Abstract
The NRF2 pathway is frequently activated in various cancer types, yet a comprehensive analysis of its effects across different malignancies is currently lacking. We developed a NRF2 activity metric and utilized it to conduct a pan-cancer analysis of oncogenic NRF2 signaling. We identified an immunoevasive phenotype where high NRF2 activity is associated with low interferon-gamma (IFNγ), HLA-I expression and T cell and macrophage infiltration in squamous malignancies of the lung, head and neck area, cervix and esophagus. Squamous NRF2 overactive tumors comprise a molecular phenotype with SOX2/TP63 amplification, TP53 mutation and CDKN2A loss. These immune cold NRF2 hyperactive diseases are associated with upregulation of immunomodulatory NAMPT, WNT5A, SPP1, SLC7A11, SLC2A1 and PD-L1. Based on our functional genomics analyses, these genes represent candidate NRF2 targets, suggesting direct modulation of the tumor immune milieu. Single-cell mRNA data shows that cancer cells of this subtype exhibit decreased expression of IFNγ responsive ligands, and increased expression of immunosuppressive ligands NAMPT, SPP1 and WNT5A that mediate signaling in intercellular crosstalk. In addition, we discovered that the negative relationship of NRF2 and immune cells are explained by stromal populations of lung squamous cell carcinoma, and this effect spans multiple squamous malignancies based on our molecular subtyping and deconvolution data.
Keywords: NRF2, KEAP1, Redox, Squamous, T cells, Interferon gamma, HLA-I, SOX2, TP63, PD-L1
Abbreviations
- LAML
Acute Myeloid Leukemia
- ACC
Adrenocortical carcinoma
- BLCA
Bladder Urothelial Carcinoma
- LGG
Brain Lower Grade Glioma
- BRCA
Breast invasive carcinoma
- CESC
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- CHOL
Cholangiocarcinoma
- LCML
Chronic Myelogenous Leukemia
- COAD
Colon adenocarcinoma
- ESCA
Esophageal carcinoma
- GBM
Glioblastoma multiforme
- HNSC
Head and Neck squamous cell carcinoma
- KICH
Kidney Chromophobe
- KIRC
Kidney renal clear cell carcinoma
- KIRP
Kidney renal papillary cell carcinoma
- LIHC
Liver hepatocellular carcinoma
- LUAD
Lung adenocarcinoma
- LUSC
Lung squamous cell carcinoma
- DLBC
Lymphoid Neoplasm Diffuse Large B-cell Lymphoma
- MESO
Mesothelioma
- OV
Ovarian serous cystadenocarcinoma
- PAAD
Pancreatic adenocarcinoma
- PCPG
Pheochromocytoma and Paraganglioma
- PRAD
Prostate adenocarcinoma
- READ
Rectum adenocarcinoma
- SARC
Sarcoma
- SKCM
Skin Cutaneous Melanoma
- STAD
Stomach adenocarcinoma
- TGCT
Testicular Germ Cell Tumors
- THYM
Thymoma
- THCA
Thyroid carcinoma
- UCS
Uterine Carcinosarcoma
- UCEC
Uterine Corpus Endometrial Carcinoma
- UVM
Uveal Melanoma
- TCGA
The Cancer Genome Atlas
- CCLE
Cancer Cell Line Encyclopedia
- IFNγ
Interferon gamma
- GTEx
Genotype Tissue Expression Portal
- SqCC
Squamous cell carcinoma
- ICB
Immune-checkpoint blockade
- TME
Tumor microenvironment
1. Introduction
Carcinogenesis – the gradual path from normal cellular behavior to malignant growth – requires a cell to acquire a set of distinct hallmark properties over time. These properties comprise functional changes in pre-malignant cells, as well as in various proximate stromal cells through intercellular crosstalk. From this standpoint, malignant tumors can be considered as complex organs harnessed to sustain neoplastic growth [1]. Due to the increase in proliferation and cellular metabolism, an inevitable consequence of malignant growth is increased oxidative stress, which renders pathways with antioxidant effects under positive selection [2]. The principal regulator of the cellular redox homeostasis is the transcription factor Nuclear factor erythroid 2-related factor 2 (NRF2, NFE2L2 gene), which is frequently hyperactive in malignant disease. Especially in non-small cell lung cancer, the oncogenic activation of NRF2 is highly frequent: somatic NRF2 activating mutations alone are among the most frequently occurring subtype specific aberrations [3]. The cytoplasmic inhibitor of NRF2, Kelch-like ECH Associated Protein 1 (KEAP1), is an E3 ubiquitin ligase substrate adaptor targeting NRF2 for proteasomal degradation under unstressed conditions. In oxidative or electrophile stress, the interaction is disrupted and de novo synthesized NRF2 is translocated to the nucleus to drive target gene expression. NRF2 target genes have antioxidant and detoxifying effects via various mechanisms including upregulation of glutathione S-transferases, as well as NAD(P)H quinone oxidoreductase, which has multiple roles in adaptive cellular responses to stress [4]. In cancer, the regulation of NRF2 is disturbed rendering NRF2 constitutively active. Mechanisms of NRF2 activation include somatic mutations and copy-number variation in NFE2L2 (gain-of-function or amplification) and KEAP1 (loss-of-function or deletion), as well as positive regulation by p62 [5,6]. Along with its antioxidant effects, NRF2 hyperactivity is known to promote cancer cell proliferation and survival via various other mechanisms, for instance by promoting anabolic metabolism and increasing chemoresistance via enhanced phase II enzyme and drug efflux transporter expression [7].
One hallmark property of cancer is its ability to evade destruction by the immune system [1]. The past decade has complemented oncogene-centric targeted therapies with treatments that modulate the antitumor immune response. However, current diagnostic approaches fail to detect the clinical responders for immune-checkpoint blockade (ICB) with reproducible precision [8]. From this viewpoint, the effects of known oncogenic events on the crosstalk between cancer cells and immune cells should be elucidated, as these events are often straightforward to identify, and if associated with altered immune responses, may serve as predictive biomarkers for ICB. It has been previously shown that NRF2 drives PD-L1 expression and leads to reduced leukocyte infiltration in mouse allograft models of melanoma and lung adenocarcinoma [9,10], but a comprehensive characterization of the effect of NRF2 overactivity on cancer immunity is currently lacking. The aim of this study was therefore to characterize NRF2 hyperactivity at a pan-cancer scale and interrogate unknown biological effects of oncogenic NRF2 activation, with a special emphasis on cancer immunity. To this end, we utilized The Cancer Genome Atlas (TCGA) and Cancer Cell Line Encyclopedia (CCLE), which are publicly available multi-omics databases of tumors and cell lines, respectively. Furthermore, the key findings of this work were validated with independent clinical cohorts.
2. Results
2.1. NRF2 activity score improves detection of NRF2 driven malignancies
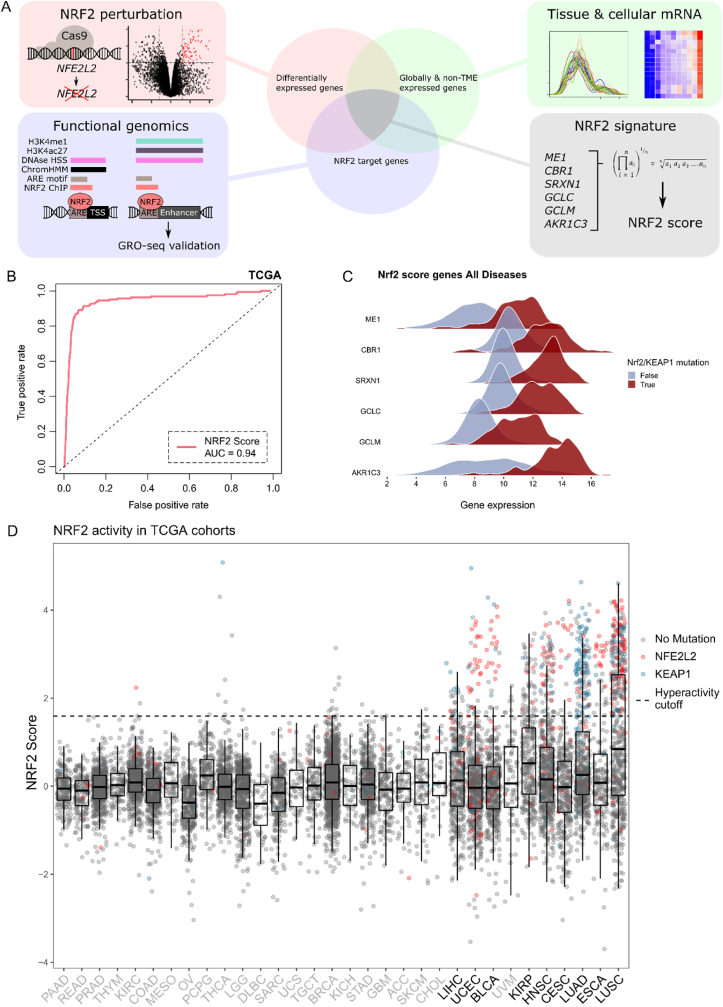
We developed a NRF2 activity scoring metric from experimentally confirmed robust NRF2 target genes (Fig. 1A). The utility of such a score is to overcome the limitations of using gold-standard mutations as a classifier, namely low sample size and the lack of statistical power, as well as ambivalent functionality of rare variants. Furthermore, alternative NRF2 activation mechanisms exist [5,6], highlighting the importance of using NRF2 target gene expression as a marker of activity instead of somatic variants. Our goal was to generate a metric that is: a) based on expression of evident target genes with robust responses to NRF2 activity; b) tissue-agnostic; and c) unbiased towards signal arising from the TME. The score was developed as follows: first, we used A549 lung adenocarcinoma cells harboring the KEAP1 inactivating mutation G333W rendering NRF2 overactive (A549-NRF2OE) and knocked out NRF2 with Cas9-sgNRF2 (comparison of A549 NRF2 overexpressed vs knockout, hereafter referred to as A549-NRF2OEvsKO) to detect NRF2 dependent differentially expressed genes (Figs. S1A and S1B, list of genes in Supplementary Table 1). Second, we used publicly available functional genomics data (See materials & methods, Functional genomics analysis of NRF2 target genes) to subset the genes that are directly regulated by NRF2 (See Supplementary Table 2 for a complete reference of candidate targets). Third, we utilized public tissue as well as stromal cell transcriptomic data from Genotype Tissue Expression portal (GTEx) and Database of Immune Cell Expression (DICE), respectively, to discard genes that are clearly tissue specific or prominently expressed in TME-populations (Fig. S1C and Fig. S1D). The final NRF2 signature comprised genes CBR1, SRXN1, GCLC, GCLM, AKR1C3 and ME1 and the final score was defined as a geometric mean of their linear TMM normalized mRNA-expression. The score values were scaled within disease entities (to density distribution peak) in TCGA-samples to decrease variance between cancer types.
Fig. 1.
NRF2 score development, performance and distribution across TCGA malignancies.
A: Schematic of the score development procedure. NRF2 perturbation, functional genomics and tissue-specific expression data were integrated together and the NRF2 score was defined as a geometric mean of the mRNA expression of overlapping genes.
B: Receiver operating characteristic (ROC) curve for the NRF2 score in classifying functionally relevant somatic variants defined in OncoKB.
C: Differential log2 CPM gene expression distribution between NFE2L2/KEAP1 mutated and non-mutated NRF2 score comprising genes in TCGA cohorts.
D: NRF2 score with NFE2L2/KEAP1 mutations shown as a boxplot across TCGA-cohorts sorted by variance. The cohorts chosen into subsequent analyses are colored in black. The hyperactivity threshold was chosen with values of TPR >0.85 and FPR <0.1.
