Abstract
Background
No effective early diagnostic biomarkers are available for colorectal cancer (CRC). Therefore, we sought to identify new biomarkers that could identify CRC from progression as a pre-cancerous lesion to its invasive form. Recent studies have shown that microRNAs (miRs) are associated with the onset of cancer invasion and progression.
Aims
We hypothesized that the identification of miRs associated with CRC might be useful to detect this disease at early stages.
Methods
We conducted an integrated analysis of 79 isolated colorectal tumor glands, including adenomas, intramucosal cancers, and invasive CRCs that showed a microsatellite stable phenotype using GeneChip miRNA 4.0 microarray assays. The colorectal tumors we examined were divided into 2 cohorts (42 in the first cohort and 37 in the second cohort).
Results
First, cluster analysis was performed to stratify expression patterns of multiple miRs that were pooled according to the following criteria: fold change in expression (< −2.0 or > 2.0), p < 0.05, and mature miRs. As a result, the expression patterns of pooled miRs were subdivided into 3 subgroups that were correlated with tumor grade. Each subgroup was characterized by specific miRs. In addition, we found that specific miRs, including miR-140-3p and miR-378i, were closely associated with cancer invasion. Finally, we analyzed paired dysregulated miRs between adenomatous and cancerous components present within the same tumor.
Discussion
We showed that several miRs were dysregulated during progression from adenoma to intramucosal cancer. Specific miRs may have key roles in progression from intramucosal tumor to invasive CRC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10620-022-07576-8.
Keywords: Adenoma, Adenoma–carcinoma sequence, Colorectal cancer, Intramucosal cancer, microRNA
Introduction
Colorectal cancer (CRC) is a common disease and one of the leading causes of death worldwide [1]. At present, most patients are diagnosed with CRC at an advanced stage. Consequently, early detection of CRC is important to achieve better patient outcomes. Colorectal genetic pathways are largely classified into 3 models: the adenoma–carcinoma sequence, the serrated pathway, and the de novo pathway [2]. Among these models, the most important model is thought to be the adenoma–carcinoma sequence because this pathway is frequently encountered [3]. Thus, researchers should focus on molecular evaluation of progression in the adenoma–carcinoma sequence model for early detection of CRC.
CRC is a heterogeneous disease that can be categorized into distinct consensus molecular subtypes (CMSs) based on gene transcription [4]; these CMSs are helpful for predicting prognosis in patients with CRC [4–6]. The CMS classification suggests clear potential for clinical use in predicting both prognosis and response to systemic therapy, which seems to be independent of the classifier used. Prospective studies are warranted to support implementation of the CMS classification in clinical practice [4–6]. According to this theory, genomic and epigenetic alterations play key roles in colorectal carcinogenesis. Recently, many studies have focused on dysregulation of epigenetic changes, including altered expression of microRNAs (miRs) [4, 7].
Over the past few years, miRs have emerged as a prominent class of gene regulators. miRs are single-stranded RNAs 18–24 nt in length and are generated by an RNase III type enzyme from endogenous transcripts [7, 8]. miRs regulate gene expression by modulating the expression of mRNA. By silencing or activating various target mRNAs, miRs play key roles in diverse regulatory signal pathways, including control of development, cell differentiation, apoptosis, and proliferation of normal cells [7, 8]. Moreover, miRs also have roles in cancer development and progression [9, 10]. Indeed, specific miRs are expressed in multiple types of cancer cells and are involved in various signal pathways [7–11]. Thus, these miRs likely contribute to colorectal tumorigenesis, and it is important to elucidate how dysregulated miRs contribute to the development and progression of CRC tumors, particularly the progression from adenoma to intramucosal cancer and ultimately to invasive CRC [11].
Here, we performed a comprehensive analysis of miRs. Our goal was to evaluate the roles of specific miRs and their modified expression during colorectal tumorigenesis. In addition, we analyzed dysregulated expression of miRs occurring in adenomatous and cancerous components present within the same intramucosal neoplasias.
Materials and Methods
Patients
A total of 79 patients, including 30 with colorectal adenoma, 16 with intramucosal cancer (IMC), and 33 with invasive CRC with a microsatellite stable (MSS) phenotype, were enrolled in the current study. The 79 patients were divided into 2 groups, constituting a first cohort (cohort 1) for analysis and a second for validation (cohort 2). Histological diagnoses of adenoma and IMC were made according to the World Health Organization (WHO; 2019) criteria, with slight modifications. The adenomas we examined included low- and high-grade adenomas [3]. Clinicopathological findings were recorded according to the General Rules for Management of the Japanese Colorectal Cancer Association [12]. The clinicopathological findings in cohorts 1 and 2 are shown in Table 1-left. In addition, we investigated molecular alterations associated with direct progression from adenomatous to cancerous components occurring within the same tumor using a carcinoma in/with adenoma sequence model. The clinicopathological variables are summarized in Table 1-right.
Table 1.
