Abstract
Introduction
To design and evaluate a deep learning model based on ultra-widefield images (UWFIs) that can detect several common fundus diseases.
Methods
Based on 4574 UWFIs, a deep learning model was trained and validated that can identify normal fundus and eight common fundus diseases, namely referable diabetic retinopathy, retinal vein occlusion, pathologic myopia, retinal detachment, retinitis pigmentosa, age-related macular degeneration, vitreous opacity, and optic neuropathy. The model was tested on three test sets with data volumes of 465, 979, and 525. The performance of the three deep learning networks, EfficientNet-B7, DenseNet, and ResNet-101, was evaluated on the internal test set. Additionally, we compared the performance of the deep learning model with that of doctors in a tertiary referral hospital.
Results
Compared to the other two deep learning models, EfficientNet-B7 achieved the best performance. The area under the receiver operating characteristic curves of the EfficientNet-B7 model on the internal test set, external test set A and external test set B were 0.9708 (0.8772, 0.9849) to 1.0000 (1.0000, 1.0000), 0.9683 (0.8829, 0.9770) to 1.0000 (0.9975, 1.0000), and 0.8919 (0.7150, 0.9055) to 0.9977 (0.9165, 1.0000), respectively. On a data set of 100 images, the total accuracy of the deep learning model was 93.00%, the average accuracy of three ophthalmologists who had been working for 2 years and three ophthalmologists who had been working in fundus imaging for more than 5 years was 88.00% and 94.00%, respectively.
Conclusion
High performance was achieved on all three test sets using our UWFI multidisease classification model with a small sample size and fast model inference. The performance of the artificial intelligence model was comparable to that of a physician with 2–5 years of experience in fundus diseases at a tertiary referral hospital. The model is expected to be used as an effective aid for fundus disease screening.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40123-022-00627-3.
Keywords: Ultra-widefield images, Deep learning, Fundus diseases, Artificial intelligence
Key Summary Points
| Why carry out this study? |
| Compared with conventional color fundus photography, ultra-widefield images (UWFIs) have the advantages of no pupil dilatation, a wide imaging range, and fast acquisition, which are suitable for fundus disease screening. |
| We aim to develop a more clinically applicable deep learning model for multidisease classification, as few studies have done so previously. |
| What was learned from the study? |
| The UWFI multidisease classification model that we designed on the basis of a small sample size performed well with fast model inference. Its performance was comparable to that of doctors with 2–5 years of experience at a tertiary referral hospital. |
| The model is expected to be applied in developing countries and remote areas to assist in fundus disease screening. |
| Further expansion of the sample size and number of diseases and optimization of the model are needed in the future to achieve higher performance. |
Introduction
Retinal and optic nerve diseases are major causes of severe vision loss and blindness worldwide and can irreversibly affect a patient’s vision [1]. Failure to diagnose and treat the disease in a timely manner can often result in reduced quality of life and increased financial burden for patients. The availability of ophthalmologists specializing in fundus diseases is often inadequate, especially in developing countries and some underdeveloped areas, which can delay treatment for patients. In recent years, deep learning has been gradually applied to screen, diagnose, classify, and guide the treatment of retinal diseases, which can greatly reduce the human and material resources required for conventional screening modalities [2, 3]. Artificial intelligence (AI) models for multidisease classification based on conventional retinal fundus images have gradually matured [4–6]. However, color fundus photography (CFP) has a small imaging range and a time-consuming pupil dilation preparation. Ultra-widefield fundus images (UWFIs) have a wide imaging range, acquiring 200° fundus images in a single scan, and has the advantages of being noncontact, pupil-dilation-free, and easy to use [7]. The technique offers the benefit of much more rapid screening of fundus diseases.
The UWFI-based deep learning model has performed well in the identification of various single diseases and lesions, such as diabetic retinopathy (DR), retinal vein occlusion (RVO), retinal hemorrhage, and glaucomatous optic neuropathy (GON) [8–12]. In particular, by taking advantage of the wide range of UWFIs, excellent performance in identifying retinal detachment (RD) and peripheral retinal degeneration, such as lattice degeneration and retinal breaks, can be achieved using deep learning models [13–16]. However, there has been relatively little research into deep learning models that better meet clinical needs for the identification of multiple fundus diseases. Antaki et al. [17] applied automatic machine learning in Google Cloud AutoML Vision to implement a multiclassification task for UWFIs. Zhang et al. [18] improved the performance of deep learning models in classifying UWFIs by optimizing image preprocessing. However, in both of these studies, only normal eyes and three fundus diseases were identified. Recently, Cao et al. [19] proposed a UWFI-based four-hierarchical interpretable eye diseases screening system that can identify 30 common fundus diseases and lesions. We also carried out some work on multidisease identification with UWFIs. In this study, we developed an end-to-end deep learning UWFI disease classification model using a small sample size to identify normal eyes and eight retinal or optic nerve diseases, which were then tested externally at two hospitals.
Methods
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Clinical Research Ethics Committee of Renmin Hospital of Wuhan University (ethics number WDRY2021-K034). The ethics committee waived informed consent, as none of the UWFIs contained any personal information about the patients included.