We assessed the score performance with ROC-analysis against OncoKB [11] defined hotspots in NFE2L2 and KEAP1 and functional variants (KEAP1-truncating aberrations) in TCGA-data (Fig. 1B). The score exhibited excellent overall discrimination (AUC = 0.94) and genes showed markedly different distributions in mRNA-expression in TCGA KEAP1/NFE2L2 mutated vs wild type samples (Fig. 1C). Individual ROC analyses for cohorts with somatic mutations in NFE2L2/KEAP1 are shown in Fig. S1E. We computed the correlations of NRF2 score to NFE2L2 mRNA expression. Based on the correlation patterns, multiple cancer types display significant positive correlations (Fig. S1F). As canonical NRF2 regulation is based on protein turnover, upregulation of NFE2L2 mRNA and its correlation with target gene expression suggests additional contribution of an upstream regulator or regulators. Score variance correlated considerably with KEAP1/NFE2L2 mutation frequency (r = 0.8, P < 0.0001), thereby predicting the oncogenicity of NRF2 irrespective of activating mechanism (Fig. S1G). By this rationale, diseases associated with >5% mutation frequency or σ2 > 0.75 were defined to harbor significant oncogenic NRF2 activity. From these malignancies, we discarded TCGA cohorts with N < 100 to maintain high statistical power across the datasets. The score distribution and cohort selection are shown in Fig. 1D (colored mutations above the specified hyperactivity cutoff of TPR >0.85 are shown in Fig. S1H). NRF2 activity scores for all TCGA samples are listed in Supplementary Table 3). Notably, malignancies in the lung, uterus, bladder, kidney and those with dominating squamous histology had a significant proportion of high scoring samples.
2.2. Enriched pathways in NRF2 hyperactive cancers reveal differential immunomodulatory association across cancer types
For TCGA cohorts meeting the inclusion criteria, as well as for the A549-NRF2OEvsKO and CCLE transcriptome and proteome data, we conducted gene set enrichment analysis (GSEA) to assess the pathways enriched with NRF2 hyperactivity (Fig. 2A. See Supplementary Table 4 for all data). Interestingly, two directly oncogenic signaling pathways, MYC and WNT, showed prominent global positive enrichment. Other global pathways associated with NRF2 hyperactivity were mainly metabolic, drug efflux and redox-regulatory processes, whereas immune microenvironment related processes were negatively enriched in NSCLC and squamous diseases in contrast to other diseases, which exhibited positive enrichment. With the curated data, cohorts clustered into two populations based on the immune milieu associated gene sets. Notably, IFNγ response, HLA- and T cell signaling gene sets enriched to the negative end in lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), esophageal carcinoma (ESCA), cervical carcinoma (CESC) as well as head and neck cancer (HNSC), while the same pathways had positive enrichment scores in kidney renal papillary cell carcinoma (KIRP), uterine corpus endometrial carcinoma (UCEC), bladder carcinoma (BLCA) and liver hepatocellular carcinoma (LIHC) (Fig. 2A–B). As the immunological gene sets were not enriched in the pure cell populations (A549-NRF2OEvsKO or CCLE), they likely emerge from the crosstalk between cancer- and TME-cell populations. Of note, downregulation of cytokines, HLA-I, IL-12, IFNγ and TCR signaling are all characteristic to ‘immune cold’ tumors with documented poor responses to ICB therapies [12]. Upon further characterization of the pathways differing between the defined disease groups, we observed further differences in TME-related enrichment terms (Fig. 2C). As the pathways comprised largely lymphoid cell associated signatures, these data suggest that oncogenic NRF2 signaling is, in the context of NSCLC and squamous diseases, associated with less lymphocyte infiltration. The data also shows a dichotomous association of NRF2 to IFNγ response and lymphocyte associated pathway enrichment between NSCLC and squamous cancers vs kidney, uterine and liver cancer.
Fig. 2.
NRF2 activity associates with disease specific immunoevasive characteristics.
A: Pan-cancer GSEA analysis. GSEA normalized enrichment scores for pathways correlated to NRF2 activity shown as a Heatmap for TCGA, A549-NRF2OEvsKO, CCLE mRNA and CCLE protein. Significance of enrichment is shown as: <0.1, * <0.05, ** <0.01, *** <0.001, **** <0.0001. For TCGA, combined p-values (Stouffer's method) are shown. Redundant pathway terms were discarded, See Table S2 for full list of pathway terms. Pathways were curated to reflect cancer hallmarks that are shown on the right.
B: Correlation of ssGSEA Hallmarks IFNγ response and NRF2 score. Cohorts in blue, red and grey have negative, positive or insignificant correlation, respectively.
C: Top enrichment terms deviating between the defined cancer groups with differential enrichment scores in the IFNγ response pathway. Normalized enrichment scores are shown on the heatmap. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.3. Squamous diseases comprise a NRF2 hyperactive subtype with SOX2 amplification, CDKN2A/B and TP53 loss and upregulated immunosuppressive genes
Since we observed similar immune cold characteristics across the TCGA squamous diseases, we proceeded to study subtype-effects within the diseases using Uniform Manifold Approximation and Projection (UMAP) and community detection-based clustering. We identified a distinct pan-squamous subtype with hyperactive NRF2 (identified communities are shown in Fig. 3A, TCGA cohorts in Fig. 3B and NRF2 activity in Fig. 3C). Associated to this subtype, we observed co-occurring copy number variation (CNV) and a characteristic mutational landscape, most notably amplified SOX2/TP63 (q-arm of chromosome 3) and loss of CDKN2A/CDKN2B (9p21) (Fig. 3D and S2 A-D), as well as mutated TP53 and CDKN2A (Figs. S2E and F). Furthermore, we identified a group of cell lines with similar genomic profiles using the CCLE/DepMap dataset, confirming the genomic determinants of this subtype (Figs. S2G, H, I, J, K, L, and M). There were no prominent peaks in chromosome 6, suggesting that HLA-I loss events do not contribute to the phenotype. To confirm this, we defined HLA-I loss as at least one shallow deletion or LoF-mutation in major HLA-I genes and did not observe an association between the two variables with Fisher's test (OR = 0.40, P = 0.09). The association of tumor mutational burden and NRF2 overexpression has been reported before [13]. Thus, we assessed the prospect of immunological effects arising from differential neoantigen load by computing mutational burden (log2 total mutation count) across the cluster comprising cohorts with respect to NRF2 activity, and did not observe a uniform trend between mutation count and NRF2 (Fig. S2N). To follow up on amplified transcription factors SOX2 and TP63, we downloaded publicly available ChIP-seq data (GSE46837) in squamous cancer cell lines and generated a list of target genes (pipeline as in Fig. 1A, functional genomics) to interrogate direct targets of these transcription factors (see Supplementary Tables 5 and 6 for a complete reference of targets). To specifically assess differentially expressed, putative direct targets in immunomodulatory genes, we studied the overlap of the differentially expressed genes in the squamous cluster and the target gene list as well as curated genes from the CellPhoneDB framework [14] and TISIDB [15] (Fig. 3E). From these results, SLC7A11, NAMPT, SLC2A1 and MPP3 were identified to confer resistance to T cell mediated killing by high-throughput screening. Moreover, the listed genes had evidence for upstream activation by NRF2. SLC7A11, NAMPT and SCL2A1 are genes attributed to metabolism, involved in cysteine uptake, NAD + biosynthesis and glucose uptake, respectively. From the literature annotated genes, while most were ambivalent, SOX2 was shown to be a) upregulated in the case of effector cell resistance in co-culture; b) downregulate IFN type I response in vitro and c) decrease T cell infiltration in a HNSC murine model [16]. Interestingly, we observed increased NFE2L2 mRNA in SOX2 amplified TCGA squamous cell carcinoma (SqCC) cases, suggesting the presence of an upstream regulator in Chr3 q2 locus (Fig. S2O). Finally, in the squamous NRF2 cluster, we identified prominent downregulation of the IFNγ response analogous to the initial GSEA analysis (Fig. 3F). Taken together, these data support the notion that downregulation of IFNγ-response and/or HLA-I genes is downstream of NRF2 or other co-expressed transcription factors (SOX2 or TP63). Based on the functional role of the identified immunomodulatory genes (Fig. 3E), modification of the tumor metabolic landscape may contribute to the immune escape of NRF2 hyperactive cancer.
Fig. 3.
NRF2 hyperactivity is associated with a distinct squamous subtype with SOX2/TP63 amplification, CDKN2A/CDKN2B loss and downregulated IFNγ-response in TCGA.
A: UMAP representation and Louvain clustering of included TCGA cohorts.
B: TCGA diseases in Louvain clusters.
C: NRF2 score in Louvain clusters.
D: Gistic2 copynumber profile of the NRF2 hyperactive cluster*. The G-score in gistic2 is CNV amplitude multiplied by CNV frequency, which measures the significance of a CNV event in a dataset.
E: A heatmap of differentially expressed immunomodulatory genes between the NRF2 hyperactive cluster and other SqCC putatively regulated by NRF2, SOX2 or TP63. From the TISIDB-database, genes colored with green indicate a literature cited effect, whereas genes colored in magenta depict a hit from CRISPR/Cas9 functional screens. Panel on the right depicts hits from a functional genomics target gene analysis on NRF2, SOX2 and TP63, mint green hits designate a direct target based on the analysis (See methods for details).
F: Volcano plot of enriched pathways in the NRF2 hyperactive cluster*.
*All analyses were conducted against squamous samples; LUSC, HNSC, ESCA and CESC. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.4. Interferon-gamma response pathway is downregulated in NRF2 hyperactive cancer cells in situ
To follow up on the identified targets, we proceeded to further explore signaling between NRF2 hyperactive cancer cells and immune cells in higher resolution in an in situ setting in a relevant cancer type. Thereby, we assigned NRF2 activity score to cells in a publicly available HNSC single-cell-RNAseq dataset (GSE103322). UMAP projection of single cells revealed distinct clustering of cancer cells with high NRF2 score across patient samples, suggesting that NRF2 activation could be linked to global shifts in cell phenotype (Fig. 4A and B). Similar to bulk tumors, the cluster exhibited high expression of SOX2 (Fig. 4C). In addition, TP63 was also overexpressed in the NRF2 hyperactive cancer cell cluster, although its expression was also present in other clusters (Fig. S3A). In further agreement with the bulk-tumor analysis, inflammatory response and IFNγ-signaling were the most prominent negatively enriched pathways in the NRF2 cluster relative to other cancer cell clusters (Fig. 4D). The single cell analysis distinguished that the response to interferon is downregulated in malignant cells with NRF2 hyperactivity (Fig. S3B). These data suggest that the negative correlation between IFNγ response and NRF2 in squamous cancer types originates from the response in cancer cells, either due to low interferon ligand or by intrinsic properties of NRF2 hyperactive cancer cells.
Fig. 4.
NRF2 hyperactive cancer cells have a dampened IFNγ response in situ and exhibit unique immunomodulatory intracellular interactions.
A: UMAP representation for cell populations.
B: UMAP representation for NRF2 score.
C: UMAP representation for SOX2 mRNA.
D: Volcano plot of GSEA enriched pathways in NRF2 hyperactive vs other cancer cells.
E: Curated interactions from the CellPhoneDB analysis. CellPhoneDB statistical significance is denoted as: *P < 0.05; *P < 0.001. Heatmap colors show the ratio of CellPhoneDB interaction mean for the clusters NRF2 hyperactive cancer cells vs other cancer cells. Panel on the left shows CellPhoneDB annotation for secreted (orange) and non-secreted (grey) factors. Panel on the right portrays hits from the functional genomics target gene analysis on NRF2, SOX2 and TP63 (light green), and shows genes included in the IFNγ signaling pathway (yellow).
F: UMAP representation for SPP1 mRNA.