Clinicopathological findings for the colorectal lesions examined in this study
| Cohort 1 (Microarray) (%) | Cohort 2 (Validation test) (%) | p value | |||
|---|---|---|---|---|---|
| Total | 42 | 37 | |||
| Sex | |||||
| Male | 29 | (69.0) | 15 | (40.5) | |
| Female | 13 | (31.0) | 22 | (59.5) | < 0.05 |
| Age, median (range), years | 67.0 | (43–81) | 66.0 | (46–85) | N.S |
| Location | |||||
| Left side/right side | 26/16 | 21/16 | N.S | ||
| Histological type | |||||
| CA | 15 | (35.7) | 15 | (40.5) | |
| IMC | 8 | (19) | 8 | (21.6) | |
| CRC with an MSS phenotype | 19 | (45.2) | 14 | (37.8) | N.S |
| Metastasis of lymph node | |||||
| Positive | 9 | (21.4) | 4 | (10.8) | |
| Negative | 33 | (78.9) | 33 | (89.2) | N.S |
| Lymphatic invasion | |||||
| Positive | 9 | (21.4) | 7 | (18.9) | |
| Negative | 33 | (78.9) | 30 | (81.1) | N.S |
| Vascular invasion | |||||
| Positive | 12 | (28.6) | 8 | (21.6) | |
| Negative | 30 | (65.2) | 29 | (78.4) | N.S |
CA, conventional adenoma; IMC, intramucosal cancer; CRC, colorectal cancer; MSS, microsatellite stable; N.S, not statistically significant
This study was approved by the local ethics committee of Iwate Medical University (approval number MH2020-066), and all patients provided informed consent.
Crypt Isolation Method
We isolated crypts from the tumors and normal mucosa (distal site of the colon for surgical or endoscopic specimens) as described elsewhere [13]. Tumor glands (adenomatous and intramucosal carcinomatous glands) were obtained from suspected target lesions as determined by magnification, whereas cancer glands in invasive CRC were taken from the invasive front. Briefly, fresh tissues were minced with a razor into small pieces and incubated at 37 °C for 30 min in calcium- and magnesium-free Hanks’ balanced salt solution (CMF) containing 30 mM EDTA. The isolated crypts were immediately fixed in 70% ethanol and stored at 4 °C until use for RNA extraction. The fixed isolated crypts were observed under a dissecting microscope (SZ60; Olympus, Tokyo, Japan). Some isolated crypts were routinely processed for histopathological analysis to confirm the histological nature of the isolated glands. Contamination, such as interstitial cells, was not evident in any of the 79 samples.
RNA Extraction
For each patient, isolation of total RNA from tumors and normal glands was performed using RNeasy Mini kits (Qiagen, Valencia, CA, USA) in accordance with the manufacturer’s instructions. The nucleic acid concentration was determined using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and the RNA purity was verified using 1.5% denaturing agarose gels.
Analysis of Microsatellite Instability (MSI)
DNA was extracted from isolated normal and tumor glands derived from each lesion. The MSI status was determined using a consensus panel of five reference microsatellite markers (BAT25, BAT26, D2S123, D3S546, and D17S250) using a previously described method [14]. When no marker was altered, the tumors were defined as MSS. When only one marker was altered, the tumors were defined as low MSI. When 2 or more markers were altered, the tumors were defined as high MSI.
MiR Microarray Analysis
For microarray analysis, 200 ng RNA was polyadenylated and labeled using a FlashTag Biotin HSR RNA Labeling kit and then treated with DNA ligase. The labeled RNA was hybridized to GeneChip miRNA 4.0 microarrays (Thermo Fisher Scientific) at 48 °C for 16 h, followed by washing and staining using a streptavidin–PE solution. The Affymetrix miRNA 4.0 microarray contains 6631 probes, including 2570 mature miR probes. The stained arrays were assessed using a GeneChip Scanner 3000 7G System (Thermo Fisher Scientific). Detailed methods have been described previously [15].
Hierarchical Clustering Based on Marker Scores
We conducted hierarchical cluster analysis to avoid arbitrary selection of pooled miRs using array-based analysis according to their quantitative levels. Next, we identified appropriate miRs for characterizing subgroups. This approach maximized homogeneity for each subgroup and assured the greatest differences between the subgroups. This was achieved with open-access clustering software (Cluster 3.0 software; bonsai.hgc.jp/ ~ mdehoon/software/cluster/software.htm). The clustering algorithm was set to centroid linkage clustering, which is the standard hierarchical clustering method used in biological studies.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
To validate the microarray results, RT-qPCR was performed. One microgram of total RNA was reverse transcribed into first-strand cDNA using a Qiagen cDNA Synthesis Kit (Qiagen). The ubiquitous β-actin gene was used as a control for constitutive gene expression. RT-qPCR was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). Relative expression levels (2−ΔCt) were calculated according to the Livak and Schmittgen method. Expression levels of each gene were compared with the expression level of RNU6B. Finally, for miR-185-5p, which is considered an advanced MiR in TaqMan Advanced miRNA Assays (Thermo Fisher, Waltham, MA, USA), miR-30e-5p was used as an internal control.
Receiver Operating Characteristic (ROC) Curve Analysis and Determination of Cutoff Values
Cutoff expression levels for each miR were determined using ROC analysis. For each cutoff expression level, the weighted mean sensitivity and specificity values for differentiating between lesion types were plotted to generate an ROC curve. The expression level closest in distance to the point on the curve with both the maximum sensitivity and specificity was selected as the cutoff, representing the expression level that correctly classified the greatest number of tumors with or without downregulation of that miR. The area under the ROC curve (AUC) was then calculated. These analyses were conducted using JMP Pro15.0 software.
Work Flow of the Current Study
The work flow of this study is shown in Supplementary Fig. 1. First, we examined significant differences in gene expression levels between isolated tumors and normal glands (adenomatous, intramucosal cancerous, and invasive cancerous glands) using paired t tests. From this analysis, 53 miRs were selected from 4603 miRs according to the following criteria: fold change in expression (< −2.0 or > 2.0), Benjamini–Hochberg method/false discovery rate (FDR) adjusted p value less than 0.05, and mature miRs using GeneChip miRNA 4.0 microarrays. In addition, hierarchical analysis (cluster analysis) was performed to avoid arbitrary assessment of the expression patterns of miRs occurring in tumor cells. We then evaluated candidate miRs that characterized subgroups stratified by hierarchical cluster analysis. Next, we examined whether these candidate miRs were correlated with colorectal tumors, including adenoma, IMC, and CRC with an MSS phenotype. Finally, candidate miRs were validated using RT-qPCR analysis in the second cohort (cohort 2).
