Abstract
Optimal conditions for two-dimensional gel electrophoresis of total cellular proteins from Myxococcus xanthus were established. Using these conditions, we analyzed protein patterns of heat-shocked M. xanthus cells. Eighteen major spots and 15 minor spots were found to be induced by heat shock. From N-terminal sequences of 15 major spots, DnaK, GroEL, GroES, alkyl hydroperoxide reductase, aldehyde dehydrogenase, succinyl coenzyme A (CoA) synthetase, 30S ribosomal protein S6, and ATP synthase α subunit were identified. Three of the 18 major spots had an identical N-terminal sequence, indicating that they may be different forms of the same protein. Although a DnaK homologue, SglK, has been identified in M. xanthus (R. M. Weimer, C. Creghton, A. Stassinopoulos, P. Youderian, and P. L. Hartzell, J. Bacteriol. 180:5357–5368, 1998; Z. Yang, Y. Geng, and W. Shi, J. Bacteriol. 180:218–224, 1998), SglK was not induced by heat shock. In addition, there were seven substitutions within the N-terminal 30-residue sequence of the newly identified DnaK. This is the first report to demonstrate that succinyl CoA synthetase, 30S ribosomal protein S6, and ATP synthase α subunit are heat shock inducible.
All organisms respond and adapt to heat shock by inducing heat shock proteins (HSPs). A number of HSPs from bacteria to animals are well conserved (14, 29). HSPs are also induced in response to carbon, nitrogen, or phosphate starvation in prokaryotes (19, 20, 21, 24). In eukaryotes, HSP100, HSP90, HSP70, HSP60, and HSP40 function as molecular chaperones, and in prokaryotes, DnaK (a homologue of HSP70) (12), GroEL (HSP60) (26), DnaJ (HSP40) (12), and GroES (27) do so. In addition, ATP-dependent proteases such as ClpP and Lon are known to be HSPs (7, 10).
Myxococcus xanthus is a gram-negative soil bacterium that feeds on microorganisms and organic debris (5, 6). Under nutrient starvation, M. xanthus cells aggregate by gliding motility and form multicellular fruiting bodies (FB) in which cells differentiate into spores. It has been shown that a number of developmental signals are coordinated during the differentiation process. Spores are metabolically dormant and resistant to desiccation, heat, and UV irradiation.
The heat shock response of M. xanthus was previously investigated by labeling M. xanthus cells with [35S] methionine during vegetative growth, glycerol-induced spore formation, and starvation-induced FB formation (17). During vegetative growth, 18 major HSPs with molecular masses of 91 to 14.5 kDa were found. When cells were heat shocked prior to starvation-induced FB and spore formation, FB and spore formation was accelerated with no effect on spore yield. During glycerol-induced spore formation, heat shock accelerated the rate of spore formation and enhanced the spore yield by fivefold (13).
Here, we reinvestigated the heat shock response by two-dimensional (2D) gel electrophoresis and N-terminal microsequencing of heat-shock-induced proteins in M. xanthus. We found that in addition to well-known molecular chaperones (DnaK, GroEL, and GroES), alkyl hydroperoxide reductase, aldehyde dehydrogenase, succinyl coenzyme A (CoA) synthetase, 30S ribosomal protein S6, and ATP synthase α subunit are induced by heat shock in vegetatively growing M. xanthus cells.
MATERIALS AND METHODS
Sample preparation.
M. xanthus DZF1 was grown in 20 ml of CYE (3) liquid medium at 30°C. For heat shock stress, cells growing exponentially at 30°C were shifted to 42°C and incubated for 30 and 60 min. Cells were harvested by centrifugation and washed with TM buffer (10 mM Tris-Cl [pH 7.6], 8 mM MgSO4). Cells were resuspended in 100 μl of lysis buffer (7 M urea, 2 M thiourea, 5% N-cyclohexyl-3-aminopropanesulfonic acid [CHAPS; DOJINDO Ltd.], 2% IPG buffer [Amersham Pharmacia Biotech], 50 mM 2-mercaptoethanol, 2.5 μg of DNase I per ml, 2.5 μg of RNase A per ml) and disrupted by sonication. After 320 μl of lysis buffer was added, samples were mixed by gentle shaking at room temperature for 1 h. Supernatants obtained after centrifugation at 100,000 × g for 30 min were used for 2D gel electrophoresis. Protein concentrations in samples were determined with a protein assay rapid kit (Wako Chemicals Co. Ltd.). All chemicals used were purchased from Wako Chemicals, unless otherwise indicated.