Image Acquisition and Data Sets
We obtained the original set for this study by reviewing the UWFIs (Optos Daytona and Optos 200TX, Dunfermline, UK) of patients who attended the Eye Center, Renmin Hospital of Wuhan University from 2016 to 2022. One trained physician excluded fundus images with a small imaging field (fundus image obscured by eyelids and eyelashes or the examiner’s finger; more than 1/3 of the image not visible), significant refractive media opacity, and obvious signs of treatment, such as laser photocoagulation, silicone oil, or gas filling. Three attending physicians with more than 5 years of fundus imaging experience classified the images separately, and images with the same classification were included directly in the data set. Images that were classified differently at first were adjudicated by two senior physicians with more than 25 years of experience. Images for which two senior physicians could not agree on a classification were directly excluded. Normal fundus and eight clinically common fundus diseases were selected for model construction, namely, DR, RVO, pathologic myopia, RD, retinitis pigmentosa (RP), age-related macular degeneration, vitreous opacity, and optic neuropathy (including possible GON, optic nerve atrophy, and papilledema or optic disc edema). The images were labeled and classified according to the diagnostic imaging features of the disease (see Table S1 in the electronic supplementary material for details).
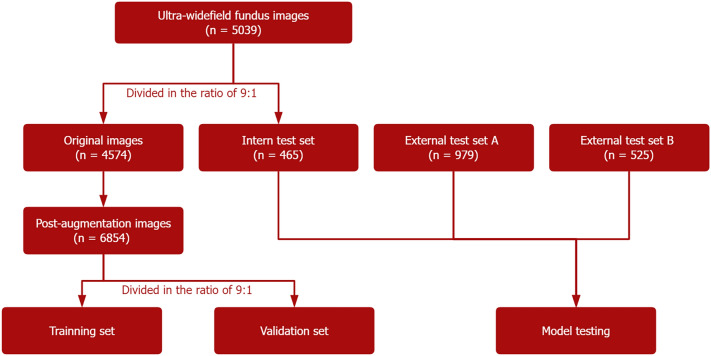
Only one image per eye was included for each patient, and images where multiple diseases were present were placed into multiple categories simultaneously. The original images were divided in a ratio of approximately 9:1 and used for model development and internal testing. Given the unbalanced amount of data across the disease species in the model development data set, random augmentation of the data was performed for a smaller number of disease species, including random horizontal and vertical flips, random rotations, and random Gaussian blurs, so that more balanced data could be used to better fit the neural network. After augmentation, the data were divided into a training set and a validation set at a ratio of 9:1. As there was no publicly available data set, we acquired UWFIs from Wuhan Optics Valley Central Hospital and UWFIs from Tianjin Medical University Eye Hospital as two external test sets sequentially: external test set A and external test set B. The distribution of images for each disease in the data set is shown in Table 1. The workflow for developing the deep learning model for multidisease recognition in UWFI is shown in Fig. 1.
Table 1.
Characteristics of the model training set and test sets of deep learning model
| Label, n (%) | Data set for model training | Test sets (n = 1969) | |||
|---|---|---|---|---|---|
| Original images (n = 4574) | Post-augmentation images (n = 6854) | Internal test set (n = 465) | External test set A (n = 979) | External test set B (n = 525) | |
| Normal fundus | 825 (18.0) | 825 (12.0) | 83 (17.8) | 280 (28.6) | 96 (18.3) |
| Referable diabetic retinopathy | 784 (17.1) | 784 (11.4) | 78 (16.8) | 123 (12.6) | 74 (14.1) |
| Retinal vein occlusion | 723 (15.8) | 723 (10.5) | 72 (15.5) | 69 (7.0) | 89 (17.0) |
| Pathologic myopia | 557 (12.2) | 776 (11.3) | 56 (12.0) | 105 (10.7) | 88 (16.8) |
| Retinal detachment | 545 (11.9) | 741 (10.8) | 55 (11.8) | 135 (13.8) | 49 (9.3) |
| Age-related macular degeneration | 239 (5.2) | 717 (10.5) | 27 (5.8) | 41 (4.2) | 33 (6.2) |
| Retinitis pigmentosa | 296 (6.5) | 761 (11.1) | 30 (6.4) | 32 (3.2) | 29 (5.5) |
| Vitreous opacity | 277 (6.1) | 755 (11.0) | 28 (6.0) | 96 (9.8) | 38 (7.2) |
| Optic neuropathy | 328 (7.2) | 772 (11.3) | 36 (7.7) | 98 (10.0) | 29 (5.5) |
Fig. 1.
Workflow for developing the deep learning model for multidisease recognition in UWFI
Deep-Learning Model Construction
Several convolutional neural networks with high performance in multilabel classification tasks, including EfficientNet-B7, DenseNet, and ResNet-101, were selected for this study, and pretrained models of ImageNet were used to initialize the model parameters. In training, we designed an end-to-end multilabel prediction model directly using the normal eye as a classification. The original UWFI image size was 3900 × 3072 pixels and was nonsquare. All input images were first preprocessed so that the model had an image input size of 224 × 224 pixels. After multiple convolution and block modules, an additional self-attentive layer was added to balance the feature dimensions of the images, mainly by means of a 1 × 1 convolution layer that rebalanced the weights of the input feature maps to achieve global perception. We set the Adam optimizer using CrossEntropy as the loss function. The training performance of each epoch was evaluated using the validation set, and the EarlyStopping strategy was set to stop training when there was no significant change in the validation set loss for 10 consecutive epochs.
