Abstract
Schizophrenia is a heterogeneous psychiatric disorder with a strong genetic basis, whose etiology and pathophysiology remain poorly understood. Exome sequencing studies have uncovered rare, loss-of-function variants that greatly increase risk of schizophrenia [1], including loss-of-function mutations in GRIN2A (aka GluN2A or NR2A, encoding the NMDA receptor subunit 2A) and AKAP11 (A-Kinase Anchoring Protein 11). AKAP11 and GRIN2A mutations are also associated with bipolar disorder [2], and epilepsy and developmental delay/intellectual disability [1, 3, 4], respectively. Accessible in both humans and rodents, electroencephalogram (EEG) recordings offer a window into brain activity and display abnormal features in schizophrenia patients. Does loss of Grin2a or Akap11 in mice also result in EEG abnormalities? We monitored EEG in heterozygous and homozygous knockout Grin2a and Akap11 mutant mice compared with their wild-type littermates, at 3- and 6-months of age, across the sleep/wake cycle and during auditory stimulation protocols. Grin2a and Akap11 mutants exhibited increased resting gamma power, attenuated auditory steady-state responses (ASSR) at gamma frequencies, and reduced responses to unexpected auditory stimuli during mismatch negativity (MMN) tests. Sleep spindle density was reduced in a gene dose-dependent manner in Akap11 mutants, whereas Grin2a mutants showed increased sleep spindle density. The EEG phenotypes of Grin2a and Akap11 mutant mice show a variety of abnormal features that overlap considerably with human schizophrenia patients, reflecting systems-level changes caused by Grin2a and Akap11 deficiency. These neurophysiologic findings further substantiate Grin2a and Akap11 mutants as genetic models of schizophrenia and identify potential biomarkers for stratification of schizophrenia patients.
Subject terms: Neuroscience, Schizophrenia, Physiology
Introduction
A severe psychiatric disorder affecting approximately 0.5% of the global population, schizophrenia is characterized by hallucinations, delusions, disorganized thoughts and behavior, social withdrawal, reduced emotional expression, and cognitive deficits. While genome-wide association studies (GWAS) have identified many genetic loci associated with schizophrenia, it is often difficult to identify the causal gene or to interpret the biological effect of the GWAS common variants, which are mainly non-coding, challenging to fine-map, and associated with only small increases in disease risk (odds ratio typically around 1.1) [5–7]. Compared with GWAS, the rare copy number variants that are associated with schizophrenia confer much higher risk but affect large regions of the genome, making it difficult to identify the relevant pathogenic gene(s) [7, 8]. As a result, the field has been hampered by lack of specific genetic models that can be brought to bear on the mechanisms of schizophrenia.
Recently, the discovery of schizophrenia risk genes has been enhanced by large-scale exome or genome sequencing of tens of thousands of cases versus controls. Such studies have the power to uncover rare loss-of-function coding variants (such as protein-truncating variants, PTVs) that have a large impact on schizophrenia risk [1, 9, 10]. In one of the largest sequencing studies to date of 24,248 cases and 97,322 controls, the Schizophrenia Exome Sequencing Meta-analysis Consortium (SCHEMA) has identified multiple rare loss-of-function genetic variants at exome-wide level of significance that confer substantial disease risk (“SCHEMA genes”; odds ratios in the range of 4-50) [1]. Because these rare variants are often PTVs occurring upstream of the last exon junction, which are predicted to lead to complete loss of function (i.e., null mutations, which presumably lead to haploinsufficiency, rather than missense mutations resulting in a reduced protein function, which presumably do not) [1, 11–14], these disease-causing mutations can be easily modeled by genetic disruption (‘knockout’) in animals such as the mouse. Moreover, these null mutations can be studied in homozygous as well as heterozygous states, the latter being more relevant to the human disease, where only one of the alleles is disrupted in patients. By systematically analyzing the phenotypes of mouse lines bearing loss-of-function mutations in SCHEMA genes (SCHEMA mouse mutants), we hope to discover convergent molecular and neurobiological mechanisms and identify potential biomarker signatures caused by these relatively highly penetrant mutations. Insights derived from these genetic animal models of schizophrenia should further our understanding of the biology of schizophrenia and could aid our efforts to develop more effective treatments [15]. In this study, we characterize by chronic electroencephalogram (EEG) the changes in brain activity caused by mutations in two SCHEMA genes, GRIN2A and AKAP11.
One of ten genes reaching exome-wide significance [1], GRIN2A (glutamate ionotropic receptor NMDA type subunit 2A) is a particularly compelling SCHEMA gene for further study because it is also a significant GWAS hit for schizophrenia [5]. GRIN2A encodes the 2A subunit of the N-methyl-D-aspartate (NMDA) receptor (also known as GluN2A or NR2A), a protein that is highly expressed in neurons in the brain and localized at postsynaptic sites of glutamatergic synapses [16, 17]. Reduced NMDA receptor function has long been proposed as a pathophysiologic mechanism underlying schizophrenia, in part because NMDA receptor antagonists at low concentrations can induce psychosis-like symptoms in humans [18]. Interestingly, GRIN2A is expressed later in brain development than the genes encoding the GRIN1 and GRIN2B subunits of the NMDA receptor; expression of GRIN2A starts postnatally and rises through juvenile and adolescent stages in humans and rodents, inviting comparison with the typical onset of schizophrenia in adolescence and early adulthood [19–21].
AKAP11 (A-Kinase Anchoring Protein 11) has been identified as a rare-variant large-effect risk gene for schizophrenia, ranking #12 in the recent SCHEMA exome meta-analysis (https://schema.broadinstitute.org/) [1]. In addition, AKAP11 is a bipolar disorder risk gene, recently identified as the top hit from Bipolar Exome (BipEx) sequencing studies of 13933 bipolar cases and 14422 controls [2]. Thus, AKAP11 is a shared risk gene for schizophrenia and bipolar disorder, underscoring the genetic overlap between these two disorders on the psychosis spectrum. AKAP11 (formerly known as AKAP220) encodes an A-kinase anchoring protein that is expressed broadly in the body, including in neurons in the brain. Biochemically, AKAP11 interacts with protein kinase A (PKA), a protein that is involved in a variety of biological processes including neuronal plasticity, and with glycogen synthase kinase 3 beta (GSK3β), one of the targets of lithium, a mainstay therapy for bipolar disorder [22–24]. However, AKAP11’s function in the brain and its biological role in psychiatric disease are uncharacterized.
Here we investigate how GRIN2A and AKAP11 loss-of-function affect brain activity by monitoring EEG in mutant mice deficient in these genes. EEG provides a non-invasive, high temporal-resolution, systems-level readout of neural activity in vivo, a neurophysiologic assay that is clinically translatable to human patients. The present work provides an in-depth characterization of EEG phenotypes across different behavioral states (sleep, wake, auditory stimulation) and ages (3 and 6 months) in Grin2a and Akap11 heterozygous (Het) and homozygous knockout (KO) mice, as compared with their wild-type littermates (WT). We found that Grin2a and Akap11 mutant mice possess several EEG features shared with human schizophrenia and bipolar disorder patients, including elevated gamma oscillations at rest [25, 26], attenuated auditory steady-state responses (ASSR) at gamma frequencies [27, 28], and changes in sleep spindle density [29, 30]. These mouse EEG phenotypes reveal systems-level abnormalities caused by (even heterozygous) mutations in Grin2a and Akap11, and provide further justification, beyond their genetic validity, that these mouse mutants can serve as useful animal models of schizophrenia/bipolar disorder.