2D gel electrophoresis.
2D gel electrophoresis was carried out by the method of Gorg et al. (8, 9) with modifications. Briefly, for the first-dimension isoelectric focusing gel, Immobiline DryStrip pH 3-10 NL (13 cm) (Amersham Pharmacia Biotech) was used. Sample solution (110 μl; 2 mg of proteins) and 110 μl of rehydration buffer (8 M urea, 0.5% CHAPS, 20 mM dithiothreitol, 0.5% IPG buffer) were mixed and poured into the gel rehydration tray (Nihon Eido Co., Ltd., Tokyo, Japan). The strips were covered with silicone oil (KF-96-10CS; Shin-Etsu Chemical Co., Ltd.) to prevent samples from evaporation and rehydrated at room temperature overnight. Rehydrated strips were placed on Kimwipes to remove silicone oil from the surface. First-dimension isoelectric focusing gel electrophoresis was carried out by using an electrophoresis apparatus from Nihon Eido at 500 V for 2 h, at 700 V for 1 h, at 1,000 V for 1 h, at 2,000 V for 1 h, and at 3,500 V for 15 to 16 h. After electrophoresis, the strips were soaked in equilibration buffer (0.05 M Tris-Cl [pH 6.8], 6 M urea, 30% glycerol, 1% sodium dodecyl sulfate [SDS], 16 mM dithiothreitol, 0.04% bromophenol blue) at room temperature for 40 min with gentle shaking.
2D SDS-polyacrylamide-gel electrophoresis was performed with a 17.5% acrylamide gel at 5 mA/gel for the first 2 h and then at 10 mA/gel for 7 h. After electrophoresis, the gels were fixed in 10% trichloroacetic acid solution for 1.5 h and then stained with Coomassie brilliant blue R (CBB).
pI standards were purchased from Daiichi Pure Chemicals, and molecular weight standards were obtained from Bio-Rad Laboratories. Protein patterns were analyzed with Gel LABII version 2.0 (Scanalytics).
Protein microsequencing.
For preparing samples for microsequencing, Immobiline DryStrip pH 3-10 NL (18 cm) was used for the first-dimension isoelectric focusing gel. After 2D gel electrophoresis, the gel was blotted onto polyvinylidene difluoride membranes (Japan Genetics Co., Ltd.) with buffer containing 10 mM Tris-Cl (pH 8), 2 mM glycine, and 1.5% methanol. The membranes were stained with CBB and washed with 50% methanol and distilled water. The membranes were dried, and spots of interest were excised with a razor blade. The excised membranes were stored at 4°C. Determination of N-terminal sequences was performed with a Shimazu PPSQ10 protein sequencer. The membranes were washed with 10% ethanol several times and dried before being applied to the sequencer. One gel spot obtained from 2 mg of total cellular proteins was enough for the determination of N-terminal sequences. Electrophoresis was carried out at least three times to obtain reproducibility.
RESULTS
Establishment of the optimal conditions for 2D gel electrophoresis for M. xanthus.
Proteome analysis is often performed by 2D gel electrophoresis with cells grown under various stress conditions, such as heat shock, oxidative stress, and nutritional starvation. A major problem in analyzing total cellular proteins is that M. xanthus cells contain a large amount of polysaccharides or slime, which interferes with protein solubility, resolution, and reproducibility in 2D gel analysis. Furthermore, the M. xanthus DZF1 genome (approximately 9,500 kbp) is approximately twice the size of the Escherichia coli genome (4). To establish optimal conditions for 2D gel electrophoresis of M. xanthus total cellular proteins, we improved the methods for sample preparation, electrophoresis, and detection of proteins for subsequent sequencing on the basis of the methods described by Gorg et al. (8, 9). For sample preparation, to prevent high proteolytic activity, M. xanthus cells were lysed in lysis buffer containing 7 M urea and 2 M thiourea as denaturants and 5% CHAPS as a detergent and immediately sonicated to disrupt cells. For the first-dimension isoelectric focusing gel, electrophoresis was conducted initially at 500 V for 2 h, and the voltage was increased to 3,500 V in a stepwise manner, as described in Materials and Methods. Note that the initial step at 500 V is important to obtain high resolution for the isoelectric focusing. For 2D SDS-polyacrylamide gel electrophoresis, electrophoresis was carried out at a low current (5 mA/gel) until the dye migrated 2 cm from the top of the gel. This procedure allowed us to avoid using stacking gels and to obtain clear spots for higher-molecular-weight proteins. Proteins were detected by CBB staining, which allowed us to analyze protein patterns quantitatively and reproducibly.