G: Kaplan-Meier survival for SPP1 high vs low in pan–HNSC–LUAD-LUSC. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.5. NRF2 hyperactive cancer cells are associated with less TME interactions via HLA-I and increased interactions via NAMPT, SPP1 and WNT5A
We used the statistical framework of CellPhoneDB to interrogate putative intercellular ligand-receptor interactions between cancer- and TME cells in the whole HNSC single-cell dataset. With integration of our NRF2 target catalogue and a priori IFNγ gene sets from MSigDB (Hallmarks and Reactome), we identified multiple differential interactions between the cancer clusters against TME clusters with either direct NRF2 targets or genes involved in IFNγ mediated signaling. The most prominent hits were downregulated HLA type I interactions with cytotoxic T cells, and upregulated NAMPT, SPP1, WNT5A (Fig. 4E and F). These hits were also differentially expressed genes in the earlier pan-SqCC bulk tumor analysis and in CCLE cell lines mRNA, while SPP1 and WNT5A proteins were also upregulated (Figure S3C, D and E). Furthermore, NAMPT and SPP1 were differentially expressed in A549OEvsKO and all of the hits were in our NRF2 target catalogue (Fig. S3F). Moreover, in line with earlier observations, PD-L1 - PD1 interaction between NRF2 hyperactive cancer cells and T cells was statistically significant, further corroborating the role of PD-L1 in NRF2 driven immune-escape and suggesting that its effect extends the previously studied melanoma [10] (Fig. S3G). From the direct targets, NAMPT is the rate-limiting enzyme in the biosynthesis of NAD+. While its interaction with P2RY6 in CellPhoneDB was inferred with protein pulldown and lacks functional data, NAMPT knockdown was recently shown to increase CD8+ T cell infiltration in murine tumors via attenuating inducible PD-L1 expression [17]. The second hit, Osteopontin (SPP1), has been shown to inhibit T cell proliferation and IFNγ production in vitro [18]. The third hit, WNT5A with frizzled receptors, also has implications in tumor immunity: WNT5a signaling in cancer cells has been attributed to immunosuppressive metabolite induction via dendritic cells through FZD [19]. To assess the clinical relevance of these findings, we downloaded a publicly available targeted mRNA-expression dataset of PD-1 inhibitor treated HNSC and NSCLC patients and established SPP1 mRNA as a negative predictive biomarker to treatment-response (Odds ratio = 0.20 for response with high SPP1 expression; P < 0.05, Fisher's test) and therapy associated progression-free survival (mPFS 2.8 vs mPFS 12.33 months, P < 0.01) in pan HNSC-LUSC-LUAD (n = 5; n = 22; n = 13, respectively) (Fig. 4G). The inferior survival may either be directly caused by effects of SPP1 or arise from NRF2 activation where SPP1 mRNA acts as a surrogate marker. This clinical observation might prove relevant in classifying non-responders to ICB, as the current biomarkers suffer from inadequate reproducibility. In summary, these data demonstrate that NRF2 hyperactive cancer cells overexpress unique TME interacting ligands NAMPT, SPP1 and WNT5A, which have been shown to cause immunoevasion in cancer.
2.6. NRF2 hyperactivity associates with reduced stromal lymphocyte and macrophage infiltration and increased inducible cancer cell PD-L1 expression in squamous cancers
Finally, to explore the relationship between NRF2 activity and the immune milieu in greater detail, we performed deconvolution for the TCGA gene expression data with CIBERSORT [20] to infer immune cell content and correlated the cell fractions to the NRF2 score. In the NRF2 hyperactive IFNγ negative malignancies (Fig. 5A, cohorts color coded as cadet blue), many lymphocyte populations and different macrophage polarization states correlated negatively with NRF2 activity, while the correlations of cell subsets varied between cancer types. In the NRF2 hyperactive IFNγ positive cancers (cohorts color coded as coral), we observed mostly positive correlations to different immune cells with variability in the correlating cell types between diseases: in KIRP, there was a strong association to macrophages and in UCEC to other antigen presenting cells (Fig. 5A). Different malignancies have variability in their immune cell contents [21]. Therefore, we compared the overall distribution of each immune cell type using Mann-Whitney U tests between the IFNγ response curated NRF2 hyperactive cancers and observed that the NRF2 hyperactive IFNγ negative malignancies often had a higher disease-intrinsic proportion of immune cells, including macrophages, lymphocytes and dendritic cells (Fig. 5A, side panel). To evaluate the relationship of NRF2 and total T lymphocyte content, we calculated the sum of T cell fractions and observed fewer bulk lymphocytes in all of the IFNγ-negative NRF2 hyperactive cancers (Fig. 5B). We proceeded to validate the result in LUSC with a different approach utilizing a publicly available TIL dataset generated from TCGA H&E images with deep learning [22] and observed a decrease in the median percentage of tumor-infiltrating lymphocytes (TILs) in the NRF2 hyperactive cases (P < 0.05, Fig. 5C). To corroborate this, we stained an independent tissue-microarray dataset of NSCLC (obtained from Auria Biobank, 117 and 211 cases of LUSC and LUAD, respectively) with NQO1 (a marker of NRF2 activity) and the universal T cell marker CD3, and observed a similar result in LUSC, that is, NQO1 positive tumors (above the upper quartile of the mean intensity distribution) harbored less CD3+ cells (P < 0.05, Fig. 5D and E). In our TMA data, the cytotoxic T cell marker CD8 showed a similar trend but did not reach statistical significance (P = 0.09, Fig. S4A), and in LUAD, the relationship was not observed (Data not shown). In addition, the contribution of NFE2L2 and KEAP1 mutations was assessed separately in TCGA LUSC cohort, and a lower median fraction of total lymphocyte infiltration was present in all groups, irrespective of mutated gene (Fig. S4B). As the earlier methods did not differentiate between stromal and epithelia infiltrating populations (defined as cells within the malign epithelial component), we next sought to assess the relationship between NRF2 activity and intraepithelial immune cells with multiplexed immunohistochemistry (mIHC, Fig. 5F) in a separate NSCLC-cohort of resected cases (obtained from Central Finland Biobank, final sample sizes after quality control filtering, 67 and 53 for LUSC and LUAD, respectively), using AKR1B10 as a marker for NRF2 activation (cutoffs for groups are presented in Fig. S4C). Surprisingly, we did not observe differences in the intraepithelial populations in either LUSC or LUAD, except AKR1B10 high LUSC harbored fewer intraepithelial granulocytes (Supplementary Figs. S4D–E). However, in LUSC, most of the stromal population densities from the total core area, that is CD8+ and CD4+ positive T cells as well as M2 macrophages and granulocytes were decreased (Fig. 5G and H). Expectedly, also tumor purity, defined as the relative tumor area to total area had a positive relationship to NRF2 activity (Fig. 5I). We validated this result with TCGA using ABSOLUTE quantifications (Fig. S4F). Similar effects on tumor purity were observed in the rest of the NRF2 immune cold TCGA cancer types, HNSC, CESC, ESCA and LUAD. In LUAD mIHC, we did not observe differences in lymphocytes, but there were fewer unspecified macrophages in the AKR1B10-high group. In addition, median tumor purity was greater in LUAD AKR1B10-high but not at a statistically significant level (Supplementary Figs. 5G–E). Interestingly, in the LUSC stromal macrophages, the ratio of PD-L1+ cells correlated significantly with mean cancer cell AKR1B10 expression (Fig. 5J and K). Given the observed effect on stromal macrophages and the earlier observations of PD-L1 as a downstream target of NRF2 [9,10], we asked whether NRF2 activity would contribute to inducible PD-L1 expression in LUSC. In cancer cells, intraepithelial CD8+ T cell infiltration (E-CD8+) had a positive correlation to PD-L1 expression, which is an expected effect from proximal proinflammatory signaling (Supplementary Fig. 5I). Given this baseline effect, we grouped the samples to E-CD8+ infiltrated and deserted cases (that is, high density of CD8+ cells adjacent to cancer cells, cutoff in red in Supplementary Fig. 5I) and observed that in infiltrated samples, mean of cancer cell AKR1B10 expression correlated positively with the fraction of PD-L1+ cancer cells (R = 0.69, P < 0.001, Fig. 5L-M). The correlation between cancer cell AKR1B10 and PD-L1 expression was not present with low E-CD8+ suggesting that proinflammatory stimulation is needed to induce NRF2 driven PD-L1 (Fig. S4J). Of note, only one of these E-CD8+ infiltrated samples exceeded our cutoff to be defined as NRF2 hyperactive (intensity of 180 A U.) and the fraction of E-CD8+ infiltrated samples was less than 25% (Fig. S4K). In order to corroborate the positive association between PD-L1 and NRF2, we computed purity corrected CD274 mRNA between the NRF2 basal and hyperactive groups of TCGA LUSC and observed increased median expression in the NRF2 hyperactive group (P < 0.0001, Fig. S4L). We did not discover similar associations between NRF2 and PD-L1 in LUAD in either our mIHC-data or TCGA (data not shown). The effects of NRF2 signaling to the immune milieu in LUAD remain largely elusive, as our data did not indicate similar differences between the groups. However, the PD-L1 association was present in other TCGA squamous cancers (Fig. S4M). Taken together, these data suggest that overactive NRF2 associates with less overall stromal cell infiltration in pan-squamous cell carcinoma, mainly lower lymphocyte and macrophage density as well as a decline in stromal cell occupied area of the tumor. The TMA data and the tumor purity associations suggest that the differences observed in TCGA deconvolution data may arise specifically from the stroma occupied populations, which is expected as large proportion of the immune cells reside in the stroma instead of epithelium. Moreover, these results indicate that overall PD-L1 expression is associated with increased cancer cell NRF2 activity, both in cancer cells and stromal macrophages, suggesting a global PD-L1 immunosuppressive milieu across the stromal and tumoral landscapes of NRF2 hyperactive LUSC. The CD274 mRNA expression distributions imply that this effect extends to other squamous malignancies (Fig. S4M).
Fig. 5.
NRF2 hyperactive tumors display negative correlations to overall lymphocytes in pan-squamous diseases.
A: Correlation matrix of NRF2 score vs CIBERSORT cell types. Right annotation panel depicts the distributions of respective immune cells in the IFNγ pathway classified diseases. Statistical significance is shown as: *P < 0.05, **P < 0.01, ***P < 0.001. Negative correlations in lymphocytes and macrophages are largely present in squamous cancers and lung adenocarcinoma.
B: Violin plots for CIBERSORT bulk T lymphocyte fractions. Less bulk T lymphocytes were observed in silico in squamous cancers and lung adenocarcinoma, while in KIRP, UCEC, BLCA and LIHC there was no association.
C: Percentage of tumor-infiltrating lymphocytes (TIL) in digitized TCGA LUSC H&E images.
D: Tissue microarray IHC staining of NQO1 (NRF2 activity marker) and CD3+ cells.
E: Representative images of the NQO1-stained groups.
F: Schematic of the mIHC workflow.
G: AKR1B10 vs stromal immune cell correlations in mIHC LUSC TMAs.
H: Representative pseudofluorescence images of mIHC LUSC tumor cores. Cancer AKR1B10, macrophages, T cells and granulocytes are shown in magenta, red, green and cyan, respectively.
I: Tumor purity by AKR1B10 groups in LUSC tumor cores.
J: Scatter plot of mean cancer AKR1B10 expression and macrophage PD-L1.
K: Representative image of macrophage PD-L1 expression from AKR1B10-high samples.
L: Scatter plot of mean cancer AKR1B10 expression and the fraction of PD-L1+ cancer cells.
M: Representative image of PD-L1 expression from a high AKR1B10 expressed CD8+ T-cell infiltrated sample. Upper image comprises PD-L1+ and CD8+, and lower shows AKR1B10 pseudofluorescence for the matching area. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3. Discussion
Cancer immunotherapies, especially ICB, has become a mainstay in cancer care. Despite its undeniable success, biomarkers to stratify responders and non-responders are currently elusive. Recently, the relationship between oncogenic pathways and the TME have been explored. For instance, the effect of MYC and WNT signaling, as well as the loss of TP53, LKB1 and PTEN on tumor immune responses have been previously characterized [23]. In this work, we identified a phenotype of high NRF2 activity accompanied with low lymphocyte content, HLA-I and IFNγ, spanning multiple cancer types. Since the negative relationship between immune infiltration and immunotherapy responses has been clearly established before, it is reasonable to assume that NRF2 hyperactive cases of the characterized malignancies would associate with weaker responses, and that alternative strategies to activate the antitumor immune response should be studied. Indeed, during the course of this study, an association between NRF2 pathway mutations and inferior ICB-response was shown in NSCLC [24]. The present study, to our understanding, is the first attempt to provide pan-cancer wide perspective to NRF2 signaling and tumor immunity, with important implications for future studies: first, immune checkpoint inhibitor treatment efficacy should be assessed prospectively across all squamous malignancies with NRF2 hyperactivity and eligibility for ICB; and second, if proven inferior, alternative first-line or effective adjuvant treatment options should be explored.