Pathway Analysis
We explored target mRNAs that may be closely associated with corresponding miRs using Ingenuity Pathway Analysis (Qiagen).
Statistical Analysis
Clinicopathological variables (sex, location, macroscopic type, tumor location, and histological type) were analyzed using Fisher’s exact test. If statistical differences among the three lesion types were detected, comparisons between two groups were performed using Fisher’s exact test. Differences in age and tumor size between two groups were evaluated using Mann–Whitney U-tests. Differences in the expression levels of miRs between isolated normal and neoplastic glands were examined using t-tests (p values) with an adjusted Benjamini–Hochberg method/FDR correction. We used statistical analysis software (JMP Pro 13.0 software package for Windows; SAS Institute Inc., Cary, NC, USA) for these analyses. Results with p values less than 0.05 were accepted as significant.
Results
miR Expression Profiling in Colorectal Tumors
We identified potential miR biomarkers in 42 colorectal neoplasias, including 15 adenomas, 8 IMCs, and 19 invasive CRCs. We used global miR expression profiling in the analysis of the 42 colorectal neoplasias and compared them with normal gland samples (Supplementary Table 1). Thus, in the colorectal tumors examined in this study, 21 upregulated miRs and 32 downregulated miRs were subjected to cluster analysis (total of 53 miRs).
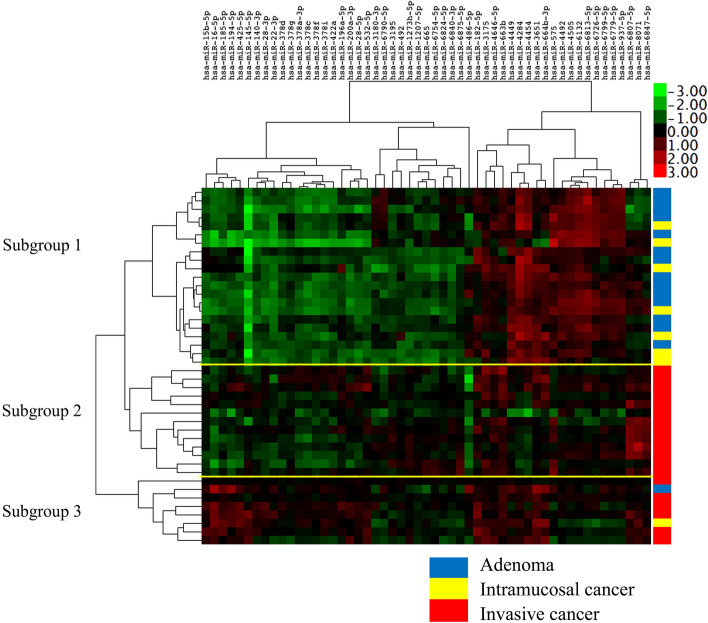
Hierarchical Clustering Based on Dysregulated miR Expression
We examined the expression patterns of miRs using hierarchical clustering. As a result, we identified 3 distinct subgroups (Fig. 1) based on their similar expression patterns. There were significant differences in the frequencies of adenoma or CRC with an MSS phenotype between subgroups 1 and 2 or 3 (p < 0.01). In addition, there were significant differences in the frequencies of IMCs between subgroups 1 and 2. Detailed clinicopathological findings of subgroups are shown in Table 2. Finally, detailed data are depicted in Supplementary Table 2.
Fig. 1.
A hierarchical cluster analysis. Expression patterns in tumors were classified into 3 subgroups
Table 2.
Clinicopathological findings for each subgroup for the colorectal lesions examined in this study (cohort 1)
| Cohort 1 (microarray) (%) | Subgroup 1 (%) | Subgroup 2 (%) | Subgroup 3 (%) | p value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 42 | 21 | 13 | 8 | |||||||||
| Sex | |||||||||||||
| Male | 29 | (69) | 15 | (71.4) | 10 | (76.9) | 4 | (50.0) | |||||
| Female | 13 | (31) | 6 | (28.6) | 3 | (23.1) | 4 | (50.0) | N.S | ||||
| Age, median (range), years | 67.0 | (43–81) | 70.0 | (52–80) | 67.0 | (43–81) | 65.5 | (58–73) | |||||
| Location | |||||||||||||
| Left side/right side | 26/16 | 11/10 | 10/3 | 4/4 | N.S | ||||||||
| ** | |||||||||||||
| Histological type | *** | ||||||||||||
| CA | 15 | (35.7) | 14 | (66.7) | 0 | (0) | 1 | (12.5) | |||||
| ** | |||||||||||||
| IMC | 8 | (19) | 7 | (33.3) | 0 | (0) | 1 | (12.5) | |||||
| *** | |||||||||||||