Analysis of protein patterns induced by heat shock.
M. xanthus cells growing exponentially at 30°C were heat shocked by shifting to 42°C. Cells were harvested 0, 15, 30, and 60 min after heat shock. Significant changes in protein patterns were observed for heat-shocked cells at 30 and 60 min after the temperature shift. Since there were many proteins at about pI 5, strips with a nonlinear pH gradient from 3 to 10 were used for the first-dimension isoelectric focusing gel.
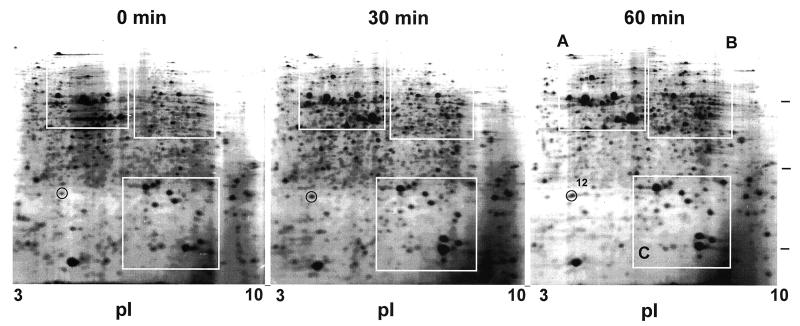
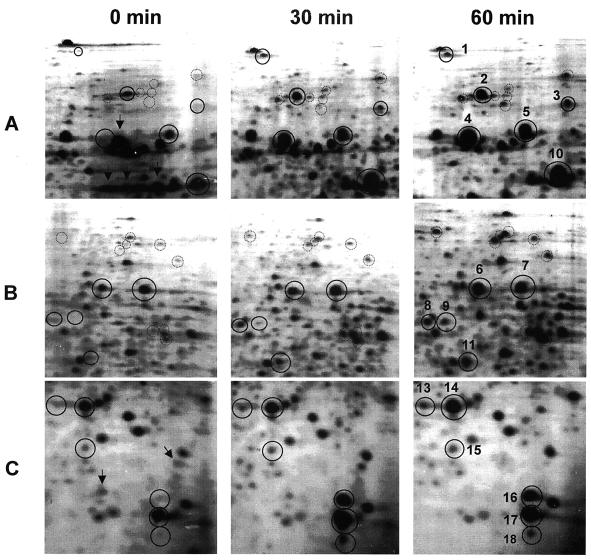
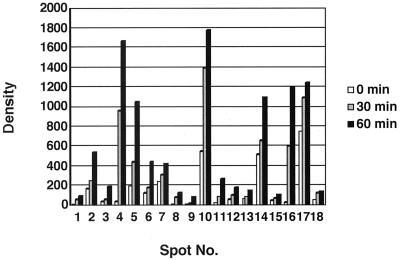
As shown in Fig. 1, about 1,000 to 1,200 protein spots were detected in each gel. It is known to be difficult to separate basic proteins with a first-dimension isoelectric focusing gel because of horizontal streaking due to incomplete focusing. Under the conditions described above, both basic and acidic proteins were well resolved as round spots. When these patterns were compared with those of E. coli in the SWISS-2DPAGE database (22), a majority of M. xanthus proteins were basic and had high molecular weights, in contrast to E. coli proteins, whose pIs were mostly less than 6. Three regions in Fig. 1 were enlarged and depicted in Fig. 2. At least 18 major spots, except for spot 12, circled and numbered in these regions, were found to be heat shock inducible. Spot 12 was detected in an acidic region in Fig. 1. Their expression levels were estimated by measuring the densities of individual spots (Fig. 3). Among them, spots 2, 5, 6, 7, 10, 12, 14, and 17 were also present before heat shock. Their molecular weights and pIs are shown in Table 1.
FIG. 1.