Materials and methods
Animals
All experiments were approved by the Broad Institute IACUC (Institutional Animal Care and Use Committee) and conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals. Mice were housed at AAALAC-approved facilities on a 12-hour light/dark cycle, with food and water available ad libitum. Grin2a (B6;129S-Grin2a < tm1Nak>; RBRC02256, Riken BioResource Center, Saitama, Japan; MGI: 1928506) and Akap11 (B6.Cg-Akap11[tm1.2Jsco/J]; #028922, Jackson Laboratory, Bar Harbor, ME; MGI:5751858) mutant mice on the C57BL/6J background were originally generated as described [31, 32]. Grin2a and Akap11 heterozygous breeding pairs were generated in-house by crossing homozygous KO with C57BL/6J wild-type mice (#000664, Jackson Laboratory). The resulting heterozygous breeding pairs were used to generate Grin2a and Akap11−/− (homozygous knockout, KO) and +/− (heterozygous, Het) mice and their wild-type (+/+, WT) littermates. Adult male and female Grin2a mice and male Akap11 mice were used for all experiments (open field testing: n = 12 Grin2a WT [6 male, 6 female], n = 13 Grin2a Het [7 male, 6 female], n = 11 Grin2a KO [5 male, 6 female]; n = 11 Akap11 WT, n = 10 Akap11 Het, n = 10 Akap11 KO; EEG recording: n = 12 Grin2a WT [6 male, 6 female], n = 12 Grin2a Het [4 male, 8 female], n = 12 Grin2a KO [11 male, 1 female]; n = 11 Akap11 WT, n = 12 Akap11 Het, n = 12 Akap11 KO). The sample size (n~10–12 mice/group) was estimated based on statistical power calculations for mouse disease models that produce mild phenotypes (e.g., 50% change in EEG power). No randomization was used to allocate animals to experimental groups. 1 male Grin2a WT mouse was removed from sleep/wake and MMN analyses at 6 months due to damaged EEG electrodes. Open field behavior and auditory EEG experiments were conducted during the light phase of the daily cycle. All experiments and analyses were conducted by investigators who were blinded to the mouse genotype.
Open field testing
11- to 14-week-old Grin2a and Akap11 mutant mice and their WT littermates (n = 10–13 animals/group) were monitored using the SuperFlex Open Field system (40 cm x 40 cm x 40 cm; Omnitech Electronics, Inc., Columbus, OH) for 60 minutes. The animals’ position was captured in real-time using Fusion system software (Omnitech Electronics, Inc.).
EEG implantation surgery
6- to 12-week-old mice (n = 11–12 mice/group) were deeply anesthetized with isoflurane. A prefabricated EEG/EMG headmount (#8201-SS, Pinnacle Technology, Lawrence, KS) was secured to the skull with four 0.10” intracranial electrode screws (#8403, Pinnacle Technology) at the following stereotactic coordinates: frontal recording electrode (+1.5 AP, 1.5 ML to Bregma), parietal recording electrode (-2 AP, 1.5 ML to Bregma), ground and reference electrodes (bilaterally −1 AP, 2 ML to Lambda). The electromyogram (EMG) electrodes were placed bilaterally in the nuchal muscles. Electrodes were soldered to the EEG/EMG headmount and dental acrylic was used to secure the connections. Animals were given at least one week of post-operative recovery before EEG recording.
EEG recording
Following recovery from EEG implantation, mice were tethered to the Pinnacle recording system, with at least 3 hours of habituation before testing. Experiments were conducted over a period of 4–5 days. EEG/EMG signals were recorded in freely moving mice across 24 hours of sleep/wake, followed by ASSR and mismatch negativity (MMN) testing (see Supplementary Information for details). Animals remained tethered to the Pinnacle system throughout the testing period with ad libitum access to food and water. All signals were digitized at a sampling rate of 1000 Hz, filtered (1–100 Hz bandpass for EEG; 10–1 kHz bandpass for EMG), and acquired using the Sirenia Acquisition program (Pinnacle Technology). EEG recordings took place at two time points, roughly corresponding to 3- and 6-months of age (mouse age at the time of Recording 1: 8–17 weeks, Recording 2: 22-33 weeks). Mice were returned to their home cage between recording sessions.
EEG analysis
Sleep state classification, power, sleep spindles, ASSR, and MMN were analyzed as described in the Supplementary Information. All analyses shown were conducted using data from the frontal EEG electrode.
Statistical analysis
All comparisons across genotypes (WT vs. Het vs. KO) were conducted using one-way ANOVAs followed by Tukey post-hoc pairwise comparisons, unless otherwise specified. Variance was similar across groups that were statistically compared.
Results
Grin2a and Akap11 mutants exhibit increased and decreased locomotor activity, respectively, in the open field test
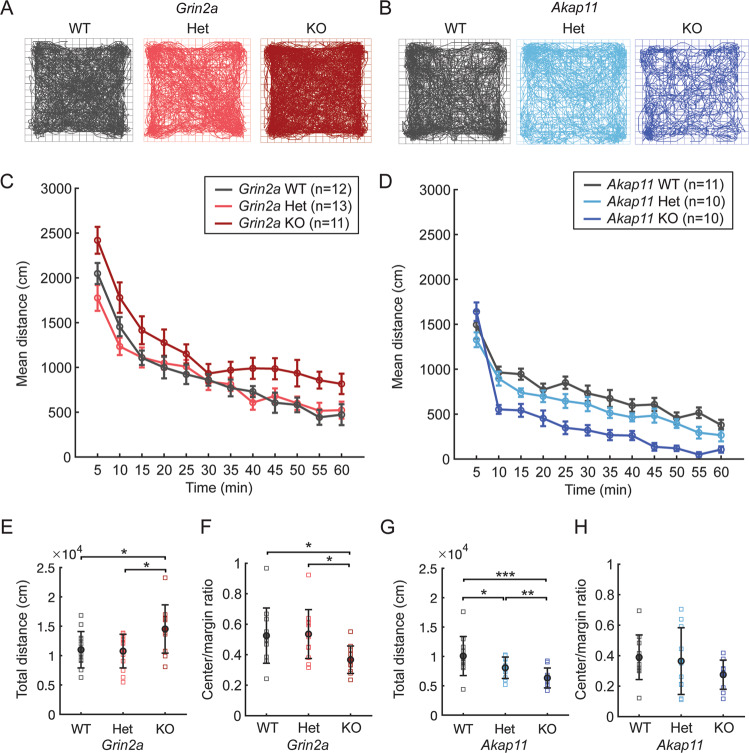
The open-field test is frequently used to measure general locomotor activity and infer anxiety-like behavior in rodents [33, 34]. At 3 months of age, Grin2a KO mice showed increased locomotor activity in the open field (Fig. 1A, C, E; ~25% increase from WT; p = 0.033), consistent with previous reports [35–37]. Grin2a+/− (Het) mice exhibited similar activity levels as WT littermates. In contrast, Akap11 Het (p = 0.0337) and KO (p = 7.49e-07) showed reduced locomotor activity ~by 25% and 50%, respectively, compared to their WT littermates (Fig. 1B, D, G), consistent with previous findings [32]. Grin2a KO (but not Het) mice had reduced ratios of center/margin distance traveled (by ~30%) relative to WT animals (p = 0.0438), suggesting anxiety-like behavior (Fig. 1F). Akap11 Het and KO had similar center/margin ratios as WT littermates (Fig. 1H).
Fig. 1. Locomotor behavior of Grin2a and Akap11 mutant mice in open field test.
A, B Representative traces of mouse movements (60-minute test). C, D Mean distance traveled, binned into 5-minute segments. E–H Total distance traveled and ratio of center/margin distance in Grin2a (left) and Akap11 (right) mutants in comparison to WT littermates. Error bars denote mean ± standard error; *p < 0.05, **p < 0.01, ***p < 0.001.
Akap11−/− mice exhibit NREM sleep deficits
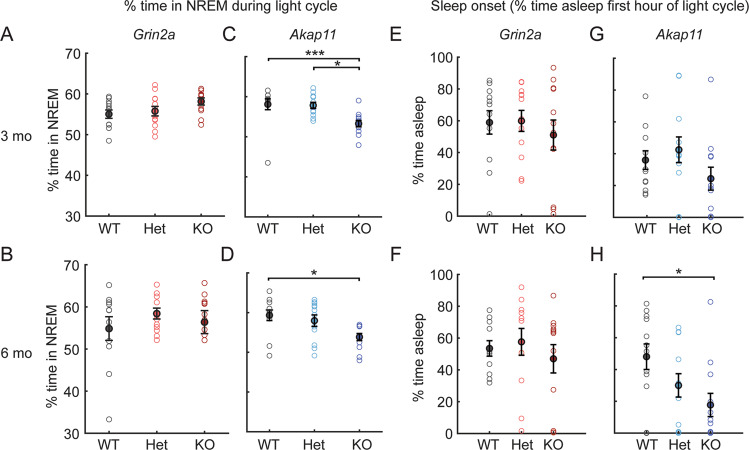
Patients with schizophrenia often show sleep abnormalities, including reduced sleep, increased sleep, delayed sleep onset, or fragmented sleep [38, 39]; altered sleep patterns (e.g., more sleep in depressive phase, less sleep in manic phase) are also found in bipolar disorder [40]. To assess sleep differences in Grin2a and Akap11 mutant mice, we recorded EEG/EMG signals across 24 hours and used a feature-based model (see Supplementary Information) to classify periods of NREM, REM, and wake. Sleep analyses were conducted during the light cycle when mice are predominantly asleep. Grin2a heterozygous and homozygous mutant mice exhibited similar sleep patterns as their WT littermates (Fig. 2A–B). Akap11-/- mice exhibited reduced (~10%) NREM sleep relative to WT littermates at 3 months (p = 0.0052) and 6 months of age (p = 0.0106), whereas Akap11+/− animals were not significantly different than WT (Fig. 2C,D). At 6-months of age, Akap11−/− mice also showed delayed sleep onset at the start of the light cycle, relative to WT littermates (p = 0.0207, Fig. 2E–H). Neither Grin2a or Akap11 mutants (heterozygous or homozygous) displayed abnormal sleep fragmentation (Figure S1), although NREM bout length was reduced in Akap11 KO (p = 0.0478) compared with WT littermates (Figure S1G).