| *** | |||||||||||||
| CRC with an MSS phenotype | 19 | (45.2) | 0 | (0) | 13 | (100) | 6 | (75.0) | < 0.001 | ||||
CA, conventional adenoma; IMC, intramucosal cancer; CRC, colorectal cancer; MSS, microsatellite stable; N.S, not statistically significant
***p < 0.0001; **p < 0.01; *p < 0.05
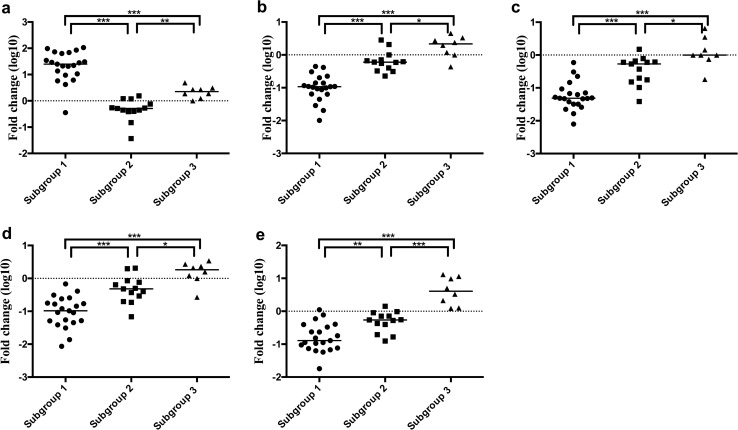
Statistically Significant Differences in the Expression Levels of Specific miRs Among Each Subgroup in the First Cohort (Cohort 1)
We then compared the expression levels of miRs between subgroups (subgroups 1 versus 2; subgroups 2 versus 3; and subgroups 1 versus 3), which were stratified by cluster analysis using 53 specific miRs (Supplementary Fig. 2). There were significant differences in the expression levels of 7 miRs (including miR-4, miR-4284, miR-140-3p, miR-185-5p, miR-22-3p, miR-378i, miR-15b-5p, and miR-378c) between subgroups (subgroups 1 versus 2; subgroups 1 versus 3; and subgroups 2 versus 3). The association of specific miRs with each subgroup is shown in Fig. 2. In addition, we examined the association of the 7 miRs with each lesion, including subgroups 1, 2, and 3. Among the 7 miRs, there were significant differences in the expression levels of 5 miRs (including miR-4284, miR-140-3p, miR-22-3p, miR-378i, and miR-185-5p) between subgroups 1 and 2, subgroups 2 and 3, and subgroups 1 and 3 (Fig. 2).
Fig. 2.
Expression levels of miRNAs in subgroups 1, 2, and 3 (cohort 1). a. miR-4284; b. miR-140-3p; c. miR-22-3p; d. miR-378i; e. miR-185-5p
Next, ROC analysis was used to determine the cutoff value of the expression level of the above-mentioned 5 miRs that best differentiated invasive CRCs from IMCs (Supplementary Fig. 2). The AUCs and optimal cutoff expression levels of 5 miRs are shown in Table 3. The cutoff values, AUCs, sensitivities, specificities, positive and negative predictive values, and positive and negative likelihood ratios discriminating between IMC and invasive CRC for each miR are shown in Table 3a.
Table 3.
Sensitivity, specificity, positive predictive value, negative predictive value, positive likelihood, and negative likelihood of the 5 miRNAs, a (cohort 1), b (cohort 2; validation cohort)
| a | ||||||||
|---|---|---|---|---|---|---|---|---|
| miRs | AUC | Cutoff value | Se | Sp | PPV | NPV | LH + | LH − |
| hsa-miR-4284 | 0.96 | 0.4298 | 1.00 | 0.96 | 0.95 | 1.00 | 23.00 | 0.00 |
| hsa-miR-140-3p | 0.91 | − 0.6435 | 1.00 | 0.78 | 0.79 | 1.00 | 4.60 | 0.00 |
| hsa-miR-185-5p | 0.80 | − 0.4031 | 0.84 | 0.74 | 0.73 | 0.85 | 3.23 | 0.21 |
| hsa-miR-22-3p | 0.85 | − 0.9886 | 0.95 | 0.74 | 0.75 | 0.94 | 3.63 | 0.07 |
| hsa-miR-378i | 0.83 | − 0.7284 | 0.95 | 0.70 | 0.72 | 0.94 | 3.11 | 0.08 |
| b | ||||||||
|---|---|---|---|---|---|---|---|---|
| miRs | AUC | Cutoff (1st cohort) | Se | Sp | PPV | NPV | LH + | LH − |
| hsa-miR-378i | 0.71 | − 0.7284 | 0.93 | 0.61 | 0.59 | 0.93 | 2.37 | 0.12 |
| hsa-miR-140-3p | 0.85 | − 0.6435 | 1.00 | 0.57 | 0.58 | 1.00 | 2.30 | 0.00 |
| hsa-miR-22-3p | 0.65 | − 0.9886 | 0.86 | 0.52 | 0.52 | 0.86 | 1.79 | 0.27 |