2D gel electrophoresis of M. xanthus total proteins before and after heat shock. Spot 12 is circled on the gels. The other heat shock-induced spots in boxes A, B, and C in the 60-min gel are assigned in Fig. 2. Bars at the right are positions of molecular mass markers, 66, 29, and 14 kDa, from the top.
FIG. 2.
Enlarged areas of Fig. 1. Heat-shock-induced spots used for determination of the N-terminal sequences are circled with a solid line, and other, minor heat-shock-induced spots are circled with a dotted line. Arrows indicate spots suppressed by heat shock.
FIG. 3.
Induction of HSPs in M. xanthus. The density of each spot was measured with Imagemaster and Gel LABII version 2.0.
TABLE 1.
Identification of heat-shock-induced proteins of M. xanthus
| Spot | MW (103) | pI | N-terminal sequencea | Proteinb |
|---|---|---|---|---|
| 1 | 81 | 5.4 | ND | |
| 2 | 71 | 5.7 | GKIIGIDLGTTNSVVAIMEGREPSVIVNPE | DnaK |
| 3 | 68 | 6.4 | ND | |
| 4 | 60 | 5.7 | AAKEIFFHQSAREAILRGVRTLSDA | GroEL1 |
| 5 | 62 | 6.1 | AKDILFDVRAREAILRGVNILADAVKVTLGPK | GroEL2 |
| 6 | 62 | 6.9 | MIYAAPNQPGSKVQFKPRYQNFIGG | AldA |
| 7 | 62 | 7.3 | MEIRADEISRIIREQIXDYGKXVT | AtpA |
| 8 | 53 | 6.6 | KRGNIVLQXRVLLEKQREEMDXL | Unknown |
| 9 | 53 | 6.7 | GNKKRGNIVLXRXVLLEKQXE | Unknown |
| 10 | 50 | 6.3 | ND | |
| 11 | 45 | 6.8 | MKIHEYQGKEIFRKYGVPTPKGIL | SucC |
| 12 | 25 | 5.4 | AETQAATRLREYETIFLVKPDLTDDNVDKL | RpsF |
| 13 | 26 | 6.5 | MLTVGDKIPNFKVKAXVSLEKGKEFQQHTN | AhpC |
| 14 | 26 | 6.7 | MLTVGDKIPNFKVKATVSLEKGKEFQQXTN | AhpC |
| 15 | 22 | 6.7 | MLTVGDKIPNFKVKATVSLEK | AhpC |
| 16 | 17 | 7.5 | MQTRNPFNSAVVVNPLMRDVDALFRELTQPPLRIA | Unknown |
| 17 | 16 | 7.5 | MKIRPLQDRLIVKRVAEENKTKGGL | GroES |
| 18 | 14 | 7.5 | MNRALQITYRGMESQEALNE | Unknown |
ND, not determined. X, unidentified amino acid residue.
Abbreviations: AldA, aldehyde dehydrogenase; AtpA, ATP synthase α subunit; SucC, succinyl CoA synthetase; RpsF, 30S ribosomal protein S6; AhpC, alkyl hydroperoxide reductase.
Identification of proteins.
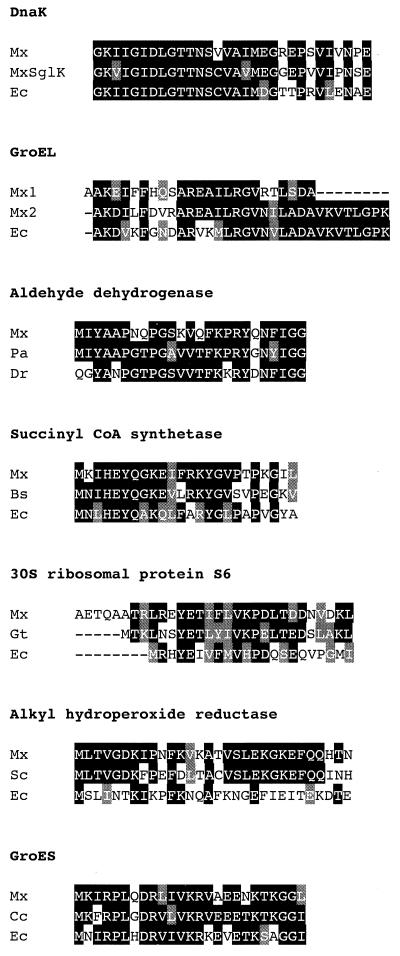
To identify the proteins, microsequence analysis was performed as described in Materials and Methods. The N-terminal sequences determined for 15 spots are shown in Table 1. The N-terminal sequences of spots 1, 3, and 10 could not be determined. Among these three spots, the N-terminal end of the spot 10 protein was considered to be blocked, as judged from the amount of the protein. For the 15 spots, 11 proteins were identified by their high homologies to known proteins (Table 1 and Fig. 4). Spot 2 shows high homology to DnaK. A DnaK homologue, SglK, has been identified in M. xanthus (25, 28). However, DnaK identified in this study was distinctly different from SglK because there were seven substitutions within the N-terminal 30-residue sequence (Fig. 4). Apparently, M. xanthus contains at least two DnaK homologues, one an HSP and the other a non-HSP. Both spots 4 and 5 exhibit high homology to GroEL. Since there are eight substitutions within the N-terminal 24-residue sequence between them, spots 4 and 5 represent two different GroEL homologues. Spots 13, 14, and 15 show high homology to alkyl hydroperoxide reductase. Because all three spots have the same N-terminal sequence, spot 13 may be shifted to the acidic side by posttranslational modification and spot 15 may be a degraded product lacking part of the C terminus. Spot 7 has the same N-terminal sequence as the ATP synthase α subunit identified by Munoz-Dorado et al. (15). Spots 6, 11, 12, and 17 show high homology to aldehyde dehydrogenase, succinyl CoA synthetase, 30S ribosomal protein S6, and GroES, respectively. The sequences determined for spots 8, 9, 16, and 18 did not show significant homology to known proteins.