Fig. 2. NREM sleep and sleep onset time of Grin2a and Akap11 mutant mice.
A–D % of time spent in NREM sleep during the light cycle in 3- and 6-month Grin2a and Akap11 mutants. E–H Sleep onset, as quantified by the % of time spent asleep in the first hour of the light cycle. Error bars denote mean ± standard error; p < 0.05, **p < 0.01, ***p < 0.001; n = 11–12 mice/group.
Elevated resting gamma power in Grin2a and Akap11 mutant mice
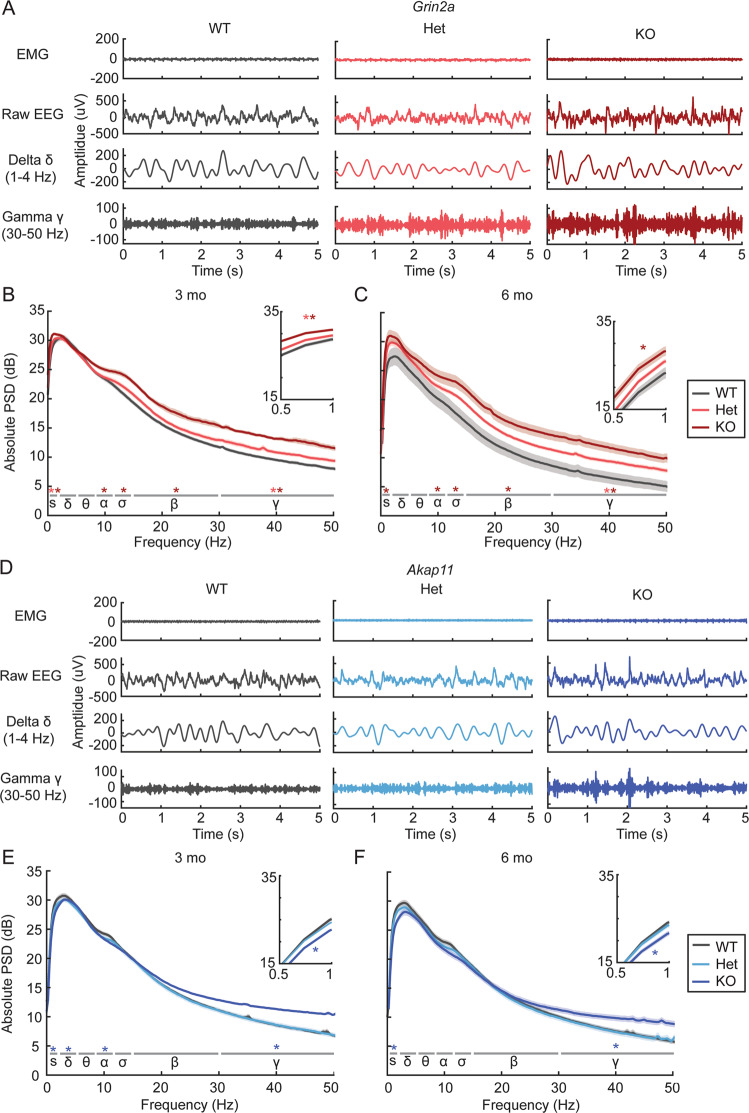
Many patients with schizophrenia exhibit altered EEG oscillatory power at rest (e.g., during sleep or in the absence of a task) especially increased gamma oscillations [41, 42]. Focusing our analysis on NREM sleep, where EEG signals are less prone to movement-related variation, we measured absolute power of brain oscillations in Grin2a and Akap11 mutants versus their WT littermates for each frequency band (slow: 0.5–1 Hz, delta: 1–4 Hz, alpha: 8–12 Hz, sigma: 12–15 Hz, beta: 15–30 Hz, gamma: 30–50 Hz). Grin2a+/− and −/− mice exhibited gene dose-dependent increases in gamma oscillations (Fig. 3A–C, S2): a 10% and ~20% increase in absolute gamma power compared with WT for heterozygous and homozygous mutants, respectively (Het, 3 months: p = 0.0187; Het, 6 months: p = 0.0385; KO, 3 months: p = 1.07e−08; KO, 6 months: p = 1.15e-04). Resting gamma power was also increased by ~10% in Akap11−/− mice at 3 months (p = 1.80e-04) and 6 months (p = 0.0112) of age, but unlike Grin2a heterozygotes, Akap11 heterozygous mutants were not significantly different from WT (Fig. 3D–F, S2). Most observed increases in resting gamma power were associated with increased spectral entropy and/or a reduced aperiodic exponent (“flatter” PSD) in the parameterized power spectrum (Table S1). Besides NREM, elevated gamma power was also found during REM sleep (Figure S3, S4) and quiet wake (Figure S5) in Grin2a and Akap11 homozygous mutants. Grin2a (but not Akap11) heterozygous mutants exhibited an intermediate phenotype of increased gamma oscillations during REM sleep (Figure S3), although this result did not reach statistical significance (3 months: p = 0.190; 6 months: p = 0.111)
Fig. 3. Power spectral analysis of brain oscillations in Grin2a and Akap11 mutant mice during NREM sleep.
A Representative EMG and EEG (raw, delta- and gamma-filtered) traces from Grin2a WT, Het, and KO animals during NREM sleep. B, C Absolute NREM power spectra for 3- and 6-month Grin2a mutants during the light cycle. Light and dark red/blue asterisks indicate oscillations for which +/− and −/− mice, respectively, differed significantly from WT littermates (p < 0.05) in each of the frequency bands: slow (s), 0.5–1 Hz; delta (δ), 1–4 Hz; theta (θ), 4–8 Hz; alpha (α), 8–12 Hz; sigma (σ), 12–15 Hz; beta (β), 15–30 Hz; gamma (γ), 30–50 Hz. Shading indicates mean ± standard error. Insets show magnified view of the graph in the slow oscillation range (0.5–1 Hz). n = 11–12 mice/group. D Representative EMG and EEG traces from Akap11 WT, Het and KO animals during NREM sleep. E, F Absolute NREM power spectra for 3- and 6-month Akap11 mutants during the light cycle.
We found changes in oscillation power outside of gamma frequencies. The EEGs of Grin2a KO exhibited increased power broadly across multiple frequency bands, including slow oscillations (3 months: p = 8.12e−08; 6 months: p = 0.0023), alpha (3 months: p = 0.0252; 6 months: p = 0.0357), sigma (3 months: p = 4.00e−05; 6 months: p = 0.0030), and beta (3 months: p = 2.71e−06; 6 months: p = 7.76e−04) (Fig. 3B, C; Fig. S2). Grin2a Hets also exhibited increased power of slow oscillations at 3 months (Figure S2; p = 0.0259). Unlike Grin2a KO mice, Akap11−/− animals had reduced power of slow oscillations (Fig. 3E, F, S2), by ~15% compared to WT (3 months: p = 2.85e−05; 6 months: p = 0.0059), as well as reduced delta (p = 0.0191) and alpha (p = 0.0375) oscillations at 3 months (Figs. 3E, S2). Heterozygous Akap11 mutants showed no significant changes in oscillation power for any frequency band.