Human Physicians Versus Deep-Learning Models
To compare the accuracy of the proposed fundus disease classification between doctors with different seniority and deep learning models, 100 images were randomly selected from the external test set of Wuhan Optics Valley Central Hospital for testing. There were 20 normal fundus images in this data set and 10 images for each of the remaining diseases. Three fundus doctors with 2 years of experience and three fundus doctors with more than 5 years of experience from the Renmin Hospital of Wuhan University (a tertiary referral hospital) were invited to participate in this comparison.
Model Performance Evaluation
The performance of the classification models on the test set was assessed using receiver operating characteristic (ROC) curves, area under the ROC curve (AUC), sensitivity, specificity, accuracy, and Matthews correlation coefficient (MCC). All data are presented as numerical values with 95% confidence intervals. In addition, the data sets were trained on ResNet-101 and DenseNet models, and the performance of EfficientNet-B7 was compared with the aforementioned two models. To obtain the logical interpretability of the models, the information learned by our deep learning models was visualized using class activation mapping (CAM) plots, an important technique in convolutional neural networks, which can be obtained by rearranging the feature maps derived from the last layer of convolution. All statistical analyses were performed using Python 3.7.11.
Results
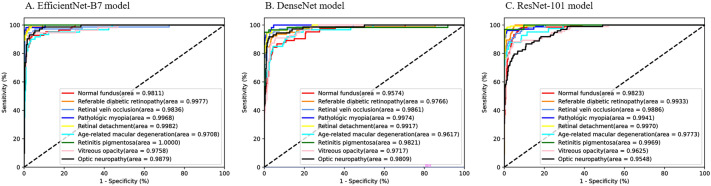
A total of 4574 original UWFIs were included in this study for training and validation of the model, which was then tested using an internal test set and two external test sets with data set volumes of 465, 979, and 525. The average AUCs of the EfficientNet-B7, ResNet-101, and DenseNet models on the internal test set were 0.9880, 0.9784, and 0.9830, respectively. EfficientNet-B7 achieved the best performance on the internal test set compared to the DenseNet and ResNet-101 models (Fig. 2, Supplementary Table S2). Therefore, the EfficientNet-B7 network was chosen for the construction of the deep learning model.
Fig. 2.
Receiver operating characteristic curves for the three deep learning models on the internal test set
Performance of the EfficientNet-B7 Model on the Test Set
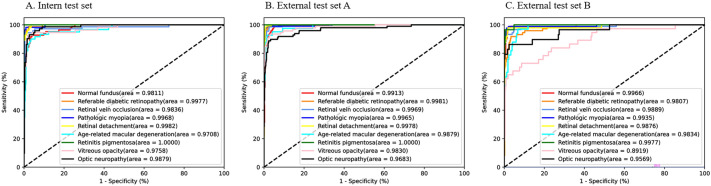
The AUCs of the EfficientNet-B7 model on the internal test set, external test set A, and external test set B were 0.9708 (0.8772, 0.9849) to 1.0000 (1.0000, 1.0000), 0.9683 (0.8829, 0.9770) to 1.0000 (0.9975, 1.0000), and 0.8919 (0.7150, 0.9055) to 0.9977 (0.9165, 1.0000); the corresponding sensitivity values were 0.8611 (0.8529, 0.8661) to 1.0000 (1.0000, 1.0000), 0.6633 (0.6452, 0.6702) to 1.0000 (1.0000, 1.0000), and 0.5946 (0.5932, 0.6226) to 0.9896 (0.9875, 0.9930); the specificity values were 0.9755 (0.9736, 0.9763) to 1.0000 (1.0000, 1.0000), 0.9861 (0.9856, 0.9870) to 0.9989 (0.9987, 1.0000), and 0.9531 (0.9522, 0.9550) to 1.0000 (1.0000, 1.0000); the accuracy values were 0.9644 (0.9637, 0.9675) to 1.0000 (1.0000, 1.0000), 0.9528 (0.9513, 0.9550) to 0.9979 (0.9988, 0.9988) and 0.9579 (0.9563, 0.9600) to 0.9923 (0.9925, 0.9938); and the MCCs were 0.8035 (0.8250, 0.8380) to 1.0000 (1.0000, 1.0000), 0.7508 (0.7714, 0.7943) to 0.9691 (0.9630, 0.9796), 0.7026 (0.7037, 0.7246) to 0.9235 (0.9263, 0.9398). The AUC, sensitivity, specificity, accuracy, and MCC for each disease on the three test sets are shown in Table 2. The ROC curves for each disease are plotted in Fig. 3.
Table 2.
