their confocal microscopes for fluorescence and fluorescence lifetime imaging. We also thank Christian Combs (National Heart Lung, and Blood Institute Light Microscopy Core), Ju-Gyeong Kang, Jay H. Chung, and Dan L. Sackett for helpful discussions and assistance.
Author contributions
M. P. M., R. P., J. R. K., and P. M. H. conceptualization; M. P. M., R. P., J. M., G. A., Y.-C. K., P.-y. W., J. R. K., and P. M. H. formal analysis; M. P. M, R. P., J. M., G. A., A. A. Y.-C. K., and P.-y. W. investigation; M. P. M., R. P., P.-y. W., J. R. K., and P. M. H. writing–original draft.
Funding and additional information
This work was supported by the National Heart, Lung, and Blood Institute—National Institutes of Health Division of Intramural Research (grant no.: HL006051) (to P. M. H.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Ruma Banerjee
Supporting information
References
- 1.Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 2.McCord J.M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J. Biol. Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 3.Wickens A.P. Ageing and the free radical theory. Respir. Physiol. 2001;128:379–391. doi: 10.1016/s0034-5687(01)00313-9. [DOI] [PubMed] [Google Scholar]
- 4.Harman D. The biologic clock: the mitochondria? J. Am. Geriatr. Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 5.Lennicke C., Cocheme H.M. Redox metabolism: ROS as specific molecular regulators of cell signaling and function. Mol. Cell. 2021;81:3691–3707. doi: 10.1016/j.molcel.2021.08.018. [DOI] [PubMed] [Google Scholar]
- 6.Hekimi S., Lapointe J., Wen Y. Taking a "good" look at free radicals in the aging process. Trends Cell Biol. 2011;21:569–576. doi: 10.1016/j.tcb.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown G.C., Borutaite V. There is no evidence that mitochondria are the main source of reactive oxygen species in mammalian cells. Mitochondrion. 2012;12:1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Zsurka G., Peeva V., Kotlyar A., Kunz W.S. Is there still any role for oxidative stress in mitochondrial DNA-dependent aging? Genes (Basel) 2018;9:175. doi: 10.3390/genes9040175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Starkov A.A. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann. N. Y Acad. Sci. 2008;1147:37–52. doi: 10.1196/annals.1427.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruyninckx W.J., Mason H.S., Morse S.A. Are physiological oxygen concentrations mutagenic? Nature. 1978;274:606–607. doi: 10.1038/274606a0. [DOI] [PubMed] [Google Scholar]
- 11.Sung H.J., Ma W., Wang P.-y., Hynes J., O'Riordan T.C., Combs C.A., et al. Mitochondrial respiration protects against oxygen-associated DNA damage. Nat. Commun. 2010;1:1–8. doi: 10.1038/ncomms1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrari M., Jain I.H., Goldberger O., Rezoagli E., Thoonen R., Cheng K.H., et al. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4241–E4250. doi: 10.1073/pnas.1621511114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain I.H., Zazzeron L., Goldberger O., Marutani E., Wojtkiewicz G.R., Ast T., et al. Leigh syndrome mouse model can Be rescued by Interventions that normalize brain hyperoxia, but not HIF activation. Cell Metab. 2019;30:824–832.e823. doi: 10.1016/j.cmet.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dmitriev R.I., Papkovsky D.B. Optical probes and techniques for O2 measurement in live cells and tissue. Cell Mol. Life Sci. 2012;69:2025–2039. doi: 10.1007/s00018-011-0914-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirabello V., Cortezon-Tamarit F., Pascu S.I. Oxygen sensing, hypoxia tracing and in vivo imaging with functional metalloprobes for the early detection of non-communicable diseases. Front. Chem. 2018;6:27. doi: 10.3389/fchem.2018.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostyuk A.I., Kokova A.D., Podgorny O.V., Kelmanson I.V., Fetisova E.S., Belousov V.V., et al. Genetically encoded tools for research of cell signaling and metabolism under brain hypoxia. Antioxidants (Basel) 2020;9:516. doi: 10.3390/antiox9060516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori M.P., Penjweini R., Knutson J.R., Wang P.Y., Hwang P.M. Mitochondria and oxygen homeostasis. FEBS J. 2021;289:6959–6968. doi: 10.1111/febs.16115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones D.P. Intracellular diffusion gradients of O2 and ATP. Am. J. Physiol. 1986;250:C663–675. doi: 10.1152/ajpcell.1986.250.5.C663. [DOI] [PubMed] [Google Scholar]
- 19.Gnaiger E., Steinlechner-Maran R., Mendez G., Eberl T., Margreiter R. Control of mitochondrial and cellular respiration by oxygen. J. Bioenerg. Biomembr. 1995;27:583–596. doi: 10.1007/BF02111656. [DOI] [PubMed] [Google Scholar]