| hsa-miR-185-5p | 0.60 | − 0.4031 | 0.69 | 0.37 | 0.43 | 0.64 | 1.10 | 0.84 |
| hsa-miR-4284 | 0.66 | 0.4298 | 0.57 | 0.26 | 0.32 | 0.50 | 0.77 | 1.64 |
Se, sensitivity; Sp, specificity; PPV, positive predictive value; NPV, negative predictive value; LH + , positive likelihood; LH − , negative likelihood
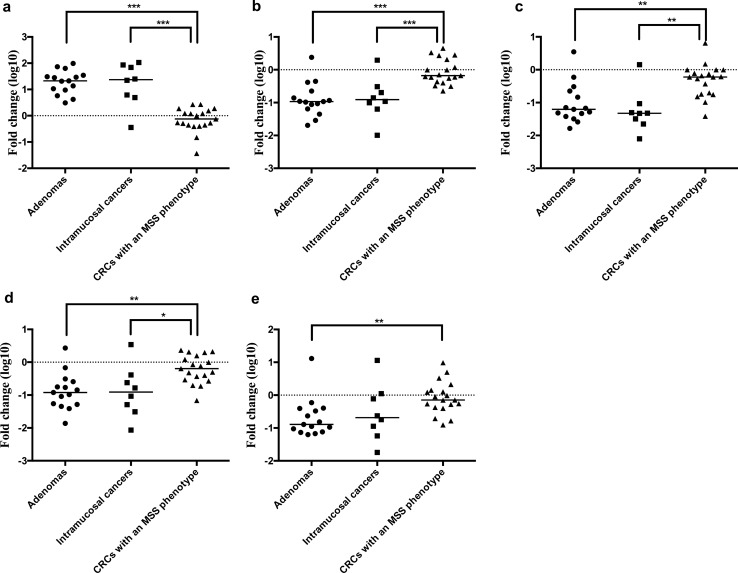
Statistically Significant Differences in the Expression Levels of Specific miRs Among Each Lesion in the Second Cohort (Cohort 2) for Validation
We selected 5 miRs (miR-378i, miR-140-3p, miR-22-3p, miR-185-5p, and miR-4284) to assess the usefulness of the candidate miRs in assessing the correlations between their expression and invasion (Fig. 3). First, we evaluated the relationships between the expression levels of the 5 candidate miRs with adenomas, IMCs, and invasive CRCs with an MSS phenotype. We found that there were statistically significant differences in the expression levels of miR-140-3p (p < 0.001) and miR-378i (p < 0.05) between adenomas and CRCs with an MSS phenotype and between IMCs and CRCs with an MSS phenotype (Fig. 4). To identify the candidate miRs differentiating IMCs from invasive CRCs, we examined the usefulness of the 5 candidate miRs to function as biological markers using the same criteria of a positive likelihood ratio greater than 2 and negative likelihood ratio less than 0.5. We found 2 specific miRs (miR-140-3p and miR-378i) for prediction of invasion beyond the submucosa (Table 3b).
Fig. 3.
Expression level of hsa-miRNAs in adenoma, intramucosal cancer, and CRC with an MSS phenotype (cohort 2). a. miR-4284; b. miR-140-3p; c. miR-22-3p; d. miR-378i; e. miR-185-5p
Fig. 4.
Expression level of each miRNA in each lesion (cohort 2). a. miR-4284; b. miR-140-3p; c. miR-22-3p; d. miR-378i; e. miR-185-5p
Association of miR-140-3p and miR-378i with Candidate mRNAs
We examined candidate connections between miRs and mRNAs according to Ingenuity Pathway Analysis (Qiagen). From this analysis, we identified 6 connections (CASP9, PDPK1, TUSC2, ODC1, IGF1R, and SUFU) of miR-378i with specific mRNAs based on confidence levels (experimentally observed, moderate prediction level; Supplementary Fig. 3). However, we could not find such connections of miR-140-3p with specific mRNAs based on the same confidence level.
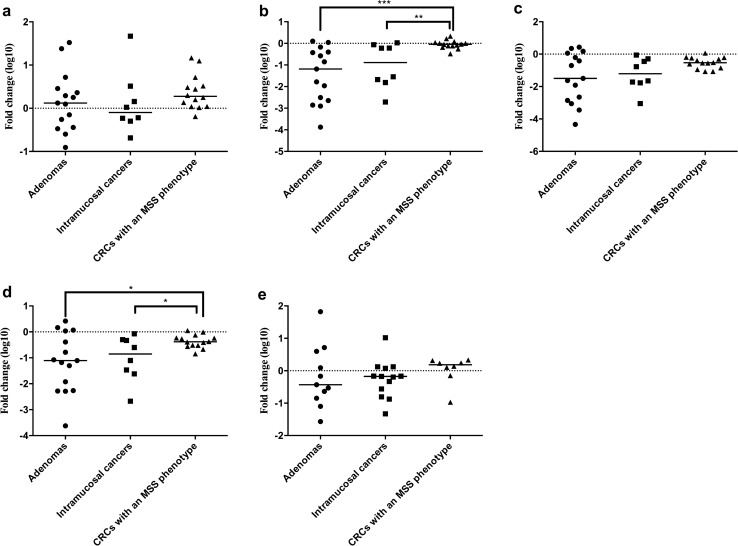
Association of miR Expression Level with the Progression of Adenomatous Lesions to IMC Lesions Within the Same Tumor
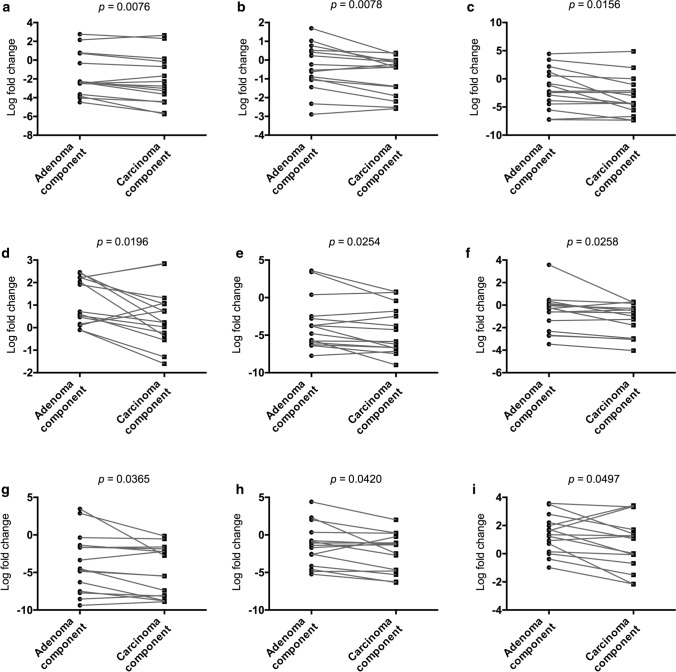
We explored specific miRs that were closely associated with the progression from an adenoma component to an IMC component within the same tumor. We found that 9 miRs (miR-6754-5p, miR-148b-3p, miR-4417, miR-4740-3p, miR-145-5p, miR-30e-5p, miR-143-3p, miR-378c, and miR-365a-5) were significantly downregulated from adenomatous components to carcinomatous components within the same tumor (using paired t tests). Thus, the expression of these miRs was significantly reduced during the progression of adenomas to IMCs within the same tumor. The associations are depicted in Fig. 5.