Characterizing cancers by mutational profiling of large datasets has been invaluable in gaining insight into the biological processes enabling and governing malignant growth. However, profiling only by mutational status has limitations: The functional consequences of individual mutations may be ambivalent, other mechanisms for perturbation of the same pathway are overlooked, or the frequency of events is too low for robust statistical inference. Relevant to NRF2 mediated oncogenic signaling, mutation-independent means of activation are common, necessitating the use of gene expression classifier to deduct NRF2 activation [25]. Herein, we introduce a robust NRF2 scoring metric comprising genes that have minimal tissue specificity and sound evidence for direct, prominent activation. The score performs well in all of the tested TCGA cohorts (Fig. S1E) and it can be applied to bulk tissue and single cell data. Moreover, the metric was constructed to support ease of use by utilizing a low number of genes and prioritizing global, robust targets. Finally, we demonstrated the utility of the score by predicting a distinct immunological phenotype in NRF2 hyperactive cases in an external dataset using IHC staining as an alternative method for NRF2 activity assessment.
Our study provided several leads towards elucidating the mechanisms by which NRF2 overactivity affects the host immune response. We ruled out the contribution of HLA-I loss and neoantigen effects. We identified a distinct squamous cell subtype that was characterized by NRF2 overactivation together with SOX2/TP63 amplification and immune cold TME. SOX2 is a key driver of malignant transformation and stemness in squamous type cancers [26]. Our analysis showed the metabolic genes SLC2A1, SLC7A11 and NAMPT that associate with functional evidence in driving resistance to T cell mediated killing as downstream candidates of NRF2. Previous work shows that the expression of SLC2A1 - a glucose transporter - is enhanced by p63 and SOX2 [27]. Our work implicates functional synergy of NRF2 and p63/SOX2 in SLC2A1 driven glucose uptake. Increased glucose uptake via SLC2A1 has been shown to result in weaker anti tumoural immune responses in a murine model [28], supporting the functional role of this gene identified by the TISIDB high-throughput CRISPR-screen. SLC7A11, a well characterized NRF2 target gene [29], is a glutamate-cystine antiporter that increases intracellular cystine while exporting glutamate. IFNγ has been reported to downregulate the glutamate-cystine antiporter members SLC7A11 and SLC3A2, lowering intracellular cystine resulting in increased T cell induced ferroptosis [30]. Thus, the higher expression of SLC7A11 in NRF2 overactive cancers may render the cells resistant to immune destruction by this mechanism. NAMPT driven NAD + metabolism is involved in tumor immunity: increased NAD + by NAMPT enables inducible PD-L1 expression by epigenetic regulation of Irf1, restricting the antitumor action of cytotoxic T cells [17]. To conclude, the results suggest metabolic crosstalk between NRF2 overexpressing and TME cells through multiple mechanisms.
SPP1 and WNT5A were hits in both the bulk tumor and single cell analysis. Both genes have implications in tumor immunity. In line with the immunoevasive phenotype, soluble SPP1 has been shown to directly inhibit T cell proliferation, decrease T cell activation and IFNγ production in vitro [18]. The observed interaction of WNT5A and frizzled receptors in the single-cell data has implications in dendritic cell (DC) function: paracrine WNT5A signaling was shown to induce IDO1 in DCs, which increased extracellular kynurenine, supporting an immunosuppressive TME [19].
Our mIHC TMA data indicated that in LUSC, the stromal populations explain the negative relationship between NRF2-signaling and multiple immune cell types. The purity associations in other squamous cohorts suggest that this effect extends to HNSC, ESCA and possibly squamous CESC. Interestingly, macrophage as well as inducible cancer cell PD-L1 correlated positively with cancer NRF2 activity. PD-L1 expression has been shown to decrease tumor infiltrating lymphocytes over time and may thus partly explain the observed phenotype [31]. Interestingly, the ICB-responses in NRF2 hyperactive NSCLC have been inferior despite of our observation of correlating inducible PD-L1 expression in LUSC, while PD-L1 serves as the gold-standard predictive marker for immunotherapy in NSCLC. Based on our findings, these seemingly conflicting observations may have multiple explanations. First, WNT signaling has been indicated to drive immunosuppression in the TME, and reversal of the WNT5A driven immunotolerant microenvironment augmented PD-1 blockade response in a murine melanoma model [19]. As we found soluble WNT5A and WNT signaling gene sets upregulated in NRF2 hyperactive SqCC, this confounding factor in therapy response might extend to squamous cancers. Of note, WNT pathway activation has been shown to upregulate PD-L1 [32], and thus paracrine WNT5A could additionally explain the upregulated PD-L1 in stromal macrophages. Secondly, the other immunosuppressive ligands identified in this study could serve as confounding factors, as, for instance, high SPP1 expressing cases had worse response rates in ICB-treatment. Finally, as the intraepithelial CD8+ infiltrated cases comprised a small subpopulation of the whole cohort and having intraepithelial CD8+ infiltration and oncogenic NRF2 hyperactivity is exceedingly rare, they may constitute a unique subcategory responsive to PD-L1 blockade, as they do harbor both CD8+ cells and PD-L1+ cancer cells [33]. To this end, currently the contributions of stromal vs intraepithelial CD8+ cells to clinical response metrics are unclear, as the spatial information is often overlooked and cannot be accounted for with the current deconvolution methods or conventional single-cell RNAseq. Due to today's predictive markers for ICB being clearly elusive and to the diversity of mechanisms in immune escape, a robust predictive metric will likely require multiple accounted factors, analogous to the immunoscore in prognostic evaluation [34]. Given the similarities between NRF2 signaling and the molecular and TME phenotypes across squamous malignancies, these questions should span all cancer types with squamous histology.
In summary, our results provide an integrated NRF2-centric resource in cancer biology, and establish a connection between immune cold tumors and NRF2 signaling across squamous carcinomas from a pan-cancer wide perspective. Finally, these data highlight multiple avenues to pursue in future studies aiming to mechanistically characterize NRF2 signaling and its effect to the tumor immune milieu, including direct regulation via immunosuppressive ligands or immunosuppressive metabolites.
4. Materials and methods
4.1. Processing of genome-wide multilevel data
4.1.1. TCGA
Processed multiomics data, sample level analysis results and clinical data, were retrieved from https://gdc.cancer.gov/about-data/publications/panimmune (Thorsson et al., 2018) and https://gdc.cancer.gov/node/905/(PanCanAtlas) for 33 cancer types available in TCGA cohort.
4.1.2. CCLE
Preprocessed CCLE data was downloaded from https://portals.broadinstitute.org/ccle/data and https://depmap.org/portal/.
4.2. Functional genomics analysis of NRF2 target genes
4.2.1. Enhancer and promoter catalogue generation
A549 and ENCODE genomic data for other cell types were used as reference to generate broad catalogue of candidate regulatory elements, that could be linked with NRF2 regulation. DNAse hotspots and ChromHMM classified genomic sites were downloaded from ENCODE for A549. In addition, DNAse clustered sites version 3 were downloaded from ENCODE for all cell types. Promoters were defined primarily based on A549 ChromHMM annotation, but if the annotation was missing from the data, promoters were defined as 500 bp upstream and 1500 bp downstream from each transcription start sites. Next, A549 DNAse hotspot sites were integrated with ENCODE DNAse clusters for various cell types, with score above 400 and cluster identified in at least 20 cell types. Promoters were intersected with DNAse hotspots to identify regulatory sites at promoters.
4.2.2. GRO-seq regression analysis of enhancer elements
eRNA can be used to detect active enhancers and it correlates with the gene expression of the target gene [35]. To infer enhancer-gene pairs, we utilized publicly available GRO-seq data curated from Gene Expression Omnibus (GEO, GSE51225, GSE51633, GSE53964, GSE60454, GSE62046, GSE62296, GSE52642, GSE66448, GSE84432, GSE67519, GSE67540, GSE101803, GSE96859, GSE102819, GSE86165, GSE91011, GSE67295, GSE154427, GSE136813, GSE118530, GSE94872, GSE92375 and GSE117086). The final dataset comprised 336 samples representing 45 cell types. Homer analyzeRepeats.pl software was used for GRO-seq quantification of nascent RNA gene expression using gene introns as coordinates, with parameters: strand + -noadj -noCondensing -pc 3. For enhancers, in case of intragenic enhancers, quantification was performed from the opposite strand and for intergenic enhancers from both strands, using the same parameters as before. Enhancers' start and end coordinates were expanded by 500bp. Gene end coordinates were expanded by 5000bp for annotating intragenic gene enhancers and account for transcription at the end of transcripts. A linear model was fitted for each gene-enhancer pair in the same topologically associating domain that had Spearman's Rho >0 and P-value <0.05. R2 value was used to estimate, how much of the gene expression could be explained by the eRNA expression. The enhancer-gene pairs were defined as features with regression P-value <0.05 and R2 > 0.1, and the maximum distance was set to 500kbp.
4.2.3. NRF2 target gene identification
To infer NRF2 bound regions at the enhancer sites, the preprocessed ENCODE ChIP-seq data for NFE2L2 was downloaded from GEO (Datasets GSE91997, GSE91894, GSE91809, GSE91565). The ARE-motif from JASPAR database (NFE2L2, MA0150.1) was queried across the ChIP-peaks with Homer annotatePeaks, and NRF2 motif score cutoff for a binding site was set to 6. From these data, the minimum amount of ChIP-seq datasets with the given peak was set to two. Using this analysis framework 3000 bidirectional and 2000 intragenic enhancers were identified for NRF2.
4.3. Development of the NRF2 score
A computational NRF2 score was developed by utilizing multiple datasets. The final gene set was derived by applying the following rationale: a) ChIP-seq peaks must be present at ChromHMM-promoters across the ENCODE NFE2L2 ChIP-seq data b) the gene must be differentially expressed in A549-NRF2OEvsKO (log2 fold-change >1.5, Bonferroni adjusted p < 1e-5); and c) mRNA-expression must be observed across tissues in GETxportal data (https://www.gtexportal.org/home/datasets) and expression must be uniform across cell types in DICE-database (https://dice-database.org/), to counter variance arising from tissue specific expression and the immune-milieu, respectively.
Differential expression analysis for A549-NRF2OEVSKO was conducted with limma. Direct NRF2 targets were defined as having a NRF2 binding in the NRF2 ChIP-seq data in the ChromHMM promoters: the peaks were narrowed down to regions that intersect all four datasets and include an ARE-sequence (motif score >6).
Genes with tissue-specific expression patterns were excluded based on the expression profiles in the GETxportal data: median tissue expression cutoff was set to 1.5 log2 TPM for binary filtering (Fig. S1A). Genes exhibiting high values or observable variance in median expression in immune-cells were excluded heuristically (Fig. S1B).
Genes passing all these steps were defined as the NRF2 signature. The samples were scored with a geometric mean from the genes’ linear mRNA-expression. For TCGA samples, the score was normalized within each disease to the peak of the score distribution (mode). Performance of the scores was evaluated with receiver operating characteristic against functional NRF2 activating somatic variants (defined in OncoKB) using the ROCR-package for R.
4.4. Gene set enrichment analysis
GSEA (Java-GSEA) was performed for the selected TCGA-cohorts and CCLE cell lines of the matching diseases using NRF2 score as a continuous feature. Pan-cancer analysis for TCGA cancer types was conducted by calculating Stouffer's statistic for each gene set across the selected cohorts. For the A549 cells, GSEA was conducted with a binary feature of NRF2−/− vs NRF2+/+. Multiple-comparison corrections were performed with the Benjamini-Hochberg (BH) –method.