Grin2a and Akap11 mutants exhibit gene dose-dependent increases and decreases in sleep spindles, respectively
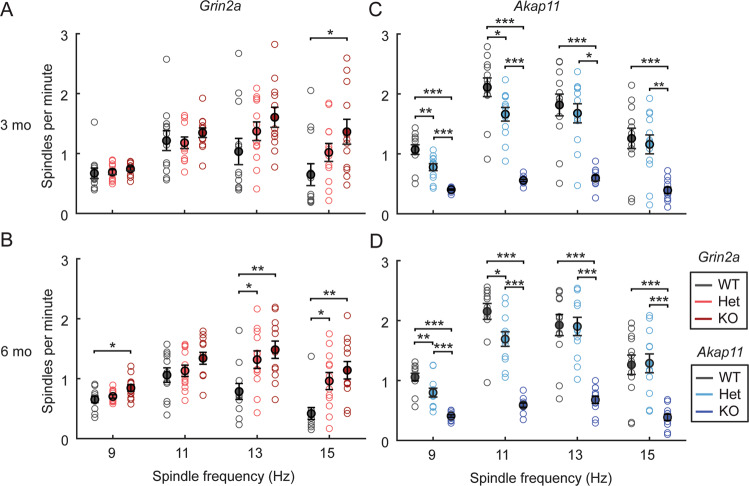
Sleep spindles, an EEG feature possibly linked to overnight memory consolidation, are commonly reduced in schizophrenia patients [30, 43, 44]. Measuring sleep spindle density during NREM sleep in the light cycle, we found striking gene dose-dependent increases in sleep spindle density in Grin2a mutants, especially at 6 months (Fig. 4A, B; ~10-40% increase in heterozygous, and 25-50% increase in homozygous mutants). This increase in spindle density occurred across a range of spindle frequencies from 9 Hz to 15 Hz (3 months, KO, 15 Hz: p = 0.0234; 6 months, Het, 13 Hz: p = 0.0340; 6 months, Het, 15 Hz: p = 0.0192; 6 months, KO, 9 Hz: p = 0.0188; 6 months, KO, 13 Hz: p = 0.0044; 6 months, KO, 15 Hz: p = 0.0016). In contrast, Akap11 mutants showed the opposite phenotype of decreased spindles (Fig. 4C, D), also in a gene dose-dependent manner (heterozygotes ~25% reduction and homozygous KO ~50–75% reduction, compared with WT littermates) (3 months, Het, 9 Hz: p = 0.0029, 3 months, Het, 11 Hz: p = 0.0193; 3 months, KO, 9 Hz: p = 5.53e−09; 3 months, KO, 11 Hz: p = 6.58e−11; 3 months, KO, 13 Hz: p = 2.27e−06; 3 months, KO, 15 Hz: p = 2.61e−04; 6 months, Het, 9 Hz: p = 0.0088; 6 months, Het, 11 Hz: p = 0.0106; 6 months, KO, 9 Hz: p = 1.03e−08; 6 months, KO, 11 Hz: p = 1.39e−11; 6 months, KO, 13 Hz: p = 1.00e−06; 6 months, KO, 15 Hz: p = 1.52e−04).
Fig. 4. Sleep spindle density in Grin2a and Akap11 mutants.
A–D Density for 9 Hz, 11 Hz, 13 Hz, and 15 Hz spindles during NREM sleep during the light cycle. Error bars show mean ± standard error; p < 0.05, **p < 0.01, ***p < 0.001; n = 11–12 mice/group.
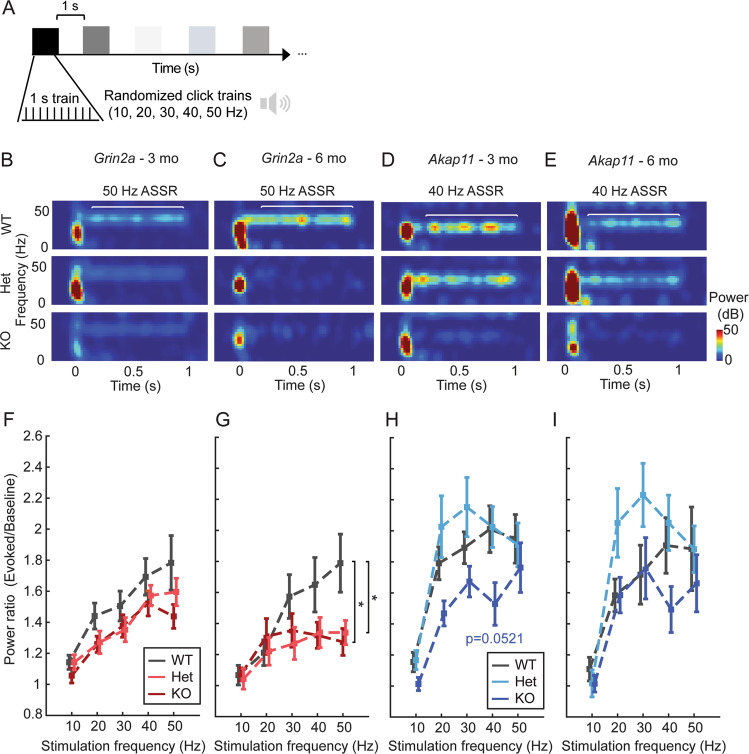
Grin2a mutants exhibit attenuated ASSR at gamma frequencies
ASSR is a highly translatable sensory processing assay that measures cortical entrainment by comparing the evoked response to click trains of different frequencies relative to pre-stimulus activity [45]. ASSR is typically reduced for gamma (40–50 Hz), but not lower frequency (<40 Hz), stimuli in schizophrenia and bipolar disorder patients relative to healthy controls, signifying impaired entrainment of cortical gamma rhythms [25, 27, 28, 45, 46]. Using an ASSR paradigm with trains of randomized 10-50 Hz stimulation frequencies (Fig. 5A), we found a significant reduction of 50 Hz entrainment (as quantified by the power ratio) for 6-month-old Grin2a Hets and KO mice, which was not present at 3 months (Fig. 5B–G). Relative to WT, homozygous (p = 0.0467) and heterozygous (p = 0.0214) Grin2a mutants showed a ~30% reduction in 50 Hz ASSR at 6 months (Fig. 5C, G). There was also a trend (p = 0.0521) towards decreased 40 Hz ASSR for Akap11 KO at 3 months (Fig. 5H), but no other consistent differences between Akap11 mutants and WT (Fig. 5D, E, H, I).
Fig. 5. Auditory steady-state responses (ASSR) in Grin2a and Akap11 mutants.
A Schematic of ASSR paradigm. B–E Representative ASSR for 3- and 6-month-old Grin2a and Akap11 animals. F–I Power ratio quantification of the evoked (0.2–1 s post-stimulus) / baseline (0.5 s pre-stimulus) response for 10-50 Hz auditory stimulation (evaluated using Welch’s method, window = 0.5 s, overlap = 0.25 s). Asterisks indicate significant differences in Het or KO vs. WT littermates (p < 0.05). Error bars denote mean ± standard error.