FIG. 4.
Sequence alignments of HSPs identified in this study and those of other bacteria. Identical amino acids are shaded in black, and conservative changes are shaded in gray. Bs, B. subtilis; Cc, Caulobacter crescentus; Dr, Deinococcus radiodurans; Ec, E. coli; Gt, Guillardia theta; Mx, M. xanthus; Pa, Pseudomonas aeruginosa; Sc, Streptomyces coelicolor. Accession numbers (GenBank) for the proteins shown are as follows: SglK from Mx, U83800; Dnak from Ec, K01298; GroEL from Ec, U14003; aldehyde dehydrogenase from Pa, AE004625; aldehyde dehydrogenase from Dr, AE001863; alkylhydroperoxide reductase from Sc, AL391754; alkyl hydroperoxide reductase from Ec, D13187; succinyl CoA synthetase from Bs, AJ000975; succinyl CoA synthetase from Ec, D90711; 30S ribosomal protein S6 from Gt, AF041468; and 30S ribosomal protein S6 from Ec, L41394; GroES from Cc, L41394; GroES from Ec, X07899.
DISCUSSION
The improved method for 2D gel analysis described here enables us to separate M. xanthus total cellular proteins at high resolution. Notably, proteins in both acidic and basic regions are detected as distinct round spots, and these protein patterns are highly reproducible. After transfer to the polyvinylidene difluoride membrane, spots of interest from one gel were sufficient for the determination of N-terminal sequences by microsequencing.
Proteins induced by heat shock have been shown to play an essential role in bacterial physiology under heat shock stress. In particular, GroEL (26), GroES (27), DnaK (12), and DnaJ (12) of E. coli are well known for their roles in protein folding as molecular chaperones. In E. coli, the heat shock response is known to be controlled by the heat shock sigma factors ς32 (RpoH) and ς24 (RpoE) (29). Interestingly, in M. xanthus, three sigma factors, SigB, SigC, and SigE, homologous to ς32 have been identified; however, none of them has been found to be essential for the induction of HSPs (23). Regulation of the heat shock response in M. xanthus is not clear at present.
In this study, at least 18 major spots were found to be induced by heat shock in vegetatively growing M. xanthus cells by 2D gel analysis. Subsequently, we determined the N-terminal sequences of the proteins extracted from 15 spots. Among them, 11 spots were identified as known proteins; 3 of these spots were found to be the same protein (Table 1). In addition to the 18 major spots, there were at least 15 minor spots (Fig. 2A and B). Therefore, M. xanthus contains more than 30 HSPs induced by heat shock during vegetative growth. It is also interesting that several proteins were repressed by heat shock (Fig. 2A and C). Although Nelson and Killeen reported that 18 major HSPs were induced in M. xanthus cells (17), it is difficult to match our spots to their 18 HSPs because of subtle differences in molecular masses.