Performance of the EfficientNet-B7 model in the classification of different fundus diseases
| Label | Internal test set | External test set A | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | MCC | Sensitivity | Specificity | Accuracy | AUC | MCC | |
| Normal fundus | 0.8916 (0.8857, 0.9010) | 0.9783 (0.9780, 0.9801) | 0.9661 (0.9650, 0.9688) | 0.9811 (0.9207, 0.9892) | 0.8332 (0.8493, 0.8633) | 0.9857 (0.9831, 0.9868) | 0.9861 (0.9856, 0.9870) | 0.9528 (0.9513, 0.9550) | 0.9913 (0.9513, 0.9939) | 0.8718 (0.8889, 0.9024) |
| Referable diabetic retinopathy | 0.9231 (0.9208, 0.9307) | 0.9980 (0.9985, 0.9986) | 0.9881 (0.9875, 0.9900) | 0.9977 (0.9307, 0.9994) | 0.8479 (0.8807, 0.8868) | 0.8943 (0.8866, 0.9011) | 0.9988 (0.9986, 1.0000) | 0.9856 (0.9850, 0.9875) | 0.9981 (0.9193, 0.999) | 0.8883 (0.9221, 0.9267) |
| Retinal vein occlusion | 0.9583 (0.9588, 0.9674) | 0.9942 (0.9943, 0.9956) | 0.9898 (0.9875, 0.9900) | 0.9836 (0.9528, 0.9997) | 0.9470 (0.9496, 0.9593) | 0.9265 (0.9167, 0.9375) | 0.9989 (0.9987, 1.0000) | 0.9938 (0.9938, 0.9950) | 0.9969 (0.9314, 0.998) | 0.9501 (0.9558, 0.9614) |
| Pathologic myopia | 0.9643 (0.9583, 0.9674) | 0.9963 (0.9959, 0.9972) | 0.9932 (0.9938, 0.9950) | 0.9968 (0.9556, 1.0000) | 0.9366 (0.9492, 0.9569) | 0.9524 (0.9467, 0.9595) | 0.9977 (0.9972, 0.9986) | 0.9928 (0.9925, 0.9938) | 0.9965 (0.9545, 0.9976) | 0.9195 (0.9348, 0.9417) |
| Retinal detachment | 0.9636 (0.9615, 0.9718) | 0.9944 (0.9932, 0.9945) | 0.9915 (0.9900, 0.9925) | 0.9982 (0.9307, 0.9994) | 0.9571 (0.9605, 0.9673) | 0.9776 (0.9754, 0.9817) | 0.9905 (0.9898, 0.9914) | 0.9887 (0.9888, 0.9900) | 0.9978 (0.9711, 0.9987) | 0.9569 (0.9647, 0.9724) |
| Age-related macular degeneration | 0.8667 (0.8636, 0.8810) | 0.9755 (0.9736, 0.9763) | 0.9644 (0.9637, 0.9675) | 0.9708 (0.8772, 0.9849) | 0.8035 (0.8250, 0.8380) | 0.9512 (0.9512, 0.9697) | 0.9861 (0.9856, 0.9870) | 0.9846 (0.9838, 0.9862) | 0.9879 (0.9351, 1.0000) | 0.8360 (0.8257, 0.8539) |
| Retinitis pigmentosa | 1.0000 (1.0000, 1.0000) | 1.0000 (1.0000, 1.0000) | 1.0000 (1.0000, 1.0000) | 1.0000 (1.0000, 1.0000) | 1.0000 (1.0000, 1.0000) | 1.0000 (1.0000, 1.0000) | 0.9979 (0.9974, 0.9987) | 0.9979 (0.9988, 0.9988) | 1.0000 (0.9975, 1.0000) | 0.9691 (0.9630, 0.9796) |
| Vitreous opacity | 0.8727 (0.8696, 0.8861) | 0.9832 (0.9822, 0.9848) | 0.9729 (0.9725, 0.9738) | 0.9758 (0.8832, 0.9827) | 0.8627 (0.8877, 0.8976) | 0.8602 (0.8533, 0.8765) | 0.9932 (0.9931, 0.9945) | 0.9805 (0.9800, 0.9825) | 0.9830 (0.8912, 0.9872) | 0.7508 (0.7714, 0.7943) |
| Optic neuropathy | 0.8611 (0.8529, 0.8661) | 0.9903 (0.9895, 0.9914) | 0.9746 (0.9738, 0.9762) | 0.9879 (0.8853, 0.9962) | 0.9472 (0.9528, 0.9593) | 0.6633 (0.6452, 0.6702) | 0.9966 (0.9959, 0.9972) | 0.9630 (0.9613, 0.9650) | 0.9683 (0.8829, 0.977) | 0.9466 (0.9556, 0.9650) |
| Label | External test set B | ||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | MCC | |
| Normal fundus | 0.9896 (0.9875, 0.9930) | 0.9531 (0.9522, 0.9550) | 0.9598 (0.9587, 0.9625) | 0.9966 (0.957, 0.9986) | 0.7030 (0.7045, 0.7356) |
| Referable diabetic retinopathy | 0.8082 (0.7944, 0.8224) | 0.9911 (0.9912, 0.9926) | 0.9655 (0.9637, 0.9663) | 0.9807 (0.854, 0.9953) | 0.8794 (0.8951, 0.9043) |