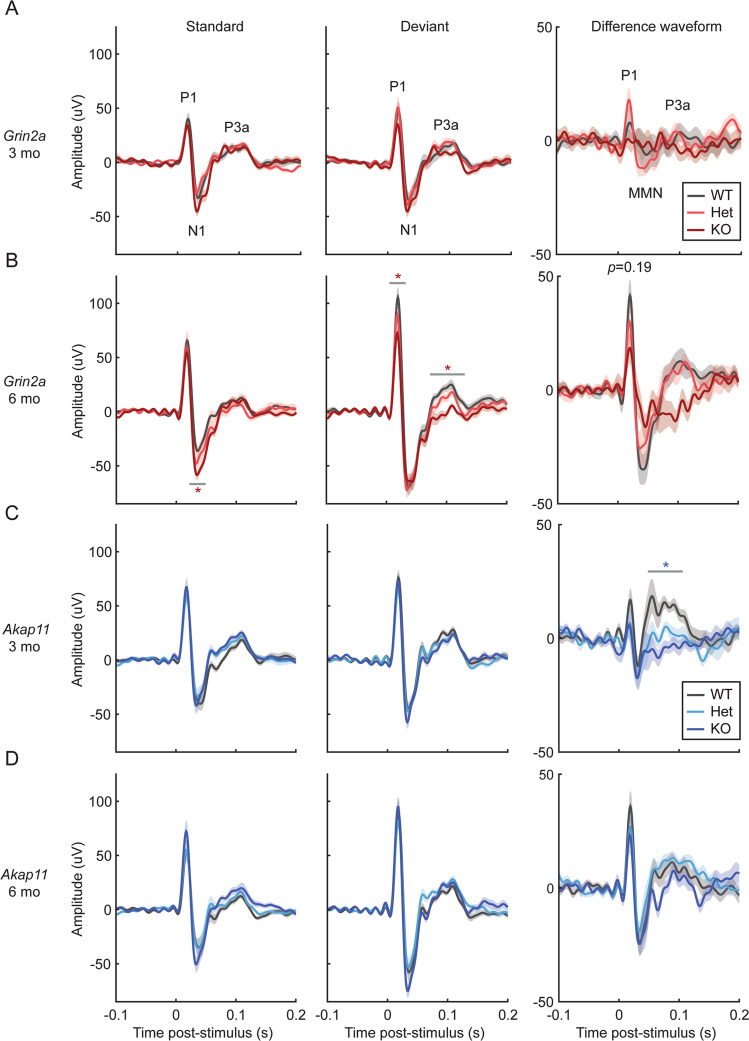
Grin2a−/− and Akap11−/− mice show reduced responses to deviant auditory stimuli in the MMN paradigm
MMN (sometimes known as the “auditory oddball” paradigm) measures novelty-related responses to unexpected stimuli by comparing the event-related potentials in response to infrequent stimuli (deviant, 10% occurrence) vs. frequent stimuli (standard, 90% occurrence) that differ by at least one parameter (e.g., frequency, duration, or amplitude) [47, 48]. Schizophrenia patients display reduced responses to unexpected auditory stimuli, as reflected by the decreased amplitude of event-related potential components MMN and P3a, compared with healthy comparison subjects [47, 49, 50]. These differential responses are thought to reflect deficits in sensory discrimination and novelty-related attention shifts, respectively. We found that Grin2a and Akap11 KO exhibited altered responses to auditory stimuli during the MMN paradigm (Fig. 6). Grin2a−/− animals at 6 months (but not 3 months) exhibited reduced P1 (p = 0.031) and reduced P3a (p = 0.0099) peak amplitudes (~30% and 40% decrease from WT levels, respectively) in response to deviant tones, while the N1 peak was increased in response to standard tones in these animals (~35% increase from WT level, p = 0.0217) (Fig. 6A-B, S6A-C). P1 and MMN amplitudes in the difference waveform (response to deviant tones minus the response to standard tones) of 6-month-old Grin2a KO mice were reduced compared to WT and Het (Fig. 6B), though these results were not statistically significant (P1: p = 0.19; MMN: p = 0.93) due to large variation in the data. Akap11−/− animals showed a strongly reduced P3a amplitude in the difference waveform at 3 months (~70% reduction vs. WT; p = 0.0192), while Akap11 heterozygous mutants had similar responses as their WT littermates (Fig. 6C, D, S6).
Fig. 6. Mismatch negativity (MMN) responses in Grin2a and Akap11 mutants.
A–D Mean response to standard (n = 900 trials), deviant (n = 100 trials) and difference waveforms [response to deviant tones minus the response to standard tones], averaged across each genotype. Shading indicates mean ± standard error. Red or blue asterisks indicate statistical differences in peak amplitude between WT and KO animals, which was assessed for the P1, N1/MMN, and P3a peaks (as indicated on the first graphs). p < 0.05, **p < 0.01, ***p < 0.001; n = 11–12 mice/group.
Discussion
Recent large-scale exome-sequencing efforts have uncovered rare genetic variants that confer a high risk for schizophrenia, including GRIN2A and AKAP11 [1, 2]. These relatively penetrant, loss-of-function mutations lend themselves to disease modeling in genetically modified mice. Besides their genetic validity, do Grin2a and Akap11 mutant mice have any neurobiological phenotype that resembles the human disease (schizophrenia and/or bipolar disorder)? Using a neurophysiological assay (EEG) that is largely translatable between rodents and humans, we report that Grin2a and Akap11 mutants show several key EEG abnormalities that are also observed in patients with schizophrenia and bipolar disorder (summarized in Table S2), including increased resting gamma power and reduced evoked gamma oscillations, altered sleep spindle density, and abnormal MMN (in particular, altered P3a event-related potentials in the MMN paradigm). These shared EEG abnormalities suggest that Grin2a and Akap11 deficiencies impact, at least in part, the same brain networks and circuits that go awry in schizophrenia patients. One limitation of our study design is that male and female Grin2a mutant mice, but only male Akap11 mutant mice, were used; therefore, we cannot exclude the possibility that female Akap11 mutant mice may exhibit a different phenotype than the male Akap11 mutants tested.
Gamma oscillations
A major finding of our study is that Grin2a and Akap11 mutants show elevated gamma power during sleep and quiet wake (Figs. 3, S3, S5), consistent with EEG findings in schizophrenia [25, 41, 42, 51] and to a lesser extent, bipolar disorder patients [51–53]. Gamma oscillations are generated by local cortical networks of fast-spiking, parvalbumin-positive (PV+) interneurons and excitatory pyramidal cells [54, 55]. Evidence of deficiency of PV+ interneuron function has been found in postmortem studies of schizophrenia patient brains [26, 56, 57], and resting gamma power is elevated in NMDAR hypofunction rodent models [58, 59], including mice lacking NMDAR transmission specifically in PV+ interneurons [60]. In the present study, Grin2a and Akap11 mutants showed increased spectral entropy and/or a reduced aperiodic exponent (indicative of a “flatter” PSD) in the parameterized power spectrum during NREM sleep (Table S1), indicating that the resting gamma increases observed in Grin2a and Akap11 mutants (Fig. 3) most likely reflect increases in asynchronous firing that manifest as increased broadband gamma [61, 62]. These increases in asynchronous neural activity may be driven by an enhanced E/I ratio [63], potentially arising from insufficient PV inhibition, as has been reported for PV-Cre/NR1 f/f mice [62]. In contrast to elevated resting gamma power, evoked gamma oscillations such as ASSR are reduced in schizophrenia [25, 27, 45, 46, 64] and bipolar disorder patients [28, 65, 66]. Similarly, we observed reduced 50 Hz ASSR in both Grin2a KO and Hets at 6 months and a strong trend towards reduced 40 Hz ASSR in Akap11 KO at 3 months (Fig. 5).
Our findings are in line with the long-standing “glutamatergic hypothesis” that NMDAR dysfunction may be an underlying cause of schizophrenia [18, 67]. Indeed, NMDA receptors, including GRIN2A, are important for the function of PV+ interneurons [68], and undergo changes during development that may make these cells particularly sensitive to perturbation in psychiatric disease [69]. Importantly, even in the heterozygous state (which is the most relevant to human schizophrenia [1]), Grin2a mutations had a significant effect on resting (particularly during NREM sleep) (Fig. 3C) and stimulus-evoked gamma power (Fig. 5G). Overall, our findings suggest that Grin2a and Akap11 deficiency affect convergent neural pathways, potentially involving circuits of PV+ interneurons and excitatory neurons, that drive gamma oscillations [55].
In our study, Grin2a−/− mice exhibited increases in spontaneous power across multiple frequency bands (Fig. 3), reminiscent of an “over-excitation” phenotype [70]. In contrast, broadband power increases (other than gamma) were not observed in Akap11 mutants, and slow oscillations were reduced. Intriguingly, mutations in GRIN2A, but not AKAP11, are associated with certain epilepsy disorders (including epilepsy-aphasia syndromes [1, 3] and severe developmental and epileptic encephalopathy [71]), and a drug that acts as a positive allosteric modulator of GRIN2A-containing NMDA receptors (GNE-0723) reduces spontaneous power (including slow oscillation power) in mouse models of Dravet syndrome and Alzheimer’s disease [35, 72]. These and our findings further substantiate a link between schizophrenia and epilepsy [73], which is affected by GRIN2A, but not AKAP11 mutation [1, 2].