It has been reported that a DnaK homologue (SglK) which is essential for FB development is induced during FB formation but not by heat shock in M. xanthus (25, 28). DnaK identified here may be also involved in FB development in M. xanthus, because it was also induced during the initiation of FB formation (data not shown). It is interesting that M. xanthus GroEL1 (spot 4) was induced earlier than GroEL2 (spot 5) after heat shock. Therefore, it is possible that GroEL1 and GroEL2 have different roles in heat shock adaptation in M. xanthus. In addition, alkyl hydroperoxide reductase, aldehyde dehydrogenase, succinyl CoA synthetase, 30S ribosomal protein S6, and ATP synthase α subunit were also identified as HSPs in M. xanthus.
Alkyl hydroperoxide reductase of Bacillus subtilis was proposed to be involved in the detoxication of organic hydroperoxides, which are produced from unsaturated fatty acids and nucleic acids under oxidative stress conditions (1). Its subunits, AhpC and AhpF, are induced not only under oxidative stress but also under heat or salt stress or glucose starvation. These results indicate that alkyl hydroperoxide reductase plays an important role under various stress conditions. Aldehyde dehydrogenase is known to be induced by a variety of stress conditions, including heat shock in Saccharomyces cerevisiae (16). 30S ribosomal protein S6 has been identified as a cold shock protein in E. coli (18) and B. subtilis (11), suggesting that 30S ribosomal protein S6 may play a unique role in sensing temperature differences to control ribosome function. Succinyl CoA synthetase is involved in the citric acid cycle and catalyzes a reaction from succinate to succinyl CoA, which is an important intermediate substance in the synthesis of various compounds. Therefore, the induction of succinyl CoA synthetase is necessary for the synthesis of the compounds required for heat shock adaptation in M. xanthus. ATP-dependent proteases, such as Lon and Clp proteases in E. coli (7, 10) and other bacteria, are known to be induced by heat shock (2). In M. xanthus, LonD, an ATP-dependent protease essential for development, has been identified as an HSP (T. Ueki and S. Inouye, unpublished data). The observed heat shock induction of ATP synthase α subunit thus may be important not only for ATP-dependent proteases but also for the synthesis of various macromolecules requiring ATP.
For the analysis of global changes in protein expression induced by various stress conditions, 2D gel electrophoresis is a powerful method. Since FB formation and sporulation in M. xanthus have been shown to be accelerated by heat shock treatment prior to FB formation (13), some of the heat-shock-induced proteins may be involved in FB formation and sporulation. In the present study, we established optimal conditions for analyzing M. xanthus HSPs. By use of the improved 2D gel electrophoresis method, it is possible to identify specific proteins induced by other stress conditions and during FB and spore formation. Identification of these proteins will provide important insights into their functions in growth under stress conditions and during M. xanthus development.
ACKNOWLEDGMENTS
We are grateful to M. Inouye for discussions and critical reading of the manuscript.
This work was supported by a grant from the Foundation of the University of Medicine and Dentistry of New Jersey.
REFERENCES
- 1.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress response in Bacillus subtilis: cloning, expression, and mutation of alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernhardt J, Volker U, Volker A, Antelmann H, Schmid R, Mach H, Heker M. Specific and general stress proteins in Bacillus subtilis—a two-dimensional protein electrophoresis study. Microbiology. 1997;143:999–1017. doi: 10.1099/00221287-143-3-999. [DOI] [PubMed] [Google Scholar]
- 3.Campos J M, Geisselsoder J, Zusman D R. Isolation of bacteriophage M×4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Keseler I M, Shimkets L J. Genome size of Myxococcus xanthus determined by pulsed-field gel electrophoresis. J Bacteriol. 1990;172:4206–4213. doi: 10.1128/jb.172.8.4206-4213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dworkin M, Kaiser D. Myxobacteria II. Washington, D.C.: ASM Press; 1993. [Google Scholar]