4.5. UMAP analysis and Louvain clustering of TCGA and CCLE data
UMAP dimensionality reduction analysis was performed for 750 most variable genes using uwot R package, with parameters set to n. neighbors = 12 and min. dist = 0.4. Community detection based clustering for the same 750 most variable genes was performed using the Louvain algorithm, with k = 4 implemented in igraph R package. Similarly, for CCLE 750 most variable genes were used with n. neighbors = 6 and min. dist = 0.3 and k = 4. Various number of most variable genes were used for UMAP to confirm robustness of sample group detection.
4.6. Functional genomics analysis of SOX2 and TP63 target genes
Target genes for SOX2 and TP63 were inferred from ChIP-seq data of the respective transcription factors (GEO dataset GSE46837). All parameters were set as in the analysis for NRF2 target genes (NRF2 target gene identification) using the enhancer and promoter catalogues defined earlier.
4.7. Differential expression analysis for the NRF2 squamous cluster
Fold-differences across the transcriptome were computed between the NRF2 squamous cluster vs other squamous samples, and the statistical significance of differentials of gene-expression distributions were assessed with two-sided Wilcoxon tests, and subsequently multiple-comparison corrected with the BH-method. The result was filtered with FDR <0.001 and a logFC threshold of 1 for absolute values. The overlap of the resulting genes was assessed with a) CCLE gene-expression and protein correlation to NRF2-score; b) genes in CellphoneDB and TISIDB (literature annotation and CRISPR-screen resistance genes); and c) the candidate target gene catalogues of NRF2, TP63 and SOX2. High-confidence hits (shown in Fig. 3E) were defined as genes that correlate in CCLE protein or mRNA (FDR <0.1, R > 0.25), are listed in CellPhoneDB or TISIDB and are listed in the target gene catalogues for at least one of the transcription factors.
4.8. In vitro experiments
4.8.1. Cell culture
Cells were incubated in 37 °C and 5% CO2 throughout the experiments and the passage number was kept under 10 over the course of this study. Cell passaging was conducted before reaching confluency.
4.8.2. CRISPR/Cas9 NRF2 knockout
A549 cells were transfected with a 20bp single-guide-RNA (CAAGCTGGTTGAGACTACCA) targeting exon 5 of NFE2L2 (ENST00000446151) and scramble-sequence including plasmid vectors (SpCas9(BB)-2 A-GFP (Addgene, PX458). NRF2 knockout and control cells were generated via CRISPR-Cas9-mediated non-homologous end-joining (NHEJ). Transfection-positive cells were sorted with fluorescence-activated cell sorting (FACS) to obtain clonal populations. Validation of the clones was conducted with NRF2 western blot (Fig. S1A), Sanger sequencing (Data not shown) and RT-qPCR (Fig. S1B). Sequencing confirmed a truncating homozygous frameshift mutation (c.562delA, p. T188fs*) in exon 5 of NFE2L2 (ENST00000446151).
4.8.3. Sanger sequencing and western blot
DNA was extracted with Genejet Genomic DNA purification kit (Thermo Scientific, catalog. no: K0702) and using the GENEWIZ Sanger Sequencing Services.
Cells were lysed and protein concentration measured using BCA kit (Pierce). 30 μg of total protein with 1X SDS-PAGE sample buffer (Biorad) was loaded in 4–20% mini-Protean TGX gels (Biorad) and the gel electrophoresis was done using Tris glycine running buffer containing SDS. Proteins were then transferred onto nitrocellulose membrane (0.2 μm, Biorad) using Owl Hep-1 semi-dry transfer system (Thermo scientific) following manufacturer's instructions. The blots were blocked with 5% milk-TBST solution for 1 h at room temperature (RT). Blots were stained overnight at 4 °C with NRF2 (1:5000 dilution, catalog no.16396-1-AP, Proteintech) and beta-actin (1:5000 dilution, catalog no. sc-47778, Santa Cruz, USA) antibodies in 5% milk-TBST solution. And secondary staining was done for 1 h at RT using Alexafluor 488- or 680- labelled anti-rabbit and anti-mouse antibodies (Invitrogen) in 2% milk-TBST solution. The blots were then visualized using Biorad documentation system.
4.9. Copy-number analysis
Copy-number analysis was conducted for firehose-derived TCGA segment-files for HNSC, LUSC, ESCA, and CESC, stratified by the NRF2 subtype (cluster 8) with Gistic2 using the following parameters: ta 0.1 -Peakpeel 1 -brlen 0.7 -cap 1.5 -conf 0.99 -td 0.1 -genegistic 1 -gcm extreme -js 4 -maxseg 2000 -qvt 0.25 -rx 0 -savegene 1 -broad 1.
4.10. Processing of single cell data
The HNSC single cell dataset was downloaded from GEO (GSE103322) and processed with Seurat R-package v3. Cells with more than 8000 detected genes were filtered out. Seurat SCTransform with 3000 variable features was used for data normalization. 25 principal components and default parameters were used for UMAP projection and Louvain clustering. SingleR 1.0.1 [36] was used for the automated cell type annotation. These annotations were manually refined according to cell types identified in Puram et al., 2017 [37].
4.11. CellPhoneDB analysis
The HNSC single-cell dataset was analyzed with CellPhoneDB statistical analysis method using 1000 iterations and an expression threshold of 0.1. To assess the differential interactions, the data was visualized as a ratio of interactions means in the NRF2 cluster vs other cancer clusters, only including interactions that were exclusively statistically significant in either the NRF2 cluster (positive ratio) or the other clusters (negative ratio). Statistically non-significant ratios were removed from the heatmap for clarity.
4.12. Survival analysis
A pan–HNSC–NSCLC-SKCM dataset was downloaded from GEO (GSE110390) and the associated clinical data was obtained from TIDE-database (http://tide.dfci.harvard.edu/). SKCM samples were excluded from the analysis, and the nCounter PanCancer Immune Profiling Panel targets were normalized to 40 housekeeping genes with NanoStringNorm R-package. Mean normalized log2 mRNA expression was used as a cutoff to dissect SPP1 high and low expressing samples. Survival analysis was performed with survival R-package. The Kaplan-Meier curve was plotted with survminer R-package.
4.13. CIBERSORT analysis for TCGA data
CIBERSORT was run for the TCGA data with the CIBERSORT R-package separately for each cancer type with the absolute method (sig.score), using 100 permutations and without quantile normalization. Correlations to NRF2 score were assessed with Pearson's method. The bulk lymphocyte analysis was conducted by taking the sum of all T lymphocyte fractions, and using a cutoff for NRF2 hyperactivity as TPR >0.85 from the ROC-analysis. The statistics were computed with Mann-Whitney U.
4.14. Conventional immunohistochemistry
NSCLC tissue microarray sections (LUAD n = 211, LUSC n = 117) were obtained from Auria Biobank, Turku University Hospital, Turku, Finland. NQO1 (Cell Signaling NQO1 A180 Mouse mAb, 1:250; catalog. no. 3187S) was stained by incubating overnight at +4C°. NQO1 signal was quantified as the mean of relative positive-stained area across replicates, and NQO1-positive samples were defined as Q3 of the mean NQO1-signal across biological replicates. For T lymphocyte immunohistochemistry, TMA sections were stained with anti-CD3 (LN10, 1:200; Novocastra) and anti-CD8 (SP16, 1:400; Thermo Scientific) using a LabVision Autostainer 480 (ImmunoVision Technologies Inc.). Antigen retrieval was done with Tris-EDTA buffer at pH 9 by microwaving the slides in 98° Celsius for 15 min. Samples were incubated with diluted antibodies for 30 min at room temperature. Diaminobenzidine (DAB) was used as a chromogen and haematoxylin as a counterstain. Positive control tissue for CD3 and CD8 immunohistochemistry was normal tonsil. The slides were digitized with a slide scanner (Nano Zoomer XR, Hamamatsu) and quantification of CD3+ and CD8+ T cells was performed using QuPath, an open-source bioimage analysis software (version 0.1.2) (Bankhead, P., Loughrey, M.B., Fernández, J.A. et al. QuPath: Open source software for digital pathology image analysis [38].
4.15. Multiplex immunohistochemistry
4.15.1. Staining
NSCLC tissue microarray sections were obtained from the Biobank of Central Finland. We designed a panel of 10 markers to a) quantify PD-L1 expression (E1L3N), and b) identify macrophages (CD68, KP1), T cells (CD3, LN10), granulocytes (CD66b, G10F5) and tumor cells (Pan-cytokeratin, BS5 & AKR1B10). T cells were further characterized to CD8+ and CD4+ cells with the respective markers (clones 4B11 and EP204, respectively), and macrophages to M1 and M2 polarization states with CD86 (E2G8P) and CD206 (E2L9N), respectively. The multiplex staining was conducted with Bond-III automated IHC stainer (Leica Biosystems) and Bond Refine Detection kit (DS9800, Leica Biosystems). ImmPACT AMEC Red (SK-4285, Vector Laboratories) was used as the chromogen, except for AKR1B10 which was stained with DAB on the final round. AKR1B10 was used instead of NQO1 based on antibody validation for Bond-III, where AKR1B10 outperformed NQO1. The slides were mounted with VectaMount AQ Aqueous Mounting Medium (H-5501, Vector Laboratories), scanned with NanoZoomer XR (Hamamatsu) with a 20x objective. De-staining was conducted with ethanol and heating was applied between all cycles to remove prior antibodies.
4.15.2. Image analysis
Digitized images of the mIHC-slides were processed with QuPath (version 0.2.3), and individual cores were recognized with TMA-dearrayer function to obtain single core images. We excluded cores that were folded or detached. Single core images were stacked into one multichannel image with pseudofluorescence colors using Fiji [39]. We further discarded cores that were misaligned in the stack images. The staining intensities were consistent between the TMA slides indicating uniform performance of the protocol.
4.15.3. Data analysis
Total sum of tissue areas and cell counts of the core replicates were used to collapse the dataset to true biological replicates. Cutoffs for markers and AKR1B10 groups were chosen based on density distributions across the patient merged TMA cores (intensities for LUSC and LUAD 180 A U. and 165 A U., respectively). Macrophage M1 and M2 polarization was assessed by normalizing the CD206 and CD86 values to percentiles and calculating the difference between the normalized values. Quantiles of this polarization index were determined as M1 and M2 populations. Cell counts were normalized to the area (mm2) of the tumor (for epithelia infiltrating densities) and total area of the core (for stromal cells occupying the core). Statistical testing between the groups were conducted with two-sided Mann-Whitney U tests. Pearson's correlations were computed for linear relationship assessment between continuous variables.
Author contributions
Conceptualization, AL.L., P.P. and J.H.; Supervision, AL.L. and M.H.; methodology, P.P., J.H. and EY.D.; Software, P.P. and J.H.; Validation, T.K., M.A., EM. T, P.T., M.K., N.T., J.P.V., S.A.V., H.E. and T.E.; Formal analysis, J.H. and P.P.; Investigation, J.H., P.P., A.J.D. I.S. and HR.T.; Resources, AL.L., M.H., M.U.K., M.K, M.H., J.P.V. and T.K.; Data curation, P.P. and J.H.; Writing - Original Draft, J.H., AL.L., and P.P.; Writing - Review & Editing, M.H., T.K., A.J.D., J.P.V. and M.U.K.; Visualization, J.H., P.P. and A.J.D.; Project administration, AL.L.; Funding acquisition, AL. L and M.H.
Declaration of competing interest
The authors declare no potential conflicts interest.
Acknowledgements
This study was supported by University of Eastern Finland Doctoral Program in Molecular Medicine, Finnish Cancer Foundation, The Academy of Finland (Grant number 332697), Sigrid Juselius Foundation, Paavo Koistinen Foundation (AL.L. lab) and Aatos Erkko Foundation (Grant number 210013) (T.K. lab). The study benefited from samples from Auria Biopankki Turku, Finland, and Central Finland Biobank, Jyväskylä, Finland.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102644.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
NGS-data has been submitted to GEO (GSE189804). Other data is either publicly available or clinical cohorts with sharing restrictions.