Sleep spindles
Sleep spindles (12-15 Hz) are another EEG feature typical of NREM sleep, generated by neurons across the cortex, thalamus, and thalamic reticular nucleus (TRN) [29]. Although rather heterogeneous, patients with schizophrenia or bipolar disorder have on average a lower sleep spindle density relative to healthy comparison subjects [30, 40, 74]. Despite sharing several other key features, the EEG phenotypes of Grin2a and Akap11 mutants showed opposite and striking changes in sleep spindles. Grin2a mutants had elevated sleep spindle density, whereas Akap11 mutants showed fewer spindles (Fig. 4). In this context, it is noteworthy that both Grin2a and Akap11 heterozygous mutants exhibited a sleep spindle phenotype intermediate between homozygous knockouts and wild-type littermates. Because Grin2a and Akap11 mutations have such disparate effects on spindle density, it is tempting to speculate that the heterogeneity of spindle density in human patients might arise in part from underlying genetic heterogeneity. The opposite effects of Grin2a and Akap11 on sleep spindles implies that these genes differentially regulate the thalamocortical circuits responsible for spindle generation [44]. Although the precise neurophysiologic mechanisms remain to be discovered, our findings suggest that sleep spindles could serve as a biomarker separating different circuit mechanisms affected by Grin2a and Akap11 loss-of-function and be potentially useful in stratifying the human disease population.
Mismatch negativity (MMN)
The MMN test is a clinically translatable assay of sensory processing of familiar versus unexpected stimuli [47, 75]. The auditory event-related potential peak components P1, N1, and P3a are thought to represent early detection, automatic sensory discrimination, and novelty-related attention shifts, respectively [76]. On average, schizophrenia [49, 77, 78] and bipolar disorder [79, 80] patients tend to exhibit reduced N1 and P3a peak amplitudes, typically measured in the difference waveform (the responses to deviant stimuli minus the responses to standard stimuli). Interestingly, Grin2a−/− (6 months) and Akap11−/− mice (3 months) exhibited reduced P3a responses to deviant stimuli (Fig. 6). The N1/MMN peak in the difference waveform is reported to be reduced on average in schizophrenia and bipolar disorder patients; however, we found no significant changes in Grin2a or Akap11 mutant mice, perhaps because of high variability in this metric [76, 77, 81]. 6-month-old Grin2a−/− mice exhibited increased N1 amplitude in response to standard tones (Fig. 6B), potentially indicating lack of habituation to the repeated stimulus.
Conclusion
The convergent and divergent EEG features identified in this study highlight systems-level functional changes in the brain which may provide insight into the neurobiological pathways affected by the loss of Grin2a and Akap11. Overall, the considerable overlap in EEG phenotypes between mouse mutants and human patients gives further credence to Akap11- and Grin2a-deficient mice as animal models of schizophrenia, particularly so as heterozygous mutants—SCHEMA mutations are found as heterozygotes in humans—tend to show abnormal phenotypes that are intermediate between knockout and WT (e.g., Grin2a+/− in gamma oscillation; Grin2a+/− and Akap11+/− in spindle density). Further investigation of the mechanistic foundations for these EEG changes, including transcriptomic and proteomic characterizations of Grin2a and Akap11 mutants, will help elucidate which brain regions, cell types, and molecular pathways contribute to the network-level dysfunctions of schizophrenia and bipolar disorder.
Supplementary information
Acknowledgements
We thank Nate Shepard for assisting with the mouse colony and Patrick Ihejirika for helping with data analysis, as well as the laboratories of Dr. Motoya Katsuki and Dr. John Scott which generated the Grin2a and Akap11 mutant mice used in this study. Research reported in this manuscript was supported by the Stanley Center for Psychiatric Research. Z.F. is supported by an Otto Hahn Fellowship of the Max Planck Society. M.S. serves on the Scientific Advisory Boards of Biogen, Cerevel, Neumora, Vanqua Bio, and ArcLight Therapeutics. L.H., L.W., E.Y., S.C., B.S., and J.Q.P. reported no biomedical financial interests or potential conflicts of interest.
Author contributions
LEH, JQP, and MS contributed to the study design. LEH contributed to data acquisition. LEH, LW, EY, SC, JQP, and MS contributed to data analysis and interpretation. LEH, LW, EY, SC, ZF, BJS, JQP, and MS contributed to writing and revising the manuscript.
Code availability
LUNA software (http://zzz.bwh.harvard.edu/luna, v0.26) was used for sleep/wake EEG analysis. The Tort lab’s script (https://github.com/tortlab/phase-amplitude-coupling, v1.0) was used for phase-coupling analysis, and the FOOOF algorithm (https://fooof-tools.github.io/fooof/index.html, v1.0.0) was used for spectral parameterization. All other analyses were conducted using custom Python and MATLAB (MathWorks, Natick, MA, RRID: SCR_001622) scripts, which the authors will provide upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Linnea E. Herzog, Email: lherzog@broadinstitute.org
Morgan Sheng, Email: msheng@broadinstitute.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-023-02393-7.
References
- 1.Singh T, Poterba T, Curtis D, Akil H, Al Eissa M, Barchas JD, et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature. 2022;604:509–16. doi: 10.1038/s41586-022-04556-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer DS, Howrigan DP, Chapman SB, Adolfsson R, Bass N, Blackwood D, et al. Exome sequencing in bipolar disorder identifies AKAP11 as a risk gene shared with schizophrenia. Nat Genet. 2022;54:541–7. doi: 10.1038/s41588-022-01034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvill GL, Regan BM, Yendle SC, O’Roak BR, Lozovaya N, Bruneau N, et al. GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nat Gene. 2013;45:1073–6. doi: 10.1038/ng.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salmi M, Bolbos R, Bauer S, Minlebaev M, Burnashev N, Szepetowski P. Transient microstructural brain anomalies and epileptiform discharges in mice defective for epilepsy and language-related NMDA receptor subunit gene Grin2a. Epilepsia. 2018;59:1919–30. doi: 10.1111/epi.14543. [DOI] [PubMed] [Google Scholar]
- 5.The Schizophrenia Working Group of the Psychiatric Genomics Consortium. Ripke S, Neale BM, Corvin A, Walters JTR, Farh K, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502–8. doi: 10.1038/s41586-022-04434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennison CA, Legge SE, Pardiñas AF, Walters JTR. Genome-wide association studies in schizophrenia: Recent advances, challenges and future perspective. Schizophr Res. 2020;217:4–12. doi: 10.1016/j.schres.2019.10.048. [DOI] [PubMed] [Google Scholar]
- 8.Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landén M, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–41. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh T, Walters JTR, Johnstone M, Curtis D, Suvisaari J, Torniainen M, et al. The contribution of rare variants to risk of schizophrenia in individuals with and without intellectual disability. Nat Genet. 2017;49:1167–73. doi: 10.1038/ng.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, et al. Impact of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–9. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–9. doi: 10.1016/S0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 13.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krey I, von Spiczak S, Johannesen K, Hikel C, Kurlemann G, Muhle H, et al. L-Serine treatment is associated with improvements in behavior, EEG, and seizure frequency in individuals with GRIN-related disorders due to null variants. Neurotherapeutics. 2022;19:334–41. doi: 10.1007/s13311-021-01173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyamoto S, Miyake N, Jarskog LF, Fleischhacker WW, Lieberman JA. Pharmacological treatment of schizophrenia: a critical review of the pharmacology and clinical effects of current and future therapeutic agents. Mol Psychiatry. 2012;17:1206–27. doi: 10.1038/mp.2012.47. [DOI] [PubMed] [Google Scholar]