| Retinal vein occlusion | 0.8989 (0.8944, 0.9014) | 0.9954 (0.9955, 0.9970) | 0.9789 (0.9775, 0.9800) | 0.9889 (0.9155, 0.9988) | 0.8469 (0.8489, 0.8657) |
| Pathologic myopia | 0.9091 (0.9055, 0.9149) | 0.9931 (0.9925, 0.9941) | 0.9789 (0.9788, 0.9812) | 0.9935 (0.9206, 0.9957) | 0.8260 (0.8571, 0.8700) |
| Retinal detachment | 0.9583 (0.9571, 0.9677) | 0.9726 (0.9698, 0.9728) | 0.9713 (0.9700, 0.9725) | 0.9876 (0.936, 0.995) | 0.9071 (0.9316, 0.9381) |
| Age-related macular degeneration | 0.8485 (0.8372, 0.8636) | 0.9652 (0.9628, 0.9659) | 0.9579 (0.9563, 0.9600) | 0.9834 (0.8442, 0.9895) | 0.7026 (0.7037, 0.7246) |
| Retinitis pigmentosa | 0.9310 (0.9333, 0.9516) | 0.9959 (0.9960, 0.9973) | 0.9923 (0.9925, 0.9938) | 0.9977 (0.9165, 1.0000) | 0.9235 (0.9263, 0.9398) |
| Vitreous opacity | 0.5946 (0.5932, 0.6226) | 0.9959 (0.9960, 0.9973) | 0.9674 (0.9675, 0.9700) | 0.8919 (0.715, 0.9055) | 0.9054 (0.9328, 0.9412) |
| Optic neuropathy | 0.7586 (0.7400, 0.7857) | 1.0000 (1.0000, 1.0000) | 0.9866 (0.9850, 0.9875) | 0.9569 (0.8001, 0.9586) | 0.8526 (0.8387, 0.8657) |
AUC area under the curve, MCC Matthews correlation coefficient
Fig. 3.
Receiver operating characteristic curves for the EfficientNet-B7 model on the internal test set and the two external test sets
Heatmap Features for Model Identification
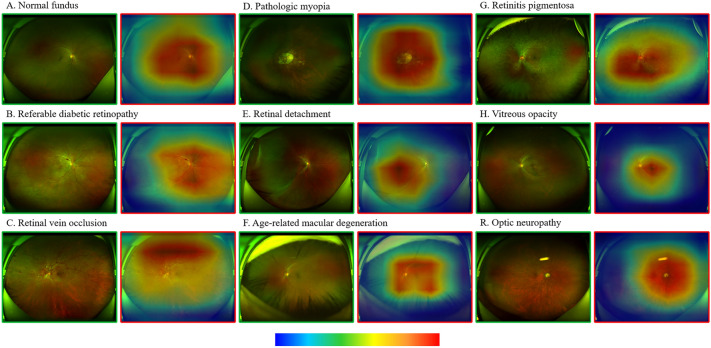
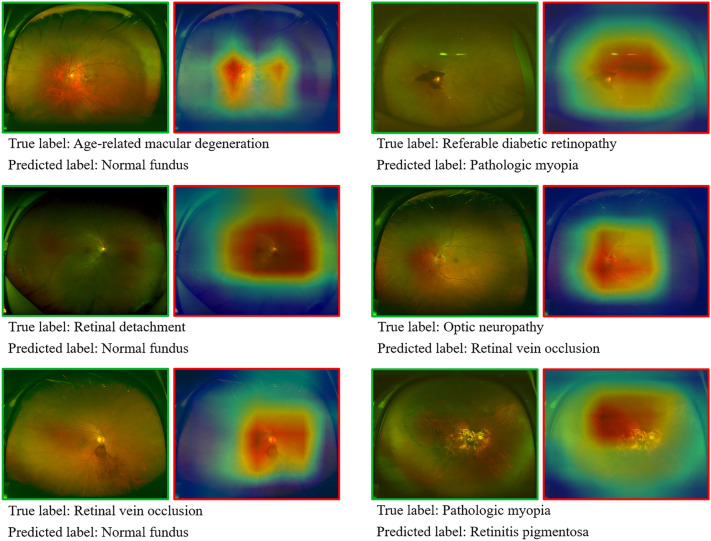
The CAM shows a heatmap of the distribution of contributions to the predicted output. Ranging from blue to red, redder colors indicate greater contributions to the network from the corresponding regions of the original image. The original images of the nine true-positive fundus disease classifications and the heatmaps overlaying the areas suggestive of contribution are shown in Fig. 4. On the basis of the CAM plot, the AI model is largely consistent with the clinician’s identification of the overall area of lesion features, while identifying lesions in more detail remains difficult. However, some images were misclassified (Fig. 5). In some of the images, the deep learning model made obvious errors in identifying areas that contribute to the diagnostic value.
Fig. 4.
True positive UWFI images of the nine labels on the internal test set (green boxes) and the corresponding CAM heatmap (red boxes). The heatmap shows a progressively higher contribution to the predicted output from blue to red (color bar below). CAM, class activation mapping
Fig. 5.
Misclassified UWFI images. The figure shows the true label and the predicted mislabel
Human Doctors Versus Deep-Learning Models
On a data set of 100 images, the total accuracy of the deep learning model was 93.00%, the average accuracy of the three ophthalmologists who had been working for 2 years was 88.00%, and the average accuracy of the three ophthalmologists who had been working in fundus imaging for more than 5 years was 94.00%. The average sensitivity and specificity of the aforementioned three groups were 92.22% and 99.07%, 88.15% and 98.52%, and 93.89% and 99.26%, respectively. The performance of the AI model ranged between the ophthalmologists who had worked in tertiary referral hospitals for 2 years and the others who had worked for more than 5 years.