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation, Cell. 2011;144 doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Hayes J.D., Dinkova-Kostova A.T., Tew K.D. Oxidative stress in cancer. Cancer Cell. 2020;38 doi: 10.1016/j.ccell.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell J.D., Alexandrov A., Kim J., Wala J., Berger A.H., Pedamallu C.S., Shukla S.A., Guo G., Brooks A.N., Murray B.A., Imielinski M., Hu X., Ling S., Akbani R., Rosenberg M., Cibulskis C., Ramachandran A., Collisson E.A., Kwiatkowski D.J., Lawrence M.S., Weinstein J.N., Verhaak R.G.W., Wu C.J., Hammerman P.S., Cherniack A.D., Getz G., Artyomov M.N., Schreiber R., Govindan R., Meyerson M. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016;48 doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross D., Siegel D. Functions of NQO1 in cellular protection and CoQ10 metabolism and its potential role as a redox sensitive molecular switch. Front. Physiol. 2017;8 doi: 10.3389/fphys.2017.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leinonen H.M., Kansanen E., Pölönen P., Heinäniemi M., Levonen A.L. Role of the keap1-Nrf2 pathway in cancer. Adv. Cancer Res. 2014 doi: 10.1016/B978-0-12-420117-0.00008-6. [DOI] [PubMed] [Google Scholar]
- 6.Pölönen P., Jawahar Deen A., Leinonen H.M., Jyrkkänen H.K., Kuosmanen S., Mononen M., Jain A., Tuomainen T., Pasonen-Seppänen S., Hartikainen J.M., Mannermaa A., Nykter M., Tavi P., Johansen T., Heinäniemi M., Levonen A.L. Nrf2 and SQSTM1/p62 jointly contribute to mesenchymal transition and invasion in glioblastoma. Oncogene. 2019;38 doi: 10.1038/s41388-019-0956-6. [DOI] [PubMed] [Google Scholar]
- 7.Rojo de la Vega M., Chapman E., Zhang D.D. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34 doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ancevski Hunter K., Socinski M.A., Villaruz L.C. PD-L1 testing in guiding patient selection for PD-1/PD-L1 inhibitor therapy in lung cancer. Mol. Diagn. Ther. 2018;22 doi: 10.1007/s40291-017-0308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Best S.A., de Souza D.P., Kersbergen A., Policheni A.N., Dayalan S., Tull D., Rathi V., Gray D.H., Ritchie M.E., McConville M.J., Sutherland K.D. Synergy between the KEAP1/NRF2 and PI3K pathways drives non-small-cell lung cancer with an altered immune microenvironment. Cell Metabol. 2018;27 doi: 10.1016/j.cmet.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Zhu B., Tang L., Chen S., Yin C., Peng S., Li X., Liu T., Liu W., Han C., Stawski L., Xu Z.X., Zhou G., Chen X., Gao X., Goding C.R., Xu N., Cui R., Cao P. Targeting the upstream transcriptional activator of PD-L1 as an alternative strategy in melanoma therapy. Oncogene. 2018;37 doi: 10.1038/s41388-018-0314-0. [DOI] [PubMed] [Google Scholar]
- 11.Chakravarty D., Gao J., Phillips S., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H., Chang M.T., Chandarlapaty S., Traina T.A., Paik P.K., Ho A.L., Hantash F.M., Grupe A., Baxi S.S., Callahan M.K., Snyder A., Chi P., Danila D.C., Gounder M., Harding J.J., Hellmann M.D., Iyer G., Janjigian Y.Y., Kaley T., Levine D.A., Lowery M., Omuro A., Postow M.A., Rathkopf D., Shoushtari A.N., Shukla N., Voss M.H., Paraiso E., Zehir A., Berger M.F., Taylor B.S., Saltz L.B., Riely G.J., Ladanyi M., Hyman D.M., Baselga J., Sabbatini P., Solit D.B., Schultz N. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017 doi: 10.1200/po.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duan Q., Zhang H., Zheng J., Zhang L. Turning cold into hot: firing up the tumor microenvironment. Trends Cancer. 2020;6 doi: 10.1016/j.trecan.2020.02.022. [DOI] [PubMed] [Google Scholar]
- 13.Liu P., Ma D., Wang P., Pan C., Fang Q., Wang J. Nrf2 overexpression increases risk of high tumor mutation burden in acute myeloid leukemia by inhibiting MSH2. Cell Death Dis. 2021;12 doi: 10.1038/s41419-020-03331-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 2020;15 doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 15.Ru B., Wong C.N., Tong Y., Zhong J.Y., Zhong S.S.W., Wu W.C., Chu K.C., Wong C.Y., Lau C.Y., Chen I., Chan N.W., Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35 doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 16.Tan Y.S., Sansanaphongpricha K., Xie Y., Donnelly C.R., Luo X., Heath B.R., Zhao X., Bellile E., Hu H., Chen H., Polverini P.J., Chen Q., Young S., Carey T.E., Nor J.E., Ferris R.L., Wolf G.T., Sun D., Lei Y.L. Mitigating SOX2-potentiated immune escape of head and neck squamous cell carcinoma with a STING-inducing nanosatellite vaccine. Clin. Cancer Res. 2018;24 doi: 10.1158/1078-0432.CCR-17-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv H., Lv G., Chen C., Zong Q., Jiang G., Ye D., Cui X., He Y., Xiang W., Han Q., Tang L., Yang W., Wang H. NAD+ metabolism maintains inducible PD-L1 expression to drive tumor immune evasion. Cell Metabol. 2021;33 doi: 10.1016/j.cmet.2020.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Klement J.D., Paschall A.v., Redd P.S., Ibrahim M.L., Lu C., Yang D., Celis E., Abrams S.I., Ozato K., Liu K. An osteopontin/CD44 immune checkpoint controls CD8+ T cell activation and tumor immune evasion. J. Clin. Invest. 2018;128 doi: 10.1172/JCI123360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao F., Xiao C., Evans K.S., Theivanthiran T., DeVito N., Holtzhausen A., Liu J., Liu X., Boczkowski D., Nair S., Locasale J.W., Hanks B.A. Paracrine wnt5a-β-catenin signaling triggers a metabolic Program that drives dendritic cell tolerization. Immunity. 2018;48 doi: 10.1016/j.immuni.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12 doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., Ziv E., Culhane A.C., Paull E.O., Sivakumar I.K.A., Gentles A.J., Malhotra R., Farshidfar F., Colaprico A., Parker J.S., Mose L.E., Vo N.S., Liu J., Liu Y., Rader J., Dhankani V., Reynolds S.M., Bowlby R., Califano A., Cherniack A.D., Anastassiou D., Bedognetti D., Rao A., Chen K., Krasnitz A., Hu H., Malta T.M., Noushmehr H., Pedamallu C.S., Bullman S., Ojesina A.I., Lamb A., Zhou W., Shen H., Choueiri T.K., Weinstein J.N., Guinney J., Saltz J., Holt R., Rabkin C.E., Caesar-Johnson S.J., Demchok J.A., Felau I., Kasapi M., Ferguson M.L., Hutter C.M., Sofia H.J., Tarnuzzer R., Wang Z., Yang L., Zenklusen J.C., Julia J., Zhang, Chudamani S., Liu J., Lolla L., Naresh R., Pihl T., Sun Q., Wan Y., Wu Y., Cho J., DeFreitas T., Frazer S., Gehlenborg N., Getz G., Heiman D.I., Kim J., Lawrence M.S., Lin P., Meier S., Noble M.S., Saksena G., Voet D., Zhang H., Bernard B., Chambwe N., Dhankani V., Knijnenburg T., Kramer R., Leinonen K., Liu Y., Miller M., Reynolds S., Shmulevich I., Thorsson V., Zhang W., Akbani R., Broom B.M., Hegde A.M., Ju Z., Kanchi R.S., Korkut A., Li J., Liang H., Ling S., Liu W., Lu Y., Mills G.B., Ng K.S., Rao A., Ryan M., Wang J., Weinstein J.N., Zhang J., Abeshouse A., Armenia J., Chakravarty D., Chatila W.K., de Bruijn I., Gao J., Gross B.E., Heins Z.J., Kundra R., La K., Ladanyi M., Luna A., Nissan M.G., Ochoa A., Phillips S.M., Reznik E., Sanchez-Vega F., Sander C., Schultz N., Sheridan R., Sumer S.O., Sun Y., Taylor B.S., Wang J., Zhang H., Anur P., Peto M., Spellman P., Benz C., Stuart J.M., Wong C.K., Yau C., Hayes D.N., Parker J.S., Wilkerson M.D., Ally A., Balasundaram M., Bowlby R., Brooks D., Carlsen R., Chuah E., Dhalla N., Jones S.J.M., Kasaian K., Lee D., Ma Y., Marra M.A., Mayo M., Moore R.A., Mungall A.J., Mungall K., Robertson A.G., Sadeghi S., Schein J.E., Sipahimalani P., Tam A., Thiessen N., Tse K., Wong T., Berger A.C., Beroukhim R., Cherniack A.D., Cibulskis C., Gabriel S.B., Ha G., Meyerson M., Schumacher S.E., Shih J., Kucherlapati M.H., Kucherlapati R.S., Baylin S., Cope L., Danilova L., Bootwalla M.S., Lai P.H., Maglinte D.T., van den Berg D.J., Weisenberger D.J., Auman J.T., Balu S., Bodenheimer T., Fan C., Hoadley K.A., Hoyle A.P., Jefferys S.R., Jones C.D., Meng S., Mieczkowski P.A., Mose L.E., Perou A.H., Perou C.M., Roach J., Shi Y., Simons J.v., Skelly T., Soloway M.G., Tan D., Veluvolu U., Fan H., Hinoue T., Laird P.W., Shen H., Zhou W., Bellair M., Chang K., Covington K., Creighton C.J., Dinh H., Doddapaneni H.V., Donehower L.A., Drummond J., Gibbs R.A., Glenn R., Hale W., Han Y., Hu J., Korchina V., Lee S., Lewis L., Li W., Liu X., Morgan M., Morton D., Muzny D., Santibanez J., Sheth M., Shinbrot E., Wang L., Wang M., Wheeler D.A., Xi L., Zhao F., Hess J., Appelbaum E.L., Bailey M., Cordes M.G., Ding L., Fronick C.C., Fulton L.A., Fulton R.S., Kandoth C., Mardis E.R., McLellan M.D., Miller C.A., Schmidt H.K., Wilson R.K., Crain D., Curley E., Gardner J., Lau K., Mallery D., Morris S., Paulauskis J., Penny R., Shelton C., Shelton T., Sherman M., Thompson E., Yena P., Bowen J., Gastier-Foster J.M., Gerken M., Leraas K.M., Lichtenberg T.M., Ramirez N.C., Wise L., Zmuda E., Corcoran N., Costello T., Hovens C., Carvalho A.L., de Carvalho A.C., Fregnani J.H., Longatto-Filho A., Reis R.M., Scapulatempo-Neto C., Silveira H.C.S., Vidal D.O., Burnette A., Eschbacher J., Hermes B., Noss A., Singh R., Anderson M.L., Castro P.D., Ittmann M., Huntsman D., Kohl B., Le X., Thorp R., Andry C., Duffy E.R., Lyadov V., Paklina O., Setdikova G., Shabunin A., Tavobilov M., McPherson C., Warnick R., Berkowitz R., Cramer D., Feltmate C., Horowitz N., Kibel A., Muto M., Raut C.P., Malykh A., Barnholtz-Sloan J.S., Barrett W., Devine K., Fulop J., Ostrom Q.T., Shimmel K., Wolinsky Y., Sloan A.E., de Rose A., Giuliante F., Goodman M., Karlan B.Y., Hagedorn C.H., Eckman J., Harr