- 16.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat Rev Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Q, Sheng M. NMDA receptors in nervous system diseases. Neuropharmacology. 2013;74:69–75. doi: 10.1016/j.neuropharm.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Moghaddam B, Javitt D. From revolution to evolution: the glutamate hypothesis of schizophrenia and its implication for treatment. Neuropsychopharmacology. 2012;37:4–15. doi: 10.1038/npp.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solmi M, Radua J, Olivola M, Croce E, Soardo L, Salazar de Pablo G, et al. Age at onset of mental disorders worldwide: large-scale meta-analysis of 192 epidemiological studies. Mol Psychiatry. 2012;27:281–95. doi: 10.1038/s41380-021-01161-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–7. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- 21.Ohi K, Shimada T, Nitta Y, Kihara H, Okubo H, Uehara T, et al. Specific gene expression patterns of 108 schizophrenia-associated loci in cortex. Schizophr Res. 2016;174:35–38. doi: 10.1016/j.schres.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 22.Deng Z, Li X, Ramirez MB, Purtell K, Choi I, Lu J, et al. Selective autophagy of AKAP11 activates cAMP/PKA to fuel mitochondrial metabolism and tumor cell growth. Proc Natl Acad Sci. 2012;118:e2020215118. doi: 10.1073/pnas.2020215118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varjosalo M, Keskitalo S, Van Drogen A, Nurkkala H, Vichalkovski A, Aebersold R, et al. The protein interaction landscape of the human CMGC kinase group. Cell Rep. 2013;3:1306–20. doi: 10.1016/j.celrep.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Freland L, Beaulieu JM. Inhibition of GSK3 by lithium, from single molecules to signaling networks. Front Mol Neurosci. 2012;5:14. doi: 10.3389/fnmol.2012.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlhaas PJ, Singer W. Abnormal neural oscillations and synchrony in schizophrenia. Nat Rev Neurosci. 2012;11:100–13. doi: 10.1038/nrn2774. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Burgos G, Lewis DA. NMDA receptor hypofunction, parvalbumin-positive neurons, and cortical gamma oscillations in schizophrenia. Schizophr Bull. 2012;38:950–7. doi: 10.1093/schbul/sbs010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thuné H, Recasens M, Uhlhaas PJ. The 40-Hz auditory steady-state response in patients with schizophrenia: a meta-analysis. JAMA Psychiatry. 2016;73:1145–53. doi: 10.1001/jamapsychiatry.2016.2619. [DOI] [PubMed] [Google Scholar]
- 28.Sugiyama S, Ohi Z, Kuramitsu A, Takai K, Muto Y, Taniguchi T, et al. The auditory steady-state response: electrophysiological index for sensory processing dysfunction in psychiatric disorders. Front Psychiatry. 2012;12:644541. doi: 10.3389/fpsyt.2021.644541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manoach DS, Pan JQ, Purcell SM, Stickgold R. Reduced sleep spindles in schizophrenia: a treatable endophenotype that links risk genes to impaired cognition? Biol Psychiatry. 2016;80:599–608. doi: 10.1016/j.biopsych.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozhemiako N, Wang J, Jiang C, Wang L, Gai G, Zou K, et al. Non-rapid eye movement sleep and wake neurophysiology in schizophrenia. eLife 10.7554/eLife.76211 (2022). [DOI] [PMC free article] [PubMed]
- 31.Kadotani H, Hirano T, Masugi M, Nakamura K, Nakao K, Katsuki M, et al. Motor discoordination results from combined gene disruption of the NMDA receptor NR2A and NR2C subunits, but not from single disruption of the NR2A or NR2C subunit. J Neurosci. 1996;16:7859–67. doi: 10.1523/JNEUROSCI.16-24-07859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiting JL, Ogier L, Forbush KA, Bucko P, Gopalan J, Seternes O, et al. AKAP220 manages apical actin networks that coordinate aquaporin-2 location and renal water reabsorption. Proc Natl Acad Sci. 2016;113:E4328–37. doi: 10.1073/pnas.1607745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh RN, Cummins RA. The Open-Field Test: a critical review. Psychol Bull. 1976;83:482–504. doi: 10.1037/0033-2909.83.3.482. [DOI] [PubMed] [Google Scholar]
- 34.Seibenhener ML, Wooten MC. Use of the Open Field Maze to measure locomotor and anxiety-like behavior in mice. J Vis Exp. 2015;96:e52434. doi: 10.3791/52434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson JE, Ma K, Elstrott J, Weber M, Saillet S, Khan AS, et al. GluN2A NMDA receptor enhancement improves brain oscillations, synchrony, and cognitive functions in Dravet Syndrome and Alzheimer’s disease models. Cell Rep. 2020;30:381–96. doi: 10.1016/j.celrep.2019.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amador A, Bostick CD, Olsen H, Peters J, Camp CR, Krizay D, et al. Modelling and treating GRIN2A developmental and epileptic encephalopathy in mice. Brain. 2020;143:2039–57. doi: 10.1093/brain/awaa147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bertocchi I, Eltokhi A, Rozov A, Chi VN, Jensen V, Bus T, et al. Voltage-independent GluN2A-type NMDA receptor Ca2+ signaling promotes audiogenic seizures, attentional and cognitive deficits in mice. Commun Biol. 2021;4:59. doi: 10.1038/s42003-020-01538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wulff K, Dijk D, Middleton B, Foster RG, Joyce EM. Sleep and circadian rhythm disruption in schizophrenia. Br J Psychiatry. 2012;200:308–16. doi: 10.1192/bjp.bp.111.096321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan M, Chung K, Yung K, Yeung W. Sleep in schizophrenia: A systematic review and meta-analysis of polysomnographic findings in case-control studies. Sleep Med Rev. 2017;32:69–84. doi: 10.1016/j.smrv.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Zangani C, Casetta C, Saunders AS, Donati F, Maggioni E, D’Agostino A. Sleep abnormalities across different clinical stages of Bipolar Disorder: A review of EEG studies. Neurosci Biobehav Rev. 2020;118:247–57. doi: 10.1016/j.neubiorev.2020.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Tekell JL, Hoffmann R, Hendrickse W, Greene RW, Rush AJ, Armitage R. High frequency EEG activity during sleep: characteristics in schizophrenia and depression. Clin EEG Neurosci. 2005;36:25–35. doi: 10.1177/155005940503600107. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka-Koshiyama K, Koshiyama D, Miyakoshi M, Joshi YB, Molina JL, Sprock J, et al. Abnormal spontaneous Gamma power is associated with verbal learning and memory dysfunction in schizophrenia. Front Psychiatry. 2020;11:832. doi: 10.3389/fpsyt.2020.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrarelli F. Sleep in patients with schizophrenia. Curr Sleep Med Rep. 2015;1:150–6. doi: 10.1007/s40675-015-0010-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manoach DS, Stickgold R. Abnormal sleep spindles, memory consolidation, and schizophrenia. Ann Rev Clin Psychol. 2019;15:451–79. doi: 10.1146/annurev-clinpsy-050718-095754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Donnell BF, Vohs JL, Krishnan GP, Rass O, Hetrick WP, Morzorati SL. The auditory steady-state response (ASSR): a translational biomarker for schizophrenia. Suppl Clin Neurophysiol. 2013;62:101–12. doi: 10.1016/B978-0-7020-5307-8.00006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun C, Zhou P, Wang C, Fan Y, Tian Q, Dong F, et al. Defects of Gamma oscillations in auditory steady-state evoked potential of schizophrenia. Shanghai Arch Psychiatry. 2018;30:27–38. doi: 10.11919/j.issn.1002-0829.217078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tada M, Kirihara K, Mizutani S, Uka T, Kunii N, Koshiyama D, et al. Mismatch negativity (MMN) as a tool for translational investigations into early psychosis: A review. Int J Psychophysiol. 2019;145:5–14. doi: 10.1016/j.ijpsycho.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 48.Herzog L, Salehi KL, Bohon KS, Wiest MC. Prestimulus frontal-parietal coherence predicts auditory detection performance in rats. J Neurophysiol. 2014;111:1986–2000. doi: 10.1152/jn.00781.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Light GA, Swerdlow NR, Thomas ML, Calkins ME, Green MF, Greenwood TA, et al. Validation of mismatch negativity and P3a for use in multi-site studies of schizophrenia: characterization of demographic, clinical, cognitive, and functional correlates in COGS-2. Schizophr Res. 2015;163:63–72. doi: 10.1016/j.schres.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaur M, Battisti RA, Ward PB, Ahmed A, Hickie IB, Hermens DF. MMN/P3a deficits in first episode psychosis: comparing schizophrenia-spectrum and affective-spectrum subgroups. Schizophr Res. 2011;130:203–9. doi: 10.1016/j.schres.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 51.Venables NC, Bernat EM, Sponheim SR. Genetic and disorder-specific aspects of resting state EEG abnormalities in schizophrenia. Schizophr Bull. 2009;35:826–39. doi: 10.1093/schbul/sbn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Narayanan B, O’Neil K, Berwise C, Stevens MC, Calhoun VD, Clementz BA. Resting state electroencephalogram oscillatory abnormalities in schizophrenia and psychotic bipolar patients and their relatives from the bipolar and schizophrenia network on intermediate phenotypes study. Biol Psychiatry. 2014;76:456–65. doi: 10.1016/j.biopsych.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arikan MK, Günver G, İlhan R, Öksüz O, Metin B. Gamma oscillations predict treatment response to aripiprazole in bipolar disorder. J Affect Disord. 2021;294:159–62. doi: 10.1016/j.jad.2021.07.044. [DOI] [PubMed] [Google Scholar]