- 6.Dworkin M. Recent advances in the social and developmental biology of myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gayad R C, Stephens P E, Hewick R, Schoemaker J M, Dreyer W J, Markovitz A. Regulatory region of the heat-shock-inducible capR (lon) gene: DNA and protein sequences. J Bacteriol. 1985;162:271–275. doi: 10.1128/jb.162.1.271-275.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorg A, Obermaier C, Boguth G, Csordas A, Diaz J J, Madjar J J. Very alkaline immobilized pH gradients for two-dimensional electrophoresis of ribosomal and nuclear proteins. Electrophoresis. 1997;18:328–337. doi: 10.1002/elps.1150180306. [DOI] [PubMed] [Google Scholar]
- 9.Gorg A, Obermaier C, Boguth G, Weiss W. Recent developments in two dimensional electrophoresis with immobilized pH gradients: wide pH gradients up to pH 12, longer separation distances and simplified procedures. Electrophoresis. 1999;20:712–717. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<712::AID-ELPS712>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S, Clark W P, de Crecy-Lagard V, Maurizi M R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993;268:22618–22626. [PubMed] [Google Scholar]
- 11.Graumann P, Schroder K, Schmid R, Marahiel M A. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol. 1996;178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrick J P, Hartl F U. Molecular chaperone function of heat shock protein. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 13.Killeen K P, Nelson D R. Acceleration of starvation- and glycerol-induced myxospore formation by prior heat shock in Myxococcus xanthus. J Bacteriol. 1988;170:5200–5207. doi: 10.1128/jb.170.11.5200-5207.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindquist S, Craig E A. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 15.Munoz-Dorado J, Inouye S, Inouye M. Identification of the Myxococcus xanthus 59-kDa membrane-associated GTP-binding protein as a proton-translocating ATPase. Gene. 1994;138:133–137. doi: 10.1016/0378-1119(94)90795-1. [DOI] [PubMed] [Google Scholar]
- 16.Navarro-Avino J P, Prasad R, Miralles V J, Benito R M, Serrano R. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast. 1999;15:829–842. doi: 10.1002/(SICI)1097-0061(199907)15:10A<829::AID-YEA423>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Nelson D R, Killeen K P. Heat shock proteins of vegetative and fruiting Myxococcus xanthus cells. J Bacteriol. 1986;168:1100–1106. doi: 10.1128/jb.168.3.1100-1106.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phadtare S, Yamanaka K, Inouye M. The cold shock response. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 35–45. [Google Scholar]
- 19.Rince A, Flahaut S, Auffray Y. Identification of general stress genes in Enterococcus faecalis. Int J Food Microbiol. 2000;55:87–91. doi: 10.1016/s0168-1605(00)00180-x. [DOI] [PubMed] [Google Scholar]
- 20.Svensater G, Sjogreen B, Hamilton I R. Multiple stress responses in Streptococcus mutants and the induction of general and stress-specific proteins. Microbiology. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- 21.Teixeira-Gomes A P, Cloechaert A, Zygmunt M S. Characterization of heat, oxidative, and acid stress responses in Brucella melitensis. Infect Immun. 2000;68:2954–2961. doi: 10.1128/iai.68.5.2954-2961.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonella L, Walsh B J, Sanchez J C, Ou K, Wlikins M R, Tyler M, Frutiger S, Gppley A A, Pescaru I, Appel R D, Yan J X, Bairoch A, Hoogland C, Morch F S, Hughes G J, Williams K L, Hochstrasser D F. '98 Escherichia coli SWISS-2DPAGE database update. Electrophoresis. 1998;19:1960–1971. doi: 10.1002/elps.1150191114. [DOI] [PubMed] [Google Scholar]
- 23.Ueki T, Inouye S. SigB, SigC, and SigE from Myxococcus xanthus homologous to ς32 are not required for heat shock response but for multicellular differentiation. J Mol Microbiol Biotechnol. 2001;3:287–293. [PubMed] [Google Scholar]
- 24.Volker U, Engelmann S, Maul B, Riethdorf S, Volker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 25.Weimer R M, Creghton C, Stassinopoulos A, Youderian P, Hartzell P L. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissman J S, Hohl C M, Kovalenko O, Kashi Y, Chen S, Braig K, Saibil H R, Fenton W A, Horwich A L. Mechanism of GroEL action: productive release of polypeptide from a sequestered position under GroES. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- 27.Weissman J S, Rye H S, Fenton W A, Beechem J M, Horwich A L. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- 28.Yang Z, Geng Y, Shi W. A DnaK homolog in Myxococcus xanthus is involved in social motility and fruiting body formation. J Bacteriol. 1998;180:218–224. doi: 10.1128/jb.180.2.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yura T, Kanemori M, Morita M T. The heat shock response: regulation and function. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 3–18. [Google Scholar]