Discussion
The UWFI-based multidisease classification model designed in this study achieved high sensitivity, specificity, and accuracy in the identification of normal fundus and the classification of eight common fundus diseases, achieving good performance on both the internal test set and the two external test sets. In addition, the model matched the capabilities of experienced fundus clinicians at tertiary referral hospitals. The model could be used in the future to assist in the screening and diagnosis of fundus diseases in developing countries and remote areas.
According to statistics, the number of ophthalmologists has increased at an average rate of 2.6% per year from 2010 to 2015. However, this rate is still lower than the 2.9% annual growth rate of the global aging population. In addition, the estimated average density of ophthalmologists varies widely across the world. Approximately 17% of the global population in 132 countries has access to less than 5% of the world’s ophthalmologists [20]. Of these, there may be even fewer ophthalmologists specializing in fundus diseases than for cataracts and refractions. At the same time, there is an imbalance in medical resources between developed and remote areas. It is more difficult to achieve early screening and referral for fundus diseases in remote areas. In recent years, there has been a rapid development in the direction of deep learning, which can correct the lack of manpower and relevant expertise as an alternative screening tool for eye diseases [21].
Conventional CFP has an imaging range of approximately 30–60° and is currently the most widely used clinical imaging modality for retinal imaging. Earlier studies have constructed recognition models for individual common fundus diseases [22, 23]. In recent years, several teams have demonstrated good performance with multiclassification task models derived from CFP [4–6, 24]. Real-world applications of these AI platforms with broader clinical applicability will greatly enhance the early screening and diagnosis of fundus diseases. Nevertheless, conventional CFP has a limited imaging range and can miss information from the peripheral retina. For instance, the detection of peripheral retinal diseases that can significantly affect vision, such as RD and RP, is limited in conventional CFP [4]. UWFI has the advantages of no pupil dilatation, a wide imaging range, and fast acquisition [25]. UWFI is now widely used in clinical work, almost replacing conventional CFP in many hospitals, and with the increasing popularity of UWFI, it will be more likely to achieve rapid screening of fundus diseases. Several teams have already tried to combine deep learning with UWFI for fundus screening. Japanese scholars have used convolutional neural networks to separately classify multiple single diseases from normal eyes [12, 26–28]. However, these models only demonstrate the feasibility of deep learning in identifying single diseases on UWFIs and are somewhat far from clinical application. Li et al. used high-quality annotated data to focus AI on the identification, classification, and localization of fundus lesions [10, 13, 15]. Good performance on diseases, such as peripheral retinal degeneration, retinal tears, RD, and GON, were achieved using their model, which takes advantage of the ultra-widefield of view of UWFIs.
There are still relatively few UWFI-based multidisease AI models for the fundus [17–19]. Recently, in parallel to our present study, Cao et al. [19] innovatively constructed a four-hierarchical interpretable eye diseases screening system using UWFIs. In the study, extensive image-level and lesion-level labeling was performed, the images were divided into 21 anatomical regions based on the anatomical location of the optic nerve head and macular fovea, and a lesion atlas mapping module was constructed to distinguish abnormal findings and determine their anatomical locations. The model is optimized by combining lesion mapping and diagnostic annotations, and different models are used to classify the lesions according to different disease types. Not only can it provide disease diagnosis but pathological and anatomical information can also be output. Ultimately, excellent performance is achieved using this model for the identification of 30 diseases and lesions and can be applied to multimorbidity beyond the training sample. In our study, an end-to-end model was applied with much lower engineering complexity, which offers significant advantages in terms of training time and model inference speed. The EfficientNet network is one of the most accurate models in the field of image classification and uses a joint adjustment technique of depth, width, and resolution of the input image with low parameters, high accuracy, and a flexible training strategy [29]. The model helps us to achieve higher accuracy with a relatively small sample size. The model achieves similar accuracy rates compared to clinicians and allows the use of computers to reduce the time required to classify diseases in the UWFI. In the future, we will continue to expand the sample size and disease categories in the hope of building a UWFI AI diagnostic model with more disease categories and greater accuracy.
There are also some limitations in this study. First, the number of images included in the training and test sets of this study is relatively small. As a result of the limited amount of data, the number of included disease classifications is still not broad enough; for example, optic papilledema, possible GON, and optic nerve atrophy were grouped under one classification of optic neuropathy because of the small number of optic nerve disease images. The number of diseases in the classification needs to be further increased in the future by using an increased sample size. Second, we actively excluded images with obvious treatment traces from the study, and the generalizability of the model may be reduced. In clinical applications, eyes with treatment traces are not rare, and the model may be incorrect for the recognition of these UWFI images. Third, the accuracy of this study is not high enough for the identification of some diseases, such as optic nerve disease and vitreous opacity. Fourth, disease labeling is primarily based on image features and relies on physician experience. Fifth, our model still cannot give multiple diagnoses for a single image, and we need to optimize or build other models in the future. Finally, the greatest advantage of UWFI is the detection of peripheral and total retinal lesions with slightly less sensitivity for less significant optic nerve or macular disease; human physicians face the same problem when reading the images.