- 54.Buzsaki G, Wang XJ. Mechanisms of gamma oscillations. Annu Rev Neurosci. 2012;35:203–25. doi: 10.1146/annurev-neuro-062111-150444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, Deisseroth K, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–7. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakazawa K, Zsiros V, Jiang Z, Nakao K, Kolata S, Zhang S, et al. GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology. 2012;62:1574–83. doi: 10.1016/j.neuropharm.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis DA, Curley AA, Glausier JR, Volk DW. Cortical parvalbumin interneurons and cognitive dysfunction in schizophrenia. Trends Neurosci. 2012;35:57–67. doi: 10.1016/j.tins.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakao K, Singh M, Sapkota K, Hagler BC, Hunter RN, Raman C, et al. GSK3beta inhibition restores cortical gamma oscillation and cognitive behavior in a mouse model of NMDA receptor hypofunction relevant to schizophrenia. Neuropsychopharmacology. 2020;45:2207–18. doi: 10.1038/s41386-020-00819-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gandal MJ, Sisti J, Klook K, Ortinski PI, Leitman V, Liang Y, et al. GABAB-mediated rescue of altered excitatory-inhibitory balance, gamma synchrony and behavioral deficits following constitutive NMDAR-hypofunction. Transl Psychiatry. 2012;2:e142. doi: 10.1038/tp.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carlén M, Meletis K, Siegle JH, Cardin JA, Futai K, Vierling-Claassen D, et al. A critical role for NMDA receptors in parvalbumin interneurons for gamma rhythm induction and behavior. Mol Psychiatry. 2011;17:537–48. doi: 10.1038/mp.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donoghue T, Haller M, Peterson EJ, Varma P, Sebastian P, Gao R, et al. Parameterizing neural power spectra into periodic and aperiodic components. Nat Neurosci. 2020;23:1655–65. doi: 10.1038/s41593-020-00744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guyon N, Zacharias LR, Fermino de Oliveira E, Kim H, Leite JP, Lopes-Aguiar C, et al. Network asynchrony underlying increased broadband gamma power. J Neurosci. 2021;41:2944–63. doi: 10.1523/JNEUROSCI.2250-20.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gao R, Peterson EJ, Voytek B. Inferring synaptic excitation/inhibition balance from field potentials. NeuroImage. 2017;158:70–78. doi: 10.1016/j.neuroimage.2017.06.078. [DOI] [PubMed] [Google Scholar]
- 64.Sun Y, Farzan F, Barr MS, Kirihara K, Fitzgerald PB, Light GA, et al. Gamma oscillations in schizophrenia: Mechanisms and clinical significance. Brain Res. 2011;1413:98–114. doi: 10.1016/j.brainres.2011.06.065. [DOI] [PubMed] [Google Scholar]
- 65.Oda Y, Onitsuka T, Tsuchimoto R, Hirano S, Oribe N, Ueno T, et al. Gamma band neural synchronization deficits for auditory steady state responses in bipolar disorder patients. PLoS One. 2021;7:e39955. doi: 10.1371/journal.pone.0039955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isomura S, Onitsuka T, Tsuchimoto R, Nakamura I, Hirano S, Oda Y, et al. Differentiation between major depressive disorder and bipolar disorder by auditory steady-state responses. J Affect Disord. 2016;190:800–6. doi: 10.1016/j.jad.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 67.Uno Y, Coyle JT. Glutamate hypothesis in schizophrenia. Psychiatry Clin Neurosci. 2019;73:204–15. doi: 10.1111/pcn.12823. [DOI] [PubMed] [Google Scholar]
- 68.Kinney JW, Davis CN, Tabarean I, Conti B, Bartfai T, Behrens MM. A specific role for NR2A-containing NMDA receptors in the maintenance of parvalbumin and GAD67 immunoreactivity in cultured interneurons. J Neurosci. 2006;26:1604–15. doi: 10.1523/JNEUROSCI.4722-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang HX, Gao WJ. Cell type-specific development of NMDA receptors in the interneurons of rat prefrontal cortex. Neuropsychopharmacology. 2009;34:2028–40. doi: 10.1038/npp.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang W. The broadband power shifts in entorhinal EEG are related to the firing of grid cells. Heliyon. 2021;7:e06087. doi: 10.1016/j.heliyon.2021.e06087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Strehlow V, Rieubland C, Gallati S, Kim S, Myers SJ, Peterson V, et al. Compound-heterozygous GRIN2A null variants associated with severe developmental and epileptic encephalopathy. Epilepsia. 2022;63:e132–e137. doi: 10.1111/epi.17394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hackos DH, Hanson JE. Diverse modes of NMDA receptor positive allosteric modulation: Mechanisms and consequences. Neuropharmacology. 2017;112:34–45. doi: 10.1016/j.neuropharm.2016.07.037. [DOI] [PubMed] [Google Scholar]
- 73.Chang Y, Chen P, Tsai I, Sung F, Chin Z, Kuo H, et al. Bidirectional relation between schizophrenia and epilepsy: A population-based retrospective cohort study. Epilepsia. 2011;52:2036–42. doi: 10.1111/j.1528-1167.2011.03268.x. [DOI] [PubMed] [Google Scholar]
- 74.Ritter PS, Schwabedal J, Brandt M, Schrempf W, Brezan F, Krupka A, et al. Sleep spindles in bipolar disorder - a comparison to healthy control subjects. Acta Psychiatr Scand. 2018;138:163–72. doi: 10.1111/acps.12924. [DOI] [PubMed] [Google Scholar]
- 75.Todd J, Harms L, Schall U, Michie PT. Mismatch negativity: translating the potential. Front Psychiatry. 2013;4:171. doi: 10.3389/fpsyt.2013.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rissling AJ, Light GA. Neurophysiological measures of sensory registration, stimulus discrimination, and selection in schizophrenia patients. Curr Top Behav Neurosci. 2010;4:283–309. doi: 10.1007/7854_2010_59. [DOI] [PubMed] [Google Scholar]
- 77.Nagai T, Kirihara K, Tada M, Koshiyama D, Koike S, Suga M, et al. Reduced mismatch negativity is associated with increased plasma level of glutamate in first-episode psychosis. Sci Rep. 2017;7:2258. doi: 10.1038/s41598-017-02267-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jahshan C, Wynn J, Mathis KI, Altshuler LL, Glahn DC, Green MF. Cross-diagnostic comparison of duration mismatch negativity and P3a in bipolar disorder and schizophrenia. Bipolar Disord. 2012;14:239–48. doi: 10.1111/j.1399-5618.2012.01008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chitty KM, Lagopoulos J, Lee RSC, Hickie IB, Hermens DF. A systematic review and meta-analysis of proton magnetic resonance spectroscopy and mismatch negativity in bipolar disorder. Eur Neuropsychopharmacol. 2013;23:1348–63. doi: 10.1016/j.euroneuro.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 80.Wada M, Kurose S, Miyazaki T, Nakajima S, Masuda F, Mimura Y, et al. The P300 event-related potential in bipolar disorder: A systematic review and meta-analysis. J Affect Disord. 2019;256:234–49. doi: 10.1016/j.jad.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 81.Umbricht D, Vyssotki D, Latanov A, Nitsch R, Lipp H. Deviance-related electrophysiological activity in mice: is there mismatch negativity in mice? Clin Neurophysiol. 2005;116:353–63. doi: 10.1016/j.clinph.2004.08.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LUNA software (http://zzz.bwh.harvard.edu/luna, v0.26) was used for sleep/wake EEG analysis. The Tort lab’s script (https://github.com/tortlab/phase-amplitude-coupling, v1.0) was used for phase-coupling analysis, and the FOOOF algorithm (https://fooof-tools.github.io/fooof/index.html, v1.0.0) was used for spectral parameterization. All other analyses were conducted using custom Python and MATLAB (MathWorks, Natick, MA, RRID: SCR_001622) scripts, which the authors will provide upon request.