Fig. 5.
Dysregulated miRNAs in adenomatous and carcinomatous components within the same tumor. A, miR-6754-5p; B. miR-148b-3p; C. miR-4417; D. miR-4740-3p; E. miR-145-5p; F. miR-30e-5p; G. miR-143-3p; H. miR-378c; I. miR-365a-5p
Association of the immunohistochemical expression of clusterin and ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (ASAP3) with miR-140-3p and miR-378i expression in CRC
Discussion
Improvements in our understanding of molecular alterations associated with tumor progression greatly enhance the diagnosis and treatment of patients with cancer [2, 3, 16]. Recent studies have shown that dysregulated expression of miRs is closely associated with tumor progression [8, 9]. In the current study, we set cutoff values to predict submucosal invasion of tumor cells. Although the cutoff value we set may be a limitation in the use of this approach in clinical practice, we suggest that this value might still be applicable.
miR-140 is encoded in the intron of the WW domain containing the E3 ubiquitin protein ligase 2 gene. The sequence produces 2 mature miRs (miR-140-5p and miR-140-3p), which have been shown to be related to breast, lung, colorectal, and ovarian cancers [17–20]. However, it is unknown whether miR-140-3p plays a role in colorectal tumors. In the current study, miR-140-3p expression was upregulated in isolated CRC glands compared with isolated adenomas and IMC glands. However, in previous studies, the expression of miR-140-3p was found to be downregulated in breast cancer, ovarian cancer, and CRC [17–20]. These studies showed that miR-140-3p inhibits tumor proliferation, migration, and invasion, suggesting this miR may behave as a suppressor miR [17–20]. In addition, Liu et al. showed that BCL9 and BCL2 are direct targets of miR‐140‐3p, suppressing the proliferation, migration, and invasion of CRC cells and perhaps mediating apoptosis [21]. They also suggested that β‐catenin nuclear accumulation resulting from Wnt signal activation may be caused by high expression of BCL9 and BCL2 through downregulation of miR-140-3p [18]. Moreover, a recent study showed that miR-140-3p suppresses growth and induces apoptosis in CRC by targeting programmed death ligand 1 in CRC [20]. By contrast, Salem et al. indicated that both the canonical miR-140-3p and its 5′ iso miR-140-3p, which are termed isomiRs based upon length and/or sequence variations, are highly expressed in tumors compared with normal breast tissue in miRNAseq data from patients with breast cancer (The Cancer Genome Atlas dataset) [17]. Importantly, the same miR can have the opposite function in different types of cells [17, 21]. Although target mRNAs of miR-140-3p were not examined in this study, we suggest that increased expression of miR-140-3p may be a novel candidate marker of cancer invasion during the progression from intramucosal neoplasia to invasive CRC.
In previous studies, the conformation of the miR-378 target gene was analyzed using bioinformatic analysis and luciferase reporter systems in melanoma cells [22, 23]. Expression of miR-378 was often increased in melanoma and has been shown to cause migration, invasion, and tumorigenicity [23]. miR-378 is thought to act, at least in part, through inhibition of the potential target gene FOXN3 and via Wnt/β-catenin pathway activation [23]. In the current study, our findings suggested that several target transcripts, including CASP9, PDPK1, TUSC2, ODC1, IGF1R, and SUFU, are suppressed by high expression of miR-378i. According to these findings, the expression of miR-378i was higher in CRC with an MSS phenotype than in adenoma and IMC, suggesting that miR-378i expression may suppress the target transcripts. Here, we found that the increasing expression of miR-378i from IMC and adenoma to CRC with an MSS phenotype may be associated with potential cancer invasion by promoting the progression of intramucosal neoplasia to invasive CRC. This miR could be a novel diagnostic biological marker for invasion and be useful for targeted endoscopic treatment. In addition, a recent study revealed that high expression of miR-378i reduced chemoresistance to cisplatin in lung adenocarcinoma cell by targeting secreted Clusterin, which is involved in tumor development and cancer progression [22]. This finding suggests that upregulation of mi-378i indicates a requirement for chemotherapy in CRC.
The clinical benefit of examining miR expression, including that of miR-140-3p and miR-378i, suggests a need for additional endoscopic treatment. If the expression levels of the miRs are higher than the cutoff values determined in this study, additional endoscopic treatment may be required. Under these circumstances, although unnecessary endoscopic treatment may be performed, examination of the target miRs may help to avoid such unnecessary endoscopic treatment. The current study may contribute to selection of the appropriate additional endoscopic treatment.