Conclusions
Overall, the UWFI multidisease classification model we designed achieves a balance of small sample size and high accuracy and has relatively fast performance. The performance of the AI model was comparable to that of a physician with 2–5 years of experience in fundus diseases at a tertiary referral hospital. The model is expected to be used as an effective aid for fundus disease screening. In the future, we will incorporate a larger volume of data and more disease types to improve the generalization of the model performance and to validate it in primary care facilities and physical examination centers.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank all of the patients who participated in this study.
Funding
This project was supported by the National Natural Science Foundation of China (Grant No. 81500744). The Rapid Service Fee was funded by Wuhan Aiyanbang Technology Co., Ltd.
Authorship
All authors conform to the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, account for the integrity of the whole work, and have approved the version to be published.
Author Contributions
Conceptualization: Gongpeng Sun and Changzheng Chen; Methodology: Gongpeng Sun, Xiaoling Wang, Lizhang Xu, and Jian Zheng; Formal analysis and investigation: Gongpeng Sun, Xiaoling Wang, Lizhang Xu, Chang Li, Wenyu Wang, Zuohuizi Yi, Huijuan Luo, Yu Su, Jian Zheng, Zhiqing Li, Zhen Chen, Hongmei Zheng; Image Classification and Adjudication: Xiaoling Wang, Chang Li, Zuohuizi Yi, Zhen Chen, Hongmei Zheng; Writing—original draft preparation: Gongpeng Sun and Lizhang Xu; Writing—review and editing: Xiaoling Wang and Changzheng Chen; Funding acquisition: Yu Su, Zhen Chen, Changzheng Chen; Resources: Zhiqing Li, Zhen Chen, Hongmei Zheng; Supervision: Changzheng Chen. All authors read and approved the final manuscript.
Disclosures
Gongpeng Sun, Xiaoling Wang, Lizhang Xu, Chang Li, Wenyu Wang, Zuohuizi Yi, Huijuan Luo, Yu Su, Jian Zheng, Zhiqing Li, Zhen Chen, Hongmei Zheng and Changzheng Chen have nothing to disclose.
Compliance with Ethics Guidelines
This study was approved by the Clinical Research Ethics Committee of Renmin Hospital of Wuhan University (ethics number WDRY2021-K034) and conducted in accordance with the tenets of the Declaration of Helsinki. Informed consent was waived by the ethics committee as none of the UWFIs contained personal information about the patients.
Data Availability
The UWFI data sets utilized during the current study are not publicly available for data protection reasons. The other data generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Footnotes
Gongpeng Sun and Xiaoling Wang contributed equally.
Contributor Information
Zhen Chen, Email: hchenzhen@163.com.
Hongmei Zheng, Email: 312930646@qq.com.
Changzheng Chen, Email: whuchenchzh@163.com.
References
- 1.GBD 2019 Blindness and Vision Impairment Collaborators; Vision Loss Expert Group of the Global Burden of Disease Study. Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. Lancet Glob Health. 2021;9:e144–e160. [DOI] [PMC free article] [PubMed]
- 2.Schmidt-Erfurth U, Sadeghipour A, Gerendas BS, Waldstein SM, Bogunović H. Artificial intelligence in retina. Prog Retin Eye Res. 2018;67:1–29. doi: 10.1016/j.preteyeres.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Li T, Bo W, Hu C, et al. Applications of deep learning in fundus images: a review. Med Image Anal. 2021;69:101971. doi: 10.1016/j.media.2021.101971. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Chen H, Zhang B, et al. Development and evaluation of a deep learning model for the detection of multiple fundus diseases based on colour fundus photography. Br J Ophthalmol. 2022;106(8):1079–86. [DOI] [PubMed]
- 5.Lin D, Xiong J, Liu C, et al. Application of comprehensive artificial intelligence retinal expert (CARE) system: a national real-world evidence study. Lancet Digit Health. 2021;3:e486–e495. doi: 10.1016/S2589-7500(21)00086-8. [DOI] [PubMed] [Google Scholar]
- 6.Dong L, He W, Zhang R, et al. Artificial intelligence for screening of multiple retinal and optic nerve diseases. JAMA Netw Open. 2022;5:e229960. doi: 10.1001/jamanetworkopen.2022.9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Silva PS, Cavallerano JD, Sun JK, Noble J, Aiello LM, Aiello LP. Nonmydriatic ultrawide field retinal imaging compared with dilated standard 7-field 35-mm photography and retinal specialist examination for evaluation of diabetic retinopathy. Am J Ophthalmol. 2012;154:549–559.e542. doi: 10.1016/j.ajo.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 8.Tang F, Luenam P, Ran AR, et al. Detection of diabetic retinopathy from ultra-widefield scanning laser ophthalmoscope images: a multicenter deep learning analysis. Ophthalmol Retina. 2021;5:1097–1106. doi: 10.1016/j.oret.2021.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Guo C, Nie D, et al. Development and evaluation of a deep learning system for screening retinal hemorrhage based on ultra-widefield fundus images. Transl Vis Sci Technol. 2020;9:3. doi: 10.1167/tvst.9.2.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Guo C, Lin D, et al. Deep learning for automated glaucomatous optic neuropathy detection from ultra-widefield fundus images. Br J Ophthalmol. 2020;105:1548–1554. doi: 10.1136/bjophthalmol-2020-317327. [DOI] [PubMed] [Google Scholar]