J., Myers J., Tucker K., Zach L.A., Deyarmin B., Hu H., Kvecher L., Larson C., Mural R.J., Somiari S., Vicha A., Zelinka T., Bennett J., Iacocca M., Rabeno B., Swanson P., Latour M., Lacombe L., Têtu B., Bergeron A., McGraw M., Staugaitis S.M., Chabot J., Hibshoosh H., Sepulveda A., Su T., Wang T., Potapova O., Voronina O., Desjardins L., Mariani O., Roman-Roman S., Sastre X., Stern M.H., Cheng F., Signoretti S., Berchuck A., Bigner D., Lipp E., Marks J., McCall S., McLendon R., Secord A., Sharp A., Behera M., Brat D.J., Chen A., Delman K., Force S., Khuri F., Magliocca K., Maithel S., Olson J.J., Owonikoko T., Pickens A., Ramalingam S., Shin D.M., Sica G., van Meir E.G., Zhang H., Eijckenboom W., Gillis A., Korpershoek E., Looijenga L., Oosterhuis W., Stoop H., van Kessel K.E., Zwarthoff E.C., Calatozzolo C., Cuppini L., Cuzzubbo S., DiMeco F., Finocchiaro G., Mattei L., Perin A., Pollo B., Chen C., Houck J., Lohavanichbutr P., Hartmann A., Stoehr C., Stoehr R., Taubert H., Wach S., Wullich B., Kycler W., Murawa D., Wiznerowicz M., Chung K., Edenfield W.J., Martin J., Baudin E., Bubley G., Bueno R., de Rienzo A., Richards W.G., Kalkanis S., Mikkelsen T., Noushmehr H., Scarpace L., Girard N., Aymerich M., Campo E., Giné E., Guillermo A.L., van Bang N., Hanh P.T., Phu B.D., Tang Y., Colman H., Evason K., Dottino P.R., Martignetti J.A., Gabra H., Juhl H., Akeredolu T., Stepa S., Hoon D., Ahn K., Kang K.J., Beuschlein F., Breggia A., Birrer M., Bell D., Borad M., Bryce A.H., Castle E., Chandan V., Cheville J., Copland J.A., Farnell M., Flotte T., Giama N., Ho T., Kendrick M., Kocher J.P., Kopp K., Moser C., Nagorney D., O'Brien D., O'Neill B.P., Patel T., Petersen G., Que F., Rivera M., Roberts L., Smallridge R., Smyrk T., Stanton M., Thompson R.H., Torbenson M., Yang J.D., Zhang L., Brimo F., Ajani J.A., Gonzalez A.M.A., Behrens C., Bondaruk J., Broaddus R., Czerniak B., Esmaeli B., Fujimoto J., Gershenwald J., Guo C., Lazar A.J., Logothetis C., Meric-Bernstam F., Moran C., Ramondetta L., Rice D., Sood A., Tamboli P., Thompson T., Troncoso P., Tsao A., Wistuba I., Carter C., Haydu L., Hersey P., Jakrot V., Kakavand H., Kefford R., Lee K., Long G., Mann G., Quinn M., Saw R., Scolyer R., Shannon K., Spillane A., onathan Stretch, Synott M., Thompson J., Wilmott J., Al-Ahmadie H., Chan T.A., Ghossein R., Gopalan A., Levine D.A., Reuter V., Singer S., Singh B., Tien N.V., Broudy T., Mirsaidi C., Nair P., Drwiega P., Miller J., Smith J., Zaren H., Park J.W., Hung N.P., Kebebew E., Linehan W.M., Metwalli A.R., Pacak K., Pinto P.A., Schiffman M., Schmidt L.S., Vocke C.D., Wentzensen N., Worrell R., Yang H., Moncrieff M., Goparaju C., Melamed J., Pass H., Botnariuc N., Caraman I., Cernat M., Chemencedji I., Clipca A., Doruc S., Gorincioi G., Mura S., Pirtac M., Stancul I., Tcaciuc D., Albert M., Alexopoulou I., Arnaout A., Bartlett J., Engel J., Gilbert S., Parfitt J., Sekhon H., Thomas G., Rassl D.M., Rintoul R.C., Bifulco C., Tamakawa R., Urba W., Hayward N., Timmers H., Antenucci A., Facciolo F., Grazi G., Marino M., Merola R., de Krijger R., Gimenez-Roqueplo A.P., Piché A., Chevalier S., McKercher G., Birsoy K., Barnett G., Brewer C., Farver C., Naska T., Pennell N.A., Raymond D., Schilero C., Smolenski K., Williams F., Morrison C., Borgia J.A., Liptay M.J., Pool M., Seder C.W., Junker K., Omberg L., Dinkin M., Manikhas G., Alvaro D., Bragazzi M.C., Cardinale V., Carpino G., Gaudio E., Chesla D., Cottingham S., Dubina M., Moiseenko F., Dhanasekaran R., Becker K.F., Janssen K.P., Slotta-Huspenina J., Abdel-Rahman M.H., Aziz D., Bell S., Cebulla C.M., Davis A., Duell R., Elder J.B., Hilty J., Kumar B., Lang J., Lehman N.L., Mandt R., Nguyen P., Pilarski R., Rai K., Schoenfield L., Senecal K., Wakely P., Hansen P., Lechan R., Powers J., Tischler A., Grizzle W.E., Sexton K.C., Kastl A., Henderson J., Porten S., Waldmann J., Fassnacht M., Asa S.L., Schadendorf D., Couce M., Graefen M., Huland H., Sauter G., Schlomm T., Simon R., Tennstedt P., Olabode O., Nelson M., Bathe O., Carroll P.R., Chan J.M., Disaia P., Glenn P., Kelley R.K., Landen C.N., Phillips J., Prados M., Simko J., Smith-McCune K., VandenBerg S., Roggin K., Fehrenbach A., Kendler A., Sifri S., Steele R., Jimeno A., Carey F., Forgie I., Mannelli M., Carney M., Hernandez B., Campos B., Herold-Mende C., Jungk C., Unterberg A., von Deimling A., Bossler A., Galbraith J., Jacobus L., Knudson M., Knutson T., Ma D., Milhem M., Sigmund R., Godwin A.K., Madan R., Rosenthal H.G., Adebamowo C., Adebamowo S.N., Boussioutas A., Beer D., Giordano T., Mes-Masson A.M., Saad F., Bocklage T., Landrum L., Mannel R., Moore K., Moxley K., Postier R., Walker J., Zuna R., Feldman M., Valdivieso F., Dhir R., Luketich J., Pinero E.M.M., Quintero-Aguilo M., Carlotti C.G., dos Santos J.S., Kemp R., Sankarankuty A., Tirapelli D., Catto J., Agnew K., Swisher E., Creaney J., Robinson B., Shelley C.S., Godwin E.M., Kendall S., Shipman C., Bradford C., Carey T., Haddad A., Moyer J., Peterson L., Prince M., Rozek L., Wolf G., Bowman R., Fong K.M., Yang I., Korst R., Rathmell W.K., Fantacone-Campbell J.L., Hooke J.A., Kovatich A.J., Shriver C.D., DiPersio J., Drake B., Govindan R., Heath S., Ley T., van Tine B., Westervelt P., Rubin M.A., il Lee J., Aredes N.D., Mariamidze A., Serody J.S., Demicco E.G., Disis M.L., Vincent B.G., Shmulevich llya. The immune landscape of cancer. Immunity. 2018;48 doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saltz J., Gupta R., Hou L., Kurc T., Singh P., Nguyen V., Samaras D., Shroyer K.R., Zhao T., Batiste R., van Arnam J., Caesar-Johnson S.J., Demchok J.A., Felau I., Kasapi M., Ferguson M.L., Hutter C.M., Sofia H.J., Tarnuzzer R., Wang Z., Yang L., Zenklusen J.C., Julia J., Zhang, Chudamani S., Liu J., Lolla L., Naresh R., Pihl T., Sun Q., Wan Y., Wu Y., Cho J., DeFreitas T., Frazer S., Gehlenborg N., Getz G., Heiman D.I., Kim J., Lawrence M.S., Lin P., Meier S., Noble M.S., Saksena G., Voet D., Zhang H., Bernard B., Chambwe N., Dhankani V., Knijnenburg T., Kramer R., Leinonen K., Liu Y., Miller M., Reynolds S., Shmulevich I., Thorsson V., Zhang W., Akbani R., Broom B.M., Hegde A.M., Ju Z., Kanchi R.S., Korkut A., Li J., Liang H., Ling S., Liu W., Lu Y., Mills G.B., Ng K.S., Rao A., Ryan M., Wang J., Weinstein J.N., Zhang J., Abeshouse A., Armenia J., Chakravarty D., Chatila W.K., de Bruijn I., Gao J., Gross B.E., Heins Z.J., Kundra R., La K., Ladanyi M., Luna A., Nissan M.G., Ochoa A., Phillips S.M., Reznik E., Sanchez-Vega F., Sander C., Schultz N., Sheridan R., Sumer S.O., Sun Y., Taylor B.S., Wang J., Zhang H., Anur P., Peto M., Spellman P., Benz C., Stuart J.M., Wong C.K., Yau C., Hayes D.N., Parker J.S., Wilkerson M.D., Ally A., Balasundaram M., Bowlby R., Brooks D., Carlsen R., Chuah E., Dhalla N., Holt R., Jones S.J.M., Kasaian K., Lee D., Ma Y., Marra M.A., Mayo M., Moore R.A., Mungall A.J., Mungall K., Robertson A.G., Sadeghi S., Schein J.E., Sipahimalani P., Tam A., Thiessen N., Tse K., Wong T., Berger A.C., Beroukhim R., Cherniack A.D., Cibulskis C., Gabriel S.B., Gao G.F., Ha G., Meyerson M., Schumacher S.E., Shih J., Kucherlapati M.H., Kucherlapati R.S., Baylin S., Cope L., Danilova L., Bootwalla M.S., Lai P.H., Maglinte D.T., van den Berg D.J., Weisenberger D.J., Auman J.T., Balu S., Bodenheimer T., Fan C., Hoadley K.A., Hoyle A.P., Jefferys S.R., Jones C.D., Meng S., Mieczkowski P.A., Mose L.E., Perou A.H., Perou C.M., Roach J., Shi Y., Simons J.v., Skelly T., Soloway M.G., Tan D., Veluvolu U., Fan H., Hinoue T., Laird P.W., Shen H., Zhou W., Bellair M., Chang K., Covington K., Creighton C.J., Dinh H., Doddapaneni H.V., Donehower L.A., Drummond J., Gibbs R.A., Glenn R., Hale W., Han Y., Hu J., Korchina V., Lee S., Lewis L., Li W., Liu X., Morgan M., Morton D., Muzny D., Santibanez J., Sheth M., Shinbrot E., Wang L., Wang M., Wheeler D.A., Xi L., Zhao F., Hess J., Appelbaum E.L., Bailey M., Cordes M.G., Ding L., Fronick C.C., Fulton L.A., Fulton R.S., Kandoth C., Mardis E.R., McLellan M.D., Miller C.A., Schmidt H.K., Wilson R.K., Crain D., Curley E., Gardner J., Lau K., Mallery D., Morris S., Paulauskis J., Penny R., Shelton C., Shelton T., Sherman M., Thompson E., Yena P., Bowen J., Gastier-Foster J.M., Gerken M., Leraas K.M., Lichtenberg T.M., Ramirez N.C., Wise L., Zmuda E., Corcoran N., Costello T., Hovens C., Carvalho A.L., de Carvalho A.C., Fregnani J.H., Longatto-Filho A., Reis R.M., Scapulatempo-Neto C., Silveira H.C.S., Vidal D.O., Burnette A., Eschbacher J., Hermes B., Noss A., Singh R., Anderson M.L., Castro P.D., Ittmann M., Huntsman D., Kohl B., Le X., Thorp R., Andry C., Duffy E.R., Lyadov V., Paklina O., Setdikova G., Shabunin A., Tavobilov M., McPherson C., Warnick R., Berkowitz R., Cramer D., Feltmate C., Horowitz N., Kibel A., Muto M., Raut C.P., Malykh A., Barnholtz-Sloan J.S., Barrett W., Devine K., Fulop J., Ostrom Q.T., Shimmel K., Wolinsky Y., Sloan A.E., de Rose A., Giuliante F., Goodman M., Karlan B.Y., Hagedorn C.H., Eckman J., Harr J., Myers J., Tucker K., Zach L.A., Deyarmin B., Hu H., Kvecher L., Larson C., Mural R.J., Somiari S., Vicha A., Zelinka T., Bennett J., Iacocca M., Rabeno B., Swanson P., Latour M., Lacombe L., Têtu B., Bergeron A., McGraw M., Staugaitis S.M., Chabot J., Hibshoosh H., Sepulveda A., Su T., Wang T., Potapova O., Voronina O., Desjardins L., Mariani O., Roman-Roman S., Sastre X., Stern M.H., Cheng F., Signoretti S., Berchuck A., Bigner D., Lipp E., Marks J., McCall S., McLendon R., Secord A., Sharp A., Behera M., Brat