- 20.Wenger R.H. Mitochondria: oxygen sinks rather than sensors? Med. Hypotheses. 2006;66:380–383. doi: 10.1016/j.mehy.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 21.Penjweini R., Andreoni A., Rosales T., Kim J., Brenner M.D., Sackett D.L., et al. Intracellular oxygen mapping using a myoglobin-mCherry probe with fluorescence lifetime imaging. J. Biomed. Opt. 2018;23:1–14. doi: 10.1117/1.JBO.23.10.107001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penjweini R., Roarke B., Alspaugh G., Gevorgyan A., Andreoni A., Pasut A., et al. Single cell-based fluorescence lifetime imaging of intracellular oxygenation and metabolism. Redox Biol. 2020;34 doi: 10.1016/j.redox.2020.101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sedlack A.J.H., Penjweini R., Link K.A., Brown A., Kim J., Park S.J., et al. Computational modeling and imaging of the intracellular oxygen gradient. Int. J. Mol. Sci. 2022;23:12597. doi: 10.3390/ijms232012597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Place T.L., Domann F.E., Case A.J. Limitations of oxygen delivery to cells in culture: an underappreciated problem in basic and translational research. Free Radic. Biol. Med. 2017;113:311–322. doi: 10.1016/j.freeradbiomed.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leary S.C., Cobine P.A., Kaufman B.A., Guercin G.H., Mattman A., Palaty J., et al. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto T., Wang P.Y., Ma W., Sung H.J., Matoba S., Hwang P.M. Polo-like kinases mediate cell survival in mitochondrial dysfunction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14542–14546. doi: 10.1073/pnas.0904229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacheu-Grau D., Bareth B., Dudek J., Juris L., Vogtle F.N., Wissel M., et al. Cooperation between COA6 and SCO2 in COX2 maturation during cytochrome c oxidase assembly links two mitochondrial cardiomyopathies. Cell Metab. 2015;21:823–833. doi: 10.1016/j.cmet.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Shmakova A., Batie M., Druker J., Rocha S. Chromatin and oxygen sensing in the context of JmjC histone demethylases. Biochem. J. 2014;462:385–395. doi: 10.1042/BJ20140754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S., Lee J., Chae S., Moon Y., Lee H.Y., Park B., et al. Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucl. Acids Res. 2017;45:11643–11657. doi: 10.1093/nar/gkx747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Batie M., Frost J., Frost M., Wilson J.W., Schofield P., Rocha S. Hypoxia induces rapid changes to histone methylation and reprograms chromatin. Science. 2019;363:1222–1226. doi: 10.1126/science.aau5870. [DOI] [PubMed] [Google Scholar]
- 31.Chakraborty A.A., Laukka T., Myllykoski M., Ringel A.E., Booker M.A., Tolstorukov M.Y., et al. Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science. 2019;363:1217–1222. doi: 10.1126/science.aaw1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Margulis L. 2 ed. W.H. Freeman and Company; NY: 1993. Symbiosis in Cell Evolution. [Google Scholar]
- 33.O'Riordan T.C., Fitzgerald K., Ponomarev G.V., Mackrill J., Hynes J., Taylor C., et al. Sensing intracellular oxygen using near-infrared phosphorescent probes and live-cell fluorescence imaging. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2007;292:R1613–1620. doi: 10.1152/ajpregu.00707.2006. [DOI] [PubMed] [Google Scholar]
- 34.Hara D., Umehara Y., Son A., Asahi W., Misu S., Kurihara R., et al. Tracking the oxygen status in the cell nucleus with a hoechst-tagged phosphorescent ruthenium complex. Chembiochem. 2018;19:956–962. doi: 10.1002/cbic.201700685. [DOI] [PubMed] [Google Scholar]
- 35.Wilson D.F., Erecinska M., Drown C., Silver I.A. The oxygen dependence of cellular energy metabolism. Arch. Biochem. Biophys. 1979;195:485–493. doi: 10.1016/0003-9861(79)90375-8. [DOI] [PubMed] [Google Scholar]
- 36.Al-Mehdi A.B., Pastukh V.M., Swiger B.M., Reed D.J., Patel M.R., Bardwell G.C., et al. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas L.W., Staples O., Turmaine M., Ashcroft M. CHCHD4 regulates intracellular oxygenation and perinuclear distribution of mitochondria. Front. Oncol. 2017;7:71. doi: 10.3389/fonc.2017.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandel N.S., McClintock D.S., Feliciano C.E., Wood T.M., Melendez J.A., Rodriguez A.M., et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 39.Chua Y.L., Dufour E., Dassa E.P., Rustin P., Jacobs H.T., Taylor C.T., et al. Stabilization of hypoxia-inducible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J. Biol. Chem. 2010;285:31277–31284. doi: 10.1074/jbc.M110.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar A., Vaish M., Karuppagounder S.S., Gazaryan I., Cave J.W., Starkov A.A., et al. HIF1alpha stabilization in hypoxia is not oxidant-initiated. eLife. 2021;10:91–106. doi: 10.7554/eLife.72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pientka F.K., Hu J., Schindler S.G., Brix B., Thiel A., Johren O., et al. Oxygen sensing by the prolyl-4-hydroxylase PHD2 within the nuclear compartment and the influence of compartmentalisation on HIF-1 signalling. J. Cell Sci. 2012;125:5168–5176. doi: 10.1242/jcs.109041. [DOI] [PubMed] [Google Scholar]
- 42.Dimauro I., Pearson T., Caporossi D., Jackson M.J. A simple protocol for the subcellular fractionation of skeletal muscle cells and tissue. BMC Res. Notes. 2012;5:513. doi: 10.1186/1756-0500-5-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patino W.D., Mian O.Y., Kang J.G., Matoba S., Bartlett L.D., Holbrook B., et al. Circulating transcriptome reveals markers of atherosclerosis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3423–3428. doi: 10.1073/pnas.0408032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dowling C.M., Walsh D., Coffey J.C., Kiely P.A. The importance of selecting the appropriate reference genes for quantitative real time PCR as illustrated using colon cancer cells and tissue. F1000Res. 2016;5:99. doi: 10.12688/f1000research.7656.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spandidos A., Wang X., Wang H., Seed B. PrimerBank: a resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucl. Acids Res. 2010;38:D792–799. doi: 10.1093/nar/gkp1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described are located in this article.