- 11.Nagasato D, Tabuchi H, Ohsugi H, et al. Deep neural network-based method for detecting central retinal vein occlusion using ultrawide-field fundus ophthalmoscopy. J Ophthalmol. 2018;2018:1875431. [DOI] [PMC free article] [PubMed]
- 12.Nagasato D, Tabuchi H, Ohsugi H, et al. Deep-learning classifier with ultrawide-field fundus ophthalmoscopy for detecting branch retinal vein occlusion. Int J Ophthalmol. 2019;12:94–99. doi: 10.18240/ijo.2019.01.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Guo C, Nie D, et al. A deep learning system for identifying lattice degeneration and retinal breaks using ultra-widefield fundus images. Ann Transl Med. 2019;7:618. doi: 10.21037/atm.2019.11.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C, He F, Li B, et al. Development of a deep-learning system for detection of lattice degeneration, retinal breaks, and retinal detachment in tessellated eyes using ultra-wide-field fundus images: a pilot study. Graefes Arch Clin Exp Ophthalmol. 2021;259:2225–2234. doi: 10.1007/s00417-021-05105-3. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Guo C, Nie D, et al. Deep learning for detecting retinal detachment and discerning macular status using ultra-widefield fundus images. Commun Biol. 2020;3:15. doi: 10.1038/s42003-019-0730-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohsugi H, Tabuchi H, Enno H, Ishitobi N. Accuracy of deep learning, a machine-learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting rhegmatogenous retinal detachment. Sci Rep. 2017;7:9425. [DOI] [PMC free article] [PubMed]
- 17.Antaki F, Coussa RG, Kahwati G, Hammamji K, Sebag M, Duval R. Accuracy of automated machine learning in classifying retinal pathologies from ultra-widefield pseudocolour fundus images. Br J Ophthalmol. 2021. 10.1136/bjophthalmol-2021-319030. [DOI] [PubMed]
- 18.Zhang W, Zhao X, Chen Y, Zhong J, Yi Z. DeepUWF: an automated ultra-wide-field fundus screening system via deep learning. IEEE J Biomed Health Inform. 2021;25:2988–2996. doi: 10.1109/JBHI.2020.3046771. [DOI] [PubMed] [Google Scholar]
- 19.Cao J, You K, Zhou J, et al. A cascade eye diseases screening system with interpretability and expandability in ultra-wide field fundus images: a multicentre diagnostic accuracy study. EClinicalMedicine. 2022;53:101633. doi: 10.1016/j.eclinm.2022.101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Resnikoff S, Lansingh VC, Washburn L, et al. Estimated number of ophthalmologists worldwide (International Council of Ophthalmology update): will we meet the needs? Br J Ophthalmol. 2020;104:588–592. doi: 10.1136/bjophthalmol-2019-314336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ting DSW, Peng L, Varadarajan AV, et al. Deep learning in ophthalmology: the technical and clinical considerations. Prog Retin Eye Res. 2019;72:100759. doi: 10.1016/j.preteyeres.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Gargeya R, Leng T. Automated identification of diabetic retinopathy using deep learning. Ophthalmology. 2017;124:962–969. doi: 10.1016/j.ophtha.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 23.Peng Y, Dharssi S, Chen Q, et al. DeepSeeNet: a deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126:565–575. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cen LP, Ji J, Lin JW, et al. Automatic detection of 39 fundus diseases and conditions in retinal photographs using deep neural networks. Nat Commun. 2021;12:4828. doi: 10.1038/s41467-021-25138-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagiel A, Lalane RA, Sadda SR, Schwartz SD. Ultra-widefield fundus imaging: a review of clinical applications and future trends. Retina. 2016;36:660–678. doi: 10.1097/IAE.0000000000000937. [DOI] [PubMed] [Google Scholar]
- 26.Nagasawa T, Tabuchi H, Masumoto H, et al. Accuracy of ultrawide-field fundus ophthalmoscopy-assisted deep learning for detecting treatment-naive proliferative diabetic retinopathy. Int Ophthalmol. 2019;39:2153–2159. doi: 10.1007/s10792-019-01074-z. [DOI] [PubMed] [Google Scholar]
- 27.Matsuba S, Tabuchi H, Ohsugi H, et al. Accuracy of ultra-wide-field fundus ophthalmoscopy-assisted deep learning, a machine-learning technology, for detecting age-related macular degeneration. Int Ophthalmol. 2019;39:1269–1275. doi: 10.1007/s10792-018-0940-0. [DOI] [PubMed] [Google Scholar]
- 28.Nagasawa T, Tabuchi H, Masurnoto H, et al. Accuracy of deep learning, a machine learning technology, using ultra-wide-field fundus ophthalmoscopy for detecting idiopathic macular holes. PeerJ. 2018;6:e5696. doi: 10.7717/peerj.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan MX, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. In: 36th International Conference on Machine Learning (ICML). Long Beach, CA; 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The UWFI data sets utilized during the current study are not publicly available for data protection reasons. The other data generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.