D.J., Chen A., Delman K., Force S., Khuri F., Magliocca K., Maithel S., Olson J.J., Owonikoko T., Pickens A., Ramalingam S., Shin D.M., Sica G., van Meir E.G., Zhang H., Eijckenboom W., Gillis A., Korpershoek E., Looijenga L., Oosterhuis W., Stoop H., van Kessel K.E., Zwarthoff E.C., Calatozzolo C., Cuppini L., Cuzzubbo S., DiMeco F., Finocchiaro G., Mattei L., Perin A., Pollo B., Chen C., Houck J., Lohavanichbutr P., Hartmann A., Stoehr C., Stoehr R., Taubert H., Wach S., Wullich B., Kycler W., Murawa D., Wiznerowicz M., Chung K., Edenfield W.J., Martin J., Baudin E., Bubley G., Bueno R., de Rienzo A., Richards W.G., Kalkanis S., Mikkelsen T., Noushmehr H., Scarpace L., Girard N., Aymerich M., Campo E., Giné E., Guillermo A.L., van Bang N., Hanh P.T., Phu B.D., Tang Y., Colman H., Evason K., Dottino P.R., Martignetti J.A., Gabra H., Juhl H., Akeredolu T., Stepa S., Hoon D., Ahn K., Kang K.J., Beuschlein F., Breggia A., Birrer M., Bell D., Borad M., Bryce A.H., Castle E., Chandan V., Cheville J., Copland J.A., Farnell M., Flotte T., Giama N., Ho T., Kendrick M., Kocher J.P., Kopp K., Moser C., Nagorney D., O'Brien D., O'Neill B.P., Patel T., Petersen G., Que F., Rivera M., Roberts L., Smallridge R., Smyrk T., Stanton M., Thompson R.H., Torbenson M., Yang J.D., Zhang L., Brimo F., Ajani J.A., Gonzalez A.M.A., Behrens C., Bondaruk J., Broaddus R., Czerniak B., Esmaeli B., Fujimoto J., Gershenwald J., Guo C., Lazar A.J., Logothetis C., Meric-Bernstam F., Moran C., Ramondetta L., Rice D., Sood A., Tamboli P., Thompson T., Troncoso P., Tsao A., Wistuba I., Carter C., Haydu L., Hersey P., Jakrot V., Kakavand H., Kefford R., Lee K., Long G., Mann G., Quinn M., Saw R., Scolyer R., Shannon K., Spillane A., onathan Stretch, Synott M., Thompson J., Wilmott J., Al-Ahmadie H., Chan T.A., Ghossein R., Gopalan A., Levine D.A., Reuter V., Singer S., Singh B., Tien N.V., Broudy T., Mirsaidi C., Nair P., Drwiega P., Miller J., Smith J., Zaren H., Park J.W., Hung N.P., Kebebew E., Linehan W.M., Metwalli A.R., Pacak K., Pinto P.A., Schiffman M., Schmidt L.S., Vocke C.D., Wentzensen N., Worrell R., Yang H., Moncrieff M., Goparaju C., Melamed J., Pass H., Botnariuc N., Caraman I., Cernat M., Chemencedji I., Clipca A., Doruc S., Gorincioi G., Mura S., Pirtac M., Stancul I., Tcaciuc D., Albert M., Alexopoulou I., Arnaout A., Bartlett J., Engel J., Gilbert S., Parfitt J., Sekhon H., Thomas G., Rassl D.M., Rintoul R.C., Bifulco C., Tamakawa R., Urba W., Hayward N., Timmers H., Antenucci A., Facciolo F., Grazi G., Marino M., Merola R., de Krijger R., Gimenez-Roqueplo A.P., Piché A., Chevalier S., McKercher G., Birsoy K., Barnett G., Brewer C., Farver C., Naska T., Pennell N.A., Raymond D., Schilero C., Smolenski K., Williams F., Morrison C., Borgia J.A., Liptay M.J., Pool M., Seder C.W., Junker K., Omberg L., Dinkin M., Manikhas G., Alvaro D., Bragazzi M.C., Cardinale V., Carpino G., Gaudio E., Chesla D., Cottingham S., Dubina M., Moiseenko F., Dhanasekaran R., Becker K.F., Janssen K.P., Slotta-Huspenina J., Abdel-Rahman M.H., Aziz D., Bell S., Cebulla C.M., Davis A., Duell R., Elder J.B., Hilty J., Kumar B., Lang J., Lehman N.L., Mandt R., Nguyen P., Pilarski R., Rai K., Schoenfield L., Senecal K., Wakely P., Hansen P., Lechan R., Powers J., Tischler A., Grizzle W.E., Sexton K.C., Kastl A., Henderson J., Porten S., Waldmann J., Fassnacht M., Asa S.L., Schadendorf D., Couce M., Graefen M., Huland H., Sauter G., Schlomm T., Simon R., Tennstedt P., Olabode O., Nelson M., Bathe O., Carroll P.R., Chan J.M., Disaia P., Glenn P., Kelley R.K., Landen C.N., Phillips J., Prados M., Simko J., Smith-McCune K., VandenBerg S., Roggin K., Fehrenbach A., Kendler A., Sifri S., Steele R., Jimeno A., Carey F., Forgie I., Mannelli M., Carney M., Hernandez B., Campos B., Herold-Mende C., Jungk C., Unterberg A., von Deimling A., Bossler A., Galbraith J., Jacobus L., Knudson M., Knutson T., Ma D., Milhem M., Sigmund R., Godwin A.K., Madan R., Rosenthal H.G., Adebamowo C., Adebamowo S.N., Boussioutas A., Beer D., Giordano T., Mes-Masson A.M., Saad F., Bocklage T., Landrum L., Mannel R., Moore K., Moxley K., Postier R., Walker J., Zuna R., Feldman M., Valdivieso F., Dhir R., Luketich J., Pinero E.M.M., Quintero-Aguilo M., Carlotti C.G., dos Santos J.S., Kemp R., Sankarankuty A., Tirapelli D., Catto J., Agnew K., Swisher E., Creaney J., Robinson B., Shelley C.S., Godwin E.M., Kendall S., Shipman C., Bradford C., Carey T., Haddad A., Moyer J., Peterson L., Prince M., Rozek L., Wolf G., Bowman R., Fong K.M., Yang I., Korst R., Rathmell W.K., Fantacone-Campbell J.L., Hooke J.A., Kovatich A.J., Shriver C.D., DiPersio J., Drake B., Govindan R., Heath S., Ley T., van Tine B., Westervelt P., Rubin M.A., il Lee J., Aredes N.D., Mariamidze A., Shmulevich I., Rao A.U.K., Sharma A., Thorsson V. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23 doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spranger S., Gajewski T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat. Rev. Cancer. 2018;18 doi: 10.1038/nrc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A., Daemen A., Nickles D., Jeon S.M., Foreman O., Sudini K., Gnad F., Lajoie S., Gour N., Mitzner W., Chatterjee S., Choi E.J., Ravishankar B., Rappaport A., Patil N., McCleland M., Johnson L., Acquaah-Mensah G., Gabrielson E., Biswal S., Hatzivassiliou G. NRF2 Activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin. Cancer Res. 2021;27 doi: 10.1158/1078-0432.CCR-20-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kansanen E., Kuosmanen S.M., Leinonen H., Levonenn A.L. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer, Redox Biol. 2013;1 doi: 10.1016/j.redox.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porter L., McCaughan F. SOX2 and squamous cancers. Semin. Cancer Biol. 2020;67 doi: 10.1016/j.semcancer.2020.05.007. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh M.H., Choe J.H., Gadhvi J., Kim Y.J., Arguez M.A., Palmer M., Gerold H., Nowak C., Do H., Mazambani S., Knighton J.K., Cha M., Goodwin J., Kang M.K., Jeong J.Y., Lee S.Y., Faubert B., Xuan Z., Abel E.D., Scafoglio C., Shackelford D.B., Minna J.D., Singh P.K., Shulaev V., Bleris L., Hoyt K., Kim J., Inoue M., DeBerardinis R.J., Kim T.H. J. whan Kim, p63 and SOX2 Dictate Glucose Reliance and Metabolic Vulnerabilities in Squamous Cell Carcinomas, Cell Rep. 2019;28 doi: 10.1016/j.celrep.2019.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ottensmeier C.H., Perry K.L., Harden E.L., Stasakova J., Jenei V., Fleming J., Wood O., Woo J., Woelk C.H., Thomas G.J., Thirdborough S.M. Upregulated glucose metabolism correlates inversely with CD8+ T-cell infiltration and survival in squamous cell carcinoma. Cancer Res. 2016;76 doi: 10.1158/0008-5472.CAN-15-3121. [DOI] [PubMed] [Google Scholar]
- 29.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism, Trends Biochem. Sci. 2014;39 doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A., Xia H., Zhou J., Li G., Li J., Li W., Wei S., Vatan L., Zhang H., Szeliga W., Gu W., Liu R., Lawrence T.S., Lamb C., Tanno Y., Cieslik M., Stone E., Georgiou G., Chan T.A., Chinnaiyan A., Zou W. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569 doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casey S.C., Tong L., Li Y., Do R., Walz S., Fitzgerald K.N., Gouw A.M., Baylot V., Gütgemann I., Eilers M., Felsher D.W. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castagnoli L., Cancila V., Cordoba-Romero S.L., Faraci S., Talarico G., Belmonte B., v Iorio M., Milani M., Volpari T., Chiodoni C., Hidalgo-Miranda A., Tagliabue E., Tripodo C., Sangaletti S., di Nicola M., Pupa S.M. WNT signaling modulates PD-L1 expression in the stem cell compartment of triple-negative breast cancer. Oncogene. 2019;38:4047–4060. doi: 10.1038/s41388-019-0700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng M.W.L., Ngiow S.F., Ribas A., Smyth M.J. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75:2139–2145. doi: 10.1158/0008-5472.CAN-15-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pagès C., Tosolini M., Camus M., Berger A., Wind P., Zinzindohoué F., Bruneval P., Cugnenc P.-H., Trajanoski Z., Fridman W.-H., Pagès F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 35.Bouvy-Liivrand M., de Sande A.H., Pölönen P., Mehtonen J., Vuorenmaa T., Niskanen H., Sinkkonen L., Kaikkonen M.U., Heinäniemi M. Analysis of primary microRNA loci from nascent transcriptomes reveals regulatory domains governed by chromatin architecture. Nucleic Acids Res. 2017;45 doi: 10.1093/nar/gkx680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., Butte A.J., Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20 doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.S V. Puram, I. Tirosh, A.S. Parikh, A.P. Patel, K. Yizhak, S. Gillespie, C. Rodman, C.L. Luo, E.A. Mroz, K.S. Emerick, D.G. Deschler, M.A. Varvares, R. Mylvaganam, O. Rozenblatt-rosen, J.W. Rocco, W.C. Faquin, D.T. Lin, A. Regev, B.E. Bernstein, single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171 doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankhead P., Loughrey M.B., Fernández J.A., Dombrowski Y., McArt D.G., Dunne P.D., McQuaid S., Gray R.T., Murray L.J., Coleman H.G., James J.A., Salto-Tellez M., Hamilton P.W. QuPath: open source software for digital pathology image analysis. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS-data has been submitted to GEO (GSE189804). Other data is either publicly available or clinical cohorts with sharing restrictions.