Colorectal tumorigenesis is thought to be a multistep process in which genetic alterations accumulate, ultimately producing a malignant phenotype [2, 3]. An adenoma–carcinoma sequence model was proposed to explain the genetic basis of colorectal neoplasia, including several salient molecular features [2, 3]. The adenoma–carcinoma sequence model, in which both adenoma and cancer components coexist within the same tumor, is a novel model in which molecular differences are simultaneously present within the same tumor [8]. Although the transition from adenoma to carcinoma has been reported to be accompanied by the acquisition of molecular alterations [2, 3], researchers have attempted to discover new molecular factors that determine the risk of progression of adenoma to adenocarcinoma. In the current study, we found that specific miRs, including miR-6754-5p, miR-148b-3p, miR-4417, miR-4740-3p, miR-145-5p, miR-30e-5p, miR-143-3p, miR-378c, and miR-365a-5p, were candidate markers that may be associated with tumor progression within the same tumor. We focused on two specific miRs (miR-143-3p and miR-378c) owing to their close association with CRC progression [23–28], including the invasion, proliferation, and migration of CRC cells in vitro. In addition, the invasive potential of CRC cells was enhanced by expression of target mRNAs, such as clusterin, FOXN3, ASAP3, and SDAD1 [23–28]. Hence, our data may suggest new molecular mechanisms for CRC progression. By identifying new mechanisms, we could provide new diagnostic targets that have clinical value.
There were some limitations to this study. First, the sample size may have been small. However, we used isolated tumors and normal glands, enabling us to conduct a comprehensive and controlled molecular analysis. This is a clear advantage of this study, compared with other molecular studies, such as those [29, 30] using fresh tissues containing interstitial cells. Second, we obtained samples from a single site of the lesion regardless of tumor sample size. The single site might not reflect all of the molecular alterations present within the same tumor. However, we used the central lesion of the tumor or the invasive front of CRC, which are thought to be representative of molecular alterations.
In conclusion, we report a comprehensive and sensitive miR expression analysis of isolated tumor glands, including adenoma, IMC, and invasive CRC with an MSS phenotype. From our analysis, miR-140-3p and miR-378i were selected as markers for the transition from intramucosal neoplasia to CRC in both the first (cohort 1) and second cohorts (cohort 2). Our findings provided insights into the possibility of targeting oncogenic miRs as invasive markers. In addition, we examined dysregulated miRs that may be associated with intramucosal progression from adenoma to IMC within the same tumor, data confirmed with a paired t test. We suggest that several miRs could be used as candiate markers of progression from adenoma to intramucosal cancer within the same tumor.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary Figure 1. Work flow in the present study. (TIF 518 kb)
Supplementary Figure 2. ROC of each miRNA. a. miR-4284; b. miR-140-3p; c. miR-22-3p; d. miR-378i; e. miR-185-5p. (TIFF 673 kb)
Supplementary Figure 3. Suggested connections of miR-378i with several transcripts, including CASP9, PDPK1, TUSC2, ODC1, IGF1R, and SUFU. (TIF 375 kb)
Acknowledgments
We gratefully acknowledge the technical assistance of members of the Department of Molecular Diagnostic Pathology, Iwate Medical University, for their support.
Abbreviations
- miR
MicroRNA
- CRC
Colorectal cancer
- PCR
Polymerase chain reaction
- ROC
Receiver operating characteristic curve
- AUC
Area under the curve
Author’s contribution
TS, who is the first and corresponding author, contributed to the preparation of the manuscript, including all aspects of the data collection and analysis. RS constructed the figures and tables and performed the statistical analyses. MO assisted the statistical analyses. NY helped in the interpretation of pathological findings. KO and AS provided clinical support during the preparation of the manuscript. ME and TM provided clinical information on endoscopic treatment. HS helped with molecular analysis.
Funding
No funding was received for this study.
Data Availability
The microarray data presented in this article have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/geo/query/XXXXXXXXXXXXX) under accession number GSE6XXXXX.
Code Availability
N/A.
Declarations
Conflict of interest
We declare that we have no conflicts of interest.
Ethical Approval
Informed consent was obtained from each patient according to institutional guidelines, and the research protocols were approved by the ethics committee of Iwate Medical University Hospital (approval number MH2020-066).
Consent of Publication
Not applicable.
Human Rights Statement and Informed Consent
All procedures were performed in accordance with the ethical standards of the Iwate Medical University and with the Declaration of Helsinki. Substitute for informed consent (approval by the institutional review board of Iwate Medical University) was obtained from all patients included in the study.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10:13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamilton SR, Sekine S. Conventional colorectal adenoma. WHO classification of tumours of the digestive system. International Agency for Research on Cancer, Lyon 2019:170–173.
- 4.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ten Hoorn S, de Back TR, Sommeijer DW, Vermeulen L. Clinical value of consensus molecular subtypes in colorectal cancer: a systematic review and meta-analysis. J Natl Cancer Inst. 2021:djab106. doi: 10.1093/jnci/djab106. [DOI] [PMC free article] [PubMed]
- 6.Ten Hoorn S, Trinh A, de Jong J, Koens L, Vermeulen L. Classification of colorectal cancer in molecular subtypes by immunohistochemistry. Methods Mol Biol. 2018;1765:179–191. doi: 10.1007/978-1-4939-7765-9_11. [DOI] [PubMed] [Google Scholar]
- 7.Strubberg AM, Madison BB. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis Model Mech. 2017;10:197–214. doi: 10.1242/dmm.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahadi A. The significance of microRNA deregulation in colorectal cancer development and the clinical uses as a diagnostic and prognostic biomarker and therapeutic agent. Noncoding RNA Res. 2020;5:125–134. doi: 10.1016/j.ncrna.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fadaka AO, Pretorius A, Klein A. Biomarkers for stratification in colorectal cancer: microRNAs. Cancer Control. 2019;26:1073274819862784. doi: 10.1177/1073274819862784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saberinia A, Alinezhad A, Jafari F, Soltany S, Akhavan Sigari R. Oncogenic miRNAs and target therapies in colorectal cancer. Clin Chim Acta. 2020;508:77–91. doi: 10.1016/j.cca.2020.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Sugai T, Osakabe M, Habano W, et al. A genome-wide analysis of the molecular alterations occurring in the adenomatous and carcinomatous components of the same tumor based on the adenoma–carcinoma sequence. Pathol Int. 2021;71:582–593. doi: 10.1111/pin.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe T, Muro K, Ajioka Y, et al. Japanese Society for Cancer of the Colon and Rectum. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2016 for the treatment of colorectal cancer. Int J Clin Oncol. 2018;23:1–34. [DOI] [PMC free article] [PubMed]
- 13.Sugai T, Habano W, Nakamura S, Uesugi N, Sasou S, Itoh C. A unique method for mutation analysis of tumor suppressor genes in colorectal carcinomas using a crypt isolation technique. Arch Pathol Lab Med. 2000;124:382–386. doi: 10.5858/2000-124-0382-AUMFMA. [DOI] [PubMed] [Google Scholar]
- 14.Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 15.Sato C, Osakabe M, Nagasawa T, et al. Genome-wide analysis of microRNA to evaluate prognostic markers in isolated cancer glands and surrounding stroma in high-grade serous ovarian carcinoma. Oncol Lett. 2020;20:338. doi: 10.3892/ol.2020.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Network Cancer Genome Atlas. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salem O, Erdem N, Jung J, et al. The highly expressed 5'isomiR of hsa-miR-140-3p contributes to the tumor-suppressive effects of miR-140 by reducing breast cancer proliferation and migration. BMC Genomics. 2016;17:566. doi: 10.1186/s12864-016-2869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong W, Yao C, Teng X, et al. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016;37:973–2985. doi: 10.1007/s13277-015-3452-9. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Zhao J, Wang H, et al. MicroRNA-140-3p enhances the sensitivity of hepatocellular carcinoma cells to sorafenib by targeting pregnenolone X receptor. Onco Targets Ther. 2018;11:5885–5894. doi: 10.2147/OTT.S179509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang W, Li T, Wang J, et al. miR-140-3p suppresses cell growth and induces apoptosis in colorectal cancer by targeting PD-L1. Onco Targets Ther. 2019;12:10275–10285. doi: 10.2147/OTT.S226465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu D, Chen C, Cui M, Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med. 2021;10:3358–3372. doi: 10.1002/cam4.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Jiang Y, Huang Z, et al. miRNA-378 reverses chemoresistance to cisplatin in lung adenocarcinoma cells by targeting secreted clusterin. Sci Rep. 2016;6:19455. doi: 10.1038/srep19455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun M, Ma X, Tu C, et al. MicroRNA-378 regulates epithelial-mesenchymal transition and metastasis of melanoma by inhibiting FOXN3 expression through the Wnt/β-catenin pathway. Cell Biol Int. 2019;43:1113–1124. doi: 10.1002/cbin.11027. [DOI] [PubMed] [Google Scholar]
- 24.Guo L, Fu J, Sun S, et al. MicroRNA-143-3p inhibits colorectal cancer metastases by targeting ITGA6 and ASAP3. Cancer Sci. 2019;110:805–816. doi: 10.1111/cas.13910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding X, Du J, Mao K, et al. MicroRNA-143-3p suppresses tumorigenesis by targeting catenin-delta1 in colorectal cancer. Onco Targets Ther. 2019;12:3255–3265. doi: 10.2147/OTT.S184118. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Zeng M, Zhu L, Li L, Kang C. miR-378 suppresses the proliferation, migration and invasion of colon cancer cells by inhibiting SDAD1. Cell Mol Biol Lett. 2017;22:12. doi: 10.1186/s11658-017-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slattery ML, Herrick JS, Pellatt DF, et al. MicroRNA profiles in colorectal carcinomas, adenomas and normal colonic mucosa: variations in miRNA expression and disease progression. Arcinogenesis. 2016;37:245–261. doi: 10.1093/carcin/bgv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagy ZB, Wichmann B, Kalmár A, et al. Colorectal adenoma and carcinoma specific miRNA profiles in biopsy and their expression in plasma specimens. Clin Epigenet. 2017;9:22. doi: 10.1186/s13148-016-0305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habano W, Sugai T, Nakamura S, Yoshida T. A novel method for gene analysis of colorectal carcinomas using a crypt isolation technique. Lab Invest. 1996;74:933–940. [PubMed] [Google Scholar]
- 30.Takahashi H, Sugai T, Habano W, Nakamura S. Application of the crypt isolation technique to the assessment of genetic alterations of colorectal carcinomas. Pathol Int. 2002;52:628–635. doi: 10.1046/j.1440-1827.2002.01395.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Work flow in the present study. (TIF 518 kb)
Supplementary Figure 2. ROC of each miRNA. a. miR-4284; b. miR-140-3p; c. miR-22-3p; d. miR-378i; e. miR-185-5p. (TIFF 673 kb)
Supplementary Figure 3. Suggested connections of miR-378i with several transcripts, including CASP9, PDPK1, TUSC2, ODC1, IGF1R, and SUFU. (TIF 375 kb)
Data Availability Statement
The microarray data presented in this article have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/geo/query/XXXXXXXXXXXXX) under accession number GSE6XXXXX.
N/A.